
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 15 November 2023
Sec. Intestinal Microbiome
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1294082
Microbiota colonize exposed body tissues (e.g., gastrointestinal tract, skin, lungs, female genital tract, and urogenital tracts) and unexposed sites (e.g., breast). Persistent bacterial infection in the host lead to the development of multiple disease. They are implicated in the pathogenesis of various complex diseases, including diabetes, atherosclerosis, autoimmune diseases, Alzheimer’s disease, and malignant diseases. Amounting studies have demonstrated the role of bacterial infection in carcinogenesis. The study of microbiota in tumorigenesis is primarily focused on lung cancer, colorectal cancer (CRC), breast cancer, gastric cancer, and gynecologic tumors, and so on. Infection of Helicobacter pylori in gastric cancer carcinogenesis is recognized as class I carcinogen by the World Health Organization (WHO) decades ago. The role of Fusobacterium nucleatum in the development of colorectal cancer is extensively investigated. Variable bacteria have been cultured from the tumor tissues. The identification of microbiota in multiple tumor tissues reveal that bacterial infection and microbiota are associated with tumor development. The microbiota affects multiple aspects of carcinogenesis and tumor development, including favoring epithelial cells proliferation, establishing inflammatory microenvironment, promoting metastasis, and causing resistance to therapy. On the other hand, microbiota can shape a tumor surveillance environment by enhancing cell activity, and sensitize the tumor cells to immune therapy. In the present review, the roles of microbiota in multiple malignancies are summarized, and unraveling the mechanisms of host-microbiota interactions can contribute to a better understanding of the interaction between microbiota and host cells, also the development of potential anti-tumor therapeutic strategies.
The link between bacterial infection and cancer development was discovered almost a century ago (Chen et al., 2023). It is estimated that 20% of malignancies are related to bacterial infection (Wong-Rolle et al., 2021). However, the role of microbiota in cancer development is not fully elucidated; hence, this topic has attracted considerable research attention worldwide. Identification of microbiota in multiple tumor types were emerging by through high through-put sequencing including 16S rRNA sequencing and Metagenomic sequencing (Fu et al., 2022). The application of bacterial infection in carcinogenesis, tumor metastasis, response to therapy were investigated in gastrointestinal tumors, breast cancer and gynecological tumors (Stein et al., 2000) (Fu et al., 2022) (Wang et al., 2022). Bacteria effect the host directly by malignant transformation of the host cells through secretion of virulence factors or causing inflammatory factors (Odenbreit et al., 2000). Bacteria can also modify the tumor microenvironment by shaping the tumor microenvironment to tumor-promotive or tumor-suppressive directions (Poutahidis and Erdman, 2016) (Dong et al., 2021). The roles of bacterial infection and microbiota in tumor biological functions and various types of tumors will be discussed in different topics in the current review.
The identification of microorganisms in tumor tissues has been conducted for almost a century (Nejman et al., 2020). Researches have investigated the relationship between microbiota and cancer in multiple solid tumors (Zhao et al., 2021) (Garrett, 2015). By examining the microbiota composition in >1,500 tumor samples (Nejman et al., 2020), Nejman et al. found that bacteria can reside both in macrophages and epithelial cells in tumor tissues (Nejman et al., 2020). Living bacteria can be cultured from tumor samples (Livyatan et al., 2020). The bacterial composition in various types of cancer (e.g., melanoma, ovarian cancer, glioblastoma, pancreatic cancer, breast cancer, and lung cancer) are examined through numerous methods, including 16S-rRNA sequencing, and lipopolysaccharide (LPS) and lipoteichoic acid immunohistochemistry (Nejman et al., 2020). The development of the third-generation sequencing technologies led to the study of the intra-tumoral microbiome (Park et al., 2022). Bacterial 16s ribosomal RNA (16S-rRNA) gene DNA sequencing is a powerful tool for the identification of the bacterial composition in cancer tissues. Costantini et al. identified the bacterial composition in breast cancer through analysis of multi hypervariable 16S-rRNA gene regions (Costantini et al., 2018). The microbiota composition of breast cancer core needle biopsies was analyzed through 16S-rRNA gene sequencing (Costantini et al., 2018). Seven hypervariable regions of the 16S-rRNA gene were simultaneously examined. The results showed that Proteobacteria were the most abundant bacteria in the breast cancer tissues (Costantini et al., 2018). Diaphorobacter, Micrococcus, Paracoccus, Phascolarctobacterium, and Ralstonia are identified to be the most abundant genera in non-small cell lung cancer and the adjacent healthy tissue (Dumont-Leblond et al., 2021). Colorectal cancer (CRC) is colonized by more Escherichia coli, E. faecalis, F. nucleatum, and Streptococcus gallolyticus (Chattopadhyay et al., 2021). The identification of bacteria in the tumor tissues gave the direct evidence that microbiota might participate in the development of carcinogenesis (Figure 1). But regarding to the role of bacterial infection in tumor tissues as bystander or as effector need further investigation.
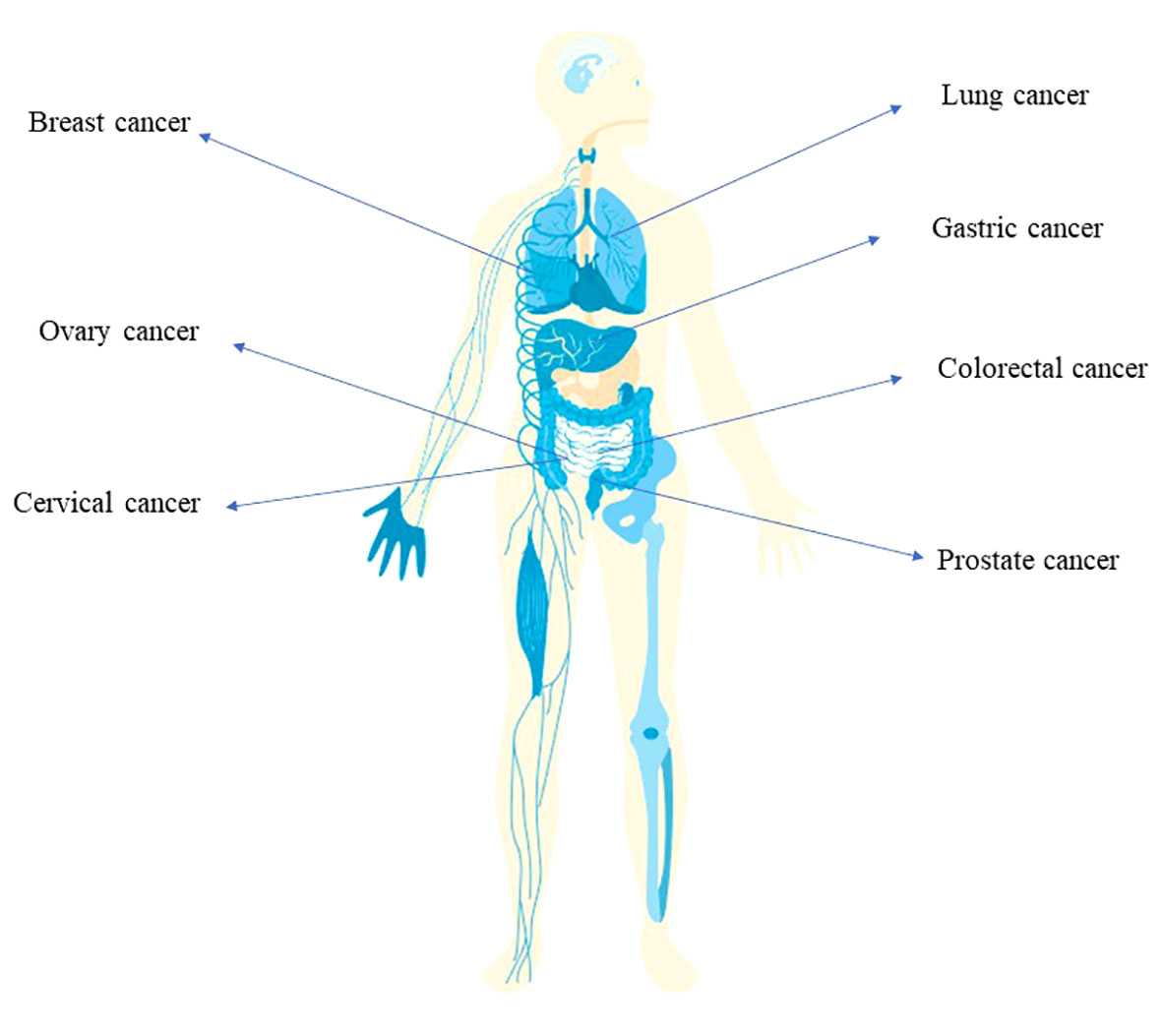
Figure 1 The implication of bacterial infection in multiple cancer types Infection of Helicobacter pylori is associated with the development of gastric cancer. The tumorigenesis of colorectal cancer is related with the infection of E. coli, Salmonella and Fusobacterium nucleatum. Also, breast cancer, ovary cancer, lung cancer, prostate cancer involves bacterial infection.
Kalaora et al. reported that, apart from antigen presenting cells that can present peptides from bacteria (Pfeifer et al., 1993; Kovacsovics-Bankowski and Rock, 1995; Bettencourt et al., 2020), tumor cells can digest bacteria inside the tumor and present them on tumor cells (Kalaora et al., 2021). Analysis of 17 melanoma metastasis samples extracted from nine patients detected 248 unique human leukocyte antigen class I (HLA-I) and 35 HLA-II peptides in 41 species of bacteria (Kalaora et al., 2021). The presented bacterial peptides can initiate the activation of T-cell response (Kalaora et al., 2021). Moreover, gentamicin protection assay and immunofluorescence staining demonstrated that Staphylococcus capitis and Staphylococcus succinus from tumors can invade melanoma cells (Kalaora et al., 2021). After co-culture of bacteria with a melanoma-derived melanoma cell line and analysis by HLA peptidomics, HLA-I and HLA-II bacterial peptides were found on cells (Kalaora et al., 2021). This study supported the antigen-presenting function of cancer cells through interaction with microbiota.
The role of bacterial infection in carcinogenesis is complex, and warrant extensive investigation on this topic. All aspects of cancer cells biological behavior can be regulated by microbiota. Bacterial infection can drive the carcinogenesis by multiple mechanisms. Malignant transformation can be induced by bacterial virulence factors. Through producing virulence factor, e.g., Helicobacter pylori secret virulence factor which participate in the tumorigenesis of gastric cancer (Kikuchi et al., 2012), Fusobacterium nucleatum can produce virulence factors (such as FadA and Fap2) which is implicated in the carcinogenesis of colon cancer (Jawad Abed et al., 2016).
Bacterial infection, especially persistent bacterial infection, cause chronic inflammation which can promote the development of malignancies (Reuter et al., 2010). Bacterial infection can cause the release of reactive oxygen species (ROS) which further activate multiple signaling pathways involving in tumor development, e.g., nuclear factor κB (NF-κB) and signal transducer and activator of transcription 3 (STAT3) (Reuter et al., 2010).
Persistent bacterial infection promotes the malignant transformation of epithelial cells (Normark et al., 2003). Free radicals released by the immune cells, including reactive oxygen species (ROS) and nitrogen oxide species, cause damage to epithelial cells and DNA (Kawanishi et al., 2017). Cytokines and chemokines released by the bacteria facilitate tumor cell growth (Fiorentini et al., 2020) (Figure 2). Bacteria remodel the tumor microenvironment (TME) to facilitate its colonization, the presence of bacteria in the TME modulates immune balance to benefit persistent bacterial infections. Bacterial LPS binds to the toll-like receptor 4 (TLR4) receptor of monocytes, thereby shifting their differentiation to the M2 phenotype (Li et al., 2019). Proinflammatory cytokines, including tumor necrosis factor-alpha (TNF-α), IL23, and IL8, can activate immune cells (Hartog et al., 2016). In addition, the accumulation of metabolites and ROS in the tumor microenvironment (TME) can mediate DNA damage and inhibit the function of CD8+ T cells (Srinivas et al., 2019).
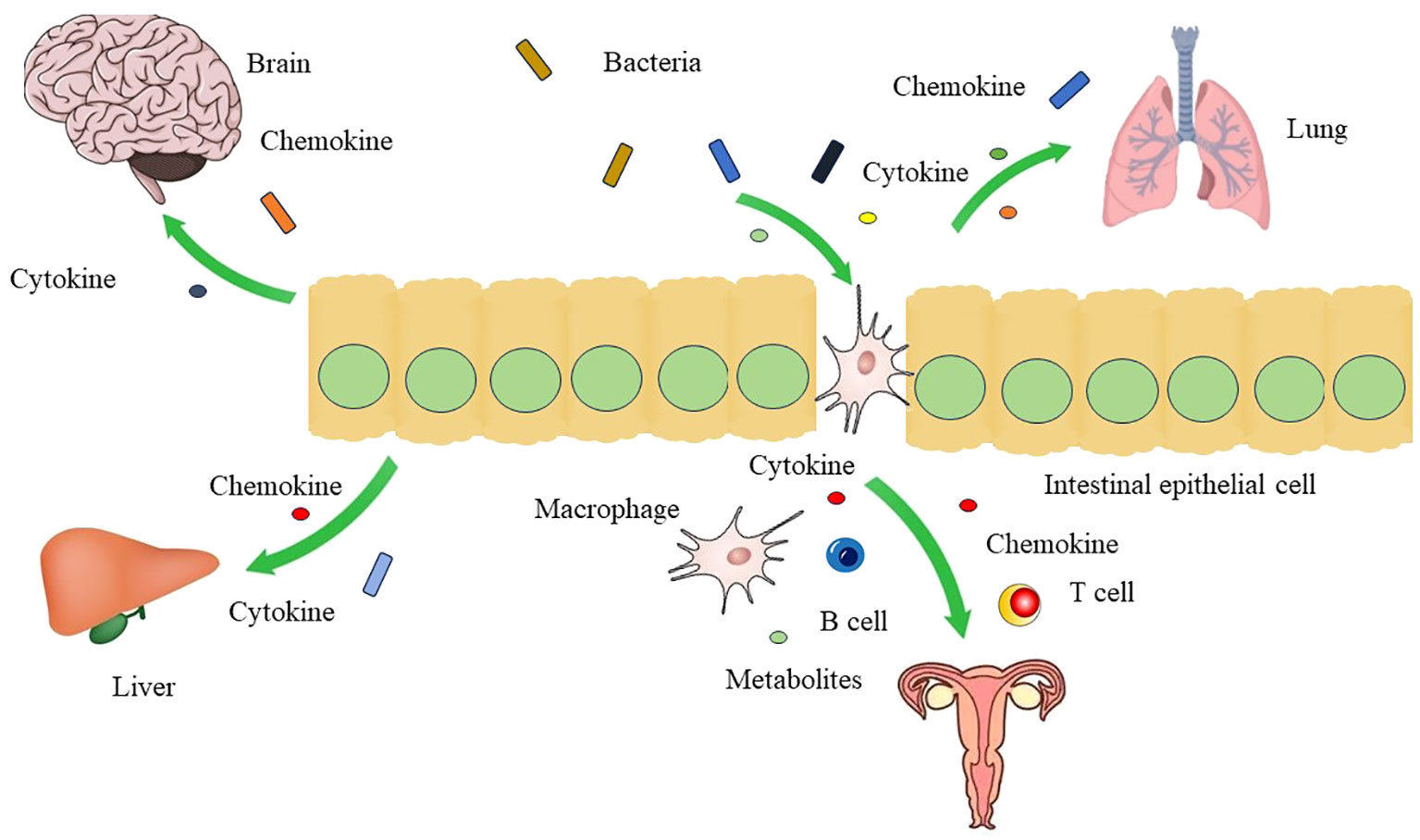
Figure 2 The crosstalk between gut microbiota and other organs Gut microbiota can affect the function of brain, liver, lung and genital tract through gut-brain axis, gut-liver axis, gut-lung axis, and gut-vagina axis by mediation of cytokines, chemokines, and metabolites.
Diet can affect the gut microbiota composition (Nogal et al., 2021). High fiber and polyphenols, low saturated fats diets will affect the gut microbiota composition (Nogal et al., 2021). High fiber diet increases the microbiota diversity (Nogal et al., 2021). Soluble inulin-type fructans (ITFs), one source of fiber, increase the level of Bifidobacterium (Nogal et al., 2021). Mediterranean diet can argument the abundance of Bifidobacterium. A ketogenic diet decreases the Bifidobacterium levels (Nogal et al., 2021).
Microbiota derived SCFAs enhance regulatory T cells and elicit immune tolerance (Nogal et al., 2021). High fat diet can change the gut microbiota composition by increasing the abundance of Alistipes, Bilophila and Bacteroides, and promote the development of gastrointestinal cancer (Tong et al., 2021).
Infection with viral pathogens can modify the microenvironment of tumor cells. The TME modulated by bacteria enhances cancer development and promotes bacterial infection (Iida N et al., 2013; Poutahidis and Erdman, 2016). Microbiota in the gastrointestinal tract can influence tumor development at remote sites (Zhao et al., 2021), e.g., the composition of gut microbiota modulates lung cancer pathogenesis (Wang et al., 2021). Bacteria from the intestinal lumen translocate to other sites, where they promote tumorigenesis (Wells and Maddaus MA, 1988). The gut microbiota influences the immune environment, for example, Akkermansia muciniphila modulates CD8+ T cell response and inhibits colitis-associated tumorigenesis of colon cancer (Wang et al., 2020). Aleksandar et al. reported that F. nucleatum modulated the tumor immune microenvironment by recruiting tumor-infiltrating myeloid cells, and promoted tumorigenesis of CRC (Kostic et al., 2013). Bullman et al. reported that F. nucleatum was transported with colon cancer cells during metastasis, and facilitated colonization of the metastasis sites (Bullman et al., 2017). Furthermore, whole-genome sequencing of Fusobacterium revealed >99.9% average nucleotide identity between the primary and metastatic tumors (Bullman et al., 2017). Fu et al. reported that tumor-resident intracellular microbiota promoted the metastasis of breast cancer cells (Fu et al., 2022); live bacteria, including Staphylococcus, Lactobacillus, Enterococcus, and Streptococcus, can be cultured from breast cancer tissues (Fu et al., 2022). The investigators traced the motility of the bacteria in vivo through fluorescent labeling (Fu et al., 2022). They found that bacteria were transported along with tumor cells in the circulation; intracellular bacteria facilitated the establishment of metastatic colonization sites by tumor cells (Fu et al., 2022).
Intratumor bacteria not only affect the metastasis of cancer cells, but also affiliate resistance to therapy of cancer cells. The microbiota in tumors modify the immune response in the TME, and regulate the response to anti-tumor therapy. Bacterial infection can shift the immune response (Kim and Covington A, 2017) against tumor cells to a tumor-permissive state. The composition of the gut microbiota can affect the response of cancer cells to immune checkpoint blockade immunotherapy (Gopalakrishnan et al., 2018). Gopalakrishnan et al. reported that, in patients with melanoma who received anti-PD-1 immunotherapy, gut microbiota varied between the responders and non-responders (Gopalakrishnan et al., 2018). Higher alpha diversity in the composition of fecal microbiota was observed in responders versus non-responders (Gopalakrishnan et al., 2018). The gut microbiome can affect the efficacy of PD-1-based immunotherapy in multiple tumor types (Long et al., 2019). Disturbance of the gut microbiome composition can increase the resistance to immune checkpoint inhibitors that target the PD-1/PD-L1 axis (Routy et al., 2018). Transplantation of fecal microbiota from responders to germ-free mice increases the therapeutic efficacy of PD-1 blockade (Routy et al., 2018). In contrast, transplantation of fecal microbiota from non-responders to germ-free mice did not ameliorate the response to therapy (Routy et al., 2018). A correlation between the abundance of Akkermansia muciniphila and response to immune checkpoint inhibitor therapy exists, as shown by a metagenomics analysis of stool samples obtained from patients (Routy et al., 2018). In colon cancer, Fusobacterium mediates chemoresistance to capecitabine (and 5-FU) by TLR4/MyD88-driven activation of autophagy (Yu et al., 2017). F. nucleatum infection induces light chain 3-II (LC3-II) expression, increases the RNA and protein expression of autophagy-related proteins, such as, unc-51 like autophagy activating kinase 1 (ULK1), phosphorylated-ULK1, and autophagy related 7 (ATG7) (Yu et al., 2017). Antibiotic treatment can reduce the tumor load of the colorectal cancer and may benefit the treatment of colorectal cancer positive with Fusobacterium (Bullman et al., 2017).
Gastric cancer is the fifth most common type of cancer worldwide, and is associated with a high incidence rate in Asian countries (Smyth et al., 2020). Gastric cancer can be divided to 2 histological types: intestinal-type and diffuse-type carcinomas (Yasui et al., 2011). Pathologically, approximately 90% of gastric cancers are adenocarcinomas. The development of gastric cancer involves a series of pathological steps (i.e., Correa’s steps), including atrophy, intestinal metaplasia, and dysplasia to adenocarcinoma (Sheh A, 2013). Compared with other body sites, the colonization of the stomach is challenging (Sheh A, 2013). The low pH 1–2 in the stomach is apparently formidable and hospitable for bacterial colonization (Sheh A, 2013). This contributes to significant differences in microbiota composition between the stomach and other sites of the gastrointestinal tract (Stearns et al., 2011). The cascade of events underlying the process from gastritis to intestinal metaplasia and gastric adenocarcinoma may require decades. Notably, the colonization by microbiota changes during this process (Alarcón and Llorca L, 2017).
The relationship of bacterial infection with tumorigenesis has been recognized since the discovery of H. pylori decades ago (Wang et al., 2014). Helicobacter pylori (H. pylori) infection in Mongolian gerbil is an established model for identifying the role of bacterial infection in the carcinogenesis and development of gastric cancer (Wang et al., 2014). H. pylori infection promotes the development of gastric cancer through multiple steps. But only 1–3% of the population infected by H. pylori will develop gastric cancer (Wang et al., 2014). Cag pathogenicity island (Cag PAI) of the bacteria is a key determinant in the malignant transformation of gastric epithelial cells by H. pylori (Odenbreit et al., 2000; Kikuchi et al., 2012). Virulence factors of the bacteria Cag PAI can be injected into the cytosol of gastric epithelial cells through the syringe-like structure type IV secretion system (Odenbreit et al., 2000).Subsequently, the tyrosine residue on four distinct glutamate-proline−isoleucine−tyrosine−alanine (EPIYA) motifs at the C-terminal region of CagA is phosphorylated (Stein et al., 2000). CagA promotes the acquisition of a ‘hummingbird’ phenotype by epithelial cells and the epithelial–mesenchymal transition of epithelial cells (Baj et al., 2020), thereby promoting the migration of cells (Kikuchi et al., 2012; Zhang et al., 2022). And increases the stem cell traits of gastric epithelial cells. Moreover, it increases the expression of gastric cancer stem cell marker CD44 (Bessède et al., 2014). CagA of H. pylori activates the Wnt signaling of the gastric epithelial cells, and drives carcinogenesis (Guo et al., 2022) as shown in Figure 3. Apart from the effect of the bacterial virulence factor, persistent inflammation induced by chronic H. pylori infection can accelerate the development of gastric cancer (Normark et al., 2003). Eliminating H. pylori infection by antibiotics proved to be beneficial to the patients (Wong et al., 2004). After 7.5 years’ follow-up, H. pylori eradiation in precancerous lesions group including gastric atrophy, intestinal metaplasia, and dysplasia group, prevent the gastric cancer carcinogenesis (Eun et al., 2014).
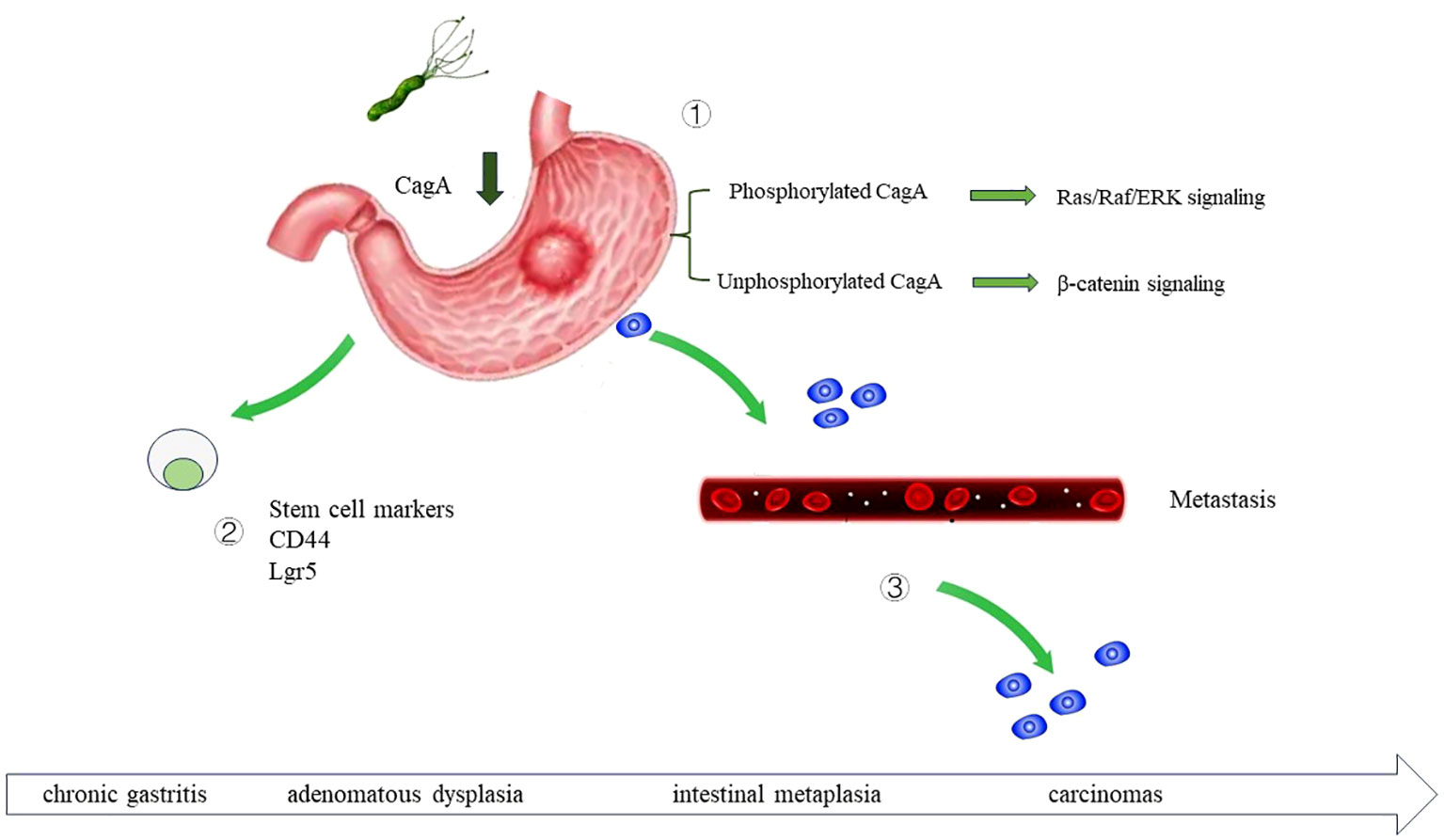
Figure 3 The role of H. pylori infection in gastric cancer carcinogenesis H. pylori infection can drive the development of gastric cancer through multiple mechanisms. By secreting virulence factor CagA, upregulating the stem cell traits of gastric epithelial cells, and promote the metastasis of gastric cancer cell. ① Once injected into the host cells, CagA is phosphorylated at glutamate-proline-isoleucine-tyrosine alanine (EPIYA) motifs. Phosphorylated CagA activate Ras/Raf/ERK signaling, and further activate the transcription of cyclin D and promote cell proliferation. Non-phosphorylated CagA activate β-catenin signaling and further upregulate cyclin D1 and c-Myc. ② H. pylori regulate the stem cell traits of gastric epithelial cell, increase CD44, Sox2, Oct4 and Nanog expression. ③ H. pylori can accelerate the metastasis of gastric cancer cell.
Except for H. pylori, other bacteria also reside in the stomach. By pyrosequencing methods, Eun et al. profiled the bacteria composition in the patients with chronic gastritis, intestinal metaplasia, and gastric cancer (Eun et al., 2014). Significantly different microbiota composition was identified in the gastric mucosa from Helicobacter colonized gastric cancer patients compared to gastritis and intestinal metaplasia patients (Eun et al., 2014). Through sequencing analysis, Dai et al. detected significant differences between gastric cancer tissues and matched non-tumor tissues (Dai et al., 2021).
The composition of the microbiota in gastric cancer were reported by different studies (Sheh A, 2013) (Bik et al., 2006) (Stewart and Wu F, 2020). It has been demonstrated that Firmicutes, Proteobacteria, and Bacteroidetes are the most abundant phyla in the microbiota in the stomach (Sheh A, 2013). Consistent results were reported by Elisabeth et al. that Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Fusobacteria phyla were the dominant bacteria in the stomach (Bik et al., 2006) (Stewart and Wu F, 2020). Ferreira et al. determined the microbiota composition of 54 patients with gastric carcinoma and 81 patients with chronic gastritis, through 16S rRNA gene next-generation sequencing (Ferreira et al., 2018). And it was found that a reduced microbial diversity was present in the gastric carcinoma microbiota (Ferreira et al., 2018). Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria and Fusobacteria were the most dominant bacteria in the stomach. And this is consistent with other studies (Bik et al., 2006) (Stewart and Wu F, 2020). Elucidating the role of microbiota in the gastric cancer will introduce a new dimension of the microbiome in the common malignancies and benefit the development of therapeutic strategies.
CRC is the third leading cause of cancer-related death worldwide (Siegel et al., 2022 2022). CRC is related to mutation of the adenomatous polyposis coli (APC) tumor suppressor gene, which activates the Wnt/β-catenin signaling pathway (Bian et al., 2020; JvP et al., 2021; Zhao et al., 2022). The translocation of β-catenin into the nucleus can activate the signaling pathways related to the development of cancer (Bian et al., 2020). The involvement of bacterial infection in colon cancer has been reported for decades. It has been shown that an imbalance in colon microbiota was tumorigenic. In addition, the occurrence of colon cancer has been linked to dietary habits (i.e., consumption of red meat, animal fat, and alcohol) (Campos et al., 2005). Notably, smoking has been associated with a high incidence of CRC (Bai et al., 2022).
Diet composition can affect the production of short-chain fatty acids (SCFAs), including acetate, propionate, and butyrate, which regulate the function of intestinal epithelial cells (Louis and Hold GL, 2014). Among them, butyrate and propionate inhibit histone deacetylases (HDACs) and regulate the function of CD8+ T cells (Bai et al., 2022) (Louis and Hold GL, 2014). The level of propionate in colorectal cancer decreases. Propionate regulate the mTORC2/PDK1/AKT signaling pathway (Louis and Hold GL, 2014). The metabolites from the microbiota modify the biological behaviors of the cancer cell. SCFAs propionate and butyrate, by-products of the intestinal bacterial fermentation, can induce autophagy of colon cancer cell and retard the cellular apoptosis due to mitochondrial dysfunction (Tang et al., 2011). Increased LC3-II and reduced p62/SQSTM1 were observed in the propionate treated colon cancer cells (Tang et al., 2011). It also indicated the therapeutic potential of SCFAs might be enhanced by autophagy inhibitor (Tang et al., 2011).
It is estimated that a load of 1010 to 1012 CFU/g bacteria is contained in the gastrointestinal tract (Korecka A, 2012; Bretin and Gewirtz AT, 2018). After sequencing nine metastatic CRC tissues and seven non-metastatic CRC tissues (Chen et al., 2020), Chen et al. found that Fusobacteriaceae is more frequently present in patients with metastatic CRC versus those with non-metastatic CRC (Chen et al., 2020). The association of Escherichia coli (E. coli) with colon cancer was reported in 1998 (Swidsinski et al., 1998). Based on quantitative polymerase chain reaction and 16S-rRNA sequencing analyses, Swidsinski et al. identified intracellular E. coli in mucous obtained from patients with colorectal carcinoma (Swidsinski et al., 1998). In addition, Long et al. reported that Peptostreptococcus anaerobius promoted carcinogenesis of CRC (Long et al., 2019).
The composition of colorectal cancer microbiota was investigated by different groups. Fusobacterium, Peptostreptococcus, Porphyromonas, Bacteroides, Parvimonas, Prevotella, Gemella, Streptococcus, Clostridium, Escherichia, Bilophila, Campylobacter, Phascolarctobacterium, Selenomonas, Ruminococcus, Shigella, Akkermansia, Desulfovibrio, Eubacterium, Leptotrichia are among the most common bacteria in colorectal cancer (Ternes et al., 2020).Zhou et al. identified “Bifidobacteria, Fusobacterium nucleatum, Geotrichum candidum, Porphyromonas asaccharolytica, Escherichia coli, Rhodococcus, Anaerostipes caccae, Enhydrobacter, Lachnoclostridiumsp.m3, Bacteroides clarus, Clostridium hathewayi, Ruminococcaceae, Bacteroides thetaiotaomicron, Culinariside, and enterotoxigenic Bacteroides fragilis (ETBF)” as biomarkers in gut microbiome as early diagnostic markers of colorectal cancer (Zhou et al., 2022). Fusobacterium nucleatum, Escherichia coli, and Bacteroides fragilis are among the most evaluated bacteria in the carcinogenesis of colorectal cancer (Zhou et al., 2022). Experimental evidence supporting the role of Fusobacterium nucleatum, Escherichia coli, and Bacteroides fragilis in colorectal cancers substantially increased (Tilg et al., 2018).
The relationship between F. nucleatum and CRC is well established (Bashir et al., 2015). F. nucleatum is an opportunistic, Gram-negative bacterium that has been associated with the metastasis of colon cancer cells. F. nucleatum reaches the gastrointestinal tract through the oral cavity or by hematogenous translocation (Ashare et al., 2009; Kostic et al., 2013). Bacterial infection modifies the phenotype of macrophage to tumor promoting phenotype (Chen et al., 2018). Monocytes differentiate toward the M1 and M2 phenotypes under different stimuli. M2 macrophages, termed alternatively activated macrophages, promote tumor development and cancer cell metastasis, and inhibit anti-tumor immunity (Mantovani et al., 2002). In vitro, F. nucleatum infection favors the polarization of macrophages toward the M2 phenotype. This process depends on the signaling of pattern-recognition receptor TLR4. It modulates the polarization of macrophages toward the CD206+ M2 phenotype in the TME, and promotes the development of colorectal tumors in a TLR4-dependent mechanism (Chen et al., 2018). Typically, the activation of LPS-TLR4 signaling lead to the polarization of macrophages toward the M1 phenotype. However, evidence revealed that bacterial infection induced the differentiation of macrophages to the M2 phenotype by IL6/STAT3/c−MYC signaling (Chen et al., 2018).
The migration and invasion of colorectal cancer can be regulated by gastrointestinal microbiota (JvP et al., 2021). Fusobacterium nucleatum drive colorectal cancer migration by inducing IL-8 and cytokine CXCL1 secretion (Casasanta et al., 2020). Han et al. reported that F. nucleatum promoted liver metastasis of CRC (Yin et al., 2022) as shown in Figure 4. Following infection of mice with F. nucleatum through the oral route, metastasis of CRC cells to the liver was increased, whereas the body weight and overall survival time of the mice were decreased (Yin et al., 2022). The bacterium modified the TME of the liver by recruiting Th17 cells and regulatory T cells (Yin et al., 2022). Furthermore, F. nucleatum binds to the inhibitory immune receptor targeting the T cell immunoglobulin and ITIM domain (TIGIT) of immune cells through Fap2 (Garrett, 2019). This leads to inhibition of the function of tumor-infiltrating lymphocytes and natural killer cells, thereby facilitating the immune evasion of tumor cells (Garrett, 2019).
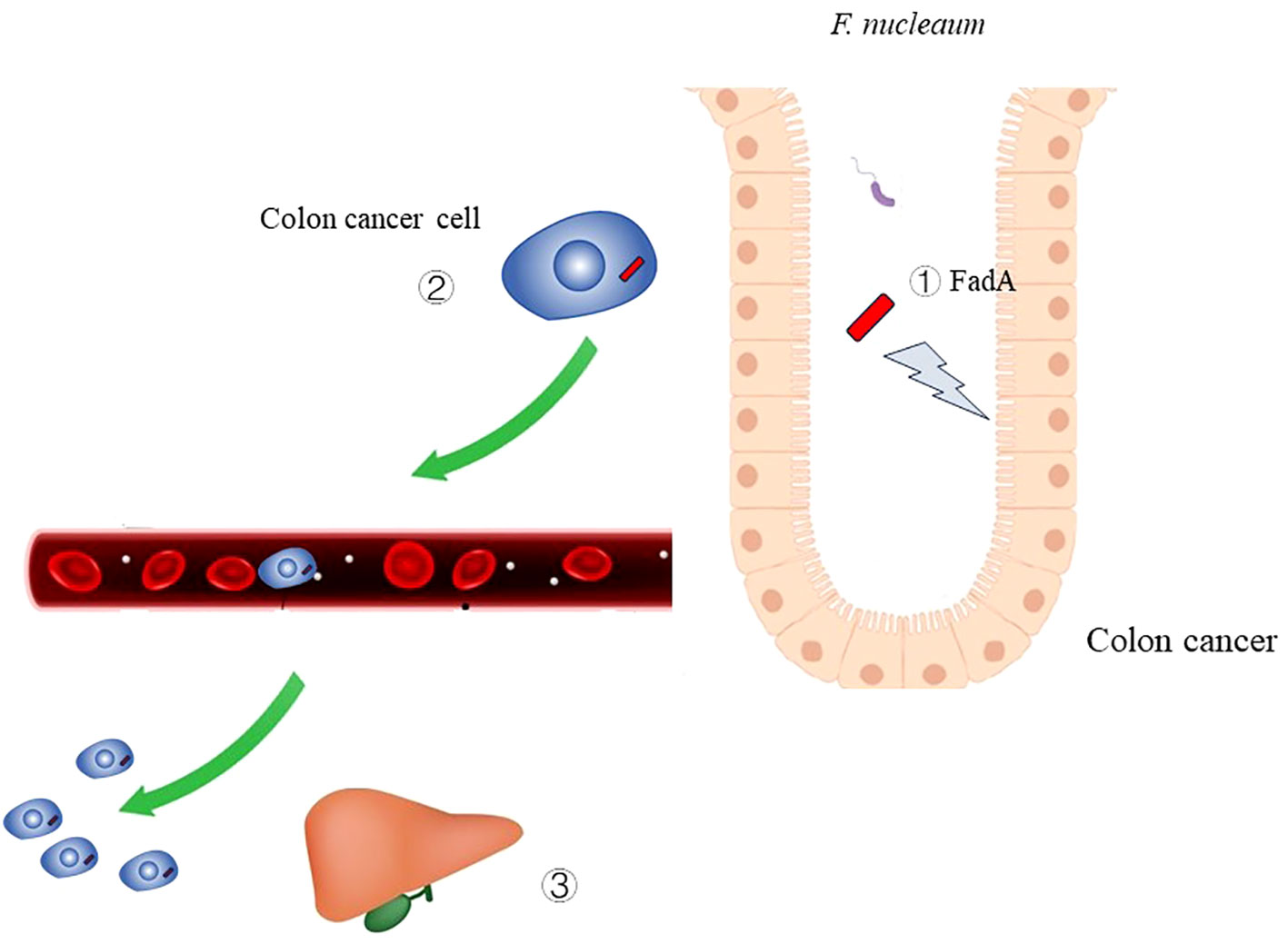
Figure 4 The role of Fusobacterium nucleatum infection in colon cancer ① Virulence factor FadA of F. nucleaum can cause damage to DNA.② F. nucleaum can metastasize along with colon cancer cell.
Activation of NF-κB signal mediated by Toll-like receptors and nucleotide-binding oligomerization domain-like receptors bind with Fusobacterium nucleatum, induce chronic inflammation (Garrett, 2015). Activation of IL-6, TNF and STAT3 signaling is also tumor promoting (Garrett, 2015).
Bacterial infection can lead to DNA damage and mutation caused by the metabolites of the bacteria. E. coli harboring the polyketide-nonribosomal peptide synthase operon (pks) island produces colibactin, thereby causing genomic instability of the host cell (Nougayrede et al., 2006). Colibactin secreted by Escherichia coli activates the senescence-associated secretory phenotype and promotes the development of colon cancer (Dalmasso et al., 2014). E. coli harboring the pks island causes DNA double-strand breaks. Pleguezuelos-Manzano et al. analyzed 5,876 human cancer genomes and reported that pks+ E. coli induced a distinct mutational signature in 20% of the healthy population, 40% of patients with inflammatory bowel disease, and 60% of patients with CRC (Pleguezuelos-Manzano et al., 2020).
Enterotoxigenic Bacteroides fragilis (ETBF) has been found in biofilms extracted from the colon of patients with familial adenomatous polyposis (Dejea et al., 2018). This bacterium secretes a metalloprotease toxin (Bacteroides fragilis toxin). The occurrence of familial adenomatous polyposis is related to mutation in the APC tumor suppressor gene. ETBF in CRC can cleave and degrade E-cadherin through the secretion of metalloproteases (Dejea et al., 2018). E. coli and ETBF are dominant bacteria in the biofilm of patients with CRC. Wu et al. reported that ETBF promoted colon cancer tumorigenesis by activating T helper type 17 (Th17) T cell responses (Wu et al., 2009). Use of antibodies against IL17 and IL23 receptor alleviated the inflammatory infiltration and hyperproliferation of colonic mucosal cells (Wu et al., 2009).
Salmonella species (Gram-negative bacteria) can cause various diseases, ranging from self-limiting gastroenteritis to typhoid fever (Bretin and Gewirtz AT, 2018). Enteric Salmonella infection, e.g., Salmonella Enteritidis, has been associated with CRC (Mughini-Gras et al., 2018). Salmonella infection in the azoxymethane/dextran sodium sulfate mouse model or APC-deficient mice model significantly increased the incidence of CRC (Lu et al., 2014). Moreover, this relationship involves the Salmonella AvrA protein, which is expressed at higher levels in tumor-adjacent versus non-cancer colorectal mucosa (Lu et al., 2014; Lu et al., 2016; Lu et al., 2017). AvrA activates the STAT3 and β-catenin signaling pathways (Lu et al., 2014; Lu et al., 2016).
Female genital tract is occupied by microbiota. Gynecologic cancer refers to a group of cancers of the female reproductive organs (e.g., cervical, endometrial, and ovarian). The association between bacterial infection and gynecologic tumors is complex. Due to its anatomy, the female genital tract is exposed to the external environment. This provides an opportunity for bacteria to enter the reproductive organs. Lactobacillus predominantly colonizes low-grade squamous intra-epithelial lesions compared with invasive cervical cancers (Mitra et al., 2015). Greater diversity in vaginal microbiota correlates with more severe cervical intra-epithelial neoplasia disease (Mitra et al., 2015). This finding indicated that Lactobacillus exerts protective effects against cervical cancer tumorigenesis, possibly by promoting human papillomavirus clearance (Mitra et al., 2015). Persistent infection with Neisseria gonorrhoeae is associated with urethritis (Unemo et al., 2019). Neisseria gonorrhoeae has been associated with malignant transformation due to the regulation of cyclin B (CCNB) expression and the induction of cell cycle arrest in the G1 phase (Jones et al., 2007). The intratumoral abundance of F. nucleatum may function as a prognostic marker for cervical carcinoma (Huang et al., 2020).
The composition of gynecologic tumors was elucidated by multiple researches. High microbial diversity and overgrowth of anaerobic bacteria were found in the vaginal microbiome of cervical cancer (Laniewski et al., 2020). Atopobium and Porphyromonas are associated with endometrial cancer (Laniewski et al., 2020). Brucella, Mycoplasma and Chlamydia, Acinetobacter are related with ovary cancer (Laniewski et al., 2020). Dysbiosis of female reproductive tract (FRT) are emerging as one of the drivers of gynaecological malignancies (Laniewski et al., 2020).
Ovarian cancer is the most common type of gynecologic cancer, and is typically diagnosed at the late stage of the disease (Lengyel, 2010). Bacteria can also colonize the ovary through the pelvic cavity, particularly in patients suffering from chronic pelvic inflammatory disease (Infectious Vaginitis, 2023). Studies reported that microbiota play multiple functions in gynecologic tumors (Routy et al., 2018). For example, the abundance of Akkermansia correlated with the response of patients to immunotherapy with immune checkpoint inhibitors targeting programmed cell death 1 (PD-1) (Routy et al., 2018). Studies analyzed ovarian cancer samples through 16S-rRNA sequencing, using distal fallopian tube samples as control (Zhou et al., 2019). The results demonstrated that Proteobacteria and Firmicutes are the predominant phyla in ovarian cancer by Lefse (LDA Effect Size) analysis (Zhou et al., 2019). Genera within Firmicutes can produce butyrate which is tumor preventive in the early stages of tumor development (Zhou et al., 2019). Higher abundance of F. nucleatum correlates with lower overall survival and progression-free survival rates, as well as enhanced characteristics of cancer stem cells (Huang et al., 2020). Wang et al. reported that transplantation of fecal microbiota from patients with ovarian cancer to a mouse model of ovarian cancer accelerated tumor development. Interestingly, Akkermansia supplementation in mice reversed this effect (Wang et al., 2022).
The female genital tract is adjacent to the urinary tract (Walther-António et al., 2016). Walther-António et al. found that the uterine microbiome contributed to the development of endometrial cancer (Walther-António et al., 2016). Through microbiome sequencing (16S-rDNA V3–V5 region), researchers identified the bacterial composition in endometrial cancer (Walther-António et al., 2016). It is found that A. vaginae and Porphyromonas sp. correlate with presence of endometrial cancer (Walther-António et al., 2016). This study proposed the potential roles of microbiota in the progression of endometrial cancer (Walther-António et al., 2016). Hawkins et al. reported that Actinobacteria, Bacteroidetes, Firmicutes, OD1, and Proteobacteria phyla were present in both benign and malignant uterine tissue specimens (Mitra et al., 2015). Comparison of the microbial profiles (at the genus level) revealed that the microbial diversity is greater in endometrial cancer versus the benign uterus (Hawkins et al., 2022).
Bladder cancer is the tenth most common type of cancer worldwide (Bray et al., 2018), and the predominant tumor type of the urological tract. Several research groups have discovered that the urinary tract is colonized by unique urinary microbiota (Parra-Grande et al., 2022) (Martin et al., 2022). In urinary carcinomas, the microbiome promotes the carcinogenesis of epithelial cells through different mechanisms (Parra-Grande et al., 2022). Persistent bacterial infection can induce DNA mutation and genomic instability (Hanahan and Weinberg, 2011; Hanahan, 2022). Moreover, ROS and metabolites, such as short-chain fatty acids, can modify the urinary microenvironment and promote tumorigenesis (Hanahan and Weinberg, 2011; Hanahan, 2022). In addition, bacteria (e.g., E. coli) can persistently survive in bladder epithelial cells (Hannan et al., 2012).
The composition of the urinary carcinomas’ microbiota was verified. In bladder cancer patients’ samples, abundant levels of Proteobacteria, Firmicutes and Actinobacteria at the phylum level are more common in the urinary microbiota of bladder cancer patients than in control patient (Karam et al., 2022). Fusobacterium, Acinetobacter, Cupriavidus, Corynebacterium, Streptococcus and Staphylococcus are higher in bladder cancer urine samples (Karam et al., 2022). In tissue samples, Firmicutes, Bacteroidetes, Proteobacteria and Actinobacteria were detected on the phyla level both in bladder cancer patients and normal samples (Karam et al., 2022). With lower abundance of Firmicutes and Bacteroidetes, higher abundance of Proteobacteria and Actinobacteria presented in bladder cancer samples (Karam et al., 2022). Streptococcus have a prominent abundance in the urine microbiota of prostate cancer patients (Alanee et al., 2019) (Karam et al., 2022).
Apart from 16S-rRNA sequencing, shotgun metagenomic sequencing and microbiome metabolomics have been used to identify the microbiota composition in bladder cancer (Zhang et al., 2023). Parra-Grande et al. compared the microbial composition between matched tumor and non-tumor samples (Parra-Grande et al., 2022). They observed lower biodiversity in the tumor samples than in the mucosa of healthy controls (Parra-Grande et al., 2022). The most abundant phyla in the tumor and non-tumor samples were Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Parra-Grande et al., 2022). Higher abundance of Actinobacteria was observed in the non-tumor mucosa samples versus the tumor samples (Parra-Grande et al., 2022). Notably, higher abundance of Enterococcus correlated with lower tumor grade (Parra-Grande et al., 2022).
Lung cancer is the most common type of cancer worldwide (Avasarala and Rickman, 2022). Small cell lung cancer and non-small cell lung cancer are the most common categories of lung cancer. The latter accounts for up to 85% of lung cancer cases. The most common types of lung cancer include adenocarcinoma, squamous cell carcinoma, and large cell carcinoma (Jackson et al., 2001).
The role of the microbiome in lung cancer has been more and more recognized by researchers (Jin et al., 2019) (Zhao et al., 2021). Bacteria can colonize the lungs and induce chronic inflammation, while various cytokines and chemokines can facilitate tumor growth. Not only the lung microbiota is involved in the carcinogenesis of lung cancer (Jin et al., 2019), but also the microbiome in the intestinal lumen can modify the TME of lung cancer from remote sites (Budden et al., 2017). Lung cancer carcinogenesis is affected by the gut microbiota (Liu et al., 2021). The crosstalk between the lungs and intestines is involved in the development of lung cancer (Budden et al., 2017) (He Zhuang et al., 2019). It has been shown that the transportation of Th17 cells from the intestines to the lungs aggregated immune imbalance in the lungs (Liu et al., 2021). Zheng et al. reported that patients with early-stage lung cancer have a specific gut microbial profile (Zheng et al., 2020). Different microbial compositions in the gut correlate with the stages and subtypes of lung cancer, indicating that a gut microbiota signature may predict the development of lung cancer (Zheng et al., 2020).
The composition of the microbiota in lung cancer were not extensively reported as gastrointestinal cancer (Xu et al., 2020). Increase of oral taxa Streptococcus and Veillonella was detected in the lower airways of lung cancer patients (Pizzo et al., 2022). It is related with ERK and PI3K signaling pathway activation (Pizzo et al., 2022). Enrichment of oral bacteria Prevotella, Veillonella, Rothia, Streptococcus, and Porphyromonas in the lower airway were reported by multiple studies (Wu et al., 2017). Lower alpha diversity of bacterial community was reported in lung cancer patients than in non-malignant lung tissues (Pizzo et al., 2022). Herbaspirillum and Sphingomonadaceae are more common in lung cancer tissues than normal lung tissues (Jin et al., 2019). Enrichment of Firmicutes, Granulicatella, Abiotrophia, and Streptococcus with a decreased bacterial community diversity were reported in lung cancer patients (Lee et al., 2016). Enrichment of Streptococcus and deficiency in Staphylococcus were evidenced in lung cancer-associated microbiota (Liu et al., 2017).
The lung microbiome also affects the response of lung cancer cells to chemotherapy (Wang et al., 2021). Bacteria colonizing the TME can suppress immune response against tumor cells (Jin et al., 2019). Notably, IL17-producing γδ T cells are associated with the development of lung cancer. Moreover, the release of IL17 and IL23 can boost inflammation in lung tissues and favor the lung cancer cells proliferation (Jin et al., 2019). Antibiotic treatment can alleviate the tumor burden in the lung (Jin et al., 2019).
Microbiota promote tumor development, modulated the tumor environment to benefit cancer cells and effect the responses to chemotherapy (Figure 5). Comprehensive research is warranted to address numerous unanswered questions in the interaction of microbiota and host cells. Part of the bacteria play a driver function in the carcinogenesis, part of the bacteria plays the passenger function (Tjalsma et al., 2012). For example, Salmonella and Citrobacter can function as the driver of colon cancer carcinogenesis (Tjalsma et al., 2012), causing malignant transformation of the intestinal epithelial cell by metabolites (Avril and DePaolo, 2021). Therapeutic strategies targeting the microbiota is merging as a potential antitumor strategy. Investigation of the role of microbiota in cancer development may provide targets for anti-tumor therapy. Moreover, skewing of the microbiota balance may prevent tumor development. Finally, the microbiota composition in tumors may be used as an alternative biomarker for predicting prognosis and response to therapy.
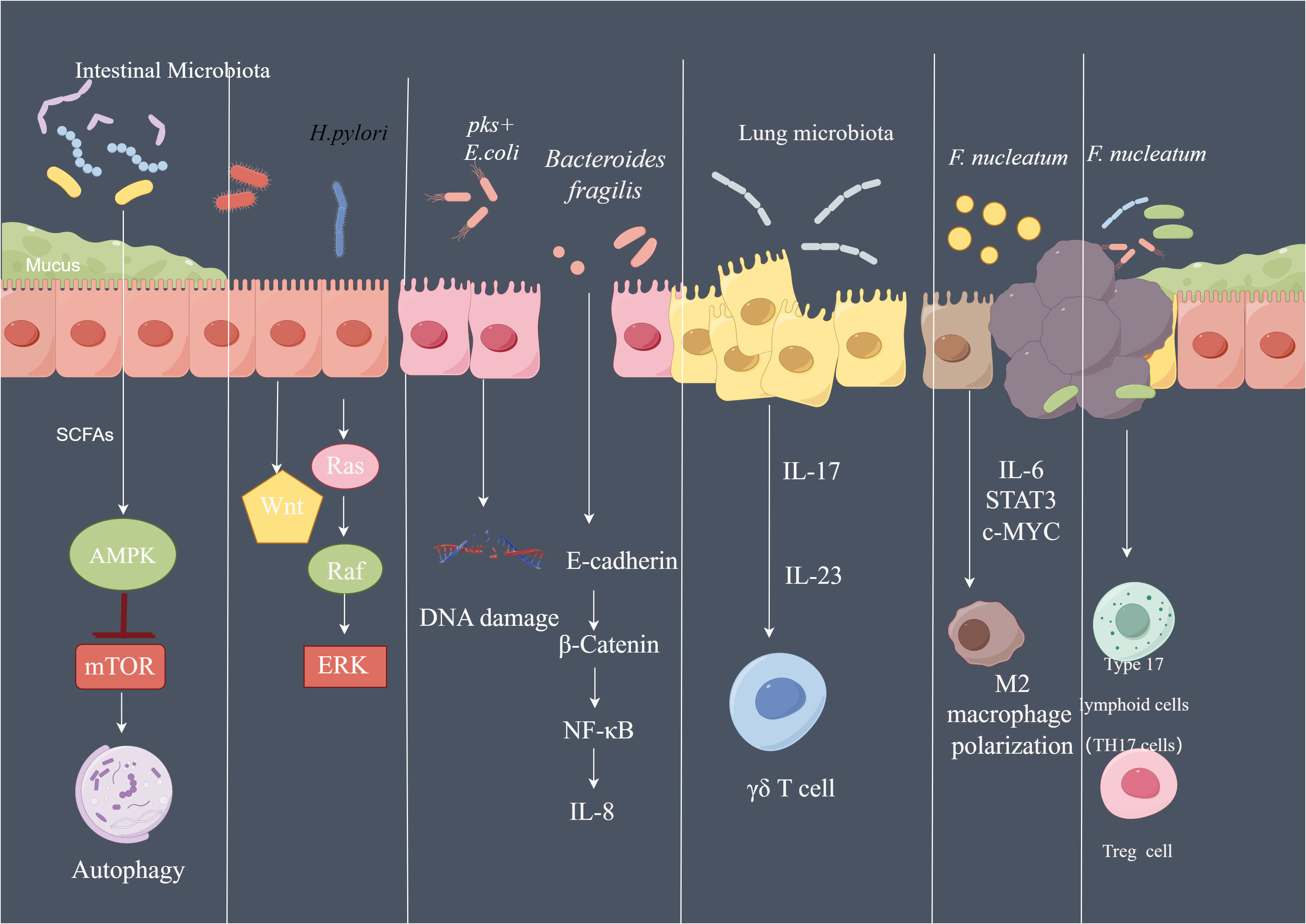
Figure 5 The pathways affected by microbiota in cancer SCFA of the microbiota can activate AMPK signaling pathway and inhibit mTOR signaling, further activate autophagy machinery. Phosphorylated CagA of H. pylori can activate Ras/Raf/ERK signaling and Wnt signaling in gastric epithelial cells. pks+ E. coli led to DNA damage of the intestinal epithelial cell. Bacteroides fragilis induce th production of IL-8 by activating E-cadherin/β-catenin/NF-κB signaling pathway. Lung microbiota promote the development of lung Cancer via γδ T Cells which can be activated by Myd88-dependent IL-1β and IL-23 induced by commensal bacteria. F.nucleatum regulate the polarization of macrophage to M2 phenotype by secreting IL-6, and activate IL-6/STAT3/c-MYC signaling. By Figdraw.
QL: Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
The support from Tongji Hospital is appreciated.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alanee, S., El-Zawahry, A., Dynda, D., Dabaja, A., McVary, K., Karr, M., et al. (2019). A prospective study to examine the association of the urinary and fecal microbiota with prostate cancer diagnosis after transrectal biopsy of the prostate using 16sRNA gene analysis. Prostate 79, 81–87. doi: 10.1002/pros.23713
Alarcón, T., Llorca L, G. P.-P. (2017). Impact of the microbiota and gastric disease development by helicobacter pylori. Curr. Top. Microbiol. Immunol. 400, 253–275. doi: 10.1007/978-3-319-50520-6_11
Ashare, A., Stanford, C., Hancock, P., Stark, D., Lilli, K., Birrer, E., et al. (2009). Chronic liver disease impairs bacterial clearance in a human model of induced bacteremia. Clin. Transl. Sci. 2, 199–205. doi: 10.1111/j.1752-8062.2009.00122.x
Avasarala, S. K., Rickman, O. B. (2022). Endobronchial therapies for diagnosis, staging, and treatment of lung cancer. Surg. Clin. North Am. 102, 393–412. doi: 10.1016/j.suc.2022.01.004
Avril, M., DePaolo, R. W. (2021). Driver-passenger” bacteria and their metabolites in the pathogenesis of colorectal cancer. Gut Microbes 13, 1941710. doi: 10.1080/19490976.2021.1941710
Bai, X., Wei, H., Liu, W., Coker, O. O., Gou, H., Liu, C., et al. (2022). Cigarette smoke promotes colorectal cancer through modulation of gut microbiota and related metabolites. Gut 71, 2439–2450. doi: 10.1136/gutjnl-2021-325021
Baj, J., Korona-Głowniak, I., Forma, A., Maani, A., Sitarz, E., Rahnama-Hezavah, M., et al. (2020). Mechanisms of the epithelial-mesenchymal transition and tumor microenvironment in helicobacter pylori-induced gastric cancer. Cells 9, 1055. doi: 10.3390/cells9041055
Bashir, A., Miskeen, A. Y., Bhat, A., Fazili, K. M., Ganai, B. A. (2015). Fusobacterium nucleatum: an emerging bug in colorectal tumorigenesis. Eur. J. Cancer Prev. 24, 373–385. doi: 10.1097/CEJ.0000000000000116
Bessède, E., Staedel, C., Acuña Amador, L. A., Nguyen, P. H., Chambonnier, L., Hatakeyama, M., et al. (2014). Helicobacter pylori generates cells with cancer stem cell properties via epithelial-mesenchymal transition-like changes. Oncogene 33, 4123–4131. doi: 10.1038/onc.2013.380
Bettencourt, P., Müller, J., Nicastri, A., Cantillon, D., Madhavan, M., Charles, P. D., et al. (2020). Identification of antigens presented by MHC for vaccines against tuberculosis. NPJ Vaccines 5, 2. doi: 10.1038/s41541-019-0148-y
Bian, J., Dannappel, M., Wan, C., Firestein, R. (2020). Transcriptional regulation of wnt/β-catenin pathway in colorectal cancer. Cells 9, 2125. doi: 10.3390/cells9092125
Bik, E. M., Eckburg, P. B., Gill, S. R., Nelson, K. E., Purdom, E. A., Francois, F., et al. (2006). Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. U.S.A. 103, 732–737. doi: 10.1073/pnas.0506655103
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Bretin, A., Gewirtz AT, B. C. (2018). Microbiota and metabolism: what’s new in 2018? Am. J. Physiol. Endocrinol. Metab. 315, E1–E6. doi: 10.1152/ajpendo.00014.2018
Budden, K. F., Gellatly, S. L., Wood, D. L., Cooper, M. A., Morrison, M., Hugenholtz, P., et al. (2017). Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 15, 55–63. doi: 10.1038/nrmicro.2016.142
Bullman, S., Pedamallu, C. S., Sicinska, E., Clancy, T. E., Zhang, X., Cai, D., et al. (2017). Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448. doi: 10.1126/science.aal5240
Campos, F. G., Logullo Waitzberg, A. G., Kiss, D. R., Waitzberg, D. L., Habr-Gama, A., Gama-Rodrigues, J. (2005). Diet and colorectal cancer: current evidence for etiology and prevention. Nutr. Hosp. 20 (1), 18–25.
Casasanta, M. A., Yoo, C. C., Udayasuryan, B., Sanders, B. E., Umaña, A., Zhang, Y., et al. (2020). Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci. Signal. 13, eaba9157. doi: 10.1126/scisignal.aba9157
Chattopadhyay, I., Dhar, R., Pethusamy, K., Seethy, A., Srivastava, T., Sah, R., et al. (2021). Exploring the role of gut microbiome in colon cancer. Appl. Biochem. Biotechnol. 193, 1780–1799. doi: 10.1007/s12010-021-03498-9
Chen, J., Gao, L., Wu, X., Fan, Y., Liu, M., Peng, L., et al. (2023). BCG-induced trained immunity: history, mechanisms and potential applications. J. Transl. Med. 21, 106. doi: 10.1186/s12967-023-03944-8
Chen, T., Li, Q., Wu, J., Wu, Y., Peng, W., Li, H., et al. (2018). Fusobacterium nucleatum promotes M2 polarization of macrophages in the microenvironment of colorectal tumours via a TLR4-dependent mechanism. Cancer Immunol. Immunother. 67, 1635–1646. doi: 10.1007/s00262-018-2233-x
Chen, Y., Chen, Y., Zhang, J., Cao, P., Su, W., Deng, Y., et al. (2020). Fusobacterium nucleatum Promotes Metastasis in Colorectal Cancer by Activating Autophagy Signaling via the Upregulation of CARD3 Expression. Theranostics 10, 323–339. doi: 10.7150/thno.38870
Costantini, L., Magno, S., Albanese, D., Donati, C., Molinari, R., Filippone, A., et al. (2018). Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions. Sci. Rep. 8, 16893. doi: 10.1038/s41598-018-35329-z
Dai, D., Yang, Y., Yu, J., Dang, T., Qin, W., Teng, L., et al. (2021). Interactions between gastric microbiota and metabolites in gastric cancer. Cell Death Dis. 12, 1104. doi: 10.1038/s41419-021-04396-y
Dalmasso, G., Cougnoux, A., Delmas, J., Darfeuille-Michaud, A., Bonnet, R. (2014). The bacterial genotoxin colibactin promotes colon tumor growth by modifying the tumor microenvironment. Gut Microbes 5, 675–680. doi: 10.4161/19490976.2014.969989
Dejea, C. M., Fathi, P., Craig, J. M., Boleij, A., Taddese, R., Geis, A. L., et al. (2018). Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597. doi: 10.1126/science.aah3648
Dong, Q., Chen, E. S., Zhao, C., Jin, C. (2021). Host-microbiome interaction in lung cancer. Front. Immunol. 12. doi: 10.3389/fimmu.2021.679829
Dumont-Leblond, N., Veillette, M., Racine, C., Joubert, P., Duchaine, C. (2021). Non-small cell lung cancer microbiota characterization: Prevalence of enteric and potentially pathogenic bacteria in cancer tissues. PloS One 16, e0249832. doi: 10.1371/journal.pone.0249832
Eun, C. S., Kim, B. K., Han, D. S., Kim, S. Y., Kim, K. M., Choi, B. Y., et al. (2014). Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 19, 407–416. doi: 10.1111/hel.12145
Ferreira, R. M., Pereira-Marques, J., Pinto-Ribeiro, I., Costa, J. L., Carneiro, F., MaChado, J. C., et al. (2018). Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut 67, 226–236. doi: 10.1136/gutjnl-2017-314205
Fiorentini, C., Carlini, F., Germinario, E. A. P., Maroccia, Z., Travaglione, S., Fabbri, A. (2020). Gut microbiota and colon cancer: A role for bacterial protein toxins? Int. J. Mol. Sci. 21, 6201. doi: 10.3390/ijms21176201
Fu, A., Yao, B., Dong, T., Chen, Y., Yao, J., Liu, Y., et al. (2022). Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 185, 1356–72.e26. doi: 10.1016/j.cell.2022.02.027
Garrett, W. S. (2019). The gut microbiota and colon cancer. Science 364, 1133–1135. doi: 10.1126/science.aaw2367
Gopalakrishnan, V., Spencer, C. N., Nezi, L., Reuben, A., Andrews, M. C., Karpinets, T. V., et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. doi: 10.1126/science.aan4236
Guo, K., Duan, J., Lu, J., Xiao, L., Han, L., Zeng, S., et al. (2022). Tumor necrosis factor-α-inducing protein of Helicobacter pylori promotes epithelial-mesenchymal transition and cancer stem-like cells properties via activation of Wnt/β-catenin signaling pathway in gastric cancer cells. Pathog. Dis. 80, ftac025. doi: 10.1093/femspd/ftac025
Hanahan, D. (2022). Hallmarks of cancer: new dimensions. Cancer Discovery 12, 31–46. doi: 10.1158/2159-8290.CD-21-1059
Hanahan, D., Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. doi: 10.1016/j.cell.2011.02.013
Hannan, T. J., Totsika, M., Mansfield, K. J., Moore, K. H., Schembri, M. A., Hultgren, S. J. (2012). Host-pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic Escherichia coli bladder infection. FEMS Microbiol. Rev. 36, 616–648. doi: 10.1111/j.1574-6976.2012.00339.x
Hartog, G. d., Chattopadhyay, R., Ablack, A., Hall, E. H., Butcher, L. D., Bhattacharyya, A., et al. (2016). Regulation of rac1 and reactive oxygen species production in response to infection of gastrointestinal epithelia. PloS Pathog. 12, e1005382. doi: 10.1371/journal.ppat.1005382
Hawkins, G. M., Burkett, W. C., McCoy, A. N., Nichols, H. B., Olshan, A. F., Broaddus, R., et al. (2022). Differences in the microbial profiles of early stage endometrial cancers between Black and White women. Gynecol Oncol. 165, 248–256. doi: 10.1016/j.ygyno.2022.02.021
He Zhuang, L. C., Wang, Y., Zhang, Y.-K., Zhao, M.-F., Liang, G.-D., Zhang, M.-C., et al. (2019). Dysbiosis of the gut microbiome in lung cancer. Front. Cell Infect. Microbiol. 18. doi: 10.3389/fcimb.2019.00112
Huang, S. T., Chen, J., Lian, L. Y., Cai, H. H., Zeng, H. S., Zheng, M., et al. (2020). Intratumoral levels and prognostic significance of Fusobacterium nucleatum in cervical carcinoma. Aging (Albany NY) 12, 23337–23350. doi: 10.18632/aging.104188
Iida N, D. A., Stewart, C. A., Smith, L., Bouladoux, N., Weingarten, R. A., Molina, D. A., et al. (2013). Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967–970. doi: 10.1126/science.1240527
Infectious Vaginitis, S. S. (2023). Cervicitis, and pelvic inflammatory disease. Med. Clin. North Am. 107, 299–315. doi: 10.1016/j.mcna.2022.10.009
Jackson, E. L., Willis, N., Mercer, K., Bronson, R. T., Crowley, D., Montoya, R., et al. (2001). Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248. doi: 10.1101/gad.943001
Jawad Abed, J. E. M. E., Zamir, G., Faroja, M., Almogy, G., Grenov, A., Sol, A., et al. (2016). Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-galNAc. Cell Host Microbe 20, 215–225. doi: 10.1016/j.chom.2016.07.006
Jin, C., Lagoudas, G. K., Zhao, C., Bullman, S., Bhutkar, A., Hu, B., et al. (2019). Commensal microbiota promote lung cancer development via γδ T cells. Cell 176, 998–1013.e16. doi: 10.1016/j.cell.2018.12.040
Jones, A., Jonsson, A.-B., Aro, H. (2007). Neisseria gonorrhoeae infection causes a G1 arrest in human epithelial cells. FASEB J. 21, 345–355. doi: 10.1096/fj.06-6675com
JvP, C. H., Olinga, P., Nagelkerke, A. (2021). The gastrointestinal microbiota in colorectal cancer cell migration and invasion. Clin. Exp. Metastasis 38, 495–510. doi: 10.1007/s10585-021-10130-x
Kalaora, S., Nagler, A., Nejman, D., Alon, M., Barbolin, C., Barnea, E., et al. (2021). Identification of bacteria-derived HLA-bound peptides in melanoma. Nature 592, 138–143. doi: 10.1038/s41586-021-03368-8
Karam, A., Mjaess, G., Albisinni, S., El Daccache, Y., Farah, M., Daou, S., et al. (2022). Uncovering the role of urinary microbiota in urological tumors: a systematic review of literature. World J. Urology. 40, 951–964. doi: 10.1007/s00345-021-03924-x
Kawanishi, S., Ohnishi, S., Ma, N., Hiraku, Y., Murata, M. (2017). Crosstalk between DNA damage and inflammation in the multiple steps of carcinogenesis. Int. J. Mol. Sci. 18 (8), 1808. doi: 10.3390/ijms18081808
Kikuchi, K., Murata-Kamiya, N., Kondo, S., Hatakeyama, M. (2012). Helicobacter pylori stimulates epithelial cell migration via CagA-mediated perturbation of host cell signaling. Microbes Infect. 14, 470–476. doi: 10.1016/j.micinf.2011.12.003
Kim, S., Covington A, E. G. P. (2017). The intestinal microbiota: Antibiotics, colonization resistance, and enteric pathogens. Immunol. Rev. 279, 90–105. doi: 10.1111/imr.1256310.1111/imr.12563
Korecka A, V. A. (2012). The gut microbiome: scourge, sentinel or spectator? J. Oral. Microbiol. 2012, 4. doi: 10.3402/jom.v4i0.9367
Kostic, A. D., Chun, E., Robertson, L., Glickman, J. N., Gallini, C. A., Michaud, M., et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor immune microenvironment. Cell Host Microbe 14, 207–215. doi: 10.1016/j.chom.2013.07.007
Kovacsovics-Bankowski, M., Rock, K. L. (1995). A phagosome-to-cytosol pathway for exogenous antigens presented on MHC class I molecules. Science 267, 243–246. doi: 10.1126/science.7809629
Laniewski, P., Ilhan, Z. E., Herbst-Kralovetz, M. M. (2020). The microbiome and gynaecological cancer development, prevention and therapy. Nat. Rev. Urol. 17, 232–250. doi: 10.1038/s41585-020-0286-z
Lee, S. H., Sung, J. Y., Yong, D., Chun, J., Kim, S. Y., Song, J. H., et al. (2016). Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. 102, 89–95. doi: 10.1016/j.lungcan.2016.10.016
Lengyel, E. (2010). Ovarian cancer development and metastasis. Am. J. Pathol. 177, 1053–1064. doi: 10.2353/ajpath.2010.100105
Li, R., Zhou, R., Wang, H., Li, W., Pan, M., Yao, X., et al. (2019). Gut microbiota-stimulated cathepsin K secretion mediates TLR4-dependent M2 macrophage polarization and promotes tumor metastasis in colorectal cancer. Cell Death Differ. 26, 2447–2463. doi: 10.1038/s41418-019-0312-y
Liu, H. X., Tao, L. L., Zhang, J., Zhu, Y. G., Zheng, Y., Liu, D., et al. (2017). Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int. J. Cancer. 142, 769–778. doi: 10.1002/ijc.31098
Liu, X., Cheng, Y., Zang, D., Zhang, M., Li, X., Liu, D., et al. (2021). The role of gut microbiota in lung cancer: from carcinogenesis to immunotherapy. Front. Oncol. 11. doi: 10.3389/fonc.2021.720842
Livyatan, I., Nejman, D., Shental, N., Straussman, R. (2020). Characterization of the human tumor microbiome reveals tumor-type specific intra-cellular bacteria. Oncoimmunology 9, 1800957. doi: 10.1080/2162402X.2020.1800957
Long, X., Wong, C. C., Tong, L., Chu, E. S.H., Szeto, C. H., Go, M. Y.Y., et al. (2019). Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat. Microbiol. 4, 2319–2330. doi: 10.1038/s41564-019-0541-3
Louis, P., Hold GL, H. J. F. (2014). The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12, 661–672. doi: 10.1038/nrmicro3344
Lu, R., Bosland, M., Xia, Y., Zhang, Y.-G., Kato, I., Sun, J. (2017). Presence of Salmonella AvrA in colorectal tumor and its precursor lesions in mouse intestine and human specimens. Oncotarget 8, 55104–55115. doi: 10.18632/oncotarget.19052
Lu, R., Wu, S., Zhang, Y.-G., Xia, Y., Liu, X., Zheng, Y., et al. (2014). Enteric bacterial protein AvrA promotes colonic tumorigenesis and activates colonic beta-catenin signaling pathway. Oncogenesis 3, e105. doi: 10.1038/oncsis.2014.20
Lu, R., Wu, S., Zhang, Y. G., Xia, Y., Zhou, Z., Kato, I., et al. (2016). Salmonella protein avrA activates the STAT3 signaling pathway in colon cancer. Neoplasia 18, 307–316. doi: 10.1016/j.neo.2016.04.001
Mantovani, A., Sozzani, S., Locati, M., Allavena, P., Sica, A. (2002). Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23, 549–555. doi: 10.1016/s1471-4906(02)02302-5
Martin, A., Woolbright, B. L., Umar, S., Ingersoll, M. A., Taylor, J. A., 3rd (2022). Bladder cancer, inflammageing and microbiomes. Nat. Rev. Urol 19, 495–509. doi: 10.1038/s41585-022-00611-3
Mitra, A., MacIntyre, D. A., Lee, Y. S., Smith, A., Marchesi, J. R., Lehne, B., et al. (2015). Cervical intraepithelial neoplasia disease progression is associated with increased vaginal microbiome diversity. Sci. Rep. 5, 16865. doi: 10.1038/srep16865
Mughini-Gras, L., Schaapveld, M., Kramers, J., Mooij, S., Neefjes-Borst, E. A., Pelt, W. V., et al. (2018). Increased colon cancer risk after severe Salmonella infection. PloS One 13, e0189721. doi: 10.1371/journal.pone.0189721
Nejman, D., Livyatan, I., Fuks, G., Gavert, N., Zwang, Y., Geller, L. T., et al. (2020). The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980. doi: 10.1126/science.aay9189
Nogal, A., Valdes, A. M., Menni, C. (2021). The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 13, 1–24. doi: 10.1080/19490976.2021.1897212
Normark, S., Nilsson, C., Normark, B. H., Hornef, M. W. (2003). Persistent infection with Helicobacter pylori and the development of gastric cancer. Adv. Cancer Res. 90, 63–89. doi: 10.1016/s0065-230x(03)90002-9
Nougayrede, J. P., Homburg, S., Taieb, F., Boury, M., Brzuszkiewicz, E., Gottschalk, G., et al. (2006). Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313, 848–851. doi: 10.1126/science.1127059
Odenbreit, S., Püls, J., Sedlmaier, B., Gerland, E., Fischer, W., Haas, R. (2000). Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287, 1497–1500. doi: 10.1126/science.287.5457.1497
Park, E. M., Chelvanambi, M., Bhutiani, N., Kroemer, G., Zitvogel, L., Wargo, J. A. (2022). Targeting the gut and tumor microbiota in cancer. Nat. Med. 28, 690–703. doi: 10.1038/s41591-022-01779-2
Parra-Grande, M., Oré-Arce, M., Martínez-Priego, L., D’Auria, G., Rosselló-Mora, R., Lillo, M., et al. (2022). Profiling the bladder microbiota in patients with bladder cancer. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.718776
Pfeifer, J. D., Wick, M. J., Roberts, R. L., Findlay, K., Normark, S. J., Harding, C. V. (1993). Phagocytic processing of bacterial antigens for class I MHC presentation to T cells. Nature 361, 359–362. doi: 10.1038/361359a0
Pizzo, F., Maroccia, Z., Hammarberg Ferri, I., Fiorentini, C. (2022). Role of the microbiota in lung cancer: insights on prevention and treatment. Int. J. Mol. Sci. 23 (11), 6138. doi: 10.3390/ijms23116138
Pleguezuelos-Manzano, C., Puschhof, J., Rosendahl Huber, A., van Hoeck, A., Wood, H. M., Nomburg, J., et al. (2020). Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nat. 580, 269–273. doi: 10.1038/s41586-020-2080-8
Poutahidis, T., Erdman, S. E. (2016). Commensal bacteria modulate the tumor microenvironment. Cancer Lett. 380, 356–358. doi: 10.1016/j.canlet.2015.12.028
Reuter, S., Gupta, S. C., Chaturvedi, M. M., Oxidative stress, B. B. A. (2010). inflammation, and cancer: how are they linked? Free Radic. Biol. Med. 49, 1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006
Routy, B., Le Chatelier, E., Derosa, L., Duong, C. P. M., Alou, M. T., Daillère, R., et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. doi: 10.1126/science.aan3706
Sheh A, J. G. F. (2013). The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes 4, 505–531. doi: 10.4161/gmic.26205
Siegel, R. L., Miller, K. D., Fuchs, H. E., Jemal, A. (2022). Cancer statistics, 2022. Cancer statistics 72 (1), 7–33. doi: 10.3322/caac.21708
Smyth, E. C., Nilsson, M., Grabsch, H. I., van Grieken, N. C. (2020). Gastric cancer. Lancet 396, 635–648. doi: 10.1016/S0140-6736(20)31288-5
Srinivas, U. S., Tan, B. W. Q., Vellayappan, B. A., Jeyasekharan, A. D. (2019). ROS and the DNA damage response in cancer. Redox Biol. 25, 101084. doi: 10.1016/j.redox.2018.101084
Stearns, J. C., Lynch, M. D., Senadheera, D. B., Tenenbaum, H. C., Goldberg, M. B., Cvitkovitch, D. G., et al. (2011). Bacterial biogeography of the human digestive tract. Sci. Rep. 1, 170. doi: 10.1038/srep00170
Stein, M., Rappuoli, R., Covacci, A. (2000). Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. U.S.A. 97, 1263–1268. doi: 10.1073/pnas.97.3.1263
Stewart, O. A., Wu F, Y. C. (2020). The role of gastric microbiota in gastric cancer. Gut Microbes 11, 1220–1230. doi: 10.1080/19490976.2020.1762520
Swidsinski, A., Khilkin, M., Kerjaschki, D., Schreiber, S., Ortner, M., Weber, J., et al. (1998). Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology, 115, 281–286. doi: 10.1016/s0016-5085(98)70194-5
Tang, Y., Chen, Y., Jiang, H., Nie, D. (2011). Short-chain fatty acids induced autophagy serves as an adaptive strategy for retarding mitochondria-mediated apoptotic cell death. Cell Death Differ. 18, 602–618. doi: 10.1038/cdd.2010.117
Ternes, D., Karta, J., Tsenkova, M., Wilmes, P., Haan, S., Letellier, E. (2020). Microbiome in colorectal cancer: how to get from meta-omics to mechanism? Trends Microbiol. 28, 401–423. doi: 10.1016/j.tim.2020.01.001
Tilg, H., Adolph, T. E., Gerner, R. R., Moschen, A. R. (2018). The intestinal microbiota in colorectal cancer. Cancer Cell. 33, 954–964. doi: 10.1016/j.ccell.2018.03.004
Tjalsma, H., Boleij, A., Marchesi, J. R., Dutilh, B. E. (2012). A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat. Rev. Microbiol. 10, 575–582. doi: 10.1038/nrmicro2819
Tong, Y., Gao, H., Qi, Q., Liu, X., Li, J., Gao, J., et al. (2021). High fat diet, gut microbiome and gastrointestinal cancer. Theranostics 11, 5889–5910. doi: 10.7150/thno.56157
Unemo, M., Seifert, H. S., Hook, E. W., 3rd, Hawkes, S., Ndowa, F., Dillon, J. R. (2019). Gonorrhoea. Nat. Rev. Dis. Primers. 5, 79. doi: 10.1038/s41572-019-0128-6
Walther-António, M. R., Chen, J., Multinu, F., Hokenstad, A., Distad, T. J., Cheek, E. H., et al. (2016). Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 8, 122. doi: 10.1186/s13073-016-0368-y
Wang, D., Cheng, J., Zhang, J., Zhou, F., He, X., Shi, Y., et al. (2021). The role of respiratory microbiota in lung cancer. Int. J. Biol. Sci. 17, 3646–3658. doi: 10.7150/ijbs.51376
Wang, F., Meng, W., Wang, B., Qiao, L. (2014). Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 345, 196–202. doi: 10.1016/j.canlet.2013.08.016
Wang, L., Tang, L., Feng, Y., Zhao, S., Han, M., Zhang, C., et al. (2020). A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8+ T cells in mice. Gut 69, 1988–1997. doi: 10.1136/gutjnl-2019-320105
Wang, Z., Qin, X., Hu, D., Huang, J., Guo, E., Xiao, R., et al. (2022). Akkermansia supplementation reverses the tumorpromoting effect of the fecal microbiota transplantation in ovarian cancer. Cell Rep. 41, 111890. doi: 10.1016/j.celrep.2022.111890
Wells, C. L., Maddaus MA, R. L. S. (1988). Proposed mechanisms for the translocation of intestinal bacteria. Rev. Infect. Dis. 10, 958–979. doi: 10.1093/clinids/10.5.958
Wong, B. C.-Y., Lam, S. K., Wong, W. M., Chen, J. S., Zheng, T. T., Feng, R. E., et al. (2004). Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. JAMA 291, 187–194. doi: 10.1001/jama.291.2.187
Wong-Rolle, A., Wei, H. K., Zhao, C. (2021). Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Protein Cell 12, 426–435. doi: 10.1007/s13238-020-00813-8
Wu, S., Rhee, K.-J., Albesiano, E., Rabizadeh, S., Wu, X., Yen, H.-R., et al. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022. doi: 10.1038/nm.2015
Wu, B. G., Segal, L. N., Britton, R. A., Cani, P. D. (2017). Lung microbiota and its impact on the mucosal immune phenotype. Microbiol. Spectrum. 5 (3), 10.1128/microbiolspec.BAD-0005-2016. doi: 10.1128/microbiolspec.BAD-0005-2016
Xu, N., Wang, L., Li, C., Ding, C., Li, C., Fan, W., et al. (2020). Microbiota dysbiosis in lung cancer: evidence of association and potential mechanisms. Trans. Lung Cancer Res. 9, 1554–1568. doi: 10.21037/tlcr-20-156
Yasui, W., Sentani, K., Sakamoto, N., Anami, K., Naito, Y., Oue, N. (2011). Molecular pathology of gastric cancer: research and practice Pathol. Res. Pract. 207, 608–612. doi: 10.1016/j.prp.2011.09.006
Yin, H., Miao, Z., Wang, L., Su, B., Liu, C., Jin, Y., et al. (2022). Fusobacterium nucleatum promotes liver metastasis in colorectal cancer by regulating the hepatic immune niche and altering gut microbiota. Aging (Albany NY). 14, 1941–1958. doi: 10.18632/aging.203914
Yu, T., Guo, F., Yu, Y., Sun, T., Ma, D., Han, J., et al. (2017). Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–63.e16. doi: 10.1016/j.cell.2017.07.008
Zhang, W., Yang, F., Mao, S., Wang, R., Chen, H., Ran, Y., et al. (2023). Bladder cancer-associated microbiota: Recent advances and future perspectives. Heliyon 9, e13012. doi: 10.1016/j.heliyon.2023.e13012
Zhang, X., Li, C., Chen, D., He, X., Zhao, Y., Bao, L., et al. (2022). H. pylori CagA activates the NLRP3 inflammasome to promote gastric cancer cell migration and invasion. Inflammation Res. 71, 141–155. doi: 10.1007/s00011-021-01522-6
Zhao, H., Ming, T., Tang, S., Ren, S., Yang, H., Liu, M., et al. (2022). Wnt signaling in colorectal cancer: pathogenic role and therapeutic target. Mol. Cancer. 21, 144. doi: 10.1186/s12943-022-01616-7
Zhao, Y., Liu, Y., Li, S., Peng, Z., Liu, X., Chen, J., et al. (2021). Role of lung and gut microbiota on lung cancer pathogenesis. J. Cancer Res. Clin. Oncol. 147, 2177–2186. doi: 10.1007/s00432-021-03644-0
Zheng, Y., Fang, Z., Xue, Y., Zhang, J., Zhu, J., Gao, R., et al. (2020). Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes 11, 1030–1042. doi: 10.1080/19490976.2020.1737487
Zhou, B., Sun, C., Huang, J., Xia, M., Guo, E., Li, N., et al. (2019). The biodiversity composition of microbiome in ovarian carcinoma patients. Sci. Rep. 9, 1691. doi: 10.1038/s41598-018-38031-2
Keywords: microbiota, cancer, tumor microenvironment, sequencing, bacteria
Citation: Li Q (2023) Bacterial infection and microbiota in carcinogenesis and tumor development. Front. Cell. Infect. Microbiol. 13:1294082. doi: 10.3389/fcimb.2023.1294082
Received: 14 September 2023; Accepted: 31 October 2023;
Published: 15 November 2023.
Edited by:
Soumya Panigrahi, Case Western Reserve University, United StatesReviewed by:
Anurag Shukla, Drexel University, United StatesCopyright © 2023 Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiao Li, bGlxaWFvZ3JhY2VAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.