- 1Faculty of Medicine, University of Zurich, Zurich, Switzerland
- 2Department of Medicine, Mount Auburn Hospital, Cambridge, MA, United States
- 3Harvard Medical School, Boston, MA, United States
- 4Mahidol University, Faculty of Medicine Siriraj Hospital, Bangkok, Thailand
Purpose: Raoultella spp. is a genus of bacteria that is known to be closely related to Klebsiella. It has been debated whether Raoultella should be reclassified as a subgroup of Klebsiella. The aim of this study is to compare clinical aspects of Raoultella and Klebsiella oxytoca, a species of Klebsiella that is known to be bacteriologically similar to Raoultella spp.
Methods: Using data collected at a tertiary care hospital in the United States, we identified 43 patients with Raoultella infection and 1173 patients with Klebsiella oxytoca infection. We compared patient demographics (age and sex), hospitalization status, isolation sites and antibiotic resistance profiles between the two species.
Results: There was no significant difference in patient demographics between the two bacteria species. The proportions of intensive care unit (ICU) admission were higher among patients with Raoultella infection (p=0.008). The most common site of isolation was urine for both species (39.5% of all patients with Raoultella spp. vs. 59.3% for K. oxytoca). The second most common site of isolation was blood stream for Raoultella spp. (23.3%) and respiratory tract for K. oxytoca (10.8%). Except for the high proportion of resistant isolates of Raoultella spp. for Trimethoprim/sulfamethoxazole, the antibiotic susceptibility profiles were similar between the two bacteria species. Both were susceptible to ciprofloxacin and meropenem.
Conclusion: While there are no significant differences in the patient demographics and antibiotic susceptibility profiles between Raoultella spp. and K. oxytoca, Raoultella may cause more serious infection requiring ICU admissions. Also, Raoultella may cause blood stream infection more frequently than K. oxytoca.
1 Introduction
Raoultella spp. are Gram-negative encapsulated aerobic rods that belong to the family Enterobacteriaceae. This organism was initially classified in cluster II of genus Klebsiella as Klebsiella ornithinolytica until 2001, when cluster II of the genus Klebsiella was renamed to its own genus, Raoultella, named after Didier Raoult, a French physician/microbiologist (Hajjar et al., 2020).
R. ornithinolytica and R. planticola can be found naturally in water environments, fish and insects and are known to cause uncommon infections in humans (Hajjar et al., 2020; Pi et al., 2020; Chen et al., 2021). There are a few recent reports of Raoultella colonization and infection in children and adults of varying health status (Naganathan and Amin, 2018; Hajjar et al., 2020; Pi et al., 2020; Chen et al., 2021). Given the rarity of its infection, data on the antimicrobial resistance profile of this organism is relatively limited. It was reported in four university hospitals in France that a significant proportion of R. ornithinolytica isolates were resistant to ceftriaxone (4%), quinolones (6%) and trimethoprim/sulfamethoxazole (13%) (Seng et al., 2016).
There have been debates as to whether Raoultella spp. should be taxonomically reclassified as a subgroup of the genus Klebsiella. A recent genetic analysis of Raoultella and Klebsiella suggested that they are genetically close enough to belong to one genus (Ma et al., 2021). Klebsiella oxytoca, an enteric flora with emerging pathogenic potential, is so similar to Raoultella in its morphology and growth pattern that R. ornithinolytica can be sometimes misclassified as Klebsiella oxytoca (Alves et al., 2006; Park et al., 2011). It remains unclear whether Raoultella spp. should be treated differently from Klebsiella in clinical settings.
The objective of this study is to compare clinical aspects of Raoultella spp. and Klebsiella oxytoca. In addition, we analyzed the antibiotic resistance profile of Raoultella spp. and compared it to that of Klebsiella oxytoca to help guide the antimicrobial management of this uncommon organism.
2 Materials and method
2.1 Data source
Our study is based on data collected at an academic tertiary care center in Boston, Massachusetts, between 2008 and 2019 (Johnson et al.). This database, MIMIC-IV, contains de-identified clinical data of patients who were seen at the Emergency Department (ED) or admitted to one of the Intensive Care Units (ICU) during the study period. Patients who were hospitalized after presenting to the ED were included in the data, but those who were hospitalized through other routes bypassing the ED (such as hospital-to-hospital transfers) were not included, unless they were admitted to the ICU. As our study is a retrospective analysis of publicly available data, an individual ethics committee approval was waived.
2.2 Patients and inclusion criteria
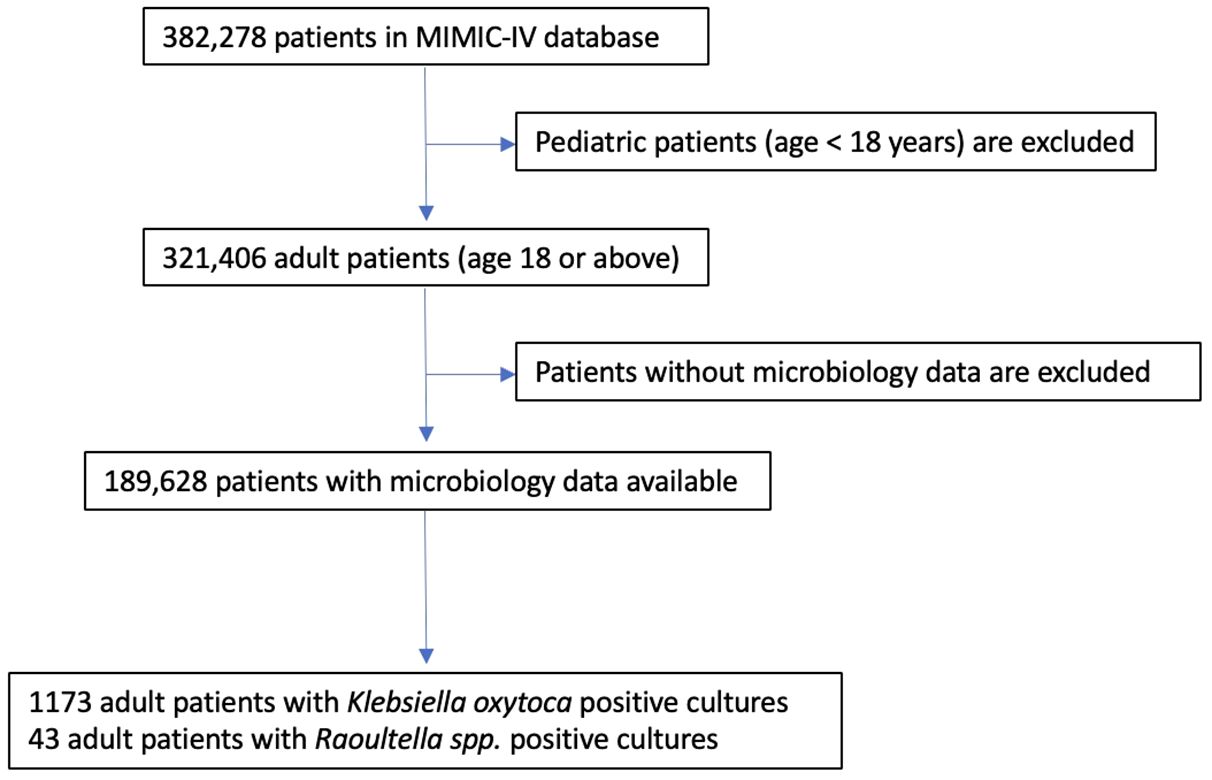
The patient list of the utilized database contains 382,278 unique individuals. Pediatric patients are excluded, and only adult patients (18 years-old or older) are included in our study. A flow chart of the inclusion criteria is shown in Figure 1.
2.3 Microbiology data
The database contains all microbiology laboratory events of the above patient population recorded in the hospital electronic health record systems. The microbiology specimens were mostly collected at the ED, during a hospitalization or during an ICU stay. In rare cases, some specimens may have been collected at an affiliated outpatient office and the results are shared in the system when patients visited ED or are admitted to ICU. The data include specimen collection time, specimen source, types of laboratory tests, organisms detected, antibiotic susceptibility studies and MIC dilution values.
We analyzed all microbiology events where Raoultella spp. or Klebsiella oxytoca were detected. The Raoultella species detected were either R. ornithinolytica or R. planticola.
2.4 MIC dilution values and antibiotic susceptibility interpretation
We re-interpreted the susceptibility studies of each antibiotic agent, using the most recent guidelines and MIC breakpoints published by the Clinical Laboratory Standards Institute (Lewis, 2022). When the MIC dilution value is missing while the original interpretation of susceptibility was resistant, we considered the new interpretation to be also resistant.
The proportions of missing values by each antibiotic agent and its original susceptibility interpretation are available in the Supplementary Material.
2.5 Statistical methods
For descriptive statistics, numbers of unique specimens and numbers of unique patients are reported separately. Descriptive statistics of patients’ age are computed at the specimen level. For example, if a patient had two specimens collected, one at age 30 and another at age 50, both ages were counted at the average age. Sex statistics are computed at the patient level. P-values are computed using two sample t-tests for continuous variables and two-sample z-tests for proportions. Two sample t-tests or two sample z-tests are performed only on the patient level but not on the specimen level.
The overall antibiotic susceptibility is defined as the number of sensitive specimens to the given antibiotic divided by the number specimens tested for the antibiotic. Each antibiotic susceptibility interpretation at the specimen level, not at the patient level, is used to compute the overall antibiotic susceptibility. In order to avoid bias caused by repeated bacterial cultures within the same patient, the 95% confidence interval for the overall antibiotic susceptibility is computed using a cluster bootstrap method. Each patient is considered to be a cluster. Five thousand bootstrap samples are drawn for each antibiotic and its susceptibility is computed for each bootstrap sample. The 95% bootstrap confidence interval is given by the 2.5th and 97.5th percentiles of the bootstrap estimates.
All statistical analyses are performed using the statistics software R.
3 Results
3.1 Patient demographics
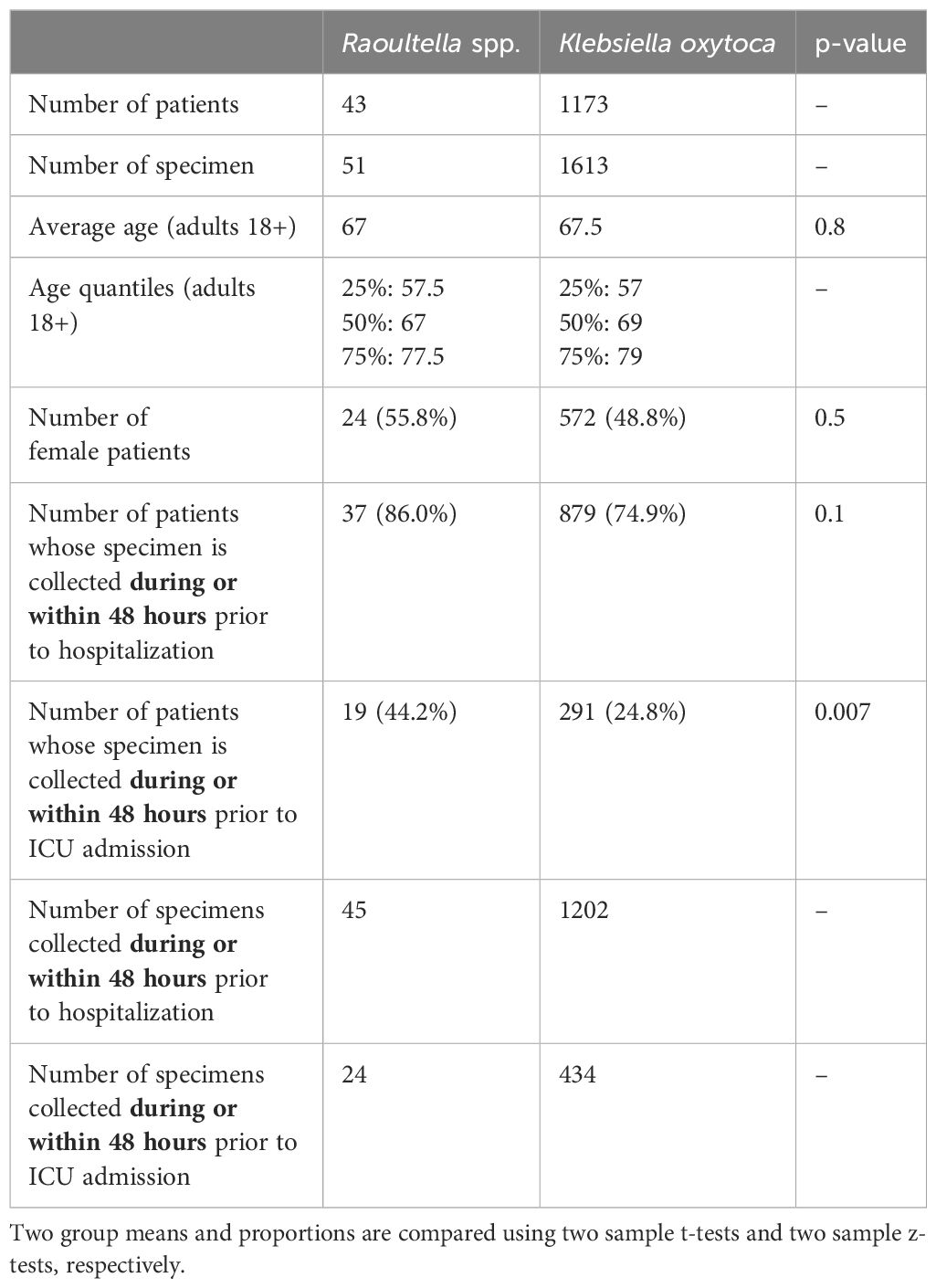
Table 1 summarizes the numbers of patients and specimens, patient demographics (age and sex) and hospitalization status. A total of 51 specimens obtained from 43 patients grew Raoultella spp. and 1613 specimens from 1173 patients grew Klebsiella oxytoca. Among adult patients (18 years old or older), the average age at the time of positive culture was 67 years for Raoultella spp. and 67.5 years for K. oxytoca (p-value = 0.8). The proportion of female patients was higher among those with Raoultella positive cultures (24 patients, 55.8%) than among those with K. oxytoca positive cultures (572 patients, 48.8%). However, this difference was not statistically significant (p-value = 0.5).
Among 43 patients with Raoultella positive cultures, 37 patients (86.0%) were hospitalized or became hospitalized within 48 hours of specimen collection. This proportion of hospitalization was lower among patients with K. oxytoca positive cultures (879 patients, 74.9%), but the difference did not reach statistical significance (p-value = 0.1). For ICU admissions, however, patients with Raoultella positive cultures were significantly more likely to be admitted to ICU at the time of or within 48 hours of specimen collection compared to patients with K. oxytoca positive cultures (44.2% vs. 24.8%, p-value = 0.007).
Results only including blood stream infection are available in the Supplementary Material.
3.2 Culture sites
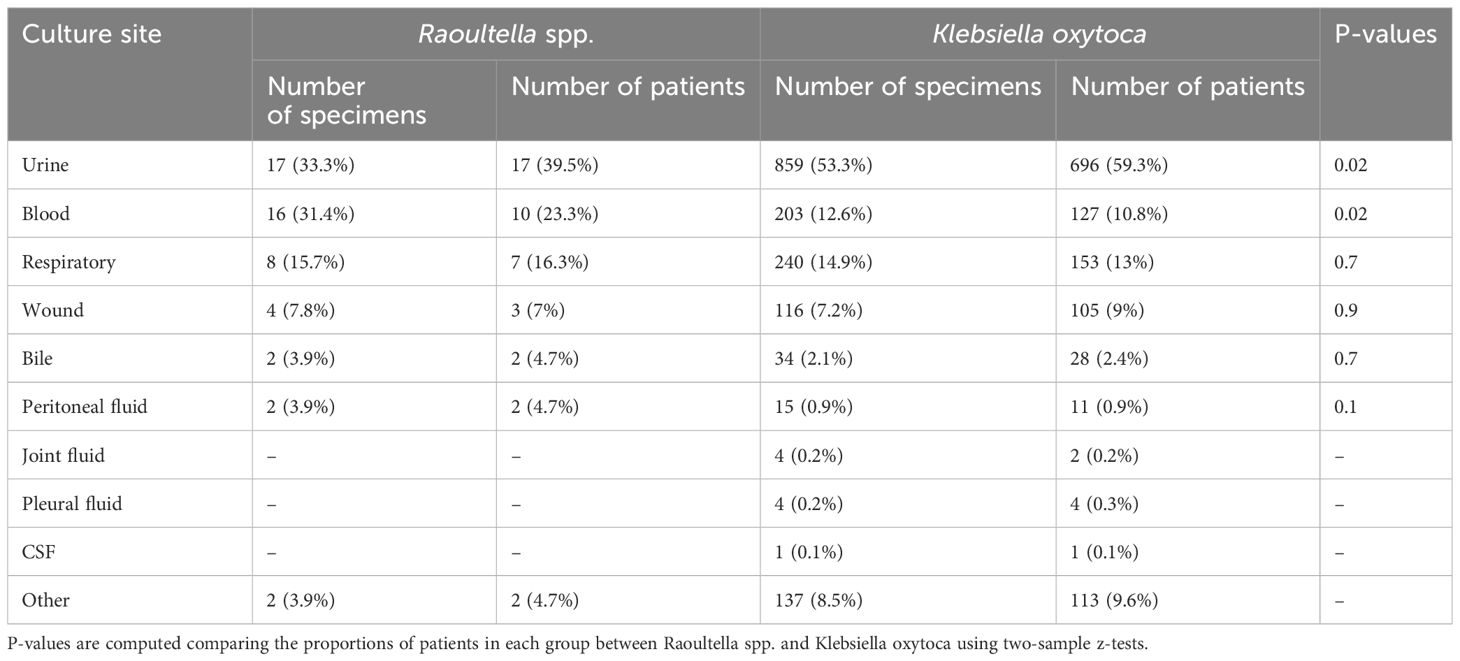
The numbers and proportions of culture sites are shown in Table 2A. For Raoultella spp., the most common culture site was urine (30.9%), closely followed by blood (29.1%) and respiratory tract (21.8%). K. oxytoca was predominantly found in urine (52.6%), followed by respiratory tract (15.8%) and blood (12.4%). The proportion of urinary tract isolation was significantly higher among patients with K. oxytoca than among patients with Raoultella (p-value = 0.01). The proportion of blood stream infection was significantly higher among Raoultella patients than among K. oxytoca patients (p-value = 0.02). Other rarer sites included wound, bile, and peritoneal fluid.
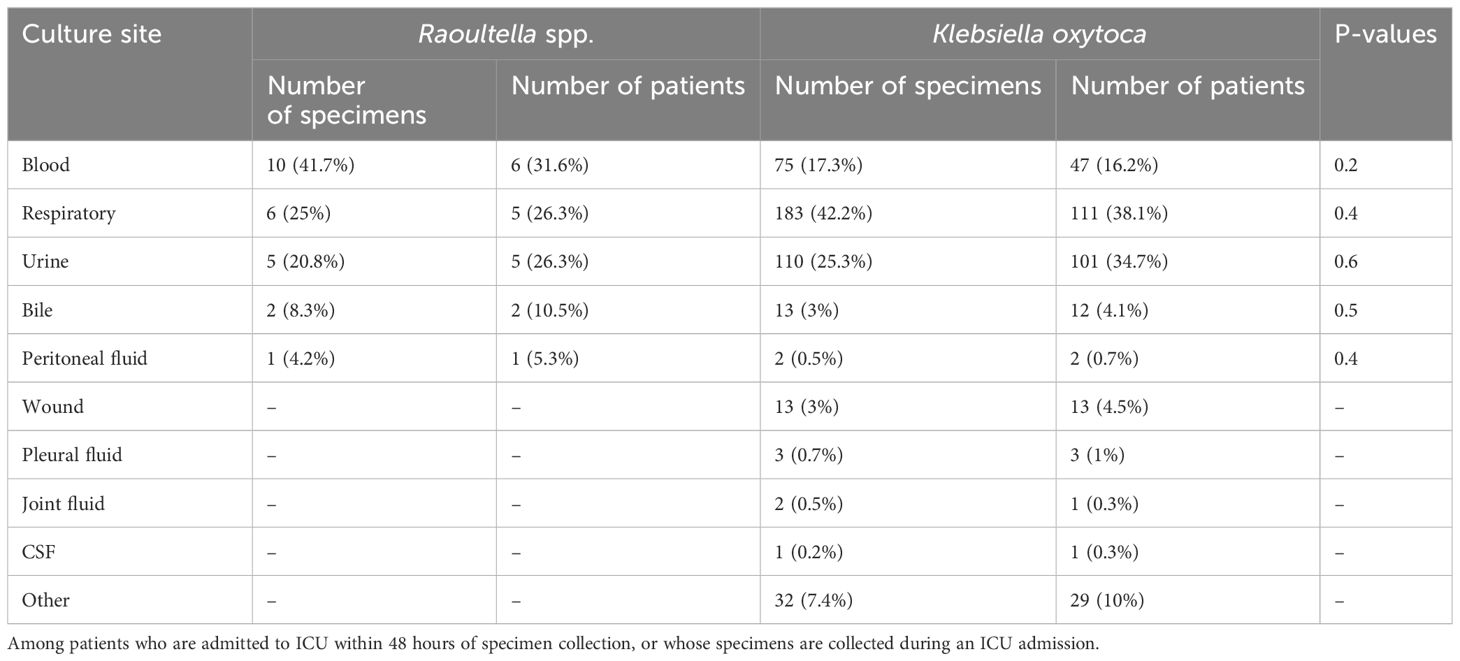
We compared the stratified results by the clinical context (ICU admission, hospitalization and no hospitalization). Table 2B shows comparison of cultures sites among patients who were admitted to ICU at the time of or within 48 hours of specimen collection. Comparison tables of culture sites among hospitalized patients and those who were not hospitalized are available in the Supplementary Material. When only severely ill patients who needed to be admitted to ICU are compared, blood stream infection accounted for the majority (6 patients, 31.6%) among Raoultella patients, while respiratory infection was most common (111 patients, 38.1%) among K. oxytoca patients. Among K. oxytoca patients admitted to ICU, the proportion of blood stream infection was lower with 16.2% than 31.6% among Raoultella patients. However, this difference was not statistically significant (p-value = 0.2).
3.3 Antibiotic resistance
MIC50, MIC 90 and antibiotic susceptibility for different antibiotics are shown in Table 3. The number of specimens tested for each antibiotic is listed also shown in Table 3. Antibiotic susceptibility and 95% confidence intervals are also shown in Figure 2. All confidence intervals for antibiotic susceptibility overlapped between Raoultella spp. and K. oxytoca except for Trimethoprim/Sulfamexthoxazole. Both Raoultella spp. and K. oxytoca were very susceptible to meropenem, piperacillin/tazobactam, and ciprofloxacin. Although the sample sizes are small, both species were also susceptible to levofloxacin and amikacin. Both Raoultella spp. and K. oxytoca were less sensitive to cefazolin (MIC50 of 4 and 8, respectively and MIC90 of 64 for both) and ampicillin/sulbactam (MIC50 of 4 and 8, respectively and MIC90 of 32 for both).
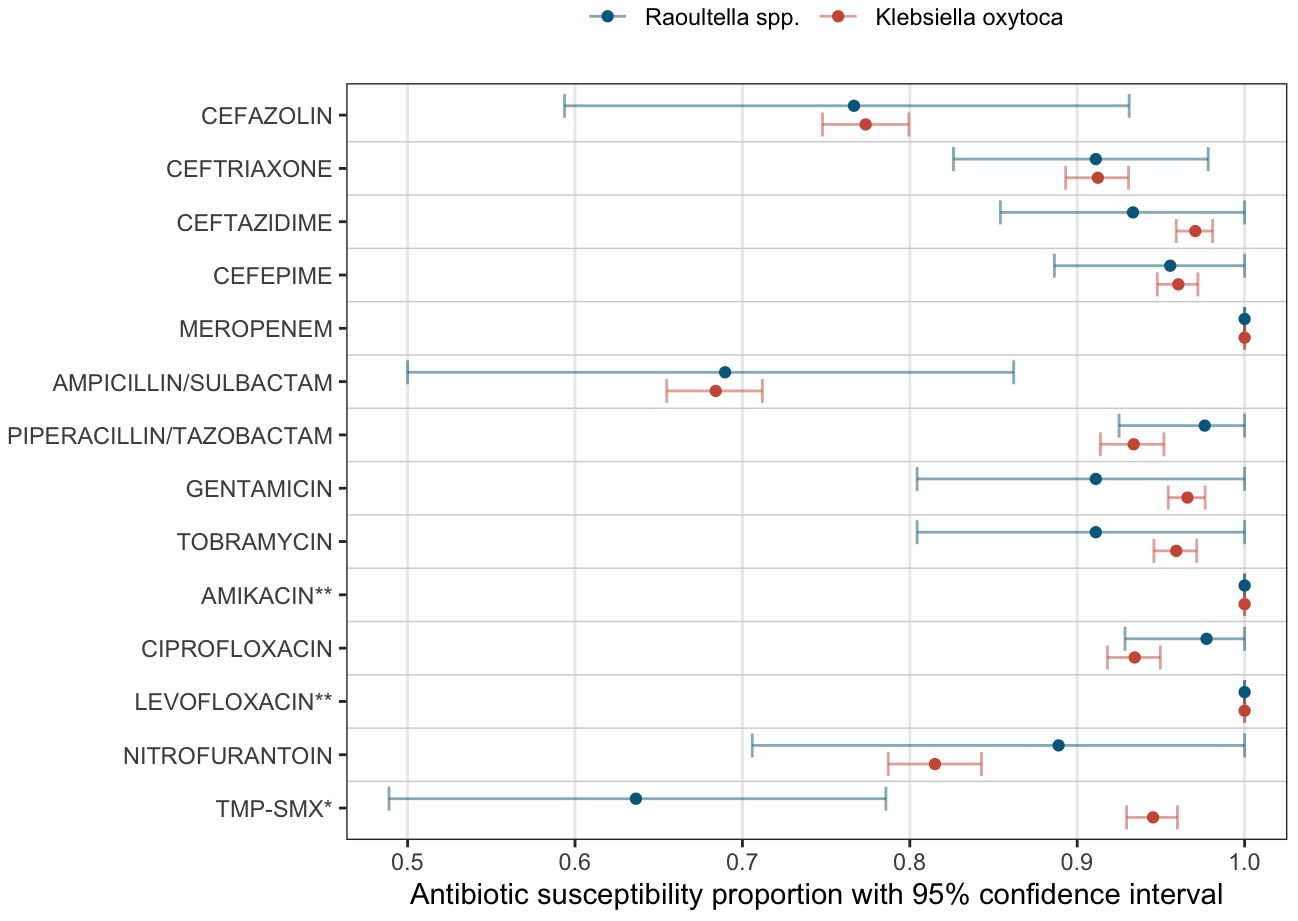
Figure 2 Antibiotic susceptibility. Proportions of susceptible specimens per each antibiotic tested are shown as dots (Blue: Raoultella spp., Red: K. oxytoca). The bars show the estimated 95% bootstrap confidence intervals for the proportion of susceptibility per each antibiotic agent. *: Trimethoprim/Sulfamethoxazole. **: Insufficient sample sizes (Amikacin: 4 samples in Raoultella spp. and 50 samples in K. oxytoca; Levofloxacin: 1 sample in Raoultella spp. and 4 samples in K. oxytoca.).
4 Discussion
To the best of our knowledge, our study is the first study to compare the clinical differences between Raoultella spp. and Klebsiella oxytoca within the same study population. We found similarities in patient demographics (age and sex) and antibiotic susceptibility studies between the two bacteria species. However, there was a significant difference in disease severity measured by the proportion of ICU admissions within 48 hours of specimen collection. Patients with Raoultella positive cultures were significantly more likely to be admitted to ICU compared to patients with Klebsiella oxytoca. There were also significant differences in the culture sites. The frequency of urinary tract isolates was significantly higher among Klebsiella oxytoca patients than among Raoultella patients. At the same time, Raoultella patients were significantly more likely to have blood stream isolates, which likely represent blood stream infection. When only ICU admissions are compared, however, these differences were statistically insignificant.
An important microbiological difference between Raoultella spp. and Klebsiella spp. is known to be the ability of Raoultella spp. to produce histamines (Appel et al., 2021). Histamine is a potent proinflammatory molecule which plays a key role in initiating the immune response and is a possible contributor to cytokine storm and shock (Dale and Laidlaw, 1919; O’Mahony et al., 2011; Eldanasory et al., 2020). The higher proportion of ICU admissions among patients with Raoultella infection may be related to the histamine producing ability of Raoultella spp. Further studies are needed to investigate the role of Raoultella-produced histamine in patients’ disease severity.
Among different techniques to identify Raoultella species, MALDI-TOF is reported to be most reliable in distinguishing Raoultella from Klebsiella species (Ponce-Alonso et al., 2016). The application of MALDI-TOF in microbiological identification is relatively new (Singhal et al., 2015) and not every microbiology laboratory has this newer technology available. This may affect our analysis in two ways: 1) The difference between Raoultella spp. and K. oxytoca species may appear smaller, because some Raoultella specimens are misclassified as K. oxytoca. 2) If there were multiple microbiology laboratories using different detection techniques and certain specimens were more likely to be analyzed using MALDI-TOF, a bias arising from such systemic difference in detection techniques cannot be ruled out in our analysis.
There were more re-infection and repeat cultures among patients with K. oxytoca than patients with Raoultella (the number of specimens divided by the number of patients, 1.38 vs. 1.25). Because Raoultella is more readily identified using newer technologies, Raoultella positive cultures are likely to be more frequent towards the end of the study period. In this case, there was less time for patients with Raoultella infection to have a re-infection and repeat cultures. This may have contributed to the larger number of repeated cultures on average in K. oxytoca than in Raoultella spp.
Multidrug resistant Raoultella spp. is uncommonly reported (Mal et al., 2019; Tayo and Nyame, 2022). In our study, Raoultella spp. had high rates of susceptibility (>85%) to most broad-spectrum antibiotics, including carbapenem, third-generation cephalosporin, fluoroquinolones, aminoglycosides and trimethoprim/sulfamethoxazole. These observed rates were comparable to a case series of 187 isolates of Raoultella ornithinolytica (Seng et al., 2016). In addition, Raoultella spp. tended to have a higher rate of cefazolin susceptibility than K. oxytoca (72% vs. 52%), although statistical significance was not reached, likely due to the relatively small sample size of Raoultella spp. resulting in limited statistical power. Studies with a larger sample size are required to further determine the resistance profile of Raoultella species.
An important limitation of our study is selection bias. The patient population is likely skewed towards sicker patients as the inclusion criteria to the database we utilized are ED visits or ICU admissions. It is possible that sites of bacterial culture are strongly affected by the selection of patient population or the context of specimen collection (e.g. at ED or at ICU). Data from the outpatient setting is limited and patients with less severe infection are likely underrepresented in our study. Another limitation of our study is as mentioned above the unstandardized identification technique. Prior to the introduction of MALDI-TOF many Raoultella species are misclassified as Klebsiella species (Park et al., 2011). As a result, the differences between the two organisms may appear smaller and no difference found in our study does not prove that there are no differences between the two organisms. Furthermore, the relatively small number of patients with Raoultella positive cultures limits the statistical power of our analysis to detect differences between the two species. Finally, because we did not have access to the full clinical records (e.g., clinician’s notes) or to the microbiology samples, the presented results are limited by the data availability. Despite these limitations, our study adds clinically important information about the differences between the newly classified bacterial species, Raoultella spp., and its closest species according to the original classification, K. oxytoca, which has not been studied previously. Our study gives justification for future studies that are dedicated to compare these two species from clinical perspectives. Future studies should focus on isolates of Raoutella and K. oxytoca identified by standardized identification techniques such as MALDI-TOF or genome-sequencing and compare patients’ clinical courses prospectively. Developing a broader dataset of the clinical characteristics of Raoultella species in comparison to Klebsiella species will help us better understand this newly classified bacteria species and guide patient care.
5 Conclusion
While we did not find any significant differences in the patient demographics between Raoultella spp. and K. oxytoca, Raoultella species may cause more serious infection requiring ICU admissions. Also, Raoultella species may cause blood stream infection more frequently than K. oxytoca. Finally, Raoultella isolates were more often resistant to Trimethoprim/Sulfamethoxazole than K. oxytoca.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: https://physionet.org/content/mimiciv/2.2/.
Author contributions
SM: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. NC: Conceptualization, Methodology, Writing – original draft, Writing – review & editing. RC: Conceptualization, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2024.1260212/full#supplementary-material
References
Alves, M. S., Dias, R. C., de Castro, A. C., Riley, L. W., Moreira, B. M. (2006). Identification of clinical isolates of indole-positive and indole-negative Klebsiella spp. J. Clin. Microbiol. 44, 3640–3646. doi: 10.1128/JCM.00940-06
Appel, T. M., Quijano-Martinez, N., de la Cadena, E., Mojica, M. F., Villegas, M. V. (2021). Microbiological and clinical aspects of Raoultella spp. Front. Public Health 9. doi: 10.3389/fpubh.2021.686789
Chen, X., Zhou, X., Cao, J., Ma, K., Xia, Z. (2021). A case report of community-acquired Raoultella ornithinolytica infection in a healthy, young individual. BMC Infect. Dis. 21, 1095. doi: 10.1186/s12879-021-06799-w
Dale, H. H., Laidlaw, P. P. (1919). Histamine shock. J. Physiol. 52, 355–390. doi: 10.1113/jphysiol.1919.sp001837
Eldanasory, O. A., Eljaaly, K., Memish, Z. A., Al-Tawfiq, J. A. (2020). Histamine release theory and roles of antihistamine in the treatment of cytokines storm of COVID-19. Travel Med. Infect. Dis. 37, 101874. doi: 10.1016/j.tmaid.2020.101874
Hajjar, R., Ambaraghassi, G., Sebajang, H., Schwenter, F., Su, S. H. (2020). Raoultella ornithinolytica: emergence and resistance. Infect. Drug Resist. 13, 1091–1104. doi: 10.2147/IDR.S191387
Johnson, A., Bulgarelli, L., Pollard, T., Horng, S., Celi, L. A., Mark, R. MIMIC-IV (version 2.1). PhysioNet. doi: 10.13026/rrgf-xw32
Lewis, J. (2022). Performance standards for antimicrobial susceptibility testing M100, 32nd Ed. (Berwyn, PA, USA: Clinical & Laboratory Standards Institute).
Ma, Y., Wu, X., Li, S., Tang, L., Chen, M., An, Q. (2021). Proposal for reunification of the genus Raoultella with the genus Klebsiella and reclassification of Raoultella electrica as Klebsiella electrica comb. nov. Res. Microbiol. 172, 103851. doi: 10.1016/j.resmic.2021.103851
Mal, P. B., Sarfaraz, S., Herekar, F., Ambreen, R. (2019). Clinical manifestation and outcomes of multi-drug resistant (MDR) Raoultella terrigena infection - A case series at Indus Health Network, Karachi, Pakistan. IDCases 18, e00628. doi: 10.1016/j.idcr.2019.e00628
Naganathan, G., Amin, N. K. (2018). Raoultella Planticola associated necrotizing appendicitis: A novel case report. Int. J. Surg. Case Rep. 44, 38–41. doi: 10.1016/j.ijscr.2018.01.021
O’Mahony, L., Akdis, M., Akdis, C. A. (2011). Regulation of the immune response and inflammation by histamine and histamine receptors. J. Allergy Clin. Immunol. 128, 1153–1162. doi: 10.1016/j.jaci.2011.06.051
Park, J. S., Hong, K. H., Lee, H. J., Choi, S. H., Song, S. H., Song, K. H., et al. (2011). Evaluation of three phenotypic identification systems for clinical isolates of Raoultella ornithinolytica. J. Med. Microbiol. 60, 492–499. doi: 10.1099/jmm.0.020768-0
Pi, D. D., Zhou, F., Bai, K., Liu, C., Xu, F., Li, J. (2020). Raoultella ornithinolytica infection in the pediatric population: A retrospective study. Front. Pediatr. 8. doi: 10.3389/fped.2020.00362
Ponce-Alonso, M., Rodriguez-Rojas, L., Del Campo, R., Canton, R., Morosini, M. I. (2016). Comparison of different methods for identification of species of the genus Raoultella: report of 11 cases of Raoultella causing bacteraemia and literature review. Clin. Microbiol. Infect. 22, 252–257. doi: 10.1016/j.cmi.2015.10.035
Seng, P., Boushab, B. M., Romain, F., Gouriet, F., Bruder, N., Martin, C., et al. (2016). Emerging role of Raoultella ornithinolytica in human infections: a series of cases and review of the literature. Int. J. Infect. Dis. 45, 65–71. doi: 10.1016/j.ijid.2016.02.014
Singhal, N., Kumar, M., Kanaujia, P. K., Virdi, J. S. (2015). MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00791
Keywords: Raoultella, Klebsiella oxytoca, antibiotic, resistance, susceptability, bacteremia, intensive care
Citation: Mettler SK, Charoenngam N and Colgrove RC (2024) Clinical differences between Raoultella spp. and Klebsiella oxytoca. Front. Cell. Infect. Microbiol. 14:1260212. doi: 10.3389/fcimb.2024.1260212
Received: 17 July 2023; Accepted: 15 May 2024;
Published: 03 June 2024.
Edited by:
Margie Morgan, Cedars Sinai Medical Center, United StatesReviewed by:
Kate McCarthy, Royal Brisbane and Women’s Hospital, AustraliaEgidia Miftode, Grigore T. Popa University of Medicine and Pharmacy, Romania
Copyright © 2024 Mettler, Charoenngam and Colgrove. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sofia K. Mettler, c29maWFreW9uaGkubWV0dGxlckB1emguY2g=
 Sofia K. Mettler
Sofia K. Mettler Nipith Charoenngam2,3,4
Nipith Charoenngam2,3,4