
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 06 March 2024
Sec. Clinical Microbiology
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1361432
 Satomi Takei1,2
Satomi Takei1,2 Kanae Teramoto2,3
Kanae Teramoto2,3 Junya Fujimura4
Junya Fujimura4 Megumi Fujiwara4
Megumi Fujiwara4 Mai Suzuki5
Mai Suzuki5 Yukiko Fukui5
Yukiko Fukui5 Yuji Sekiguchi6
Yuji Sekiguchi6 Takaaki Kawakami7
Takaaki Kawakami7 Masayoshi Chonan7
Masayoshi Chonan7 Mitsuru Wakita7
Mitsuru Wakita7 Yuki Horiuchi1
Yuki Horiuchi1 Takashi Miida1
Takashi Miida1 Toshio Naito2,5
Toshio Naito2,5 Teruo Kirikae2,8
Teruo Kirikae2,8 Tatsuya Tada9*
Tatsuya Tada9* Yoko Tabe1,2
Yoko Tabe1,2Wickerhamiella is a genus of budding yeast that is mainly isolated from environmental samples, and 40 species have been detected. The yeast isolated from human clinical samples usually only contain three species: W. infanticola, W. pararugosa and W. sorbophila. In this study, we isolated W. tropicalis from a blood sample of a six-year-old female with a history of B-cell precursor lymphoblastic leukemia in Japan in 2022. Though the strain was morphologically identified as Candida species by routine microbiological examinations, it was subsequently identified as W. tropicalis by sequencing the internal transcribed spacer (ITS) of ribosomal DNA (rDNA). The isolate had amino acid substitutions in ERG11 and FKS1 associated with azole and echinocandin resistance, respectively, in Candida species and showed intermediate-resistant to fluconazole and micafungin. The patient was successfully treated with micafungin. Furthermore, matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) detected three novel peaks that are specific for W. tropicalis, indicating that MALDI-MS analysis is useful for rapid detection of Wickerhamiella species in routine microbiological examinations.
Yeast-like fungi and yeasts often cause catheter associated bloodstream infections in immunocompromised patients who receive immunosuppressants, chemotherapies, or organ transplants (Papp et al., 2018). Due to the increase of these therapies, non-Candida albicans species have been increasingly identified recently (Kumar et al., 2022). Since these organisms have different susceptibility to antifungal agents, it is critical to identify the species correctly and initiate the appropriate therapies as soon as possible to improve patients’ prognoses.
Wickerhamiella is a genus of budding yeast in the family Trichomonascaceae that was redescribed in 1973 (van der Walt and Liebenberg, 1973). Wickerhamiella species consist of more than 40 species which are mostly environmental organisms. For instance, W. vanderwaltii PYCC 3671T (GenBank accession no. GCA_022577775) was isolated from winery equipment in South Africa, W. sorbophila DS02T (= NBRC 1583T) (GenBank accession no. GCF_002251995) was isolated from industrial water in South Korea in 2015, and W. spandovensis PYCC 8431 (GenBank accession no. GCA_022577695) was isolated from alcoholic beverages in Germany. To date, there are only several Wickerhamiella species that were isolated from human samples. For example, W. pararugosa (Candida pararugosa) PH2204 (GenBank accession no. JAMAJR000000000) was isolated from human feces in China in 2019, oral cavity in Italy (Giammanco et al., 2004), and saliva in Japan (Nakagawa et al., 2004). Furthermore, W. sorbophila was isolated from human feces in China in 2019 (GenBank accession no. GCA_023629035), and W. infanticola from an ear of a baby in Germany in 1995 (GenBank accession no. GCA_004125145). Previous studies have reported that W. pararugosa causes bloodstream infections. These species were detected in a six-month-old male with intrauterine growth in Qatar in 2006 (Taj-Aldeen et al., 2014), a three-year-old female with acute lymphoblastic leukemia in Greece in 2008 (Noni et al., 2020), a five-year-old male in Qatar in 2010 (Taj-Aldeen et al., 2014), a 39-year-old female with morbid obesity in the United States in 2017 (El Helou and Palavecino, 2017), and a three-year-old male with acute myeloid leukemia in Iran in 2022 (Nasri et al., 2023). Though Wickerhamiella tropicalis was first isolated from environmental samples in 2020 (Sakpuntoon et al., 2020), it has never been isolated from human samples. In this case report, we first isolated W. tropicalis in blood cultures that caused severe infections.
Although, gram-staining and fungal isolation media are routinely used to identify yeast-like fungi in clinical laboratories, these methods are insufficient to accurately differentiate fungal species (Pravin Charles et al., 2015). In contrast, genetic sequencing of the internal transcribed spacer (ITS) region of rDNA is an excellent method to accurately differentiate strains. Indeed, predicted taxonomic thresholds for identifying yeast species have been reported to be 98.41% for the ITS-rDNA (Vu et al., 2016). However, sequencing is not suitable for routine microbiological examinations since a short turnaround time is required. Therefore, matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) has been developed to identify species quickly and accurately (Eigner et al., 2009). Despite using technology, detection of W. tropicalis is currently difficult due to the lack of library database. In this study, we attempted to identify MALDI-MS peaks specific to Wickerhamiella species and found three new peaks related to the Wickerhamiella species.
A six-year-old female who was suffering from B-cell precursor lymphoblastic leukemia was hospitalized from May 2022 at a university hospital for a remission induction therapy. She had repeated fevers and showed high levels of C-reactive protein (CRP) with cervical lymphadenopathies. The treatment with piperacillin/tazobactam (3.0 g/day) and linezolid (450 mg/day) was started and improved her symptoms in June. Though blood cultures were tested 5 times at the time of fever, which were negative, a set of blood culture was positive after 62 h of incubation in July. The isolate was initially identified as Candida species. A similar specimen was obtained three days later. A treatment of micafungin (3 mg/kg/day) was started. The blood culture became negative eight days after the treatment.
A W. tropicalis JUWT001 strain was isolated from a patient treated at Juntendo University Hospital in Japan in July 2022. The type strain of W. tropicalis TBRC 11426T was obtained from Thailand Bioresource Research Center (TBRC, Thailand) and the type strains of W. sorbophila NBRC 1583T and W. spandovensis NBRC 10249T (= PYCC 8431T) were obtained from the Biological Resource Center, National Institute of Technology and Evaluation (NITE) (Tokyo, Japan). W. tropicalis TBRC 11426T was imported into Japan with permission from the Minister of Agriculture, Forestry and Fisheries under the Plant Protection Act. The isolates were cultured in Sabouraud dextrose agar (SDA) plate (Becton, Dickinson-Diagnostic Systems, Sparks, MD, USA), Potato Dextrose Agar (PDA) (Becton, Dickinson-Diagnostic Systems), and/or CHROM agar Candida medium (Kanto Chemical Co., Tokyo, Japan) at 25°C or 35°C under aerobic conditions for 48 hours. The minimum inhibitory concentrations (MICs) of the isolate were determined using the broth microdilution method, as described by the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2017).
Drug susceptibility was tested by microdilution assay according to the guidelines of the Clinical and Laboratory Standard Institute (CLSI) (CLSI, 2017) using the Yeast-Like Fungus DP (Eiken Chemical Co., Ltd., Tokyo). The ranges of antibiotic concentrations tested were: 5-fluorocytosine 0.12 to 64 μg/mL, amphotericin B 0.03 to 16 μg/mL, fluconazole 0.12 to 64 μg/mL, itraconazole 0.015 to 8 μg/mL, micafungin 0.015 to 16 μg/mL, miconazole 0.03 to 16 μg/mL, and voriconazole 0.015 to 8 μg/mL (Table 1).
DNA of the clinical isolate for sanger sequencing was extracted using DNeasy UltraClean Microbial kit (QIAGEN, Tokyo, Japan). The ITS region of ribosomal DNA (rDNA) was amplified using fungal universal primers Its1 (5’- TCCGTAGGTGAACCTGCGG -3’) and Its4 (5’- TCCTCCGCTTATTGATATGC -3’) (Zhai et al., 2021). The PCR products were sequenced using 3500 XL genetic analyzer (ABI-Hitachi, Tokyo, Japan). Sequence data were analyzed using BLAST on the National Center for Biotechnology Information (NCBI) database.
Genomic DNA of the clinical isolate for whole genome sequencing was extracted as described previously (Mizukoshi et al., 2021). The genome was sequenced by Illumina MiSeq platform using v3 chemistry (600 cycles). Raw reads were trimmed and assembled using CLC Genomic Workbench version 10.0.1 (CLC bio, Aarhus, Denmark). The genome completeness and contamination were assessed using Eukcc v2.1.0 with default settings (Saary et al., 2020). Species identification of the isolate was also determined by ITS using BLAST. Amino acid substitutions in BCR1, BRG1, CBK1, EFG1, and IRE1 (Talapko et al., 2021) associated with virulence, in ERG3, ERG11, PDR1, and TAC1 associated with azole resistance, and in FKS1, FKS2, and GSC1 (Spettel et al., 2019) associated with echinocandin resistance in Candida species were analyzed based on whole genome sequencing.
Phylogenetic trees were constructed based on concatenated single-copy marker protein sequences predicted from genomes using GTDB-Tk v2.2.6 software (Chaumeil et al., 2022) and visualized using iTol ver.6. The following type strains were used for a phylogenic tree among Wickerhamiella species: W. alocasiicola PYCC 8427T (GenBank accession number GCA_022577715), W. azyma PYCC 8333T (GCA_022577855), W.cacticola NRRL Y-27362T (GCA_003705615), W. dianesei PYCC 8330T (GCA_022577725), W. domercqiae JCM 9478T (GCA_001599275), W. galacta NRRL Y-17645T (GCA_003045245), W. hasegawae JCM 12559T (GCA_004125105), W. infanticola NRRL Y-17858T (GCA_004125145), W. kurtzmanii PYCC 8437T (GCA_022577765), W. nectarea PYCC 8436T (GCA_022577815), W. occidentalis NRRL Y-27364T (GCA_004125095), W. pararugosa PX1910T (GCA_023628975), W. parazyma PYCC 8426T (GCA_022577825), W. sorbophila NBRC 1583T (GCF_002251995), W. spandovensis NBRC 10249T (GCA_022577695), W. vanderwaltii PYCC 3671T (GCA_022577775), and W. versatilis JCM 5958T (GCA_001600375).
The whole-genome sequences of W. tropicalis JUWT001 have been deposited in GenBank as accession number DRR488519 (https://www.ncbi.nlm.nih.gov/sra/DRR488519).
Alpha-cyano-4-hydroxycinnamic acid (CHCA) was used as a matrix. Ten mg of 4-CHCA was dissolved in 1 mL of the solvent consisting of 1% (v/v) trifluoroacetic acid, 35% (v/v) ethanol, 15% (v/v) acetonitrile, and milliQ water. For the analysis using cell lysate, a full loop of fungal cells was dispersed to 200 μL of distilled water in a microtube and mixed with 800 μL of ethanol with zirconia beads. The suspensions were vortexed briefly and centrifuged at 15,000 g for 2 min. The pellets were dried for 5 min. After freezing the tubes at -80 °C, the pellets were suspended in 100 μL of 70% formic acid, crushed using a micro tube mixer (MT-400, Tomy Seiko, Tokyo, Japan) for 5 min, suspended in 100 μL of acetonitrile, and centrifuged at 15,000 g for 2 min. The analysis using protein fraction was performed as described previously (Teramoto et al., 2019). Briefly, a full loop of fungal cells was dispersed in 500 μL of distilled water in a microtube with zirconia beads. After freezing the tubes at -80 °C, the suspensions were centrifuged at 10,000 g for 2 min, 200 μL of supernatants were removed, crushed using a Fast Prep 24 apparatus (Funakoshi Co., Ltd.) for a total of 3 min (9 times for 20 s), and centrifuged at 10,000 g for 2 min. The suspensions were washed in milliQ water and centrifuged at 15,000 g for 5 min. The supernatants were ultrafiltered using Amicon® Ultra-0.5ml (Merck Millipore, Germany), with cut-off points at 100 kDa, centrifuged at 14,000 g for 5 min, and the devices set upside down at 2,000 g for 2 min to remove any residual solutions contained in the filter. Supernatants were analyzed by MALDI-MS according to the manufacturer’s instruction.
MALDI-MS measurements were performed in positive linear mode using MALDI-8020 RUO (Shimadzu Corporation, Kyoto, Japan) equipped with a 200 Hz Nd : YAG laser (355 nm). Mass calibration was performed using 6 peaks with m/z 4365.4, 5381.4, 6411.6, 7274.0, 8369.8, and 10300.1 from Escherichia coli DH5α. Five individual mass spectra were acquired for each fungal extract in the range from m/z 2,000 to 20,000. Biomarker search and peak matching were carried out using eMSTAT Solution™ software (Shimadzu Corp., Kyoto, Japan).
Samples of the positive blood culture were Gram stained and revealed the presence of yeast-like cells (Figure 1). The positive blood culture samples were inoculated on Sabouraud Dextrose Agar (SDA) plate, Potato Dextrose Agar (PDA), and CHROM agar Candida medium at 35°C. After 48h of incubation, a large number of monomorphic, cream-colored smooth colonies were observed on the SDA and PDA. Purple-colored colonies were observed on the CHROM agar Candida medium (Figure 2). The smear of the colony showed yeasts that were observed by the direct smear of the blood culture specimen. The isolate was identified as Candida species. The MICs of the isolate to fluconazole and micafungin were 4 μg/mL and 0.5 μg/mL, respectively (Table 1). According to the breakpoints of fluconazole and micafungin in Candida species, the isolate showed intermediate resistance.
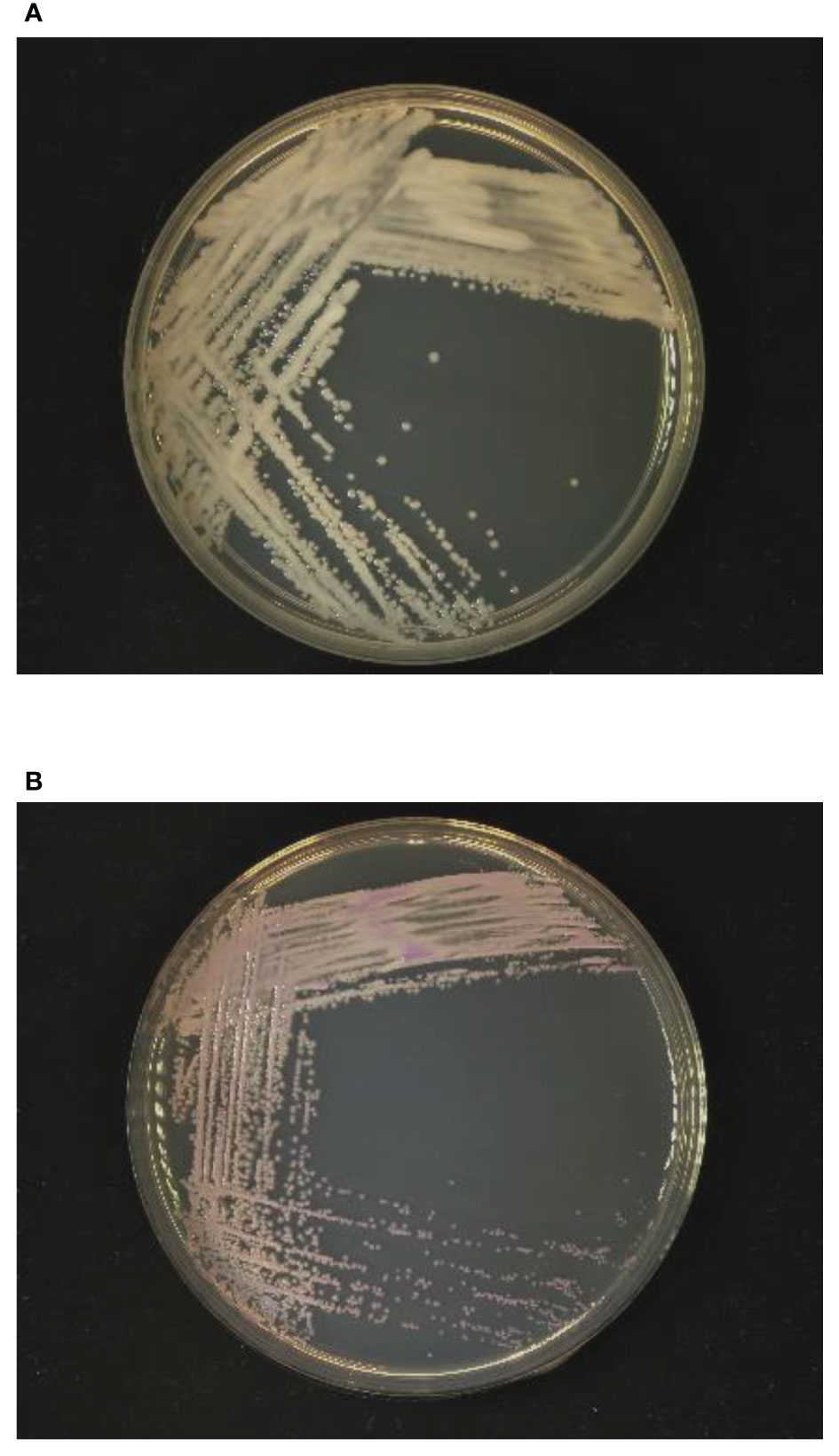
Figure 2 Growth of yeast-like colonies after 48h incubation in (A) Sabouraud dextrose agar and (B) CHROM agar Candida medium.
The ITS sequence of the isolate was 98.8% identical to the sequence of W. tropicalis (GenBank accession number MN218416). Therefore, the isolate was confirmed to be W. tropicalis and designated as W. tropicalis JUWT001.
The W. tropicalis JUWT001 harbored two virulence genes encoding protein kinase regulators, IRE1 and CBK1, that cause pathogenic fungal infections (Talapko et al., 2021). The IRE1 protein had 69.4% and 94.5% identity to that in C. albicans SC 5314 (GenBank accession no. GCF_000182965) and W. sorbophila NBRC 1583T (accession no. GCF_002251995), respectively. The CBK1 protein had 40.3% and 90.7% identity to that in C. albicans SC 5314 (accession no. GCF_000182965) and W. sorbophila NBRC 1583T (accession no. GCF_002251995), respectively.
The isolate had amino acid substitutions in ERG11 and FKS1 associated with azole and echinocandin resistance, respectively. Twenty-one amino-acid substitutions in ERG11 and 9 amino-acid substitutions in FKS1 were detected in hot spot regions which were highly conserved antimicrobial-binding domain in fungi (Favre et al., 1999; Papp et al., 2018) (Figure 3). Of the nine amino acid substitutions of FKS1, an amino acid substitution from Met to Leu at the 696 position was reported in C. parapsilosis in 2018 (Papp et al., 2018; Spettel et al., 2019). There is no report on the remaining mutations in these resistant factors in the hot spot regions (Figure 3).

Figure 3 Amino acid sequence alignments of hot spot regions of ERG11 and FKS1 in C. albicans SC 5314 (GenBank accession no. GCF_000182965), W. sorbophila NBRC 1583T (accession no. GCF_002251995) and W. tropicalis JUWT001. Amino acid substitutions are shaded in black, compared with C. albicans SC 5314. The arrow indicates an amino acid substitution of FKS1 from Met to Leu at the position of 696.
As shown in Figure 4, the phylogenetic tree revealed three clades: A, B, and C. W. tropicalis JUWT001 belonged to clade A, along with W. sorbophila (GenBank accession no. GCA_023629035) and W. infanticola (accession no. GCA_004125145), which have already been reported to be isolated from humans. Another previously reported clinical isolate, W. pararugosa (accession no. JAMAJR000000000), belonged to clade C. The other species were mostly isolated from plant (Lachance et al., 1998; Nakase et al., 2007; Barata et al., 2008; Wang et al., 2008; Lachance et al., 2010; de Vega et al., 2017).
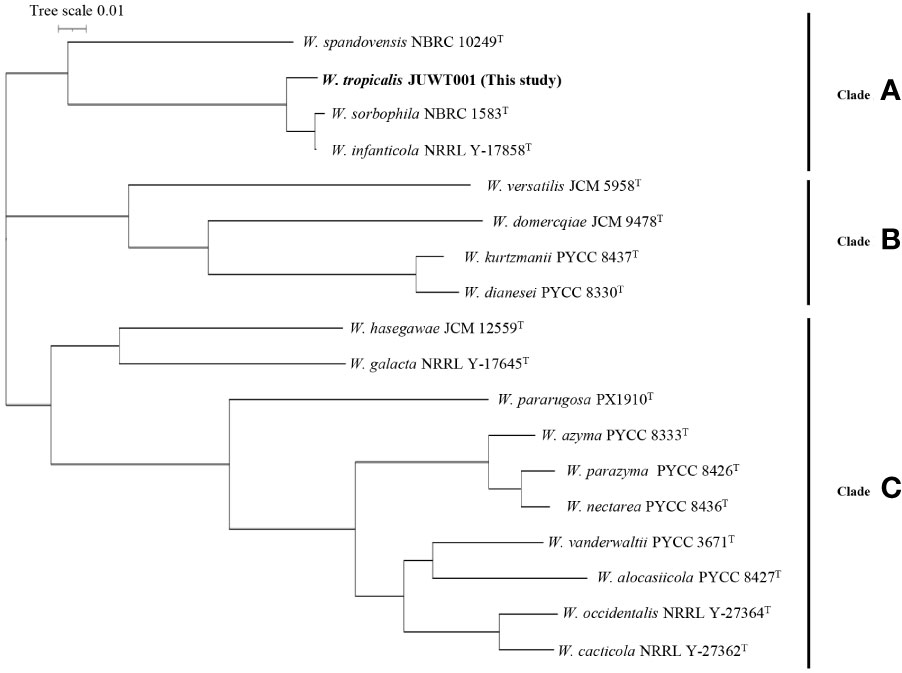
Figure 4 Phylogenetic tree of a clinical isolate and 17 type strains of Wickerhamiella species. The phylogenetic tree revealed three clades: Clades A, B, and C.
We investigated the feasibility in identifying W. tropicalis using MALDI-MS. The protein fraction and cell lysate mass spectra of W. tropicalis JUWT001 were examined by MALDI-MS along with the type strains of W. tropicalis, W. sorbophila, and W. spandovensis. The numbers of mass peaks detected in the protein fractions were higher than in the cell lysates for all strains tested (Supplementary Figure 1). Therefore, the protein fractions were used in the following study. As shown in Figure 5, we observed that the protein fraction mass spectra of W. tropicalis JUWT001 were almost identical to W. tropicalis type strain, but different from W. sorbophila and W. spandovensis type strains. In order to find useful biomarker peaks, we used the eMSTAT Solution software. Biomarker peaks were selected from those with a relative peak intensity ratio of 0.5% or higher which was detected in all five replicates. The number of major peaks were 19 for the W. tropicalis type strain, 18 for W. tropicalis JUWT001, 17 for W. sorbophila type strain and 16 for W. spandovensis type strain (data not shown). The 19 peaks of the W. tropicalis type strain were then used as biomarker peaks, and the matching peaks of the observed masses to the biomarker masses were judged from a tolerance within 500 ppm. Then, the homology to the reference peaks of W. tropicalis type strain were evaluated for W. tropicalis JUWT001, W. sorbophila type strain, and W. spandovensis type strain (Figure 5). As shown in Table 2, 15 mass peaks (79%) of W. tropicalis JUWT001, 6 mass peaks (32%) of W. sorbophila type strain, and 2 mass peaks (11%) of W. spandovensis type strain were detected at the same mass as the 19 mass peaks of W. tropicalis type strain. Compared with the W. tropicalis type strain and JUWT001, W. sorbophila and W. spandovensis type strain had unique peaks shown in Figure 5 and Table 2. The same peak (less than 500 ppm) at m/z 6,040 (peak No. 2) were observed in W. tropicalis JUWT001 and W. tropicalis type strain, whereas the different peaks at m/z 6,024 (peak No. 2-1) and m/z 6,031 (peak No. 2-2) were observed in W. sorbophila and W. spandovensis type strains, respectively. The same peaks at m/z 6,128 (peak No. 3) were observed in W. tropicalis and W. sorbophila, whereas the different peaks of m/z 6,157 (peak number 3-1) was observed in W. spandovensis. The same peaks at m/z 6,399 (Peak No. 4) were observed in W. tropicalis, whereas the different peaks at m/z 6,426 (peak No. 4-1) and m/z 6,487 (No. 4-2) were observed in W. sorbophila and W. spandovensis, respectively. These specific peaks were observed not only in the protein fraction, but also in the mass spectra of cell lysates cultured on SDA, PDA, and CHROM agar Candida medium (data not shown).
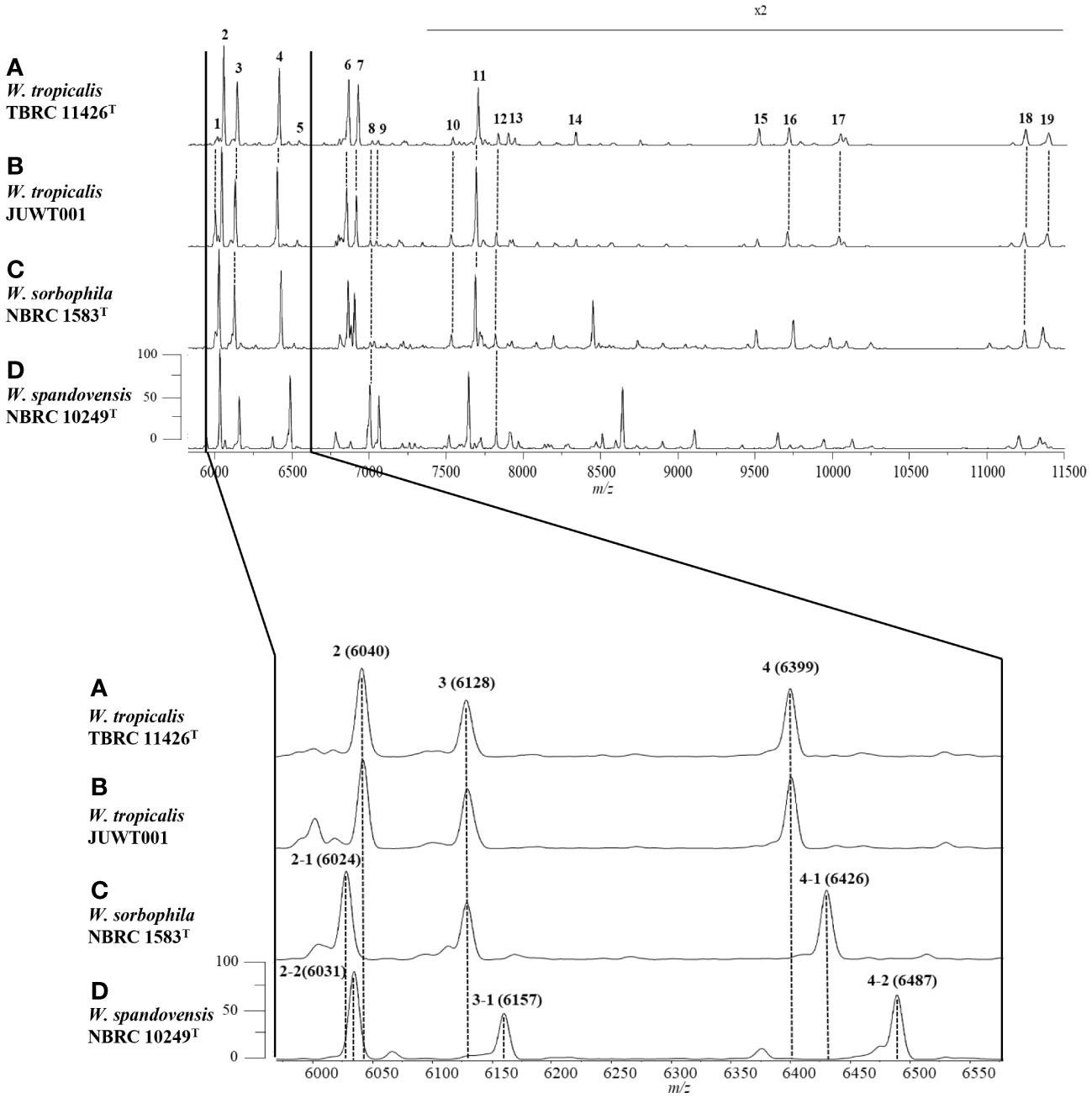
Figure 5 Representative mass spectra of (A) W. tropicalis TBRC 11426T, (B) JUWT001, (C) W. sorbophila NBRC 1583T, and (D) W. spandovensis NBRC 10249T in the protein fractions. Upper figure indicates mass spectra from m/z 6,000 to 11,500. Peak numbers correspond to those detailed in Table 2. Amplification of the m/z 6,000 to 6600 section of mass spectra results (lower figure) reveals small variations of m/z values for peaks 2, 3 and 4 for W. sorbophila and W. spandovensis, with reference to those of W. tropicalis strains, as detailed in Table 2.
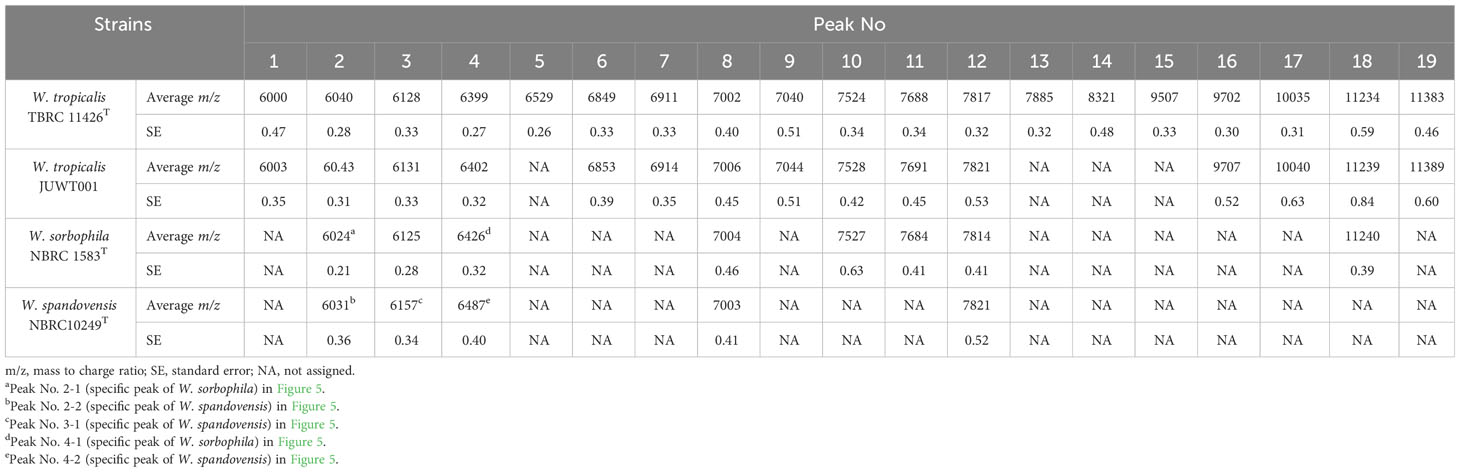
Table 2 Characteristics of the peaks of the protein fractions in the MALDI-MS analysis in the isolates of Wickerhamiella species.
To date, W. tropicalis strains were only isolated from a grease trap obtained in Thailand in 2017 and from sea surface microlayer in Taiwan in 2005 (Sakpuntoon et al., 2020). To our knowledge, this is the first report that W. tropicalis can cause human infectious disease.
The immunocompromised patients have possibilities of severe infections caused by environmental fungi including Wickerhamiella species. The mortality of non-Candida albicans species in pediatric patients with malignancies was 26.3% between 2015 and 2019 (Vasileiou et al., 2020). The previous studies reported that the 5 patients with W. pararugosa bloodstream infections were immunocompromised because of malignancy, sarcoma, surgery, and leukemia, and 2 of the 5 patients could not be cured (Nasri et al., 2023). Similarly for W. tropicalis infections, accurate and rapid diagnosis of infection in immunocompromised patients is critical.
Wickerhamialla species show intrinsically intermediate resistance/resistance to fluconazole. W. pararugosa clinical isolates causing bloodstream infections had MICs of 4 to 8 μg/mL against fluconazole (Taj-Aldeen et al., 2014; El Helou and Palavecino, 2017; Noni et al., 2020; Nasri et al., 2023). W. infanticola isolate from environmental sample had MICs of 4 μg/mL against fluconazole (Maciel et al., 2019). However, there are no MIC breakpoints for azole and echinocandin in Wickerhamiella species, and the breakpoints for Candida species in CLSI are used in clinical laboratories in many countries. Therefore, antifungal agents should be used with caution in the treatment of Wickerhamiella infections.
Furthermore, the identification of non-Candida species is important for initiating early effective antifungal therapy because these species, such as the Cryptococcus and Wickerhamiella species, have the potential to develop antifungal resistance. Previous studies on the identification of yeast isolates using MALDI-MS have shown that the identification accuracy of yeasts by MALDI-MS is not sufficient, with an identification accuracy of 96.8% for Candida species and 84.2% for non-Candida species (Zhang et al., 2014). On the other hand, the identification accuracy of Cryptococcus species by MALDI-MS was reported to have improved from 76.0% using a commercial database, to 100% by detecting five new biomarker peaks (Zvezdanova et al., 2020). Thus, it is essential for clinical laboratories to establish biomarker peaks and fingerprinting libraries for rare yeast species, including Wickerhamiella and Cryptococcus. The three MALDI-MS peaks specific for W. tropicalis obtained in this study may serve as an indicator for the identification of rare fungal species, including Wickerhamiella species, in routine microbiological examinations.
This study has the following limitations. First, this study was conducted at a single institution. The results should be evaluated and discussed at other study sites in the future. Second, only one sample was obtained in this study because W. tropicalis is rare to obtain from humans. Further data on clinical isolates of Wickerhamiella species should be collected. Third, the lack of genomic data on W. tropicalis precluded the use of theoretical protein masses in this study. It is hoped that genomic data will be constructed in the future.
In conclusion, MALDI-MS analysis has the potential for rapid identification of Wickerhamiella species as well as other fungus infections. It is important to establish the database using MALDI-MS for identification of yeast-like fungi causing severe infections in clinical laboratories.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving humans were approved by the Ethical Committee of Juntendo University. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by- product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements. Written informed consent was obtained from the minor(s)’ legal guardian/next of kin for the publication of any potentially identifiable images or data included in this article.
ST: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. KT: Methodology, Visualization, Writing – original draft, Writing – review & editing. JF: Resources, Writing – original draft. MF: Resources, Writing – original draft. MS: Resources, Writing – original draft. YF: Resources, Writing – original draft. YS: Writing – original draft. TaK: Resources, Writing – original draft. MC: Resources, Writing – original draft. MW: Resources, Writing – original draft. YH: Writing – original draft. TM: Writing – original draft. TN: Writing – original draft. TeK: Writing – original draft. TT: Conceptualization, Funding acquisition, Project administration, Supervision, Visualization, Writing – original draft, Writing – review & editing. YT: Project administration, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the Japan Society for the Promotion of Science (Grant number: 21K07031 to TT and 22K15675 to ST).
The authors would like to thank the staff of Juntendo University Hospital for their contribution in collecting data.
The Department of MALDI-TOF MS Practical Application Research at Juntendo University Graduate School of Medicine has been endowed partially by Shimadzu Corp. Kyoto, Japan to develop and validate new diagnostic technology and to conduct academic research through collaborations. ST, KT, TN, TK, and YT belong to the Department of MALDI-TOF MS Practical Application Research. Shimadzu Corp. provided MALDI-8020 and reagents for MALDI-MS analysis free of cost to YT. Author KT is employed by Shimadzu Corp. The study was performed by scientifically proper methods without any bias.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1361432/full#supplementary-material
Barata, A., Seborro, F., Belloch, C., Malfeito-Ferreira, M., Loureiro, V. (2008). Ascomycetous yeast species recovered from grapes damaged by honeydew and sour rot. J. Appl. Microbiol. 104, 1182–1191. doi: 10.1111/j.1365-2672.2007.03631.x
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., Parks, D. H. (2022). GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics 38, 5315–5316. doi: 10.1093/bioinformatics/btac672
CLSI (2017). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard-4th ed CLSI Document M27-A4. (Wayne, PA: Clinical and Laboratory Standards).
de Vega, C., Albaladejo, R. G., Guzmán, B., Steenhuisen, S. L., Johnson, S. D., Herrera, C. M., et al. (2017). Flowers as a reservoir of yeast diversity: description of Wickerhamiella nectarea f.a. sp. nov., and Wickerhamiella natalensis f.a. sp. nov. from South African flowers and pollinators, and transfer of related Candida species to the genus Wickerhamiella as new combinations. FEMS Yeast. Res. 17. doi: 10.1093/femsyr/fox054
Eigner, U., Holfelder, M., Oberdorfer, K., Betz-Wild, U., Bertsch, D., Fahr, A. M. (2009). Performance of a matrix-assisted laser desorption ionization-time-of-flight mass spectrometry system for the identification of bacterial isolates in the clinical routine laboratory. Clin. Lab. 55, 289–296.
El Helou, G., Palavecino, E. (2017). Candida pararugosa: first reported bloodstream infection in an adult. Cureus 9, e1283. doi: 10.7759/cureus.1283
Favre, B., Didmon, M., Ryder, N. S. (1999). Multiple amino acid substitutions in lanosterol 14alpha-demethylase contribute to azole resistance in Candida albicans. Microbiol. (Reading). 145, 2715–2725. doi: 10.1099/00221287-145-10-2715
Giammanco, G. M., Melilli, D., Pizzo, G. (2004). Candida pararugosa isolation from the oral cavity of an Italian denture wearer. Res. Microbiol. 155, 571–574. doi: 10.1016/j.resmic.2004.04.003
Kumar, S., Kumar, A., Roudbary, M., Mohammadi, R., Černáková, L., Rodrigues, C. F. (2022). Overview on the infections related to rare candida species. Pathogens 11. doi: 10.3390/pathogens11090963
Lachance, M. A., Dobson, J., Wijayanayaka, D. N., Smith, A. M. (2010). The use of parsimony network analysis for the formal delineation of phylogenetic species of yeasts: Candida apicola, Candida azyma, and Candida parazyma sp. nov., cosmopolitan yeasts associated with floricolous insects. Antonie. Van. Leeuwenhoek. 97, 155–170. doi: 10.1007/s10482-009-9399-3
Lachance, M. A., Rosa, C. A., Starmer, W. T., Schlag-Edler, B., Barker, J. S., Bowles, J. M. (1998). Wickerhamiella australiensis, Wickerhamiella cacticola, Wickerhamiella occidentalis, Candida drosophilae and Candida lipophila, five new related yeast species from flowers and associated insects. Int. J. Syst. Bacteriol. 48 Pt 4, 1431–1443. doi: 10.1099/00207713-48-4-1431
Maciel, N. O., Johann, S., Brandão, L. R., Kucharíková, S., Morais, C. G., Oliveira, A. P., et al. (2019). Occurrence, antifungal susceptibility, and virulence factors of opportunistic yeasts isolated from Brazilian beaches. Mem. Inst. Oswaldo. Cruz. 114, e180566. doi: 10.1590/0074-02760180566
Mizukoshi, F., Kobayashi, N., Kirikae, F., Ohta, K., Tsuyuguchi, K., Yamada, N., et al. (2021). Molecular epidemiology of drug-resistant mycobacterium tuberculosis in Japan. mSphere 6, e0097820. doi: 10.1128/mSphere.00978-20
Nakagawa, Y., Robert, V., Kawarazaki, J., Epping, W., Smith, M. T., Poot, G. A., et al. (2004). Recurrent isolation of an uncommon yeast, Candida pararugosa, from a sarcoma patient. Med. Mycol. 42, 267–271. doi: 10.1080/13693780310001597674
Nakase, T., Jindamorakot, S., Limtong, S., Am-in, S., Kawasaki, H., Imanishi, Y., et al. (2007). Candida kazuoi sp. nov. and Candida hasegawae sp. nov., two new species of ascomycetous anamorphic yeasts isolated from insect frass in Thailand. J. Gen. Appl. Microbiol. 53, 239–245. doi: 10.2323/jgam.53.239
Nasri, E., Vaezi, A., Shelerangkon, M., Rizi, M. H., Ghafel, S., Abbasi, K., et al. (2023). Catheter-associated blood stream infections due to Candida pararugosa in a patient with acute myeloid leukemia: A case report. Braz. J. Microbiol. 54, 891–895. doi: 10.1007/s42770-023-00985-5
Noni, M., Stathi, A., Velegraki, A., Malamati, M., Kalampaliki, A., Zachariadou, L., et al. (2020). Rare invasive yeast infections in greek neonates and children, a retrospective 12-year study. J. Fungi. (Basel). 6. doi: 10.3390/jof6040194
Papp, C., Kocsis, K., Tóth, R., Bodai, L., Willis, J. R., Ksiezopolska, E., et al. (2018). Echinocandin-induced microevolution of candida parapsilosis influences virulence and abiotic stress tolerance. mSphere 3. doi: 10.1128/mSphere.00547-18
Pravin Charles, M. V., Kali, A., Joseph, N. M. (2015). Performance of chromogenic media for Candida in rapid presumptive identification of Candida species from clinical materials. Pharmacognosy. Res. 7, S69–S73. doi: 10.4103/0974-8490.150528
Saary, P., Mitchell, A. L., Finn, R. D. (2020). Estimating the quality of eukaryotic genomes recovered from metagenomic analysis with EukCC. Genome Biol. 21, 244. doi: 10.1186/s13059-020-02155-4
Sakpuntoon, V., Angchaun, J., Boonmak, C., Chang, C. F., Liu, S. M., Lee, C. F., et al. (2020). Wickerhamiella osmotolerans sp. nov. and Wickerhamiella tropicalis sp. nov., novel ascomycetous yeast in the family Wickerhamiellaceae. Int. J. Syst. Evol. Microbiol. 70, 2596–2601. doi: 10.1099/ijsem.0.004075
Spettel, K., Barousch, W., Makristathis, A., Zeller, I., Nehr, M., Selitsch, B., et al. (2019). Analysis of antifungal resistance genes in Candida albicans and Candida glabrata using next generation sequencing. PloS One 14, e0210397. doi: 10.1371/journal.pone.0210397
Taj-Aldeen, S. J., AbdulWahab, A., Kolecka, A., Deshmukh, A., Meis, J. F., Boekhout, T. (2014). Uncommon opportunistic yeast bloodstream infections from Qatar. Med. Mycol. 52, 552–556. doi: 10.1093/mmycol/myu016
Talapko, J., Juzbašić, M., Matijević, T., Pustijanac, E., Bekić, S., Kotris, I., et al. (2021). Candida albicans-the virulence factors and clinical manifestations of infection. J. Fungi. (Basel). 7. doi: 10.3390/jof7020079
Teramoto, K., Okubo, T., Yamada, Y., Sekiya, S., Iwamoto, S., Tanaka, K. (2019). Classification of Cutibacterium acnes at phylotype level by MALDI-MS proteotyping. Proc. Jpn. Acad. Ser. B. Phys. Biol. Sci. 95, 612–623. doi: 10.2183/pjab.95.042
van der Walt, J. P., Liebenberg, N. V. (1973). The yeast genus Wickerhamiella gen. nov. (Ascomycetes). Antonie. Van. Leeuwenhoek. 39, 121–128. doi: 10.1007/bf02578847
Vasileiou, E., Paisiou, A., Tsipou, C., Pourtsidis, A., Galani, V., Katzilakis, N., et al. (2020). Candidemia in children with Malignancies: report from the infection working group of the hellenic society of pediatric hematology-oncology. J. Fungi. (Basel). 6. doi: 10.3390/jof6040276
Vu, D., Groenewald, M., Szöke, S., Cardinali, G., Eberhardt, U., Stielow, B., et al. (2016). DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 85, 91–105. doi: 10.1016/j.simyco.2016.11.007
Wang, S. A., Jia, J. H., Bai, F. Y. (2008). Candida alocasiicola sp. nov., Candida hainanensis sp. nov., Candida heveicola sp. nov. and Candida musiphila sp. nov., novel anamorphic, ascomycetous yeast species isolated from plants. Antonie. Van. Leeuwenhoek. 94, 257–265. doi: 10.1007/s10482-008-9238-y
Zhai, L., Zhou, Y., Wu, Y., Jin, Y., Zhu, Q., Gao, S., et al. (2021). Isolation and identification of Candida tropicalis in sows with fatal infection: a case report. BMC Vet. Res. 17, 108. doi: 10.1186/s12917-021-02821-0
Zhang, L., Xiao, M., Wang, H., Gao, R., Fan, X., Brown, M., et al. (2014). Yeast identification algorithm based on use of the Vitek MS system selectively supplemented with ribosomal DNA sequencing: proposal of a reference assay for invasive fungal surveillance programs in China. J. Clin. Microbiol. 52, 572–577. doi: 10.1128/jcm.02543-13
Zvezdanova, M. E., Arroyo, M. J., Méndez, G., Guinea, J., Mancera, L., Muñoz, P., et al. (2020). Implementation of MALDI-TOF mass spectrometry and peak analysis: application to the discrimination of cryptococcus neoformans species complex and their interspecies hybrids. J. Fungi. (Basel). 6. doi: 10.3390/jof6040330
Keywords: Wickerhamiella tropicalis, blood culture, yeast, MALDI-MS, whole-genome
Citation: Takei S, Teramoto K, Fujimura J, Fujiwara M, Suzuki M, Fukui Y, Sekiguchi Y, Kawakami T, Chonan M, Wakita M, Horiuchi Y, Miida T, Naito T, Kirikae T, Tada T and Tabe Y (2024) Isolation and identification of Wickerhamiella tropicalis from blood culture by MALDI-MS. Front. Cell. Infect. Microbiol. 14:1361432. doi: 10.3389/fcimb.2024.1361432
Received: 26 December 2023; Accepted: 20 February 2024;
Published: 06 March 2024.
Edited by:
Alin Laurentiu Tatu, Dunarea de Jos University, RomaniaReviewed by:
Maria Dolores Moragues, University of the Basque Country, SpainCopyright © 2024 Takei, Teramoto, Fujimura, Fujiwara, Suzuki, Fukui, Sekiguchi, Kawakami, Chonan, Wakita, Horiuchi, Miida, Naito, Kirikae, Tada and Tabe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tatsuya Tada, dC10YWRhQGp1bnRlbmRvLmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.