- Centre for Host-Microbiome Interactions, Faculty of Dental, Oral & Craniofacial Sciences, King’s College London, London, United Kingdom
Biofilm formation is an important survival strategy commonly used by bacteria and fungi, which are embedded in a protective extracellular matrix of organic polymers. They are ubiquitous in nature, including humans and other animals, and they can be surface- and non-surface-associated, making them capable of growing in and on many different parts of the body. Biofilms are also complex, forming polymicrobial communities that are difficult to eradicate due to their unique growth dynamics, and clinical infections associated with biofilms are a huge burden in the healthcare setting, as they are often difficult to diagnose and to treat. Our understanding of biofilm formation and development is a fast-paced and important research focus. This review aims to describe the advancements in clinical biofilm research, including both in vitro and in vivo biofilm models, imaging techniques and techniques to analyse the biological functions of the biofilm.
1 Introduction
Biofilms are described as colonies of bacterial and fungal cells that adhere to and proliferate on both biological and non-biological materials (Costerton et al., 1995). Biofilms were first discovered centuries ago by Anton Von Leeuwenhoek, who used his microscope to examine tooth scrapings, but the first biofilm literature was not published until the 1970s, with the examination of “microbial films” taken from environmental water sources (Mack et al., 1975). However, the term “biofilm” was only introduced in the 1980s by Bill Costeron and collegues, which was used to describe microbial growth within medical samples (Lam et al., 1980; Marrie et al., 1982). More than 40 years on, we now realise that biofilms are the primary mode of growth for many bacterial and fungal species, which often form multikingdom communites with other organisms including protozoa and viruses (Burmølle et al., 2014). Therefore, understanding how biofilms form both inside and outide of the body has become an important aspect of modern medical research, such that new strategies can be developed to control environmental resevoirs of pathogens, and treat infectious diseases.
Biofilms go through a generalised life cycle. The classic model of biofilm development is a five-step process where biofilms resemble a mushroom-type shape. Planktonic cells (1) adhere, irreversibly, to surfaces which are primed with a conditioning film, followed by (2) proliferation of bacterial cells and an increase in biomass. This mass of cells then undergoes (3) maturation and (4) excretion of extracellular matrix. The final step is (5) dispersal of cells within the biofilm, allowing the dispersed planktonic cells the opportunity to adhere at other sites (Sauer et al., 2002; Stoodley et al., 2002). However, it is now thought that not all bacterial species grow in this classical way, especially within a native environment, and this concept has therefore been expanded on in recent years. A new conceptual model has been presented (Sauer et al., 2022), which instead suggests that there are 3 main stages of biofilm development; (1) aggregation and/or attachment, (2) growth and accumulation, and (3) disaggregation and/or detachment, with the possibility to aggregate and detach at any point and that these phases are affected by the environment and different conditions. This new model also accounts for non-surface-associated biofilms, particularly those observed in medicine, such as biofilms that form in the airways of patients with impaired mucociliary clearance (Sauer et al., 2022). Most importantly though, both models show that the maturation of biofilms, and the difference between simple aggregation and true biofilms, is defined by the excretion of extracellular polymeric substances (EPS), composed of carbohydrates, proteins, extracellular DNA (eDNA) and lipids (Di Martino, 2018).
Within the body a diverse microbial flora, arranged as biofilms, is associated with the skin and mucosal membranes, and these are highly beneficial to the host, for example within the gut (Macfarlane and Dillon, 2007), vagina (Leccese Terraf et al., 2016) and oral cavity (Robertson and McLean, 2015). However, dysbiosis can lead to changes within these biofilm communities, and can cause diseases such as inflammatory bowel disease (Baumgartner et al., 2021), bacterial vaginosis (Castro et al., 2019), and dental caries (Marsh, 2010), respectively. Bacteria can also cause a range of infections by forming biofilms on almost any indwelling medical device, including prosthetic joints, cardiac pacemakers, prosthetic heart valves, urinary catheters, and intravenous catheters (El Rafei et al., 2016; Beaver et al., 2021; Vilchez et al., 2021). These infections are often healthcare acquired (i.e. as a direct result of surgical contamination) but can also result from transient haematogenous spread of bacteria in the bloodstream (Honkanen et al., 2019). In addition, non-surface-associated bacterial aggregate biofilm infections are becoming more well-characterised (Alhede et al., 2011; Pabst et al., 2016; Cai, 2020), which are often chronic and low-grade, such as lung infections in cystic fibrosis patients (Høiby et al., 2010), chronic wound infections (Wei et al., 2019), and chronic otitis media (Akyıldız et al., 2013).
The type of infection being studied is important when choosing a biofilm model and screening technique, and some methods will be more appropriate than others. This review aims to discuss the different modelling techniques and the methods available for assessing functionality of biofilms.
2 Modelling biofilm growth
In vitro models for biofilm investigation can be highly tailored to the system that is being modelled. A multitude of variables can be chosen from, including different substrates on which biofilms can grow, mono- and poly-microbial seeding, seeding with bodily fluids, human-derived or artificial growth medium, chemically defined growth medium, and different culture conditions. Models can range from very simple systems to highly advanced biological simulations (Table 1).
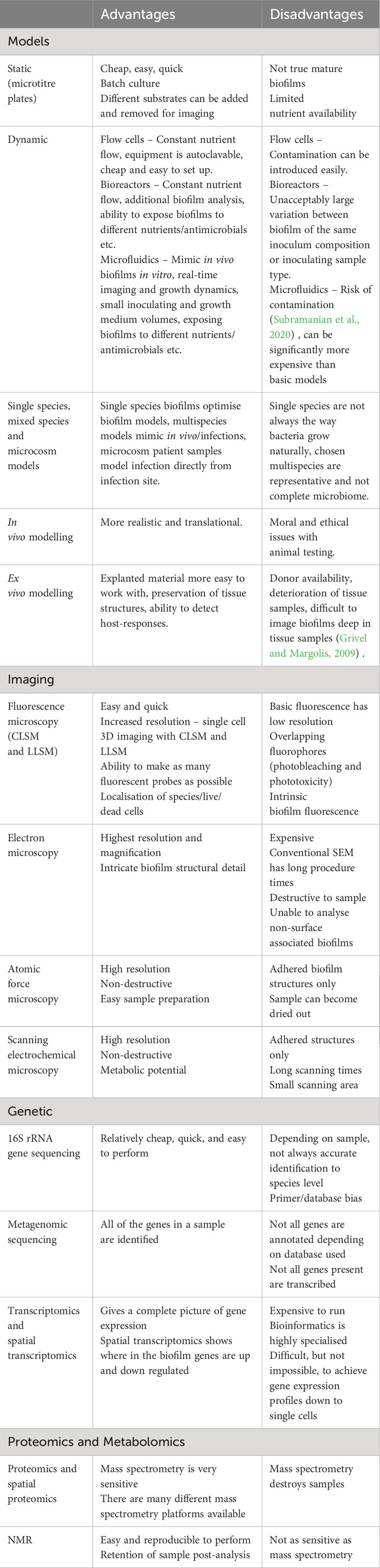
Table 1 An overview of the analysis techniques included in this review, including the advantages and disadvantages of the different methods.
2.1 Static growth
The most simple biofilm experiment is the microtitre plate assay where biofilms are formed on the bottom of a microtitre plate (O’Toole, 2011) and are typically stained with crystal violet to assay biofilm biomass (Figure 1A). Additionally, different substrates can be added to the bottom of the well for biofilms to adhere to and grow on, such as glass beads, titanium discs and hydroxyapatite discs (Figure 1B), which can be removed for imaging or for other types of quantative analysis. Another simple model is using an 8 chamber glass slide, where biofilms can be grown and directly imaged (Jurcisek et al., 2011). Although these methods are quick, cheap, and easy, these are typically not considered to be true mature biofilms as they are statically grown sedimentation cultures and are not exposed to sheer forces that are typical at many in vivo sites (e.g., urinary tract, gut, oral cavity). Furthermore, static cultures also limit the availability of nutrients and build up of bioproducts, as the culture medium is generally only changed manually every 12-48 hours.
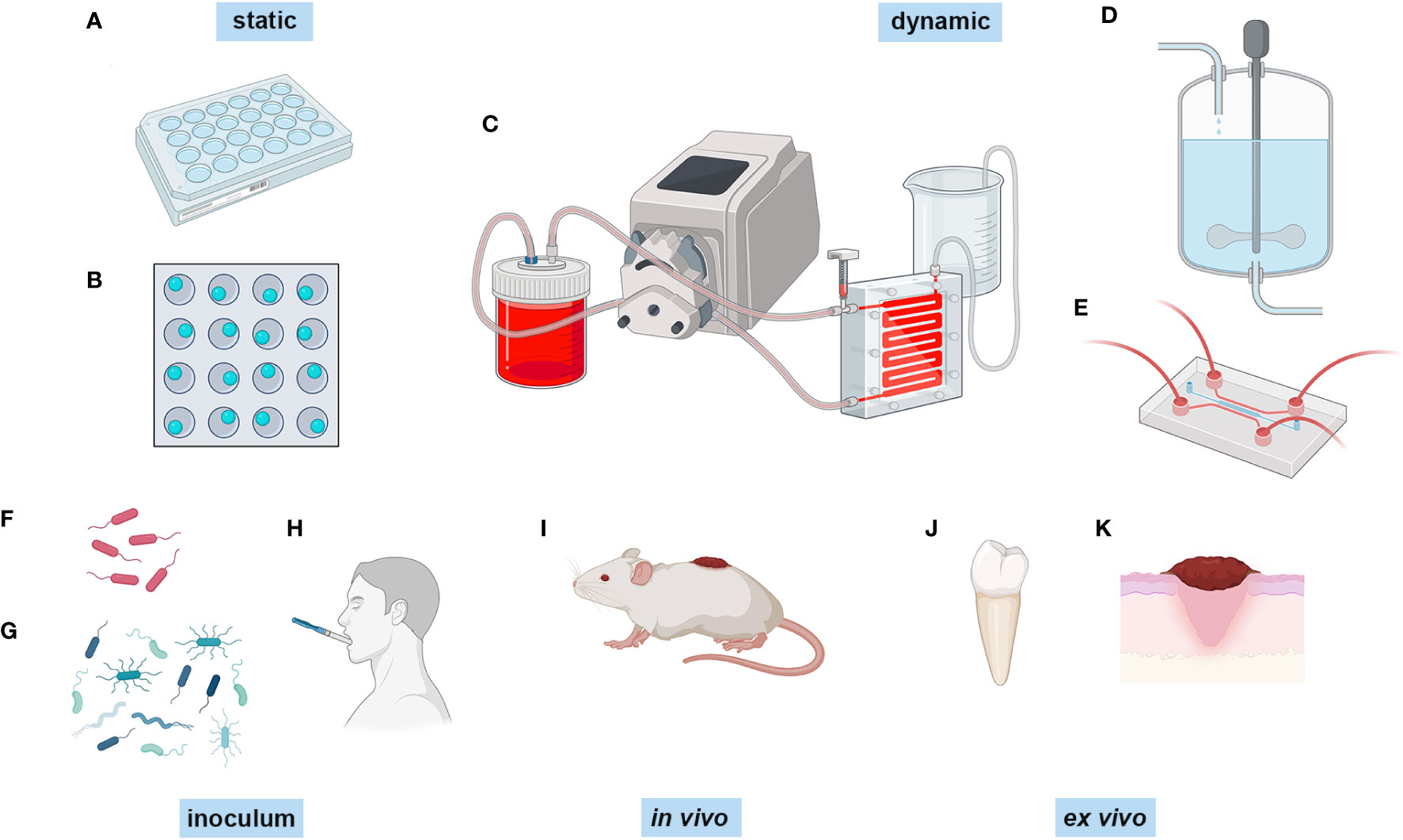
Figure 1 Different methods of biofilm growth. Biofilms can be grown in a number of different ways. Biofilms can be grown statically in (A) microtitre plates and (B) microtitre plates with glass beads, titanium discs or hydroxyapatite discs in the bottom of the well. They can be grown dynamically in (C) flow cells combined wuth peristaltic pumps, (D) in a constant depth film fermenter, and (E) in a microfluidic system such as the BioFlux. The inoculum can range from (F) single and (G) mixed species polymicrobial consortia, and inoculating collected samples from participants, such as (H) collected saliva. In vivo modelling is generally peformed on (I) live animals, and ex vivo modelling is performed on extracted samples such as (J) teeth and (K) skin samples, and also skin models. Created with BioRender.com.
2.2 Dynamic growth
2.2.1 Flow cells
A more advanced system for growing biofilms, that overcomes the limited nutrient availability and allows them to reach maturation, is the use of flow cell systems or constant flow chambers (Peters and Wimpenny, 1988) (Figure 1C). This is a method of growing and evaluating biofilms under hydrodynamic conditions. This comprises a basic set up with a flask for growth media which is fed to the biofilm that grows within a chamber by a peristaltic pump and syringe bubble trap, followed by a waste bottle (Nielsen et al., 2011; Crusz et al., 2012) . The growth medium can be replaced with the dye of choice for imaging, the flow cell can then be removed from the system and biofilm images can be captured directly using fluorescence and confocal microscopy (Pamp et al., 2009; Nielsen et al., 2011) . One benefit of the flow cell system is that the chambers and the equipment can be sterilised by autoclave, therefore facilitating multiple reuse and subsequently reducing laboratory costs. However, this is also one of the disadvantages as this can quickly introduce contamination to the system if inadequate sterilisation has been performed (Palmer, 1999). Nonetheless, flow cells are a satisfactory choice if the user wants to analyse biofilm growth in real-time.
2.2.2 Bioreactor systems
Another method of dynamic growth of biofilms is by using systems such as the constant depth film fermentor (CDFF) (Figure 1D). These have been used for decades to grow biofilms, and consist of ports for nutrient and gas exchange, housing for substrates which are inserted to a known depth, and a turntable with a scraper which keeps the biofilm at a constant depth, if required (Pratten, 2007). Biofilms can be pulsed with nutrients or inhibitors (e.g. antimicrobials) as there are additional ports, and the substrates upon which the biofilms have formed can be removed for additional analysis. These types of biofilm model benefit from constant nutrient availability, and are well-suited for use in oral biofilm modelling as the constant flow mimics the saliva coating the tooth surface (Wilson, 1999; Hope and Wilson, 2006; Pratten, 2007; Sousa et al., 2022) and for wound biofilms which grow at the air-liquid interface and are constantly nourished from the exudate beneath (Duckworth et al., 2018). However, there are issues with heterogeneity of the biofilms that are formed due to either minor changes in the number of bacteria in a defined consortium inoculum, or the variability of human samples that are inoculated (Hope et al., 2012) which can affect the final biofilm composition.
2.2.3 Microfluidic systems
In recent years, the state of the art in experimental biofilm modelling and analysis is through the use of microfluidics (Figure 1E), which includes devices such as Bioflux (Fluxion Biosciences) (Pouget et al., 2021) and BiofilmChip (Blanco-Cabra et al., 2021). Microfluidics is the use of small channels, microns in thickness, and a constant, low volume flow of fluid that facilitates small-scale experiments, not only for biofilm development but also for other applications such as organ-on-a-chip and cancer studies (Li et al., 2016). In the case of the Bioflux system, the set-up allows for growth at different temperatures using a heating plate, different flow rates dependent on dyne pressure, different air requirements by the addition of gas tanks, and the double inlet of the Bioflux allows for switching between growth media. Biofilms can be grown either by binding and forming within the glass chamber alone or by seeding the channels with monolayers of cells (Tremblay et al., 2015). The inlets can also be used to pulse biofilms with antimicrobials (Naudin et al., 2019; Zhang et al., 2019a; Czechowicz et al., 2022). The BiofilmChip is a similar concept to the Bioflux, except it does not have the same plate-type technology. However, it does allow for the analysis of biofilm growth using electrochemical impedance spectroscopy (EIS) technology (Blanco-Cabra et al., 2021), which is the same technology used in the closed-system microtitre plate biofilm growth dynamic analysis xCELLigence Real Time Cell Analyzer (RTCA) (Acea Biosciences) (van Duuren et al., 2017). EIS uses electrical currents applied to a sample and measures the response using sensors to detect biofilm growth at the solid-liquid interphase (McGlennen et al., 2023). This allows for dynamic growth analysis without having to use a confocal microscope, although this can still be utilised if required. Arguably, one of the major advantages of microfluidic devices is the ability to image the biofilm directly in situ in real-time (Brown et al., 2019; Straub et al., 2020; Corsini et al., 2021) using a number of different probes and stains at single-cell resolution using confocal microscopy to generate complex images of the biofilm structure and its components. Whilst one advantage is being able to use low inoculation and culture volumes, therefore accommodating small clinical samples, this also comes with small output volumes for downstream sampling. However, one study using a robotic sampler for non-destructive sampling of the biofilm has shown that very small 10 μl samples can be retrieved and used directly for 16S rRNA gene sequencing (Hansen et al., 2019), along with high performance liquid chromatography (HPLC). Microfluidic systems have also been integrated with downstream applications in situ by pairing real-time biofilm growth in a vacuum compatible microfluidic reactor and time-of-flight secondary ion mass spectrometry (ToF-SIMS) (Hua et al., 2015) to assess spatial chemical mapping within the biofilm.
One of the disadvantages of using these biofilm models is that the biofilms must be adhered to a surface, therefore not considering non-surface associated biofilms. However, this can be overcome by using droplet microfluidics, a method of using small, encapsulated droplets to grow bacteria in biofilms. Specialised platforms are required to carry out these experiments, therefore they may not currently be accessible to most laboratories. This method is in a phase of optimisation for biofilm development, with two studies using Bacillus subtilis and P. aeruginosa to develop biofilms in droplets (Chang et al., 2015; Jin et al., 2018). Both studies have shown that biofilm development is highly dependent on the size of the droplet, and that uniformity of droplet size to be consistent across the samples is one major limitation of this method of biofilm growth. However, this method is very high throughput, with the ability to assess thousands, even millions, of droplet biofilms at one time.
2.3 Single, mixed species and microcosm biofilm models
Traditionally, biofilm investigations focused on single species of bacteria and characterised their ability to form biofilms as a mono-species model (Figure 1F). This has been hugely beneficial, and continues to increase our understanding of bacterial biofilm development immensely. Growing biofilms as single species allows for the optimisation and assessment of different biofilm growth models and assessing antimicrobials (Sena et al., 2006; Wang et al., 2019), as variability is lower in single species, therefore a more reliable comparison can be made. However, it has been shown that growth as single-species is not the prefered way that bacteria thrive and survive in natural biofilms, such as in dental plaque (Periasamy and Kolenbrander, 2010). As mentioned previously, biofilms grow in a state of symbiosis, sharing nutrients and antimicrobial resistance, which makes them far more resilient to environmental stresses (Lee et al., 2014). The in vitro growth of multispecies biofilms is essentially an effort to replicate the environment in which the biofilms grow. Dual-species and multispecies microcosm biofilms (Figure 1G) have been used to investigate biofilms related to different infections or body locations, such as dual-species S. aureus and Candida aureus infection (Tran et al., 2022), a nine species supragingival model (Bradshaw et al., 1989), a four species catheter-associated infection model (Hou et al., 2022) , a dual species E. coli and Enterococcus faecalis gut biofilm model (Govindarajan et al., 2022). In vitro biofilm models can also be generated using samples collected from patients/participants as the inoculating fluid, as opposed to a defined consortium of bacteria. The most common is retrievied saliva (Figure 1H) and plaque from the oral cavity (Signori et al., 2016), but studies have also used vaginal fluid from patients with chronic candidiasis (Cordeiro et al., 2020).
2.4 Modelling host-microbe interactions
In vitro modelling of biofilm infections is limited in that it is purely a prediction of what will happen in situ, and does not take into account the host response to biofilm infection. In vivo animal models have been used to grow biofilms in a more realistic and translational manner, when compared to in vitro biofilm modelling. For wound biofilm modelling (reviewed in (Ganesh et al., 2015)) pigs, mice, rats and even drosophila were inflicted with a wound (Figure 1I) which was then challenged with differing combinations of bacteria that are common in wound infections (P. aeruginosa, S. aureus, and Acinetobacter baumanii for example). In vivo modelling has also been utilised to study prosthetic joint infections, such as in mouse models where the animals receive a prosthetic joint and are then given an intra-articular injection of bacteria (Carli et al., 2017), or using horse or pig synovial fluid to mimic non-surface attached biofilms in the joint space (Gilbertie et al., 2019) . Host-microbe interactions can be modelled in vivo in animal models, although this is falling out of favour because of the obvious moral and ethical issues associated with these types of experiments. Ex vivo modelling using mock tissue systems or collected human tissues as the substrate is also gaining traction as an acceptable replacement (Corzo-León et al., 2019).
Whilst extracted teeth have been used for decades to grow oral biofilms (Figure 1J), ex vivo biofilm modelling is advancing rapidly with the advent of new technologies. Simpler models include P. aeruginosa biofilms that have been successfully modelled using synthetic cystic fibrosis sputum medium (Davies et al., 2017; De Bleeckere et al., 2023). Most commonly, ex vivo biofilm experiments use explanted material such as explanted porcine skin (Yang et al., 2013; Wilkinson et al., 2018) to study wound biofilms (Figure 1K), explanted corneal samples from rabbits and humans to study biofilms that contribute to keratitis (Pinnock et al., 2017) , explanted rat bone to study osteomyelitis (Junka et al., 2017), explanted porcine bronchiolar tissue to study cystic fibrosis biofilms (Harrison and Diggle, 2016), and explanted porcine heart valves (Chuang-Smith et al., 2010). More recently, reconstructed human epidermis (RHE) from stem cells known as Labskin has been used to model host-microbe interactions (Larson et al., 2021), therefore overcoming the requirement to harvest live tissue from animals or humans.
3 Image analysis techniques
As with biofilm modelling, there are basic techniques which offer low-resolution imaging, graduating to more advanced techniques which offer high-resolution information on the overall biofilm structure and its components (Table 1). Light microscopy is the most basic biofilm imaging technique and is useful as a way of visually confirming if biofilm material is present on the substrate being analysed (Bakke and Olsson, 1986). It is cheap and easy to perform – almost all microbiology laboratories will have a light microscope. However, this microscopy method is limited in biofilm analysis, as the resolution is far too low to elucidate the complex structures within these models.
3.1 Fluorescence-based microscopy
A step-up from the light microscope is the use of fluorescence microscopes, coupled with fluorescence staining. A number of fluorophores can be used, such as SYTO-9 and propidium iodide for live/dead staining, or probes using DNA tags for specific species staining (i.e., fluorescence in situ hybridisation (FISH) staining). Fluorescence microscopy is often paired with higher-resolution microscopy as a way of identifying live and dead bacteria at the surface of the biofilm prior to electron microscopy (Hassan et al., 2010). Fluorescence microscopy has been assessed in comparison to confocal laser scanning microscopy (CLSM) in an in vitro CDFF biofilm model, and was shown to image to greater depths than CLSM without a loss of resolution in this biofilm type (Vroom et al., 1999) . Not only can fluorescence microscopy be used to assess in vitro biofilms, but also removed medical devices to look for residual biofilm contamination (Wong et al., 2020). One of the main limitations to fluorescence microscopy is the limited time that fluorescence remains active – fluorophores are easily bleached by excitation under the microscope, limiting the time to analyse. Additionally, fluorescence staining overlap is a limitation here, with some probes having overlapping excitation/emission wavelengths which can cause issues with acquiring and analysing images.
Fluorescence microscopy and CLSM share similar attributes, as fluorescent probes and stains can also be used in CLSM. Although CLSM uses fluorescence to visualise samples, it is different from fluorescence microscopy due to the pinpoint laser which improves resolution and the scanning of the laser through the sample on the x, y and z planes which provides z-stack images which can be reconstructed using open-source software such as COMSTAT2 (Vorregaard, 2008; Samarian et al., 2014) and BiofilmQ (Hartmann et al., 2021) . The information obtained from z-stack image analysis can provide information on biofilm depth, biomass, and surface area which is useful to compare biofilm characteristics when exposed to different molecules, along with live/dead staining (Figures 2A, B) which can show the distribution of bacterial viability within the biofilm (Bridier et al., 2010; Morris et al., 2020; Cleaver et al., 2021). Specific dyes and probes can be used, such as FM 1-43, fluorescent in situ hybridisation (FISH) probes (Thurnheer et al., 2004; Malic et al., 2009; Lebeer et al., 2011), and carboxy-SNARF (Corsini et al., 2021) for the monitoring of pH within the biofilm. However, CLSM suffers from photobleaching of samples, therefore the sampling of biofilms over a long period of time becomes a challenge. Lattice light sheet microscopy (LLSM) has been adapted to overcome this difficulty (Zhang et al., 2021a; Zhang et al., 2021b). LLSM has high resolution, both spatially and temporally, and it has low phototoxicity and photobleaching. Its benefits for biofilm experiments have been limited until recently, due to the water-immersion objectives on the microscope, however this microscopy methodology has been adapted to be used for biofilm dynamic analysis by pairing with hermetically sealed flow cells (Zhang et al., 2021b). An open-source analytical pipeline has also been developed to analyse 3D images captured using LLSM (Zhang et al., 2022). To achieve good quality images, samples must be grown on transparent media, or kept at a thin depth, which is a current limitation of this method.
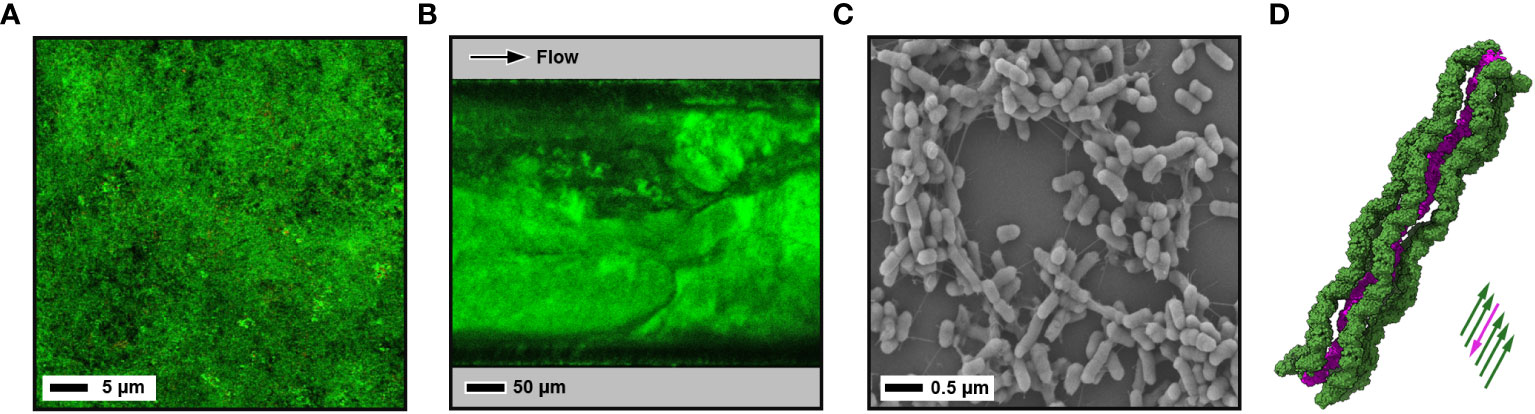
Figure 2 Images of biofilms using different techniques. (A) CLSM image of a 10-day old microcosm biofilm grown on a hydroxyapatite disc from a saliva sample inoculum. Biofilm is stained with LIVE/DEAD™ BacLight™ Bacterial Viability Kit (green - live; red - dead). (B) CLSM image of a mature 4-day old E. coli W strain biofilm grown under shear flow with a BioFlux microfluidic system. Bacterial membranes are stained with FilmTracer™ FM™ 1-43 dye. (C) Negative stain SEM image of a 1-day old enterotixogenic E. coli (ETEC) H10407 strain biofilm showing microcolony formation. (D) Cryo-EM derived atomic structure of bundled in situ archaeal bundling pili from Pyrobaculum calidifontis at 4 Å resolution (PDB ID: 7ueg) (Wang et al., 2022) . One extended pilus (purple) is surrounded by five other pili running in the opposite direction (green).
3.2 Electron microscopy
The most advanced methods for microscopic analyses of biofilm samples are high-resolution scanning electron microscopy (SEM) and transmission electron microscopy (TEM). The main difference between the two methods is that in SEM, 3D surface images are produced through the detection of secondary electrons emitted from the sample surface after excitation from a primary electron beam (Garcez et al., 2013; Hung et al., 2013; Nishitani et al., 2015). On the otherhand, in TEM the primary beam passes through a sample, is diffracted, and then refocussed to produce an image as a 2D projection (Richardson et al., 2009; Sugimoto et al., 2016).
SEM is a well-established method that has been used extensively to assess the relationship between bacteria within a biofilm and with the substrate surface (Figure 2C). Conventional negative stain SEM has been expanded upon to include field emission SEM (FE-SEM), variable pressure SEM (VP-SEM) and cryo-SEM. Although negative stain SEM is relatively low in resolution (up to ~100 nm) it has high magnification (up to x30,000) (Relucenti et al., 2021), and the preparation and preservation of samples allows the intricate water channels and the biofilm matrix to be clearly visualised. However, preparative steps can lead to loss of the sample through fixation and drying, and the use of metal coating. Ionic coating of biofilms has overcome this to some extent (Asahi et al., 2015), but cryo-SEM is now considerd much more superior, involving freeze fracture of the sample which enhances the retention of the inner structures (Hrubanova et al., 2016; Hrubanova et al., 2018) (Figure 2D). Although SEM alone can only provide a surface image, focussed ion beam (FIB) milling can be used to remove ~10 nm thick sections from biofilms, and when coupled iteratively with SEM imaging (FIB-SEM), this can be used to investigate the subsurface structure and create 3D tomographical biofilm reconstructrions (Alhede et al., 2012). SEM is an incredibly useful tool in the investigation of biofilms and has been used in a range of different ways. For example, it has been used to characterise biofilm structure and formation (Fleeman et al., 2023), to assess the effects of antimicrobials on biofilms formed on different surfaces (Gomes and Mergulhão, 2017), and also clinically to assess the biofilm removal efficiency of different disruption techniques (Vyas et al., 2016) and for the detection of biofilms in ureteral stents in renal transplant patients (Barajas-García et al., 2023).
TEM has been used much less-frequently than SEM for the analysis of biofilms due to thin samples being required, and it has generally only been useful for imaging surface and internal structures of bacterial cells. However, TEM has extremely high spatial resolution (~1 Å) and with the emergence of cryo-TEM as a powerful tool for structural biology, cryo-TEM approaches have the potential to deliver in situ atomic resolution details within biofilms (Cossa and Trépout, 2022). Although not mature biofilms, Pyrobaculum calidifontis archaea, aggregated through intertwined intercellular pili, have been imaged by cryo-TEM, and using helical reconstruction the structure of these bundled pili has been modelled at 4 Å resolution (Wang et al., 2022). Likewise, using FIB milling with cryo-electron tomography (cryo-ET) to examine biofilm-like floccules of P. aeruginosa cells has shown how the surface protein CdrA localises at the intercellular interface and mediates bacterial aggregation (Melia et al., 2021). The ability to section mature biofilms in a non-disruptive fashion is a current draw back in being able to study their high resolution features in situ. However, by growing biofilms directly onto EM grids and/or using advanced sectioning approaches such as FIB milling, it may be possible in the future to overcome these barriers.
3.3 Atomic force microscopy
Another method for the microscopic analysis of biofilms is atomic force microscopy (AFM), which can produce an image by scanning a small cantilever over the surface of a sample, and measuring the force between the two (Hiesgen and Friedrich, 2011). This can be achieved through tapping mode where the cantilever is intermittently oscilated up and down, and contact mode where the cantilever is dragged across the sample and is in constant contact with the biofilm. Although resolution is generally low in the lateral plane (~2-30 nm), atomic resolution (~1 Å) can be achieved in the vertical direction. AFM has been used in biofilm analysis to study the growth of different bacteria on different substrates, namely investigating bacterial adhesion and morphology, the interaction of bacteria within polymicrobial biofilms, and the effect of surface treatments and antimicrobials (Beech et al., 1996; Chaw et al., 2005; Núñez et al., 2005; Ahimou et al., 2007; Oh et al., 2009; Germano et al., 2013). There are many advantages to using AFM in biofilm studies, including the ability to image nanoscale structures, simplified sample preparation, and its non-destructive nature (Vahabi et al., 2013). One disadvantage to AFM is that biofilms are often dried-out during the process, therefore often losing the integral structures and topography of biofilms, but one study has overcome this by creating a moist bioreactor that inhibits this drying process (Ahimou et al., 2007). Another disadvantage is that the sample must be well-adhered to the surface, which makes studying cell-cell interactions within non-adhered biofilms impossible with this technique.
3.4 Scanning electrochemical microscopy
Another method of imaging of biofilms is by using scanning electrochemical microscopy (SECM). This method of microscopy assesses bioelectric currents that are produced by bacteria within biofilms using small microelectrodes, similar to those used in EIS (Revsbech, 2005; Huang et al., 2018; Caniglia and Kranz, 2020) . The probe scans the biofilm surface and retrieves information regarding the redox processes around the sample and can provide a micron-scale 3D map of the environment (Darch and Koley, 2018). This methodology can be used to assess the growth and metabolic potential of biofilms (reviewed extensively by (Zhou et al., 2022)), but also to measure the antimicrobial effects of electrical stimulation on the biofilm (Del Pozo et al., 2009; Darvishi et al., 2021; Darvishi and Girault, 2023). SECM can be combined with other microscopic techniques, such as AFM (Huang et al., 2018) and CLSM (Cannan et al., 2002). One of the major advantages of SECM is that it is non-destructive and can be used in real-time to assess different molecules at the surface of the biofilm, such as virulence factors including pyocyanin in P. aeruginosa (Koley et al., 2011). There are some limitations to SECM, mainly the long scanning times, the inability to scan large surfaces and the inability to scan rough surfaces (Santana Santos et al., 2023), however the hybridisation of this method with others has the potential to overcome these limitations.
4 Genetic and molecular analysis techniques
4.1 Sequencing analysis
Molecular biology techniques range from simple targeted polymerase chain reactions (PCR) to advanced metatranscriptomic analysis (Table 1). All techniques have their place within the scope of the experiment being carried out, and some answer “what bacteria are there?” and others “what are those bacteria doing?”. All molecular biology investigations require a solid research hypothesis before being performed, as the vast amount of data derived, particularly from high throughput sequencing techniques, can be difficult to decipher without one. Their application in biofilm research is extensive, particularly in dental biofilm samples where 26% of the oral microbiome is unculturable (https://www.homd.org/; last accessed 20th October 2023), therefore allowing the detection of bacteria that would normally be missed in conventional microbiology techniques (Chen et al., 2010; Dewhirst et al., 2010).
Sequencing of the variable 16S rRNA gene is a relatively simple method of identifying bacteria in biofilms down to the genus level confidently (Dempsey et al., 2007; Swearingen et al., 2016; Cleaver et al., 2019). However, 16S rRNA gene sequencing can be limited by several variables, such as the bias of primer pairs that are used for amplification (specifically those that are universal primers (Morales and Holben, 2009)), the variable region that is targeted (Johnson et al., 2019) , and also the curated database that is used to identify sequences to species level. A study by Johnson and colleagues (Johnson et al., 2019) also suggested that the most accurate method to discriminate down to species level is to sequence the entire 16S rRNA gene using long sequence reads, which can be produced by platforms such as Oxford Nanopore (Tianyuan et al., 2023). Molecular identification of fungal species within a biofilm requires sequencing of the nuclear ribosomal internal transcribed spacer (ITS) region (Nilsson et al., 2009), and cannot be performed concurrently with 16S rRNA gene sequencing.
To overcome these limitations, one might choose to use metagenomic next generation sequencing to sequence all the genes present within a polymicrobial biofilm sample. A study comparing the two showed that metagenomic sequencing has a much higher level of taxonomic diversity and specificity, and that 16S rRNA gene sequencing showed broader community composition and is less specific (Poretsky et al., 2014). Metagenomic sequencing also gives information on antimicrobial resistance within a biofilm sample, and also allows for predicting functionality of the biofilm using the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Matsumoto et al., 2021; Yen and Johnson, 2021). Whole genome sequencing has its place in biofilm analysis, however, the presence of a gene within the genome does not necessarily mean that it is being transcribed.
4.2 Transcriptomics
Transcriptomics is the analysis of the complete mRNA transcripts of cells and provides information on differential gene expression of single cell types or polymicrobial communities and is an advancing method of identifying the functionality and activity of biofilms (Table 1). Sequencing the mRNA transcripts of cells allows both dual sequencing of bacterial and fungal biofilms, however, the database used to blast sequences must include both bacterial and fungal genes. The switch from planktonic to sessile growth is tightly regulated by bacterial gene expression, and this has been shown in different biofilm models (Oggioni et al., 2006; Shemesh et al., 2007; Sánchez et al., 2019) and biofilm growth dynamics are different from planktonic cell growth dynamics (Resch et al., 2005; Robertsson et al., 2021; Zheng et al., 2022). Metatranscriptomics is a powerful tool in microbiology research for the analysis of in vitro biofilm growth models (Zhang et al., 2019b), but also for examination of clinical samples, such as dental plaque (Edlund et al., 2018; Huang et al., 2021), prosthetic joint infections (Goswami et al., 2021), and host-microbiome interactions in diabetic wound biofilms (Malone et al., 2022).
One of the limitations of biofilm metatranscriptomics has been the inability to resolve spatial functionality within the biofilm structure. However, spatial transcriptomics is now a fast-developing method which is being used to understand how microbes behave in different topographical locations within the biofilm (Dar et al., 2021a). A range of different methodologies to achieve this have been reported in the literature. One group have developed RAINBOW-seq, a method where fluorescently labelled dyes are used to label bacteria spatially within a biofilm (surface, middle, interior) during growth within a microfluidic system (Figure 3A). These are then sorted by fluorescence-activated cell sorting (FACS), after which RNA-seq is performed (Wang et al., 2023). Another group has grown biofilms in a drip-flow chamber on stainless steel coupons, which were then cryo-sectioned and slices from the surface, middle and interior of the biofilm and then analysed with microarrays (Heacock-Kang et al., 2017). Sequencing of RNA transcripts from colony biofilms has also been carried out, whereby filter membranes seeded with E. coli were grown on solid agar plates at regular intervals from 12-72 hours and the colonies at these time periods were then subject to transcriptomics, adaptive microscopy, metabolomics using ultra-high performance liquid chromatography (UPLC), and oxygen measurements (Díaz-Pascual et al., 2021). Although technically not a true biofilm, this methodology gives a good picture of colony biofilm growth and functionality over time. Agar block biofilm assays (ABBA) have also been used to grow P. aeruginosa biofilms in chambered coverslips, which were then hybridised with fluorescent probes targeted to genes of interest and imaged using CLSM (Livingston et al., 2022) to visualise gene expression within the biofilms. Furthermore, a set of 105 unique genes, representing physiology and virulence, from P. aeruginosa biofilms grown in coverslip chambers have been spatially imaged using par-seqFISH, which is a new method of parallel sequential fluorescence in situ hybridisation CLSM (Dar et al., 2021).
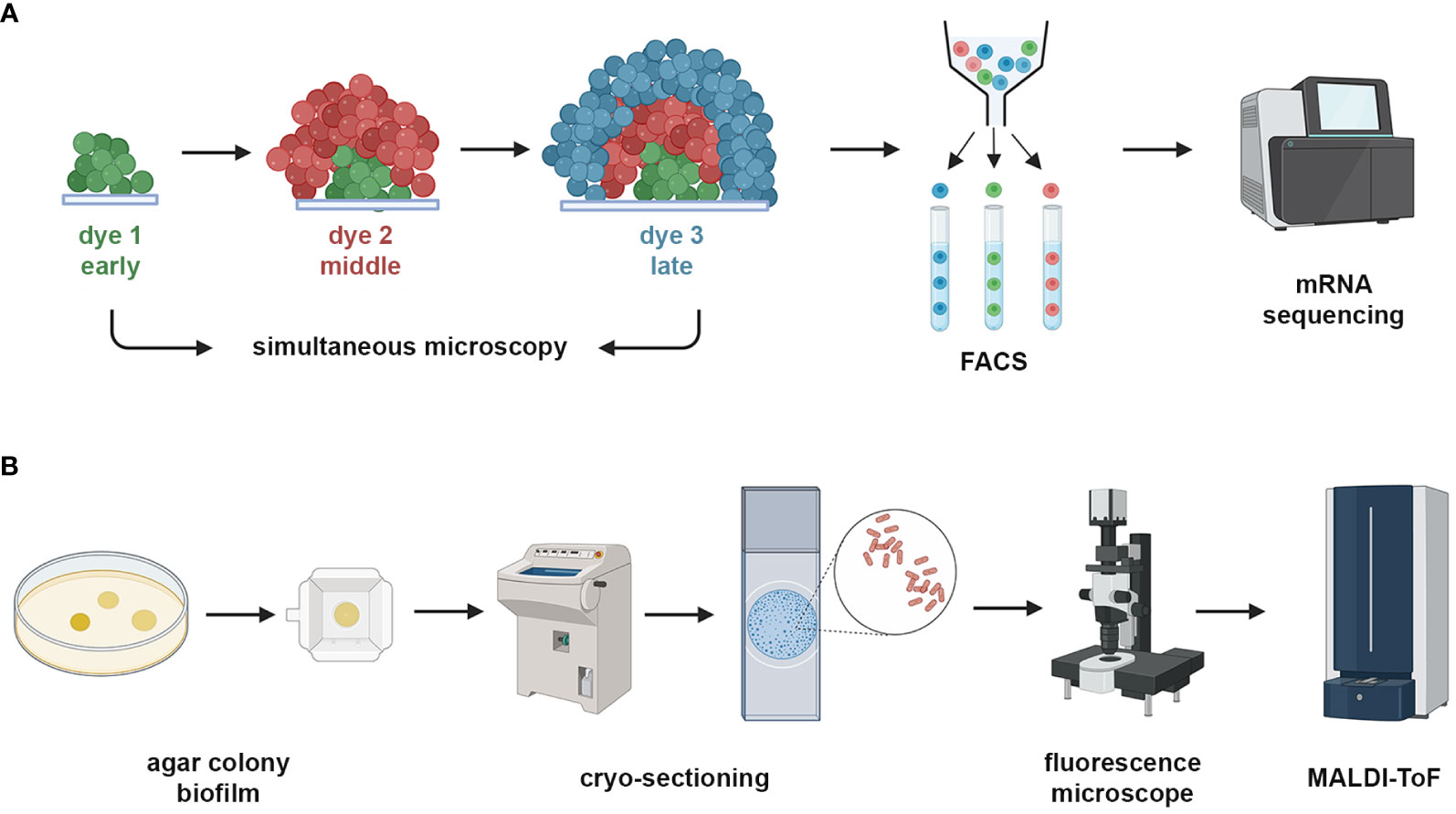
Figure 3 Spatial and functional analysis of biofilms. Imaging can be coupled with transcriptomics and mass spectrometry to assess the functionality of biofilm samples. (A) A schematic view of RAINBOW-seq, where different fluorescent dyes are sequentially added to a biofilm which is imaged at regular intervals, after which the biofilm is subjected to FACS cell sorting and then low quantity mRNA sequencing is performed which is mapped back to the image using bioinformatic gating parameters (Dar et al., 2021) . (B) A schematic view of imaging mass spectroscopy, where agar grown colony biofilms that are eGFP fluorescence tagged are grown and cryosectioned into thin slices, which are imaged and then subjected to MALDI-ToF mass spectrometry with post-processing mapping to the image. Created with BioRender.com.
However, these tehniques so far can only resolve regions within biofilms rather than at the single cell level. The development of single cell transcriptomics from bacterial cells has been challenging, as bacterial cells contain low levels of mRNA, the mRNA does not have a poly(A) tail making some sequencing techniques unsuitable, and bacterial mRNA is notoriously unstable (Homberger et al., 2022). Overcoming these challenges will allow significant advancements in our insight into bacterial functionality within these complex biofilm communities.
4.3 Proteomics and metabolomics
Sequencing bacterial mRNA from biofilm samples can give information on which genes are being expressed but does not necessarily equate to translation of those genes and how the molecules that are translated are utilised, especially in polymicrobial biofilm samples (Table 1). However, analysing the proteome and metabolome of biofilms, with or without gene expression data, is able to provide a much more complete picture of the biofilm environment.
Bacterial proteomics is the analysis of all the proteins that are present in a sample and can be used to analyse the degradation of nutrients by bacteria. Proteomic analysis of biofilms has been used to identify key proteins that are essential in complex biofilm networks, including; in the EPS (Egorova et al., 2022) , required for temperature adaptation stress (Lee and Wang, 2020) , required for adaptation to antimicrobial treatment (MaChado and Coquet, 2016) , and that change with biofilm development (Serra et al., 2008; Suriyanarayanan et al., 2018) and aging (Rahman et al., 2022), and between planktonic and biofilm conditions (Khan et al., 2019; Suryaletha et al., 2019; Llama-Palacios et al., 2020; Tang et al., 2020). These studies use a range of different mass spectrometry methodologies, including tandem liquid chromatography (LC-MS/MS) alone (Lee and Wang, 2020) or coupled with nano-high performance liquid chromatography (HPLC) (Egorova et al., 2022), lab-on-a-chip and XCT mass spectrometry (MaChado and Coquet, 2016), tandem mass tag (TMT) mass spectrometry (Rahman et al., 2022), fourier transform infrared spectroscopy (FT-IR) (Serra et al., 2008), nano-LC/MS coupled with quadrupole-ToF (Q-ToF) (Suryaletha et al., 2019) , iTRAQ labelling plus LC-MS/MS for quantitative proteomics, and matrix-assisted laser desorption ionisation time-of-flight (MALDI-ToF) mass spectrometry (Llama-Palacios et al., 2020; Tang et al., 2020). MALDI-ToF analysis of bacteria is routinely used in clinical diagnostic laboratories to identify bacterial species, and this method is increasingly being developed to discriminate between isolates and their status as biofilm-producers or non-producers (Caputo et al., 2018), which will aid in establishing effective antimicrobial therapy.
Proteomics has also been coupled with imaging analysis to investigate the proteome spatially within the biofilm. One of the most utilised methods of achieving this is with MALDI-ToF imaging mass spectrometry (MALDI-ToF IMS) (Floyd et al., 2015; Rivera et al., 2022), whereby biofilms are cryo-sectioned and imaged with a fluorescent microscope prior to MALDI-ToF (Rivera et al., 2022) (Figure 3B) or biofilms are grown on slides and subjected to electron microscopy and MALDI-ToF (Floyd et al., 2015). Another method is laser ablation sample transfer (LAST), where biofilms can be grown on transwell membranes which are then laid on a microscope slide on the anoxic or oxic side and subjected to LC-MS/MS (Pulukkody et al., 2021). The latter method does not employ image analysis but rather proteomic analysis based on oxygen availability within the biofilm.
Bacterial metabolomics is the identification and quantification of metabolites within a sample that have either been produced by bacteria (metabolised) or are a result of the breakdown of a substrate into other downstream products (catabolised) by bacteria. This can be; (1) targeted, where the absolute concentration of a metabolite is quantified using labelled isotopes, or (2) untargeted, where metabolites are semi-quantitatively quantified using a control sample. Mass spectrometry is used regularly to undertake metabolomic analysis of biofilm samples, however nuclear magnetic resonance (NMR) spectroscopy has been used more frequently in recent biofilm metabolomic studies (Renslow et al., 2013; Cleaver et al., 2019; Cleaver et al., 2021; Czajkowska et al., 2021; Leggett et al., 2022). Whereas mass spectrometry assesses the mass to charge ratio of charged molecules to determine molecular identity (Emonet et al., 2010) , NMR detects changes in the local electronic environment and can provide fingerprint spectra for specific molecular structures (Singh and Singh, 2022). Although NMR is more reproducible than mass spectrometry, does not destroy the sample, and is more efficient to run, one of its limitations is that it is less sensitive, where mass spectrometry approaches tend to identify a higher range of metabolites (Emwas et al., 2013).
5 Conclusions
Biofilms are incredibly complex and dynamic structures, and cause a range of difficult to treat medical infections. Advancements in biofilm research is fast-paced and new ways to study biofilms are in constant development. This review has detailed the numerous ways to grow and image biofilms, and investigate the multifaceted dynamics of biofilm behaviour. It is important to use a technique that is appropriate for the biofilm infection being studied and the methods that are chosen ultimately depend on the hypotheses and questions that are aiming to be answered, and these should be carefully considered.
Author contributions
LC: Conceptualization, Writing – original draft, Writing – review & editing. JG: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by MRC grant MR/W000814/1.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahimou, F., Semmens, M. J., Novak, P. J., Haugstad, G. (2007). Biofilm cohesiveness measurement using a novel atomic force microscopy methodology. Appl. Environ. Microbiol. 73, 2897–2904. doi: 10.1128/AEM.02388-06
Akyıldız, I., Take, G., Uygur, K., Kızıl, Y., Aydil, U. (2013). Bacterial biofilm formation in the middle-ear mucosa of chronic otitis media patients. Indian J. Otolaryngol Head Neck Surg. 65, 557–561. doi: 10.1007/s12070-012-0513-x
Alhede, M., Kragh, K. N., Qvortrup, K., Allesen-Holm, M., van Gennip, M., Christensen, L. D., et al. (2011). Phenotypes of non-attached Pseudomonas aeruginosa aggregates resemble surface attached biofilm. PloS One 6, e27943. doi: 10.1371/journal.pone.0027943
Alhede, M., Qvortrup, K., Liebrechts, R., Høiby, N., Givskov, M., Bjarnsholt, T. (2012). Combination of microscopic techniques reveals a comprehensive visual impression of biofilm structure and composition. FEMS Immunol. Med. Microbiol. 65, 335–342. doi: 10.1111/j.1574-695X.2012.00956.x
Asahi, Y., Miura, J., Tsuda, T., Kuwabata, S., Tsunashima, K., Noiri, Y., et al. (2015). Simple observation of Streptococcus mutans biofilm by scanning electron microscopy using ionic liquids. AMB Express 5, 6. doi: 10.1186/s13568-015-0097-4
Bakke, R., Olsson, P. Q. (1986). Biofilm thickness measurements by light microscopy. J. Microbiol. Methods 5, 93–98. doi: 10.1016/0167-7012(86)90005-9
Barajas-García, C. C., Guerrero-Barrera, A. L., Arreola-Guerra, J. M., Avelar-González, F. J., Ramírez-Castillo, F. Y. (2023). Detection of microbial biofilms inside the lumen of ureteral stents: two case reports. J. Med. Case Rep. 17, 174. doi: 10.1186/s13256-023-03849-6
Baumgartner, M., Lang, M., Holley, H., Crepaz, D., Hausmann, B., Pjevac, P., et al. (2021). Mucosal biofilms are an endoscopic feature of irritable bowel syndrome and ulcerative colitis. Gastroenterology 161, 1245–1256.e20. doi: 10.1053/j.gastro.2021.06.024
Beaver, M., Lagundzin, D., Thapa, I., Lee, J., Ali, H., Kielian, T., et al. (2021). Cutibacterium acnes central nervous system catheter infection induces long-term changes in the cerebrospinal fluid proteome. Infect. Immun. 89. doi: 10.1128/IAI.00531-20
Beech, I. B., Cheung, C. W. S., Johnson, D. B., Smith, J. R. (1996). Comparative studies of bacterial biofilms on steel surfaces using atomic force microscopy and environmental scanning electron microscopy. Biofouling 10, 65–77. doi: 10.1080/08927019609386271
Blanco-Cabra, N., López-Martínez, M. J., Arévalo-Jaimes, B. V., Martin-Gómez, M. T., Samitier, J., Torrents, E. (2021). A new BiofilmChip device for testing biofilm formation and antibiotic susceptibility. NPJ Biofilms Microbiomes 7, 62. doi: 10.1038/s41522-021-00236-1
Bradshaw, D. J., Mckee, A. S., Marsh, P. D. (1989). Effects of Carbohydrate Pulses and pH on Population Shifts within Oral Microbial Communities in vitro. J. Dent. Res. 68, 1298–1302. doi: 10.1177/00220345890680090101
Bridier, A., Dubois-Brissonnet, F., Boubetra, A., Thomas, V., Briandet, R. (2010). The biofilm architecture of sixty opportunistic pathogens deciphered using a high throughput CLSM method. J. Microbiol. Methods 82, 64–70. doi: 10.1016/j.mimet.2010.04.006
Brown, J. M., Blunk, B., Williams, P., Hardie, K. R. (2019). Microfluidic-based growth and imaging of bacterial biofilms. Bio Protoc. 9, e3460. doi: 10.21769/BioProtoc.3460
Burmølle, M., Ren, D., Bjarnsholt, T., Sørensen, S. J. (2014). Interactions in multispecies biofilms: Do they actually matter? Trends Microbiol. 22, 84–91. doi: 10.1016/j.tim.2013.12.004
Cai, Y.-M. (2020). Non-surface attached bacterial aggregates: A ubiquitous third lifestyle. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.557035
Caniglia, G., Kranz, C. (2020). Scanning electrochemical microscopy and its potential for studying biofilms and antimicrobial coatings. Anal. Bioanal Chem. 412, 6133–6148. doi: 10.1007/s00216-020-02782-7/
Cannan, S., Douglas Macklam, I., Unwin, P. R. (2002). Three-dimensional imaging of proton gradients at microelectrode surfaces using confocal laser scanning microscopy. Electrochem Commun. 4, 886–892. doi: 10.1016/S1388-2481(02)00482-4
Caputo, P., Di Martino, M. C., Perfetto, B., Iovino, F., Donnarumma, G. (2018). Use of MALDI-TOF MS to discriminate between biofilm-producer and non-producer strains of staphylococcus epidermidis. Int. J. Environ. Res. Public Health 15. doi: 10.3390/ijerph15081695
Carli, A. V., Bhimani, S., Xu, Y., Shirley, M. B., De Mesy Bentley, K. L., Ross, F. P., et al. (2017). Quantification of peri-implant bacterial load and in vivo biofilm formation in an innovative, clinically representative mouse model of periprosthetic joint infection. J. Bone Joint Surg. - Am. Volume 99, e25(1)–e25(8). doi: 10.2106/JBJS.16.00815
Castro, J., MaChado, D., Cerca, N. (2019). Unveiling the role of Gardnerella vaginalis in polymicrobial Bacterial Vaginosis biofilms: the impact of other vaginal pathogens living as neighbors. ISME J. 13, 1306–1317. doi: 10.1038/s41396-018-0337-0
Chang, C. B., Wilking, J. N., Kim, S.-H., Shum, H. C., Weitz, D. A. (2015). Monodisperse emulsion drop microenvironments for bacterial biofilm growth. Small 11, 3954–3961. doi: 10.1002/smll.201403125
Chaw, K. C., Manimaran, M., Tay, F. E. H. (2005). Role of silver ions in destabilization of intermolecular adhesion forces measured by atomic force microscopy in Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 49, 4853–4859. doi: 10.1128/aac.49.12.4853-4859.2005
Chen, T., Yu, W.-H., Izard, J., Baranova, O. V., Lakshmanan, A., Dewhirst, F. E. (2010). The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010. doi: 10.1093/database/baq013
Chuang-Smith, O. N., Wells, C. L., Henry-Stanley, M. J., Dunny, G. M. (2010). Acceleration of Enterococcus faecalis biofilm formation by aggregation substance expression in an Ex Vivo model of cardiac valve colonization. PloS One 5. doi: 10.1371/journal.pone.0015798
Cleaver, L. M., Moazzez, R., Carpenter, G. H. (2019). Mixed aerobic-anaerobic incubation conditions induce proteolytic activity from in vitro salivary biofilms. J. Oral. Microbiol. 11, 1643206. doi: 10.1080/20002297.2019.1643206
Cleaver, L. M., Moazzez, R. V., Carpenter, G. H. (2021). Evidence for proline utilization by oral bacterial biofilms grown in saliva. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.619968
Cordeiro, R., de, A., de Andrade, A. R. C., Portela, F. V. M., Pereira, L. M. G., Moura, S. G. B., et al. (2020). Proposal for a microcosm biofilm model for the study of vulvovaginal candidiasis. Biofouling 36, 610–620. doi: 10.1080/08927014.2020.1785435
Corsini, P., Wang, S., Rehman, S., Fenn, K., Sagar, A., Sirovica, S., et al. (2021). Molecular and cellular insight into Escherichia coli SslE and its role during biofilm maturation. NPJ Biofilms Microbiomes 8 (1). doi: 10.1101/2021.02.07.430137
Corzo-León, D. E., Munro, C. A., MacCallum, D. M. (2019). An ex vivo human skin model to study superficial fungal infections. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01172
Cossa, A., Trépout, S. (2022). “Cryo-electron Microscopy to Analyze the Structure of Bacterial Amyloids In Vitro,” in Bacterial Amyloids: Methods and Protocols. Eds. Arluison, V., Wien, F., Marcoleta, A. (New York, NY: Springer US), 13–23. doi: 10.1007/978-1-0716-2529-3_2
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R., Lappin-Scott, H. M. (1995). Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745. doi: 10.1146/annurev.mi.49.100195.003431
Crusz, S. A., Popat, R., Rybtke, M. T., Cámara, M., Givskov, M., Tolker-Nielsen, T., et al. (2012). Bursting the bubble on bacterial biofilms: A flow cell methodology. Biofouling 28, 835–842. doi: 10.1080/08927014.2012.716044
Czajkowska, J., Junka, A., Hoppe, J., Toporkiewicz, M., Pawlak, A., Migdał, P., et al. (2021). The co-culture of staphylococcal biofilm and fibroblast cell line: the correlation of biological phenomena with metabolic NMR1 footprint. Int. J. Mol. Sci. 22. doi: 10.3390/ijms22115826
Czechowicz, P., Nowicka, J., Neubauer, D., Chodaczek, G., Krzyżek, P., Gościniak, G. (2022). Activity of Novel Ultrashort Cyclic Lipopeptides against Biofilm of Candida albicans Isolated from VVC in the Ex Vivo Animal Vaginal Model and BioFlux Biofilm Model—A Pilot Study. Int. J. Mol. Sci. 23. doi: 10.3390/ijms232214453
Dar, D., Dar, N., Cai, L., Newman, D. K. (2021). Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Sci. (1979) 373. doi: 10.1126/science.abi4882
Darch, S. E., Koley, D. (2018). Quantifying microbial chatter: Scanning electrochemical microscopy as a tool to study interactions in biofilms. Proc. R. Soc. A: Mathematical Phys. Eng. Sci. 474. doi: 10.1098/rspa.2018.0405
Darvishi, S., Girault, H. H. (2023). Revealing the Effects of Three Different Antimicrobial Agents on E. coli Biofilms by Using Soft-Probe Scanning Electrochemical Microscopy. Revealing the Effects of Three Different Antimicrobial Agents on E. coli Biofilms by Using Soft-Probe Scanning Electrochemical Microscopy. Appl. Nano 4, 260–279. doi: 10.3390/applnano
Darvishi, S., Pick, H., Oveisi, E., Girault, H. H., Lesch, A. (2021). Soft-probe-scanning electrochemical microscopy reveals electrochemical surface reactivity of E. coli biofilms. Sens Actuators B Chem. 334. doi: 10.1016/j.snb.2021.129669
Davies, E. V., James, C. E., Brockhurst, M. A., Winstanley, C. (2017). Evolutionary diversification of Pseudomonas aeruginosa in an artificial sputum model. BMC Microbiol. 17. doi: 10.1186/s12866-016-0916-z
De Bleeckere, A., Van den Bossche, S., De Sutter, P. J., Beirens, T., Crabbé, A., Coenye, T. (2023). High throughput determination of the biofilm prevention concentration for Pseudomonas aeruginosa biofilms using a synthetic cystic fibrosis sputum medium. Biofilm 5. doi: 10.1016/j.bioflm.2023.100106
Del Pozo, J. L., Rouse, M. S., Mandrekar, J. N., Steckelberg, J. M., Patel, R. (2009). The electricidal effect: Reduction of Staphylococcus and Pseudomonas biofilms by prolonged exposure to low-intensity electrical current. Antimicrob. Agents Chemother. 53, 41–45. doi: 10.1128/AAC.00680-08
Dempsey, K. E., Riggio, M. P., Lennon, A., Hannah, V. E., Ramage, G., Allan, D., et al. (2007). Identification of bacteria on the surface of clinically infected and non-infected prosthetic hip joints removed during revision arthroplasties by 16S rRNA gene sequencing and by microbiological culture. Arthritis Res. Ther. 9, R46. doi: 10.1186/ar2201
Dewhirst, F. E., Chen, T., Izard, J., Paster, B. J., Tanner, A. C. R., Yu, W. H., et al. (2010). The human oral microbiome. J. Bacteriol 192, 5002–5017. doi: 10.1128/JB.00542-10
Díaz-Pascual, F., Lempp, M., Nosho, K., Jeckel, H., Jo, J. K., Neuhaus, K., et al. (2021). Spatial alanine metabolism determines local growth dynamics of escherichia coli colonies. Elife 10, 1–29. doi: 10.7554/eLife.70794
Di Martino, P. (2018). Extracellular polymeric substances, a key element in understanding biofilm phenotype. AIMS Microbiol. 4, 274–288. doi: 10.3934/MICROBIOL.2018.2.274
Duckworth, P. F., Rowlands, R. S., Barbour, M. E., Maddocks, S. E. (2018). A novel flow-system to establish experimental biofilms for modelling chronic wound infection and testing the efficacy of wound dressings. Microbiol. Res. 215, 141–147. doi: 10.1016/j.micres.2018.07.009
Edlund, A., Yang, Y., Yooseph, S., He, X., Shi, W., McLean, J. S. (2018). Uncovering complex microbiome activities via metatranscriptomics during 24 hours of oral biofilm assembly and maturation. Microbiome 6, 217. doi: 10.1186/s40168-018-0591-4
Egorova, D. A., Solovyev, A. I., Polyakov, N. B., Danilova, K. V., Scherbakova, A. A., Kravtsov, I. N., et al. (2022). Biofilm matrix proteome of clinical strain of P. aeruginosa isolated from bronchoalveolar lavage of patient in intensive care unit. Microb. Pathog. 170, 105714. doi: 10.1016/j.micpath.2022.105714
El Rafei, A., Desimone, D. C., Sohail, M. R., Desimone, C. V., Steckelberg, J. M., Wilson, W. R., et al. (2016). Cardiovascular implantable electronic device infections due to propionibacterium species. Pacing Clin. Electrophysiol 39, 522–530. doi: 10.1111/pace.12845
Emonet, S., Shah, H. N., Cherkaoui, A., Schrenzel, J. (2010). Application and use of various mass spectrometry methods in clinical microbiology. Clin. Microbiol. Infection 16, 1604–1613. doi: 10.1111/j.1469-0691.2010.03368.x
Emwas, A. H. M., Salek, R. M., Griffin, J. L., Merzaban, J. (2013). NMR-based metabolomics in human disease diagnosis: Applications, limitations, and recommendations. Metabolomics 9, 1048–1072. doi: 10.1007/s11306-013-0524-y
Fleeman, R. M., Mikesh, M., Davies, B. W. (2023). Investigating Klebsiella pneumoniae biofilm preservation for scanning electron microscopy. Access Microbiol. 5. doi: 10.1099/acmi.0.000470.v3
Floyd, K. A., Moore, J. L., Eberly, A. R., Good, J. A. D., Shaffer, C. L., Zaver, H., et al. (2015). Adhesive fiber stratification in uropathogenic escherichia coli biofilms unveils oxygen-mediated control of type 1 pili. PloS Pathog. 11, 1–26. doi: 10.1371/journal.ppat.1004697
Ganesh, K., Sinha, M., Mathew-Steiner, S. S., Das, A., Roy, S., Sen, C. K. (2015). Chronic wound biofilm model. Adv. Wound Care (New Rochelle) 4, 382–388. doi: 10.1089/wound.2014.0587
Garcez, A. S., Núñez, S. C., Azambuja, N., Fregnani, E. R., Rodriguez, H. M. H., Hamblin, M. R., et al. (2013). Effects of photodynamic therapy on gram-positive and gram-negative bacterial biofilms by bioluminescence imaging and scanning electron microscopic analysis. Photomed Laser Surg. 31, 519–525. doi: 10.1089/pho.2012.3341
Germano, F., Ennio, B., Arcuri, C., Cecchetti, F., Cicciù, M. (2013). Atomic force microscopy of bacteria from periodontal subgingival biofilm: Preliminary study results. Eur. J. Dent. 07, 152–158. doi: 10.4103/1305-7456.110155
Gilbertie, J. M., Schnabel, L. V., Hickok, N. J., Jacob, M. E., Conlon, B. P., Shapiro, I. M., et al. (2019). Equine or porcine synovial fluid as a novel ex vivo model for the study of bacterial free-floating biofilms that form in human joint infections. PloS One 14. doi: 10.1371/journal.pone.0221012
Gomes, L. C., Mergulhão, F. J. (2017). SEM analysis of surface impact on biofilm antibiotic treatment. Scanning 2017, 2960194. doi: 10.1155/2017/2960194
Goswami, K., Shope, A., Wright, J., Purtill, J., Lamendella, R., Parvizi, J. (2021). Shotgun metatranscriptomics for periprosthetic joint infection diagnosis: A novel prospective investigation. Orthopaedic Proc. 103-B, 85. doi: 10.1302/1358-992X.2021.15.085
Govindarajan, D. K., MeGhanathan, Y., Sivaramakrishnan, M., Kothandan, R., Muthusamy, A., Seviour, T. W., et al. (2022). Enterococcus faecalis thrives in dual-species biofilm models under iron-rich conditions. Arch. Microbiol. 204. doi: 10.1007/s00203-022-03309-7
Grivel, J. C., Margolis, L. (2009). Use of human tissue explants to study human infectious agents. Nat. Protoc. 4, 256–269. doi: 10.1038/nprot.2008.245
Hansen, S. H., Kabbeck, T., Radtke, C. P., Krause, S., Krolitzki, E., Peschke, T., et al. (2019). Machine-assisted cultivation and analysis of biofilms. Sci. Rep. 9, 8933. doi: 10.1038/s41598-019-45414-6
Harrison, F., Diggle, S. P. (2016). An ex vivo lung model to study bronchioles infected with Pseudomonas aeruginosa biofilms. Microbiol. (United Kingdom) 162, 1755–1760. doi: 10.1099/mic.0.000352
Hartmann, R., Jeckel, H., Jelli, E., Singh, P. K., Vaidya, S., Bayer, M., et al. (2021). Quantitative image analysis of microbial communities with BiofilmQ. Nat. Microbiol. 6, 151–156. doi: 10.1038/s41564-020-00817-4
Hassan, A. N., Anand, S., Avadhanula, M. (2010). Microscopic observation of multispecies biofilm of various structures on whey concentration membranes1. J. Dairy Sci. 93, 2321–2329. doi: 10.3168/jds.2009-2800
Heacock-Kang, Y., Sun, Z., Zarzycki-Siek, J., McMillan, I. A., Norris, M. H., Bluhm, A. P., et al. (2017). Spatial transcriptomes within the Pseudomonas aeruginosa biofilm architecture. Mol. Microbiol. 106, 976–985. doi: 10.1111/mmi.13863
Hiesgen, R., Friedrich, K. A. (2011). Atomic force microscopy. PEM Fuel Cell Diagn. Tools 395–421. doi: 10.4011/shikizai.93.321
Høiby, N., Ciofu, O., Bjarnsholt, T. (2010). Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 5, 1663–1674. doi: 10.2217/fmb.10.125
Homberger, C., Barquist, L., Vogel, J. (2022). Ushering in a new era of single-cell transcriptomics in bacteria. microLife 3, 1–9. doi: 10.1093/femsml/uqac020
Honkanen, M., Jämsen, E., Karppelin, M., Huttunen, R., Eskelinen, A., Syrjänen, J. (2019). Periprosthetic joint infections as a consequence of bacteremia. Open Forum Infect. Dis. 6. doi: 10.1093/ofid/ofz218
Hope, C. K., Bakht, K., Burnside, G., Martin, G. C., Burnett, G., de Josselin de Jong, E., et al. (2012). Reducing the variability between constant-depth film fermenter experiments when modelling oral biofilm. J. Appl. Microbiol. 113, 601–608. doi: 10.1111/j.1365-2672.2012.05368.x
Hope, C. K., Wilson, M. (2006). Biofilm structure and cell vitality in a laboratory model of subgingival plaque. J. Microbiol. Methods 66, 390–398. doi: 10.1016/j.mimet.2006.01.003
Hou, J., Wang, L., Alm, M., Thomsen, P., Monsen, T., Ramstedt, M., et al. (2022). Enhanced antibiotic tolerance of an in vitro multispecies uropathogen biofilm model, useful for studies of catheter-associated urinary tract infections. Microorganisms 10. doi: 10.3390/microorganisms10061207
Hrubanova, K., Nebesarova, J., Ruzicka, F., Krzyzanek, V. (2018). The innovation of cryo-SEM freeze-fracturing methodology demonstrated on high pressure frozen biofilm. Micron 110, 28–35. doi: 10.1016/j.micron.2018.04.006
Hrubanova, K., Skoupy, R., Nebesarova, J., Ruzicka, F., Krzyzanek, V. (2016). “The sample preparation for cryo-SEM: the real ultrastructure of microbial biofilm or just artifacts?,” in European Microscopy Congress 2016: Proceedings (Wiley-VCH Verlag GmbH & Co. KGaA). 203–204. doi: 10.1002/9783527808465.EMC2016.6907
Hua, X., Marshall, M. J., Xiong, Y., Ma, X., Zhou, Y., Tucker, A. E., et al. (2015). Two-dimensional and three-dimensional dynamic imaging of live biofilms in a microchannel by time-of-flight secondary ion mass spectrometry. Biomicrofluidics 9, 31101. doi: 10.1063/1.4919807
Huang, L., Li, Z., Lou, Y., Cao, F., Zhang, D., Li, X. (2018). Recent advances in scanning electrochemical microscopy for biological applications. Materials 11. doi: 10.3390/ma11081389
Huang, Y., Zhao, X., Cui, L., Huang, S. (2021). Metagenomic and metatranscriptomic insight into oral biofilms in periodontitis and related systemic diseases. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.728585
Hung, C., Zhou, Y., Pinkner, J. S., Dodson, K. W., Crowley, J. R., Heuser, J., et al. (2013). Escherichia coli biofilms have an organized and complex extracellular matrix structure. mBio 4. doi: 10.1128/mBio.00645-13
Jin, Z., Nie, M., Hu, R., Zhao, T., Xu, J., Chen, D., et al. (2018). Dynamic sessile-droplet habitats for controllable cultivation of bacterial biofilm. Small 14, 1800658. doi: 10.1002/smll.201800658
Johnson, J. S., Spakowicz, D. J., Hong, B.-Y., Petersen, L. M., Demkowicz, P., Chen, L., et al. (2019). Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10, 5029. doi: 10.1038/s41467-019-13036-1
Junka, A., Szymczyk, P., Ziółkowski, G., Karuga-Kuzniewska, E., Smutnicka, D., Bil-Lula, I., et al. (2017). Bad to the bone: On in vitro and ex vivo microbial biofilm ability to directly destroy colonized bone surfaces without participation of host immunity or osteoclastogenesis. PloS One 12. doi: 10.1371/journal.pone.0169565
Jurcisek, J. A., Dickson, A. C., Bruggeman, M. E., Bakaletz, L. O. (2011). In vitro biofilm formation in an 8-well chamber slide. J. Vis. Exp 47. doi: 10.3791/2481
Khan, M. M., Chattagul, S., Tran, B. Q., Freiberg, J. A., Nita-Lazar, A., Shirtliff, M. E., et al. (2019). Temporal proteomic profiling reveals changes that support Burkholderia biofilms. Pathog. Dis. 77. doi: 10.1093/femspd/ftz005
Koley, D., Ramsey, M. M., Bard, A. J., Whiteley, M. (2011). Discovery of a biofilm electrocline using real-time 3D metabolite analysis. Proc. Natl. Acad. Sci. U.S.A. 108, 19996–20001. doi: 10.1073/pnas.1117298108
Lam, J., Chan, R., Lam, K., Costerton, J. W. (1980). Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28, 546–556. doi: 10.1128/iai.28.2.546-556.1980
Larson, P. J., Chong, D., Fleming, E., Oh, J. (2021). Challenges in developing a human model system for skin microbiome research. J. Invest. Dermatol. 141, 228–231.e4. doi: 10.1016/j.jid.2020.05.096
Lebeer, S., Verhoeven, T. L. A., Claes, I. J. J., De Hertogh, G., Vermeire, S., Buyse, J., et al. (2011). FISH analysis of Lactobacillus biofilms in the gastrointestinal tract of different hosts. Lett. Appl. Microbiol. 52, 220–226. doi: 10.1111/j.1472-765X.2010.02994.x
Leccese Terraf, M. C., Juárez Tomás, M. S., Rault, L., Le Loir, Y., Even, S., Nader-Macías, M. E. F. (2016). Biofilms of vaginal Lactobacillus reuteri CRL 1324 and Lactobacillus rhamnosus CRL 1332: kinetics of formation and matrix characterization. Arch. Microbiol. 198, 689–700. doi: 10.1007/s00203-016-1225-5
Lee, K. W. K., Periasamy, S., Mukherjee, M., Xie, C., Kjelleberg, S., Rice, S. A. (2014). Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 8, 894–907. doi: 10.1038/ismej.2013.194
Lee, Y.-J., Wang, C. (2020). Proteomic analysis reveals the temperature-dependent presence of extracytoplasmic peptidases in the biofilm exoproteome of Listeria monocytogenes EGD-e. J. Microbiol. 58, 761–771. doi: 10.1007/s12275-020-9522-8
Leggett, A., Li, D.-W., Bruschweiler-Li, L., Sullivan, A., Stoodley, P., Brüschweiler, R. (2022). Differential metabolism between biofilm and suspended Pseudomonas aeruginosa cultures in bovine synovial fluid by 2D NMR-based metabolomics. Sci. Rep. 12, 17317. doi: 10.1038/s41598-022-22127-x
Li, Y., Gao, A., Yu, L. (2016). Monitoring of TGF-β 1-induced human lung adenocarcinoma A549 cells epithelial-mesenchymal transformation process by measuring cell adhesion force with a microfluidic device. Appl. Biochem. Biotechnol. 178, 114–125. doi: 10.1007/s12010-015-1862-1
Livingston, J., Spero, M. A., Lonergan, Z. R., Newman, D. K. (2022). Visualization of mRNA Expression in Pseudomonas aeruginosa Aggregates Reveals Spatial Patterns of Fermentative and Denitrifying Metabolism. Appl. Environ. Microbiol. 88. doi: 10.1128/aem.00439-22
Llama-Palacios, A., Potupa, O., Sánchez, M. C., Figuero, E., Herrera, D., Sanz, M. (2020). Proteomic analysis of Fusobacterium nucleatum growth in biofilm versus planktonic state. Mol. Oral. Microbiol. 35, 168–180. doi: 10.1111/omi.12303
Macfarlane, S., Dillon, J. F. (2007). Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol. 102, 1187–1196. doi: 10.1111/j.1365-2672.2007.03287.x
MaChado, I., Coquet, L. (2016). Proteomic Changes in Pseudomonas aeruginosa Biofilm Cells after Adaptive Resistance Development. J. Proteomics Bioinform. 09, 58–62. doi: 10.4172/jpb.1000390
Mack, W. N., Mack, J. P., Ackerson, A. O. (1975). Microbial film development in a trickling filter. Microb. Ecol. 2, 215–226. doi: 10.1007/BF02010441
Malic, S., Hill, K. E., Hayes, A., Percival, S. L., Thomas, D. W., Williams, D. W. (2009). Detection and identification of specific bacteria in wound biofilms using peptide nucleic acid fluorescent in situ hybridization (PNA FISH). Microbiol. (N Y) 155, 2603–2611. doi: 10.1099/mic.0.028712-0
Malone, M., Radzieta, M., Peters, T. J., Dickson, H. G., Schwarzer, S., Jensen, S. O., et al. (2022). Host-microbe metatranscriptome reveals differences between acute and chronic infections in diabetes-related foot ulcers. APMIS 130, 751–762. doi: 10.1111/apm.13200
Marrie, T. J., Nelligan, J., Costerton, J. W. (1982). A scanning and transmission electron microscopic study of an infected endocardial pacemaker lead. Circulation 66, 1339–1341. doi: 10.1161/01.cir.66.6.1339
Marsh, P. D. (2010). Microbiology of dental plaque biofilms and their role in oral health and caries. Dent. Clin. North Am. 54, 441–454. doi: 10.1016/j.cden.2010.03.002
Matsumoto, K., Sakami, T., Watanabe, T., Taniuchi, Y., Kuwata, A., Kakehi, S., et al. (2021). Metagenomic analysis provides functional insights into seasonal change of a non-cyanobacterial prokaryotic community in temperate coastal waters. PloS One 16. doi: 10.1371/journal.pone.0257862
McGlennen, M., Dieser, M., Foreman, C. M., Warnat, S. (2023). Using electrochemical impedance spectroscopy to study biofilm growth in a 3D-printed flow cell system. Biosens Bioelectron X 14. doi: 10.1016/j.biosx.2023.100326
Melia, C. E., Bolla, J. R., Katharios-Lanwermeyer, S., Mihaylov, D. B., Hoffmann, P. C., Huo, J., et al. (2021). Architecture of cell-cell junctions in situ reveals a mechanism for bacterial biofilm inhibition. PNAS 118. doi: 10.1073/pnas.2109940118/-/DCSupplemental
Morales, S. E., Holben, W. E. (2009). Empirical testing of 16S rRNA gene PCR primer pairs reveals variance in target specificity and efficacy not suggested by in silico analysis. Appl. Environ. Microbiol. 75, 2677–2683. doi: 10.1128/AEM.02166-08
Morris, A. J., Li, A., Jackson, L., Yau, Y. C. W., Waters, V. (2020). Quantifying the Effects of Antimicrobials on In vitro Biofilm Architecture using COMSTAT Software. J. Vis. Exp 166. doi: 10.3791/61759
Naudin, B., Heins, A., Pinhal, S., Dé, E., Nicol, M. (2019). BioFluxTM 200 Microfluidic System to Study A. baumannii Biofilm Formation in a Dynamic Mode of Growth BT - Acinetobacter baumannii: Methods and Protocols. Eds. Biswas, I., Rather, P. N. (New York, NY: Springer New York), 167–176. doi: 10.1007/978-1-4939-9118-1_16
Nielsen, M. W., Sternberg, C., Molin, S., Regenberg, B. (2011). Pseudomonas aeruginosa and saccharomyces cerevisiae biofilm in flow cells. J. Visualized Experiments 47. doi: 10.3791/2383
Nilsson, R. H., Ryberg, M., Abarenkov, K., Sjökvist, E., Kristiansson, E. (2009). The ITS region as a target for characterization of fungal communities using emerging sequencing technologies. FEMS Microbiol. Lett. 296, 97–101. doi: 10.1111/j.1574-6968.2009.01618.x
Nishitani, K., Sutipornpalangkul, W., De Mesy Bentley, K. L., Varrone, J. J., Bello-Irizarry, S. N., Ito, H., et al. (2015). Quantifying the natural history of biofilm formation in vivo during the establishment of chronic implant-associated Staphylococcus aureus osteomyelitis in mice to identify critical pathogen and host factors. J. Orthopaedic Res. 33, 1311–1319. doi: 10.1002/jor.22907
Núñez, M. E., Martin, M. O., Chan, P. H., Spain, E. M. (2005). Predation, death, and survival in a biofilm: Bdellovibrio investigated by atomic force microscopy. Colloids Surf B Biointerfaces 42, 263–271. doi: 10.1016/j.colsurfb.2005.03.003
Oggioni, M. R., Trappetti, C., Kadioglu, A., Cassone, M., Iannelli, F., Ricci, S., et al. (2006). Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol. Microbiol. 61, 1196–1210. doi: 10.1111/j.1365-2958.2006.05310.x
Oh, Y. J., Lee, N. R., Jo, W., Jung, W. K., Lim, J. S. (2009). Effects of substrates on biofilm formation observed by atomic force microscopy. Ultramicroscopy 109, 874–880. doi: 10.1016/j.ultramic.2009.03.042
Pabst, B., Pitts, B., Lauchnor, E., Stewart, P. S. (2016). Gel-entrapped staphylococcus aureus bacteria as models of biofilm infection exhibit growth in dense aggregates, oxygen limitation, antibiotic tolerance, and heterogeneous gene expression. Antimicrob. Agents Chemother. 60, 6294–6301. doi: 10.1128/AAC.01336-16
Palmer, R. J. (1999). Microscopy flowcells: Perfusion chambers for real-time study of biofilms. Methods Enzymol. 310, 160–166. doi: 10.1016/S0076-6879(99)10014-4
Pamp, S. J., Sternberg, C., Tolker-Nielsen, T. (2009). Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. in. Cytometry Part A 75A, 90–103. doi: 10.1002/cyto.a.20685
Periasamy, S., Kolenbrander, P. E. (2010). Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J. Bacteriol 192, 2965–2972. doi: 10.1128/JB.01631-09
Peters, A. C., Wimpenny, J. W. (1988). A constant-depth laboratory model film fermentor. Biotechnol. Bioeng 32, 263–270. doi: 10.1002/bit.260320302
Pinnock, A., Shivshetty, N., Roy, S., Rimmer, S., Douglas, I., MacNeil, S., et al. (2017). Ex vivo rabbit and human corneas as models for bacterial and fungal keratitis. Graefe’s Arch. Clin. Exp. Ophthalmol. 255, 333–342. doi: 10.1007/s00417-016-3546-0
Poretsky, R., Rodriguez-R, L. M., Luo, C., Tsementzi, D., Konstantinidis, K. T. (2014). Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PloS One 9. doi: 10.1371/journal.pone.0093827
Pouget, C., Dunyach-Remy, C., Pantel, A., Schuldiner, S., Sotto, A., Lavigne, J.-P. (2021). New adapted in vitro technology to evaluate biofilm formation and antibiotic activity using live imaging under flow conditions. Diagnostics (Basel) 11. doi: 10.3390/diagnostics11101746
Pratten, J. (2007). Growing oral biofilms in a constant depth film fermentor (CDFF). Curr. Protoc. Microbiol. doi: 10.1002/9780471729259.mc01b05s6
Pulukkody, A. C., Yung, Y. P., Donnarumma, F., Murray, K. K., Carlson, R. P., Hanley, L. (2021). Spatially resolved analysis of Pseudomonas aeruginosa biofilm proteomes measured by laser ablation sample transfer. PloS One 16, 1–18. doi: 10.1371/journal.pone.0250911
Rahman, M. A., Amirkhani, A., Chowdhury, D., Mempin, M., Molloy, M. P., Deva, A. K., et al. (2022). Proteome of staphylococcus aureus biofilm changes significantly with aging. Int. J. Mol. Sci. 23. doi: 10.3390/ijms23126415
Relucenti, M., Familiari, G., DonFrancesco, O., Taurino, M., Li, X., Chen, R., et al. (2021). Microscopy methods for biofilm imaging: focus on SEM and VP-SEM pros and cons. Biol. (Basel) 10. doi: 10.3390/biology10010051
Renslow, R. S., Babauta, J. T., Majors, P. D., Mehta, H. S., Ewing, R. J., Ewing, T. W., et al. (2013). A biofilm microreactor system for simultaneous electrochemical and nuclear magnetic resonance techniques. Water Sci. Technol. 69, 966–973. doi: 10.2166/wst.2013.802
Resch, A., Rosenstein, R., Nerz, C., Götz, F. (2005). Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 71, 2663–2676. doi: 10.1128/AEM.71.5.2663-2676.2005
Revsbech, N. P. (2005). Analysis of microbial communities with electrochemical microsensors and microscale biosensors. Methods Enzymol. 397, 147–166. doi: 10.1016/S0076-6879(05)97009-2
Richardson, N., Mordan, N. J., Figueiredo, J. A. P., Ng, Y. L., Gulabivala, K. (2009). Microflora in teeth associated with apical periodontitis: A methodological observational study comparing two protocols and three microscopy techniques. Int. Endod. J. 42, 908–921. doi: 10.1111/j.1365-2591.2009.01594.x
Rivera, E. S., Weiss, A., Migas, L. G., Freiberg, J. A., Djambazova, K. V., Neumann, E. K., et al. (2022). Imaging mass spectrometry reveals complex lipid distributions across Staphylococcus aureus biofilm layers. J. Mass Spectrometry Adv. Clin. Lab. 26, 36–46. doi: 10.1016/j.jmsacl.2022.09.003
Robertson, S., McLean, R. (2015). Beneficial biofilms. AIMS Bioeng 2, 437–448. doi: 10.3934/bioeng.2015.4.437
Robertsson, C., Svensäter, G., Blum, Z., Jakobsson, M. E., Wickström, C. (2021). Proteomic response in Streptococcus gordonii DL1 biofilm cells during attachment to salivary MUC5B. J. Oral. Microbiol. 13, 1967636. doi: 10.1080/20002297.2021.1967636
Samarian, D. S., Jakubovics, N. S., Luo, T. L., Rickard, A. H. (2014). Use of a high-throughput in vitro microfluidic system to develop oral multi-species biofilms. JoVE 94, e52467. doi: 10.3791/52467
Sánchez, M. C., Romero-Lastra, P., Ribeiro-Vidal, H., Llama-Palacios, A., Figuero, E., Herrera, D., et al. (2019). Comparative gene expression analysis of planktonic Porphyromonas gingivalis ATCC 33277 in the presence of a growing biofilm versus planktonic cells. BMC Microbiol. 19, 58. doi: 10.1186/s12866-019-1423-9
Santana Santos, C., Jaato, B. N., Sanjuán, I., Schuhmann, W., Andronescu, C. (2023). Operando scanning electrochemical probe microscopy during electrocatalysis. Chem. Rev. 123, 4972–5019. doi: 10.1021/acs.chemrev.2c00766
Sauer, K., Camper, A. K., Ehrlich, G. D., Costerton, J. W., Davies, D. G. (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol 184, 1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002
Sauer, K., Stoodley, P., Goeres, D. M., Hall-Stoodley, L., Burmølle, M., Stewart, P. S., et al. (2022). The biofilm life cycle: expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 20, 608–620. doi: 10.1038/s41579-022-00767-0
Sena, N. T., Gomes, B. P. F. A., Vianna, M. E., Berber, V. B., Zaia, A. A., Ferraz, C. C. R., et al. (2006). In vitro antimicrobial activity of sodium hypochlorite and chlorhexidine against selected single-species biofilms. Int. Endod. J. 39, 878–885. doi: 10.1111/j.1365-2591.2006.01161.x
Serra, D. O., Lücking, G., Weiland, F., Schulz, S., Görg, A., Yantorno, O. M., et al. (2008). Proteome approaches combined with Fourier transform infrared spectroscopy revealed a distinctive biofilm physiology in Bordetella pertussis. Proteomics 8, 4995–5010. doi: 10.1002/pmic.200800218
Shemesh, M., Tam, A., Steinberg, D. (2007). Differential gene expression profiling of Streptococcus mutans cultured under biofilm and planktonic conditions. Microbiol. (N Y) 153, 1307–1317. doi: 10.1099/mic.0.2006/002030-0
Signori, C., Van De Sande, F. H., Maske, T. T., De Oliveira, E. F., Cenci, M. S. (2016). Influence of the inoculum source on the cariogenicity of in vitro microcosm biofilms. Caries Res. 50, 97–103. doi: 10.1159/000443537
Singh, M. K., Singh, A. (2022). “Chapter 14 - Nuclear magnetic resonance spectroscopy,” in Characterization of Polymers and Fibres. The Textile Institute Book Series. Eds. Singh, M. K., Singh, A. (Elsevier Ltd), 321–339. doi: 10.1016/B978-0-12-823986-5.00011-7
Sousa, V., Spratt, D., Davrandi, M., Mardas, N., Beltrán, V., Donos, N. (2022). Oral microcosm biofilms grown under conditions progressing from peri-implant health, peri-implant mucositis, and peri-implantitis. Int. J. Environ. Res. Public Health 19. doi: 10.3390/ijerph192114088
Stoodley, P., Sauer, K., Davies, D. G., Costerton, J. W. (2002). Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209. doi: 10.1146/annurev.micro.56.012302.160705
Straub, H., Eberl, L., Zinn, M., Rossi, R. M., Maniura-Weber, K., Ren, Q. (2020). A microfluidic platform for in situ investigation of biofilm formation and its treatment under controlled conditions. J. Nanobiotechnol 18, 166. doi: 10.1186/s12951-020-00724-0
Subramanian, S., Huiszoon, R. C., Chu, S., Bentley, W. E., Ghodssi, R. (2020). Microsystems for biofilm characterization and sensing – A review. Biofilm 2, 100015. doi: 10.1016/j.bioflm.2019.100015
Sugimoto, S., Okuda, K. I., Miyakawa, R., Sato, M., Arita-Morioka, K. I., Chiba, A., et al. (2016). Imaging of bacterial multicellular behaviour in biofilms in liquid by atmospheric scanning electron microscopy. Sci. Rep. 6. doi: 10.1038/srep25889
Suriyanarayanan, T., Qingsong, L., Kwang, L. T., Mun, L. Y., Truong, T., Seneviratne, C. J. (2018). Quantitative proteomics of strong and weak biofilm formers of enterococcus faecalis reveals novel regulators of biofilm formation*. Mol. Cell. Proteomics 17, 643–654. doi: 10.1074/mcp.RA117.000461
Suryaletha, K., Narendrakumar, L., John, J., Radhakrishnan, M. P., George, S., Thomas, S. (2019). Decoding the proteomic changes involved in the biofilm formation of Enterococcus faecalis SK460 to elucidate potential biofilm determinants. BMC Microbiol. 19, 146. doi: 10.1186/s12866-019-1527-2
Swearingen, M. C., DiBartola, A. C., Dusane, D., Granger, J., Stoodley, P. (2016). 16S rRNA analysis provides evidence of biofilms on all components of three infected periprosthetic knees including permanent braided suture. Pathog. Dis. 74. doi: 10.1093/femspd/ftw083
Tang, T., Chen, G., Guo, A., Xu, Y., Zhao, L., Wang, M., et al. (2020). Comparative proteomic and genomic analyses of Brucella abortus biofilm and planktonic cells. Mol. Med. Rep. 21, 731–743. doi: 10.3892/mmr.2019.10888
Thurnheer, T., Gmür, R., Guggenheim, B. (2004). Multiplex FISH analysis of a six-species bacterial biofilm. J. Microbiol. Methods 56, 37–47. doi: 10.1016/j.mimet.2003.09.003
Tianyuan, Z., Hanzhou, L., Silin, M., Jian, C., Hao, L., Qiaoyun, H., et al. (2023). The newest Oxford Nanopore R10.4.1 full-length 16S rRNA sequencing enables the accurate resolution of species-level microbial community profiling. Appl. Environ. Microbiol. 0, e00605-23. doi: 10.1128/aem.00605-23
Tran, V. N., Khan, F., Han, W., Luluil, M., Truong, V. G., Yun, H. G., et al. (2022). Real-time monitoring of mono- and dual-species biofilm formation and eradication using microfluidic platform. Sci. Rep. 12. doi: 10.1038/s41598-022-13699-9
Tremblay, Y. D. N., Vogeleer, P., Jacques, M., Harel, J. (2015). High-throughput microfluidic method to study biofilm formation and host-pathogen interactions in pathogenic escherichia coli. Appl. Environ. Microbiol. 81, 2827–2840. doi: 10.1128/AEM.04208-14
Vahabi, S., Nazemi Salman, B., Javanmard, A. (2013). Atomic force microscopy application in biological research: a review study. Iran J. Med. Sci. 38, 76–83.
van Duuren, J. B. J. H., Müsken, M., Karge, B., Tomasch, J., Wittmann, C., Häussler, S., et al. (2017). Use of single-frequency impedance spectroscopy to characterize the growth dynamics of biofilm formation in pseudomonas aeruginosa. Sci. Rep. 7, 5223. doi: 10.1038/s41598-017-05273-5
Vilchez, H. H., Escudero-Sanchez, R., Fernandez-Sampedro, M., Murillo, O., Auñón, Á., Rodríguez-Pardo, D., et al. (2021). Prosthetic shoulder joint infection by Cutibacterium acnes: Does rifampin improve prognosis? A retrospective, multicenter, observational study. Antibiotics 10. doi: 10.3390/antibiotics10050475
Vorregaard, M. (2008). Comstat2 - a modern 3D image analysis environment for biofilms, in Informatics and Mathematical Modelling. (Kongens Lyngby, Denmark: Technical University of Denmark) Available at: https://www.comstat.dk/#:~:text=What%20is%20Comstat2%3F,contain%20any%20statistical%20calculation%20methods.Kongens Lyngby, Denmark: Technical University of Denmark.
Vroom, J. M., Grauw, K. J., De, Gerritsen, H. C., Bradshaw, D. J., Marsh, P. D., Watson, G. K., et al. (1999). Depth penetration and detection of pH gradients in biofilms by two-photon excitation microscopy. Appl. Environ. Microbiol. 65, 3502–3511. doi: 10.1128/AEM.65.8.3502-3511.1999
Vyas, N., Sammons, R. L., Addison, O., Dehghani, H., Walmsley, A. D. (2016). A quantitative method to measure biofilm removal efficiency from complex biomaterial surfaces using SEM and image analysis. Sci. Rep. 6, 32694. doi: 10.1038/srep32694
Wang, F., Cvirkaite-Krupovic, V., Krupovic, M., Egelman, E. H. (2022). Archaeal bundling pili of Pyrobaculum calidifontis reveal similarities between archaeal and bacterial biofilms. Proc. Natl. Acad. Sci. 119, e2207037119. doi: 10.1073/pnas.2207037119
Wang, T., Shen, P., He, Y., Zhang, Y., Liu, J. (2023). Spatial transcriptome uncovers rich coordination of metabolism in E. coli K12 biofilm. Nat. Chem. Biol. doi: 10.1038/s41589-023-01282-w
Wang, L., Xie, X., Qi, M., Weir, M. D., Reynolds, M. A., Li, C., et al. (2019). Effects of single species versus multispecies periodontal biofilms on the antibacterial efficacy of a novel bioactive Class-V nanocomposite. Dental Materials 35, 847–861. doi: 10.1016/J.DENTAL.2019.02.030
Wei, D., Zhu, X.-M., Chen, Y.-Y., Li, X.-Y., Chen, Y.-P., Liu, H.-Y., et al. (2019). Chronic wound biofilms: diagnosis and therapeutic strategies. Chin. Med. J. (Engl), 132 (22), 2737–2744. doi: 10.1097/CM9.0000000000000523
Wilkinson, H. N., Iveson, S., Catherall, P., Hardman, M. J. (2018). A novel silver bioactive glass elicits antimicrobial efficacy against Pseudomonas aeruginosa and Staphylococcus aureus in an ex vivo skin wound biofilm model. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01450
Wilson, M. (1999). Use of constant depth film fermentor in studies of biofilms of oral bacteria. Methods Enzymol. 310, 264–279. doi: 10.1016/s0076-6879(99)10023-5
Wong, M., Wang, Y., Wang, H., Marrone, A. K., Haugen, S. P., Kulkarni, K., et al. (2020). RESEARCH: fluorescence microscopy-based protocol for detecting residual bacteria on medical devices. Biomed. Instrumentation \& Technol. 54, 397–409. doi: 10.2345/0899-8205-54.6.397
Yang, Q., Phillips, P. L., Sampson, E. M., Progulske-Fox, A., Jin, S., Antonelli, P., et al. (2013). Development of a novel ex vivo porcine skin explant model for the assessment of mature bacterial biofilms. Wound Repair Regeneration 21, 704–714. doi: 10.1111/wrr.12074
Yen, S., Johnson, J. S. (2021). Metagenomics: a path to understanding the gut microbiome. Mamm. Genome 32, 282–296. doi: 10.1007/s00335-021-09889-x
Zhang, Y., Shi, W., Song, Y., Wang, J. (2019b). Metatranscriptomic analysis of an in vitro biofilm model reveals strain-specific interactions among multiple bacterial species. J. Oral. Microbiol. 11, 1599670. doi: 10.1080/20002297.2019.1599670
Zhang, X.-Y., Sun, K., Abulimiti, A., Xu, P.-P., Li, Z.-Y. (2019a). Microfluidic system for observation of bacterial culture and effects on biofilm formation at microscale. Micromachines (Basel) 10. doi: 10.3390/mi10090606
Zhang, J., Wang, Y., Donarski, E. D., Toma, T. T., Miles, M. T., Acton, S. T., et al. (2022). BCM3D 2.0: accurate segmentation of single bacterial cells in dense biofilms using computationally generated intermediate image representations. NPJ Biofilms Microbiomes 8, 99. doi: 10.1038/s41522-022-00362-4
Zhang, J., Wang, Y., Zhang, M., Achimovich, A., Wang, J., Acton, S., et al. (2021a). High-resolution imaging of single-cell behaviors in 3D bacterial biofilms using lattice-light sheet microscopy and deep learning-based image processing. Microscopy Microanalysis 27, 3038–3040. doi: 10.1017/S1431927621010540
Zhang, J., Zhang, M., Wang, Y., Donarski, E., Gahlmann, A. (2021b). Optically accessible microfluidic flow channels for noninvasive high-resolution biofilm imaging using lattice light sheet microscopy. J. Phys. Chem. B 125, 12187–12196. doi: 10.1021/acs.jpcb.1c07759
Zheng, L., Zhang, X., Lu, Z., Ma, W., Hu, A., Zhou, H., et al. (2022). Transcriptome sequencing reveals the difference in the expression of biofilm and planktonic cells between two strains of Salmonella Typhimurium. Biofilm 4, 100086. doi: 10.1016/j.bioflm.2022.100086
Keywords: biofilm, biofilm model, biofilm imaging, biofilm analysis, infection, host-microbe interactions
Citation: Cleaver L and Garnett JA (2023) How to study biofilms: technological advancements in clinical biofilm research. Front. Cell. Infect. Microbiol. 13:1335389. doi: 10.3389/fcimb.2023.1335389
Received: 08 November 2023; Accepted: 30 November 2023;
Published: 13 December 2023.
Edited by:
Jinsong Feng, Zhejiang University, ChinaReviewed by:
Tahereh Navidifar, Shoushtar Faculty of Medical Sciences, IranJens Kreth, Oregon Health and Sciences University, United States
Copyright © 2023 Cleaver and Garnett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James A. Garnett, amFtZXMuZ2FybmV0dEBrY2wuYWMudWs=; Leanne Cleaver, bGVhbm5lLmNsZWF2ZXJAa2NsLmFjLnVr
 Leanne Cleaver
Leanne Cleaver James A. Garnett
James A. Garnett