
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 12 February 2024
Sec. Parasite and Host
Volume 14 - 2024 | https://doi.org/10.3389/fcimb.2024.1270060
This article is part of the Research TopicAdvancement in Malaria TreatmentView all 6 articles
Background: Malaria has always been a serious infectious disease prevalent in the world. Antimalarial drugs such as chloroquine and artemisinin have been the main compounds used to treat malaria. However, the massive use of this type of drugs accelerates the evolution and spread of malaria parasites, leading to the development of resistance. A large number of related data have been published by researchers in recent years. CiteSpace software has gained popularity among us researchers in recent years, because of its ability to help us obtain the core information we want in a mass of articles. In order to analyze the hotspots and develop trends in this field through visual analysis, this study used CiteSpace software to summarize the available data in the literature to provide insights.
Method: Relevant literature was collected from the Web of Science Core Collection (WOSCC) from 1 January 2015 to 29 March 2023. CiteSpace software and Microsoft Excel were used to analyze and present the data, respectively.
Results: A total of 2,561 literatures were retrieved and 2,559 literatures were included in the analysis after the removal of duplicates. An irrefutable witness of the ever-growing interest in the topic of antimalarial drug resistance could be expressed by the exponentially increased number of publications and related citations from 2015 to 2022, and its sustained growth trend by 2023. During the past 7 years, USA, Oxford University, and David A Fidock are the country, institution, and author with the most publications in this field of research, respectively. We focused on the references and keywords from literature and found that the research and development of new drugs is the newest hotspot in this field. A growing number of scientists are devoted to finding new antimalarial drugs.
Conclusion: This study is the first visual metrological analysis of antimalarial drug resistance, using bibliometric methods. As a baseline information, it is important to analyze research output published globally on antimalarial drug resistance. In order to better understand the current research situation and future research plan agenda, such baseline data are needed accordingly.
Malaria is a vector-borne infectious disease caused by Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, Plasmodium ovale, and Plasmodium knowlesi and transmitted by Anopheles mosquitoes. There are two proven effective strategies to combat malaria (White, 2004). One is to block the Anopheles mosquitoes from approaching humans, by blocking the transmission route of Anopheles mosquitoes and using insecticide-equipped mosquito nets. The other is to use antimalarial drugs for case management. In 2006, the World Health Organization (WHO) recommended the use of artemisinin-based combination therapies (ACTs) as first-line treatment for falciparum malaria (Eastman and Fidock, 2009). If ACTs fail in Southeast Asia and other regions, there will be no fully effective drug capable of completely replacing them in the future. As the new antimalarial drugs are unlikely to become generally available within the next few years, active measures should be given the highest priority to preserve the current antimalarials.
Widespread drug use leads to antimalarial drug resistance, which is an inevitable risk factor in malaria treatment. This risk cannot be disregarded, particularly with unregulated drug use. Early detection and proper medical intervention can effectively lower malaria mortality rates and prevent the emergence of Plasmodium resistance (Landier et al., 2018; Schultz et al., 2023). Over the last few years, antimalarial drug resistance has posed a significant challenge to worldwide malaria control initiatives, especially in the Greater Mekong subregion (Ménard et al., 2018). There are alarming early reports of disturbing mutations in other regions too (Anantabotla et al., 2019; Hussien et al., 2020; Narh et al., 2020; Uwimana et al., 2020; Balikagala et al., 2021; Uwimana et al., 2021; Conrad et al., 2023; Fola et al., 2023; Mihreteab et al., 2023). The global prevalence of Plasmodium infection and deaths declined prior to the outbreak, and healthcare services were compromised due to the public health emergencies that exacerbated the common health crisis (Bhatt et al., 2015; McLean et al., 2018; Kiarie et al., 2022). In accordance with the prognosis of the WHO, the insufficient prevention, diagnosis, and treatment of the illness is expected to lead to the doubling of malaria-related fatalities during the pandemic (Norman et al., 2022). According to the 2022 World Malaria Report, there were 247 million cases of malaria globally in 2021, an increase from 245 million cases in 2020. The number of deaths attributed to the disease decreased from 625,000 in 2020 to 619,000 in 2021 (Fact sheet about malaria, n.d). Among available antimalarials, factors that contribute include parasite mutation rates, total parasite load, drug strength, adherence to treatment guidelines, dosing appropriateness, pharmacokinetic properties, inadequate drug contact due to falsified drugs, and poor-quality antimalarials that may lead to or eliminate resistance (Shibeshi et al., 2020). In 2021, amid antimalarial drug resistance, the African region has contributed to 95% of the total global malaria cases and 96% of all deaths related to malaria, with children under five accounting for 80% of all malaria-related deaths in Africa. While artemisinin-based malaria treatment continues to be highly effective in Africa with no evidence of complete resistance, the mounting genetic mutations linked to artemisinin resistance may ultimately result in enhanced overall artemisinin resistance as well as selective pressure on partner drugs (Owoloye et al., 2021). Therefore, continuous monitoring of treatment efficacy and genetic surveillance across Africa is imperative.
Regular monitoring of drug efficacy is necessary to enhance treatment strategies in malaria-endemic countries and ensure early detection and response to drug resistance. The WHO recommends surveillance of antimalarial drug resistance every 2 years to control the spread of antimalarial drug resistance. The predominant techniques employed to oversee antimalarial drug resistance encompass drug efficacy studies, in vitro/in vivo studies of malaria parasites, and molecular studies that evaluate recognized markers of antimalarial drug resistance (Ariey et al., 2014; Ménard et al., 2016; Chen et al., 2021). To identify antimalarial drug resistance, specific gene mutations and molecular markers associated with antimalarial drug resistance are utilized. Detection of antimalarial drug resistance serves as a useful monitoring mechanism to enhance the control of Plasmodium susceptibility. Bibliometrics is a qualitative and quantitative visualization study of the retrieved literature, involving the application of mathematical and statistical methods to scholarly publications. Researchers evaluate research trends and research priorities within the field by analyzing literature databases, which have extensive applications in the pharmaceutical field. This study aimed to visually analyze the research status and development trends of the antimalarial drug resistance, by means of bibliometrics and knowledge mapping.
The study employed Web of Science Core Collection (WOSCC) as the literature search database. The retrieved literature was analyzed for research trends, references, and keywords with the aid of CiteSpace and Microsoft Excel. Relevant literature from 1 January 2015 to 29 March 2023 was included, with the date of publication as the selection criterion. The search terms used were “(TS= (Antimalarial Drug) AND TS=(Resistance))”.
CiteSpace (6.2.R1) was utilized for bibliometric analysis, a tool created by Professor Chen Chaomei. The most current version of CiteSpace can be downloaded from the official website and no longer requires the JAVA environment. Its primary function is to visually and analytically represent information found in the academic literature within a given field. The primary objective of visualizing knowledge domains is to identify and track their development by illustrating scientific collective networks within scientific domains, such as scientific networks, social networks of co-authors, citation networks, and networks of collaborators. CiteSpace achieves this by representing relations between scholars or publications through network nodes and connectivity, with the size of a node proportional to the publication output. The publication date is indicated by the color of each node. Technical terms are explained upon their first use. Mediator centrality is assigned to every node in a network, determining the probability of each shortest path passing through it. Mediated centrality is defined for each node in the network to determine the probability that any shortest path in the network passes via the node. Mediator nodes with high centrality may be found at the center of two significant clusters or sub-networks, hence the term mediator. CiteSpace illustrates nodes with high mediator centrality as purple rings, with the thickness representing the magnitude of the value of mediator centrality. The co-cited literature and keywords were clustered using the log-likelihood ratio (LLR) method. The silhouette (S) value denotes the mean contour value of the cluster. It is generally accepted that clustering is justifiable when S > 0.5 and persuasive when S > 0.7.
For this research, CiteSpace was used with the following specific parameters: time slice spanned January 2015 to March 2023; year slice interval was 1 year; term source used included title, abstract, author, and keywords; node type encompassed author, institution, country, keyword, literature, cited author, and cited journal; and the top 50 articles were compiled based on the selection criteria.
Microsoft Excel was utilized to construct tables that illustrate annual national publication trends.
A total of 2,561 papers related to antimalarial drug resistance in the WOSCC were searched, 2,559 papers were shortlisted after excluding duplicates using CiteSpace, and all papers were included in the analysis after excluding duplicates. From 1 January 2015 to 29 March 2023, a total of 2,561 articles with the subject terms “antimalarial drug” and “resistance” were retrieved from the WOSCC, consisting of the following article types: Article (1,979), Article; Book Chapter (6), Article; Early Access (33), Article; Proceedings Paper (7), Correction (2), Editorial Material (31), Meeting Abstract (31), News Item (1), Review (462), Review; Book Chapter (5), and Review; Early Access (4).
There were 386 papers in 2021, compared to 363 in 2020, 343 in 2022, and approximately 300 in other years, with the largest amount of literature published (Figure 1). In general, the rise in the number of academic papers on malaria may be linked to growing apprehension about the emergence of resistance genes in Africa over the past few years.
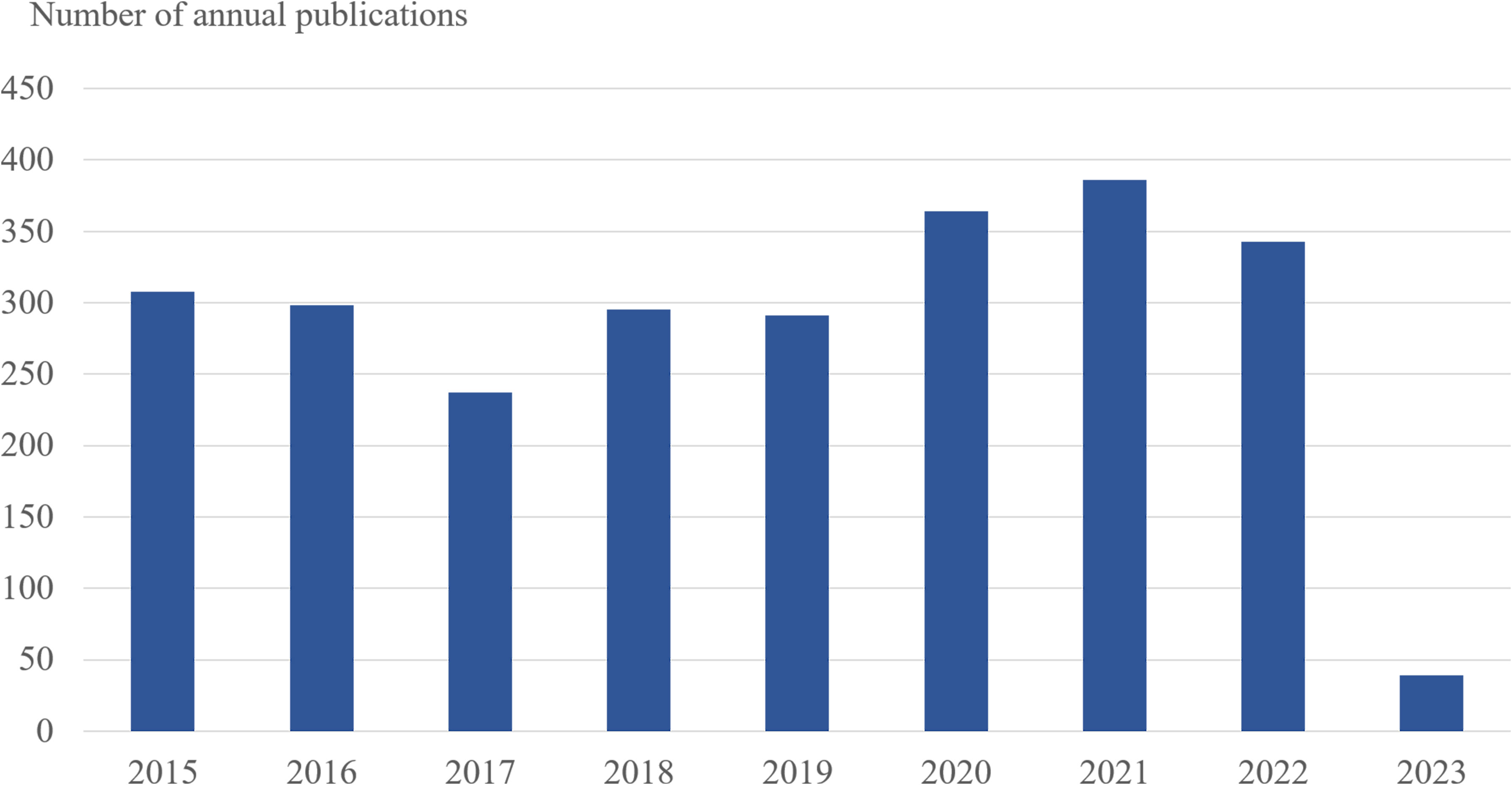
Figure 1 The number of annual publications on antimalarial drug resistance research between 2015 and 2023. The horizontal coordinates represent the year of publication, and the vertical coordinates represent the number of publications.
In order to understand the research hotspots in the analysis of antimalarial drug resistance in various countries, visual analysis was carried out in each country. As Table 1 and Figure 2 indicate, the top 10 countries with published papers were USA (744), India (403), England (382), Australia (232), China (217), Thailand (199), Switzerland (197), France (176), South Africa (170), and Germany (154). For centrality, the top ranking is Germany (0.14) followed by Switzerland (0.12), Spain (0.12), Sudan (0.08), and Bangladesh (0.07) (Table 1).
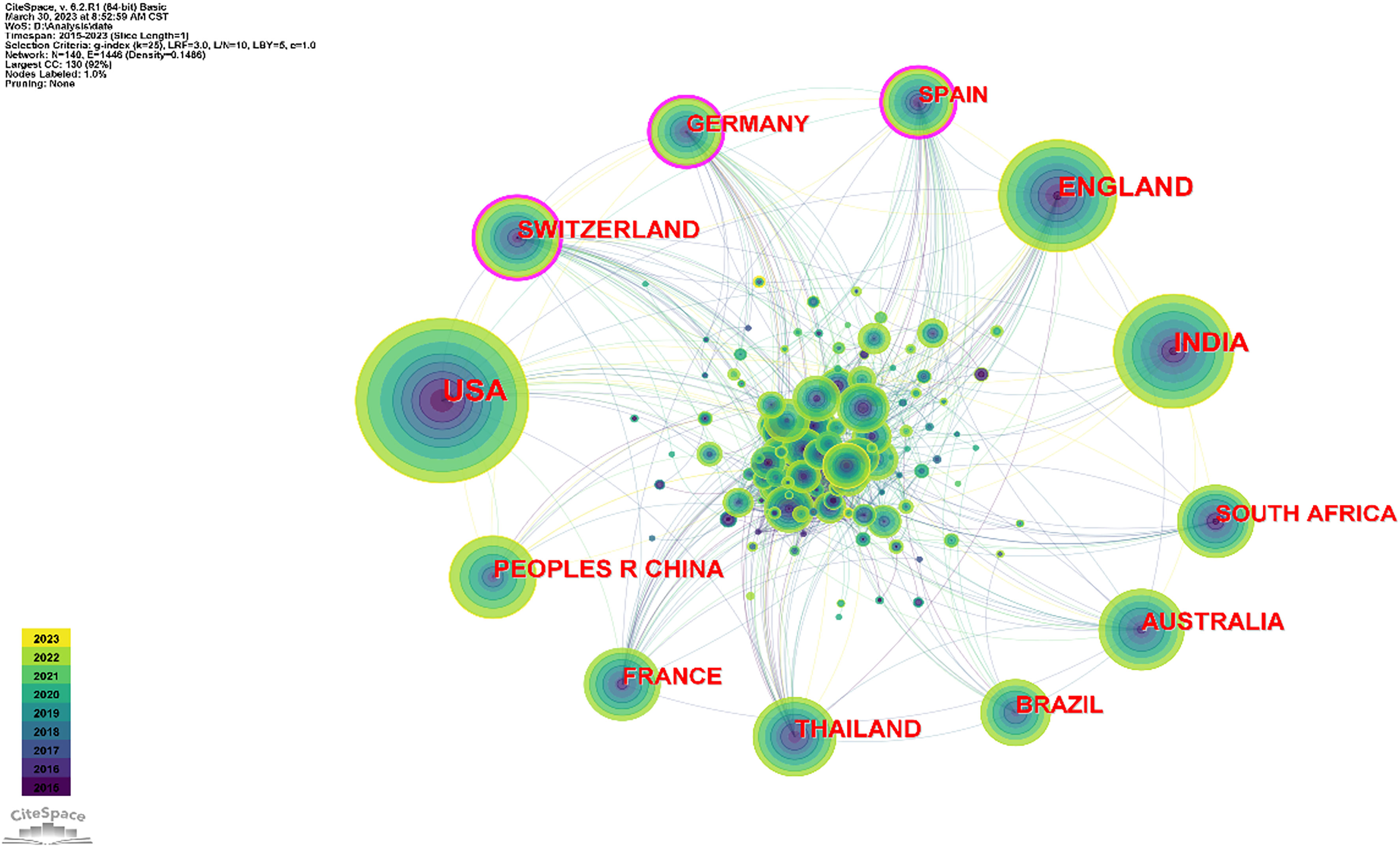
Figure 2 Map illustrating networks of cooperation between countries. Labels with purple circles denote centrality values >0.1, revealing Switzerland, Germany, and Spain as the three countries with the highest centrality. The size of each circle corresponds to the number of countries with which a country cooperates. The largest circle in the diagram represents the USA, indicating its high level of cooperation with other countries.
Many institutions and universities from different countries around the world have contributed to this research field. As shown in Table 2 and Figure 3, the top 5 institutions were the University of Oxford (England, 186), Mahidol University (Thailand, 168), the University of London (England, 127), the University of California (USA, 122), and Mahidol Oxford Tropical Medicine Research Unit (MORU) (Thailand, 122). The top 5 institutions with the highest centrality were Columbia University (USA, 0.09), Harvard University (USA, 0.08), Swiss Tropical & Public Health Institute (Switzerland, 0.07), Mahidol University (Thailand, 0.05), and the University of Basel (Switzerland, 0.05).
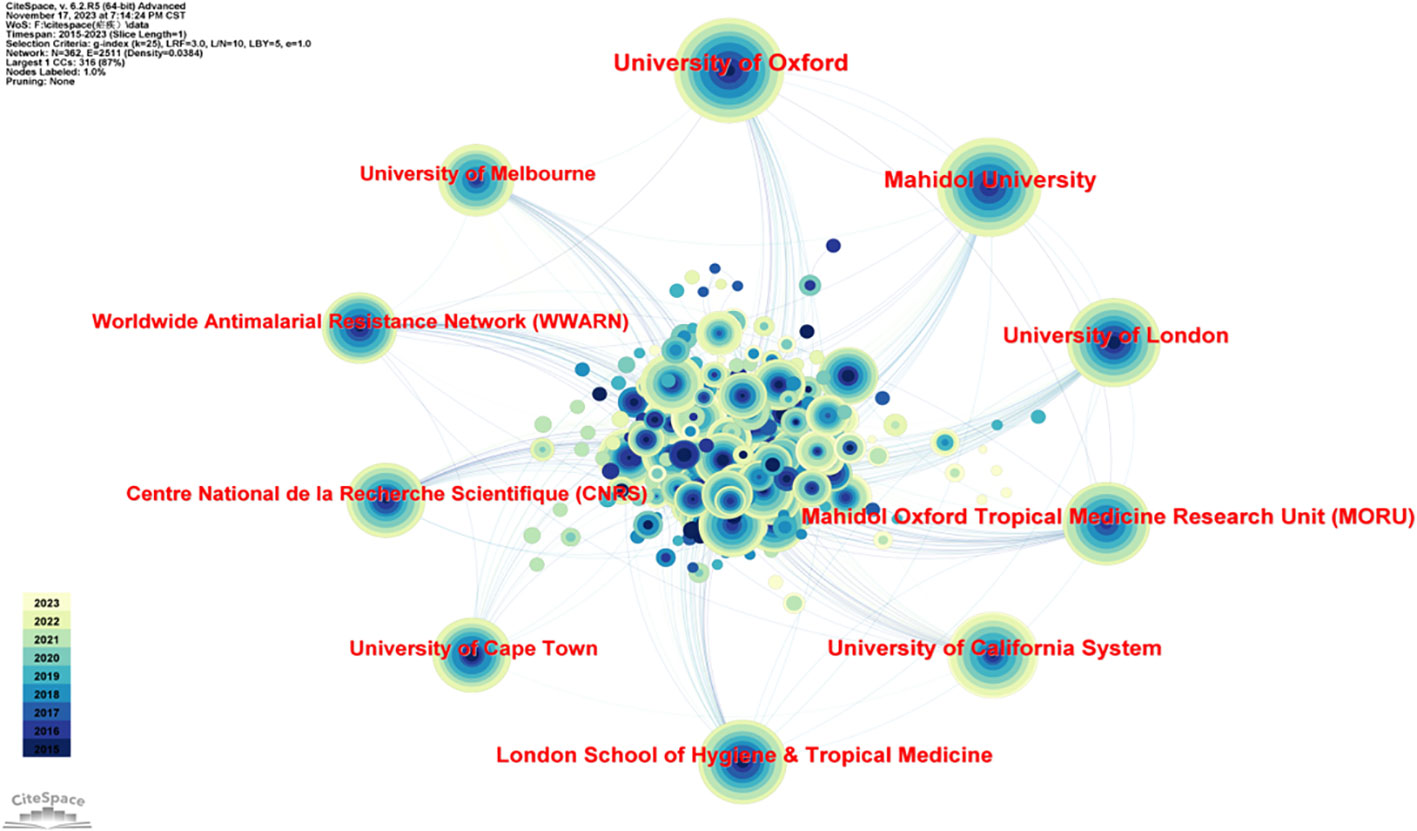
Figure 3 Diagram of the network of cooperation between institutions. The five most prominent institutions are the University of Oxford (England, 186), Mahidol University (Thailand, 168), the University of London (England, 127), the University of California (USA, 122), and Mahidol Oxford Tropical Medicine Research Unit (MORU) (Thailand, 122).
The author partnership network was analyzed to provide information for finding industry giants and seeking partners. As Table 3 and Figure 4 show, the top 10 authors were David A. Fidock (74), Nicholas J. White (44), Elizabeth A. Winzeler (42), Philip J. Rosenthal (41), Sergio Wittlin (32), Arjen M. Dondorp (31), François Nosten (30), Kelly Chibale (28), Bruno Pradines (27), and Liwang Cui (22). The top 5 authors in terms of centrality size were David A. Fidock (0.16), Sergio Wittlin (0.14), Didier Menard (0.13), Ric N. Price (0.13), and Elizabeth A. Winzeler (0.10).
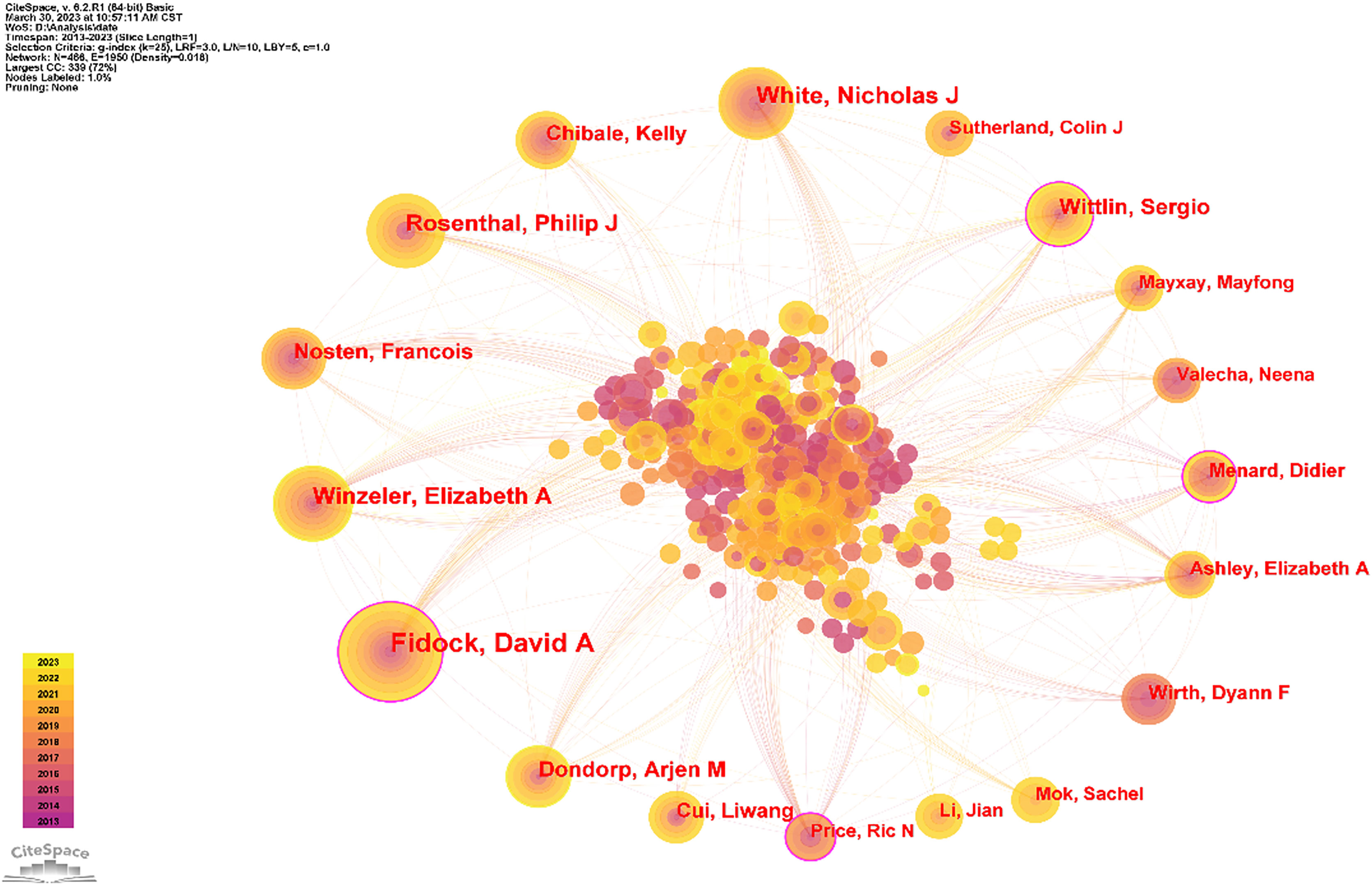
Figure 4 Author collaboration network diagram. As shown in the figure, David A. Fidock is the most collaborative and the most central author. Secondly, Wittlin Serglo’s article may be helpful for research in this direction.
The software defaults the WHO as author, and this study does not consider the WHO to be among the authors; thus, it was excluded. Some anonymous authors appeared during the analysis, and because such appearance affected the analysis, this part was excluded as well. The top 5 cited authors were Arjen M. Dondorp (634), Elizabeth A. Ashley (564), Nicholas J. White (554), Frédéric Ariey (463), and W. Trager (356). The top 5 co-cited authors for centrality were Timothy J Egan (0.07), Paul M O'Neill (0.06), David A Fidock (0.05), Judith Straimer (0.05), and Christin Sisowath (0.05), (Table 4 and Figure 5).
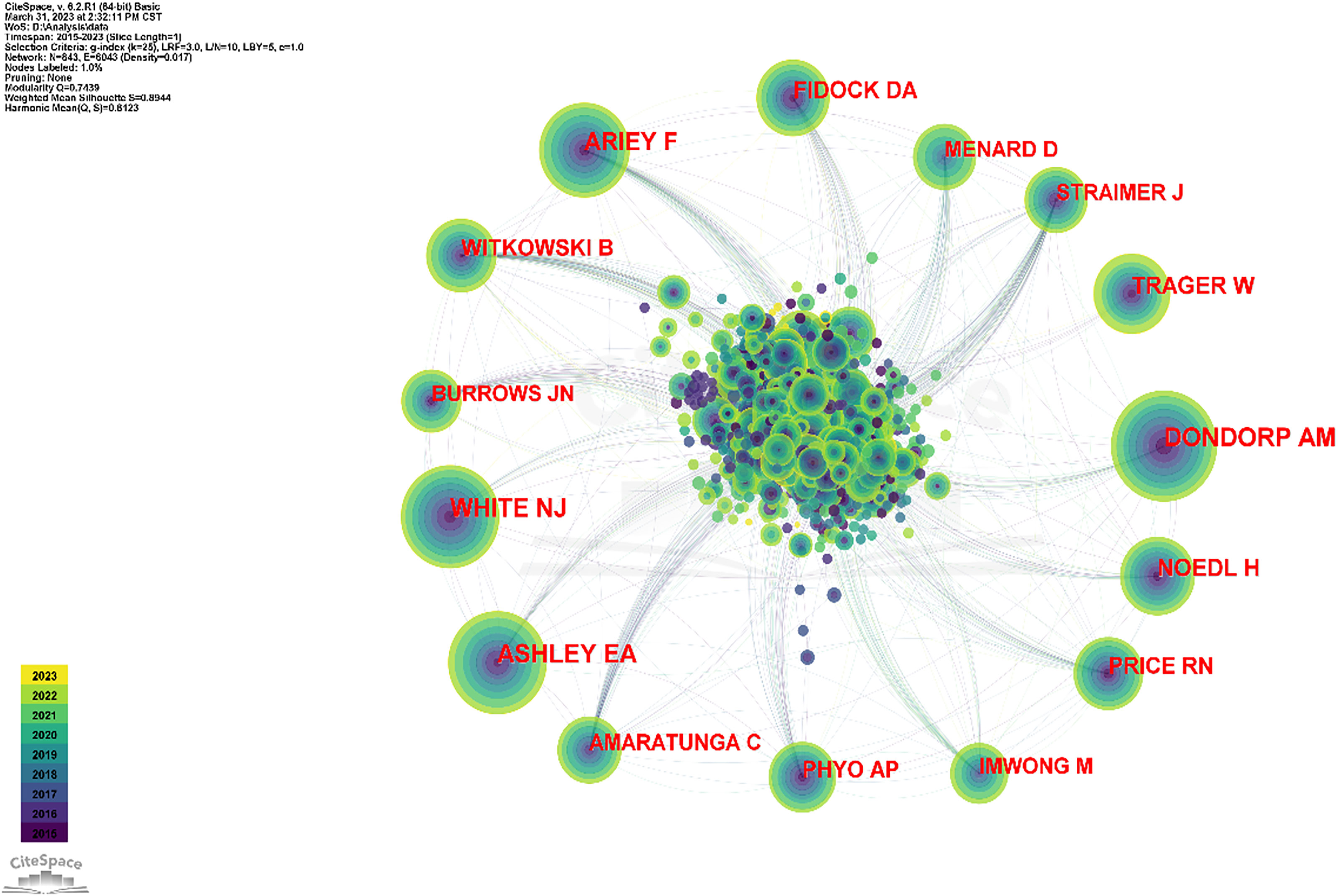
Figure 5 The collaboration network of co-cited authors. The articles published by Dondorp AM, White NJ, and Ashley EA are the most commonly cited.
In the analysis of co-cited journals, the top 5 journals were Antimicrobial Agents and Chemotherapy (1,784), Malaria Journal (1,784), PLOS One (1,495), Nature (1,361), and Proceedings of National Academy of Sciences of the United States of America (1,284). The top 5 journals for centrality were Carcinogenesis (0.36), International Journal of Nanomedicine (0.29), Frontiers in Microbiology (0.24), Dalton Transactions (0.22), and Journal of Chemical Information and Modeling (0.21) (Table 5, Figure 6).
As shown in Table 6, Ashley et al. (2014), Ariey et al. (2014), World Health Organization (2013), Straimer et al. (2015), and World Health Organization (2015) were the top 5 references cited. The S values of all clusters were greater than 0.7, indicating that the clustering results are convincing. The first 10 clustering tags (shown in Table 7, Figure 7) were “ISPD”, “sulfadoxine-pyrimethamine”, “drug discovery”, “ozonide”, “Plasmodium”, “artemisinin resistance”, “ferrocene”, “drug transport”, “medicinal plants”, and “animal models”. The timeline graph of co-cited references showed that “drug discovery”, “ozonide”, and “Plasmodium “ were the current research hotspots. Figure 8 shows the top 25 most frequently cited articles.
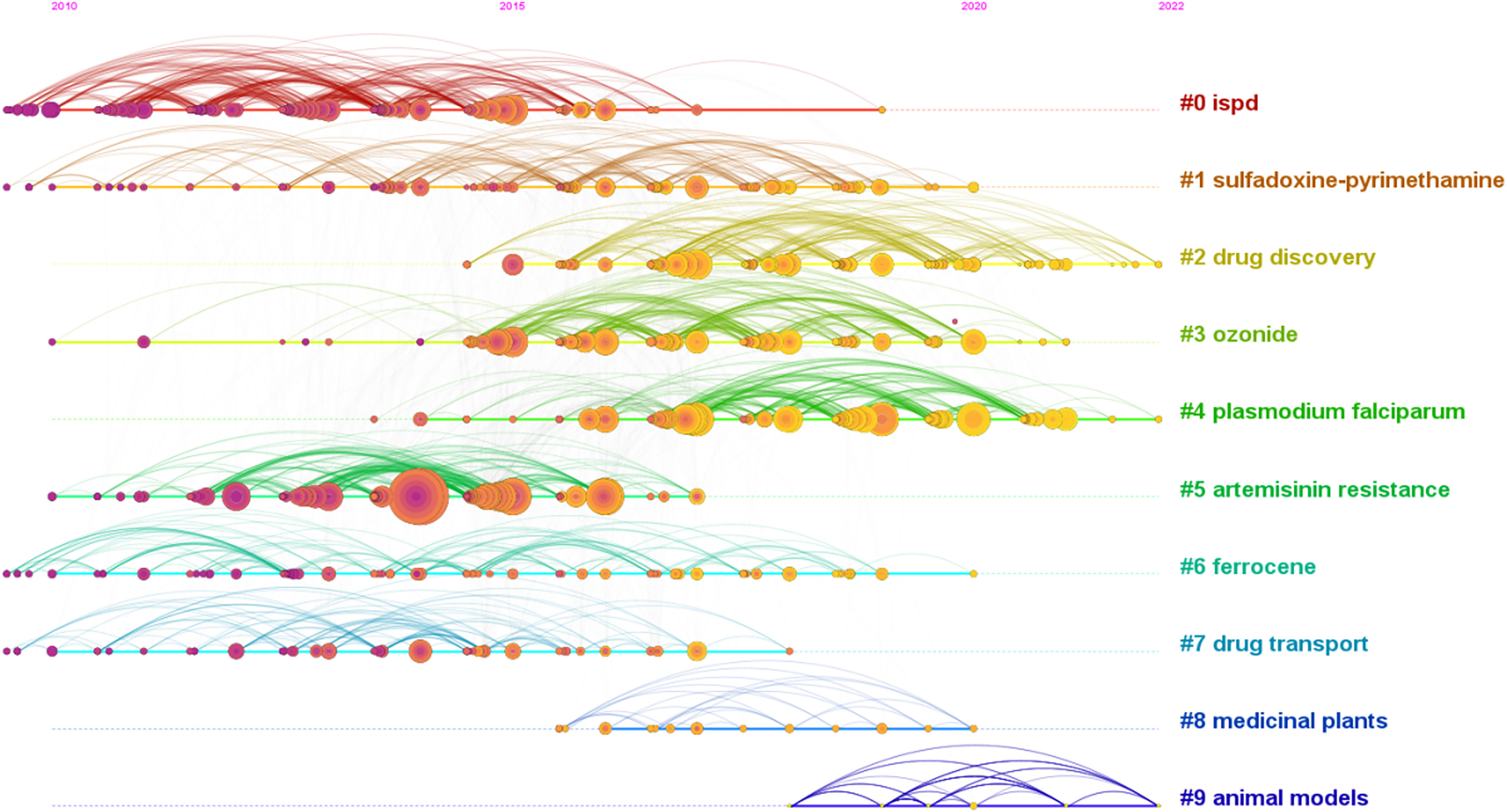
Figure 7 The timeline of co-cited literature showed that the current and future research hotspots in this field may be “new drug development” and “Plasmodium falciparum”.
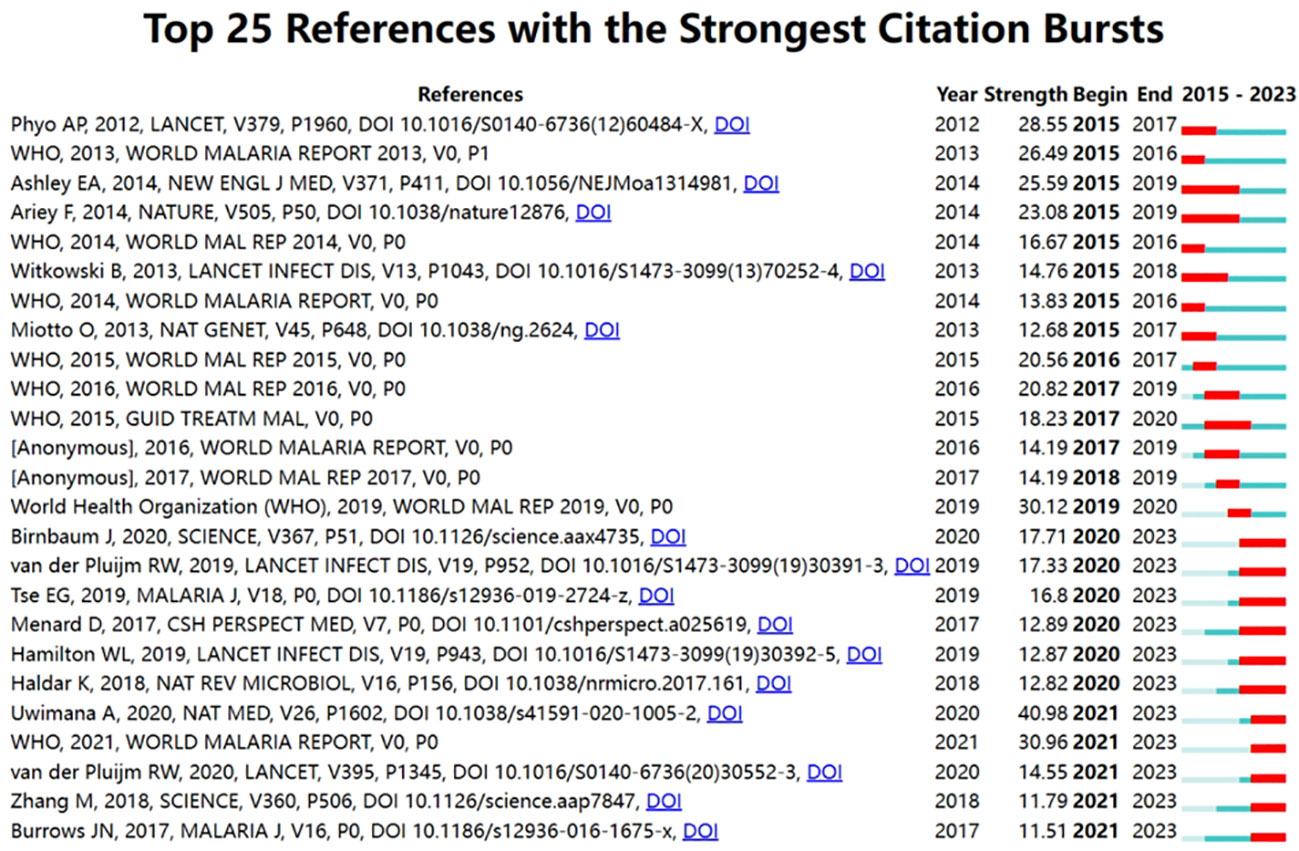
Figure 8 The top 25 most cited references in the field are presented, with the most cited reference being DOI: 10.1016/S0140-6736(12)60484-X.
The research hotspots in this field were the keywords that appear most frequently in the web co-occurrences. “Plasmodium falciparum”, “malaria”, “drug resistance”, “artemisinin resistance”, and “in vitro analysis” were high-frequency keywords (Table 8, Figure 9). The top 5 largest clusters of keywords were antimalarial activity, malaria parasite, artesunate, drug resistance, and artemisia annua (shown in Table 9).
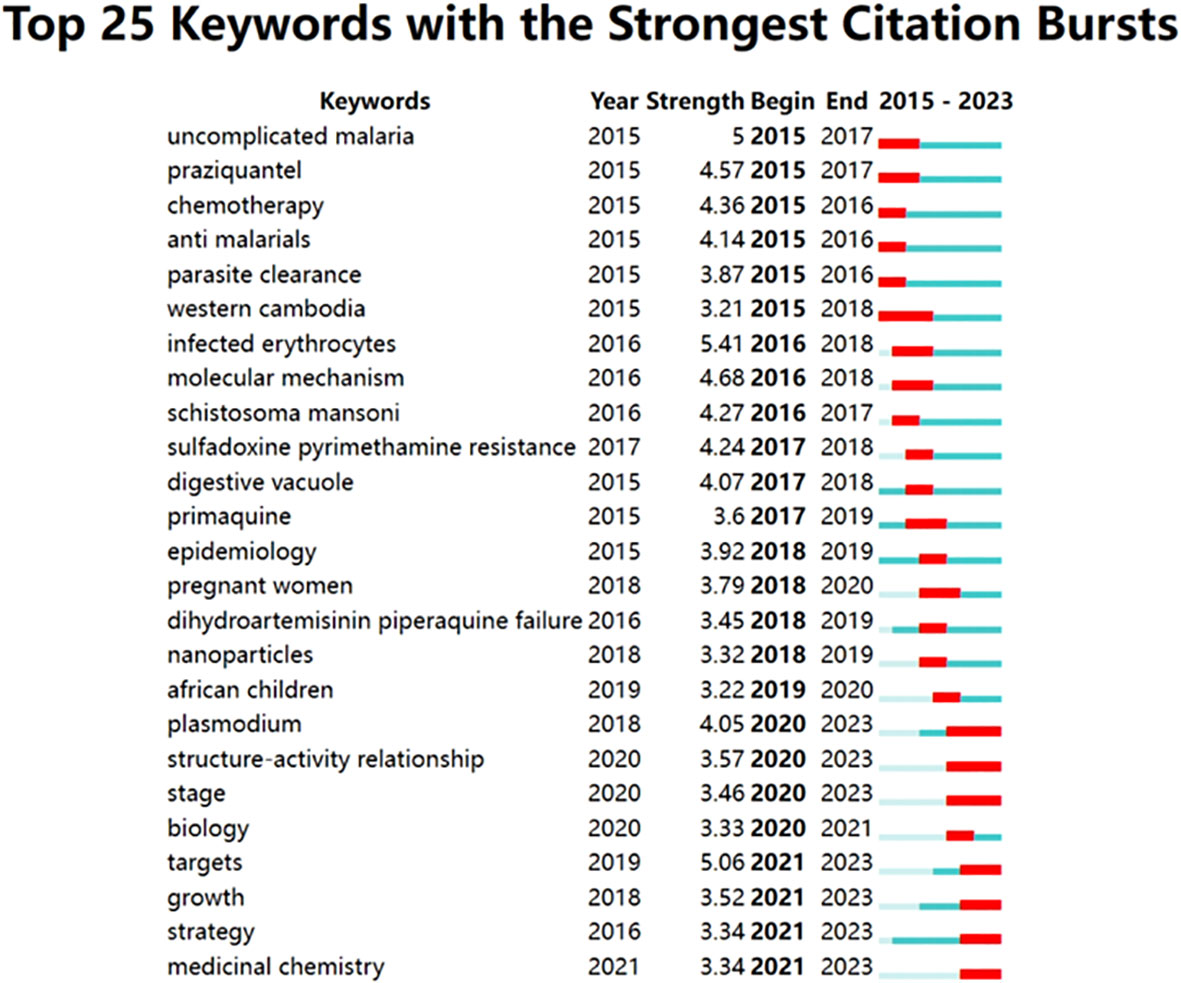
Figure 9 The chart presents the 25 most prominent keywords, their respective duration and intensity of prominence.
Between 2015 and 2023, a variable upward trend was observed in the literature pertaining to antimalarial medication. The USA boasts the highest number of published papers in this area. Alassane Mbengue and colleagues’ research, which has received 384 citations, stands as the most commonly cited paper in this field within the USA. Their publication explicates that PI3P serves as a critical mechanism regarding artemisinin resistance at the cellular and biochemical levels in P. falciparum and also identifies PfPI3K as a key target in the fight against malaria (Mbengue et al., 2015). Following this, India, UK, Australia, and other countries are listed.
Oxford University achieved the highest number of publications and the highest centrality ranking, with a total of 184 papers published. These papers delve into the evaluation of resistance activity in various regions, as well as the exploration of resistance mechanisms in P. falciparum and P. vivax (Pulcini et al., 2015; Yogavel et al., 2018). The study found that Fidock A. David, affiliated with Columbia University Medical Center in the USA, had published the most papers and had the strongest influence among the co-cited authors. He primarily worked on detecting malaria resistance genes in areas susceptible to the disease (Ross et al., 2018), researching drug resistance mechanisms resulting from P. falciparum gene mutations (Veiga et al., 2016), and developing novel antimalarial medications through diverse methods (Baragaña et al., 2015).
Moreover, the journal Antimicrobial Agents and Chemotherapy was the most co-cited publication. One of its most cited papers provided an overview of the essential characteristics and recent advances in understanding the biology, epidemiology, and clinical significance of malaria (Cowman et al., 2016). Leah S. has discovered a novel tyrosine amide compound, (S)-SW228703 [(S)-SW703]. Imlay et al. (2023) have demonstrated activity in both the asexual blood and hepatic stages (Imlay et al., 2023).
Through an examination of relevant keywords and significant literature, this paper presents a synthesis of the latest research trends concerning the topic of antimalarial drug resistance:
Artemisinin-based combination therapies (ACTs) are currently the primary treatment for malaria. The treatment mechanism involves swiftly eliminating both young and mature trophozoites with artemisinin, which is crucial in managing severe conditions and bringing about a cure (Dondorp et al., 2010), while the partner drug kills residual parasites by other means within a few weeks. Furthermore, ACTs exhibit multiple antimalarial mechanisms that contribute significantly towards their effectiveness. Wicht et al. (2020) discovered that the distinctive antimalarial impact of ACTs results mainly from the over-activation of their peroxide bridge by heme, an unavoidable by-product of the parasite’s hemoglobin metabolism in red blood cells. An extensive amount of free radicals could alkylate numerous Plasmodium proteins and biomolecules, ultimately causing rapid death of the parasite (Wicht et al., 2020). Artesunate, derived from artemisinin, targets several essential proteins in multiple P. falciparum pathways, including REDOX homeostasis, lipid metabolism, and protein synthesis. As such, it exhibits antimalarial effects (Gao, 2023).
Slow clearance of malaria parasites in vivo poses a new challenge for ACT treatment. It may jeopardize future global malaria control and elimination efforts. However, resistance to ACT drugs is not entirely comprehensive, the 3-day treatment is variable, and it is unclear whether extending the duration of treatment would boost the clearance rate of the malaria parasite. Presently, there are many ongoing efforts to tackle this predicament, with a primary focus on the resistance mechanism of P. falciparum against ACTs drugs. Blood samples were collected from areas where malaria is endemic, and samples were sequenced to obtain the drug-resistant mutant gene of P. falciparum using whole genome sequencing (Miotto et al., 2015). Numerous studies have indicated that the P. falciparum Kelch13 mutation is a significant factor in the slow parasite clearance rate in ACTs. Aline Uwimana et al. collected cases of pediatric malaria from three locations in Rwanda and observed delayed parasite clearance in over 10% of patients. The prevalence of the Pfkelch13 R561H mutation was significantly greater in patients with recurrent infection than in those who had cleared the infection (Woodrow and White, 2017). Yang et al. discovered that reduced K13 levels hindered hemoglobin breakdown and reduced artemisinin activation (Yang et al., 2019). This aligns with the conclusion by Wicht et al. that heme activation of artemisinin results in a distinct antimalarial mechanism.
To overcome the emergence of antimalarial drug resistance, there is an urgent need to develop antimalarial drugs with broad therapeutic potential and new modes of action. In conducting a keyword search, we have discovered that numerous new drugs possess certain antimalarial effects. A study has found that DDD107498 has effective and novel antimalarial activities against multiple life cycle stages of Plasmodium parasites, and its molecular target is extension factor 2 (eEF2) (Baragaña et al., 2015); Tanya Paquet et al. found that MMV390048 was effective against multiple stages of the Plasmodium life cycle using a whole-cell screen and that its molecular target was the Plasmodium parasite kinase phosphatidylinositol 4-kinase (Paquet et al., 2017). The chemical drug DSM265, which has entered the stage of clinical pharmacology research, is a novel dihydroorotate dehydrogenase inhibitor and has the prospect of collaborating with antimalarial drugs for drug development (McCarthy et al., 2017); 2-c-methyl-methyl-erythrin 4-phosphosidal transferase (ispd), an essential and effective antimalarial target, is active against both P. falciparum and P. vivax (Saggu et al., 2018). MMV008138 was found to selectively target ispd (Ghavami et al., 2018).
Ozonides are one of the most advanced drug classes in antimalarial drug development, aiming to improve the limitations associated with current artemisinin-based first-line therapies, in which peroxide bonds are a key factor in efficacy (Giannangelo et al., 2019). Artefenomel (OZ439), a novel synthetic trioxolane modification, has better pharmacokinetic properties than other antimalarial artemisinin pharmacophores (Phyo et al., 2016), and OZ439 remains effective against most parasite strains expressing mutant K13 in the case of parasite resistance (Straimer et al., 2017). Studies have shown that peroxide antimalarials disproportionately alkylate proteins involved in REDOX homeostasis and that the mechanism of action of these important antimalarials is related to the disruption of REDOX processes. Finding new antimalarial targets is one of the new ways to deal with resistance to existing antimalarial drugs (Siddiqui et al., 2022). These drugs have good pharmacokinetic profiles and acceptable safety profiles, which may become one of the antimalarial drugs of choice in the future.
Over the past 5 years, research into new antimalarial targets and the development of novel drugs have been the primary focus. As antimalarial drug resistance poses a significant challenge, emphasis is being placed on future development towards tackling this issue. The identification of new drug targets and the subsequent research into discovering effective antimalarial drugs have generated considerable interest within the field. Whole gene scanning and untargeted metabolomics research are currently the popular research methods (Challis et al., 2022). Co-occurrence analysis of the highest-ranked citations shows that “Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study” (Hamilton et al., 2019) indicates that drug resistance genes in Southeast Asia have garnered significant attention in this research area.
This paper presents the inaugural bibliometric visualization of antimalarial drug resistance. However, noteworthy limitations exist. The study solely comprises pertinent literature from the Web of Science (WOS) core database, excluding relevant content in other search engines such as PubMed and Google Scholar. As CiteSpace predominantly supports the WOS database, the core database acted as the data source to accommodate the software requirements. Other databases were seldom utilized for the analysis of the software. The software’s limitation to automatically eliminate duplicate entries from distinct databases prohibits its ability to conduct simultaneous analyses on two or more databases. In addition, the study’s merging of near-synonyms cannot completely eliminate possible analysis errors due to subjectivity.
The volume of literature has consistently shown an upward trend every year, signifying a persistent interest within the field. The visual analysis of authors originating from diverse nations, institutions, and journals displays their academic contributions without subjectivity while offering potential collaboration for future research. An overview of literature and keywords within the field reveals research priorities and patterns in antimalarial drug resistance. For instance, developing new drugs represents a current priority, while screening new drug targets using network models could emerge as a future trend. Scholarly peers can reference this analysis to gain insights into the direction of research in this field.
JZ: Visualization, Data curation, Writing – original draft. MS: Writing – review & editing. MI: Writing – review & editing. HZ: Writing – review & editing, Conceptualization, Visualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (No. 82004037), Introduce innovative team projects of Jinan (No. 202228029), Traditional Chinese Medicine Science and Technology Project of Shandong Province (M-2022173), and Academician Workstation of Jinnan. All sponsors provided software support, as well as data download support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Anantabotla, V. M., Antony, H. A., Parija, S. C., Rajkumari, N., Kini, J. R., Manipura, R., et al. (2019). Polymorphisms in genes associated with drug resistance of Plasmodium vivax in India. Parasitol. Int. 70, 92–97. doi: 10.1016/j.parint.2019.03.001
Ariey, F., Witkowski, B., Amaratunga, C., Beghain, J., Langlois, A.-C., Khim, N., et al. (2014). A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505, 50–55. doi: 10.1038/nature12876
Ashley, E. A., Dhorda, M., Fairhurst, R. M., Amaratunga, C., Lim, P., Suon, S. (2014). Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 371 (5), 411–423. doi: 10.1056/NEJMoa1314981
Balikagala, B., Fukuda, N., Ikeda, M., Katuro, O. T., Tachibana, S.-I., Yamauchi, M., et al. (2021). Evidence of artemisinin-resistant malaria in Africa. N. Engl. J. Med. 385, 1163–1171. doi: 10.1056/NEJMoa2101746
Baragaña, B., Hallyburton, I., Lee, M. C. S., Norcross, N. R., Grimaldi, R., Otto, T. D., et al. (2015). A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature 522, 315–320. doi: 10.1038/nature14451
Bhatt, S., Weiss, D. J., Cameron, E., Bisanzio, D., Mappin, B., Dalrymple, U., et al. (2015). The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 526, 207–211. doi: 10.1038/nature15535
Challis, M. P., Devine, S. M., Creek, D. J. (2022). Current and emerging target identification methods for novel antimalarials. Int. J. Parasitol. Drugs Drug Resist. 20, 135–144. doi: 10.1016/j.ijpddr.2022.11.001
Chen, Y., Li, Y., Guo, L., Hong, J., Zhao, W., Hu, X., et al. (2021). Bibliometric analysis of the inflammasome and pyroptosis in brain. Front. Pharmacol. 11. doi: 10.3389/fphar.2020.626502
Conrad, M. D., Asua, V., Garg, S., Giesbrecht, D., Niaré, K., Smith, S., et al. (2023). Evolution of partial resistance to artemisinins in malaria parasites in Uganda. N. Engl. J. Med. 389, 722–732. doi: 10.1056/NEJMoa2211803
Cowman, A. F., Healer, J., Marapana, D., Marsh, K. (2016). Malaria: biology and disease. Cell 167, 610–624. doi: 10.1016/j.cell.2016.07.055
Dondorp, A. M., Fanello, C. I., Hendriksen, I. C. E., Gomes, E., Seni, A., Chhaganlal, K. D., et al. (2010). Artesunate versus quinine in the treatment of severe falciparum malaria in African children (AQUAMAT): an open-label, randomised trial. Lancet Lond. Engl. 376, 1647–1657. doi: 10.1016/S0140-6736(10)61924-1
Eastman, R. T., Fidock, D. A. (2009). Artemisinin-based combination therapies: a vital tool in efforts to eliminate malaria. Nat. Rev. Microbiol. 7, 864–874. doi: 10.1038/nrmicro2239
Fact sheet about malaria. Available at: https://www.who.int/news-room/fact-sheets/detail/malaria (Accessed July 3, 2023).
Fola, A. A., Feleke, S. M., Mohammed, H., Brhane, B. G., Hennelly, C. M., Assefa, A., et al. (2023). Plasmodium falciparum resistant to artemisinin and diagnostics have emerged in Ethiopia. Nat. Microbiol. 8, 1911–1919. doi: 10.1038/s41564-023-01461-4
Gao, P. (2023). Profiling the Antimalarial Mechanism of Artemisinin by Identifying Crucial Target Proteins. doi: 10.1016/j.eng.2023.06.001
Ghavami, M., Merino, E. F., Yao, Z.-K., Elahi, R., Simpson, M. E., Fernández-Murga, M. L., et al. (2018). Biological studies and target engagement of the 2- C-methyl-d-erythritol 4-phosphate cytidylyltransferase (IspD)-targeting antimalarial agent (1 R,3 S)-MMV008138 and analogs. ACS Infect. Dis. 4, 549–559. doi: 10.1021/acsinfecdis.7b00159
Giannangelo, C., Fowkes, F. J. I., Simpson, J. A., Charman, S. A., Creek, D. J. (2019). Ozonide antimalarial activity in the context of artemisinin-resistant malaria. Trends Parasitol. 35, 529–543. doi: 10.1016/j.pt.2019.05.002
Hamilton, W. L., Amato, R., van der Pluijm, R. W., Jacob, C. G., Quang, H. H., Thuy-Nhien, N. T., et al. (2019). Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect. Dis. 19, 943–951. doi: 10.1016/S1473-3099(19)30392-5
Hussien, M., Abdel Hamid, M. M., Elamin, E. A., Hassan, A. O., Elaagip, A. H., Salama, A. H. A., et al. (2020). Antimalarial drug resistance molecular makers of Plasmodium falciparum isolates from Sudan during 2015-2017. PloS One 15, e0235401. doi: 10.1371/journal.pone.0235401
Imlay, L. S., Lawong, A. K., Gahalawat, S., Kumar, A., Xing, C., Mittal, N., et al. (2023). Fast-killing tyrosine amide ((S)-SW228703) with blood- and liver-stage antimalarial activity associated with the cyclic amine resistance locus (PfCARL). ACS Infect. Dis. 9, 527–539. doi: 10.1021/acsinfecdis.2c00527
Kiarie, H., Temmerman, M., Nyamai, M., Liku, N., Thuo, W., Oramisi, V., et al. (2022). The COVID-19 pandemic and disruptions to essential health services in Kenya: a retrospective time-series analysis. Lancet Glob. Health 10, e1257–e1267. doi: 10.1016/S2214-109X(22)00285-6
Landier, J., Parker, D. M., Thu, A. M., Lwin, K. M., Delmas, G., Nosten, F. H., et al. (2018). Effect of generalised access to early diagnosis and treatment and targeted mass drug administration on Plasmodium falciparum malaria in Eastern Myanmar: an observational study of a regional elimination programme. Lancet 391, 1916–1926. doi: 10.1016/S0140-6736(18)30792-X
Mbengue, A., Bhattacharjee, S., Pandharkar, T., Liu, H., Estiu, G., Stahelin, R. V., et al. (2015). A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature 520, 683–687. doi: 10.1038/nature14412
McCarthy, J. S., Lotharius, J., Rückle, T., Chalon, S., Phillips, M. A., Elliott, S., et al. (2017). Safety, tolerability, pharmacokinetics, and activity of the novel long-acting antimalarial DSM265: a two-part first-in-human phase 1a/1b randomised study. Lancet Infect. Dis. 17, 626–635. doi: 10.1016/S1473-3099(17)30171-8
McLean, A. R. D., Wai, H. P., Thu, A. M., Khant, Z. S., Indrasuta, C., Ashley, E. A., et al. (2018). Malaria elimination in remote communities requires integration of malaria control activities into general health care: an observational study and interrupted time series analysis in Myanmar. BMC Med. 16, 183. doi: 10.1186/s12916-018-1172-x
Ménard, D., Clain, J., Ariey, F. (2018). Multidrug-resistant Plasmodium falciparum malaria in the Greater Mekong subregion. Lancet Infect. Dis. 18, 238–239. doi: 10.1016/S1473-3099(18)30071-9
Ménard, D., Khim, N., Beghain, J., Adegnika, A. A., Shafiul-Alam, M., Amodu, O., et al. (2016). A worldwide map of plasmodium falciparum K13-propeller polymorphisms. N. Engl. J. Med. 374, 2453–2464. doi: 10.1056/NEJMoa1513137
Mihreteab, S., Platon, L., Berhane, A., Stokes, B. H., Warsame, M., Campagne, P., et al. (2023). Increasing prevalence of artemisinin-resistant HRP2-negative malaria in Eritrea. N. Engl. J. Med. 389, 1191–1202. doi: 10.1056/NEJMoa2210956
Miotto, O., Amato, R., Ashley, E. A., MacInnis, B., Almagro-Garcia, J., Amaratunga, C., et al. (2015). Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet. 47, 226–234. doi: 10.1038/ng.3189
Narh, C. A., Ghansah, A., Duffy, M. F., Ruybal-Pesántez, S., Onwona, C. O., Oduro, A. R., et al. (2020). Evolution of antimalarial drug resistance markers in the reservoir of plasmodium falciparum infections in the upper east region of Ghana. J. Infect. Dis. 222, 1692–1701. doi: 10.1093/infdis/jiaa286
Norman, F. F., Treviño-Maruri, B., Ruiz Giardín, J. M., Gullón-Peña, B., Salvador, F., Serre, N., et al. (2022). Trends in imported malaria during the COVID-19 pandemic, Spain (+Redivi Collaborative Network). J. Travel Med. 29, taac083. doi: 10.1093/jtm/taac083
Owoloye, A., Olufemi, M., Idowu, E. T., Oyebola, K. M. (2021). Prevalence of potential mediators of artemisinin resistance in African isolates of Plasmodium falciparum. Malar. J. 20, 451. doi: 10.1186/s12936-021-03987-6
Paquet, T., Le Manach, C., Cabrera, D. G., Younis, Y., Henrich, P. P., Abraham, T. S., et al. (2017). Antimalarial efficacy of MMV390048, an inhibitor of Plasmodium phosphatidylinositol 4-kinase. Sci. Transl. Med. 9, eaad9735. doi: 10.1126/scitranslmed.aad9735
Phyo, A. P., Jittamala, P., Nosten, F. H., Pukrittayakamee, S., Imwong, M., White, N. J., et al. (2016). Antimalarial activity of artefenomel (OZ439), a novel synthetic antimalarial endoperoxide, in patients with Plasmodium falciparum and Plasmodium vivax malaria: an open-label phase 2 trial. Lancet Infect. Dis. 16, 61–69. doi: 10.1016/S1473-3099(15)00320-5
Pulcini, S., Staines, H. M., Lee, A. H., Shafik, S. H., Bouyer, G., Moore, C. M., et al. (2015). Mutations in the Plasmodium falciparum chloroquine resistance transporter, PfCRT, enlarge the parasite’s food vacuole and alter drug sensitivities. Sci. Rep. 5, 14552. doi: 10.1038/srep14552
Ross, L. S., Dhingra, S. K., Mok, S., Yeo, T., Wicht, K. J., Kümpornsin, K., et al. (2018). Emerging Southeast Asian PfCRT mutations confer Plasmodium falciparum resistance to the first-line antimalarial piperaquine. Nat. Commun. 9, 3314. doi: 10.1038/s41467-018-05652-0
Saggu, G. S., Garg, S., Pala, Z. R., Kochar, S. K., Saxena, V. (2018). Deciphering the role of IspD (2−C−methyl−D−erythritol 4−phosphate cytidyltransferase) enzyme as a potential therapeutic drug target against Plasmodium vivax. Gene 675, 240–253. doi: 10.1016/j.gene.2018.06.084
Schultz, J. S., Mace, K. E., Tan, K. R. (2023). Return to travel in the coronavirus disease 2019 pandemic recovery period and implications for imported malaria: reinforcing prevention, early diagnosis, and appropriate treatment of malaria. Clin. Infect. Dis. 76 (7), 1161–1163. doi: 10.1093/cid/ciad061
Shibeshi, M. A., Kifle, Z. D., Atnafie, S. A. (2020). Antimalarial drug resistance and novel targets for antimalarial drug discovery. Infect. Drug Resist. 13, 4047–4060. doi: 10.2147/IDR.S279433
Siddiqui, G., Giannangelo, C., De Paoli, A., Schuh, A. K., Heimsch, K. C., Anderson, D., et al. (2022). Peroxide antimalarial drugs target redox homeostasis in plasmodium falciparum infected red blood cells. ACS Infect. Dis. 8, 210–226. doi: 10.1021/acsinfecdis.1c00550
Straimer, J., Gnädig, N. F., Stokes, B. H., Ehrenberger, M., Crane, A. A., Fidock, D. A. (2017). Plasmodium falciparum K13 mutations differentially impact ozonide susceptibility and parasite fitness in vitro. mBio 8, e00172–e00117. doi: 10.1128/mBio.00172-17
Straimer, J., Gnädig, N. F., Witkowski, B., Amaratunga, C., Duru, V., Pramundita Ramadani, A. (2015). Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347 (6220), 428–31. doi: 10.1126/science.1260867
Uwimana, A., Legrand, E., Stokes, B. H., Ndikumana, J.-L. M., Warsame, M., Umulisa, N., et al. (2020). Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 26, 1602–1608. doi: 10.1038/s41591-020-1005-2
Uwimana, A., Umulisa, N., Venkatesan, M., Svigel, S. S., Zhou, Z., Munyaneza, T., et al. (2021). Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: an open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect. Dis. 21, 1120–1128. doi: 10.1016/S1473-3099(21)00142-0
Veiga, M. I., Dhingra, S. K., Henrich, P. P., Straimer, J., Gnädig, N., Uhlemann, A.-C., et al. (2016). Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat. Commun. 7, 11553. doi: 10.1038/ncomms11553
White, N. J. (2004). Antimalarial drug resistance. J. Clin. Invest. 113, 1084–1092. doi: 10.1172/JCI21682
Wicht, K. J., Mok, S., Fidock, D. A. (2020). Molecular mechanisms of drug resistance in plasmodium falciparum malaria. Annu. Rev. Microbiol. 74, 431–454. doi: 10.1146/annurev-micro-020518-115546
Woodrow, C. J., White, N. J. (2017). The clinical impact of artemisinin resistance in Southeast Asia and the potential for future spread. FEMS Microbiol. Rev. 41, 34–48. doi: 10.1093/femsre/fuw037
World Health Organization. (2013). World malaria report. Available at: https://www.who.int/publications/i/item/9789241564694 [Accessed December 09, 2023].
World Health Organization (2015). Guidelines for the treatment of malaria, 3rd ed. Available at: https://iris.who.int/handle/10665/162441 [Accessed December 09, 2023].
Yang, T., Yeoh, L. M., Tutor, M. V., Dixon, M. W., McMillan, P. J., Xie, S. C., et al. (2019). Decreased K13 abundance reduces hemoglobin catabolism and proteotoxic stress, underpinning artemisinin resistance. Cell Rep. 29, 2917–2928.e5. doi: 10.1016/j.celrep.2019.10.095
Yogavel, M., Nettleship, J. E., Sharma, A., Harlos, K., Jamwal, A., Chaturvedi, R., et al. (2018). Structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase–dihydropteroate synthase from Plasmodium vivax sheds light on drug resistance. J. Biol. Chem. 293, 14962–14972. doi: 10.1074/jbc.RA118.004558
Keywords: bibliometric analysis, antimalarial drug, resistance, anti-malarial, CiteSpace
Citation: Zhang J, Shahbaz M, Ijaz M and Zhang H (2024) Bibliometric analysis of antimalarial drug resistance. Front. Cell. Infect. Microbiol. 14:1270060. doi: 10.3389/fcimb.2024.1270060
Received: 05 September 2023; Accepted: 02 January 2024;
Published: 12 February 2024.
Edited by:
Martin Craig Taylor, University of London, United KingdomReviewed by:
Jyoti Chhibber-Goel, International Centre for Genetic Engineering and Biotechnology, IndiaCopyright © 2024 Zhang, Shahbaz, Ijaz and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huimin Zhang, aHVpbWluemhhbmdAMTYzLmNvbQ==; Muhammad Ijaz, ZHJtdWhhbW1hZGlqYXoxQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.