- 1Chemical Engineering Research Center, School of Chemical Engineering and Technology, Tianjin University, Tianjin, China
- 2R&D Department, Dynamiker Biotechnology (Tianjin) Co., Ltd, Tianjin, China
- 3Tianjin Key Laboratory of Membrane Science and Desalination Technology, Tianjin University, Tianjin, China
- 4State Key Laboratory of Chemical Engineering, Tianjin University, Tianjin, China
- 5Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Tianjin, China
With advances in medicine, increasing medical interventions have increased the risk of invasive fungal disease development. (1-3)-β-D glucan (BDG) is a common fungal biomarker in serological tests. However, the scarcity of Limulus resources for BDG detection poses a challenge. This study addresses the need for an alternative to Limulus amebocyte lysate by using BDG mutant antibody for chemiluminescence detection. The wild-type BDG antibody was obtained by immunizing rabbits. An optimal V52HI/N34L Y mutant antibody, which has increased 3.7-fold of the testing efficiency compared to the wild-type antibody, was first achieved by mutating “hot-spot” residues that contribute to strong non-covalent bonds, as determined by alanine scanning and molecular dynamics simulation. The mutant was then applied to develop the magnetic particle chemiluminescence method. 574 clinical samples were tested using the developed method, with a cutoff value of 95 pg/mL set by Limulus amebocyte lysate. The receiver operating characteristic curve demonstrated an area under the curve of 0.905 (95% CI: 0.880–0.929). Chemiluminescence detected an antigen concentration of 89.98 pg/mL, exhibiting a sensitivity of 83.33% and specificity of 89.76%. In conclusion, the results showed a good agreement with Limulus amebocyte lysate and demonstrated the feasibility of using BDG mutant antibodies for invasive fungal disease diagnosis. The new method based on chemiluminescence for detecting BDG could shorten the sample-to-result time to approximately 30 min, rescue Limulus from being endangered and is resource efficient in terms of equipment and the non-use of a skilled technician.
1 Introduction
The increasing use of invasive tests, antibiotics, immunosuppressants, glucocorticoids, and chemotherapeutic agents has led to a rise in fungal infections, which poses a serious threat to public health. Each year, fungal diseases cause over 1.6 million death and severely affect more than 1 billion people worldwide (Almeida et al., 2019; Fisher et al., 2022). Fungi are diverse and adaptable organisms. They usually exist on human mucosal surfaces, but they can invade tissues and cause invasive fungal disease (IFD) in immunocompromised individuals (Salazar et al., 2022). IFD has typical clinical symptoms and is difficult to diagnose, leading to delayed treatment and severe consequences (Terrero-Salcedo and Powers-Fletcher, 2020). However, traditional diagnostic methods such as culture and histopathology have limitations in sensitivity and speed, making rapid and accurate microbiological diagnosis crucial (Ricna et al., 2019; Lass-Flörl et al., 2021). Serological diagnosis has become a popular auxiliary clinical diagnostic method for IFD with the advantages of high sensitivity, strong specificity, and convenience (Hage et al., 2019; Azap, 2020).
(1–3)-β-D glucan (BDG) is one of the fungal biomarkers widely used in serological detection (Mercier et al., 2019; Finkelman, 2021). Fungal Glucan Detection (G test) was then developed for testing the concentration of BDG in serum, which could help to diagnose IFD infection (Yoshida, 2021). The G test for the measurement of BDG uses the coagulation cascade mediated by factor G in the Limulus amebocyte lysate test (Yoshida, 2021). Unfortunately, the raw materials of Limulus amebocyte lysate come from natural Limulus resources, which are widely killed for their medicinal and edible value. In addition, the artificial cultivation of limulus is difficult, and large-scale cultivation has never been achieved (Kwan et al., 2018). With the increasing scarcity of Limulus resources, it is necessary to find a way to replace Limulus amebocyte lysate.
Chemiluminescence immunoassay, which is based on a fast “mix-and-measure” protocol, provides a promising automatic alternative for testing analytical procedures (Calero et al., 2022). Although it highly relies on instrument, chemiluminescence immunoassay still has significant advantages over other non-isotopic ELISA such as the high sensitivity and a wide linear range. In addition, compared with the Limulus amebocyte lysate method, the chemiluminescence method is free of endotoxin contamination. Most importantly, it has a sustainable raw material source, which offers a long-term development for the testing method. Therefore, this study aimed to develop a magnetic particle-based chemiluminescence immunoassay for the measurement of fungal BDG. The process was as follows (1): The wild-type antibody (WT-BDG-Ab) was obtained by immunizing rabbits and the full sequence plasmid expression vector was constructed (2). Alanine scanning of the antibody CDR region revealed seven residues (Trp33H, Val52H, Asp54H, Phe58H, Tyr99H, Pro105H, and Tyr106H) in the heavy chain and five residues (Tyr31L Asn33L, Asn34L, Gly52L, and Arg55L) in the light chain as critical residues for antigen–antibody binding (3). The optimal mutant antibody V52HI/N34LY was obtained by combined mutation screening (4). The V52HI/N34LY mutant was applied to the chemiluminescence platform, and the results of 574 clinical samples showed a good correlation with the Limulus amebocyte lysate. This study provides a reasonable basis for the modification of WT-BDG-Ab to improve its potency and an effective detection method for the early diagnosis of IFD, which broadens the technical field of Limulus amebocyte lysate.
2 Materials and methods
2.1 Immunization and specificity analysis
Owing to the poor water solubility of standard BDG, carboxymethylated BDG was chosen as the immunogen, which was purchased from Megazyme (carboxymethylated substitution degree of 20%) and purified. It was mixed with Freund’s adjuvant complete in equal volumes. After full emulsification, New Zealand large-eared rabbits were injected subcutaneously at multiple points, and the immunization dose was controlled at 0.1–0.2 mg/time per rabbit. Ear blood was taken 3 days before immunization and the serum was isolated for negative control. After the initial immunization, the rabbits were immunized every 2 weeks in the same way. A total of eight immunizations were performed and the spleen was taken after the last immunization.
Enzyme-linked immunosorbent assay (ELISA) was used to analyze the specificity of the BDG antibody (BDG-Ab). Enzyme plates were coated with 50 ng/well of BDG, candida mannan, trehalose galactomannan, or cryptococcal polysaccharide, and the directly blocked plates were used as the negative control. The wells were blocked and incubated at 37°C for 1 h, with 100 μL of the serum which 16-fold diluted by 0.01 M PBS. They were washed and placed in 100 μL of goat anti-rabbit-HRP antibody for 30 min at 37°C, then washed and developed with 100 μL of tetraethylbenzidine (TMB) substrate for 15 min.
2.2 Sequence comparison and model construction of BDG-Ab
BDG-Ab sequencing was commissioned from GenScript (Supplementary Table 1, GenBank OR729008-OR729009). The IgBLAST system was used to compare the antibody libraries (Sayers et al., 2021). The heavy chain (BDG-Ab-H) and light chain (BDG-Ab-L) were submitted to the Kabat database, respectively, and the framework regions (FRs) and complementarity-determining regions (CDRs) in the variable region of BDG-Ab were identified.
The three-dimensional structure of BDG-Ab was constructed based on homology modeling and the machine learning method by trRosetta and Uni-Fold software, respectively (Du et al., 2021; Su et al., 2021). Since antibodies consist of two heavy and two light chains, homology modeling can only build single chains and then match the constructed single chains together. The machine learning method directly predicts the atomic coordinates of the antibody using a combination of its amino acid sequence, multiple sequence alignment, and solved homologous structures (Li et al., 2022).
2.3 Molecular docking and molecular dynamics simulations
The BDG model built by AmberTools was assigned the Amber ff99SB force field (Tian et al., 2020) and docked to the CDR region of BDG-Ab by Autodock vina (Eberhardt et al., 2021). GROMACS 2019.5 software was used for molecular dynamics (MD) simulations (Abraham et al., 2015). The simulation process was as follows (1): The water molecules and ions were modeled and inserted into the box to make the system charge equilibrium (2). The 2,000 energy minimization steps and 40-ns production simulations were applied to achieve the minimum energy point (3). The simulations were carried out in an isothermal–isobaric ensemble with periodic boundary conditions. The simulation parameters were a 25-fs step length, a 1-atm pressure, and a 298 K temperature, with the coupling constants of 12.0 and 1.0 ps, respectively. The simulation results were drawn and analyzed using Gromacs utility and VMD software (Humphrey et al., 1996). The binding energy of antibody and BDG was calculated by gmx_mmpbsa.py python script (Valdés-Tresanco et al., 2021).
2.4 Detection of the relative binding capacity of BDG-Ab mutant
The sequences of BDG-Ab-H and BDG-Ab-L were ligated to the AbVec2.0 plasmid by Genewiz Biotech Co., Ltd (Suzhou, China) (Supplementray Figures 1 and 2). The non-alanine sequence in the CDR region of the BDG-Ab-H and BDG-Ab-L plasmids was sequentially point mutated to the alanine sequence. The mutated plasmids were provided by Genewiz Biotech Co., Ltd (Suzhou, China).
The mutated BDG-Ab-H/L plasmids paired with unmutated BDG-Ab-L/H plasmids were transfected into 293T cells. Cell supernatants were harvested after 48 h for two separate ELISA assays, in which enzyme plates were coated with BDG or mouse anti-rabbit lgG Fab antibody. The plates were added to the BDG-Ab separately, incubated at 37°C for 1 h, washed and placed in mouse anti-rabbit lgG Fc-HRP antibody for 30 min at 37°C, and then washed and developed with TMB substrate for 15 min at 37°C. The relative binding capacity (RBC) of BDG-Ab was calculated as:
where A and B are the result of ELISA assay with BDG and mouse anti-rabbit lgG Fab antibody-coated enzyme plates, respectively, and WT is the wild-type BDG-Ab.
2.5 Potency testing of BDG-Ab mutants
BDG-Ab mutants were purified by the AmMagTM ProteinA Magnetic Beads (GenScript) kit after the plasmids were transfected into 293T cells. Antibody potency was verified by the indirect ELISA method. The ELISA plates were coated with 20 ng/well of BDG, and BDG-Ab mutants were diluted at concentrations of 1,000, 500, 250, 125, 62.5, 31.25, 15.625, 7.8125, and 3.90625 ng/mL, respectively. The plates were added to the dilutions separately, incubated at 37°C for 1 h, washed and placed in goat anti-rabbit-HRP antibody for 30 min at 37°C, and then washed and developed with TMB substrate for 15 min at 37°C. We defined the BDG-Ab potency as the reciprocal of the BDG-Ab concentration at OD = 0.
2.6 Fungi and bacteria recognizing ability comparison of wild-type and mutant BDG-Ab
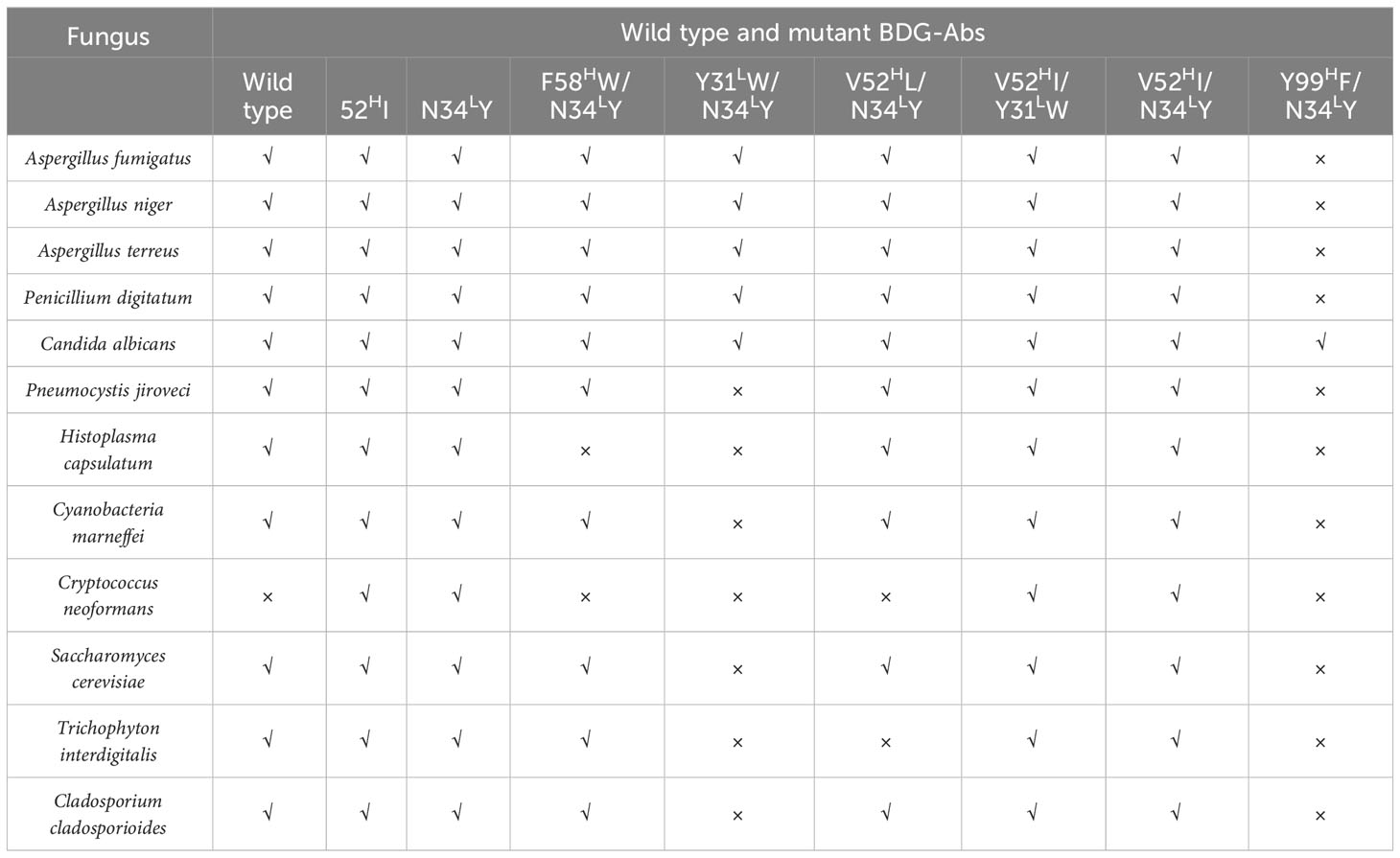
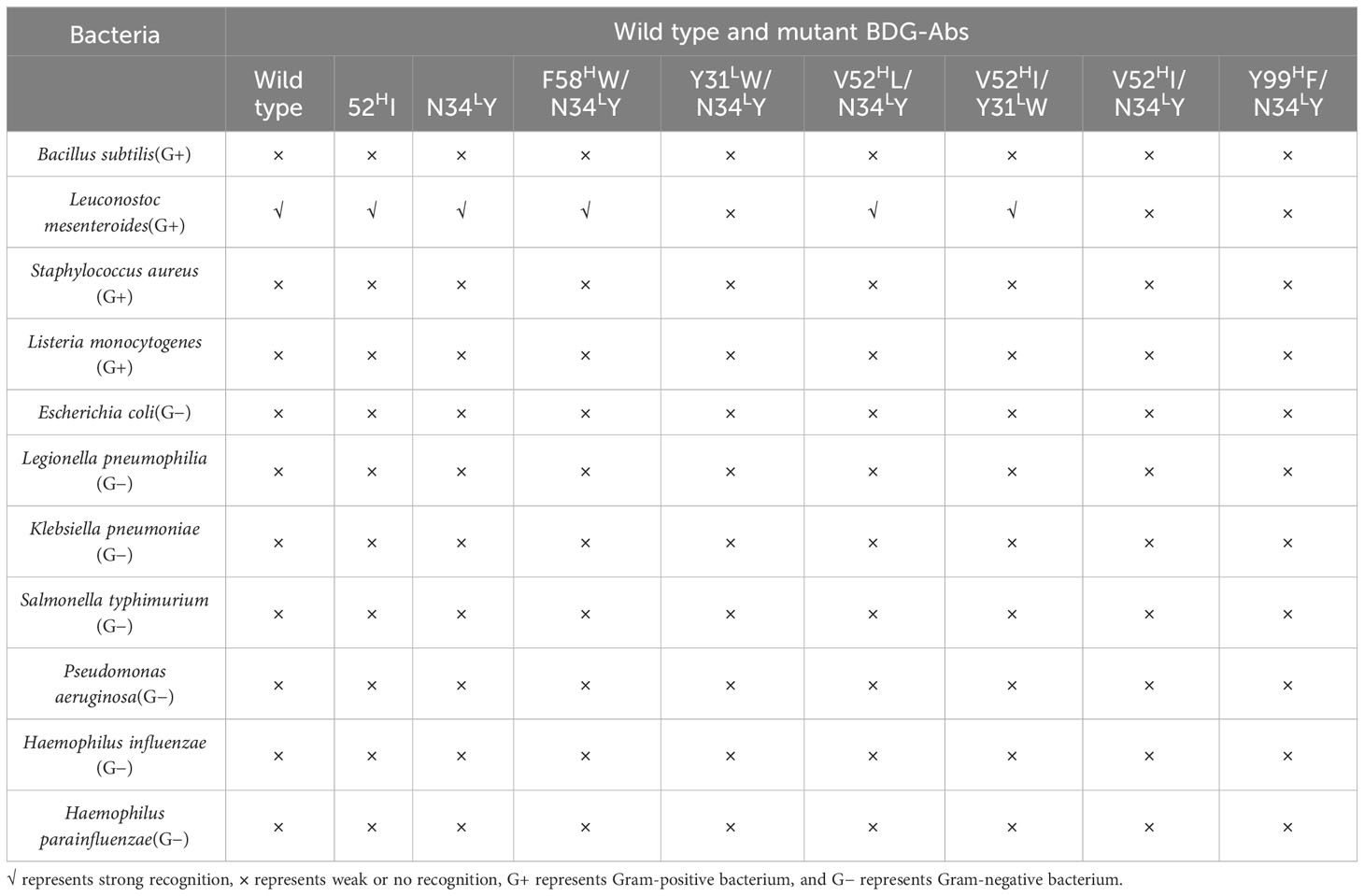
Microplates were coated with filtered ultrasonic crushing solutions of different fungi and bacteria. Then, 100 ng/mL BDG-Ab mutants and wild-type BDG-Ab were added into the coated microplates to validate the recognition capability of different fungi and bacteria. In addition, the microplates coated with fungi- and bacteria-free ultrasonic crushing solution were used as negative control. Aspergillus fumigatus, Aspergillus niger, Aspergillus terreus, Penicillium digitatum, Candida albicans, Pneumocystis jiroveci, Histoplasma capsulatum, Cyanobacteria marneffei, Cryptococcus neoformans, Saccharomyces cerevisiae, Trichophyton interdigitalis, and Cladosporium cladosporioides were used in this research.
2.7 Comparison of the analytical capability of the Limulus amebocyte lysate method and the chemiluminescence method
According to EP 17 A2, the limit of blank (LoB), limit of detection (LoD) of the Limulus amebocyte lysate method, and the chemiluminescence method were compared. Precision comparison was conducted based on EP05 A3. The linearity evaluation was conducted referring to EP06 A. Long-term stability was tested according to EP25 A.
The chemiluminescence testing system was constructed using the following procedure. The BDG-Ab mutant diluent (1 mg/mL) was configured with 0.02 M PB (pH = 7.2) buffer, Sulfo-NHS-LC-Biotin (10 mg/mL) was added to 14.85 μL/mL, mixed for 2 h in the dark, and then dialyzed in 0.02 M PB (pH = 7.2) and labeled with biotin. Meanwhile, the BDG-Ab mutant diluent (1 mg/mL) was configured with 0.05 M CBS buffer, NSP-SA-NHS (2 mg/mL) was added to 45.5 μL/mL, mixed for 2 h in the dark, and then dialyzed in 0.02 M PB (pH 7.2) and labeled with acridine ester. A SMART 6500 automatic chemiluminescence detector was used for chemiluminescence detection. The clinical sample size was set to 100 μL and the amount of streptavidin-magnetic particles (0.4 mg/mL) was set to 40 μL. According to the requirement of antibody gradient concentration, the amount of two kinds of antibody master mixes that were added was adjusted. The fungal (1–3)-β-D-glucan assay kit [Dynamiker (Tianjin) Biotechnology Co. Ltd] was used for the Limulus amebocyte lysate method. The detailed operation method referred to protocol of the BDG testing kit. The LoB, LoD, linear range, and stability of the Limulus amebocyte lysate method and chemiluminescence method were compared. The sample volume for both BDG detention methods is 20 μL.
2.8 Clinical sample testing
A total of 574 serum samples were collected from Shandong Chest Hospital. A total of 52 samples were from fungal infection patients, 140 samples were from probable fungal infection patients, and 382 samples were from the fungal infection-free or healthy cohort. The samples were tested using the chemiluminescence method and the Limulus amebocyte lysate method simultaneously. The BDG testing method was performed in duplicate for each sample. The fungal (1–3)-β-D-glucan assay kit (chromogenic method) [Dynamiker (Tianjin) Biotechnology Co. Ltd] was used for the Limulus amebocyte lysate method. The chemiluminescence testing system is described in Section 2.7. ROC was calculated using SPSS, which was used for consistency evaluation for both the Limulus amebocyte lysate method and the chemiluminescence method. The sensitivity of both BDG testing method was compared (sensitivity is calculated as the ratio of positive sample amount tested by the Limulus amebocyte lysate method or the chemiluminescence method to a sample amount of fungal infection patients).
3 Results and discussion
3.1 Immunization with single-targeted antibody
Standardized carboxymethylated pachyman (CP-BDG) was used as the immunogen to obtain WT-BDG-Ab by immunizing rabbits. Pachyman, a type of BDG from Poria cocos, was carboxymethylated to overcome the limitation of water solubility (Liu et al., 2021). Since the purity of the antigen will directly affect the quality of antibody, standardized antigen was prepared by semi-preparative high performance liquid chromatography (Supplementray Figure 3). The specificity analysis of WT-BDG-Ab (Figure 1) suggested that it did not produce immune responses to candida mannoprotein, aspergillus galactomannan, and cryptococcal polysaccharide, which was comparable to the background.
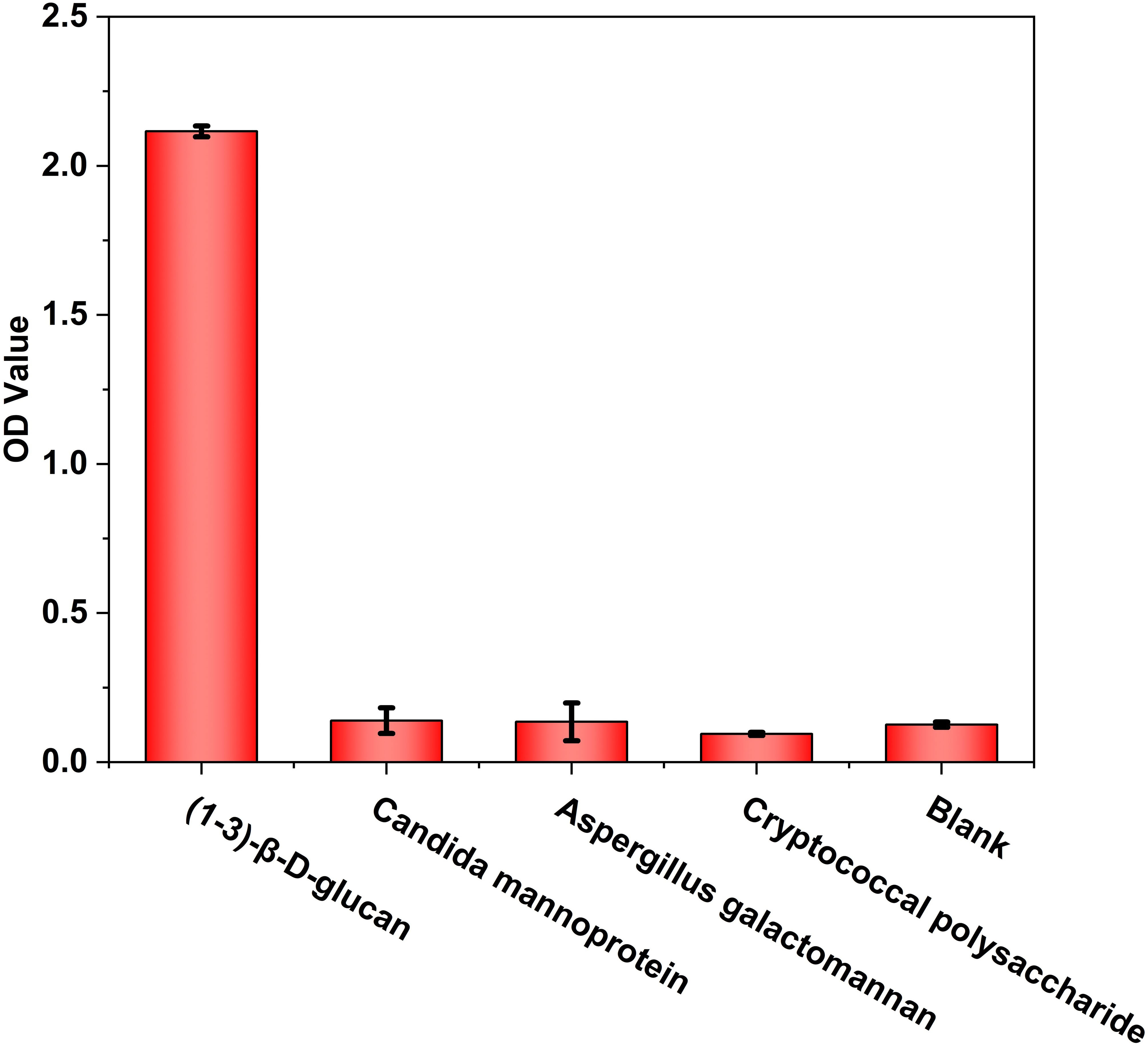
Figure 1 The specificity analysis of WT-BDG-Ab to different antigens using enzyme-linked immunosorbent assay (ELISA). In the blank control, the equal volume of endotoxin-free water was used instead of antigen.
3.2 Analysis of the properties of key amino acids
Endogenous proteins are autonomously produced by the organism to meet physiological functions, which is difficult to meet the needs of clinical treatment. Therefore, studying the antigen–antibody binding mechanism is conducive to the development of BDG-Ab in clinical diagnosis. Antibody specificity is determined by the antigen binding sites in the CDR region. We obtained a large number of single-point mutant antibodies in the CDR region by alanine scanning to discuss the key sites, which are the main contributors of non-covalent bonds. They induce interactions at other sites. Therefore, mutation of key sites causes loss of non-covalent bonds through broad structural change of the interface (Fernández Quintero et al., 2020). In this study, the AI tetra-chain modeling was selected (Supplementray Figures 4, 5). The variable regions of one heavy chain (chain B) and one light chain (chain C) were selected for the calculated alanine scan. The general trend of the experimental alanine scans was consistent with the calculated results.
The relative binding and mutation energy results reveal that Trp33H, Val52H, Asp54H, Phe58H, Tyr99H, Pro105H, and Tyr106H residues in chain B and Tyr31L, Asn33L, Asn34L, Gly52L, and Arg55L residues (the superscripts H and L indicate the heavy chain and the light chain of BDG-Ab, respectively) in chain C are the key sites in the antigen–antibody binding (Figures 2A, B). Antigen recognition is primarily governed by several types of non-covalent bond interactions: electrostatic interactions, van der Waals forces, hydrogen bonds, and hydrophobic interactions, while the latter is dominant (Yoshida et al., 2019). The Y106HA mutant had a high mutation energy of 1.22 kcal/mol and an RBC value of −29.5%, indicating that the Tyr106 residue is a “hot spot” residue in the interaction. Similarly, the mutant binding affinity of V52HA, F58HA, and P105HA mutants was significantly reduced (RBC values were −98.4%, −69.3%, and −29.9%, respectively). The “hot spot” residues Val52H, Phe58H, Pro105H, and Tyr106H mostly play the hydrophobic function in the analysis of the docking data. In light chain, Y31LA and N34LA mutants had a significant decrease in binding affinity (RBC values were −101.8% and −92.7%, respectively) and had a mutation energy of 0.61 and 0.34 kcal/mol, respectively. The four “hot spot” residues Tyr31L, Asn33L, Asn34L, and Arg55L bind the antigen mainly through hydrogen bonds, and the mutation energy also reveals that the hydrogen bonding force is weaker than the hydrophobic interaction. As shown in Supplementray Figure 6, Asn34L has a hydrogen bond connection length of 3.4 Å, which is close to the hydrogen bond limit connection length. Moreover, the Trp33H, Asp54H, and Tyr94L residues did not interact directly with the antigen, but their mutants also had a significant decrease in binding affinity. The positions of these residues were mainly distributed near “hot spot” residues (Figure 3), indicating that these mutants generate new interactions at the periphery of the interface to affect the antibody affinity. Although experimental and computational results for the Y99HA mutant show the same trend, the effects imposed by the mutation vary widely. Experiments found that the Tyr99H residue interacts directly with the antigen and that mutation results in loss of antibody function, but the model of the antibody protein at this position deviated from the actual structure and produced inconsistent results.
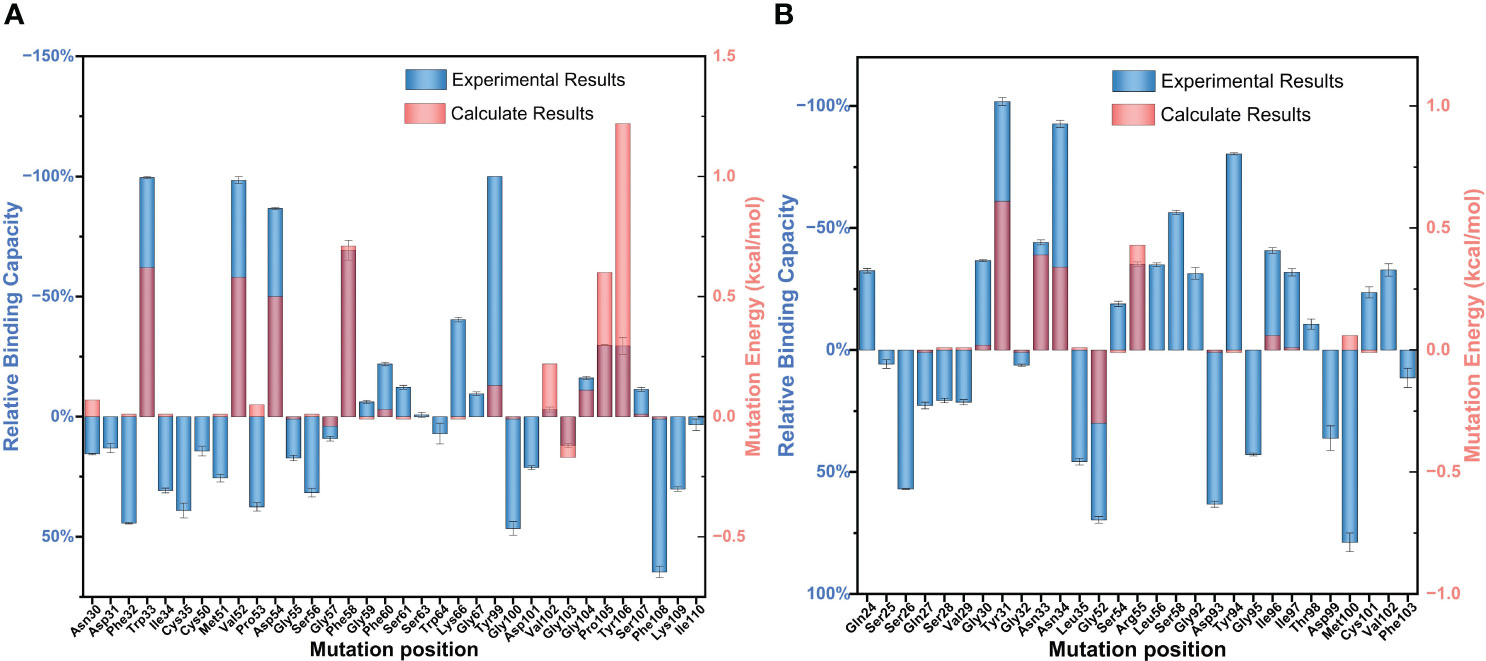
Figure 2 Partial results of the identification of BDG-Ab complementarity-determining region (CDR) residues involved in antigen binding via alanine scanning mutagenesis. The relative antigen binding of the BDG-Abs alanine substitution mutations on (A) heavy chain and (B) light chain was evaluated by indirect ELISA method and molecular dynamics simulation. Error bars represent standard deviations from the means for three independent experiments.
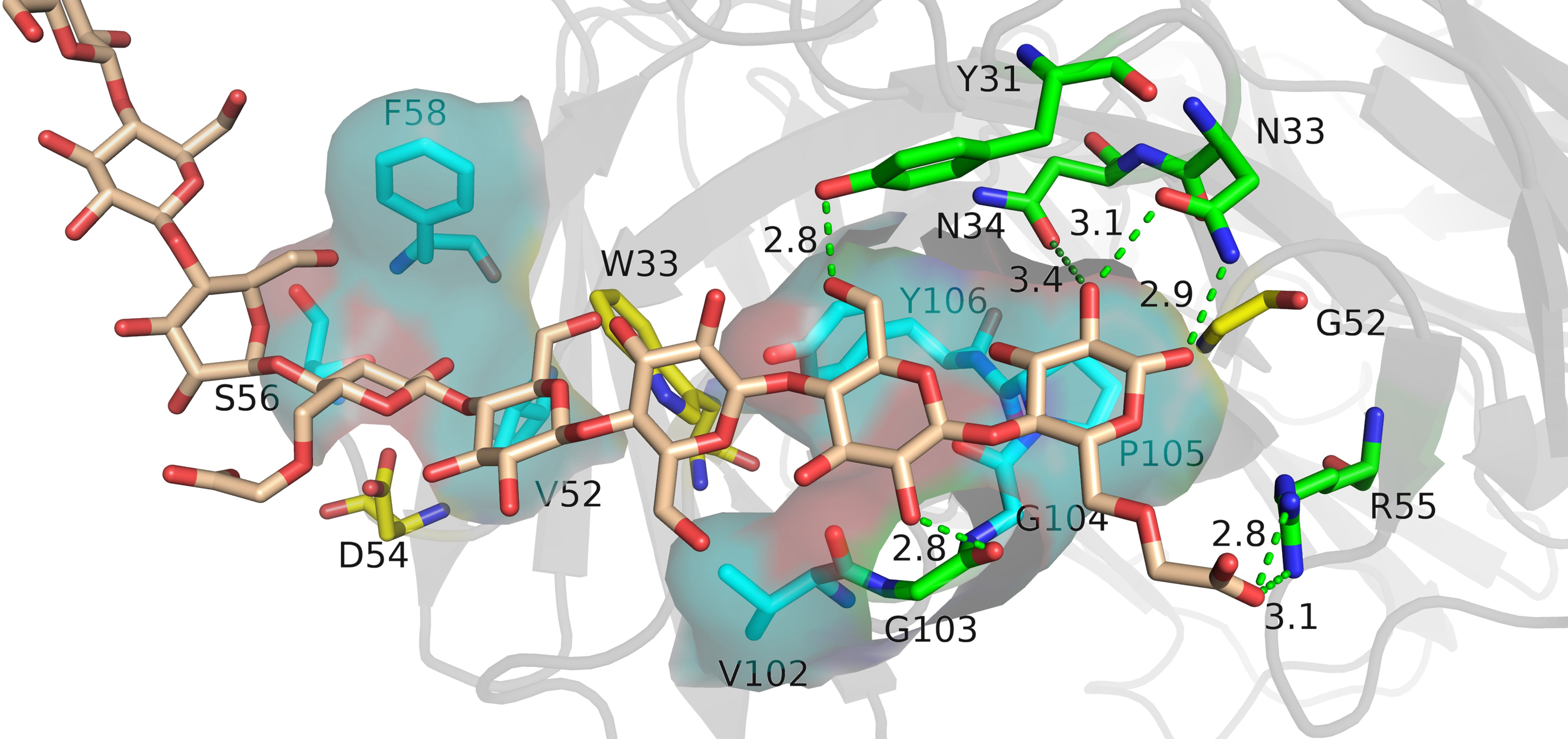
Figure 3 The analysis diagram of the key residues of antigen–antibody binding. Indigo represents the residues that interact with antigens through hydrophobic interaction, green represents the residues that bind to antigens through hydrogen bonds, and yellow represents residues that play an indirect role in the binding process between antigens and antibodies.
3.3 Rational design of BDG-Ab
The antigen–antibody binding was dependent on the role played by the side chains of residues in the interaction (Akiba et al., 2019). Amino acid residues of the same type have similar functions. Therefore, we calculated saturation mutations for the “hot spot” residues with the same type of amino acids to identify better combinations. At most, two residues were predicted to mutate simultaneously. More mutation sites would affect the antibody’s tertiary structure more significantly, which would bias the simulation results. Meanwhile, the mutation energy of the double point mutant antibody was calculated by pairwise combination. The mutation energies of 944 combinations were calculated by mutating the 11 “hot spot” residues to the same type residues (Supplementray Figure 7). Most mutations lead to increased energy, exacerbating the instability of antigen–antibody binding. Therefore, we selected the eight mutation combinations with the lowest energy for antibody expression, purification, and potency testing from 16 results that had mutation energy below −0.5 kcal/mol.
Most of the mutants outperformed the WT-BDG-Ab at different antibody concentrations (Figure 4A). However, the detection ability varied widely at higher antibody concentrations. We used the antibody concentration that corresponded to an absorbance of 0.5 as the measure of antibody potency in this work (Figure 4B). The combined mutation of F58HW/N34LY and V52HL/N34LY was comparable in potency to the WT-BDG-Ab, while the V52HI, N34LY, V52HI/Y31LW, and V52HI/N34LY mutants were superior to the WT-BDG-Ab. However, antibodies with the Tyr99H mutation had low binding capacity to the antigen, consistent with the experimental alanine scan results. The calculation failed to predict this mutation accurately. Secondly, the Y31LW/N34LY double mutation also reduced the antigen–antibody affinity substantially. The Tyr31L and Asn34L residues are spatially close, and the mutation may have altered the conformation of the light chain domain. The experimental results showed that the V52HI/N34LY mutant had the highest potency, which was 3.7-fold higher than that of WT-BDG-Ab.
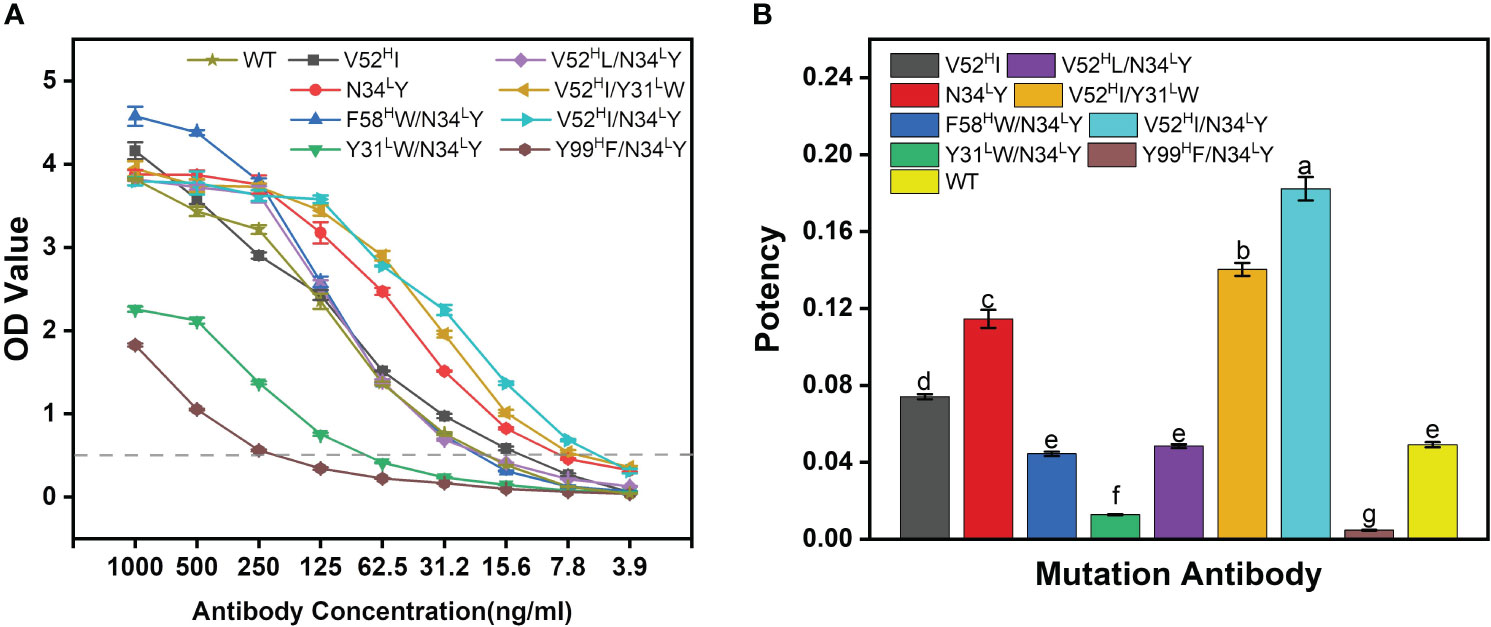
Figure 4 The potency comparison of BDG-Abs. (A) Potency graph for WT-BDG-Ab and its mutants. The antibodies were multiple proportion diluted (1,000–3.90625 ng/mL), then added to the plates coated with BDG. Then, the indirect ELISA was operated (detailed procedure is presented in Section 2.5). The potency of BDG-Abs was compared according to OD value (n = 3). (B) Comparison of the potency of WT-BDG-Ab and its mutants. The BDG-Ab potency was defined as the reciprocal of the BDG-Ab concentration at OD = 0.5. Error bars represent the standard deviations of three independent experiments. Different letters above the bars indicate statistically significant differences between the samples (one-way ANOVA followed by a post hoc Tukey test, p< 0.05).
3.4 Recognition ability of wild-type and mutant BDG-Ab to different fungi and bacteria
Wild-type BDG-Ab and mutant BDG-Ab display different recognition ability to different fungi and bacteria. As shown in Tables 1 and 2, apart from C. neoformans, all the testing fungi could be recognized by wild-type BDG-Ab; meanwhile, 52HI, N34LY, F58HW/N34LY, V52HI/Y31LW, and V52HI/N34LY mutant BDG-Ab could recognize all the fungi being tested. Other mutants have weaker recognizing abilities to the tested fungi. All the BDG-Abs could not recognize Gram-negative bacteria, whereas wild-type, 52HI, N34LY, F58HW/N34LY, and V52HI/Y31LW mutant BDG-Ab could recognize Leuconostoc mesenteroides (a kind of Gram-negative bacterium). This may be due to the fact that L. mesenteroides could also produce BDG. The bacterial BDG could not be recognized by V52HI/N34LY mutant BDG-Ab, indicating its increase in specificity of BDG-Ab. N34LY single-point mutation could increase Ab potency as well. However, when combined with the Y31LW or Y99HF mutant, the antigen–antibody binding ability of Abs could be decreased significantly. The thickness of capsule of C. neoformans may lead to the incomplete release of BDG; thus, C. neoformans could be poorly recognized by BDG-Abs. Gram-negative bacterium could not be significantly recognized either by wild-type BDG-Ab or by mutant BDG-Abs, which makes a significant contribution to improve the testing specificity of BDG. The result has settled the main shortcoming of the G test, which broadens the horizon of BDG detection. Recognition of L. mesenteroides by wild type and some of the mutant BDG-Ab may be due to BDG production by L. mesenteroides. Nevertheless, V52HI/N34LY mutant BDG-Ab could not recognize L. mesenteroides; the reason for this has to be further discussed.
3.5 Analytical capability comparison
The BDG testing ability of the chemiluminescence method was further evaluated, and the Limulus amebocyte lysate method was used as the control. The result is shown in Table 3. The LoB and LoD of the Limulus amebocyte lysate method are 15 pg/mL and 5 pg/mL, respectively, while the LoB and LoD of the chemiluminescence method were decreased to 5 pg/mL and 10 pg/mL for LoB and LoD, respectively. This may be due to the fact that the chemiluminescence method is an endotoxin contamination-free BDG testing method. Moreover, the chemiluminescence method could widen the linear range from 37.5–600 pg/mL to 200–1,000 pg/mL. Both two BDG testing methods share similar precision and stability.
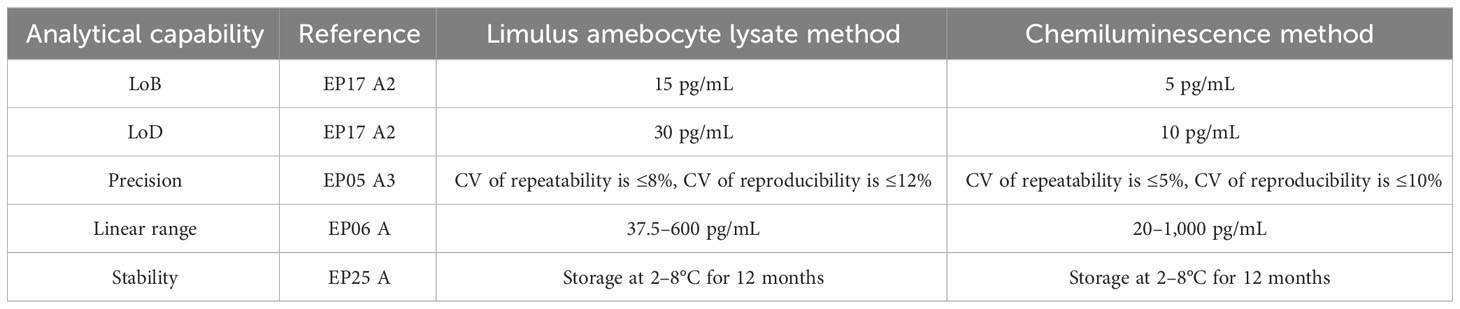
Table 3 Analytical capability comparison data of the Limulus amebocyte lysate method and the chemiluminescence method.
3.6 Clinical application of the V52HI/N34LY mutant
The V52HI/N34LY mutant was applied to a chemiluminescent platform. Chemiluminescence assay does not require immobilizing the primary antibody on the ELISA plate, unlike the ELISA double-antibody sandwich assay. This enables more complete interaction between the free antibodies and the antigen, resulting in better detection. Both the biotin-labeled and acridine ester-labeled antibodies target the same antigen. Firstly, the optimal concentration of the biotin-labeled antibody was determined. The concentration of the acridine ester-labeled antibody was fixed at 0.5 μg/mL, then different concentrations of the biotin-labeled antibody were used to detect the gradient concentration of antigen, and the standard curve was plotted. Figure 5A has shown that the increase of the biotin-labeled antibody occupied a large number of the epitopes on BDG, and the light value of 1 μg/mL biotin antibody decreased by about half at the same BDG concentration. The maximum light value gradually entered the plateau period as the biotin-labeled antibody addition decreased, and basically ceased to change at 0.4 μg/mL.
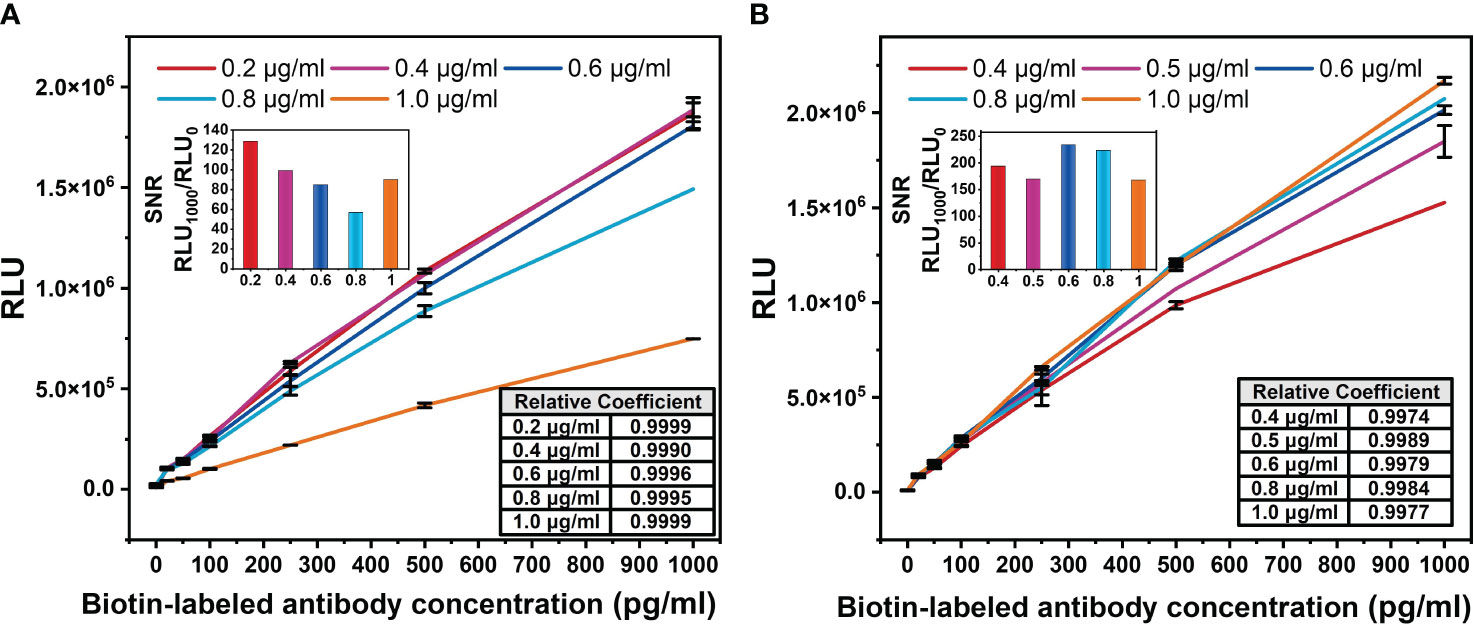
Figure 5 Optimization diagram of antibody concentration in chemiluminescence method. (A) Biotin-labeled antibody concentration. The concentration of acridine ester-labeled antibody was fixed at 0.5 μg/mL. (B) Acridine ester-labeled antibody concentration. The concentration of biotin-labeled antibody was fixed at 0.2 μg/mL. The signal-to-noise ratio (SNR) of the standard curve of biotin-labeled antibody with different concentration was defined as the ratio of the light value of 1,000 pg/mL antigen to the background light value. The correlation coefficients between the logarithm of BDG concentrations and light values were calculated.
In order to accurately detect the actual concentration of antigen and increase the threshold of detectable light value, the signal-to-noise ratio (SNR) of the standard curve of the biotin-labeled antibody with different concentrations was calculated. We defined the SNR as the ratio of the light value of 1,000 pg/mL antigen to the background light value. Among the five concentrations, the biotin-labeled antibody at 0.2 μg/mL had the highest SNR (Figure 5A), which was used for the subsequent study. The light value increased with the concentration of the acridine ester-labeled antibody (Figure 5B), and we selected the concentration of 0.6 μg/mL with the highest SNR for the subsequent study. The correlation coefficients between the logarithm of BDG concentrations and the light values were greater than 0.99, indicating a strong correlation.
A total of 574 clinical samples were tested using the developed method and the Limulus amebocyte lysate method, respectively. The Limulus amebocyte lysate method can accurately identify patients with BDG concentrations above 95 pg/mL as positive and below 70 pg/mL as negative, but not those with BDG concentrations between 70 and 95 pg/mL. The ROC curves of the chemiluminescence method (Figures 6A, B) were plotted using the Limulus amebocyte lysate method as the cutoff values for negativity and positivity. With a cutoff value of 70 pg/mL, the chemiluminescence method had an area under the curve (AUC) of 0.903 (95% CI: 0.876–0.930) and a maximum Youden index of 0.7409. The antigen concentration detected by the chemiluminescence method was 89.98 pg/mL, which was within the 70–95 pg/mL range. The sensitivity and specificity of the chemiluminescence method were 86.72% and 87.37%, respectively, using this antigen concentration as the cutoff value. With a cutoff value of 95 pg/mL, the AUC of the chemiluminescence method was 0.905 (95% CI: 0.880–0.929), which was higher than the cutoff value of 70 pg/mL, with a maximum Youden index of 0.7309, and the antigen concentration was 89.98 pg/mL. The sensitivity and specificity were 83.33% and 89.76%, respectively, with the antigen concentration cutoff value of 89.98 pg/mL. In addition, compared with the Limulus amebocyte lysate method, the chemiluminescence method has lower sensitivity (Table 4).
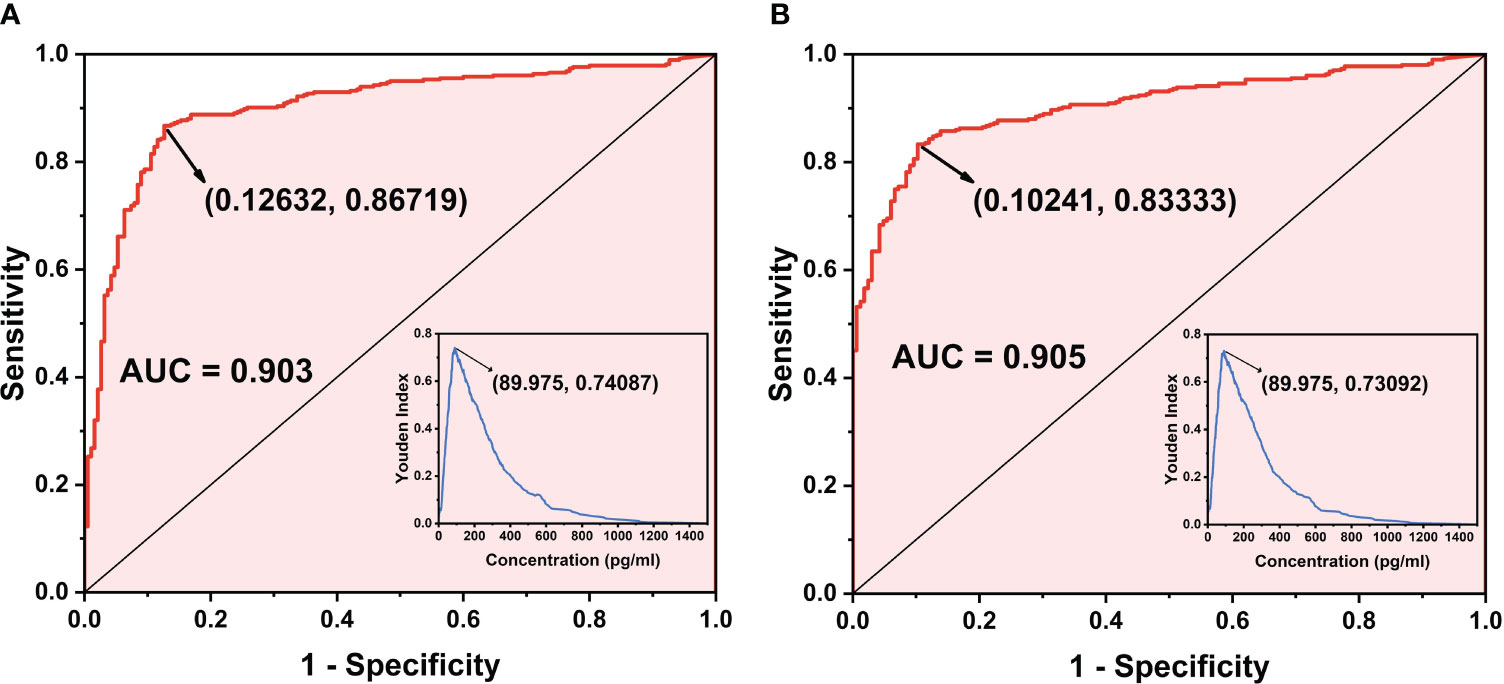
Figure 6 ROC curve of the chemiluminescence method. (A) ROC curve when the cutoff value of the Limulus amebocyte lysate detection method is 70 pg/mL as the standard. (B) ROC curve when the cutoff value of the Limulus amebocyte lysate detection method is 95 pg/mL as the standard.
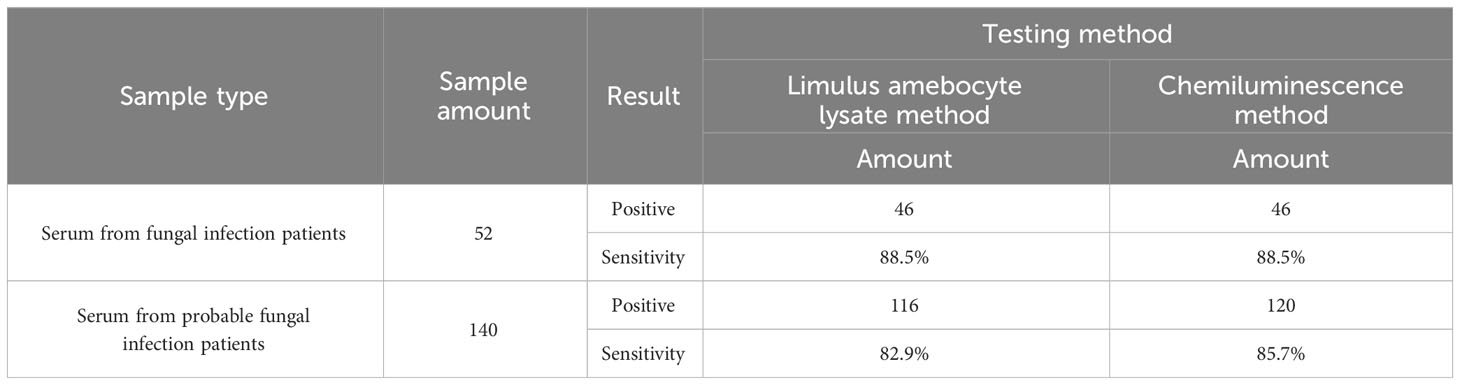
Table 4 Sensitivity comparison between the chemiluminescence method and the Limulus amebocyte lysate method.
The existing technologies for the quantitation of BDG is limited. The Limulus amebocyte lysate method relies on cells derived from the blood of horseshoe crabs, which may lead to the population decline of horseshoe crabs. Compared to the Limulus amebocyte lysate method, chemiluminescence is free of endotoxin contamination. Last but not least, such method has a sustainable raw material source, which offers the testing method a long-term development.
In this study, a V52HI/N34LY mutant BDG-Ab was discovered for the construction of a chemiluminescence-based BDG testing system. According to the clinical sample testing result, the chemiluminescence-based method possesses a similar clinical compliance rate with the Limulus amebocyte lysate method and better sensitivity and specificity. The operation protocol is practically simple since the chemiluminescence-based method is a direct “mix-to-test” method without a washing procedure. Based on such reasons, the chemiluminescence-based method is a better choice for BDG testing.
4 Conclusion
In the study, a high-performance mutant antibody was generated by immunizing rabbits with purified CP-BDG and experimental as well as computational alanine scanning on the CDR region of BDG-Ab. The V52HI/N34LY mutant exhibited a 3.7-fold higher potency than WT-BDG-Ab, which was labeled with biotin and acridine ester for the chemiluminescence assay. Then, a novel detection method for BDG based on a magnetic particle chemiluminescence platform using V52HI/N34LY mutant was developed. Compared to the Limulus amebocyte lysate method, the chemiluminescence method showed a lower LoB and LoD, a wider linear range, and better sensitivity, which poses accurate quantitation for clinical sample testing. The assay demonstrated high sensitivity and specificity (83.33% and 89.76%, respectively) for detecting BDG in serum, with an antigen concentration cutoff value of 89.98 pg/mL. The results indicated that the developed method could serve as an alternative to the Limulus amebocyte lysate assay and enable a rapid and accurate diagnosis of IFD.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The study was approved by the Medical Ethics Committee of Shandong Chest Hospital under the registration number 2020XKYYEC-48. The animal study was approved by Animal Ethics Committee of Tianjin Fifth Central Hospital under the registration number TJWZX2018048. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
CL: Conceptualization, Methodology, Validation, Writing – original draft. LZ: Investigation, Methodology, Visualization, Writing – review & editing. JZ: Conceptualization, Methodology, Software, Writing – review & editing. MW: Formal analysis, Methodology, Writing – review & editing. SY: Conceptualization, Writing – review & editing. RS: Resources, Supervision, Writing – review & editing. WQ: Project administration, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2022YFC2106300), The National Natural Science Foundation of China (No. 21621004, 22278314, 22078239), The Beijing-Tianjin-Hebei Basic Research Cooperation Project (B2021210008), Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-004), and Enterprise horizontal entrustment 2021GKF-0559, 2021GKF-0562.
Conflict of interest
Author CL was employed by the company Dynamiker Biotechnology Tianjin Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1322264/full#supplementary-material
References
Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., et al. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SOFTWAREX 1-2(C), 19–25. doi: 10.1016/j.softx.2015.06.001
Akiba, H., Tamura, H., Caaveiro, J. M. M., Tsumoto, K. (2019). Computer-guided library generation applied to the optimization of single-domain antibodies. Protein Engineering Design Selection 32 (9), 423–431. doi: 10.1093/protein/gzaa006
Almeida, F., Rodrigues, M. L., Coelho, C. (2019). The still underestimated problem of fungal diseases worldwide. Front. Microbiol. 10, 214. doi: 10.3389/fmicb.2019.00214
Azap, A. (2020). Role of non-culture tests for the diagnosis of invasive fungal infections. Klimik Dergisi/Klimik J. 32 (Supp2), 131–134. doi: 10.5152/kd.2019.56
Calero, A. L., Alonso, R., Gadea, I., Vega, M. D. M., García, M. M., Muñoz, P., et al. (2022). Comparison of the performance of two galactomannan detection tests: platelia aspergillus ag and aspergillus galactomannan ag virclia monotest. Microbiol. Spectr. 10 (2), e0262621–e0262621. doi: 10.1128/spectrum.02626-21
Du, Z., Su, H., Wang, W., Ye, L., Wei, H., Peng, Z., et al. (2021). The trRosetta server for fast and accurate protein structure prediction. Nat. Protoc. 16 (12), 5634–5651. doi: 10.1038/s41596-021-00628-9
Eberhardt, J., Santos-Martins, D., Tillack, A. F., Forli, S. (2021). AutoDock vina 1.2.0: new docking methods, expanded force field, and python bindings. J. Chem. INF Model. 61 (8), 3891–3898. doi: 10.1021/acs.jcim.1c00203
Fernández Quintero, M. L., Hoerschinger, V. J., Lamp, L. M., Bujotzek, A., Georges, G., Liedl, K. R. (2020). VH -VL interdomain dynamics observed by computer simulations and NMR. Proteins: Structure Function Bioinf. 88 (7), 830–839. doi: 10.1002/prot.25872
Finkelman, M. A. (2021). Specificity influences in (1→3)-β-d-glucan-supported diagnosis of invasive fungal disease. J. FUNGI 7 (1), 14. doi: 10.3390/jof7010014
Fisher, M. C., Alastruey-Izquierdo, A., Berman, J., Bicanic, T., Bignell, E. M., Bowyer, P., et al. (2022). Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol. 20 (9), 557–571. doi: 10.1038/s41579-022-00720-1
Hage, C. A., Carmona, E. M., Epelbaum, O., Evans, S. E., Gabe, L. M., Haydour, Q., et al. (2019). Microbiological laboratory testing in the diagnosis of fungal infections in pulmonary and critical care practice. An official american thoracic society clinical practice guideline. Am. J. Resp. Crit. Care 200 (5), 535–550. doi: 10.1513/AnnalsATS.201908-582CME
Humphrey, W., Dalke, A., Schulten, K. (1996). VMD: Visual molecular dynamics. J. Mol. Graphics 14 (1), 33–38. doi: 10.1016/0263-7855(96)00018-5
Kwan, B. K. Y., Chan, H. K., Cheung, S. G. (2018). Habitat use of globally threatened juvenile Chinese horseshoe crab,Tachypleus tridentatus under the influence of simulated intertidal oyster culture structures in Hong Kong. Aquat. Conservation: Mar. Freshw. Ecosyst. 28 (1), 124–132. doi: 10.1002/aqc.2811
Lass-Flörl, C., Samardzic, E., Knoll, M. (2021). Serology anno 2021—fungal infections: from invasive to chronic. Clin. Microbiol. Infect. 27 (9), 1230–1241. doi: 10.1016/j.cmi.2021.02.005
Li, Z., Liu, X., Chen, W., Shen, F., Bi, H., Ke, G., et al. (2022). Uni-fold: an open-source platform for developing protein folding models beyond alphaFold (Cold Spring Harbor: Cold Spring Harbor Laboratory Press).
Liu, F., Liu, Y., Feng, X., Ibrahim, S. A., Huang, W. (2021). Structure characterization and in vitro immunomodulatory activities of carboxymethyl pachymaran. Int. J. Biol. MACROMOL 178, 94–103. doi: 10.1016/j.ijbiomac.2021.02.046
Mercier, T., Guldentops, E., Patteet, S., Beuselinck, K., Lagrou, K., Maertens, J. (2019). Beta-d -glucan for diagnosingPneumocystis pneumonia: a direct comparison between the wako β-glucan assay and the fungitell assay. J. Clin. Microbiol. 57 (6), e00322–19. doi: 10.1128/JCM.00322-19
Ricna, D., Lengerova, M., Bezdicek, M., Kocmanova, I., Drgona, L., Weinbergerova, B., et al. (2019). Detection and identification of fungi in bronchoalveolar lavage fluid from immunocompromised patients using panfungal PCR. Folia Microbiol. 64 (3), 421–428. doi: 10.1007/s12223-018-00669-w
Salazar, F., Bignell, E., Brown, G. D., Cook, P. C., Warris, A. (2022). Pathogenesis of respiratory viral and fungal coinfections. Clin. Microbiol. Rev. 35 (1), e21–e94. doi: 10.1128/CMR.00094-21
Sayers, E. W., Beck, J., Bolton, E. E., Bourexis, D., Brister, J. R., Canese, K., et al. (2021). Database resources of the national center for biotechnology information. Nucleic Acids Res. 49 (D1), D10–D17. doi: 10.1093/nar/gkaa892
Su, H., Wang, W., Du, Z., Peng, Z., Gao, S. H., Cheng, M. M., et al. (2021). Improved protein structure prediction using a new multi-scale network and homologous templates. Adv. Sci. 8 (24), e2102592. doi: 10.1002/advs.202102592
Terrero-Salcedo, D., Powers-Fletcher, M. V. (2020). Updates in laboratory diagnostics for invasive fungal infections. J. Clin. Microbiol. 58 (6), e01487–19. doi: 10.1128/JCM.01487-19
Tian, C., Kasavajhala, K., Belfon, K. A. A., Raguette, L., Huang, H., Migues, A. N., et al. (2020). ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J. Chem. Theory Comput. 16 (1), 528–552. doi: 10.1021/acs.jctc.9b00591
Valdés-Tresanco, M. S., Valdés-Tresanco, M. E., Valiente, P. A., Moreno, E. (2021). gmx_MMPBSA: A new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 17 (10), 6281–6291. doi: 10.1021/acs.jctc.1c00645
Yoshida, M. (2021). Development of a method of measuring β-D-glucan and its use in preemptive therapy for invasive fungal infections. Int. J. Mol. Sci. 22 (17), 9265. doi: 10.3390/ijms22179265
Yoshida, K., Kuroda, D., Kiyoshi, M., Nakakido, M., Nagatoishi, S., Soga, S., et al. (2019). Exploring designability of electrostatic complementarity at an antigen-antibody interface directed by mutagenesis, biophysical analysis, and molecular dynamics simulations. Sci. REP-UK 9 (1), 4482. doi: 10.1038/s41598-019-40461-5
Keywords: invasive fungal infections, (1-3)-β-D glucan, antibody rational design, rapid detection, limulus, chemiluminescence
Citation: Liu C, Zhang L, Zhang J, Wang M, You S, Su R and Qi W (2024) Rational design of antibodies and development of a novel method for (1–3)-β-D glucan detection as an alternative to Limulus amebocyte lysate assay. Front. Cell. Infect. Microbiol. 14:1322264. doi: 10.3389/fcimb.2024.1322264
Received: 16 October 2023; Accepted: 02 January 2024;
Published: 24 January 2024.
Edited by:
Ying Zhao, Peking Union Medical College Hospital (CAMS), ChinaReviewed by:
Sandra Jesus, University of Coimbra, PortugalAhmad Mobed, Tabriz University of Medical Sciences, Iran
Chunhui Xu, Chinese Academy of Medical Sciences and Peking Union Medical College, China
Copyright © 2024 Liu, Zhang, Zhang, Wang, You, Su and Qi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengping You, eXNwQHRqdS5lZHUuY24=; Wei Qi, cWl3ZWlAdGp1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Chunlong Liu
Chunlong Liu Lin Zhang
Lin Zhang Jiaxing Zhang
Jiaxing Zhang Mengfan Wang1
Mengfan Wang1 Shengping You
Shengping You Rongxin Su
Rongxin Su Wei Qi
Wei Qi