
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 31 October 2023
Sec. Bacteria and Host
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1286842
This article is part of the Research TopicRole of Microbes in Ocular Surface Health and DiseasesView all 8 articles
 Robert M. Q. Shanks1*
Robert M. Q. Shanks1* Sarah Atta1
Sarah Atta1 Nicholas A. Stella1
Nicholas A. Stella1 Chollapadi V. Sundar-Raj1
Chollapadi V. Sundar-Raj1 John E. Romanowski1
John E. Romanowski1 Arman S. Grewal1
Arman S. Grewal1 Hazel Q. Shanks1
Hazel Q. Shanks1 Sonya M. Mumper1
Sonya M. Mumper1 Deepinder K. Dhaliwal1
Deepinder K. Dhaliwal1 Alex Mammen1
Alex Mammen1 Jake D. Callaghan1
Jake D. Callaghan1 Rachel C. Calvario1
Rachel C. Calvario1 Eric G. Romanowski1
Eric G. Romanowski1 Regis P. Kowalski1
Regis P. Kowalski1 Michael E. Zegans2,3
Michael E. Zegans2,3 Vishal Jhanji1
Vishal Jhanji1Introduction: Pseudomonas aeruginosa causes vision threatening keratitis. The LasR transcription factor regulates virulence factors in response to the quorum sensing molecule N-3-oxo-dodecanoyl-L-homoserine lactone. P. aeruginosa isolates with lasR mutations are characterized by an iridescent high sheen phenotype caused by a build-up of 2-heptyl-4-quinolone. A previous study demonstrated 22% (n=101) of P. aeruginosa keratitis isolates from India between 2010 and 2016 were sheen positive lasR mutants, and the sheen phenotype correlated with worse clinical outcomes for patients. In this study, a longitudinal collection of P. aeruginosa keratitis isolates from Eastern North America were screened for lasR mutations by the sheen phenotype and sequencing of the lasR gene.
Methods: Keratitis isolates (n=399) were classified by sheen phenotype. The lasR gene was cloned from a subset of isolates, sequenced, and tested for loss of function or dominant-negative status based on an azocasein protease assay. A retrospective chart review compared outcomes of keratitis patients infected by sheen positive and negative isolates.
Results: A significant increase in sheen positive isolates was observed between 1993 and 2021. Extracellular protease activity was reduced among the sheen positive isolates and a defined lasR mutant. Cloned lasR alleles from the sheen positive isolates were loss of function or dominant negative and differed in sequence from previously reported ocular lasR mutant alleles. Retrospective analysis of patient information suggested significantly better visual outcomes for patients infected by sheen positive isolates.
Discussion: These results indicate an increase in lasR mutations among keratitis isolates in the United States and suggest that endemic lasR mutants can cause keratitis.
The bacterium Pseudomonas aeruginosa is the most frequent cause of contact lens associated microbial keratitis and is of concern because keratitis caused by P. aeruginosa has rapid progression and poor clinical outcomes (Shen et al., 2014; Vazirani et al., 2015). P. aeruginosa keratitis isolates resistant to fluoroquinolones and other antibiotics typically used to treat keratitis have been reported (Alexandrakis et al., 2000; Rhee et al., 2004; Bispo et al., 2022; Khan et al., 2023). Microbial keratitis caused by antibiotic resistant P. aeruginosa isolates correlates with worse clinical outcomes including an increase in corneal perforations from 12% of cases with normal P. aeruginosa to 52% with multidrug-resistant (MDR) P. aeruginosa (Vazirani et al., 2015; Fernandes et al., 2016). Beyond resistance, P. aeruginosa has numerous virulence factors associated with establishing corneal infections (Pearlman et al., 2013; Hilliam et al., 2020). These include pathogen-associated molecular pattern (PAMP) such as lipopolysaccharide and flagellin (Fleiszig and Evans, 2002; Willcox, 2007), a variety of proteases including elastases (LasA and LasB) and Pseudomonas aeruginosa Small Protease (PASP) (O'Callaghan et al., 2019; Hilliam et al., 2020), and the type III secretion system that are important for virulence in experimental models (Zhu et al., 2006; Evans and Fleiszig, 2013; Pearlman et al., 2013; Hilliam et al., 2020). These virulence factors are highly regulated through multiple transcriptional regulators including the LasR quorum sensing master regulator that mediates population density collective responses (Lee and Zhang, 2015; Moradali et al., 2017). Likely the most studied among these in P. aeruginosa is the LasR transcription factor. LasR responds to quorum sensing molecule N-(3-oxododecanoyl) homoserine lactone, controls a large portion of the P. aeruginosa genome, and is an important regulator of pathogenesis in lung and burn infection models (Rumbaugh et al., 1999; Pearson et al., 2000; Lesprit et al., 2003), as it positively regulates a number of pro-virulence factors including elastase proteases LasA and LasB, and rhamnolipids (Kariminik et al., 2017; Moradali et al., 2017).
A prior study on the keratitis isolates of P. aeruginosa from the Steroids for Corneal Ulcers Trial (SCUT) (Hammond et al., 2016) reported a colony iridescent sheen positive phenotype in 22 of the 101 isolates taken during the course of the study from India (Hammond et al., 2016). This sheen phenotype correlated with significantly worse visual outcomes. These included significantly reduced visual acuity and infiltrate/scar size for patients infected sheen isolates compared to typical P. aeruginosa (Hammond et al., 2016). The basis for the sheen phenotype has been shown to be due to mutation of the lasR transcription factor gene (D'Argenio et al., 2007). LasR is a positive regulator of the gene pqsH, which codes for an enzyme that converts 2-heptyl-4-quinolone (HHQ) to heptyl-3-hydroxy-4(1H)-quinone (PQS) (D'Argenio et al., 2007). In the absence of LasR function, HHQ builds up in the cell and creates the sheen phenotype. PQS is an important signaling molecule known as Pseudomonas quinolone signal (Garcia-Reyes et al., 2020). Surprisingly only two nonsynonymous mutations in the lasR gene were detected in 21 out of 22 sequenced sheen positive isolates suggesting that the mutations were already present in strains endemic to the country (India). By contrast, in chronic lung infections, such as those associated with cystic fibrosis, P. aeruginosa are frequently observed to gain mutations in lasR (Smith et al., 2006; D'Argenio et al., 2007). In the airway, patients are thought to be initially infected by wild-type (WT) P. aeruginosa, and lasR mutants can then increase over time (Kohler et al., 2009). Like keratitis patients, cystic fibrosis patients infected with lasR mutants have been recorded to experience worse disease progression compared to patients infected by wild-type P. aeruginosa (Hoffman et al., 2009). Together these prior studies suggest that sheen isolates are associated with worse clinical outcomes.
In the SCUT study, all of the sheen isolates tested were isolated between 2006-2010 and caused largely by isolates with one of two lasR mutations (Hammond et al., 2016). This study sought to determine whether sheen isolates were a general phenomenon among P. aeruginosa keratitis isolates or a geographically isolated observation and, if wide-spread, whether the same mutant alleles of lasR were present and associated with worse visual outcomes. Here we found a concerning increase in lasR mutants among the keratitis isolates taken in a tertiary care hospital in the Eastern United States. Data suggests that the mutations were highly variable with one exception, being an insertion element present in several strains; moreover, retrospective analysis suggests that patients with lasR mutations had better visual outcomes contrary to the former study.
De-identified P. aeruginosa strains isolated from the corneas of patients with keratitis were retrieved from a clinical tissue bank which is used for validation of diagnostic testing and antibiotic evaluation. The P. aeruginosa isolates were collected from 1993 through 2021 by The Charles T. Campbell Ophthalmic Microbiology Laboratory at the University of Pittsburgh School of Medicine and stored at -80°C. P. aeruginosa isolates were plated on tryptic soy agar with 5% sheep’s red blood cells and incubated for 18-20 hours at 37°C and sheen phenotype was established visually.
The lasR gene was deleted from strain PaC (Kowalski et al., 2001) using allelic exchange with plasmid pMQ30, as previously described (Shanks et al., 2006). The plasmid was modified with a ΔlasR allele cloned from PA14 ΔlasR (Hammond et al., 2016) to generate pMQ767. Primers to amplify the lasR region (approximately 500 bp upstream and downstream of the ORF) were 4835 and 4836 and listed in Table 1. The resultant strain was verified by PCR and whole genome sequencing. The lasR ORF was amplified by PCR and cloned into shuttle vector pMQ132 under control of the Escherichia coli lac promoter using yeast homologous recombination as previously described (Shanks et al., 2009). Primers to amplify the lasR ORF were 3217 and 3218 (Table 1). Plasmids were sequenced at the University of Pittsburgh Genomics Core or PlasmidSaurus.
Milk agar plates were made with brain heart infusion and skim milk as previously described (Sokol et al., 1979). Bacteria were incubated at 37°C for 24 hours and zones of clearing were measured on at least three separate occasions. For more quantitative analysis azocasein was used as previously described (Shanks et al., 2013).
The minimum inhibitory concentrations (MICs) of Pseudomonas aeruginosa keratitis isolates were determined to ciprofloxacin (CIP), tobramycin (TOB), and ceftazidime (CAZ) using MIC strips (Fisher Scientific, LIOFILCHEM, MA) on Mueller-Hinton II agar (BD BBL) agar as previously described (Harshaw et al., 2021). The keratitis isolates tested, taken from the cornea, were chosen arbitrarily out of the deidentified strain bank. The isolates in question were collected anonymously from 1993 to 2021 by the Charles T. Campbell Ophthalmic Microbiology Laboratory and stored at -80°C. The antibiotic susceptibility was determined by comparing the MIC of each to the Clinical and Laboratory Standards Institute breakpoints (Humphries et al., 2021).
Retrospective review of medical records of all patients diagnosed with culture-positive P. aeruginosa keratitis at the University of Pittsburgh Medical Center between 2017 and 2021 was performed. The study was approved by the Institutional Review Board of the University of Pittsburgh and followed the tenets of the Declaration of Helsinki. Clinical data were collected for each patient, including clinical features, treatment, and outcomes. Demographic features were recorded, including gender and age. Visual acuity was recorded at presentation and after resolution. The visual outcomes were BCVA on the Snellen chart and converted to LogMAR.
Graph-pad Prism was used to perform Mann-Whitney and ANOVA analysis with Tukey’s post-test, chi-square, and Fisher’s exact tests. P<0.05 was considered significant.
P. aeruginosa keratitis isolates between 1993-2021 were evaluated for sheen phenotypes on blood agar plates (Figure 1A). The isolate collection and storage approach remained consistent over that time period. A notable increase in sheen positive isolates was observed over the time frame going from 0% between 1993-1997 to 26.2% between 2018-2021 (Figure 1B). All time frames tested were significantly different from 1993-1997 by Fisher’s Exact and chi-square Test and (p<0.01).
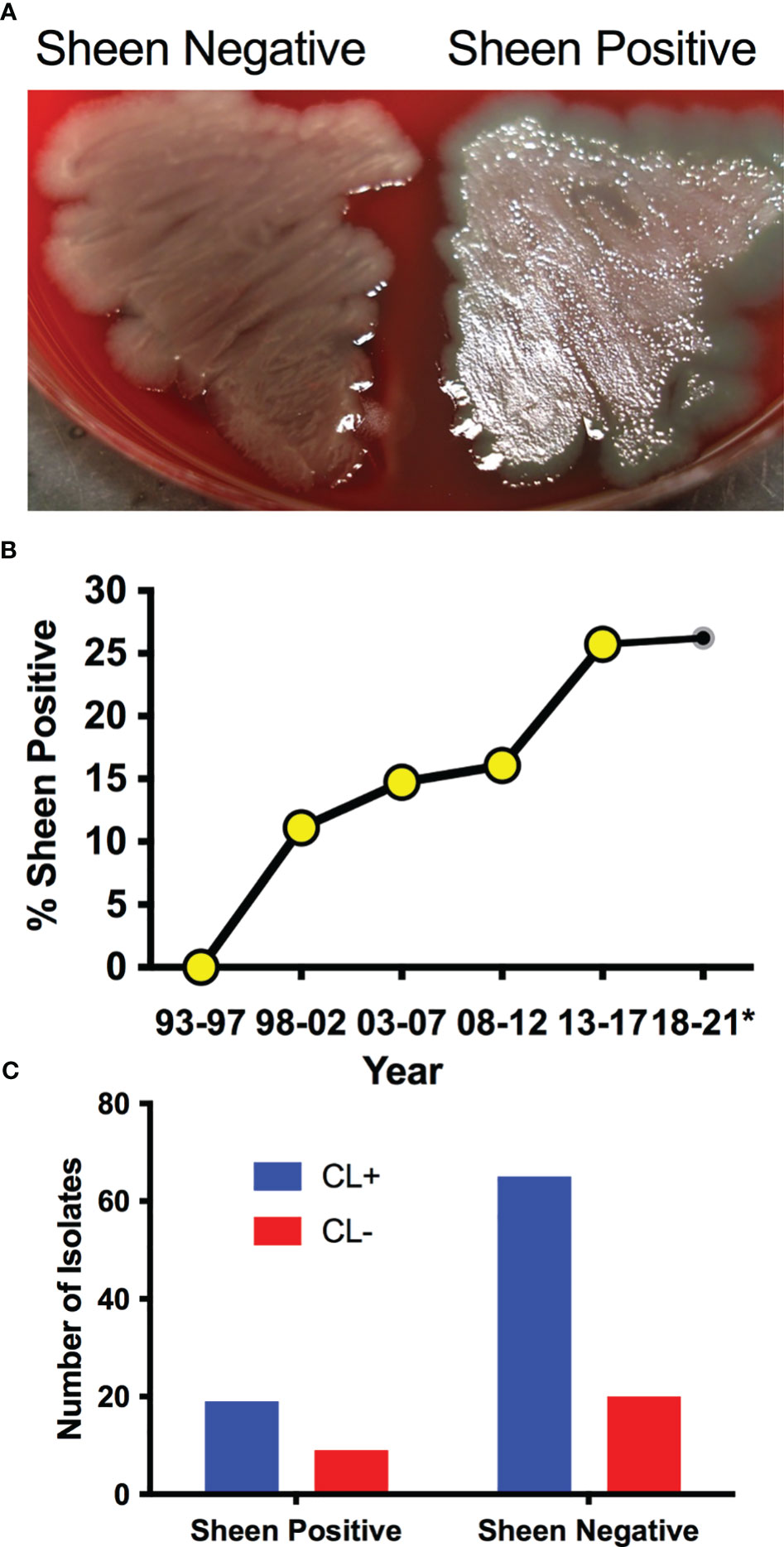
Figure 1 P. aeruginosa keratitis isolates with a sheen phenotype are increasing over the last two decades. (A) Appearance of sheen negative and positive P. aeruginosa keratitis isolates on blood agar. (B) Frequency of PA keratitis isolates with sheen positive phenotype. 5 year periods are shown except for 2018-2021 as denoted by the asterisk. n=399, n≥50 per time period. (C) Correlation between contact lens (CL) use and sheen status. p=0.455.
Contact lens use is the major risk factor for P. aeruginosa keratitis (Green et al., 2019; Enzor et al., 2021; Ung and Chodosh, 2021). Where possible, the contact lens use status of the patient was correlated with the sheen phenotype and no significant difference was observed (p=0.45 by Fisher’s Exact Test) with 23% sheen positive isolates from contact lens associated keratitis (19/84) and 31% sheen positive isolates from non-contact lens associated keratitis (9/29) (Figure 1C).
We also screened a small library of fluoroquinolone resistant isolates from keratitis patients in the New York City area obtained before 2001 (Kowalski et al., 2001) and found that two out of six where sheen positive (33.3%), further suggesting that sheen positive ocular clinical isolates are a general rather than geographically isolated phenomenon.
The lasR allele from a subset of arbitrarily chosen strains were cloned into a shuttle vector and sequenced. The prior study with patients from India reported multiple independent isolates with two specific LasR variants I215S (14/22) and P117L (7/22), and one having a missense mutation yielding V221L (1/22) (Hammond et al., 2016). By contrast, our study did not find these mutations and found a wider variety of alterations in the lasR sequence. We cloned and sequenced the lasR open reading frame (ORF) from 27 strains including 4 from sheen negative isolates and 23 from sheen positive isolates (Table 2). Of the sheen positive strains, 1 had a wild-type lasR sequence, others had amino acid substitutions, deletions of insertions that created out of frame mutations, premature stop codons, and insertion elements. Strikingly, 4 of 6 with insertion elements had identical insertions at base pair 126 despite being found over 20 years.
It has been reported that lasR isolates have altered susceptibility to a variety of antibiotics (Hoffman et al., 2010; Qi et al., 2016; Abisado-Duque et al., 2023). To test whether sheen status had an impact on susceptibility to the major topical antibiotics for P. aeruginosa keratitis (Lin et al., 2019), we evaluated minimum inhibitory concentrations from keratitis isolates (Table 3). Drugs from three different antibiotic classes were evaluated. There was no difference in the percent susceptible to the fluoroquinolone ciprofloxacin for sheen positive versus negative isolates (92.5% compared to 93.5% respectively, p=0.29 Fisher’s Exact test). For the aminoglycoside tobramycin, there was a non-significant trend toward a higher percentage of susceptible isolates in the sheen negative group which was 10% higher than the sheen positive group (p=0.23). While the cephalosporin ceftazidime is not used as commonly for keratitis, it has been used successfully used to treat keratitis and has been suggested as an alternative for treatment of aminoglycoside and fluoroquinolone resistant isolates (Rhee et al., 2004; Dehghani et al., 2009; Subedi et al., 2018). For ceftazidime, the sheen negative isolates had a higher frequency of susceptibility than the sheen positive isolates (98.1% vs 85.3% respectively, p=0.01).
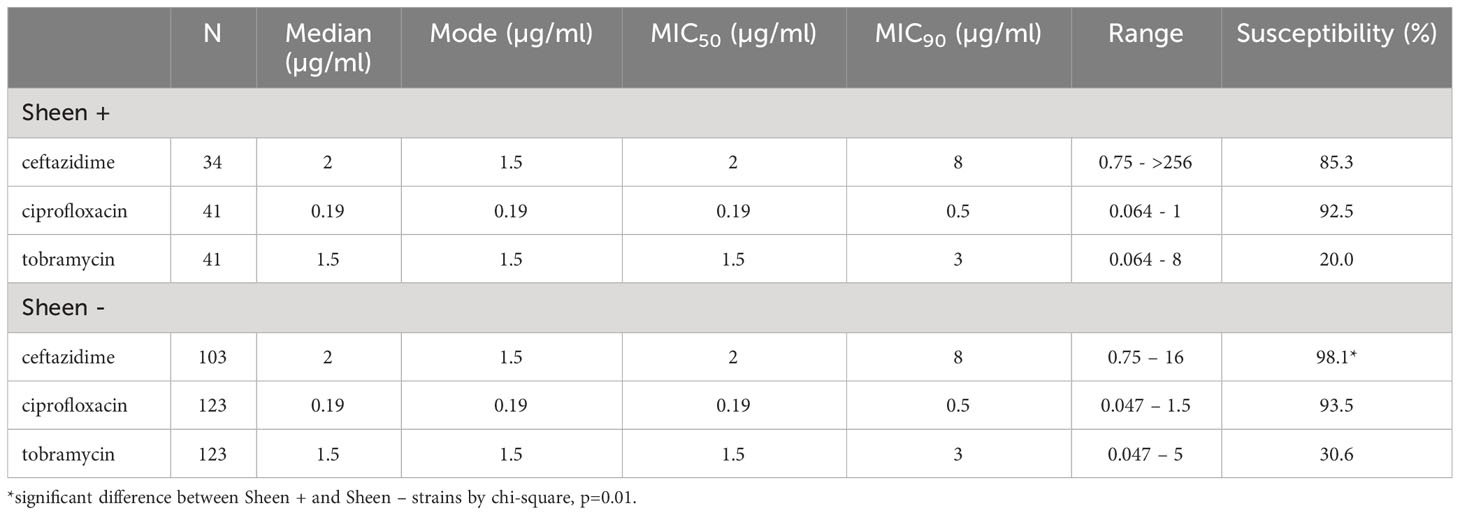
Table 3 Descriptive statistics of minimum inhibitory concentrations (MICs) for Pseudomonas aeruginosa keratitis isolates to ceftazadime, ciprofloxacin, and tobramycin.
LasR is a known regulator of elastase B (lasB) and other proteases thought to aid the bacteria in microbial keratitis (Gambello and Iglewski, 1991). The protease activity of arbitrarily chosen keratitis isolates (14 sheen positive and 60 sheen negative) was assessed by measuring the zone of clearance on milk agar plates (Figure 2A). The sheen negative isolates had a zone of 5.6 mm versus 2.7 mm for the sheen positive isolate (p<0.0001, Mann-Whitney test).
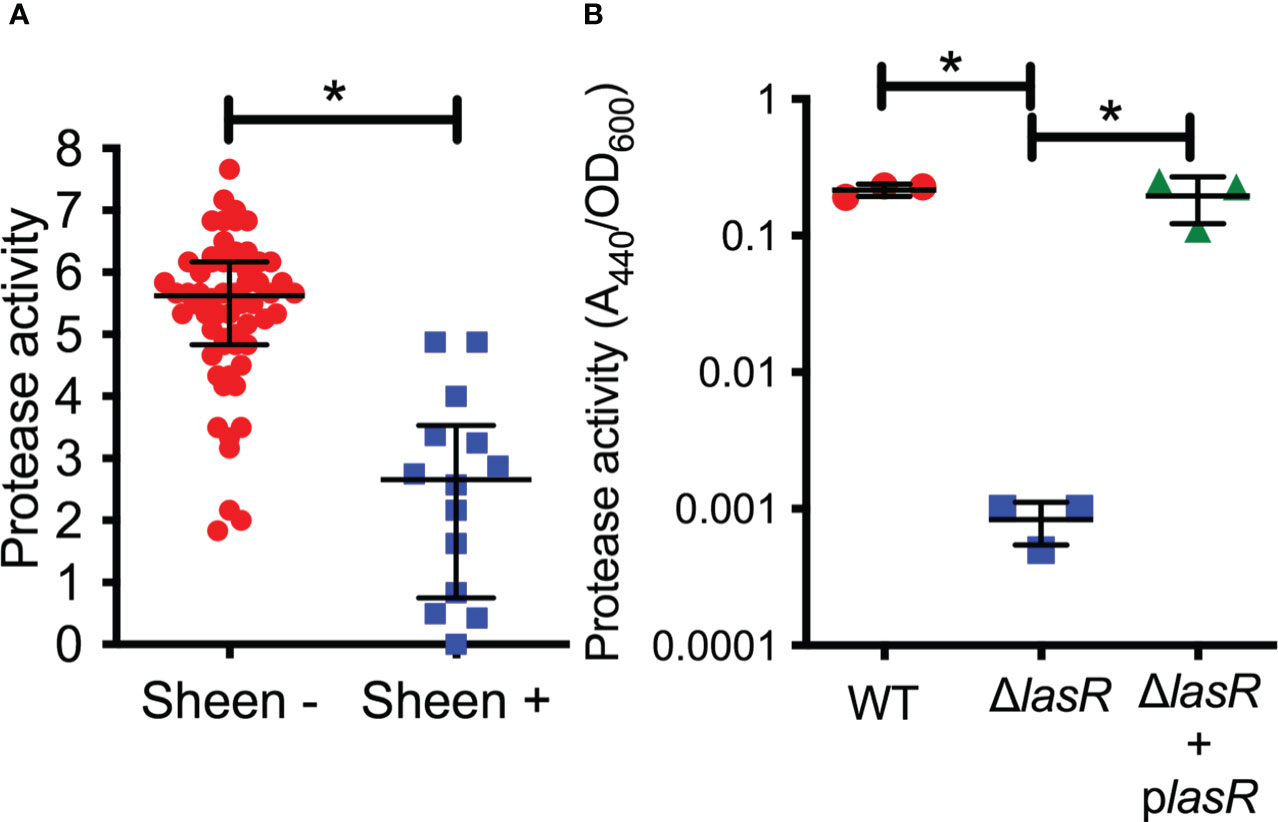
Figure 2 Secreted protease activity is reduced among sheen positive PA keratitis isolates and can be complemented. (A) Secreted protease activity by PA keratitis isolates. The zone of clearance (mm) on milk plate assay is shown. Each data point indicates the mean zone of clearance for an individual isolate. Medians and IQ ranges are shown. *, p<0.001 by Mann-Whitney. (B) Secreted protease activity measured by azocasein and normalized by bacterial density from sterile culture filtrates from the clinical isolate PAC (WT) and isogenic ΔlasR mutant and the mutant with wild-type lasR on a plasmid. The bacteria were grown in LB medium and were harvested at OD600 = 2. Asterisks indicate p<0.01 between indicated groups by ANOVA with Tukey’s post-test, n=3, mean and SD are shown.
To test this further, we generated a lasR deletion mutation in strain PaC (Kowalski et al., 2001) which is a fluoroquinolone resistant keratitis isolate. The PaC ΔlasR mutant was more than 100-fold reduced in protease activity compared to the wild type as measured using azocasein and this defect could be complemented by adding the wild-type lasR gene back on a plasmid (Figure 2B).
While the sheen phenotype is linked to lasR loss of function mutations, the sheen phenotype can occur due to other mutations such as mutation of the pqsH gene which converts HHQ to PQS (Diggle et al., 2006; O'Connor et al., 2022). Several of the cloned lasR mutants were tested for function by expression in the PaC ΔlasR mutant followed by protease evaluation as a surrogate for LasR function (Figure 3A). Wild-type alleles from PA14 and K2361 restored wild-type levels of protease activity, whereas the lasR alleles cloned from sheen positive strains had highly reduced protease activity (Figure 3A).
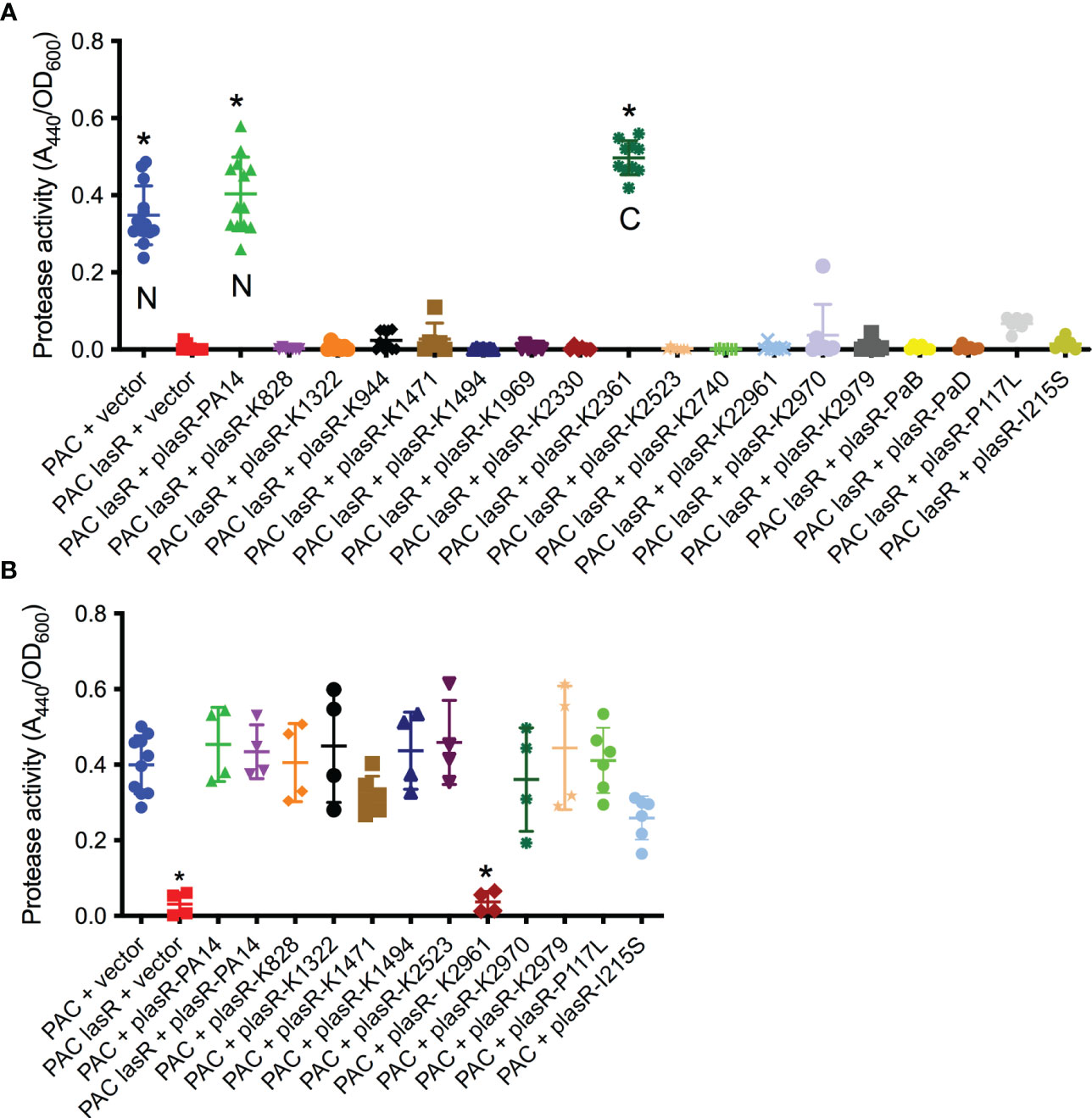
Figure 3 Tested lasR alleles from sheen positive isolates were loss of function and generally not dominant negative. (A, B) Protease activity in supernatants from overnight cultures (18-20h) grown in LB medium was measured using azocasein and normalized by culture density. n≥6, median and standard deviation is shown. (A) The vector alone negative control (pMQ132), wild-type lasR plasmid (plasR-PA14) positive control, and candidate plasmids were expressed in the wild-type PaC to establish base level or PaC ΔlasR to determine lasR function. “N” indicates a lasR allele from a sheen negative strain. “C” indicates lasR from a sheen positive isolate with no amino acid changes in the ORF transcript. Asterisk indicates p<0.001 by ANOVA with Tukey’s post-test. (B) As A, but plasmids were tested in the wild-type PaC to detect dominant negative activity. Asterisk indicates difference from PaC + vector by ANOVA, p<0.01.
Genes for the major two LasR variants I215S (14/22) and P117L (7/22) from the SCUT isolates were also cloned as above, moved into the PaC ΔlasR mutant and based on protease activity, both were loss of function mutations (Figure 3A). The PI117L mutant appeared to maintain some activity maintaining 16.6% of wild-type protease activity and 17-fold higher than the vector alone negative control, but was not significantly different than the vector alone negative control. The I215S allele allowed only 3.6-fold higher protease levels than the vector alone negative control.
The dominant negative status of the lasR alleles was also tested by expression in the wild-type PaC strain (Figure 3B). Notably, the K2961 allele strongly reduced protease activity in the PaC wild type, whereas the other tested lasR alleles did not significantly alter protease activity. The PaC strain with the lasR_K2961 allele expressed on a plasmid was remade to ensure that the effect was not artifactual and the reduced protease phenotype was again observed. A similar, although less severe reduction in protease activity was measured when the lasR_K2961 allele was expressed in the PA14 wild-type strain (Figure 3B). When the previously described lasR alleles from the India isolates were expressed at multicopy in the wild type bacteria, the P117L allele had no effect, but the I215S may have a dominant negative effect with a 43% reduction compared to the wild type expressing the PA14 lasR allele (Figure 3B).
Where possible the clinical outcomes of patients were determined from clinical records with n=49 for sheen negative isolates and n=19 for sheen positive. Of all of the evaluated variables only final average vision was significantly different (p=0.0106) (Table 4). Surprisingly, unlike the prior study evaluating lasR mutant associated keratitis (Hammond et al., 2016), the average visual outcomes were favorable for patients infected with the sheen positive isolates. Though not significantly different, the most severe outcomes, corneal transplants and enucleation, were absent in the sheen positive infected eyes, further suggesting reduced virulence by the sheen positive isolates.
This study has demonstrated an increase in sheen positive P. aeruginosa keratitis isolates in a tertiary care hospital in the Eastern United States. This suggests that the abundance of sheen positive isolates is a general rather than a regional phenomenon. The reason for the increase observed in this study was not clear. The clinical microbiologist that collected the samples maintained the same collection protocol over the period of isolate collection, so differences in this would not account for the increase in sheen isolates.
Another consideration was whether the sheen positive isolates from our study are lasR mutants. The majority (22 out of 24) of sheen isolates that were sequenced had changes in the lasR sequence and those tested did not code for functional proteins. Because only the ORFs were cloned, other mutations that render the strain LasR-deficient could be missed, for example, promoter mutations. However, it is formally possible that in a small subset of the keratitis isolates, LasR-independent changes could be responsible for a subset of the sheen isolates. Therefore, we conclude that the majority if not all sheen positive keratitis isolates have defects in LasR function.
Whereas the sheen phenotype appears to be a good predictor of lasR mutant status, it has been observed that some lasR mutants do not manifest the sheen phenotype (Feltner et al., 2016; Mould et al., 2022). In this study the lasR gene was sequenced from three sheen negative isolates and all three had WT sequences; two were used to complement a ΔlasR strain for protease production and showed functional complementation. However, the protease levels from 7 out of 60 sheen negative isolates were close to the median for the sheen positive isolates suggesting that ~10% of the sheen negative isolates are defective in LasR function. Therefore, our data suggest that the sheen negative phenotype may have around a 10% error rate for predicting a functional LasR protein.
The Hammond study indicated worse visual outcomes for patients with sheen positive isolates (Hammond et al., 2016), by contrast patients in this study had strikingly better visual acuity, as well as no incidence of the severe outcomes of enucleation and corneal transplantation that were present in the sheen negative infected patients. The reason for this discrepancy is not clear, but could possibly be due to the different strains or the specific mutations associated with strains isolated in the SCUT study. Moreover, the SCUT study used a standard protocol for timing and methodology for obtaining visual acuity measurements, which lends more weight to that analysis.
Nevertheless, the reduced severity observed in the clinical data from this study were consistent with a recent paper using a rabbit corneal infection model that demonstrated reduced corneal perforation and bacterial proliferation of a lasR deletion mutant of strain PA14 compared to an isogenic wild type suggesting that LasR promotes keratitis severity (Romanowski et al., 2023). Studies with mice show mixed results with strain PA01 with lasR deletion mutations. The Pier group reported that C3H/HeN mice with scarified corneas required fewer lasR mutant bacteria to cause keratitis compared to the wild type (Preston et al., 1997). Whereas the Willcox group used the same bacterial strains with BALB/c mice and found indistinguishable infection frequencies for both bacterial strains, but reported that bacterial proliferation and severity scores were reduced in eyes infected with the lasR mutant (Preston et al., 1997).
Protease activity was used in this study as a reporter for LasR function; however, the reduction of protease production by the lasR mutants may contribute to reduced virulence. Several studies have linked extracellular proteases to the ability of P. aeruginosa to spread and proliferate in the cornea and to alter the immune response (Matsumoto, 2004; Willcox, 2007; O'Callaghan et al., 2019; Hilliam et al., 2020). For example, elastase injected into rabbit corneas causes corneal melting that can be prevented by elastase inhibitors (Pallares et al., 2021), though proteases are not absolutely necessary for P. aeruginosa to cause corneal inflammation (Hobden, 2002).
In conclusion, lasR mutant P. aeruginosa appear to be increasing among keratitis patients and this may be a world-wide phenomenon. The highly variable nature of the lasR mutations among isolates in our study does not unequivocally indicate whether the strains mutated during infection or prior to infection in general. The exception being identification of multiple isolates with a mutation at base pair 126 which suggests the existence of a regional endemic strain which is in agreement with a study by Hammond, et al (Hammond et al., 2016), which postulated that strains with existing lasR mutations initiate ocular infections. Interestingly, the identical insertion element in the same location was reported in the lasR gene from a P. aeruginosa isolated from a bean plant in Spain suggesting an environmental source (Ruiz-Roldan et al., 2021). Insertion elements interrupting the lasR ORF have also been observed among sheen positive isolates from young cystic fibrosis patients and other sources (Hoffman et al., 2009). Whether these lasR mutant strains are more or less virulent in keratitis is in question and more research is needed to determine the level of concern, however, our study suggests that the sheen positive lasR isolates are less pathogenic.
The original contributions presented in the study are included in the article/supplementary materials, further inquiries can be directed to the corresponding author.
The studies involving humans were approved by University of Pittsburgh Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
RS: Data curation, Formal Analysis, Funding acquisition, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing. SA: Data curation, Formal Analysis, Methodology, Writing – review & editing. NS: Formal Analysis, Investigation, Writing – review & editing. CS: Data curation, Formal Analysis, Investigation, Writing – review & editing. JR: Data curation, Investigation, Writing – review & editing. AG: Data curation, Formal Analysis, Investigation, Writing – review & editing. HS: Data curation, Formal Analysis, Investigation, Writing – review & editing. SM: Data curation, Formal Analysis, Investigation, Writing – review & editing. DD: Conceptualization, Methodology, Writing – review & editing. AM: Conceptualization, Investigation, Writing – review & editing. JC: Formal Analysis, Investigation, Methodology, Resources, Writing – review & editing. RC: Data curation, Formal Analysis, Methodology, Writing – review & editing. ER: Conceptualization, Formal Analysis, Investigation, Writing – review & editing. RK: Conceptualization, Funding acquisition, Resources, Writing – review & editing. MZ: Conceptualization, Formal Analysis, Methodology, Writing – original draft. VJ: Conceptualization, Data curation, Formal Analysis, Investigation, Writing – original draft.
The authors declare financial support was received for the research, authorship, and/or publication of this article. NEI provided funds for the study EY032517, EY08098. The Campbell Family foundation provided funds for experiments and publication fees. Research to Prevent Blindness and the Eye and Ear Foundation of Pittsburgh provided department funds to pay for infrastructure.
The authors thank Deborah Hogan at Dartmouth Medical School for strains.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abisado-Duque, R. G., Townsend, K. A., McKee, B. M., Woods, K., Koirala, P., Holder, A. J., et al. (2023). An amino acid substitution in elongation factor EF-G1A alters the antibiotic susceptibility of Pseudomonas aeruginosa lasR-null mutants. J. Bacteriol 205 (6), e0011423. doi: 10.1128/jb.00114-23
Alexandrakis, G., Alfonso, E. C., Miller, D. (2000). Shifting trends in bacterial keratitis in south Florida and emerging resistance to fluoroquinolones. Ophthalmology 107 (8), 1497–1502. doi: 10.1016/s0161-6420(00)00179-2
Bispo, P. J. M., Sahm, D. F., Asbell, P. A. (2022). A systematic review of multi-decade antibiotic resistance data for ocular bacterial pathogens in the United States. Ophthalmol. Ther. 11 (2), 503–520. doi: 10.1007/s40123-021-00449-9
D'Argenio, D. A., Wu, M., Hoffman, L. R., Kulasekara, H. D., Deziel, E., Smith, E. E., et al. (2007). Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 64 (2), 512–533. doi: 10.1111/j.1365-2958.2007.05678.x
Dehghani, A. R., Fazel, F., Akhlaghi, M. R., Ghanbari, H., Ilanloo, M. R., Ahmadi-Azad, D. (2009). Cefazolin-gentamicin versus vancomycin-ceftazidime eye drops for bacterial corneal ulcers;a randomized clinical trial. J. Ophthalmic Vis. Res. 4 (1), 19–23.
Diggle, S. P., Cornelis, P., Williams, P., Camara, M. (2006). 4-quinolone signalling in Pseudomonas aeruginosa: old molecules, new perspectives. Int. J. Med. Microbiol. 296 (2-3), 83–91. doi: 10.1016/j.ijmm.2006.01.038
Enzor, R., Bowers, E. M. R., Perzia, B., Perera, C., Palazzolo, L., Mammen, A., et al. (2021). Comparison of clinical features and treatment outcomes of Pseudomonas aeruginosa keratitis in contact lens and non-contact lens wearers. Am. J. Ophthalmol. 227, 1–11. doi: 10.1016/j.ajo.2021.02.024
Evans, D. J., Fleiszig, S. M. (2013). Why does the healthy cornea resist Pseudomonas aeruginosa infection? Am. J. Ophthalmol. 155 (6), 961–970.e2. doi: 10.1016/j.ajo.2013.03.001
Feltner, J. B., Wolter, D. J., Pope, C. E., Groleau, M. C., Smalley, N. E., Greenberg, E. P., et al. (2016). LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in pseudomonas aeruginosa. mBio 7 (5), e01513-16. doi: 10.1128/mBio.01513-16
Fernandes, M., Vira, D., Medikonda, R., Kumar, N. (2016). Extensively and pan-drug resistant Pseudomonas aeruginosa keratitis: clinical features, risk factors, and outcome. Graefes Arch. Clin. Exp. Ophthalmol. 254 (2), 315–322. doi: 10.1007/s00417-015-3208-7
Fleiszig, S. M., Evans, D. J. (2002). The pathogenesis of bacterial keratitis: studies with Pseudomonas aeruginosa. Clin. Exp. Optom 85 (5), 271–278. doi: 10.1111/j.1444-0938.2002.tb03082.x
Gambello, M. J., Iglewski, B. H. (1991). Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol 173 (9), 3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991
Garcia-Reyes, S., Soberon-Chavez, G., Cocotl-Yanez, M. (2020). The third quorum-sensing system of Pseudomonas aeruginosa: Pseudomonas quinolone signal and the enigmatic PqsE protein. J. Med. Microbiol. 69 (1), 25–34. doi: 10.1099/jmm.0.001116
Green, M., Sara, S., Hughes, I., Apel, A., Stapleton, F. (2019). Trends in contact lens microbial keratitis 1999 to 2015: a retrospective clinical review. Clin. Exp. Ophthalmol. 47 (6), 726–732. doi: 10.1111/ceo.13484
Hammond, J. H., Hebert, W. P., Naimie, A., Ray, K., Van Gelder, R. D., DiGiandomenico, A., et al. (2016). Environmentally Endemic Pseudomonas aeruginosa Strains with Mutations in lasR Are Associated with Increased Disease Severity in Corneal Ulcers. mSphere 1 (5), e00140-16. doi: 10.1128/mSphere.00140-16
Harshaw, N. S., Stella, N. A., Lehner, K. M., Romanowski, E. G., Kowalski, R. P., Shanks, R. M. Q. (2021). Antibiotics used in empiric treatment of ocular infections trigger the bacterial Rcs stress response system independent of antibiotic susceptibility. Antibiotics 10 (9), 1033. doi: 10.3390/antibiotics10091033
Hilliam, Y., Kaye, S., Winstanley, C. (2020). Pseudomonas aeruginosa and microbial keratitis. J. Med. Microbiol. 69 (1), 3–13. doi: 10.1099/jmm.0.001110
Hobden, J. A. (2002). Pseudomonas aeruginosa proteases and corneal virulence. DNA Cell Biol. 21 (5-6), 391–396. doi: 10.1089/10445490260099674
Hoffman, L. R., Kulasekara, H. D., Emerson, J., Houston, L. S., Burns, J. L., Ramsey, B. W., et al. (2009). Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J. Cyst Fibros 8 (1), 66–70. doi: 10.1016/j.jcf.2008.09.006
Hoffman, L. R., Richardson, A. R., Houston, L. S., Kulasekara, H. D., Martens-Habbena, W., Klausen, M., et al. (2010). Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog. 6 (1), e1000712. doi: 10.1371/journal.ppat.1000712
Humphries, R., Bobenchik, A. M., Hindler, J. A., Schuetz, A. N. (2021). Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100, 31st edition. J. Clin. Microbiol. 59 (12), e0021321. doi: 10.1128/JCM.00213-21
Kariminik, A., Baseri-Salehi, M., Kheirkhah, B. (2017). Pseudomonas aeruginosa quorum sensing modulates immune responses: An updated review article. Immunol. Lett. 190, 1–6. doi: 10.1016/j.imlet.2017.07.002
Khan, M., Ma, K., Wan, I., Willcox, M. D. (2023). Ciprofloxacin resistance and tolerance of Pseudomonas aeruginosa ocular isolates. Cont Lens Anterior Eye 46 (3), 101819. doi: 10.1016/j.clae.2023.101819
Kohler, T., Buckling, A., van Delden, C. (2009). Cooperation and virulence of clinical Pseudomonas aeruginosa populations. Proc. Natl. Acad. Sci. U.S.A. 106 (15), 6339–6344. doi: 10.1073/pnas.0811741106
Kowalski, R. P., Pandya, A. N., Karenchak, L. M., Romanowski, E. G., Husted, R. C., Ritterband, D. C., et al. (2001). An in vitro resistance study of levofloxacin, ciprofloxacin, and ofloxacin using keratitis isolates of Staphylococcus aureus and Pseudomonas aeruginosa. Ophthalmology 108 (10), 1826–1829. doi: 10.1016/s0161-6420(01)00724-2
Lee, J., Zhang, L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6 (1), 26–41. doi: 10.1007/s13238-014-0100-x
Lesprit, P., Faurisson, F., Join-Lambert, O., Roudot-Thoraval, F., Foglino, M., Vissuzaine, C., et al. (2003). Role of the quorum-sensing system in experimental pneumonia due to Pseudomonas aeruginosa in rats. Am. J. Respir. Crit. Care Med. 167 (11), 1478–1482. doi: 10.1164/rccm.200207-736BC
Lin, A., Rhee, M. K., Akpek, E. K., Amescua, G., Farid, M., Garcia-Ferrer, F. J., et al. (2019). American academy of ophthalmology preferred practice pattern and P. External disease: bacterial keratitis preferred practice pattern(R). Ophthalmology 126 (1), P1–P55. doi: 10.1016/j.ophtha.2018.10.018
Matsumoto, K. (2004). Role of bacterial proteases in pseudomonal and serratial keratitis. Biol. Chem. 385 (11), 1007–1016. doi: 10.1515/BC.2004.131
Moradali, M. F., Ghods, S., Rehm, B. H. (2017). Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00039
Mould, D. L., Stevanovic, M., Ashare, A., Schultz, D., Hogan, D. A. (2022). Metabolic basis for the evolution of a common pathogenic Pseudomonas aeruginosa variant. Elife 11, e76555. doi: 10.7554/eLife.76555
O'Callaghan, R., Caballero, A., Tang, A., Bierdeman, M. (2019). Pseudomonas aeruginosa keratitis: protease IV and PASP as corneal virulence mediators. Microorganisms 7 (9), 281. doi: 10.3390/microorganisms7090281
O'Connor, K., Zhao, C. Y., Mei, M., Diggle, S. P. (2022). Frequency of quorum-sensing mutations in Pseudomonas aeruginosa strains isolated from different environments. Microbiol. (Reading) 168 (12), 0001265. doi: 10.1099/mic.0.001265
Pallares, R. M., An, D. D., Hebert, S., Faulkner, D., Loguinov, A., Proctor, M., et al. (2021). Multidimensional genome-wide screening in yeast provides mechanistic insights into europium toxicity. Metallomics 13 (12), mfab061. doi: 10.1093/mtomcs/mfab061
Pearlman, E., Sun, Y., Roy, S., Karmakar, M., Hise, A. G., Szczotka-Flynn, L., et al. (2013). Host defense at the ocular surface. Int. Rev. Immunol. 32 (1), 4–18. doi: 10.3109/08830185.2012.749400
Pearson, J. P., Feldman, M., Iglewski, B. H., Prince, A. (2000). Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect. Immun. 68 (7), 4331–4334. doi: 10.1128/iai.68.7.4331-4334.2000
Preston, M. J., Seed, P. C., Toder, D. S., Iglewski, B. H., Ohman, D. E., Gustin, J. K., et al. (1997). Contribution of proteases and LasR to the virulence of Pseudomonas aeruginosa during corneal infections. Infect. Immun. 65 (8), 3086–3090. doi: 10.1128/iai.65.8.3086-3090.1997
Qi, Q., Toll-Riera, M., Heilbron, K., Preston, G. M., MacLean, R. C. (2016). The genomic basis of adaptation to the fitness cost of rifampicin resistance in Pseudomonas aeruginosa. Proc. Biol. Sci. 283 (1822), 20152452. doi: 10.1098/rspb.2015.2452
Rhee, M. K., Kowalski, R. P., Romanowski, E. G., Mah, F. S., Ritterband, D. C., Gordon, Y. J. (2004). A laboratory evaluation of antibiotic therapy for ciprofloxacin-resistant Pseudomonas aeruginosa. Am. J. Ophthalmol. 138 (2), 226–230. doi: 10.1016/j.ajo.2004.03.016
Romanowski, E. G., Stella, N. A., Brazile, B. L., Lathrop, K. L., Franks, J. M., Sigal, I. A., et al. (2023). Predatory bacteria can reduce Pseudomonas aeruginosa induced corneal perforation and proliferation in a rabbit keratitis model. Ocul Surf 28, 254–261. doi: 10.1016/j.jtos.2023.05.002
Ruiz-Roldan, L., Rojo-Bezares, B., Lozano, C., Lopez, M., Chichon, G., Torres, C., et al. (2021). Occurrence of Pseudomonas spp. in raw vegetables: molecular and phenotypical analysis of their antimicrobial resistance and virulence-related traits. Int. J. Mol. Sci. 22 (23), 12626. doi: 10.3390/ijms222312626
Rumbaugh, K. P., Griswold, J. A., Iglewski, B. H., Hamood, A. N. (1999). Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect. Immun. 67 (11), 5854–5862. doi: 10.1128/IAI.67.11.5854-5862.1999
Shanks, R. M., Caiazza, N. C., Hinsa, S. M., Toutain, C. M., O'Toole, G. A. (2006). Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 72 (7), 5027–5036. doi: 10.1128/AEM.00682-06
Shanks, R. M., Kadouri, D. E., MacEachran, D. P., O'Toole, G. A. (2009). New yeast recombineering tools for bacteria. Plasmid 62 (2), 88–97. doi: 10.1016/j.plasmid.2009.05.002
Shanks, R. M., Stella, N. A., Arena, K. E., Fender, J. E. (2013). Mutation of crp mediates Serratia marcescens serralysin and global secreted protein production. Res. Microbiol. 164 (1), 38–45. doi: 10.1016/j.resmic.2012.10.006
Shen, E. P., Hsieh, Y. T., Chu, H. S., Chang, S. C., Hu, F. R. (2014). Correlation of Pseudomonas aeruginosa genotype with antibiotic susceptibility and clinical features of induced central keratitis. Invest. Ophthalmol. Vis. Sci. 56 (1), 365–371. doi: 10.1167/iovs.14-15241
Smith, E. E., Buckley, D. G., Wu, Z., Saenphimmachak, C., Hoffman, L. R., D'Argenio, D. A., et al. (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U.S.A. 103 (22), 8487–8492. doi: 10.1073/pnas.0602138103
Sokol, P. A., Ohman, D. E., Iglewski, B. H. (1979). A more sensitive plate assay for detection of protease production by Pseudomanas aeruginosa. J. Clin. Microbiol. 9 (4), 538–540. doi: 10.1128/jcm.9.4.538-540.1979
Subedi, D., Vijay, A. K., Willcox, M. (2018). Overview of mechanisms of antibiotic resistance in Pseudomonas aeruginosa: an ocular perspective. Clin. Exp. Optom 101 (2), 162–171. doi: 10.1111/cxo.12621
Ung, L., Chodosh, J. (2021). Foundational concepts in the biology of bacterial keratitis. Exp. Eye Res. 209, 108647. doi: 10.1016/j.exer.2021.108647
Vazirani, J., Wurity, S., Ali, M. H. (2015). Multidrug-resistant Pseudomonas aeruginosa keratitis: risk factors, clinical characteristics, and outcomes. Ophthalmology 122 (10), 2110–2114. doi: 10.1016/j.ophtha.2015.06.007
Willcox, M. D. (2007). Pseudomonas aeruginosa infection and inflammation during contact lens wear: a review. Optom Vis. Sci. 84 (4), 273–278. doi: 10.1097/OPX.0b013e3180439c3e
Zhu, H., Conibear, T. C., Bandara, R., Aliwarga, Y., Stapleton, F., Willcox, M. D. (2006). Type III secretion system-associated toxins, proteases, serotypes, and antibiotic resistance of Pseudomonas aeruginosa isolates associated with keratitis. Curr. Eye Res. 31 (4), 297–306. doi: 10.1080/02713680500536746
Keywords: keratitis, Pseudomonas aeruginosa, ocular infection, quorum sensing, transcription factor, protease
Citation: Shanks RMQ, Atta S, Stella NA, Sundar-Raj CV, Romanowski JE, Grewal AS, Shanks HQ, Mumper SM, Dhaliwal DK, Mammen A, Callaghan JD, Calvario RC, Romanowski EG, Kowalski RP, Zegans ME and Jhanji V (2023) A rise in the frequency of lasR mutant Pseudomonas aeruginosa among keratitis isolates between 1993 and 2021. Front. Cell. Infect. Microbiol. 13:1286842. doi: 10.3389/fcimb.2023.1286842
Received: 31 August 2023; Accepted: 16 October 2023;
Published: 31 October 2023.
Edited by:
Paulo J. M. Bispo, Harvard Medical School, United StatesReviewed by:
Ajai Dandekar, University of Washington, United StatesCopyright © 2023 Shanks, Atta, Stella, Sundar-Raj, Romanowski, Grewal, Shanks, Mumper, Dhaliwal, Mammen, Callaghan, Calvario, Romanowski, Kowalski, Zegans and Jhanji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert M. Q. Shanks, c2hhbmtzcm1AdXBtYy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.