- 1Department of General Surgery, The First Affiliated Hospital of Dalian Medical University, Dalian, China
- 2Laboratory of Integrative Medicine, The First Affiliated Hospital of Dalian Medical University, Dalian, China
- 3Departments of Gastrointestinal Surgery, The Second Affiliated Hospital of Dalian Medical University, Dalian, China
- 4Institute (College) of Integrative Medicine, Dalian Medical University, Dalian, China
Colorectal cancer (CRC) is a common malignancy of the gastrointestinal tract, accounting for the second most common cause of gastrointestinal tumors. As one of the intestinal barriers, gut bacteria form biofilm, participate in intestinal work, and form the living environment of intestinal cells. Metagenomic next-generation sequencing (mNGS) of the gut bacteria in a large number of CRC patients has been established, enabling specific microbial signatures to be associated with colorectal adenomato-carcinoma. Gut bacteria are involved in both benign precursor lesions (polyps), in situ growth and metastasis of CRC. Therefore, the term tumorigenic bacteria was proposed in 2018, such as Escherichia coli, Fusobacterium nucleatum, enterotoxigenic Bacteroides fragilis, etc. Meanwhile, bacteria toxins (such as cytolethal distending toxin (CDT), Colibactin (Clb), B. fragilis toxin) affect the tumor microenvironment and promote cancer occurrence and tumor immune escape. It is important to note that there are differences in the bacteria of different types of CRC. In this paper, the role of tumorigenic bacteria in the polyp-cancer transformation and the effects of their secreted toxins on the tumor microenvironment will be discussed, thereby further exploring new ideas for the prevention and treatment of CRC.
1 Introduction
Colorectal cancer (CRC) is a malignant tumor of the colon or rectum that usually originates from mucosal epithelial cells. It is a common type of cancer with high incidence rates worldwide. Several risk factors, such as age, family history, dietary habits, intestinal polyps, and inflammatory bowel disease, are associated with the development of CRC (Center et al., 2009). The intestinal microbiome is a multifaceted ecosystem consisting of a rich array of bacteria, viruses, and fungi. It harbors a vast reservoir of genetic diversity, surpassing that which resides within an individual’s own DNA, making it a profoundly intricate and unique entity. The intricate interplay between bacteria and the host leads to multifaceted impacts of intestinal microbiota and their metabolites on the initiation and progression of CRC, as well as the modulation of the immune microenvironment. Intestinal colonizing bacteria secrete metabolites and enter the blood circulation, thereby affecting important physiological processes such as nutrient absorption, material metabolism, and immune defense (Sun et al., 2023). Moreover, the oncogenic flora promotes the occurrence of CRC by inducing DNA damage in epithelial cells, which in turn promotes the proliferation of bacteria that have a growth advantage in the tumor microenvironment (Tjalsma et al., 2012; Clavenna et al., 2023). The definition of intestinal microbiome is becoming more and more clear, and it is related to countless health conditions. These interactions are now understood to occur locally and throughout the body through changes in the immune system and other mechanisms. The local proximity of intestinal microbiome to the colon led many early researchers to study its effect on CRC, making CRC a frontier for studying the response of microbiome to cancer development, progression and treatment.
2 The occurrence and development of CRC
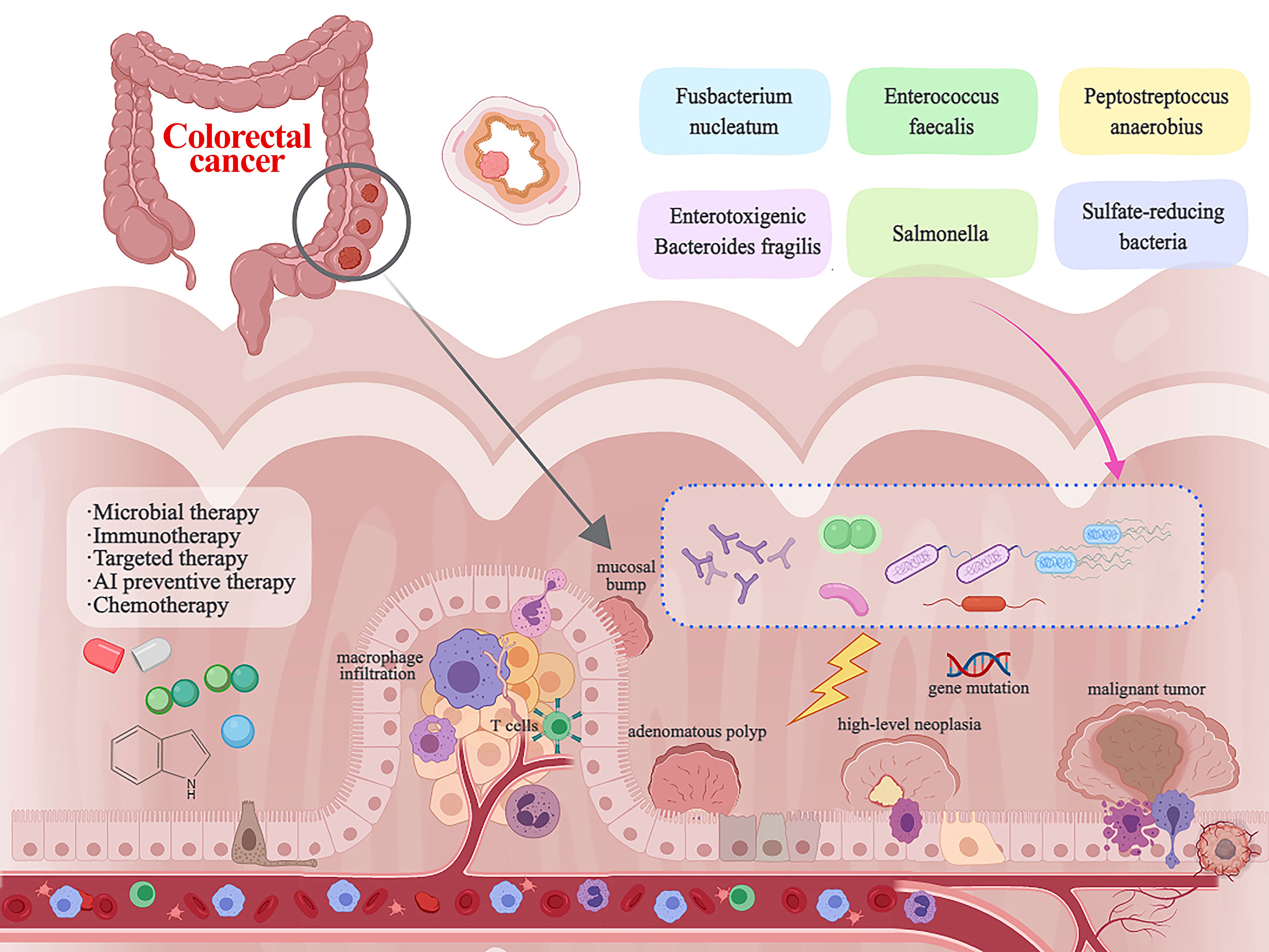
CRC originates from the mucosal epithelial cells in the colorectal mucosa layer. Clinically, CRC is mainly secondary to intestinal polyps and inflammatory bowel disease (Dyson and Rutter, 2012; Wolf et al., 2023). Novel ideas about CRC progression course are that normal mucosa after mucosal bump, small adenomatous polyp, large adenoma, high-level neoplasia, eventually into malignant tumor. The types of polyp tissue prone to cancer include tubular adenoma, villous adenoma, tubular-villous adenoma (mixed adenoma), and serrated adenoma (Knudsen et al., 2023). In a recent investigation, researchers delved into the composition of “mucosal-associated metabolites” in low-grade versus high-grade dysplastic polyps. Notably, they observed an enrichment of the genus Pelomonas, a member of the Proteobacteria phylum, in the low-grade dysplastic polyps. Conversely, microbiota analyses of high-grade dysplastic adenomas unveiled an elevated presence of the genus Anaerococcus, a taxon that has been notably abundant in CRC tissues (Clavenna et al., 2023). In a clinical study of Chinese patients, it was found that Bifidobacterium bifidum, Candida albicans, and Saccharomyces cerevisiae in the feces of CRC patients were more prevalent than those of healthy population (Li X. et al., 2023). In research conducted among individuals diagnosed with familial adenomatous polyposis (FAP), the colonic biofilms were observed to harbor oncogenic bacteria, primarily Escherichia coli and Bacteroides fragilis (Dejea et al., 2018).
In approximately 85% of colon cancers, the adenomatous polyposis coli (APC) gene, a critical tumor suppressor, undergoes deletion or inactivation (Grivennikov et al., 2012). APC gene is not only associated with FAP, but also plays an important role in the occurrence of CRC. NOTUM retains tumor suppressor activity in APC-ineffective adenomas. However, NOTUM becomes a specific oncogene when it develops into adenocarcinoma with p53 deletion (Tian et al., 2023). Oncogenic microbial communities wield the ability to reshape the entire gut microbiota’s composition, inciting pro-inflammatory reactions and incipient cellular metamorphosis, culminating in carcinogenesis (Yan et al., 2023). Furthermore, oncogenic microbiota catalyze CRC progression through the instigation of DNA damage within the epithelial cells. Epithelial barrier damage may be a consequence of β-catenin activation as well as loss of APC, microbial products drive IL-23/IL-17-mediated tumor growth (Grivennikov et al., 2012).
As early as 2012, the bacterial driver–passenger model was proposed (Tjalsma et al., 2012). Certain driver bacteria, such as E. faecalis, produce extracellular superoxide, which causes cellular DNA damage (Table 1). In a 16s RNA sequencing discovery, 7 bacterial genera were identified as potential drivers (e.g., unclassified Pseudomonadaceae and Neissenaceae) and 12 bacterial genera as potential passengers (e.g., Staphylococcus and Veillonella) (Geng et al., 2014). Some studies have also proposed the “Alpha-bug” model (Sears and Pardoll, 2011; Avril and DePaolo, 2021), enterotoxigenic Bacteroides fragilis induces colon tumors in mice (Sears and Pardoll, 2011; Yu and Fang, 2015).
3 Gut bacterial products associated with CRC
Bacteria can obtain the ability to penetrate the intestinal mucosal barrier through flagella, pili, and adhesins, as well as adhere to and invade intestinal epithelial cells, produce endotoxin or exotoxin, and then form pathogenicity (Perez-Lopez et al., 2016). Common pathogenic bacteria have been mentioned before and will not be repeated.
A recent study has suggested that an analysis of the microbial community in tumors holds the potential to identify distinct prognostic subtypes of CRC. This classification system delineates three principal subtypes: OCS1, predominantly associated with Fusobacteria and oral pathogens; OCS2, characterized by a prevalence of Firmicutes and Bacteroidetes; and OCS3, featuring an abundance of Escherichia, Pseudomonas and Shigella (Mouradov et al., 2023). OCS1 tumors mostly occur in the right colon and have high pathological grade. In contrast, OCS2 and OCS3 tumors are mostly located in the left colon and rectum with low pathological grade (Mouradov et al., 2023). There was no significant difference in clinical features between OCS2 and OCS3 (Mouradov et al., 2023). It has been found that the expression of Gal‐GalNAc (recognized by Fusobacterium Fap2) may promote the binding of Fusobacterium to CRC (Abed et al., 2016). F. nucleatum utilizes the non-lectin structure of Clostridium Fap2 to achieve tumor-promoting effects (Alon-Maimon et al., 2022). Additionally, in a pathological context, F. nucleatum augments its virulence through the secretion of an amyloid-like adhesin called FadA, utilizing a Fap2-like autotransporter (Meng et al., 2021). In addition, F. nucleatum can enhance drug resistance of tumor cells, inhibit neutrophil infiltration, and ultimately change the tumor immune microenvironment (Alon-Maimon et al., 2022; Garcia-Serrano et al., 2023). F. nucleatum is involved in tumor initiation or progression before cancer formation, which regulating the tumor immune microenvironment and promoting the proliferation of tumor-infiltrating immune cells (Kostic et al., 2013). F. nucleatum pro-inflammatory genes are characterized by upregulation of PTGS2 (Kostic et al., 2013). Nevertheless, certain experiments have revealed that F. nucleatum is not an unequivocal instigator of cancer (Nawab et al., 2023). Instead, its carcinogenic potential hinges on the particular dietary context in which it operates.
E. coli is involved in the development of CRC through the induction of inflammation and genotoxic host responses by bacteria-derived virulence factors. Some strains of E. coli produce a secondary metabolite called colibactin (Clb), and bacteria carrying pks genomic islands have DNA-damaging properties associated with CRC (Dougherty et al., 2023; Harnack et al., 2023). Blocking bacterial adhesion attenuates colibactin-mediated genotoxicity and CRC exacerbations (Jans et al., 2023). Pks+ E. coli can opportunistically enter the epithelium and promote existing mucosal damage, while mice colonized with pks+ E. coli cannot reestablish functional barriers (Harnack et al., 2023). Grotesquely, it has also been found that about half of colibactin-producing E. coli (CoPEC) can encode cytotoxic necrotizing factor-1 (CNF1) which induces CRC in mice by reducing CoPEC (Chat et al., 2023). The influence of microorganisms such as F. nucleatum, E. coli, enterotoxigenic B. fragilis, and Faecalibacterium prausnitzii on miRNAs is well-established, and this microbial impact leads to the stimulation of tumor growth and exacerbates inflammatory responses (Xing et al., 2022). Microbiota reprograms mouse intestinal lipid metabolism by suppressing expression of lncRNA Snhg9 in small intestinal epithelial cells (Tian et al., 2023).
Lostridium sporogenes is responsible for breaking down tryptophan and secreting the metabolite indole propionic acid (IPA), which has been shown to help strengthen the intestinal barrier and interact with the immune system, then change the biological characteristics of the intestine (Dodd et al., 2017). The gut microbiota metabolizes tryptophan to generate Indole-3-acetic acid (3-IAA), which effectively downregulates the expression of TNF-α. This reduction in TNF-α expression is attributed to the enzymatic conversion of tryptophan, highlighting the microbiota’s significant role in modulating inflammatory responses (Tomii et al., 2023). Furthermore, the metabolization of tryptophan by the bacterial flora results in the production of indole, which exerts regulatory control over mucosal immunity by activating receptors associated with polycyclic aromatic hydrocarbons (Lavelle and Sokol, 2020; Hezaveh et al., 2022). Bacteroides thetaiotaomicron inhibits tumor growth by producing short-chain fatty acids (SCFAs) such as propionate (Xu et al., 2023). Elevating the abundance of species such as Ruminococcaceae, Parabacterium, and Blautellae known for their capacity to generate SCFAs, Zearalenone (ZEA) exhibits a notable capacity to effectively suppress the development of colorectal tumors (Leung et al., 2023). The initiation of AhR signaling is triggered by microbiome-derived formate, which subsequently leads to the expansion of Th17 cells and promotes CRC tumor invasion (Ternes et al., 2022).
The occurrence and progression of CRC are influenced by DNA mismatch repair (MMR). In a recent examination of DNA mismatch repair deficiencies (dMMR) versus proficient DNA mismatch repair (pMMR), researchers investigated the impact of microbial-driven metabolic reconfiguration (Hale et al., 2018; Li J. et al., 2023). In the realm of dMMR, a total of 211 distinct species thrived, with noteworthy representatives including F. nucleatum, A. muciniphila and O. splanchnicus (Hsueh et al., 2022; Li J. et al., 2023). In stark contrast, a mere 2 species displayed a deficiency in dMMR, as exemplified by F. plautii. Furthermore, the dMMR environment boasted 13 metabolites in abundance, with retinoic acid being a prominent member, while on the opposite end of the spectrum, 77 metabolites experienced a significant depletion in the dMMR context, encompassing lactic acid, succinic acid, and 2,3-dihydroxyvaleric acid (Li J. et al., 2023).
The improved prognosis of colon cancer can be attributed to specific mucosal biota, namely Faecalibacterium prausnitzii and Ruminococcus gnavus. These microorganisms play a pivotal role by producing metabolites that encompass a spectrum of fatty acid species, including medium chain (MCFAs), long-chain (LCFAs), and very long-chain (VLCFAs) fatty acids, alongside ceramides and lysophospholipids (Alexander et al., 2023).
Similarly, gut bacteria can also produce substances that reverse CRC progression. In a study of female CRC patients, it was found that Carnobacterium maltaromaticum was missing (Li Q. et al., 2023). Intestinal colonization of C. maltaromaticum is influenced by estrogen and increases the abundance of vitamin D-related metabolites in colon tissue (Li Q. et al., 2023). Remarkably, the progression of CRC has been observed to be exacerbated by alterations in the male gut microbiome (Wang L. et al., 2023). This includes an augmentation in the presence of the pathogenic bacterium Akkermansia muciniphila and a reduction in the levels of the beneficial probiotic Parabacterium kingeri (Wang L. et al., 2023).
4 Gut bacteria regulate the tumor microenvironment
The CRC tumor microenvironment (TME) constitutes a multifaceted and intricate ecosystem, and plays a pivotal role in tumor growth, metastasis, and treatment response. TME comprises a diverse array of cellular components and molecular elements. It encompasses tumor cells, immune cell populations, vascular networks, fibroblasts, intestinal flora and the extracellular matrix (ECM) (Zhang et al., 2023).
It is currently believed that the TME of CRC mainly consists of the intestinal bacteria microenvironment, the inflammatory microenvironment and the hypoxic microenvironment, which work together and coordinate with each other (Wang et al., 2017). This article mainly describes the impact of intestinal bacteria on TME. Bifidobacterium adolescentis is a probiotic found in the human intestine. It can inhibit the proliferation of patholgen in the intestine and maintain the homeostasis of the bacterial microenviroment. It has been experimentally confirmed that B. adolescentis inhibits tumorigenesis by inducing a new CD143+ cancer-associated fibroblasts through Wnt signaling-regulated GAS1 (Chen et al., 2023). In addition, B. adolescentis inhibits colorectal carcinogenesis through TLR2 induction of decorin+ macrophages (Lin et al., 2023). In AOM/DSS-induced mice, B. thetaiotaomicron suppresses tumorigenesis of colitis-associated CRC and MC38 allograft tumors (Xu et al., 2023). Not only in CRC, but other experiments have shown that in melanoma, Eubacterium rectale significantly improves the efficacy of anti-PD1 treatment and the overall survival rate of tumor-bearing mice (Liu et al., 2023). Eubacterium rectale consumes l-serine to enhance NK cell function and anti-PD1 therapeutic effect, leading to activation of NK cell activity through the FOS/FOSL2 signaling pathway (Liu et al., 2023).
In an in vitro study, F. nucleatum infection was found to induce a significant increase in the production of neutrophil extracellular traps (NETs) (Kong et al., 2023). This demonstrates that F. nucleatum-induced NETs indirectly accelerate malignant tumor growth through angiogenesis and promote tumor metastasis. This is exemplified by cellular migration linked to the process of epithelial-mesenchymal transition (EMT), the breakdown of basement membrane proteins facilitated by matrix metalloproteinases (MMPs), and the entrapment of CRC cells (Kong et al., 2023). In research, exposure of peripheral blood mononuclear cells (PBMCs) to LPS derived from these microorganisms revealed that F. periodonticum triggers cytokine synthesis in PBMCs, whereas both B. fragilis and P. asaccharolytica exerted a suppressive influence (Sulit et al., 2023). In a study of intratumoral bacteria, elevated autophagy induced by F. nucleatum led to increased resistance to reactive oxygen species (ROS) in CRC, this resistance was alleviated, ultimately promoting apoptosis in cancer cells, and apoptosis was triggered by intracellular redox imbalance caused by the interaction with BSA-Cu SAN (Wang X. et al., 2023).
5 Metastasis and immune escape of CRC cells
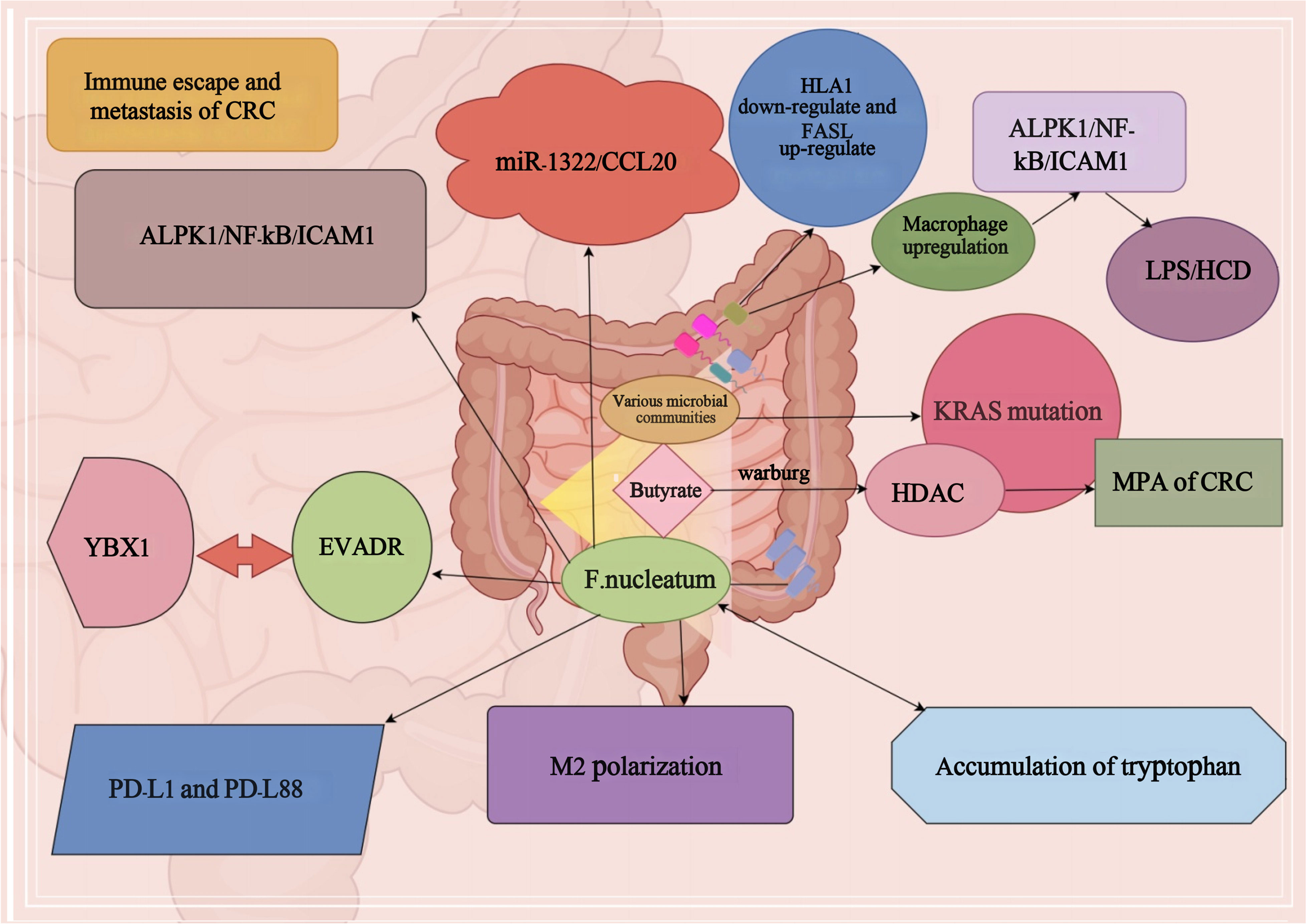
Studies have shown that relevant DNA analysis of CRC patients and fecal microorganisms found that KRAS gene mutations have a significant impact on distant metastasis of CRC (Sui et al., 2020). At the same time, in CRC, the abundance of different bacterial groups is also influencing the mutation of KRAS gene, which affects the metastasis and progression of CRC (Sui et al., 2020). Microorganisms such as Rosella, Paramecium, Post-Rosella, Staphylococcaceae and Bacillariophyta in the mutant group significantly affected distant metastasis of CRC through KRAS gene mutation, and their prevalence and metastasis were significantly higher than those in the non-mutant group (Liu et al., 2021). Furthermore, butyrate, a prominent component among SCFAs, plays a pivotal role in the metabolic processes of normal colorectal epithelial cells (Yan et al., 2024). Remarkably, a substantial portion of butyrate remains unmetabolized, largely attributed to the fact that colon cells have a Warburg effect pathway (Eslami et al., 2020). Butyrate serves as a potent histone deacetylase (HDAC) inhibitor, influencing the intricate orchestration of tumor cell metabolism, proliferation, and apoptosis (Korsten et al., 2023). Consequently, these multifaceted interactions exert a significant impact on the metastatic potential of CRC (Li et al., 2021). At the same time, it was shown that F. nucleatum was found to be highly abundant in CRC and promote CRC metastasis by affecting the miR-1322/CCL20 axis and M2 polarization (Xu et al., 2021). The ALPK1/NF-κB/ICAM1 pathway can be induced by F. nucleatum, leading to enhanced adhesion of CRC cells to intestinal endothelial cells, as well as increased infiltration and distant metastasis (Zhang et al., 2022). Additionally, EVADR induction has the potential to facilitate CRC metastasis through YBX1-dependent translation processes (Lu et al., 2022). It has been reported that sustained F. nucleatum exposure reduces the diversity of the intestinal microbiota in mice, leading to an imbalance of the intestinal bacteria, and a reorganization of the associated bacteria, which intricately affects colorectal carcinogenesis and progression through the secretion of pro-inflammatory cytokines (Yin et al., 2022).
F. nucleatum promotes CRC progression and upregulates PD-L1 protein expression in CRC cell lines, thereby promoting immune escape from the tumor (Gao et al., 2023). Furthermore, studies have shown that the accumulation of tryptophan derivatives in the gut promotes the formation of suitable targets for immune escape (Puccetti et al., 2015). Simultaneously, the oncogenic bacteria in the gut, or the metabolites they generate, stimulate the generation of macrophages. The presence of LPS or HCD-induced macrophage infiltration notably triggers the activation of the macrophage-derived CCL5-p65/STAT3-CSN5-PD-L1 signaling pathway, which plays a crucial role in facilitating immune evasion in CRC (Liu et al., 2020). F. nucleatum can also lead to tumor subclones with PD-L1 mutations, nonsense-mediated RNA decay in PD-L1 K1fs, and protein degradation in PD-L162 L1S, thereby promoting its immune escape and tumor metastasis (Stein et al., 2021). It has also been shown that metabolites associated with F. nucleatum can affect up to 50% of dMMR/high microsatellite instability (MSI-H) advanced cancer patients who progress after PD-1 blockade, leading to a high probability of immune escape (Cohen et al., 2020). F. nucleatum has the capacity to promote CRC immune escape by influencing the depletion of human leukocyte antigen class I (HLA-I) (Anderson et al., 2021). In addition, F. nucleatum can also help colon cancer evade immune surveillance and immune elimination by influencing Fas expression (O’Connell et al., 2000). Simultaneously, it can bolster the resistance of CRC to the immune system through the upregulation of FasL expression (Zhu et al., 2005). In summary, as mentioned in Figure 1 CRC immune escape and distant metastasis can be caused by the joint action of intestinal carcinogenic flora and their metabolites.
6 Microbiological therapy for CRC
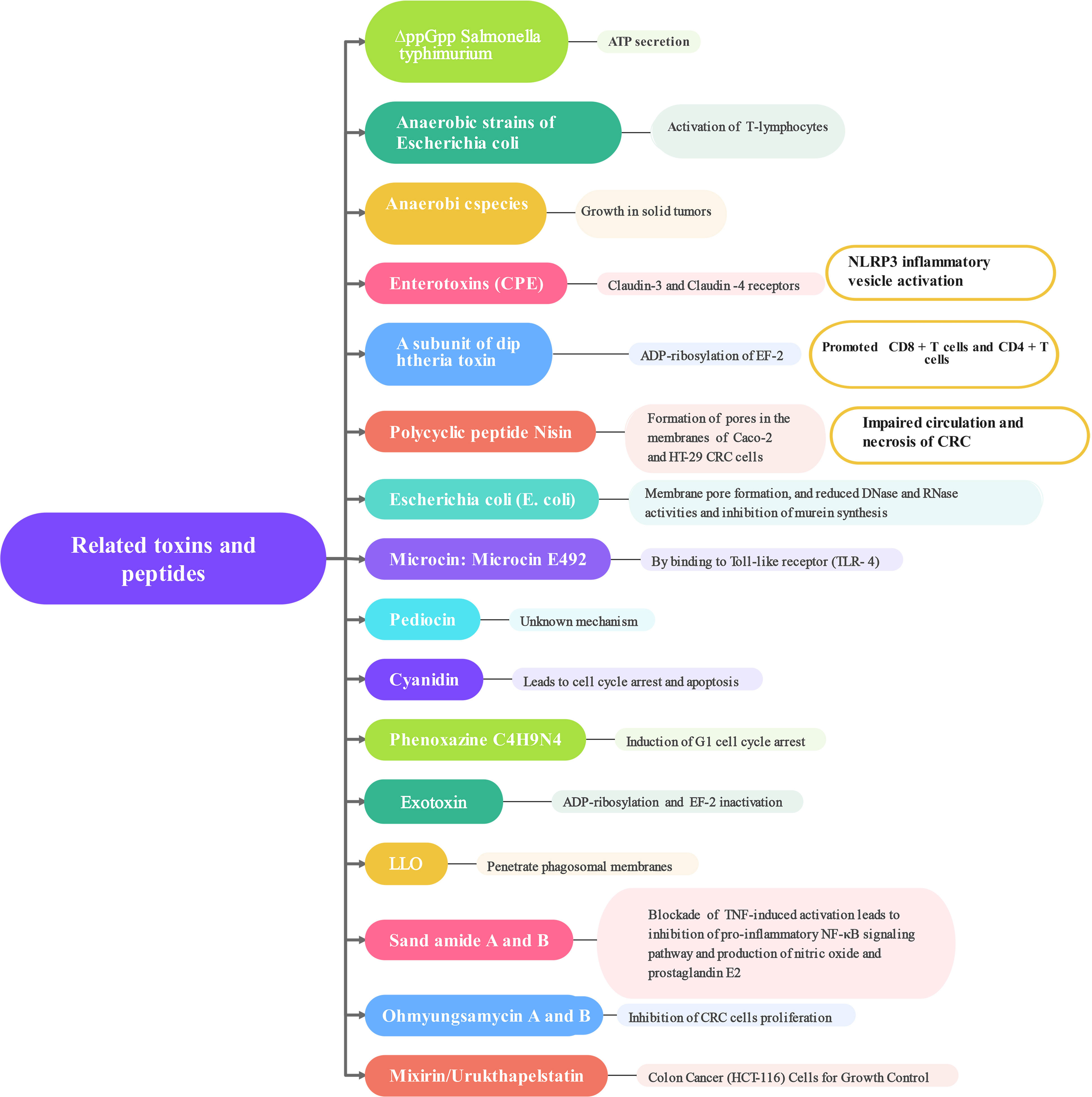
The connection between CRC and the gut microbiota is strong. While we still don’t fully understand how the microbiota impacts the development and progression of CRC, there is increasing proof that it plays a direct role in influencing signaling pathways, anti-tumor immune responses, and cell growth (Montalban-Arques and Scharl, 2019). It has been shown that the gut microbiota immune system kills the bacterial flora through specific receptors (Toll-like receptors) and related metabolites (Figure 2). Clostridium nucleatum, Escherichia coli, and Mimicronium fragilis play a crucial role in the development of CRC. Increasing dietary fiber, including fructans and oligogalactans, has an inhibitory effect on CRC, but it also affects the abundance of Bifidobacteria and Lactobacillus, which increases fecal butyrate concentrations (Rebersek, 2021). It has been reported that intestinal flora plays an anti-cancer role in the efficacy of PD-L1 immune checkpoint inhibitor blockade (Yu, 2018). F. nucleatum has been shown to induce different immune responses in CRCs with varying microsatellite instability (MSI) states. F. nucleatum could induce PD-L8 expression by activating STING signaling during PD-L1 blockade therapy and increase the interferon-gamma (IFN-γ) CD1 tumor-infiltrating lymphocytes (TILs), which increases tumor sensitivity to PD-L1 blockade (Gao et al., 2021). It has also been reported that inhibition of F. nucleatum and reduction of its abundance modulate the TLR-4-mediated pathway and MyD88-induced cellular autophagy, which may enhance the chemotherapeutic effect of CRC (Mima et al., 2015; Yu et al., 2017). Simultaneously, the restoration of the gut microbiota composition can lead to the augmentation of regulatory T cell populations within the colonic mucosa (Routy et al., 2018; Shi et al., 2023). According to recent studies, the anticancer effects of microbial therapies such as bacterial therapies are mainly manifested in the form of bacterial-related biologics, including toxins and peptides (Mueller et al., 2022). These compounds produce regulatory cytokines, like TNF-α, which leads to the activation or blocking of NF-κB, and they also activate pro-apoptotic proteins (Bcl-1, Bad, Bax, Bak), combine cytochrome C with caspase-9 to form an apoptotic complex, and ultimately promote CRC cells apoptosis. Apoptosis is a key target of cancer therapy and is characterized by an imbalance between cell proliferation and death, resulting in autophagy in CRC cells (Mueller et al., 2022). Next, how the following related strains and their metabolites combat CRC was explored (Figure 3). According to some studies, timulation of the inflammatory vesicle pathway triggered by bacteria can activate the immune system, and ΔppGpp Salmonella typhimurium inhibits primary and even metastatic CRC by secreting ATP, which causes activation of the NLRP3 inflammatory vesicle in macrophages (Mengesha et al., 2007; Nguyen et al., 2010). It has also been shown that the anaerobic strain of E. coli counteracts CRC cells by activating the production of T-lymphocytes, thereby greatly contributing to the tumor-protective activity of CD8+ and CD4+ T-cells (Azadi et al., 2021). At the same time, anaerobic bacterial species can invade and grow in solid tumors, allowing impaired circulation and necrosis of CRC (Fox et al., 1996; Zhao et al., 2005; Agrawal et al., 2017; Kasper et al., 2020). The antagonistic effect of related toxins on CRC was also investigated. Based on relevant reports and experiments, it has been shown that Clostridium perfringens enterotoxin (CPE) produced by Clostridium perfringens can bind to Claudin-3 and -4 receptors on the surface of CRC, leading to the breakdown of cellular osmotic homeostasis and the lysis of cancer cells (Pahle et al., 2017; Sasaki et al., 2020). The subunit derived from Gram-positive Corynebacterium diphtheriae can halt protein production by ADP-ribosylating cytoplasmic elongation factor 2 (EF-2), eventually resulting in the demise of CRC cells (Vallera et al., 2002; Martarelli et al., 2009). The polycyclic peptide Nisin secreted by Lactococcus lactis strains enables the formation of pores in the membranes of Caco-2 and HT-29 CRC cells ultimately leading to membrane depolarization and apoptosis in CRC cells (Ahmadi et al., 2017). Cytotoxic effects of colistin on CRC cells include membrane pore formation, reduced DNase and RNase activities, and inhibition of murein synthesis (Kohoutova et al., 2020). Microcin/Microcin E492 causes apoptosis by enabling pore formation in CRC cell membranes and ultimately by binding to Toll-like receptor 4 (Hetz et al., 2002; Lagos et al., 2009). Pediocin has been observed to trigger apoptosis through a mechanism that remains unidentified (Mueller et al., 2022). Proteins capable of entering CRC cells and inducing cell cycle arrest and apoptosis by aspyrins (Mueller et al., 2022). Phenazine, a nitrogen-containing metabolite, is produced by various bacterial strains, with notable secretion observed in numerous Pseudomonas aeruginosa strains. This compound includes phenazine 1-carboxylic acid and phenazine 1,6-dicarboxylic acid (PDC) (Wolf and Elsässer-Beile, 2009). Crucially, it induces G1 cell-cycle arrest, consequently prompting apoptosis, while also negatively impacting CRC cell viability and hampering DNA synthesis (Iglewski and Kabat, 1975; Wolf and Elsässer-Beile, 2009). Recall antigens delivered via Listeria might serve as a viable option for cancer immunotherapy beyond neoantigens (Selvanesan et al., 2022). Listeriolysin O (LLO), a poisonous compound produced by the anaerobic microorganism Listeria monocytogenes, possesses the ability to infiltrate the cytoplasm of antigen-presenting cells and rupture the phagosome membranes (Mueller et al., 2022).
Following that, non-ribosomal peptides are discussed, which constitute an alternative group of peptides produced by bacteria, fungi, and cyanobacteria. These peptides play a role in combatting CRC. Lucentamycins, Arenamides, Ohmyungsamycins, Mixirins, and Urukthapelstatin A possess the ability to engage with CRC cells, either through direct interactions or indirect mechanisms (Sacks et al., 2018). For instance, sarcosamides A and B have demonstrated their potential in inhibiting the pro-inflammatory NF-κB signaling pathway by effectively blocking TNF-induced activation, ultimately leading to a reduction in inflammation (Byun et al., 2020). Consequently, this decrease in inflammation hinders the production of NO and PGE2, effectively opposing the activities of CRC cells (Byun et al., 2020). Cyclic depsipeptides, specifically Ohmyungsamycin A and B, display a discerning ability to impede the proliferation of CRC cells (Um et al., 2013; Byun et al., 2020). Mixirin, derived from Bacillus marinus, is a cyclic thiopeptide that can exhibit cytotoxicity against the HCT-116 (human colon cancer cell line) (Yamamoto et al., 2015). Urukthapelstatin A is a cyclic sulfur peptide produced by Mechercharimyces asporophorigenens, a marine microorganism affiliated with the Thermoactinobacteriaceae family (Matsuo et al., 2007). This compound exerts inhibitory effects on the proliferation of HCT-116 cell line through its biological activity (Mueller et al., 2022).
7 Conclusion
Intestinal microorganisms constitute a rich ecosystem, with more than 1000 species of bacteria belonging to 50 genera and 17 families. Their composition depends largely on environmental conditions, and there are differences among individuals. With the in-depth study of intestinal bacteria, we can find that intestinal bacteria and their metabolites have many effects on CRC, such as inflammatory transformation, malignant transformation of intestinal polyps, tumor escape, treatment and so on. According to relevant studies, it can be reported that apoptosis of CRC cells can be induced by inhibiting the activity of glutamate dehydrogenase, regulating the MAPK signaling pathway, PI3K/AKT, and other related pathway mechanisms, which are crucial for the development of CRC (Chang and Kang, 2023; Yang et al., 2023).
In this paper, we reviewed that intestinal bacteria can participate in adenoma-adenocarcinoma transformation through their metabolites and affect the DNA coding of intestinal cells. It is believed that in the initial stage of CRC, “driver” bacteria are dominant in the intestine, which leads to adenoma and even malignant tumor with the increase of DNA damage and chromosome instability in intestinal cells. In addition, intestinal flora can directly induce tumor-associated immune cell infiltration and promote the formation of tumor microenvironment. In some familial hereditary adenomatous polyposis, specific intestinal bacteria often play a role in promoting the carcinogenesis of adenomas. No matter which kind of colon cancer patients, the determination of intestinal flora and its metabolites has great clinical significance, because it may early warn the occurrence of colorectal cancer and adenoma, or improve the prognosis of patients with CRC. Tailoring the regulation of gut microbiota on an individual basis is poised to emerge as a focal point and innovative strategy in the realm of preventing and supporting the treatment of CRC.
Author contributions
JH: Conceptualization, Writing – original draft, Writing – review & editing. BZ: Conceptualization, Writing – original draft, Writing – review & editing. YZ: Conceptualization, Writing – original draft, Writing – review & editing. TY: Conceptualization, Writing – original draft, Writing – review & editing. YC: Conceptualization, Writing – original draft, Writing – review & editing. JL: Conceptualization, Writing – original draft, Writing – review & editing. YY: Conceptualization, Writing – original draft, Writing – review & editing. HS: Conceptualization, Writing – original draft, Writing – review & editing. DS: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abed, J., Emgård, J. E. M., Zamir, G., Faroja, M., Almogy, G., Grenov, A., et al. (2016). Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-galNAc. Cell Host Microbe 20 (2), 215–225. doi: 10.1016/j.chom.2016.07.006
Agrawal, S., Acharya, D., Adholeya, A., Barrow, C. J., Deshmukh, S. K. (2017). Nonribosomal peptides from marine microbes and their antimicrobial and anticancer potential. Front. Pharmacol. 8, 828. doi: 10.3389/fphar.2017.00828
Ahmadi, S., Ghollasi, M., Hosseini, H. M. (2017). The apoptotic impact of nisin as a potent bacteriocin on the colon cancer cells. Microb. Pathog. 111, 193–197. doi: 10.1016/j.micpath.2017.08.037
Alexander, J. L., Posma, J. M., Scott, A., Poynter, L., Mason, S. E., Doria, M. L., et al. (2023). Pathobionts in the tumour microbiota predict survival following resection for colorectal cancer. Microbiome 11 (1), 100. doi: 10.1186/s40168-023-01518-w
Alon-Maimon, T., Mandelboim, O., Bachrach, G. (2022). Fusobacterium nucleatum and cancer. Periodontol 2000 89 (1), 166–180. doi: 10.1111/prd.12426
Anderson, P., Aptsiauri, N., Ruiz-Cabello, F., Garrido, F. (2021). HLA class I loss in colorectal cancer: implications for immune escape and immunotherapy. Cell Mol. Immunol. 18 (3), 556–565. doi: 10.1038/s41423-021-00634-7
Avril, M., DePaolo, R. W. (2021). “Driver-passenger” bacteria and their metabolites in the pathogenesis of colorectal cancer. Gut Microbes 13 (1), 1941710. doi: 10.1080/19490976.2021.1941710
Azadi, A., Golchini, A., Delazar, S., Abarghooi Kahaki, F., Dehnavi, S. M., Payandeh, Z., et al. (2021). Recent advances on immune targeted therapy of colorectal cancer using bi-specific antibodies and therapeutic vaccines. Biol. Proced Online 23 (1), 13. doi: 10.1186/s12575-021-00147-7
Biarc, J., Nguyen, I. S., Pini, A., Gossé, F., Richert, S., Thiersé, D., et al. (2004). Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S.bovis). Carcinogenesis 25 (8), 1477–1484. doi: 10.1093/carcin/bgh091
Byun, W. S., Kim, S., Shin, Y. H., Kim, W. K., Oh, D. C., Lee, S. K. (2020). Antitumor activity of ohmyungsamycin A through the regulation of the Skp2-p27 axis and MCM4 in human colorectal cancer cells. J. Nat. Prod 83 (1), 118–126. doi: 10.1021/acs.jnatprod.9b00918
Center, M. M., Jemal, A., Smith, R. A., Ward, E. (2009). Worldwide variations in colorectal cancer. CA: Cancer J. Clin. 59 (6), 366–378. doi: 10.3322/caac.20038
Chang, S. N., Kang, S. C. (2023). Decursinol angelate inhibits glutamate dehydrogenase 1 activity and induces intrinsic apoptosis in MDR-CRC cells. Cancers (Basel) 15 (14), 3541. doi: 10.3390/cancers15143541
Chat, H., Dalmasso, G., Godfraind, C., Bonnin, V., Beyrouthy, R., Bonnet, M., et al. (2023). Cytotoxic necrotizing factor 1 hinders colon tumorigenesis induced by colibactin-producing Escherichia coli in ApcMin/+ mice. Gut Microbes 15 (1), 2229569. doi: 10.1080/19490976.2023.2229569
Chen, S., Fan, L., Lin, Y., Qi, Y., Xu, C., Ge, Q., et al. (2023). Bifidobacterium adolescentis orchestrates CD143+ cancer-associated fibroblasts to suppress colorectal tumorigenesis by Wnt signaling-regulated GAS1. Cancer Commun. (Lond). 43 (9), 1027–1047. doi: 10.1002/cac2.12469
Clavenna, M. G., La Vecchia, M., Sculco, M., Joseph, S., Barberis, E., Amede, E., et al. (2023). Distinct signatures of tumor-associated microbiota and metabolome in low-grade vs. High-grade dysplastic colon polyps: inference of their role in tumor initiation and progression. Cancers (Basel) 15 (12), 3065. doi: 10.3390/cancers15123065
Cohen, R., Rousseau, B., Vidal, J., Colle, R., Diaz, L. A., Jr., André, T. (2020). Immune checkpoint inhibition in colorectal cancer: microsatellite instability and beyond. Target Oncol. 15 (1), 11–24. doi: 10.1007/s11523-019-00690-0
Dejea, C. M., Fathi, P., Craig, J. M., Boleij, A., Taddese, R., Geis, A. L., et al. (2018). Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359 (6375), 592–597. doi: 10.1126/science.aah3648
Dodd, D., Spitzer, M. H., Van Treuren, W., Merrill, B. D., Hryckowian, A. J., Higginbottom, S. K., et al. (2017). A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551 (7682), 648–652. doi: 10.1038/nature24661
Dougherty, M. W., Valdés-Mas, R., Wernke, K. M., Gharaibeh, R. Z., Yang, Y., Brant, J. O., et al. (2023). The microbial genotoxin colibactin exacerbates mismatch repair mutations in colorectal tumors. Neoplasia 43, 100918. doi: 10.1016/j.neo.2023.100918
Dyson, J. K., Rutter, M. D. (2012). Colorectal cancer in inflammatory bowel disease: what is the real magnitude of the risk? World J. Gastroenterol. 18 (29), 3839–3848. doi: 10.3748/wjg.v18.i29.3839
Eslami, M., Sadrifar, S., Karbalaei, M., Keikha, M., Kobyliak, N. M., Yousefi, B. (2020). Importance of the microbiota inhibitory mechanism on the warburg effect in colorectal cancer cells. J. Gastrointest Cancer 51 (3), 738–747. doi: 10.1007/s12029-019-00329-3
Evans, M. D., Dizdaroglu, M., Cooke, M. S. (2004). Oxidative DNA damage and disease: induction, repair and significance. Mutat. Res. 567 (1), 1–61. doi: 10.1016/j.mrrev.2003.11.001
Fox, M. E., Lemmon, M. J., Mauchline, M. L., Davis, T. O., Giaccia, A. J., Minton, N. P., et al. (1996). Anaerobic bacteria as a delivery system for cancer gene therapy: in vitro activation of 5-fluorocytosine by genetically engineered clostridia. Gene Ther. 3 (2), 173–178.
Gao, Y., Bi, D., Xie, R., Li, M., Guo, J., Liu, H., et al. (2021). Fusobacterium nucleatum enhances the efficacy of PD-L1 blockade in colorectal cancer. Signal Transduct Target Ther. 6 (1), 398. doi: 10.1038/s41392-021-00795-x
Gao, Y., Zou, T., Xu, P., Wang, Y., Jiang, Y., Chen, Y. X., et al. (2023). Fusobacterium nucleatum stimulates cell proliferation and promotes PD-L1 expression via IFIT1-related signal in colorectal cancer. Neoplasia 35, 100850. doi: 10.1016/j.neo.2022.100850
Garcia-Serrano, A., Mukhedkar, D., Hultin, E., Rudsander, U., Wettergren, Y., Ure, A. E., et al. (2023). Assessment of bacterial and viral gut communities in healthy and tumoral colorectal tissue using RNA and DNA deep sequencing. Cancer Med. 12 (18), 19291–19300. doi: 10.1002/cam4.6483
Geng, J., Song, Q., Tang, X., Liang, X., Fan, H., Peng, H., et al. (2014). Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathog. 6, 26. doi: 10.1186/1757-4749-6-26
Grivennikov, S. I., Wang, K., Mucida, D., Stewart, C. A., Schnabl, B., Jauch, D., et al. (2012). Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491 (7423), 254–258. doi: 10.1038/nature11465
Hale, V. L., Jeraldo, P., Chen, J., Mundy, M., Yao, J., Priya, S., et al. (2018). Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers. Genome Med. 10 (1), 78. doi: 10.1186/s13073-018-0586-6
Harnack, C., Berger, H., Liu, L., Mollenkopf, H.-J., Strowig, T., Sigal, M. (2023). Short-term mucosal disruption enables colibactin-producing E. coli to cause long-term perturbation of colonic homeostasis. Gut Microbes 15 (1), 2233689. doi: 10.1080/19490976.2023.2233689
Hetz, C., Bono, M. R., Barros, L. F., Lagos, R. (2002). Microcin E492, a channel-forming bacteriocin from Klebsiella pneumoniae, induces apoptosis in some human cell lines. Proc. Natl. Acad. Sci. U.S.A. 99 (5), 2696–2701. doi: 10.1073/pnas.052709699
Hezaveh, K., Shinde, R. S., Klötgen, A., Halaby, M. J., Lamorte, S., Ciudad, M. T., et al. (2022). Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity 55 (2), 324–340.e8. doi: 10.1016/j.immuni.2022.01.006
Hsueh, C.-Y., Lau, H.-C., Huang, Q., Gong, H., Sun, J., Cao, P., et al. (2022). Fusobacterium nucleatum impairs DNA mismatch repair and stability in patients with squamous cell carcinoma of the head and neck. Cancer 128 (17), 3170–3184. doi: 10.1002/cncr.34338
Iglewski, B. H., Kabat, D. (1975). NAD-dependent inhibition of protein synthesis by Pseudomonas aeruginosa toxin. Proc. Natl. Acad. Sci. U.S.A. 72 (6), 2284–2288. doi: 10.1073/pnas.72.6.2284
Jans, M., Kolata, M., Blancke, G., Ciers, M., Dohlman, A. B., Kusakabe, T., et al. (2023). Colibactin-induced genotoxicity and colorectal cancer exacerbation critically depends on adhesin-mediated epithelial binding. bioRxiv. doi: 10.1101/2023.08.16.553526
Kasper, S. H., Morell-Perez, C., Wyche, T. P., Sana, T. R., Lieberman, L. A., Hett, E. C. (2020). Colorectal cancer-associated anaerobic bacteria proliferate in tumor spheroids and alter the microenvironment. Sci. Rep. 10 (1), 5321. doi: 10.1038/s41598-020-62139-z
Knudsen, M. D., Wang, K., Wang, L., Polychronidis, G., Berstad, P., Wu, K., et al. (2023). Development and validation of a risk prediction model for post-polypectomy colorectal cancer in the USA: a prospective cohort study. EClinicalMedicine 62, 102139. doi: 10.1016/j.eclinm.2023.102139
Kohoutova, D., Forstlova, M., Moravkova, P., Cyrany, J., Bosak, J., Smajs, D., et al. (2020). Bacteriocin production by mucosal bacteria in current and previous colorectal neoplasia. BMC Cancer 20 (1), 39. doi: 10.1186/s12885-020-6512-5
Kong, X., Zhang, Y., Xiang, L., You, Y., Duan, Y., Zhao, Y., et al. (2023). Fusobacterium nucleatum-triggered neutrophil extracellular traps facilitate colorectal carcinoma progression. J. Exp. Clin. Cancer Res. 42 (1), 236. doi: 10.1186/s13046-023-02817-8
Korsten, S. G. P. J., Vromans, H., Garssen, J., Willemsen, L. E. M. (2023). Butyrate protects barrier integrity and suppresses immune activation in a caco-2/PBMC co-culture model while HDAC inhibition mimics butyrate in restoring cytokine-induced barrier disruption. Nutrients 15 (12), 2760. doi: 10.3390/nu15122760
Kostic, A. D., Chun, E., Robertson, L., Glickman, J. N., Gallini, C. A., Michaud, M., et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14 (2), 207–215. doi: 10.1016/j.chom.2013.07.007
Lagos, R., Tello, M., Mercado, G., García, V., Monasterio, O. (2009). Antibacterial and antitumorigenic properties of microcin E492, a pore-forming bacteriocin. Curr. Pharm. Biotechnol. 10 (1), 74–85. doi: 10.2174/138920109787048643
Lavelle, A., Sokol, H. (2020). Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 17 (4), 223–237. doi: 10.1038/s41575-019-0258-z
Leung, H. K. M., Lo, E. K. K., Chen, C., Zhang, F., Felicianna, Ismaiah, M. J., et al. (2023). Zearalenone attenuates colitis associated colorectal tumorigenesis through Ras/Raf/ERK pathway suppression and SCFA-producing bacteria promotion. BioMed. Pharmacother. 164, 114973. doi: 10.1016/j.biopha.2023.114973
Li, Q., Chan, H., Liu, W.-X., Liu, C.-A., Zhou, Y., Huang, D., et al. (2023). Carnobacterium Maltaromaticum boosts intestinal vitamin D production to suppress colorectal cancer in female mice. Cancer Cell 41 (8), 1450–1465.e8. doi: 10.1016/j.ccell.2023.06.011
Li, X., Feng, J., Wang, Z., Liu, G., Wang, F. (2023). Features of combined gut bacteria and fungi from a Chinese cohort of colorectal cancer, colorectal adenoma, and post-operative patients. Front. Microbiol. 14, 1236583. doi: 10.3389/fmicb.2023.1236583
Li, J., Guo, Y., Liu, J., Guo, F., Du, L., Yang, Y., et al. (2023). Depicting the landscape of gut microbial-metabolic interaction and microbial-host immune heterogeneity in deficient and proficient DNA mismatch repair colorectal cancers. J. Immunother. Cancer 11 (8), e007420. doi: 10.1136/jitc-2023-007420
Li, W., Lu, Y., Ye, C., Ouyang, M. (2021). The regulatory network of microRNA in the metabolism of colorectal cancer. J. Cancer 12 (24), 7454–7464. doi: 10.7150/jca.61618
Lin, Y., Fan, L., Qi, Y., Xu, C., Jia, D., Jiang, Y., et al. (2023). Bifidobacterium adolescentis induces Decorin+ macrophages via TLR2 to suppress colorectal carcinogenesis. J. Exp. Clin. Cancer Res. 42 (1), 172. doi: 10.1186/s13046-023-02746-6
Liu, N., Chen, L., Yan, M., Tao, Q., Wu, J., Chen, J., et al. (2023). Eubacterium rectale Improves the Efficacy of Anti-PD1 Immunotherapy in Melanoma via l-Serine-Mediated NK Cell Activation. Res. (Wash D C) 6, 0127. doi: 10.34133/research.0127
Liu, C., Yao, Z., Wang, J., Zhang, W., Yang, Y., Zhang, Y., et al. (2020). Macrophage-derived CCL5 facilitates immune escape of colorectal cancer cells via the p65/STAT3-CSN5-PD-L1 pathway. Cell Death Differ 27 (6), 1765–1781. doi: 10.1038/s41418-019-0460-0
Liu, W., Zhang, X., Xu, H., Li, S., Lau, H. C.-H., Chen, Q., et al. (2021). Microbial community heterogeneity within colorectal neoplasia and its correlation with colorectal carcinogenesis. Gastroenterology 160 (7), 2395–2408. doi: 10.1053/j.gastro.2021.02.020
Lu, X., Xu, Q., Tong, Y., Zhang, Z., Dun, G., Feng, Y., et al. (2022). Long non-coding RNA EVADR induced by Fusobacterium nucleatum infection promotes colorectal cancer metastasis. Cell Rep. 40 (3), 111127. doi: 10.1016/j.celrep.2022.111127
Martarelli, D., Pompei, P., Mazzoni, G. (2009). Inhibition of adrenocortical carcinoma by diphtheria toxin mutant CRM197. Chemotherapy 55 (6), 425–432. doi: 10.1159/000264689
Matsuo, Y., Kanoh, K., Imagawa, H., Adachi, K., Nishizawa, M., Shizuri, Y. (2007). Urukthapelstatin A, a novel cytotoxic substance from marine-derived Mechercharimyces asporophorigenens YM11-542. II. Physico-chemical properties and structural elucidation. J. Antibiot (Tokyo) 60 (4), 256–260. doi: 10.1038/ja.2007.30
Meng, Q., Gao, Q., Mehrazarin, S., Tangwanichgapong, K., Wang, Y., Huang, Y., et al. (2021). Fusobacterium nucleatum secretes amyloid-like FadA to enhance pathogenicity. EMBO Rep. 22 (7), e52891. doi: 10.15252/embr.202152891
Mengesha, A., Dubois, L., Chiu, R. K., Paesmans, K., Wouters, B. G., Lambin, P., et al. (2007). Potential and limitations of bacterial-mediated cancer therapy. Front. Biosci. 12, 3880–3891. doi: 10.2741/2357
Mima, K., Sukawa, Y., Nishihara, R., Qian, Z. R., Yamauchi, M., Inamura, K., et al. (2015). Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 1 (5), 653–661. doi: 10.1001/jamaoncol.2015.1377
Montalban-Arques, A., Scharl, M. (2019). Intestinal microbiota and colorectal carcinoma: Implications for pathogenesis, diagnosis, and therapy. EBioMedicine 48, 648–655. doi: 10.1016/j.ebiom.2019.09.050
Mouradov, D., Greenfield, P., Li, S., In, E.-J., Storey, C., Sakthianandeswaren, A., et al. (2023). Oncomicrobial community profiling identifies clinicomolecular and prognostic subtypes of colorectal cancer. Gastroenterology 165 (1), 104–120. doi: 10.1053/j.gastro.2023.03.205
Mueller, A. L., Brockmueller, A., Fahimi, N., Ghotbi, T., Hashemi, S., Sadri, S., et al. (2022). Bacteria-mediated modulatory strategies for colorectal cancer treatment. Biomedicines 10 (4), 832. doi: 10.3390/biomedicines10040832
Nawab, S., Bao, Q., Ji, L.-H., Luo, Q., Fu, X., Fan, S., et al. (2023). The pathogenicity of fusobacterium nucleatum modulated by dietary fibers-A possible missing link between the dietary composition and the risk of colorectal cancer. Microorganisms 11 (8), 2004. doi: 10.3390/microorganisms11082004
Nguyen, V. H., Kim, H. S., Ha, J. M., Hong, Y., Choy, H. E., Min, J. J. (2010). Genetically engineered Salmonella typhimurium as an imageable therapeutic probe for cancer. Cancer Res. 70 (1), 18–23. doi: 10.1158/0008-5472.CAN-09-3453
O’Connell, J., Bennett, M. W., Nally, K., Houston, A., O’Sullivan, G. C., Shanahan, F. (2000). Altered mechanisms of apoptosis in colon cancer: Fas resistance and counterattack in the tumor-immune conflict. Ann. N Y Acad. Sci. 910, 178–92. doi: 10.1111/j.1749-6632.2000.tb06708.x
Pahle, J., Menzel, L., Niesler, N., Kobelt, D., Aumann, J., Rivera, M., et al. (2017). Rapid eradication of colon carcinoma by Clostridium perfringens Enterotoxin suicidal gene therapy. BMC Cancer 17 (1), 129. doi: 10.1186/s12885-017-3123-x
Perez-Lopez, A., Behnsen, J., Nuccio, S. P., Raffatellu, M. (2016). Mucosal immunity to pathogenic intestinal bacteria. Nat. Rev. Immunol. 16 (3), 135–148. doi: 10.1038/nri.2015.17
Puccetti, P., Fallarino, F., Italiano, A., Soubeyran, I., MacGrogan, G., Debled, M., et al. (2015). Accumulation of an endogenous tryptophan-derived metabolite in colorectal and breast cancers. PloS One 10 (4), e0122046. doi: 10.1371/journal.pone.0122046
Rebersek, M. (2021). Gut microbiome and its role in colorectal cancer. BMC Cancer 21 (1), 1325. doi: 10.1186/s12885-021-09054-2
Routy, B., Gopalakrishnan, V., Daillère, R., Zitvogel, L., Wargo, J. A., Kroemer, G. (2018). The gut microbiota influences anticancer immunosurveillance and general health. Nat. Rev. Clin. Oncol. 15 (6), 382–396. doi: 10.1038/s41571-018-0006-2
Sacks, D., Baxter, B., Campbell, B. C. V., Carpenter, J. S., Cognard, C., Dippel, D., et al. (2018). Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int. J. stroke: Off. J. Int. Stroke Soc. 13 (6), 612–632. doi: 10.1016/j.jvir.2017.11.026
Sasaki, T., Mori, S., Kishi, S., Fujiwara-Tani, R., Ohmori, H., Nishiguchi, Y., et al. (2020). Effect of proton pump inhibitors on colorectal cancer. Int. J. Mol. Sci. 21 (11), 3877. doi: 10.3390/ijms21113877
Sears, C. L., Pardoll, D. M. (2011). Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J. Infect. Dis. 203 (3), 306–311. doi: 10.1093/jinfdis/jiq061
Selvanesan, B. C., Chandra, D., Quispe-Tintaya, W., Jahangir, A., Patel, A., Meena, K., et al. (2022). Listeria delivers tetanus toxoid protein to pancreatic tumors and induces cancer cell death in mice. Sci. Transl. Med. 14 (637), eabc1600. doi: 10.1126/scitranslmed.abc1600
Shi, Z., Li, H., Song, W., Zhou, Z., Li, Z., Zhang, M. (2023). Emerging roles of the gut microbiota in cancer immunotherapy. Front. Immunol. 14, 1139821. doi: 10.3389/fimmu.2023.1139821
Stein, A., Simnica, D., Schultheiß, C., Scholz, R., Tintelnot, J., Gökkurt, E., et al. (2021). PD-L1 targeting and subclonal immune escape mediated by PD-L1 mutations in metastatic colorectal cancer. J. Immunother. Cancer 9 (7), e002844. doi: 10.1136/jitc-2021-002844
Sui, X., Chen, Y., Liu, B., Li, L., Huang, X., Wang, M., et al. (2020). The relationship between KRAS gene mutation and intestinal flora in tumor tissues of colorectal cancer patients. Ann. Transl. Med. 8 (17), 1085. doi: 10.21037/atm-20-5622
Sulit, A. K., Daigneault, M., Allen-Vercoe, E., Silander, O. K., Hock, B., McKenzie, J., et al. (2023). Bacterial lipopolysaccharide modulates immune response in the colorectal tumor microenvironment. NPJ Biofilms Microbiomes 9 (1), 59. doi: 10.1038/s41522-023-00429-w
Sun, J., Chen, F., Wu, G. (2023). Potential effects of gut microbiota on host cancers: focus on immunity, DNA damage, cellular pathways, and anticancer therapy. ISME J. 17 (10), 1535–1551. doi: 10.1038/s41396-023-01483-0
Ternes, D., Tsenkova, M., Pozdeev, V. I., Meyers, M., Koncina, E., Atatri, S., et al. (2022). The gut microbial metabolite formate exacerbates colorectal cancer progression. Nat. Metab. 4 (4), 458–475. doi: 10.1038/s42255-022-00558-0
Tian, Y., Wang, X., Cramer, Z., Rhoades, J., Estep, K. N., Ma, X., et al. (2023). APC and P53 mutations synergise to create a therapeutic vulnerability to NOTUM inhibition in advanced colorectal cancer. Gut. 72 (12), 2294–2306. doi: 10.1136/gutjnl-2022-329140
Tjalsma, H., Boleij, A., Marchesi, J. R., Dutilh, B. E. (2012). A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat. Rev. Microbiol. 10 (8), 575–582. doi: 10.1038/nrmicro2819
Tomii, A., Higa, M., Naito, K., Kurata, K., Kobayashi, J., Takei, C., et al. (2023). Activation of the TLR4-JNK but not the TLR4-ERK pathway induced by indole-3-acetic acid exerts anti-proliferative effects on Caco-2 cells. Biosci. Biotechnol. Biochem. 87 (8), 839–849. doi: 10.1093/bbb/zbad055
Um, S., Choi, T. J., Kim, H., Kim, B. Y., Kim, S.-H., Lee, S. K., et al. (2013). Ohmyungsamycins A and B: cytotoxic and antimicrobial cyclic peptides produced by Streptomyces sp. volcanic island. J. Org Chem. 78 (24), 12321–12329. doi: 10.1021/jo401974g
Vallera, D. A., Li, C., Jin, N., Panoskaltsis-Mortari, A., Hall, W. A. (2002). Targeting urokinase-type plasminogen activator receptor on human glioblastoma tumors with diphtheria toxin fusion protein DTAT. J. Natl. Cancer Inst 94 (8), 597–606. doi: 10.1093/jnci/94.8.597
Wang, X., Chen, Q., Zhu, Y., Wang, K., Chang, Y., Wu, X., et al. (2023). Destroying pathogen-tumor symbionts synergizing with catalytic therapy of colorectal cancer by biomimetic protein-supported single-atom nanozyme. Signal Transduction Targeted Ther. 8 (1), 277. doi: 10.1038/s41392-023-01491-8
Wang, L., Tu, Y.-X., Chen, L., Zhang, Y., Pan, X.-L., Yang, S.-Q., et al. (2023). Male-biased gut microbiome and metabolites aggravate colorectal cancer development. Adv. Sci. (Weinh) 10 (25), e2206238. doi: 10.1002/advs.202206238
Wang, M., Zhao, J., Zhang, L., Wei, F., Lian, Y., Wu, Y., et al. (2017). Role of tumor microenvironment in tumorigenesis. J. Cancer 8 (5), 761–773. doi: 10.7150/jca.17648
Wolf, P., Elsässer-Beile, U. (2009). Pseudomonas exotoxin A: from virulence factor to anti-cancer agent. Int. J. Med. Microbiol. 299 (3), 161–176. doi: 10.1016/j.ijmm.2008.08.003
Wolf, T., Lewis, A., Beaugerie, L., Svrcek, M., Kirchgesner, J. (2023). Risk of colorectal neoplasia according to histologic disease activity in patients with inflammatory bowel disease and colonic post-inflammatory polyps. Aliment Pharmacol. Ther. 57 (12), 1445–1452. doi: 10.1111/apt.17495
Xing, J., Liao, Y., Zhang, H., Zhang, W., Zhang, Z., Zhang, J., et al. (2022). Impacts of microRNAs induced by the gut microbiome on regulating the development of colorectal cancer. Front. Cell Infect. Microbiol. 12, 804689. doi: 10.3389/fcimb.2022.804689
Xu, C., Fan, L., Lin, Y., Shen, W., Qi, Y., Zhang, Y., et al. (2021). Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes 13 (1), 1980347. doi: 10.1080/19490976.2021.1980347
Xu, Y., Wang, F., Mi, K., Wang, X., Wang, D., Zhao, Q., et al. (2023). Biglycan regulated colorectal cancer progress by modulating enteric neuron-derived IL-10 and abundance of Bacteroides thetaiotaomicron. iScience 26 (9), 107515. doi: 10.1016/j.isci.2023.107515
Yamamoto, S., Shiraishi, S., Suzuki, S. (2015). Are cyclic lipopeptides produced by Bacillus amyloliquefaciens S13-3 responsible for the plant defence response in strawberry against Colletotrichum gloeosporioides? Lett. Appl. Microbiol. 60 (4), 379–386. doi: 10.1111/lam.12382
Yan, J., Duan, W., Gao, Q., Mao, T., Wang, M., Duan, J., et al. (2023). ENPP2 inhibitor improves proliferation in AOM/DSS-induced colorectal cancer mice via remodeling the gut barrier function and gut microbiota composition. Pharmacol. Res. 195, 106877. doi: 10.1016/j.phrs.2023.106877
Yan, J., Xiao, L., Feng, D., Chen, B., Yang, T., Tong, B., et al. (2024). Vitamin A deficiency suppresses CEACAM1 to impair colonic epithelial barrier function via downregulating microbial-derived short-chain fatty acids. Genes Dis. 11 (2), 1066–1081. doi: 10.1016/j.gendis.2023.03.032
Yang, Y., Jin, Y., Yin, L., Liu, P., Zhu, L., Gao, H. (2023). Sertaconazole nitrate targets IDO1 and regulates the MAPK signaling pathway to induce autophagy and apoptosis in CRC cells. Eur. J. Pharmacol. 942, 175515. doi: 10.1016/j.ejphar.2023.175515
Yin, H., Miao, Z., Wang, L., Su, B., Liu, C., Jin, Y., et al. (2022). Fusobacterium nucleatum promotes liver metastasis in colorectal cancer by regulating the hepatic immune niche and altering gut microbiota. Aging (Albany NY) 14 (4), 1941–1958. doi: 10.18632/aging.203914
Yu, L. C. (2018). Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis. J. BioMed. Sci. 25 (1), 79. doi: 10.1186/s12929-018-0483-8
Yu, Y.-N., Fang, J.-Y. (2015). Gut microbiota and colorectal cancer. Gastrointest Tumors 2 (1), 26–32. doi: 10.1159/000380892
Yu, T., Guo, F., Yu, Y., Sun, T., Ma, D., Han, J., et al. (2017). Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170 (3), 548–563.e516. doi: 10.1016/j.cell.2017.07.008
Zhang, B., Liu, J., Li, H., Huang, B., Zhang, B., Song, B., et al. (2023). Integrated multi-omics identified the novel intratumor microbiome-derived subtypes and signature to predict the outcome, tumor microenvironment heterogeneity, and immunotherapy response for pancreatic cancer patients. Front. Pharmacol. 14. doi: 10.3389/fphar.2023.1244752
Zhang, Y., Zhang, L., Zheng, S., Li, M., Xu, C., Jia, D., et al. (2022). Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-κB/ICAM1 axis. Gut Microbes 14 (1), 2038852. doi: 10.1080/19490976.2022.2038852
Zhao, M., Yang, M., Li, X. M., Jiang, P., Baranov, E., Li, S., et al. (2005). Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc. Natl. Acad. Sci. U.S.A. 102 (3), 755–760. doi: 10.1073/pnas.0408422102
Keywords: colorectal cancer, gut bacteria, tumor microbial microenvironment, immune escape, therapy
Citation: Han J, Zhang B, Zhang Y, Yin T, Cui Y, Liu J, Yang Y, Song H and Shang D (2023) Gut microbiome: decision-makers in the microenvironment of colorectal cancer. Front. Cell. Infect. Microbiol. 13:1299977. doi: 10.3389/fcimb.2023.1299977
Received: 23 September 2023; Accepted: 20 November 2023;
Published: 12 December 2023.
Edited by:
Wei Wang, Nanjing Medical University, ChinaCopyright © 2023 Han, Zhang, Zhang, Yin, Cui, Liu, Yang, Song and Shang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanfei Yang, MTMyOTYzOTYwMDdAMTM5LmNvbQ==; Huiyi Song, aHVpeWlzb25nbWFpbEAxNjMuY29t; Dong Shang, c2hhbmdkb25nQGRtdS5lZHUuY24=
†These authors have contributed equally to this work
 Jingrun Han
Jingrun Han Biao Zhang
Biao Zhang Yongnian Zhang
Yongnian Zhang Tianyi Yin
Tianyi Yin Yuying Cui1,2,4
Yuying Cui1,2,4 Jinming Liu
Jinming Liu Yanfei Yang
Yanfei Yang Huiyi Song
Huiyi Song Dong Shang
Dong Shang