- 1Yunnan Tropical and Subtropical Animal Viral Disease Laboratory, Yunnan Animal Science and Veterinary Institute, Kunming, China
- 2The Aquaculture Workstation of Yuanyang County Agriculture, Rural Affairs, and Science and Technology Bureau, Yuanyang, China
Introduction: Culicoides plays a crucial role as an insect vector in the field of veterinary medicine. The transmission of significant viruses such as bluetongue virus (BTV) and African horse sickness virus (AHSV) by this insect poses a substantial threat, leading to the development of severe diseases in domestic animals. This study aimed to explore the Culicoides species, identify their blood-meal sources, and assess the presence of BTV and AHSV carried by Culicoides in Yuanyang County, Yunnan Province. The aim was to gain insights into the potential vectors of these two viruses and elucidate their potential roles in the transmission of pathogens.
Methods: The midges were collected from cattle (Bos indicus), pig (Sus scrofa), and goat (Capra hircus) pens in Yuanyang County, Yunnan Province in June 2020. Initial identification of midges was conducted through morphological characteristics, followed by molecular identification using the cytochrome C oxidase subunit I (COI) gene. The determination of Culicoides blood-meal sources was accomplished using specific primers targeting the cytochrome b (Cyt b) gene from potential hosts. BTV and AHSV RNA were identified in Culicoides pools through the application of reverse transcriptase PCR and quantitative real-time PCR. Nucleotide homology and phylogenetic analysis were performed using MegAlign (DNAStar) and Mega 6.0 software.
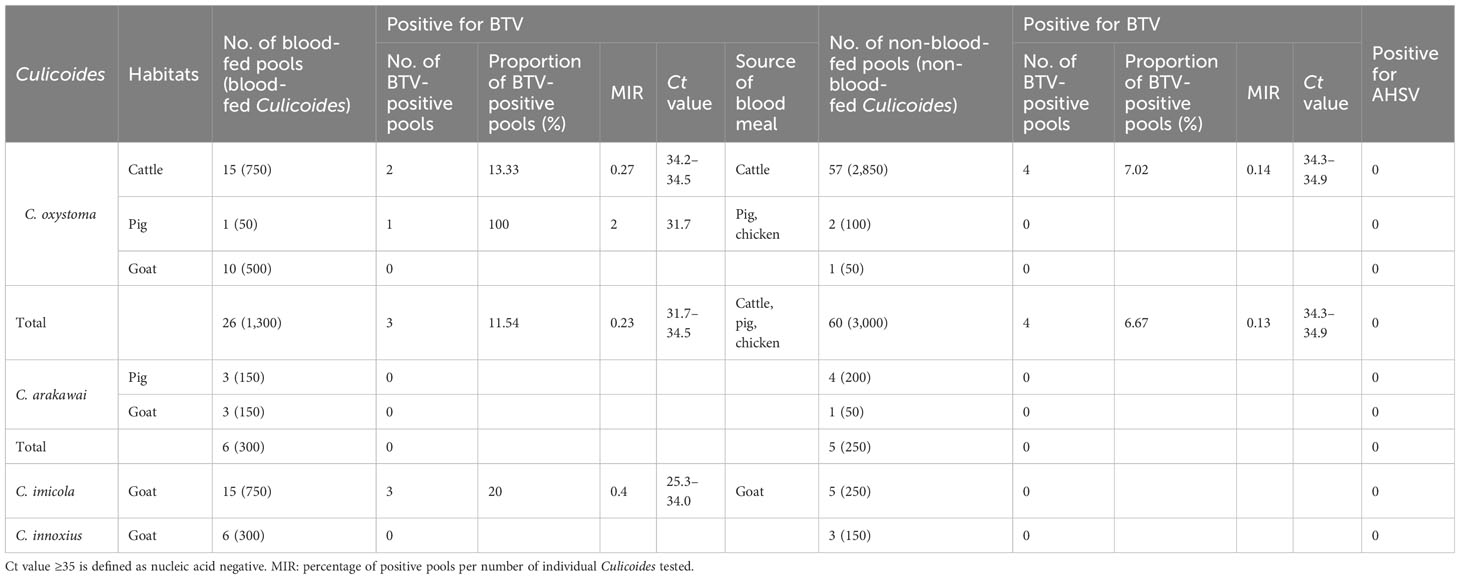
Results: A total of 6,300 Culicoides, consisting of C. oxystoma, C. arakawai, C. imicola, and C. innoxius, were collected from cattle, pigs, and goat pens. The engorgement rates for these species were 30.2%, 54.6%, 75%, and 66.7%, respectively. In the cattle pen, the prevailing species is C. oxystoma (100%). In the pig pen, C. arakawai dominates (70%), with C. oxystoma following at 30%. In the goat pen, C. imicola holds the majority (45.45%), trailed by C. oxystoma (25%), C. innoxius (20.45%), and C. arakawai (9.09%). These Culicoides species were identified as feeding on cattle, pigs, goats, chickens (Gallus gallus), and humans (Homo sapiens). The positivity rates for BTV were 20.00% and 11.54% in blood-fed specimens of C. imicola and C. oxystoma, respectively. Conversely, the positivity rates for BTV in non-blood-fed specimens were 0.00% and 6.67% for C. imicola and C. oxystoma, respectively. BTV was not detected in C. arakawai and C. innoxius. The specimens (YY86) from C. imicola that tested positive for BTV had the closest genetic relationship to YTS-4 isolated from Mangshi, Yunnan Province in 1996. All test results for the nucleic acid of AHSV were negative.
Conclusion: The study reveals variations in the species distribution, community composition, blood sucking rate, and blood-feeding sources of Culicoides across different habitats. Notably, C. imicola and C. oxystoma emerge as potential vectors for the transmission of BTV in local animals. Accordingly, this investigation provides crucial insights that can serve as a valuable reference for the prevention and control of BTV in local animals, particularly from the perspective of vector management.
1 Introduction
Bluetongue virus (BTV) and African horse sickness virus (AHSV) are globally recognized as two distinct orbivirus that bear the closest association with animal diseases and have a significant impact on animal husbandry and veterinary health. These orbiviruses pose a serious threat to economically valuable livestock species, including cattle (Bos indicus), goat (Capra hircus), pig (Sus scrofa), and horses, often leading to high mortality rates in affected animals. For example, the death rate of bluetongue (BT) in goat and African horse sickness (AHS) in horses can reach as high as 75% and 90%, respectively (Coetzer and Guthrie, 2004; Najarnezhad and Rajae, 2013; Carpenter et al., 2017). Initially confined to Africa and parts of Europe, these two animal diseases have witnessed a surge in prevalence in recent years. This escalation is attributed to factors such as social development, increased transportation networks, and rise in livestock exchanges and trade. Consequently, there has been a significant upswing in the transmission and spread of Culicoides, the vector responsible for the BTV and AHSV in recent years. Not only has the intensity of these epidemics increased, but their geographical scope has also expanded continuously, often leading to trans-regional and trans-continental outbreak and a large number of animal fatalities and resulting in significant economic losses (Alkhamis et al., 2020).
Culicoides plays an important role in the transmission of BTV and AHSV, and the occurrence and spread of BT are closely linked to the species composition and distribution of potential Culicoides. The genus Culicoides comprises a global total of 1,399 species (Borkent and Dominiak, 2020), but only a limited number have been implicated with the transmission of BTV and AHSV. Notable species proven to be vectors of BTV include C. imicola, C. obsoletus, C. schultzei, C. wadai, C. fulvus, C. brevitarsis, C. variipennis, C. insignis, C. actoni, C. oxystoma, C. orientalis, and C. peregrines (Sendow et al., 1996; Mellor et al., 2000; Venter et al., 2000; Dadawala et al., 2012). Additionally, species such as C. oxystoma, C. imicola, and C. bolitinos have been identified as potential vectors of AHSV(Meiswinkel and Paweska, 2003; Moussa et al., 2015). The distribution and ecosystem of these species of Culicoides significantly influence the occurrence, spread, and severity of both BT and AHS. A comprehensive understanding of the species composition, distribution, blood-meal host of Culicoides, and the arbovirus they carry is the key to effectively prevent and control the epidemic and transmission of BT and AHS.
The initial report of a BT outbreak in goats dates back to 1979 in Yunnan Province, China. Since then, BT has been identified in animals, including cattle, across various regions of China. Up to now, BTV has been detected in infected livestock in 29 provinces, including Guangdong, Guangxi, and Xinjiang. Ten regions have reported the isolation of 16 BTV serotypes (BTV-1, BTV-2, BTV-3, BTV-4, BTV-5, BTV-7, BTV-9, BTV-11, BTV-12, BTV-14, BTV-15, BTV-16, BTV-17, BTV-21, BTV-24, and BTV-29) from infected goat or sentinel animals (Ma et al., 2017; Qin et al., 2018; Duan et al., 2019; Li et al., 2020; Li et al., 2021). The diverse array of BTV serotypes poses a significant threat to the country’s cattle and goat breeding industry. AHSV primarily affects regions in Africa and the Middle East (King et al., 2020). Notably, C. imicola, the primary vector of AHSV, is presented in the southern regions of China, heightening the risk of AHS being introduced to China. Despite this potential risk, there is a notable absence of research on the species composition, blood-meal sources, and arbovirus-carrying vectors associated with both BT and AHS. The aim of this study was to identify Culicoides collected from different habitats in Yuanyang, Yunnan Province, utilizing morphological and molecular identification methods. Additionally, the investigation seeks to analyze the blood-meal sources and arbovirus-carrying potential of Culicoides.
2 Materials and methods
2.1 Collection of Culicoides specimens
In June 2020, Culicoides specimens were collected from feedlots at three different sites using light traps (12 V, 300 mA; Wuhan Lucky Star Environmental Protection, Hubei, China) situated in Yuanyang County, Honghe Prefecture, Yunnan Province. The sites chosen included goat, cattle, and pig feedlots. The goat feedlot (23°12′N; 102°55′E) and cattle feedlot (23°13′N; 102°50′E) are situated in Wubang and Nansha Village, Nansha Town, at altitudes of 230 m and 242 m, respectively. In contrast, the pig feedlot (23°6′N; 102°45′E) is located in Quanfuzhuang Village, Xinjie Town, at a higher altitude of 1,700 m above sea level. Culicoides specimens were collected overnight, spanning from 7:00 pm to 7:00 am. The collected specimens were promptly frozen in a −20°C freezer for 20 min to sacrifice. Subsequently, they were preliminarily classified and identified on ice according to their morphological characteristics following the keys described by Yu et al. (2005). Fifty Culicoides per tube were dispensed into cell-freezing tubes, recorded and labeled, and then stored in liquid nitrogen. Finally, those samples were transported to our laboratory for further analysis.
2.2 Molecular identification of Culicoides
Genomic DNA was extracted from Culicoides pools using the TIANamp Micro DNA kit (TIANGEN, Beijing) in accordance with the manufacturer’s recommendation and subsequently stored at −20°C. A fragment of the COI gene was amplified using the primers C1-J-1718 and C1-N-2191 (Dallas et al., 2003) and TaKaRa Ex Taq® DNA Polymerase (TaKaRa, Beijing). PCR reactions were conducted in a total volume of 25 µL, comprising 2 µL of DNA template, 2.5 µL of 10×Ex Taq buffer, 2.5 µL of 2.5 mmol/L dNTP mixture, 0.3 µL of Ex Taq, 0.5 µL of 20 µM of each primer, and 16.7 µL of ddH2O. The COI amplification process included an initial denaturation of 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 54°C for 30 s, 72°C for 40 s, and a final elongation step of 72°C for 10 min. The amplified products were subjected to detection via 1% agarose gel electrophoresis, purification, and Sanger sequencing by Tsingke Biotechnology Co., Ltd. (Beijing, China).
2.3 Identification of blood-meal sources
DNA was extracted from engorged Culicoides and used as a template, and the fragments of the Cyt b region of the mitochondrion were amplified. This amplification was achieved through the utilization of universal vertebrate-specific primer Cytb-F and Cytb-R (Townzen et al., 2008), mammalian-specific primer Mammal-F and Mammal-R (Ngo and Kramer, 2003), and species-specific primer Cow121-F and UNREV1025 (Kent and Norris, 2005). Each 25 µL of PCR reaction comprised 2 µL of DNA template, 2.5 µL of 10× Ex Taq buffer, 2.5 µL of 2.5 mmol/L dNTP mixture, 0.3 µL of Ex Taq, 0.5 µL of 20 µM of each primer, and 16.7 µL of ddH2O. The PCR protocol involved an initial heating phase of 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 30 s; annealing at 50°C (Cytb-F and Cytb-R), 55°C (Mammal-F and Mammal-R), and 58°C (Cow121-F and UNREV1025); and elongation at 72°C for 45~60 s. A final elongation step was conducted at 72°C for 10 min. The amplified products were subjected to detection via 1% agarose gel electrophoresis, purification, and Sanger sequencing by Tsingke Biotechnology Co., Ltd. (Beijing, China).
2.4 Detection of BTV and AHSV in Culicoides
Total RNA was extracted from the grinding supernatant of Culicoides pools using RNAiso Plus (TaKaRa), following the manufacturer’s recommendations, and stored at −80°C. To detect BTV and AHSV in Culicoides pools, specific primer pairs and TaqMan probes for reverse transcription quantitative PCR (RT-qPCR) were employed (Hofmann et al., 2008; Guthrie et al., 2013). The reaction mix comprised of 2 µL of RNA template, 10 µL of 2× One Step RT-PCR buffer III;, 0.4 µL of TaKaRa Ex Taq HS (5 U/μL), 0.4 µL of PrimeScript RT Enzyme Mix II, 0.4 µL of ROX Reference Dye II (50×), 0.4 µL of 10 µM of each primer and probe, and 5.6 µL of ddH2O. The reactions were conducted using One Step PrimeScript™ RT-PCR Kit (TaKaRa) on a Fast 7500 Real-Time PCR machine (Applied Biosystems, Carlsbad, CA, USA) under the following conditions: 42°C for 5 min, 95°C for 10 s, and 35 cycles of 95°C for 3 s and 60°C for 30 s.
2.5 Sequence amplification of BTV Seg-3
For RNA samples positive for BTV nucleic acid, reverse transcriptase M-MLV (RNase H−) (TaKaRa) was employed to synthesize the first-strand cDNA in accordance with the manufacturer’s recommendations. Subsequent PCR reactions were conducted using the primer pair BTV L3-1 and BTV L3-2 (Ohashi et al., 2004). Each 25 µL of PCR reaction included 2 µL of cDNA template, 2.5 µL 10× of Ex Taq buffer, 2.5 µL of 2.5 mmol/L dNTP mixture, 0.3 µL of Ex Taq, 0.5 µL of 20 µM of each primer, and 16.7 µL of ddH2O. The PCR protocol involved an initial heating phase of 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 30 s, and elongation at 72°C for 40 s. A final elongation step was performed at 72°C for 10 min. The amplified products were subjected to detection via 1% agarose gel electrophoresis, purification, and subsequent Sanger sequencing by Tsingke Biotechnology Co., Ltd. (Beijing, China). All nucleotide positions were confirmed by independent sequencing reactions in both directions.
2.6 Sequence analysis
The sequences of the Culicoides COI gene and BTV Seg-3 were analyzed using the MegAlign program within the DNAStar software package. The phylogenetic trees for the COI gene and Seg-3 were conducted using the neighbor-joining (NJ) method through MEGA 6.0 software, with a bootstrap value of 1,000 to ensure robustness. Additionally, the nucleotide sequences of the Cyt b gene were compared using Nucleotide BLAST in GenBank to analyze the blood-meal sources of Culicoides.
2.7 Statistical analysis
The minimum infection rate (MIR) value was expressed as a percentage of the number of positive pools per number of individual Culicoides tested (Larska et al., 2013). The differences in the composition of Culicoides from different environments, the engorgement rate, and the positive rate of BTV detection in different Culicoides species were analyzed by chi-square analyses carried out with the SPSS Statistics 26.0 software package. p ≤0.05 was considered statistically significant.
3 Results
3.1 Trapping and identification of Culicoides species
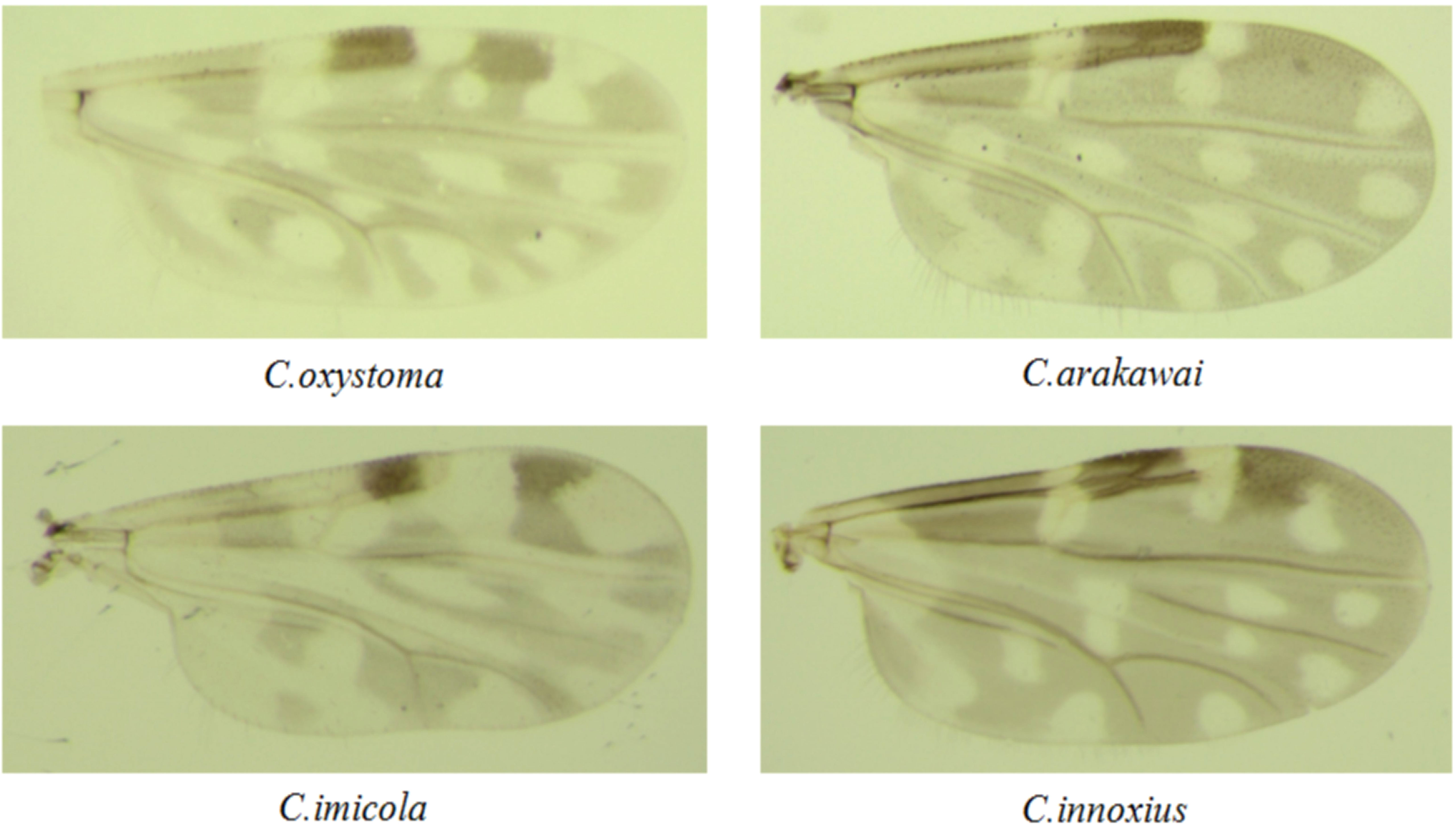
After collecting the specimens of midges, the collected midges were frozen to death in a refrigerator at −20°C, and then the miscellaneous insects were picked out. Under a stereomicroscope, the morphology of the midges was preliminarily identified according to the morphology of the wing spot, size, and color of the midges. Fifty Culicoides of the same species collected in the same site were placed in a tube as a pool. They were recorded, marked, stored in liquid nitrogen, and finally transported to our laboratory.
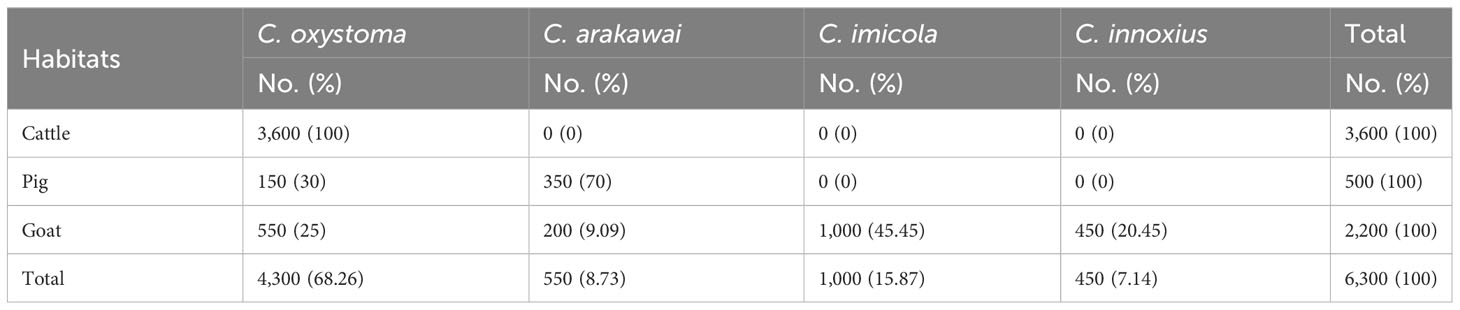
A comprehensive collection of approximately 6,300 specimens from the Culicoides genus was obtained through light traps set in goat, cattle, and pig feedlots in Yuanyang County, Honghe Prefecture, Yunnan Province. Utilizing both morphological and molecular identification methods, four distinct Culicoides species were identified: C. oxystoma (68.26%), C. arakawai (8.73%), C. imicola (15.87%), and C. innoxius (7.14%) (Figures 1, 2). The sequences of these Culicoides species have been deposited in GenBank with accession numbers (OR472938–OR472948). There were significant differences in the species composition of Culicoides collected in cattle, sheep, and pig habitats (χ² = 6,408.703, p < 0.01). Culicoides oxystoma emerged as the predominant species in the cattle habitat (100%). In the pig habitat, the dominant species was C. arakawai (70%). Within the goat habitat, C. imicola (45.45%) was the most abundant species (Table 1).
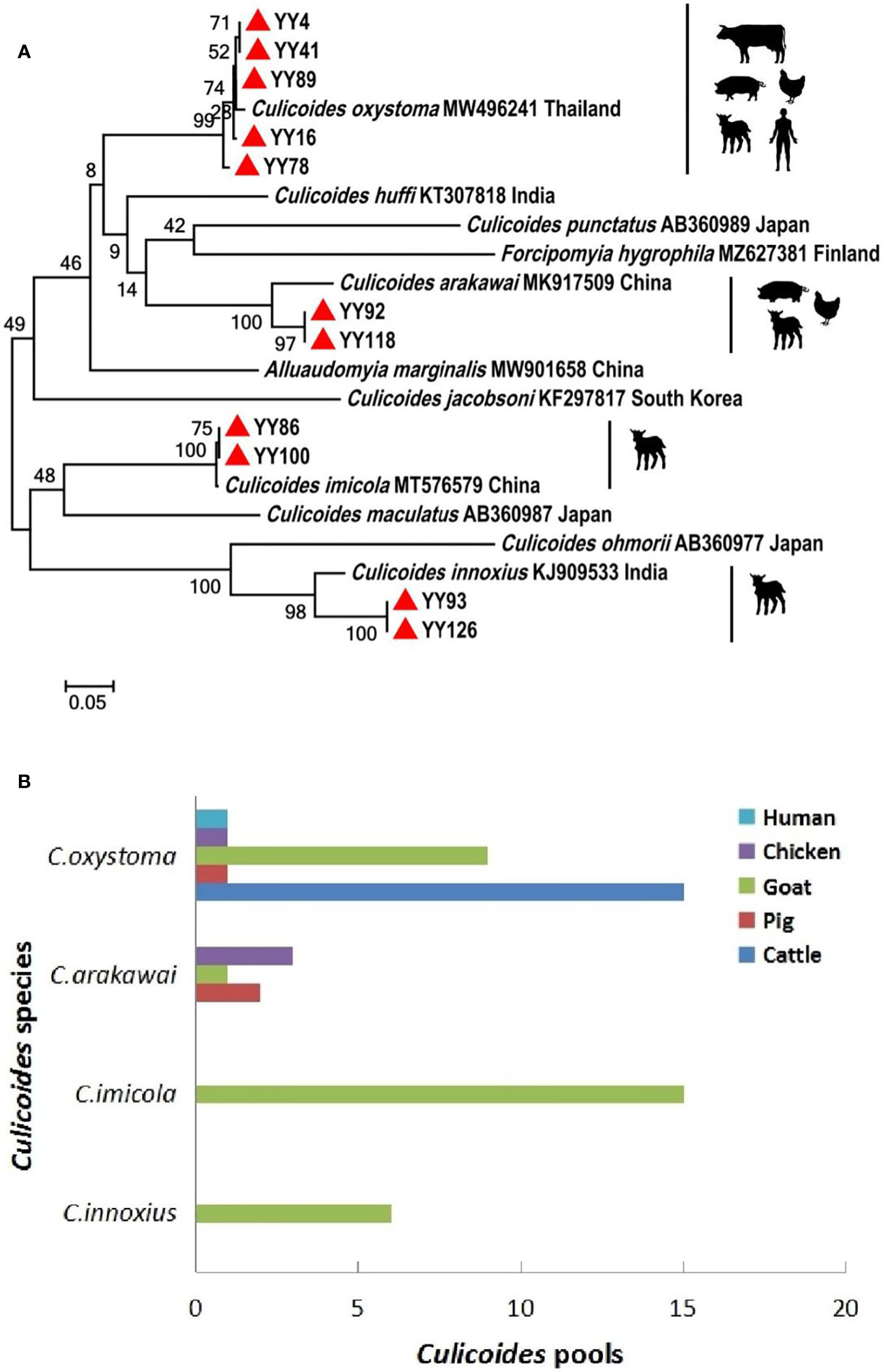
Figure 2 Phylogenetic tree of COI sequences and molecular identification of blood-meal hosts for Culicoides species. (A) Phylogenetic tree of COI sequences of Culicoides and potential hosts of different Culicoides species. The specimens created in this study are labeled by solid red triangles. (B) Blood-meal hosts were identified from different Culicoides species.
3.2 Blood-meal source analysis
In total, 6,300 specimens representing Culicoides species were collected, with 2,650 of them classified as blood-engorged (42.06%). The collection encompassed various feedlots, including cattle (20.83% blood-fed rate, n = 750), pig (40% blood-fed rate, n = 200), and goat (77.27% blood-fed rate, n = 1,700). There were significant differences in the engorgement rate of Culicoides collected in these three environments (χ² = 712.825, p < 0.001). The engorgement rates of C. oxystoma, C. arakawai, C. imicola, and C. innoxius were 30.23%, 54.55%, 75%, and 66.67%, respectively. The engorgement rates of different Culicoides species were also significantly different (Fisher’s precision probability test p < 0.001).
An effort was made to identify vertebrate hosts for 53 pools of the blood-fed specimens (n = 2,650). Initially, all samples were screened with a universal vertebrate primer pair. Subsequently, screening transitioned to a mammalian-specific primer pair and a species-specific primer pair (Bos taurus L.) to enhance the identification of specimens. As a result, the total blood-meal host results were obtained through the utilization of several primer pairs. All DNA sequences from blood-engorged specimens showed ≥94% identity matches to vertebrate hosts in GenBank. The origins of the blood-meal host of four Culicoides species were identified at the species level. Culicoides oxystoma fed on five different vertebrate species: cattle, pig, goat, chicken (Gallus gallus), and human (Homo sapiens). Culicoides arakawai fed on three different vertebrate species: pig, goat, and chicken. In contrast, C. imicola and C. innoxius exclusively fed on goat (Table 2 and Figure 2).
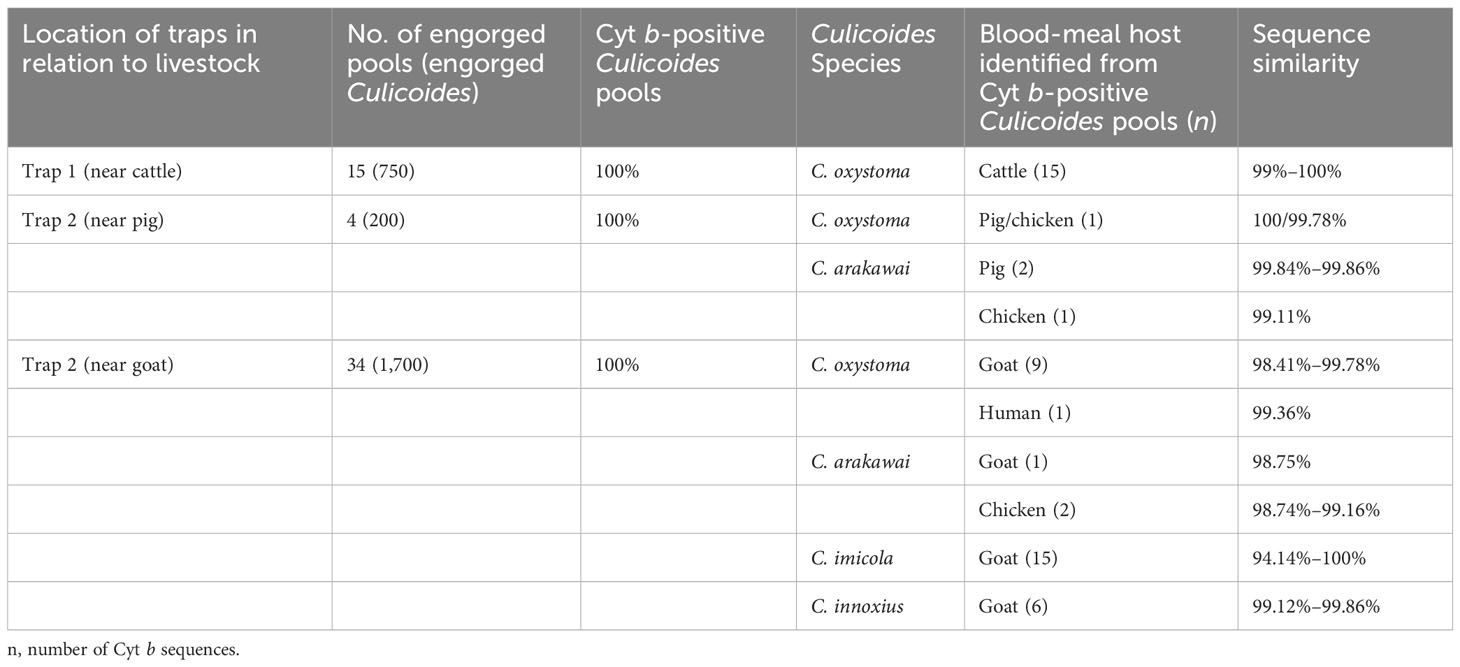
Table 2 Results of blood-meal source analyses in the different Culicoides species, according to source (cattle, pig, goat).
3.3 BTV and AHSV detection
A total of 126 pools, compromising specimens from four Culicoides species collected at three distinct sites, underwent testing with BTV- and AHSV-specific RT-qPCR assay. The results of the BTV and AHSV RT-qPCRs for these specimens are summarized in Table 3. Among the 86 pools of C. oxystoma tested, 26 blood-fed and 60 non-blood-fed specimens were examined, revealing that 3 (11.54%; MIR 0.23%) blood-fed and 4 (6.67%; MIR 0.13%) non-blood-fed pools were identified as infected with BTV, respectively. In the case of C. imicola, 15 blood-fed and 5 non-blood-fed pools from the 20 tested pools showed that only 3 (20.00%; MIR 0.4%) blood-fed pools were positive for BTV infection. For C. arakawai and C. innoxius, 6 blood-fed and 5 non-blood-fed and 6 blood-fed and 3 non-blood-fed specimens from 11 pools and 9 pools, respectively, were found to be negative for BTV infection. Additionally, no specimens from the four Culicoides species tested positive for AHSV infection.
3.4 Phylogenetic analysis of BTV
A Seg-3 sequence, spanning 688 bp, was derived from the BTV-infected C. imicola specimen (YY86) obtained from a goat feedlot. The obtained fragment was utilized to construct phylogenetic trees alongside sequences downloaded from NCBI. As shown in Figure 3, YY86 was positioned in the same branch as YTS-4 isolated from Mangshi, Yunnan Province, in 1996, displaying a 96.4% homogeneity (Yang et al., 2012). The data presented here unequivocally affirm that the virus carried by C. imicola (YY86) is indeed BTV. The sequence of the BTV Seg-3 has been deposited in GenBank with accession number OR481010.
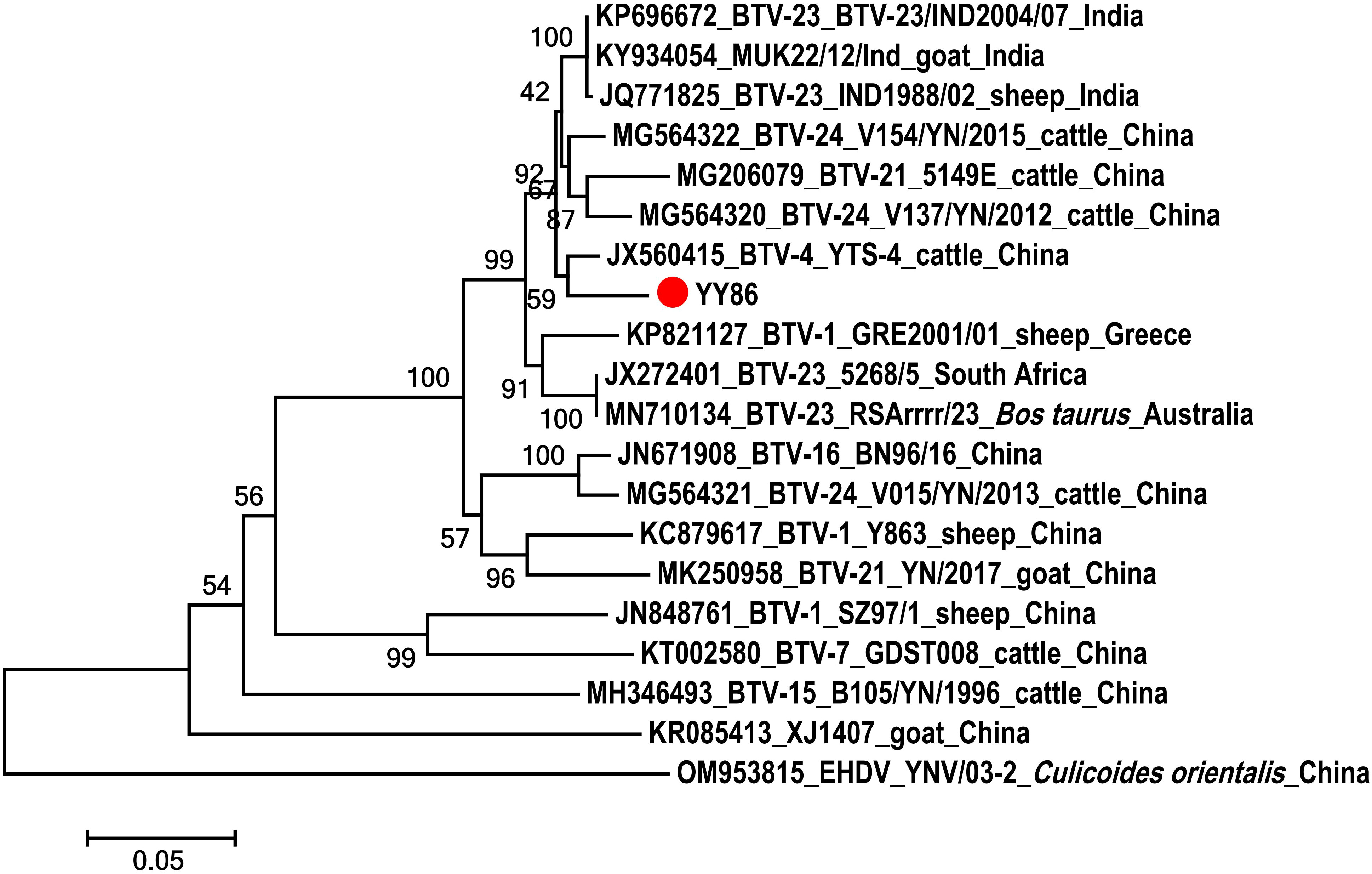
Figure 3 Phylogenetic analysis of the sequences of segment 3 of BTV. The specimens created in this study are labeled by a solid red circle.
4 Discussion
BT and AHS stand out as the two most severe Culicoides-borne livestock diseases affecting ruminants and horses, respectively. BTV and AHSV naturally persist through a series of alternative cycles of replication between Culicoides vectors and susceptible hosts. Culicoides imicola has been confirmed in previous studies as a principal vector for both BT and AHS, exhibiting a high transmission efficiency for both BTV and AHSV (Braverman et al., 1985; Venter et al., 2000; Paweska et al., 2002; Guichard et al., 2014; de Waal et al., 2016). The widespread distribution of C. imicola, spanning the African continent, southern Europe, and extending eastward to South China, poses a significant risk for the transmission and escalation of BT and AHS epidemics (Guichard et al., 2014). In a laboratory setting, C. imicola has demonstrated susceptibility to various BTV strains, and it has been established that this vector can also be infected with AHSV (Venter et al., 2004; Venter et al., 2005; Venter et al., 2006a; Venter et al., 2006b; Venter and Paweska, 2007b; Venter et al., 2007a; Venter et al., 2009). South Africa, where C. imicola is highly abundant, has reported BTV isolations from field-collected C. imicola (Nevill et al., 1992; Meiswinkel et al., 2004). In addition, in Italy, BTV has been detected in C. imicola (Goffredo et al., 2015). The presence of BTV in C. imicola has also been confirmed in western Thailand and southern Yunnan, indicating the potential significant role of C. imicola in BTV transmission (Duan et al., 2021; Fujisawa et al., 2021). Apart from C. imicola, C. oxystoma is considered a potential vector for arboviruses such as BTV, Akabane virus (AKAV), and epizootic hemorrhagic disease virus (EHDV). This species is geographically widespread in East Asia, Southeast Asia, Australia, and the Middle East (Kitaoka, 1984; Wirth and Hubert, 1989), playing a crucial role in the transmission and distribution of these viruses (Yanase et al., 2005; Dadawala et al., 2012; Bakhoum et al., 2013; Harsha et al., 2020). Dadawala et al. reported the isolation of BTV-1 from non-engorged C. oxystoma in India, underscoring the direct involvement of this vector in BTV transmission (Dadawala et al., 2012). Additionally, AKAV has been isolated and detected from C. oxystoma in Japan and South Korea (Oem et al., 2013; Kato et al., 2016).
Yunnan Province, situated in the southwest of China, boasts a tropical and subtropical monsoon climate conducive to the survival and reproduction of vector insects and host animals. BTV has been isolated from Culicoides or hosts in tropical and subtropical regions of southern Yunnan Province, particularly in epidemic-prone regions (Kirkland et al., 2002; Yang et al., 2016). Yuanyang County, located in the southern part of Yunnan Province, exhibits significant differences in altitude. High-density feedlots in this region house a substantial number of cattle, goats, and other livestock, fostering an environment more conducive to the propagation and transmission of arboviruses. Culicoides were collected from three distinct sites with varying elevations in Yuanyang County, encompassing goat (230 m), cattle (242 m), and pig (1,700 m) feedlots, and their composition of Culicoides was identified. Four Culicoides species (C. imicola, C. oxystoma, C. arakawai, and C. innoxius) were identified. Morphological and molecular identification results aligned, confirming C. imicola and C. oxystoma as potential vectors of BTV. Culicoides arakawai has been associated with the transmission of significant livestock pathogens, including BTV, AHSV, AKAV, Chuzan virus (CHUV), and bovine ephemeral fever virus (BEFV) (Yanase et al., 2005; Yang et al., 2018; Kim et al., 2021). The role of C. innoxius in disease transmission remains unclear. Culicoides activity may be influenced by host distribution. In this study, C. imicola was exclusively present in goat habitat, indicating a heightened attraction to goat in the local area. Additionally, topography and climate may impact the abundance and active period of C. imicola, favoring hot days in lower-altitude sites. The species’ confinement to low-altitude goat habitats aligns with previous findings (Torina et al., 2004; Conte et al., 2007; Barceló et al., 2021). The highest number of C. oxystoma specimens was found across all three sites, with a significant presence in cattle habitats, suggesting an enhanced attraction to cattle. Understanding the local Culicoides species and their distribution is pivotal for elucidating their transmission ability and the pathogens they carry. The climate and temperature conditions in Yuanyang County provide an ideal ecosystem for Culicoides propagation, coupled with the abundance of susceptible animals, likely facilitating the transmission and spread of BTV.
Locally, livestock serves as the main blood-meal sources of insect vectors, acting as the continuous suppliers of arboviruses. Understanding Culicoides’ selection of susceptible hosts is crucial for studying potential vector species (Hadj-Henni et al., 2015; Riddin et al., 2019). Identifying the blood-meal hosts of Culicoides is essential to clarify the range of hosts for vectors (Lassen et al., 2011; Leta et al., 2019; Mcgregor et al., 2019). However, compared with mosquitoes and ticks, the blood-meal hosts of Culicoides have received relatively less attention (Börstler et al., 2016; Shahhosseini et al., 2018). We employed PCR to amplify the vertebrate host DNA in a blood meal from Culicoides collected at various sites, successfully analyzing 53 pools (42.06%) of the blood-fed Culicoides. The findings revealed that these four Culicoides species mainly feed on mammals, with most blood-meal analysis results aligning with the livestock present at the three sites. Culicoides occasionally fed on chickens or humans, suggesting that, in addition to cattle, pigs, and goats, nearby livestock and humans may also contribute to their blood meal. Culicoides oxystoma exhibited a diverse range of blood-meal hosts, including cattle, pigs, goats, chickens, and humans. The observation of feeding on susceptible cattle and goats indicated that C. oxystoma is likely a potential vector species for BTV and other arboviruses. Furthermore, its broad host preference enhances the potential transmission of pathogens between hosts and domestic animals.
Moreover, significant variations were observed in the species composition and blood-fed rates of Culicoides collected at different sites. Notably, the Culicoides species collected in goat feedlot exhibited the highest abundance, featuring four Culicoides species and a blood-fed rate of 77.27%. In the pig feedlot, two Culicoides species were identified, with a blood-fed rate of 40%. Conversely, the cattle feedlot only yielded C. oxystoma, with the lowest blood-fed rate at 20.83%. These findings imply that Culicoides are more attracted to goats than other domestic animals, such as cattle and pigs. It is noteworthy that despite the relatively small total number of C. imicola, it boasted the highest blood-fed rate at 75%. Following closely were C. innoxius and C. arakawai, with blood-fed rates of 66.67% and 54.55%, respectively. This suggests that C. imicola may exhibit stronger vector competence than the other three species. The blood-meal source of the arthropod plays a pivotal role in pathogen transmission and maintenance in natural systems. Identifying the blood-meal sources from engorged Culicoides indirectly provides more host information related to Culicoides, elucidating their potential host range and activity information. This information is crucial for understanding pathogen transmission and disease risk.
The study was conducted to determine the significant presence of Culicoides around livestock, particularly cattle and goats. These Culicoides exhibited a preference for feeding on cattle and goats, which are susceptible to BTV, suggesting a potential risk of pathogen transmission and spread among susceptible species. Further BTV tests demonstrated that the positive rates of BTV were 15.00% and 8.14% in C. imicola and C. oxystoma, respectively. This confirmed the indirect involvement of these two Culicoides species in BTV transmission among local domestic animals. Notably, BTV was detected in blood-fed C. imicola, and RT-PCR amplification and sequence analysis of YY86 indicated that this BTV had the closest genetic relationship with YTS-4 isolated from Mangshi, Yunnan Province in 1996. This study detected BTV in C. imicola and C. oxystoma, suggesting that they are potential vectors of BTV in this region. Hence, local livestock should be vaccinated using several licensed commercial vaccines, including modified live (live-attenuated) and inactivated products. However, all test results for AHSV were negative, indicating that, despite the presence of suitable insect vectors, no introduction of AHSV was identified in the local area at the moment.
The presence of BTV RNA detected in Culicoides in this study has been previously demonstrated in Yunnan Province (Duan et al., 2021). Here, we identified four species of Culicoides and used DNA sequencing to directly link Culicoides species with blood-feeding on hosts in Yuanyang. Further analysis of blood-meal hosts and virus detection data was conducted, linking C. imicola and C. oxystoma with the transmission of BTV among local livestock. These findings offer crucial insights for the prevention and control of BTV in the region. It will be very meaningful for the prediction and prevention of BTV in other endemic areas in China. We recommend future surveillance studies of potential vectors related to BTV in other parts of China.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OR472938–OR472948; https://www.ncbi.nlm.nih.gov/genbank/, OR481010.
Author contributions
NL: Conceptualization, Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. JM: Data curation, Investigation, Software. YH: Investigation. WW: Investigation. JW: Conceptualization, Funding acquisition, Resources, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the earmarked fund for National Natural Science Foundation of China (32260896), National Key Research and Development Program of China (2022YFC2601603), The Central Government to Guide Local Scientific and Technological Development (202207AB110006), Basic Research Projects of Yunnan Province (202201AS070062, 202301AT070028), Foreign Talents Introduction Project of Yunnan Province (202305AO350020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alkhamis, M. A., Aguilar-Vega, C., Fountain-Jones, N. M., Lin, K., Perez, A. M., Sánchez-Vizcaíno, J. M. (2020). Global emergence and evolutionary dynamics of bluetongue virus. Sci. Rep. 10 (1), 21677. doi: 10.1038/s41598-020-78673-9
Bakhoum, M. T., Fall, M., Fall, A. G., Bellis, G. A., Gottlieb, Y., Labuschagne, K., et al. (2013). First record of Culicoides oxystoma Kieffer and diversity of species within the Schultzei group of Culicoides Latreille (Diptera: Ceratopogonidae) biting midges in Senegal. PLoS One 8 (12), e84316. doi: 10.1371/journal.pone.0084316
Barceló, C., Purse, B. V., Estrada, R., Lucientes, J., Miranda, M.Á., Searle, K. R. (2021). Environmental drivers of adult seasonality and abundance of biting midges Culicoides (Diptera: Ceratopogonidae), bluetongue vector species in Spain. J. Med. Entomol. 58 (1), 350–364. doi: 10.1093/jme/tjaa160
Borkent, A., Dominiak, P. (2020). Catalog of the biting midges of the world (diptera: ceratopogonidae). Zootaxa 4787 (1), 1–377. doi: 10.11646/zootaxa.4787.1.1
Börstler, J., Jöst, H., Garms, R., Krüger, A., Tannich, E., Becker, N., et al. (2016). Host-feeding patterns of mosquito species in Germany. Parasit Vectors. 9 (1), 318. doi: 10.1186/s13071-016-1597-z
Braverman, Y., Barzila, i E., Frish, K., Rubina, M. (1985). Bluetongue virus isolation from pools of Culicoides spp. in Israel during the years 1981 to 1983. Prog. Clin. Biol. Res. 178, 191–193.
Carpenter, S., Mellor, P. S., Fall, A. G., Garros, C., Venter, G. J. (2017). African horse sickness virus: history, transmission, and current status. Annu. Rev. Entomol. 62 (1), 343–358. doi: 10.1146/annurev-ento-031616-035010
Coetzer, J. A. W., Guthrie, A. J. (2004). “African horse sickness,” in Infectious Diseases of Livestock, 2nd. Eds. Coetzer, J. A. W., Tustin, R. C. (Southern Africa: Oxford University Press), 1231–1246.
Conte, A., Goffredo, M., Ippoliti, C., Meiswinkel, R. (2007). Influence of biotic and abiotic factors on the distribution and abundance of Culicoides imicola and the Obsoletus Complex in Italy. Vet. Parasitol. 150 (4), 333–344. doi: 10.1016/j.vetpar.2007.09.021
Dadawala, A. I., Biswas, S. K., Rehman, W., Chand, K., De, A., Mathapati, B. S., et al. (2012). Isolation of bluetongue virus serotype 1 from Culicoides vector captured in livestock farms and sequence analysis of the viral genome segment-2. Transbound Emerg. Dis. 59 (4), 361–368. doi: 10.1111/j.1865-1682.2011.01279.x
Dallas, J. F., Cruickshank, R. H., Linton, Y.-M., Nolan, D. V., Patakakis, M., Braverman, Y., et al. (2003). Phylogenetic status and matrilineal structure of the biting midge, Culicoides imicola, in Portugal, Rhodes and Israel. Med. Vet. Entomol. 17 (4), 379–387. doi: 10.1111/j.1365-2915.2003.00454.x
de Waal, T., Liebenberg, D., Venter, G. J., Mienie, C. M., Van Hamburg, H. (2016). Detection of African horse sickness virus in Culicoides imicola pools using RT-qPCR. J. Vector Ecol. 41 (1), 179–185. doi: 10.1111/jvec.12210
Duan, Y. L., Bellis, G., Li, L., Li, H. C., Miao, H. S., Kou, M. L., et al. (2019). Potential vectors of bluetongue virus in high altitude areas of Yunnan Province, China. Parasit Vectors. 12 (1), 464. doi: 10.1186/s13071-019-3736-9
Duan, Y. L., Li, L., Bellis, G., Yang, Z. X., Li, H. C. (2021). Detection of bluetongue virus in Culicoides spp. in southern Yunnan Province, China. Parasit Vectors. 14 (1), 68. doi: 10.1186/s13071-020-04518-z
Fujisawa, Y., Homat, T., Thepparat, A., Changbunjong, T., Sutummaporn, K., Kornmatitsuk, S., et al. (2021). DNA barcode identification and molecular detection of bluetongue virus in Culicoides biting midges (Diptera: Ceratopogonidae) from western Thailand. Acta Trop. 224, 106147. doi: 10.1016/j.actatropica.2021.106147
Goffredo, M., Catalani, M., Federici, V., Portanti, O., Marini, V., Mancini, G., et al. (2015). Vector species of Culicoides midges implicated in the 2012−2014 Bluetongue epidemics in Italy. Vet. Ital. 51 (2), 131–138. doi: 10.12834/VetIt.771.3854.1
Guichard, S., Guis, H., Tran, A., Garros, C., Balenghien, T., Kriticos, D. J. (2014). Worldwide niche and future potential distribution of Culicoides imicola, a major vector of bluetongue and african horse sickness viruses. PLoS One 9 (11), e112491. doi: 10.1371/journal.pone.0119323
Guthrie, A. J., Maclachlan, N. J., Joone, C., Lourens, C. W., Weyer, C. T., Quan, M., et al. (2013). Diagnostic accuracy of a duplex real-time reverse transcription quantitative PCR assay for detection of African horse sickness virus. J. Virol. Methods 189 (1), 30–35. doi: 10.1016/j.jviromet.2012.12.014
Hadj-Henni, L., De Meulemeester, T., Depaquit, J., Noël, P., Germain, A., Helder, R., et al. (2015). Comparison of vertebrate cytochrome b and prepronociceptin for blood meal analyses in Culicoides. Front. Vet. Sci. 2. doi: 10.3389/fvets.2015.00015
Harsha, R., Mazumdar, S. M., Mazumdar, A. (2020). Abundance, diversity and temporal activity of adult Culicoides spp. associated with cattle in West Bengal, India. Med. Vet. Entomol. 34 (3), 327–343. doi: 10.1111/mve.12446
Hofmann, M., Griot, C., Chaignat, V., Perler, L., Thür, B. (2008). Bluetongue disease reaches Switzerland. Schweiz Arch. Tierheilkd. 150 (2), 49–56. doi: 10.1024/0036-7281.150.2.49
Kato, T., Shirafuji, H., Tanaka, S., Sato, M., Yamakawa, M., Tsuda, T., et al. (2016). Bovine arboviruses in Culicoides biting midges and sentinel cattle in southern Japan from 2003 to 2013. Transbound Emerg. Dis. 63 (6), e160–e172. doi: 10.1111/tbed.12324
Kent, R. J., Norris, D. E. (2005). Identification of mammalian blood meals in mosquitoes by a multiplexed polymerase chain reaction targeting cytochrome B. Am. J. Trop. Med. Hyg 73 (2), 336–342. doi: 10.4269/ajtmh.2005.73.336
Kim, M. S., Kim, H. C., Bellis, G. A., Chong, S. T., Kim, H. S., Klein, T. A. (2021). Seasonal abundance of Culicoides at Yongsan US Army Garrison (USAG) and Camp Humphreys USAG, Republic of Korea 2010-2013 and 2014-2017. Korean J. Parasitol. 59 (3), 273–280. doi: 10.3347/kjp.2021.59.3.273
King, S., Rajko-Nenow, P., Ashby, M., Frost, L., Carpenter, S., Batten, C. (2020). Outbreak of African horse sickness in Thailand, 2020. Transbound Emerg. Dis. 67 (5), 1764–1767. doi: 10.1111/tbed.13701
Kirkland, P. D., Zhang, N., Hawkes, R. A., Li, Z., Zhang, F., Davis, R. J., et al. (2002). Studies on the epidemiology of bluetongue virus in China. Epidemiol. Infect. 128 (2), 257–263. doi: 10.1017/s0950268801006525
Kitaoka, S. (1984). Japanese Culicoides (Diptera: Cerato-pogonidae) and keys for the species. Bull. Natl. Inst. Anim. Health 87, 73–108.
Larska, M., Lechowski, L., Grochowska, M., Żmudziński, J. F. (2013). Detection of the Schmallenberg virus in nulliparous Culicoides obsoletus/scoticus complex and C. punctatus–the possibility of transovarial virus transmission in the midge population and of a new vector. Vet. Microbiol. 166 (3-4), 467–473. doi: 10.1016/j.vetmic.2013.07.015
Lassen, S. B., Nielsen, S. A., Skovgård, H., Kristensen, M. (2011). Molecular identification of bloodmeals from biting midges (Diptera: Ceratopogonidae: Culicoides Latreille) in Denmark. Parasitol. Res. 108 (4), 823–829. doi: 10.1007/s00436-010-2123-4
Leta, S., Fetene, E., Mulatu, T., Amenu, K., Jaleta, M. B., Beyene, T. J., et al. (2019). Modeling the global distribution of Culicoides imicola: an Ensemble approach. Sci. Rep. 9 (1), 14187. doi: 10.1038/s41598-019-50765-1
Li, M. L., Zhang, Q. J., Xu, Q. Y. (2021). Global distribution of bluetongue. Acta Veterinaria Zootechnica Sin. 52 (04), 881–890. doi: 10.11843/j.issn.0366-6964.2021.04.004
Li, Z. R., Zhu, J. B., Liao, D. F., Xiao, L., Wang, J. P., Gao, L., et al. (2020). Genetic characteristics of”historical strains”of bluetongue virus isolated from Yunnan Province. China Anim. Health Inspection 37 (08), 26–35+42. doi: 10.3969/j.issn.1005-944X.2020.08.007
Ma, J. G., Zhang, X. X., Zheng, W. B., Xu, Y. T., Zhu, X. Q., Hu, G. X., et al. (2017). Seroprevalence and risk factors of bluetongue virus infection in tibetan goat and yaks in tibetan plateau, China. BioMed. Res. Int. 2017, 5139703. doi: 10.1155/2017/5139703
Mcgregor, B. L., Stenn, T., Sayler, K. A., Blosser, E. M., Blackburn, J. K., Wisely, S. M., et al. (2019). Host use patterns of Culicoides spp. biting midges at a big game preserve in Florida, U.S.A. and implications for the transmission of orbiviruses. Med. Vet. Entomol. 33 (1), 110–120. doi: 10.1111/mve.12331
Meiswinkel, R., Paweska, J. T. (2003). Evidence for a new field Culicoides vector of African horse sickness in South Africa. Prev. Vet. Med. 60 (3), 243–253. doi: 10.1016/s0167-5877(02)00231-3
Meiswinkel, R., Venter, G. J., Nevill, E. M. (2004). “Vectors: Culicoides spp,” in Infectious Diseases of Livestock, 2nd edn, vol. 1 . Eds. Coetzer, J. A. W., Tustin, R. C. (Cape Town: Oxford University Press), 93–136.
Mellor, P. S., Boorman, J., Baylis, M. (2000). Culicoides biting midges: their role as arbovirus vectors. Annu. Rev. Entomol. 45 (1), 307–340. doi: 10.1146/annurev.ento.45.1.307
Moussa, F., Maryam, D., Assane, G. F., Thomas, B., Momar, T. S., Jérémy, B., et al. (2015). Culicoides (Diptera: Ceratopogonidae) midges, the vectors of African horse sickness virus–a host/vector contact study in the Niayes area of Senegal. Parasit Vectors. 8 (1), 39. doi: 10.1186/s13071-014-0624-1
Najarnezhad, V., Rajae, M. (2013). Seroepidemiology of bluetongue disease in small ruminants of north-east of Iran. Asian Pac J. Trop. Biomed. 3 (6), 492–495. doi: 10.1016/S2221-1691(13)60102-1
Nevill, E. M., Erasmus, B. J., Venter, G. J. (1992). “A six-year study of viruses associated with Culicoides biting midges throughout South Africa (Diptera: Ceratopogonidae),” in Bluetongue, African Horse Sickness and Related Orbiviruses. Eds. Walton, T. E., Osburn, B. I. (Boca Raton, FL: CRC Press), 314–319.
Ngo, K. A., Kramer, L. D. (2003). Identification of mosquito bloodmeals using polymerase chain reaction (PCR) with order-specific primers. J. Med. Entomol. 40 (2), 215–222. doi: 10.1603/0022-2585-40.2.215
Oem, J. K., Chung, J. Y., Kwon, M. S., Kim, T. K., Lee, T. U., Bae, Y. C. (2013). Abundance of biting midge species (Diptera: Ceratopogonidae, Culicoides spp.) on cattle farms in Korea. J. Vet. Sci. 14 (1), 91–94. doi: 10.4142/jvs.2013.14.1.91
Ohashi, S., Yoshida, K., Yanase, T., Kato, T., Tsuda, T. (2004). Simultaneous detection of bovine arboviruses using single-tube multiplex reverse transcription-polymerase chain reaction. J. Virol. Methods 120 (1), 79–85. doi: 10.1016/j.jviromet.2004.04.006
Paweska, J. T., Venter, G. J., Mellor, P. S. (2002). Vector competence of South African Culicoides species for bluetongue virus serotype 1 (BTV-1) with special reference to the effect of temperature on the rate of virus replication in C. imicola and C. bolitinos. Med. Vet. Entomol. 16 (1), 10–21. doi: 10.1046/j.1365-2915.2002.00334.x
Qin, S. M., Yang, H., Zhang, Y. X., Li, Z. H., Lin, J., Gao, L., et al. (2018). Full genome sequence of the first bluetongue virus serotype 21 (BTV-21) isolated from China: evidence for genetic reassortment between BTV-21 and bluetongue virus serotype 16 (BTV-16). Arch. Virol. 163 (5), 1379–1382. doi: 10.1007/s00705-018-3718-9
Riddin, M. A., Venter, G. J., Labuschagne, K., Villet, M. H. (2019). Bloodmeal analysis in Culicoides midges collected near horses, donkeys and zebras in the Eastern Cape, South Africa. Med. Vet. Entomol. 33 (4), 467–475. doi: 10.1111/mve.12381
Sendow, I., Sukarsih, S. E., Soleha, E., Pearce, M., Bahri, S., Daniels, P. W. (1996). Bluetongue virus research in Indonesia.In: In: St George, T. D., Kegao, P., editors. Bluetongue Disease in Southeast Asia and the Pacific (Canberra: Australian Centre for International Agricultural Research), 28–32.
Shahhosseini, N., Friedrich, J., Moosa-Kazemi, S. H., Sedaghat, M. M., Kayedi, M. H., Tannich, E., et al. (2018). Host-feeding patterns of Culex mosquitoes in Iran. Parasit Vectors. 11 (1), 669. doi: 10.1186/s13071-018-3237-2
Torina, A., Caracappa, S., Mellor, P. S., Baylis, M., Purse, B. V. (2004). Spatial distribution of bluetongue virus and its Culicoides vectors in Sicily. Med. Vet. Entomol. 18 (2), 81–89. doi: 10.1111/j.0269-283X.2004.00493.x
Townzen, J. S., Brower, A. V. Z., Judd, D. D. (2008). Identification of mosquito bloodmeals using mitochondrial cytochrome oxidase subunit I and cytochrome b gene sequences. Med. Vet. Entomol. 22 (4), 386–393. doi: 10.1111/j.1365-2915.2008.00760.x
Venter, G. J., Gerdes, G. H., Mellor, P. S., Paweska, J. T. (2004). Transmission potential of South African Culicoides species for live-attenuated bluetongue virus. Vet. Ital. 40 (3), 198–202.
Venter, G. J., Graham, S. D., Hamblin, C. (2000). African horse sickness epidemiology: vector competence of South African Culicoides species for virus serotypes 3, 5 and 8. Med. Vet. Entomol. 14 (3), 245–250. doi: 10.1046/j.1365-2915.2000.00245.x
Venter, G. J., Koekemoer, J. J. O., Paweska, J. T. (2006b). Investigations on outbreaks of African horse sickness in the surveillance zone in South Africa. Rev. Sci. Tech. 25 (3), 1097–1109.
Venter, G. J., Mellor, P. S., Paweska, J. T. (2006a). Oral susceptibility of South African stock-associated Culicoides species to bluetongue virus. Med. Vet. Entomol. 20 (3), 329–934. doi: 10.1111/j.1365-2915.2006.00635.x
Venter, G. J., Mellor, P. S., Wright, I., Paweska, J. T. (2007a). Replication of live-attenuated vaccine strains of bluetongue virus in orally infected South African Culicoides species. Med. Vet. Entomol. 21 (3), 239–247. doi: 10.1111/j.1365-2915.2007.00687.x
Venter, G. J., Paweska, J. T. (2007b). Virus recovery rates for wild-type and live-attenuated vaccine strains of African horse sickness virus serotype 7 in orally infected South African Culicoides species. Med. Vet. Entomol. 21 (4), 377–383. doi: 10.1111/j.1365-2915.2007.00706.x
Venter, G. J., Paweska, J. T., Lunt, H., Mellor, P. S., Carpenter, S. (2005). An alternative method of blood-feeding Culicoides imicola and other haematophagous Culicoides species for vector competence studies. Vet. Parasitol. 131 (3-4), 331–335. doi: 10.1016/j.vetpar.2005.05.002
Venter, G. J., Wright, I. M., van der Linde, T. C., Paweska, J. T. (2009). The oral susceptibility of South African field populations of Culicoides to African horse sickness virus. Med. Vet. Entomol. 23 (4), 367–378. doi: 10.1111/j.1365-2915.2009.00829.x
Wirth, W. W., Hubert, A. A. (1989). The Culicoides of southeast Asia (Diptera: Ceratopogonidae). Mem Am. Entomol. Inst. 44, 1–508.
Yanase, T., Kato, T., Kubo, T., Yoshida, K., Ohashi, S., Yamakawa, M., et al. (2005). Isolation of bovine arboviruses from Culicoides biting midges (Diptera: Ceratopogonidae) in southern Japan: 1985–2002. J. Med. Entomol. 42 (1), 63–67. doi: 10.1093/jmedent/42.1.63
Yang, H., Xiao, L., Wang, J., Meng, J., Lv, M., Liao, D., et al. (2016). Phylogenetic characterization genome segment 2 of bluetongue virus strains belonging to serotypes 5, 7 and 24 isolated for the first time in China during 2012 to 2014. Transbound Emerg. Dis. 64 (4), 1317–1321. doi: 10.1111/tbed.12479
Yang, D., Yang, M. S., Rhim, H., Han, J. I., Oem, J. K., Kim, Y. H., et al. (2018). Analysis of five arboviruses and Culicoides distribution on cattle farms in Jeollabuk-do, Korea. Korean J. Parasitol. 56 (5), 477–485. doi: 10.3347/kjp.2018.56.5.477
Yang, H., Zhu, J., Li, H., Xiao, L., Wang, J., Li, N., et al. (2012). Full genome sequence of bluetongue virus serotype 4 from China. J. Virol. 86 (23), 13122–13123. doi: 10.1128/jvi.02393-12
Keywords: Culicoides, molecular identification, blood-meal source, Bluetongue virus, African horse sickness virus
Citation: Li N, Meng J, He Y, Wang W and Wang J (2024) Potential roles of Culicoides spp. (Culicoides imicola, Culicoides oxystoma) as biological vectors of bluetongue virus in Yuanyang of Yunnan, P. R. China. Front. Cell. Infect. Microbiol. 13:1283216. doi: 10.3389/fcimb.2023.1283216
Received: 25 August 2023; Accepted: 18 December 2023;
Published: 11 January 2024.
Edited by:
Hong Liu, Shandong University of Technology, ChinaReviewed by:
Benjamin Cull, University of Minnesota Twin Cities, United StatesSusana Remesar, University of Santiago de Compostela, Spain
Xiaolong Li, University of Florida, United States
Copyright © 2024 Li, Meng, He, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinglin Wang, V2FuZ2psMTA3QDE2My5jb20=
 Nan Li
Nan Li Jinxin Meng
Jinxin Meng Yuwen He1
Yuwen He1 Jinglin Wang
Jinglin Wang