
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 19 April 2024
Sec. Fungal Pathogenesis
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1295841
This article is part of the Research Topic Invasive Fungal Diseases: Pathogen Detection and Diagnosis Development View all 10 articles
 Faezeh Rouhi1†
Faezeh Rouhi1† Sajedeh Soltani1†
Sajedeh Soltani1† Somayeh Sadeghi2
Somayeh Sadeghi2 Elahe Nasri2
Elahe Nasri2 Mahnaz Hosseini3
Mahnaz Hosseini3 Safiyeh Ghafel3
Safiyeh Ghafel3 Shima Aboutalebian1,3
Shima Aboutalebian1,3 Hamed Fakhim2
Hamed Fakhim2 Hossein Mirhendi1,3*‡
Hossein Mirhendi1,3*‡Introduction: Although the existence of Candida species in the respiratory tract is often considered commensal, it is crucial to recognize the significance of Candida colonization in immunocompromised or COVID-19 patients. The emergence of Candida auris as an emerging pathogen further emphasizes the importance of monitoring yeast infection/colonization, particularly in COVID-19 patients.
Methods: In this study, respiratory samples mainly from COVID-19 patients, primarily those suspected of having a fungal infection, were cultured on Sabouraud dextrose agar plates and the yeast colonies were identified using a two-step multiplex PCR method. The samples suspected of C. auris underwent specific nested PCR followed by sequence analysis.
Results: A total of 199 respiratory samples were collected from 73 women and 126 men, ranging in age from 1.6 to 88 years. Among the patients, 141 had COVID-19, 32 had cancer, 5 were hospitalized in ICU, 2 had chronic obstructive pulmonary disease)COPD(, and others were patients with combination diseases. From these samples, a total of 334 yeast strains were identified. C. albicans (n=132, 39.52%) was the most common species, followed by C. tropicalis (n=67, 20%), C. glabrata (n=56, 16.76%), C. krusei (n=18, 5.4%), C. parapsilosis (n=17, 5.08%), Saccharomyces cerevisiae (n=10, 3%), C. kefyr (n=9, 2.6%), C. dubliniensis (n=7, 2.1%), C. lusitaniae (n=5, 1.5%), C. auris (n=3, 0.9%), C. guilliermondii (n=2, 0.6%), C. rugosa (n=1, 0.3%), C. intermedia (n=1, 0.3%), and Trichosporon spp. (n=1, 0.3%). C. auris was detected in a patient in ICU and two COVID-19 patients. While its presence was confirmed through sequence analysis, our extensive efforts to isolate C. auris were unsuccessful.
Conclusion: While C. albicans colonization remains prevalent, our study found no evidence of Candida lung infection. Since the role of Candida colonization in airway secretions remains ambiguous due to limited research, further studies are imperative to shed light on this matter.
Fungal infection and colonization emerged as significant clinical challenges in recent decades (Delisle et al., 2008). Among nosocomial infections, fungi account for 8-10% of cases, with Candida species responsible for 80% of them (Pendleton et al., 2017), particularly in the intensive care unit (ICU) (El-Ebiary et al., 1997). As of October 25, 2022, the World Health Organization (WHO) issued a list of 19 fungal priority pathogens identified as posing the greatest threat to public health, with six Candida species (C. albicans, C. parapsilosis, C. glabrata, C. auris, C. tropicalis, and C. krusei) included in this prioritization (Parums, 2022). Candida species are commonly found in the normal microbial flora of the human body and have historically been considered part of the oral microbiota in healthy individuals. They can also be present in the sputum of 20-55% of healthy subjects (Terraneo et al., 2016). Studies have indicated that the presence of Candida species in respiratory specimens of immunocompetent patients without concurrent infection is clinically insignificant, as it typically represents colonization rather than infection (Azoulay et al., 2006; Terraneo et al., 2016). However, in immunocompromised patients, Candida isolation from sputum, endotracheal aspirates, bronchoscopic samples, percutaneous lung needle aspirates, and lung tissue may rarely indicate invasive pneumonia instead of colonization (El-Ebiary et al., 1997). Factors such as mucus plugging in the lungs, prolonged antibiotic treatment, corticosteroid use, surgically implanted catheters, and longer hospital stays for immunocompromised patients predispose to Candida colonization (Baradkar et al., 2009; Muthig et al., 2010).
Although bronchial Candida colonization does not directly lead to increased mortality, it is associated with longer durations of ICU and hospital stays, elevated management costs, and poorer outcomes (Azoulay et al., 2006). Furthermore, Candida colonization in the lower respiratory tract (RT) or hypersensitivity reactions in different areas have been identified as independent risk factors for the presence of multidrug-resistant bacteria and the development of ventilator-associated pneumonia caused by Pseudomonas aeruginosa (Terraneo et al., 2016). Therefore, it is crucial to consider the potential significance of these commensal fungi when they are repeatedly and clearly detected at infection sites. Despite controversies surrounding the diagnosis of pulmonary candidiasis, the definitive diagnosis still relies on the histological presentation of yeast in lung tissue accompanied by inflammation (El-Ebiary et al., 1997), and the value of culturing respiratory specimens for diagnosing Candida pneumonia remains unclear. The Clinical Practice Guideline for the Management of Candidiasis does not recommend treatment when Candida species are isolated from RT secretions (Pappas et al., 2016). However, it is proposed that the presence of Candida colonization in the bronchial area, along with colonization of multiple body sites, should be considered as a risk factor for systemic candidiasis (Azoulay et al., 2006).
Candida auris, an emerging pathogen categorized in the WHO critical priority group (Parums, 2022), has been associated with varying levels of antifungal resistance and rapid spread in intensive care settings, leading to widespread nosocomial outbreaks worldwide. Its global presence and recent cases reported in Iran have been demonstrated (Abastabar et al., 2019; Mirhendi et al., 2022; Safari et al., 2022). Accurately identifying this organism poses challenges, which have hindered our understanding of its scope and created uncertainty about its prevalence, transmission, and environmental niches. This highlights the urgent need for ongoing research to address its impact on mortality and healthcare systems. During the recent viral pandemic, the isolation of C. auris in COVID-19 patients with acute respiratory distress syndrome (ARDS) has raised worldwide concern. This association has been linked to stays in the ICU, mechanical ventilation, bacterial infections, and the use of immunomodulators such as corticosteroids as risk factors for colonization or infection. It is crucial to identify C. auris, even as a colonizer, as candidemia can arise from colonization. Reports of colonization or co-infection by C. auris have been documented in COVID-19 patients in several countries, including Italy, Lebanon, the USA, China, Brazil, Colombia, and Spain (Pan American Health Organization/World Health Organization, 2016; Chowdhary et al., 2020; Allaw et al., 2021; Corcione et al., 2022).
There is a dearth of studies specifically examining the importance and role of Candida colonization in airway secretions, thus leaving this matter unresolved. Consequently, additional research is required to elucidate the role of Candida species, which are generally considered to have a low pathogenicity. The objectives of this study were to assess the frequency of pulmonary yeast colonization in respiratory samples, to identify the yeast isolated from pulmonary specimens, particularly from COVID-19 patients, and to gain insights into their colonization profile. C. auris colonization in the patients was also emphasized owing to its clinical significance.
Between March 2020 and March 2023, respiratory samples including tracheal aspiration (TA), sputum, and bronchoalveolar lavage (BAL) fluid, were collected from patients hospitalized in referral hospitals in Isfahan, Iran. The majority of the samples were from COVID-19 patients who exhibited pulmonary symptoms suggestive of fungal infections, and a smaller number of samples were obtained from immunocompromised patients without COVID-19 infection. Among the patients, there were cases of COVID-19 (n=141), cancer (n=32), ICU admission (n=5), COPD (n=2), and the others were patients with combination diseases (Table 1). The study protocol was approved by the ethics committee of the Isfahan University of Medical Science (IR.MUI.MED.REC.1400.448).
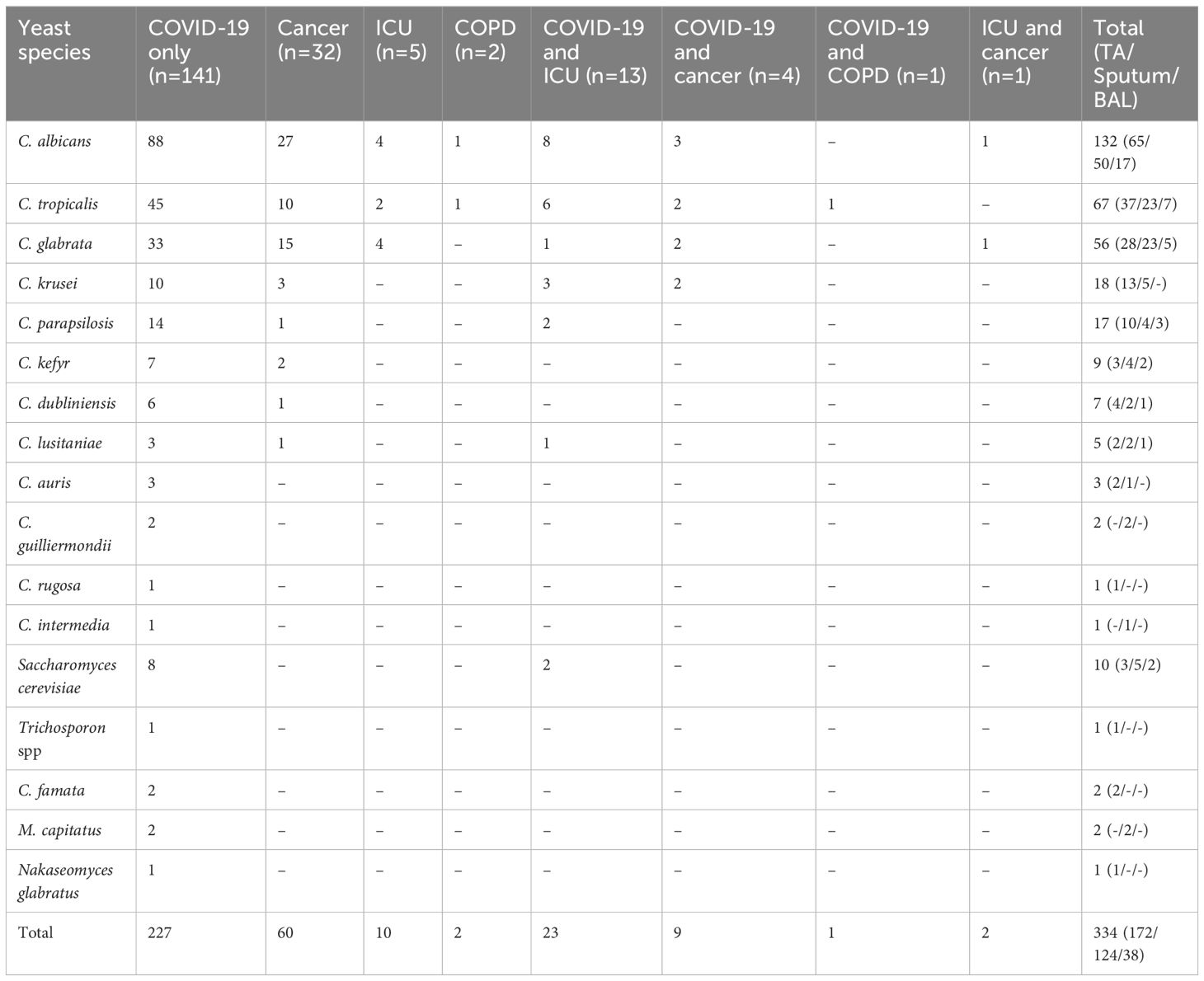
Table 1 Distribution of the yeast species in respiratory specimens (BAL, sputum, and TA) among diverse patient groups, including patients with COVID-19, cancer, ICU-hospitalized, and COPD.
As part of the routine diagnostic procedure, all samples were plated onto two Sabouraud dextrose agar plates supplemented with 0.5% chloramphenicol. The plates were then incubated at both 25°C and 37°C and monitored daily for significant fungal growth. For this study, yeast colonies that developed on the plates were harvested and subcultured into a fresh medium. The resulting pure colonies were suspended in a 30% glycerol solution and stored in a -20°C freezer until further analysis.
The DNA extraction from the yeast isolates was carried out using the boiling method (Silva et al., 2012; Salehipour et al., 2021). In summary, colonies were submerged in 50 µl of sterile distilled water and boiled for 20 minutes in a water bath. After centrifugation at 5000 rpm for 10 minutes, the supernatants were stored at -20°C until being used as DNA templates for PCR.
For species identification of the yeast isolates, a stepwise multiplex PCR known as YEAST PLEX was performed as previously described (Aboutalebian et al., 2022). Accordingly, Tube A was utilized for identifying C. albicans, C. dubliniensis, C. parapsilosis, C. glabrata (Nakaseomyces glabratus), C. tropicalis, C. krusei (Pichia kudriavzevii), C. kefyr (Kluyveromyces marxianus), and C. auris; Tube B was used for identifying C. guilliermondii (Meyerozyma guilliermondii), C. rugose (Diutina rugosa), C. intermedia, C. lusitaniae (Clavispora lusitaniae), and C. norvegensis (Pichia norvegensis); and Tube C was employed for identifying Cryptococcus neoformans, Rhodotorula mucilaginosa, Trichosporon spp., and Saccharomyces cerevisiae. To identify the rare yeasts that were not covered by YEAST PLEX, a pan-fungal PCR reaction was conducted amplifying the entire internal transcribed spacer (ITS1-5.8SrDNA-ITS2) by using well-known universal primers ITS1 and ITS4, as described previously (White et al., 1990), and the resulting PCR products were purified and sequenced (Core Facilities Research Laboratory, Isfahan, Iran) followed by analysis using the BLAST platform (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for species identification.
To confirm the presence of C. auris observed as the faint specific bands in the YEAST PLEX test of certain yeast isolates, a PCR test was performed using universal ITS1 and ITS4 primers, revealing bands of approximately 400 bp, suggesting the possibility of C. auris presence. Furthermore, a nested PCR targeting a part of the ITS region was employed. In the first stage of amplification, the external primers 5’-ATTTTGCATACACACTGATTTGG-3’ and 5’-CGACAACAAAACGAAAAAAAGCG-3’ were used to amplify a 312 bp fragment. In the second stage, the internal primers 5’-AACTAACCCAACGTTAAGTTCAAC-3’ and 5’-AACGCCACCGCGAAGATT-3’ were used, yielding a final fragment of 220 bp (Aboutalebian et al., 2021; Aboutalebian et al., 2022) (Figure 1A). The first round of PCR included 7.5 µl of 2x premix (Master Mix Red, Ampliqon, Denmark), 0.5 µM primers, and 2 µl DNA template in a total volume of 15 µl, and the thermal conditions were 95°C for 5 min, followed by 35 cycles of 95°C 15 s, 60°C 30 s, and 72°C 30 s. The second PCR consisted of 7.5 µl of 2x premix, 0.33 µM primers, and 2 µl of 1/50 diluted first PCR product, to a final volume of 15 µl, and the reaction conditions included 95°C 5 min, followed by 30 cycles of 95°C 15 s, 60°C 30 s, and 72°C 20 s. Five microliters of the PCR product were electrophoresed, stained, and visualized under ultraviolet light. For further confirmation, the PCR products from positive samples underwent sequencing and BLAST analysis.
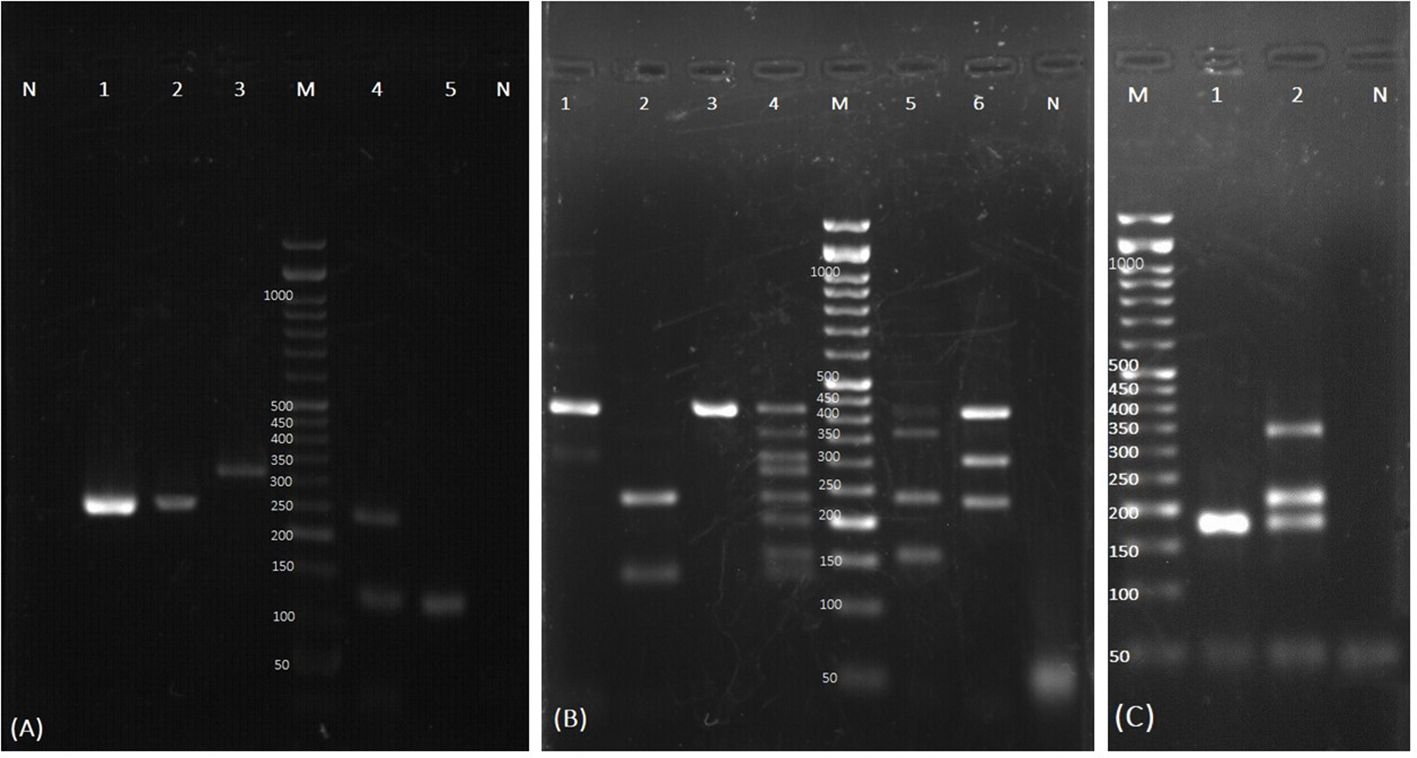
Figure 1 Examples of agarose gel electrophoresis of PCR products for identification of the yeast colonies isolated from respiratory samples. (A) Test tube specific for detection of C. auris. Lane N: negative control of nested PCR; Lane 1 and 2: C. auris with specific final product (220 bp); Lane 3: first round of nested PCR product of C. auris (312 bp); Lane M: 50-bp size marker. The yeasts identified by Tube C. are shown in Lane 4,5. Lane 4: Trichosporon spp., Saccharomyces cerevisiae; Lane 5: Saccharomyces cerevisiae; Lane N: negative control of Tube C. (B) The yeasts identified by Tube A: Lane 1: C. albicans and C. parapsilosis; Lane 2: C. glabrata and C. tropicalis; Lane 3: C. albicans; Lane 4: Home made size marker for yeast identification; Lane M: commercial 50-bp size marker; Lane 5: C. albicans, C. dubliniensis, C. glabrata, and C. krusei; Lane 6: C. albicans, C. parapsilosis, and C. glabrata; Lane N: negative control. (C) The yeasts identified by Tube B. Lane M: 50-bp size marker; Lane 1: C. lusitaniae; Lane 2: C. guilliermondii, C. intermedia, and C. lusitaniae; Lane N: negative control.
In an attempt to isolate C. auris colonies from those colonies that were positive for C. auris in molecular tests, selective mediums were used: 1) Sabouraud broths (Merck. Germany) with 10% sodium chloride and 2% mannitol was incubated at 42°C and shacked at 80 rpm for 24 h; 2) Sabouraud broths with 2% of Dulcitol and 4, 8, and 16 μg/mL fluconazole (Rossi et al., 2023) was incubated at 42°C for up to 10 days without agitation; 3) and yeast extract peptone dextrose agar with 12.5% sodium chloride and 9 ml of 9 mM/liter Iron (II) sulfate solution was mixed and incubated for one week at 35°C (Das et al., 2021).
A total of 199 respiratory samples, including 95 TA, 76 sputum, and 28 BAL were included in this study. The patients consisted of 73 women and 126 men, with ages ranging from 1.6 to 88 years and an average age of 56.
A total of 334 yeast strains were identified from the cultures, predominantly from TA samples, and subjected to PCR-based identification. The most frequently identified species were: C. albicans (n=132, 39.52%), C. tropicalis (n=67, 20%), and C. glabrata (n=56, 16.76%), followed by C. krusei (n=18, 5.4%), C. parapsilosis (n=17, 5.08%), Saccharomyces cerevisiae (n=10, 3%), C. kefyr (n=9, 2.6%), C. dubliniensis (n=7, 2.1%), C. lusitaniae (n=5, 1.5%), C. auris (n=3, 0.9%), C. guilliermondii (n=2, 0.6%), C. rugosa (n=1, 0.3%), C. intermedia (n=1, 0.3%), and Trichosporon spp. (n=1, 0.3%) (Figures 1A–C). Five yeast isolates tested negative with the YEAST PLEX assay, and therefore they were subjected to DNA amplification using ITS universal primers followed by sequencing. They were identified as C. famata (Debaryomyces hansenii) (n=2, 0.6%), Magnusiomyces capitatus (Geotrichum capitatum) (n=2, 0.6%) (accession numbers in GenBank is OR600243), and Nakaseomyces glabratus (n=1, 0.3%). Although the multiplex PCR system used in this study includes specific primers for the identification of C. norvegensis, Cryptococcus neoformans, and Rhodotorula mucilaginosa, none of these isolates were detected. The distribution of Candida species in different clinical samples and the underlying conditions of the patients are presented in Table 1.
Within our samples, 109 patients were colonized by one species of Candida, 56 patients by two species, and 27 patients by three species. Interestingly, in seven cases four different Candida species were identified. Additionally, as shown in Table 2, We observed nine cases (4.5%) of co-existence of common Candida species (C. albicans, C. glabrata, C. tropicalis, C. krusei, C. dubliniensis) with other fungi. Among these cases, A. niger (n=2), A. flavus (n=2), and A. fumigatus (n=1) were isolated in 5 samples, confirmed through direct microscopy, culture, and PCR identification. Also, Among the patients with Candida species colonization, 5 cases had co-existence with Pneumocystis, as documented by specific nested PCR (Matouri et al., 2023). Out of 9 patients with co-occurrence infections with other fungi, one of the patients had a three-fungi isolation of A. fumigatus, Pneumocystis, and C. albicans.
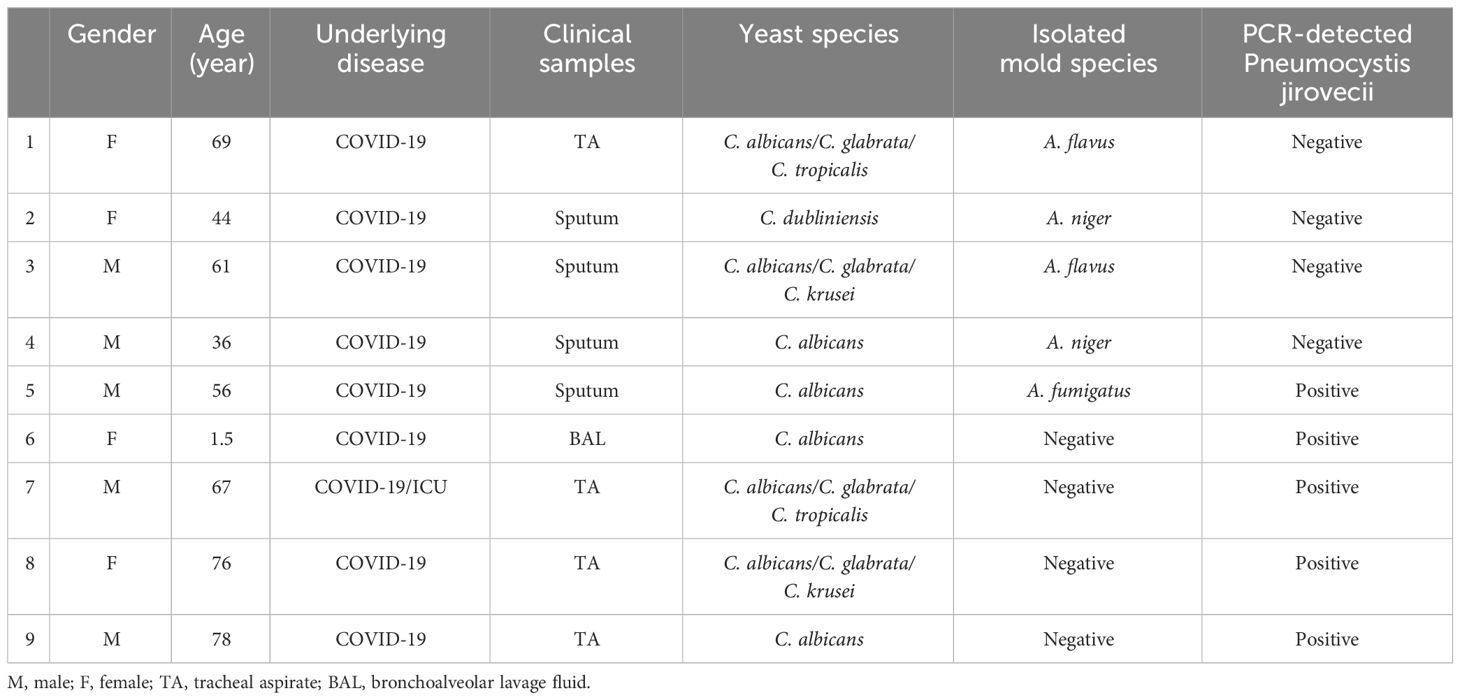
Table 2 Concurrent pulmonary fungal infection along with yeast colonization in some respiratory samples examined in this study.
By utilizing C. auris-specific nested PCR, we were able to detect C. auris in one ICU-hospitalized patient and two COVID-19 patients, as outlined in Table 3. Sequence analysis of PCR products confirmed the presence of C. auris (accession numbers in GenBank are: OR600362, OR600363, OR600282), however, in spite of multiple mycological attempts made to isolate the colony, none of the selective cultures yielded positive results for C. auris.
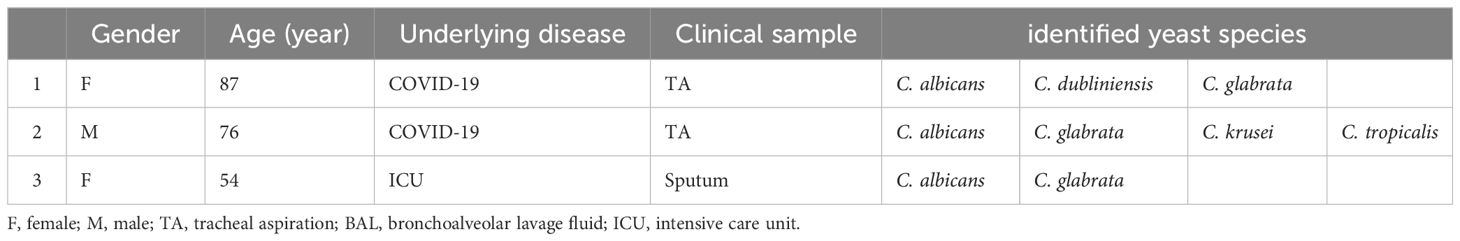
Table 3 Yeast species profiles of three patients in whom Candida auris was molecularly detected as a concurrent species.
As a member of the mycobiome in the human body, Candida species can colonize various sites, including the mucosal surfaces of the mouth and vagina, RT, skin folds, and gastrointestinal tract in both healthy and immunocompromised individuals (Caramalac et al., 2007). Microbial colonization plays a significant role in the development of secondary infections that can complicate pulmonary disorders. In immunocompromised patients, asymptomatic colonization of Candida species in the RT may be associated with poor outcomes and could potentially alter the antibiotic resistance patterns of pathogenic bacteria through the formation of polymicrobial biofilms. However, the significance of Candida colonization in the RT remains a subject of controversy (Liu et al., 2021; Froidefond et al., 2023). In this study, we conducted an analysis of the RT yeast mycobiome by culturing respiratory samples from patients with COVID-19, COPD, and cancer. The results revealed that Candida species were the dominant members in the heterogeneous patient populations, with C. albicans being the most frequently identified species (39.52%), highlighting the common occurrence of C. albicans in hospitalized patients. The recently developed WHO Fungal Priority Pathogens List now identifies critical, high, and medium-priority fungal pathogens ranked based on their public health impact and/or risk of emerging antifungal resistance. For Candida species, the WHO critical priority group currently includes C. albicans and C. auris. Also, C. glabrata, C. parapsilosis, and C. tropicalis are classified as high-priority pathogens, while C. krusei is listed in the medium-priority group (Parums, 2022).
There have been reports of an increasing emergence of non-albicans Candida species, possibly due to the development of antifungal resistance (Wang et al., 2022) and improved differentiation methods. In our study, C. tropicalis (20%), followed by C. glabrata (16.76%), C. krusei (5.4%), and C. parapsilosis (5.08%) were the most prominent non-C. albicans species in the respiratory specimens. Other studies have also shown that Candida species, particularly C. albicans, are commonly isolated from respiratory specimens of the patients. In a study conducted on 100 sputum samples from pulmonary tuberculosis in Mumbai, India, C. albicans was identified in 24 out of 26 isolates (92.31%), while C. tropicalis and C. parapsilosis were isolated from only one case each (Baradkar et al., 2009). In Michigan, C. albicans was the most prevalent (78%) species, followed by C. glabrata (11%) (Pendleton et al., 2018). Similarly, in Massachusetts, among patients with cystic fibrosis (CF), C. albicans (44.6%), C. tropicalis (6%), C. glabrata (5%), and C. parapsilosis (4%) were the most commonly isolated yeasts in respiratory secretions (Azoulay et al., 2006). Therefore, it seems that composition of the species colonizing the respiratory system is consistent with the species responsible for infection leading to hypothesis that Candida colonization may serve as a marker/source of patient deterioration.
In COVID-19 patients, immune dysfunction, lung injury, invasive managements including mechanical ventilation, and treatment with various drugs such as antibiotics and corticosteroids contribute to a high risk of colonization or secondary fungal infections (Erami et al., 2023) including invasive candidiasis (Avkan-Oğuz et al., 2022; Froidefond et al., 2023). In the present study, C. albicans (n=99, 29.64%) was the most commonly isolated yeast from the RT of COVID-19 patients followed by C. tropicalis (n=54, 16.16%) and C. glabrata (n=36, 10.77%). Similarly, Avkan-Oguz et al. reported that C. albicans (45.5%), C. glabrata (15.9%), and C. parapsilosis (13.6%) were the most frequently isolated yeasts from critically ill COVID-19 patients who had fungal infection/colonization (Avkan-Oğuz et al., 2022). Likewise, in COVID-19 patients admitted to ICUs with severe SARS-CoV-2 infection in France, C. albicans (74%), C. tropicalis (8%), C. glabrata (6%), C. dubliniensis (3%), and C. parapsilosis (3%) were most commonly isolated yeast species in respiratory sample cultures (Froidefond et al., 2023). In a study by Erami et al. conducted in Kashan, Iran, among patients with COVID-19 pneumonia who were on mechanical ventilation in ICUs, C. albicans (79.7%) was the most prevalent species, followed by C. glabrata (17.4%) and C. africana (2.9%) (Erami et al., 2022). In our study, 18 cases of co-colonization of C. albicans and C. glabrata were observed in patients with COVID-19 who received corticosteroid therapy. While previous reports have highlighted the cases of C. glabrata pneumonia with candidemia in patients suffering from COPD (Yazici et al., 2016), our study found that C. tropicalis was the most common species among COPD patients.
In our study, the most common yeast species identified from 37 hospitalized cancer patients who received anti-cancer therapy was C. albicans, which confirms the previous reports (Yu and Liu, 2022). Other Candida species, including C. glabrata, C. tropicalis, C. krusei, C. kefyr, C. dubliniensis, C. lusitaniae, and C. parapsilosis were also identified in these patients. C. albicans, C. tropicalis, and C. glabrata were similar across all other groups of patients (i.e., COVID-19, cancer, and ICU patients), except for the COPD group, in which the only species were C. albicans and C. tropicalis.
Candida auris, a global nosocomial pathogen with varying antifungal resistance, poses identification challenges, especially in ICU settings. In our study, C. auris colonization was detected in three COVID-19 and ICU patients who were hospitalized in the ICU. These patients displayed immunosuppression due to extended ICU stays, multiple complications including severe acute respiratory syndrome coronavirus infection, and the frequent use of high-dose corticosteroid therapy. The presence of C. auris appears to correlate with patients’ vulnerability arising from comorbidities, invasive treatments, and prolonged hospitalization. Among the reviewed studies, we encountered a single case of C. auris detected in the RT of an ICU-admitted patient in Italy (Corcione et al., 2022). Therapies involving steroids, immunomodulatory agents, and consistent use of broad-spectrum antibiotics might pose risk factors for C. auris acquisition, aligning with known risks for invasive Candida species infections (Snyder and Wright, 2019). The occurrence of three cases of C. auris colonization COVID-19 ICU-admitted in our patients notably emphasizes that immunosuppressed patients with extended hospital stays and multiple complications may present concurrent conditions.
Two strains of M. capitatus were isolated from our patient and classified as colonization. M. capitatus, previously known as Geotrichum capitatum or Blastoschizomyces capitatus, is a rare cause of systemic fungal infection in immunocompromised individuals. However, its true frequency is often underestimated, as it is an emerging opportunistic pathogen (Zhu et al., 2022). The prevalence of M. capitatus infections has been documented in European countries like Italy, Spain, and France (Mazzocato et al., 2015), and there have been reports highlighting its potential to infect immunocompetent patients as well (Tanabe and Patel, 2018). M. capitatus has been found in household dishwashers due to its thermophilic nature, suggesting a potential source of contamination (Zalar et al., 2011). However, there is limited information available from Asia, as there are only a few case reports (Subramanya Supram et al., 2015).
We observed non-Candida colonization including Saccharomyces cerevisiae, Trichosporon spp, M. capitatus, and Nakaseomyces glabratus in the COVID-19 patients. Saccharomyces cerevisiae colonization was found in 3.52% of the COVID-19 patients and 8.69% of ICU/COVID-19 patients, suggesting that ICU hospitalization may provide favorable conditions for Saccharomyces cerevisiae growth. Trichosporon spp, M. capitatus, and Nakaseomyces glabratus colonization were observed in approximately 2.5% of the COVID-19 patients.
Yeast isolation from RT samples, such as sputum, BAL, and TA, is a common occurrence, and distinguishing between colonization and infection can be challenging. In our study, none of the colonized patients were proven to have Candida lung infection. The significance of a positive culture for Candida in severely immunosuppressed patients remains unclear. Currently, the widely accepted criterion for the definitive diagnosis of Candida pneumonia is the histologic demonstration of the fungus in lung tissue, accompanied by inflammation (Baum, 1960; De Pascale and Antonelli, 2014).
This study has limitations: 1) The absence of demographic data, patients’ mortality status, duration of hospitalization, cause of death, and short-term follow-up of the patients, primarily due to the critical conditions and high workload during the COVID-19 epidemics. 2) Some yeast colonies had delayed molecular identification, resulting in missing several isolates in subcultures over the course of several months. 3) Antifungal susceptibility testing was not performed for the isolates. In conclusion, Candida colonization in the respiratory tracts is a complex issue with varying implications depending on the context. While some studies suggest that it may be associated with worse outcomes in immunocompromised patients and an increased risk of pneumonia, other studies propose that it may be a normal part of the respiratory flora and contribute to community-acquired pneumonia (Pendleton et al., 2017). Candida colonization in the RT might act as an independent risk factor promoting ventilator-associated pneumonia and even altering the antibiotic resistance patterns of pathogenic bacteria through polymicrobial biofilm formation (Liu et al., 2021). Airway Candida colonization is associated with pulmonary inflammation and subsequent cellular immune dysfunction (De Pascale and Antonelli, 2014). Further research is needed to fully understand the significance of Candida colonization in the RT. Although Candida pneumonia is rare in both immunocompetent and immunocompromised patients, it is important not to underestimate the clinical implications of Candida isolation from the RT, especially in immunodeficient patients.
The data presented in the study are deposited in the NCBI (GenBank) repository, accession numbers OR600362, OR600363, OR600282, and OR600243. The original contributions presented in the study are included in the article/supplementary materials. Further inquiries can be directed to the corresponding author.
The studies involving humans were approved by The ethics committee of the Isfahan University of Medical Science IR.MUI.MED.REC.1400.448. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
FR: Writing – review & editing, Writing – original draft, Visualization, Software, Methodology, Investigation, Data curation. SaS: Writing – review & editing, Writing – original draft, Visualization, Software, Methodology, Investigation, Data curation. SoS: Writing – review & editing, Resources. EN: Writing – review & editing, Resources. MH: Writing – review & editing, Investigation. SG: Writing – review & editing, Investigation. SA: Writing – review & editing, Visualization, Methodology, Investigation. HF: Writing – review & editing, Resources. HM: Writing – review & editing, Visualization, Supervision, Resources, Project administration, Methodology, Funding acquisition, Data curation, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Isfahan University of Medical Science (Grant number: 1400180).
Also, the authors are grateful to the staff at Al-Zahra and Sayyed Al-Shohada Hospital, Isfahan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abastabar, M., Haghani, I., Ahangarkani, F., Rezai, M. S., Taghizadeh Armaki, M., Roodgari, S., et al. (2019). Candida auris otomycosis in Iran and review of recent literature. Mycoses. 62, 101–105. doi: 10.1111/myc.12886
Aboutalebian, S., Ahmadikia, K., Fakhim, H., Chabavizadeh, J., Okhovat, A., Nikaeen, M., et al. (2021). Direct detection and identification of the most common bacteria and fungi causing otitis externa by a stepwise multiplex PCR. Front. Cell. infection Microbiol. 11, 644060. doi: 10.3389/fcimb.2021.644060
Aboutalebian, S., Mahmoudi, S., Charsizadeh, A., Nikmanesh, B., Hosseini, M., Mirhendi, H. (2022). Multiplex size marker (YEAST PLEX) for rapid and accurate identification of pathogenic yeasts. J. Clin. Lab. Analysis. 36, e24370. doi: 10.1002/jcla.24370
Allaw, F., Kara Zahreddine, N., Ibrahim, A., Tannous, J., Taleb, H., Bizri, A. R., et al. (2021). First Candida auris outbreak during a COVID-19 pandemic in a tertiary-care center in Lebanon. Pathogens. 10, 157. doi: 10.3390/pathogens10020157
Avkan-Oğuz, V., Çelİk, M., Eren-Kutsoylu, OÖ, Nazli, A., Uğur, Y. L., Taylan, A., et al. (2022). Fungal colonization and infections in patients with COVID-19 in intensive care units: A real-life experience at a tertiary-care hospital. Respir. Med. Res. 82, 100937. doi: 10.1016/j.resmer.2022.100937
Azoulay, E., Timsit, J.-F., Tafflet, M., de Lassence, A., Darmon, M., Zahar, J.-R., et al. (2006). Candida colonization of the respiratory tract and subsequent pseudomonas ventilator-associated pneumonia. Chest. 129, 110–117. doi: 10.1378/chest.129.1.110
Baradkar, V., Mathur, M., Wanjari, K., Kumar, S. (2009). Candida in pulmonary tuberculosis. Bombay Hosp. J. 51, 52–53.
Baum, G. L. (1960). The significance of candida albicans in human sputum. New Engl. J. Med. 263, 70–73. doi: 10.1056/NEJM196007142630204
Caramalac, D. A., da Silva Ruiz, L., de Batista, G. C. M., Birman, E. G., Duarte, M., Hahn, R., et al. (2007). Candida isolated from vaginal mucosa of mothers and oral mucosa of neonates: occurrence and biotypes concordance. Pediatr. Infect. Dis. J. 26, 553–557. doi: 10.1097/INF.0b013e31806166d7
Chowdhary, A., Tarai, B., Singh, A., Sharma, A. (2020). Multidrug-resistant Candida auris infections in critically ill coronavirus disease patients, India, April–July 2020. Emerging Infect. diseases. 26, 2694. doi: 10.3201/eid2611.203504
Corcione, S., Montrucchio, G., Shbaklo, N., De Benedetto, I., Sales, G., Cedrone, M., et al. (2022). First cases of candida auris in a referral intensive care unit in piedmont region, Italy. Microorganisms. 10, 1521. doi: 10.3390/microorganisms10081521
Das, S., Singh, S., Tawde, Y., Chakrabarti, A., Rudramurthy, S. M., Kaur, H., et al. (2021). A selective medium for isolation and detection of Candida auris, an emerging pathogen. J. Clin. Microbiol. 59 (2), 10–1128. doi: 10.1128/JCM.00326-20
Delisle, M.-S., Williamson, D. R., Perreault, M. M., Albert, M., Jiang, X., Heyland, D. K. (2008). The clinical significance of Candida colonization of respiratory tract secretions in critically ill patients. J. Crit. Care 23, 11–17. doi: 10.1016/j.jcrc.2008.01.005
De Pascale, G., Antonelli, M. (2014). Candida colonization of respiratory tract: to treat or not to treat, will we ever get an answer? Intensive Care Med. 40, 1381–1384. doi: 10.1007/s00134-014-3364-y
El-Ebiary, M., Torres, A., Fabregas, N., de la BELLACASA, J. P., Gonzalez, J., Ramirez, J., et al. (1997). Significance of the isolation of Candida species from respiratory samples in critically ill, non-neutropenic patients: an immediate postmortem histologic study. Am. J. Respir. Crit. Care Med. 156, 583–590. doi: 10.1164/ajrccm.156.2.9612023
Erami, M., Mirhendi, H., Momen-Heravi, M., Sharif, A., Hashemi Hezaveh, S. J., Matini, A. H., et al. (2023). Case report: COVID-19-associated mucormycosis co-infection with Lomentospora prolificans: The first case and review on multiple fungal co-infections during COVID-19 pandemic. Front. Med. 10, 1078970. doi: 10.3389/fmed.2023.1078970
Erami, M., Raiesi, O., Momen-Heravi, M., Getso, M. I., Fakhrehi, M., Mehri, N., et al. (2022). Clinical impact of Candida respiratory tract colonization and acute lung infections in critically ill patients with COVID-19 pneumonia. Microbial Pathogenesis. 166, 105520. doi: 10.1016/j.micpath.2022.105520
Froidefond, M., Sevestre, J., Chaudet, H., Ranque, S. (2023). COVID-19 is a confounder of increased candida airway colonisation. Pathogens. 12, 463. doi: 10.3390/pathogens12030463
Liu, J., Yu, Y.-T., Xu, C.-H., Chen, D.-C. (2021). Candida colonization in the respiratory tract: what is the significance? Front. Med. 7, 598037. doi: 10.3389/fmed.2020.598037
Matouri, R., Aboutalebian, S., Nasri, E., Sadeghi, S., Rostami, S., Fakhim, H., et al. (2023). Molecular and microscopy detection of Pneumocystis jirovecii in hospitalized patients during the COVID-19 pandemic. Front. Med. 10, 1148320. doi: 10.3389/fmed.2023.1148320
Mazzocato, S., Marchionni, E., Fothergill, A., Sutton, D., Staffolani, S., Gesuita, R., et al. (2015). Epidemiology and outcome of systemic infections due to Saprochaete capitata: case report and review of the literature. Infection. 43, 211–215. doi: 10.1007/s15010-014-0668-3
Mirhendi, H., Charsizadeh, A., Aboutalebian, S., Mohammadpour, M., Nikmanesh, B., de Groot, T., et al. (2022). South Asian (Clade I) Candida auris meningitis in a paediatric patient in Iran with a review of the literature. Mycoses. 65, 134–139. doi: 10.1111/myc.13396
Muthig, M., Hebestreit, A., Ziegler, U., Seidler, M., Müller, F.-M. C. (2010). Persistence of Candida species in the respiratory tract of cystic fibrosis patients. Med. Mycology. 48, 56–63. doi: 10.3109/13693780802716532
Pan American Health Organization, World Health Organization. (2016). Epidemiological alert: Candida auris outbreaks in health care services. Regional Office for the Americas of the World Health Organization. www.PAHO.org.
Pappas, P. G., Kauffman, C. A., Andes, D. R., Clancy, C. J., Marr, K. A., Ostrosky-Zeichner, L., et al. (2016). Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Diseases. 62, e1–e50. doi: 10.1093/cid/civ933
Parums, D. V. (2022). Editorial: the world health organization (WHO) fungal priority pathogens list in response to emerging fungal pathogens during the COVID-19 pandemic. Med. Sci. Monit. 28, e939088. doi: 10.12659/MSM.939088
Pendleton, K. M., Dickson, R. P., Newton, D. W., Hoffman, T. C., Yanik, G. A., Huffnagle, G. B. (2018). Respiratory tract colonization by candida species portends worse outcomes in immunocompromised patients. Clin. Pulm Med. 25, 197–201. doi: 10.1097/CPM.0000000000000279
Pendleton, K. M., Huffnagle, G. B., Dickson, R. P. (2017). The significance of Candida in the human respiratory tract: our evolving understanding. Pathog. Dis. 75 (3), ftx029. doi: 10.1093/femspd/ftx029
Rossi, A., Chavez, J., Iverson, T., Hergert, J., Oakeson, K., LaCross, N., et al. (2023). Candida auris discovery through community wastewater surveillance during healthcare outbreak, Nevada, USA, 2022. Emerging Infect. Diseases. 29 (2), 422. doi: 10.3201/eid2902.221523
Safari, F., Madani, M., Badali, H., Kargoshaie, A.-A., Fakhim, H., Kheirollahi, M., et al. (2022). A chronic autochthonous fifth clade case of Candida auris otomycosis in Iran. Mycopathologia. 187, 121–127. doi: 10.1007/s11046-021-00605-6
Salehipour, K., Aboutalebian, S., Charsizadeh, A., Ahmadi, B., Mirhendi, H. (2021). Differentiation of Candida albicans complex species isolated from invasive and non-invasive infections using HWP1 gene size polymorphism. Curr. Med. Mycol. 7, 34–38. doi: 10.18502/cmm.7.2.7034
Silva, G., Bernardi, T. L., Schaker, P. D. C., Menegotto, M., Valente, P. (2012). Rapid yeast DNA extraction by boiling and freeze-thawing without using chemical reagents and DNA purification. Braz. Arch. Biol. technology. 55, 319–327. doi: 10.1590/S1516-89132012000200020
Snyder, G. M., Wright, S. B. (2019). The epidemiology and prevention of candida auris. Curr. Infect. Dis. Rep. 21, 19. doi: 10.1007/s11908-019-0675-8
Subramanya Supram, H., Gokhale, S., Chakrabarti, A., Rudramurthy, S. M., Gupta, S., Honnavar, P. (2015). Emergence of Magnusiomyces capitatus infections in Western Nepal. Sabouraudia. 54, 103–110. doi: 10.1093/mmy/myv075
Tanabe, M., Patel, S. (2018). Blastoschizomyces capitatus pulmonary infections in immunocompetent patients: case report, case series and literature review. Epidemiol. Infection. 146, 58–64. doi: 10.1017/S0950268817002643
Terraneo, S., Ferrer, M., Martín-Loeches, I., Esperatti, M., Di Pasquale, M., Giunta, V., et al. (2016). Impact of Candida spp. isolation in the respiratory tract in patients with intensive care unit-acquired pneumonia. Clin. Microbiol. Infection. 22, 94.e1–94.e8. doi: 10.1016/j.cmi.2015.09.002
Wang, Q., Cai, X., Li, Y., Zhao, J., Liu, Z., Jiang, Y., et al. (2022). Molecular identification, antifungal susceptibility, and resistance mechanisms of pathogenic yeasts from the China antifungal resistance surveillance trial (CARST-fungi) study. Front. Microbiol. 13, 1006375. doi: 10.3389/fmicb.2022.1006375
White, T. J., Bruns, T., Lee, S., Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: guide to Methods applications. 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Yazici, O., Cortuk, M., Casim, H., Cetinkaya, E., Mert, A., Benli, A. R. (2016). Candida glabrata pneumonia in a patient with chronic obstructive pulmonary disease. Case Rep. Infect. Diseases. 2016, 4737321. doi: 10.1155/2016/4737321
Yu, D., Liu, Z. (2022). The research progress in the interaction between Candida albicans and cancers. Front. Microbiol. 13, 988734. doi: 10.3389/fmicb.2022.988734
Zalar, P., Novak, M., de Hoog, G. S., Gunde-Cimerman, N. (2011). Dishwashers–a man-made ecological niche accommodating human opportunistic fungal pathogens. Fungal Biol. 115, 997–1007. doi: 10.1016/j.funbio.2011.04.007
Keywords: yeast species, respiratory mycobiome, COVID-19, Candida auris, colonization
Citation: Rouhi F, Soltani S, Sadeghi S, Nasri E, Hosseini M, Ghafel S, Aboutalebian S, Fakhim H and Mirhendi H (2024) Yeast species in the respiratory samples of COVID-19 patients; molecular tracking of Candida auris. Front. Cell. Infect. Microbiol. 14:1295841. doi: 10.3389/fcimb.2024.1295841
Received: 17 September 2023; Accepted: 29 March 2024;
Published: 19 April 2024.
Edited by:
Ying Zhao, Peking Union Medical College Hospital (CAMS), ChinaReviewed by:
Ludmila Baltazar, Federal University of Minas Gerais, BrazilCopyright © 2024 Rouhi, Soltani, Sadeghi, Nasri, Hosseini, Ghafel, Aboutalebian, Fakhim and Mirhendi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hossein Mirhendi, cy5oLm1pcmhlbmRpQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
‡ORCID: Hossein Mirhendi, orcid.org/0000-0002-1006-4169
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.