- 1School of Life Sciences, Faculty of Science, University of Technology Sydney, Ultimo, NSW, Australia
- 2Faculty of Science, University of Technology Sydney, Ultimo, NSW, Australia
Chlamydia trachomatis is a strict intracellular human pathogen. It is the main bacterial cause of sexually transmitted infections and the etiologic agent of trachoma, which is the leading cause of preventable blindness. Despite over 100 years since C. trachomatis was first identified, there is still no vaccine. However in recent years, the advancement of genetic manipulation approaches for C. trachomatis has increased our understanding of the molecular pathogenesis of C. trachomatis and progress towards a vaccine. In this mini-review, we aimed to outline the factors related to the developmental cycle phase and specific pathogenesis activity of C. trachomatis in order to focus priorities for future genetic approaches. We highlight the factors known to be critical for developmental cycle stages, gene expression regulatory factors, type III secretion system and their effectors, and individual virulence factors with known impacts.
1 Introduction
The sexually transmitted pathogen Chlamydia trachomatis (CT) is the leading cause of preventable blindness worldwide (Taylor et al., 2014) and the most common bacterial sexually transmitted infection (STIs) in humans, with approximately 131 million cases each year and rising (Newman et al., 2015). The majority of chlamydial infections are asymptomatic. The infection when diagnosed can be treated with antibiotics (Scidmore, 2009). Complications of CT infections are known to include reproductive tract impacts which can cause considerable morbidity and cost (Davies et al., 2016).
The Chlamydiae are a ubiquitous family of Gram-negative pathogens, with a unique lifecycle. Chlamydiae species causes infection in specific animals, such as Chlamydia muridarum in mice and hamsters (Mishkin et al., 2022), while Chlamydia psittaci commonly infects birds and rarely humans (Hogerwerf et al., 2020). Chlamydia pneumoniae and CT are endemic to humans, they infect different anatomical sites, respiratory and oculogenital, respectively (Whittum-Hudson and Hudson, 2005). CT is further divided into serovars based on the hypervariable region of the major outer membrane protein (MOMP) (Hepler et al., 2018). CT serovars exhibit different tissue tropisms. A-C infects ocular tissue and causes trachoma. D-K primarily infects the genital tract resulting in the STI ‘chlamydia’ but in newborns can also infect respiratory and ocular conjunctiva tissues causing pneumonia and conjunctivitis. L1-L3 infects genital tissues and macrophages resulting in the invasive STI ‘lymphogranuloma venereum (LGV)’ (Abdelsamed et al., 2013).
The Chlamydia developmental cycle is characterised as biphasic, relying on specific interactions with the host cell for nutrients in order to survive and replicate due to the reduced chlamydial genome (Gitsels et al., 2019). There are two main morphological bacterial phases; the extracellular infectious elementary body (EB) and the intracellular, non-infectious, replicative reticulate body (RB) [reviewed in (Hafner et al., 2008; Elwell et al., 2016; Witkin et al., 2017)] (Figure 1). The infectious EBs are robust formations with spore-like structures that consist of disulphide cross-linked outer membrane protein complex on the surface. The development cycle of CT can be impacted by environmental factors and stresses. Known stressors include nutrient deprivation, exposure to host cytokines, and cell wall synthesis-targeting antibiotics (Gitsels et al., 2019). Under these conditions, RBs transition to altered morphological forms of non-culturable, non-dividing, aberrantly enlarged “persistent” forms, called aberrant bodies (ABs) (Panzetta et al., 2018). These forms remain viable and can revert into an active infection once conditions are favourable (Schoborg, 2011).
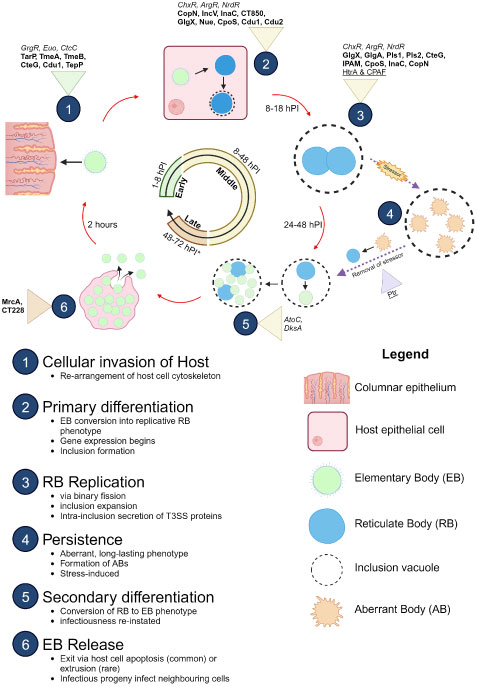
Figure 1 Developmental cycle of C. trachomatis. Important proteins expressed at different stages of the development cycle are shown with established transcription factors (italicised), T3SS effectors (bolded), and proteases (underlined). Infectious EBs (coloured green) adhere to and enter the host cell (coloured pink) using various adhesins (Ctad1, Pmps) and the T3SS. Once inside, EBs begin differentiation into replicative RBs (coloured blue). During the RB replication phase, host cell metabolites are used to facilitate the pathogen’s survival. After several rounds of replication, the RBs then asynchronously re-differentiate into EBs for exit via extrusion of the inclusion vacuole from the host cell or lysis of the host cell. The released EB progeny then restarts the infection cycle. Release of EBs (late stage) varies from 48 hours to 72 hours, depending on the serovar (asterisk). In atypical cycles, stressors trigger persistence, in which RBs (coloured green) pause replication and differentiate into long-surviving, stable ABs (coloured orange) and enter a stasis-like state until the stressor is removed. The ABs “detect” this change and re-differentiate back into replicative RBs to resume development. Figure produced with BioRender.
As an obligate intracellular organism, virulence and pathogenic features interplay critically with host cellular factors and the host immune system which can also contribute to disease pathology [reviewed in (Geisler, 2010; Menon et al., 2015; Murthy et al., 2018; Chen et al., 2019)]. The intracellular nature and unusual bi-phasic developmental cycle has, until recently, hampered progress on biological investigations and genetic manipulation of Chlamydia. Recently, genetic methods have rapidly progressed and accordingly our understanding of pathogenic mechanisms has increased (Fisher and Beare, 2023). However, each genetic approach has their advantages and limitations [reviewed in, (Banerjee and Nelson, 2021; Wan et al., 2023)]. Previously, Chlamydia was thought to be genetically intractable due to its intracellular development, vacuole niche, evasive persistence mechanism, serovar genetic variability, and the thick wall of EB absorbing external DNA (Andersen et al., 2021). As such, specialised techniques were developed to better understand this clinically significant pathogen. Whole genome sequencing (Stephens et al., 1998), allelic exchange mutagenesis (Mueller et al., 2016), CRISPRi knockdown (Ouellette, 2018; Ouellette et al., 2021) and intron insertion gene inactivation (O'Neill et al., 2021) assist in associating genes with chlamydial phenotypic traits, bypassing impractical traditional approaches in genome studies [reviewed in (Read and Massey, 2014; Fisher and Beare, 2023; Luu et al., 2023)]. Nonetheless, many of these genetic manipulation approaches for Chlamydia are still relatively time consuming and requires expert culture skills, hence we set out to review what is known about potential virulence factors to guide priorities for future genetic experiments. We do not cover metabolic factors, although it is arguable that these are related to pathogenesis for an obligate intracellular pathogen. We also do not cover unknown hypothetical proteins.
2 Transcription and regulatory factors as a target to understand pathogenesis
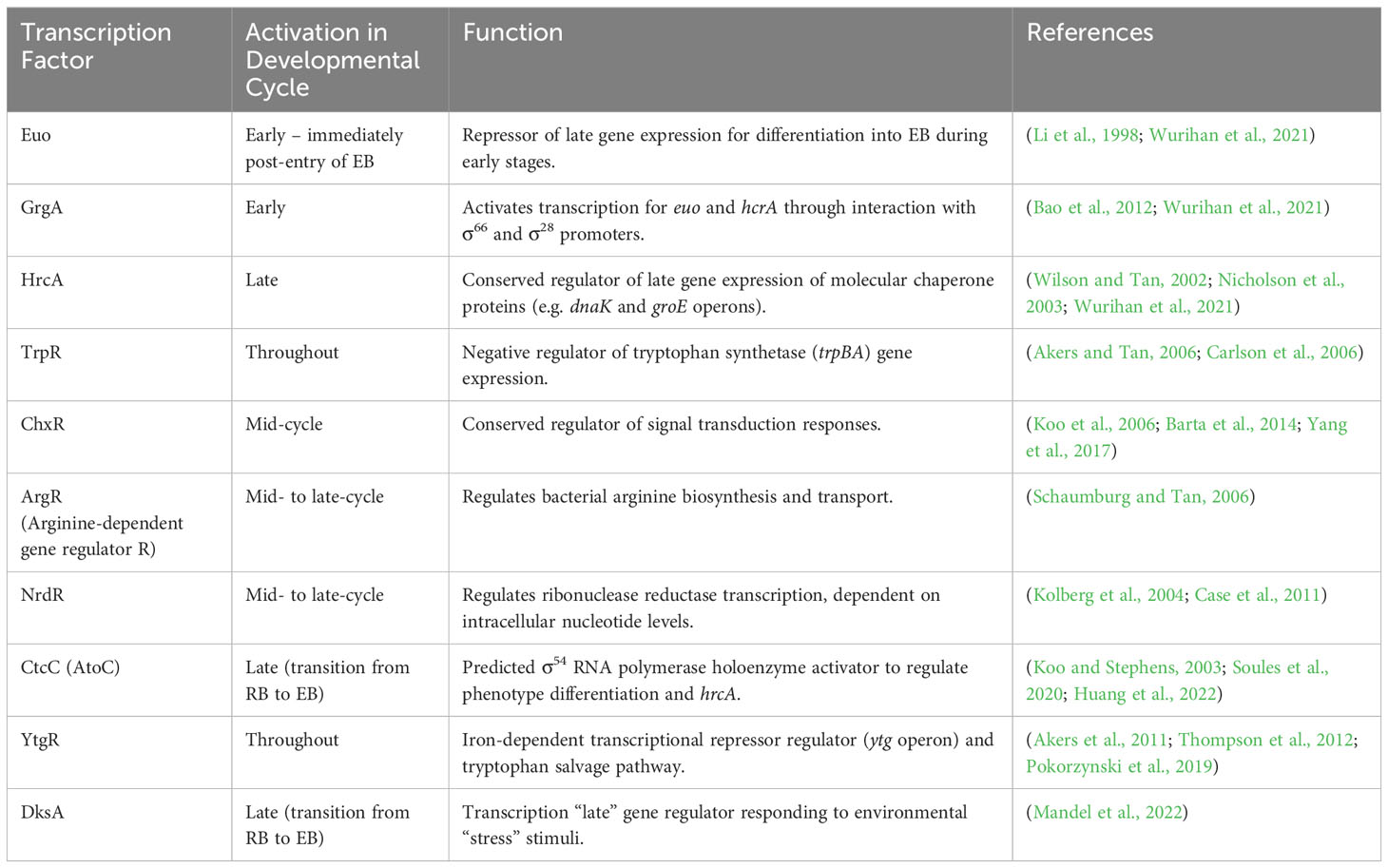
The bi-phasic development is regulated by transcription factors resulting in distinct gene expression profiles characterised as early, mid-, or late response. Chlamydia’s genome encodes a small number of known transcription factors (outlined in Table 1A) and sigma factors (σ) (Akers et al., 2011). The trio of chlamydial sigma factors (σ66, σ28, and σ54) serve as RNA polymerases to various promotor genes (Nicholson et al., 2003; Wurihan et al., 2021). Belland et al. (2003) detected σ28 and σ54 mRNA mid-development at 8 h PI (hours post infection), whilst σ66 was detected earliest at 3 h PI. σ66 is responsible for activating the majority of CT promoters, whilst select late genes rely on σ28- or σ54-dependent promoters (Mathews et al., 1999). These sigma factors are regulated by specific transcription factors (Table 1A) and anti-sigma factors. A comprehensive review of transcriptional regulation is available from Nicholson et al. (2003) and Belland et al. (2003).
To date, only two anti-sigma factors which inhibit σ66 activity have been characterised: CT663, a T3SS Scc4 chaperone, and the switch-protein Regulator of Sigma B W (RsbW) kinase (Rao et al., 2009) (Thompson et al., 2015). RsbW forms a regulatory network with its own anti-anti-sigma factor RsbV1 and the phosphatase RsbU. The activity of RsbW is driven by the levels unphosphorylated RsbV1, where a deficit of RsbV1 drives the switch-kinase towards antagonizing σ66, altering transcription of σ66 development-related genes involved in metabolism and growth (Hanson et al., 2015; Thompson et al., 2015).
Recently, transcriptional profiles for the σ54 regulon was generated using CtcC (an ATP-hydrolyzing response regulator for σ54) mutants which further revealed the role of this sigma factor in membrane remodelling and incorporating T3SS effectors into EBs during RB differentiation into infectious EB progeny (Soules et al., 2020). These observations were supported by recent findings from Hatch and Ouellette (2023) which employed CRISPR interference (CRISPRi) gene knockdown of σ54. Hatch and Ouellette (2023) also used CRISPRi to knockdown σ28 and identified this sigma-factor to be epistatic of σ54 and also involved in secondary RB differentiation.
The use of CRISPRi to knockdown and investigate sigma factors has provided new insights into how RB to EB differentiation is regulated (Swoboda et al., 2023). This approach should be adopted to investigate other transcriptional factors in CT (listed in Table 1A). It is likely that many regulators will be essential and cannot be inactivated, thus utilising CRISPRi to knockdown will clarify how critical their role is in the CT development cycle as well as determine their regulon.
3 Type III secretion system and effector proteins
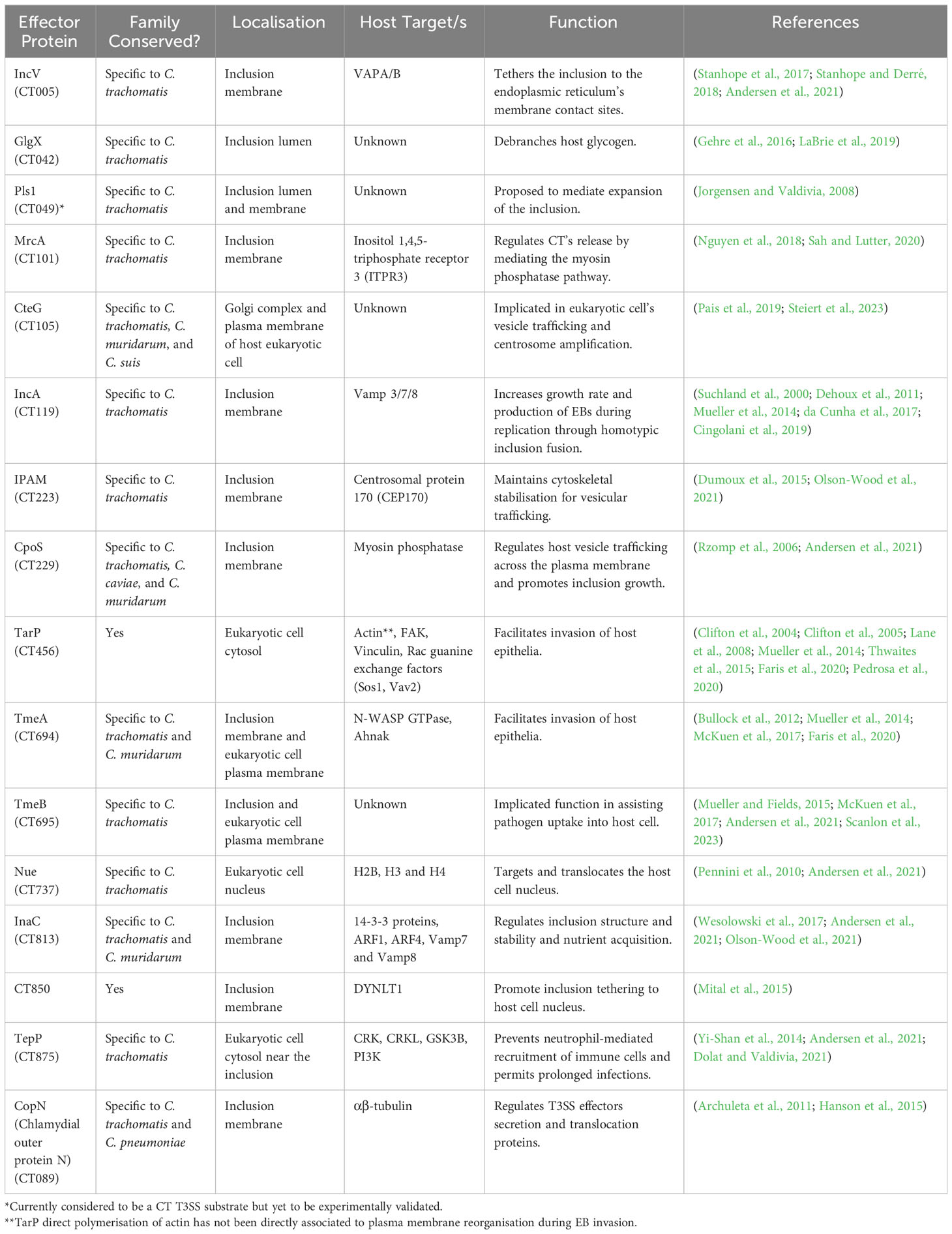
A well-known pathogenic feature of C. trachomatis is the T3SS and their associated effector proteins (Bugalhão and Mota, 2019). The T3SS is composed of a needle-like injectosome apparatus, which secretes effector proteins directly from the chlamydial cytoplasm into the host cell. Specific chlamydial chaperone 4 (Scc4; formerly CT663) precisely regulates T3SS function through gene expression and effector networks (Gao et al., 2020). There are 80 speculated anti-host T3SS-secreted effectors (Betts-Hampikian and Fields, 2010).
The secreted effectors have a variety of functions, including; disrupting the cytoskeletal structure, evasion of host defences, and prevention of host cell apoptosis, described in Table 1B (Coburn et al., 2007). Evidence shows the T3SS not only translocates bacterial-encoded proteins into the eukaryotic host cell’s cytoplasm, but also into the inclusion’s lumen (da Cunha et al., 2017). Some effectors (e.g. TarP and TepP) confer protection against cellular immune response triggers, TLR2, NOD1, and STING (stimulator of interferon genes) (Murray and McKay, 2021). The stimulation of IFN-γ from STING results in activation of indoleamine 2,3-dioxygenase (IDO), an integral element of host defence by reducing tryptophan available for the organism. Tryptophan is crucial to CT pathogenesis and genital CT serovars are able to synthesise tryptophan if indole is present through a functional tryptophan synthase (trypA) gene while ocular strains cannot as trpA is inactivated (McClarty et al., 2006).
Currently, many proposed effectors remain unconfirmed and even more remain uncharacterised, with the only secretion evidence provided using heterologous T3SS expression systems. By utilising knockout genetics to disable the T3SS, followed by phenotypic and proteomic comparison of wildtype and T3SS-null strains will be important in confirming established and discovering new effectors as well as unravelling their functions.
3.1 Tarp
The extracellular translocated actin-recruiting phosphoprotein (TarP, CT456) is required in the early developmental stages (Table 1B) (Clifton et al., 2004). TarP is Slc-1-dependent and involved in chlamydial internalisation, and invasion of host cell (Mueller et al., 2014; Faris et al., 2020). TarP binds to the host cell’s major cytoskeletal component, actin. Phosphorylated TarP interacts with a multitude of host cell signalling molecules, as well as phosphatidylinositol 3-kinase (PI3K) and SHC-transforming protein 1 (SHC1), involved in diverse cell functions (e.g. growth, differentiation, motility, survival) (Shehat et al., 2021). TarP was also discovered to impair the highly conserved Hippo signalling pathway in infected epithelial cells, a previously unknown chlamydia-affected pathway involved in cell proliferation and death (Shehat et al., 2021).
3.2 TepP
The early translocator phosphoprotein, TepP, is the second Slc-1-dependent effector and one of the few T3SS effectors where null mutants have been characterised (Chen et al., 2014). TepP is secreted by the T3SS to enhance CT infectivity and dampen host immune activation. TepP is localised near the inclusion in the eukaryotic cell’s cytosol, where it is phosphorylated by the host kinases (Carpenter et al., 2017). It then targets host factors CRK, PI3K and GSK3B to locally synthesise phosphoinositide-(3,4,5)-triphosphate (PIP3) and modulate host cell signalling in nascent infections. Specifically, PI3K is linked to inhibiting IFN-induced gene transcription within early inclusions. A TepP null-mutant displayed defected growth and altered host IFN-associated gene expression required to activate early immune responses (Chen et al., 2014). Recruitment of PI3K also suggests a role for TepP in modulating membrane vesicle trafficking events (Carpenter et al., 2017).
3.3 Inc proteins
Inc proteins or inclusion membrane proteins, share bilobed hydrophobic domains enabling anchoring to the inclusion membrane. Some Incs contain the presence of vesicle-targeting SNARE-like motifs in their coding sequence (e.g. IncA), which may facilitate host-pathogen interactions (Cingolani et al., 2019; Paumet et al., 2009).CT genome is predicted to encode 55 putative Inc proteins, with 37 confirmed (Weber et al., 2016). There is divergent Inc content across the Chlamydiaceae family, each possessing a distinct distribution pattern along the chlamydial inclusion membrane (Mital et al., 2013). IncA is required for inclusion vacuole fusion (Table 1B), other functions of Inc proteins include; a possible role in maintaining host cell viability (IncG) (Scidmore and Hackstadt, 2001; Verbeke et al., 2006) and as an early Inc, IncD may be important for establishing the inclusion as a replicative niche (Shaw et al., 2000), although further studies are required to confirm this. Chlamydial T3SS-secreted inclusion factors have been studied in greater detail by da Cunha et al. (2017), and Dehoux et al. (2011). Here, we focus on IncM, CTL0390 and CpoS which have been recently characterised.
The IncM effector targets host cell microtubules, as exhibited by its action in host cell multinucleation, centrosome positioning and Golgi distribution. This was found to impact inclusion morphology stabilisation, as IncM-null mutants exhibited defects in these areas, compared to wildtype strains (Luís et al., 2023).
The CTL0390 Inc effector is thought to play a role in mediating EB exit via host-cell lysis during the late stages. A CTL0390 mutant was observed to have reduced Golgi translocation of STING, which is likely required to regulate host-cell lysis for bacterial exit (Bishop and Derré, 2022).
Lastly, CpoS (chlamydial promoter of survival) suppresses apoptosis and necrosis of host cells to prolong chlamydial infection and increase infectivity (Sixt et al., 2017). CpoS affords protection from within the inclusion, thus can only counteract nearby pro-death host signalling (e.g. STING). CpoS-null mutants exhibited attenuated infections, with expedited clearance from murine genital tract models and reduce propagation in cell cultures. Recently, CpoS was found to block Rab GTPases required for lipid transport regulation, as well as the formation of inclusion microdomains of many Incs needed for pathogen-host cell interactions (i.e. IPAM, IncD and CT222) (Meier et al., 2023).
3.4 ChlaDUB
CT868 (ChlaDUB1/Cdu1) and CT867 (ChlaDUB2/Cdu2) are both deubiquitinases which are delivered to the host cytosol, but have differing impacts on CT infectivity. ChlaDUB1 is solely localised to the inclusion membrane where it deubiquinates the anti-apoptosis protein MCL-1. This prevents MCL-1 depletion and apoptosis, thus inhibiting premature host-cell death to allow EB progeny release and increasing infectivity (Fischer et al., 2017). ChlaDUB1 also prevents degradation of host glucose-transporter-1 (GLUT-1) proteins (Wang et al., 2017). This likely supports growth as knockdown of host GLUT-1 impairs C. trachomatis infection (Wang et al., 2017). Inactivation of ChlaDUB1 in CT also enhanced sensitivity to IFN-γ and reduced infectivity in murine infections (Fischer et al., 2017). Alternatively, inactivation of ChlaDUB2, which localises at the inclusion membrane and host cytosol, did not impair infection (Pruneda et al., 2018). However, both ChlaDUBs were found to be involved in Golgi fragmentation (Pruneda et al., 2018).
4 Adhesins
4.1 Ctad1
C. trachomatis adhesin 1 (Ctad1) is a highly conserved 47 kDa invasin encoded by CT017. It contains two SH3 (src Homology-3) domains located near the N-terminus. These domains are responsible for Ctad1 binding to integrin β1 subunits on human epithelial cells (Stallmann and Hegemann, 2016). Ctad1 is expressed on the surface of EBs and mediates C. trachomatis invasion. Upon binding to integrin β1, Ctad1 triggers activation of the host mitogen-activated protein kinase (MAPK) pathway leading to extracellular signal-regulated kinase 1/2 (ERK1/2) phosphorylation and internalisation of EBs (Stallmann and Hegemann, 2016). Blocking of Ctad1 receptors with recombinant Ctad1 was shown to significantly reduce EB adhesion and internalisation, illustrating its role in virulence (Stallmann and Hegemann, 2016).
4.2 PMPs
Polymorphic membrane proteins (Pmps) are a group of type V autotransporter proteins unique to Chlamydiaceae, with variable numbers of Pmps encoded on the genomes. In CT, there are 9 Pmps and all nine are involved in adhesion (Becker and Hegemann, 2014).
Pmps consist of a signal peptide, an N-terminus passenger domain and a C-terminus β channel domain. The C-terminus domain allows translocation of the passenger domain through the outer membrane. After translocation through the outer membrane, Pmps can remain surface bound or be further cleaved and secreted into the inclusion. It has been suggested that the different cleavage of PmpD may lead to fragments with different functions (Swanson et al., 2009).
The expression of Pmps are temporally and developmentally driven. For instance, pmpA and pmpI expression peaked earliest at 12–18 hours post infection and were only detected in RBs (Nunes et al., 2007; Carrasco et al., 2011; Saka et al., 2011). This suggests that both proteins play an important role in RB development. In contrast, pmpD peaks mid-cycle at 12-24 hours post infection, whereas expression of other pmps (pmpB-H) occurred later (Kiselev et al., 2009). This indicates the latter’s involvement in the late stages (such as RB to EB transition) or early stages of infection (EB attachment) (Kiselev et al., 2009; Carrasco et al., 2011; Saka et al., 2011). Remarkably, the expression of the same pmp can vary drastically between different strains (Tan et al., 2009) and even in different inclusions for the same culture (Tan et al., 2010). Besides pmpG, the regulatory mechanism behind Pmp variation is unknown. The flexibility to alter Pmps expression may allow CT to evade antibody recognition targeted against a specific Pmp.
The importance of Pmps as adhesins in CT pathogenesis is best characterised in PmpD. Pre-incubation with recombinant PmpD was found to inhibit CT attachment (Becker and Hegemann, 2014). CT PmpD null mutants also had a 70% reduced host cell attachment and decreased RBs attached to the inclusion membrane, suggesting an additional role for PmpD in maintaining RB-inclusion interactions (Kari et al., 2014). Infection in non-human primates with these PmpD null mutants showed decreased chlamydial burden illustrating its role in pathogenesis (Kari et al., 2014). Although all Pmps are involved in adhesion, it remains unclear what the relative importance of each Pmp to CT is. pmpA, pmpD and pmpI are the most conserved CT Pmp genes and their expression are also unaffected by penicillin-induced stress, suggesting a greater selection pressure to maintain those genes relative to other Pmps (Carrasco et al., 2011).
Pmps may also play an important role in modulating the immune response. In C. pneumoniae and C. psittaci, the N-terminal Pmp21/PmpD activates Toll-like receptor 2 (TLR2), myeloid differentiation factor 88 (MYD88) and nuclear factor κB (NF-κB) signalling leading to Th2 polarised macrophages and upregulation of various cytokines/chemokines including IL-8, IL-6, IL-10 and monocyte chemoattractant protein-1 (MCP-1) (Wehrl et al., 2004). Importantly, Th2 polarised macrophages were associated with reduced nitric oxide (NO) production and Chlamydia killing (Chu et al., 2020). Whether CT Pmps also can modulate the host immune response remains to be seen.
Finally, genomic evidence has suggested that Pmps may be associated with tissue tropism and adaptation. Phylogenetic typing using 6 pmp genes (pmpB, pmpC, pmpF, pmpG, pmpH and pmpI) were able to separate CT strains into their respective biovars and serotypes (Gomes et al., 2006) These genes were under positive selection in one or more niches (Borges et al., 2012), suggesting that Pmps are involved in driving specific host-cell interactions or disease outcomes. However, the mechanisms of how these unique Pmp mutations contribute to phenotypic or virulence differences in CT are yet to be elucidated and are likely to benefit from fluorescence-reported allelic exchange mutagenesis (FRAEM) approaches now available (Mueller et al., 2016).
5 Cytotoxin
The human CT urogenital serovars (D-K) encode a partial 73 kDa cytotoxin (CT166) (Carlson et al., 2004). This cytotoxin was found to contain a functional glycosyltransferase DXD domain and UDP-glucose binding domain with significant homology to the large cytotoxins (LCT) from Clostridioides difficile (Belland et al., 2001). The chlamydial cytotoxin is responsible for the cytopathic ballooning effects seen in infected host cells by glucosylating the Rho-GTPase protein, Rac1. This inactivates Rac1 leading to actin remodelling (Thalmann et al., 2010). In addition to Rac1, the cytotoxin also glucosylates other small GTPases including H-Ras, K-Ras and N-Ras. This was found to inhibit ERK1/2 and P13K/Akt signalling pathways leading to reduced cell cycle progression, division, and migration (Bothe et al., 2015). Crucial to Rac1 and Ras glucosylation is the DXD motif with mutations in this motif shown to abrogate glucosylation (Thalmann et al., 2010; Bothe et al., 2015). However, DXD mutated cytotoxin was still capable of reducing cellular migration suggesting that there may be other toxin effects independent to the DXD motif (Bothe et al., 2015).
Currently, the importance and role of the cytotoxin in CT pathogenesis is unclear. CT166 is present in EBs at the start of infection but is rapidly degraded after 2 hours post-infection (Belland et al., 2001). This suggests potential importance in the initial stages of infection. Ectopic overexpression of CT166 in HeLa cells reduced CT uptake. It was suggested that the chlamydial cytotoxin controls Chlamydia uptake and functions to limit excessive actin polymerisation from other virulence factors such as Tarp (Thalmann et al., 2010). The cytotoxin has also been hypothesised to preserve host energy and nutrients for Chlamydia by limiting energy intensive processes such as cell division and migration (Bothe et al., 2015).
Only urogenital serovars encode a functional cytotoxin. In ocular serovars, the glycosyltransferase domain is deleted while in LGV serovars, both domains are absent (Carlson et al., 2004). It has been argued that the progressive loss of the cytotoxin gene in CT compared to other Chlamydia species like C. muridarum, which encodes 3 paralogous copies, may reflect host adaptation and that the cytotoxin is redundant for human infection.
6 Plasmid
Chlamydia retains a highly conserved ~7.5 kb virulence plasmid that was found to be significant in vivo (Zhong, 2016). The plasmid encodes 8 glycoproteins (pGP1-8) that have a variety of functions including promoting infection ascension, inducing pro-inflammatory responses, and promoting extrusion processes. pGP4, is a putative transcriptional regulator of both plasmid and chromosomal genes (Turman et al., 2023). Virulence plasmid-deficient or pGP3-deficient strains resulted in attenuated infections (Carlson et al., 2008). Under stressful conditions, CT plasmid copy numbers increase, presumably to upregulate T3SS-secreted substrates and ensure in vivo survival (Dirks et al., 2021).
Expression of pGP3 is regulated by pGP4, which similarly is involved in regulating expression of chromosomal glycogen synthase (GlgA), and other proteins with currently unknown functions in pathogenesis, CT050, CT143, and CT144 (Lei et al., 2021). pGP4 was proposed to serve as a plasmid pGP4-dependent secretion system, imperative for delivering pGP3 and GlgA to the host cytosol to increase host glycogen stores (Lei et al., 2021). Contrary to previous results from (da Cunha et al., 2017) this secretion was independent of the T2SS and T3SS (Lei et al., 2021). pGP4-deficient CT strains demonstrate varied transcription of chromosomal and plasmid genes, responsible for various duties (e.g. glycogen synthesis and inclusion morphology) within the developmental cycle that impact virulence (Song et al., 2013). Additionally, pGP4 has been implicated in boosting Euo’s ability to bind and repress Euo-dependent transcription promoters (Table 1A), thus temporally altering late gene expression necessary for RB differentiation into infectious progeny (Zhang et al., 2020). However, plasmid-deficient CT is also capable of inducing sequelae as severe as wild-type strains containing the plasmid (Qu et al., 2015). Thus, the exact role of chlamydial plasmid’s function in pathogenesis remains unknown but can be investigated using new techniques such as FRAEM to create gene deletions (Mueller et al., 2016; Ghosh et al., 2020; Fields et al., 2022).
7 Other characterised chlamydial virulence factors
CT’s pathogenesis is additionally mediated by various proteases and membrane proteins. HtrA (DegP) is a periplasmic serine protease with chaperone functions, including proteolysis of abnormal or misfolded proteins, and serves as a stress response protein that is upregulated in some persistence and stress models (e.g. high temperatures, antibiotics) (Huston et al., 2007; Huston et al., 2008). Little was known about the exact role HtrA played in Chlamydia’s pathogenesis prior to inhibition studies with JO146 (Patel et al., 2014). This inhibitor was most effective in CT’s mid-replication cycle, where administration of the drug resulted in a significant loss in overall vacuole size and in viable infectious EB progeny (Gloeckl et al., 2013; Ong et al., 2013; Lawrence et al., 2016).
Chlamydia Protease-like Activity Factor (CPAF) is a secreted protease that was initially thought to degrade dozens of host proteins involved in Golgi reorganisation, apoptosis, cytoskeleton remodelling and immune regulation. However, these initial CPAF targets identified were disproven as artefacts from in vitro post-lysis degradation, as CPAF is not inhibited by standard protease inhibitors (Chen et al., 2012). To clarify the role of CPAF, a reverse genetics approach re-established CPAF’s likely functions in targeting specific host cell proteins (i.e. vimentin and lamin-associated protein 1) impacting on late stages of infection and development of infectious progeny, however CPAF is not essential for either process (Snavely et al., 2014). Additional experiments also revealed CPAF as a key factor for evading host immune response by paralysing early recruitment of polymorphonuclear cells. Secreted CPAF cleaves FPR2 on the surface of neutrophils, inhibiting their activation, oxidative bursts and production of neutrophil extracellular traps thus establishing longer infections (Rajeeve et al., 2018).
Other important CT proteases include Ptr which is localised to the inclusion lumen and is expressed throughout CT’s development. A CT LGV-L2 ptr-null mutant demonstrated impaired genome replication after IFN-γ removal, along with decreased progeny generation (Panzetta et al., 2019). This links Ptr’s role in recovering from IFN-γ-induced persistence. The periplasmic Tail-specific protease, Tsp, has also been identified as critical for the differentiation from RB to EB via genetic approaches which alter Tsp expression levels (Swoboda et al., 2023).
Lastly, the major outer membrane protein (MOMP) is an immunodominant antigen. It functions as a porin, a cytoadhesin, and is the dominant component of the outer membrane complex that maintains EB stability (Wang et al., 2006; Guifeng et al., 2007).
8 Discussion and conclusion
Genetic approaches to investigate CT are still time consuming and technically difficult. Genome wide genetic manipulation and screening approaches remain challenging. Therefore, selective approaches to genetic manipulation guided by current knowledge of virulence functions, homolog functions, etc. has been used to target loci for genetic manipulation. These genetic approaches in CT have already confirmed the role of several established virulence factors while also overturning or disputing other widely-held understandings in Chlamydia pathogenesis such as the role of the T3SS in secreting plasmid regulated chromosomally-encoded proteins and the role of CPAF (Snavely et al., 2014; Lei et al., 2021). While significant advances in genetic approaches have been made in CT, the progress in other Chlamydia species remains behind. Application or modification of genetic approaches in CT like CRISPRi to other species will allow comparative studies to reveal important determinants of host tropism. Moreover, the recent development of a multiplexed CRISPRi knockdown system in CT is an exciting advancement which will allow interactions between two or more genes to be investigated, providing a more wholistic systems understanding of chlamydia pathogenesis (Hatch and Ouellette, 2023). Ultimately, rapidly expanding the arsenal of genetically investigated virulence factors in CT could inform novel treatment approaches or future attempts to develop a live attenuated vaccine strain.
Author contributions
BJ: Visualization, Writing – original draft, Writing – review & editing. CF: Supervision, Writing – review & editing. WH: Conceptualization, Supervision, Writing – review & editing. LL: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. LL is funded by a University of Technology Sydney Chancellor’s Research Fellowship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CT, Chlamydia trachomatis; RB, Reticulate body; EB, Elementary body; AB, Aberrant body; T3SS, Type 3 secretion system.
References
Abdelsamed, H., Peters, J., Byrne, G. I. (2013). Genetic variation in Chlamydia trachomatis and their hosts: impact on disease severity and tissue tropism. Future Microbiol. 8 (9), 1129–1146. doi: 10.2217/fmb.13.80
Akers, J. C., HoDac, H., Lathrop, R. H., Tan, M. (2011). Identification and functional analysis of CT069 as a novel transcriptional regulator in chlamydia. J. Bacteriol. 193 (22), 6123–6131. doi: 10.1128/JB.05976-11
Akers, J. C., Tan, M. (2006). Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis. J. Bacteriol. 188 (12), 4236–4243. doi: 10.1128/JB.01660-05
Andersen, S. E., Bulman, L. M., Steiert, B., Faris, R., Weber, M. M. (2021). Got mutants? How advances in chlamydial genetics have furthered the study of effector proteins. Pathog. Dis. 79 (2), ftaa078. doi: 10.1093/femspd/ftaa078
Archuleta, T. L., Du, Y., English, C. A., Lory, S., Lesser, C., Ohi, M. D., et al. (2011). The chlamydia effector chlamydial outer protein N (CopN) sequesters tubulin and prevents microtubule assembly. J. Biol. Chem. 286 (39), 33992–33998. doi: 10.1074/jbc.M111.258426
Banerjee, A., Nelson, D. E. (2021). The growing repertoire of genetic tools for dissecting chlamydial pathogenesis. Pathog. Dis. 79 (5), ftab025. doi: 10.1093/femspd/ftab025
Bao, X., Nickels, B. E., Fan, H. (2012). Chlamydia trachomatis protein GrgA activates transcription by contacting the nonconserved region of σ66. Proc. Natl. Acad. Sci. 109 (42), 16870–16875. doi: 10.1073/pnas.1207300109
Barta, M. L., Hickey, J. M., Anbanandam, A., Dyer, K., Hammel, M., Hefty, P. S. (2014). Atypical response regulator ChxR from Chlamydia trachomatis is structurally poised for DNA binding. PloS One 9 (3), e91760–e91760. doi: 10.1371/journal.pone.0091760
Becker, E., Hegemann, J. H. (2014). All subtypes of the Pmp adhesin family are implicated in chlamydial virulence and show species-specific function. MicrobiologyOpen 3 (4), 544–556. doi: 10.1002/mbo3.186
Belland, R. J., Scidmore, M. A., Crane, D. D., Hogan, D. M., Whitmire, W., McClarty, G., et al. (2001). Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. 98 (24), 13984–13989. doi: 10.1073/pnas.241377698
Belland, R. J., Zhong, G., Crane, D. D., Hogan, D., Sturdevant, D., Sharma, J., et al. (2003). Genomic transcriptional profiling of the developmental cycle of chlamydia trachomatis. Proc. Natl. Acad. Sci. 100 (14), 8478–8483. doi: 10.1073/pnas.1331135100
Betts-Hampikian, H. J., Fields, K. A. (2010). The chlamydial type III secretion mechanism: revealing cracks in a tough nut. Front. Microbiol. 1. doi: 10.3389/fmicb.2010.00114
Bishop, R. C., Derré, I. (2022). The Chlamydia trachomatis Inclusion Membrane Protein CTL0390 Mediates Host Cell Exit via Lysis through STING Activation. Infect. Immun. 90 (6), e0019022–e0019022. doi: 10.1128/iai.00190-22
Borges, V., Nunes, A., Ferreira, R., Borrego, M. J., Gomes, J. P. (2012). Directional Evolution of Chlamydia trachomatis towards Niche-Specific Adaptation. J. Bacteriol. 194 (22), 6143–6153. doi: 10.1128/JB.01291-12
Bothe, M., Dutow, P., Pich, A., Genth, H., Klos, A. (2015). DXD motif-dependent and -independent effects of the chlamydia trachomatis cytotoxin CT166. Toxins 7 (2), 621–637. doi: 10.3390/toxins7020621
Bugalhão, J. N., Mota, L. J. (2019). The multiple functions of the numerous Chlamydia trachomatis secreted proteins: the tip of the iceberg. Microbial Cell 6 (9), 414–449. doi: 10.15698/mic2019.09.691
Bullock, H. D., Hower, S., Fields, K. A. (2012). Domain analyses reveal that chlamydia trachomatis CT694 protein belongs to the membrane-localized family of type III effector proteins. J. Biol. Chem. 287 (33), 28078–28086. doi: 10.1074/jbc.M112.386904
Carlson, J. H., Hughes, S., Hogan, D., Cieplak, G., Sturdevant, D. E., McClarty, G., et al. (2004). Polymorphisms in the chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect. Immun. 72 (12), 7063–7072. doi: 10.1128/IAI.72.12.7063-7072.2004
Carlson, J. H., Whitmire, W. M., Crane, D. D., Wicke, L., Virtaneva, K., Sturdevant, D. E., et al. (2008). The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect. Immun. 76 (6), 2273–2283. doi: 10.1128/iai.00102-08
Carlson, J. H., Wood, H., Roshick, C., Caldwell, H. D., McClarty, G. (2006). In vivo and in vitro studies of Chlamydia trachomatis TrpR: DNA interactions. Mol. Microbiol. 59 (6), 1678–1691. doi: 10.1111/j.1365-2958.2006.05045.x
Carpenter, V., Chen, Y.-S., Dolat, L., Valdivia, R. H. (2017). The effector tepP mediates recruitment and activation of phosphoinositide 3-kinase on early chlamydia trachomatis vacuoles. mSphere 2 (4), e00207–17. doi: 10.1128/mSphere.00207-17
Carrasco, J. A., Tan, C., Rank, R. G., Hsia, R.c., Bavoil, P. M. (2011). Altered developmental expression of polymorphic membrane proteins in penicillin-stressed Chlamydia trachomatis. Cell. Microbiol. 13 (7), 1014–1025. doi: 10.1111/j.1462-5822.2011.01598.x
Case, E. D. R., Akers, J. C., Tan, M. (2011). CT406 encodes a chlamydial ortholog of nrdR, a repressor of ribonucleotide reductase. J. Bacteriol. 193 (17), 4396–4404. doi: 10.1128/JB.00294-11
Chen, A. L., Johnson, K. A., Lee, J. K., Sütterlin, C., Tan, M. (2012). CPAF: a Chlamydial protease in search of an authentic substrate. PLoS Pathog. (8), e1002842. doi: 10.1371/journal.ppat.1002842
Chen, Y.-S., Bastidas, R. J., Saka, H. A., Carpenter, V. K., Richards, K. L., Plano, G. V., et al. (2014). The chlamydia trachomatis type III secretion chaperone slc1 engages multiple early effectors, including tepP, a tyrosine-phosphorylated protein required for the recruitment of crkI-II to nascent inclusions and innate immune signaling. PloS Pathog. 10 (2), e1003954. doi: 10.1371/journal.ppat.1003954
Chen, H., Wen, Y., Li, Z. (2019). Clear victory for chlamydia : the subversion of host innate immunity. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01412
Chu, J., Li, X., Qu, G., Wang, Y., Li, Q., Guo, Y., et al. (2020). Chlamydia psittaci pmpD-N exacerbated chicken macrophage function by triggering th2 polarization and the TLR2/myD88/NF-κB signaling pathway. Int. J. Mol. Sci. 21 (6), 2003. doi: 10.3390/ijms21062003
Cingolani, G., McCauley, M., Lobley, A., Bryer, A. J., Wesolowski, J., Greco, D. L., et al. (2019). Structural basis for the homotypic fusion of chlamydial inclusions by the SNARE-like protein IncA. Nat. Commun. 10 (1), 2747–2712. doi: 10.1038/s41467-019-10806-9
Clifton, D. R., Dooley, C. A., Grieshaber, S. S., Carabeo, R. A., Fields, K. A., Hackstadt, T. (2005). Tyrosine phosphorylation of the chlamydial effector protein Tarp is species specific and not required for recruitment of actin. Infect. Immun. 73 (7), 3860–3868. doi: 10.1128/IAI.73.7.3860-3868.2005
Clifton, D. R., Fields, K. A., Grieshaber, S. S., Dooley, C. A., Fischer, E. R., Mead, D. J., et al. (2004). A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. 101 (27), 10166–10171. doi: 10.1073/pnas.0402829101
Coburn, B., Sekirov, I., Finlay, B. B. (2007). Type III secretion systems and disease. Clin. Microbiol. Rev. 20 (4), 535–549. doi: 10.1128/CMR.00013-07
da Cunha, M., Pais, S. V., Bugalhão, J. N., Mota, L. J. (2017). The Chlamydia trachomatis type III secretion substrates CT142, CT143, and CT144 are secreted into the lumen of the inclusion. PloS One 12 (6), e0178856–e0178856. doi: 10.1371/journal.pone.0178856
Davies, B., Turner, K. M., Frølund, M., Ward, H., May, M. T., Rasmussen, S., et al. (2016). Risk of reproductive complications following chlamydia testing: a population-based retrospective cohort study in Denmark. Lancet Infect. Dis. 16 (9), 1057–1064. doi: 10.1016/S1473-3099(16)30092-5
Dehoux, P., Flores, R., Dauga, C., Zhong, G., Subtil, A. (2011). Multi-genome identification and characterization of chlamydiae-specific type III secretion substrates: the Inc proteins. BMC Genomics 12 (1), 109–109. doi: 10.1186/1471-2164-12-109
Dirks, J. A. M. C., Janssen, K., Hoebe, C. J. P. A., Geelen, T. H. B., Lucchesi, M., Dukers-Muijrers, N. H. T. M., et al. (2021). Chlamydia trachomatis intra-bacterial and total plasmid copy number in clinical urogenital samples. Sci. Rep. 11 (1), 259–256. doi: 10.1038/s41598-020-80645-y
Dolat, L., Valdivia, R. H. (2021). An endometrial organoid model of interactions between Chlamydia and epithelial and immune cells. J. Cell Sci. 134 (5), jcs252403. doi: 10.1242/jcs.252403
Dumoux, M., Menny, A., Delacour, D., Hayward, R. D. (2015). A Chlamydia effector recruits CEP170 to reprogram host microtubule organization. J. Cell Sci. 128 (18), 3420–3434. doi: 10.1242/jcs.169318
Elwell, C., Mirrashidi, K., Engel, J. (2016). Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 14 (6), 385–400. doi: 10.1038/nrmicro.2016.30
Faris, R., McCullough, A., Andersen, S. E., Moninger, T. O., Weber, M. M. (2020). The Chlamydia trachomatis secreted effector TmeA hijacks the N-WASP-ARP2/3 actin remodeling axis to facilitate cellular invasion. PloS Pathog. 16 (9), e1008878–e1008878. doi: 10.1371/journal.ppat.1008878
Fields, K. A., Bodero, M. D., Scanlon, K. R., Jewett, T. J., Wolf, K.. (2022). A minimal replicon enables efficacious, species-specific gene deletion in chlamydia and extension of gene knockout studies to the animal model of infection using chlamydia muridarum. Infect. Immun. 90 (12), e0045322. doi: 10.1128/iai.00453-22
Fischer, A., Harrison, K. S., Ramirez, Y., Auer, D., Chowdhury, S. R., Prusty, B. K., et al. (2017). Chlamydia trachomatis -containing vacuole serves as deubiquitination platform to stabilize Mcl-1 and to interfere with host defense. eLife 6, e21465. doi: 10.7554/eLife.21465
Fisher, D. J., Beare, P. A. (2023). Recent advances in genetic systems in obligate intracellular human-pathogenic bacteria. Front. Cell. Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1202245
Gao, L., Cong, Y., Plano, G. V., Rao, X., Gisclair, L. N., Schesser Bartra, S., et al. (2020). Context-dependent action of scc4 reinforces control of the type III secretion system. J. Bacteriol. 202 (15), 1. doi: 10.1128/JB.00132-20
Gehre, L., Gorgette, O., Perrinet, S., Prevost, M.-C., Ducatez, M., Giebel, A. M., et al. (2016). Sequestration of host metabolism by an intracellular pathogen. eLife 5, e12552–e12552. doi: 10.7554/eLife.12552
Geisler, W. M. (2010). Duration of untreated, uncomplicated Chlamydia trachomatis genital infection and factors associated with chlamydia resolution: a review of human studies. J. Infect. Dis. 201 Suppl 2, S104–S113. doi: 10.1086/652402
Gitsels, A., Sanders, N., Vanrompay, D. (2019). Chlamydial infection from outside to inside. Front. Microbiol. 10 (2329). doi: 10.3389/fmicb.2019.02329
Gloeckl, S., Ong, V. A., Patel, P., Tyndall, J. D., Timms, P., Beagley, K. W., et al. (2013). Identification of a serine protease inhibitor which causes inclusion vacuole reduction and is lethal to Chlamydia trachomatis. Mol. Microbiol. 89 (4), 676–689. doi: 10.1111/mmi.12306
Ghosh, S., Ruelke, E. A., Ferrell, J. C., Bodero, M. D., Fields, K. A., Jewett, T. J. (2020). Fluorescence-reported allelic exchange mutagenesis-mediated gene deletion indicates a requirement for chlamydia trachomatis tarp during in vivo infectivity and reveals a specific role for the C terminus during cellular invasion. Infect. Immun. 88 (5), e00841–19. doi: 10.1128/IAI.00841-19
Gomes, J. P., Nunes, A., Bruno, W. J., Borrego, M. J., Florindo, C., Dean, D. (2006). Polymorphisms in the Nine Polymorphic Membrane Proteins of Chlamydia trachomatis across All Serovars: Evidence for Serovar Da Recombination and Correlation with Tissue Tropism. J. Bacteriol. 188 (1), 275–286. doi: 10.1128/JB.188.1.275-286.2006
Guifeng, S. U. N., Pal, S., Sarcon, A. K., Kim, S., Sugawara, E., Nikaido, H., et al. (2007). Structural and functional analyses of the major outer membrane protein of chlamydia trachomatis. J. Bacteriol. 189 (17), 6222–6235. doi: 10.1128/JB.00552-07
Hafner, L., Beagley, K., Timms, P. (2008). Chlamydia trachomatis infection: host immune responses and potential vaccines. Mucosal Immunol. 1 (2), 116–130. doi: 10.1038/mi.2007.19
Hanson, B. R., Slepenkin, A., Peterson, E. M., Tan, M. (2015). Chlamydia trachomatis type III secretion proteins regulate transcription. J. Bacteriol. 197 (20), 3238–3244. doi: 10.1128/JB.00379-15
Hatch, N. D., Ouellette, S. P. (2023). Identification of the alternative sigma factor regulons of Chlamydia trachomatis using multiplexed CRISPR interference. mSphere, e0039123. doi: 10.1128/msphere.00391-23
Hepler, R. W., Nahas, D. D., Lucas, B., Kaufhold, R., Flynn, J. A., Galli, J. D., et al. (2018). Spectroscopic analysis of chlamydial major outer membrane protein in support of structure elucidation: Biophysical Characterization of MOMP. Protein Sci. 27 (11), 1923–1941. doi: 10.1002/pro.3501
Hogerwerf, L., Roof, I., de Jong, M. J. K., Dijkstra, F., van der Hoek, W. (2020). Animal sources for zoonotic transmission of psittacosis: a systematic review. BMC Infect. Dis. 20 (1), 192–192. doi: 10.1186/s12879-020-4918-y
Huang, Y., Wurihan, W., Lu, B., Zou, Y., Wang, Y., Weldon, K., et al. (2022). Robust heat shock response in chlamydia lacking a typical heat shock sigma factor. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.812448
Huston, W. M., Swedberg, J. E., Harris, J. M., Walsh, T. P., Mathews, S. A., Timms, P. (2007). The temperature activated HtrA protease from pathogen Chlamydia trachomatis acts as both a chaperone and protease at 37°C. FEBS Lett. 581 (18), 3382–3386. doi: 10.1016/j.febslet.2007.06.039
Huston, W. M., Theodoropoulos, C., Mathews, S. A., Timms, P. (2008). Chlamydia trachomatis responds to heat shock, penicillin induced persistence, and IFN-gamma persistence by altering levels of the extracytoplasmic stress response protease HtrA. BMC Microbiol. 8 (1), 190–190. doi: 10.1186/1471-2180-8-190
Jorgensen, I., Valdivia, R. H. (2008). Pmp-Like Proteins Pls1 and Pls2 Are Secreted into the Lumen of the Chlamydia trachomatis Inclusion. Infect. Immun. 76 (9), 3940–3950. doi: 10.1128/IAI.00632-08
Kari, L., Southern, T. R., Downey, C. J., Watkins, H. S., Randall, L. B., Taylor, L. D., et al. (2014). Chlamydia trachomatis polymorphic membrane protein D is a virulence factor involved in early host-cell interactions. Infect. Immun. 82 (7), 2756–2762. doi: 10.1128/IAI.01686-14
Kiselev, A. O., Skinner, M. C., Lampe, M. F. (2009). Analysis of pmpD expression and PmpD post-translational processing during the life cycle of Chlamydia trachomatis serovars A, D, and L2. PloS One 4 (4), e5191–e5191. doi: 10.1371/journal.pone.0005191
Kolberg, M., Strand, K. R., Graff, P., Kristoffer Andersson, K. (2004). Structure, function, and mechanism of ribonucleotide reductases. BBA - Proteins Proteomics 1699 (1), 1–34. doi: 10.1016/j.bbapap.2004.02.007
Koo, I. C., Stephens, R. S. (2003). A developmentally regulated two-component signal transduction system in chlamydia. J. Biol. Chem. 278 (19), 17314–17319. doi: 10.1074/jbc.M212170200
Koo, I. C., Walthers, D., Hefty, P. S., Kenney, L. J., Stephens, R. S. (2006). ChxR is a transcriptional activator in chlamydia. Proc. Natl. Acad. Sci. - PNAS 103 (3), 750–755. doi: 10.1073/pnas.0509690103
LaBrie, S. D., Dimond, Z. E., Harrison, K. S., Baid, S., Wickstrum, J., Suchland, R. J., et al. (2019). Transposon mutagenesis in chlamydia trachomatis identifies CT339 as a comEC homolog important for DNA uptake and lateral gene transfer. mBio 10 (4), e01343–19. doi: 10.1128/mBio.01343-19
Lane, B. J., Mutchler, C., Al Khodor, S., Grieshaber, S. S., Carabeo, R. A. (2008). Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PloS Pathog. 4 (3), e1000014. doi: 10.1371/journal.ppat.1000014
Lawrence, A., Fraser, T., Gillett, A., Tyndall, J. D. A., Timms, P., Polkinghorne, A., et al. (2016). Chlamydia Serine Protease Inhibitor, targeting HtrA, as a New Treatment for Koala Chlamydia infection. Sci. Rep. 6 (1), 31466. doi: 10.1038/srep31466
Lei, L., Yang, C., Patton, M. J., Smelkinson, M., Dorward, D., Ma, L., et al. (2021). A chlamydial plasmid-dependent secretion system for the delivery of virulence factors to the host cytosol. mBio 12 (3), e0117921–e0117921. doi: 10.1128/mBio.01179-21
Li, Z., Douglas, A. L., Hatch, T. P. (1998). Characterization of a Chlamydia psittaci DNA binding protein (EUO) synthesized during the early and middle phases of the developmental cycle. Infect. Immun. 66 (3), 1167–1173. doi: 10.1128/iai.66.3.1167-1173.1998
Luís, M. P., Pereira, I. S., Bugalhão, J. N., Simões, C. N., Mota, C., Romão, M. J., et al. (2023). The chlamydia trachomatis incM protein interferes with host cell cytokinesis, centrosome positioning, and golgi distribution and contributes to the stability of the pathogen-containing vacuole. Infect. Immun. 91 (4), e0040522–e0040522. doi: 10.1128/iai.00405-22
Luu, L. D. W., Kasimov, V., Phillips, S., Myers, G. S., Jelocnik, M. (2023). Genome organization and genomics in Chlamydia: whole genome sequencing increases understanding of chlamydial virulence, evolution, and phylogeny. Front. Cell. Infect. Microbiol. 13, 1178736. doi: 10.3389/fcimb.2023.1178736
Mandel, C., Yang, H., Buchko, G. W., Abendroth, J., Grieshaber, N., Chiarelli, T., et al. (2022). Expression and structure of the Chlamydia trachomatis DksA ortholog. Pathog. Dis. 80 (1), ftac007. doi: 10.1093/femspd/ftac007
Mathews, S. A., Volp, K. M., Timms, P. (1999). Development of a quantitative gene expression assay for Chlamydia trachomatis identified temporal expression of σ factors. FEBS Lett. 458 (3), 354–358. doi: 10.1016/S0014-5793(99)01182-5
McClarty, G., Caldwell, H. D., Nelson, D. E. (2006). Chlamydial interferon gamma immune evasion influences infection tropism. Curr. Opin. Microbiol. 10 (1), 47–51. doi: 10.1016/j.mib.2006.12.003
McKuen, M. J., Mueller, K. E., Bae, Y. S., Fields, K. A. (2017). Fluorescence-reported allelic exchange mutagenesis reveals a role for chlamydia trachomatis tmeA in invasion that is independent of host AHNAK. Infect. Immun. 85 (12), e00640–17. doi: 10.1128/IAI.00640-17
Meier, K., Jachmann, L. H., Türköz, G., Babu Sait, M. R., Pérez, L., Kepp, O., et al. (2023). The Chlamydia effector CpoS modulates the inclusion microenvironment and restricts the interferon response by acting on Rab35. mBio 14 (4), e0319022. doi: 10.1128/mbio.03190-22
Menon, S., Timms, P., Allan, J. A., Alexander, K., Rombauts, L., Horner, P., et al. (2015). Human and pathogen factors associated with chlamydia trachomatis-related infertility in women. Clin. Microbiol. Rev. 28 (4), 969–985. doi: 10.1128/CMR.00035-15
Mishkin, N., Ricart Arbona, R. J., Carrasco, S. E., Lawton, S., Henderson, K. S., Momtsios, P., et al. (2022). Reemergence of the murine bacterial pathogen chlamydia muridarum in research mouse colonies. Comp. Med. 72 (4), 230–242. doi: 10.30802/AALAS-CM-22-000045
Mital, J., Lutter, E. I., Barger, A. C., Dooley, C. A., Hackstadt, T. (2015). Chlamydia trachomatis inclusion membrane protein CT850 interacts with the dynein light chain DYNLT1 (Tctex1). Biochem. Biophys. Res. Commun. 462 (2), 165–170. doi: 10.1016/j.bbrc.2015.04.116
Mital, J., Miller, N. J., Dorward, D. W., Dooley, C. A., Hackstadt, T. (2013). Role for chlamydial inclusion membrane proteins in inclusion membrane structure and biogenesis. PloS One 8 (5), e63426–e63426. doi: 10.1371/journal.pone.0063426
Mueller, K. E., Fields, K. A. (2015). Application of β-lactamase reporter fusions as an indicator of effector protein secretion during infections with the obligate intracellular pathogen Chlamydia trachomatis. PloS One 10 (8), e0135295–e0135295. doi: 10.1371/journal.pone.0135295
Mueller, K. E., Plano, G. V., Fields, K. A. (2014). New frontiers in type III secretion biology: the Chlamydia perspective. Infect. Immun. 82 (1), 2–9. doi: 10.1128/IAI.00917-13
Mueller, K. E., Wolf, K., Fields, K. A. (2016). Gene deletion by fluorescence-reported allelic exchange mutagenesis in chlamydia trachomatis. mBio 7 (1), e01817–e01815. doi: 10.1128/mBio.01817-15
Murray, S. M., McKay, P. F. (2021). Chlamydia trachomatis: Cell biology, immunology and vaccination. Vaccine 39 (22), 2965–2975. doi: 10.1016/j.vaccine.2021.03.043
Murthy, A. K., Li, W., Ramsey, K. H. (2018). Immunopathogenesis of chlamydial infections. Curr. topics Microbiol. Immunol. 412, 183–215. doi: 10.1007/82_2016_18
Newman, L., Rowley, J., Vander Hoorn, S., Wijesooriya, N. S., Unemo, M., Low, N., et al. (2015). Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PloS One 10 (12), e0143304–e0143304. doi: 10.1371/journal.pone.0143304
Nguyen, P. H., Lutter, E. I., Hackstadt, T. (2018). Chlamydia trachomatis inclusion membrane protein MrcA interacts with the inositol 1,4,5-trisphosphate receptor type 3 (ITPR3) to regulate extrusion formation. PloS Pathog. 14 (3), e1006911. doi: 10.1371/journal.ppat.1006911
Nicholson, T. L., Olinger, L., Chong, K., Schoolnik, G., Stephens, R. S. (2003). Global Stage-Specific Gene Regulation during the Developmental Cycle of Chlamydia trachomatis. J. Bacteriol. 185 (10), 3179–3189. doi: 10.1128/JB.185.10.3179-3189.2003
Nunes, A., Gomes, J. P., Mead, S., Florindo, C., Correia, H., Borrego, M. J., et al. (2007). Comparative expression profiling of the Chlamydia trachomatis pmp gene family for clinical and reference strains. PloS One 2 (9), e878–e878. doi: 10.1371/journal.pone.0000878
O'Neill, C. E., Skilton, R. J., Forster, J., Cleary, D. W., Pearson, S. A., Lampe, D. J., et al. (2021). An inducible transposon mutagenesis approach for the intracellular human pathogen Chlamydia trachomatis. Wellcome Open Res. 6, 312. doi: 10.12688/wellcomeopenres.16068.1
Olson-Wood, M. G., Jorgenson, L. M., Ouellette, S. P., Rucks, E. A. (2021). Inclusion membrane growth and composition are altered by overexpression of specific inclusion membrane proteins in chlamydia trachomatis L2. Infect. Immun. 89 (7), e0009421–e0009421. doi: 10.1128/IAI.00094-21
Ong, V. A., Marsh, J. W., Lawrence, A., Allan, J. A., Timms, P., Huston, W. M. (2013). The protease inhibitor JO146 demonstrates a critical role for CtHtrA for Chlamydia trachomatis reversion from penicillin persistence. Front. Cell. Infect. Microbiol. 3. doi: 10.3389/fcimb.2013.00100
Ouellette, S. P. (2018). Feasibility of a conditional knockout system for chlamydia based on CRISPR interference. Front. Cell. Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00059
Ouellette, S. P., Blay, E. A., Hatch, N. D., Fisher-Marvin, L. A. (2021). CRISPR interference to inducibly repress gene expression in chlamydia trachomatis. Infect. Immun. 89 (7), e0010821. doi: 10.1128/iai.00108-21
Pais, S. V., Key, C. E., Borges, V., Pereira, I. S., Gomes, J. P., Fisher, D. J., et al. (2019). CteG is a Chlamydia trachomatis effector protein that associates with the Golgi complex of infected host cells. Sci. Rep. 9 (1), 6133–6133. doi: 10.1038/s41598-019-42647-3
Panzetta, M. E., Luján, A. L., Bastidas, R. J., Damiani, M. T., Valdivia, R. H., Saka, H. A. (2019). Ptr/CTL0175 is required for the efficient recovery of chlamydia trachomatis from stress induced by gamma-interferon. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00756
Panzetta, M. E., Valdivia, R. H., Saka, H. A. (2018). Chlamydia persistence: A survival strategy to evade antimicrobial effects in-vitro and in-vivo. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.03101
Patel, P., De Boer, L., Timms, P., Huston, W. M. (2014). Evidence of a conserved role for Chlamydia HtrA in the replication phase of the chlamydial developmental cycle. Microbes infection 16 (8), 690–694. doi: 10.1016/j.micinf.2014.07.003
Paumet, F., Wesolowski, J., Garcia-Diaz, A., Delevoye, C., Aulner, N., Shuman, H. A., et al. (2009). Intracellular bacteria encode inhibitory SNARE-like proteins. PloS One 4 (10), e7375.
Pedrosa, A. T., Murphy, K. N., Nogueira, A. T., Brinkworth, A. J., Thwaites, T. R., Aaron, J., et al. (2020). A post-invasion role for Chlamydia type III effector TarP in modulating the dynamics and organization of host cell focal adhesions. J. Biol. Chem. 295 (43), 14763–14779. doi: 10.1074/jbc.RA120.015219
Pennini, M. E., Perrinet, S., Dautry-Varsat, A., Subtil, A. (2010). Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PloS Pathog. 6 (7), e1000995–e1000995. doi: 10.1371/journal.ppat.1000995
Pokorzynski, N. D., Brinkworth, A. J., Carabeo, R. (2019). A bipartite iron-dependent transcriptional regulation of the tryptophan salvage pathway in Chlamydia trachomatis. eLife 8, e42295. doi: 10.7554/eLife.42295
Pruneda, J. N., Bastidas, R. J., Bertsoulaki, E., Swatek, K. N., Santhanam, B., Clague, M. J., et al. (2018). A Chlamydia effector combining deubiquitination and acetylation activities induces Golgi fragmentation. Nat. Microbiol. 3 (12), 1377–1384. doi: 10.1038/s41564-018-0271-y
Qu, Y., Frazer, L. C., O'Connell, C. M., Tarantal, A. F., Andrews, J. C. W., O'Connor, S. L., et al. (2015). Comparable genital tract infection, pathology, and immunity in rhesus macaques inoculated with wild-type or plasmid-deficient chlamydia trachomatis serovar D. Infect. Immun. 83 (10), 4056–4067. doi: 10.1128/IAI.00841-1510.1128/IAI.00841-15
Rajeeve, K., Das, S., Prusty, B. K., Rudel, T. (2018). Chlamydia trachomatis paralyses neutrophils to evade the host innate immune response. Nat. Microbiol. 3 (7), 824–835. doi: 10.1038/s41564-018-0182-y
Rao, X., Deighan, P., Hua, Z., Hu, X., Wang, J., Luo, M., et al. (2009). A regulator from Chlamydia trachomatis modulates the activity of RNA polymerase through direct interaction with the beta subunit and the primary sigma subunit. Genes Dev. 23 (15), 1818–1829. doi: 10.1101/gad.1784009
Read, T. D., Massey, R. C. (2014). Characterizing the genetic basis of bacterial phenotypes using genome-wide association studies: A new direction for Bacteriol. Genome Med. 6 (11), 109–109. doi: 10.1186/s13073-014-0109-z
Rzomp, K. A., Moorhead, A. R., Scidmore, M. A. (2006). The GTPase rab4 interacts with chlamydia trachomatis inclusion membrane protein CT229. Infect. Immun. 74 (9), 5362–5373. doi: 10.1128/IAI.00539-06
Sah, P., Lutter, E. I. (2020). Hijacking and use of host kinases by chlamydiae. Pathogens 9 (12), 1034. doi: 10.3390/pathogens9121034
Saka, H. A., Thompson, J. W., Chen, Y.-S., Kumar, Y., Dubois, L. G., Moseley, M. A., et al. (2011). Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms: Quantitative proteomics analysis of C. trachomatis developmental forms. Mol. Microbiol. 82 (5), 1185–1203. doi: 10.1111/j.1365-2958.2011.07877.x
Scanlon, K. R., Keb, G., Wolf, K., Jewett, T. J., Fields, K. A. (2023). Chlamydia trachomatis TmeB antagonizes actin polymerization via direct interference with Arp2/3 activity. Front. Cell. Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1232391
Schaumburg, C. S., Tan, M. (2006). Arginine-dependent gene regulation via the argR repressor is species specific in chlamydia. J. Bacteriol. 188 (3), 919–927. doi: 10.1128/JB.188.3.919-927.2006
Schoborg, R. V. (2011). Chlamydia persistence – a tool to dissect chlamydia–host interactions. Microbes infection 13 (7), 649–662. doi: 10.1016/j.micinf.2011.03.004
Scidmore, M. A. (2009). "Chlamydia," in Encyclopedia of Microbiology (Third Edition). Ed. Schaechter, M. (Oxford: Academic Press), 74–86.
Scidmore, M., Hackstadt, T. (2001). Mammalian 14-3-3β associates with the chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 39 (6), 1638–1650.
Shehat, M. G., Aranjuez, G. F., Kim, J., Jewett, T. J. (2021). The Chlamydia trachomatis Tarp effector targets the Hippo pathway. Biochem. Biophys. Res. Commun. 562, 133–138. doi: 10.1016/j.bbrc.2021.05.057
Sixt, B. S., Bastidas, R. J., Finethy, R., Baxter, R. M., Carpenter, V. K., Kroemer, G., et al. (2017). The chlamydia trachomatis inclusion membrane protein cpoS counteracts STING-mediated cellular surveillance and suicide programs. Cell Host Microbe 21 (1), 113–121. doi: 10.1016/j.chom.2016.12.002
Shaw, E., Dooley, C., Fischer, E., Scidmore, M., Fields, K., Hackstadt, T. (2000). Three temporal classes of gene expression during the chlamydia trachomatis developmental cycle. Mol. Microbiol. 37 (4), 913–925.
Snavely, E. A., Kokes, M., Dunn, J. D., Saka, H. A., Nguyen, B. D., Bastidas, R. J., et al. (2014). Reassessing the role of the secreted protease CPAF in Chlamydia trachomatis infection through genetic approaches. Pathog. Dis. 71 (3), 336–351. doi: 10.1111/2049-632X.12179
Song, L., Carlson, J. H., Whitmire, W. M., Kari, L., Virtaneva, K., Sturdevant, D. E., et al. (2013). Chlamydia trachomatis plasmid-encoded pgp4 is a transcriptional regulator of virulence-associated genes. Infect. Immun. 81 (3), 636–644. doi: 10.1128/IAI.01305-12
Soules, K. R., LaBrie, S. D., May, B. H., Hefty, P. S. (2020). Sigma 54-Regulated Transcription Is Associated with Membrane Reorganization and Type III Secretion Effectors during Conversion to Infectious Forms of Chlamydia trachomatis. mBio 11 (5), e01725–20. doi: 10.1128/mBio.01725-20
Stallmann, S., Hegemann, J. H. (2016). The Chlamydia trachomatis Ctad1 invasin exploits the human integrin β1 receptor for host cell entry: Chlamydia trachomatis Ctad1 binds to integrin β1. Cell. Microbiol. 18 (5), 761–775. doi: 10.1111/cmi.12549
Stanhope, R., Derré, I. (2018). Making contact: VAP targeting by intracellular pathogens. Contact 1, 251525641877551. doi: 10.1177/2515256418775512
Stanhope, R., Flora, E., Bayne, C., Derré, I. (2017). IncV, a FFAT motif-containing Chlamydia protein, tethers the endoplasmic reticulum to the pathogen-containing vacuole. Proc. Natl. Acad. Sci. 114 (45), 12039–12044. doi: 10.1073/pnas.1709060114
Steiert, B., Icardi, C. M., Faris, R., McCaslin, P. N., Smith, P., Klingelhutz, A. J., et al. (2023). The Chlamydia trachomatis type III-secreted effector protein CteG induces centrosome amplification through interactions with centrin-2. Proc. Natl. Acad. Sci. 120 (20), e2303487120. doi: 10.1073/pnas.2303487120
Stephens, R. S., Kalman, S., Lammel, C., Fan, J., Marathe, R., Aravind, L., et al. (1998). Genome sequence of an obligate intracellular pathogen of humans: chlamydia trachomatis. Science 282 (5389), 754–759. doi: 10.1126/science.282.5389.754
Suchland, R. J., Rockey, D. D., Bannantine, J. P., Stamm, W. E. (2000). Isolates of chlamydia trachomatis that occupy nonfusogenic inclusions lack incA, a protein localized to the inclusion membrane. Infect. Immun. 68 (1), 360–367. doi: 10.1128/IAI.68.1.360-367.2000
Swanson, K. A., Taylor, L. D., Frank, S. D., Sturdevant, G. L., Fischer, E. R., Carlson, J. H., et al. (2009). Chlamydia trachomatis polymorphic membrane protein D is an oligomeric autotransporter with a higher-order structure. Infect. Immun. 77 (1), 508–516. doi: 10.1128/IAI.01173-08
Swoboda, A. R., Wood, N. A., Saery, E. A., Fisher, D. J., Ouellette, S. P. (2023). The periplasmic tail-specific protease, tsp, is essential for secondary differentiation in chlamydia trachomatis. J. Bacteriol. 205 (5), e0009923–e0009923. doi: 10.1128/jb.00099-23
Tan, C., Hsia, R.-c., Shou, H., Carrasco, J. A., Rank, R. G., Bavoil, P. M. (2010). Variable expression of surface-exposed polymorphic membrane proteins in in vitro-grown Chlamydia trachomatis. Cell. Microbiol. 12 (2), 174–187. doi: 10.1111/j.1462-5822.2009.01389.x
Tan, C., Hsia, R.-c., Shou, H., Haggerty, C. L., Ness, R. B., Gaydos, C. A., et al. (2009). Chlamydia trachomatis-infected patients display variable antibody profiles against the nine-member polymorphic membrane protein family. Infect. Immun. 77 (8), 3218–3226. doi: 10.1128/IAI.01566-08
Taylor, H. R., Burton, M. J., Haddad, D., West, S., Wright, H. (2014). Trachoma. Lancet 384 (9960), 2142–2152. doi: 10.1016/S0140-6736(13)62182-0
Thalmann, J., Janik, K., May, M., Sommer, K., Ebeling, J., Hofmann, F., et al. (2010). Actin re-organization induced by Chlamydia trachomatis serovar D–evidence for a critical role of the effector protein CT166 targeting Rac. PloS One 5 (3), e9887–e9887. doi: 10.1371/journal.pone.0009887
Thompson, C. C., Griffiths, C., Nicod, S. S., Lowden, N. M., Wigneshweraraj, S., Fisher, D. J., et al. (2015). The Rsb Phosphoregulatory Network Controls Availability of the Primary Sigma Factor in Chlamydia trachomatis and Influences the Kinetics of Growth and Development. PloS Pathog. 11 (8), e1005125. doi: 10.1371/journal.ppat.1005125
Thompson, C. C., Nicod, S. S., Malcolm, D. S., Grieshaber, S. S., Carabeo, R. A. (2012). Cleavage of a putative metal permease in Chlamydia trachomatis yields an iron-dependent transcriptional repressor. Proc. Natl. Acad. Sci. - PNAS 109 (26), 10546–10551. doi: 10.1073/pnas.1201398109
Thwaites, T. R., Pedrosa, A. T., Peacock, T. P., Carabeo, R. A. (2015). Vinculin interacts with the Chlamydia effector TarP via a tripartite vinculin binding domain to mediate actin recruitment and assembly at the plasma membrane. Front. Cell. Infect. Microbiol. 5, 88. doi: 10.3389/fcimb.2015.00088
Turman, B. J., Darville, T., O'Connell, C. M. (2023). Plasmid-mediated virulence in chlamydia. Front. Cell. Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1251135
Verbeke, P., Welter-Stahl, L., Ying, S., Hansen, J., Häcker, G., Darville, T., et al. (2006). Recruitment of BAD by the chlamydia trachomatis vacuole correlates with host-cell survival. PloS Pathog. 2 (5), e45–e45. doi: 10.1371/journal.ppat.0020045
Wan, W., Li, D., Li, D., Jiao, J. (2023). Advances in genetic manipulation of Chlamydia trachomatis. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1209879
Wang, Y., Berg, E. A., Feng, X., Shen, L., Smith, T., Costello, C. E., et al. (2006). Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci. 15 (1), 122–134. doi: 10.1110/ps.051616206
Wang, X., Hybiske, K., Stephens, R. S. (2017). Orchestration of the mammalian host cell glucose transporter proteins-1 and 3 by Chlamydia contributes to intracellular growth and infectivity. Pathog. Dis. 75 (8), ftx108. doi: 10.1093/femspd/ftx108
Weber, M. M., Noriea, N. F., Bauler, L. D., Lam, J. L., Sager, J., Wesolowski, J., et al. (2016). A functional core of incA is required for chlamydia trachomatis inclusion fusion. J. Bacteriol. 198 (8), 1347–1355. doi: 10.1128/JB.00933-1510.1128/JB.00933-15
Wehrl, W., Brinkmann, V., Jungblut, P. R., Meyer, T. F., Szczepek, A. J. (2004). From the inside out – processing of the Chlamydial autotransporter PmpD and its role in bacterial adhesion and activation of human host cells. Mol. Microbiol. 51 (2), 319–334. doi: 10.1046/j.1365-2958.2003.03838.x
Wesolowski, J., Weber, M. M., Nawrotek, A., Dooley, C. A., Calderon, M., St Croix, C. M., et al. (2017). Chlamydia hijacks ARF GTPases to coordinate microtubule posttranslational modifications and golgi complex positioning. mBio 8 (3), e02280–16. doi: 10.1128/mBio.02280-16
Whittum-Hudson, J. A., Hudson, A. P. (2005). Human chlamydial infections : persistence, prevalence, and outlook for the future. Natures Sci. sociétés 13 (4), 371–382. doi: 10.1051/nss:2005057
Wilson, A. C., Tan, M. (2002). Functional analysis of the heat shock regulator HrcA of Chlamydia trachomatis. J. Bacteriol. 184 (23), 6566–6571. doi: 10.1128/JB.184.23.6566-6571.2002
Witkin, S. S., Minis, E., Athanasiou, A., Leizer, J., Linhares, I. M. (2017). Chlamydia trachomatis: the persistent pathogen. Clin. Vaccine Immunol. 24 (10), e00203–e00217. doi: 10.1128/CVI.00203-17
Wurihan, W., Zou, Y., Weber, A. M., Weldon, K., Huang, Y., Bao, X., et al. (2021). Identification of a grgA-euo-hrcA transcriptional regulatory network in chlamydia. mSystems 6 (4), e0073821–e0073821. doi: 10.1128/mSystems.00738-21
Yang, C., Kari, L., Sturdevant, G. L., Song, L., Patton, M. J., Couch, C. E., et al. (2017). Chlamydia trachomatis ChxR is a transcriptional regulator of virulence factors that function in in vivo host–pathogen interactions. Pathog. Dis. 75 (3), ftx035. doi: 10.1093/femspd/ftx035
Yi-Shan, C., Robert, J. B., Hector, A. S., Victoria, K. C., Kristian, L. R., Gregory, V. P., et al. (2014). The chlamydia trachomatis type III secretion chaperone slc1 engages multiple early effectors, including tepP, a tyrosine-phosphorylated protein required for the recruitment of crkI-II to nascent inclusions and innate immune signaling. PloS Pathog. 10 (2), e1003954. doi: 10.1371/journal.ppat.1003954
Zhang, Q., Rosario, C. J., Sheehan, L. M., Rizvi, S. M., Brothwell, J. A., He, C., et al. (2020). The repressor function of the chlamydia late regulator EUO is enhanced by the plasmid-encoded protein pgp4. J. Bacteriol. 202 (8), e00793–19. doi: 10.1128/JB.00793-19
Keywords: Chlamydia trachomatis, pathogenesis, virulence, immune evasion, type III secretion system, proteases, cytotoxin, polymorphic membrane proteins
Citation: Jury B, Fleming C, Huston WM and Luu LDW (2023) Molecular pathogenesis of Chlamydia trachomatis. Front. Cell. Infect. Microbiol. 13:1281823. doi: 10.3389/fcimb.2023.1281823
Received: 23 August 2023; Accepted: 03 October 2023;
Published: 18 October 2023.
Edited by:
Scot P. Ouellette, University of Nebraska Medical Center, United StatesReviewed by:
Catherine Mary O’Connell, University of North Carolina at Chapel Hill, United StatesHector Alex Saka, Consejo Nacional de Investigaciones Científicas y Técnicas, Córdoba, Argentina
Copyright © 2023 Jury, Fleming, Huston and Luu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laurence Don Wai Luu, bGF1cmVuY2UubHV1QHV0cy5lZHUuYXU=
 Brittany Jury1
Brittany Jury1 Wilhelmina M. Huston
Wilhelmina M. Huston Laurence Don Wai Luu
Laurence Don Wai Luu