- 1Department of Laboratory Medicine, Huadong Hospital Affiliated to Fudan University, Shanghai, China
- 2Shanghai Key Laboratory of Clinical Geriatric Medicine, Huadong Hospital Affiliated to Fudan University, Shanghai, China
- 3Research Center on Aging and Medicine, Fudan University, Shanghai, China
- 4Department of Endoscopy, Huadong Hospital Affiliated to Fudan University, Shanghai, China
- 5Department of Research and Development, Ningbo HEALTH Gene Technologies Co., Ltd, Ningbo, China
- 6Department of Gerontology, Huadong Hospital Affiliated to Fudan University, Shanghai, China
Aim: This study established a high-throughput multiplex genetic detection assay (HMGA) for rapid identification, semi-quantification and virulence analysis of Helicobacter pylori directly from the clinical non-invasive oral samples.
Methods: The gastric mucosa and oral samples were collected from 242 patients in Shanghai from 2021 to 2022. All the samples were detected by routine clinical tests for H. pylori and Sanger sequenced for inconsistent results. A new multiplex PCR assay providing results within 4 hours was designed and optimized involving fluorescent dye-labeled specific primers targeted 16S rRNA gene, semi-quantitative gene ureC and 10 virulence genes of H. pylori. Semi-quantification was carried out by simulating the serial 10-fold dilutions of positive oral samples, and the H. pylori loads in different clinical samples were further compared. The mixed plasmids of virulence genes vacA s1, vacA m1 and vacA m2 were used to evaluate the performance on different genotypes. The consistency of 10 virulence genes in gastric mucosa, saliva, mouthwash and dental plaque of H. pylori-positive patients was compared.
Results: The non-invasive HMGA was highly specific for detection of all 12 targets of H. pylori and human internal reference gene β-globin, and the sensitivity to all target genes could reach 10 copies/μL. Compared with routine clinical tests and sequencing, non-invasive HMGA has a high level (>0.98) of sensitivity, specificity, accuracy, PPV, NPV and kappa coefficient for direct detection of H. pylori in oral samples. Moreover, by detecting peak area levels of ureC, it was confirmed that the H. pylori loads in gastric mucosa were significantly higher than those of the three kinds of oral samples (p<0.05). We also found that 45.0% (91/202) of patients had different H. pylori virulence genes in different oral samples. The concordance of positive detection rates of each virulence gene between saliva and gastric mucosa was more than 78% (p<0.05).
Conclusion: The non-invasive HMGA proved to be a reliable method for the rapid H. pylori identification, semi-quantification and detection of 10 virulence genes directly in oral samples, providing a new idea for non-invasive detection of H. pylori.
1 Introduction
Helicobacter pylori is one of the most common causes of cancer-related deaths and is thought to infect half of the world’s population (Kesharwani et al., 2023). The common clinical detection methods of H. pylori are Urea breath test (non-invasive test), histochemical method and culture (invasive test), all of which have certain advantages and disadvantages (Kismat et al., 2022). 13C/14C-urea breath test, as the gold standard for H. pylori detection, is mainly applicable to screen H. pylori in the stomach during physical examination. Although there is no physical pain to the patients, its results may be affected by drugs and endogenous production of CO2 depending much on basal metabolic rate (Eisdorfer et al., 2018; Chen et al., 2023). Histochemistry has high sensitivity and specificity, but the operation is complicated and brings greater pain to patients due to gastroscopy biopsy (Kocsmár and Lotz, 2022). Even worse, the harsh culture conditions of H. pylori result in a low positive detection rate (McNulty, 2023).
The oral cavity is an important storage location for microbial population and a potential reservoir of H. pylori, which may cause the recurrence of H. pylori infection in the stomach (Nagata et al., 2023). In addition, it has been reported that the H. pylori in the oral cavity is highly homologous to that in stomach and closely related to the occurrence and development of gastric diseases(Siavoshi et al., 2005; Chen et al., 2022). However, some researchers have indicated that the genotypes of H. pylori in the oral cavity and stomach are different (Sohrabi et al., 2021). Therefore, the correlation between oral and gastric H. pylori infection is controversial (Mao et al., 2021). Moreover, the positive detection rates of H. pylori infection in oral samples vary greatly among different detection methods, ranging from 0% to 100% (Wang et al., 2014; Zhou et al., 2016; Xu et al., 2018). Therefore, there is an urgent need to develop a non-invasive detection technology for H. pylori in oral samples, providing a reliable etiological diagnosis basis for H. pylori infection.
Our team has previously established the HMGS platform for direct detection of H. pylori in gastric mucosa (Zhang et al., 2016a; Zhang et al., 2016b; Zhou et al., 2016). However, sampling methods have drawbacks such as invasion, complex sampling process and are only applicable to specialized laboratories. In the present study, we established a non-invasive detection technology with high accuracy, specificity and sensitivity for H. pylori in oral samples. In gastric mucosa, culture, histology, rapid urease test (RUT) and quantitative real-time PCR (qRT-PCR) were used as the comparators for the non-invasive HMGA platform. In oral samples, only RUT and qRT-PCR were used as the comparators for the non-invasive HMGA platform. And we further explored the consistency and difference between oral and gastric H. pylori, providing reliable etiological diagnosis basis for clinically rapid and convenient detection of H. pylori.
2 Methods
2.1 Ethics statement
This study was conducted in accordance with the Declaration of Helsinki and approved by Medical Ethics Committee of Huadong Hospital Affiliated to Fudan University. All participants have signed the informed consent forms (The Ethics Approval Number: 2020K080).
2.2 Inclusion & exclusion criteria
Patients who underwent upper gastrointestinal endoscopy due to recurrent upper gastrointestinal symptoms of abdominal pain, acid reflux, vomiting or abdominal distention, and chronic gastritis, peptic ulcer or gastric cancer with family history, were included(Kotilea et al., 2023). Patients who received anti-H. pylori therapy within 4 weeks preceding the study, experienced subtotal gastrostomy, suffered from severe cardiovascular and respiratory diseases or recently consumed NSAID or alcohol, were excluded(Kotilea et al., 2023). In addition, all included patients did not brush their teeth or drink water in the morning.
2.3 Collection of gastric specimens and oral specimens
From February 2021 to January 2022, 242 patients with oral samples and gastric mucosa who met the inclusion criteria and received gastroscopy in Huadong Hospital Affiliated to Fudan University were recruited into the final cohort. The gastric biopsy specimens were taken from two parts of antrum of the patients undergoing endoscopy. The two biopsies were homogenized, combined together, and then divided into four portions for the following tests: culture, RUT, qRT-PCR and non-invasive HMGA. Among 242 patients, there were 126 (52.1%, 126/242) males with mean age of 48.0 ± 18.8 years (age range: 15-93 years) and 116 (47.9%, 116/242) female patients (age range: 14-89 years, mean age: 50.0 ± 19.0 years). In addition, to further verify no interference in target genes, oral samples were obtained from 30 healthy volunteers.
The oral samples including saliva, mouthwash and dental plaque were collected. Firstly, 1 mL of the saliva from patients who did not drink, eat or brush teeth were collected into a disposable sterile sputum cup. Then, patients drink 1mL of 0.9% saline to rinse their mouths for 1 minute. The collected mouthwash was placed in a disposable sterile sputum cup. We then used the disposable probe to scrape the subgingival plaque from the patient’s first and second molars gently to avoid gums bleeding and transferred the scraped plaque into a sterile tube containing 1 mL of 0.9% saline.
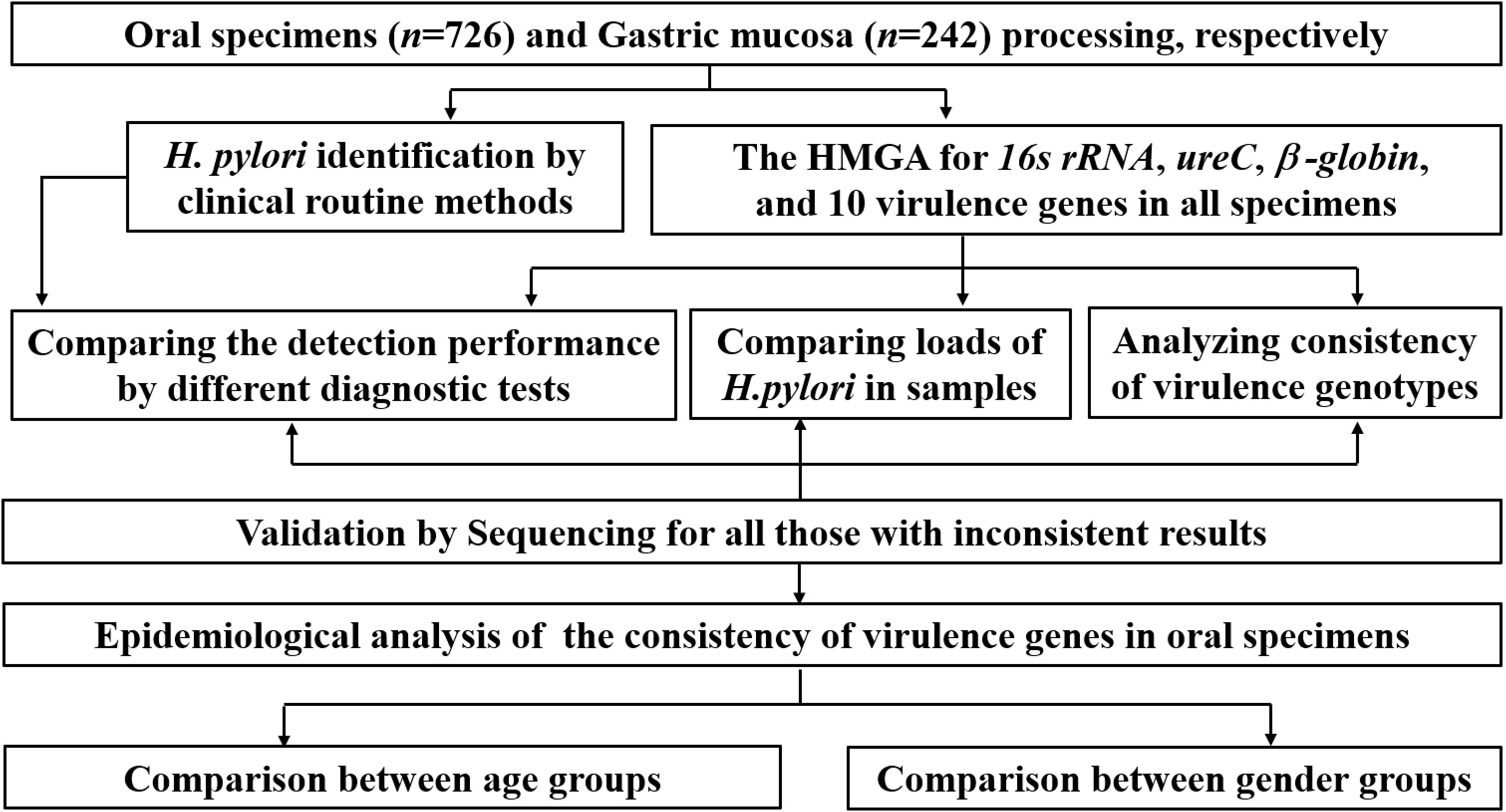
All samples were stored for clinical performance verification, comparison of H. pylori loads and consistency analysis of virulence genes. The main experimental and analyzing procedure is shown in Figure 1. Firstly, the non-invasive HMGA was used to detect H. pylori in clinical samples, and further compare the detection performance of different diagnostic methods. Then, the differences of H. pylori loads in clinical samples were compared. And epidemiological characteristics of the consistency of virulence genes in oral samples were further analyzed. In addition, the consistency of virulence genotypes was compared between gastric mucosa and oral samples.
2.4 Clinical routine methods
Gastric mucosa specimens for histological examination were fixed in 4.5% buffered formalin and embedded in paraffin. A modified Giemsa stain was used to detect H. pylori. An experienced histopathologist reviewed each biopsy without knowing clinical data or H. pylori status. The gastric biopsies sent to the microbiology laboratory were homogenized and then inoculated on Columbia agar medium (OXOID Microbiology Products, Thermo Fisher Scientific Inc., MA, USA) containing 8% sterile defibrinated sheep blood and 0.5% selective antibiotics supplement. The cultures were kept at 35°C under microaerophilic conditions of 5% O2, 10% CO2 and 85% N2 for 3-7 days. Colonies with typical H. pylori morphology were selected and identified by Gram staining, RUT, oxidase and catalase tests. Notably, RUT was used to detect H. pylori in each oral and gastric biopsy specimen according to the instructions (H. pylori rapid detection kit, Huiyi Biology Companies, Shanghai, China). The brief operation was as follow. The gastric mucosa biopsy and oral samples were placed into semisolid 2% agar with a sterile needle and then incubated at room temperature. It was positive for H. pylori when the indicator light changes from yellow to red after 30min. The operation was performed as the instruction (Siavoshi et al., 2015). Moreover, qRT-PCR performed by H. pylori Nucleic Acid Detection Kit (Da’an Gene Co., Ltd, Guangzhou, China) was used to identify H. pylori in each sample by ABI Prism®7500 Real‐Time PCR Instrument (Thermo-Fisher, MA, USA). Specific primers and fluorescent probes were designed with the highly conserved region of ureA of H. pylori as the target. The CT value ≤ 27.02 indicated a positive H. pylori infection status.
2.5 Establishment of the non-invasive HMGA
2.5.1 Primer design
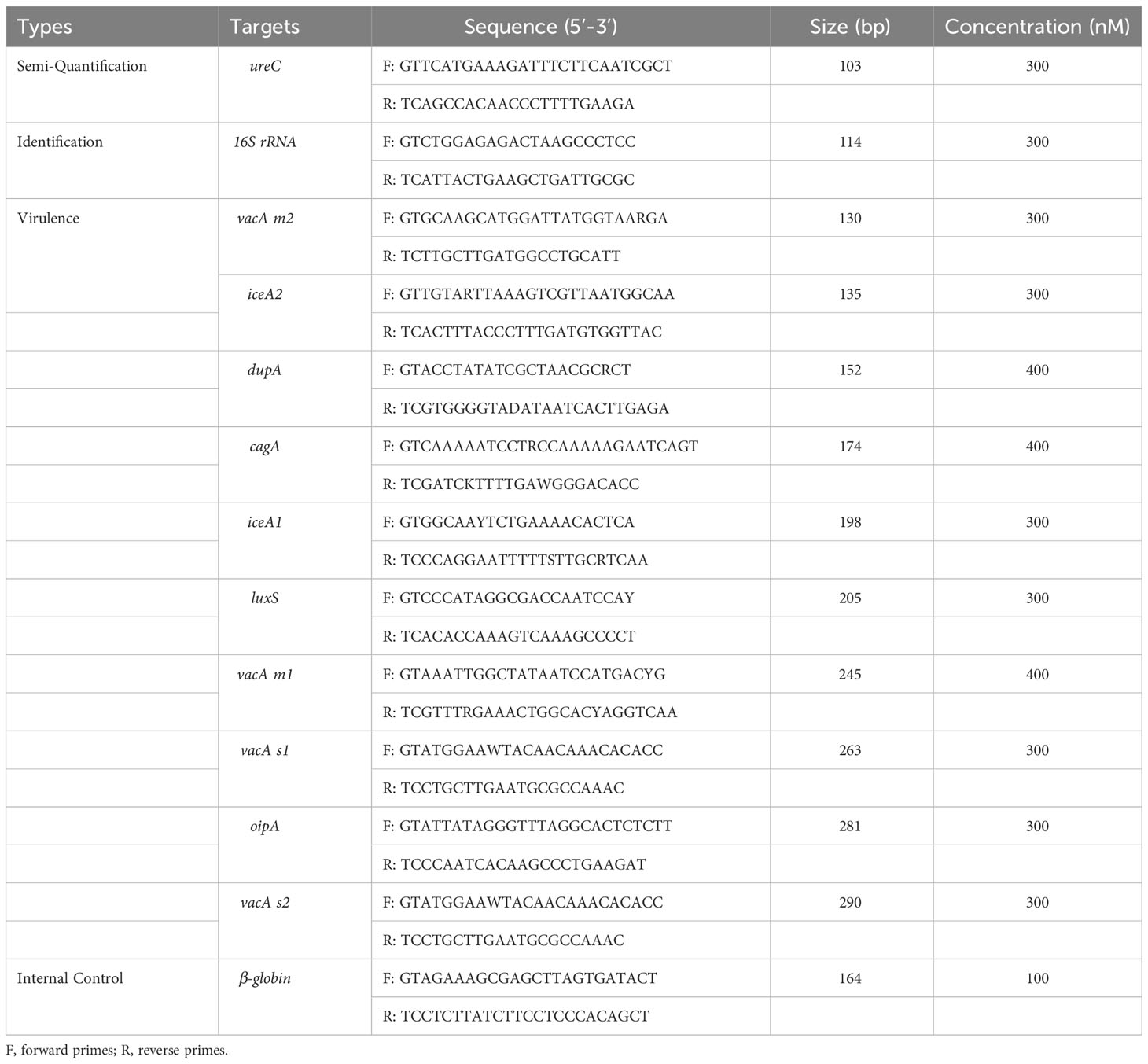
The gene sequences of H. pylori strain-specific identification gene 16S rRNA, single-copy semi-quantitative gene ureC, 10 virulence genes (vacA s1, vacA s2, vacA m2, vacA m1, cagA, oipA, luxS, iceA2, iceA1 and dupA) and one human internal reference gene β-globin were downloaded from the National Center of Biotechnology Information (NCBI). Vector NTI (Invitrogen, Carlsbad, USA) was used for multiple sequence alignment, and highly conserved specific sequences were selected to design primers (Sun et al., 2021). Next, primers were designed to amplify each of the highly conserved regions using DNASTAR (DNASTAR Inc., Madison, WI, USA) and Primer Premier 6.0 (Premier Biosoft International, Palo Alto, CA, USA)(Sun et al., 2021). The primers are detailed in Table 1.
2.5.2 DNA extraction and multiplex PCR
Total DNA from the strains and clinical samples was extracted by an automatic nucleic acid extraction instrument with magnetic beads (Smart LabAssist, Taiwan Advanced Nanotech, Taiwan, China). Concentration of each extracted DNA was determined using the Nanodrop spectrophotometer (Thermo Fisher Scientific Inc., MA, USA). DNA was kept in -20°C for further analysis.
Multiplex PCR mixture with the 15µL final volume included 6µL of 2.5*NuHi SU11 PCR Mix (NuHigh Biotechnologies Co., Ltd, Suzhou, China), 1.5µL of pooled primers, 2.5µL of ddH2O, and 5µL of template. The primer pool consisted of one pair of primer for identification gene, one pair for semi-quantitation gene, ten pairs for virulence genes and one pair for the quality control gene. The primers were mixed in different proportions to achieve optimum sensitivity for all targets, and the final concentration of each primer in the pool was listed in Table 1. The PCR was performed using the Veriti 96-well Thermal Cycler (Applied Biosystems, California, USA): 50°C for 5 min; 95°C for 10 min; 32 cycles of 95°C for 30 seconds, 60°C for 30 seconds and 72°C for 1 min; and 72°C for 15 min.
2.5.3 Capillary electrophoresis and fragment analysis
The separation by capillary electrophoresis and fragment analysis using the Applied Biosystems 3500DX Genetic Analysis System (Applied Biosystems, California, USA) were same as our previous study (Sun et al., 2021). In this study, when the detection peak height of the target gene product fragment >500 relative fluorescence units (rfu), it was judged positive for the target gene (Figure 2A). The ddH20 and seven negative control pathogens, including Candida albicans (C. albicans), Streptococcus salivarius (S. salivarius), Acinetobacter baumannii (A. baumannii), Staphylococcus aureus (S.aureus), Pseudomonas aeruginosa (P. aeruginosa), Escherichia coli (E.coli) and Klebsiella pneumoniae (K. pneumoniae) were used as negative quality controls throughout the detection. All results from the non-invasive HMGA were further verified by Sanger sequencing, as the gold standard for gene identification in this study (Drevinek et al., 2023). The identification gene for H. pylori with Sanger sequencing is 576 bp-16S rRNA referred to previously reported paper (Forward primer: (5′-TCTAACGAATAAGCACCGGCTA-3′), Reverse primer: (5′-GTGCAGCACCTGTTTTCAAGG-3′)(Hu et al., 2016).
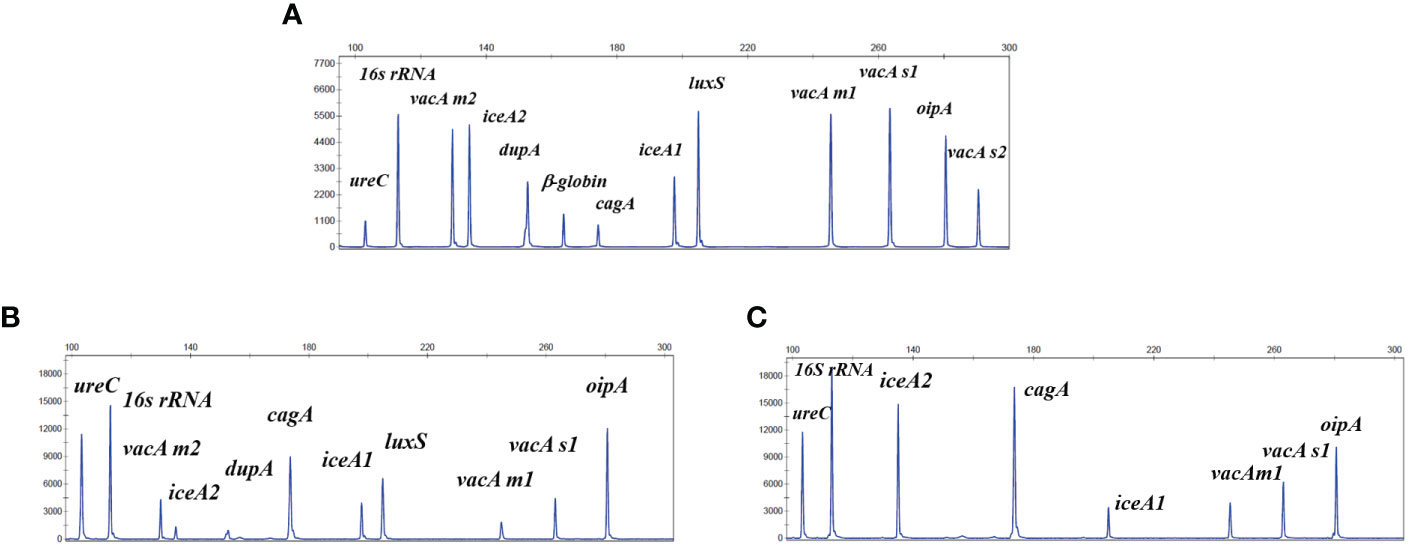
Figure 2 The non-invasive HMGA could produce specific amplification signals for 13 targets. The horizontal coordinate indicated the actual PCR product size(bp), and the vertical coordinate indicated the dye signal (rfu). (A) The specific amplification signals arising from the mixed plasmids of the 12 target genes of H. pylori and one β-globin gene at the same concentration of 20 copies/μL and 7 negative control pathogens DNA all at the concentration of 105 copies/μL. (B, C) The specific amplification signals arising from the amplification of the traditional clinical isolates. Note that all gene targets were specifically amplified without non-specific amplification by the non-invasive HMGA.
2.5.4 Optimization of the non-invasive HMGA
The 13 targets detection for the non-invasive high-throughput multiplex genetic detection assay (HMGA) were optimized as following principle: primer sequences were optimized so that each signature of detection target could be amplified specifically without cross-interaction; other reaction parameters of PCR components and procedures, such as buffer, enzyme, and reaction time, were also systematically optimized. Additionally, detections of β-globin in the clinical samples indicated that no significant nucleic acid degradation had occurred during specimen handling/storage. In addition, we mixed the plasmids of the 12 target genes of H. pylori and β-globin at the same concentration of 20 copies/μL and added seven negative control pathogen DNA, including Candida albicans, Streptococcus salivarius, Acinetobacter baumannii, Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli and Klebsiella pneumoniae all at the concentration of 105 copies/μL. The H. pylori negative results were validated in saliva (n=30), mouthwash (n=30) and dental plaque samples (n=30) from healthy people using RUT and qRT-PCR. Then the non-invasive HMGA was used to detect the extracted DNA of H. pylori negative oral samples from healthy people, respectively.
2.6 Evaluation of the semi-quantification of non-invasive HMGA
In order to prove whether non-invasive HMGA can semi-quantitatively detect H. pylori, the single copy gene ureC of H. pylori was selected as the detection target. Firstly, the standard strain ATCC26695 was fully mixed with the saliva of healthy volunteers to extract DNA from the simulated H. pylori positive saliva. The concentration of ureC in the standard strain was quantified by Droplet Digital PCR (dd-PCR), and the ureC gene was gradually diluted to 1×104, 1×103, 1×102, 1×101 and 1×100 copies/μL, respectively. The non-invasive HMGA was used to detect the ureC gene at each gradient concentration for three times. The gradient dilution concentration of H. pylori was used as the horizontal coordinate, and the peak areas of the three assays corresponding to different gradient concentrations were as the vertical coordinate.
2.7 Statistical analysis
The true positive is defined as consistent positive results of both the detection methods and the gold standard. The true negative is defined as consistent negative results of both the detection methods and the gold standard. GraphPad Prism 8.0.2 (San Diego, CA, USA) was used for statistical analysis. The positive rates of H. pylori virulence genes from saliva, mouthwash and dental plaque samples in genders and ages were compared by the Chi-squared test. The Wilcoxon rank sum test was used for comparison of ureC peak area levels in different samples and above. For all figures, * means p <0.05, ** means p <0.01, *** means p <0.001, **** means p <0.0001, and n.s. means not significant (p >0.05). Differences were considered statistically significant for p <0.05.
3 Results
3.1 The non-invasive HMGA is specific for H. pylori identification
The results showed that 13 target genes had specific peaks without non-specific products amplified by their corresponding primers. Subsequently, As shown in Figure 2A, 13 specific amplification peaks were observed clearly. Two traditional clinical isolates were used to evaluate the accuracy of simultaneous detection of target genes of the non-invasive HMGA. All the specific amplification signals were observed clearly (Figures 2B, C). In addition, seven negative control pathogens did not produce any specific amplification peaks (Supplementary Figures S2A–G). As shown, only β-globin was found in the electrophoresis maps of saliva (Supplementary Figure S2H), mouthwash (Supplementary Figure S2I) and dental plaque (Supplementary Figure S2J), indicating that the detection of H. pylori target genes by non-invasive HMGA was not interfered by complex microorganisms in oral samples. The above results demonstrated that non-invasive HMGA was highly specific to all targets. Additionally, the minimum detection limit of non-invasive HMGA was as low as 10 copies/μL for simultaneous detection of 12 target genes using serial 10-fold dilution.
3.2 Positive detection rate and performance evaluation of non-invasive HMGA for H. pylori in oral samples
The results of non-invasive HMGA showed that H. pylori was detected in 208 (86.0%, 208/242) gastric mucosa, 194 (80.2%, 194/242) saliva samples, 167 (69.0%, 167/242) mouthwash samples and 128 (52.9%, 128/242) dental plaque samples. Furthermore, 202 patients (83.5%, 202/242) tested positive for more than one kind of oral samples, 169 patients (69.8%, 169/242) tested positive for more than two kinds of oral samples, and 115 patients (47.5%, 115/242) tested positive for all three oral samples. Additionally, the results of non-invasive HMGA showed that H. pylori was all negative in oral samples from 30 healthy volunteers.
Comparing with sequencing, the non-invasive HMGA system for detecting H. pylori in gastric mucosa (Table 2) and oral samples (Table 3) was evaluated. The sensitivities of non-invasive HMGA to H. pylori in saliva, gargle and dental plaque samples were 1.00, 1.00 and 0.99. respectively. The specificities of non-invasive HMGA detections were all 1.00 in oral samples. The kappa values measuring the consistency values of non-invasive HMGA and sequencing in saliva, mouthwash and dental plaque samples were 1.000, 1.000 and 0.992, respectively (Tables 2, 3).
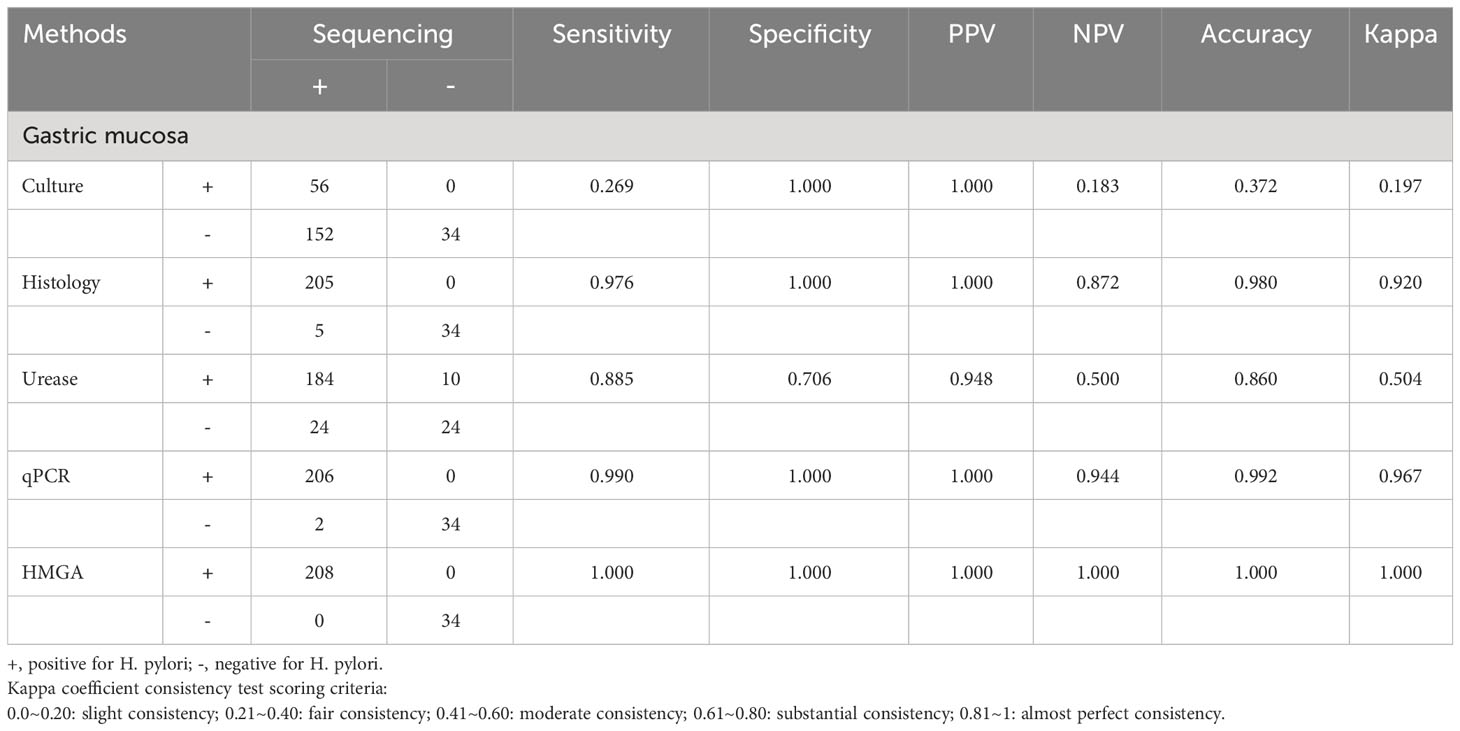
Table 2 Non-invasive HMGA exhibited high levels of accuracy for the identification of H. pylori in gastric biopsies when compared to conventional methods.
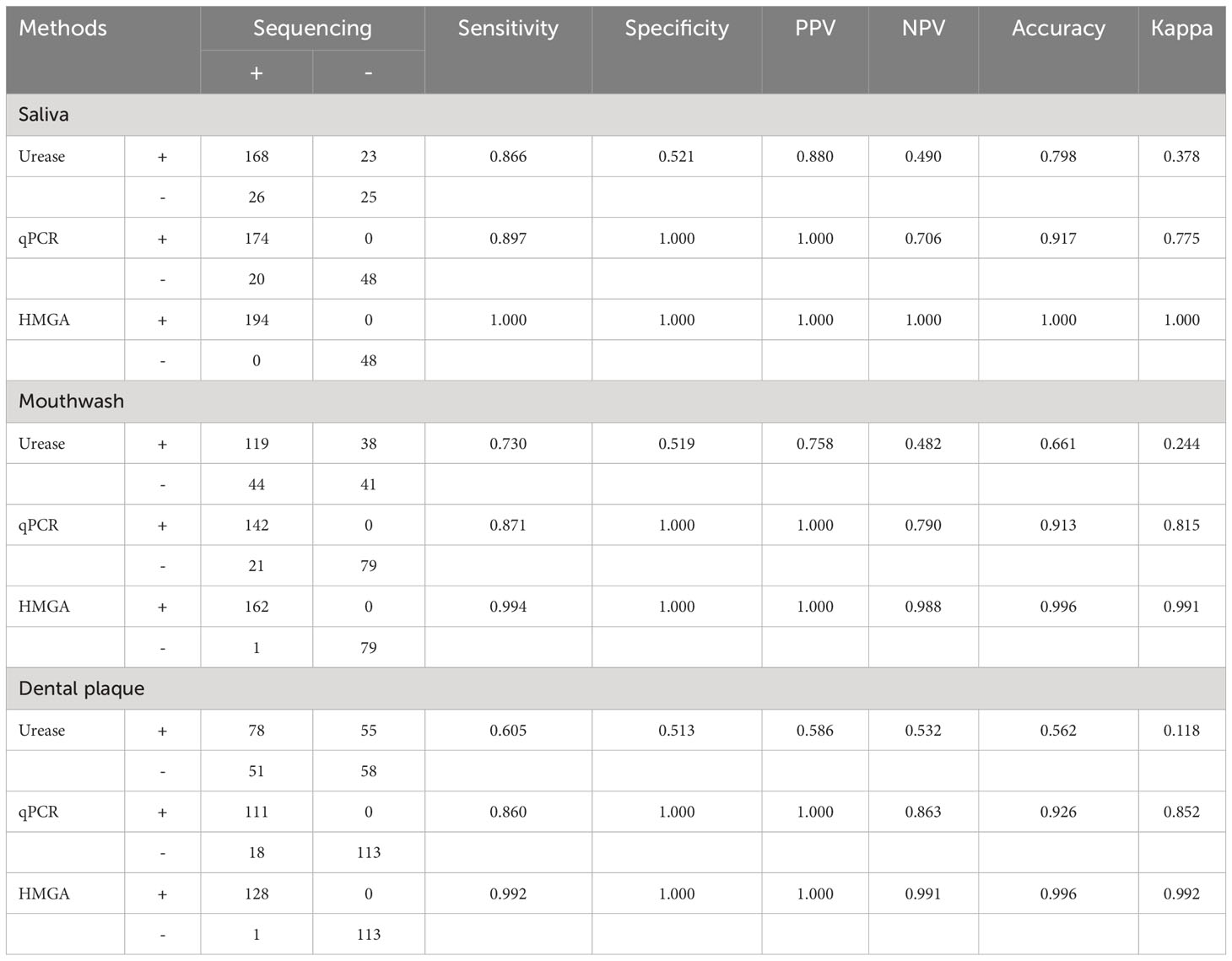
Table 3 Non-invasive HMGA exhibited high levels of accuracy for the identification of H. pylori in oral specimens when compared to conventional methods.
3.3 Relative quantitative analysis of H. pylori infection in oral samples
Our data showed that the detection peak area of non-invasive HMGA increased with the increase of ureC concentration. The correlation coefficient between ureC gene concentration and peak area was 0.989 (Figure 3A). We further used non-invasive HMGA to detect the peak area of ureC gene in gastric mucosa and oral samples of 242 patients and drew violin graphs based on the distribution and probability density of peak area of ureC in different samples. As shown, the peak area of ureC in gastric mucosa samples was relatively high (from 150000 to 300000), and significantly higher than that of three oral samples (p <0.001) (Figures 3B–D). The ureC peak area of saliva samples was distributed in the relatively middle position (from 100 000 to 250 000), and the peak area level was significantly higher than that of mouthwash samples (p <0.001) and dental plaque samples (p <0.001) (Figure 3E). Furthermore, we calculated the copy numbers of H. pylori corresponding to the relatively concentrated detection peak area of ureC distribution in clinical samples. We found, more than 60% of the ureC peak areas of gastric mucosa concentrated around 250 000, and the corresponding copy numbers of H. pylori were about 100 copies/μL. More than 70% of the ureC peak areas of saliva samples ranged from 50 000 to 250 000, and the corresponding copy numbers of H. pylori were ranged from 10 to 100 copies/μL. About 50% of the ureC peak areas of mouthwash samples ranged from 50 000 to 250 000, and the corresponding copy numbers were ranged from 10 to 100 copies/μL. About 50% of the ureC peak areas of dental plaque samples below 50 000, and the corresponding to the copy numbers of H. pylori were less than 10 copies/μL. H. pylori loads in gastric mucosa (p<0.05) were significantly higher than those in three kinds of oral samples by detecting ureC peak area levels. Furthermore, according to the semi-quantitative standard analysis, when the peak areas of H. pylori DNA in different clinical samples concentrated around 5 000, the corresponding mean copy numbers was 10 copies/μL.
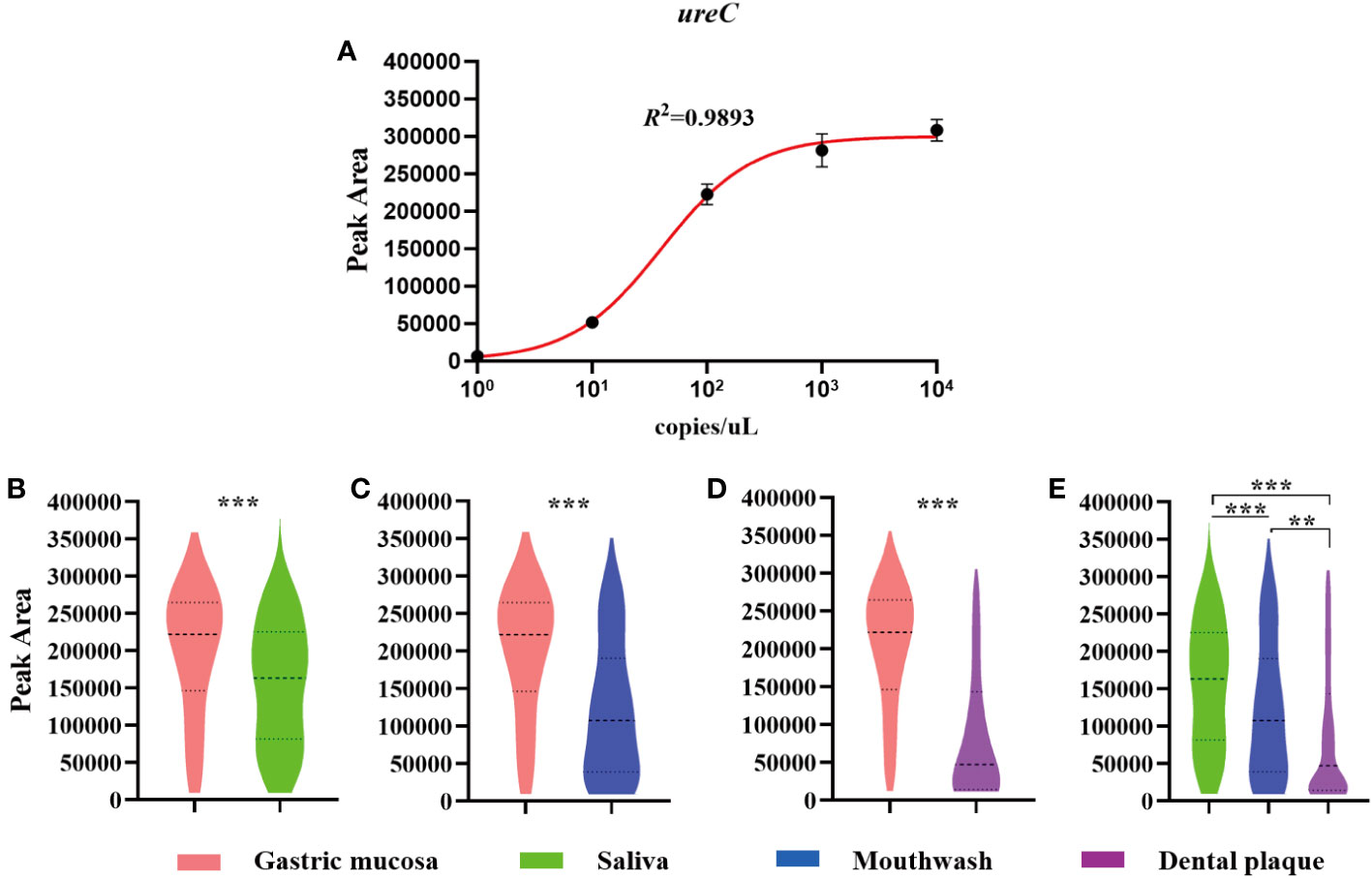
Figure 3 The semi-quantitation of H. pylori in clinical samples by detecting the peak area of ureC using non-invasive HMGA. The violin plot was drawn by GraphPad Prism 8.0.2. (A) The peak areas of ureC at gradient concentration of 1×100~1×104 copies/µL in simulated positive saliva were detected by non-invasive HMGA to evaluate the semi-quantitation for H. pylori. (B–D) The peak area levels of ureC in gastric mucosa samples were significantly higher than those in the three oral samples (p<0.001). (E) The peak area levels of ureC in saliva were significantly higher than those in mouthwash (p<0.001) and dental plaque (p < 0.001). **denotes p < 0.01, ***denotes p < 0.001.
3.4 The non-invasive HMGA was able to distinguish different virulence genotypes
To evaluate whether non-invasive HMGA could accurately identify and distinguish different vacA genotypes, plasmids of virulence genes vacA m1 and vacA m2 were mixed at final concentrations of 102 and 103copies/μL to form mixed templates with seven negative control pathogens DNA. As shown in Figures 4A, B, the position and signal intensity of the target peaks of the two virulence genes were consistent whether seven negative control pathogens DNA existed. As shown in Figures 4C, D, the position and signal intensity of the target peaks of the two virulence gene plasmids were basically consistent with those of the mixed detection, indicating that when non-invasive HMGA was used to detect different vacA genotypes, the position and signal intensity of the target peaks were not interfered.
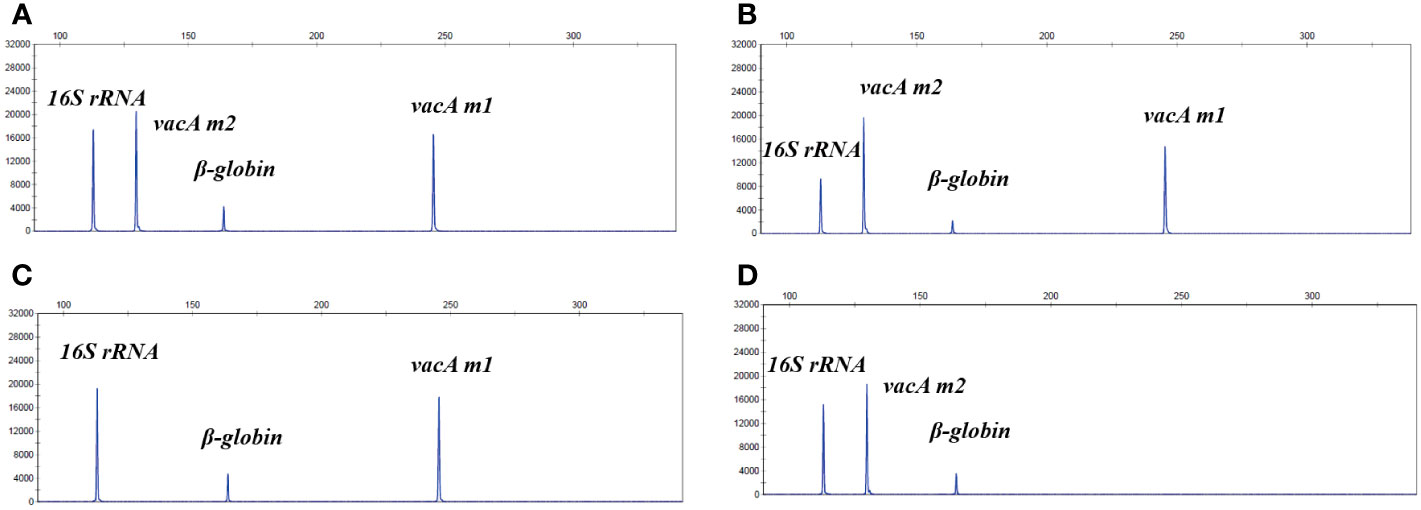
Figure 4 The non-invasive HMGA accurately distinguished different virulence genotypes. (A) The system was used to detect mixed plasmids of vacA m1 (102 copies/μL) and vacA m2 (103 copies/μL), and the mixed plasmids of 16S rRNA and β-globin. The vacA m1 and vacA m2 were detected specific peaks of about 16000 rfu and 20000 rfu at 245 bp and 130 bp, respectively. (B) The system was used to detect the mixed plasmids of vacA m1 (102 copies/μL), vacA m2 (103 copies/μL) and negative controls (105 copies/μL). (C, D) The position and signal intensity of the target peaks were basically consistent with those in (A), when vacA m1 (102 copies/μL) and vacA m2 (103 copies/μL) plasmids detected separately.
3.5 Detection and clinical correlation analysis of consistency of virulence genes in oral samples by non-invasive HMGA
Non-invasive HMGA was used to detect H. pylori in saliva, mouthwash and dental plaque samples of 242 patients. The results showed that H. pylori was positive in at least one kind of oral sample of 202 patients (83.5%, 202/242). Clinical analysis showed that the proportion of male patients with oral H. pylori positive was 51.5% (104/202), which was higher than that of female patients (48.5%, 98/202). Consistent with the loads of H. pylori in oral samples, we found that the positive detection rates of H. pylori virulence genotypes in saliva were higher than those in mouthwash and dental plaque. The positive detection rates of H. pylori virulence genotypes in mouthwash were higher than those in dental plaque. Among them, the positive detection rates of cagA, iceA1, luxS and oipA in saliva were significantly higher than those in mouthwash and dental plaque (p <0.05). The positive detection rates of vacA s1m2, cagA, iceA2 and oipA in mouthwash were significantly higher than those in dental plaque (p <0.05) (Figures 5A–D).
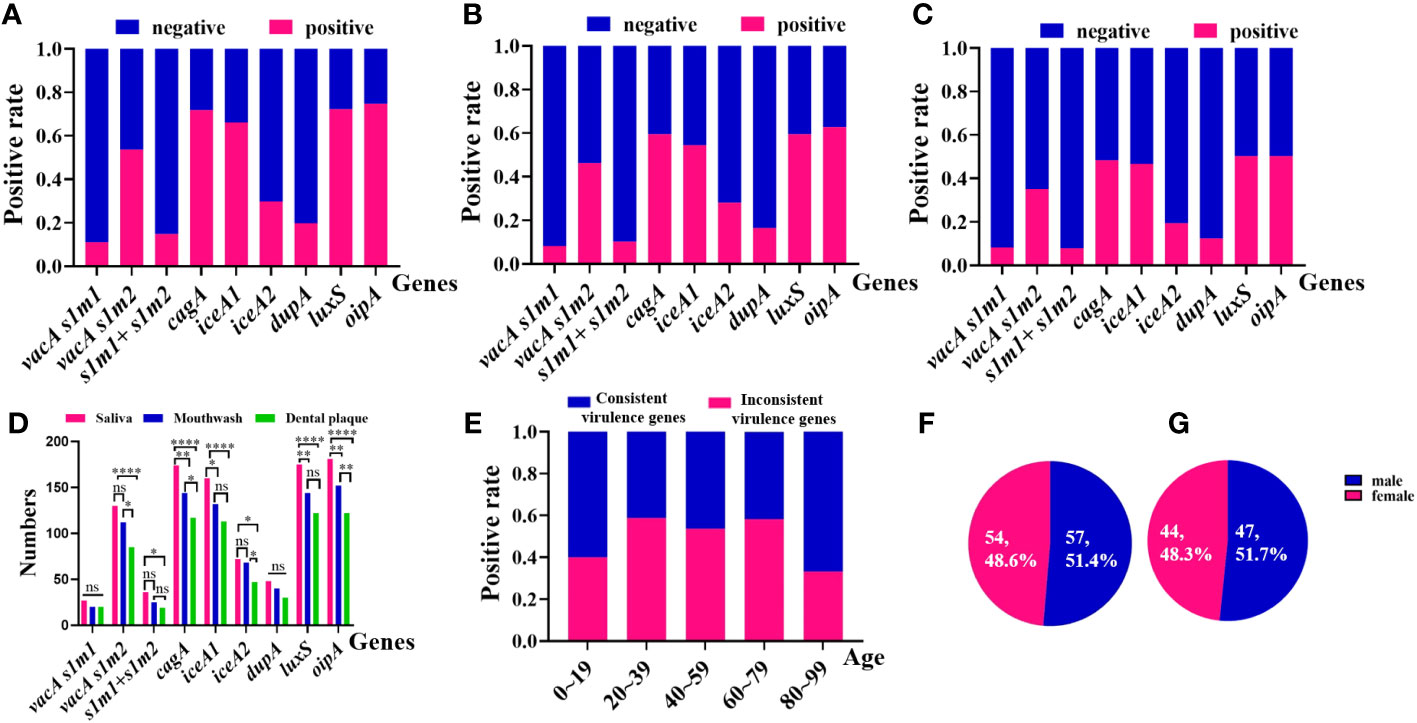
Figure 5 Positive rates of the consistency of virulence genes of H. pylori from 202 oral samples in different groups by non-invasive HMGA. (A) The virulence genotypes of H. pylori in saliva samples. From left to right were vacA s1m1(11.2%), meaning vacA s1 and vacA m1, vacA s1m2(53.7%), meaning vacA s1 and vacA m2, vacA s1m1+vacA s1m2(14.9%), meaning vacA s1, vacA m1 and vacA m2, cagA (71.9%), iceA1(66.1%), iceA2(29.8%), dupA(19.8%), luxS(72.3%) and oipA(74.8%). (B) The virulence genotypes of H. pylori in mouthwash samples. From left to right were vacA s1m1(8.3%), vacA s1m2(46.3%), vacA s1m1+vacA s1m2(10.3%), cagA (59.5%), iceA1(54.6%), iceA2(28.1%), dupA(16.5%), luxS(59.5%) and oipA(62.8%). (C) The virulence genotypes of H. pylori in dental plaque samples. From left to right vacA s1m1(8.3%), vacA s1m2(35.1%), vacA s1m1+vacA s1m2(7.9%), cagA (48.4%), iceA1(46.7%), iceA2(19.4%), dupA(12.4%), luxS(50.4%) and oipA(50.4%). (D) The numbers of virulence genes in saliva were higher than those in mouthwash and dental plaque, and the numbers of virulence genes in mouthwash were higher than those in dental plaque. (E) The distributions of the consistency of virulence genes of H. pylori from oral samples in different ages were compared. The proportion of consistent virulence genes of H. pylori in age groups 0-19, 20-39, 40-59, 60-79 and 80-99 were 40.0%, 58.8%, 53.7%, 58.3% and 33.3%, respectively. (F) The distributions of consistent virulence genes of H. pylori in different genders. (G) The distributions of inconsistent virulence genes of H. pylori in different genders.
By comparing the consistency of 10 virulence gene combinations of H. pylori in different oral samples from 202 patients, we found that in 55.0% (111/202) of patients, 10 virulence genes of H. pylori were completely consistent among different oral samples. And 45.0% (91/202) of patients were detected different virulence genes in different oral samples. In addition, we analyzed the epidemiological characteristics of H. pylori in 202 oral samples. We found that, the positive rates of consistent virulence genes of H. pylori were higher than those of the inconsistent virulence genes in patients aged 20-79 years old (p<0.05) (Figure 5E). The proportion of consistent virulence genes in males and females in oral samples was 51.4% (57/111) and 48.6% (54/111), and the ratio of males to females was close to 1.1:1 (Figure 5F). The proportion of inconsistent virulence genes in male and female patients was 51.7% (47/91) and 48.3% (44/91). The sex ratio of inconsistent virulence genes in oral samples was close to 1.1:1 (p >0.05) (Figure 5G).
3.6 Consistency of detecting virulence genes of H. pylori in oral and gastric mucosa samples by non-invasive HMGA
Firstly, we compared the consistency of 10 virulence gene combinations between saliva and gastric mucosa of each patient. The results showed that 10 virulence genes combinations in 47.5% (115/242) saliva samples were completely consistent with that in gastric mucosa, 32.6% (79/242) were different, and 19.8% (48/242) were H. pylori negative (Table 4). Secondly, we found that 10 virulence gene combinations in 38.8% (94/242) mouthwash samples were completely consistent with that in the gastric mucosa (Supplementary Table S1). Finally, 10 virulence gene combinations between dental plaque and gastric mucosa were completely consistent in 36.8% (89/242) patients (Supplementary Table S2). Furthermore, the consistency of positive detection rates of seven virulence genes (vacA s1, vacA m2, vacA m1, cagA, oipA, luxS and dupA) in saliva and gastric mucosa was > 84% (kappa coefficient > 0.61) (Table 5). The kappa values of positive detection rates of iceA1 and iceA2 in saliva and gastric mucosa were 0.555 and 0.426, respectively.
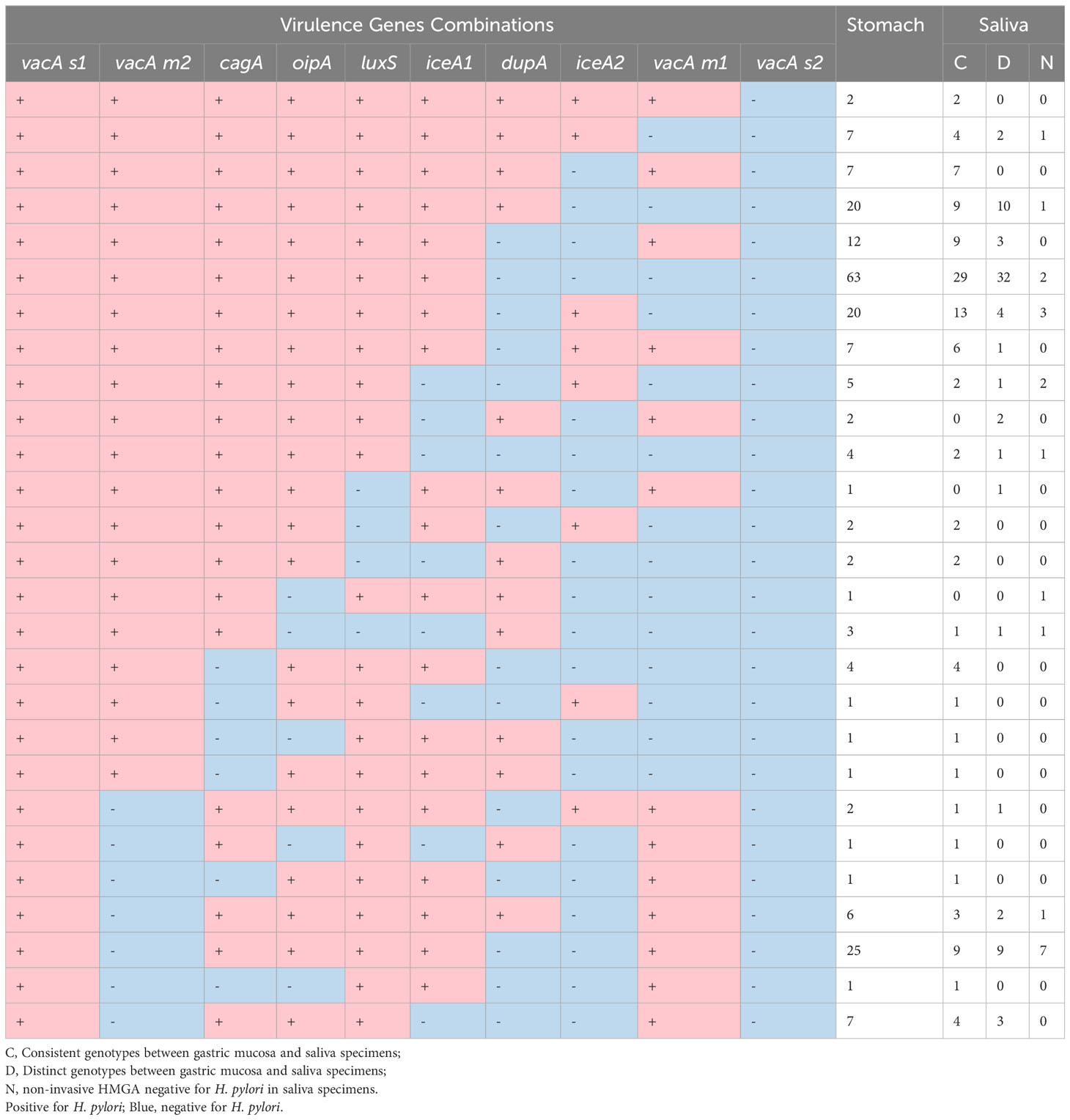
Table 4 Frequencies of virulence genes combinations of H. pylori from gastric mucosa and saliva specimens in 242 patients. Pink.
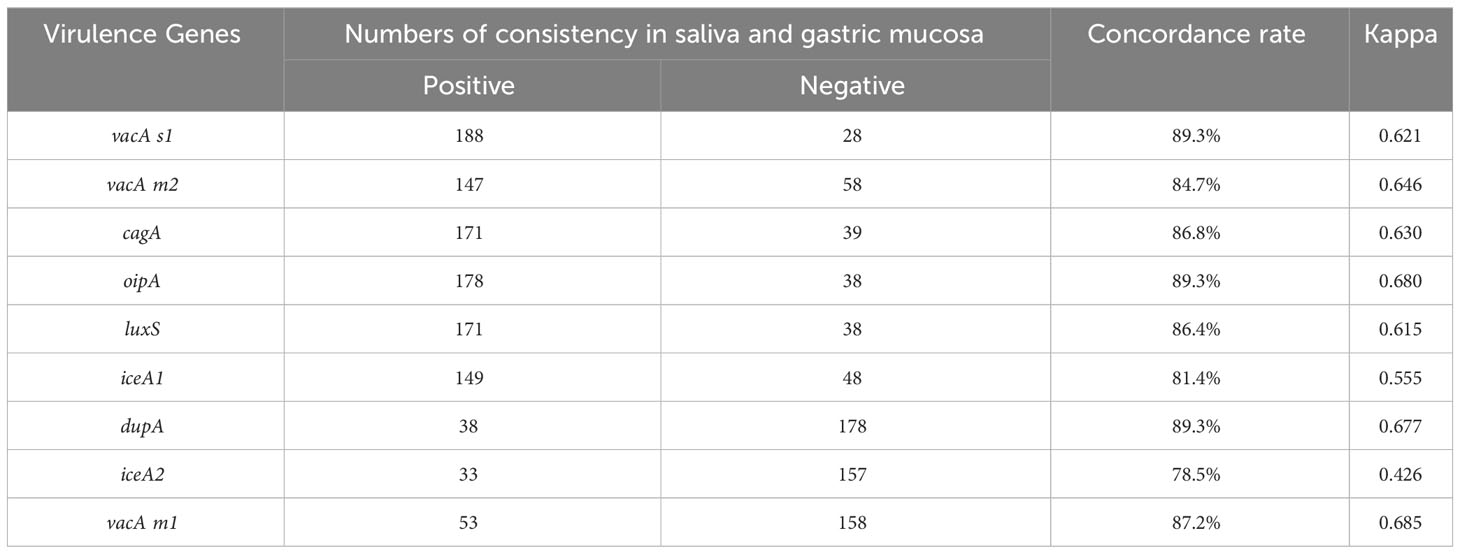
Table 5 Comparison of consistency between each virulence gene of H. pylori in saliva and gastric mucosa samples based on Kappa coefficient.
4 Discussion
4.1 The HMGA provided a new direction and idea for non-invasive detection of H. pylori
H. pylori is a gram-negative bacillus closely related to the occurrence and development of gastric-related diseases (Zang et al., 2023). Effective and timely detection of H. pylori is important to prevent and treat gastric cancer and other gastric-related diseases (Ebell, 2023). The clinical traditional detection methods of H. pylori, such as culture, rapid urease test, immunohistochemical staining, are all invasive methods, which seriously affect the timely diagnosis of H. pylori (Idowu et al., 2022). By referring to the reported high detection rates of virulence genes oipA, iceA and dupA, etc. (Xue et al., 2021), we developed and optimized a non-invasive HMGA directly from the clinical oral specimens. It could identify 12 targets of H. pylori, including identification, semi-quantification and virulence genes, directly from one oral sample within 4 hours. Synchronous detection in one tube is easy to operate, and the detection cost is low for each target ($1.3). Namely, the non-invasive HMGA was fast, convenient and cost-effective. It broke through the bottleneck of invasive sampling of conventional detection methods and made up for the limitations and deficiencies of current methods.
4.2 The detection performance of H. pylori in oral samples was reliable
The sensitivity, specificity, accuracy and consistency of the non-invasive HMGA for the detection of H. pylori in oral samples were all greater than 0.99, confirming that it had excellent detection performance for the identification of H. pylori from three kinds of oral samples. The gold standard is Sanger sequencing, and other clinical methods are used to detect H. pylori in parallel. The sensitivities of RUT to detect H. pylori gradually decreased in gastric mucosa, saliva, mouthwash and dental plaque samples, which may be corelated with the less H. pylori loads in oral samples than those in gastric mucosa samples (Liu et al., 2019). Considering RUT is fast, simple and easy to operate.(Dahlén et al., 2018), it was used in this study for detecting H. pylori in oral samples, although its results maybe doubtful due to the existence of urease producing organisms such as Haemophilus, Actinomyces and Streptococcus sp. present in the human oral cavity. And our study also confirmed the lower specificity of RUT in oral samples (0.51), than that in gastric mucosa (0.71). In addition, the sensitivity, specificity, accuracy and consistency of qPCR kit for H. pylori detection in gastric mucosa were all greater than 0.95, which showed reliable detection performance. However, the detection sensitivity of this method for H. pylori in oral samples decreased to about 0.87, which may be related to the fact that the kit did not specifically improve the limit detection with relatively low loads of H. pylori in oral samples. The specificity of culture in gastric mucosa samples was as high as 1.00, but the sensitivity was low, only 0.27, due to the harsh and time-consuming culture conditions of H. pylori. In addition, we tried to use three kinds of oral samples for H. pylori culture, but it failed due to too many colonization bacteria in the oral cavity. Furthermore, the negative results of oral samples from 30 healthy volunteers by non-invasive HMGA indicated that oral microflora did not affect the detection performance of the HMGA.
4.3 Non-invasive HMGA could be used to detect the consistency of virulence genes in oral samples of H. pylori
The inconsistent virulence genes of H. pylori in the oral cavity may be related to the open environment in oral cavity (Momtaz et al., 2012). Some studies have found significant differences of H. pylori genotypes in gastric mucosa, saliva and fecal samples, and different H. pylori genotypes may exist in the different site of digestive tract in the same patient (Momtaz et al., 2012; Sepúlveda et al., 2012; Wang et al., 2020). The findings suggested that H. pylori were not consistently in saliva and dental plaque samples, which may be the result of occasional gastroesophageal reflux (Momtaz et al., 2012; Mao et al., 2021). Some researchers have indicated that H. pylori in oral cavity may serve as the important source of gastric reinfection (Abdul et al., 2023). Our results of non-invasive HMGA showed 45.0% patients with inconsistent virulence genes of H. pylori in oral samples, which might because humans can be simultaneously infected with two or more H. pylori genotypes (Momtaz et al., 2010; Momtaz et al., 2012).
4.4 The consistency of H. pylori genotypes in saliva and gastric mucosa by non-invasive HMGA was high
Previous studies have shown that H. pylori in oral cavity was closely related to H. pylori in stomach (Wang et al., 2014). Eradication of H. pylori in oral cavity may effectively reduce the positive rate of gastric H. pylori infection (Wang et al., 2014). It was of great clinical significance to accurately detect and analyze the genotypes consistency of H. pylori in oral samples and gastric mucosa. In this study, we analyzed the consistency of 10 virulence genes of H. pylori in saliva, mouthwash and dental plaque, as well as that in gastric mucosa. The results showed that the combination genotypes of identification gene and 10 virulence genes in saliva and gastric mucosa was completely consistent in 47.5% (115/242) patients (Table 4). In addition, the consistency of each virulence gene in saliva and gastric mucosa samples was more than 0.78, which confirmed that H. pylori from oral samples was highly consistent with gastric mucosa. Our results were consistent with the findings reported by Wang et al, which showed 64% homology between saliva and gastric samples from the same patients (Wang et al., 2002). These findings supported the notion that saliva was a possible source of H. pylori infection (Momtaz et al., 2012). Based on the non-invasive HMGA, we first realized the simultaneous detection of 10 important virulence genes of H. pylori in oral samples and analyzed the consistency of H. pylori in oral samples and gastric tissues, providing a more comprehensive, detailed and accurate diagnosis basis for clinical H. pylori infection.
5 Conclusion
In this study, based on oral samples, a non-invasive HMGA was established and optimized for the identification, semi-quantitation and virulence genes of H. pylori. In addition, the different distribution of H. pylori virulence genes in oral and gastric mucosa samples of Shanghai population were analyzed by non-invasive HMGA. This study developed a new method for simple, non-invasive, rapid and accurate detection of H. pylori, and provided a timelier and more comprehensive diagnosis basis for H. pylori infection. It was expected to become an effective assay for clinical identification and monitoring of H. pylori. Considering the frequent update of H. pylori virulence genes, we would develop and simplify new versions of non-invasive HMGA system, for the better, easier and more robust diagnosis of H. pylori infection.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by Medical Ethics Committee of Huadong Hospital Affiliated to Fudan University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
WC: Writing – original draft, Writing – review & editing, Formal Analysis, Software, Data curation, Methodology. SuW: Writing – review & editing, Writing – original draft, Formal Analysis, Validation. TL: Methodology, Writing – review & editing. WJ: Data curation, Writing – review & editing. LD: Data curation, Writing – review & editing. YM: Data curation, Writing – review & editing. FY: Software, Writing – review & editing. JZ: Software, Writing – review & editing. DJ: Methodology, Writing – review & editing. ZX: Methodology, Writing – review & editing. HWZ: Resources, Writing – review & editing. YW: Resources, Writing – review & editing. ZB: Project administration, Writing – review & editing, Supervision. HuZ: Project administration, Writing – review & editing, Funding acquisition, Investigation, Validation, Supervision. ShW: Supervision, Writing – review & editing, Funding acquisition, Investigation, Project administration, Validation.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (No. 82272987, 82172933, 81902380), the Scientific Research Project of Shanghai Science and Technology Commission (No. 21ZR1421700), Shanghai Technological Innovation Action Projects (No. 21Y11900900), the Shanghai health and family planning commission youth project of scientific research subject (No. 20204Y0296), the Shanghai Rising-Star Program (No. 23QA1403000), the Key Subject of Huadong Hospital (No. ZDXK2212, GZRPY010Y), and the Clinical Research Project of Huadong Hospital (No. HDLC2022007).
Conflict of interest
Authors HWZ and YW were employed by the company Ningbo HEALTH Gene Technologies Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1267288/full#supplementary-material
References
Abdul, N. S., Khalid Alkhelaiwi, A., Awadh Alenazi, A., Fehaid Alrashidi, R., Ghaleb Salma, R. (2023). The association of helicobacter pylori in the oral cavity with dental caries in patients with and without gastric infection: A systematic review. Cureus 15 (5), e38398. doi: 10.7759/cureus.38398
Chen, X., Wang, N., Wang, J., Liao, B., Cheng, L., Ren, B. (2022). The interactions between oral-gut axis microbiota and Helicobacter pylori. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.914418
Chen, Y., Yang, C., You, N., Zhang, J. (2023). Relationship between Helicobacter pylori and glycated hemoglobin: a cohort study. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1196338
Dahlén, G., Hassan, H., Blomqvist, S., Carlén, A. (2018). Rapid urease test (RUT) for evaluation of urease activity in oral bacteria in vitro and in supragingival dental plaque ex vivo. BMC Oral. Health 18 (1), 89. doi: 10.1186/s12903-018-0541-3
Drevinek, P., Hollweck, R., Lorenz, M. G., Lustig, M., Bjarnsholt, T. (2023). Direct 16S/18S rRNA gene PCR followed by Sanger sequencing as a clinical diagnostic tool for detection of bacterial and fungal infections: a systematic review and meta-analysis. J. Clin. Microbiol., e0033823. doi: 10.1128/jcm.00338-23
Ebell, M. H. (2023). H. pylori Eradication: Effective for Cure or Improvement of Functional Dyspepsia, Especially if Eradication Is Confirmed. Am. Fam Physician 107 (6), Online.
Eisdorfer, I., Shalev, V., Goren, S., Chodick, G., Muhsen, K. (2018). Sex differences in urea breath test results for the diagnosis of Helicobacter pylori infection: a large cross-sectional study. Biol. Sex Differ 9 (1), 1. doi: 10.1186/s13293-017-0161-7
Hu, B., Zhao, F., Wang, S., Olszewski, M. A., Bian, H., Wu, Y., et al. (2016). A high-throughput multiplex genetic detection system for Helicobacter pylori identification, virulence and resistance analysis. Future Microbiol. 11, 1261–1278. doi: 10.2217/fmb-2016-0023
Idowu, S., Bertrand, P. P., Walduck, A. K. (2022). Gastric organoids: Advancing the study of H. pylori pathogenesis and inflammation. Helicobacter 27 (3), e12891. doi: 10.1111/hel.12891
Kesharwani, A., Dighe, O. R., Lamture, Y. (2023). Role of helicobacter pylori in gastric carcinoma: A review. Cureus 15 (4), e37205. doi: 10.7759/cureus.37205
Kismat, S., Tanni, N. N., Akhtar, R., Roy, C. K., Rahman, M. M., Molla, M. M. A., et al. (2022). Diagnosis and comparison of three invasive detection methods for helicobacter pylori infection. Microbiol. Insights 15, 11786361221133947. doi: 10.1177/11786361221133947
Kocsmár, É., Lotz, G. (2022). Comment on skrebinska et al. who could be blamed in the case of discrepant histology and serology results for helicobacter pylori detection? Diagnostics. Diagnostics (Basel) 1212 (6), 133. doi: 10.3390/diagnostics12061424
Kotilea, K., Iliadis, E., Nguyen, J., Salame, A., Mahler, T., Miendje Deyi, V. Y., et al. (2023). Antibiotic resistance, heteroresistance, and eradication success of Helicobacter pylori infection in children. Helicobacter 28 (5), e13006. doi: 10.1111/hel.13006
Liu, Q., Li, X., Zhang, Y., Song, Z., Li, R., Ruan, H., et al. (2019). Orally-administered outer-membrane vesicles from Helicobacter pylori reduce H. pylori infection via Th2-biased immune responses in mice. Pathog. Dis. 77 (5), ftz050. doi: 10.1093/femspd/ftz050
Mao, X., Jakubovics, N. S., Bächle, M., Buchalla, W., Hiller, K. A., Maisch, T., et al. (2021). Colonization of Helicobacter pylori in the oral cavity - an endless controversy? Crit. Rev. Microbiol. 47 (5), 612–629. doi: 10.1080/1040841x.2021.1907740
McNulty, C. A. M. (2023). The first 5 years of Helicobacter pylori research-With an emphasis on the United Kingdom. Helicobacter 28 (4), e12982. doi: 10.1111/hel.12982
Momtaz, H., Souod, N., Dabiri, H. (2010). Comparison of the virulence factors of Helicobacter pylori isolated in stomach and saliva in Iran. Am. J. Med. Sci. 340 (5), 345–349. doi: 10.1097/MAJ.0b013e3181d94fbc
Momtaz, H., Souod, N., Dabiri, H., Sarshar, M. (2012). Study of Helicobacter pylori genotype status in saliva, dental plaques, stool and gastric biopsy samples. World J. Gastroenterol. 18 (17), 2105–2111. doi: 10.3748/wjg.v18.i17.2105
Nagata, R., Sato, H., Takenaka, S., Yokoyama, J., Terai, S., Mimuro, H., et al. (2023). Analysis of genetic relatedness between gastric and oral helicobacter pylori in patients with early gastric cancer using multilocus sequence typing. Int. J. Mol. Sci. 24 (3), 2211. doi: 10.3390/ijms24032211
Sepúlveda, E., Moreno, J., Spencer, M. L., Quilodrán, S., Brethauer, U., Briceño, C., et al. (2012). [Comparison of Helicobacter pylori in oral cavity and gastric mucosa according to virulence genotype (cagA and vacA m 1)]. Rev. Chil. Infectol 29 (3), 278–283. doi: 10.4067/s0716-10182012000300005
Siavoshi, F., Salmanian, A. H., Akbari, F., Malekzadeh, R., Massarrat, S. (2005). Detection of Helicobacter pylori-specific genes in the oral yeast. Helicobacter 10 (4), 318–322. doi: 10.1111/j.1523-5378.2005.00319.x
Siavoshi, F., Saniee, P., Khalili-Samani, S., Hosseini, F., Malakutikhah, F., Mamivand, M., et al. (2015). Evaluation of methods for H. pylori detection in PPI consumption using culture, rapid urease test and smear examination. Ann. Transl. Med. 3 (1), 11. doi: 10.3978/j.issn.2305-5839.2014.11.16
Sohrabi, A., Franzen, J., Tertipis, N., Zagai, U., Li, W., Zheng, Z., et al. (2021). Efficacy of loop-mediated isothermal amplification for H. pylori detection as point-of-care testing by noninvasive sampling. Diagnostics (Basel) 11 (9), 1538. doi: 10.3390/diagnostics11091538
Sun, Z., Liu, W., Zhang, J., Wang, S., Yang, F., Fang, Y., et al. (2021). The direct semi-quantitative detection of 18 pathogens and simultaneous screening for nine resistance genes in clinical urine samples by a high-throughput multiplex genetic detection system. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.660461
Wang, J., Chi, D. S., Laffan, J. J., Li, C., Ferguson, D. A., Jr., Litchfield, P., et al. (2002). Comparison of cytotoxin genotypes of Helicobacter pylori in stomach and saliva. Dig Dis. Sci. 47 (8), 1850–1856. doi: 10.1023/a:1016417200611
Wang, Y. H., Wang, F. F., Gong, X. L., Yan, L. L., Zhao, Q. Y., Song, Y. P., et al. (2020). Genotype profiles of Helicobacter pylori from gastric biopsies and strains with antimicrobial-induced resistance. Therap Adv. Gastroenterol. 13, 1756284820952596. doi: 10.1177/1756284820952596
Wang, X. M., Yee, K. C., Hazeki-Taylor, N., Li, J., Fu, H. Y., Huang, M. L., et al. (2014). Oral Helicobacter pylori, its relationship to successful eradication of gastric H. pylori and saliva culture confirmation. J. Physiol. Pharmacol. 65 (4), 559–566.
Xu, Y., Song, Y., Wang, X., Gao, X., Li, S., Yee, J. K. (2018). A clinical trial on oral H. pylori infection of preschool children. Ann. Clin. Lab. Sci. 48 (6), 751–756.
Xue, Z., Yang, H., Su, D., Song, X., Deng, X., Yu, C., et al. (2021). Geographic distribution of the cagA, vacA, iceA, oipA and dupA genes of Helicobacter pylori strains isolated in China. Gut Pathog. 13 (1), 39. doi: 10.1186/s13099-021-00434-4
Zang, H., Wang, J., Wang, H., Guo, J., Li, Y., Zhao, Y., et al. (2023). Metabolic alterations in patients with Helicobacter pylori-related gastritis: The H. pylori-gut microbiota-metabolism axis in progression of the chronic inflammation in the gastric mucosa. Helicobacter 28 (4), e12984. doi: 10.1111/hel.12984
Zhang, Y., Wang, S., Hu, B., Zhao, F., Xiang, P., Ji, D., et al. (2016a). Direct detection of Helicobacter pylori in biopsy specimens using a high-throughput multiple genetic detection system. Future Microbiol. 11, 1521–1534. doi: 10.2217/fmb-2016-0149
Zhang, Y., Zhao, F., Kong, M., Wang, S., Nan, L., Hu, B., et al. (2016b). Validation of a high-throughput multiplex genetic detection system for helicobacter pylori identification, quantification, virulence, and resistance analysis. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01401
Keywords: Helicobacter pylori, high-throughput multiplex genetic detection assay (HMGA), oral samples, performance evaluation, semi-quantification, virulence genes
Citation: Chi W, Wang S, Liu T, Jiang W, Ding L, Miao Y, Yang F, Zhang J, Ji D, Xiao Z, Zhu H, Wu Y, Bao Z, Zhao H and Wang S (2023) A rapid and high-throughput multiplex genetic detection assay for detection, semi-quantification and virulence genotyping of Helicobacter pylori in non-invasive oral samples. Front. Cell. Infect. Microbiol. 13:1267288. doi: 10.3389/fcimb.2023.1267288
Received: 26 July 2023; Accepted: 11 September 2023;
Published: 29 September 2023.
Edited by:
Marie Hallin, Université libre de Bruxelles, BelgiumReviewed by:
Eng Guan Chua, University of Western Australia, AustraliaMarkus Gerhard, Technical University of Munich, Germany
Copyright © 2023 Chi, Wang, Liu, Jiang, Ding, Miao, Yang, Zhang, Ji, Xiao, Zhu, Wu, Bao, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijun Bao, emhpanVuYmFvQGZ1ZGFuLmVkdS5jbg==; Hu Zhao, aHViZXJ0emhhb0AxNjMuY29t; Shiwen Wang, d2FuZ3NoaXdlbjE5ODZAYWxpeXVuLmNvbQ==
†These authors have contributed equally to this work
 Wenjing Chi
Wenjing Chi Su Wang
Su Wang Tao Liu1,2,3
Tao Liu1,2,3 Li Ding
Li Ding Yingxin Miao
Yingxin Miao Feng Yang
Feng Yang Jinghao Zhang
Jinghao Zhang Zili Xiao
Zili Xiao Haowei Zhu
Haowei Zhu Zhijun Bao
Zhijun Bao Hu Zhao
Hu Zhao Shiwen Wang
Shiwen Wang