- 1Emergency Department, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China
- 2Emergency and Critical Care Center, Intensive Care Unit, Zhejiang Provincial People's Hospital (Affiliated People's Hospital), Hangzhou Medical College, Hangzhou, Zhejiang, China
Introduction: Pneumonia are the leading cause of death worldwide, and antibiotic treatment remains fundamental. However, conventional sputum smears or cultures are still inefficient for obtaining pathogenic microorganisms.Metagenomic next-generation sequencing (mNGS) has shown great value in nucleic acid detection, however, the NGS results for lower respiratory tract microorganisms are still poorly studied.
Methods: This study dealt with investigating the efficacy of mNGS in detecting pathogens in the lower respiratory tract of patients with pulmonary infections. A total of 112 patients admitted at the First Affiliated Hospital of Zhengzhou University between April 30, 2018, and June 30, 2020, were enrolled in this retrospective study. The bronchoalveolar lavage fluid (BALF) was obtained from lower respiratory tract from each patient. Routine methods (bacterial smear and culture) and mNGS were employed for the identification of pathogenic microorganisms in BALF.
Results: The average patient age was 53.0 years, with 94.6% (106/112) obtaining pathogenic microorganism results. The total mNGS detection rate of pathogenic microorganisms significantly surpassed conventional methods (93.7% vs. 32.1%, P < 0.05). Notably, 75% of patients (84/112) were found to have bacteria by mNGS, but only 28.6% (32/112) were found to have bacteria by conventional approaches. The most commonly detected bacteria included Acinetobacter baumannii (19.6%), Klebsiella pneumoniae (17.9%), Pseudomonas aeruginosa (14.3%), Staphylococcus faecium (12.5%), Enterococcus faecium (12.5%), and Haemophilus parainfluenzae (11.6%). In 29.5% (33/112) of patients, fungi were identified using mNGS, including 23 cases of Candida albicans (20.5%), 18 of Pneumocystis carinii (16.1%), and 10 of Aspergillus (8.9%). However, only 7.1 % (8/112) of individuals were found to have fungi when conventional procedures were used. The mNGS detection rate of viruses was significantly higher than the conventional method rate (43.8% vs. 0.9%, P < 0.05). The most commonly detected viruses included Epstein–Barr virus (15.2%), cytomegalovirus (13.4%), circovirus (8.9%), human coronavirus (4.5%), and rhinovirus (4.5%). Only 29.4% (33/112) of patients were positive, whereas 5.4% (6/112) of patients were negative for both detection methods as shown by Kappa analysis, indicating poor consistency between the two methods (P = 0.340; Kappa analysis).
Conclusion: Significant benefits of mNGS have been shown in the detection of pathogenic microorganisms in patients with pulmonary infection. For those with suboptimal therapeutic responses, mNGS can provide an etiological basis, aiding in precise anti-infective treatment.
Introduction
Pulmonary infection stands as a major global cause of mortality, with antibiotic treatment remaining its cornerstone (Magill et al., 2014). In recent times, there has been a shift toward precise anti-infective therapy, emphasizing the early acquisition of an etiological basis to enable the selection of appropriate antibiotics, enhance therapeutic outcomes, and mitigate antibiotic abuse and drug resistance in bacteria (Caliendo et al., 2013). It has been proven that conventional approaches for detecting pathogenic microorganisms, including bacterial smears of sputum or bronchoalveolar lavage fluid (BALF), culture, or nucleic acid detection of pharyngeal swabs, have only a limited level of effectiveness. Numerous studies indicate that these approaches are only capable of identifying pathogenic microorganisms in between 30 and 40% of patients, with even lower detection rates in those who have previously used antibiotics, posing a challenge to clinical requirements (Jain et al., 2015; Torres et al., 2019).
Metagenomic next-generation sequencing (mNGS), as an advanced molecular method of disease diagnosis, provides a high throughput and sensitivity while also being less impacted by exposure to antibiotics. All microbial fragments in a sample may be directly identified by mNGS (Gu et al., 2019; McCombie et al., 2019; Duan et al., 2021). Recently, mNGS has found increased application in clinical settings, proving valuable both in non-infectious diseases (Kamps et al., 2017; Mellis et al., 2018; Cainap et al., 2021) and infectious diseases, particularly in detecting unique infections and emerging pathogens (Afshinnekoo et al., 2017; Simner et al., 2018; Ai et al., 2020; Chen et al., 2020). Studies indicate that the diagnostic performance of mNGS for infectious diseases is 50.7% in sensitivity and 85.7% in specificity (Miao et al., 2018). Despite this, there is still a paucity of mNGS data concerning pathogenic microorganisms in the lower respiratory tract of patients suffering from pulmonary infections, particularly in BALF obtained through bronchoscopy. The focus of this study is to examine mNGS’s efficacy in detecting pathogens in the lower respiratory tracts of individuals with pulmonary infections, in comparison with conventional detection techniques. It seeks to delineate the characteristics of the pathogen spectrum, providing novel insights into precise anti-infective treatment.
Methods
Study participants
Patients admitted due to pulmonary infections between April 30, 2018, and June 30, 2020, at the First Affiliated Hospital of Zhengzhou University were included retrospectively. The inclusion criteria were as follows: (1) definite diagnosis according to the criteria outlined in the “Diagnosis and Treatment of Adults With Community-acquired Pneumonia 2016” (Metlay et al., 2019) and “Management of Adults With Hospital-acquired and Ventilator-associated Pneumonia:2016 Clinical Practice Guidelines by the Infectious Diseases Society of America and the American Thoracic Society” (Kalil et al., 2016) and (2) During the patient’s stay in the hospital, BALF bacterial smear, culture, and mNGS (BALF) to identify pathogenic microorganisms in the respiratory tract. The exclusion criterion comprised complications with other infections and incomplete clinical data. Low immune function was identified as occurring when at least one of the following conditions was present: hematopathy, use of chemotherapeutic drugs and glucocorticoids for autoimmune disorders, immunosuppressant use for solid organ transplantation, neutropenia or chemotherapy for solid tumors in the past 3 months, and impaired immunity owing to other factors, including hereditary or congenital factors.
Microbiological testing and pathogenic analysis
The bronchoalveolar lavage was performed to obtain BALF. Pathogenic microorganisms were identified using the routine methods of bacterial smear and culture. Simultaneously, mNGS was employed to identify pathogenic microorganisms in BALF. mNGS testing involved immediate storage of 5mL BALF at 4°C, with submission for inspection within two hours. The mNGS procedure for BALF samples includes nucleic acid extraction, library construction, sequencing, and information analysis. mNGS testing is completed by two third party companies(BGI-Huada Genomics Institute, Shenzhen, China;Capital Bio Technology, Beijing, China).
Acquisition of clinical data and antibiotic treatments
Medical records were queried for clinical data, which included demographic information, lab test outcomes, intensive care unit (ICU) special treatment records, and antibiotic treatment particulars. The following information was also obtained: antibiotic treatment before ICU admission, initial antibiotics administered upon ICU admission, and adjustments made based on mNGS pathogen data.
Statistical analysis
Data were analyzed statistically utilizing the SPSS 22.0 software. The t-test was used to examine measurement data that conformed to a normal distribution, which were presented as mean ± standard deviation (x ± s). Measurement data that did not conform to a normal distribution were presented as a median (M) and an interquartile range (IQR), with the Mann-Whitney rank sum test applied for comparison. Categorical data were presented as the number of cases and percentages [n (%)], with comparisons conducted using the χ2 test or Fisher’s exact test. Statistical significance was set at P < 0.05.
Results
Participants’ general characteristics
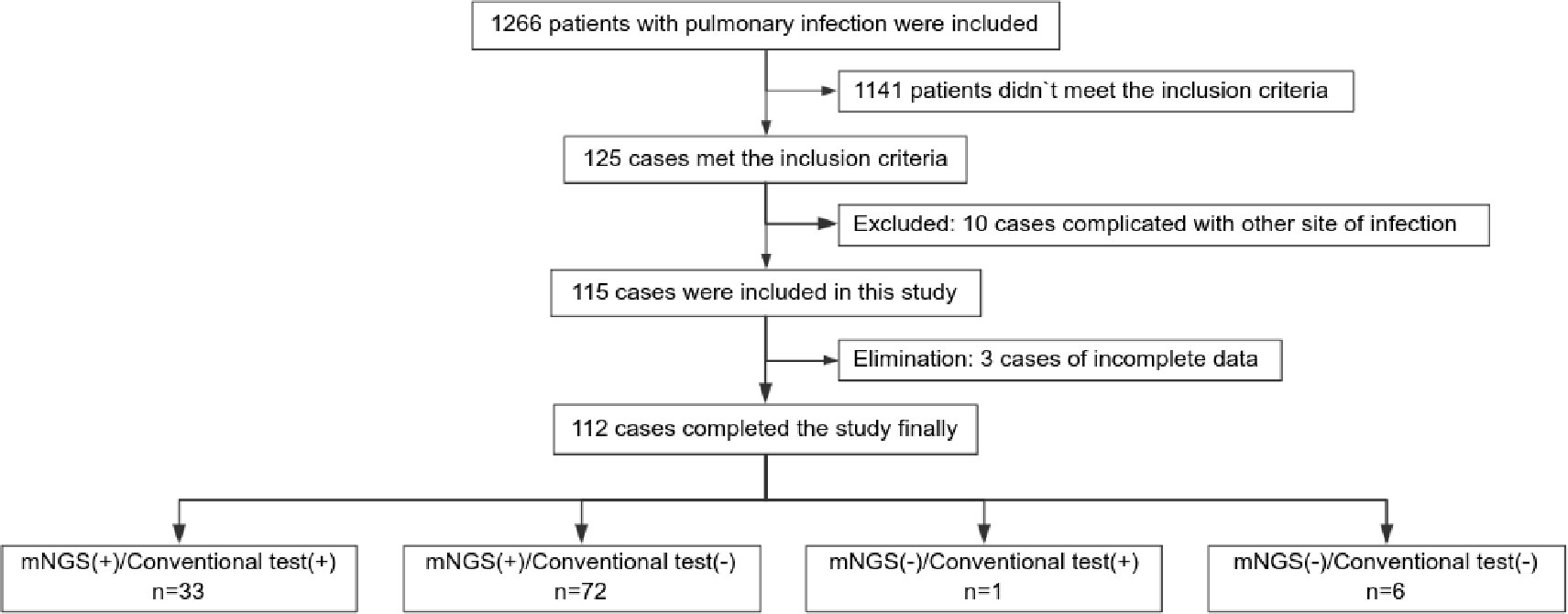
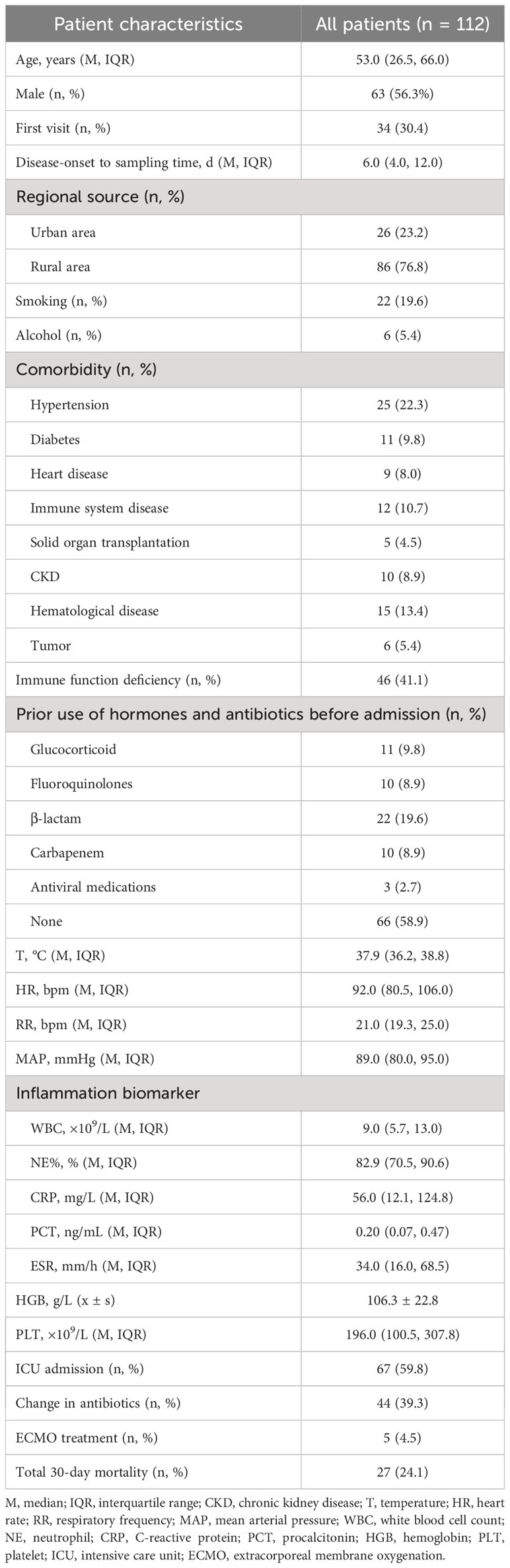
A total of 112 individuals successfully completed the study out of 1,266 patients who received treatment for pulmonary infection between April 30, 2018, and June 30, 2020. Of the total number of participants, 34.0% (34/112) were diagnosed with pulmonary infection for the first time. The mean age of the patients was 53.0 years. Coughing was the most common respiratory symptom, affecting 58.0% of patients (66/112), followed by chest tightness, which affected 39.3% (44/112) of patients. A significant proportion (41.1%) of patients exhibited low immune function. Additionally, 9.8% (11/112) of patients had used glucocorticoids, and 41.1% (46/112) had used antibiotics before admission Furthermore, 39.3% of patients adjusted their antibiotic prescriptions based on pathogenic microorganism results, 4.5% received extracorporeal membrane oxygenation treatment, 59.8% of those patients were admitted to the ICU, and the 30-day hospital mortality rate was 24.1% (Figure 1; Table 1).
Difference between conventional tests and mNGS in detecting pathogenic microorganisms
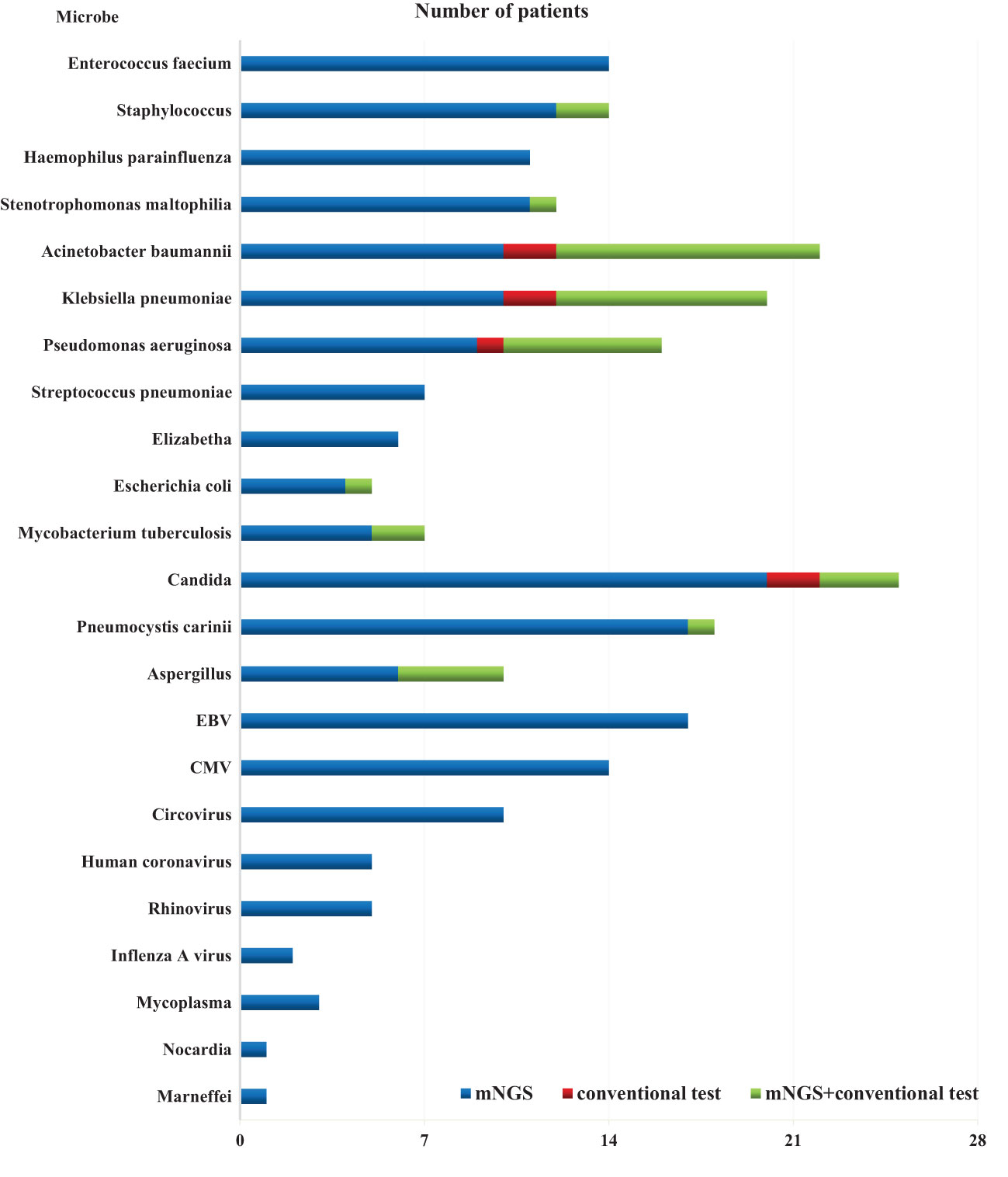
The overall detection rate of mNGS for pathogenic microorganisms demonstrated a significant superiority over conventional methods(93.7% vs. 32.1%, P < 0.05) (Figures 2, 3). mNGS identified bacteria in 75.0% (84/112) of patients, whereas conventional methods detected bacteria in only 28.6% (32/112) of patients. Predominantly identified bacteria included Acinetobacter baumannii (19.6%), Klebsiella pneumoniae (17.9%), Pseudomonas aeruginosa (14.3%), Enterococcus faecium (12.5%), Staphylococcus faecium (12.5%), and Haemophilus parainfluenzae (11.6%). Additionally, mNGS detected fungi in 29.5% (33/112) of patients, encompassing Candida (23 patients, 20.5%), Pneumocystis carinii (18 patients, 16.1%), and Aspergillus (10 patients, 8.9%). In contrast, the conventional method identified fungi in only 7.1% (8/112) of patients. The detection rate of viruses by mNGS significantly surpassed that of conventional methods (43.8% vs. 0.9%).The most commonly detected viruses included Epstein–Barr virus (15.2%), cytomegalovirus (12.5%), circovirus (8.9%), rhinovirus (4.5%), and human coronavirus (4.5%). Notably, mNGS detected two cases of Influenza A virus. It is worth mentioning that rare pathogens Nocardia and cyanobacteria Marneffei were identified in two patients by mNGS.
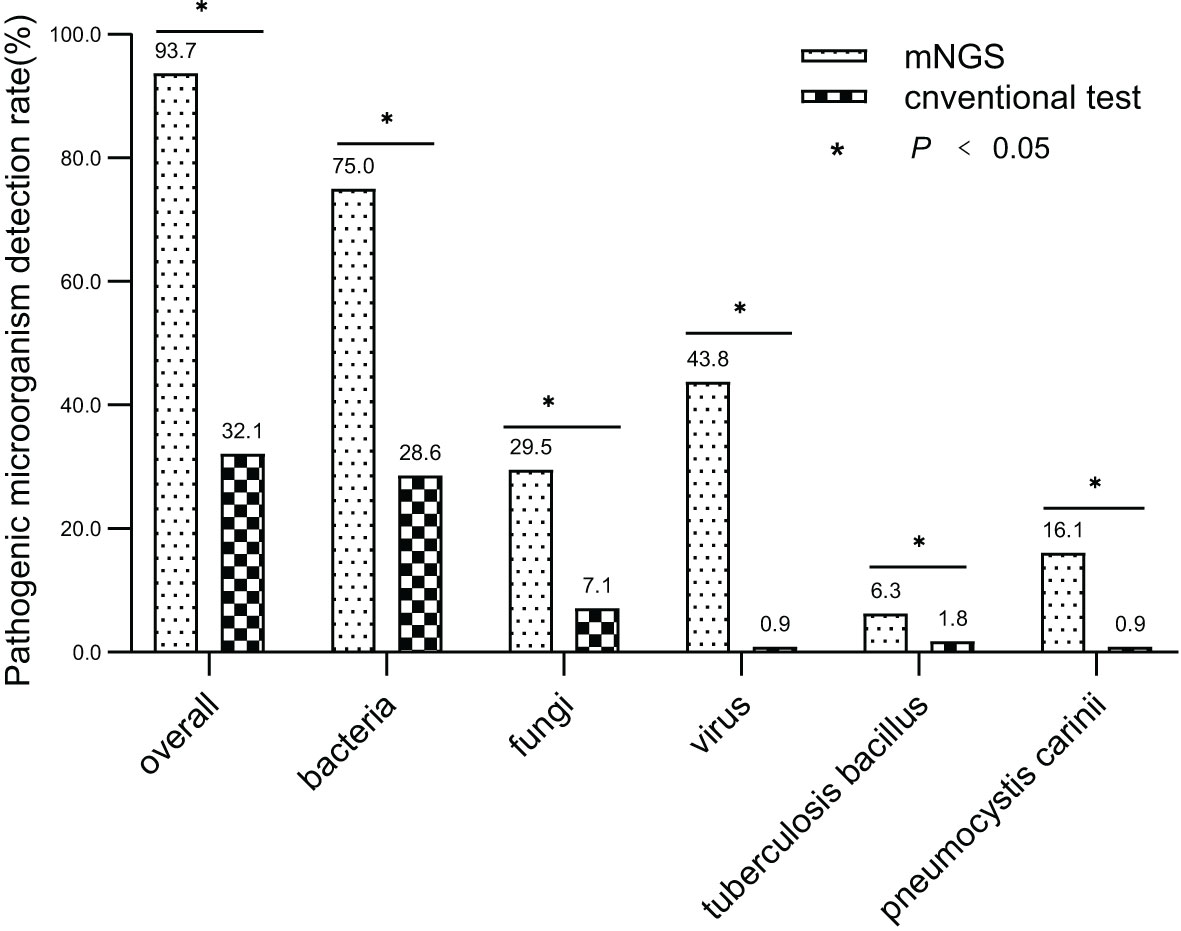
Figure 2 Comparing mNGS-based pathogen detection with conventional detection techniques. (* P<0.05).
Consistency between mNGS and conventional approaches in detecting pathogenic microorganisms
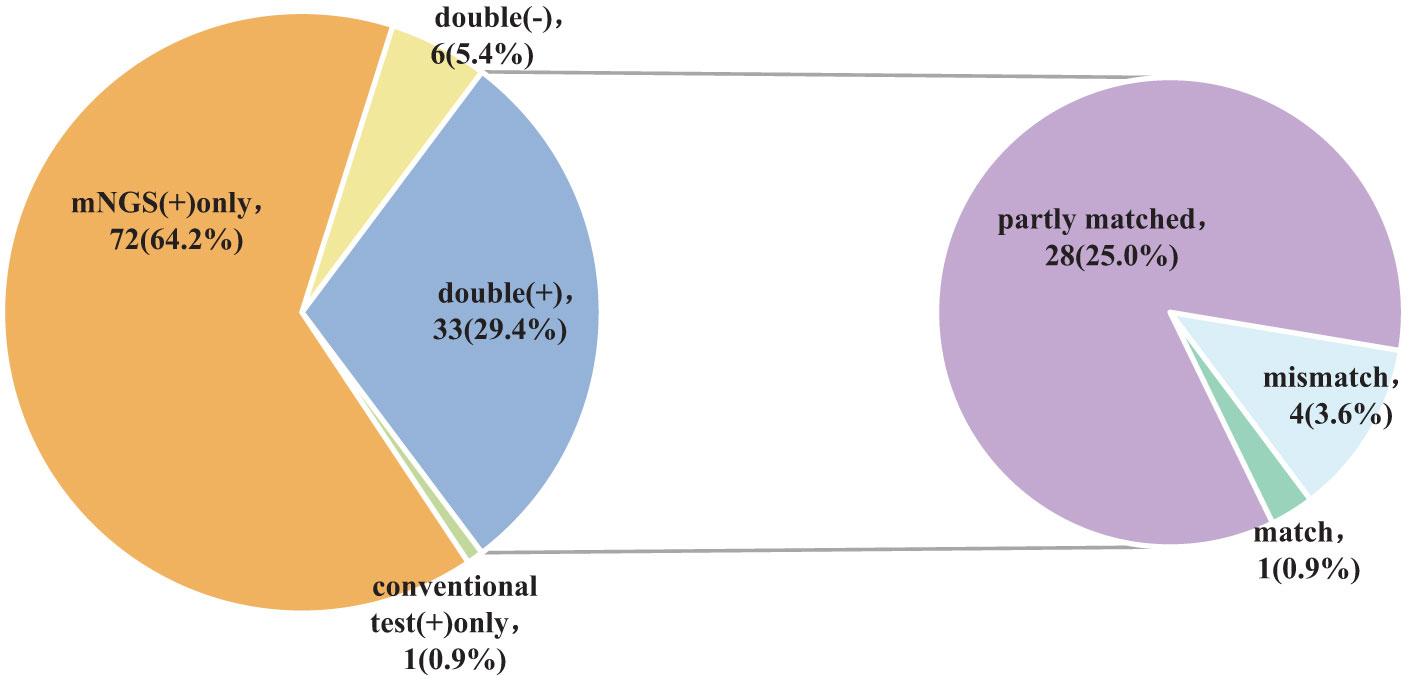
The findings of the Kappa analysis suggested that there was poor consistency between the two methods (P = 0.340). Notably, both methods yielded positive results in 29.4% (33/112) of cases, while 5.4% (6/112) were negative for both methods. Additionally, 64.2% (72/112) of participants tested positive for mNGS only, and a minimal 0.9% (1/112) showed positivity solely for conventional approaches. Out of the 33 cases who tested positive for both methods, only one (0.9%) exhibited complete matching, whereas 4 cases (3.6%) were entirely mismatched and 28 (25.0%) demonstrated partial matching (Figure 4; Table 2).
Comparative analysis of clinical features between patients with positive and negative results detected by routine methods
Among the 105 patients whose mNGS findings were positive, 33 were positive according to the conventional techniques, whereas 72 were negative as per the conventional techniques. Notably, the negative group exhibited a significantly lower proportion of smokers, tumor history, prior use of fluoroquinolones before admission, and admissions to the ICU than the positive group. Additionally, Comparisons between the positive and negative groups showed that patients in the negative group had remarkably lower proportion of neutrophils in peripheral blood(all P < 0.05) (Table 3).
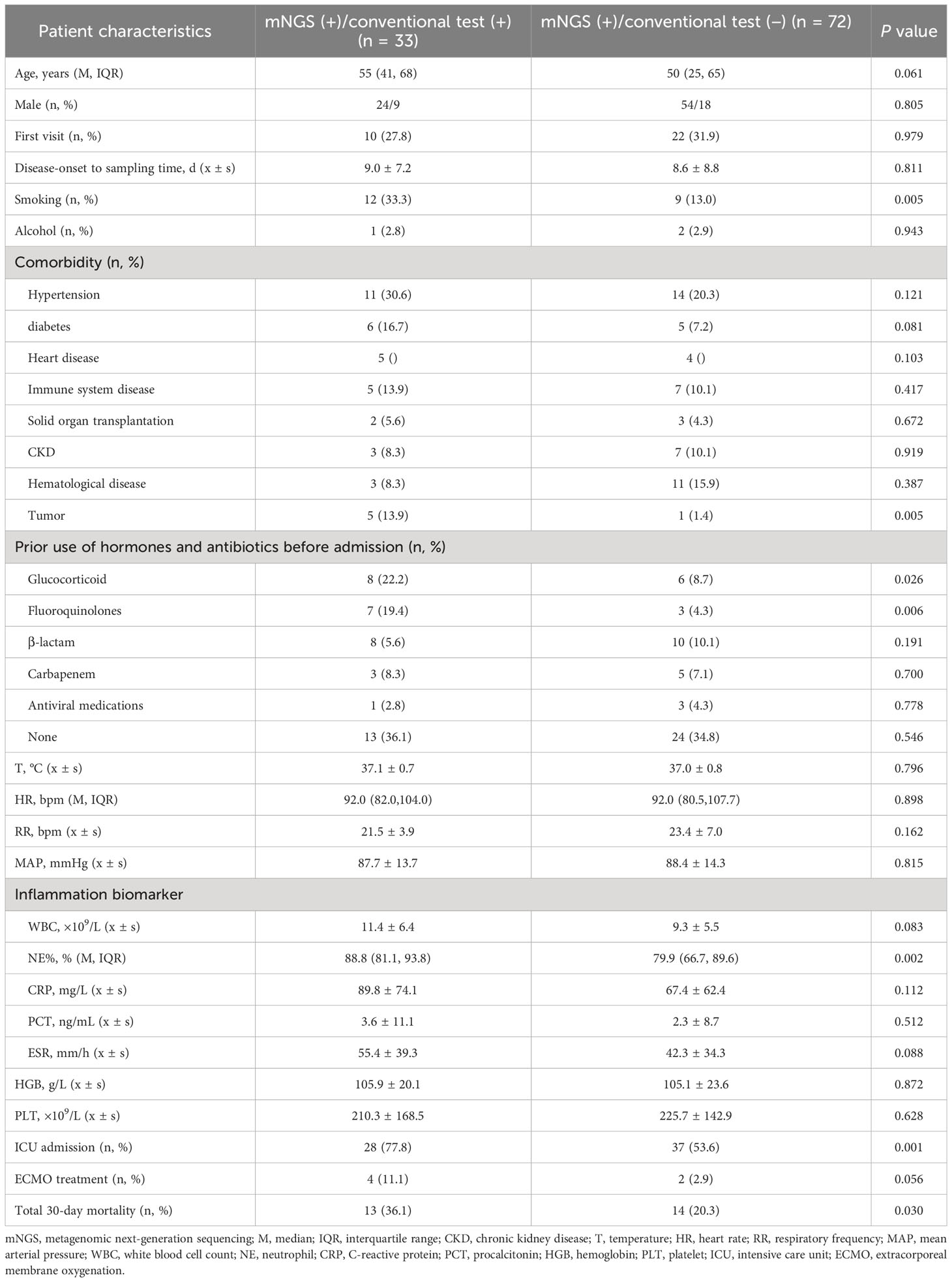
Table 3 Comparative analysis of the clinical features of mNGS (+) patients who obtained positive and negative results outcomes as detected by conventional approaches (n = 105).
Discussion
Comparative analysis of mNGS and conventional detection techniques were performed in this retrospective research to detect pathogens in the BALF of patients with pulmonary infection. The findings highlight the exceptional detection capability of mNGS, identifying pathogens even in cases where routine detection methods yielded negative results, offering significant support for antibiotic selection.
In total, 1,266 patients presenting with pulmonary infection were considered for this study, with 112 patients meeting the inclusion criteria (Figure 1). Notably, 41.1% (46/112) of these patients had immunodeficiency, 59.8% (67/112) were admitted to the ICU, and 39.3% (44/112) altered their antibiotic regimen based on pathogenic microbiological results (Table 1). The study observed that 94.6% of those diagnosed with pulmonary infection obtained pathogenic microorganism results through mNGS or conventional tests (Figure 2), aligning with previous research by Wu et al. (Wu et al., 2020). Further analysis revealed that the conventional approaches identified bacteria in only 28.6% of cases, falling significantly short of clinical needs. In contrast, bacteria were detected in 75% of patients using mNGS (Figure 2), providing crucial insights for application in clinical settings. The most commonly detected bacteria included A. baumannii, K. pneumoniae, P. aeruginosa, S. faecium, E. faecium, and H. parainfluenzae (Figure 3), conforming to typical bacteria prevalent in both hospital- and community-acquired pneumonia (Huang et al., 2020; Wu et al., 2020). With its superior microbial detection rate, mNGS shows great promise for directing antibiotic selection, improving the detection of pathogenic microorganisms, and monitoring the lower respiratory tract in patients who do not respond well to empirical therapy (Chen et al., 2021).
Fungal infections are significant contributors to pneumonia, particularly in patients with risk factors such as immune deficiency, immunosuppressant use, diabetes, organ transplantation, chemotherapy or hormonal therapy, antibiotic misuse, and others (Kauffman, 2015). The study revealed that in comparison to conventional approaches, which only had a 7.1% success rate in detecting fungus, the mNGS detection rate was 29.5% (33/112) (Figure 2). Notably, mNGS significantly outperformed conventional approaches in detecting Pneumocystis carinii and Aspergillus. Given the challenges in detecting fungi, especially with the need for advanced wall-breaking technology and the potential for false negatives, clinicians should interpret results cautiously and consider repeated testing if necessary (Sanmiguel, 2011; Biswas et al., 2017). The study emphasizes the high incidence of fungal infection in patients with immunosuppression, reaching 41.1% (46/112). Conventional methods often face challenges in obtaining an etiological basis for fungal infections in these patients, leading to difficulties in follow-up antifungal treatment. Lack of etiological basis may result in premature discontinuation of antifungal treatment and subsequent recurrence of infection. mNGS proves to be a valuable tool in addressing these challenges. The study reveals that the detection rate of fungi is dramatically increased by mNGS, going from 7.1% (8/112) to 29.5% (33/112), providing clinicians with more references for accurate diagnosis and treatment. Nevertheless, this highlights the need for thorough assessment by clinicians, taking into consideration clinical symptoms, laboratory indicators, imaging results, and the possibility of normal pulmonary fungal colonization, particularly for pulmonary Candida (Zeng et al., 2019). The study emphasizes the importance of considering fungal infections when managing aggravated infections in patients with immunosuppression.
Virus infection indeed plays a crucial role in respiratory tract infections, and conventional methods struggle with low detection rates, even with gene amplification like PCR (Capobianchi et al., 2013; Parker and Chen, 2017). This study underscores that the conventional method’s virus detection rate is a mere 0.9%, while mNGS achieves an impressive 43.8% (Figure 2). The most prevalent viruses identified are still Epstein–Barr virus and cytomegalovirus, followed by parvovirus, rhinovirus, and human coronavirus, in agreement with extensive research findings (Chen et al., 2021). Detecting viruses in BALF holds vital clinical significance, though further investigation is needed to determine its clinical relevance in conjunction with specific epidemiology.
In summary, the use of mNGS results in a significant increase in the detection rate of pathogenic microorganisms in BALF, providing an important point of reference for medical practitioners. When the two approaches are compared, it is found that there is a remarkable lack of consistency in the detection rates of pathogenic microorganisms (Kappa value = 0.030, P = 0.340) (Table 2). Only 29.4% (33/112) of the patients got positive results at the same time,and of the 33 double positive patients, only 1 case (0.9%) was completely matched, 4 cases (3.6%) were completely mismatched, and 28 cases (25.0%) were partially matched (Figure 4). This indicates that mNGS can detect pathogenic microorganisms not identified by conventional methods. However, considering the lung’s non-sterile environment and the presence of numerous microorganisms in healthy individuals, clinicians should apply caution when interpreting mNGS data, as they need to distinguish between colonization and infection to determine the most appropriate antibiotic regimen (Erb-Downward et al., 2011; Dickson et al., 2017; Langelier et al., 2018). As observed by Wang et al. (Wang et al., 2019), 26 out of 55 patients with mixed pulmonary infection tested positive using both mNGS and routine approaches, and only 3 (5.5%) cases exhibited matching results, aligning with the current study’s findings. For rare pathogens, conventional methods often struggle with detection, while mNGS proves effective. In this study, 18 cases of Pneumocystis carinii, 7 cases of Mycobacterium tuberculosis, 1 case of Nocardia, and 1 case of cyanobacteria Marneffei were detected by mNGS (Figure 3). With this knowledge, clinicians may choose appropriate antibiotic treatment plans, particularly when conventional methods encounter obstacles such as low gene load, antibiotic exposures, or deficiencies in routine methods for the detection, particularly culture (Chan et al., 2020). Based on the results of this study, physicians should quickly conduct mNGS to identify the types of pathogenic microorganisms in patients who have not responded well to empirical therapy. According to microbial results, 39.3% (44/112) of patients in this study were able to modify their antibiotic prescription, leading to effective treatment outcomes and precise anti-infective medication. The anti-infective treatment was adjusted depending on microbiological findings when mNGS was used to identify respiratory microbes in 21 patients presenting with severe community-acquired pneumonia and poor immune response, as well as in 23 patients with normal immune function. According to these findings, compared with patients with normal immune function, patients with low immune function received fewer full-coverage anti-infective regimens before obtaining pathogenic microbiological results (14.7% vs. 57.1%, P = 0.022) and more adjusted anti-infective regimens after obtaining microbiological results (87.0% vs. 57.1%, P = 0.027), with >50% of the patients receiving a downgrade in antibiotics.
The analysis of patients who have varying testing results provides valuable insights into the clinical characteristics affecting the detection of pathogenic microorganisms. Patients who had negative findings in the mNGS test showed a significantly lower proportion of smokers, tumor history, prior use of fluoroquinolones before admission and admissions to the ICU than the positive group, and decreased proportion of neutrophils in peripheral blood (Table 3). This shows that clinicians may have a more difficult time finding pathogenic microorganisms in patients who do not smoke, patients with a history of cancer, patients exposured to fluoroquinolones before admission, patients who are treated in general wards, and patients whose inflammatory indices are low. For such patients, the study recommends timely mNGS detection of BALF, especially if the initial anti-infective treatment proves ineffective. The implementation of mNGS in these cases is highlighted as a crucial step to significantly improve the detection rate of pathogenic microorganisms. Critically ill patients in the ICU, with established artificial airways, find it relatively easier to obtain BALF through bedside tracheoscopy. However, patients in general wards with milder illnesses face challenges due to the discomfort caused by tracheoscopy as an invasive procedure (Wu et al., 2020).
At present, new nucleic acid detection techniques have been used to detect pathogenic microorganisms. Droplet digital PCR (ddPCR) is a method of absolute nucleic acid quantification by using Poisson statistical analysis of the number of positive and negative droplets and has been proven to achieve higher accuracy and sensitivity than qPCR (Shirai et al., 2022). It has been used in a lot of clinical fields, such as liquid biopsies for cancer monitoring and noninvasive prenatal testing for genetic abnormality detection(Cainap et al., 2021; Boldrin et al., 2022). However, due to the testing of specimens, it is currently limited to the detection and diagnosis of bloodstream infections (Lin et al., 2023). It has great clinical value in patients with lung infections combined with bloodstream infections, especially because ddPCR detects the same pathogen in the blood as the lower respiratory tract secretions (Wu et al., 2022).
Acknowledging the study’s limitations is crucial. First, the presentation solely focuses on the findings of the examination into the presence of pathogenic microorganisms in the lower respiratory tract, leaving the judgment of whether they are pathogenic or colonized bacteria to clinicians. Second, considering that it was conducted in a single site retrospectively, there is a possibility that it was subjected to selection bias. Prospective and multicenter studies are necessary for further confirmation. Third, mNGS technology has some inherent flaws, with detection results prone to contamination by the kit-related background microorganisms and nucleic acids. Advancements and updates in mNGS technology are expected to address this issue.
Conclusions
In conclusion, mNGS demonstrates significant advantages in detecting pathogenic microorganisms in the lower respiratory tract of individuals suffering from pulmonary infections, offering valuable references for clinicians. As a novel molecular diagnostic tool, the ongoing innovation in mNGS technology is anticipated to provide even more substantial assistance to clinicians.
Data availability statement
The original contributions presented in the study are included in the article/supplementary materials. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Scientific Research and Clinical Trial Ethics Committee of the First Affiliated Hospital of Zhengzhou University (approval number: 2021-KY-0952-003). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
J-ZW: Writing – original draft. DY: Data curation, Investigation, Writing – review & editing. X-HY: Funding acquisition, Project administration, Writing – review & editing. C-HS: Methodology, Resources, Writing – review & editing. L-LH: Data curation, Resources, Writing – review & editing. YZ: Data curation, Formal analysis, Writing – review & editing. Y-XG: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Sub-projects of major National Science and Technology Projects (2017ZX10103005-009), The Key Scientific Research Projects of Institutions of Higher Learning in Henan Province (20A320046), Zhengzhou Science and Technology Program for Public Wellbeing (2020KJHM0001), The Project of Primary Health Care Foundation of China (YLGX-JZ-2020001), and Peking Union Medical Foundation-Rui E (Rui Yi) Emergency Medical Research Special Fund (R2020014).
Acknowledgments
We would like to thank all participants for their contributions to this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Afshinnekoo, E., Chou, C., Alexander, N., Ahsanuddin, S., Schuetz, A. N., Mason, C. E. (2017). Precision metagenomics: rapid metagenomic analyses for infectious disease diagnostics and public health surveillance. J. Biomol Tech. 28 (1), 40–45. doi: 10.7171/jbt.17-2801-007
Ai, J. W., Zhang, Y., Zhang, H. C., Xu, T., Zhang, W. H. (2020). Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned. Emerg. Microbes Infect. 9 (1), 597–600. doi: 10.1080/22221751.2020.1738905
Biswas, C., Chen, S. C., Halliday, C., Martinez, E., Rockett, R. J., Wang, Q., et al. (2017). Whole genome sequencing of candida glabrata for detection of markers of antifungal drug resistance. J. Vis. Exp. 28(130), 56714. doi: 10.3791/56714
Boldrin, E., Mazza, M., Piano, M. A., Alfieri, R., Montagner, I. M., Magni, G., et al. (2022). Putative Clinical Potential of ERBB2 Amplification Assessment by ddPCR in FFPE-DNA and cfDNA of Gastroesophageal Adenocarcinoma Patients. Cancers (Basel). 14 (9), 2180. doi: 10.3390/cancers14092180
Cainap, C., Balacescu, O., Cainap, S. S., Pop, L. A. (2021). Next generation sequencing technology in lung cancer diagnosis. Biol. (Basel). 10 (9), 864. doi: 10.3390/biology10090864
Caliendo, A. M., Gilbert, D. N., Ginocchio, C. C., Hanson, K. E., May, L., Quinn, T. C., et al. (2013). Better tests, better care: improved diagnostics for infectious diseases. Clin. Infect. Dis. 57 Suppl 3, S139–S170. doi: 10.1093/cid/cit578
Capobianchi, M. R., Giombini, E., Rozera, G. (2013). Next-generation sequencing technology in clinical virology. Clin. Microbiol. Infect. 19 (1), 15–22. doi: 10.1111/1469-0691.12056
Chan, W. S., Au, C. H., Leung, S. M., Ho, D. N., Wong, E. Y. L., To, M. Y., et al. (2020). Potential utility of targeted Nanopore sequencing for improving etiologic diagnosis of bacterial and fungal respiratory infection. Diagn. Pathol. 15 (1), 41. doi: 10.1186/s13000-020-00960-w
Chen, L., Liu, W., Zhang, Q., Xu, K., Ye, G., Wu, W., et al. (2020). RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 9 (1), 313–319. doi: 10.1080/22221751.2020.1725399
Chen, Y., Feng, W., Ye, K., Guo, L., Xia, H., Guan, Y., et al. (2021). Application of metagenomic next-generation sequencing in the diagnosis of pulmonary infectious pathogens from bronchoalveolar lavage samples. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.541092
Dickson, R. P., Erb-Downward, J. R., Freeman, C. M., McCloskey, L., Falkowski, N. R., Huffnagle, G. B., et al. (2017). Bacterial topography of the healthy human lower respiratory tract. mBio 8 (1), e02287-16. doi: 10.1128/mBio.02287-16
Duan, H., Li, X., Mei, A., Li, P., Liu, Y., Li, X., et al. (2021). The diagnostic value of metagenomic next rectanglegeneration sequencing in infectious diseases. BMC Infect. Dis. 21 (1), 62. doi: 10.1186/s12879-020-05746-5
Erb-Downward, J. R., Thompson, D. L., Han, M. K., Freeman, C. M., McCloskey, L., Schmidt, L. A., et al. (2011). Analysis of the lung microbiome in the "healthy" smoker and in COPD. PloS One 6 (2), e16384. doi: 10.1371/journal.pone.0016384
Gu, W., Miller, S., Chiu, C. Y. (2019). Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. 14, 319–338. doi: 10.1146/annurev-pathmechdis-012418-012751
Huang, J., Jiang, E., Yang, D., Wei, J., Zhao, M., Feng, J., et al. (2020). Metagenomic next-generation sequencing versus traditional pathogen detection in the diagnosis of peripheral pulmonary infectious lesions. Infect. Drug Resist. 13, 567–576. doi: 10.2147/idr.S235182
Jain, S., Self, W. H., Wunderink, R. G., Fakhran, S., Balk, R., Bramley, A. M., et al. (2015). Community-acquired pneumonia requiring hospitalization among U.S. Adults. N Engl. J. Med. 373 (5), 415–427. doi: 10.1056/NEJMoa1500245
Kalil, A. C., Metersky, M. L., Klompas, M., Muscedere, J., Sweeney, D. A., Palmer, L. B., et al. (2016). Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the infectious diseases society of America and the American thoracic society. Clin. Infect. Dis. 63 (5), e61–e111. doi: 10.1093/cid/ciw353
Kamps, R., Brandão, R. D., Bosch, B. J., Paulussen, A. D., Xanthoulea, S., Blok, M. J., et al. (2017). Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int. J. Mol. Sci. 18 (2), 308. doi: 10.3390/ijms18020308
Kauffman, C. A. (2015). Pulmonary and invasive fungal infections. Semin. Respir. Crit. Care Med. 36 (5), 639–640. doi: 10.1055/s-0035-1562923
Langelier, C., Zinter, M. S., Kalantar, K., Yanik, G. A., Christenson, S., O'Donovan, B., et al. (2018). Metagenomic sequencing detects respiratory pathogens in hematopoietic cellular transplant patients. Am. J. Respir. Crit. Care Med. 197 (4), 524–528. doi: 10.1164/rccm.201706-1097LE
Lin, K., Zhao, Y., Xu, B., Yu, S., Fu, Z., Zhang, Y., et al. (2023). Clinical diagnostic performance of droplet digital PCR for suspected bloodstream infections. Microbiol. Spectr. 11 (1), e0137822. doi: 10.1128/spectrum.01378-22
Magill, S. S., Edwards, J. R., Bamberg, W., Beldavs, Z. G., Dumyati, G., Kainer, M. A., et al. (2014). Multistate point-prevalence survey of health care-associated infections. N Engl. J. Med. 370 (13), 1198–1208. doi: 10.1056/NEJMoa1306801
McCombie, W. R., McPherson, J. D., Mardis, E. R. (2019). Next-generation sequencing technologies. Cold Spring Harb. Perspect. Med. 9 (11), a036798. doi: 10.1101/cshperspect.a036798
Mellis, R., Chandler, N., Chitty, L. S. (2018). Next-generation sequencing and the impact on prenatal diagnosis. Expert Rev. Mol. Diagn. 18 (8), 689–699. doi: 10.1080/14737159.2018.1493924
Metlay, J. P., Waterer, G. W., Long, A. C., Anzueto, A., Brozek, J., Crothers, K., et al. (2019). Diagnosis and treatment of adults with community-acquired pneumonia. An official clinical practice guideline of the American thoracic society and infectious diseases society of America. Am. J. Respir. Crit. Care Med. 200 (7), e45–e67. doi: 10.1164/rccm.201908-1581ST
Miao, Q., Ma, Y., Wang, Q., Pan, J., Zhang, Y., Jin, W., et al. (2018). Microbiological diagnostic performance of metagenomic next-generation sequencing when applied to clinical practice. Clin. Infect. Dis. 67 (suppl_2), S231–s240. doi: 10.1093/cid/ciy693
Parker, J., Chen, J. (2017). Application of next generation sequencing for the detection of human viral pathogens in clinical specimens. J. Clin. Virol. 86, 20–26. doi: 10.1016/j.jcv.2016.11.010
Sanmiguel, P. (2011). Next-generation sequencing and potential applications in fungal genomics. Methods Mol. Biol. 722, 51–60. doi: 10.1007/978-1-61779-040-9_4
Shirai, R., Osumi, T., Sato-Otsubo, A., Nakabayashi, K., Ishiwata, K., Yamada, Y., et al. (2022). Quantitative assessment of copy number alterations by liquid biopsy for neuroblastoma. Genes Chromosomes Cancer. 61 (11), 662–669. doi: 10.1002/gcc.23073
Simner, P. J., Miller, S., Carroll, K. C. (2018). Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin. Infect. Dis. 66 (5), 778–788. doi: 10.1093/cid/cix881
Torres, A., Chalmers, J. D., Dela Cruz, C. S., Dominedò, C., Kollef, M., Martin-Loeches, I., et al. (2019). Challenges in severe community-acquired pneumonia: a point-of-view review. Intensive Care Med. 45 (2), 159–171. doi: 10.1007/s00134-019-05519-y
Wang, J., Han, Y., Feng, J. (2019). Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm Med. 19 (1), 252. doi: 10.1186/s12890-019-1022-4
Wu, X., Li, Y., Zhang, M., Li, M., Zhang, R., Lu, X., et al. (2020). Etiology of severe community-acquired pneumonia in adults based on metagenomic next-generation sequencing: A prospective multicenter study. Infect. Dis. Ther. 9 (4), 1003–1015. doi: 10.1007/s40121-020-00353-y
Wu, J., Tang, B., Qiu, Y., Tan, R., Liu, J., Xia, J., et al. (2022). Clinical validation of a multiplex droplet digital PCR for diagnosing suspected bloodstream infections in ICU practice: a promising diagnostic tool. Crit. Care 26 (1), 243. doi: 10.1186/s13054-022-04116-8
Zeng, Z. R., Tian, G., Ding, Y. H., Yang, K., Liu, J. B., Deng, J. (2019). Surveillance study of the prevalence, species distribution, antifungal susceptibility, risk factors and mortality of invasive candidiasis in a tertiary teaching hospital in Southwest China. BMC Infect. Dis. 19 (1), 939. doi: 10.1186/s12879-019-4588-9
Keywords: pulmonary infection, antibiotics, etiology, metagenomic next-generation sequencing, lower respiratory tract
Citation: Wang J-z, Yuan D, Yang X-h, Sun C-h, Hou L-l, Zhang Y and Gao Y-x (2024) Etiology of lower respiratory tract in pneumonia based on metagenomic next-generation sequencing: a retrospective study. Front. Cell. Infect. Microbiol. 13:1291980. doi: 10.3389/fcimb.2023.1291980
Received: 10 September 2023; Accepted: 13 December 2023;
Published: 09 January 2024.
Edited by:
Beiwen Zheng, Zhejiang University, ChinaReviewed by:
Yu-Lei Gao, Tianjin Medical University General Hospital, ChinaChengqi Xin, Dalian Medical University, China
Copyright © 2024 Wang, Yuan, Yang, Sun, Hou, Zhang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-xia Gao, Z2FveWFueGlhenp1QDE2My5jb20=
 Jin-zhu Wang
Jin-zhu Wang Ding Yuan1
Ding Yuan1