
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 18 January 2024
Sec. Clinical Microbiology
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1351814
 Shuang Chao1†
Shuang Chao1† Yuejiao Wang1†
Yuejiao Wang1† Baolei Wu2†
Baolei Wu2† Runqing Li3
Runqing Li3 Jingxiao Dong3
Jingxiao Dong3 Lina Ji1
Lina Ji1 Xuejun Li1
Xuejun Li1 Ran Li1
Ran Li1 Xiaomei Yin1
Xiaomei Yin1 Xiuying Zhao3*
Xiuying Zhao3* Wannian Liang2*
Wannian Liang2*Introduction: Respiratory viral infection (RVI) is of very concern after the outbreak of COVID-19, especially in pediatric departments. Learning pathogen spectrum of RVI in children previous the epidemic of COVID-19 could provide another perspective for understanding RVI under current situation and help to prepare for the post COVID-19 infection control.
Methods: A nucleic acid sequence-based amplification (NASBA) assay, with 19 pairs of primers targeting various respiratory viruses, was used for multi-pathogen screening of viral infections in children presenting influenza-like illness (ILI) symptoms. Children with ILI at the outpatient department of Beijing Tsinghua Changgung Hospital during the influenza epidemic from 12/2018 to 01/2019 were included. Throat swabs were obtained for both the influenza rapid diagnostic test (IRDT) based on the colloidal gold immunochromatographic assay and the NASBA assay, targeting various respiratory viruses with an integrated chip technology.
Results and discussion: Of 519 patients, 430 (82.9%) were positive in the NASBA assay. The predominant viral pathogens were influenza A H1N1 pdm1/2009 (pH1N1) (48.4%) and influenza A (H3N2) (18.1%), followed by human metapneumovirus (hMPV) (8.8%) and respiratory syncytial virus (RSV) (6.1%). Of the 320 cases identified with influenza A by NASBA, only 128 (40.0%) were positive in the IRDT. The IRDT missed pH1N1 significantly more frequently than A (H3N2) (P<0.01). Influenza A pH1N1 and A (H3N2) were the major pathogens in <6 years and 6-15 years old individuals respectively (P<0.05). In summary, influenza viruses were the major pathogens in children with ILI during the 2018-2019 winter influenza epidemic, while hMPV and RSV were non-negligible. The coexistence of multiple pathogen leading to respiratory infections is the normalcy in winter ILI cases.
Currently, respiratory infections caused by immune debt post the spreading of COVID-19 have attracted attention from both academic and medical communities (Principi et al., 2023). Seasonal respiratory viral infections (RVI) mainly affect the nose, throat, and airways, bring to nasal congestion, a runny nose, sore throat, cough, and to extent with fever, generally with influenza-like illness (Lafond et al., 2016). Numerous viruses, including influenza virus A (FluA) sub-strains A H1N1 pdm1/2009 (pH1N1), A (H3N2), and two lineages of influenza virus B (FluB) have been implicated in the pathogen spectrum of pediatric RVI (Paul Glezen et al., 2013; Bedford et al., 2015). Patients with RVI may develop severe symptoms or even life-threatening complications, particularly children, the elderly, and individuals with underlying chronic conditions (Al-Awaidy et al., 2015; Kwong et al., 2018). Upon introduction in poultry, other members of the Orthomyxoviridae family, such as the subtypes of H5 and H7, may lead to outbreaks of highly pathogenic avian influenza, or even the acute respiratory distress syndrome (ARDS) (Tang et al., 2019; Zhong et al., 2019). On the other hand, pathogenic microorganisms other than influenza viruses, like the respiratory syncytial virus (RSV) (Takashita et al., 2021); the members of the coronavirus family including the novel SARS-CoV-2, the human rhinovirus (HRV), adenoviruses (ADV), and mycoplasma pneumonia (MP), can contribute to influenza-like illness (ILI) symptoms (Adam et al., 2017); leading to confusion in both diagnosis and disease managing. After emergence of SARS-CoV-2, from the early spring of 2020 to late 2021, reported influenza cases declined considerably due to implementation of quarantine and improved hygiene behavioral changes (Cowling et al., 2020; Feng et al., 2021). While increases in influenza activity were observed with the FluB/Victoria lineage as major pathogen in the beginning of 2022. Cocirculation of SARS-CoV-2 with influenza viruses and other else co-infections has been paid more attention in winter of 2023 (Wolters et al., 2021; Yun et al., 2021).
According to recent guidelines (General Office of National Health Commission of People's Republic of China, 2018; Uyeki et al., 2019b), administration of oseltamivir is recommended for selected patients with severe influenza. Therefore, timely differentiation of influenza viruses, as well as other respiratory pathogens, is essential for clinical management. Influenza rapid diagnostic tests (IRDTs) targeting the antigens of FluA and FluB viruses are widely used for timely diagnosis (Chartrand et al., 2012). Unfortunately, given the high false-negative rates of IRDTs (Uyeki et al., 2009), they were removed from priority detection tools in recent guidelines (Miller et al., 2015). Instead, nucleic acid-based tests including real-time RT-PCR are recommended to assist in the diagnosis of respiratory viral infection (Uyeki et al., 2019a; Losier and Dela Cruz, 2022). Respiratory panel assays based on automated multiplex RT-PCR method or other isothermal amplification techniques that integrates all reagents required for common respiratory viruses have been promoted (Couturier et al., 2013; Takashita et al., 2021). One of them featured by detecting 17 respiratory viruses and 3 atypical pathogens, had been proved to be helpful in the diagnosis of acute respiratory infections (Loeffelholz et al., 2011), with good performance in the diagnosis of community-acquired pneumonia (CAP) and hospital-acquired infections (Busson et al., 2019; Thongpan et al., 2019). A high-throughput, multi-index isothermal amplification platform for rapid detection of 19 types of common respiratory viruses or subspecies, including SARS-CoV-2 had been reported in 2020, the assays based on nucleic acid sequence-based amplification (NASBA) and previously reported micro/nanofluidic chip platform (MNCP) (Xing et al., 2020). Other multiple assay platforms based on real-time nucleic acid assay have been incorporated into routine diagnosis during and post the COVID-19 epidemic, broaden the understanding of multiple viral pathogenic infections (Li et al., 2019).
Under such background, to clarify the pathogenic characteristics previous the COVID-19 pandemic would help us prepare for the post-COVID-19 era. The present study concerns on a NASBA platform and 19 pairs of primers for multiple respiratory viruses, focus on its application in screening and identifying viral infections in pediatric outpatients presenting ILI symptoms during the winter of 2018-2019. The results were summarized based on demographic and clinical characterization.
The study population included outpatients who sought medical attention at the Department of Pediatrics of Beijing Tsinghua Changgung Hospital during a severe influenza epidemic from December 2018 to January 2019. All patients were children aged 0-15 years whose cases met the standard definition of ILI, including sudden onset of fever (≥38°C) and a cough and/or a sore throat without a known cause other than influenza (National Health and Family Planning Commission of the People's Republic of the China, 2018). They were classified into four age groups: babies ≤12 months (≤12m, infant period), children >12 months but <3 years (12m-3y, toddler period), children ≥3 years but <6 years (3-6y, preschool age), and children ≥6 years but ≤15 years (6-15y, school age and adolescence) (Table 1). Two throat swabs were obtained from each patient, one of which was used for immediate IRDT and the other was stored in 500 µl of the viral transport medium (VTM) at -80°C for RNA extraction within 7 days.
The throat swabs were vortexed for 30 s in the VTM (Yocon Biological Pharmacy, Beijing, China). Then, 300 μl of the medium was used for RNA extraction with TRIzol™ LS Reagent (Life Technologies Co., Grand Island, NY, USA) as recommended by the manufacturer. The RNA solution was obtained by adding 50 μl of RNase-free H2O.
The NASBA assay is an isothermal reaction coupled with the micro/nanofluidic chip (MNCP) platform. The data were analyzed with the software coupled with the RTisochip-A detector (CapitalBio Corp., Hong Kong, China) (Zhang et al., 2018). Briefly, 19.5 µl of RNA solution was mixed with amplification reagents, and the mixture was loaded into an MNCP, in which 24 reaction chambers are designed, with each chamber connected to a sine-shaped infusing channel by a micro-channel. The 19 pairs of primers targeted to the Flu A matrix protein (MP) gene, pH1N1 hemagglutinin (HA) gene, seasonal A H1N1 (sH1N1) HA gene, Flu A H3 HA gene, Flu A H7 HA gene (Chen et al., 2019), Flu B MP gene for general influenza B virus, RSV fusion protein (FP) gene, human entero-rhinovirus (HRV) 5’-UTR, enterovirus EV71 subtype (EV71) VP gene, coxsackievirus CA16 subtype (CA16) VP gene, coxsackievirus CA6 subtype (CA6) VP gene, ADV terminal protein gene, parainfluenza-1 (PIV1) hemagglutinin-neuraminidase (HN) gene, PIV2 HN gene, PIV3 HN gene, PIV4 HN gene, human metapneumovirus (hMPV) fusion protein (F) gene, universal primers for coronavirus OC43 and HKU1 (OC43/HKU1) orf1ab polyprotein gene, and universal primers for coronavirus NL63 and 229E (NL63/229E) orf1ab polyprotein gene. Totally 18 pathogens, the specific viruses causing respiratory symptoms, or its sub-species were included.
The system had been validated before it being used for the assay. To evaluate the sensitivity of the platform, the synthesized RNA templates were used to make sure that the sensitivity are below 200 copies/ul for all the pathogens. RNA from pH1N1 was used for the system repeatability test at concentration of 500 copies/µL (Xing et al., 2020). As a qualitative test, two positive controls, two negative controls were included. Assessment of the internal control (IC) glyceraldehyde 3-phosphate dehydrogenase (GAPDH) confirmed that swab samples were collected successfully, and nucleic acid extraction performed correctly. Only results with adequate positive, negative, and IC controls were used for analysis. The subtypes of Flu A sH1N1, pH1N1, A (H3N2), and A (H7N9) were identified with the specific influenza HA gene being positive. Influenza A (IA) was identified with only the Flu A MP gene being positive but the specific Flu A HA genes were negative.
The Clearview Exact Influenza A&B by Alear (Abon Biopharm Co., Ltd., Shanghai, China) was used at Beijing Tsinghua Changgung Hospital for influenza antigen detection. The reagent is coated with anti-influenza A and anti-influenza B NP in one strip, enabling differentiation between Flu A and Flu B. Undiluted swabs from the 519 patients were used for detection immediately after collection. All specimens were tested in a single experiment according to the manufacturer’s instructions.
Whole blood samples collected with EDTA.2K anticoagulant were tested for the 519 cases on a Sysmex XN1000 automatic blood cell analyzer (Sysmex Corp., Kobe, Japan).
The data were analyzed with SPSS 24.0 for Windows (IBM, Armonk, NY, USA). Continuous variables with normal distribution (Kolmogorov-Smirnov test) were presented as mean ± standard deviations (SD) and analyzed by analysis of variance; otherwise, continuous data were presented as median (interquartile ranges) and analyzed by the Wilcoxon test. Categorical variables were presented as frequency (percentage) and analyzed by the chi-square test or Fisher’s exact test. Two-sided (except for the chi-square test) P<0.05 was considered statistically significant.
The study population included 519 outpatients (269 males and 250 females; 4.4 ± 3.2 years) who sought medical attention during the severe influenza epidemic of 2018-2019 (Table 1).
Of the 519 patients, 430 (82.9%) were tested positive for at least one pathogen, and the remaining 89 (17.2%) cases were tested negative. In the 430 positive cases, the predominant single virus infection was due to Flu A (293/430, 68.1%), including 208 (48.4%) cases of pH1N1 and 78 (18.1%) of A (H3N2), followed by hMPV (38/430, 8.8%), RSV (26/430, 6.1%), and OC43/HKU1 (13/430, 3.0%). The results indicated that Flu A mainly accounted for the ILI symptoms during this epidemic season. Cases with two or more pathogens were defined as co-infection cases (41/430, 9.5%), of which the most common combination was pH1N1 with RSV (10/430, 2.3%) (Table 2).
Children aged 3-6y (161/191, 84.3%) and 12m-3y (173/210, 82.4%) represented the majority of the viral positive cases, followed by the 6-15y (81/99, 81.8%) and ≤12m (15/19, 79.0%) groups; the frequency of the general positivity showed no significant difference among the tested age groups (χ2 = 0.82, P=0.84).
The detection rate was significantly different among age groups for pH1N1, A (H3N2) and RSV. Among the 430 positive cases, pH1N1 was the major pathogen in the 12m-3y and 3-6y groups, showing a significant difference between the 3-6y and 6-15y groups (χ2 = 8.18, P<0.001). Compared with the other groups, children aged 6-15y were mainly infected by A (H3N2) (P=0.01, <0.001, <0.001, respectively). Children aged 12m-3y were found to be infected with RSV more frequently than other viruses, differences between ≤12m vs. 3-6y, ≤12m vs. 6-15y, 12m-3y vs. 3-6y, and 12m-3y vs. 6-15y were statistically significant (P=0.04, 0.02, <0.001, and <0.001, respectively). Meanwhile, hMPV was mainly identified in the 12m-3y and 3-6y groups, but there were no significant differences among groups (P>0.05) (Figure 1).
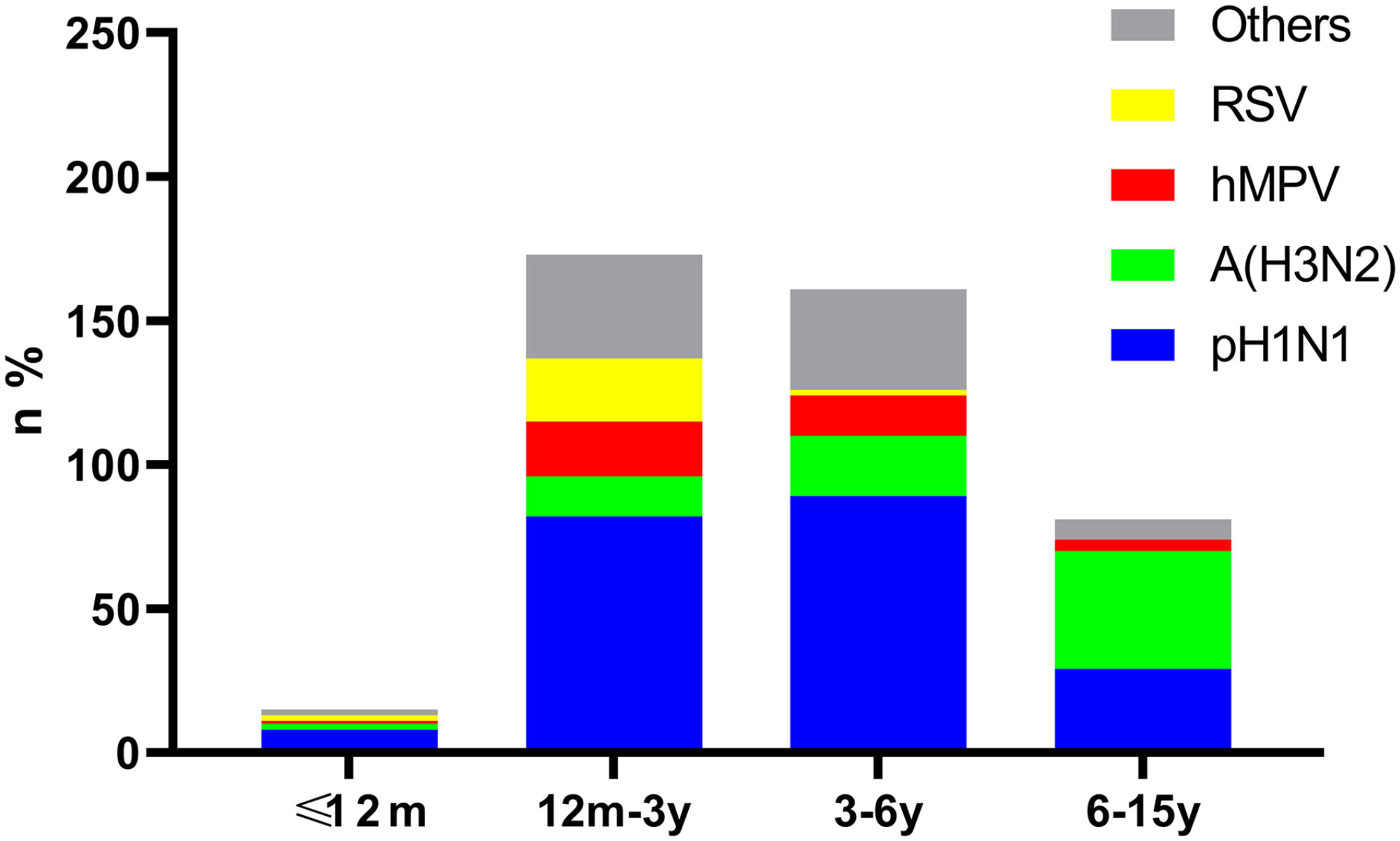
Figure 1 Age distribution of cases positive for pH1N1, A (H3N2), hMPV and RSV. Bars show the age distribution of case numbers and rates for pH1N1, H3N2, hMPV, RSV and other viruses in the 430 positive cases.
There were 330 positive cases of Flu A (76.7%, 330/430) and 3 positive cases of Flu B (0.7%, 3/430). Sub-typing of Flu A indicated that 232 (232/330, 70.3%) were pH1N1, 88 (88/330, 26.7%) were A (H3N2), 8 (8/330, 2.4%) with IA positive were classified to Flu A group but could not be differentiated, and 2 (2/330, 0.6%) were co-infected by pH1N1 and A (H3N2).
Considering that the IRDT is still commonly used as a point-of-care test (POCT) for influenza diagnosis, we compared the results in 320 individuals positive for Flu A by the NASBA (232 tested positive for pH1N1, 88 tested positive for A (H3N2)) with IRDT results. In general, 128 of the 320 cases were positive for influenza A by the IRDT (40.0%). When analysis was performed by subtypes, the IRDT’s positivity rate in patients with pH1N1 was 27.6% (64/232), and that of patients with A (H3N2) was 72.7% (64/88). Compared with NASBA, the detection of A (H3N2) infection by the IRDT was significantly more efficient than that of pH1N1 infection (χ2 = 54.17, P<0.001) (Table 3). These results strongly suggested that the IRDT was likely to miss Flu A, especially as the epidemic was caused by the pH1N1 virus. From the perception, NASBA and other nucleic based assays could significantly improve the efficiency of influenza diagnosis versus IRDTs, especially when infections are caused by pH1N1.
We compared white blood cell (WBC) counts, neutrophil (NE) counts, thermal spikes, frequencies of cough, and frequencies of other clinical characteristics in the four virus-positive groups with total cases number more than 25. Individuals who were hMPV-positive and RSV-positive were more likely to present with higher WBC counts compared with patients infected by pH1N1 or A(H3N2) (P-values were <0.001, 0.003, <0.001, and 0.006, respectively). No significant differences in WBC counts were found between pH1N1 and A (H3N2) cases (P=0.93), or between hMPV and RSV cases (P=0.55). In addition, children infected by hMPV tended to show higher NE amounts compared with those infected by the remaining three viruses (P=0.01, 0.01, 0.02, respectively). NE counts in children infected by pH1N1, A (H3N2) and RSV showed no significant differences (all P>0.05). There were significant differences in frequency of cough between A (H1N1)-positive (79.8%) and hMPV-positive (94.7%) individuals (χ2 = 4.88, P=0.03) (Table 4).
Compared with the A (H3N2)-positive and RSV-positive individuals, pH1N1-positive patients had higher mean thermal spike (P=0.01 and <0.001, respectively) (Figure 2A). Children aged 12m-3y and 3-6y were more likely to have high fever compared with those aged ≤12m and 6-15y (Figure 2B).
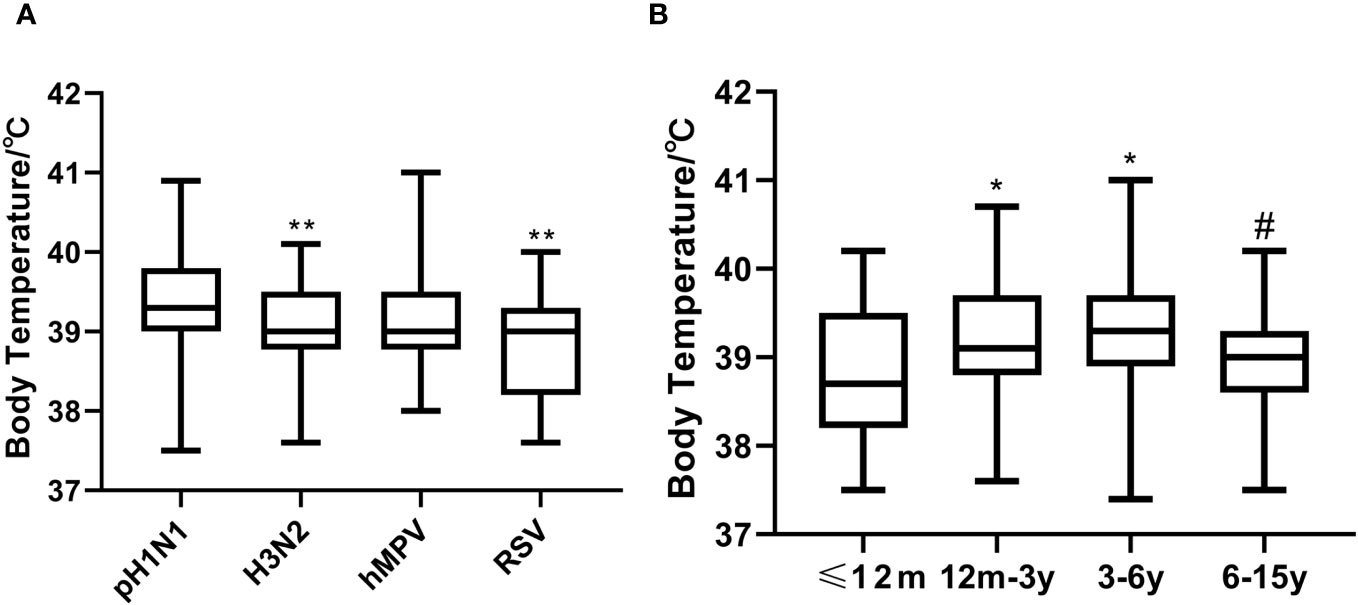
Figure 2 Thermal spikes among the viruses-positive groups and age groups. (A) Compared A (H3N2)-positive and RSV-positive individuals, pH1N1-positive patients had higher mean thermal spike (**P<0.01). (B) Thermal spike differences between the ≤12m and 12m-3y groups, and the ≤12m and 3-6y groups were significant (*P<0.05), as well as between the 12m-3y and 6-15y groups (#P<0.001). However, thermal spikes between the ≤12m and 6-15y groups, and the 12m-3y and 3-6y groups were not significant (all P>0.05).
In this study, we employed a multiplex nucleic acid assay to characterize 18 respiratory viruses or their subspecies in pediatric cases during Dec. 2018-Jan. 2019, one year previous the COVID-19 epidemic. The method had been validated before it was incorporated in the study and could detect 18 common pathogens or subtypes within 45 minutes after loading the nucleic acid sample. Based on the isothermal amplification technology and the combined MNCP platform, the assay platform had been applied to detect ILI cases with influenza virus infection during the 2017-2018 seasonal influenza epidemic, with improved performance compared with a commercially available rRT-PCR test (Li et al., 2019). After emergence of COVID-19, SARS-CoV-2 spike and nucleocapsid genes were incorporated into the system and the sequences of all the primers and probes can be found from published references (Xing et al., 2020).
From the 519 non-repeat pediatric cases, the NASBA assay revealed 430 viral infections, with a positivity rate of 82.9%. The results corroborate other studies based on rRT-PCR methods, with a positivity rate of up to 72%-95% in symptomatic ILI children, depending on age, and diagnostic and detection methods (Upadhyay et al., 2018; Busson et al., 2019; Li et al., 2019). Further analysis indicated that influenza virus A was the predominant virus (68.1%), with pH1N1 being the leading pathogen, but A (H3N2) consisted of 26.6% of Flu A positive cases. Pathogens besides influenza viruses were identified in 31.9% of the 430 cases, with hMPV at 8.8%, RSV at 6.1%, and OC43/HKU1 at 3.0% etc. The results indicate that medical professionals should be aware of pathogens other than influenza in ILI patients even during the influenza epidemic season. Given that oseltamivir therapy is widely adopted by clinicians (Lee et al., 2017), screening assays are necessary for antiviral therapy or in patients without significant improvement after oseltamivir administration. The results add value to multi-pathogen assays in pediatric clinic.
The current study also indicates significant differences in age of children infected with various pathogens. Indeed, pH1N1 was the major pathogen accounting for ILI in children of 3-6 years old, consistent with other studies showing that children in 3-5 years comprised the greatest proportion of pH1N1 cases in 2018/19 (Skowronski et al., 2019). A (H3N2) was more frequently identified in school-attending children (Wei et al., 2013). Meanwhile, hMPV was frequently detected in children between 1 and 6 years old, and RSV was detected more frequently in children younger than 3 years (Salimi et al., 2016; Thongpan et al., 2019). Only 19 cases under 1 year old included in the study, two reasons account for this, one was indeed very few baby cases present clinic with ILI in this time period, second was increased difficulty in interpreting and obtaining informed consent. We estimate that normal babies still have maternally derived antibodies that protect them from pathogenic infection (Zhang et al., 2013; Langel et al., 2022). The reasons why different ages were susceptible to variable infection might be attributed to acquired immunity such as vaccine injection, respiratory tract receptor for certain pathogen, and scope of social activities (Mansbach et al., 2021; WHO, 2022). There were 89 cases who were tested negative for pathogens listed in the study, but they could not be ruled out the possibility of mycoplasma pneumoniae or other community-source bacterial infections. In addition, all patients were sampled once, possibility of improper sampling or collection time could not be ruled out (Zhu et al., 2013; Liu et al., 2015; Zhu et al., 2019; Zhu et al., 2021).
We compared the two influenza A results of NASBA to those of the IRDT, which is not priority recommended in current guidelines but had been widely used as a POCT due to its convenience. The positivity rate (40.0%) by the IRDT was significantly lower than that obtained by the NASBA method. We compared IRDT-positive cases between the pH1N1 and A(H3N2) groups, and A(H3N2) was more frequently detected by IRDT (72.7%) than pH1N1 (27.6%). IRDT missed pH1N1 cases more than the A (H3N2) infections (P<0.01). This might be attributed to the absence of a specific antibody toward pH1N1 in the IRDT reagent. pH1N1 emerged in 2009 and replaced the sH1N1 virus as the prevalent strain. There were reports about IRDT sensitivity and its correlation with viral loads of Flu A or Flu B viruses, while our study indicates viral subtypes was a significant factor associated with sensitivity, whether it was due to viral load deserve further analysis (Li et al., 2019). The results suggested that IRDT manufacturers should keep renewing reagents even if the product has been approved for marketing. From this aspect, rRT-PCR or NASBA assays based on specific sequences may avoid this situation. Above all, the quality of swabs was fundamentally important in both the IRDT and NASBA. In the NABSA method, GAPDH was included as the internal control for swabs and RNA extraction, and the results were much more reliable than IRDT data when the reference is correct.
We analyzed clinical characteristics, including WBC, NE, fever and other symptoms among cases infected by the four viruses. The results indicated that compared with A (H3N2) -positive and RSV-positive individuals, pH1N1-positive patients were more likely to have higher temperatures. Individuals who were hMPV- and RSV-positive were more likely to present increased WBC counts compared with pH1N1-positive and A (H3N2)-positive patients (P<0.05). Children infected by hMPV tended to have higher WBC and NE counts than those infected by the other three viruses and were more likely to present cough than pH1N1-positive cases. Given that children with hMPV infection tended to get severe symptoms and were easily considered severe influenza cases or administered Oseltamivir without pathogen detection assay, attention should be paid to those not improving after antiviral therapy. Mechanism behind hMPV could cause severe diseases needs further study.
However, this study had limitations. It was a cross sectional study using random samples collected in one center from Northern China, and the sample size was relatively small, especially in those under 1 year old. In addition, the sampling time limited in 2 months, therefore, the pathogens spectrum reported in the study may be different from others (Huang et al., 2018; Yu et al., 2019). The study provides an opportunity to look at the distribution of 18 viral pathogens or their subtypes one year previous the COVID-19 epidemic. pH1N1 was the main pathogen in ILI cases in the winter of 2018-2019, but other pathogens existed as well. Other pathogens should not be neglected during the influenza season, and appropriate prevention, treatment and assay strategies are necessary for proper clinical management, in case the COVID-19 related quarantine policy being removed.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
The studies involving humans were approved by ethics committee of Beijing Tsinghua Changgung Hospital (No. 17120-0-01). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
SC: Formal analysis, Investigation, Writing – original draft. YW: Methodology, Data curation, Investigation, Writing – original draft. BW: Data curation, Investigation, Writing – original draft. RL: Data curation, Writing – original draft, Software, Validation. JD: Investigation, Validation, Writing – original draft. LJ: Data curation, Writing – original draft, Methodology. XL: Investigation, Writing – original draft, Methodology. RaL: Data curation, Writing – original draft, Investigation. XY: Data curation, Investigation, Supervision, Writing – original draft. XZ: Writing – review & editing, Conceptualization, Funding acquisition, Methodology, Project administration, Resources. WL: Writing – review & editing, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Beijing Municipal Science & Technology Commission Program of China [grant no. Z181100001718148, 2018 and Beijing High-level Public Health Technical Personnel Project 2023-03-03]. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Dr. Yan Zhang, Mr. Yanjing Chen and Miss Li Feng from National Engineering Research Center for Beijing Biochip Technology for useful discussion and excellent technical assistant.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
RVI, Respiratory viral infection; ILI, influenza-like illness; IRDT, influenza rapid diagnostic test; NASBA, nucleic acid sequence-based amplification; hMPV, human metapneumovirus; RSV, respiratory syncytial virus; Flu, Influenza; FluA, influenza virus A; FluB, influenza virus B; ARDS, acute respiratory distress syndrome; HRV, human rhinovirus; ADV, adenoviruses; MP, mycoplasma pneumonia; FA-RP, FilmArray respiratory panel; CAP, community-acquired pneumonia; NASBA, nucleic acid sequence-based amplification; MNCP, micro/nanofluidic chip platform; VTM, viral transport medium; HA, hemagglutinin; sH1N1, seasonal A H1N1; FP, fusion protein; HRV, human entero-rhinovirus; PIV1, parainfluenza-1; HN, hemagglutinin-neuraminidase; IC, internal control; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IA, Influenza A; SD, standard deviations; POCT, point-of-care test; WBC, white blood cell; NE, neutrophil.
Adam, K., Pangesti, K. N., Setiawaty, V. (2017). Multiple viral infection detected from influenza-like illness cases in Indonesia. BioMed. Res. Int. 2017, 9541619. doi: 10.1155/2017/9541619
Al-Awaidy, S., Hamid, S., Al Obaidani, I., Al Baqlani, S., Al Busaidi, S., Bawikar, S., et al. (2015). The burden of influenza-associated hospitalizations in Oman, january 2008-june 2013. PloS One 10, e0144186. doi: 10.1371/journal.pone.0144186
Bedford, T., Riley, S., Barr, I. G., Broor, S., Chadha, M., Cox, N. J., et al. (2015). Global circulation patterns of seasonal influenza viruses vary with antigenic drift. Nature 523, 217–220. doi: 10.1038/nature14460
Busson, L., Bartiaux, M., Brahim, S., Konopnicki, D., Dauby, N., Gérard, M., et al. (2019). Contribution of the FilmArray Respiratory Panel in the management of adult and pediatric patients attending the emergency room during 2015-2016 influenza epidemics: An interventional study. Int. J. Infect. Dis. 83, 32–39. doi: 10.1016/j.ijid.2019.03.027
Chartrand, C., Leeflang, M. M., Minion, J., Brewer, T., Pai, M. (2012). Accuracy of rapid influenza diagnostic tests: a meta-analysis. Ann. Intern. Med. 156, 500–511. doi: 10.7326/0003-4819-156-7-201204030-00403
Chen, L., Ruan, F., Sun, Y., Chen, H., Liu, M., Zhou, J., et al. (2019). Establishment of sandwich ELISA for detecting the H7 subtype influenza A virus. J. Med. Virol. 91, 1168–1171. doi: 10.1002/jmv.25408
Couturier, M. R., Barney, T., Alger, G., Hymas, W. C., Stevenson, J. B., Hillyard, D., et al. (2013). Evaluation of the FilmArray® Respiratory Panel for clinical use in a large children's hospital. J. Clin. Lab. Anal. 27, 148–154. doi: 10.1002/jcla.21576
Cowling, B. J., Ali, S. T., Ng, T. W. Y., Tsang, T. K., Li, J. C. M., Fong, M. W., et al. (2020). Impact assessment of non-pharmaceutical interventions against coronavirus disease 2019 and influenza in Hong Kong: an observational study. Lancet Public Health 5, e279–e288. doi: 10.1016/s2468-2667(20)30090-6
Feng, L., Zhang, T., Wang, Q., Xie, Y., Peng, Z., Zheng, J., et al. (2021). Impact of COVID-19 outbreaks and interventions on influenza in China and the United States. Nat. Commun. 12, 3249. doi: 10.1038/s41467-021-23440-1
General Office of National Health Commission of People's Republic of China National influenza surveillance program. Available at: http://www.nhc.gov.cn/jkj/s3577/201704/ed1498d9e64144738cc7f8db61a39506.shtml (Accessed December 24, 2023).
Huang, H. S., Tsai, C. L., Chang, J., Hsu, T. C., Lin, S., Lee, C. C. (2018). Multiplex PCR system for the rapid diagnosis of respiratory virus infection: systematic review and meta-analysis. Clin. Microbiol. Infect. 24, 1055–1063. doi: 10.1016/j.cmi.2017.11.018
Kwong, J. C., Schwartz, K. L., Campitelli, M. A., Chung, H., Crowcroft, N. S., Karnauchow, T., et al. (2018). Acute myocardial infarction after laboratory-confirmed influenza infection. N. Engl. J. Med. 378, 345–353. doi: 10.1056/NEJMoa1702090
Lafond, K. E., Nair, H., Rasooly, M. H., Valente, F., Booy, R., Rahman, M., et al. (2016). Global role and burden of influenza in pediatric respiratory hospitalizations 1982-2012: A systematic analysis. PloS Med. 13, e1001977. doi: 10.1371/journal.pmed.1001977
Langel, S. N., Blasi, M., Permar, S. R. (2022). Maternal immune protection against infectious diseases. Cell Host Microbe 30, 660–674. doi: 10.1016/j.chom.2022.04.007
Lee, J., Park, J. H., Jwa, H., Kim, Y. H. (2017). Comparison of efficacy of intravenous peramivir and oral oseltamivir for the treatment of influenza: systematic review and meta-analysis. Yonsei Med. J. 58, 778–785. doi: 10.3349/ymj.2017.58.4.778
Li, R., Gai, W., Zhu, D., Lok, C., Song, C., Dong, J., et al. (2019). Evaluation of a novel micro/nanofluidic chip platform for the detection of influenza A and B virus in patients with influenza-like illness. AMB Express 9, 77. doi: 10.1186/s13568-019-0791-8
Liu, Y., Song, S., Wang, W., Geng, X., Liu, W., Han, D., et al. (2015). Status of acute upper respiratory infection, influenza-like illness, and influenza vaccination coverage among community residents in Jinan. Chin. J. Prev. Med. 49, 1032–1035. doi: 10.3760/cma.j.issn.0253-9624.2015.12.003
Loeffelholz, M. J., Pong, D. L., Pyles, R. B., Xiong, Y., Miller, A. L., Bufton, K. K., et al. (2011). Comparison of the FilmArray Respiratory Panel and Prodesse real-time PCR assays for detection of respiratory pathogens. J. Clin. Microbiol. 49, 4083–4088. doi: 10.1128/jcm.05010-11
Losier, A., Dela Cruz, C. S. (2022). New testing guidelines for community-acquired pneumonia. Curr. Opin. Infect. Dis. 35, 128–132. doi: 10.1097/qco.0000000000000824
Mansbach, J. M., Geller, R. J., Hasegawa, K., Piedra, P. A., Avadhanula, V., Gern, J. E., et al. (2021). Detection of respiratory syncytial virus or rhinovirus weeks after hospitalization for bronchiolitis and the risk of recurrent wheezing. J. Infect. Dis. 223, 268–277. doi: 10.1093/infdis/jiaa348
Miller, M. R., Peters, T. R., Suerken, C. K., Snively, B. M., Poehling, K. A. (2015). Predictors of influenza diagnosis among patients with laboratory-confirmed influenza. J. Infect. Dis. 212, 1604–1612. doi: 10.1093/infdis/jiv264
National Health and Family Planning Commission of the People′s Republic of China (2018). Chinese guidelines for diagnosis and treatment of influenza, (2018 version). Chin. J. Viral Dis. 8, 81–85. doi: 10.16505/j.2095-0136.2018.0021
Paul Glezen, W., Schmier, J. K., Kuehn, C. M., Ryan, K. J., Oxford, J. (2013). The burden of influenza B: a structured literature review. Am. J. Public Health 103, e43–e51. doi: 10.2105/ajph.2012.301137
Principi, N., Autore, G., Ramundo, G., Esposito, S. (2023). Epidemiology of respiratory infections during the COVID-19 pandemic. Viruses 15, 1160. doi: 10.3390/v15051160
Salimi, V., Tavakoli-Yaraki, M., Yavarian, J., Bont, L., Mokhtari-Azad, T. (2016). Prevalence of human respiratory syncytial virus circulating in Iran. J. Infect. Public Health 9, 125–135. doi: 10.1016/j.jiph.2015.05.005
Skowronski, D. M., Leir, S., De Serres, G., Murti, M., Dickinson, J. A., Winter, A. L., et al. (2019). Children under 10 years of age were more affected by the 2018/19 influenza A(H1N1)pdm09 epidemic in Canada: possible cohort effect following the 2009 influenza pandemic. Euro Surveill 24, 1900104. doi: 10.2807/1560-7917.Es.2019.24.15.1900104
Takashita, E., Kawakami, C., Momoki, T., Saikusa, M., Shimizu, K., Ozawa, H., et al. (2021). Increased risk of rhinovirus infection in children during the coronavirus disease-19 pandemic. Influenza Other Respir. Viruses 15, 488–494. doi: 10.1111/irv.12854
Tang, J., Zhang, J., Zhou, J., Zhu, W., Yang, L., Zou, S., et al. (2019). Highly pathogenic avian influenza H7N9 viruses with reduced susceptibility to neuraminidase inhibitors showed comparable replication capacity to their sensitive counterparts. Virol. J. 16, 87. doi: 10.1186/s12985-019-1194-9
Thongpan, I., Suntronwong, N., Vichaiwattana, P., Wanlapakorn, N., Vongpunsawad, S., Poovorawan, Y. (2019). Respiratory syncytial virus, human metapneumovirus, and influenza virus infection in Bangkok 2016-2017. PeerJ 7, e6748. doi: 10.7717/peerj.6748
Upadhyay, B. P., Banjara, M. R., Shrestha, R. K., Tashiro, M., Ghimire, P. (2018). Etiology of coinfections in children with influenza during 2015/16 winter season in Nepal. Int. J. Microbiol. 2018, 8945142. doi: 10.1155/2018/8945142
Uyeki, T. M., Bernstein, H. H., Bradley, J. S., Englund, J. A., File, T. M., Fry, A. M., et al. (2019a). Clinical practice guidelines by the infectious diseases society of America: 2018 update on diagnosis, treatment, chemoprophylaxis, and institutional outbreak management of seasonal influenzaa. Clin. Infect. Dis. 68, e1–e47. doi: 10.1093/cid/ciy866
Uyeki, T. M., Bernstein, H. H., Bradley, J. S., Englund, J. A., File, T. M., Fry, A. M., et al. (2019b). Clinical practice guidelines by the infectious diseases society of america: 2018 update on diagnosis, treatment, chemoprophylaxis, and institutional outbreak management of seasonal influenzaa. Clin. Infect. Dis. 68, 895–902. doi: 10.1093/cid/ciy874
Uyeki, T. M., Prasad, R., Vukotich, C., Stebbins, S., Rinaldo, C. R., Ferng, Y. H., et al. (2009). Low sensitivity of rapid diagnostic test for influenza. Clin. Infect. Dis. 48, e89–e92. doi: 10.1086/597828
Wei, M., Yan, Z., Wang, C., Liu, W., Cao, W. (2013). Eight-hospital based influenza like illness surveillance from April 2009 to March 2011 in China. Influenza Other Respir. Viruses 7, 997–998. doi: 10.1111/irv.12064
Who (2022). Vaccines against influenza:WHO position paper – May 2022. Wkly. Epidemiol. Rec. No 19. 97, 185–208. Available at: https://iris.who.int/bitstream/handle/10665/354264/WER9719-eng-fre.pdf?sequence=1 (Accessed December 24,2023).
Wolters, F., Grünberg, M., Huber, M., Kessler, H. H., Prüller, F., Saleh, L., et al. (2021). European multicenter evaluation of Xpert® Xpress SARS-CoV-2/Flu/RSV test. J. Med. Virol. 93, 5798–5804. doi: 10.1002/jmv.27111
Xing, W., Liu, Y., Wang, H., Li, S., Lin, Y., Chen, L., et al. (2020). A high-throughput, multi-index isothermal amplification platform for rapid detection of 19 types of common respiratory viruses including SARS-coV-2. Engineering 6, 1130–1140. doi: 10.1016/j.eng.2020.07.015
Yu, J., Liu, C., Xiao, Y., Xiang, Z., Zhou, H., Chen, L., et al. (2019). Respiratory syncytial virus seasonality, Beijing, China 2007-2015. Emerg. Infect. Dis. 25, 1127–1135. doi: 10.3201/eid2506.180532
Yun, J., Park, J. H., Kim, N., Roh, E. Y., Shin, S., Yoon, J. H., et al. (2021). Evaluation of three multiplex real-time reverse transcription PCR assays for simultaneous detection of SARS-coV-2, influenza A/B, and respiratory syncytial virus in nasopharyngeal swabs. J. Korean Med. Sci. 36, e328. doi: 10.3346/jkms.2021.36.e328
Zhang, G., Zheng, G., Zhang, Y., Ma, R., Kang, X. (2018). Evaluation of a micro/nanofluidic chip platform for the high-throughput detection of bacteria and their antibiotic resistance genes in post-neurosurgical meningitis. Int. J. Infect. Dis. 70, 115–120. doi: 10.1016/j.ijid.2018.03.012
Zhang, C., Zhu, N., Xie, Z., Lu, R., He, B., Liu, C., et al. (2013). Viral etiology and clinical profiles of children with severe acute respiratory infections in China. PloS One 8, e72606. doi: 10.1371/journal.pone.0072606
Zhong, G., Fan, S., Lopes, T. J. S., Le, M. Q., Van Bakel, H., Dutta, J., et al. (2019). Isolation of highly pathogenic H5N1 influenza viruses in 2009-2013 in Vietnam. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01411
Zhu, D., Lok, C., Chao, S., Chen, L., Li, R., Zhao, Z., et al. (2019). Detection and characterization of type B influenza virus from influenza-like illness cases during the 2017-2018 winter influenza season in Beijing, China. Arch. Virol. 164, 995–1003. doi: 10.1007/s00705-019-04160-w
Zhu, G., Xu, D., Zhang, Y., Wang, T., Zhang, L., Gu, W., et al. (2021). Epidemiological characteristics of four common respiratory viral infections in children. Virol. J. 18, 10. doi: 10.1186/s12985-020-01475-y
Keywords: respiratory viral infection (RVI), influenza (flu), nucleic acid sequence-based amplification (NASBA), influenza rapid diagnostic test (IRDT), influenza-like illness (ILI), pediatrics
Citation: Chao S, Wang Y, Wu B, Li R, Dong J, Ji L, Li X, Li R, Yin X, Zhao X and Liang W (2024) Characterization of viral infections in children with influenza-like-illness during December 2018–January 2019. Front. Cell. Infect. Microbiol. 13:1351814. doi: 10.3389/fcimb.2023.1351814
Received: 07 December 2023; Accepted: 26 December 2023;
Published: 18 January 2024.
Edited by:
Peng Li, Chinese Center for Disease Control and Prevention, ChinaReviewed by:
Kun Qin, National Institute for Viral Disease Control and Prevention (China CDC), ChinaCopyright © 2024 Chao, Wang, Wu, Li, Dong, Ji, Li, Li, Yin, Zhao and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiuying Zhao, emhhb3hpdXlpbmcyMDAxQDE2My5jb20=; Wannian Liang, bGlhbmd3bkBtYWlsLnRzaW5naHVhLmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.