- 1Faculty of Public Health, Kasetsart University, Sakon Nakhon, Thailand
- 2Department of Bacteriology I, National Institute of Infectious Diseases, Tokyo, Japan
- 3Japan-Thailand Research Collaboration Center on Emerging and Re-emerging Infections, Research Institute for Microbial Diseases, Osaka University, Osaka, Japan
- 4Department of Bacterial Infections, Research Institute for Microbial Diseases, Osaka University, Osaka, Japan
- 5Center for Infectious Disease Education and Research, Osaka University, Osaka, Japan
Colistin is widely used for the prophylaxis and treatment of infectious disease in humans and livestock. However, the global food chain may actively promote the dissemination of colistin-resistant bacteria in the world. Mobile colistin-resistant (mcr) genes have spread globally, in both communities and hospitals. This study sought to genomically characterize mcr-mediated colistin resistance in 16 Escherichia coli strains isolated from retail meat samples using whole genome sequencing with short-read and long-read platforms. To assess colistin resistance and the transferability of mcr genes, antimicrobial susceptibility testing and conjugation experiments were conducted. Among the 16 isolates, 11 contained mcr-1, whereas three carried mcr-3 and two contained mcr-1 and mcr-3. All isolates had minimum inhibitory concentration (MIC) for colistin in the range 1–64 μg/mL. Notably, 15 out of the 16 isolates demonstrated successful transfer of mcr genes via conjugation, indicative of their presence on plasmids. In contrast, the KK3 strain did not exhibit such transferability. Replicon types of mcr-1-containing plasmids included IncI2 and IncX4, while IncFIB, IncFII, and IncP1 contained mcr-3. Another single strain carried mcr-1.1 on IncX4 and mcr-3.5 on IncP1. Notably, one isolate contained mcr-1.1 located on a chromosome and carrying mcr-3.1 on the IncFIB plasmid. The chromosomal location of the mcr gene may ensure a steady spread of resistance in the absence of selective pressure. Retail meat products may act as critical reservoirs of plasmid-mediated colistin resistance that has been transmitted to humans.
1 Introduction
Antimicrobial resistance (AMR) is a serious global health issue that affects both humans and animals. Misuse and overuse of antibiotics in clinical treatment and with livestock contribute to the growth and spread of antimicrobial resistance bacteria (Liu et al., 2021). Colistin is a common antibiotic that has been utilized in the veterinary field for decades, not only for prophylaxis and treatment, but also for growth promotion. It is now regarded as a last resort against antimicrobial agents in therapy infections caused by multidrug-resistant, Gram-negative bacteria in humans (Zając et al., 2019). Inevitably, the increasing use of colistin in humans and animals has led to the emergence of colistin resistance in Gram-negative bacteria, with the rates of resistance continuing to increase (Kempf et al., 2016).
Normally, colistin resistance is thought to be chromosomally mediated (Meletis and Skoura, 2018). In 2016, a mobile colistin resistance (mcr) gene was identified in the IncI2-type plasmid of E. coli isolated from food animals and raw meat in China (Liu et al., 2016). The mcr gene encodes a membrane of phosphoethanolamine transferase that catalyzes the addition of phosphoethanolamine into lipid A to modify lipopolysaccharide (LPS), leading to a reduced negative charge on the outer membrane of Gram-negative bacteria (Vu Thi Ngoc et al., 2022). The mcr genes (mcr-1 to mcr-10) have been successively reported globally; the mcr-1 gene has been widely detected in animals, food products, humans and the general environment (Martiny et al., 2022). To date, the predominate plasmid carrying mcr has been reported in different plasmids based on their replicon types including IncX3, IncX4, IncI2, IncHI1, InHI2, IncK, IncP, IncF, IncFII, IncFIB, IncP, IncY, ColE10, and ColE10-like (Sun et al., 2018; Lu et al., 2020; Tang et al., 2022). Epidemiological studies have indicated that the spread of colistin-resistant mcr-carrying bacteria is not only a problem in hospitals, but also a major concern for environmental and food safety. In this regard, environmental sources, food-producing animals, foreign travel, and food trading have expedited the worldwide spread of mcr genes at the human-animal-environment interface (Liu and Song, 2019; Liu et al., 2021; Lopes et al., 2021; Nakano et al., 2021).
Escherichia coli (E. coli) is a commensal pathogen that causes gastrointestinal tract disease in humans and animals. This bacterium has been isolated at various sites in the food chain, typically usually due to fecal contamination (Macori et al., 2021). Often, it is used as an indicator organism to monitor AMR in livestock, food products, and humans (Singh et al., 2018; Pormohammad et al., 2019; Puangseree et al., 2022) and represents a major reservoir of antimicrobial resistance genes, mostly acquired through horizontal gene transfer (Puangseree et al., 2022). The spreading of mcr-carrying E. coli along the food chain is the most common transmission mode of colistin resistance, the mcr-1 gene could have been disseminated to human populations through two primary pathways: the food supply chain and direct contact between animals and humans. Additionally, contamination of both freshwater and seawater systems likely played a role in the dissemination, subsequently leading to the contamination of vegetables and seafood (Chen et al., 2017; Lu et al., 2020). In Thailand, colistin-resistant mcr-carrying E. coli strains have been identified in humans ((Khine et al., 2020; Khanawapee et al., 2021; Paveenkittiporn et al., 2021; Trongjit and Chuanchuen, 2021; Nguyet et al., 2022; Phuadraksa et al., 2022). In our previous study, mcr genes were detected in 7.8% of slaughtered pork in Thailand (Khanawapee et al., 2021). Consequently, to comprehend the genomic characteristics in mcr-harboring E. coli strains in food animals in Thailand, we characterized the genetic features of mcr-harboring plasmids in E. coli that had been isolated from food animal products using whole-genome sequencing.
2 Materials and methods
2.1 Ethics statement
Ethical review and approval were not required because no human or animal specimens or data were used in the current study.
2.2 Bacterial isolates and identification
In this study, a total of 16 E. coli isolates carrying mcr genes were analyzed. Ten of these isolates were collected from slaughtered pigs during 2014–2015, as reported in a previous study (Khanawapee et al., 2021). Furthermore, we identified mcr genes in six out of 40 E. coli isolates obtained from 63 retail meat samples collected between 2020–2021, (unpublished data) by using a polymerase chain reaction (PCR) methods as described elsewhere (Khanawapee et al., 2021). The geographical locations of sample collection are illustrated on the map in Figure 1.
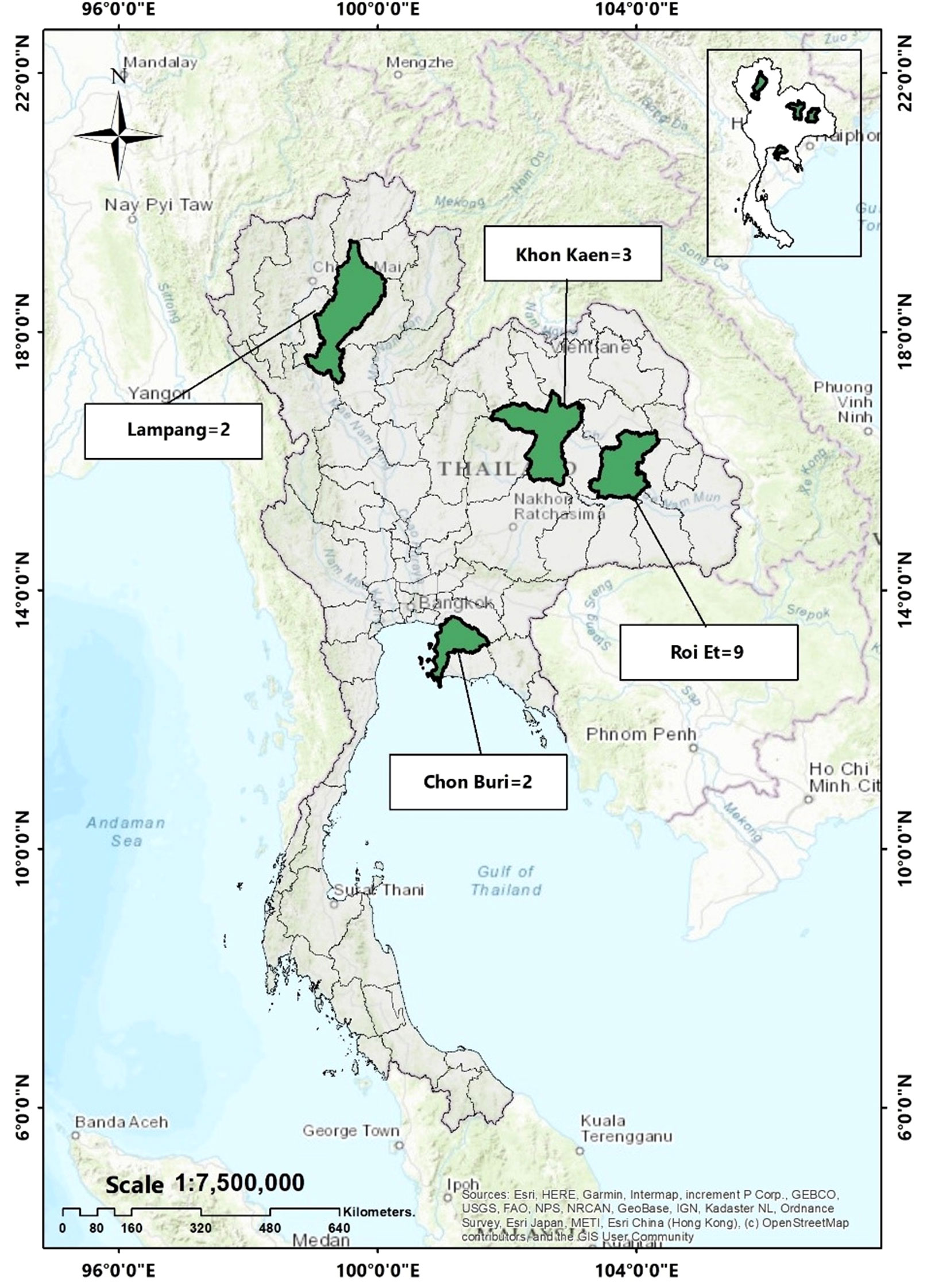
Figure 1 Geographical location of mcr-harboring E. coli in this study. (Khon Kaen = 3 isolates, Roi Et = 9 isolates, Lampang = 2 isolates and Chon Buri = 2 isolates).
2.3 Antimicrobial susceptibility testing
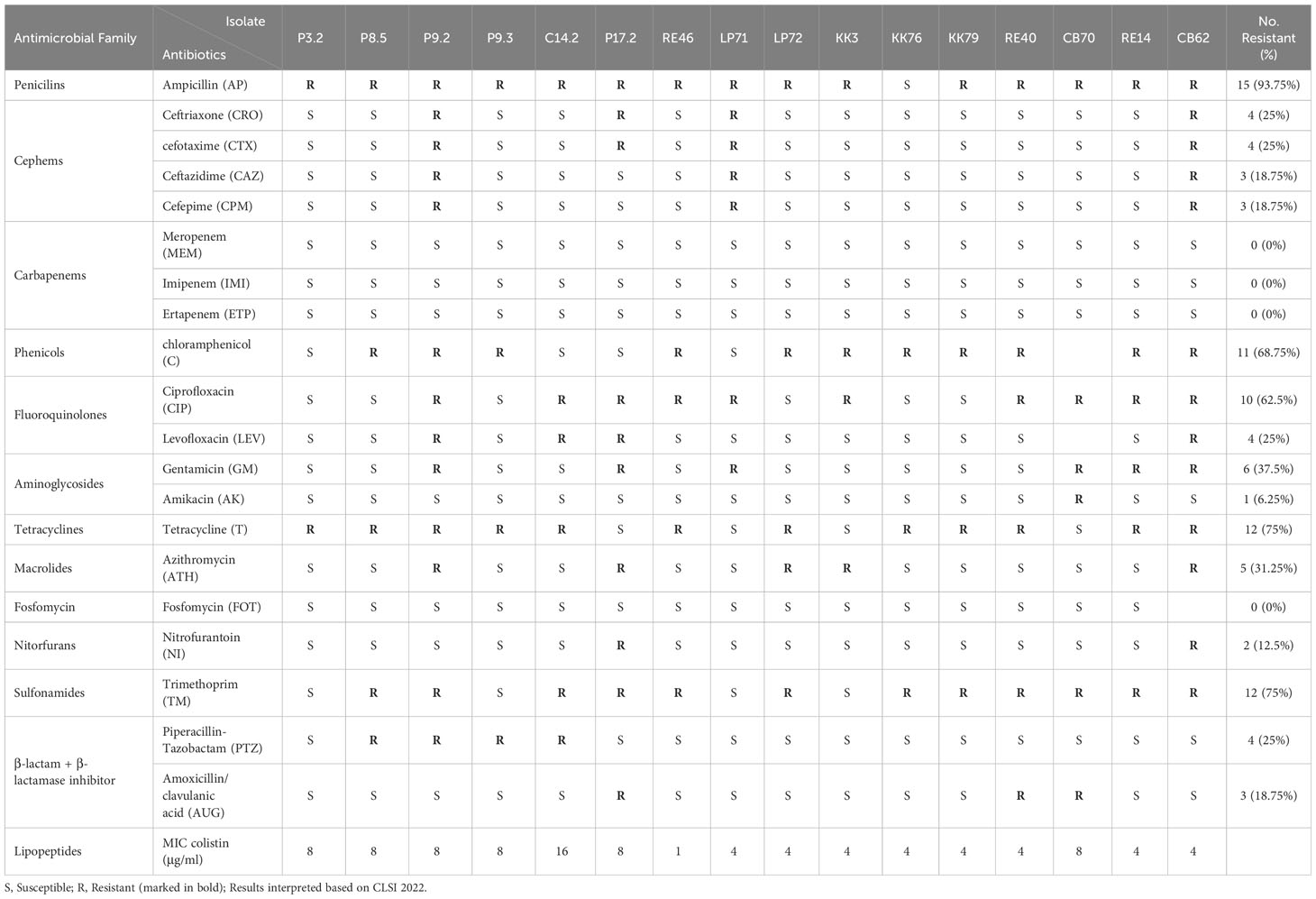
The six of E. coli isolates carrying mcr genes (P3.2, P8.5, P9.2, P9.3, P17.2 and C14.2) were subjected disk diffusion and broth microdilution (only colistin was performed with MIC) and interpreted according to the 2022 Clinical and Laboratory Standards Institute guidelines (Clinical and Laboratory Standards Institute (CLSI), 2022) subsequently, they were used for susceptibility testing of mcr-harboring E. coli isolates. E. coli ATCC 25922 was used as the control. While ten E. coli isolates carrying mcr genes have been reported in previous study (Khanawapee et al., 2021). The disk diffusion method was used to determine the susceptibility of clinical routine antimicrobial agents including ampicillin (AP; 10 µg), gentamicin (GM; 10 µg), amikacin (AK; 30 µg), amoxicillin/clavulanic acid (AUG; 30 µg), piperacillin-tazobactam (PTZ; 110 µg), cefepime (CPM; 30 µg), cefotaxime (CTX; 30 µg), ceftriaxone (CRO; 30 µg), ciprofloxacin (CIP; 5 µg), levofloxacin (LEV; 5 µg), ertapenem (ETP; 10 µg), imipenem (IMI; 10 µg), meropenem (MEM; 10 µg), ceftazidime (CAZ; 30 µg), chloramphenicol (C; 30 µg), tetracycline (T; 30 µg), fosfomycin (FOT; 200 µg), nitrofurantoin (NI; 300 µg), azithromycin (ATH; 5 µg), and trimethoprim (TM; 5µg). The MIC of colistin (colistin sulfate; Sigma-Aldrich, USA) were determined based on broth microdilution (Khanawapee et al., 2021). Multidrug-resistance status was assigned to isolates that were resistant to at least one agent from three or more different antimicrobial categories (Magiorakos et al., 2012).
2.4 Conjugation experiment
The transferability of mcr was assessed through a conjugation assay, utilizing mcr-harboring E. coli strains as donors and streptomycin-resistant E. coli UB1637 as a recipient. Briefly, both donor and recipient strains were culture in Luria-Bertani (LB) broth with shaking at 200 rpm at 37°C, then a 1:25 ration of donor to recipient strain was mixed in 1 ml of LB broth and incubated overnight at 37°C with gentle shaking 50 rpm. Transconjugants were selected on MacConkey agar plates containing streptomycin (3200 µg/mL) and colistin (4 µg/mL). The presence of mcr genes in transconjugants was detected via PCR method, and colistin MIC were determined to confirm the transconjugants as previously described (Phetburom et al., 2021).
2.5 Whole genome sequencing and bioinformatics analyses
Genomic DNA samples of the 16 mcr-1 harboring E. coli strains were extracted using a MagMAX DNA Multi-Sample Ultra 2.0 Kit (Appliedbiosystems by Thermo Fisher Scientific; Vilnius, Lithuania). DNA concentrations were determined using a Qubit dsDNA HS assay kit (Thermo Fisher Scientific; Waltham, MA, USA). Whole-genome sequencing was performed using the HiSeq 3000 (Illumina; CA, USA) and MinION (Oxford Nanopore Technologies; Oxford, UK) platforms. The DNA library was generated using the NEBNex Ultra II DNA Library Prep Kit for Illumina (New England Biolabs; MA, USA) according to the manufacturer. The Illumina HiSeq platform was prepared with 150-bp paired-end reads. We applied the Fastp v0.19.5 program (Chen et al., 2018) with default parameters for quality filtering of the Illumina reads and adapter trimming for the sequences using the Skewer v0.2.2 program (Jiang et al., 2014). The FastQC v0.118 program1 was used for quality checking of the Illumina reads.
Library preparation for ONT sequencing followed the rapid barcoding DNA sequencing protocol using the SQK-RBK004 kit (Oxford Nanopore Technologies; Oxford, UK), and the libraries were sequenced using a single R9 version/FLO-MIN106D flow cell on a MinION Mk1B sequencer (Oxford Nanopore Technologies; Oxford, UK). We base called and demultiplexed the raw data using the Guppy v5.0.11 (ONT) program. The ONT adapters were trimmed using the Porechop v0.2.4 program2 (Wick et al., 2019). Quality control of the ONT reads was undertaken using the NanoPlot v1.28.1 program3 (De Coster and Rademakers, 2023). Hybrid assemblies with the ONT and Illumina data were performed using Unicycler v0.48 (Wick et al., 2017) and the genome sequences were checked for quality using the QUAST v5.0.2 program (Gurevich et al., 2013). The genome sequences were processed using the DDBJ Fast Annotation and Submission tool (Tanizawa et al., 2018) for annotation. Default parameters were used for all software programs unless otherwise specified.
Genomes were in silico typed using the following databases: sequence types (MLST 2.0) (Larsen et al., 2012), serotypes (SerotypeFinder 2.0) (Joensen et al., 2015), subtypes (CHtyper 1.0) (Camacho et al., 2009), while FimH types were identified using the FimTyper 1.0 program (Roer et al., 2017). Plasmid replicon types were identified using the PlasmidFinder 2.1 program (Carattoli et al., 2014) from the Center for Genomic Epidemiology (CGE). Clermont phylotyping4 was performed. Acquired resistance genes and chromosomal mutation-mediated antimicrobial resistance genes were identified using the ResFinder 4.1 program (Zankari et al., 2012) from CGE and the resistance gene identifier from the Comprehensive Antibiotic Resistance Database (Alcock et al., 2023). A SNP-based phylogenetic tree was constructed to compare twelve E. coli strains isolated from patients in Thailand (Paveenkittiporn et al., 2021; Boueroy et al., 2022) (Supplementary Table 1) with the sixteen mcr-1 harboring E. coli strains from food-animal products in this study. The construction of the tree was carried out using REALPHY online tool (Bertels et al., 2014). Phylogenetic analysis of mcr carrying plasmids was applied using Mashtree (Katz et al., 2019) and visualized using the Interactive Tree of Life (iTOL) tool (Letunic and Bork, 2021). The Easyfig software (version 2.2.5) was used for the comparative and visualize the region of plasmids carrying mcr genes. (Sullivan et al., 2011). The sequence similarity search was performed using BLAST against the NCBI nucleotide database. In order to examine the distribution and relationships of other plasmid carrying mcr, plasmid sequences were downloaded from NCBI and compared (Supplementary Table 2).
2.6 Data availability statement
The complete genomes were deposited in GenBank under Bioproject Accession No. PRJNA525849.
3 Results
3.1 Antimicrobial susceptibility of mcr-carrying E. coli strains
In total, of the 16 E. coli carrying mcr isolates, 11 contained mcr-1, 3 isolates harbored mcr-3, and 2 isolates contained mcr-1 and mcr-3. The antimicrobial susceptibility results are shown in Table 1. All strains were defined as multidrug resistance (MDR) based on resistance to at least three classes of antibiotics (Magiorakos et al., 2012). Of the 16 isolates, 15 (93.75%) were resistant to ampicillin, 12 (75%) were resistant to trimethoprim and tetracycline, 11 (68.75%) were resistant to chloramphenicol, and 10 (62.5%) to ciprofloxacin, whereas there was no resistance to fosfomycin and carbapenem was detected. The colistin MIC of these isolates was in the range 1–16 µg/mL, as shown in Table 1. We detected only one isolate (RE46) did not resist to colistin (MIC = 1 ug/ml), whereas the rest isolates were resistant. We detected only one isolate (RE46) did not show resistance to colistin (MIC = 1 ug/ml), whereas the remaining isolates were resistant.
3.2 Conjugation
Of the 16 mcr harboring E. coli isolates, 15 successfully transferred the colistin resistant gene (mcr) by conjugation to the recipient strain (E. coli UB1637), with the KK3 strain being the exception. All transconjugants had harboring mcr-1 and mcr-3 depend on donor and colistin MIC values in the range 8–16 µg/mL, suggesting the transferred mcr genes were located on conjugative plasmids.
3.3 Genotypic profile of mcr- harboring E. coli isolates
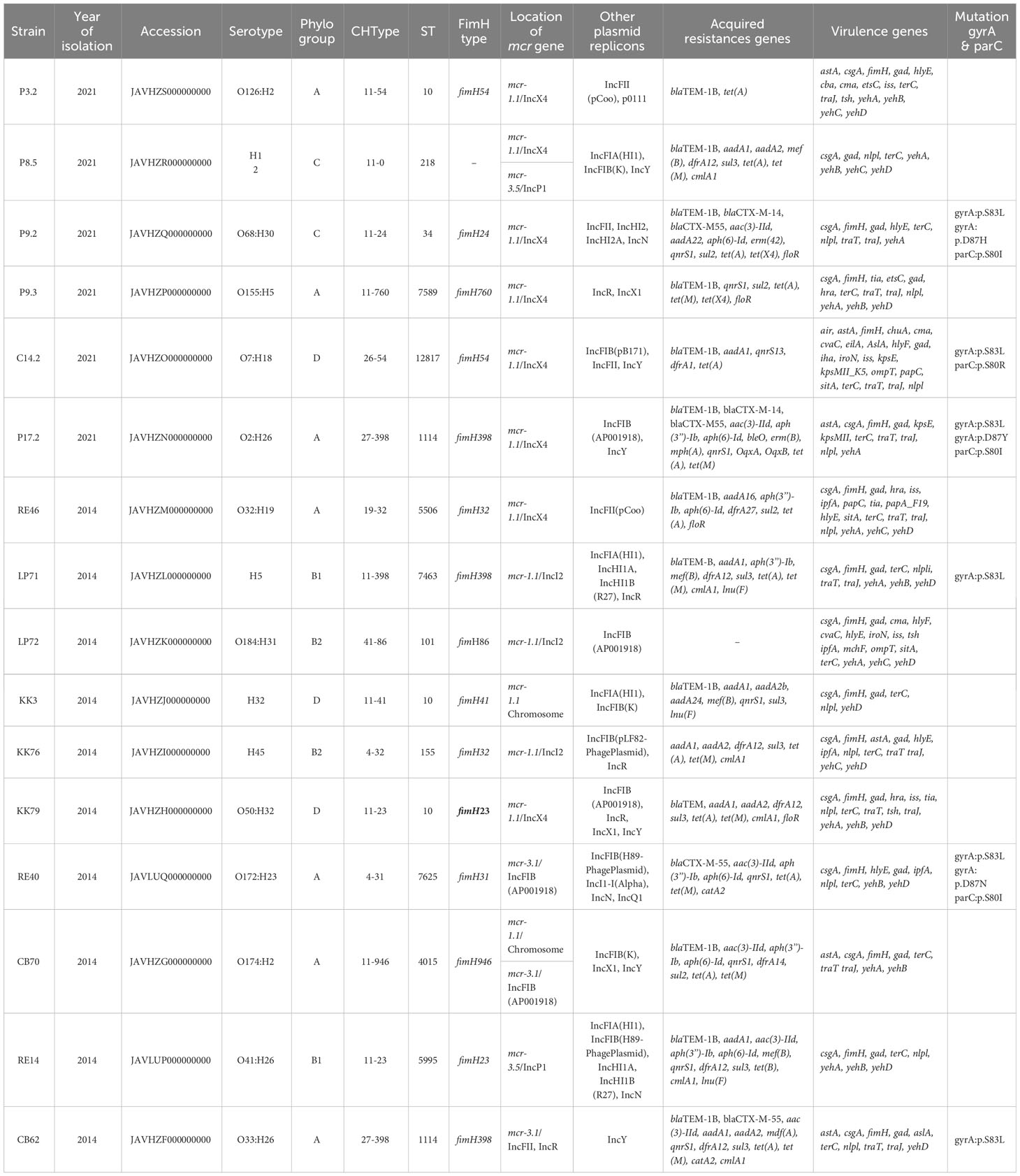
Whole-genome sequencing (WGS) of the mcr-harboring E. coli (Supplementary Table 3) including Clermont phylogroup, sequence types (ST), serotype, CHType, FimH type, plasmids replicon types, antibiotic resistance and virulence genes was shown in Table 2. The Clermont phylotyping identified 7 isolates (43.75%, 7/16) belonging to group A, 3 to group D, and 2 for each of groups C, B1, and B2 (Table 2). FimH398 was detected in 4 isolates (25%, 4/16). Two isolates had FimH54 and FimH23, while the remaining isolates belonged to individual FimH types. For the CHType, 27-398 was found in 2 isolates, with different CHType represented by the remaining isolates. The MLST revealed 13 STs among these isolates (Table 2). ST10 was found in 3 isolates, with 2 isolates belong to ST1114 and 11 STs.
To analyze the genetic relationship of the mcr-carrying E. coli isolated in this study, we conducted a phylogenomic analysis using available genomes of mcr-carrying E. coli (n = 12) isolated from patients in Thailand in another study (Boueroy et al., 2022). The phylogenetic tree had two main distinct clades. The mcr-carrying E. coli isolates from our study had a related genetic background in a cluster, except for C14.2 that was closely related to Thai human stains 54881, 54715, and 60000 (Figure 2). The strains KK76, RE40, RE46, and LP72 were located on the same cluster as the Thai human strains 53037, 2117, 62122, 58967, 56511, 53360, and V417 (Figure 2).
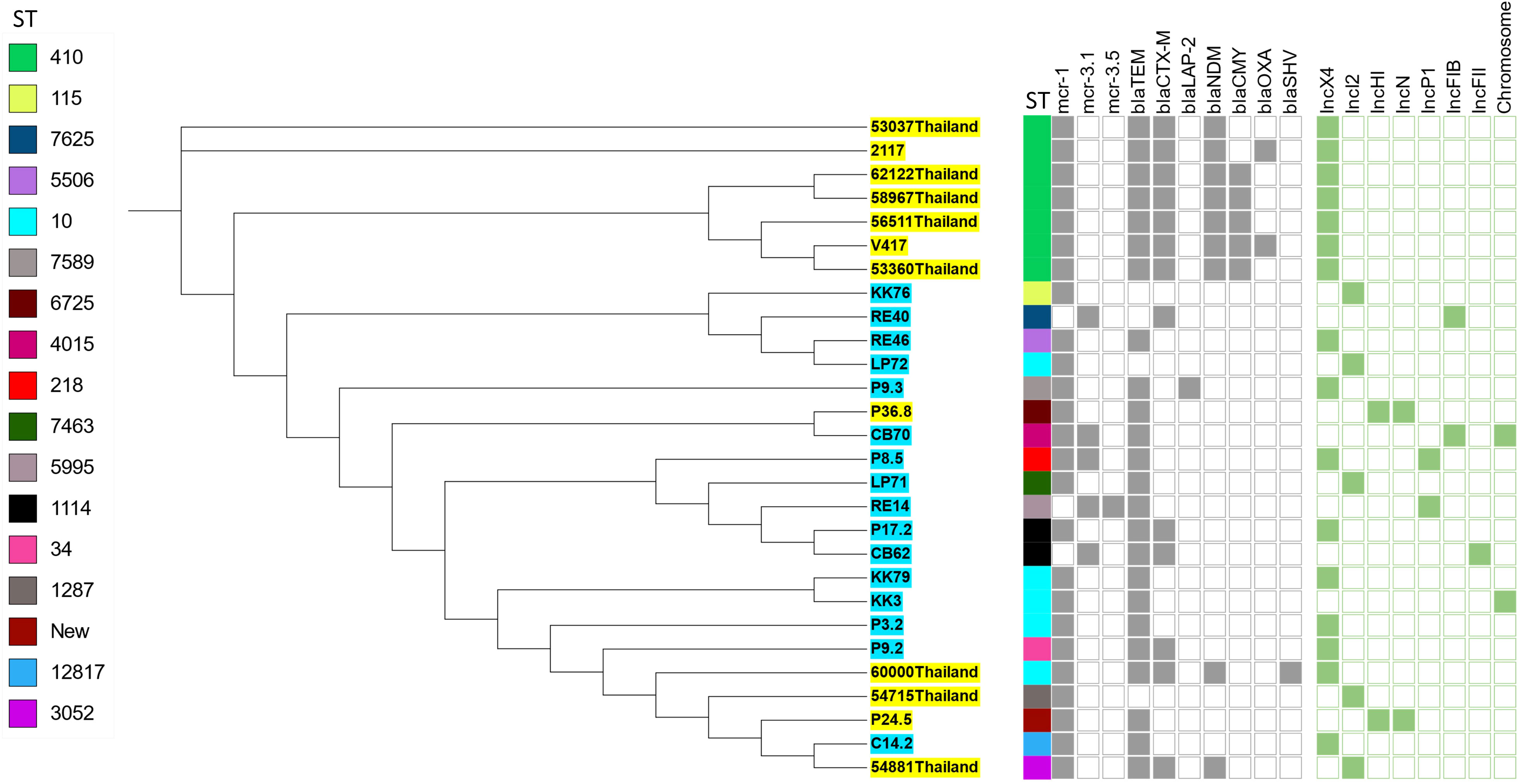
Figure 2 Phylogenetic tree of mcr-harboring E. coli, where blue color represents isolates from meat (pork and chicken) in this study, while yellow represents isolates from human specimens.
The genomic analysis of mcr-harboring E. coli revealed a high number of resistance genes, with up to 37 different antibiotic resistance genes that conferred resistance to 8 antibiotic classes, consisting of β-lactam resistance genes, aminoglycoside, tetracycline, macrolide, phenicol, fluoroquinolones, folate pathway antagonist, and sulfonamide genes. Additionally, mutations in the gyrA (S83L, D87H, D87Y, D87N) and parC (S80I, S80R) genes were identified in the C14.2, LP71, CB62, P9.2, P17.2, and RE40 strains, as displayed in Table 2, which confer high-level resistance to fluoroquinolones.
A total of 35 distinct virulence genes were identified within genomes of mcr positive E. coli isolates. All 16 mcr containing E. coli isolates carried the virulence genes gad (glutamate decarboxylase), terC (tellurium ion resistance protein, and csgA (curlin major subunit). Furthermore, 15 mcr bearing E. coli isolates carried fimH (Type 1 fimbriae), 14 mcr bearing E. coli isolates carried nlpl (lipoprotein NlpI precursor), and 11 mcr bearing E. coli isolates carried traJ (Protein TraJ (a positive regulator of conjugal transfer operon)) and yehA (outer membrane lipoprotein, YHD fimbriael cluster), as shown in Table 2.
3.4 Comparison of plasmids containing mcr in E. coli
The WGS analysis of the 16 mcr-harboring E. coli isolates revealed that 11 mcr-1 genes and 4 mcr-3 genes were located on the plasmids; however, the 2 mcr-1 genes (KK3 and CB70 strains) were located on the chromosome (Table 2). Among the 16 mcr-positive isolates identified, two strains harbored both mcr-1 and mcr-3 genes (CB70 and P8.5; Table 2). Seven isolates contained mcr-1.1 genes were located on IncX4 plasmids, while 3 isolates harbored mcr-1.1 genes on IncI2 plasmids. The mcr-3 genes were detected on IncFIB (n=2), IncP1 (n=2), and IncFII (n=1) plasmids. Notably, strain KK3 exhibited chromosomal carriage of mcr-1.1, while strain CB70 harbored mcr-1.1 on its chromosome and mcr-3.1 on an IncFIB plasmid (Table 2).
Analysis of genetic context of the 8 plasmid IncX4 samples containing mcr-1 showed the conserved mcr-1.1-pap2 segment (Figure 3A), a putative pap2 protein, a pap2 membrane-associated lipid phosphatase, which have transmembrane domains and are involved in the phosphatidic acid pathway and lipid metabolism and signal (Fan et al., 2014; Wang et al., 2018; Gaballa et al., 2023). No other antibiotic resistance genes were detected in the whole IncX4 plasmid, while 3 mcr-1.1 were carried on the IncI2 plasmid that revealed the genetic environment of mcr-1 to be relaxase-ISApl1- mcr-1.1-pap2 (Figure 3B). Furthermore, 3 mcr-3.1 genes showed arrangements of the IS3 family transposase-transposase-dgkA-mcr-3.1-Tn3family transposase (Figure 3C). The plasmids carried mcr-3.5 containing diacylglycerol kinase (dgkA) downstream and Tn3 transposase and a conjugal protein upstream (Figure 3D). Notably, the genetic features of mcr-1.1 on the chromosomes of CB70 and P8.5 contained ISApl1-pap2-mcr-1.1-ISApl1 (Figure 3E).
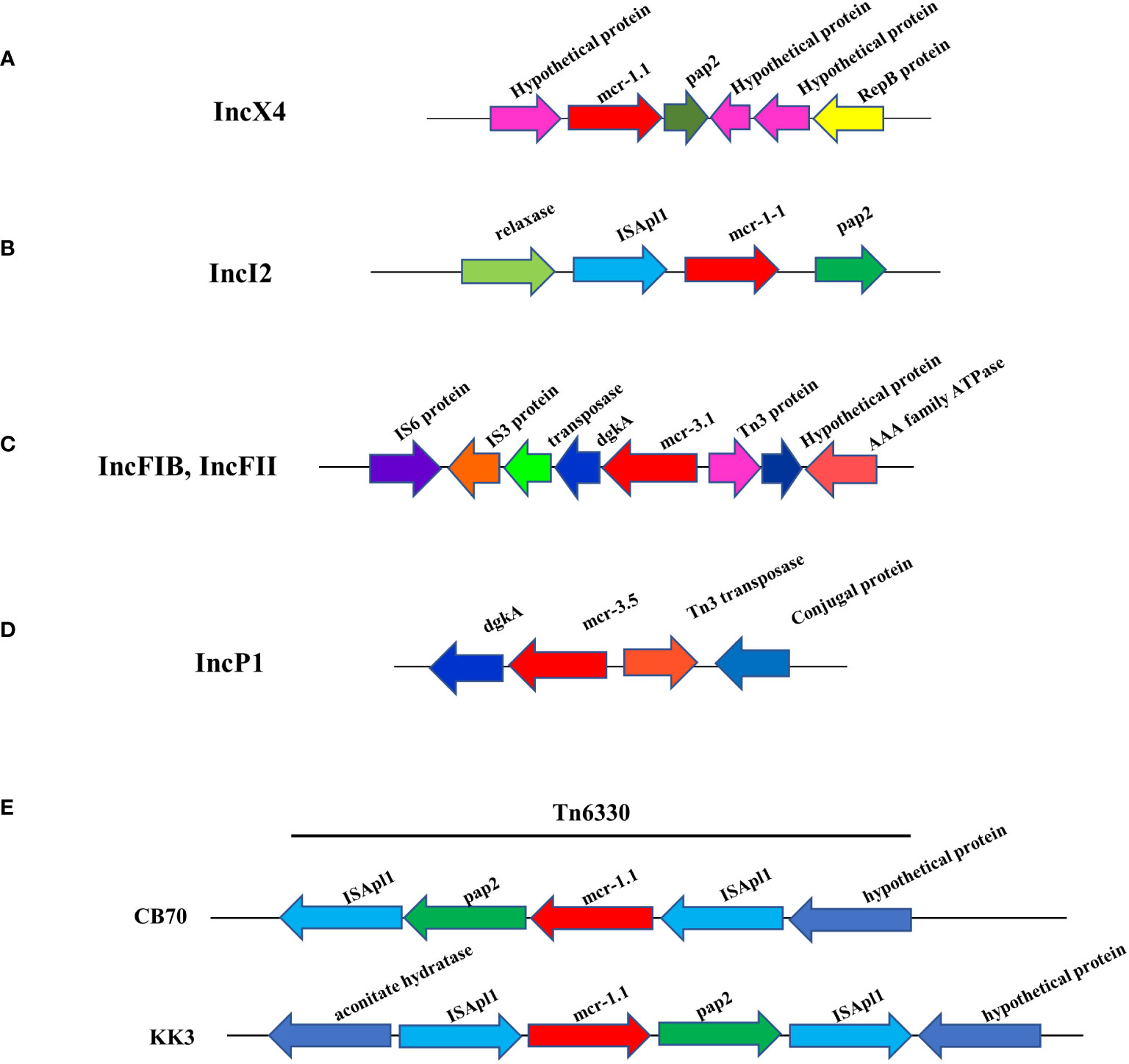
Figure 3 Schematic partial representation of genetic surrounding of mcr-harboring plasmid replicon types of E. coli, where blue color represents isolates from meat (pork and chicken) in this study and yellow represents isolates from human specimens. (A) IncX4 containing mcr-1.1, (B) IncI2 containing mcr-1.1, (C) IncF containing mcr-1.1, (D) IncP1 containing mcr-1.1 and (E) showing mcr-1.1 located on a chromosome.
The phylogenetic relationships of the plasmid replicon type IncX4 and IncI2 carrying mcr-1 in E. coli in our study were closely related with the plasmids IncX4 and IncI2 harboring mcr-1 human E. coli. (Figure 4). The comparative analysis of the IncI2 and IncX4 types carrying mcr-1 in E. coli showed that both the human and meat strains were similar and contained mcr-1.1-pap2. (Figure 5).
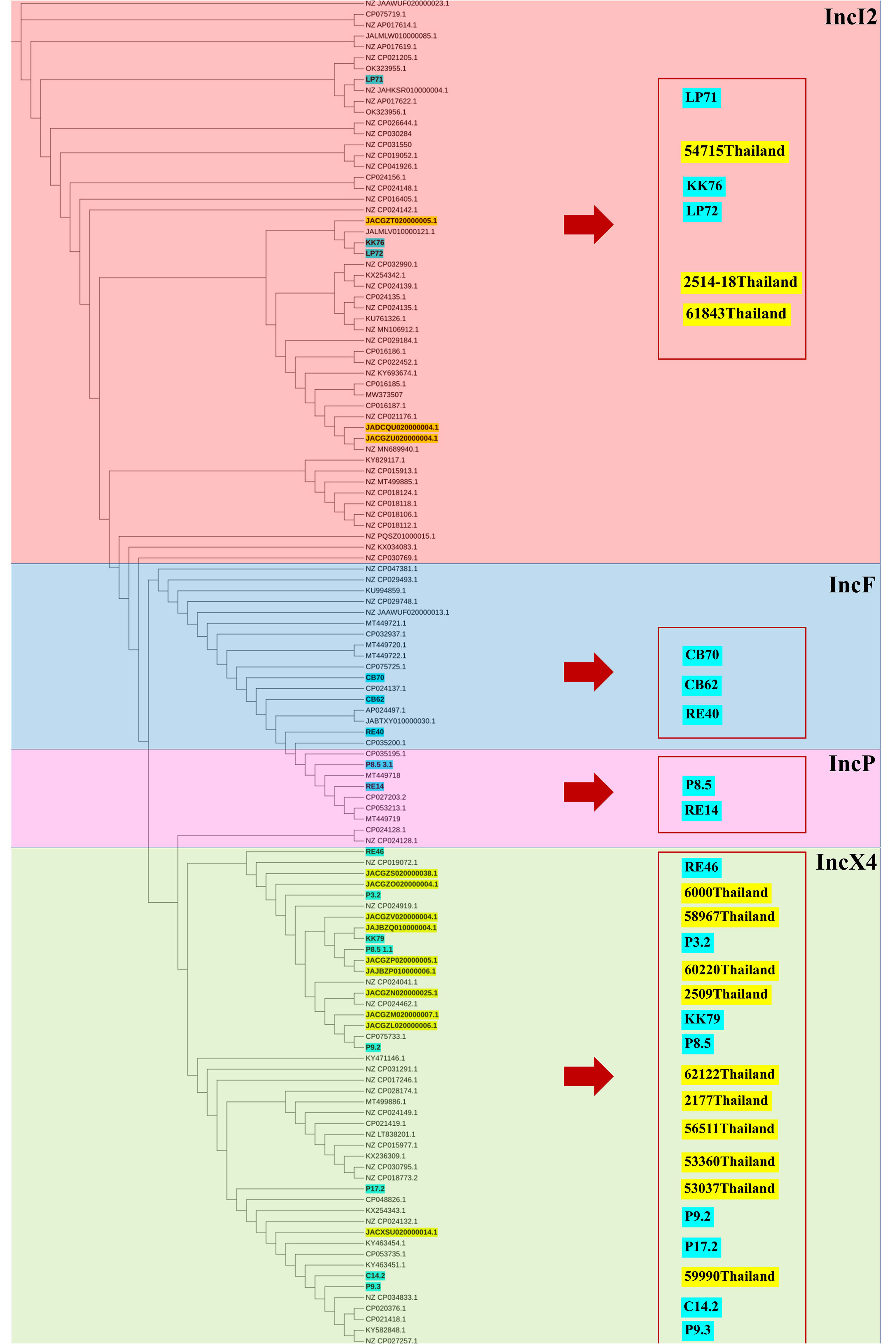
Figure 4 Phylogenetic tree of mcr-harboring plasmid replicon types of E. coli, where blue color represents isolates from meat (pork and chicken) in this study and yellow represents isolates from human specimens.
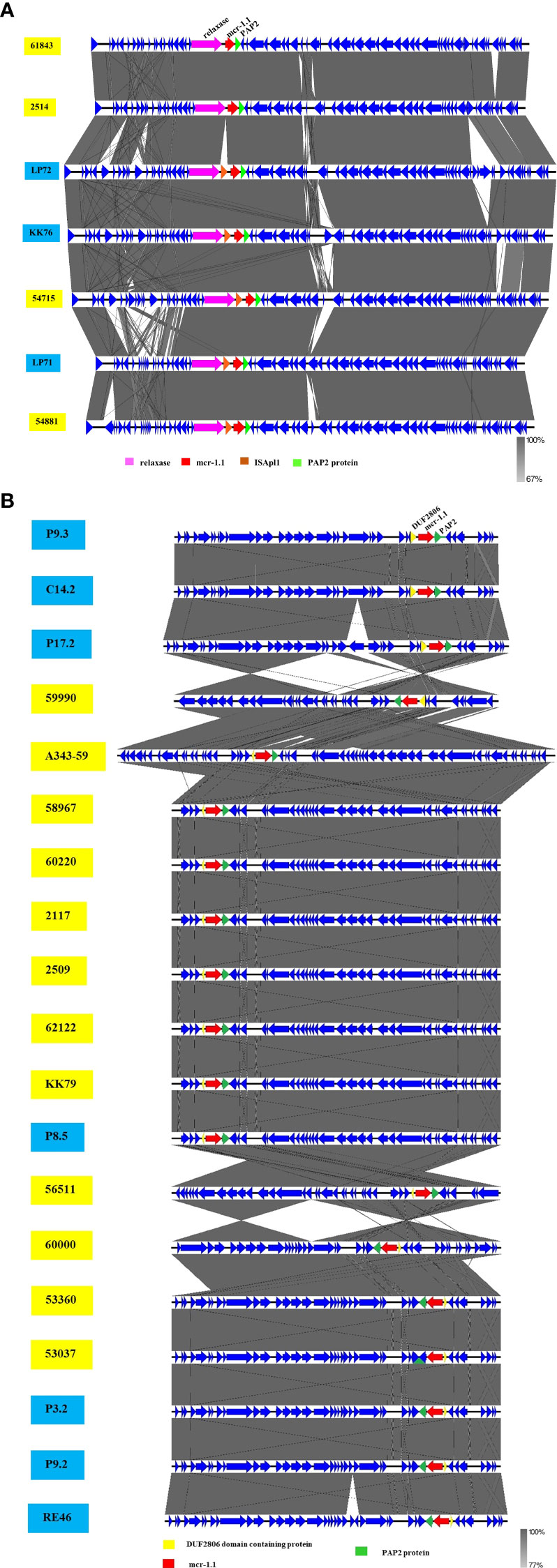
Figure 5 Schematic representation of sequence alignment analysis of IncI2 (A) and IncX4 (B) plasmids carrying mcr-1.1, comparing meat and human specimens. Boxed arrows represent position and transcriptional direction of ORFs, with mobile genetic elements (mcr-1.1) in red, pap2 protein in green, relaxase protein in pink, ISApl1 in orange, and other genes or hypothetical ones in blue. The ID strain in light blue box represents isolates from meat (pork and chicken) in this study and yellow box represents isolates from human specimens.
4 Discussion
There have been reports of the prevalence of plasmid-mediated colistin resistance in E. coli in a wide range of Enterobacterales, especially in E. coli from various sources, environments (Lopes et al., 2021; Mohsin et al., 2021; Nakano et al., 2021) food-producing animals (Huang et al., 2017), livestock (Migura-Garcia et al., 2020), and humans (Li B. et al., 2018; Yamaguchi et al., 2020). Other studies have shown that the plasmid type is important in mcr dissemination due to horizontal gene transfer in various sources. In Thailand, the mcr gene has been identified in clinical samples (Paveenkittiporn et al., 2021; Boueroy et al., 2022) and animals (Khine et al., 2020; Khanawapee et al., 2021; Trongjit and Chuanchuen, 2021). Here, we characterized the genetic feature and accessory gene of 16 mcr-1 and mcr-3 carrying E. coli strains isolated from meat samples in the food supply chain. Almost all isolates exhibited resistance to colistin, however, only one isolate did not resist. This result is consistent with other studies that reported colistin MIC values below 2 µg/ml but present mcr gene ((Khanawapee et al., 2021; Pungpian et al., 2021).
In recent decades, the incidence of extended-spectrum β-lactamases (ESBL)-producing E. coli, contributing to a growing incidence of community-acquired extra-intestinal infections in humans, animals and wide range of food animals product, including beef, pork and chicken meat globally (Shafiq et al., 2019; Shafiq et al., 2021; Shafiq et al., 2022). Our study, E. coli harboring mcr-1 and mcr-3 were detected together with ESBL genes (blaTEM-1B, blaCTX-M-14 and blaCTX-M-55). Among them, blaTEM-1B was the most common found in thirteen mcr positive E. coli, these results correlated with previous findings in Thailand that demonstrated the concurrent carriage of mcr-1.1 with blaCTX-M-14, blaCTX-M-55, and blaTEM-1B (Leangapichart et al., 2023). Moreover, the coexistence of mcr with ESBL genes in E.coli was report from other studies such as China and Pakistan, E. coli co-harboring mcr with ESBL genes such as blaCTX-M-14, blaCTX-M-55, and blaTEM-1B (Shafiq et al., 2019; Shafiq et al., 2022).
Our study, identified 12 different STs, containing 3 ST10 strains and 1 ST34 strain, which could be considered as a broad range of hosts to carry mcr that are found in humans, animals, vegetables, and wastewater (Shen et al., 2020) that have been reported in humans and animals in Thailand (Khanawapee et al., 2021; Paveenkittiporn et al., 2021; Boueroy et al., 2022). These STs were most common among the mcr-1 positive isolates (Li et al., 2022). The majority of phylogroups in our studies belonged to group A (43.75%), similar with other reports (Bosák et al., 2019; García-Meniño et al., 2019).
Horizontal gene transfer (HGT) via plasmids plays a critical role in the dissemination of antibiotic resistance determinants among Gram-negative bacteria (Sun et al., 2017). Notably, the mcr genes, responsible for colistin resistance, are located on diverse plasmids, encompassing more than 20 incompatibility groups identified as carriers of mcr-1 (Liu et al., 2023). The IncX4, IncH2 and IncI2 replicon represent the majority, exceeding 90% of plasmids that have been identified globally. While IncX4 was the predominant plasmid group carrying the mcr-1 gene in Europe and IncI2 in Asia, recently indicates a global shift towards IncX4 prevalence (Sismova et al., 2023). Our analysis of mcr carrying plasmid sequences, revealing a significant predominance of mcr-1 on IncX4 replicons, with additional presence on IncI2, IncP1, IncFIB and IncFII plasmids. These finding resonate with our previously published data, demonstrating mcr-1 carriage by IncX4 and IncI2 replicon in E. coli in humans in Thailand (Boueroy et al., 2022). This supports the notion of mcr gene emergence within specific plasmid groups in Enterobacterales from various reservoirs, including animals, food, environment, and humans (Boonyasiri et al., 2023; Leangapichart et al., 2023). Additionally, our experiments revealed the conjugative transfer capacity of IncX4 plasmids harboring mcr-1. Although this observation is limited to our laboratory conditions, it may offer a partial explanation for the increasing dominance of IncX4 in mcr-1 dissemination. Its high self-transferable at high frequencies (Sun et al., 2017; Bai et al., 2018). The high efficiency of horizontal transfer via this plasmid family poses a significant risk, considering that colistin is considered a last-line antibiotic for the treating life-threatening infections in humans (Sismova et al., 2023).
The pap2 superfamily protein was detected upstream but was not detected in the insertion sequence ISApl1 of any of the mcr-1- bearing InX4 plasmids, which indicated that the mcr-1 located in IncX4 was stable.
The genetic context analysis surrounding the mcr genes in our studies was hampered by the inherent ambiguity of the conjugation assay for determining the origin of transferred genes. The conjugation assay inability to definitively differentiate between plasmid and chromosomal transfer could lead to misinterpretations. This ambiguity stems from two scenarios. Firstly, mcr genes may reside on plasmids that integrated into the chromosome prior to conjugation. In such instances, the transferred genes would appear chromosomal despite their initial plasmid origin (Rafii and Crawford, 1988; Willett, 1988).
Furthermore, the ISAPl1 insertion sequence has been implicated as a crucial element facilitating the translocation of mcr-1 into diverse plasmid types. This sequence has been localized on the chromosome through the formation of circular intermediates (Trongjit and Chuanchuen, 2021). The close genetic association between ISApl1 and mcr-1 suggests that ISApl1 may play a pivotal role in the dissemination of mcr-1 (Li et al., 2017). The three isolates of mcr-1 located on the IncI2 plasmid contained ISAPl1-mcr-1-pap2, which is one pattern of the transposon Tn6330 (Snesrud et al., 2018). In our study, the mcr-3.1 gene was located on the IncFIB and IncFII plasmids, while mcr-3.5 was located on the IncP1 plasmid, to a study in China (Li J. et al., 2018). Both plasmids contained a diacylglycerol kinase (dgkA) upstream and this genomic context similar to another study (Phuadraksa et al., 2022). In addition, almost all the mcr-carrying plasmids could transfer to the recipient E. coli strain, which was in agreement with another study that concluded these plasmids were conjugative and dissemination could easily occur (Trongjit and Chuanchuen, 2021).
Other reports noted that the presence of the chromosomally encoded mcr gene is very rare (3.5–4%) (Li R. et al., 2018; Shen et al., 2020); however, a recent study reported E. coli strains harboring chromosomally encoded mcr-1 were found in 26-37% isolates from humans and animals in Vietnam (Vu Thi Ngoc et al., 2022). Recently, co-harboring of mcr-2 and mcr-3 on chromosomes in E. coli isolated from a healthy human was documented in Thailand (Phuadraksa et al., 2022). The current study revealed 2 mcr-1 harboring E. coli strains (KK3 and CB70) contained the segment of transposase ISApl1-mcr-1-pap2-ISApl1 (Tn6330) in the chromosome that would increase its stability and be highly mobilizable in spreading mcr genes due to vertical transfer.
5 Conclusion
Retail meat products (primarily pork and chicken), as a main food type and an important component of the food chain, were contaminated by mcr genes. Such meat products may serve as reservoirs of this antimicrobial-resistant gene that can affect humans. This finding supports the potential for the distribution and transmission of the colistin-resistant mcr gene is mediated by plasmids. Therefore, the mcr gene can spread via food chains, with the source of the strains suggesting that mcr-1 may be a public health risk by spreading to humans through farm to fork processes. Therefore, ongoing monitoring and investigations of mcr genes in agricultural sector are required to control and prevent their spread.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
PC: Conceptualization, Funding acquisition, Resources, Writing – original draft, Writing – review & editing. PB: Formal Analysis, Writing – review & editing. MM: Formal analysis, Investigation, Methodology, Software, Writing – review & editing. TI: Investigation, Methodology, Software, Writing – review & editing. YA: Conceptualization, Supervision, Writing – review & editing. SH: Supervision, Writing – review & editing. AK: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Office of the Ministry of Higher Education, Science, Research and Innovation; and the Thailand Science Research and Innovation through the Kasetsart University Reinventing University Program 2021.
Acknowledgments
The Kasetsart University Research and Development Institute (KURDI), Bangkok, Thailand provided English-editing assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1289134/full#supplementary-material
Footnotes
- ^ https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- ^ https://github.com/rrwick/Porechop
- ^ https://github.com/wdecoster/NanoPlot
- ^ http://clermontyping.iame-research.center/index.php
References
Alcock, B. P., Huynh, W., Chalil, R., Smith, K. W., Raphenya, A. R., Wlodarski, M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 51, D690–D699. doi: 10.1093/NAR/GKAC920
Bai, F., Li, X., Niu, B., Zhang, Z., Malakar, P. K., Liu, H., et al. (2018). A mcr-1-Carrying Conjugative IncX4 Plasmid in Colistin-Resistant Escherichia coli ST278 Strain Isolated From Dairy Cow Feces in Shanghai, China. Front. Microbiol. 9. doi: 10.3389/FMICB.2018.02833/BIBTEX
Bertels, F., Silander, O. K., Pachkov, M., Rainey, P. B., Van Nimwegen, E. (2014). Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 31, 1077–1088. doi: 10.1093/MOLBEV/MSU088
Boonyasiri, A., Brinkac, L. M., Jauneikaite, E., White, R. C., Greco, C., Seenama, C., et al. (2023). Characteristics and genomic epidemiology of colistin-resistant Enterobacterales from farmers, swine, and hospitalized patients in Thailand 2014–2017. BMC Infect. Dis. 23, 1–14. doi: 10.1186/S12879-023-08539-8/FIGURES/4
Bosák, J., Hrala, M., Pirková, V., Micenková, L., Čížek, A., Smola, J., et al. (2019). Porcine pathogenic Escherichia coli strains differ from human fecal strains in occurrence of bacteriocin types. Vet. Microbiol. 232, 121–127. doi: 10.1016/J.VETMIC.2019.04.003
Boueroy, P., Wongsurawat, T., Jenjaroenpun, P., Chopjitt, P., Hatrongjit, R., Jittapalapong, S., et al. (2022). Plasmidome in mcr-1 harboring carbapenem-resistant enterobacterales isolates from human in Thailand. Sci. Rep. 12 (1), 19051. doi: 10.1038/S41598-022-21836-7
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Carattoli, A., Zankari, E., Garciá-Fernández, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chen, K., Chan, E. W. C., Xie, M., Ye, L., Dong, N., Chen, S. (2017). Widespread distribution of mcr-1-bearing bacteria in the ecosystem 2015 to 2016. Eurosurveillance 22, 17–00206. doi: 10.2807/1560-7917.ES.2017.22.39.17-00206/CITE/PLAINTEXT
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/BIOINFORMATICS/BTY560
Clinical and Laboratory Standards Institute (CLSI) (2022). Performance standards for antimicrobial susceptibility testing. 32nd ed (USA: PA, U.S.: Clinical and Laboratory Standards Institute).
De Coster, W., Rademakers, R. (2023). NanoPack2: population-scale evaluation of long-read sequencing data. Bioinformatics 39 (5), btad311. doi: 10.1093/BIOINFORMATICS/BTAD311
Fan, J., Jiang, D., Zhao, Y., Liu, J., Zhang, X. C. (2014). Crystal structure of lipid phosphatase Escherichia coli phosphatidylglycerophosphate phosphatase B. Proc. Natl. Acad. Sci. U. S. A. 111, 7636–7640. doi: 10.1073/PNAS.1403097111/-/DCSUPPLEMENTAL
Gaballa, A., Wiedmann, M., Carroll, L. M. (2023). More than mcr: canonical plasmid- and transposon-encoded mobilized colistin resistance genes represent a subset of phosphoethanolamine transferases. Front. Cell. Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1060519
García-Meniño, I., Díaz-Jiménez, D., García, V., de Toro, M., Flament-Simon, S. C., Blanco, J., et al. (2019). Genomic Characterization of Prevalent mcr-1, mcr-4, and mcr-5 Escherichia coli Within Swine Enteric Colibacillosis in Spain. Front. Microbiol. 10. doi: 10.3389/FMICB.2019.02469
Gurevich, A., Saveliev, V., Vyahhi, N., Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/BIOINFORMATICS/BTT086
Huang, X., Yu, L., Chen, X., Zhi, C., Yao, X., Liu, Y., et al. (2017). High Prevalence of Colistin Resistance and mcr-1 Gene in Escherichia coli Isolated from Food Animals in China. Front. Microbiol. 8. doi: 10.3389/FMICB.2017.00562
Jiang, H., Lei, R., Ding, S. W., Zhu, S. (2014). Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinf. 15, 182. doi: 10.1186/1471-2105-15-182
Joensen, K. G., Tetzschner, A. M. M., Iguchi, A., Aarestrup, F. M., Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Katz, L., Griswold, T., Morrison, S., Caravas, J., Zhang, S., Bakker, H., et al. (2019). Mashtree: a rapid comparison of whole genome sequence files. J. Open Source Softw. 4, 10.21105/joss.01762. doi: 10.21105/joss.01762
Kempf, I., Jouy, E., Chauvin, C. (2016). Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 48, 598–606. doi: 10.1016/J.IJANTIMICAG.2016.09.016
Khanawapee, A., Kerdsin, A., Chopjitt, P., Boueroy, P., Hatrongjit, R., Akeda, Y., et al. (2021). Distribution and Molecular Characterization of Escherichia coli Harboring mcr Genes Isolated from Slaughtered Pigs in Thailand. Microb. Drug Resist. 27, 971–979. doi: 10.1089/MDR.2020.0242
Khine, N. O., Lugsomya, K., Kaewgun, B., Honhanrob, L., Pairojrit, P., Jermprasert, S., et al. (2020). Multidrug Resistance and Virulence Factors of Escherichia coli Harboring Plasmid-Mediated Colistin Resistance: mcr-1 and mcr-3 Genes in Contracted Pig Farms in Thailand. Front. Vet. Sci. 7. doi: 10.3389/FVETS.2020.582899
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Leangapichart, T., Stosic, M. S., Hickman, R. A., Lunha, K., Jiwakanon, J., Angkititrakul, S., et al. (2023). Exploring the epidemiology of mcr genes, genetic context and plasmids in Enterobacteriaceae originating from pigs and humans on farms in Thailand. J. Antimicrob. Chemother. 78, 1395–1405. doi: 10.1093/JAC/DKAD097
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/NAR/GKAB301
Li, F., Cheng, P., Li, X., Liu, R., Liu, H., Zhang, X. (2022). Molecular Epidemiology and Colistin-Resistant Mechanism of mcr-Positive and mc r-Negative Escherichia coli Isolated From Animal in Sichuan Province, China. Front. Microbiol. 13. doi: 10.3389/FMICB.2022.818548
Li, J., Hulth, A., Nilsson, L. E., Börjesson, S., Chen, B., Bi, Z., et al. (2018). Occurrence of the mobile colistin resistance gene mcr-3 in Escherichia coli from household pigs in rural areas. J. Antimicrob. Chemother. 73, 1721–1723. doi: 10.1093/JAC/DKY038
Li, B., Ke, B., Zhao, X., Guo, Y., Wang, W., Wang, X., et al. (2018). Antimicrobial Resistance Profile of mcr-1 Positive Clinical Isolates of Escherichia coli in China From 2013 to 2016. Front. Microbiol. 9. doi: 10.3389/FMICB.2018.02514
Li, R., Xie, M., Zhang, J., Yang, Z., Liu, L., Liu, X., et al. (2017). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 72, 393–401. doi: 10.1093/JAC/DKW411
Li, R., Yu, H., Xie, M., Chen, K., Dong, N., Lin, D., et al. (2018). Genetic basis of chromosomally-encoded mcr-1 gene. Int. J. Antimicrob. Agents 51, 578–585. doi: 10.1016/J.IJANTIMICAG.2017.11.015
Liu, J. H., Liu, Y. Y., Shen, Y. B., Yang, J., Walsh, T. R., Wang, Y., et al. (2023). Plasmid-mediated colistin-resistance genes: mcr. Trends Microbiol. S0966-842X(23)00295-0. Advance online publication. doi: 10.1016/J.TIM.2023.10.006
Liu, Z., Liu, Y., Xi, W., Liu, S., Liu, J., Mu, H., et al. (2021). Genetic Features of Plasmid- and Chromosome-Mediated mcr-1 in Escherichia coli Isolates From Animal Organs With Lesions. Front. Microbiol. 12. doi: 10.3389/FMICB.2021.707332
Liu, B. T., Song, F. J. (2019). Emergence of two Escherichia coli strains co-harboring mcr-1 and bla NDM in fresh vegetables from China. Infect. Drug Resist. 12, 2627–2635. doi: 10.2147/IDR.S211746
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lopes, R., Furlan, J. P. R., dos Santos, L. D. R., Gallo, I. F. L., Stehling, E. G. (2021). Colistin-Resistant mcr-1-Positive Escherichia coli ST131- H 22 Carrying bla CTX-M-15 and qnrB19 in Agricultural Soil. Front. Microbiol. 12. doi: 10.3389/FMICB.2021.659900
Lu, X., Xiao, X., Liu, Y., Huang, S., Li, R., Wang, Z. (2020). Widespread Prevalence of Plasmid-Mediated Colistin Resistance Gene mcr-1 in Escherichia coli from Père David’s Deer in China. mSphere 5 (6), e01221-20. doi: 10.1128/MSPHERE.01221-20
Macori, G., Nguyen, S. V., Naithani, A., Hurley, D., Bai, L., Garch, F., et al. (2021). Characterisation of Early Positive mcr-1 Resistance Gene and Plasmidome in Escherichia coli Pathogenic Strains Associated with Variable Phylogroups under Colistin Selection. Antibiot. (Basel Switzerland) 10, 1041. doi: 10.3390/ANTIBIOTICS10091041
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/J.1469-0691.2011.03570.X
Martiny, H.-M., Munk, P., Brinch, C., Szarvas, J., Aarestrup, F. M., Petersen, T. N. (2022). Global distribution of mcr gene variants in 214K metagenomic samples. mSystems 7 (2), e0010522. doi: 10.1128/MSYSTEMS.00105-22/SUPPL_FILE/REVIEWER-COMMENTS.PDF
Meletis, G., Skoura, L. (2018). Polymyxin resistance mechanisms: from intrinsic resistance to Mcr genes. Recent Pat. Antiinfect. Drug Discovery 13, 198–206. doi: 10.2174/1574891X14666181126142704
Migura-Garcia, L., González-López, J. J., Martinez-Urtaza, J., Aguirre Sánchez, J. R., Moreno-Mingorance, A., Perez de Rozas, A., et al. (2020). mcr-colistin resistance genes mobilized by IncX4, IncHI2, and IncI2 plasmids in Escherichia coli of pigs and white stork in Spain. Front. Microbiol. 10. doi: 10.3389/FMICB.2019.03072
Mohsin, M., Hassan, B., Martins, W. M. B. S., Li, R., Abdullah, S., Sands, K., et al. (2021). Emergence of plasmid-mediated tigecycline resistance tet(X4) gene in Escherichia coli isolated from poultry, food and the environment in South Asia. Sci. Total Environ. 787, 147613. doi: 10.1016/J.SCITOTENV.2021.147613
Nakano, A., Nakano, R., Nishisouzu, R., Suzuki, Y., Horiuchi, S., Kikuchi-Ueda, T., et al. (2021). Prevalence and Relatedness of mcr-1-Mediated Colistin-Resistant Escherichia coli Isolated From Livestock and Farmers in Japan. Front. Microbiol. 12. doi: 10.3389/FMICB.2021.664931
Nguyet, L. T. Y., Keeratikunakorn, K., Kaeoket, K., Ngamwongsatit, N. (2022). Antibiotic resistant Escherichia coli from diarrheic piglets from pig farms in Thailand that harbor colistin-resistant mcr genes. Sci. Rep. 12 (1), 9083. doi: 10.1038/S41598-022-13192-3
Paveenkittiporn, W., Kamjumphol, W., Ungcharoen, R., Kerdsin, A. (2021). Whole-genome sequencing of clinically isolated carbapenem-resistant enterobacterales harboring mcr genes in Thailand 2016-2019. Front. Microbiol. 11. doi: 10.3389/FMICB.2020.586368
Phetburom, N., Boueroy, P., Chopjitt, P., Hatrongjit, R., Akeda, Y., Hamada, S., et al. (2021). Klebsiella pneumoniae Complex Harboring mcr-1, mcr-7, and mcr-8 Isolates from Slaughtered Pigs in Thailand. Microorganisms 9, 2436. doi: 10.3390/MICROORGANISMS9122436
Phuadraksa, T., Wichit, S., Arikit, S., Songtawee, N., Yainoy, S. (2022). Co-occurrence of mcr-2 and mcr-3 genes on chromosome of multidrug-resistant Escherichia coli isolated from healthy individuals in Thailand. Int. J. Antimicrob. Agents 60 (4), 106662. doi: 10.1016/J.IJANTIMICAG.2022.106662
Pormohammad, A., Nasiri, M. J., Azimi, T. (2019). Prevalence of antibiotic resistance in Escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: a systematic review and meta-analysis. Infect. Drug Resist. 12, 1181–1197. doi: 10.2147/IDR.S201324
Puangseree, J., Prathan, R., Srisanga, S., Angkittitrakul, S., Chuanchuen, R. (2022). Plasmid profile analysis of Escherichia coli and Salmonella enterica isolated from pigs, pork and humans. Epidemiol. Infect. 150, e110. doi: 10.1017/S0950268822000814
Pungpian, C., Lee, S., Trongjit, S., Sinwat, N., Angkititrakul, S., Prathan, R., et al. (2021). Colistin resistance and plasmid-mediated mcr genes in Escherichia coli and Salmonella isolated from pigs, pig carcass and pork in Thailand, Lao PDR and Cambodia border provinces. J. Vet. Sci. 22, 1–15. doi: 10.4142/JVS.2021.22.E68
Rafii, F., Crawford, D. L. (1988). Transfer of conjugative plasmids and mobilization of a nonconjugative plasmid between Streptomyces strains on agar and in soil. Appl. Environ. Microbiol. 54, 1334–1340. doi: 10.1128/AEM.54.6.1334-1340.1988
Roer, L., Tchesnokova, V., Allesoe, R., Muradova, M., Chattopadhyay, S., Ahrenfeldt, J., et al. (2017). Development of a Web Tool for Escherichia coli Subtyping Based on fimH Alleles. J. Clin. Microbiol. 55, 2538–2543. doi: 10.1128/JCM.00737-17
Shafiq, M., Huang, J., Rahman, S. U., Shah, J. M., Chen, L., Gao, Y., et al. (2019). High incidence of multidrug-resistant Escherichia coli coharboring mcr-1 and bla CTX-M-15 recovered from pigs. Infect. Drug Resist 12, 2135–2149. doi: 10.2147/IDR.S209473
Shafiq, M., Huang, J., Shah, J. M., Ali, I., Rahman, S. U., Wang, L. (2021). Characterization and resistant determinants linked to mobile elements of ESBL-producing and mcr-1-positive Escherichia coli recovered from the chicken origin. Microb. Pathog. 150, 104722. doi: 10.1016/J.MICPATH.2020.104722
Shafiq, M., Rahman, S. U., Bilal, H., Ullah, A., Noman, S. M., Zeng, M., et al. (2022). Incidence and molecular characterization of ESBL-producing and colistin-resistant Escherichia coli isolates recovered from healthy food-producing animals in Pakistan. J. Appl. Microbiol. 133, 1169–1182. doi: 10.1111/JAM.15469
Shen, C., Zhong, L. L., Yang, Y., Doi, Y., Paterson, D. L., Stoesser, N., et al. (2020). Dynamics of mcr-1 prevalence and mcr-1-positive Escherichia coli after the cessation of colistin use as a feed additive for animals in China: a prospective cross-sectional and whole genome sequencing-based molecular epidemiological study. Lancet Microbe 1, e34–e43. doi: 10.1016/S2666-5247(20)30005-7
Singh, A. K., Das, S., Singh, S., Gajamer, V. R., Pradhan, N., Lepcha, Y. D., et al. (2018). Prevalence of antibiotic resistance in commensal Escherichia coli among the children in rural hill communities of Northeast India. PloS One 13:e0199179. doi: 10.1371/JOURNAL.PONE.0199179
Sismova, P., Sukkar, I., Kolidentsev, N., Palkovicova, J., Chytilova, I., Bardon, J., et al. (2023). Plasmid-mediated colistin resistance from fresh meat and slaughtered animals in the Czech Republic: nation-wide surveillance 2020–2021. Microbiol. Spectr. 11, e0060923. doi: 10.1128/SPECTRUM.00609-23/SUPPL_FILE/SPECTRUM.00609-23-S0001.PDF
Snesrud, E., McGann, P., Chandler, M. (2018). The birth and demise of the IS Apl1- mcr-1-IS Apl1 composite transposon: the vehicle for transferable colistin resistance. MBio 9. doi: 10.1128/MBIO.02381-17
Sullivan, M. J., Petty, N. K., Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/BIOINFORMATICS/BTR039
Sun, J., Fang, L. X., Wu, Z., Deng, H., Yang, R. S., Li, X. P., et al. (2017). Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci. Rep. 71, 1–9. doi: 10.1038/s41598-017-00095-x
Sun, J., Zhang, H., Liu, Y. H., Feng, Y. (2018). Towards understanding MCR-like colistin resistance. Trends Microbiol. 26, 794–808. doi: 10.1016/J.TIM.2018.02.006
Tang, B., Wang, J., Zheng, X., Chang, J., Ma, J., Wang, J., et al. (2022). Antimicrobial resistance surveillance of Escherichia coli from chickens in the Qinghai Plateau of China. Front. Microbiol. 13. doi: 10.3389/FMICB.2022.885132
Tanizawa, Y., Fujisawa, T., Nakamura, Y. (2018). DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34, 1037–1039. doi: 10.1093/BIOINFORMATICS/BTX713
Trongjit, S., Chuanchuen, R. (2021). Whole genome sequencing and characteristics of Escherichia coli with co-existence of ESBL and mcr genes from pigs. PloS One 16, e0260011. doi: 10.1371/JOURNAL.PONE.0260011
Vu Thi Ngoc, B., Le Viet, T., Nguyen Thi Tuyet, M., Nguyen Thi Hong, T., Nguyen Thi Ngoc, D., Le Van, D., et al. (2022). Characterization of Genetic Elements Carrying mcr-1 Gene in Escherichia coli from the Community and Hospital Settings in Vietnam. Microbiol. Spectr. 10, e0135621. doi: 10.1128/SPECTRUM.01356-21
Wang, R., Van Dorp, L., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 91(9), 1–9 doi: 10.1038/s41467-018-03205-z
Wick, R. R., Judd, L. M., Gorrie, C. L., Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/JOURNAL.PCBI.1005595
Wick, R. R., Judd, L. M., Holt, K. E. (2019). Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 20 (1), 129. doi: 10.1186/S13059-019-1727-Y
Willetts, N. (1988). 3 Conjugation. Methods Microbiol. 21, 49–77. doi: 10.1016/S0580-9517(08)70070-9
Yamaguchi, T., Kawahara, R., Hamamoto, K., Hirai, I., Khong, D. T., Nguyen, T. N., et al. (2020). High Prevalence of Colistin-Resistant Escherichia coli with Chromosomally Carried mcr-1 in Healthy Residents in Vietnam. mSphere 5, e00117-20. doi: 10.1128/MSPHERE.00117-20
Zając, M., Sztromwasser, P., Bortolaia, V., Leekitcharoenphon, P., Cavaco, L. M., Zitek-Barszcz, A., et al. (2019). Occurrence and Characterization of mcr-1-Positive Escherichia coli Isolated From Food-Producing Animals in Poland 2011-2016. Front. Microbiol. 10. doi: 10.3389/FMICB.2019.01753
Keywords: colistin-resistant E. col, mcr gene, plasmid replicon typing, foods, IncX4, IncP1, IncI2
Citation: Chopjitt P, Boueroy P, Morita M, Iida T, Akeda Y, Hamada S and Kerdsin A (2024) Genetic characterization of multidrug-resistant Escherichia coli harboring colistin-resistant gene isolated from food animals in food supply chain. Front. Cell. Infect. Microbiol. 14:1289134. doi: 10.3389/fcimb.2024.1289134
Received: 05 September 2023; Accepted: 12 January 2024;
Published: 02 February 2024.
Edited by:
Laura Maria Andrade De Oliveira, Federal University of Rio de Janeiro, BrazilReviewed by:
Adam Valcek, Vrije University Brussel, BelgiumClaudia Cortimiglia, Università Cattolica del Sacro Cuore, Italy
Muhammad Shafiq, Shantou University, China
Copyright © 2024 Chopjitt, Boueroy, Morita, Iida, Akeda, Hamada and Kerdsin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peechanika Chopjitt, cGVlY2hhbmlrYS5jQGt1LnRo
 Peechanika Chopjitt
Peechanika Chopjitt Parichart Boueroy1
Parichart Boueroy1 Masatomo Morita
Masatomo Morita Tetsuya Iida
Tetsuya Iida Yukihiro Akeda
Yukihiro Akeda Sihigeyuki Hamada
Sihigeyuki Hamada Anusak Kerdsin
Anusak Kerdsin