- 1State Key Laboratory for Animal Disease Control and Prevention, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, Gansu, China
- 2Department of Zoology, Abdul Wali Khan University Mardan, Khyber Pakhtunkhwa, Pakistan
- 3Department of Parasitology, Faculty of Veterinary and Animal Sciences, The Islamia University of Bahawalpur, Bahawalpur, Punjab, Pakistan
Multiple ticks (Acari: Ixodoidea) carrying Rickettsiales bacteria have significant importance for both human and animal health. Thus, the purpose of this work was to genetically analyze tick species and their associated Rickettsiales bacteria in animal hosts. In order to achieve these objectives, various animals (including camels, cattle, goats, sheep, dogs, and mice) were inspected in four districts (Mardan, Peshawar, Kohat, and Karak) of Khyber Pakhtunkhwa to collect ticks, while blood samples were collected from all the symptomatic and asymptomatic cattle in all four districts. A total of 234 ticks were obtained from 86 out of 143 (60.14%) host animals, which were morphologically identified as Rhipicephalus turanicus, Rhipicephalus microplus, Haemaphysalis cornupunctata, and Hyalomma asiaticum. Among these, their representative ticks (126/234, 53.85%) were processed for molecular confirmation using cytochrome c oxidase (cox1) gene. Obtained cox1 sequences of four different tick species showed 99.72%–100% maximum identity with their corresponding species reported from Pakistan, China, India, and Kazakhstan and clustered phylogenetically. This study presented the first genetic report of Hy. asiaticum ticks in Pakistan. Moreover, genetically confirmed tick species were molecularly analyzed by PCR for detection of Rickettsiales DNA using partial fragments of 16S rDNA, 190-kDa outer membrane protein A (ompA), and 120-kDa outer membrane protein B (ompB) genes. In addition, blood samples were analyzed to identify Rickettsiales bacteria using the aforementioned genes. Rickettsiales bacteria were found in 24/126 (19.05%) ticks and 4/16 (25.00%) in symptomatic cattle’s blood. The obtained ompA and ompB sequences from Hy. asiaticum ticks showed 99.73%–99.87% with Candidatus Rickettsia shennongii and unidentified Rickettsia sp., whereas the obtained 16S rDNA sequences from cattle’s blood and ticks (Hae. cornupunctata) showed 99.67% highest identity with Anaplasma phagocytophilum. The 16S rDNA sequence of Rickettsiales DNA from Rh. turanicus ticks showed 100% identity with Ehrlichia canis and unidentified Ehrlichia sp. Obtained sequences of Rickettsiales bacteria were grouped along with their respective species in phylogenetic trees, which were previously reported in Greece, Cuba, Iraq, Turkey, Pakistan, South Korea, and China (mainland and Taiwan). This extensive study explores the wide range of damaging ticks and their corresponding tick-borne bacteria in the area, suggesting a possible danger to both livestock and human communities.
Introduction
Ticks (Acari: Ixodoidea) play a significant role in the maintenance and spread of numerous zoonotic protozoans, bacteria, and tick-borne viruses, posing a significant risk to the health of people and animals worldwide (Bekloo et al., 2018). The infections caused by these tick-borne pathogens (TBPs) lead to significant losses in livestock production in tropical and subtropical regions. This has substantial economic consequences since it reduces farmers’ revenue and poses a challenge to the cattle industry in global markets (Jongejan and Uilenberg, 2004). In addition, the presence of theileriosis has a significant effect on livestock farms in Pakistan, leading to a decrease in milk output and an increase in animal deaths (Rashid et al., 2018).
Several prominent tick species from different genera, such as Rhipicephalus, Hyalomma, Haemaphysalis, Ixodes, Argas, Ornithodoros, Nosomma, and Carios have been found in diverse locations in Pakistan, infesting both humans and animals (Karim et al., 2017; Ali et al., 2019; Aiman et al., 2022; Numan et al., 2022; Zahid et al., 2023). Various tick species are responsible for the transmission of numerous bacterial pathogens including Rickettsia spp., Anaplasma spp., Ehrlichia spp., and Coxiella spp. to different host animals like cattle, buffaloes, dogs, camels, mice, goats, sheep, domestic fowls, and equids (Khan et al., 2022; Ali et al., 2023a; Ali et al., 2023b; Shehla et al., 2023a). In addition, many TBPs such as Babesia spp., Theileria spp., Hepatozoon spp., Rickettsia spp., Anaplasma spp., and Ehrlichia spp. have been identified in blood samples taken from various domestic animals, including goats, sheep, dogs, cattle, and buffaloes (Ahmad et al., 2018; Hassan et al., 2018; Malik et al., 2018; Iqbal et al., 2019; Khan et al., 2020; Iatta et al., 2021).
The order Rickettsiales comprises the genera Rickettsia, Anaplasma, and Ehrlichia, which are obligate intracellular gram-negative bacteria that replicate inside the host's cells of host's body (Schmutzhard and Helbok, 2014). Rickettsia possesses a diverse spectrum of arthropod vectors and vertebrate hosts for their transmission. The scope and influence of the recognized tick-borne Rickettsiales pathogens have significantly expanded in the last 25 years, making this intricate category of diseases an ideal model for studying emerging and reemerging infections. Many tick-borne rickettsiae that were formerly thought to be harmless are now believed to be associated with human illnesses. Additionally, new species of Rickettsia, with unknown levels of virulence, are constantly being discovered in ticks worldwide (Parola et al., 2013). Based on the biological and genetic characteristics, rickettsiae are classified into four groups: the Spotted Fever Group (SFG), Typhus Group (TG), Transitional Group (TRG), and Ancestral Group (AG) (Gillespie et al., 2007). Tick-borne Spotted Fever Group Rickettsiae (SFGR) are globally distributed and can cause life-threatening illnesses, such as Rocky Mountain spotted fever and Mediterranean spotted fever (Paddock and Alvarez-Hernández, 2018). In Pakistan, different Rickettsia species belonging to the SFG including Rickettsia massiliae, Rickettsia hoogstraalii, Candidatus Rickettsia shennongii, and Rickettsia spp. have been molecularly documented in various tick species (Ali et al., 2023b; Shehla et al., 2023a).
In addition, many species of Anaplasma are accountable for inducing anaplasmosis in both people and animals, which is a tick-borne disease (TBD) and characterized by the destruction of red blood cells (Kocan et al., 2003). The illness is becoming an escalating and significant menace to the animal breeding system. It causes an extra burden on animal healthcare due to its infection, which results in decreased body weight, reduced milk supply, and frequent abortions in animals, ultimately leading to death (Stuen et al., 2002; Stuen et al., 2003). Anaplasma species, such as Anaplasma phagocytophilum, Anaplasma centrale, Anaplasma capra, Anaplasma platys, and Anaplasma marginale have been genetically analyzed in numerous arthropod vectors and host animals worldwide (Byaruhanga et al., 2018; Iqbal et al., 2019; Ali et al., 2023b). Specifically, the A. phagocytophilum is regarded as an important zoonotic pathogen, causing disease in humans, domestic ruminants, horses, dogs, and rarely in cats, while wild mammals act as its reservoir host (Dugat et al., 2015). This obligate intracellular Rickettsiales pathogen replicates in neutrophilic granulocytes and leads to thrombocytopenia, leukopenia, anemia, and immunosuppression associated with variable clinical signs (Dumler et al., 2005). Similarly, the genus Ehrlichia is also another highlighted TBP, and its different species have been molecularly documented in tick species, humans, domestic animals (cattle, buffaloes, horses, goats, sheep, and dogs), and wild animals including coyote, white-footed mice, and white-tailed deer (Dumler et al., 2001; Ali et al., 2023b).
Main TBDs, often referred to as blood-borne diseases (BBDs) such as anaplasmosis, babesiosis, theileriosis, and ehrlichiosis have a significant impact on bovines in Pakistan, particularly in terms of economic losses (Jabbar et al., 2015; Malik et al., 2018). Livestock plays a crucial role in the animal production sector, making a significant contribution of approximately 11.8% to Pakistan’s yearly gross domestic product (GDP). The agricultural sector is primarily run by small-scale farmers who cultivate essential nutrients and proteins to provide food security and generate revenue for the nation (Jabbar et al., 2015). Nevertheless, the growing number of TBDs has a negative impact on the agricultural and animal industries in Pakistan. Hence, this study was aimed for molecular identification of various tick species and their associated Rickettsiales pathogens in Khyber Pakhtunkhwa (KPK), Pakistan.
Materials and methods
Study area description
Khyber Pakhtunkhwa is one of the colder province in Pakistan and various districts including Mardan (34.1986°N, 72.0404°E), Peshawar (34.0398°N, 71.5668°E), Kohat (33.5828°N, 71.4627°E), and Karak (33.1335°N, 71.0365°E) of KPK were selected to cover the current study. These districts are comprised of both lower and upper highlands as well as plain areas. The study region exhibits a climate that ranges from dry to semi-arid, with an average annual temperature fluctuating between 8.5°C and 33.4°C. The average humidity levels vary from 28% to 82% throughout the duration of the year. Likewise, the annual precipitation in the given regions varies from 15 mm to 200 mm (https://en.climate-data.org/asia/pakistan/khyber-pakhtunkhwa-2239; accessed on October 16, 2023). Different factors like temperature, humidity, and precipitation have major influence on the growth and distribution of various ticks (Karim et al., 2017). The geographical coordinates of the study locations were obtained using global positioning system (GPS) and inserted into Microsoft Excel 2003 (Microsoft 365®) to design a study map using the ArcGIS V. 10.3.1 (ESRI, Redlands, CA, USA) (Figure 1). Three sampling points/locations in each district are represented by a drop-like structure in Figure 1.
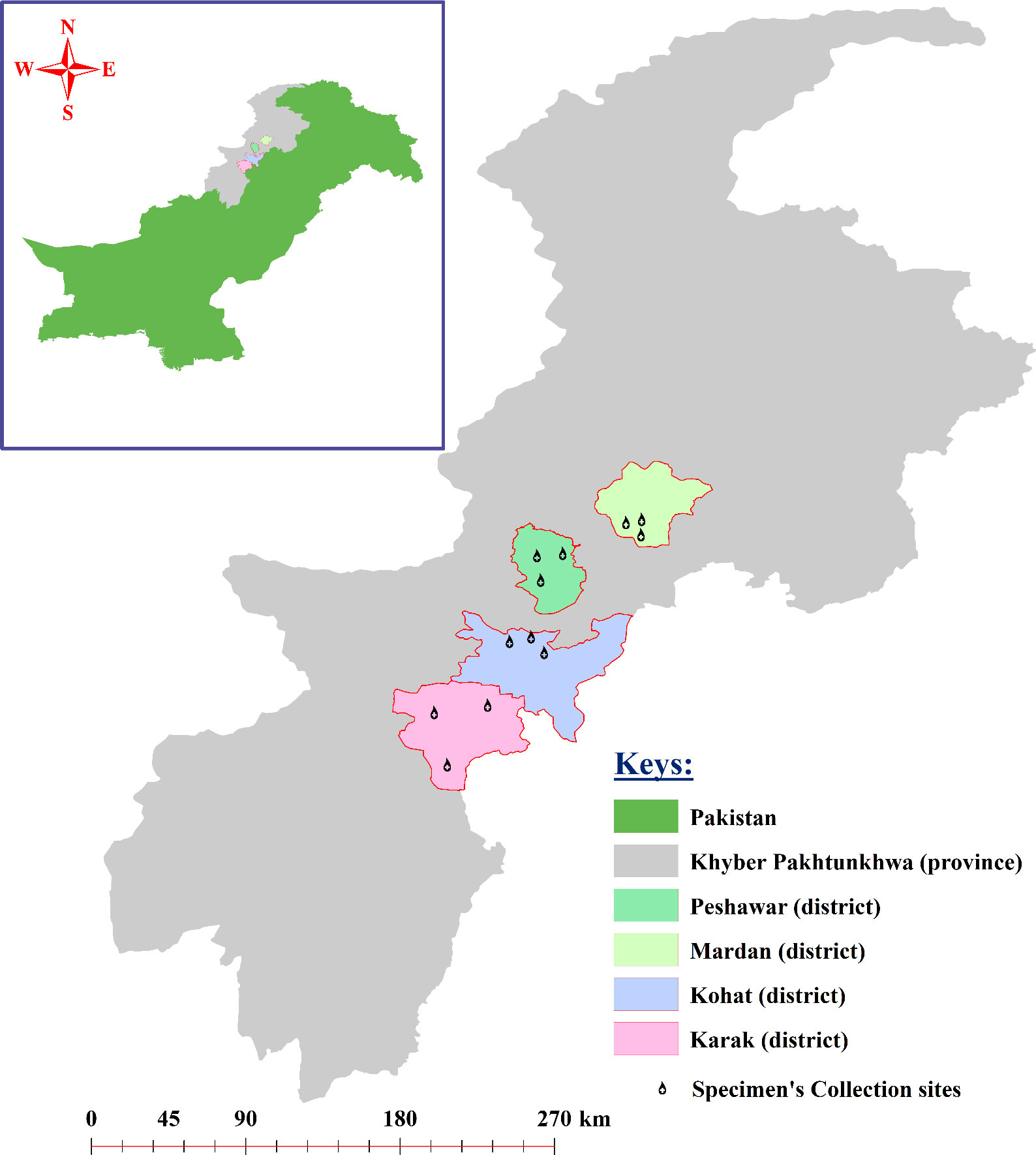
Figure 1 Study map of the four districts of the study area where the specimen collections were performed.
Collection and morphological identification of tick specimens
From March to August 2023, a total of 12 animal farms (three from each district) in the aforementioned districts were surveyed to collect ticks from different hosts such as camels, cattle, goats, sheep, and dogs. In addition, a substantial number of live mice were seized irregularly, to collect ticks from them, using plastic snap traps, which were installed/placed at different places in the studied livestock farms. Ticks were collected from various body parts like the sternum, belly, anal region, udder, legs, head, and neck region, using forceps that were applied as close as possible to the attachment point on the host’s body to pull the tick. Verbal and written permission (consent) was also taken from the animal farm’s owners before the sample collection. Ticks were collected once from each infested animal host in each farm in various districts. Ticks collected from each specific host in each district were placed separately in a labeled Eppendorf tube and were brought to the Laboratory of Molecular Parasitology, Department of Parasitology, Faculty of Veterinary and Animal Sciences, The Islamia University of Bahawalpur, Pakistan. Subsequently, all the gathered tick specimens underwent a cleansing process using 70% ethanol, followed by a final rinse with distilled water to remove any exterior contamination. Various morphological features were observed under the stereomicroscope (SZ61, Olympus Corporation, Tokyo, Japan) according to the standard identification keys (Hoogstraal and Varma, 1962; Kaiser and Hoogstraal, 1963; Dhanda and Kulkarni, 1969; Walker et al., 2005; Hosseini-Chegeni et al., 2013) to confirm the morphology of ticks. The ticks that were recognized based on their morphology were stored in 100% ethanol in the laboratory until they were ready to be processed for the extraction of genomic DNA (gDNA) and molecular analysis.
Blood collection and microscopic smear examination
In addition to inspections of tick infestation, veterinary experts undertook additional examinations of all the cattle hosts in all four districts. However, in Kohat District, the tick-infested cattle hosts were considered as suspected or positive based only on their observable signs and symptoms associated with any common TBDs, such as fever, loss of appetite, diarrhea, fatigue, coughing, and decreased milk output, as determined by a veterinary official. Therefore, blood samples were obtained from the jugular vein of each cattle in all four districts and stored in labeled vacutainers coated with ethylenediaminetetraacetic acid (EDTA). A total of 16 blood samples were promptly taken and delivered to the above-stated laboratory for the purpose of microscopic examination of a thin blood smear using Giemsa staining. Briefly, a drop of blood was poured on a glass slide for preparation of blood smear. These smears were then fixed with methanol and stained with a Giemsa (10%) for 30 min. Finally, the smears were inspected under a microscope by following the procedure outlined by Aktas and Özübek (2015) to detect the bacteria inside the cytoplasm. The positive blood samples were further subjected to gDNA extraction using the standard method of phenol–chloroform (Sambrook et al., 1989), followed by molecular analyses.
Molecular analyses of ticks and pathogens
The tick species of different life stages were identified based on their morphology and were then subjected to molecular analyses. Each tick was individually cleansed after preservation using 70% ethanol, followed by distilled water in order to remove any further contaminants from the tick’s body. Subsequently, each tick was dried separately on sterile filter paper. Afterward, each chosen tick species, including various life stages, was individually crushed using sterile scissors or blades to extract the gDNA. The gDNA from each morphologically identified tick species and collected blood samples was extracted using the standard method of phenol–chloroform (Sambrook et al., 1989). The extracted gDNA of tick species as well as blood specimens was quantified using NanoDrop (Nano-Q, Optizen, Daejeon, South Korea) and then stored at −20°C until further molecular analyses.
After that, each tick’s extracted gDNA sample was used for the amplification of the partial fragment of cox1 gene using a pair of primers in conventional PCR (PCR: GE-96G, BIOER, Hangzhou, China) for their molecular confirmation. Similarly, the gDNA of each tick species and each blood specimen were used individually in PCR for the amplification of partial fragments of the bacterial 16S rDNA gene. In addition, a set of primers targeting the 190-kDa outer membrane protein A (ompA) and the 120-kDa outer membrane protein B (ompB) genes were used to amplify the gDNA of each tick species and blood sample in order to determine the presence of Rickettsiales DNA. The primers and PCR cycling conditions of each specific gene are provided in Table 1. Each specific PCR was performed in a total volume of 25 µL, comprising 1 µL of each forward and reverse primer (concentration of 10 pmol/µL), 8.5 µL of PCR water, 2 µL (100 ng/µL) of gDNA, and 12.5 µL of DreamTaq MasterMix (2×) (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The PCR water was used as a negative control, while Hyalomma anatolicum tick species and R. massiliae were used as a positive control in each PCR. After successful amplification, almost 2 µL of each amplified product was examined by Gel Documentation System (BioDoc-It TM Imaging Systems, UVP, LLC, Upland, CA, USA) on 1.5% (w/v) agarose gel to validate the amplification efficiency.
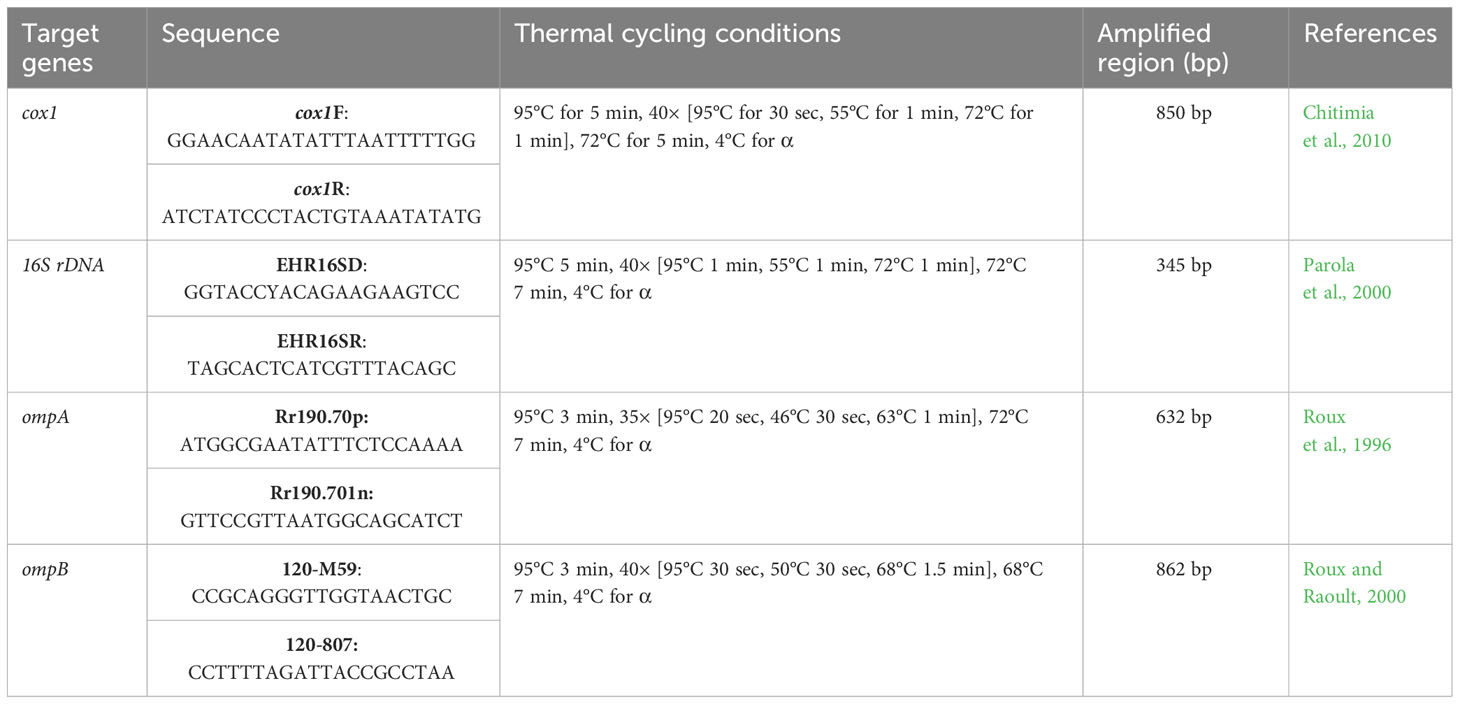
Table 1 List of primer pairs used in the current study for the molecular identification of different tick species as well as for the detection of bacterial pathogens in ticks and blood samples.
Genotyping and phylogenetic analyses of the obtained sequences
Prior to sequencing, the amplified products (126 for ticks and 35 for bacterial pathogens) underwent purification using the GeneClean II DNA purification Kit (QBIOgene, Illkirch, France) following the manufacturer’s given methodology. Subsequently, all the purified products were sequenced bidirectionally. Subsequently, the obtained sequences underwent trimming and purification using SeqMan v. 5 (DNASTAR, Inc., Madison, WI, USA). Finally, nine consensus sequences (four for different tick spp., two for Ehrlichia spp., one for Anaplasma sp., and two for Rickettsia sp.) were generated after the successful purification of all the obtained sequences. All the consensus sequences were individually subjected to the Basic Local Alignment Search Tool (BLAST) (Altschul et al., 1990) at the National Center for Biotechnology Information (NCBI) to download the high-identity sequences in FASTA format to obtain the phylogenetic relationship of our obtained sequences with already submitted sequences. The downloaded sequences along with specific selected outgroups were aligned in BioEdit Sequence Alignment Editor V.7.0.5 (Raleigh, NC, USA) (Hall et al., 2011) using ClustalW multiple alignments (Thompson et al., 1994). Separate phylogenetic trees were constructed for each specific gene using the neighbor-joining method with a p-distance model/method by applying 1,000 bootstraps in Molecular Evolutionary Genetic Analysis (MEGA-X) software (Kumar et al., 2018).
Results
Description regarding hosts and ticks
The tick collection was conducted on several host species, such as camels (20/143, 13.99%), cattle (25/143, 17.48%), sheep (28/143, 19.58%), goats (29/143, 20.28%), dogs (26/143, 18.18%), and mice (15/143, 10.49%) in the four demonstrated regions. However, only 86/143 (60.14%) host animals including the highest number of sheep (18/86, 20.93%) were found infested, followed by goats and dogs (17/86, 19.77%), cattle (16/86, 18.60%), and camels (13/86, 15.12%), while the least number of infested animals were mice (5/86, 5.81%) in this study. The infestation rate of the aforementioned animals was found the highest in Peshawar district (21/34, 61.76%), followed by Karak (24/39, 61.53%) and Kohat (22/37, 59.46%), while the least infestation rate of various host animals was recorded in Mardan district (19/33, 57.58%), as mentioned in Figure 2 and Table 2. Overall, almost 234 partially and fully engorged ticks including the highest number of nymph stages (128, 54.70%), followed by female stage (85, 36.32%), and males (21, 8.97%) were removed from the aforementioned infested host animals. The highest number of ticks were collected in Karak district (65, 27.78%), followed by Kohat (61, 26.07%) and Peshawar (57, 24.36%), and the least were collected from Mardan district (51, 21.80%). Additionally, the highest number of ticks were removed from cattle (56, 23.93%), followed by goats (47, 20.09%), sheep (46, 19.66%), dogs (39, 16.67%), camels (38, 16.24%), and mice (8, 3.42%) (Table 2).
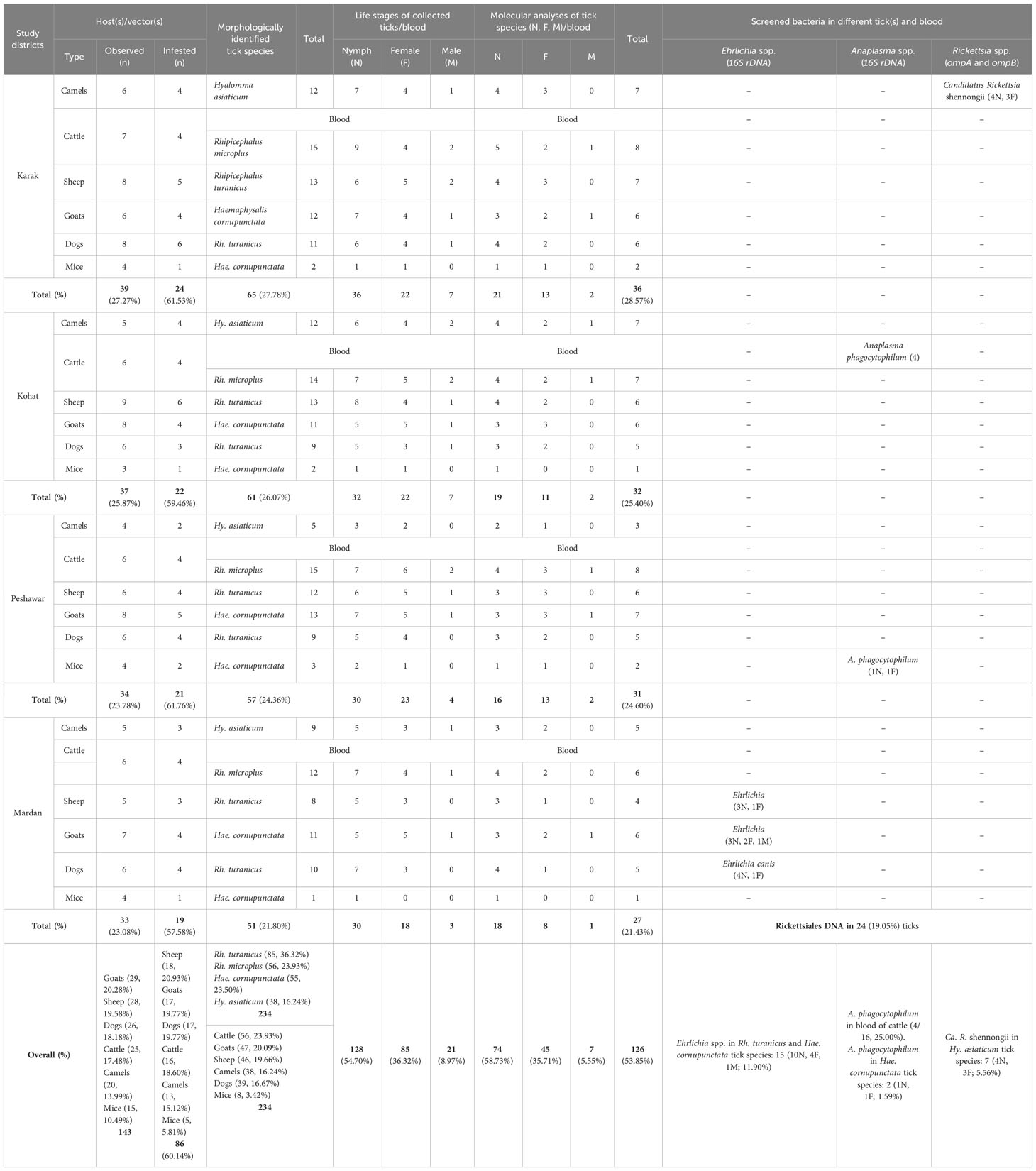
Table 2 Molecular detection of bacterial species in cattle’s blood and various tick species collected from different hosts in different districts of Khyber Pakhtunkhwa.
Morpho-molecular analyses of collected specimens
The 234 tick specimens were classified into three different genera based on their morphological characteristics: Hyalomma, Rhipicephalus, and Haemaphysalis. The largest number of ticks belonged to the specific species of Rhipicephalus turanicus (85, 36.32%), followed by Rhipicephalus microplus (56, 23.93%), Haemaphysalis cornupunctata (55, 23.50%), and Hyalomma asiaticum (38, 16.24%). Out of a total of 234 tick species, 126 (53.85%) representative tick species were molecularly confirmed using partial fragments of cox1 gene (Table 2).
Microscopic examination of blood
Intra-cytoplasmic inclusion bodies were observed in the prepared smears of almost 4/16 (25.00%) of cattle from Kohat district (Supplementary Figure 1); however, all the remaining blood samples of 12/16 (75.00%) were found negative by blood smear examination. Genomic DNA were extracted from all the positive blood samples.
Phylogenetic analyses of the obtained tick sequences
All the amplified products of partial fragments of cox1 gene from each tick species were sequenced. Since all the sequences acquired from each morphologically identifiable tick species were similar, hence a single consensus sequence was used for each tick species in their phylogenetic analysis. In conclusion, a total of four consensus and purified sequences were found for the four tick species stated before, which belong to three different genera: Rh. turanicus, Rh. microplus, Hae. cornupunctata, and Hy. asiaticum.
The consensus sequence of Rh. turanicus (723 bp) showed 99.72%–100% identity with the corresponding species by the BLAST result and clustered in the clade of the same species reported from Pakistan in the phylogenetic tree. The BLAST result of the consensus sequence of Rh. microplus (761 bp) tick species had maximum identity (99.74%–100%) with Rh. microplus tick species reported from China, Pakistan, and India, showing a close phylogenetic relationship with Rh. microplus in its phylogenetic analyses. Similarly, the consensus sequence of each Hae. cornupunctata (633 bp) and Hy. asiaticum (758 bp) showed 100% and 99.21%–99.34% identities with their corresponding species, respectively, and clustered phylogenetically with their corresponding related species reported from Kazakhstan and China (Figure 3).
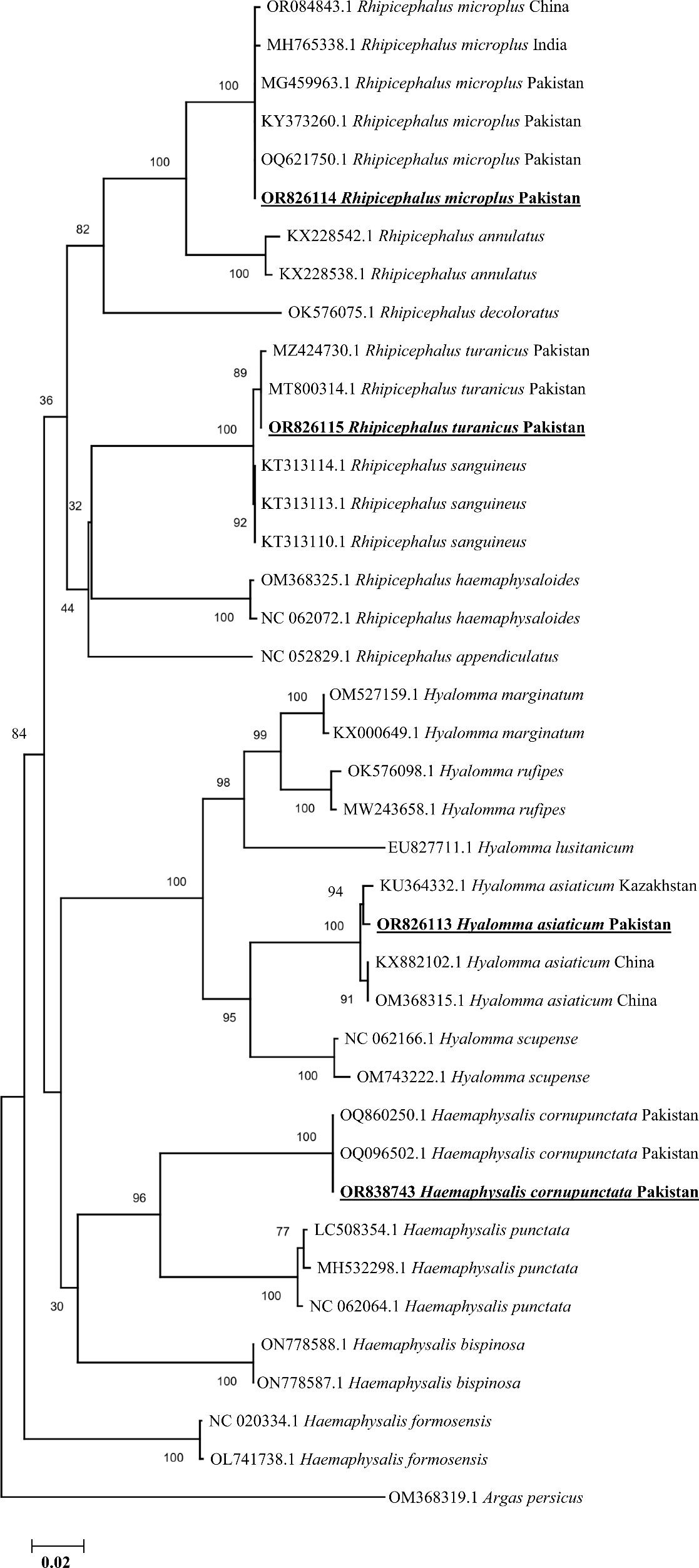
Figure 3 Phylogenetic analyses of tick species through the partial fragment of cox1 gene using neighbor-joining method and p-distance model. The sequences of the obtained tick species in the current study are marked as bold and underlined fonts (OR826113, OR826114, OR826115, and OR838743). The soft tick specie, Argus persicus, (OM368319.1) was chosen as an outgroup.
Detection of bacterial DNA and their phylogenetic analyses
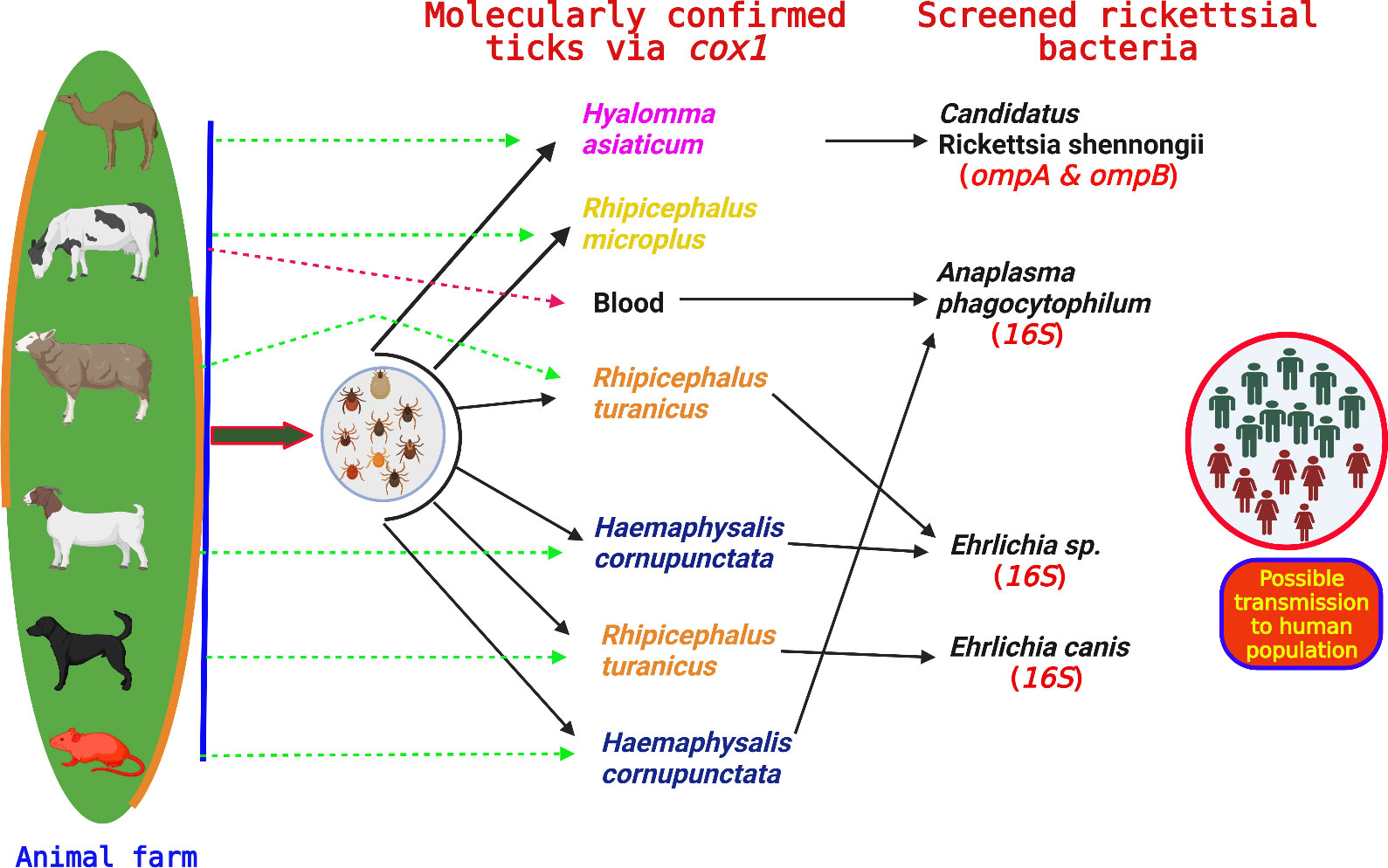
Consequently, the extracted DNA from the selected tick species and blood was tested for the detection of different bacterial pathogens including the highest existing rate of Ehrlichia spp. (15/126, 11.90% in ticks), followed by Anaplasma spp. (4/16, 25.00% in blood and 2/126, 1.59% in ticks) and Rickettsia spp. (7/126, 5.56% in ticks). However, no Rickettsiales DNA belonging to the genus Rickettsia were detected in the DNA of all the collected blood specimens.
Rickettsia spp. and phylogenetic analysis
Based on partial fragments of ompA and ompB, the Ca. R. shennongii was identified in Hy. asiaticum tick species (7/126, 5.56%) collected from camels in Karak district, and all the amplified products of each gene (ompA and ompB) were sequenced for their phylogenetic analyses. Due to the homologous/identical nature of all the obtained sequences, a single consensus sequence of each gene (ompA and ompB) was used to retrieve highly similar sequences to obtain their phylogenies. By the BLAST results, our obtained single consensus ompA sequence showed 99.29%–100% identity with the Ca. R. shennongii and Rickettsia sp. reported from Pakistan and Taiwan, respectively, that clustered with the corresponding species in the phylogenetic tree (Figure 4).
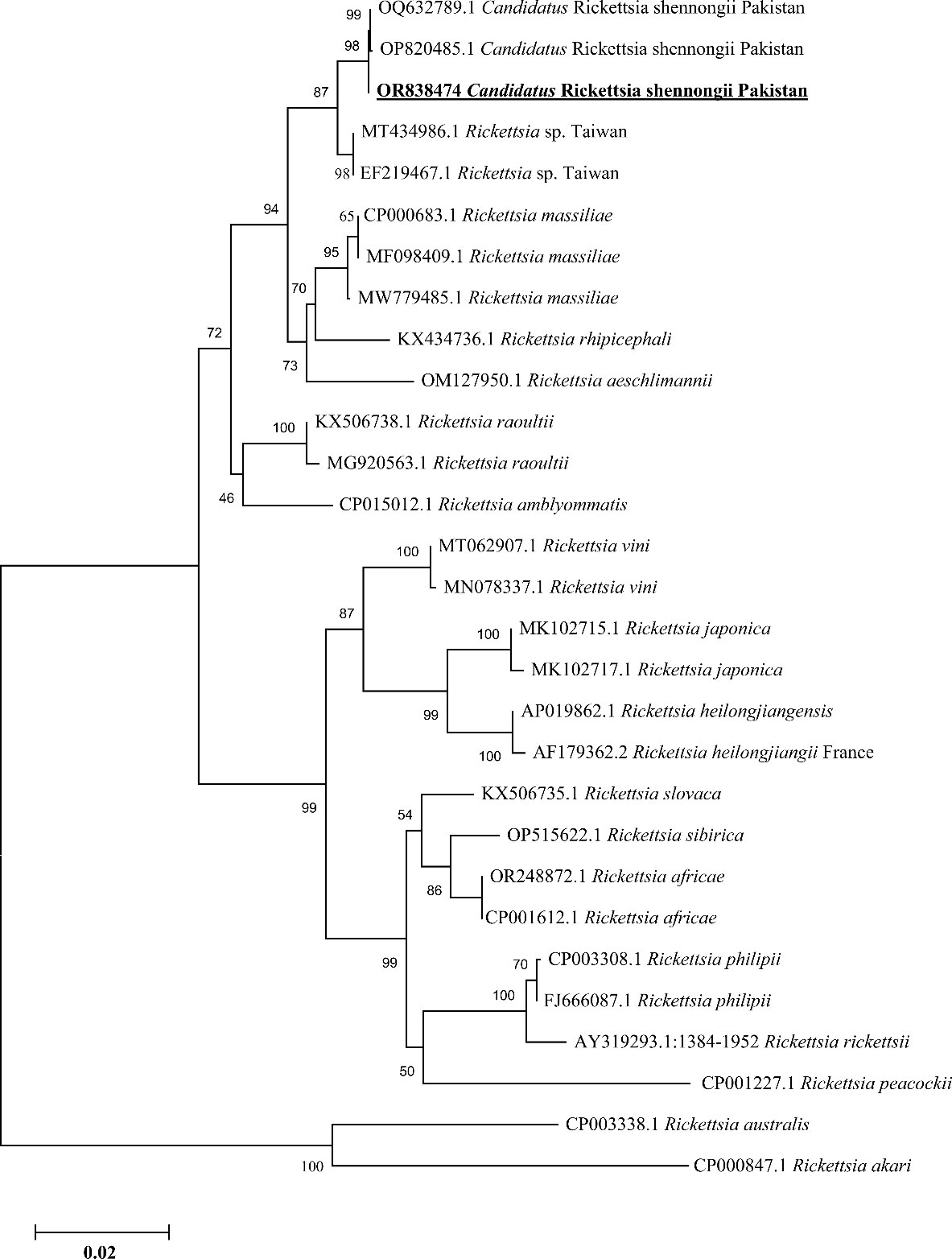
Figure 4 Phylogenetic analyses of Candidatus Rickettsia shennongii through the partial fragment of ompA gene using neighbor-joining method and p-distance model. The sequence of the obtained Ca. R. shennongii in the current study are marked as bold and underlined fonts (OR838474). Rickettsia australis (CP003338.1) and Rickettsia akari (CP000847.1) were chosen as outgroup sequences.
Similarly, the BLAST results of the obtained single consensus ompB sequence showed the maximum identity in the range of 99.73%–99.87% with the Ca. R. shennongii and Rickettsia sp. that were clustered with their corresponding species reported from Pakistan, China (the mainland and Taiwan), and Sri Lanka in the phylogenetic tree (Figure 5).
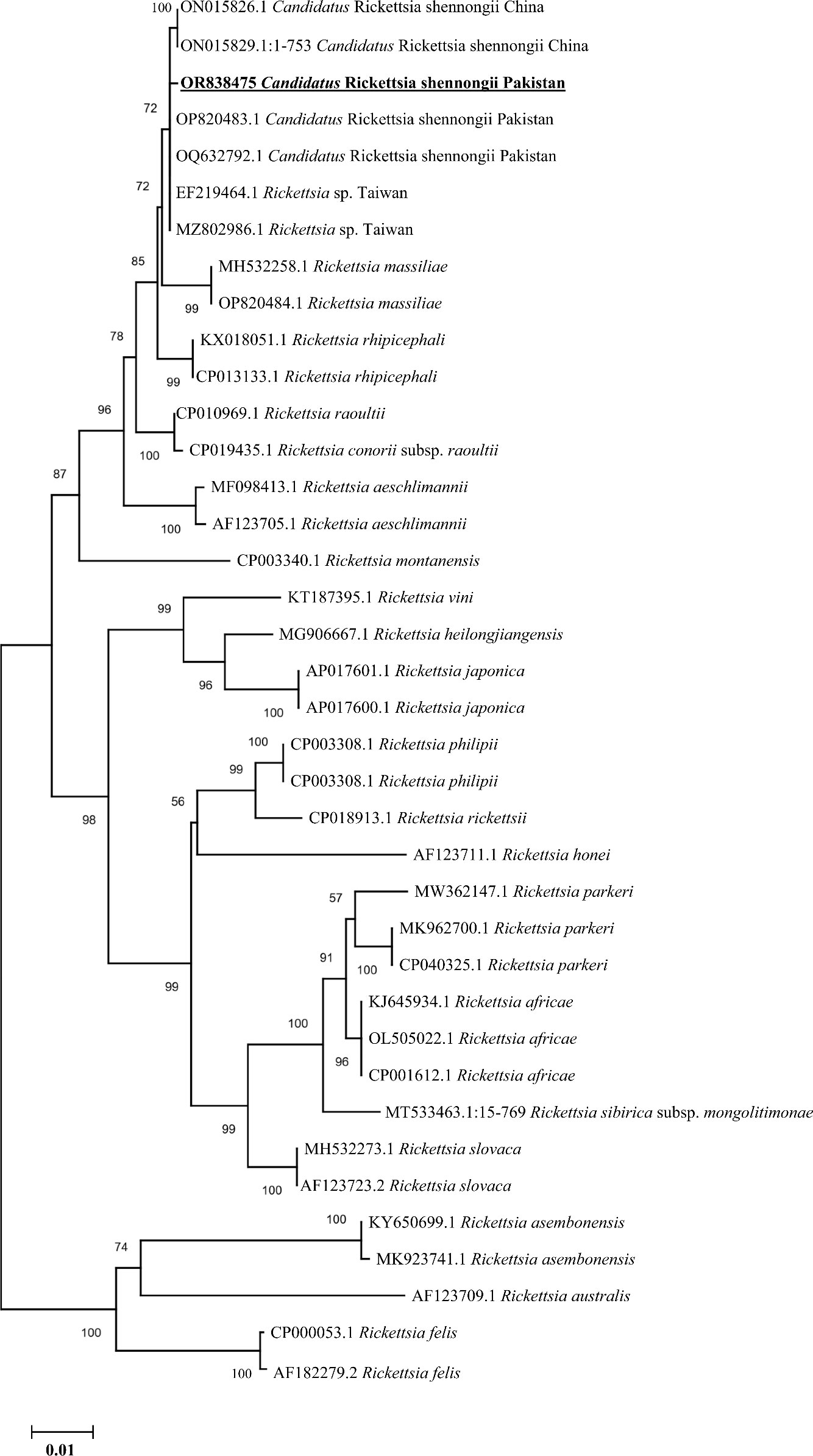
Figure 5 Phylogenetic analyses of Candidatus Rickettsia shennongii through the partial fragment of ompB gene using neighbor-joining method and p-distance model. The sequence of the obtained Ca. R. shennongii in the current study is marked as bold and underlined fonts (OR838475). Rickettsia felis (CP000053.1 and AF182279.2) were chosen as outgroup sequences.
Phylogenetic analysis of Anaplasma phagocytophilum and Ehrlichia spp.
Based on consensus sequences of 16S rDNA partial fragments, the A. phagocytophilum and Ehrlichia spp. (Ehrlichia canis and Ehrlichia sp.) were confirmed. The obtained consensus sequence of 16S rDNA partial fragment was obtained from the DNA of cattle’s blood and Hae. cornupunctata and was used to download the high-identity sequences from the NCBI using the BLAST, and our obtained consensus sequence showed a maximum of 99.67% identity with A. phagocytophilum reported from South Korea and clustered phylogenetically in a clade with the same corresponding species (Figure 6).
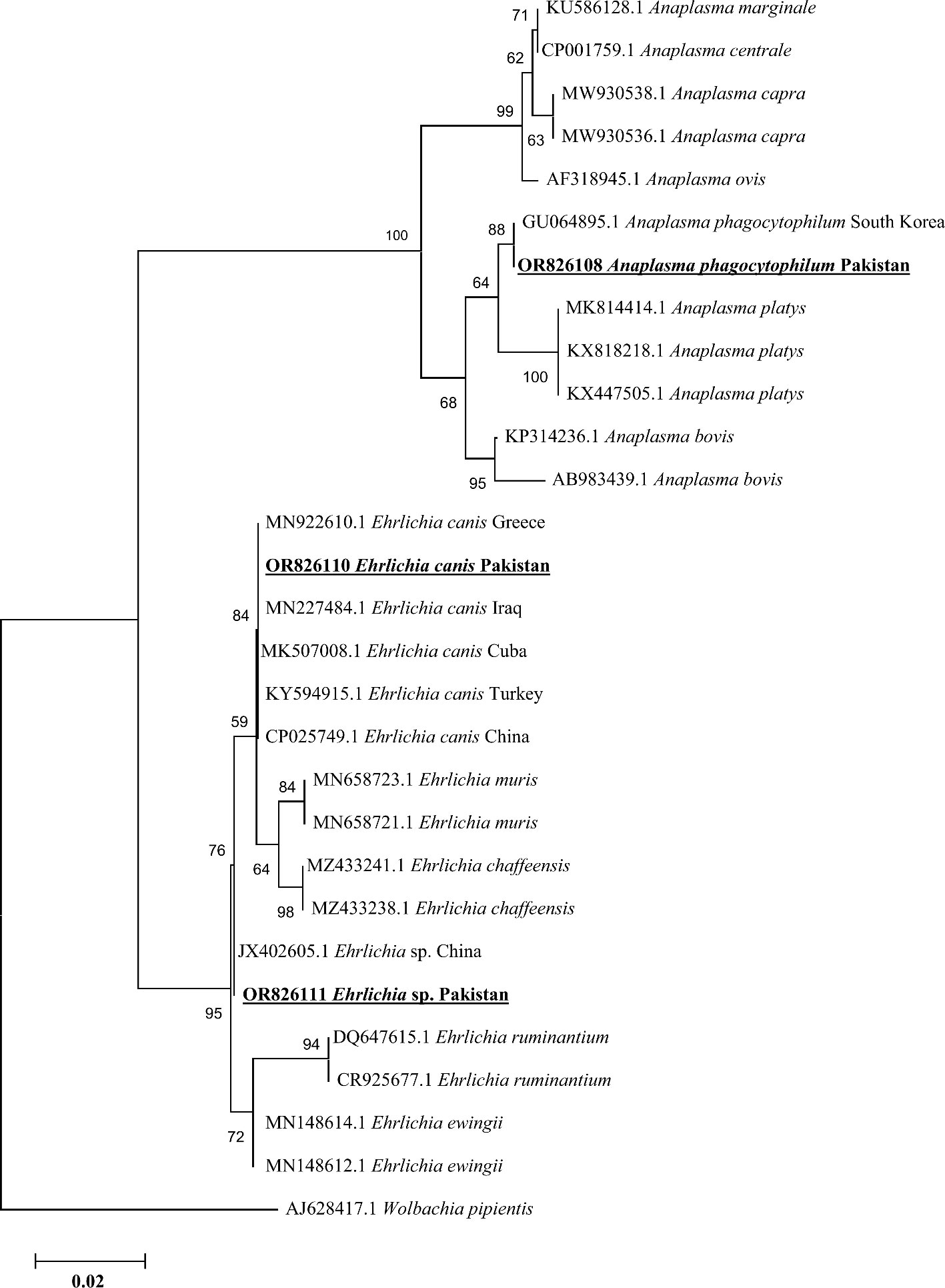
Figure 6 Phylogenetic analyses of Anaplasma phagocytophilum, Ehrlichia canis, and Ehrlichia sp. through the partial fragment of 16S rDNA gene using neighbor-joining method and p-distance model. The sequence of the obtained A. phagocytophilum, E. canis, and Ehrlichia sp. in the current study are marked as bold and underlined fonts (OR826108, OR826110, and OR826111), respectively. Wolbachia pipientis (AJ628417.1) was chosen as an outgroup sequence.
In addition, we obtained two distinct consensus sequences for Ehrlichia spp., which were then subjected to BLAST search in the NCBI to obtain the corresponding homologous sequences for their phylogenetic analysis. One consensus sequence of Ehrlichia spp. showed 100% identity with the E. canis species and clustered in the same clade with the same corresponding species reported from Greece, Cuba, Iraq, Turkey, and China. Another consensus sequence had the highest identity (99.71%) with the same unidentified species of Ehrlichia reported from China, which was clustered with the corresponding Ehrlichia sp. in the phylogenic tree (Figure 6).
Discussion
In Pakistan, small-scale animal husbandry is prevalent in rural and economically impoverished communities, supporting both domestic and commercial purposes. In order to accomplish this target, diverse animals such as camels, cattle, buffaloes, sheep, and goats are imported from neighboring countries due to their better milk production capabilities. As a result, farmers have embraced commercial farming methods that use advanced technology. However, a wide range of Ixodid ticks and TBPs are prevalent in distinct ecological and geographical areas of Pakistan (Karim et al., 2017; Ghafar et al., 2020b; Atif et al., 2022), as exotic animals are core affected and vulnerable to ticks and their associated TBPs even in a controlled environment as described by Young et al. (1988), significantly impacting the farming community in Pakistan. In recent years, numerous TBPs belonging to different genera including Rickettsia, Ehrlichia, Anaplasma, Coxiella, and Bartonella have been documented in a variety of ticks and their animal hosts from various regions of Pakistan (Iqbal et al., 2019; Ali et al., 2023b; Khan et al., 2023; Rizwan et al., 2023; Shehla et al., 2023b), showing considerable public health interest. Their phylogenic affiliations have evolved much and are still evolving with the passage of time. Hence, this study was aimed for the molecular identification of various tick species and their Rickettsiales bacteria in camels, cattle, goats, sheep, dogs, and mice from the specified targeted area because the aforementioned host animals except mice are considered as companions of humans, which might play an important role in the natural transmission cycle and dispersal of different ticks and their associated TBPs. Herein, four tick species, namely, Rh. turanicus, Rh. microplus, Hae. cornupunctata, and Hy. asiaticum, were genetically identified successfully using cox1 partial sequences. Additionally, the molecular identification of bacterial pathogens including Ca. R. shennongii (based on ompA and ompB), E. canis and unidentified Ehrlichia sp. (based on 16S rDNA), and A. phagocytophilum (based on 16S rDNA) was achieved. Moreover, this is the first-ever genetic evidence of Hy. asiaticum tick species carrying the Ca. R. shennongii from Pakistan.
The majority of the examined animals in this study were camels, although the infestation rate of ticks was found higher in sheep as compared to other host animals, which aligns with the prior research findings documented from North Africa (Khamassi-Khbou et al., 2021), and the high infection rate may be attributed to the abundance of a variety of sheep breeds and the presence of long wool on their bodies, which enables the ticks to thrive in the specific research area.
Four tick species belonging to three genera including Rhipicephalus, Hyalomma, and Haemaphysalis were found on the animal host bodies in the current study because the targeted study locations are comprised of arid to semi-arid regions, and their climatic conditions vary due to having four seasons (spring, winter, summer, and fall). Climate variability and global warming significantly influence the proliferation and distribution of tick species (Leger et al., 2013; Gilbert et al., 2014). The three aforementioned genus species exhibited dominance in the research region, perhaps attributed to the aggregation of various animals in overcrowded shelters/farms and the simultaneous grazing of diverse host animals in the studied areas. In the current study, the targeted region predominantly exhibited ticks belonging to the genus Rhipicephalus, and these findings were in accordance with previously published data from Pakistan (Obaid et al., 2023). More specifically, the Rh. turanicus tick species was predominant as compared to other tick species that were infesting both sheep and dogs in the targeted study locations, and these findings are parallel to the previous findings of Ceylan et al. (2021) and Alam et al. (2022).
Due to the presence of similarities and juvenile stages, it is inadequate to achieve the accurate morphological identification of tick species. Therefore, genetic markers such as mitochondrial cox1 gene have been successfully proven as reliable for the molecular confirmation of different ticks (Filipe et al., 2021; Zhao et al., 2020). In the current study, we report the Hy. asiaticum as the first molecular report from Pakistan, as it was reported morphologically prior to this investigation (Afridi et al., 2023), and there was no molecular evidence from Pakistan. However, the remaining three species including Rh. turanicus, Rh. microplus, and Hae. cornupunctata were already morpho-molecularly reported from various regions of Pakistan. Phylogenetically, the obtained partial sequences of cox1 gene from the aforementioned tick species revealed a close evolutionary relationship with their corresponding species reported from Kazakhstan, Pakistan, India, and China, and the accuracy regarding their topology was supported by previously published data.
The results of our investigation confirmed the presence of Rickettsiales DNA in the investigated tick species, and it is worth noting that this kind of bacterial DNA has been previously detected in other tick species globally (Yang et al., 2016; Shao et al., 2021; Hu et al., 2022; Ali et al., 2023b). More specifically, the Ca. R. shennongii based on ompA and ompB genes were presented for the first time from the Hy. asiaticum tick species collected from camels in the current study, and these findings were parallel to the previous findings that have presented the carrier nature of Hy. asiaticum for Rickettsia spp. collected from camels (Batu et al., 2020). Therefore, these considerations suggest that camels are likely reservoirs for Rickettsia species in the study area, although the pathogenicity of Ca. R. shennongii is still undetermined (Shehla et al., 2023a). Hence, extensive research work is needed to update the scientific community regarding its pathogenicity.
The A. phagocytophilum bacterium has a broad range of susceptible hosts and may induce illness in ruminants, horses, dogs, and cats. Moreover, it is recognized as a new human pathogen of growing significance worldwide (Stuen et al., 2002; Strle, 2004; Dumler, 2005). In the past, Anaplasma spp. have been genetically identified in the blood of numerous animals including cattle in different regions of the globe, using different genetic markers (Dugat et al., 2015; Bekloo et al., 2018; Byaruhanga et al., 2018; Iqbal et al., 2019; Iatta et al., 2021; Hu et al., 2022). Using the highly conserved 16S rDNA marker, we have molecularly detected A. phagocytophilum in the blood of cattle. These results provide compelling evidence that cattle might serve as a reservoir for the Anaplasma bacterium. In addition, we molecularly identified A. phagocytophilum from ticks (Hae. cornupunctata) collected from mice in livestock farms in Peshawar district, and these results are consistent with a recently published work (Santos et al., 2009) A study indicated that mice may have significant involvement in maintaining the active cycles of this bacterial pathogen. The presence of mice in animal farms in Peshawar district may be a contributing factor in the transmission of Anaplasma bacteria to livestock animals from mice through the ticks, as previous research has shown the presence of Anaplasma species in ticks collected from cattle in the same district (Khan et al., 2022).
Sheep, goats, and dogs roam freely outdoors in both rural and urban areas of the study locations to find food and hunt. This raises the risk of tick infestation and the spread of various TBPs such as Ehrlichia spp. This bacterium has been found worldwide in the blood of animals and in ticks that infest host animals including cattle, sheep, goats, dogs, and horses (Dumler et al., 2001; Bekloo et al., 2018; Malik et al., 2018; Hu et al., 2022). Our findings declared the existence of Ehrlichia genus especially E. canis and unidentified Ehrlichia sp. in Rh. turanicus and Hae. cornupunctata ticks, collected from sheep, dogs, and goats. These findings are in accordance with the previous research works performed by Ali et al. (2023b); Malik et al. (2018), and Iatta et al. (2021) in Pakistan.
Phylogenetically, the topology of various Rickettsiales bacteria including Ca. R. shennongii, A. phagocytophilum, and Ehrlichia spp. (E. canis and Ehrlichia sp.) was supported by previous studies (Yang et al., 2016; Iqbal et al., 2019; Shao et al., 2021; Ali et al., 2023b; Shehla et al., 2023a). The occurrence of Rickettsiales DNA in different animals suggests that there is a wide range of both identified and unidentified bacteria in Pakistan that may be transmitted between animals and humans. Therefore, it is crucial to carry out more extensive research studies to record these pathogenic bacteria and investigate their pathogenic roles in the natural environment. Moreover, it is crucial to conduct in-depth “One-Health” research to better understand the epidemiology, transmission, and pathogenicity of Rickettsiales bacteria in the country. The One-Health approach is especially important for devising strategies to manage tick infestations and associated TBDs for the aforementioned objectives.
Conclusion
This work presents the first genetic analysis of the Hy. asiaticum tick species in Pakistan using the cox1 genetic marker. In addition, this work reveals the genetic confirmation of tick species such as Rh. turanicus, Rh. microplus, and Hae. cornupunctata. Rickettsiales bacteria including Ca. R. shennongii, A. phagocytophilum, E. canis, and Ehrlichia sp. are molecularly reported in the aforementioned tick species. Moreover, the Ca. R. shennongii was reported for the first time in Hy. asiaticum tick species from Pakistan. Furthermore, the A. phagocytophilum was also genetically isolated from the DNA of cattle’s blood. The presence of diverse tick species and detection of Rickettsiales pathogen in ticks as well as cattle’s blood might suggest a lack of long-term surveillance that highlights the need for additional comprehensive studies regarding the epidemiology and evolutionary status of various ticks and their associated TBPs in Pakistan.
Data availability statement
The datasets presented in this study can be found in online repositories (https://www.ncbi.nlm.nih.gov/genbank/). The names of the repository/repositories and accession number(s) can be found below: Hy. asiaticum (OR826113), Rh. microplus (OR826114), Rh. turanicus (OR826115), Hae. cornupunctata (OR838743), A. phagocytophilum (OR826108), Ca. R. shennongii (ompA; OR838474, ompB; OR838475), E. canis (OR826110), and Ehrlichia sp. (OR826111).
Ethics statement
The requirement of ethical approval was waived by taking the verbal and written informed consent/General permission letter from the Animal owner’s before the sampling of ticks and blood. Additionally, all the blood samples from cattle were collected by veterinary officials, and animal owner’s showed their full trust and satisfaction. This study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MKO: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Writing – original draft, Writing – review & editing. ShS: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Writing – original draft, Writing – review & editing. GG: Formal analysis, Investigation, Funding acquisition, Methodology, Project administration, Resources, Software, Validation, Visualization, Writing – review & editing. MR: Funding acquisition, Project administration, Resources, Software, Validation, Writing – review & editing, Data curation, Supervision. SuS: Data curation, Writing – review & editing, Formal analysis, Methodology.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was carried out under the financial support of National Academy of Higher Education (NAHE), Higher Education Commission, Pakistan (Ref. No. 286/IPFP-II (Batch-I)/SRGP/NAHE/HEC/2020/120). The Science Fund for Creative Research Groups (22JR5RA024) and Special Project (22CX8NA011) of Gansu Province; National Parasitic Resources Center (NPRC-2019-194-30); the Agricultural Science and Technology Innovation Program (ASTIP, CAAS-ASTIP-2016-LVRI); NBCITS (CARS-37).
Acknowledgments
We acknowledge all the animal owners for providing consent. We acknowledge the veterinary officials for the collection of blood samples from the suspected cattle.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1346595/full#supplementary-material
References
Afridi, K. K., Habibullah, F. Z., Ahmad, F., Khan, M. T., Ahmad, T., Ullah, N., et al. (2023). Diversity and species composition of ixodid ticks in district Khyber, Khyber Pakhtunkhwa, Pakistan. Int. J. Agric. Biol. 30, 161–168.
Ahmad, A. S., Saeed, M. A., Rashid, I., Ashraf, K., Shehzad, W., Traub, R. J., et al. (2018). Molecular characterization of Hepatozoon canis from farm dogs in Pakistan. Parasitol. Res. 117, 1131–1138. doi: 10.1007/s00436-018-5790-1
Aiman, O., Ullah, S., Chitimia-Dobler, L., Nijhof, A. M., Ali, A. (2022). First report of Nosomma monstrosum ticks infesting Asian water buffaloes (Bubalus bubalis) in Pakistan. Ticks Tick Borne Dis. 13 (2), 101899. doi: 10.1016/j.ttbdis.2022.101899
Aktas, M., Özübek, S. (2015). Bovine anaplasmosis in Turkey: First laboratory confirmed clinical cases caused by Anaplasma phagocytophilum. Vet. Microb. 178 (3-4), 246–251. doi: 10.1016/j.vetmic.2015.05.021
Alam, S., Khan, M., Alouffi, A., Almutairi, M. M., Ullah, S., Numan, M., et al. (2022). Spatio-temporal patterns of ticks and molecular survey of Anaplasma marginale, with notes on their phylogeny. Microorganisms 10 (8), 1663. doi: 10.3390/microorganisms10081663
Ali, A., Khan, M. A., Zahid, H., Yaseen, P. M., Qayash Khan, M., Nawab, J., et al. (2019). Seasonal dynamics, record of ticks infesting humans, wild and domestic animals and molecular phylogeny of Rhipicephalus microplus in Khyber Pakhtunkhwa Pakistan. Front. Physiol. 10, 793. doi: 10.3389/fphys.2019.00793
Ali, A., Obaid, M. K., Almutairi, M. M., Alouffi, A., Numan, M., Ullah, S., et al. (2023a). Molecular detection of Coxiella spp. in ticks (ixodidae and argasidae) infesting domestic and wild animals: with notes on the epidemiology of tick-borne Coxiella burnetii in Asia. Front. Microbial., 14. doi: 10.3389/fmicb.2023.1229950
Ali, A., Ullah, S., Numan, M., Almutairi, M. M., Alouffi, A., Tanaka, T. (2023b). First report on tick-borne pathogens detected in ticks infesting stray dogs near butcher shops. Front. Vet. Sci. 10. doi: 10.3389/fvets.2023.1246871
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 (3), 403–410. doi: 10.1016/S0022-2836(05)80360-2
Atif, F. A., Abbas, R. Z., Mehnaz, S., Qamar, M. F., Hussain, K., Nazir, M. U., et al. (2022). First report on molecular surveillance based on duplex detection of Anaplasma marginale and Theileria annulata in dairy cattle from Punjab, Pakistan. Trop. Anim. Health Prod. 54 (2), 155. doi: 10.1007/s11250-022-03158-y
Batu, N., Wang, Y., Liu, Z., Huang, T., Bao, W., He, H., et al. (2020). Molecular epidemiology of Rickettsia sp. and Coxiella burnetii collected from Hyalomma asiaticum in Bactrian camels (Camelus bactrianus) in inner Mongolia of China. Vector Borne Zoonotic Dis. 11, 6, 101548. doi: 10.1016/j.ttbdis.2020.101548
Bekloo, J. A., Ramzgouyan, M. R., Shirian, S., Faghihi, F., Bakhshi, H., Naseri, F., et al. (2018). Molecular characterization and phylogenetic analysis of Anaplasma spp. and Ehrlichia spp. isolated from various ticks in southeastern and northwestern regions of Iran. Vector Borne Zoonotic Dis. 18, 5, 252–257. doi: 10.1089/vbz.2017.2219
Byaruhanga, C., Collins, N. E., Knobel, D. L., Khumalo, Z. T., Chaisi, M. E., Oosthuizen, M. C. (2018). Molecular detection and phylogenetic analysis of Anaplasma marginale and Anaplasma centrale amongst transhumant cattle in north-eastern Uganda. Ticks.Tick. Borne. Dis. 9, 3, 580–588. doi: 10.1016/j.ttbdis.2018.01.012
Ceylan, O., Uslu, A., Ceylan, C., Sevinc, F. (2021). Predominancy of rhipicephalus turanicus in tick-infested sheep from Turkey: a large-scale survey. Pak. Vet. J. 41 (3). doi: 10.29261/pakvetj/2021.036
Chitimia, L., Lin, R. Q., Cosoroaba, I., Wu, X. Y., Song, H. Q., Yuan, Z. G., et al. (2010). Genetic characterization of ticks from southwestern Romania by sequences of mitochondrial cox1 and nad 5 genes. Exp. Appl. Acarol. 52, 305–311. doi: 10.1007/s10493-010-9365-9
Dhanda, V., Kulkarni, S. M. (1969). Immature stages of Haemaphysalis cornupunctata Hoogstraal and Varma, (Acarina: Ixodidae) with new host and locality records, and notes on its ecology. Orient. Insects. 3, 15–21. doi: 10.1080/00305316.1969.10433891
Dugat, T., Lagrée, A. C., Maillard, R., Boulouis, H. J., Haddad, N. (2015). Opening the black box of Anaplasma phagocytophilum diversity: current situation and future perspectives. Front. Cell. Infect. Microbiol. 5, 61. doi: 10.3389/fcimb.2015.00061
Dumler, J. S. (2005). Anaplasma and ehrlichia infection. Ann. N Y Acad. Sci. 1063, 361–373. doi: 10.1196/annals.1355.069
Dumler, J. S., Barbet, A. F., Bekker, C. P., Dasch, G. A., Palmer, G. H., Ray, S. C., et al. (2001). Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and 'HGE agent'as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. 51, 6, 2145–2165. doi: 10.1099/00207713-51-6-2145
Dumler, J. S., Choi, K. S., Garcia-Garcia, J. C., Barat, N. S., Scorpio, D. G., Garyu, J. W., et al. (2005). Human granulocytic anaplasmosis and Anaplasma phagocytophilum. Emerg. Infect. Dis. 11 (12), 1828. doi: 10.3201%2Feid1112.050898
Filipe, D., Parreira, R., Pereira, A., Galvão, N., Cristóvão, J. M., Nunes, M., et al. (2021). Preliminary comparative analysis of the resolving power of COX1 and 16S-rDNA as molecular markers for the identification of ticks from Portugal. Veterinary Parasitology. Reg. Stud. Rep. 24, 100551. doi: 10.1016/j.vprsr.2021.100551
Ghafar, A., Gasser, R. B., Rashid, I., Ghafoor, A., Jabbar, A. (2020b). Exploring the prevalence and diversity of bovine ticks in five agro-ecological zones of Pakistan using phenetic and genetic tools. Ticks.Tick. Borne. Dis. 11, 5, 101472. doi: 10.1016/j.ttbdis.2020.101472
Gilbert, l., Aungier, J., Tomkins, Jl. (2014). Climate of origin affects tick (Ixodes ricinus) host-seeking behavior in response to temperature: implications for resilience to climate change? Ecol. Evol. 4, 1186–1198. doi: 10.1002/ece3.1014
Gillespie, J. J., Beier, M. S., Rahman, M. S., Ammerman, N. C., Shallom, J. M., Purkayastha, A., et al. (2007). Plasmids and Rickettsial evolution: insight from Rickettsia felis. PloS One 2 (3), e266. doi: 10.1371/journal.pone.0000266
Hall, T., Biosciences, I., Carlsbad, C. (2011). BioEdit: an important software for molecular biology. GERF Bull. Biosc. 2 (1), 60–61.
Hassan, M. A., Liu, J., Rashid, M., Iqbal, N., Guan, G., Yin, H., et al. (2018). Molecular survey of piroplasm species from selected areas of China and Pakistan. Parasites Vectors 11, 1–7. doi: 10.1186/s13071-018-3035-x
Hoogstraal, H., Varma, M. (1962). Haemaphysalis cornupunctata sp. n. and H. kashmirensis sp. n. from Kashmir, with Notes on H. sundrai Sharif and H. sewelli Sharif of India and Pakistan (Ixodoidea, Ixodidae). J. Parasitol. 48, 185–194. doi: 10.2307/3275561
Hosseini-Chegeni, A., Hosseini, R., Tavakoli, M., Telmadarraiy, Z., Abdigoudarzi, M. (2013). The Iranian Hyalomma (Acari: Ixodidae) with a key to the identification of male species 2, 3. doi: 10.22073/pja.v2i3.10046
Hu, H., Liu, Z., Fu, R., Liu, Y., Ma, H., Zheng, W. (2022). Detection and phylogenetic analysis of tick-borne bacterial and protozoan pathogens in a forest province of eastern China. Acta Tropica 235, 106634. doi: 10.1016/j.actatropica.2022.106634
Iatta, R., Sazmand, A., Nguyen, V. L., Nemati, F., Ayaz, M. M., Bahiraei, Z., et al. (2021). Vector-borne pathogens in dogs of different regions of Iran and Pakistan. Parasitol. Res. 120, 1–10. doi: 10.1007/s00436-020-06992-x
Iqbal, N., Mukhtar, M. U., Yang, J., Sajid, M. S., Niu, Q., Guan, G., et al. (2019). First molecular evidence of Anaplasma bovis and Anaplasma phagocytophilum in bovine from central Punjab, Pakistan. Pathogens 8 (3), 155. doi: 10.3390/pathogens8030155
Jabbar, A., Abbas, T., Sandhu, Z. U. D., Saddiqi, H. A., Qamar, M. F., Gasser, R. B. (2015). Tick-borne diseases of bovines in Pakistan: major scope for future research and improved control. Parasites Vectors 8 (1), 1–13. doi: 10.1186/s13071-015-0894-2
Jongejan, F., Uilenberg, G. (2004). The global importance of ticks. Parasitology 129 (S1), S3–S14. doi: 10.1017/S0031182004005967
Kaiser, M. N., Hoogstraal, H. (1963). The Hyalomma ticks (Ixodoidea, ixodidae) of Afghanistan 130–139. doi: 10.2307/3275691
Karim, S., Budachetri, K., Mukherjee, N., Williams, J., Kausar, A., Hassan, M. J., et al. (2017). A study of ticks and tick-borne livestock pathogens in Pakistan. PloS Negl. Trop. Dis. 11 (6), e0005681. doi: 10.1371/journal.pntd.0005681
Khamassi Khbou, M., Rouatbi, M., Romdhane, R., Sassi, L., Jdidi, M., Haile, A., et al. (2021). Tick infestation and piroplasm infection in barbarine and queue fine de l’ouest autochthonous sheep breeds in Tunisia, North Africa. Animals 11 (3), 839. doi: 10.3390/ani11030839
Khan, S. M., Khan, M., Alouffi, A., Almutairi, M. M., Numan, M., Ullah, S., et al. (2023). Phylogenetic Position of Haemaphysalis kashmirensis and Haemaphysalis cornupunctata, with Notes on Rickettsia spp. Genes 14 (2), 360. doi: 10.3390/genes14020360
Khan, A., Niaz, S., Khan, A., Ahmed, H., Khattak, I., Zeb, J., et al. (2020). Molecular detection of small ruminant piroplasmosis and first report of Theileria luwenshuni (Apicomplexa: Theileridae) in small ruminants of Pakistan. Exp. Parasitol. 212, 107872. doi: 10.1016/j.exppara.2020.107872
Khan, Z., Shehla, S., Alouffi, A., Kashif Obaid, M., Zeb Khan, A., Almutairi, M. M., et al. (2022). Molecular survey and genetic characterization of Anaplasma marginale in ticks collected from livestock hosts in Pakistan. Animals 12 (13), p.1708. doi: 10.3390/ani12131708
Kocan, K. M., de la Fuente, J., Guglielmone, A. A., Meléndez, R. D. (2003). Antigens and alternatives for control of Anaplasma marginale infection in cattle. Clin. Microbiol. Rev. 16 (4), pp.698–pp.712. doi: 10.1128/cmr.16.4.698-712.2003
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), p.1547. doi: 10.1093/molbev/msy096
Leger, E., Vourc’h, G., Vial, L., Chevillon, C., McCoy, K. D. (2013). Changing distributions of ticks: causes and consequences. Exp. Appl. Acarol. 59, 219–244. doi: 10.1007/s10493-012-9615-0
Malik, M. I., Qamar, M., Ain, Q., Hussain, M. F., Dahmani, M., Ayaz, M., et al. (2018). Molecular detection of Ehrlichia canis in dogs from three districts in Punjab (Pakistan). Vet. Med. Sci. 4 (2), 126–132. doi: 10.1002/vms3.94
Numan, M., Islam, N., Adnan, M., Zaman Safi, S., Chitimia-Dobler, L., Labruna, M. B., et al. (2022). First genetic report of Ixodes kashmiricus and associated Rickettsia sp. Parasites Vectors 15 (1), 1–12. doi: 10.1186/s13071-022-05509-y
Obaid, M. K., Almutairi, M. M., Alouffi, A., Safi, S. Z., Tanaka, T., Ali, A. (2023). Assessment of cypermethrin and amitraz resistance and molecular profiling of voltage-gated sodium channel and octopamine tyramine genes of Rhipicephalus microplus. Front. Front. Cell. Infect. Microbial. 13, 1176013. doi: 10.3389/fcimb.2023.1176013
Paddock, C. D., Alvarez-Hernández, G. (2018). “Rickettsia rickettsii (Rocky Mountain spotted fever),” in Principles and Practice of Pediatric Infectious Diseases (Mexico: Elsevier Inc), 952–957.
Parola, P., Paddock, C. D., Socolovschi, C., Labruna, M. B., Mediannikov, O., Kernif, T., et al. (2013). Update on tick-borne rickettsioses around the world: a geographic approach. Clin. Microbiol. Rev. 26 (4), 657–702. doi: 10.1128/CMR.00032-13
Parola, P., Roux, V., Camicas, J. L., Baradji, I., Brouqui, P., Raoult, D. (2000). Detection of Ehrlichiae in African ticks by polymerase chain reaction. Trans. R. Soci. Trop. Med. Hyg. 94 (6), 707–708. doi: 10.1016/S0035-9203(00)90243-8
Rashid, M., Akbar, H., Rashid, I., Saeed, K., Ahmad, L., Ahmad, A. S., et al. (2018). Economic significance of tropical theileriosis on a Holstein Friesian dairy farm in Pakistan. J. Parasitol. Res. 104 (3), 310–312. doi: 10.1645/16-179
Rizwan, M., Ali, S., Javid, A., Von Fricken, M. E., Rashid, M. I. (2023). Molecular epidemiology of Bartonella species from sympatric mammals collected in urban and rural areas of Punjab, Pakistan. Acta tropica. 243, 106940. doi: 10.1016/j.actatropica.2023.106940
Roux, V., Fournier, P. E., Raoult, D. (1996). Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rompA. J. Clin. Microbiol. 34 (9), 2058. doi: 10.1128/jcm.34.9.2058-2065.1996
Roux, V., Raoult, D. (2000). Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rompB (ompB). Int. J. Syst. Evol. Microbiol. 50, 4, 1449–1455. doi: 10.1099/00207713-50-4-1449
Sambrook, J., Fritsch, E. F., Maniatis, T. E. (1989). Molecular Cloning: A Laboratory Manual. Ed. 2 (New York, NY, United States: Cold Spring Harbor Laboratory Press).
Santos, A. S., Amaro, F., Santos-Silva, M. M., De Sousa, R., Mathias, M. L., Ramalhinho, M. G., et al. (2009). Detection of antibodies against Anaplasma phagocytophilum in Algerian mice (Mus spretus), Portugal. Vector-Borne Zoonotic Dis. 9 (6), 663–669. doi: 10.1089/vbz.2008.0066
Schmutzhard, E., Helbok, R. (2014). Rickettsiae, protozoa, and opisthokonta/metazoa. Handb. Clin. Neurol. 121, 1403–1443. doi: 10.1016/B978-0-7020-4088-7.00096-1
Shao, J. W., Yao, X. Y., Song, X. D., Li, W. J., Huang, H. L., Huang, S. J., et al. (2021). Molecular detection and genetic diversity of Rickettsia spp. in pet dogs and their infesting ticks in Harbin, northeastern China. BMC Vet. Res. 17, 1–9. doi: 10.1186/s12917-021-02823-y
Shehla, S., Almutairi, M. M., Alouffi, A., Tanaka, T., Chang, S. C., Chen, C. C., et al. (2023b). Molecular survey of Rickettsia raoultii in ticks infesting livestock from Pakistan with notes on pathogen distribution in palearctic and oriental regions. Vet. Sci. 10 (11), 636. doi: 10.3390/vetsci10110636
Shehla, S., Ullah, F., Alouffi, A., Almutairi, M. M., Khan, Z., Tanaka, T., et al. (2023a). Association of SFG Rickettsia massiliae and Candidatus Rickettsia shennongii with Different Hard Ticks Infesting Livestock Hosts. Pathogens 12 (9), 1080. doi: 10.3390/pathogens12091080
Strle, F. (2004). Human granulocytic ehrlichiosis in Europe. Int. J. Med. Microbiol. 293 (Suppl 37), 27–35. doi: 10.1016/S1433-1128(04)80006-8
Stuen, S., Bergström, K., Palmer, E. (2002). Reduced weight gain due to subclinical Anaplasma phagocytophilum (formerly Ehrlichia phagocytophila) infection. Exp. Appl. Acarol. 28, 209–215. doi: 10.1023/A:1025350517733
Stuen, S., Nevland, S., Moum, T. (2003). Fatal cases of tick-borne fever (TBF) in sheep caused by several 16S rRNA gene variants of Anaplasma phagocytophilum. Ann. N. Y. Acad. Sci. 990 (1), 433–434. doi: 10.1111/j.1749-6632.2003.tb07407.x
Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 (22), 4673–4680. doi: 10.1093/nar/22.22.4673
Walker, J. B., Keirans, J. E., Horak, I. G. (2005). The genus Rhipicephalus (Acari, Ixodidae): a guide to the brown ticks of the world (UK: Cambridge University Press).
Yang, J., Liu, Z., Niu, Q., Liu, J., Xie, J., Chen, Q., et al. (2016). Evaluation of different nested PCRs for detection of Anaplasma phagocytophilum in ruminants and ticks. BMC Vet. Res. 12 (1), 1–6. doi: 10.1186/s12917-016-0663-2
Young, A. S., Groocock, C. M., kariuki, D. P. (1988). Integrated control of ticks and tick-borne diseases of cattle in Africa. Parasitology 96, 403–432. doi: 10.1017/s0031182000058388
Zahid, H., Alouffi, A., Almutairi, M. M., Ateeq, M., Tanaka, T., Chang, S.-C., et al. (2023). Ticks infesting ducks, domestic fowls and bats in Pakistan: first report on molecular survey and phylogenetic position of Borrelia anserina. Vet. Sci. 10, 628.
Zhao, L., Lv, J., Li, F., Li, K., He, B., Zhang, L., et al. (2020). Identification and molecular analysis of ixodid ticks (Acari: Ixodidae) infesting domestic animals and tick-borne pathogens at the Tarim Basin of Southern Xinjiang, China. Korean J. Parasitol. 58 (1), 37. doi: 10.3347/kjp.2020.58.1.37
Keywords: Anaplasma phagocytophilum, Hyalomma asiaticum, Candidatus Rickettsia shennongii, Ehrlichia, blood-borne pathogens
Citation: Obaid MK, Shehla S, Guan G, Rashid M and Shams S (2024) Genotyping of ticks: first molecular report of Hyalomma asiaticum and molecular detection of tick-borne bacteria in ticks and blood from Khyber Pakhtunkhwa, Pakistan. Front. Cell. Infect. Microbiol. 14:1346595. doi: 10.3389/fcimb.2024.1346595
Received: 29 November 2023; Accepted: 22 January 2024;
Published: 12 March 2024.
Edited by:
Ala E. Tabor, The University of Queensland, AustraliaReviewed by:
Benjamin Cull, University of Minnesota Twin Cities, United StatesLudovic Pruneau, Université des Antilles, Guadeloupe
Copyright © 2024 Obaid, Shehla, Guan, Rashid and Shams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Kashif Obaid, a2FzaGlmb2JhaWRrYW56QGdtYWlsLmNvbQ==; Guiquan Guan, Z3Vhbmd1aXF1YW5AY2Fhcy5jbg==
 Muhammad Kashif Obaid
Muhammad Kashif Obaid Shehla Shehla
Shehla Shehla Guiquan Guan
Guiquan Guan Muhammad Rashid
Muhammad Rashid Sumaira Shams2
Sumaira Shams2