
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 18 March 2024
Sec. Bacteria and Host
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1360438
Background: The Philippines bears health and economic burden caused by high dengue cases annually. Presently, the Philippines still lack an effective and sustainable vector management. The use of Wolbachia, a maternally transmitted bacterium, that mitigate arbovirus transmission has been recommended. Cytoplasmic incompatibility and viral blocking, two characteristics that make Wolbachia suitable for vector control, depend on infection prevalence and density. There are no current Wolbachia release programs in the Philippines, and studies regarding the safety of this intervention. Here, we screened for Wolbachia in Aedes aegypti collected from Metropolitan Manila, Philippines. We designed location-specific primers for qPCR to test whether this improved Wolbachia detection in Ae. aegypti. We explored if host sex and Wolbachia strain could be potential factors affecting Wolbachia density.
Methods: Ae. aegypti mosquitoes (n=429) were screened for natural Wolbachia by taqman qPCR using location-specific Wolbachia surface protein primers (wspAAML) and known 16S rRNA primers. Samples positive for wspAAML (n=267) were processed for Sanger sequencing. We constructed a phylogenetic tree using IQ-TREE 2 to further characterize Wolbachia present in the Philippine Ae. aegypti. We then compared Wolbachia densities between Wolbachia groups and host sex. Statistical analyses were done using GraphPad Prism 9.0.
Results: Wolbachia prevalence for 16S rRNA (40%) and wspAAML (62%) markers were high. Wolbachia relative densities for 16S rRNA ranged from −3.84 to 2.71 and wspAAML from −4.02 to 1.81. Densities were higher in male than female mosquitoes. Wolbachia strains detected in Ae. aegypti clustered into supergroup B. Some 54% (123/226) of these sequences clustered under a group referred to here as “wAegML,” that belongs to the supergroup B, which had a significantly lower density than wAegB/wAlbB, and wAlbA strains.
Conclusion: Location-specific primers improved detection of natural Wolbachia in Ae. aegypti and allowed for relative quantification. Wolbachia density is relatively low, and differed between host sexes and Wolbachia strains. An economical way of confirming sporadic or transient Wolbachia in Ae. aegypti is necessary while considering host sex and bacterial strain.
The Philippines continues to experience the health burden caused by high dengue cases, ranking number one in Asia with 17,630 deaths last March 2021 (Edillo et al., 2015, 2022; Ong et al., 2022). The persistent high cases annually prompted the Philippine government to establish the National Dengue Prevention and Control Program with vector surveillance and management as one of the key targets (Dengue Prevention and Control Program | Department of Health website, n.d.). Aedes aegypti mosquito is the primary vector of Dengue in the Philippines owing to its adaptability in changing environments (Edillo et al., 2022). To mitigate infections, recommendations for vector management including the reduction of breeding sites, and improvement of water systems have been proposed. More importantly, Ong et al. (2022) highlighted the need for a more sustainable approach involving the use of the bacterium Wolbachia (Ong et al., 2022). Currently, the Philippines still lacks local studies that could provide a baseline information for assisting future mass release implementations (Ong et al., 2022).
Wolbachia was discovered in Culex pipiens (Hertig and Wolbach, 1924) and may induce cytoplasmic incompatibility (CI), wherein gametes fail to produce viable offspring owing to incompatible Wolbachia infection (Yen and Barr, 1973). Wolbachia also naturally infect arthropods such as Drosophila spp (Min and Benzer, 1997; Brownlie et al., 2009; McMeniman et al., 2009; Osborne et al., 2009), Anopheles spp (Walker et al., 2021), and Aedes albopictus (Dutton and Sinkins, 2004; Yang et al., 2021). Different Wolbachia strains, i.e. wMelpop, wMel, and wAlbB, from these natural hosts can exert life-shortening effects (Min and Benzer, 1997; McMeniman et al., 2009), or confer nutritional benefits (Brownlie et al., 2009), efficient maternal transmission (Liu and Li, 2021), and antiviral protection (Osborne et al., 2009; Liu and Li, 2021; Reyes et al., 2021). CI, maternal transmission, and the antiviral effects of Wolbachia against arthropod-borne viruses all contribute to the efficiency of the mass release of Wolbachia-transinfected Ae. aegypti (Indriani et al., 2020; Ahmad et al., 2021). Currently, only D. melanogaster-derived wMel and Ae. albopictus-derived wAlbB are used in release programs (Indriani et al., 2020; Ahmad et al., 2021; Ross et al., 2022). Release programs can either result in mosquito population suppression or replacement (Ross et al., 2020). The former involves the release of Wolbachia-infected males that induce CI with wild female mosquitoes, whereas both CI-inducing male mosquitoes and females carrying a pathogen-blocking strain that is maternally transmitted are released in the latter (Ross et al., 2020).
Unlike other host species, studies on Ae. aegypti have mostly revealed the absence of natural Wolbachia (Gloria-Soria et al., 2018; Goindin et al., 2018). Novel Wolbachia transinfection in mosquitoes facilitates population suppression and replacement (Ross et al., 2020). However, the occasional presence of Wolbachia in Ae. aegypti populations without any preceding Wolbachia have been reported. These were found in the USA (Coon et al., 2016), Mexico (Kulkarni et al., 2019), Panama (Bennett et al., 2019), India (Balaji et al., 2019), Malaysia (Teo et al., 2017), Thailand (Thongsripong et al., 2018), China (Zhang et al., 2022), Taiwan (Chao and Shih, 2023), and Philippines (Carvajal et al., 2019; Regilme et al., 2021; Muharromah et al., 2023). Thus, understanding biological factors that may influence the variable presence of natural Wolbachia in Ae. aegypti is necessary. More so, finding an economical way to improve detection of Wolbachia could help initial surveillance in countries where mass release programs could be implemented. This is particularly important in low-income countries like the Philippines, gravely affected by arboviral diseases.
Natural Wolbachia hosts are characterized by consistently high Wolbachia prevalence (Inácio da Silva et al., 2021). Culex pipiens, Culex quinquefasciatus, Ae. albopictus, Anopheles moucheti, and Anopheles demeilloni can exhibit 90% to 100% Wolbachia prevalence (Bergman and Hesson, 2021; Inácio da Silva et al., 2021). In Anopheles gambiae and Ae. aegypti, prevalence is relatively low and can vary from 8–24% and 4.3–58%, respectively (Inácio da Silva et al., 2021). The detection of natural Wolbachia in Ae. aegypti remains inconclusive. The reported presence of Wolbachia in the said mosquito host seems to be sporadic, transient, and low in prevalence making it difficult to quantify. Thus, more studies regarding prevalence, biological factors, and mechanisms are needed to understand the nature of natural Wolbachia sporadically found in different populations of Ae. aegypti.
In general, Wolbachia prevalence and density influence maternal transmission fidelity and pathogen blocking extent (Ikeda et al., 2003; Unckless et al., 2009; Hoffmann et al., 2014; López-Madrigal and Duarte, 2019; Liu and Li, 2021; Shropshire et al., 2021; Walker et al., 2021). Achieving high and stable Wolbachia introgression into communities via transinfected Ae. aegypti depends on the number of mosquitoes that become infected with the endosymbiont (Indriani et al., 2020; Ahmad et al., 2021). Other hosts like Drosophila spp. and the moth Ephestia kuehniella exhibit varying CI levels relative to Wolbachia density; i.e., mostly high CI with high density (Hoffmann et al., 1986; Bourtzis et al., 1996; Ikeda et al., 2003; Unckless et al., 2009; Turelli et al., 2018). However, one study found that high wMel density in Drosophila simulans did not translate into elevated CI but strengthened host immune expression (Shropshire et al., 2021). Density also has been linked to virus inhibition (Bian et al., 2010; Frentiu et al., 2010; Lu et al., 2020). For example, higher wAlbB and wMelpop-CLA densities in Ae. albopictus was correlated with stronger blocking in vitro (Frentiu et al., 2010; Lu et al., 2020). Likewise, Ae. aegypti mosquitoes transinfected with wAlbB inhibited dengue replication and transmission by regulating immunity and longevity, respectively (Bian et al., 2010).
Wolbachia density is host sex-dependent. Density is reported higher in adult female than male hosts, as observed in D. simulans as well as the planthoppers Laodelphax striatellus and Sogatella furcifera (Noda et al., 2001; Correa and Ballard, 2012). Wolbachia densities are also more stable in female than male insect hosts and decline with age in males (Noda et al., 2001; Unckless et al., 2009; Correa and Ballard, 2012). However, males exhibit higher Wolbachia density than females in some insects such as Diaphorina citri (Hoffmann et al., 2014). Wolbachia density and prevalence in host sex also vary with strain in Ae. albopictus (Tortosa et al., 2010; Yang et al., 2022). wAlbB density was higher in males than females, whereas wAlbA density was higher in females than males. Wolbachia wAlbA density in male Ae. albopictus also decreased with age (Tortosa et al., 2010). Further, a higher prevalence of wAlbA and wAlbB coinfection was observed in females than males, in which infection prevalence varied according to wAlb strain (Tortosa et al., 2010; Yang et al., 2022).
Wolbachia density is also determined by Wolbachia strain. Different Wolbachia strains vary in density (Dutton and Sinkins, 2004; Hu et al., 2020), viral inhibition (Osborne et al., 2009; Martinez et al., 2017), and CI (Liang et al., 2020) strength in hosts. For instance, wAlbB resides in Ae. albopictus at a higher density than wAlbA (Dutton and Sinkins, 2004; Hu et al., 2020). Further, Wolbachia strains wMel, wAu, wRi, and wNo that exist at high density in D. simulans confer viral protection, unlike wHa (Osborne et al., 2009). Lastly, strains wMelpop, wMel, and wAlbB exhibit stable and high-density infection in transinfected Ae. aegypti, despite being transferred from their natural hosts D. melanogaster and Ae. albopictus (McMeniman and O’Neill, 2010; Fraser et al., 2017; Ross, 2021).
Currently, information on the presence of natural Wolbachia in Ae. aegypti varies and knowledge on density in relation to biological factors is limited (Teo et al., 2017; Bennett et al., 2019; Carvajal et al., 2019). Detection of Wolbachia also differs among states and cities within a country (Chen et al., 2016; Schuler et al., 2018; Zhang et al., 2022). Previous studies on prevalence have used general primers based on Wolbachia sequences from established natural hosts (Teo et al., 2017; Bennett et al., 2019; Carvajal et al., 2019; Kulkarni et al., 2019); thus primers currently used may not detect rare and low-density strains (Marcon et al., 2011; Simões et al., 2011). These limitations suggest that validation methods are necessary to perform initial surveillance targeting specific local Wolbachia populations. Lastly, Wolbachia density in naturally infected hosts is regulated by multiple factors; e.g., host genotype (Mouton et al., 2006), host sex (Tortosa et al., 2010; Mejia et al., 2022), and Wolbachia strain (Dutton and Sinkins, 2004; Hu et al., 2020). However, no study has clarified the biological factors that affect Wolbachia density in Ae. aegypti.
The present study aims to investigate if locally designed primers for qPCR could help validate the presence and density of natural Wolbachia in local mosquito populations. We also determined if density could be influenced by host sex and Wolbachia strain. We did this by utilizing locally designed Wolbachia surface protein (wsp) primers suitable for Ae. aegypti collected from Metro Manila, Philippines. We then used these primers to quantify Wolbachia density in 429 individual mosquitoes and compared against a known general primer used for conventional PCR. Next, we examined Wolbachia density between Wolbachia host sex and strains in Ae. aegypti populations. We hypothesized that the presence of sporadic, low density natural Wolbachia in Ae. aegypti can be detected and quantified by using primers designed for local mosquito populations and that Wolbachia density differs depending on Wolbachia host sex and strain. An economical way to conduct initial surveillance of Wolbachia in Ae. aegypti is necessary especially in low-income countries like the Philippines, where an effective vector control strategy is needed.
We used DNA samples extracted from Ae. aegypti adult mosquitoes collected in the National Capital Region (Manila) of the Philippines; these samples were previously used for Wolbachia detection via conventional PCR (Carvajal et al., 2019) and ddRAD-Seq (Muharromah et al., 2023). Wolbachia was detected in 11.9% (80/672) of these samples by PCR (Carvajal et al., 2019) and sequence reads of 26,299 (wAlbA) and 43,778 (wAlbB) were mapped across the entire Wolbachia genome out of 146,239,637 filtered reads obtained by ddRAD-Seq (Muharromah et al., 2023). Thus, the samples were considered suitable for validating primer incompatibility. Each individual mosquito was previously screened and processed for DNA extraction as described by Carvajal et al. (2019). The total DNA of individual mosquitoes was extracted using a Blood and Tissue DNEasy Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol with slight modifications (Crane, 2013). All samples were stored as DNA at −80°C for long term preservation. Of 672 individual mosquitoes used by Carvajal et al. (2019), we selected 429 samples based on sufficient volume and DNA concentration for downstream assays.
Data on conventional PCR results were obtained from the study of Carvajal et al. (2019) (Supplementary Data) and used as a baseline reference in the present study in relation to Wolbachia prevalence and density detected with the newly designed primers. Briefly, Carvajal et al. (2019) used two known markers targeting the 16S rRNA gene, which has a slow evolutionary rate, and another marker targeting the highly variable Wolbachia surface protein (wsp), which is suitable for strain identification (O’Neill et al., 1992; Zhou et al., 1998). The sequences of 16S rRNA and wsp Wolbachia-specific primers were as follows: Wspecf (5′-GAA GAT AAT GAC GGT ACT CAC-3′) and Wspecr (5′-AGC TTC GAG TGA AAC CAA TTC-3′) (O’Neill et al., 1992); wsp 81F (5′-TGG TCC AAT AAG TGA TGA AGA AAC-3′) and wsp 691R (5′-AAA AAT TAA ACG CTA CTC CA-3′) (Zhou et al., 1998). PCR thermocycling conditions were conducted according to the published protocol (Carvajal et al., 2019).
Most wsp primers designed for Wolbachia detection are strain-specific (Zhou et al., 1998; Dutton and Sinkins, 2004). Thus, we developed new primers specific for our local samples to consider for primer incompatibility and to quantify Wolbachia density. First, we obtained 118 wsp sequences from the Ae. aegypti samples of Carvajal et al. (2019) (GenBank popset 1712729902). Next, a multiple sequence alignment was performed using MUSCLE, and the results were visualized in Codon Code Aligner version 1.2.4 (https://www.codoncode.com/aligner/). The consensus sequence produced from the alignment was then inputted into Primer-BLAST (Ye et al., 2012) to design wsp primers targeting the Ae. aegypti samples. Primer-BLAST generated five primer pairs (Supplementary Table 1), which were first validated via gradient conventional PCR using a known Cx. quinquefasciatus positive sample. Among the primer pairs, primers wspAAML 01 and wspAAML 05 were selected for further optimization because they exhibited the correct band size of target markers without nonspecific binding in the sample (Supplementary Figure 1). To select the most suitable wspAAML primer pair for downstream analysis, we determined the optimized annealing temperature and primer concentration for both pairs (Supplementary Figure 2). We selected wspAAML 05, given that its PCR efficiency (Supplementary Figure 3) fell within the standard MIQE guideline of ≥90% (Bustin et al., 2009).
To quantify Wolbachia density, Taqman qPCR targeting both 16S rRNA and wspAAML was conducted. Wolbachia quantification was performed using a well-established primer set targeting the 16S rRNA marker (16SF 5′-AGT GAA GA A GGC CTT TGG G-3′; 16SR 5′-CAC GGA GTT AGC CAG GAC TTC-3′) but with a modification of the fluorescent dye of the probe (Fraser et al., 2020). Instead of LC640, we used TET as the reporter dye and BHQ1 as its quencher (5′TET-CTG TGA GTA CCG TCA TTA TCT TCC TCA CT-BHQ13′). Wolbachia was also confirmed using the newly designed Wolbachia surface protein (wsp) primers (wspAAML F 5′-AGC ATC TTT TAT GGC TGGT GG-3′; wspAAML R 5′- AAT GCT GCC ACA CTG TTT GC-3′; wspAAML probe 5′FAM-ACG ACG TTG GTG GTG CAA CAT TTG C-TAMRA3′) with the Ae. aegypti ribosomal protein S17 (RPS17) gene as a reference gene (17SF 5′-TCC GTG GTA TCT CCA TCA AGC T-3′; 17SR 5′- CAC TTC CGG CAC GTA GTT GTC-3′; 17S probe 5′HEX- CAG GAG GAG GAA CGT GAG CGC AG-BHQ13′) (Frentiu et al., 2010). In total, 429 individual mosquitoes were screened for the presence of Wolbachia using qPCR with a cut-off Cq value of 35. The cut-off value was set from a qPCR experiment of Cx. quinquefasciatus samples representing true natural Wolbachia infection and three replicates of the no-template control for each target gene; negative detections of no template controls were confirmed using gel electrophoresis showing no bands, hence no true amplification (Supplemental Figure 4).
All singleplex PCR reactions were performed in a final volume of 10 µl containing 5 µl of 2x iTaq Universal Probes Supermix (Bio Rad) with 0.3 µM RPS17 primers, 0.2 µM 16S rRNA primers, or 0.5 µM wsp primers and 0.2 µM, 0.15 µM, or 0.3 µM of their corresponding TaqMan probes, respectively, with nuclease-free water added to reach the final volume. The following thermal profile was used for both RPS17 and 16S rRNA with a CFX96 touch deep well real-time PCR detection system (Bio-Rad Tokyo, Japan): an initial polymerase activation at 95°C for 30 s followed by 40 cycles of denaturation at 95°C for 5 s, and combined annealing/extension at 60°C for 10 s. The wsp thermal profile included an initial polymerase activation at 95°C for 2 min followed by 40 cycles of denaturation at 95°C for 5 s, and combined annealing/extension at 58.8°C for 30 s. Each PCR amplification included a Wolbachia-infected Cx. quinquefasciatus positive control and a no-template control. Wolbachia density was derived from the Cq values expressed as the relative density of wspAAML normalized to single-copy RPS17 (Livak and Schmittgen, 2001). This was used to compare density between host sex and Wolbachia strains. Relative densities reported in this study were log-transformed (log10). Following detection, qPCR-confirmed wspAAML-positive products were cleaned using a mixture of alkaline phosphatase (TaKaRa) and exonuclease I (TaKaRa). The cleaned samples were subjected to Sanger sequencing for strain identification.
Wolbachia phylogeny was inferred using the maximum-likelihood criterion. For this analysis, we used Ae. aegypti samples in which the wsp gene was detected via qPCR (n = 226). We also obtained additional wsp sequences from other host species, e.g., Aedes spp., Anopheles spp., Culex spp., and others indicated as reference sequences (n = 511), from NCBI GenBank (Table 1). In total, 737 wsp sequences were aligned using MUSCLE in CodonCode Aligner version 1.2.4 (https://www.codoncode.com/aligner/) and then trimmed to a final length of 103 nucleotide bases. The sequences containing 103 nucleotide bases correspond to the homologous region across all sequences. Using DNASp version 6.12.03 (Rozas et al., 2017), we obtained 102 haplotypes, and we subjected the representative sequences of each haplotype to phylogenetic analysis. Tree reconstruction was conducted using only wsp given the high evolutionary rate of the gene; i.e., its suitability for strain identification (Zhou et al., 1998; Ren et al., 2020). IQ-TREE 2 (http://iqtree.org) (Minh et al., 2020) was used where the appropriate substitution model was first identified through ModelFinder implemented as a function of the software (Kalyaanamoorthy et al., 2017), from which TPM2+G4 was selected as the best-fit model. We set the ultrafast bootstrap approximation (UFBoot) in IQ-TREE to 1,000 iterations, the minimum correlation coefficient to 0.99, and the other parameters to their default settings (Hoang et al., 2018). For visualization and annotation, we used iTOL (https://itol.embl.de/) (Letunic and Bork, 2021).
Mann Whitney tests were used to determine statistical differences between Wolbachia densities of male and female mosquitoes as well as those between the densities of samples detected as positive or negative using conventional PCR. To compare densities between Wolbachia strains, Dunn’s multiple comparison test was performed. Statistical calculations were conducted in GraphPad Prism version 9.2.0 for Windows (www.graphpad.com), and p -values of ≤0.01 or ≤0.0001 were considered statistically significant.
Screening of Wolbachia in Ae. aegypti using 16S rRNA and wsp qPCR revealed an overall prevalence of 40% (172/429) and 62% (267/429) in the mosquito population, respectively (Table 2). Wolbachia density expressed in logarithmic scale refers to the relative abundance of the target gene normalized to the reference gene, RPS17 (Livak and Schmittgen, 2001). Thus, the median relative Wolbachia density was −1.99 and −2.09 for 16S rRNA and wspAAML qPCR assays, respectively. The relative Wolbachia densities of 16S rRNA- and wspAAML-positive samples ranged from −3.84 to 2.17 and −4.02 to 1.81, respectively.
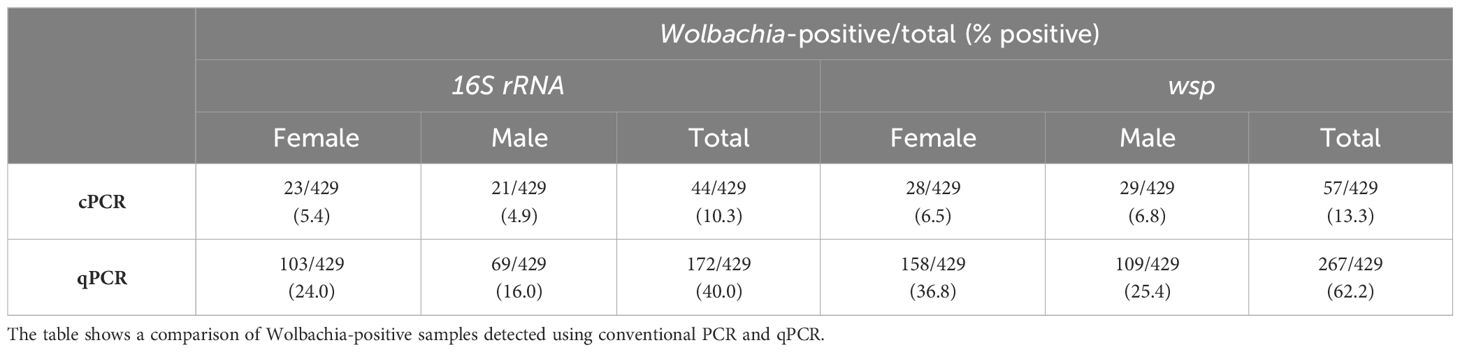
Table 2 Infection prevalence of natural Wolbachia in Ae. aegypti based on conventional PCR (cPCR) and qPCR.
Comparing our qPCR results with the conventional PCR results of Carvajal et al. (2019), we found that 91% (40/44) of the mosquitoes positive for 16S rRNA in conventional PCR showed the same result in our qPCR, whereas only 9% equivalent to 4 out of 44 of the mosquito samples previously detected as positive by standard PCR were detected as negative via qPCR. Of the 385 samples confirmed as negative for 16S rRNA via conventional PCR, 34% (132/385) were positive according to qPCR, whereas 66% (253/385) were consistent with negative conventional PCR detection. Regarding wsp, 100% (57/57) of mosquito samples that were positive according to conventional PCR were also positive in our qPCR. Conventional PCR wsp-negative samples exhibited an infection prevalence of 57% positive (213/372) and 43% negative (159/372) detection according to qPCR.
In an attempt to explain the contrasting negative conventional PCR and positive qPCR Wolbachia detection results, we compared the relative Wolbachia densities expressed in logarithmic scales, between samples found to be either Wolbachia-negative or positive via conventional PCR by Carvajal et al. (2019) (Figure 1). We found an approximately 30-fold higher relative Wolbachia density in 16S rRNA (median = −0.8250) and wspAAML (median = −0.8550) Wolbachia-positive mosquitoes compared with the conventional PCR-negative samples for 16S rRNA (p < 0.001; median = −2.325) and wsp (p < 0.001; median = −2.250). Finally, it is worth mentioning that in qPCR alone, only 21% (91/429) of samples were positive for both 16S rRNA and wspAAML markers.
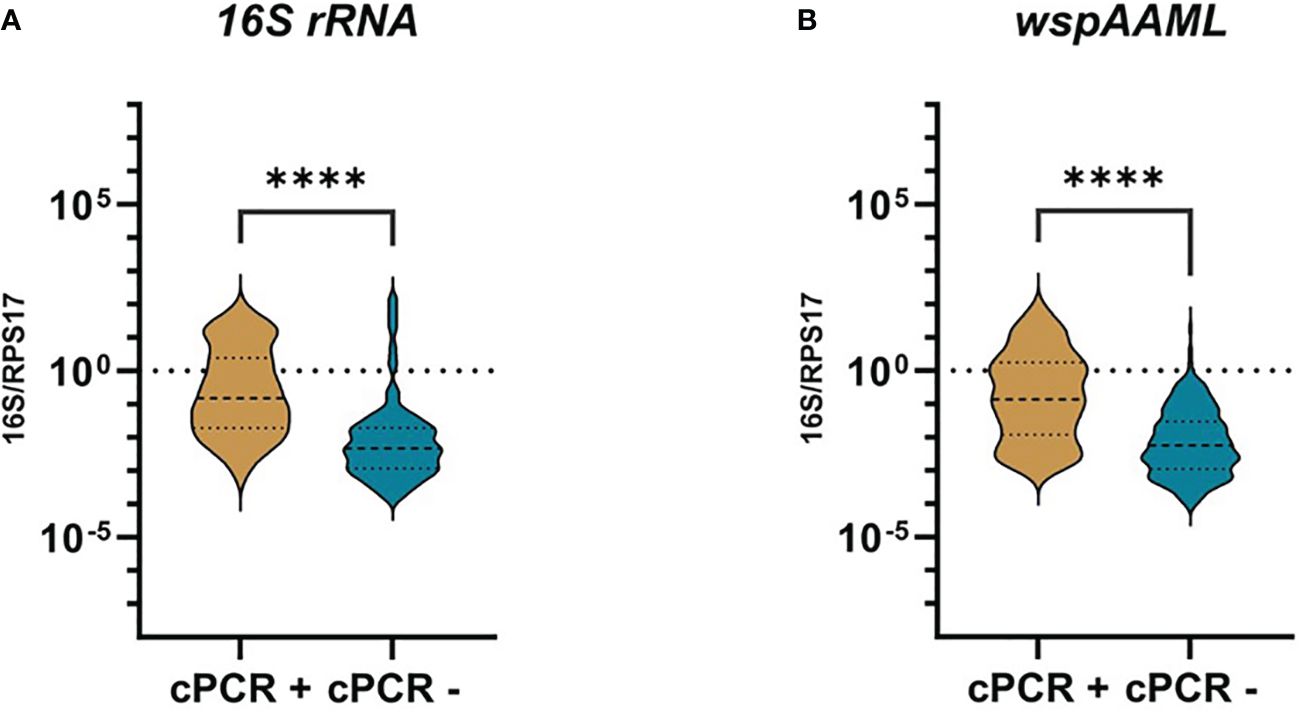
Figure 1 Wolbachia density of qPCR-positive Ae. aegypti grouped according to conventional PCR results. Individual mosquitoes detected as either positive for 16S rRNA (n =172) or wspAAML (n = 267) via qPCR were grouped based on Wolbachia detection results according to conventional PCR (cPCR). Relative Wolbachia density is expressed as the ratio of the target gene to single-copy RPS17 in logarithmic scale. (A) Using 16S rRNA primers, cPCR positive (n = 40) and negative (n = 132) samples had a median relative Wolbachia density of −0.8250 and −2.325, respectively. (B) Using wspAAML primers, cPCR positive (n = 54) and negative (n = 213) samples had a median relative Wolbachia density of −0.8550 and −2.250, respectively. **** indicates a significant difference between cPCR-positive and cPCR-negative Ae. aegypti at p ≤ 0.0001..
For both 16S rRNA and wspAAML qPCR assays, Wolbachia densities were ≥10-fold higher in male than female Ae. aegypti (p ≤ 0.01 for both; Figure 2). In the 16S rRNA marker, male mosquitoes (n = 69) exhibited relative Wolbachia densities between −3.57 and 2.17 with a median value of −1.67, whereas females (n = 103) exhibited a relative Wolbachia density range from −3.84 to 1.82 and a median value of −2.31. Regarding the wspAAML marker, male mosquitoes (n = 109) exhibited relative Wolbachia densities between −3.53 and 1.81 with a median value of −1.88, whereas females (n = 158) exhibited a range from −4.02 to 1.28 and a median value of −2.24. Although Wolbachia density was higher in male Ae. aegypti than in females, the prevalence of infection was higher among females (16S rRNA = 24%, wspAAML = 36.8%) than males (16S rRNA = 16%, wspAAML = 25%), regardless of the marker used for detection via qPCR (Table 2).
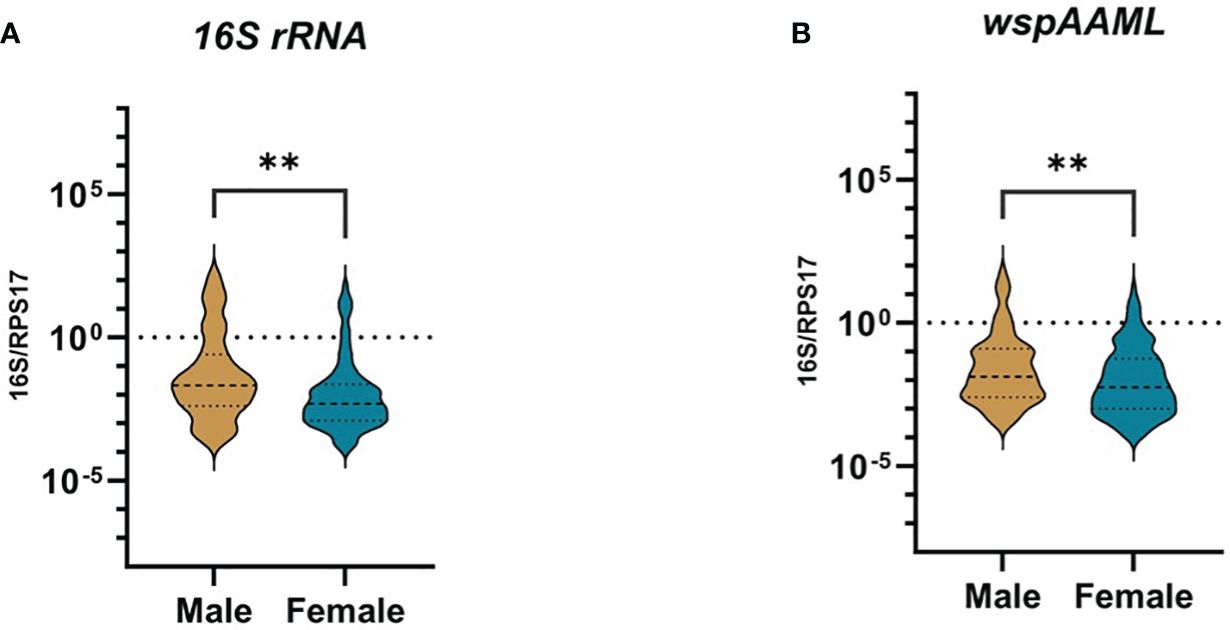
Figure 2 Presence of natural Wolbachia between male and female Ae. aegypti. Mosquitoes that were Wolbachia-positive according to TaqMan qPCR were classified as either male (16S rRNA = 69, wspAAML = 109) or female (16S rRNA = 103, wspAAML = 158). Relative Wolbachia density is shown as the ratio of the target gene to RPS17 in logarithmic scale. The 95% confidence interval of the median is indicated by the blue and violet lines. Relative densities are shown for (A) 16S rRNA and (B) wspAAML markers. In both (A, B), Wolbachia density differed significantly according to host sex. **p ≤ 0.01.
For phylogenetic analysis, we used 226 wspAAML sequences obtained from 267 wsp-positive Ae. aegypti samples collected from Metro Manila (AAML). We excluded 41 samples due to low sequencing quality and an inability to repeat sequencing owing to an inadequate volume of DNA. According to the maximum-likelihood phylogenetic tree, 83% (187/226) of wspAAML sequences in the AAML samples clustered into supergroup B, whereas only 15% (34/226) of wsp sequences clustered into supergroup A (Figure 3). The bootstrap values (≥75%; indicated on the external tree branches) supported the divergence of three clusters between supergroups A and B.
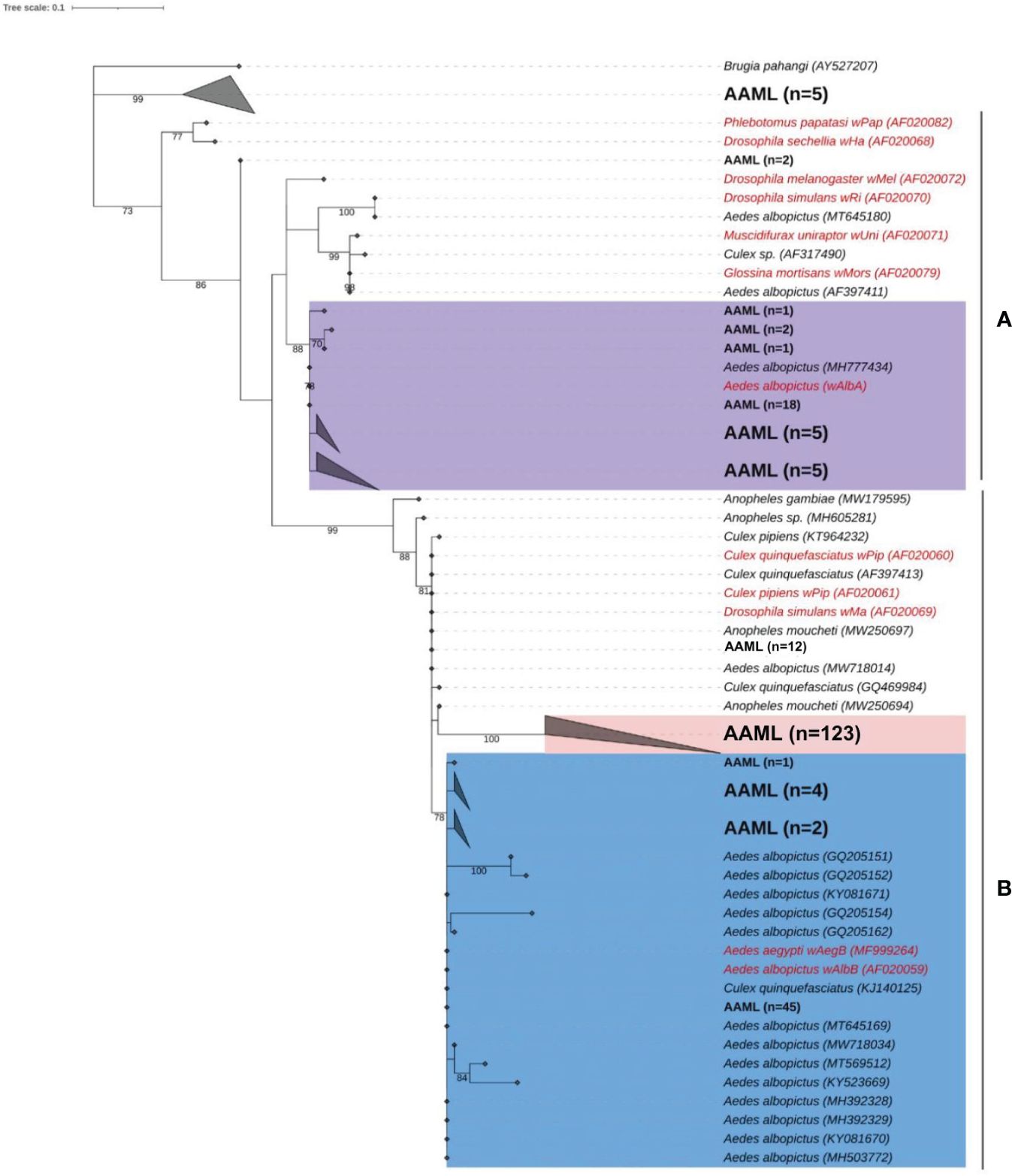
Figure 3 Phylogenetic analysis of Wolbachia according to wsp. Maximum-likelihood tree showing wspAAML sequences from Ae. aegypti samples collected in Metro Manila, Philippines (AAML in black bold text) and other wsp sequences obtained from other mosquito hosts (data from NCBI Blast; shown in black italicized text). Reference sequences are shown in red with an indication of the corresponding Wolbachia strains. Numbers on branches reflect the bootstrap support estimated with 1000 replications. The gray triangles are collapsed clades, each representing a node class. Ninety-eight percent of the wspAAML sequences clustered into either supergroup A or B. The tree shows two Wolbachia strains, namely wAlbA (violet) under supergroup A, wAlbB/wAegB (blue), and a group AAML under supergroup B (red). Scale bar indicates a phylogenetic distance of 0.1 nucleotide substitutions per site.
Under supergroup A, a clear cluster similar to the strain wAlbA was observed. Two wsp sequences of Ae. aegypti did not exhibit a clear delineation with any Wolbachia reference sequence. Numerous weak bootstrap support values (≤74%) were observed in deep nodes within supergroup A, indicating high genetic diversity. Under supergroup B, the wspAAML sequences of Ae. aegypti samples were nested within Wolbachia cluster wAegB/wAlbB together with other wsp sequences derived from Ae. albopictus or Culex spp. We found 12 wspAAML sequences of Ae. aegypti samples that fall under the wPip strain. However, another cluster was solely composed of wspAAML sequences found in the AAML samples (n = 123). For clarity, we hereafter refer to this AAML group (shaded in red in Figure 3) as the “wAegML” cluster. The two Wolbachia strains under supergroup B, i.e., wAegB/wAlbB and wAegML, share the same branch as wPip, which was consistent with the grouping previously established by Zhou et al (Zhou et al., 1998). The 12 sequences clustering with wPip share similarity with wAegML group and wAegB/wAlbB strain. We also noted that five wsp sequences from the Ae. aegypti samples formed a distinct cluster that did not fall under any of the supergroups considered; thus, these samples possibly belong to supergroups other than A and B.
Lastly, Wolbachia density of the wAegML group was less than either wAegB/wAlbB or wAlbA in Ae. aegypti (Dunn’s test, p <0.0001; Figure 4). However, Wolbachia density did not differ between wAegB/wAlbB and wAlbA strains in Ae. aegypti (Dunn’s test, p >0.0001).
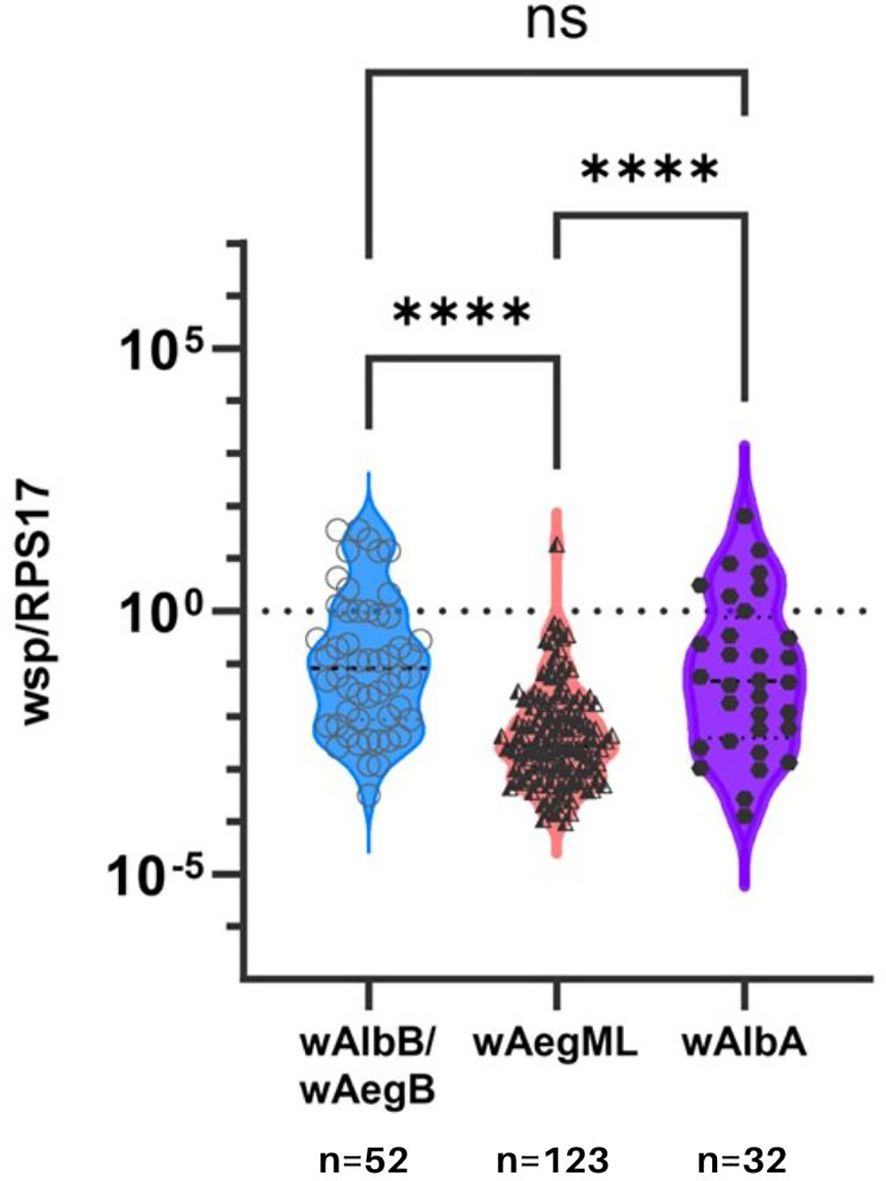
Figure 4 Wolbachia density in Ae. aegypti according to three identified Wolbachia clusters. Relative Wolbachia density of strains wAlbB/wAegB, wAegML, and wAlbA in Ae. aegypti. Relative density was considered the ratio of the target gene to RPS17. Values are shown on a logarithmic scale. Median Wolbachia relative density is indicated by a dark grey line. **** indicates p ≤0.001. ns indicates no significance between two groups.
The presence of natural Wolbachia in Ae. aegypti has been investigated in different countries. Based on these reports, Ae. aegypti is either absent or when present, exists sporadically and in low prevalence (Coon et al., 2016; Teo et al., 2017; Gloria-Soria et al., 2018; Goindin et al., 2018; Thongsripong et al., 2018; Balaji et al., 2019; Bennett et al., 2019; Carvajal et al., 2019; Zhang et al., 2022; Muharromah et al., 2023). Particularly in the Philippines, Wolbachia has been detected in cities within Metropolitan Manila finding a relatively low prevalence for both 16S rRNA (13.2%) and wsp markers (16.8%) by conventional PCR (Carvajal et al., 2019). More recently, natural Wolbachia detected in Ae. aegypti from the same mosquito population has been validated by ddRAD-sequencing. Sequence reads of 26,299 (wAlbA) and 43,778 (wAlbB) were mapped across the entire Wolbachia genome out of 146,239,637 filtered reads obtained (Muharromah et al., 2023). There are currently no Wolbachia mass release programs being implemented in the Philippines but its potential as a vector control method has been proposed (Ong et al., 2022). Hence, an economical way of conducting an initial surveillance to characterize not just the presence but also the density of natural Wolbachia in Ae. aegypti is warranted. In the present study, we used locally designed primers (wspAAML) to validate the presence of natural Wolbachia in Ae. aegypti that resulted to a higher prevalence of Wolbachia infection in wsp compared to the conventional PCR method used by Carvajal et al. We also found a high detection rate of Wolbachia, although to a lesser extent, using an established 16S rRNA primers. We found that relative Wolbachia density varied between host sex and Wolbachia phylogenetic groups in natural Ae. aegypti populations. The Wolbachia strains found present in Ae. aegypti were closely related to strains found in Ae. albopictus. We identified one cluster in the phylogenetic tree referred to as wAegML that was present in lower density but in higher prevalence in Ae. aegypti.
Wolbachia is ubiquitous in numerous host species (Noda et al., 2001; Kyei-Poku et al., 2005; Ros et al., 2009; Hughes et al., 2011) but its presence in mosquitoes vary. Presence of natural Wolbachia is usually high among Culex spp. and Ae. albopictus which are hosts regarded as naturally infected. On the other hand, Anopheles gambiae (8%-24%) and Ae. aegypti (4.3%-58%) exhibit variable prevalence results that differs according to geographical location (Inácio da Silva et al., 2021). In the present study, Ae. aegypti collected from Metro Manila, Philippines demonstrated natural Wolbachia prevalence of 40.0% and 62.2% when targeting 16S rRNA and wsp, respectively. This finding is consistent with other studies that detected natural Wolbachia in Ae. aegypti (Coon et al., 2016; Teo et al., 2017; Thongsripong et al., 2018; Balaji et al., 2019; Bennett et al., 2019; Carvajal et al., 2019; Kulkarni et al., 2019; Muharromah et al., 2023). Given the high prevalence rate of natural Wolbachia detected here, we suggest that the use of location-specific primers for qPCR increased the sensitivity of our detection method.
In a previous study, our laboratory performed conventional PCR-based detection of Wolbachia infection in Ae. aegypti collected from the current study location, finding a relatively low prevalence considering both 16S rRNA (13.2%) and wsp markers (16.8%). Contrastingly, a study conducted by Gloria-Soria et al. (2018) revealed the absence of Wolbachia in 117 Ae. aegypti mosquitoes collected from Cebu province, Philippines (Gloria-Soria et al., 2018). In both studies, conventional PCR was used. Although the primers used can amplify multiple Wolbachia strains, they were not designed using natural Wolbachia populations from Ae. aegypti mosquitoes in the local regions. Additionally, the difference in the prevalence of natural Wolbachia in Metropolitan Manila (Carvajal et al., 2019; Muharromah et al., 2023) and Cebu, Philippines (Gloria-Soria et al., 2018) further supports the fact that the presence of natural Wolbachia is variable and sporadic. The same situation occurs in China where infection prevalence between prefectures differed from 0 to 41.7% (Zhang et al., 2022).
Different Wolbachia primers used for PCR assays vary in terms of efficiency and coverage. The performance of 13 Wolbachia primer pairs was previously assessed using samples from a wide range of hosts representing supergroups A-F. The results varied, even among primers targeting the same gene, and only two primer sets yielded identical results, with other primers resulting in incorrectly sized amplifications (Simões et al., 2011). In the present study, to address the potential issue of primer incompatibility, we designed location-specific primers for wsp based on published sequences of Wolbachia populations in the same regions (Carvajal et al., 2019). In order to avoid bias, we included 16S rRNA marker which we did not design and has already been established (Fraser et al., 2020). Using either marker demonstrated a high detection rate with wspAAML being 22% higher than 16S rRNA. This suggests that the qPCR method led to an improved natural Wolbachia detection rate in Ae. aegypti owing to primer compatibility.
Notably, the qPCR method requires the use of primers that yield short amplicons (150 bp), which may have contributed to an increase in sensitivity. Additionally, qPCR has a lower limit of detection and higher sensitivity relative to conventional PCR, which could also explain the higher detection rate in the current study (Mee et al., 2015; Xia et al., 2018). When we compared Wolbachia density between samples found to be Wolbachia-negative or positive using conventional PCR, we found a 30-fold higher median Wolbachia density in the Wolbachia-positive samples by qPCR. We also found low Wolbachia density. Therefore, it is likely that both the newly designed primers and qPCR helped increase the natural Wolbachia detection rate relative to the detection performance of conventional PCR. These results suggest that location-specific primers may help validate the presence and determine Wolbachia density in Ae. aegypti samples with transient, sporadic, and/or low density natural Wolbachia infection.
The use of Ae. aegypti mosquitoes previously subjected to Wolbachia detection (Carvajal et al., 2019), allowed us to define potential reasons (e.g., primer incompatibility) for the sporadic detection of natural Wolbachia in Ae. aegypti. However, the possibility of contamination of Wolbachia from other mosquito host species in the larval stage cannot be completely avoided and therefore should be carefully considered (Ross et al., 2020). It is recommended that a comprehensive approach including imaging-based technique (FISH, IFA), demonstration of maternal transmission, reproductive manipulation e.g., CI, and antibiotic treatment of Wolbachia can be used to confirm natural infection in Ae. aegypti. Nevertheless, an economical way to improve detection for initial surveillance is also needed considering the sporadic and low-density presence of Wolbachia in Philippine Ae. aegypti.
Meanwhile, the present study adds to the existing evidence that Wolbachia (wsp) sequences found in Ae. aegypti belong to either supergroup A or B. Most wsp sequences found in Ae. aegypti collected from Metro Manila, Philippines were categorized under supergroup B consistent with the previous studies that utilized known Wolbachia primers and ddRAD-Seq, respectively (Carvajal et al., 2019; Muharromah et al., 2023). Both supergroups are widespread among arthropods and belong to a single monophyletic lineage (Gerth et al., 2014; Zug and Hammerstein, 2015). Supergroup B is likely to be the dominant supergroup found in naturally infected Ae. aegypti, as reported in most previous studies (Coon et al., 2016; Balaji et al., 2019; Carvajal et al., 2019; Kulkarni et al., 2019). Wolbachia strains under supergroup B usually reside in their hosts at high density (Hu et al., 2020; Walker et al., 2021) and exhibit resilience to cyclical heat stress, allowing them to persist in host populations (Ross et al., 2017). In our study, Wolbachia density is found to be relatively low. Balaji et al. (2019) reported 1.01 and 1.76 wsp/Rps17 of Wolbachia in male and female, respectively whereas we found an overall range of -4.02 to 1.81 regardless of gender. Natural Wolbachia density in Ae. aegypti differs throughout the developmental stage where significantly low density commonly manifests from the adult stage (Balaji et al., 2019). Our study only used random field-collected adult samples without considering age of adult mosquitoes that could account for the difference.
Interestingly, we detected three clusters representing the Wolbachia strains wAlbA, wAegB/wAlbB, and wAegML. Due to the inability to conduct further experiments to validate strain, we refer to another cluster as merely wAegML group. The bacterial density of wAegML was significantly lower than that of wAlbA, wAegB/wAlbB, and 62.8% of the wsp sequences in our Ae. aegypti samples belong to the wAegML. It is important to recognize that our study only used wsp for phylogenetic analysis which somehow limit our capacity to establish relatedness. Therefore, it is important that future studies utilize other markers and incorporate MLST data for a more accurate strain differentiation (Wang et al., 2020). Further experiments are needed to provide conclusive evidence on what impact sporadic and low-density natural Wolbachia in Ae. aegypti have.
Lastly, previous studies have revealed sex-specific Wolbachia density differences in natural populations of planthoppers S. furcifera and L. striatellus (Noda et al., 2001), as well as D. citri (Hoffmann et al., 1986), Drosophila spp (Correa and Ballard, 2012), Cx. Pipiens (Echaubard et al., 2010), Ae. albopictus (Tortosa et al., 2010; Calvitti et al., 2015), and Ae. aegypti (Mejia et al., 2022). We found that natural Wolbachia density in Ae. aegypti males was higher than in females, which is consistent with observations in D. citri (Hoffmann et al., 1986) and Ae. albopictus (Tortosa et al., 2010). Such sex-specific variation was only previously observed in Ae. albopictus mosquitoes carrying the wAlbB strain (Tortosa et al., 2010). A recent study on Wolbachia density in Ae. aegypti also reported the consistent densities in females throughout their lifetime whereas males demonstrate relatively higher and variable Wolbachia densities, in testes (Mejia et al., 2022). Although the biology underlying sex-specific differences in Wolbachia density is unknown, biological differences between the host sexes could explain our finding. For instance, female mosquitoes tend to have an expanded microbial composition relative to that of males, resulting in more bacterial competition (Zouache et al., 2011). Female mosquitoes are hematophagous, and the composition of bacterial microbiota in mosquitoes largely depends on nutrient intake (Gaio et al., 2011; Minard et al., 2013). Thus, the digestion process of female mosquitoes may act as a barrier to the survival of some symbionts, including Wolbachia.
Other studies have found that females exhibit higher Wolbachia densities than males in S. furcifera, L. striatellus (Noda et al., 2001), Drosophila spp (Correa and Ballard, 2012), Cx. pipiens (Echaubard et al., 2010), and Ae. albopictus (Tortosa et al., 2010; Calvitti et al., 2015). Thus, further investigation on gender-specific effects considering other coexisting factors is warranted. Our study only explored differences of relative Wolbachia density between male and female adult mosquitoes. It is important to further characterize how Wolbachia density is affected by host sex in different stages of the mosquito life cycle. Determining an absolute Wolbachia density rather than a relative one will provide more accurate information. Additionally, supplementing this with microscopic evidence will demonstrate the changes in Wolbachia density occurring between male and female mosquitoes during the insects’ life cycle.
Host species in which Wolbachia is naturally found usually exhibit stable high prevalence, high density, and heritable infection. Nevertheless, this is not the case for Ae. aegypti. Our study proposes that presence of natural Wolbachia in Ae. aegypti does not mean absolute presence, rather natural Wolbachia in this host is sporadic, and low-density. In the present study, we focused on investigating the factors that affect such occurrence. The use of Ae. aegypti samples previously subjected to Wolbachia detection by conventional PCR and ddRAD-Seq (Carvajal et al., 2019; Muharromah et al., 2023) allowed us to explore the biological (Wolbachia strain and host sex) and methodological (method of detection, primer incompatibility) factors affecting Wolbachia prevalence rate and density. This potentially explains the varying presence of natural Wolbachia in this host. Our findings suggest that (i) location-specific primers can improve Wolbachia density detection; (ii) relative Wolbachia densities are influenced by host sex and bacterial strain; (iii) majority of Wolbachia sequences clustered into a group referred to here wAegML that was present at a low density in Ae. aegypti. Overall, designing location-specific primers is economical for initial validation considering limited resources in other countries. These factors must be accounted for when conducting initial surveillance in places where no mass release programs have been conducted (e.g., Philippines). Moving forward, a comprehensive detection of natural Wolbachia infection in Ae. aegypti using multiple methods is still warranted.
The original contributions presented in the study are included in the article/Supplementary Material. The generated wspAAML sequences are available in GenBank database under the Accession Numbers PP278103-PP278328. Further inquiries can be directed to the corresponding author.
JR: Conceptualization, Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. TS: Data curation, Formal Analysis, Investigation, Methodology, Writing – review & editing. YS: Supervision, Writing – review & editing. KW: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the JSPS Core-to-Core Program B. Asia-Africa science platforms and the Sumitomo Electric Industries Group Corporate Social Responsibility (CSR) Foundation, Endowed Chair program.
The authors would like to thank Dr. Thaddeus Carvajal for his efforts in the field collection of Ae. aegypti mosquitoes. Finally, we are grateful to Dr. Christopher Robinson (Eawag - Swiss Federal Institute of Aquatic Science and Technology) for revising the final manuscript and sharing his insights to improve it further.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1360438/full#supplementary-material
Ahmad, N. A., Mancini, M.-V., Ant, T. H., Martinez, J., Kamarul, G. M. R., Nazni, W. A., et al. (2021). Wolbachia strain wAlbB maintains high density and dengue inhibition following introduction into a field population of Aedes aEgypti. Philos. Trans. R Soc. Lond B Biol. Sci. 376, 20190809. doi: 10.1098/rstb.2019.0809
Balaji, S., Jayachandran, S., Prabagaran, S. R. (2019). Evidence for the natural occurrence of Wolbachia in Aedes aEgypti mosquitoes. FEMS Microbiol. Lett. 366, fnz055. doi: 10.1093/femsle/fnz055
Bennett, K. L., Gómez-Martínez, C., Chin, Y., Saltonstall, K., McMillan, W. O., Rovira, J. R., et al. (2019). Dynamics and diversity of bacteria associated with the disease vectors Aedes aEgypti and Aedes albopictus. Sci. Rep. 9, 12160. doi: 10.1038/s41598-019-48414-8
Bergman, A., Hesson, J. C. (2021). Wolbachia prevalence in the vector species Culex pipiens and Culex torrentium in a Sindbis virus-endemic region of Sweden. Parasites Vectors 14, 428. doi: 10.1186/s13071-021-04937-6
Bian, G., Xu, Y., Lu, P., Xie, Y., Xi, Z. (2010). The endosymbiotic bacterium wolbachia induces resistance to dengue virus in aedes aEgypti. PloS Pathog. 6, e1000833. doi: 10.1371/journal.ppat.1000833
Bourtzis, K., Nirgianaki, A., Markakis, G., Savakis, C. (1996). Wolbachia infection and cytoplasmic incompatibility in Drosophila species. Genetics 144, 1063–1073. doi: 10.1093/genetics/144.3.1063
Brownlie, J. C., Cass, B. N., Riegler, M., Witsenburg, J. J., Iturbe-Ormaetxe, I., McGraw, E. A., et al. (2009). Evidence for Metabolic Provisioning by a Common Invertebrate Endosymbiont, Wolbachia pipientis, during Periods of Nutritional Stress. PloS Pathog. 5, e1000368. doi: 10.1371/journal.ppat.1000368
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Calvitti, M., Marini, F., Desiderio, A., Puggioli, A., Moretti, R. (2015). Wolbachia density and cytoplasmic incompatibility in Aedes albopictus: concerns with using artificial Wolbachia infection as a vector suppression tool. PloS One 10, e0121813. doi: 10.1371/journal.pone.0121813
Carvajal, T. M., Hashimoto, K., Harnandika, R. K., Amalin, D. M., Watanabe, K. (2019). Detection of Wolbachia in field-collected Aedes aEgypti mosquitoes in metropolitan Manila, Philippines. Parasites Vectors 12, 361. doi: 10.1186/s13071-019-3629-y
Chao, L.-L., Shih, C.-M. (2023). First detection and genetic identification of wolbachia endosymbiont in field-caught aedes aEgypti (Diptera: culicidae) mosquitoes collected from southern Taiwan. Microorganisms 11, 1911. doi: 10.3390/microorganisms11081911
Chen, Y.-T., Zhang, Y.-K., Du, W.-X., Jin, P.-Y., Hong, X.-Y. (2016). Geography has a greater effect than Wolbachia infection on population genetic structure in the spider mite, Tetranychus pueraricola. Bull. Entomol Res. 106, 685–694. doi: 10.1017/S0007485316000444
Coon, K. L., Brown, M. R., Strand, M. R. (2016). Mosquitoes host communities of bacteria that are essential for development but vary greatly between local habitats. Mol. Ecol. 25, 5806–5826. doi: 10.1111/mec.13877
Correa, C. C., Ballard, J. W. O. (2012). Wolbachia gonadal density in female and male Drosophila vary with laboratory adaptation and respond differently to physiological and environmental challenges. J. Invertebr Pathol. 111, 197–204. doi: 10.1016/j.jip.2012.08.003
Dengue Prevention and Control Program | Department of Health website. Available online at: https://doh.gov.ph/national-dengue-prevention-and-control-program (Accessed December 22, 2023).
Dutton, T. J., Sinkins, S. P. (2004). Strain-specific quantification of Wolbachia density in Aedes albopictus and effects of larval rearing conditions. Insect Mol. Biol. 13, 317–322. doi: 10.1111/j.0962-1075.2004.00490.x
Echaubard, P., Duron, O., Agnew, P., Sidobre, C., Noël, V., Weill, M., et al. (2010). Rapid evolution of Wolbachia density in insecticide resistant Culex pipiens. Heredity 104, 15–19. doi: 10.1038/hdy.2009.100
Edillo, F. E., Halasa, Y. A., Largo, F. M., Erasmo, J. N. V., Amoin, N. B., Alera, M. T. P., et al. (2015). Economic cost and burden of dengue in the Philippines. Am. J. Trop. Med. Hyg 92, 360–366. doi: 10.4269/ajtmh.14-0139
Edillo, F., Ymbong, R. R., Bolneo, A. A., Hernandez, R. J., Fuentes, B. L., Cortes, G., et al. (2022). Temperature, season, and latitude influence development-related phenotypes of Philippine Aedes aEgypti (Linnaeus): Implications for dengue control amidst global warming. Parasites Vectors 15, 74. doi: 10.1186/s13071-022-05186-x
Fraser, J. E., Bruyne, J. T. D., Iturbe-Ormaetxe, I., Stepnell, J., Burns, R. L., Flores, H. A., et al. (2017). Novel Wolbachia-transinfected Aedes aEgypti mosquitoes possess diverse fitness and vector competence phenotypes. PloS Pathog. 13, e1006751. doi: 10.1371/journal.ppat.1006751
Fraser, J. E., O’Donnell, T. B., Duyvestyn, J. M., O’Neill, S. L., Simmons, C. P., Flores, H. A., et al. (2020). Novel phenotype of Wolbachia strain wPip in Aedes aEgypti challenges assumptions on mechanisms of Wolbachia-mediated dengue virus inhibition. PloS Pathog. 16:e1008410. doi: 10.1371/journal.ppat.1008410
Frentiu, F. D., Robinson, J., Young, P. R., McGraw, E. A., O’Neill, S. L. (2010). Wolbachia-mediated resistance to dengue virus infection and death at the cellular level. PloS One 5, e13398. doi: 10.1371/journal.pone.0013398
Gaio, A., de, O., Gusmão, D. S., Santos, A. V., Berbert-Molina, M. A., Pimenta, P. F., et al. (2011). Contribution of midgut bacteria to blood digestion and egg production in Aedes aEgypti (diptera: culicidae) (L.). Parasites Vectors 4, 105. doi: 10.1186/1756-3305-4-105
Gerth, M., Gansauge, M.-T., Weigert, A., Bleidorn, C. (2014). Phylogenomic analyses uncover origin and spread of the Wolbachia pandemic. Nat. Commun. 5, 5117. doi: 10.1038/ncomms6117
Gloria-Soria, A., Chiodo, T. G., Powell, J. R. (2018). Lack of evidence for natural wolbachia infections in aedes aEgypti (Diptera: culicidae). J. Med. Entomol 55, 1354–1356. doi: 10.1093/jme/tjy084
Goindin, D., Cannet, A., Delannay, C., Ramdini, C., Gustave, J., Atyame, C., et al. (2018). Screening of natural Wolbachia infection in Aedes aEgypti, Aedes taeniorhynchus and Culex quinquefasciatus from Guadeloupe (French West Indies). Acta Trop. 185, 314–317. doi: 10.1016/j.actatropica.2018.06.011
Hertig, M., Wolbach, S. B. (1924). Studies on rickettsia-like micro-organisms in insects. J. Med. Res. 44, 329–374.7.
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hoffmann, A. A., Iturbe-Ormaetxe, I., Callahan, A. G., Phillips, B. L., Billington, K., Axford, J. K., et al. (2014). Stability of the wMel Wolbachia Infection following Invasion into Aedes aEgypti Populations. PloS Negl. Trop. Dis. 8, e3115. doi: 10.1371/journal.pntd.0003115
Hoffmann, A. A., Turelli, M., Simmons, G. M. (1986). Unidirectional incompatibility between populations of drosophila simulans. Evolution 40, 692–701. doi: 10.2307/2408456
Hu, Y., Xi, Z., Liu, X., Wang, J., Guo, Y., Ren, D., et al. (2020). Identification and molecular characterization of Wolbachia strains in natural populations of Aedes albopictus in China. Parasites Vectors 13, 28. doi: 10.1186/s13071-020-3899-4
Hughes, G. L., Koga, R., Xue, P., Fukatsu, T., Rasgon, J. L. (2011). Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles Gambiae. PloS Pathog. 7, e1002043. doi: 10.1371/journal.ppat.1002043
Ikeda, T., Ishikawa, H., Sasaki, T. (2003). Regulation of Wolbachia density in the Mediterranean flour moth, Ephestia kuehniella, and the almond moth, Cadra cautella. Zoolog Sci. 20, 153–157. doi: 10.2108/zsj.20.153
Inácio da Silva, L. M., Dezordi, F. Z., Paiva, M. H. S., Wallau, G. L. (2021). Systematic review of wolbachia symbiont detection in mosquitoes: an entangled topic about methodological power and true symbiosis. Pathogens 10, 39. doi: 10.3390/pathogens10010039
Indriani, C., Tantowijoyo, W., Rancès, E., Andari, B., Prabowo, E., Yusdi, D., et al. (2020). Reduced dengue incidence following deployments of Wolbachia-infected Aedes aEgypti in Yogyakarta, Indonesia: a quasi-experimental trial using controlled interrupted time series analysis. Gates Open Res. 4, 50. doi: 10.12688/gatesopenres.13122.1
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kulkarni, A., Yu, W., Jiang, J., Sanchez, C., Karna, A. K., Martinez, K. J. L., et al. (2019). Wolbachia pipientis occurs in Aedes aEgypti populations in New Mexico and Florida, USA. Ecol. Evol. 9, 6148–6156. doi: 10.1002/ece3.5198
Kyei-Poku, G. K., Colwell, D. D., Coghlin, P., Benkel, B., Floate, K. D. (2005). On the ubiquity and phylogeny of Wolbachia in lice. Mol. Ecol. 14, 285–294. doi: 10.1111/j.1365-294X.2004.02409.x
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Liang, X., Liu, J., Bian, G., Xi, Z. (2020). Wolbachia inter-strain competition and inhibition of expression of cytoplasmic incompatibility in mosquito. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01638
Liu, X.-C., Li, Z.-X. (2021). Transmission of the wMel Wolbachia strain is modulated by its titre and by immune genes in Drosophila melanogaster (Wolbachia density and transmission). J. Invertebr Pathol. 181, 107591. doi: 10.1016/j.jip.2021.107591
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López-Madrigal, S., Duarte, E. H. (2019). Titer regulation in arthropod-Wolbachia symbioses. FEMS Microbiol. Lett. 366, fnz232. doi: 10.1093/femsle/fnz232
Lu, P., Sun, Q., Fu, P., Li, K., Liang, X., Xi, Z. (2020). Wolbachia inhibits binding of dengue and zika viruses to mosquito cells. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01750
Marcon, H. S., Coscrato, V. E., Selivon, D., Perondini, A. L. P., Marino, C. L. (2011). Variations in the sensitivity of different primers for detecting Wolbachia in Anastrepha (diptera: tephritidae). Braz. J. Microbiol. 42, 778–785. doi: 10.1590/S1517-83822011000200046
Martinez, J., Tolosana, I., Ok, S., Smith, S., Snoeck, K., Day, J. P., et al. (2017). Symbiont strain is the main determinant of variation in Wolbachia-mediated protection against viruses across Drosophila species. Mol. Ecol. 26, 4072–4084. doi: 10.1111/mec.14164
McMeniman, C. J., Lane, R. V., Cass, B. N., Fong, A. W. C., Sidhu, M., Wang, Y.-F., et al. (2009). Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aEgypti. Science 323, 141–144. doi: 10.1126/science.1165326
McMeniman, C. J., O’Neill, S. L. (2010). A Virulent Wolbachia Infection Decreases the Viability of the Dengue Vector Aedes aEgypti during Periods of Embryonic Quiescence. PloS Negl. Trop. Dis. 4, e748. doi: 10.1371/journal.pntd.0000748
Mee, P. T., Weeks, A. R., Walker, P. J., Hoffmann, A. A., Duchemin, J.-B. (2015). Detection of low-level cardinium and wolbachia infections in culicoides. Appl. Environ. Microbiol. 81, 6177–6188. doi: 10.1128/AEM.01239-15
Mejia, A. J., Dutra, H. L. C., Jones, M. J., Perera, R., McGraw, E. A. (2022). Cross-tissue and generation predictability of relative Wolbachia densities in the mosquito Aedes aEgypti. Parasites Vectors 15, 128. doi: 10.1186/s13071-022-05231-9
Min, K.-T., Benzer, S. (1997). Wolbachia, normally a symbiont of Drosophila, can be virulent, causing degeneration and early death. Proc. Natl. Acad. Sci. U.S.A. 94, 10792–10796. doi: 10.1073/pnas.94.20.10792
Minard, G., Mavingui, P., Moro, C. V. (2013). Diversity and function of bacterial microbiota in the mosquito holobiont. Parasites Vectors 6, 146. doi: 10.1186/1756-3305-6-146
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mouton, L., Henri, H., Bouletreau, M., Vavre, F. (2006). Effect of temperature on Wolbachia density and impact on cytoplasmic incompatibility. Parasitology 132, 49–56. doi: 10.1017/S0031182005008723
Muharromah, A. F., Reyes, J. I. L., Kagia, N., Watanabe, K. (2023). Genome-wide detection of Wolbachia in natural Aedes aEgypti populations using ddRAD-Seq. Front. Cell. Infection Microbiol. 13. doi: 10.3389/fcimb.2023.1252656
Noda, H., Koizumi, Y., Zhang, Q., Deng, K. (2001). Infection density of Wolbachia and incompatibility level in two planthopper species, Laodelphax striatellus and Sogatella furcifera. Insect Biochem. Mol. Biol. 31, 727–737. doi: 10.1016/S0965-1748(00)00180-6
O’Neill, S. L., Giordano, R., Colbert, A. M., Karr, T. L., Robertson, H. M. (1992). 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. U.S.A. 89, 2699–2702. doi: 10.1073/pnas.89.7.2699
Ong, E. P., Obeles, A. J. T., Ong, B. A. G., Tantengco, O. A. G. (2022). Perspectives and lessons from the Philippines’ decades-long battle with dengue. Lancet Regional Health – Western Pacific 24, 100505. doi: 10.1016/j.lanwpc.2022.100505
Osborne, S. E., Leong, Y. S., O’Neill, S. L., Johnson, K. N. (2009). Variation in antiviral protection mediated by different wolbachia strains in drosophila simulans. PloS Pathog. 5, e1000656. doi: 10.1371/journal.ppat.1000656
Regilme, M. A. F., Inukai, T., Watanabe, K. (2021). Detection and phylogeny of Wolbachia in field-collected Aedes albopictus and Aedes aEgypti from Manila City, Philippines. bioRxiv 2021–08. doi: 10.1101/2021.08.24.457456
Ren, W., Wei, H., Yang, Y., Shao, S., Wu, H., Chen, X., et al. (2020). Molecular detection and phylogenetic analyses of Wolbachia in natural populations of nine galling Aphid species. Sci. Rep. 10, 12025. doi: 10.1038/s41598-020-68925-z
Reyes, J. I., Suzuki, Y., Carvajal, T., Muñoz, M. N., Watanabe, K. (2021). Intracellular interactions between arboviruses and wolbachia in Aedes aegypt. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.690087
Ros, V. I. D., Fleming, V. M., Feil, E. J., Breeuwer, J. A. J. (2009). How diverse is the genus Wolbachia? Multiple-gene sequencing reveals a putatively new Wolbachia supergroup recovered from spider mites (Acari: Tetranychidae). Appl. Environ. Microbiol. 75, 1036–1043. doi: 10.1128/AEM.01109-08
Ross, P. (2021). Designing effective wolbachia release programs for mosquito and arbovirus control. Acta Tropica 222, 106045. doi: 10.1016/j.actatropica.2021.106045
Ross, P. A., Callahan, A. G., Yang, Q., Jasper, M., Arif, M. A. K., Afizah, A. N., et al. (2020). An elusive endosymbiont: Does Wolbachia occur naturally in Aedes aEgypti? Ecol. Evol. 10, 1581–1591. doi: 10.1002/ece3.6012
Ross, P. A., Robinson, K. L., Yang, Q., Callahan, A. G., Schmidt, T. L., Axford, J. K., et al. (2022). A decade of stability for wMel Wolbachia in natural Aedes aEgypti populations. PloS Pathog. 18, e1010256. doi: 10.1371/journal.ppat.1010256
Ross, P. A., Wiwatanaratanabutr, I., Axford, J. K., White, V. L., Endersby-Harshman, N. M., Hoffmann, A. A. (2017). Wolbachia infections in aedes aEgypti differ markedly in their response to cyclical heat stress. PloS Pathog. 13, e1006006. doi: 10.1371/journal.ppat.1006006
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Schuler, H., Egan, S. P., Hood, G. R., Busbee, R. W., Driscoe, A. L., Ott, J. R. (2018). Diversity and distribution of Wolbachia in relation to geography, host plant affiliation and life cycle of a heterogonic gall wasp. BMC Evolutionary Biol. 18, 37. doi: 10.1186/s12862-018-1151-z
Shropshire, J. D., Hamant, E., Cooper, B. S. (2021). Male age and wolbachia dynamics: investigating how fast and why bacterial densities and cytoplasmic incompatibility strengths vary. mBio 12, e02998–e02921. doi: 10.1128/mbio.02998-21
Simões, P. M., Mialdea, G., Reiss, D., Sagot, M.-F., Charlat, S. (2011). Wolbachia detection: an assessment of standard PCR Protocols. Mol. Ecol. Resour. 11, 567–572. doi: 10.1111/j.1755-0998.2010.02955.x
Teo, C. H. J., Lim, P. K. C., Voon, K., Mak, J. W. (2017). Detection of dengue viruses and Wolbachia in Aedes aEgypti and Aedes albopictus larvae from four urban localities in Kuala Lumpur, Malaysia. Trop. BioMed. 34, 583–597.
Thongsripong, P., Chandler, J. A., Green, A. B., Kittayapong, P., Wilcox, B. A., Kapan, D. D., et al. (2018). Mosquito vector-associated microbiota: Metabarcoding bacteria and eukaryotic symbionts across habitat types in Thailand endemic for dengue and other arthropod-borne diseases. Ecol. Evol. 8, 1352–1368. doi: 10.1002/ece3.3676
Tortosa, P., Charlat, S., Labbé, P., Dehecq, J.-S., Barré, H., Weill, M. (2010). Wolbachia age-sex-specific density in aedes albopictus: A host evolutionary response to cytoplasmic incompatibility? PloS One 5, e9700. doi: 10.1371/journal.pone.0009700
Turelli, M., Cooper, B. S., Richardson, K. M., Ginsberg, P. S., Peckenpaugh, B., Antelope, C. X., et al. (2018). Rapid Global Spread of wRi-like Wolbachia across Multiple Drosophila. Curr. Biol. 28, 963–971.e8. doi: 10.1016/j.cub.2018.02.015
Unckless, R. L., Boelio, L. M., Herren, J. K., Jaenike, J. (2009). Wolbachia as populations within individual insects: causes and consequences of density variation in natural populations. Proc. Biol. Sci. 276, 2805–2811. doi: 10.1098/rspb.2009.0287
Walker, T., Quek, S., Jeffries, C. L., Bandibabone, J., Dhokiya, V., Bamou, R., et al. (2021). Stable high-density and maternally inherited Wolbachia infections in Anopheles moucheti and Anopheles demeilloni mosquitoes. Curr. Biol. 31, 2310–2320.e5. doi: 10.1016/j.cub.2021.03.056
Wang, X., Xiong, X., Cao, W., Zhang, C., Werren, J. H., Wang, X. (2020). Phylogenomic analysis of wolbachia strains reveals patterns of genome evolution and recombination. Genome Biol. Evol. 12, 2508–2520. doi: 10.1093/gbe/evaa219
Xia, Z., Johansson, M. L., Gao, Y., Zhang, L., Haffner, G. D., MacIsaac, H. J., et al. (2018). Conventional versus real-time quantitative PCR for rare species detection. Ecol. Evol. 8, 11799–11807. doi: 10.1002/ece3.4636
Yang, Q., Chung, J., Robinson, K. L., Schmidt, T. L., Ross, P. A., Liang, J., et al. (2022). Sex-specific distribution and classification of Wolbachia infections and mitochondrial DNA haplogroups in Aedes albopictus from the Indo-Pacific. PloS Negl. Trop. Dis. 16, e0010139. doi: 10.1371/journal.pntd.0010139
Yang, Y., He, Y., Zhu, G., Zhang, J., Gong, Z., Huang, S., et al. (2021). Prevalence and molecular characterization of Wolbachia in field-collected Aedes albopictus, Anopheles sinensis, Armigeres subalbatus, Culex pipiens and Cx. tritaeniorhynchus in China. PloS Negl. Trop. Dis. 15, e0009911. doi: 10.1371/journal.pntd.0009911
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., Madden, T. L. (2012). Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 13, 134. doi: 10.1186/1471-2105-13-134
Yen, J. H., Barr, A. R. (1973). The etiological agent of cytoplasmic incompatibility in Culex pipiens. J. Invertebrate Pathol. 22, 242–250. doi: 10.1016/0022-2011(73)90141-9
Zhang, H., Gao, J., Ma, Z., Liu, Y., Wang, G., Liu, Q., et al. (2022). Wolbachia infection in field-collected Aedes aEgypti in Yunnan Province, southwestern China. Front. Cell. Infection Microbiol. 12. doi: 10.3389/fcimb.2022.1082809
Zhou, W., Rousset, F., O’Neil, S. (1998). Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 265, 509–515. doi: 10.1098/rspb.1998.0324
Zouache, K., Raharimalala, F. N., Raquin, V., Tran-Van, V., Raveloson, L. H. R., Ravelonandro, P., et al. (2011). Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aEgypti, from different geographic regions of Madagascar. FEMS Microbiol. Ecol. 75, 377–389. doi: 10.1111/fem.2011.75.issue-3
Keywords: Wolbachia, aedes aegypti, vector control, arbovirus, mosquito
Citation: Reyes JIL, Suzuki T, Suzuki Y and Watanabe K (2024) Detection and quantification of natural Wolbachia in Aedes aegypti in Metropolitan Manila, Philippines using locally designed primers. Front. Cell. Infect. Microbiol. 14:1360438. doi: 10.3389/fcimb.2024.1360438
Received: 23 December 2023; Accepted: 04 March 2024;
Published: 18 March 2024.
Edited by:
Natarajaseenivasan Kalimuthusamy, Bharathidasan University, IndiaReviewed by:
Amit Sinha, New England Biolabs, United StatesCopyright © 2024 Reyes, Suzuki, Suzuki and Watanabe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kozo Watanabe, d2F0YW5hYmUua296by5takBlaGltZS11LmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.