- Fraunhofer Institute for Cell Therapy and Immunology, Department of Vaccines and Infection Models, Leipzig, Germany
Introduction: West Nile Virus (WNV) is a zoonotic flavivirus transmitted by mosquitoes. Especially in the elderly or in immunocompromised individuals an infection with WNV can lead to severe neurological symptoms. To date, no human vaccine against WNV is available. The Envelope (E) protein, located at the surface of flaviviruses, is involved in the invasion into host cells and is the major target for neutralizing antibodies and therefore central to vaccine development. Due to their close genetic and structural relationship, flaviviruses share highly conserved epitopes, such as the fusion loop domain (FL) in the E protein, that are recognized by cross-reactive antibodies. These antibodies can lead to enhancement of infection with heterologous flaviviruses, which is a major concern for potential vaccines in areas with co-circulation of different flaviviruses, e.g. Dengue or Zika viruses.
Material: To reduce the potential of inducing cross-reactive antibodies, we performed an immunization study in mice using WNV E proteins with either wild type sequence or a mutated FL, and WNV E domain III which does not contain the FL at all.
Results and discussion: Our data show that all antigens induce high levels of WNV-binding antibodies. However, the level of protection against WNV varied, with the wildtype E protein inducing full, the other antigens only partial protection. On the other hand, serological cross-reactivity to heterologous flaviviruses was significantly reduced after immunization with the mutated E protein or domain III as compared to the wild type version. These results have indications for choosing antigens with the optimal specificity and efficacy in WNV vaccine development.
1 Introduction
The human pathogenic and mosquito-transmitted West-Nile Virus (WNV), belongs to the family Flaviviridae, which are enveloped viruses that contain a single stranded, positive sense RNA genome with a length of 11kb. It encodes three structural proteins (Capsid protein, Envelope protein, Premembrane protein) which form the virion, and seven nonstructural proteins, which are translated as a single polyprotein that is co- and post-translationally cleaved by viral and host proteases. (Hayes et al., 2005; Welsch et al., 2009). Flaviviridae also include a number of other human pathogens, such as Dengue (DENV), Japanese encephalitis virus (JEV), Zika (ZIKV), Tick-borne encephalitis (TBEV) or Yellow fever (YFV) viruses. WNV is currently the most widely spread vector-borne flavivirus (Chancey et al., 2015) and is endemic in regions of the Americas (Hadfield et al., 2019), Europe (Giesen et al., 2023), Africa (García-Carrasco et al., 2023), Australia (Prow et al., 2016) and Asia (Bassal et al., 2017). WNV primarily circulates between birds, the natural reservoir, and mosquitoes (Kilpatrick et al., 2007). The vectors can transmit WNV also to mammals including horses and humans, however, these represent dead-end hosts and do not re-infect mosquitoes (Sewgobind et al., 2023). Nevertheless, transmission between humans may occur via organ or blood transplantation (Centers for Disease Control and Prevention (CDC), 2009; Byas and Ebel, 2020).
Infections in humans mainly lead to mild, flu-like symptoms or remain asymptomatic (Hayes et al., 2005). However, approx. 1% of infections are affecting the central nervous system (CNS) causing encephalitis, meningitis and muscle paralysis with a possible fatal outcome (Bai et al., 2019). The risk for severe, neuroinvasive WNV infections increases with age (Montgomery, 2016) or is correlated to an immunocompromised immune system (Sejvar, 2016). To date, there is no specific treatment for WNV disease, hence clinical management is purely supportive. An infection with WNV leads to a long-lasting immunity to the virus. Several vaccine approaches have been developed for the prevention of WNV induced disease in humans, although until now none has progressed beyond phase 2 clinical studies (Gould et al., 2023).
The key antigen in almost all WNV vaccine candidates is the envelope (E) protein, which is targeted by a variety of virus neutralizing antibodies. These antibodies are critical for the induction of a protective immunity (Pierson et al., 2008). The E protein consists of an ectodomain that is anchored in the viral envelope and is divided into the major domains I, II, and III. Domain II (EDII) contains the fusion loop (FL), and domain III (EDIII) binds to the host cell receptor(s) (Zhang et al., 2017). For immunization studies, the E protein has been administered in different forms: as recombinant protein, as part of virus-like particles (VLP) or via inactivated viruses and via different viral vectors, and in general, protective immune responses were induced (Davis et al., 2001; Bonafé et al., 2009; Lieberman et al., 2009; Brandler et al., 2012; Dayan et al., 2013; Pinto et al., 2013; Magnusson et al., 2014). However, the close genetic and structural relationship of flaviviruses leads to highly conserved epitopes within the E protein, and as a consequence a major problem in flavivirus immunology remains the cross-reactivity of immune responses, especially the induction of cross-reacting antibodies (Rey et al., 2018). Such cross-reacting antibodies have been linked to the phenomenon of antibody dependent enhancement of infection (ADE). Although not yet understood in detail, ADE was associated to non- or sub-neutralizing antibodies that bind the virus and can lead to increased viral entry into host cells due to Fc-receptor mediated endocytosis (Sarker et al., 2023). ADE is most problematic in areas where closely related flaviviruses co-circulate and has been documented clinically between infections of the different DENV serotypes, but also in subsequent infections of ZIKV and DENV (Rothman, 2011; Katzelnick et al., 2017; Katzelnick et al., 2020). In addition, ADE between different flaviviruses has been demonstrated in preclinical models, e.g. WNV antibodies enhanced the infection with ZIKV (Bardina et al., 2017). Importantly, ADE is of concern for the development of flavivirus vaccines, as vaccine-induced antibodies might constitute a risk for enhanced disease upon infection with another flavivirus or between different serotypes within one flavivirus (Halstead, 2018; Ulbert, 2019).
A large proportion of cross-reactive flavivirus antibodies target the highly conserved fusion loop (FL) of the E protein, which contains stretches of amino acids that are almost identical in many human pathogenic flaviviruses (Crill and Chang, 2004; Rey et al., 2018). Modifying the FL by mutations or eliminating the FL by using only EDIII leads to greatly reduced binding of cross-reactive antibodies from human infections (Roberson et al., 2007; Rockstroh et al., 2019). Using FL-deleted or FL-mutated VLPs of ZIKV, the induction of ADE was decreased in animal models, but a decrease in efficacy was also reported (Richner et al., 2017; Yang et al., 2017).
The recombinant WNV E protein is one of the few vaccine candidates that have entered early phase clinical testing (Kaaijk and Luytjes, 2018). We therefore tested a recombinant E protein with four mutations in and near the FL (termed Equad protein) as a vaccine antigen to prevent WNV induced disease in a mouse model. The induction of cross-reactive flavivirus antibodies and the protective efficacy were compared to the wildtype version of E (Ewt) and to recombinant EDIII.
2 Results
2.1 Analysis of recombinant vaccine antigens
We performed an immunization study in mice using recombinant WNV ectodomains of E proteins having either the wildtype sequence (Ewt) or four mutations in or near the FL (Equad). Additionally, the domain III (EDIII) was included. Ewt and Equad were expressed in Drosophila S2 cells and EDIII in E. coli. To analyze the structural conformation of the proteins before immunization, serological analysis was carried out by ELISA. A monoclonal antibody (mAb) binding to an epitope at the lateral ridge of domain III with virus neutralizing activity (Oliphant et al., 2006) comparably recognized Ewt, Equad and to a slightly lesser extend EDIII (Figure 1A). The proportion of protein recognized by the mAb E16 was lower on EDIII, considering that equal amounts of protein were coated and EDIII has a smaller molecular weight than Ewt and Equad (indicating a potential loss of correctly folded EDIII in the bacterial expression system). Analysis using the FL-specific monoclonal antibody 4G2 (Nawa et al., 2001; Crill and Chang, 2004) confirmed that the FL is mutated or absent in Equad and EDIII, respectively (Figure 1B).
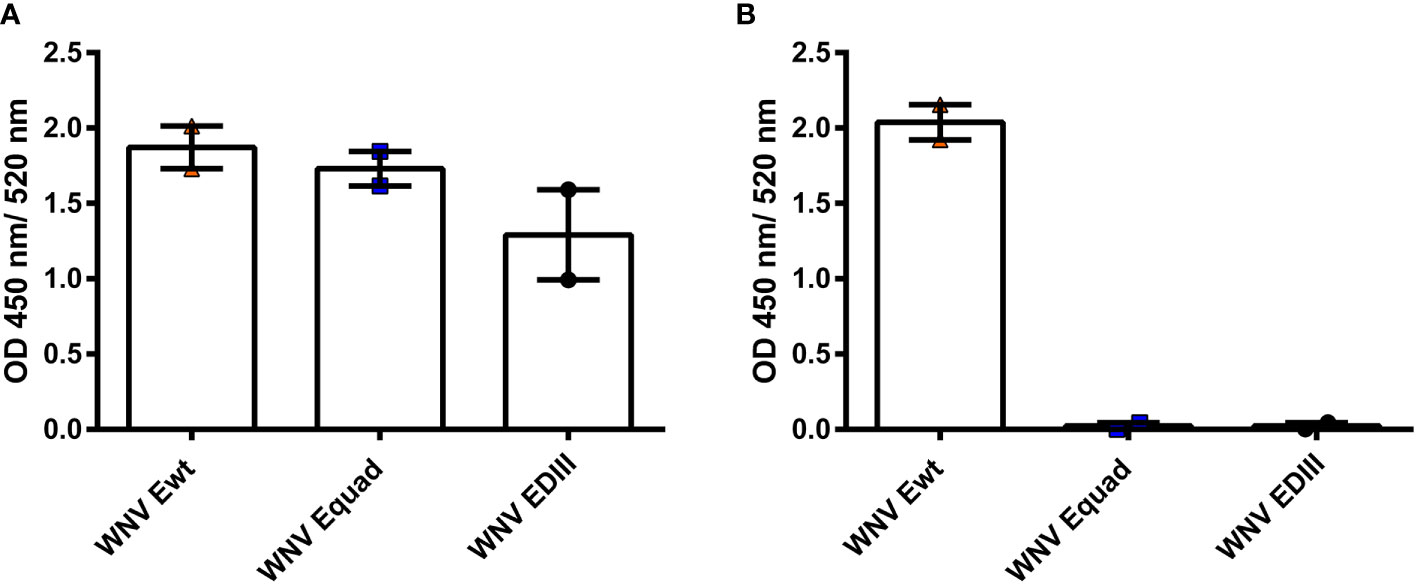
Figure 1 Structural assessment of recombinant WNV proteins with monoclonal antibodies. ELISAs with anti-WNV antibody E16, recognizing an epitope within EDIII (A) and anti-flavivirus antibody 4G2, recognizing the FL (B) to recombinant WNV E proteins (200 ng protein coating per microtiter well). The Data derive from two independent measurements with each sample measured in duplicates. Indicated are mean values ± standard error (SEM) for each group.
2.2 Humoral immune response
Mice were immunized two times with a four weeks interval with 20 µg of either Ewt, Equad, EDIII or buffer (vehicle control), all with Alhydrogel as adjuvant (Figure 2). Using ELISAs with the respective antigen, animals were assessed for the induction of a specific IgG response post vaccination (Figure 3). After the prime immunization mice vaccinated with WNV Ewt displayed the highest signals. After boost immunization, all three groups showed high titers of binding antibodies, and the titers induced by Equad were more heterogenous than in the other groups. In contrast, no antibodies were detectable in the control group measured on Ewt protein (which includes the protein sequence of EDIII) as coating antigen.
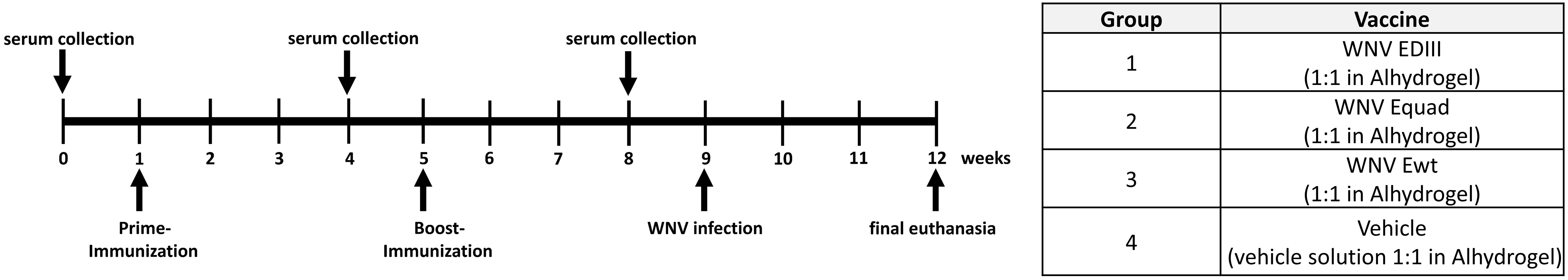
Figure 2 Immunization experiment and serum collection. Female BALB/c mice (n = 6 per group) were immunized two times with variants of the recombinant West Nile virus envelope Protein (WNV E) or with vehicle solution with adjuvant (Vehicle) as control.
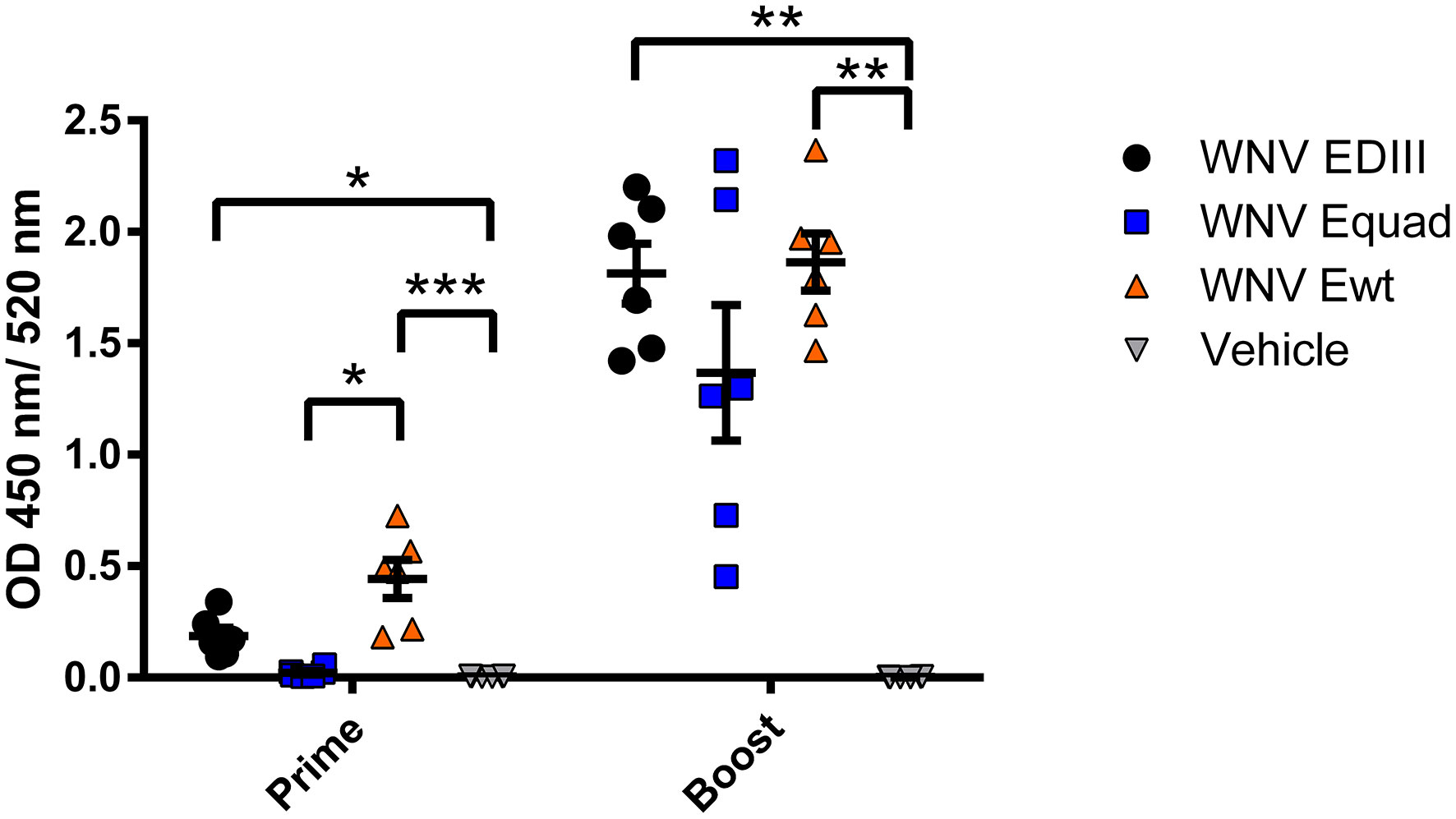
Figure 3 Humoral immune responses after vaccination with variants of the recombinant WNV envelope protein and vehicle immunization. Binding of serum antibodies three weeks after prime and boost immunization was measured in an IgG-ELISA using homologous recombinant proteins. Each data point represents the mean values of one individual mouse from two independent measurements with each sample measured in duplicates. Indicated are mean values ± standard error (SEM) for each group. The data were analyzed with Kruskal-Wallis Test followed by Dunn’s multiple comparisons test (*p<0.05; **p<0.01; ***p<0.001).
To determine the cross-reactivity of serum antibodies after boost immunization, sera were analyzed using whole virions of WNV or the related flaviviruses TBEV, ZIKV and Usutu virus (USUV) (Figure 4). Antibodies from all three immunized groups bound to WNV. For TBEV, only minimal antibody binding was detected, with no statistically significant differences between the protein immunized groups and the control group. When tested on ZIKV the signals of EDIII and Equad immunized mice were in the range of the control group. In contrast, sera from the WNV Ewt group showed a high signal on ZIKV, significantly higher than the Equad or EDIII groups. A similar pattern was observed for binding to USUV. This suggests that immunization with Ewt induces a higher amount of cross-reacting flavivirus antibodies compared to Equad or EDIII.
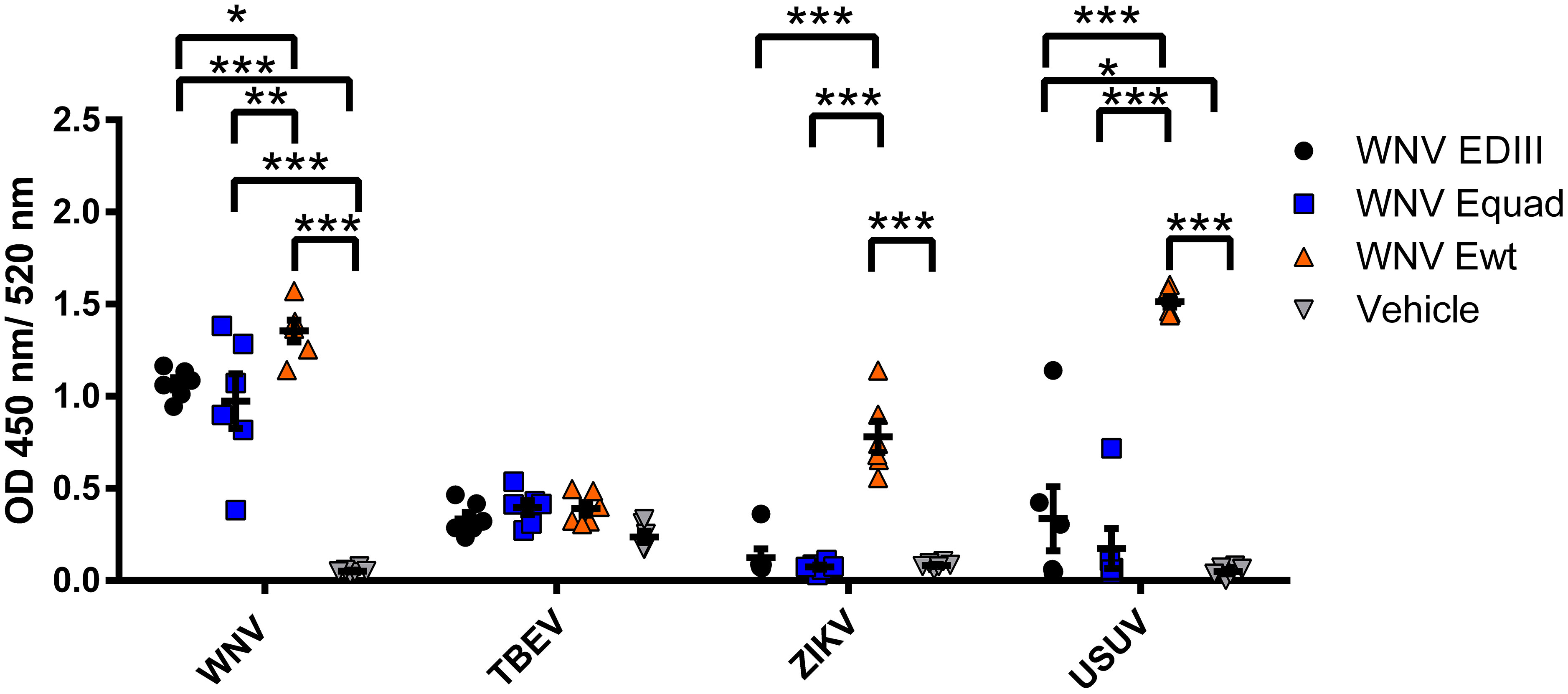
Figure 4 Cross-reactivity of mouse sera after vaccination with variants of the recombinant WNV envelope protein and vehicle immunization. Binding of serum antibodies (three weeks after boost vaccination) to full virions of flaviviruses was analyzed in an IgG-ELISA using purified TBEV, ZIKV, USUV and WNV as coating antigens. Each data point represents the mean values of one individual mouse from two independent measurements with each sample measured in duplicates. Indicated are mean values ± standard error (SEM) for each group. The data were analyzed with RM two-way ANOVA followed by Dunn’s multiple comparisons test (*p<0.05; **p<0.01; ***p<0.001).
2.3 In vivo protection assay
Next, the protective efficacy of the induced antibodies was analyzed by cellular virus neutralization tests (Figure 5). Vaccination with EDIII and Equad elicited neutralizing titers at a mean of 715 and 160, respectively. However, immunization with Ewt induced a mean neutralizing titer of 7,131, which was significantly and more than 10-fold higher than those obtained with Equad. No neutralizing antibodies were detectable in the vehicle control group.
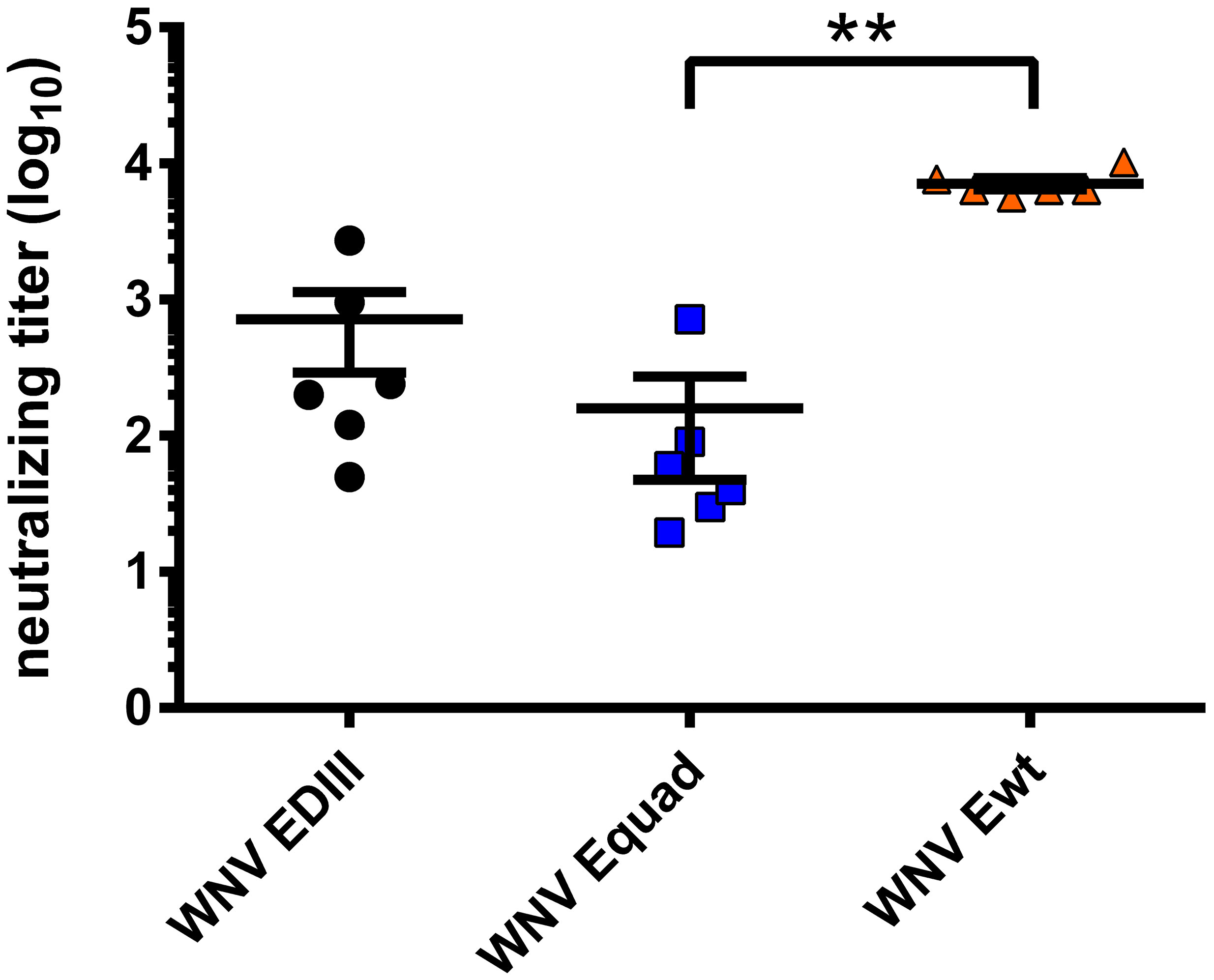
Figure 5 WNV-neutralizing capacity of mouse sera in vitro. Neutralizing antibody titers of sera three weeks after boost vaccination were measured for each mouse through focus reduction neutralization tests (FRNT50). Each data point equals the mean value for every individual mouse serum from two independent measurements. Shown are mean ± standard error (SEM) for each group. The data were analyzed with Kruskal-Wallis Test followed by Dunn’s multiple comparisons test (**p<0.01).
To assess protection from lethal infection, the weight (Figure 6A), clinical score (Figure 6B) and survival (Figure 6C), of immunized mice were monitored for three weeks after challenge with WNV strain Ita09 (104 FFU/mouse) and viral load in brain and spleen was assessed by RT-qPCR (Figure 6D). All animals in the vehicle control group had to be euthanized according to humane endpoints between day 5 and day 8 post infection. Four animals in the control group had detectable viral RNA in the brain and five in the spleen. While all animals that were immunized with Ewt survived, two animals in the Equad group and one animal in the EDIII group succumbed to the infection. In those animals, viral RNA was detected in the brain, but not in the spleen. None of the surviving mice had detectable WNV RNA in the organs investigated. The differences in survival were statistically significant between the vehicle control group and the groups immunized with Ewt (p<0.001), EDIII (p<0.001) and Equad (p<0.01).
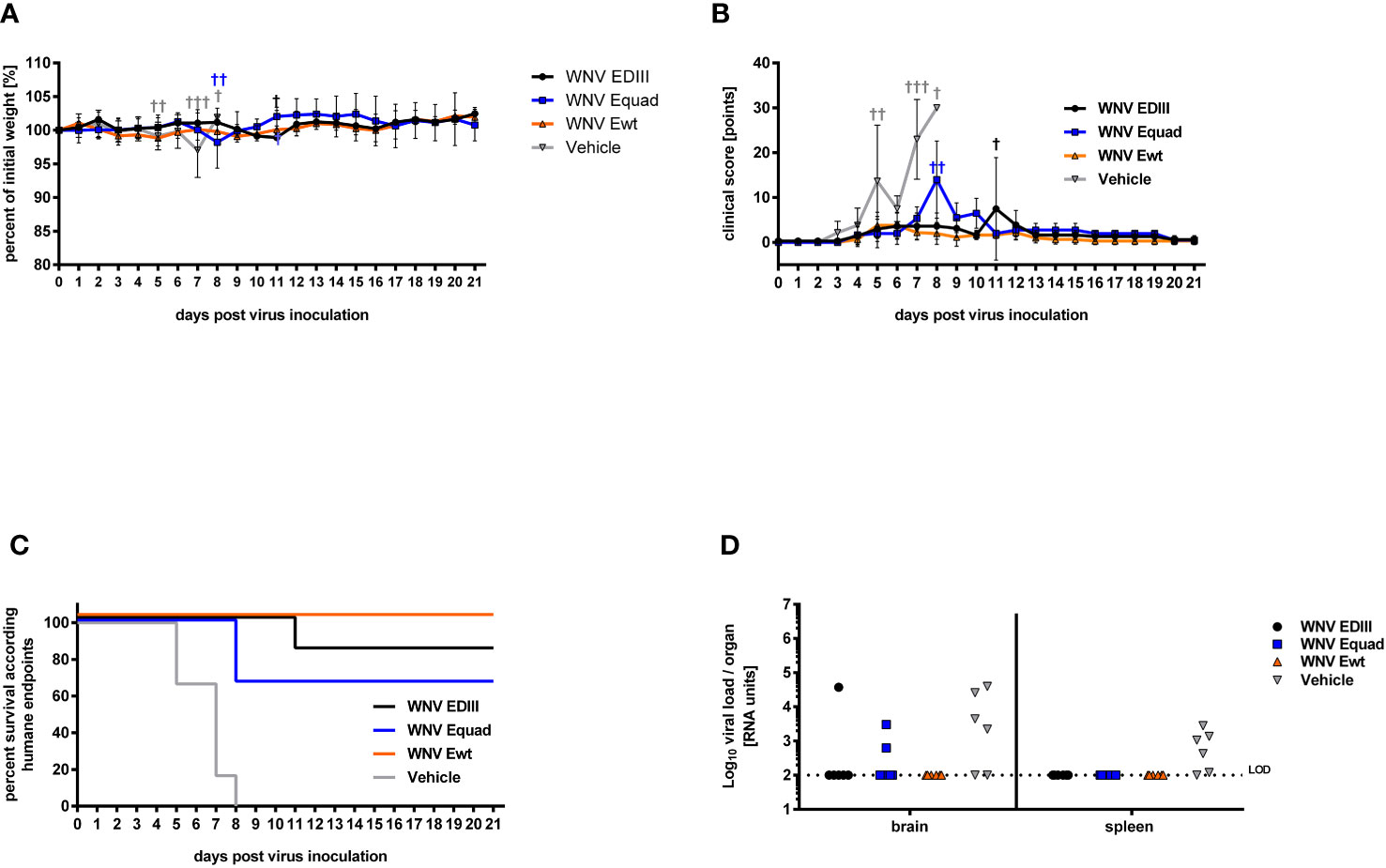
Figure 6 Protective efficacy of protein-based vaccine in mice against lethal WNV infection. Four weeks after boost immunization, female BALB/c mice (n = 6) were infected with a lethal dose of West Nile virus via intraperitoneal injection. All animals were monitored daily for body weight (A) and clinical score (B). Animals reaching humane endpoints were euthanized and are marked by a cross (†). Data are presented as means ± standard errors. The percentages of surviving animals according to humane endpoints are shown (C). Overlapping lines are partially offset for better readability. After euthanasia, brain and spleen tissues were homogenized and their viral load was quantified by RT-qPCR. Shown are viral RNA units of individual animals (D). Limit of detection (LOD) is indicated by a dotted line.
3 Discussion
Vaccine development against WNV focuses on the E protein, which plays multiple critical roles in the viral life cycle and is therefore considered the primary target for neutralizing antibodies. We conducted an immunization study using different recombinantly expressed versions of the WNV E protein to investigate the role of the FL domain in cross-reactivity and efficacy. Mice were vaccinated in a homologous prime-boost regime with the full E ectodomain either in its wildtype form or with a mutated FL, or with EDIII, which completely lacks the FL. IgG antibody titers to the homologous protein antigens remained low after prime, except for Ewt, but strongly increased after the boost immunization. These antibodies also recognized WNV virions.
When analyzing the cross-reactivity of the antibodies to other flaviviruses, the differences between the groups were significant. Mice immunized with Ewt displayed high antibody titers against ZIKV and USUV, whereas the Equad or EDIII immunized groups showed no or only minimal cross-reactive signals. This antibody cross-reactivity induced by Ewt was in accordance with the phylogenetic relationship of the analyzed flaviviruses: it was most prominent with USUV, the flavivirus most closely related to WNV and part of the same JEV serocomplex, followed by ZIKV (Dutta and Langenburg, 2023). No cross-reactivity was observed with TBEV, which is most distantly related to WNV. These results confirm the critical role of the FL in the induction of cross-reactive immune responses (Rey et al., 2018). In WNV serology, a modified FL has been demonstrated to abolish the binding of antibodies induced by heterologous flavivirus infections (Chabierski et al., 2014). In addition, FL-mutated E proteins have been used as parts of virus-like particles (VLPs) in vaccine development against ZIKV, DENV and JEV, resulting in a strong reduction of cross-reactive antibodies (Crill et al., 2012; Richner et al., 2017; Kotaki et al., 2022). In contrast to these previous vaccine studies, we have used recombinant E ectodomains rather than VLPs, and demonstrate that the WNV E protein with mutated FL significantly reduces the induction of antibodies to heterologous flaviviruses.
The phenomenon of ADE due to cross-reactivity of flavivirus-induced immune responses is best known from infections with different DENV serotypes, although the exact molecular mechanism is still not fully understood (Rothman, 2011). Recently, evidence of ADE was also reported from infections with DENV and ZIKV (Katzelnick et al., 2020). It remains uncertain to what extent cross-reactive antibodies induced by WNV infection or vaccination could lead to similar problems. While there is reported evidence that sera from WNV infected individuals can enhance ZIKV infections in vitro and in vivo (Bardina et al., 2017), this question has not yet been addressed in clinical studies. The continuous spread of WNV into areas where USUV is circulating, e.g. in Italy, increases the likelihood of subsequent infections, and to date neither cross-protection nor enhancement of infections can be excluded (Sinigaglia et al., 2019). In addition, the (re-) emergence of yet unknown or so far neglected flaviviruses should be considered when assessing the safety of flavivirus vaccines. As a prominent example, ZIKV had not been considered a major problem until its emergence in Micronesia (Duffy et al., 2009). To date, the virus is globally spread, and coinfections with DENV are commonly observed (Rodriguez-Barraquer et al., 2019). The development of WNV vaccines should therefore address the phenomenon of antibody cross-reactivity, even though there is currently no clinical evidence supporting ADE due to WNV infections.
When the immunized animals were analyzed for the induction of protective immune responses, clear differences were observed between Ewt on one side, and Equad and EDIII on the other. Ewt resulted in high titers of virus neutralizing antibodies, whereas lower titers were measured in the Equad and EDIII groups. These lower titers of neutralizing antibodies do only poorly correspond to the higher levels of binding antibodies induced by the immunizations. This reflects the finding that neutralizing antibodies only constitute a minor fraction of the humoral immune response to WNV antigens (Throsby et al., 2006). Likewise, all animals immunized with Ewt survived a lethal WNV infection, whereas two and one animals immunized with Equad or EDIII, respectively, did not. Neutralizing antibodies were induced in all immunized animals, but animals succumbing to the infection did not display the lowest titers in the individual groups. This indicates that neutralizing antibodies alone were poorly predictable for protection in the chosen immunization setup. The control animals had WNV RNA detectable in brain and spleen. In contrast, the immunized mice that did not survive the challenge had WNV RNA detectable only in the brain, suggesting a reduction of viral spreading due to the vaccines.
This study represents the first investigation of a recombinant WNV E protein with mutated FL as a vaccine. Nevertheless, vaccination studies using ZIKV VLPs with E proteins containing a mutant FL also reported a decrease in protective capacity when compared to the wildtype version (Richner et al., 2017; Thompson et al., 2022). These studies suggested impaired structures of the VLP due to the FL mutations, leading to altered quaternary epitopes. Unlike these studies we used single E ectodomains rather than VLPs. Hence, alteration of critical epitopes due to FL mutations can only affect the E protein monomer. Interestingly, the binding of the neutralizing antibody E16, which recognizes an epitope at the lateral ridge of EDIII (Oliphant et al., 2007), remained unaffected by the FL mutations (Figure 1). Therefore, impairment or loss of neutralizing epitopes apparently affects other regions of the protein. This question could be addressed by a detailed scan of the Equad protein with monoclonal antibodies against structural epitopes. In addition, it cannot be excluded that the absence or mutation of the FL itself leads to a decrease in the overall protection from WNV, as neutralizing capacity of some FL antibodies has been described for flaviviruses, although with varying efficacy (Vogt et al., 2011; Dai et al., 2016).
A potential approach to increase the protective efficacy of FL-mutant E proteins might involve additional boost immunizations, as repeated boosting is well established to enhance and broaden the neutralizing antibody repertoire and protection (Burckhardt et al., 2022). In this study only one boost was administered, similar to the previous studies reporting a lower protective response with FL mutations (Richner et al., 2017; Thompson et al., 2022). However, we have previously demonstrated for an Equad protein of ZIKV that after three immunizations no differences were observed compared to the wildtype version in the induction of neutralizing antibodies (Berneck et al., 2020). Likewise, investigations using three doses of DNA vaccines coding for VLPs with a single point mutation in the FL reported no decrease in neutralizing antibodies for WNV or DENV when compared to the wildtype VLPs (Crill et al., 2012; Yamanaka et al., 2022).
Unlike the Equad protein, recombinant EDIII from different protein expression systems has been used in previous WNV vaccination studies. Depending on the exact study design, these investigations reported the induction of highly protective or only partially protective immune responses (Chu et al., 2007; Martina et al., 2008; Zlatkovic et al., 2011; Friedrich et al., 2016; Lai et al., 2018). Similar to the antigens with a mutant FL, immunization schedules including at least two boosters generally resulted in higher efficacy. Therefore, increasing the number of booster immunizations with Equad or EDIII might be a way to increase vaccine efficacy and at the same time minimize the induction of flavivirus cross-reactive immune responses.
In summary, our results confirm that the ectodomain of the WNV E protein is a potent vaccine antigen. However, its original sequence induces cross-reacting antibodies to related flaviviruses. Although there is no clinical evidence yet that pre-existing WNV immunity could lead to disease enhancement upon infection with known heterologous flaviviruses, this concern should be taken in account in WNV vaccine development. Our findings demonstrate that the Equad or the EDIII antigens induce significantly less cross-reactive antibodies than the wildtype E ectodomain. However, the efficacy of both antigens needs to be improved through optimization of immunization schedules before WNV vaccines based on these proteins can progress to clinical development.
4 Materials and methods
4.1 Cells and viruses
Vero E6 cells (DSMZ, Braunschweig, Germany) were propagated in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, Carlsbad, USA) supplemented with 10% heat inactivated fetal calf serum (FCS, Gibco) and 1% penicillin/streptomycin (Gibco) at 37°C and 5% CO2.
Viruses used in this study were West Nile Virus (genetic lineage 1 strain WNV-Ita09, kindly provided by Luisa Barzon, Padova University), Zika Virus (Dominican Republic/2016/PD1, kindly provided by Luisa Barzon), Usutu Virus (strain 3345, isolate Arb276, provided by the European Viral Archive Global EVAg), Tick-borne encephalitis Virus (strain Hypr 9BMP U39292.1, kindly provided by Uwe Liebert, Leipzig University). Viruses were propagated in Vero E6 cells and purified from culture supernatant by ultracentrifugation. Virus titration in focus forming units (FFU) was performed as previously described (Berneck et al., 2020). In short, serial dilutions of virus were incubated on Vero E6 monolayers for 1 h at 37°C. After removal of the supernatant, cells were overlaid with 1% methylcellulose in DMEM supplemented with 2% FCS and 1% penicillin/streptomycin. Cells were fixed after 16 – 22 h with 4% formaldehyde (Roth, Karlsruhe, Germany) in phosphate buffered saline (PBS). Perm Wash buffer (0.1% BSA (Roth) and 0.1% Saponin (Roth) in PBS) was used for permeabilization, blockage and washing of cells. The primary anti-flavivirus antibody 4G2 (absolute antibody, Oxford, UK, 1:2,000), an anti-mouse IgG HRP-conjugated secondary antibody (Dako, Denmark, 1:1,500) and TrueBlue peroxidase substrate (SeraCare, Milford, USA) were used for immunostaining. Spots were counted automatically with an Immunospot Universal Analyzer (Cellular Technology Limit, Cleveland, USA).
For analyzing virus-binding antibodies, viruses were coated on ELISA plates (see below), except for WNV, which was inactivated before by overnight treatment with 0.3% H2O2 (Roth, Karlsruhe, Germany) in Sucrose-PBS at 37°C followed by dialysis against PBS.
4.2 Expression and purification of recombinant proteins
The wildtype and quadruple mutant E‐proteins (Equad) from WNV (isolate NY2000 ‐ crow3356, E‐protein amino acid residues 1‐400 bearing the mutations T76A, M77G, W101R, L107R) have been described previously (Rockstroh et al., 2019).
The expression vector encoding domain III of the WNV E-protein (isolate NY2000 ‐ crow3356 E‐protein amino acid residues 299-400) as a fusion protein with the maltose binding protein (MBP) was described previously (Schneeweiss et al., 2011). The fusion protein was expressed in E. coli strain Rosetta 2 DE3 (Merck, Darmstadt, Germany) by induction with 1 mM isopropyl-beta-D-thiogalactopyranoside (IPTG, Janssen Pharmaceuticals, Beerse, Belgium). Bacteria were harvested by centrifugation (4,500 x g, 15 min, 4°C) and lysed in buffer containing 20 mM Tris pH 7.4 (Roth), 200 mM NaCl (Roth), 1 mM EDTA, 10 mM Imidazol (Roth), 10% Glycerol (Roth), 1 mM DTT (PanReac Applichem ITW Reagents, Darmstadt, Germany) and protease-inhibitor (Sigma Aldrich, St. Louis, USA) using a high-pressure homogenizer (Constant Systems LTD, Daventry, UK). Insoluble cell debris was separated from the soluble fraction containing WNV EDIII by centrifugation (15,000 x g, 30 min, 4°C). The recombinant MBP-fusion protein was purified on Amylose resin columns (New England Biolabs, Frankfurt, Germany). Subsequently the MBP-tag was removed by Factor Xa Protease (New England Biolabs, Frankfurt, Germany) cleavage and the resulting WNV EDIII His-tagged protein was further purified on His60 Ni Superflow Resin columns (TaKaRa Bio, USA). The protein was then dialyzed against PBS, and aliquots were stored at - 80°C until use.
Bradford assay and SDS-gel electrophoresis were performed for measuring protein amount and assessment of protein purity, respectively (data not shown).
4.3 Mouse immunization and challenge experiment
The mouse experiment was carried out in accordance with the EU Directive 2010/63/EU for animal experiments and was approved by local authorities (Landesdirektion Sachsen). Female 8-week-old BALB/c mice were purchased from Charles River (Sulzfeld, Germany) and randomly assigned into groups of 6 mice. Mice were housed in a specific pathogen-free environment in individually ventilated cages, 12 h/12 h-light/dark cycle, and water and mouse chow were provided ad libitum.
For vaccine preparation, purified proteins (EDIII, Equad, and Ewt) were diluted in PBS before mixing gently with Alhydrogel (aluminum hydroxide gel adjuvant, 10 mg/mL aluminum, InvivoGen, Toulouse, France) at a 1:1 volume ratio to administer 20 µg protein in 100 µL per mouse. The adjuvant was chosen based on the wide experience available with aluminium hydroxide and recombinant protein immunizations (Zlatkovic et al., 2011). For the immunization, inhaled light isoflurane anesthesia was applied, and each vaccine was injected in the musculus gastrocnemius of each hind limb (50 µL each). Control mice were sham-immunized with vehicle solution (1:1 mixture of Alhydrogel and PBS). All mice were immunized twice at four-week interval.
Blood was sampled from the retro-bulbar venous sinus one week prior to the prime and boost immunization, as well as one week before the challenge. Collected blood samples were incubated at room temperature for 30 min and centrifuged at 8,000 x g for 10 min to obtain serum for antibody analysis. Four weeks after the boost immunization, mice were challenged by intraperitoneal (i.p.) injection of 104 FFU of purified WNV-Ita09 in a total volume of 100 µL per mouse. The infection and all following work were carried out under biosafety level 3 (BSL3) conditions. Clinical development of disease was monitored daily for 21 days post-infection and score points were given according to the following criteria: body weight loss (0 points= no weight loss, 5 points= 8-10%, 10 points= 11-19%, 20 points= weight loss ≥20% of initial weight); fur condition (0 points = shiny and clean coat, 2 points = piloerection, 5 points = ruffled fur); eye appearance (0 points= open healthy eyes, 5 points= mildly inflamed, 10 points = highly inflamed and closed); gastrointestinal symptoms due to distention of the intestine (0 points= no symptoms, 5 points= mild, 10 points= moderate abdominal swelling); body posture (0 points= normal posture, 20 points= hunched body posture); activity level and motor function (5 points= slightly reduced activity and reaction, 10 points= coordination disorder and reduced activity, 20 points= apathy and morbidity). Score points of 0-9 were defined as mild, 10-19 as moderate, and ≥20 as severe. Humane endpoints requiring euthanasia were defined as reaching a cumulative score points of 20 for a period of 24 h. Animals acquiring cumulative score points greater than 20 were immediately euthanized. Surviving mice were euthanized 21 days after the challenge.
Spleens and brains were isolated and homogenized in gentleMACS™ M Tubes (Miltenyi Biotec, Bergisch Gladbach, Germany) containing 2 mL of ice-cold PBS using gentleMACS Dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany). Homogenized tissues were cleared of cell debris by centrifugation for 5 min at 2,000 × g and 4°C. Homogenates were stored at - 80°C until viral isolation.
4.4 Viral RNA extraction and RT-qPCR
Viral RNA was isolated from 140 μL cell-free supernatant of organ homogenate using QIAamp-Viral-RNA-Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Briefly, 5 μL of isolated RNA was reverse transcribed and amplified using QuantiTect probe RT-PCR kit (Qiagen, Hilden, Germany) with sense primer (5′- GTGATCCATGTAAGCCCTCAGAA -3′), antisense primer (5′- GTCTGACATTGGGCTTTGAAGTTA-3′) and a TaqMan probe labeled with fluorophore and Quencher (5′-[6-FAM] AGGACCCCACATGTT [MGB CDPI3-BMN-Q535]-3′ (Jiménez-Clavero et al., 2006)). All oligonucleotides were ordered from Biomers, Ulm, Germany. The reaction was carried out in LightCycler® 480 instrument (Roche, Penzberg, Germany). 10-fold serial dilutions of viral RNA isolated from purified WNV-Ita09 with defined titer served as standards for the quantification of viral genome copy numbers in mouse samples.
4.5 ELISA analysis
Microtiter plates (Nunc Polysorp®, ThermoScientific, Roskilde, Denmark) were incubated either with recombinant WNV proteins (WNV Ewt, WNV Equad, WNV EDIII) or viruses in coating buffer (35 mM Na2HCO3/15 mM Na2CO3, pH 9.6) in a total volume of 100 µL per well over night at 4°C. The coated amounts of proteins per well were 200 ng for the analysis with monoclonal antibodies and 300 ng for the analysis with mouse sera. The optimal coating amounts for recombinant proteins and virus particles were evaluated experimentally (data not shown). 105 FFUs of virus particles were coated for the analysis with mouse sera. After three washing steps with 350 µL/well PBS 0.05% Tween20, plates were blocked for 2 h at room temperature (RT) with 5% non-fat milk powder in PBS (milk). After another wash step either mouse sera or monoclonal antibodies diluted in milk were incubated on the plates for 1.5 h at RT. Mouse sera were diluted 1:1,000 or 1:100 for binding to recombinant proteins or virus particles, respectively. The mouse monoclonal antibodies 4G2 (absolute antibody, Oxford, UK) and E16 (Millipore Merck, Darmstadt, Germany) were diluted 1:1,000 and 1:2,000, respectively. Following the third wash step, the anti-mouse IgG-HRP-conjugated antibody diluted in milk was incubated on the plates for another 1 h at RT. Subsequently to the final wash step, TMB substrate (Biozol, Hamburg, Germany) was incubated on plates for 30 min at RT in darkness, and the reaction was stopped with 1 M H2SO4. Absorbance was measured at 450 nm with 520 nm as reference in a microplate ELISA reader (TECAN, infinite M200 Basic, TECAN Austria GmbH, Grödig, Austria). Each sample was measured in duplicates in two independent experiments.
4.6 Focus reduction neutralization test
Focus Reduction Neutralization Test (FRNT) was performed as previously described (Finkensieper et al., 2022). In short, mouse sera taken three weeks after the second immunization were heat inactivated (56°C for 30 min), serially diluted in microplates and incubated with 75 FFU of purified WNV Ita09 for 1h at 37°C. The serum-virus-mixture was then added to Vero E6 cell monolayers in 96 well microwell plates and again incubated for 1 h at 37°C. Finally, the mix was removed and the cells were overlaid with 1% methylcellulose in DMEM with 2% FCS and 1% penicillin/streptomycin. After 16-18 h incubation at 37°C cells were fixed with 4% formaldehyde-PBS and Perm Wash buffer was used for permeabilization, blockage and washing of cells. The primary anti-flavivirus antibody 4G2 (absolute antibody, Oxford, UK, 1:2,000), an anti-mouse IgG HRP-conjugated secondary antibody (Dako, Denmark, 1:1,500) and TrueBlue peroxidase substrate (SeraCare, Milford, USA) were used for immunostaining. Spots were analyzed using the Immunospot Universal Analyzer (CTL, Cleveland, USA). The neutralizing antibody titer was defined as the reciprocal of the highest serum dilution that showed a minimal reduction in number of WNV foci of 50% compared to sera from the control group. Each serum was measured once in two independent experiments.
4.7 Statistics
Statistical analysis was performed with GraphPadPrism6 (Version 6.07, 2015). The ELISA data of sera on homologous recombinant proteins and the data of FRNT50 (arithmetic mean) assays were analyzed with Kruskal-Wallis Test. Data of ELISA assays using boost sera on virions were analyzed using two way ANOVA. All tests were followed by Dunn’s multiple comparisons test with *=p<0.05; **=p<0.01; ***=p<0.001. The survival rates were compared using Log-rank (Mantel-Cox) analysis.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was approved by Landesdirektion Sachsen, Germany. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
RW: Investigation, Resources, Writing – original draft, Writing – review & editing, Data curation, Validation, Visualization. LI: Investigation, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. AR: Investigation, Writing – review & editing, Conceptualization. TG: Conceptualization, Investigation, Writing – review & editing, Funding acquisition, Resources, Supervision. JF: Conceptualization, Funding acquisition, Resources, Supervision, Writing – original draft. SU: Conceptualization, Funding acquisition, Resources, Supervision, Writing – original draft, Formal Analysis, Investigation, Project administration, Writing – review & editing, JF: writing - review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported as a Fraunhofer FLAGSHIP PROJECT (RNAuto).
Acknowledgments
We thank Julia Finkensieper for technical discussion and Ulrike Ehlert, Steffen Jakob and Isabell Schulz for excellent technical assistance.
Conflict of interest
SU is co-author on the patent US 10060924 which describes the Equad protein of WNV.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bai, F., Thompson, E. A., Vig, P. J. S., Leis, A. A. (2019). Current understanding of west nile virus clinical manifestations, immune responses, neuroinvasion, and immunotherapeutic implications. Pathogens 8, 193. doi: 10.3390/pathogens8040193
Bardina, S. V., Bunduc, P., Tripathi, S., Duehr, J., Frere, J. J., Brown, J. A., et al. (2017). Enhancement of Zika virus pathogenesis by preexisting antiflavivirus immunity. Science 356, 175–180. doi: 10.1126/science.aal4365
Bassal, R., Shohat, T., Kaufman, Z., Mannasse, B., Shinar, E., Amichay, D., et al. (2017). The seroprevalence of West Nile Virus in Israel: A nationwide cross sectional study. PloS One 12, e0179774. doi: 10.1371/journal.pone.0179774
Berneck, B. S., Rockstroh, A., Fertey, J., Grunwald, T., Ulbert, S. (2020). A recombinant zika virus envelope protein with mutations in the conserved fusion loop leads to reduced antibody cross-reactivity upon vaccination. Vaccines 8, 603. doi: 10.3390/vaccines8040603
Bonafé, N., Rininger, J. A., Chubet, R. G., Foellmer, H. G., Fader, S., Anderson, J. F., et al. (2009). A recombinant West Nile virus envelope protein vaccine candidate produced in Spodoptera frugiperda expresSF+ cells. Vaccine 27, 213–222. doi: 10.1016/j.vaccine.2008.10.046
Brandler, S., Marianneau, P., Loth, P., Lacôte, S., Combredet, C., Frenkiel, M.-P., et al. (2012). Measles vaccine expressing the secreted form of West Nile virus envelope glycoprotein induces protective immunity in squirrel monkeys, a new model of West Nile virus infection. J. Infect. Dis. 206, 212–219. doi: 10.1093/infdis/jis328
Burckhardt, R. M., Dennehy, J. J., Poon, L. L. M., Saif, L. J., Enquist, L. W. (2022). Are COVID-19 vaccine boosters needed? The science behind boosters. J. Virol. 96, e01973–e01921. doi: 10.1128/jvi.01973-21
Byas, A. D., Ebel, G. D. (2020). Comparative pathology of west nile virus in humans and non-human animals. Pathog. Basel Switz. 9, 48. doi: 10.3390/pathogens9010048
Centers for Disease Control and Prevention (CDC) (2009). West Nile virus transmission via organ transplantation and blood transfusion - Louisiana 2008. MMWR Morb. Mortal. Wkly. Rep. 58, 1263–1267.
Chabierski, S., Barzon, L., Papa, A., Niedrig, M., Bramson, J. L., Richner, J. M., et al. (2014). Distinguishing West Nile virus infection using a recombinant envelope protein with mutations in the conserved fusion-loop. BMC Infect. Dis. 14, 246. doi: 10.1186/1471-2334-14-246
Chancey, C., Grinev, A., Volkova, E., Rios, M. (2015). The global ecology and epidemiology of west nile virus. BioMed. Res. Int. 2015, 1–20. doi: 10.1155/2015/376230
Chu, J.-H. J., Chiang, C.-C. S., Ng, M.-L. (2007). Immunization of flavivirus west nile recombinant envelope domain III protein induced specific immune response and protection against west nile virus infection. J. Immunol. 178, 2699–2705. doi: 10.4049/jimmunol.178.5.2699
Crill, W. D., Chang, G.-J. J. (2004). Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. J. Virol. 78, 13975–13986. doi: 10.1128/JVI.78.24.13975-13986.2004
Crill, W. D., Hughes, H. R., Trainor, N. B., Davis, B. S., Whitney, M. T., Chang, G.-J. J. (2012). Sculpting humoral immunity through dengue vaccination to enhance protective immunity. Front. Immunol. 3. doi: 10.3389/fimmu.2012.00334
Dai, L., Song, J., Lu, X., Deng, Y.-Q., Musyoki, A. M., Cheng, H., et al. (2016). Structures of the zika virus envelope protein and its complex with a flavivirus broadly protective antibody. Cell Host Microbe 19, 696–704. doi: 10.1016/j.chom.2016.04.013
Davis, B. S., Chang, G. J., Cropp, B., Roehrig, J. T., Martin, D. A., Mitchell, C. J., et al. (2001). West Nile virus recombinant DNA vaccine protects mouse and horse from virus challenge and expresses in vitro a noninfectious recombinant antigen that can be used in enzyme-linked immunosorbent assays. J. Virol. 75, 4040–4047. doi: 10.1128/JVI.75.9.4040-4047.2001
Dayan, G. H., Pugachev, K., Bevilacqua, J., Lang, J., Monath, T. P. (2013). Preclinical and clinical development of a YFV 17 D-based chimeric vaccine against West Nile virus. Viruses 5, 3048–3070. doi: 10.3390/v5123048
Duffy, M. R., Chen, T.-H., Hancock, W. T., Powers, A. M., Kool, J. L., Lanciotti, R. S., et al. (2009). Zika virus outbreak on Yap Island, Federated States of Micronesia. N. Engl. J. Med. 360, 2536–2543. doi: 10.1056/NEJMoa0805715
Dutta, S. K., Langenburg, T. (2023). A perspective on current flavivirus vaccine development: A brief review. Viruses 15, 860. doi: 10.3390/v15040860
Finkensieper, J., Issmail, L., Fertey, J., Rockstroh, A., Schopf, S., Standfest, B., et al. (2022). Low-energy electron irradiation of tick-borne encephalitis virus provides a protective inactivated vaccine. Front. Immunol. 13. doi: 10.3389/fimmu.2022.825702
Friedrich, B. M., Beasley, D. W. C., Rudra, J. S. (2016). Supramolecular peptide hydrogel adjuvanted subunit vaccine elicits protective antibody responses against West Nile virus. Vaccine 34, 5479–5482. doi: 10.1016/j.vaccine.2016.09.044
García-Carrasco, J.-M., Muñoz, A.-R., Olivero, J., Segura, M., Real, R. (2023). An African West Nile virus risk map for travellers and clinicians. Travel Med. Infect. Dis. 52, 102529. doi: 10.1016/j.tmaid.2022.102529
Giesen, C., Herrador, Z., Fernandez-Martinez, B., Figuerola, J., Gangoso, L., Vazquez, A., et al. (2023). A systematic review of environmental factors related to WNV circulation in European and Mediterranean countries. One Health 16, 100478. doi: 10.1016/j.onehlt.2022.100478
Gould, C. V., Staples, J. E., Huang, C. Y.-H., Brault, A. C., Nett, R. J. (2023). Combating west nile virus disease — Time to revisit vaccination. N. Engl. J. Med. 388, 1633–1636. doi: 10.1056/NEJMp2301816
Hadfield, J., Brito, A. F., Swetnam, D. M., Vogels, C. B. F., Tokarz, R. E., Andersen, K. G., et al. (2019). Twenty years of West Nile virus spread and evolution in the Americas visualized by Nextstrain. PloS Pathog. 15, e1008042. doi: 10.1371/journal.ppat.1008042
Halstead, S. B. (2018). Safety issues from a Phase 3 clinical trial of a live-attenuated chimeric yellow fever tetravalent dengue vaccine. Hum. Vaccines Immunother. 14, 2158–2162. doi: 10.1080/21645515.2018.1445448
Hayes, E. B., Sejvar, J. J., Zaki, S. R., Lanciotti, R. S., Bode, A. V., Campbell, G. L. (2005). Virology, pathology, and clinical manifestations of west nile virus disease. Emerg. Infect. Dis. 11, 1174–1179. doi: 10.3201/eid1108.050289b
Jiménez-Clavero, M. A., Agüero, M., Rojo, G., Gómez-Tejedor, C. (2006). A new fluorogenic real-time RT-PCR assay for detection of lineage 1 and lineage 2 west nile viruses. J. Vet. Diagn. Invest. 18, 459–462. doi: 10.1177/104063870601800505
Kaaijk, P., Luytjes, W. (2018). Are we prepared for emerging flaviviruses in Europe? Challenges for vaccination. Hum. Vaccines Immunother. 14, 337–344. doi: 10.1080/21645515.2017.1389363
Katzelnick, L. C., Gresh, L., Halloran, M. E., Mercado, J. C., Kuan, G., Gordon, A., et al. (2017). Antibody-dependent enhancement of severe dengue disease in humans. Science 358, 929–932. doi: 10.1126/science.aan6836
Katzelnick, L. C., Narvaez, C., Arguello, S., Lopez Mercado, B., Collado, D., Ampie, O., et al. (2020). Zika virus infection enhances future risk of severe dengue disease. Science 369, 1123–1128. doi: 10.1126/science.abb6143
Kilpatrick, A. M., LaDeau, S. L., Marra, P. P. (2007). Ecology of west nile virus transmission and its impact on birds in the western hemisphere. Auk 124, 1121–1136. doi: 10.1093/auk/124.4.1121
Kotaki, T., Nagai, Y., Yamanaka, A., Konishi, E., Kameoka, M. (2022). Japanese encephalitis DNA vaccines with epitope modification reduce the induction of cross-reactive antibodies against dengue virus and antibody-dependent enhancement of dengue virus infection. Vaccines 10, 1411. doi: 10.3390/vaccines10091411
Lai, H., Paul, A. M., Sun, H., He, J., Yang, M., Bai, F., et al. (2018). A plant-produced vaccine protects mice against lethal West Nile virus infection without enhancing Zika or dengue virus infectivity. Vaccine 36, 1846–1852. doi: 10.1016/j.vaccine.2018.02.073
Lieberman, M. M., Nerurkar, V. R., Luo, H., Cropp, B., Carrion, R., de la Garza, M., et al. (2009). Immunogenicity and protective efficacy of a recombinant subunit West Nile virus vaccine in rhesus monkeys. Clin. Vaccine Immunol. CVI 16, 1332–1337. doi: 10.1128/CVI.00119-09
Magnusson, S. E., Karlsson, K. H., Reimer, J. M., Corbach-Söhle, S., Patel, S., Richner, J. M., et al. (2014). Matrix-MTM adjuvanted envelope protein vaccine protects against lethal lineage 1 and 2 West Nile virus infection in mice. Vaccine 32, 800–808. doi: 10.1016/j.vaccine.2013.12.030
Martina, B. E., Koraka, P., Van Den Doel, P., Van Amerongen, G., Rimmelzwaan, G. F., Osterhaus, A. D. M. E. (2008). Immunization with West Nile virus envelope domain III protects mice against lethal infection with homologous and heterologous virus. Vaccine 26, 153–157. doi: 10.1016/j.vaccine.2007.10.055
Montgomery, R. R. (2016). Age-related alterations in immune responses to West Nile virus infection. Clin. Exp. Immunol. 187, 26–34. doi: 10.1111/cei.12863
Nawa, M., Takasaki, T., Yamada, K.-I., Akatsuka, T., Kurane, I. (2001). Development of dengue IgM-capture enzyme-linked immunosorbent assay with higher sensitivity using monoclonal detection antibody. J. Virol. Methods 92, 65–70. doi: 10.1016/S0166-0934(00)00274-3
Oliphant, T., Nybakken, G. E., Austin, S. K., Xu, Q., Bramson, J., Loeb, M., et al. (2007). ). Induction of epitope-specific neutralizing antibodies against west nile virus. J. Virol. 81, 11828–11839. doi: 10.1128/JVI.00643-07
Oliphant, T., Nybakken, G. E., Engle, M., Xu, Q., Nelson, C. A., Sukupolvi-Petty, S., et al. (2006). Antibody recognition and neutralization determinants on domains I and II of west nile virus envelope protein. J. Virol. 80, 12149–12159. doi: 10.1128/JVI.01732-06
Pierson, T. C., Fremont, D. H., Kuhn, R. J., Diamond, M. S. (2008). Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: implications for vaccine development. Cell Host Microbe 4, 229–238. doi: 10.1016/j.chom.2008.08.004
Pinto, A. K., Richner, J. M., Poore, E. A., Patil, P. P., Amanna, I. J., Slifka, M. K., et al. (2013). A hydrogen peroxide-inactivated virus vaccine elicits humoral and cellular immunity and protects against lethal West Nile virus infection in aged mice. J. Virol. 87, 1926–1936. doi: 10.1128/JVI.02903-12
Prow, N. A., Edmonds, J. H., Williams, D. T., Setoh, Y. X., Bielefeldt-Ohmann, H., Suen, W. W., et al. (2016). Virulence and evolution of west nile virus, Australia 1960–2012. Emerg. Infect. Dis. 22, 1353–1362. doi: 10.3201/eid2208.151719
Rey, F. A., Stiasny, K., Vaney, M., Dellarole, M., Heinz, F. X. (2018). The bright and the dark side of human antibody responses to flaviviruses: lessons for vaccine design. EMBO Rep. 19, 206–224. doi: 10.15252/embr.201745302
Richner, J. M., Himansu, S., Dowd, K. A., Butler, S. L., Salazar, V., Fox, J. M., et al. (2017). Modified mRNA Vaccines Protect against Zika Virus Infection. Cell 168, 1114–1125.e10. doi: 10.1016/j.cell.2017.02.017
Roberson, J. A., Crill, W. D., Chang, G.-J. J. (2007). Differentiation of west nile and st. Louis encephalitis virus infections by use of noninfectious virus-like particles with reduced cross-reactivity. J. Clin. Microbiol. 45, 3167–3174. doi: 10.1128/JCM.01143-07
Rockstroh, A., Moges, B., Berneck, B. S., Sattler, T., Revilla-Fernández, S., Schmoll, F., et al. (2019). Specific detection and differentiation of tick-borne encephalitis and West Nile virus induced IgG antibodies in humans and horses. Transbound Emerg. Dis. 66, tbed.13205. doi: 10.1111/tbed.13205
Rodriguez-Barraquer, I., Costa, F., Nascimento, E. J. M., Nery, N., Castanha, P. M. S., Sacramento, G. A., et al. (2019). Impact of preexisting dengue immunity on Zika virus emergence in a dengue endemic region. Science 363, 607–610. doi: 10.1126/science.aav6618
Rothman, A. L. (2011). Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat. Rev. Immunol. 11, 532–543. doi: 10.1038/nri3014
Sarker, A., Dhama, N., Gupta, R. D. (2023). Dengue virus neutralizing antibody: a review of targets, cross-reactivity, and antibody-dependent enhancement. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1200195
Schneeweiss, A., Chabierski, S., Salomo, M., Delaroque, N., Al-Robaiy, S., Grunwald, T., et al. (2011). A DNA vaccine encoding the E protein of West Nile Virus is protective and can be boosted by recombinant domain DIII. Vaccine 29, 6352–6357. doi: 10.1016/j.vaccine.2011.04.116
Sejvar, J. J. (2016). West nile virus infection. Microbiol. Spectr. 4, 4.3.20. doi: 10.1128/microbiolspec.EI10-0021-2016
Sewgobind, S., McCracken, F., Schilling, M. (2023). JMM Profile: West Nile virus. J. Med. Microbiol. 72. doi: 10.1099/jmm.0.001730
Sinigaglia, A., Pacenti, M., Martello, T., Pagni, S., Franchin, E., Barzon, L. (2019). West Nile virus infection in individuals with pre-existing Usutu virus immunity, northern Italy 2018. Eurosurveillance 24, 2–7. doi: 10.2807/1560-7917.ES.2019.24.21.1900261
Thompson, D., Guenther, B., Manayani, D., Mendy, J., Smith, J., Espinosa, D. A., et al. (2022). Zika virus-like particle vaccine fusion loop mutation increases production yield but fails to protect AG129 mice against Zika virus challenge. PloS Negl. Trop. Dis. 16, e0010588. doi: 10.1371/journal.pntd.0010588
Throsby, M., Geuijen, C., Goudsmit, J., Bakker, A. Q., Korimbocus, J., Kramer, R. A., et al. (2006). Isolation and characterization of human monoclonal antibodies from individuals infected with west nile virus. J. Virol. 80, 6982–6992. doi: 10.1128/JVI.00551-06
Ulbert, S. (2019). West Nile virus vaccines - current situation and future directions. Hum. Vaccines Immunother. 15, 2337–2342. doi: 10.1080/21645515.2019.1621149
Vogt, M. R., Dowd, K. A., Engle, M., Tesh, R. B., Johnson, S., Pierson, T. C., et al. (2011). Poorly Neutralizing Cross-Reactive Antibodies against the Fusion Loop of West Nile Virus Envelope Protein Protect In Vivo via Fcγ Receptor and Complement-Dependent Effector Mechanisms. J. Virol. 85, 11567–11580. doi: 10.1128/JVI.05859-11
Welsch, S., Miller, S., Romero-Brey, I., Merz, A., Bleck, C. K. E., Walther, P., et al. (2009). Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5, 365–375. doi: 10.1016/j.chom.2009.03.007
Yamanaka, A., Rattanaamnuaychai, P., Matsuda, M., Suzuki, R., Matsuura, Y., Tatsumi, M., et al(2022)Engineered flavivirus vaccines control induction of crossreactive infection-enhancing and -neutralizing antibodies. Vaccine. 40, 6004–6011 .doi: 10.1016/j.vaccine.2022.09.012
Yang, M., Lai, H., Sun, H., Chen, Q. (2017). Virus-like particles that display Zika virus envelope protein domain III induce potent neutralizing immune responses in mice. Sci. Rep. 7, 7679. doi: 10.1038/s41598-017-08247-9
Zhang, X., Jia, R., Shen, H., Wang, M., Yin, Z., Cheng, A. (2017). Structures and functions of the envelope glycoprotein in flavivirus infections. Viruses 9, 338. doi: 10.3390/v9110338
Keywords: West Nile virus, vaccine, recombinant proteins, fusion loop, cross-reactivity, flavivirus
Citation: Weiß R, Issmail L, Rockstroh A, Grunwald T, Fertey J and Ulbert S (2023) Immunization with different recombinant West Nile virus envelope proteins induces varying levels of serological cross-reactivity and protection from infection. Front. Cell. Infect. Microbiol. 13:1279147. doi: 10.3389/fcimb.2023.1279147
Received: 17 August 2023; Accepted: 30 October 2023;
Published: 15 November 2023.
Edited by:
Richard T. Marconi, Virginia Commonwealth University, United StatesReviewed by:
Brian D. Carey, National Institute of Allergy and Infectious Diseases (NIH), United StatesSubodh Kumar Samrat, University of Arizona, United States
Copyright © 2023 Weiß, Issmail, Rockstroh, Grunwald, Fertey and Ulbert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastian Ulbert, c2ViYXN0aWFuLnVsYmVydEBpemkuZnJhdW5ob2Zlci5kZQ==
 Rebecca Weiß
Rebecca Weiß Leila Issmail
Leila Issmail Alexandra Rockstroh
Alexandra Rockstroh Thomas Grunwald
Thomas Grunwald Jasmin Fertey
Jasmin Fertey Sebastian Ulbert
Sebastian Ulbert