- 1Department of Botany and Microbiology, College of Science, King Saud University, Riyadh, Saudi Arabia
- 2Department of Biology, College of Science, University of Jeddah, Jeddah, Saudi Arabia
- 3Microbiology Department, Riyadh Regional Laboratory, Ministry of Health, Riyadh, Saudi Arabia
- 4College of Medicine, Al-Faisal University, Riyadh, Saudi Arabia
- 5Computational Bioscience Research Center, Biological and Environmental Sciences and Engineering, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia
- 6Department of Microbiology, Food and Drug Authority, Jeddah, Saudi Arabia
- 7Reference Laboratory for Microbiology, Executive Department for Reference Laboratories, Research and Laboratories Sector, Food and Drug Authority, Riyadh, Saudi Arabia
Introduction: Staphylococcus aureus is a significant human pathogen that poses a threat to public health due to its association with foodborne contamination and a variety of infections. The factors contributing to the pathogenicity of S. aureus include virulence, drug resistance, and toxin production, making it essential to monitor their prevalence and genetic profiles. This study investigated and compared the genomic characteristics of S. aureus isolates from retail meat and patients in Saudi Arabia.
Methods: A total of 136 S. aureus isolates were obtained between October 2021 and June 2022:84 from patients and 53 from meat samples in Riyadh, Saudi Arabia. S. aureus isolates were identified using conventional methods and MALDI-TOF MS, and methicillin-resistant S. aureus (MRSA) was identified using VITEK2 and BD Phoenix systems. MRSA was confirmed phenotypically using chromogenic agar, and genotypically by detecting mecA. Genomic data were analyzed using BactopiaV2 pipeline, local BLAST, and MLST databases.
Results: Antibiotic resistance genes were prevalent in both meat and patient S. aureus isolates, with high prevalence of tet38, blaZ, and fosB. Notably, all S. aureus isolates from patients carried multidrug-resistant (MDR) genes, and a high percentage of S. aureus isolates from meat also harbored MDR genes. Phenotypically, 43% of the S. aureus isolates from meat and 100% of the patients’ isolates were MDR. Enterotoxin genes, including selX, sem, and sei, exhibited high compatibility between meat and patient S. aureus isolates. Virulence genes such as cap, hly/hla, sbi, and isd were found in all S. aureus isolates from both sources.
Conclusion: Our study established a genetic connection between S. aureus isolates from meat and patients, showing shared antibiotic resistance and virulence genes. The presence of these genes in meat derived isolates underscores its role as a reservoir. Genomic relatedness also suggests potential transmission of resistance between different settings. These findings emphasize the necessity for a comprehensive approach to monitor and control S. aureus infections in both animals and humans.
1 Introduction
Staphylococcus aureus is an opportunistic bacterium found in humans and animals, especially food-producing animals (Kadariya et al., 2014). These bacteria can enter the food chain during collection, slaughter, processing, packaging, and storage (Wu et al., 2018). It has been detected in processed cheese, raw beef, camel meat, and dairy products in Saudi Arabia (El Sheikha, 2016). Moreover, it is a major pathogen that causes a wide range of infections in community and hospital settings (Turner et al., 2019). S. aureus has a variety of virulence factors that significantly influence its clinical manifestations (Jenul and Horswill, 2019). For instance, surface adhesins, including clumping factors and fibronectin-binding proteins, promote tissue adherence and contribute to skin and soft tissue infections (Algammal et al., 2020). Other virulence factors, including toxins such as hemolysins, leukocidins [e.g., panton-valentine leukocidin (PVL)], enterotoxins, and exfoliative toxins, lead to diverse clinical outcomes, ranging from severe skin infections to food poisoning and scalded skin syndrome (Algammal et al., 2020). Staphylococcal protein A (SPA) interferes with the host immune response and increases the severity of staphylococcal infection (Algammal et al., 2020). Proteins that promote biofilm production, such as polysaccharide intercellular adhesins, contribute to chronic infections and antibiotic resistance, particularly in cases involving medical devices (Singh et al., 2009). These virulence factors contribute to S. aureus pathogenicity (Jenul and Horswill, 2019). Enterotoxigenic S. aureus strains produce heat-stable staphylococcal enterotoxins (SEs), which are one of the main causes of global outbreaks of food poisoning (Pinchuk et al., 2010). To date, more than 20 SEs have been identified (Sankomkai et al., 2020). Classically known enterotoxins include the five enterotoxins encoded by sea, seb, sec, sed, and see (Sankomkai et al., 2020). Beyond food poisoning, these proteins can contribute to a broad range of symptoms ranging from mild toxin-mediated illnesses to life-threatening systemic diseases (Ahmad-Mansour et al., 2021). According to a recent report by the European Food Safety Authority, more than 40 staphylococcal food poisoning (SFP) outbreaks, 400 cases, and 32 hospitalizations have been linked to S. aureus. aureus in the European Union by 2020 (The European Union One Health 2020 Zoonoses Report, 2021). SFP accounts for approximately 241,000 hospitalizations annually due to illness in the United States (Scallan et al., 2018). As SFP are becoming more prevalent, food safety is increasingly considered a serious public health issue that affects all other safety aspects (Kadariya et al., 2014). The extensive use of antibiotics has led to the emergence of multidrug-resistant (MDR) bacteria, including genetically distinguished methicillin-resistant S. aureus (MRSA) strains associated with communities and animals, such as community-associated MRSA (CA-MRSA) and livestock-associated MRSA (LA-MRSA) (Fetsch et al., 2021). S. aureus strains exhibit antibiotic resistance due to specific genes that render them resistant to various antimicrobials, such as penicillins, tetracyclines, cephalosporins, aminoglycosides, and fluoroquinolones (Urban-Chmiel et al., 2022). The global increase in MDR is recognized as a public health threat (Algammal et al., 2022a). Recent studies have highlighted the emergence of MDR bacterial pathogens from various sources, emphasizing the importance of routine antimicrobial susceptibility testing to identify effective antibiotics and screen for emerging MDR strains (Algammal et al., 2020; Elmoslemany et al., 2021; Gordon et al., 2021; Algammal et al., 2022b). Despite sporadic reports of S. aureus toxins and drug resistance genes in Saudi retail foods, information on the prevalence, toxigenicity, and antibiotic resistance of S. aureus remains limited (Raji et al., 2016; Alghizzi and Shami, 2021; Alghizzi et al., 2021). Molecular genomic methods are essential tools for investigating the virulence factors, antimicrobial resistance, and toxin genes in S. aureus. aureus. In addition, it aids in elaborating the toxigenic and drug resistance potential of this microorganism in retail meats and hospitals. Meat and meat products have been found to be important S. aureus reservoirs and have been linked to various foodborne bacterial outbreaks (van Loo et al., 2007; Boost et al., 2013; Ge et al., 2017; Wu et al., 2018; Thwala et al., 2021). In Saudi Arabia, data on S. aureus. aureus in retail meat from different regions was limited (Abulreesh and Organji, 2011; Kay et al., 2013; El Sheikha, 2016; Raji et al., 2016; Shawish and Al-Humam, 2016; El-Ghareeb et al., 2019). Therefore, this study aimed to investigate and compare the genomic relatedness and distribution of resistance and virulence genes among clinical and meat S. aureus isolates. The genomic investigation of S. aureus isolates from meat and their relationship with clinical S. aureus isolates may be an initial step in establishing an effective strategy in Saudi Arabia for monitoring the emergence and spread of S. aureus.
2 Material and methods
2.1 Sample collection
From October 2021 to June 2022, 84 non-duplicate isolates of S. aureus were obtained from inpatients at the bacteriology department of the Riyadh Regional Laboratory and Blood Bank. A wide range of specimens, including urine midstream, tissue culture, wound swab, nasal swab, pus swab, groin swab, axilla swab, burn swab, eye swab, bronchial wash, sputum, ascites (peritoneal) fluid, tracheal aspirate, and blood culture samples, were collected for analysis in this study. For additional details on the specimens, please refer to Supplementary Table 4 in Supplementary Material. Additionally, 250 raw meat samples, including camels, beef, chicken, fish, and lamb, were collected from various meat retailers in Riyadh, Saudi Arabia. All meat samples were transported in sealed plastic wrap or original packaging in insulated coolers maintained at a stable temperature of 4°C with adequate ice packs to sustain a stable temperature throughout the transportation process to the Reference Laboratory of Microbiology at the Saudi Food and Drug Authority. Microbiological analysis was conducted within 24 h of sampling.
2.2 Ethical approval
Ethical approval (approval number H-01-R-053) for the study was obtained from the Institutional Review Board committee of King Saud Medical City, Riyadh, Saudi Arabia and the Institutional Biosafety and Bioethics Committee of King Abdullah University of Science and Technology (approval number 22IBEC051).
2.3 Staphylococcus aureus isolation and identification
Meat samples were homogenized in a stomacher using a sterile plastic bag. This involved 25 g of each meat sample and 225 mL of buffered peptone water with 6.5% NaCl. Petrifilm™ Staph Express count plate (3M™, St. Paul, MN, USA), a modified chromogenic Baird-Parker medium, was used as a selective and differential medium for S. aureus detection. The Petrifilm™ Staph Express disk contained toluidine blue O and DNA. DNase-positive organisms degrade DNA that reacts with toluidine blue O to form pink zones. This allowed for a distinction between S. aureus colonies and other staphylococci. Single S. aureus colonies were transferred to mannitol salt agar (Neogen; Lansing, MI, USA). All S. aureus isolates were identified and confirmed using the classical method, which included Gram staining, catalase testing, and coagulase testing (PROLEX™, Neston, UK). All S. aureus isolates were further identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF) (Bruker, Bremen, Germany) using the 70% formic acid protein extraction method.
2.4 Phenotypic and genotypic detection of methicillin-resistant Staphylococcus aureus
The identification of MRSA strains was carried out using VITEK2 (bioMérieux, Craponne, France) and BD Phoenix (BD Diagnostics, Franklin Lakes, NJ, USA). The instruments’ systems classify any S. aureus isolate as MRSA based on both oxacillin and cefoxitin minimum inhibitory concentrations (MICs) breakpoints, interpreted according to Laboratory Standards Institute (CLSI). For oxacillin, isolates were considered resistant with MICs ≥4 μg/ml, whereas for cefoxitin, isolates were considered resistant with MICs ≥8 μg/ml. Individual colonies of the S. aureus isolates were transferred to Harlequin MRSA chromogenic agar for the selective and differential detection of MRSA (Neogen, Lansing, MI, USA). This medium allowed MRSA to grow because it includes a cefoxitin (Fox) supplement. The α-glucosidase enzyme produced by S. aureus cleaves the chromogenic substrate, resulting in blue colonies.
DNA was extracted from the bacterial culture using the QIAGEN DNeasy Blood and Tissue Kit (Qiagen, Manchester, UK), following the manufacturer’s instructions. DNA purity was checked using a QIAxpert spectrophotometer (Qiagen, Manchester, UK), and DNA concentration was determined using a Qubit™ Flex Fluorometer device (Thermo Fisher Scientific, Waltham, MA, USA).
The presence of methicillin resistance was determined by detecting the mecA gene, which encodes for penicillin’s binding protein 2a (PBP2a). This was performed using a forward primer (5′-GTAGAAATGACTGAACGTCCGATAA-3′) and a reverse primer (5′-CCAATTCCACATTGTTTCCGTCTAA-3′), which amplified a 310 bp fragment (Parvin et al., 2021). The reaction was performed in 25 µL using DreamTaq™ 2X Green PCR Master Mix (Thermo Scientific, Waltham, MA, USA). The reaction setup included 1 µL of each (10 pmol) primer and 2 µL of DNA template (20 ng/µL). S. aureus (ATCC43300) was used as a positive control (ATCC, Manassas, Virginia, USA). The amplification process involved an initial denaturation step at 95°C for 4 min, followed by 30 cycles at 95°C for 45 s, 56°C for 1 min, and 72°C for 1 min. This was followed by a final extension step at 72°C for 4 min. The final PCR product was analyzed on a 1.6% agarose gel stained with ethidium bromide and run for 60 min at 80 volts in 1X tris borate buffer (TBE) (BIOBasic, Markham, ON, Canada). Gel visualization was performed using the Image Lab™ 6.1 Software (Bio-Rad, Hercules, CA, USA).
2.5 Antimicrobial susceptibility testing
The antimicrobial susceptibility testing (AST) of S. aureus isolates was evaluated using both the VITEK 2 System and BD Phoenix System. Testing followed manufacturers’ instructions and adhered to CLSI guidelines (Wayne, 2018). Shared antimicrobial agents between the two systems encompassed 15 classes, including β-lactam: cefoxitin (FOX) and oxacillin (OXA); aminoglycosides: gentamicin (GEN); fluoroquinolones: levofloxacin (LEV) and moxifloxacin (MOX); macrolide: erythromycin (ERY); lincosamides: clindamycin (CLI); oxazolidinone: linezolid (LIN); glycopeptide: teicoplanin (TEI) and vancomycin (VAN); tetracycline: tetracycline (TET); glycycline: tigecycline (TIG); phosphonic acids: fosfomycin (FOS); nitroheterocyclics: nitrofurantoin (NIT); fusidanes: fusidic acid (FUS); monoxycarbolic acids: mupirocin (MUP); ansamycin: rifampin (RIF); and folate pathway antagonists: sulphamethoxazole/trimethoprim (SXT). However, the BD Phoenix System lacked benzylpenicillin (PEN) and tobramycin (TOB) present in the VITEK 2 system. Additionally, the BD Phoenix System included more antimicrobial agents such as β-lactam: ampicillin (AMP), cefotaxime (CTX), ceftaroline (CPT), and penicillin G (PEN); fluoroquinolones: ciprofloxacin (CIP); and lipopeptides: daptomycin (DAP). The reference strain (ATCC33400) was used, and the results, including the MICs, were interpreted following CLSI guidelines and MIC breakpoints for S. aureus (Clinical and Laboratory Standards Institute, 2023). S. aureus isolates resistant to three or more antimicrobial classes or to oxacillin/cefoxitin were classified as MDR (Magiorakos et al., 2012).
2.6 Whole-genome sequencing
Whole-genome sequencings (WGS) libraries were constructed using a QIAseq DNA FX library preparation kit (Qiagen, Manchester, UK). The input concentration was 100 ng of DNA, following the manufacturer’s instructions (Qiagen, Manchester, UK). The library was size-selected to have a 300–350 bp insert size. Sequencing was performed using the Illumina NovaSeq 6000 platform with two SP flow cells (Illumina, San Diego, CA, USA). Data were quality-checked prior to analysis, with a Phred score cut-off of Q30. Bioinformatics analysis was conducted using the BactopiaV2 pipeline, focusing on the S. aureus-specific workflow (Petit and Read, 2020). Screening for pvl was carried out using local BLAST database identification of the two genes, lukS and lukF, accessions (YP_002268030.1 and YP_002268029.1), respectively. For sequence type (ST) assignment, the public MLST database for S. aureus was used. The clonal lineages of all S. aureus strains isolated from the patients and meat samples were identified using MLST.
2.7 Statistical analysis
To investigate the association between antibiotic resistance, toxins, and virulence genes in S. aureus isolates obtained from meat and patients, data were analyzed using GraphPad Prism 10.0 software (GraphPad Software, Boston, Massachusetts, USA). The two-tailed chi-square test (χ2) was used to determine the significance of the differences, and p values < 0.05 were considered statistically significant. The correlation coefficient (r) was calculated among the demonstrated antimicrobial resistance genes in the S. aureus strains and different antimicrobial agents. The Pearson correlation coefficient in a correlation matrix was estimated using the “corrplot” package in R, (version 4.2.1) (Wei and Simko, 2021).
3 Results
3.1 Prevalence of Staphylococcus aureus isolates
In the analysis of 250 meat samples, 53 isolates of S. aureus were detected, comprising 21.2% of the total meat samples (53/250). Further analysis revealed 136 S. aureus isolates, of which 83 (61%) were obtained from patients and 53 (39%) from meat samples. The breakdown of S. aureus isolates across the various meat types was as follows: camel meat (18/53; 34%), beef (12/53; 23%), lamb (8/53; 15%), fish (8/53; 15%), and chicken (7/53; 13%).
3.2 Phenotypic characteristics of the Staphylococcus aureus isolates
All S. aureus isolates recovered in this study were positive for several key phenotypic tests, such as Gram staining and catalase and coagulase tests. The isolates showed beta-hemolysis on blood agar, indicating the production of hemolysins and fermented mannitol on mannitol salt agar, leading to a color change from pink to yellow. Antibiotic susceptibility testing provided specific profiles for the sensitivity and resistance to various antibiotics. Using VITEK2, 56 S. aureus isolates from patients and 11 from meat were identified as MRSA, exhibiting positivity for cefoxitin and an oxacillin MIC ≥4 mg/L. The 42 methicillin-sensitive S. aureus (MSSA) isolates from meat were cefoxitin screen-negative, with oxacillin MICs measuring 0.5 mg/L. Additionally, BD Phoenix™ identified 26 S. aureus isolates from patients as MRSA, all demonstrating cefoxitin MIC ≥8 mg/L and oxacillin MIC >2 mg/L.
3.3 Phenotypic and genotypic resistance characteristics of S. aureus isolates
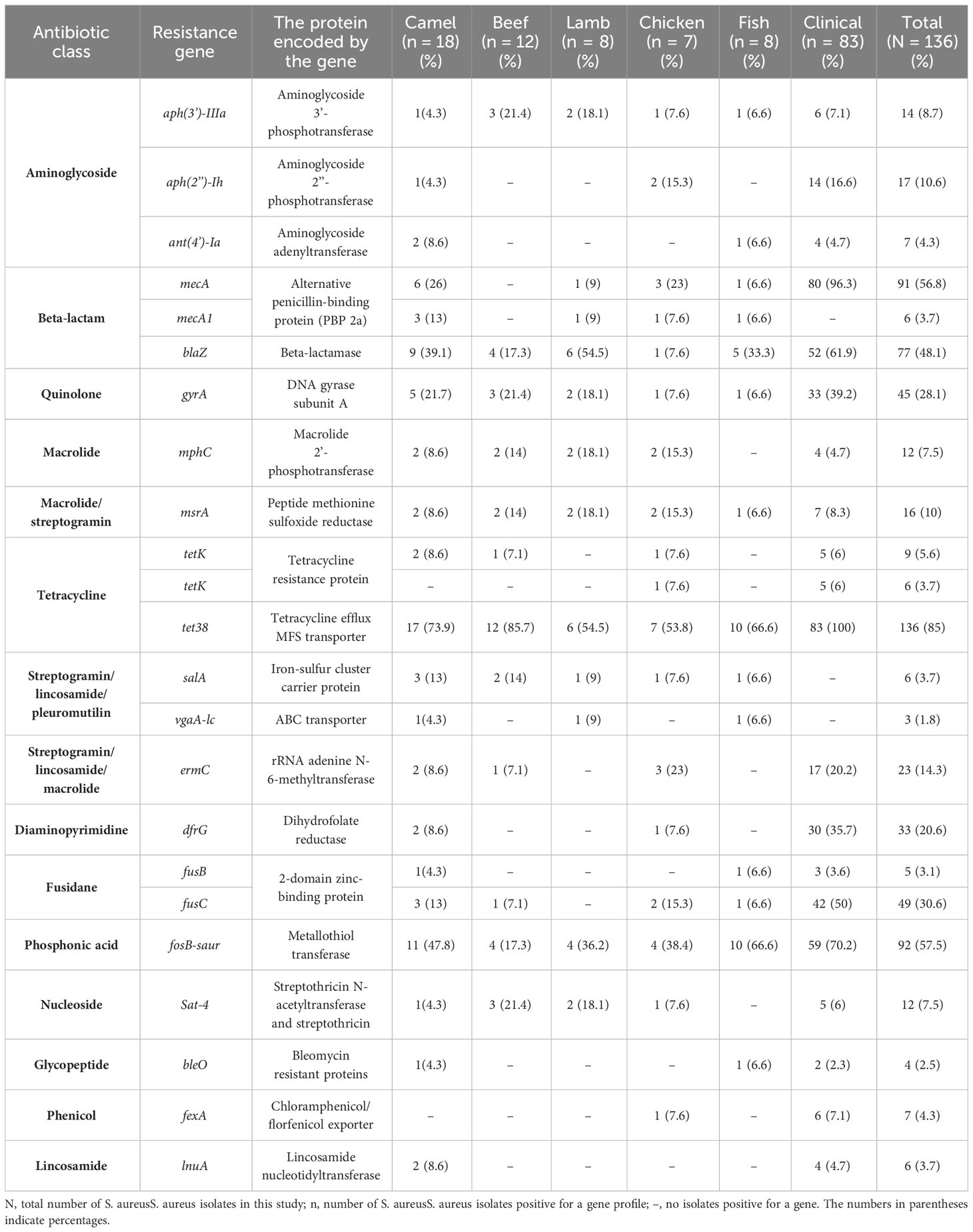
The antimicrobial susceptibility of the pooled S. aureus isolates were resistance to penicillin (97.7%), cefoxitin (68,3%), oxacillin (64,6%), levofloxacin and moxifloxacin (33.8%), erythromycin (27.9%), and trimethoprim/sulfamethoxazole (22%). Statistical analysis indicated no significant variation in susceptibility patterns among the S. aureus strains to different tested antimicrobial classes (p = 0.09). Among the meat isolates, 20.7% (11/53) demonstrated sensitivity to all antibiotics, while 35.8% (19/53) exhibited resistance (R) to penicillin (PEN) in the presence of blaZ, fosB-Saur, and tet38 genes, coupled with a MAR index of 0.05. Furthermore, 43% (23/53) of the meat isolates were categorized as MDR, displaying resistance to at least one antimicrobial agent in a minimum of three categories, according to Magiorakos et al.’s criteria (Magiorakos et al., 2012). These MDR strains harbored aph(3’)-IIIa, blaZ, mphC, msrA, sat4,and tet38 genes, and had a MAR index ranging from 0.15 to 0.3. Clinical S. aureus isolates exhibited diverse resistance patterns, with 100% demonstrating MDR and MAR indices ranging from 0.15 to 0.8, as shown in Table 1. The study revealed positive correlations between: tet gene with PEN (r=1); mecA with CFX (r=0.85); and dfrG with SXT (r=0.75), as depicted in Figure 1.
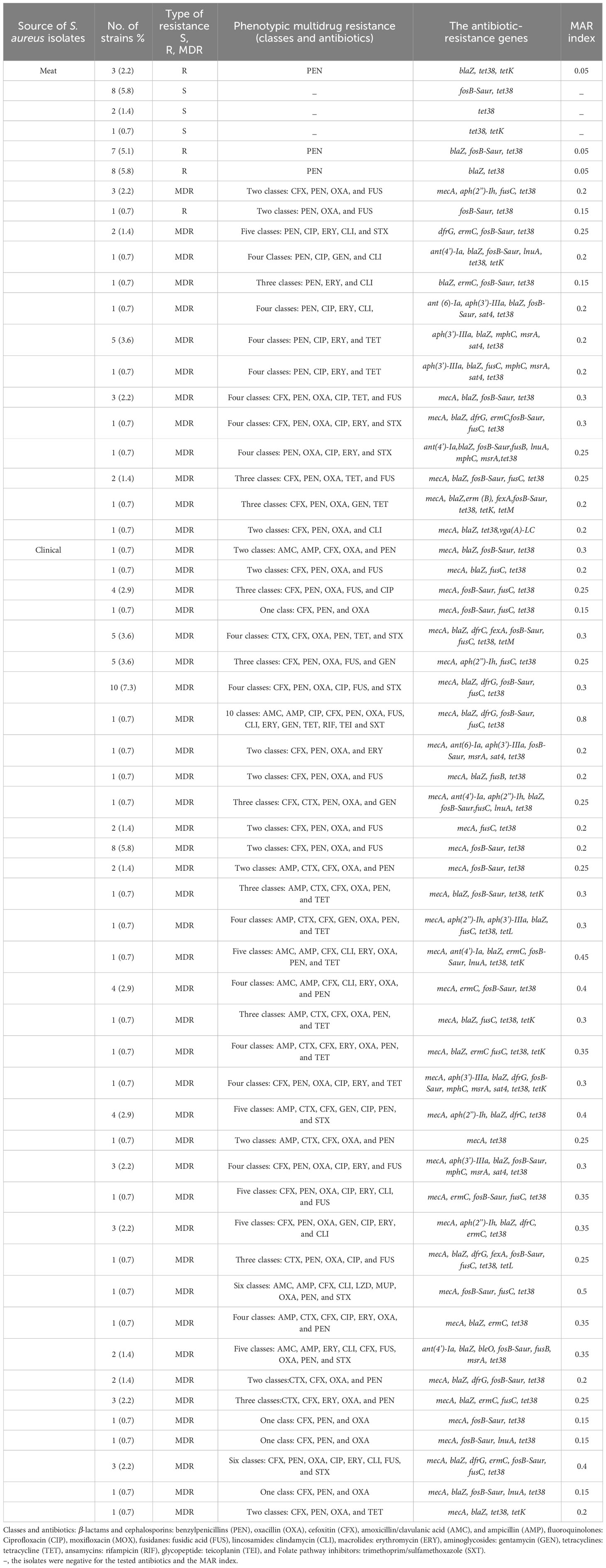
Table 1 The Prevalence of Multidrug-Resistance Profiles and the Resistance Genes Among the isolated S. aureus (n=136).
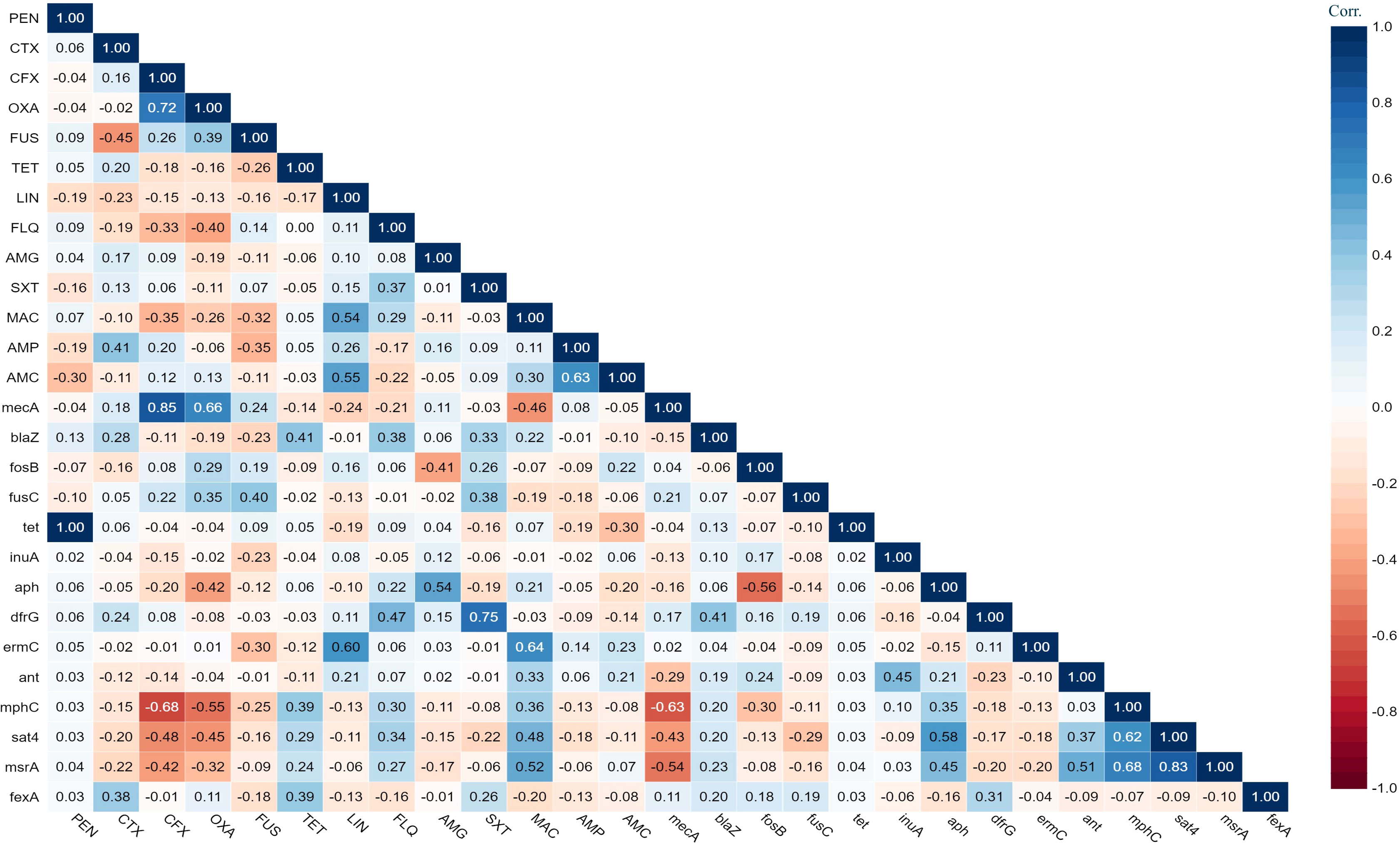
Figure 1 The heat map illustrates the correlation coefficient (r) between the demonstrated antimicrobial resistance genes in the S. aureus strains and different antimicrobial agents.
3.4 Molecular typing of Staphylococcus aureus isolates
Multiple S. aureus clones were found in both patients and meat, with clonal complex CC5 being the most prevalent in both groups. CC5 comprised (38/80, 47.5%) clones from patients and (5/11, 45.45%) clones from meat. CC97 was the second most common clone in meat, making up (4/11, 36%) of the MRSA clones, while in patients it was only (6/80, 8%). CC361 was represented by (2/11, 18%) of the meat MRSA isolates and (5/80, 6%) of patients’ MRSA isolates. The clonal lineages of all S. aureus strains isolated from the patients and meat samples were identified using MLST (Figure 2). Accessory gene regulator (agr) alleles were identified in 136 isolates by WGS. agr I was the most prevalent (58/136; 43%), followed by agr II (53/136; 39%) and agr III (22/136; 16%). Only one clinical S. aureus isolate harbored agr IV. The most common agr type of S. aureus isolated from meat was agr II, whereas agr I was the most common agr type isolated from the patients (Figure 2).
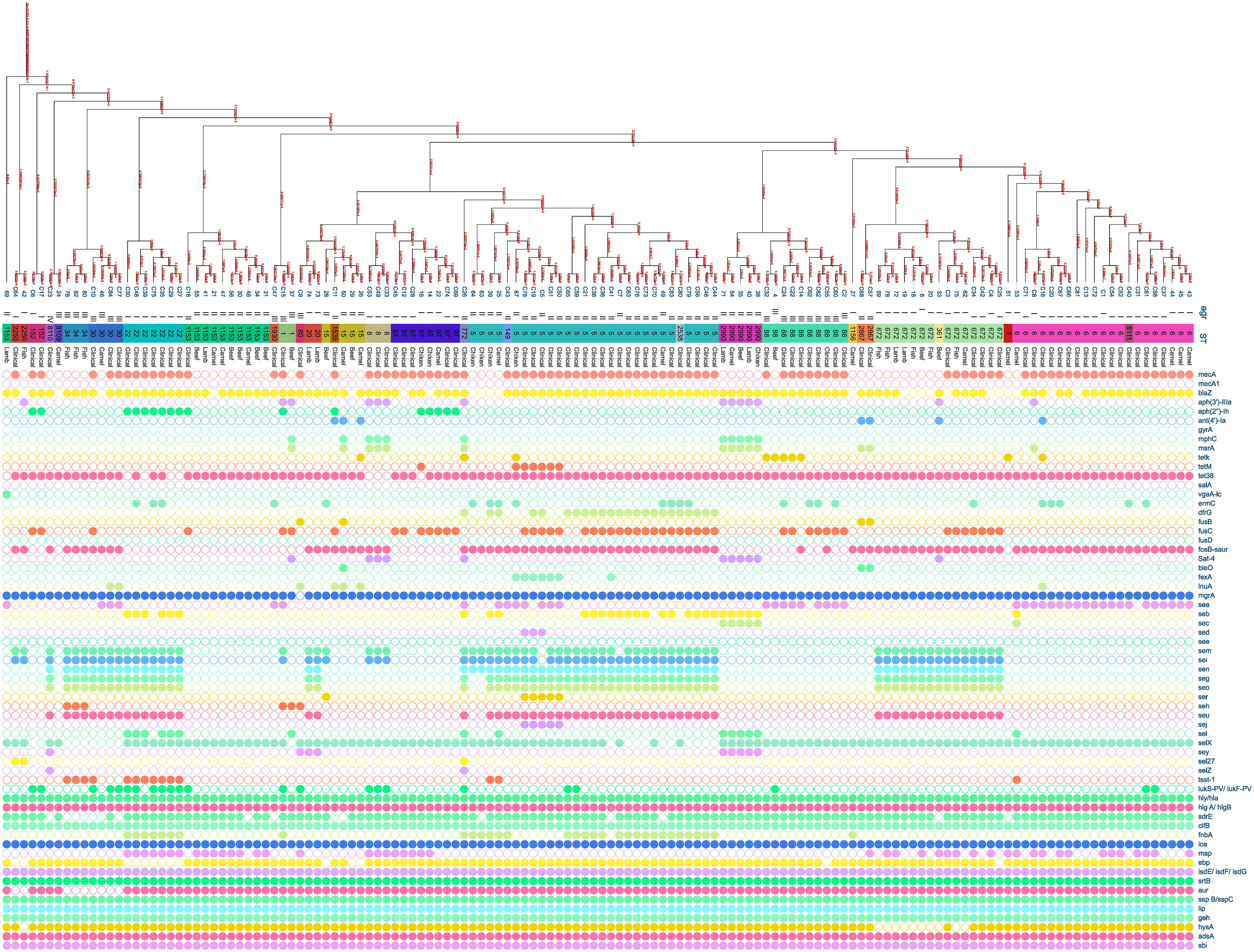
Figure 2 Distribution of antibiotic resistance genes, enterotoxin genes, and virulence genes plotted against core genome phylogeny of S. aureus isolates based on single nucleotide polymorphisms (SNPs). The agr, sequence types, and sources of the isolates were aligned with the tree. The fully colored circle indicates the presence of the target gene. White circles indicate the absence of investigated genes.
3.5 Antibiotic-resistance genes in Staphylococcus aureus strains
Staphylococcus aureus isolates from patients and meat samples demonstrated similar antimicrobial resistance gene profiles. Of the 53 meat isolates, 11 (21%) were mecA gene-positive and 42 (79%) were mecA gene-negative. Overall, the majority of S. aureus isolates from meat and patients contained β -lactam, tetracycline, and phosphonic acid resistance genes (Figures 3A, B). Meat isolates displayed a high prevalence of resistance genes harboring tet38 (69%), blaZ (70%), and fosB (58%), whereas patient isolates harbored tet38 (100%), blaZ (62%), and fosB (70%) (Figures 3A, B). A significant difference was observed between the prevalence of antibiotic resistance genes in S. aureus isolates from meat and patients (p < 0.0001). Meat isolates had multidrug resistant genes (33/53; 62.2%), specifically 100% MRSA and 52% MSSA, containing antibiotic resistance genes for three or more drug classes. All patient isolates (100%) harbored several drug resistance genes (Table 2).
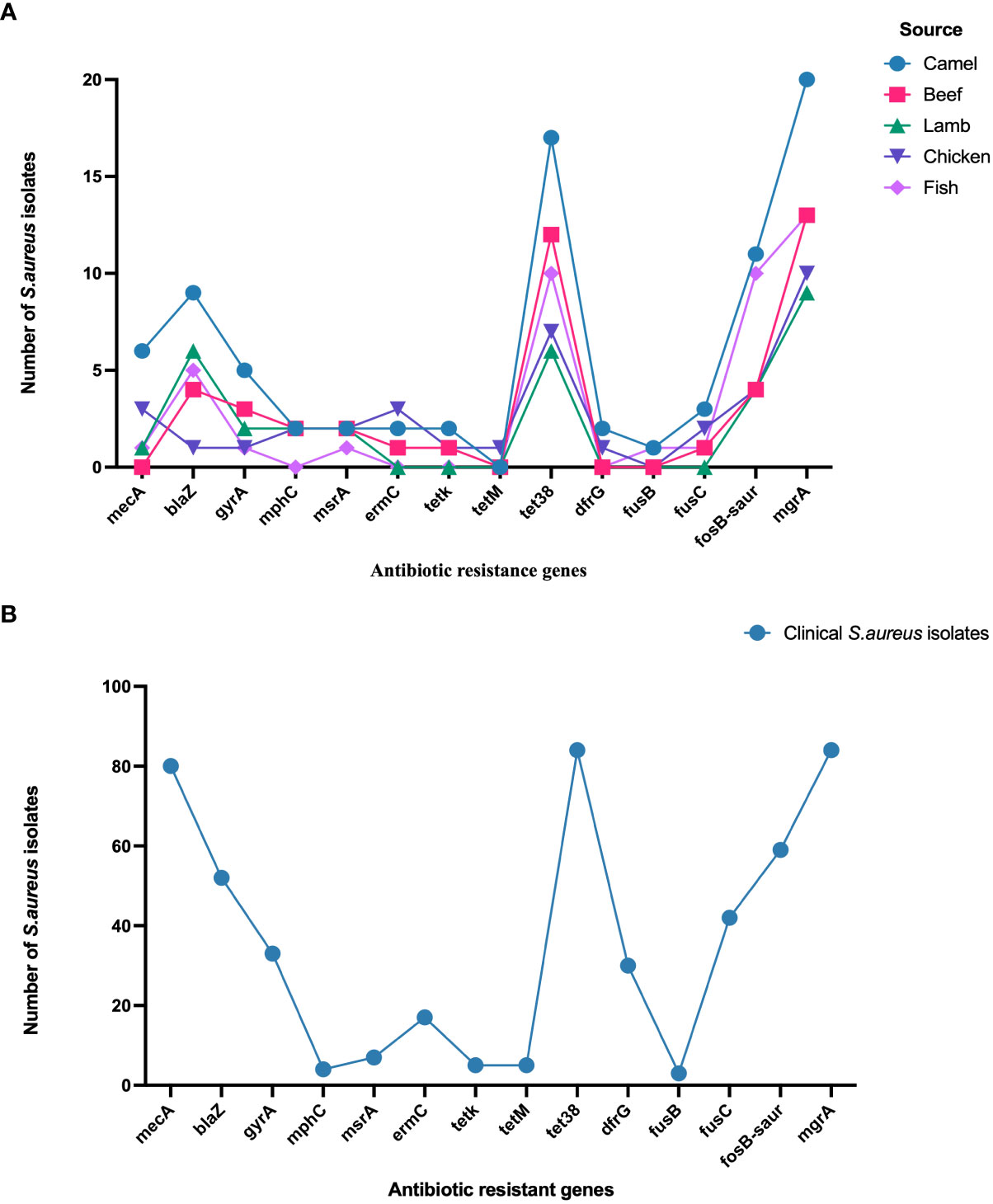
Figure 3 (A) Distribution of antibiotic resistance genes in S. aureus isolated from meat. (B). Distribution of antibiotic resistance genes in S. aureus isolated from patients.
3.6 Prevalence of enterotoxin genes in Staphylococcus aureus strains
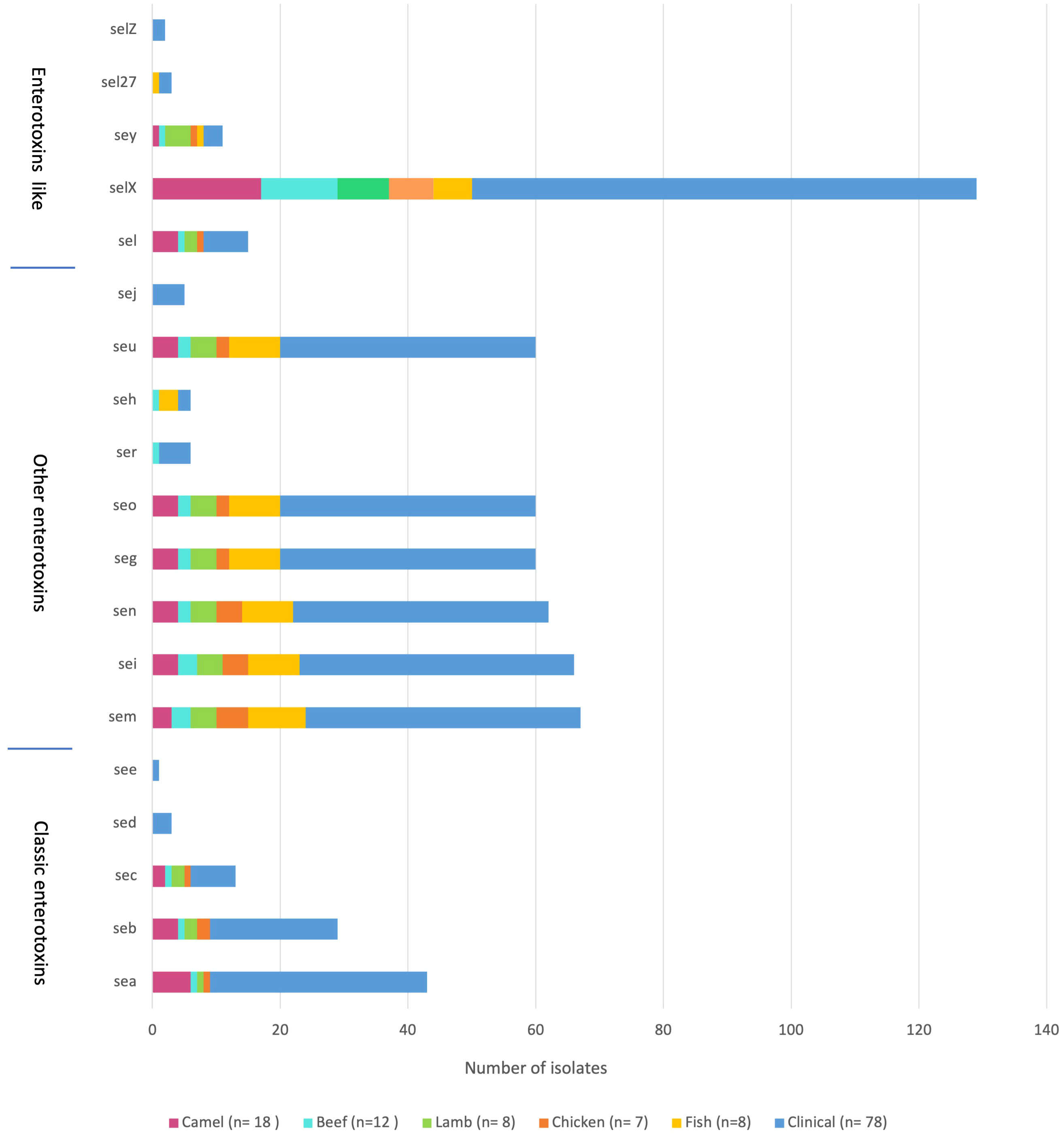
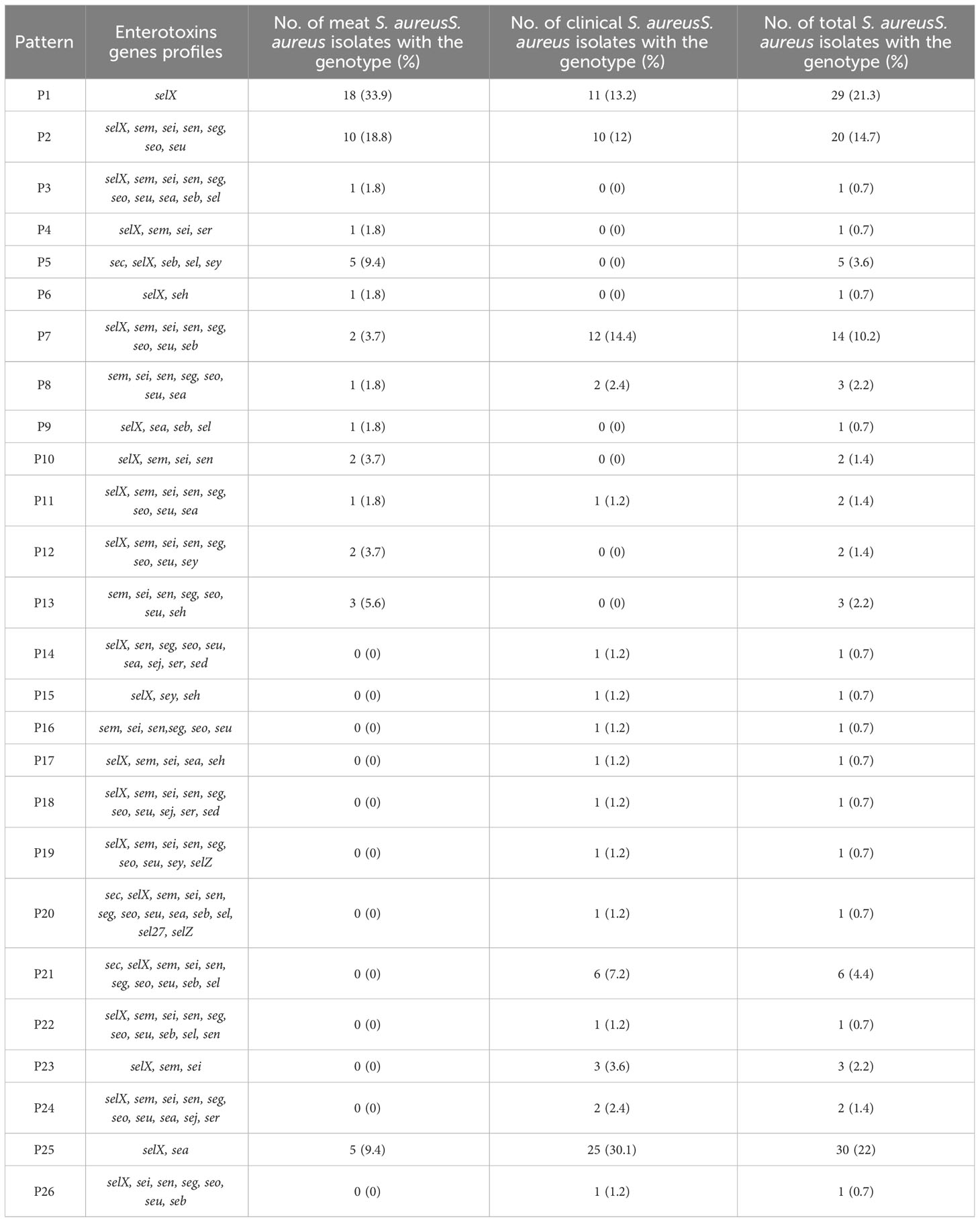
The sequencing results revealed 131 S. aureus isolates harboring different enterotoxin genotypes (Figure 4). The 18 enterotoxin genotypes detected in this study included the classic enterotoxin genes (sea, seb, sec, sed, and see), enterotoxin-like genes (sel, selX, sel27, selz, and sey), and other enterotoxin genes (sej, seh, ser, seu, seg, seo, sen, sem, and sei). MSSA and MRSA isolates from meat and patients harbored enterotoxin genes, which varied from 0 to 12. The enterotoxin-like gene selX was detected in almost all S. aureus isolates (129/136; 94.8%), followed by the sem and sei genes (66/136; 48.5%) and sen (62/136; 45.5%). The classic genes see and sed were absent from meat isolates but were present in three patient isolates. All other enterotoxin genes (seu, seg, seo, sen, sem, and sei) were observed more frequently in all the isolates. The prevalence of different enterotoxin genes in patient isolates was higher than that in the meat isolates (p =0.0239). A large proportion of the S. aureus isolates (104/136, 76.4%) harbored more than one enterotoxin gene (Table 3). The highest prevalence was found in isolates with the genotype selX, sea (23%) followed by genotype selX (22%). More than 67% of the meat and patient isolates had more than five different antibiotic resistance genes and more than 45% of these isolates had more than five enterotoxins (Figure 5).
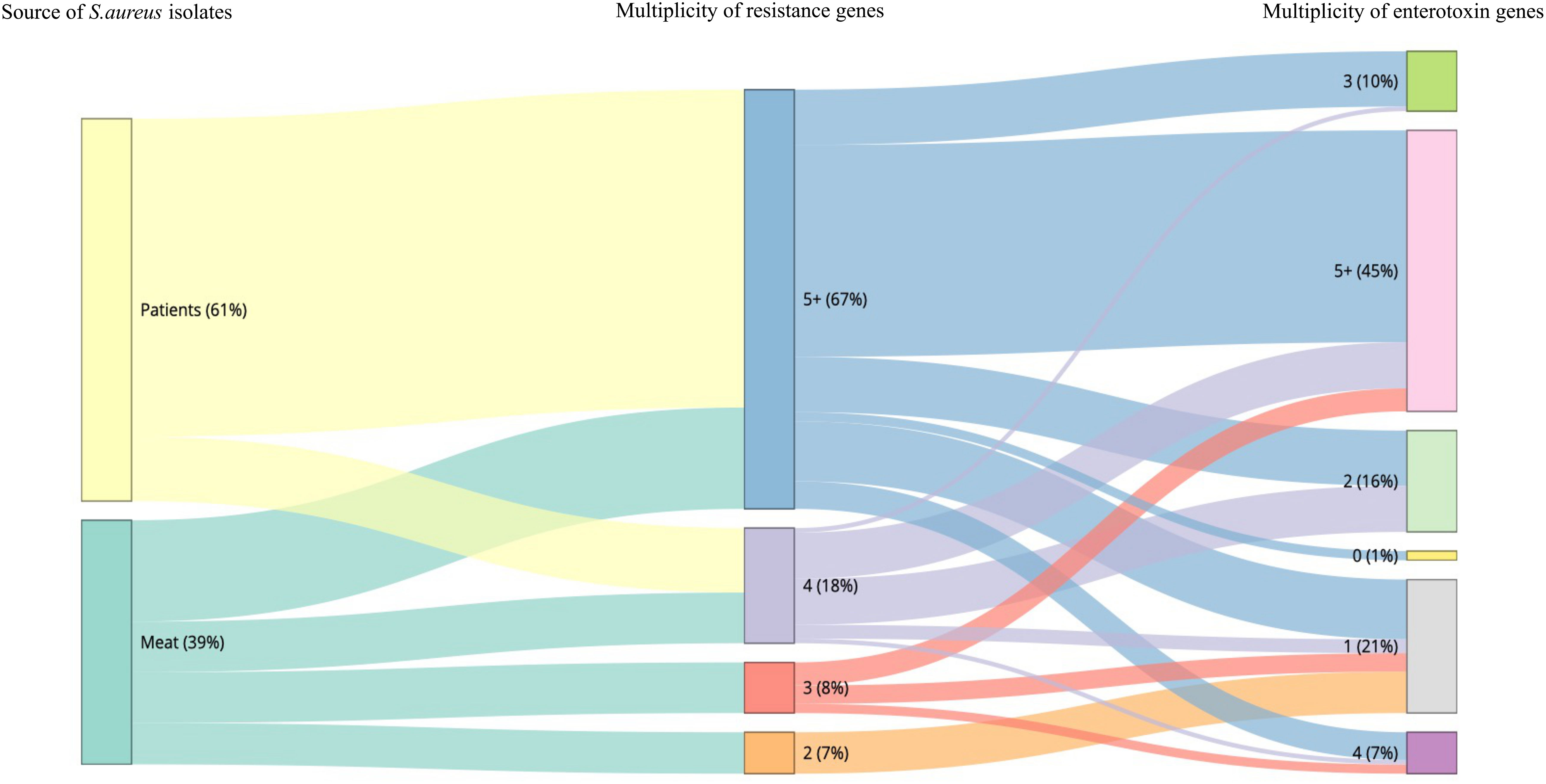
Figure 5 Snakey diagram illustrating the multiplicity of resistance and enterotoxin genes present in S. aureus isolates from both meat samples and patients.
3.7 Prevalence of virulence genes in Staphylococcus aureus strains
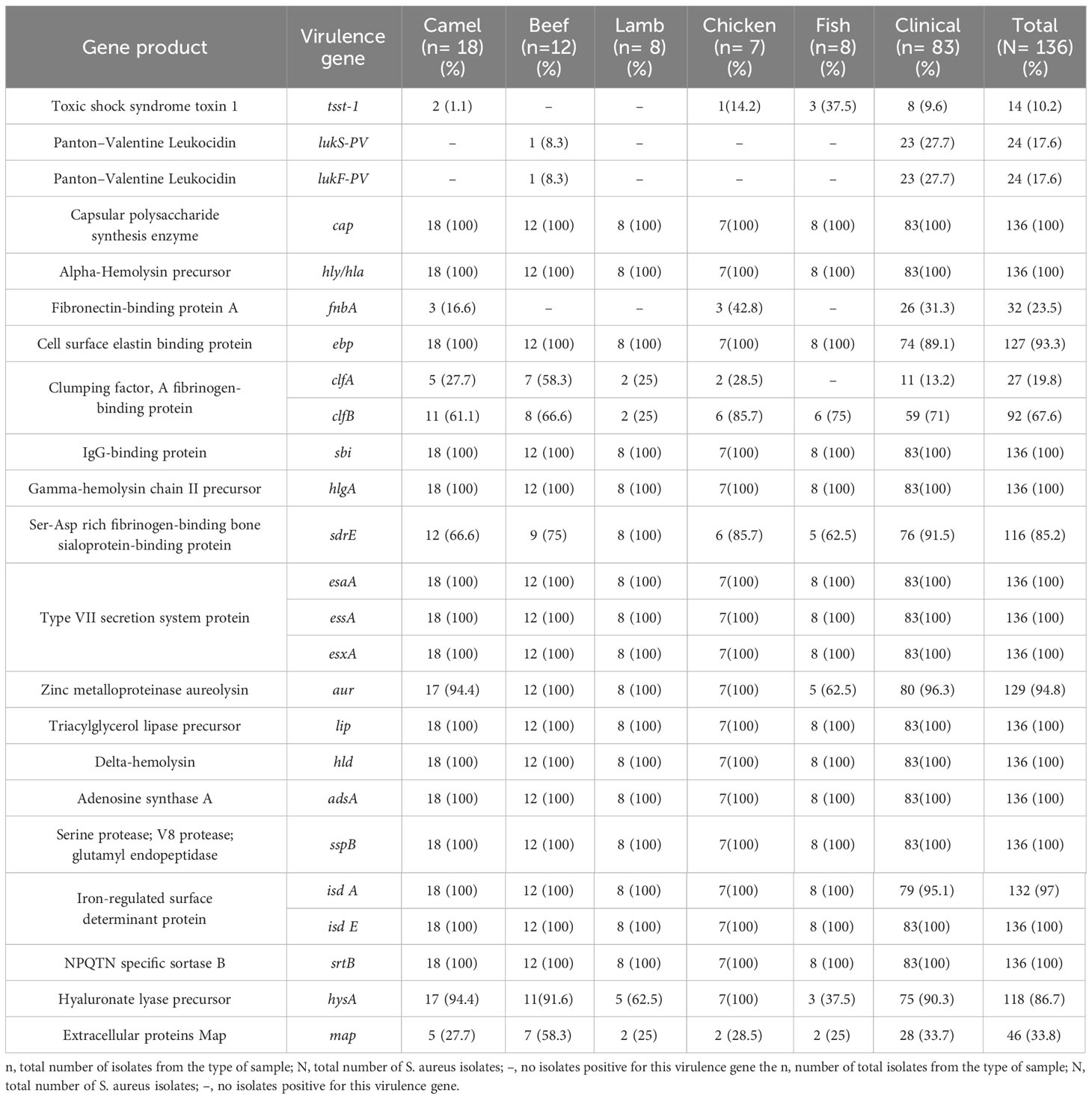
Toxin genes were distributed in a similar pattern in S. aureus isolates (Table 4). The virulence genes cap, hly/hla, sbi, geh, hlgA/hlgB, esaA/esaB, essA/essB, esxA, ica, lip, hld, adsA, sspB/sspC, isdE/isdF/isdG, and srtB were detected in all isolates. Almost all isolates harbored sspA, isdB/isdC/isdD (134/136; 99%), hlgC (133/136; 98%), isdA (132/136; 97%), aur (129/136; 95%), and ebp (127/136; 93%) genes. Other virulence genes were also prevalent among the isolates, including hysA (118/136; 87%), sdrE (116/136; 85%), sdrC (113/136; 83%), sdrD (112/136; 82%), esaC and esxB (107/136; 79%), essC (106/136; 78%), and clfB (92/136; 68%). Other virulence genes were also present: map (46/136; 34%), fnbA (32/136; 24%), clfA (27/136; 20%), pvl (lukS-PV/lukF-PV) (24/136; 18%), sell (15/136; 11%), and tst-1 (14/136; 10%). Six isolates of S. aureus carrying the toxin gene tst-1 were identified in meat, including two camels, one chicken, three fish, and eight from patients. Furthermore, a single S. aureus isolate from beef and 23 isolates obtained from patients harbored the pvl genes (lukS-PV/lukF-PV). A significant difference in the distribution of virulence genes was observed between S. aureus isolates from meat and patients (p < 0.0001).
The prevailing resistance gene patterns observed among MRSA isolates derived from both meat sources and human patients encompass mecA, aph(2’’)-Ih, fusC, and tet38 which were identified in three meat samples associated with the MRSA Clonal Complex 97 (CC97). In the patients, they were present in MRSA isolates belonging to CC97, CC152, and CC153. In the context of virulence genes, the distribution patterns of genes exhibited nearly uniform prevalence across various clonal complexes.
4 Discussion
In this study, S. aureus isolates were found in 21.2% of retail meat samples. Similarly, several previous studies have reported comparable levels of S. aureus contamination in raw meat: 27.8% prevalence in the United States (Thapaliya et al., 2017), 24.5% in various African countries (Thwala et al., 2021), 23.8% in Iran (Safarpoor Dehkordi et al., 2017), 35% in China, 33.3% in the Netherlands (van Loo et al., 2007), and 22% in Bangladesh (Pu et al., 2009). Similarly, several earlier investigations have reported comparable levels of S. aureus contamination in raw meat: 27.8% prevalence in the United States (Thapaliya et al., 2017), 24.5% in various African countries (Thwala et al., 2021), 23.8% in Iran (Safarpoor Dehkordi et al., 2017), 35% in China, 33.3% in the Netherlands (van Loo et al., 2007), and 22% in Bangladesh (Pu et al., 2009). Various factors, such as hygiene practices, storage conditions, and the use of antibiotics in animal husbandry can differ between regions and impact the presence of S. aureus in meat (Dorjgochoo et al., 2023). These regional variations are essential for understanding and addressing the food safety concerns associated with S. aureus contamination.
In addition, 136 S. aureus isolates had diverse genetic backgrounds as demonstrated by MLST and agr typing. The most prevalent agr type in patient isolates in the present study was agr I. This finding aligns with previous reports indicating the predominance of agr I among MRSA isolates associated with clinical infections in Riyadh (54.21% and 42.4%, respectively) (Monecke et al., 2012; Senok et al., 2019). In contrast, the predominant agr type in meat in this study was agr II. A previous study reported the prevalence of agr types I, II, and III in S. aureus isolated from retail meat in Riyadh, all of which were evenly distributed across isolates (Raji et al., 2016). Although agr IV has previously been identified in other clinical S. aureus isolates, only one clinical isolate in the present study harbored the same agr type (Monecke et al., 2012; Senok et al., 2019). The incompatibility of agr types over time indicates the flexibility of S. aureus isolates for adaptation. The clinical S. aureus isolates harboring agr IV in this study shared the same clonal lineage (CC121) as those from retail meat, carrying the same agr type, thereby demonstrating genetic relatedness (Raji et al., 2016).
The high prevalence of antibiotic resistance genes in both patient and meat isolates is concerning because it underscores the potential for the transmission of resistance traits from the food supply to the clinical setting. tet38, which encodes tetracycline resistance, was found in 100% and 69% of patients and meat isolates, respectively. A previous study reported a tetK prevalence of 4% among S. aureus isolates from retail meat (Raji et al., 2016). Additionally, in a hospital in Riyadh, S. aureus isolates had tetK and tetM prevalence rates of 10.2% and 20.5%, respectively (Monecke et al., 2012). The increase in the prevalence of tet gene may indicate the excessive use of tetracyclines over the past few years. Moreover, β -lactam resistance genes, including blaZ, were highly prevalent in 70% and 62% of meat and patients, respectively. Comparatively, these rates slightly decreased compared to previous studies in Saudi Arabia, where isolates from retail meat in Riyadh had a blaZ prevalence rate of 88% (Raji et al., 2016) and those from hospitals in Riyadh were 93.46% (Monecke et al., 2012). Furthermore, the gene encoding phosphonic acid resistance, fosB, had prevalence rates of 70% and 58% in the patient and meat isolates in the present study, respectively. This is consistent with a previous study on retail meat, wherein 52% of S. aureus isolates harbored fosB (Raji et al., 2016). The high prevalence of antibiotic -resistance genes is most likely a result of the overuse of β-lactams and phosphonic acid antibiotics to treat S. aureus infections in hospitals and livestock farms.
The distribution of antibiotic resistance genes among MRSA strains in a tertiary care facility in Riyadh was as follows: ermC (28.8%), msrA (10.4%), aphA3 and sat (18.4%), fusC (43.2%), tetK (17.6%), tetM (7.2%), and fosB (56.8%) (Senok et al., 2019). According to Snoussi et al. (2023), the genes associated with antimicrobial resistance in S. aureus isolates included fusC (50%), fosB, tetK, tet45, sat-4, aph(3′)-IIIa (12%), aac(6′)-aph(2″), and ermC (25%). In the present study, resistance genes in meat isolates included aph(3’)-IIIa (15%), gyrA (23%), tetK (7%), tetM (2%), ermC (9%), fosB (62%), msrA, fusC, and sat-4 (13%), whereas those in patient isolates included aph(3’)-IIIa (7%), gyrA (40%), msrA (8%), ermC (20%), fusC (51%), fosB (71%), tetK, and sat-4 (6%). Although some antibiotic resistance genes had higher prevalence levels in meat isolates, such as aph(3’)-IIIa, tetK, msrA, and sat-4, patient isolates harbored more antimicrobial resistance genes. This might due to the environment in which patient isolates are genetically adapted to thrive and flourish in hospitals. The fact that all patient isolates harbored multidrug-resistant genes is particularly alarming, highlighting the importance of prudent antibiotic use in healthcare. Furthermore, the presence of multidrug-resistant S. aureus strains in meat isolates raises concerns regarding the role of foodborne transmission in the dissemination of antibiotic resistance, highlighting the importance of surveillance in food production and distribution.
The high prevalence of some antibiotic genes in this study, such as fosB, in MRSA isolates raises concerns regarding the potential compromise of fosfomycin as a last-resort therapy for staphylococcal infections. Fosfomycin is used to treat MRSA and MDR infections because it can penetrate biofilms and exert intracellular bactericidal activity because of its low molecular weight (Chen et al., 2022). Fosfomycin is often used in combination with other antibiotics to reach targets more efficiently, thereby improving the therapeutic effects of the combined antibiotics (Chen et al., 2022). The high prevalence of fosB gene, highlights the risk of emerging resistance, which affects the efficacy of fosfomycin in the treatment MRSA infections. In the present study, the MAR index values (0.2–0.5) of most MDR S. aureus strains revealed multiple resistance patterns, suggesting that the S. aureus strains were derived from high-risk contamination (Krumperman, 1983). This result emphasizes the potential public health implications of antimicrobial resistance in S. aureus isolates.
Staphylococcus aureus enterotoxins are gastrointestinal exotoxins that can cause SFP when consumed in certain amounts (Argudín et al., 2010). Staphylococcal enterotoxin A (sea) is the most frequently identified cause of SFP, and classic enterotoxins (sea, seb, sec, sed, and see) are associated with the most reported food poisoning outbreaks (Cha et al., 2006). In Kuwait, a study of 200 isolates obtained from food handlers in various restaurants with suspected SFP revealed that the majority of isolates were resistant to various antibacterial agents, and that 71% of the isolates harbored genes for SEs, with sei being the most prevalent (Udo et al., 2009). In the present study, the enterotoxin-like gene selX was found in 95% of the patient isolates, followed by the sem and sei genes, and sen. Meat isolates exhibited comparable prevalence rates. These rates differ depending on the source of isolation and area. For instance, in Riyadh, a previous study isolated S. aureus from dairy products and identified 14 enterotoxin genes, of which seh had the highest ratio (51%). Furthermore, classic enterotoxins have a lower prevalence, except for the see gene (27.5%) (Alghizzi and Shami, 2021). In another study, the prevalence of seh gene in S. aureus isolates from processed meat was the highest at 49% (Alghizzi et al., 2021). In 2014, 165 S. aureus isolates were obtained from food handlers in Makkah, and PCR analysis of classic SEs genes revealed that sea was the most prevalent enterotoxin in MRSA (36%) and MSSA (30%) isolates (Ahmed and Mashat, 2014). Similarly, the most prevalent classic enterotoxin in S. aureus isolates in the present study was sea. The prevalence of classic SEs genes in MRSA isolates from patients in Riyadh in 2012 was < 9% (Monecke et al., 2012). In the early 2000s, 129 S. aureus isolates were identified from food handlers in Makkah during the Hajj season, and sec and sea were observed in 15.5% and 12.4% of the S. aureus isolates, respectively (Dablool and Al-Ghamdi, 2011). In the present study, sec was detected in 10% of the S. aureus isolates, and the prevalence of the sea gene was 32%. The high compatibility of enterotoxin genotypes in S. aureus isolates from meat and patients indicates a strong genomic association between the isolates from both sources. Furthermore, the higher prevalence of enterotoxin genes in the patient isolates suggests their role in the clinical manifestations of S. aureus infections, potentially leading to more severe symptoms.
Every toxin-encoding gene is equally important; the more genes an organism possesses, the more virulent it becomes (Tam and Torres, 2019). In Saudi Arabia, information regarding the prevalence and molecular characteristics of these virulence genes is limited. However, our analysis revealed a consistent distribution pattern of virulence genes across S. aureus isolates from meat samples and patients, indicating a common genetic structure among pathogens from both sources. Notably, key virulence genes, including cap, hly/hla, sbi, geh, hlgA/hlgB, esaA/esaB, essA/essB, esxA, ica, lip, hld, adsA, sspB/sspC, isdE/isdF/isdG, and srtB were present in all S. aureus isolates. This consistency suggests that these genes play essential roles in the pathogenesis of S. aureus from various sources. Although a significant difference in the distribution of virulence genes was observed between S. aureus isolates from meat samples and patients (p < 0.0001), it is essential to note the different sample sizes in each group. Nevertheless, this significance underscores the robust association between virulence gene distribution and the source of S. aureus isolates.
S. aureus produces the cytotoxin PVL, which destroys leukocytes and results in tissue necrosis (Otto, 2014). Previously, pvl gene was not abundant, except in some cases, and was often associated with CA-MRSA strains (Alghizzi and Shami, 2021; Alghizzi et al., 2021). For example, this gene was not detected in 50 S. aureus isolates obtained from patients in Makkah in 2017 (Abulreesh et al., 2017). In addition, it is absent in S. aureus isolated from dairy products, milk, and processed foods in Riyadh (Alghizzi and Shami, 2021; Alghizzi et al., 2021). In this study, 28% of S. aureus isolates from patients harbored pvl, and only one S. aureus isolate from beef that belonged to LA-MRSA CC361 harbored this gene. A previous study of 125 MRSA isolates linked to clinical infections in Riyadh reported a pvl prevalence of 30% (Senok et al., 2019). The high prevalence of pvl-carrying S. aureus clones in hospitals, which is known for its association with CA-MRSA, suggests that CA-MRSA isolates outnumber HA-MRSA in hospitals and that there is an over-time shift in the distribution of MRSA types.
Toxic shock syndrome (TSS) exotoxin is a superantigen that is generally resistant to heat and proteolysis and induces T cell-dependent shock syndrome with significant mortality by boosting the excessive release of cytokines (Dinges et al., 2000). Although S. aureus isolated from meat had a slightly higher prevalence of the tst gene in this study, it did not vary greatly from that of S. aureus isolated from patients. These numbers are slightly lower than the national tst gene prevalence rates, which range from 11.2% to 13.2% in human infections (Alkharsah et al., 2018; Senok et al., 2019). Evaluation of the frequency of virulence genes in S. aureus strains serves as a fundamental basis for formulating effective infection control strategies and anti-virulence interventions to suppress S. aureus with hypervirulent genes (Dinges et al., 2000). A limitation of this study was the small sample size, which was determined by the city’s landscape within a confined geographical area. Moreover, this study was constrained by the comparison of MRSA isolates from symptomatic humans with S. aureus and MRSA isolated from meat products. It is important to note, however, that our findings align with previous research conducted in this region (Raji et al., 2016; Aljeldah, 2020; Alghizzi et al., 2021).
The observed correlation in genetic lineages, antibiotic resistance profiles, enterotoxin genes, and virulence genes between the two sources of S. aureus has significant implications for public health. Further research is recommended to explore the genetic factors involved in S. aureus pathogenicity and their potential impact on clinical outcomes and food safety. Additionally, protocols to monitor and limit the transmission of S. aureus between clinical and foodborne settings should be developed to mitigate the potential risks to public health.
5 Conclusion
Our findings provide a comprehensive exploration of the genomic properties of the clinical and foodborne S. aureus strains. The results strongly support the close genetic association between S. aureus isolates from meat and patients, revealing shared antibiotic resistance and virulence gene profiles. Notably, the detection of a diverse array of these genes in meat-derived S. aureus, which is highly compatible with that found in patient isolates, underscores the significant genomic relatedness observed in our study. The high prevalence of specific genes such as selX, tet38, blaZ, fosB, and mecA, highlights potential challenges associated with virulence and antibiotic resistance. Our study indicates that the misuse of antibiotics may contribute to the dissemination of multidrug resistance across S. aureus isolates from different settings, with meat serving as a potential reservoir for MDR dissemination. In light of these findings, a comprehensive approach is essential for estimating the prevalence, prevent, and control S. aureus infections, thereby improving both animal and human health.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The study protocol was approved by the Institutional Review Board (IRB) committee of King Saud Medical City, Riyadh, Saudi Arabia (H-01-R-053) and the Institutional Biosafety and Bioethics Committee of King Abdullah University of Science and Technology (22IBEC051). The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from primarily isolated as part of your previous study for which ethical approval was obtained. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
DA: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. MA: Conceptualization, Investigation, Project administration, Supervision, Validation, Visualization, Writing – review & editing. AB: Resources, Supervision, Writing – review & editing. MAla: Data curation, Formal Analysis, Funding acquisition, Resources, Software, Supervision, Writing – review & editing. MM: Investigation, Writing – review & editing. SA: Methodology, Resources, Writing – review & editing. MAls: Methodology, Resources, Writing – review & editing. TG: Funding acquisition, Resources, Supervision, Writing – review & editing. SA: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was partially funded by the Saudi Food and Drug Authority (SFDA), Riyadh, Saudi Arabia, and King Abdullah University of Science and Technology (KAUST), Thwal, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The views expressed in this study are those of the authors and do not necessarily reflect those of the SFDA or its stakeholders. Guaranteeing the accuracy and validity of the data was the sole responsibility of the research team.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1339339/full#supplementary-material
References
Abulreesh, H. H., Organji, S. R. (2011). The prevalence of multidrug-resistant staphylococci in food and the environment of Makkah, Saudi Arabia. Res. J. Microbiol. 6 (6), 510–523. doi: 10.3923/jm.2011.510.523
Abulreesh, H. H., Organji, S. R., Osman, G. E. H., Elbanna, K., Almalki, M. H. K., Ahmad, I. (2017). Prevalence of antibiotic resistance and virulence factors encoding genes in clinical Staphylococcus aureus isolates in Saudi Arabia. Clin. Epidemiol. Glob Health 5 (4), 196–202. doi: 10.1016/j.cegh.2016.08.004
Ahmad-Mansour, N., Loubet, P., Pouget, C., Dunyach-Remy, C., Sotto, A., Lavigne, J. P., et al. (2021). Staphylococcus aureus toxins: an update on their pathogenic properties and potential treatments. Toxins (Basel) 13 (10), 677. doi: 10.3390/toxins13100677
Ahmed, B., Mashat, B. H. (2014). Prevalence of classical enterotoxin genes in staphylococcus aureus isolated from food handlers in Makkah city kitchens. Asian J. Sci. Tech. 5 (11), 727–731.
Algammal, A. M., Hashem, M. E. A., Alfifi, K. J., Al-Otaibi, A. S., Alatawy, M., Eltarabili, R. M., et al. (2022a). Sequence analysis, antibiogram profile, virulence and antibiotic resistance genes of XDR and MDR gallibacterium anatis isolated from layer chickens in Egypt. Infect. Drug Resist. 15, 4321–4334. doi: 10.2147/IDR.S377797
Algammal, A. M., Hetta, H. F., Elkelish, A., Alkhalifah, D. H. H., Hozzein, W. N., Batiha, G. E. S., et al. (2020). Methicillin-resistant staphylococcus aureus (MRSA): One health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect. Drug Resist. 13, 3255–3265. doi: 10.2147/IDR.S272733
Algammal, A. M., Ibrahim, R. A., Alfifi, K. J., Ghabban, H., Alghamdi, S., Kabrah, A., et al. (2022b). A First Report of Molecular Typing, Virulence Traits, and Phenotypic and Genotypic Resistance Patterns of Newly Emerging XDR and MDR Aeromonas veronii in Mugil seheli. Pathogens 11 (11), 1262. doi: 10.3390/pathogens11111262
Alghizzi, M. J., Alansari, M., Shami, A. (2021). The prevalence of Staphylococcus aureus and methicillin resistant staphylococcus aureus in processed food samples in Riyadh, Saudi Arabia. J. Pure Appl. Microbiol. 15 (1), 91–99. doi: 10.22207/JPAM.15.1.03
Alghizzi, M., Shami, A. (2021). The prevalence of Staphylococcus aureus and methicillin resistant Staphylococcus aureus in milk and dairy products in Riyadh, Saudi Arabia. Saudi J. Biol. Sci. 28 (12), 7098–7104. doi: 10.1016/j.sjbs.2021.08.004
Aljeldah, M. M. (2020). Prevalence of Methicillin-Resistant Staphylococcus aureus (MRSA) in Saudi Arabia: A systematic review. J. Pure Appl. Microbiol. 14 (1), 37–46. doi: 10.22207/JPAM.14.1.07
Alkharsah, K. R., Rehman, S., Alkhamis, F., Alnimr, A., Diab, A., Al-Ali, A. K. (2018). Comparative and molecular analysis of MRSA isolates from infection sites and carrier colonization sites. Ann. Clin. Microbiol. Antimicrob. 17 (1), 7. doi: 10.1186/s12941-018-0260-2
Argudín, M. Á., Mendoza, M. C., Rodicio, M. R. (2010). Food poisoning and staphylococcus aureus enterotoxins. Toxins (Basel) 2 (7), 1751–1773. doi: 10.3390/toxins2071751
Boost, M. V., Wong, A., Ho, J., O’Donoghue, M. (2013). Isolation of Methicillin-Resistant Staphylococcus aureus (MRSA) from retail meats in Hong Kong. Foodborne Pathog. Dis. 10 (8), 705–710. doi: 10.1089/fpd.2012.1415
Cha, J. O., Lee, J. K., Jung, Y. H., Yoo, J. I., Park, Y. K., Kim, B. S., et al. (2006). Molecular analysis of Staphylococcus aureus isolates associated with staphylococcal food poisoning in South Korea. J. Appl. Microbiol. 101 (4), 864–871. doi: 10.1111/j.1365-2672.2006.02957.x
Chen, T., Zhao, L., Liu, Y., Wang, Y., Jian, Y., Zhao, N., et al. (2022). Mechanisms of high-level fosfomycin resistance in Staphylococcus aureus epidemic lineage ST5. J. Antimicrobial Chemotherapy. 77 (10), 2816–2826. doi: 10.1093/jac/dkac236
Clinical and Laboratory Standards Institute. (2023). “M100 Performance Standards for Antimicrobial Susceptibility Testing A CLSI supplement for global application,” 33rd. Ed. Wayne, P. A. (Clinical and Laboratory Standards Institute (CLSI)). Available at: www.clsi.org.
Dablool, A. S., Al-Ghamdi, S. S. (2011). Enterotoxigenicity of Staphylococcus aureus Isolated from Food Handlers during Hajj Season in Saudi Arabia. Open J. Saf. Sci. Technology. 01 (02), 75–78. doi: 10.4236/ojsst.2011.12007
Dinges, M. M., Orwin, P. M., Schlievert, P. M. (2000). Exotoxins of staphylococcus aureus. Clin. Microbiol. Rev. 13 (1), 16–34. doi: 10.1128/CMR.13.1.16
Dorjgochoo, A., Batbayar, A., Tsend-Ayush, A., Erdenebayar, O., Byambadorj, B., Jav, S., et al. (2023). Detection of virulence genes of Staphylococcus aureus isolated from raw beef for retail sale in the markets of Ulaanbaatar city, Mongolia. BMC Microbiol. 23 (1), 372. doi: 10.1186/s12866-023-03122-2
El-Ghareeb, W., Almathen, F., Fayez, M., Alsultan, R. (2019). Methicillin resistant staphylococcus aureus (mrsa) in camel meat: prevalence and antibiotic susceptibility. Slov. Vet. Res. 56 (22-Suppl), 249–256. doi: 10.26873/SVR-764-2019
Elmoslemany, A., Elsohaby, I., Alorabi, M., Alkafafy, M., Al-Marri, T., Aldoweriej, A., et al. (2021). Diversity and risk factors associated with multidrug and methicillin-resistant staphylococci isolated from cats admitted to a veterinary clinic in eastern province, Saudi Arabia. Antibiotics 10 (4), 367. doi: 10.3390/antibiotics10040367
El Sheikha, A. F. (2016). Food safety issues in Saudi Arabia. Nutrition and Food Technology 1 (1), 1-4. doi: 10.16966/nftoa.103
Fetsch, A., Etter, D., Johler, S. (2021). Livestock-associated meticillin-resistant staphylococcus aureus-current situation and impact from a one health perspective. Curr. Clin. Microbiol. Rep. 8 (3), 103–113. doi: 10.1007/s40588-021-00170-y
Ge, B., Mukherjee, S., Hsu, C. H., Davis, J. A., Tran, T. T. T., Yang, Q., et al. (2017). MRSA and multidrug-resistant Staphylococcus aureus in U.S. retail meats, 2010-2011. Food Microbiol. 62, 289–297. doi: 10.1016/j.fm.2016.10.029
Gordon, L. G., Elliott, T. M., Forde, B., Mitchell, B., Russo, P. L., Paterson, D. L., et al. (2021). Budget impact analysis of routinely using whole-genomic sequencing of six multidrug-resistant bacterial pathogens in Queensland, Australia. BMJ Open 11 (2), e041968. doi: 10.1136/bmjopen-2020-041968
Jenul, C., Horswill, A. R. (2019). Regulation of Staphylococcus aureus virulence. Gram-Positive Pathogens. 3 (12), 948–958. doi: 10.1128/microbiolspec.GPP3-0031-2018
Kadariya, J., Smith, T. C., Thapaliya, D. (2014). Staphylococcus aureus and staphylococcal food-borne disease: an ongoing challenge in public health. BioMed. Res. Int 2014, 9. doi: 10.1155/2014/827965
Kay, G., Gamaleldin, M. S., Ahmed, S., Abdullah, A., Mohammad, K. (2013). Frequency of antibiotic resistant Salmonella, Escherichia coli, Enterococcus, and Staphylococcus aureus in meat in Saudi Arabia. Afr J. Microbiol. Res. 7 (4), 309–316. doi: 10.5897/AJMR12.930
Krumperman, P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46 (1), 165–170. doi: 10.1128/aem.46.1.165-170.1983
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infection. 18 (3), 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Monecke, S., Skakni, L., Hasan, R., Ruppelt, A., Ghazal, S. S., Hakawi, A., et al. (2012). Characterisation of MRSA strains isolated from patients in a hospital in Riyadh, Kingdom of Saudi Arabia. BMC Microbiol. 12 (1), 1–9. doi: 10.1186/1471-2180-12-146
Otto, M. (2014). Staphylococcus aureus toxins. Curr. Opin. Microbiol. 17, 32–37. doi: 10.1016/j.mib.2013.11.004
Parvin, M., Ali, M., Talukder, S., Nahar, A., Chowdhury, E. H., Rahman, M., et al. (2021). Prevalence and multidrug resistance pattern of methicillin resistant S. aureus Isolated from Frozen Chicken Meat in Bangladesh. Microorganisms. 9 (3), 636. doi: 10.3390/microorganisms9030636
Petit, R. A., Read, T. D. (2020). Bactopia: a flexible pipeline for complete analysis of bacterial genomes. mSystems 5 (4). doi: 10.1128/msystems.00190-20
Pinchuk, I. V., Beswick, E. J., Reyes, V. E. (2010). Staphylococcal enterotoxins. Toxins (Basel). 2 (8), 2177–2197. doi: 10.3390/toxins2082177
Pu, S., Han, F., Ge, B. (2009). Isolation and characterization of methicillin-resistant Staphylococcus aureus strains from louisiana retail meats. Appl. Environ. Microbiol. 75 (1), 265–267. doi: 10.1128/AEM.01110-08
Raji, M. A., Garaween, G., Ehricht, R., Monecke, S., Shibl, A. M., Senok, A. (2016). Genetic characterization of Staphylococcus aureus isolated from retail meat in Riyadh, Saudi Arabia. Front. Microbiol. 7 (JUN). doi: 10.3389/fmicb.2016.00911
Safarpoor Dehkordi, F., Gandomi, H., Basti, A. A., Misaghi, A., Rahimi, E. (2017). Phenotypic and genotypic characterization of antibiotic resistance of methicillin-resistant Staphylococcus aureus isolated from hospital food. Antimicrob. Resist. Infect. Control. 6 (1), 104. doi: 10.1186/s13756-017-0257-1
Sankomkai, W., Boonyanugomol, W., Kraisriwattana, K., Nutchanon, J., Boonsam, K., Kaewbutra, S., et al. (2020). Characterisation of classical enterotoxins, virulence activity, and antibiotic susceptibility of staphylococcus aureus isolated from Thai fermented pork sausages, clinical samples, and healthy carriers in Northeastern Thailand. J. Vet. Res. 64 (2), 289. doi: 10.2478/jvetres-2020-0036
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2018). Foodborne illness acquired in the United States-Major pathogens. Emerg. Infect. Dis. 17 (1), 7–15. doi: 10.3201/EID1701.P11101
Senok, A., Somily, A. M., Nassar, R., Garaween, G., Kim Sing, G., Müller, E., et al. (2019). Emergence of novel methicillin-resistant Staphylococcus aureus strains in a tertiary care facility in Riyadh, Saudi Arabia. Infect. Drug Resist. 12, 2739–2746. doi: 10.2147/IDR.S218870
Shawish, R. R., Al-Humam, N. A. (2016). Contamination of beef products with staphylococcal classical enterotoxins in Egypt and Saudi Arabia. GMS Hyg Infect. Control 11, Doc08. doi: 10.3205/DGKH000268
Singh, R., Ray, P., Das, A., Sharma, M. (2009). Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: an in vitro study. J. Med. Microbiol. 58 (8), 1067–1073. doi: 10.1099/jmm.0.009720-0
Snoussi, M., Noumi, E., Bouali, N., Bazaid, A. S., Alreshidi, M. M., Altayb, H. N., et al. (2023). Antibiotic susceptibility profiling of human pathogenic staphylococcus aureus strains using whole genome sequencing and genome-scale annotation approaches. Microorganisms. 11 (5), 1124. doi: 10.3390/microorganisms11051124
Tam, K., Torres, V. J. (2019). Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol. Spectr. 7 (2). doi: 10.1128/microbiolspec.GPP3-0039-2018
Thapaliya, D., Forshey, B. M., Kadariya, J., Quick, M. K., Farina, S., O’ Brien, A., et al. (2017). Prevalence and molecular characterization of Staphylococcus aureus in commercially available meat over a one-year period in Iowa, USA. Food Microbiol. 65, 122–129. doi: 10.1016/j.fm.2017.01.015
The European Union One Health 2020 Zoonoses Report (2021) EFSA journal. Available at: https://www.efsa.europa.eu/en/efsajournal/pub/6971.
Thwala, T., Madoroba, E., Basson, A., Butaye, P. (2021). Prevalence and characteristics of staphylococcus aureus associated with meat and meat products in African countries: A review. Antibiotics 10 (9), 1108. doi: 10.3390/antibiotics10091108
Turner, N. A., Sharma-Kuinkel, B. K., Maskarinec, S. A., Eichenberger, E. M., Shah, P. P., Carugati, M., et al. (2019). Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat. Rev. Microbiol. 17 (4), 203–218. doi: 10.1038/s41579-018-0147-4
Udo, E. E., Al-Mufti, S., Albert, M. J. (2009). The prevalence of antimicrobial resistance and carriage of virulence genes in Staphylococcus aureus isolated from food handlers in Kuwait City restaurants. BMC Res. Notes. 2 (1), 108. doi: 10.1186/1756-0500-2-108
Urban-Chmiel, R., Marek, A., Stępień-Pyśniak, D., Wieczorek, K., Dec, M., Nowaczek, A., et al. (2022). Antibiotic resistance in bacteria—A review. Antibiotics. 11 (8), 1079. doi: 10.3390/antibiotics11081079
van Loo, I. H. M., Diederen, B. M. W., Savelkoul, P. H. M., Woudenberg, J. H. C., Roosendaal, R., van Belkum, A., et al. (2007). Methicillin-resistant Staphylococcus aureus in meat products, the Netherlands. Emerg. Infect. Dis. 13 (11), 1753–1755. doi: 10.3201/eid1311.070358
P. A., Wayne (Ed.) (2018). “Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically,” in CLSI standard M07, 11th (Clinical and Laboratory Standards Institute (CLSI)).
Wei, T., Simko, V. (2021). corrplot: Visualization of a Correlation Matrix (Version 0.92). Available at: https://github.com/taiyun/corrplot.
Keywords: Staphylococcus aureus, MRSA, antibiotic resistance genes, virulence factors, Staphylococcal toxins
Citation: Alkuraythi DM, Alkhulaifi MM, Binjomah AZ, Alarwi M, Mujallad MI, Alharbi SA, Alshomrani M, Gojobori T and Alajel SM (2024) Comparative genomic analysis of antibiotic resistance and virulence genes in Staphylococcus aureus isolates from patients and retail meat. Front. Cell. Infect. Microbiol. 13:1339339. doi: 10.3389/fcimb.2023.1339339
Received: 16 November 2023; Accepted: 26 December 2023;
Published: 12 January 2024.
Edited by:
Laura Fernandez-Garcia, Institute of Biomedical Research of A Coruña (INIBIC), SpainReviewed by:
Abdelazeem Algammal, Suez Canal University, EgyptMadubuike Umunna Anyanwu, University of Nigeria, Nigeria
Copyright © 2024 Alkuraythi, Alkhulaifi, Binjomah, Alarwi, Mujallad, Alharbi, Alshomrani, Gojobori and Alajel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dalal M. Alkuraythi, RGFsa3JpdGhpQHVqLmVkdS5zYQ==
 Dalal M. Alkuraythi
Dalal M. Alkuraythi Manal M. Alkhulaifi
Manal M. Alkhulaifi Abdulwahab Z. Binjomah
Abdulwahab Z. Binjomah Mohammed Alarwi
Mohammed Alarwi Mohammed I. Mujallad
Mohammed I. Mujallad Saleh Ali Alharbi
Saleh Ali Alharbi Mohammad Alshomrani3
Mohammad Alshomrani3 Takashi Gojobori
Takashi Gojobori Sulaiman M. Alajel
Sulaiman M. Alajel