
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 13 May 2024
Sec. Intestinal Microbiome
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1323157
This article is part of the Research TopicImpact of landscape and feeding on the bees’ gut microbiome shaping and pathogens developmentView all 4 articles
 Chiara Braglia1
Chiara Braglia1 Daniele Alberoni1*
Daniele Alberoni1* Paula Melisa Garrido2,3
Paula Melisa Garrido2,3 Martin Pablo Porrini2,3
Martin Pablo Porrini2,3 Loredana Baffoni1
Loredana Baffoni1 Dara Scott4
Dara Scott4 Martin Javier Eguaras2,3
Martin Javier Eguaras2,3 Diana Di Gioia1
Diana Di Gioia1 David Mifsud5
David Mifsud5The genus Serratia harbors opportunistic pathogenic species, among which Serratia marcescens is pathogenic for honeybees although little studied. Recently, virulent strains of S. marcescens colonizing the Varroa destructor mite’s mouth were found vectored into the honeybee body, leading to septicemia and death. Serratia also occurs as an opportunistic pathogen in the honeybee’s gut with a low absolute abundance. The Serratia population seems controlled by the host immune system, but its presence may represent a hidden threat, ready to arise when honeybees are weakened by biotic and abiotic stressors. To shed light on the Serratia pathogen, this research aims at studying Serratia’s development dynamics in the honeybee body and its interactions with the co-occurring fungal pathogen Vairimorpha ceranae. Firstly, the degree of pathogenicity and the ability to permeate the gut epithelial barrier of three Serratia strains, isolated from honeybees and belonging to different species (S. marcescens, Serratia liquefaciens, and Serratia nematodiphila), were assessed by artificial inoculation of newborn honeybees with different Serratia doses (104, 106, and 108 cells/mL). The absolute abundance of Serratia in the gut and in the hemocoel was assessed in qPCR with primers targeting the luxS gene. Moreover, the absolute abundance of Serratia was assessed in the gut of honeybees infected with V. ceranae at different development stages and supplied with beneficial microorganisms and fumagillin. Our results showed that all tested Serratia strains could pass through the gut epithelial barrier and proliferate in the hemocoel, with S. marcescens being the most pathogenic. Moreover, under cage conditions, Serratia better proliferates when a V. ceranae infection is co-occurring, with a positive and significant correlation. Finally, fumagillin and some of the tested beneficial microorganisms could control both Serratia and Vairimorpha development. Our findings suggest a correlation between the two pathogens under laboratory conditions, a co-occurring infection that should be taken into consideration by researches when testing antimicrobial compounds active against V. ceranae, and the related honeybees survival rate. Moreover, our findings suggest a positive control of Serratia by the environmental microorganism Apilactobacillus kunkeei in a in vivo model, confirming the potential of this specie as beneficial bacteria for honeybees.
The decline in bee populations and the arising consequences are well-known to the agricultural sector. Climate changes and the widespread use of herbicides reduce the feed availability for pollinators due to a decreased flower production and delay in flowering time (Le Conte and Navajas, 2008; Carpenter et al., 2020). The stress derived from the reduced sources of foraging may find a synergy with other environmental or anthropogenic factors in the agroecosystem (Brandt et al., 2016; Raymann et al., 2017), leading to a further weakening of honeybees. For instance, it was recently demonstrated that the gut microbial community of honeybees can be altered by exposure to herbicides (Motta et al., 2018), insecticides (Alberoni et al., 2021b), and antibiotics (Baffoni et al., 2021), downregulating the expression of host-produced antimicrobial peptides, therefore altering the insect immune system (Motta et al., 2022) and pathogen susceptibility (Li et al., 2017; Castelli et al., 2020).
Among bees, honeybees are the most studied pollinators because of their economic relevance, in terms of not only their pollination services but also their colony products. A great number of honeybee microbial pathogens, whose virulence can be increased by environmental stressors, are known, including Vairimorpha ceranae (or syn. Nosema ceranae) (Higes et al., 2013) and viruses such as the chronic bee paralysis virus (Martin et al., 2013; Zheng et al., 2015; Ullah et al., 2021). In addition, in recent years, opportunistic bacterial pathogens such as Hafnia, Klebsiella, Serratia, and Enterobacter have drawn attention. These bacteria co-exist with their host and normally do not cause a disease, but they can become pathogenic or increase their relative abundance when the host’s defense system is impaired (Brown et al., 2012; Raymann and Moran, 2018) or in honeybees exposed to pesticides (Al Naggar et al., 2022; Al Naggar and Wubet, 2024). For example, some of them produce chitinases, which can seriously damage the exoskeleton of invertebrates or help them under certain conditions (Gooday, 1990). Iverson et al. (1984) found that Serratia liquefaciens and Serratia marcescens can penetrate in the pupae of Tetanops myopaeformis, establishing a symbiotic association that helps the beet root maggot emergence. Conversely, in some crab species, Serratia causes serious exoskeleton damage (Baross et al., 1978). However, the role of opportunistic bacteria is poorly investigated despite their potential impact on the host, especially Serratia.
The interaction between quality and abundance of feed sources and susceptibility to diseases has been observed in many organisms (Dolezal and Toth, 2018; Dolezal et al., 2019). Although mechanisms involved are not yet well known, it was postulated that pollen and nectar contain proteins, lipids, carbohydrates, and a variety of phytochemicals and micronutrients that may affect immune response. In addition, feed quality has been found to alter the proportion of some core gut microorganisms in honeybees, for both artificial (Alberoni et al., 2021a; Ricigliano et al., 2022) and natural nutrition (Castelli et al., 2020). Consequently, a possible reduction or loss of specific gut microbiome taxa can alter the protective functions of the immune system (Kwong et al., 2017) and potentially induce the expression of pathogenicity traits by opportunistic microorganisms (Raymann et al., 2018; Horak et al., 2020) like Serratia.
Serratia is a Gram-negative opportunistic environmental pathogen that may colonize the hemolymph of larvae and adult honeybees causing septicemia (Grimont and Grimont, 1978; Burritt et al., 2016). In adults, the bacterium causes a reduction of motility up to paralysis (Burritt et al., 2016), while larvae die and appear whitish, sticky, and smelly (El Sanousi et al., 1987) with a symptomatology like foulbrood. Serratia can be present in low amounts as a non-core bacterial species in the honeybee gut microbiota (Raymann et al., 2017, 2018); it is recognized that, when Serratia gut strains colonize the hemolymph and body cavity (hemocoel), it becomes highly virulent (Burritt et al., 2016; Raymann et al., 2018) as an opportunistic pathogen. However, a well-performing core gut microbiome is correlated with the production of antimicrobial compounds and host immune stimulation that leads to the control of pathogen proliferation in the gut, as documented for S. marcescens (Steele et al., 2017).
Concerning S. marcescens, a particularly virulent strain was isolated by Burritt et al. (2016) and named strain Sicaria (Ss1). This strain was responsible for atypical winter losses, and it was discovered to colonize the mouthpart of Varroa destructor. Moreover, the Sicaria strain has proven its virulence when vectored by V. destructor and injected directly into the honeybee hemocoel (Burritt et al., 2016). After this first isolation, Raymann et al. (2018) isolated other S. marcescens strains showing a similar mode of infection. The presence of Varroa mites associated with gut microbiota perturbations appear to be optimal conditions for opportunistic pathogens widespread in adult honeybees (Hubert et al., 2016). Therefore, conditions leading to honeybee stress could be determinant for Serratia proliferation and pathogenesis. For this reason, the aim of this research is to establish a possible connection between an immune depressing and tissue damaging honeybee gut parasite such as V. ceranae and Serratia colonization. In fact, V. ceranae is well known to cause severe gut epithelial damages (Martin et al., 2013). In this work, the pathogenicity of three different Serratia species isolated in Italy from honeybee gut was assessed together with their ability to colonize hemocoel through the gut epithelial cell wall under laboratory conditions. Moreover, the synergy with the gut pathogen V. ceranae was assessed and evaluated after the administration of a feed supplement based on microorganisms, pollen, and the antibiotic fumagillin already tested against Vairimorpha (Alberoni et al., 2018; Garrido et al., 2024). In particular, the presence and amount of the co-occurring and opportunistic pathogen Serratia were taken into consideration in a sample subset produced in Garrido et al. (2024) and a possible correlation with V. ceranae was considered.
The isolation was performed both from the gut content of honeybees and from hive debris. Approximately 15 worker honeybees were collected from an experimental apiary (Valsamoggia, Bologna, Italy) in 2014 and promptly transferred to the laboratory. All bees were sacrificed by freezing, and their gut content was extracted and mixed to obtain 1 g. One gram of colony debris was also collected. Both isolation matrixes were serially diluted in a 0.85% NaCl solution and plated on GYC Agar (50 g/L dextrose, 10 g/L yeast extract, 5 g/L calcium carbonate, and a 10 g/L agar medium with no addition of antibiotics or ethanol). Plates were incubated for 5 days at 30°C under aerobic conditions. Strains were selectively isolated based on morphology, growth rate, and color. Isolates were characterized with a PCR-dependent fingerprinting technique based on the enterobacterial repetitive intergenic consensus (ERIC) sequence. PCR was carried out with primers ERIC-1 (5’-ATGTAAGCTCCTGGGGATTCAC-3’) and ERIC-2 (5’-AAGTAAGTGACTGGGGTGAGCG-3’) as described in Alberoni et al. (2019). Fingerprinting profiles were analyzed with GelCompar II 6.6 (Applied Maths, Kor-trijk, Belgium) using the DICE coefficient and the UPMGA clustering algorithm. DNA amplification of the 16S rRNA gene was performed for samples with an ERIC unique profiles, with primers 27f (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492r (5’-GGTTACCTTGTTACGACT-3’) according to Alberoni et al. (2019). Amplicons were purified and sent to a commercial sequencing facility (Eurofins MWG, Ebersberg, Germany). Sequence chromatograms were analyzed, manually edited, and classified using BLAST tool from NCBI. The honeybee survival test using the Serratia isolated strains was performed according to Garrido et al. (2024), whereas the mortality and diet consumption information are reported in the Supplementary Materials of Garrido et al. (2024). Briefly, 50 newly emerged honeybees were caged and fed with Serratia inoculum of 1.2 × 108 CFU/mL, 1.2 × 106 CFU/mL, or 1.2 × 104 CFU/mL incorporated in sugar syrup (sucrose:water 1:1 w/v). Each experimental condition was repeated in triplicate and was performed with three different Serratia strains belonging to the species S. liquefaciens, S. marcescens, and S. nematodiphila. Dead honeybees were registered and removed daily as well as the amount of feed consumed.
Two- to three-day-old honeybees were manually collected from a mother colony located in Valsamoggia (Bologna district, Italy, 44°25’54.0”N 11°02’55.2”E). In the laboratory, honeybees were individually infected with 1.13 × 104, 1.13 × 106, and 1.13 × 108 cells of S. liquefaciens; 1.8 × 104, 1.8 × 106, and 1.8 × 108 cells of S. marcescens; and 2.5 × 104, 2.5 × 106, and 2.5 × 108 cells of S. nematodiphila. The prepared bacterial suspensions were administered with sugar syrup (sucrose:water 1:1 w/v) at a final volume of 10 μL/bee. For each Serratia strain dilution, 50 honeybees were caged and maintained at 29°C ± 2 and 60% RH. Fresh tap water and sugar syrup were supplied ad libitum and replaced every day. Every 2 days, three honeybees per cage were sacrificed and prepared for the DNA extraction: gut was carefully separated from the abdomen, and guts and the fat bodies with hemocoel processed separately although attached to the abdomen. DNA extraction was performed as described in Section 2.5, and Serratia was quantified as described in Section 2.5. The experimental conditions and abbreviations are defined in Table 1, section 1.
To obtain V. ceranae fresh spores necessary to perform the controlled infections, forager honeybees were collected from diseased hives [experimental apiary of Social Bees Research Center (CIAS), Buenos Aires Province, Argentina, 38°10’06”S, 57°38’10”W]. When spores were unavailable, to obtain fresh and infective spores, V. ceranae was multiplied by infecting 35 newly emerged and caged honeybees, allowing pathogen proliferation into the gut epithelium and sporulation for 14 days.
To reach honeybee genetic homogeneity, newly emerged honeybees were obtained from brood frames picked from multiple colonies and incubated until honeybee emergence at 32°C and 60% RH. The newly emerged honeybees were maintained under controlled conditions (29°C; 60% RH) for 3 days until individual inoculation with freshly prepared V. ceranae spores. Briefly, according to Porrini et al. (2013), each single starved honeybee was put in an infection panel and fed with 10 μL of V. ceranae spores mixed with sugar syrup.
In each laboratory experiment, homogeneous cohorts of worker honeybees from healthy hives were used. Groups of 50 newly emerged honeybees were placed into cages (12 cm × 8 cm × 6 cm) per treatment per replicate according to Porrini et al. (2020). They were fed daily with fresh tap water, freshly obtained beebread when it was necessary, and syrup to act as a vehicle for the respective treatments. Individuals were maintained at 29°C ± 2 (40% RH) during the assays.
In all cases, DNA was extracted from the gut content and used for the quantification of Serratia as described below. Some DNA samples were stored as part of wider studies according to Braglia et al. (2021) and Garrido et al. (2024), and others were obtained in this study (Table 1).
The test aimed at determining whether different infective doses of V. ceranae affect endogenous Serratia proliferation. Honeybees were inoculated with 5 × 102, 5 × 103, and 5 × 104 V. ceranae spores according to Garrido et al. (2024). At the same time, the antibiotic fumagillin [F] was also assessed for the indirect control of Serratia. The experimental conditions and abbreviations are shown in Table 1, section 2.
Three assays were performed exclusively with bacterial strains isolated from honeybee midgut. The first one aimed at comparing the effect of six single bacterial taxa and a mixture of them in the Serratia control, in the presence of V. ceranae infection. Bifidobacterium asteroides DSM20431, Bifidobacterium coryneforme C155, Bifidobacterium indicum DSM20214, Apilactobacillus kunkeei Dan39, Lactiplantibacillus plantarum Dan91, and Lactobacillus johnsonii Dan92, previously described in Baffoni et al. (2016), were grown according to Alberoni et al. (2019). Incubation conditions were standardized for each strain at 35 ± 2°C for 24 h for Lactobacillaceae and 72 h for Bifidobacteriaceae. Microbial cultures were centrifuged, and pelleted cells were resuspended in sugar syrup. When a suspension of all these bacteria was used for honeybee feeding, it is referred to as bacterial mixture [B]. This was prepared in sugar syrup suspending grown bacterial cells in order to have 5 × 107 CFU/mL of each bacterial strain except for B. coryneforme C155 that showed growth problems, and a concentration of 2.1 × 106 CFU/mL was obtained in the sugar syrup. The six single bacterial strains and their mixture were administered orally to honeybees infected with 5 × 104 V. ceranae spores according to Garrido et al. (2024). The experimental conditions and abbreviations are shown in Table 1, section 3. The samples obtained were analyzed by qPCR to obtain the number of copies of Serratia according to Section 2.5 in V. ceranae-positive samples.
The assay was carried out, as described by Cintas et al. (1995), on soft agar inoculated with indicator strains. The indicator strains used were S. marcescens AC8, S. liquefaciens AC4, and Serratia nematodiphila 1A. Fumagillin dicyclohexylamine salt was used at the following concentrations: 2.59 mM, 1.30 mM, 0.648 mM, 0.324 mM, 0.162 mM, and no antibiotic. Fumagillin was added in the wells and the sensitivity was assessed controlling the formation of inhibition halos. The assay was performed in triplicate.
Honeybees for each experimental condition per replicate were sacrificed as described in Garrido et al. (2024), and the guts were individually collected and processed according to Braglia et al. (2021) and Garrido et al. (2024). Briefly, guts were manually crumbled with plastic micro pestles in 200 μL of buffer, and DNA extraction of single honeybee guts was performed with a High Pure PCR template preparation kit (Roche Diagnostic; Buenos Aires, Argentina) following the manufacturer’s protocol. Fluorometric quantification of every sample was performed with a Qubit Flex Fluorometer (Thermo Fisher Scientific), and finally, extracted DNA was stored at −20°C until further analysis. The quorum-sensing luxS gene was selected as a molecular marker to perform Serratia quantitative PCR. The primers used were as follows: luxS1: 5’-TGCCTGGAAAGCGGCGATGG-3’; luxS2: 5’-CGCCAGCTCGTCGTTGTGGT-3’, specifically designed for Serratia’s quorum-sensing autoinducer-2 (AI-2) luxS gene by Joyner et al. (2014). The standard curve was prepared according to Baffoni et al (2012, 2016). Briefly, standard curves were constructed using PCR products of the luxS gene for the target microbial genera. The PCR products were purified with NucleoSpin® Gel and PCR Clean-up (Macherey-Nagel), quantified with Qubit high sensitivity, converted in total amount of copies per microliter and then serially diluted to obtain standards ranging from 5×103 to 5×108 gene copies.
The rRNA small ribosomal subunit gene was selected to perform V. ceranae-specific qPCR with primers Nc841f: 5’-GAGAGAACGGTTTTTTGTTTGAGA-3’ and Nc980r: 5’-ATCCTTTCCTTCCTACACTGATTG-3’ (amplicon size ~140 bp) designed by Huang and Solter (2013). Standard curve was prepared as described above. Reactions were carried out on a StepOne thermal cycler (Applied Biosystems) with the standard two-step PCR method using Fast SYBR Green PCR Master Mix (Life Technologies, Milan, Italy), according to Garrido et al. (2024).
The survival/mortality test was carried out by comparing different groups by Log-Rank survival curves and Mann–Whitney tests, respectively, unless otherwise noted, according to Garrido et al. (2024). Data were analyzed with R software (de Micheaux et al., 2013). Shapiro and Levene’s tests were performed for every dataset to test normality and homoscedasticity distribution of data. The dataset was corrected with Cook’s Distance multivariate method only for cage tests, to identify outliers based on regression analysis comparison (Cook, 1979). Datasets were analyzed with ANOVA when data were normal and homoscedastic, while GLM was applied for non-normal homoscedastic data. The Bonferroni p-value correction model was used for multiple comparisons in every independent assay. Correlation analyses were performed with the R packages devtools and ggpubr relaying on Pearson’s correlation test. For the correlation on the second test, CV2 and CV3 were deleted from the analysis since the opposite trend of CV2 and CV3 was caused by the higher Vairimorpha inoculum. Finally, boxplots and average plots were generated with ggpubr and ggplot2 packages, and where necessary, they overlapped with Adobe Illustrator. Serratia quantification was expressed as luxS gene copies/gut.
The understanding of honeybees’ pathogen dynamics, in particular the synergies among different pathogens, as well as their interactions with environmental or anthropogenic factors (climate changes, feed resources, and pesticides), is crucial for the beekeeping sector. Knowledge of the synergies between pathogen(s) and stressors may allow the design of target strategies to support honeybees’ health. A previous work (Alberoni et al., 2023) has shown that the administration of a bacterial mixture of different Lactobacillaceae and Bifidobacterium strains, tested to counteract N. ceranae, seemed to reduce the natural proliferation of Serratia in the gut. In the same work, when honeybees were not treated with the bacterial mixture, Serratia was found to proliferate, together with other gut microorganisms such as Citrobacter, Cosenzaea, and Morganella, and therefore, it was suggested that the selected mixture of microorganisms could control Serratia under cage conditions. In the present work we aimed at investigating the impact of different beneficial bacterial and their mixture, on Serratia, in case of co-infection with N. ceranae.
A total of 18 strains were isolated from the honeybee gut on GYC (Supplementary Table S1). Among these, after the 16S rRNA gene sequencing according to Alberoni et al. (2019), three bacterial species were identified as Serratia: S. marcescens AC8, S. liquefaciens AC4, and S. nematodiphila 1A (NCBI accession numbers MG649995, MG649996, and MG650059, respectively), and they were used for a survival assay to test their potential pathogenicity and virulence on honeybees. The results of the survival test showed a high pathogenicity of the S. marcescens strain when administered through feed. S. liquefaciens AC4 and S. nematodiphila 1A also caused a statistically significant mortality starting from the concentration of 1.2 × 106 (p < 0.01; Figure 1). Although a high infective dose in the environment is unlikely to be found, this test confirms that S. marcescens may be highly pathogenic when encountered by honeybee. Moreover, a lower infective dose selected such as 104 CFU/mL of sugar syrup led to high mortality, whereas S. liquefaciens and S. nematodiphila caused a honeybee mortality only at the highest doses (106–108 CFU/mL), confirming the higher pathogenicity of S. marcescens.
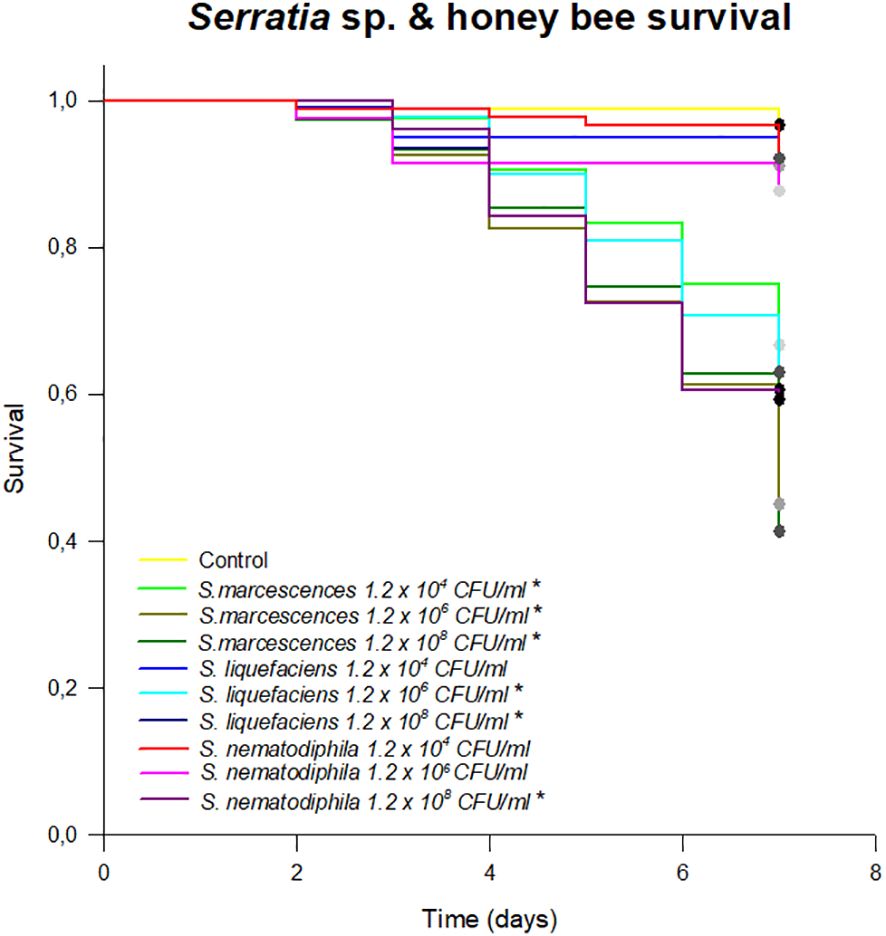
Figure 1 Survival test expressing the % of honeybees survived in an 8-day test following the chronic exposure to different species of Serratia. Asterisk (*) indicates statistical differences with the control treatment (syrup without Serratia). *p < 0.01.
Previous research works (Burritt et al., 2016; Raymann et al., 2018) have shown that particularly virulent strains of Serratia can proliferate in the hemocoel and cause a rapid death of the honeybees. Burritt et al. (2016) suggested that the transmission of the pathogen to the hemocoel was triggered by V. destructor, which acted as a vector, exactly as it happens for viruses such as Deformed Wing Virus (DWV) and Acute Bee Paralysis Virus (ABPV) (reviewed in Rosenkranz et al., 2010). However, this transmission mechanism may not be the only one (reviewed in Vallet-Gely et al., 2008). Serratia could also cross the intestinal epithelial barrier and colonize the hemocoel, and, from there, over-proliferate, causing septicemia. The test described in the present work, in which honeybees were infected with different strains of Serratia since the beginning of the test, aims at validating this proliferation path, which might easily escape the treatments for the control of Varroa mites. Our work shows that, in honeybee not inoculated with Serratia, the basal Serratia amount did not vary between the gut and the hemocoel and resulted below the detection limit (1 gene copy) for all the sampling times T1 (3 days), T2 (6 days), and T3 (9 days), respectively (Figure 2A). However, following the infection with S. marcescens at different concentrations, the number of S. marcescens cells in both gut and hemocoel proportionally increased according to the inoculum but did not significantly vary between the sampling times, as shown in Figure 2B; Supplementary Table S2. Moreover, our results show that the increase of S. marcescens load in the gut corresponded to a proportional infection in the hemocoel, during the first 6 days after Serratia inoculation. Then, S. marcescens counts decreased in the gut, while they increased in the hemocoel. The total number of cells in the hemocoel confirmed that the presence of this pathogen in the gut may act as a reservoir, and it may spread, causing septicemia. S. liquefaciens and S. nematodiphila strain infections did not follow the same growing trend linked to the inoculation dose (Figures 2C, D). The growth and colonization ability of S. liquefaciens and S. nematodiphila was therefore different from S. marcescens (except for the highest dose), and this confirms the lower virulence observed in the survival test. Even when the higher Serratia cell number was observed for S. liquefaciens after 6 days from inoculation, this development of the bacteria in the gut did not lead to a higher mortality.
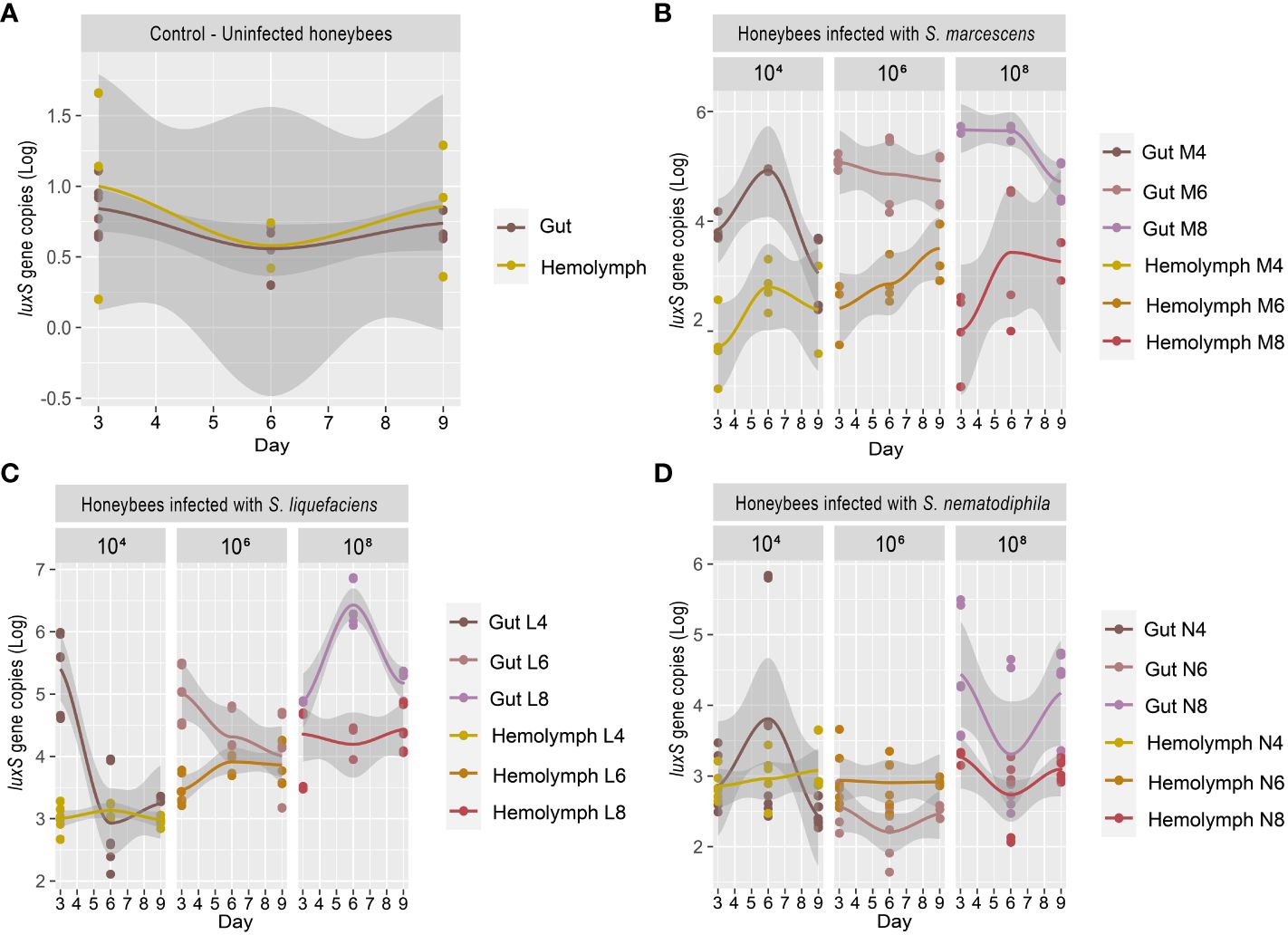
Figure 2 (A–D) Experiment 1: Serratia absolute quantification in the gut and hemocoel. (A) Scatter plot with regression curve of Serratia luxS gene copies in both gut and hemocoel in uninfected honeybees [C]. (B) Scatter plot with regression curve of Serratia luxS gene copies in both gut and hemocoel in honeybees infected with S. marcescens AC8 at different infection doses of 104, 106, and 108 [M4, M6, M8]. (C) Scatter plot with the regression curve of Serratia luxS gene copies in both gut and hemocoel in honeybees infected with S. liquefaciens AC4 at different infection doses of 104, 106, and 108 [L4, L6, L8]. (D) Scatter plot with regression curve of Serratia luxS gene copies in both gut and hemocoel in honeybees infected with S. nematodiphila 1A at different infection doses of 104, 106, and 108 [N4, N6, N8]. Sampling times at day 3, day 6, and day 9.
Our results therefore confirm that when Serratia is present in the honeybee gut, the infection can spread to hemocoel. Furthermore, depending on the initial inoculum, Serratia significantly increased the mortality of the host in the first couple of days after infection, with S. marcescens as the most virulent of the tested strains. Our results also suggest that Serratia can pass through the gut epithelial barrier, and it can colonize the hemocoel where it can also reach higher cell numbers compared to the gut. This dynamic could lead to septicemia, and the number of Serratia cells that actually developed in the gut, and then spilled to other insect organs, like hemocoel, remains undetermined. Since Serratia can overcome the gut epithelial barrier, the presence of a co-occurring infection of V. ceranae, a pathogen able to damage the gut epithelium (Tadei et al., 2020), may facilitate the transfer of Serratia in the hemolymph and its proliferation in the hemocoel.
The effect of different V. ceranae spore loads on Serratia naturally infected honeybees was therefore investigated (Table 1, Section 2). Honeybees infected with different spore doses showed a significant increase in Serratia absolute abundance when compared to uninfected control (p < 0.01 for C vs. CV1, CV2 and CV3), regardless of the number of inoculated spores. In contrast, fumagillin-treated honeybees infected with different V. ceranae doses did not show a significant increase in Serratia (C vs. FV1, FV2, and FV3) (Figure 3A). In addition to these results, our findings highlight a positive correlation between Vairimorpha infection (expressed as V. ceranae units—VcU) and the level of Serratia (correlation +0.81, p < 0.01), suggesting that the epithelial gut disruption by the Vairimorpha parasite contributes to this pathogen spread (Figure 3B). However, further studies are required to confirm this interaction among the two pathogens.
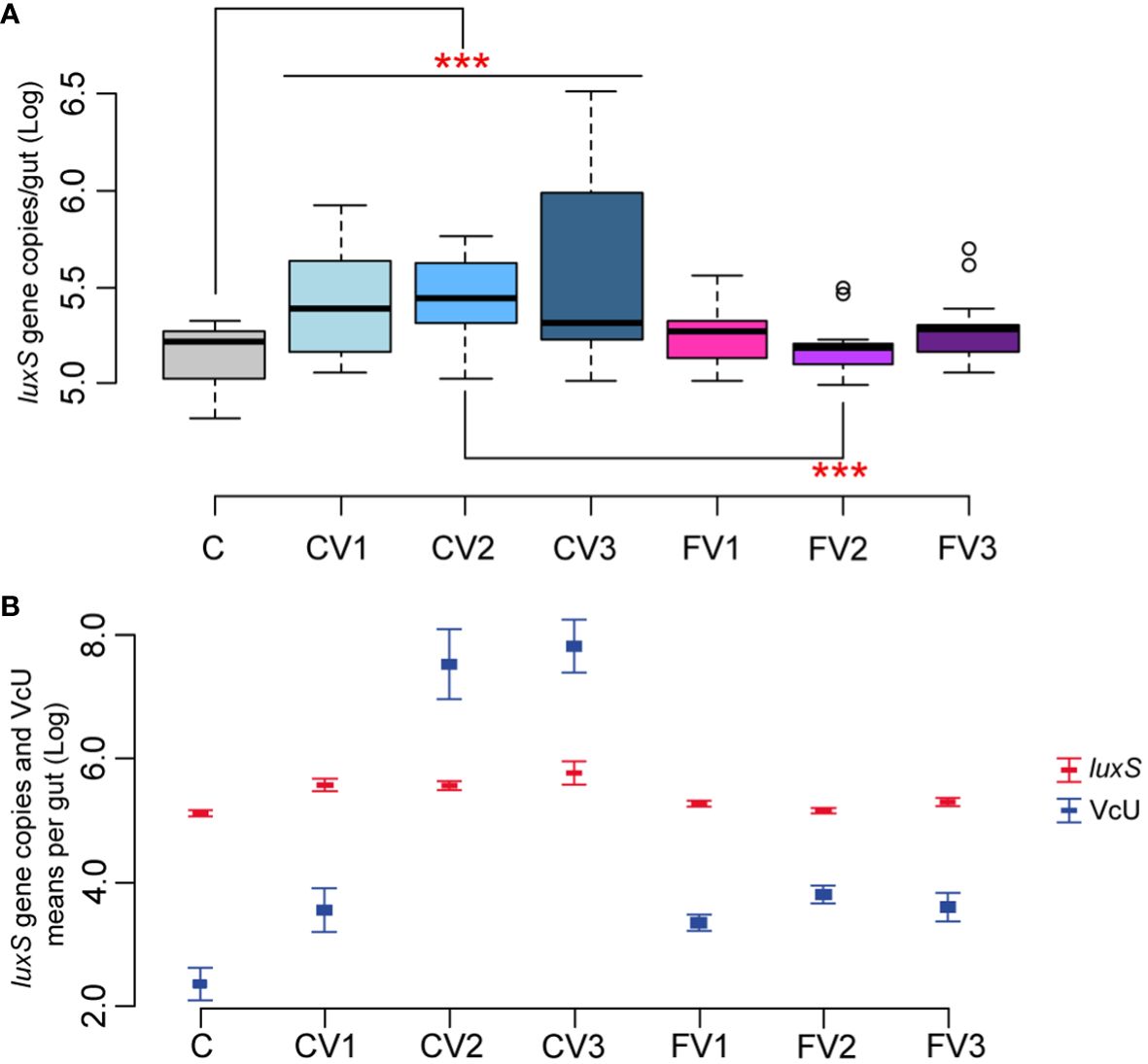
Figure 3 (A, B) Experiment 2: Serratia absolute quantification—Different doses of V. spores. (A) Box plots from experiment A showing the Log of luxS gene copies/intestine per honeybee gut obtained at 14 days post-inoculation. (B) Plot reporting the mean values of V. ceranae units (VcU) from Garrido et al. (2024) and S. marcescens (luxS) for the different experimental conditions. Bees were inoculated with 5×102 spores of V. ceranae [CV1], 5×103 spores of V. ceranae [CV2], 5×104 spores of V. ceranae [CV3], 5×102 spores of V. ceranae and Fumagillin [FV1], 5×103 spores of V. ceranae and fumagillin [FV2], and 5×104 spores of V. ceranae and Fumagillin [FV3]. [C] represents the non-infected control. ***p < 0.01.
Serratia did not proliferate when the antibiotic fumagillin was supplied, confirming that the control of V. ceranae infection also limits the proliferation of Serratia, contributing to improving honeybee health. Fumagillin is known to have an anti-amebic and anti-microsporidia action, but no anti-bacterial activity. This was confirmed in our work with an assay targeted to detect any possible antimicrobial activity of fumagillin, at different doses, on the three isolated Serratia strains. Fumagillin did not show any direct antimicrobial activity against Serratia in vitro in a well diffusion assay, at all tested concentrations (see Section 2.4), suggesting an indirect action of the antimycotic agent on Serratia proliferation. This effect is particularly evident in the honeybees infected with 5×103 Vairimorpha spores and treated with fumagillin (Figures 3A, B) where the comparison CV2 vs. FV2 was found significant (p < 0.01), but the reduction, although not significant, is also present in honeybees infected with 5×102 and 5×104 Vairimorpha spores and treated with the antibiotic fumagillin (FV1 and FV3).
In Garrido et al. (2024), the oral administration of the single bacterial strains composing a previously selected beneficial bacterial mixture aimed at controlling V. ceranae showed that only A. kunkeei significantly reduced the load of the parasite in the gut. In the present work, similar results were obtained in the reduction of Serratia, whose absolute abundance was significantly reduced in the samples in which A. kunkeei was administered (p < 0.05) (Serratia counts from 4.92 [CV] to 4.52 [AKV] Log luxS gene copies/gut; Figure 4A). Serratia amount in all samples has the same trend of V. ceranae counts previously reported (Figure 4B), showing a positive Pearson correlation of + 0.89 (p < 0.01). Given the properties characterizing several Lactobacillaceae members, the inhibition of Serratia colonization could be due to the production of antimicrobial peptides (Forsgren et al., 2010; Palmer-Young et al., 2019) and/or acidification of the gut lumen. On the other hand, it is interesting to note that the administration of the bacterial mixture, although not statistically significant, led to a reduction of Serratia as obtained for Vairimorpha.
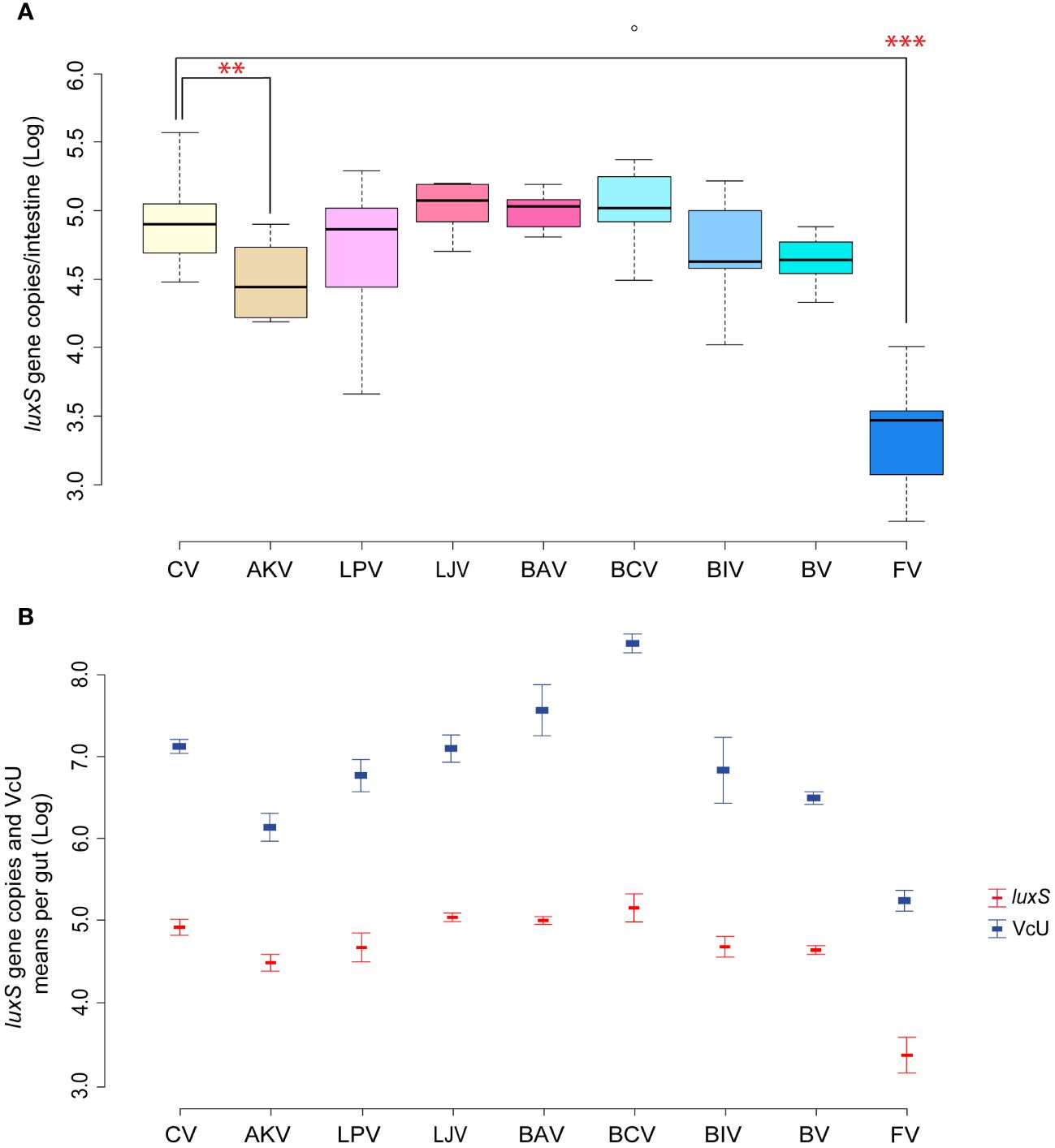
Figure 4 (A, B) Experiment 3: Serratia inhibition—Single strains. (A) Box plots from experiment 3a reporting the Log of luxS gene copies/intestine per honeybee gut. (B) Plot reporting the mean values of V. ceranae units (VcU) from Garrido et al. (2024) and S. marcescens (luxS) for the different experimental conditions. Honeybees were inoculated with 5×104 spores of V. ceranae (marked with “N”) for every treatment with the administration of sugar syrup (1:1 w/v) mixed with the following dietary ingredients: [CV] infected control, [AKV] A. kunkeei, [LPV] L. plantarum, [LJV] L. johnsonii, [BAV] B. asteroides, [BCV] B. coryneforme, [BIV] B. indicum, [BV] Bacteria Mixture, and [FV] fumagillin. **p < 0.05; ***p < 0.01.
As reported by Olofsson et al. (2016) and Kačániová et al. (2019), different strains of A. kunkeii such as Fhon2, Anhmro10, and Yubipro16 possess a high antimicrobial activity in vitro against S. marcescens. Furthermore, the tendency to reduce the infection of Serratia when B. indicum DSM 20214 or L. plantarum Dan91 are administered is evident, although not statistically significant. It is interesting to note that the two best-performing strains against Serratia are exogenous to the honeybee gut. The first one (A. kunkeei) is an environmental microorganism that is usually found in flowers and in honeybee and is usually limited to the crop. The second (B. indicum) is a typical commensal microorganism of Apis cerana (Asian honeybee) where it was isolated for the first time. It is therefore plausible that these two microorganisms triggered an immune response in the tested honeybee, as was shown by Garrido et al. (2024). Indeed, it was possible to correlate specific responses to the pathogen with target microbial strains (Steele et al., 2017; Horak et al., 2020). On the other hand, according to our results, the use of two native bifidobacteria species (B. asteroides and B. coryneforme) increases both Serratia and V. ceranae counts. This is in line with recent research showing a strict connection between V. ceranae and Bifidobacterium increase in the gut lumen (Zhang et al., 2019), probably due to a triggered gut dysbiosis, which might explain the synergy between some bifidobacteria, Nosemosis, and Serratia negatively affecting honeybee health.
Among the different Serratia species isolated from honeybee gut, the S. marcescens strain was the most virulent on honeybees compared to the S. liquefaciens and S. nemathodiphila species. The results obtained allowed us to conclude that S. marcescens can overcome the epithelial gut barrier, spread to the hemocoel, and proliferate in the hemolymph. The presence of an active Vairimorpha infection that disrupts gut epithelial cells also facilitates the Serratia development. Under cage conditions, a strong correlation between V. ceranae infection and Serratia development has been highlighted. The presence of fumagillin, active against Vairimorpha but with no direct activity against Serratia, seems to control the development of both pathogens, as well as some bacterial strain supplied as feed additive active against Vairimorpha. The microbial strains active against Vairimorpha, such as A. kunkeii, can reduce also Serratia, confirming the correlation between the two pathogens. The detection of a connection between these two chronic pathogens in certain circumstances is of concern for the researcher working under laboratory conditions on honeybees. However, field conditions may mitigate or enhance the pathogens’ spread and synergy in the honeybee colonies. Further investigations are required in order to better understand these two pathogens’ dynamics considering environmental variables as well.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. Raw data can be found in Mendeley Data repository at the following DOI: 10.17632/mjc2xky9s7.2.
Ethical approval was not required for the studies involving animals in accordance with the local legislation and institutional requirements because according to the Italian and Argentinian law, ethical approval is not necessary for insects. Written informed consent was obtained from the owners for the participation of their animals in this study.
CB: Formal analysis, Investigation, Methodology, Validation, Writing – original draft. DA: Data curation, Investigation, Writing – original draft, Conceptualization, Formal analysis, Supervision, Validation, Visualization. PG: Funding acquisition, Investigation, Methodology, Writing – review & editing. MP: Funding acquisition, Investigation, Methodology, Writing – review & editing. LB: Data curation, Resources, Visualization, Writing – review & editing. DM: Funding acquisition, Writing – review & editing. DS: Conceptualization, Funding acquisition, Methodology, Writing – review & editing. ME: Funding acquisition, Supervision, Writing – review & editing. DD: Funding acquisition, Project administration, Validation, Writing – original draft.
The research was partially funded by the EU project “NOurishingPROBiotics to bees to Mitigate Stressors” (NO PROBleMS), H2020-MSCA RISE 2017, GA 777760, 2018–2022; PICT-03969-18 and PICT 2169-17, FONCyT, Agencia Nacional de Promoción de la Investigación, el Desarrollo Tecnológico y la Innovación.
Author DS is/was employed by the company Advance Science Forbairt Eolaiochta Teo Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1323157/full#supplementary-material
Alberoni, D., Baffoni, L., Braglia, C., Gaggìa, F., Di Gioia, D. (2021b). Honeybees exposure to natural feed additives: how is the gut microbiota affected? Microorganisms 9, 1009. doi: 10.3390/microorganisms9051009
Alberoni, D., Baffoni, L., Gaggìa, F., Ryan, P. M., Murphy, K., Ross, P. R., et al. (2018). Impact of beneficial bacteria supplementation on the gut microbiota, colony development and productivity of Apis mellifera L. Benef. Microbes 9, 269–278. doi: 10.3920/BM2017.0061
Alberoni, D., Di Gioia, D., Baffoni, L. (2023). Alterations in the microbiota of caged honeybees in the presence of Nosema ceranae infection and related changes in functionality. Microb. Ecol. 86 (1), 601–616. doi: 10.1007/s00248-022-02050-4
Alberoni, D., Favaro, R., Baffoni, L., Angeli, S., Di Gioia, D. (2021a). Neonicotinoids in the agroecosystem: in-field long-term assessment on honeybeecolony strength and microbiome. Sci. Total Environ. 762, 144116. doi: 10.1016/j.scitotenv.2020.144116
Alberoni, D., Gaggìa, F., Baffoni, L., Modesto, M. M., Biavati, B., Di Gioia, D. (2019). Bifidobacterium xylocopae sp. nov. and Bifidobacterium aemilianum sp. nov., from the carpenter bee (Xylocopa violacea) digestive tract. Syst. Appl. Microbiol. 42, 205–216. doi: 10.1016/j.syapm.2018.11.005
Al Naggar, Y., Singavarapu, B., Paxton, R. J., Wubet, T. (2022). Bees under interactive stressors: the novel insecticides flupyradifurone and sulfoxaflor along with the fungicide azoxystrobin disrupt the gut microbiota of honey bees and increase opportunistic bacterial pathogens. Sci. Total Environ. 849, 157941. doi: 10.1016/j.scitotenv.2022.157941
Al Naggar, Y., Wubet, T. (2024). Chronic exposure to pesticides disrupts the bacterial and fungal co-existence and the cross-kingdom network characteristics of honey bee gut microbiome. Sci. Total Environ. 906, 167530. doi: 10.1016/j.scitotenv.2023.167530
Baffoni, L., Alberoni, D., Gaggìa, F., Braglia, C., Stanton, C., Ross, P. R., et al. (2021). Honeybee exposure to veterinary drugs: how is the gut microbiota affected? Microbiol. Spectr. 9, e00176–e00121. doi: 10.1128/Spectrum.00176-21
Baffoni, L., Gaggìa, F., Alberoni, D., Cabbri, R., Nanetti, A., Biavati, B., et al. (2016). Effect of dietary supplementation of Bifidobacterium and Lactobacillus strains in Apis mellifera L. against Nosema ceranae. Benef. Microbes 7, 45–51. doi: 10.3920/BM2015.0085
Baffoni, L., Gaggìa, F., Di Gioia, D., Santini, C., Mogna, L., Biavati, B. (2012). A Bifidobacterium based synbiotic product to reduce the transmission of C. jejuni along the poultry food chain. Int. J. Food Microbiol. 157, 156–161. doi: 10.1016/j.ijfoodmicro.2012.04.024
Baross, J. A., Tester, P. A., Morita, R. Y. (1978). Incidence, microscopy, and etiology of exoskeleton lesions in the tanner crab, Chionoecetes tanneri. J. Fish. Res. 35, 1141–1149. doi: 10.1139/f78-179
Braglia, C., Alberoni, D., Porrini, M. P., Garrido, P. M., Baffoni, L., Di Gioia, D. (2021). Screening of dietary ingredients against the honey bee parasite Nosema ceranae. Pathogens 10, 1117. doi: 10.3390/pathogens10091117
Brandt, A., Gorenflo, A., Siede, R., Meixner, M., Büchler, R. (2016). The neonicotinoids thiacloprid, imidacloprid, and clothianidin affect the immuno- competence of honey bees (Apis mellifera L.). J. Insect. Physiol. 86, 40–47. doi: 10.1016/j.jinsphys.2016.01.001
Brown, S. P., Cornforth, D. M., Mideo, N. (2012). Evolution of virulence in opportunistic pathogens: generalism, plasticity, and control. Trends Microbiol. 20, 336–342. doi: 10.1016/j.tim.2012.04.005
Burritt, N. L., Foss, N. J., Neeno-Eckwall, E. C., Church, J. O., Hilger, A. M., Hildebrand, J. A., et al. (2016). Sepsis and hemocyte loss in honey bees (Apis mellifera) infected with Serratia marcescens strain sicaria. PloS One 11, e0167752. doi: 10.1371/journal.pone.0167752
Carpenter, D. J., Mathiassen, S. K., Boutin, C., Strandberg, B., Casey, C. S., Damgaard, C. (2020). Effects of herbicides on flowering. Environ. Toxicol. Chem. 39, 1244–1256. doi: 10.1002/etc.4712
Castelli, L., Branchiccela, B., Garrido, M., Invernizzi, C., Porrini, M., Romero, H., et al. (2020). Impact of nutritional stress on honeybee gut microbiota, immunity, and Nosema ceranae infection. Microb. Ecol. 80, 908–919. doi: 10.1007/s00248-020-01538-1
Cintas, L. M., Rodriguez, J. M., Fernandez, M. F., Sletten, K., Nes, I. F., Hernandez, P. E., et al. (1995). Isolation and characterization of pediocin L50, a new bacteriocin from pediococcus acidilactici with a broad inhibitory spectrum. Appl. Environ. Microbiol. 61 (7), 2643. doi: 10.1128/aem.61.7.2643-2648.1995
Cook, R. D. (1979). Influential observations in linear regression. JASA 74, 169–174. doi: 10.1080/01621459.1979.10481634
de Micheaux, P. L., Drouilhet, R., Liquet, B. (2013). The r software (Fundamentals of Programming and Statistical Analysis), 978–971.
Dolezal, A. G., Carrillo-Tripp, J., Judd, T. M., Allen Miller, W., Bonning, B. C., Toth, A. L. (2019). Interacting stressors matter: diet quality and virus infection in honeybee health. R. Soc Open Sci. 6, e181803. doi: 10.1098/rsos.181803
Dolezal, A. G., Toth, A. L. (2018). Feedbacks between nutrition and disease in honey bee health. Curr. Opin. Insect Sci. 26, 114–119. doi: 10.1016/j.cois.2018.02.006
El Sanousi, S. M., El Sarag, M. S. A., Mohamed, S. E. (1987). Properties of Serratia marcescens isolated from diseased honeybee (Apis mellifera) larvae. Microbiology 133, 215–219. doi: 10.1099/00221287-133-1-215
Forsgren, E., Olofsson, T. C., Vásquez, A., Fries, I. (2010). Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 41, 99–108. doi: 10.1051/apido/2009065
Garrido, P. M., Porrini, M. P., Alberoni, D., Baffoni, L., Scott, D., Mifsud, D., et al. (2024). Beneficial bacteria and plant extracts promote honey bee health and reduce Nosema ceranae infection. Probiotics Antimicro., 1–16. doi: 10.1007/s12602-022-10025-7
Gooday, G. W. (1990). “The ecology of chitin degradation,” in Advances in microbial ecology (Vol.11). Ed. Marshall, K. C. (Plenum Press, New York and London), 387–430.
Grimont, P. A., Grimont, F. (1978). The genus serratia. Annu. Rev. Microbiol. 32, 221–248. doi: 10.1146/annurev.mi.32.100178.001253
Higes, M., Meana, A., Bartolomé, C., Botías, C., Martín-Hernández, R. (2013). Nosema ceranae (Microsporidia), a controversial 21st century honey bee pathogen. Environ. Microbiol. Rep. 5, 17–29. doi: 10.1111/1758-2229.12024
Horak, R. D., Leonard, S. P., Moran, N. A. (2020). Symbionts shape host innate immunity in honeybees. Proc. R. Soc B.: Biol. Sci. 287, e20201184. doi: 10.1098/rspb.2020.1184
Huang, W. F., Solter, L. F. (2013). Comparative development and tissue tropism of Nosema apis and Nosema ceranae. J. Invertebr. Pathol. 113, 35–41. doi: 10.1016/j.jip.2013.01.001
Hubert, J., Kamler, M., Nesvorna, M., Ledvinka, O., Kopecky, J., Erban, T. (2016). Comparison of Varroa destructor and worker honeybee microbiota within hives indicates shared bacteria. Microb. Ecol. 72, 448–459. doi: 10.1007/s00248-016-0776-y
Iverson, K. L., Bromel, M. C., Anderson, A. W., Freeman, T. P. (1984). Bacterial symbionts in the sugar beet root maggot, Tetanops myopaeformis (von Röder). A.E.M. 47, 22–27. doi: 10.1128/aem.47.1.22-27.1984
Joyner, J., Wanless, D., Sinigalliano, C. D., Lipp, E. K. (2014). Use of quantitative real-time PCR for direct detection of Serratia marcescens in marine and other aquatic environments. A.E.M. 80, 1679–1683. doi: 10.1128/AEM.02755-13
Kačániová, M., Kunová, S., Ivanišová, E., Terentjeva, M., Gasper, J. (2019). Antimicrobial effect of Lactobacillus kunkeei against pathogenic bacteria isolated from bees’ gut. J. Anim. Sci. Biotechnol. 52, 100–104.
Kwong, W. K., Mancenido, A. L., Moran, N. A. (2017). Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 4, e170003. doi: 10.1098/rsos.170003
Le Conte, Y., Navajas, M. (2008). Climate change: impact on honey bee populations and diseases. Rev. Sci. Tech. Off. Int. Epiz. 27, 499–510.
Li, J. H., Evans, J. D., Li, W. F., Zhao, Y. Z., DeGrandi-Hoffman, G., Huang, S. K., et al. (2017). New evidence showing that the destruction of gut bacteria by antibiotic treatment could increase the honeybee’s vulnerability to Nosema infection. PloS One 12, e0187505. doi: 10.1371/journal.pone.0187505
Martin, S. J., Hardy, J., Villalobos, E., Martín-Hernández, R., Nikaido, S., Higes, M. (2013). Do the honeybee pathogens Nosema ceranae and deformed wing virus act synergistically? Environ. Microbiol. Rep. 5, 506–510. doi: 10.1111/1758-2229.12052
Motta, E. V., Raymann, K., Moran, N. A. (2018). Glyphosate perturbs the gut microbiota of honey bees. PNAS 115, 10305–10310. doi: 10.1073/pnas.1803880115
Motta, E. V., Powell, J. E., Moran, N. A. (2022). Glyphosate induces immune dysregulation in honey bees. Anim. Microbiome 4 (1), 16. doi: 10.1186/s42523-022-00165-0
Olofsson, T. C., Butler, È., Markowicz, P., Lindholm, C., Larsson, L., Vásquez, A. (2016). Lactic acid bacterial symbionts in honeybees - an unknown key to honey’s antimicrobial and therapeutic activities. Int. Wound J. 13, 668–679. doi: 10.1111/iwj.12345
Palmer-Young, E. C., Raffel, T. R., McFrederick, Q. S. (2019). pH-mediated inhibition of a bumble bee parasite by an intestinal symbiont. Parasitol 146, 380–388. doi: 10.1017/S0031182018001555
Porrini, M. P., Garrido, P. M., Eguaras, M. J. (2013). Individual feeding of honey bees: modification of the Rinderer technique. J. Apicult. Res. 52 (5), 194–195. doi: 10.3896/IBRA.1.52.5.04
Porrini, M. P., Garrido, P. M., Umpiérrez, M. L., Porrini, L. P., Cuniolo, A., Davyt, B., et al. (2020). Effects of synthetic acaricides and Nosema ceranae (Microsporidia: Nosematidae) on molecules associated with chemical communication and recognition in honey bees. Vet. Sci. 7, 199. doi: 10.3390/vetsci7040199
Raymann, K., Coon, K. L., Shaffer, Z., Salisbury, S., Moran, N. A. (2018). Pathogenicity of Serratia marcescens strains in honey bees. mBio 9, e01649–e01618. doi: 10.1128/mBio.01649-18
Raymann, K., Moran, N. A. (2018). The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect. Sci. 26, 97–104. doi: 10.1016/j.cois.2018.02.012
Raymann, K., Shaffer, Z., Moran, N. A. (2017). Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PloS Biol. 15, e2001861. doi: 10.1371/journal.pbio.2001861
Ricigliano, V. A., Williams, S. T., Oliver, R. (2022). Effects of different artificial diets on commercial honey bee colony performance, health biomarkers, and gut microbiota. BMC Vet. Res. 18, 1–14. doi: 10.1186/s12917-022-03151-5
Rosenkranz, P., Aumeier, P., Ziegelmann, B. (2010). Biology and control of Varroa destructor. J. Invertebr. Pathol. 103, S96–S119. doi: 10.1016/j.jip.2009.07.016
Steele, M. I., Kwong, W. K., Whiteley, M., Moran, N. A. (2017). Diversification of type VI secretion system toxins reveals ancient antagonism among bee gut microbes. MBio 8, e01630–e01617. doi: 10.1128/mBio.01630-17
Tadei, R., Menezes-Oliveira, V. B., Silva-Zacarin, E. C. (2020). Silent effect of the fungicide pyraclostrobin on the larval exposure of the non-target organism Africanized Apis mellifera and its interaction with the pathogen Nosema ceranae in adulthood. Environ. pollut. 267, 115622. doi: 10.1016/j.envpol.2020.115622
Ullah, A., Gajger, I. T., Majoros, A., Dar, S. A., Khan, S., Shah, A. H., et al. (2021). Viral impacts on honey bee populations: a review. Saudi J. Biol. Sci. 28, 523–530. doi: 10.1016/j.sjbs.2020.10.037
Vallet-Gely, I., Lemaitre, B., Boccard, F. (2008). Bacterial strategies to overcome insect defences. Nat. Rev. Microbiol. 6, 302–313. doi: 10.1038/nrmicro1870
Zhang, Y., Lu, X., Huang, S., Zhang, L., Su, S., Huang, W. F. (2019). Nosema ceranae infection enhances bifidobacterium spp. abundances in the honey bee hindgut. Apidologie 50, 353–362. doi: 10.1007/s13592-019-00644-5
Keywords: Serratia, nosemosis, Nosema ceranae, fumagillin, beneficial bacteria, Apilactobacillus
Citation: Braglia C, Alberoni D, Garrido PM, Porrini MP, Baffoni L, Scott D, Eguaras MJ, Di Gioia D and Mifsud D (2024) Vairimorpha (Nosema) ceranae can promote Serratia development in honeybee gut: an underrated threat for bees? Front. Cell. Infect. Microbiol. 14:1323157. doi: 10.3389/fcimb.2024.1323157
Received: 17 October 2023; Accepted: 04 April 2024;
Published: 13 May 2024.
Edited by:
Cedric Nicolas Berger, Cardiff University, United KingdomReviewed by:
Yahya Al Naggar, Tanta University, EgyptCopyright © 2024 Braglia, Alberoni, Garrido, Porrini, Baffoni, Scott, Eguaras, Di Gioia and Mifsud. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniele Alberoni, ZGFuaWVsZS5hbGJlcm9uaUB1bmliby5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.