
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 29 September 2023
Sec. Biofilms
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1266674
Introduction: Enterococcus faecium is a leading cause of hospital-acquired infections, which has become a serious public health concern. The increasing incidence of vancomycin-resistant E. faecium (VRE-fm) raises an urgent need to find new antimicrobial agents as a complement to traditional antibiotics. The study aimed to evaluate the antimicrobial and antibiofilm activity of essential fatty acids (EFAs) against VRE-fm, and further explore the molecular mechanism of the antibiofilm activity of EFAs.
Method: The microdilution broth method was used for antimicrobial susceptibility testing with traditional antibiotics and EFAs, including α-linolenic acid (ALA), eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA), linoleic acid (LOA), γ-linolenic acid (GLA), and arachidonic acid (AA). The effect of EFAs on cell morphology of VRE-fm was investigated by scanning electron microscopy. The crystal violet method was used to evaluate the antibiofilm activities of EFAs against VRE-fm. Furthermore, the expression of biofilm-related genes (acm, atlA, esp, and sagA) of VRE-fm isolates under the action of GLA was analyzed using quantitative reverse transcription PCR (qRT-PCR) assay.
Results: VRE-fm isolates were highly resistant to most traditional antibiotics, only highly susceptible to quinupristin-dalfopristin (90.0%), tigecycline (100%), and linezolid (100%). EPA, DHA, and GLA exhibited excellent antimicrobial activity. The MIC50/90 of EPA, DHA, and GLA were 0.5/1, 0.25/0.5, and 0.5/1 mM, respectively. SEM imaging showed that strain V27 adsorbed a large number of DHA molecules. Furthermore, all EFAs exhibited excellent inhibition and eradication activities against VRE-fm biofilms. The biofilm inhibition rates of EFAs ranged from 45.3% to 58.0%, and eradication rates ranged from 54.1% to 63.4%, against 6 VRE-fm isolates with moderate biofilm formation ability. GLA exhibited remarkable antibiofilm activity against VRE-fm isolates. The qRT-PCR analysis showed that GLA could significantly down-regulate the expression of the atlA gene (P < 0.01) of VRE-fm.
Conclusion: DHA showed the strongest antibacterial activity, while GLA showed the strongest antibiofilm effect among the EFAs with antibacterial activity. Our novel findings indicate that the antibiofilm activity of GLA may be through down-regulating the atlA gene expression in VRE-fm. Therefore, DHA and GLA had the potential to be developed as therapeutic agents to treat infections related to multiple antimicrobial-resistant E. faecium.
Enterococcus faecium (E. faecium) is a Gram-positive member of the gut microbiota in humans. Commensal intestinal E. faecium is not pathogenic in healthy hosts but can cause infection in susceptible ones (Ch’ng et al., 2019). E. faecium is a leading cause of hospital-acquired infections that are often difficult to treat due to multiple antimicrobial resistances (Santajit and Indrawattana, 2016; Panda et al., 2022). In recent years, new variants of E. faecium have emerged that are better adapted to the healthcare environment, which are prone to outbreaks of infection (O’Toole et al., 2023). Infections caused by vancomycin-resistant E. faecium (VRE-fm) have further reduced treatment options due to increased resistance to a wide range of antibiotics (Lagatolla et al., 2022; Cairns et al., 2023). In addition, the ability of E. faecium to form biofilms is its remarkable pathogenicity characteristic, and the biofilm greatly enhances antibiotic resistance compared to planktonic cells (Ch’ng et al., 2019; Patil et al., 2021). Therefore, the infection caused by VRE-fm is a serious challenge worldwide, and it is urgent to develop new antimicrobial agents to combat VRE-fm, as well as to inhibit and eradicate the biofilm formed by VRE-fm.
Unsaturated fatty acids are well known to display antimicrobial activity (Desbois, 2012; Yoon et al., 2018). However, due to the discovery and development of traditional antibiotics, research on the antimicrobial properties of unsaturated fatty acids has been neglected for a long time. With the increasing severity of traditional antibiotic resistance, unsaturated fatty acids have been reconsidered as antimicrobial agents. Recently, numerous studies indicated that unsaturated fatty acids could also inhibit or eradicate biofilms formed by various microbial pathogens (Kim et al., 2018; Ramanathan et al., 2018; Cui et al., 2019; Wang et al., 2022). Consequently, unsaturated fatty acids could represent the next generation of antimicrobial agents to treat and prevent biofilm-associated infections.
Essential fatty acids (EFAs) that the body needs but cannot make and must get from food, belong to unsaturated fatty acids and play an important role in the body. EFAs are based on α-linolenic acid (ω-3 group) and linoleic acid (ω-6 group), and correlated with infant development, reduction of cardiovascular morbidity and mortality, cancer prevention, brain and vision functioning, etc. (Kaur et al., 2014) EFAs are not only nontoxic, but also essential nutrients for the human body. Therefore, it is a good model to carry out research on antimicrobial and antibiofilm using EFAs as representative of novel antimicrobial agents.
In the context, three ω-3 group including α-linolenic acid (ALA, 18:3), eicosapentaenoic acid (EPA, 20:5), and docosahexaenoic acid (DHA, 22:6), and three ω-6 group members including linoleic acid (LOA, 18:2), γ-linolenic acid (GLA; 18:3) and arachidonic acid (AA, 20:4) were used to evaluate the antimicrobial and antibiofilm effects against VRE-fm in vitro. In addition, for the first time, we study the antibiofilm mechanism of GLA against VRE-fm. Our study indicates that GLA may be developed as a novel therapeutic agent to treat VRE-fm and biofilm-related infections.
The antibiotic resistance profile of 20 isolates of VRE-fm is shown in Table 1 and summarized in Figure 1. The VRE-fm isolates in the study were highly resistant to penicillin (100%), ampicillin (100%), ciprofloxacin (100%), levofloxacin (100%), nitrofurantoin (66.7%), erythromycin (63.6%), and tetracycline (60.0%), and were moderately resistant to teicoplanin (35.0%), high-level streptomycin (30.0%), and high-level gentamicin (30.0%), while they were only highly susceptible to quinupristin-dalfopristin (90.0%), tigecycline (100%), and linezolid (100%).
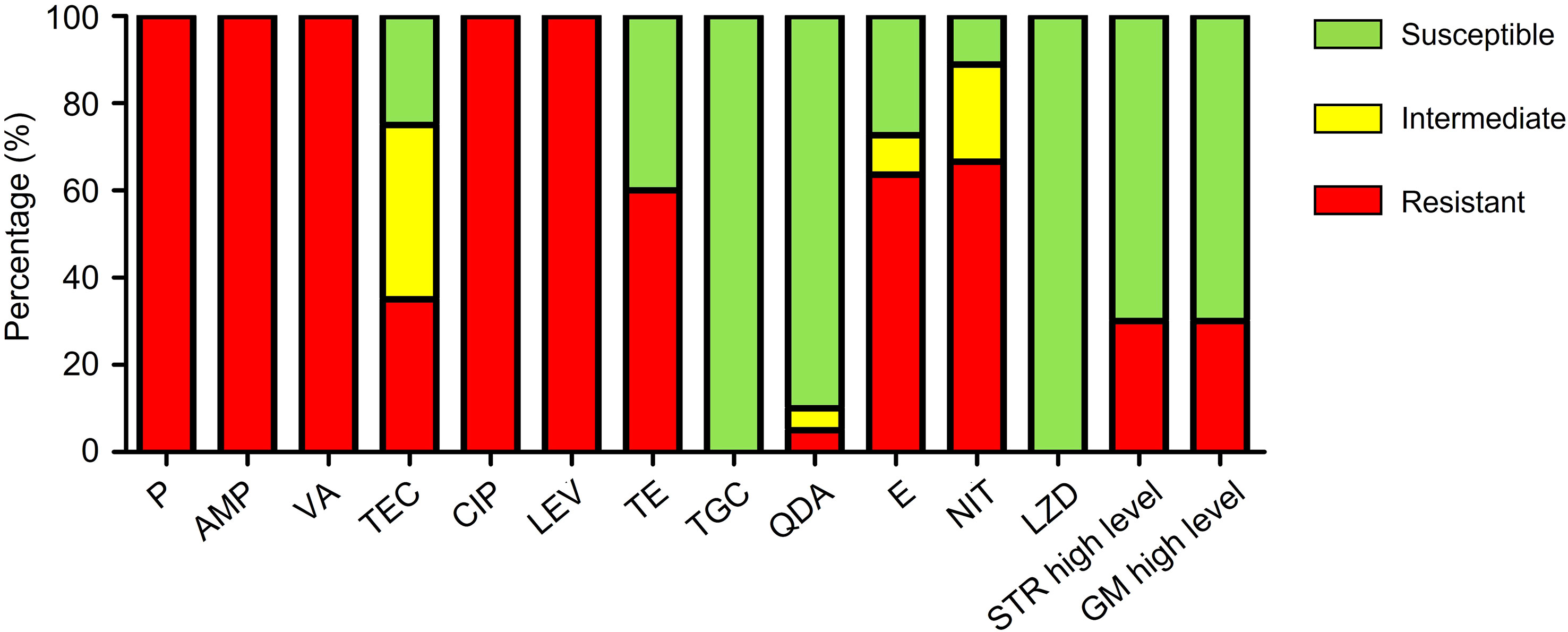
Figure 1 Antibiotic resistance patterns of VRE-fm isolates (n = 20) to 14 antibiotics. P, penicillin; AMP, ampicillin; VA, vancomycin; TEC, teicoplanin; CIP, ciprofloxacin; LEV, levofloxacin; TE, tetracycline; TGC, tigecycline; QDA, quinupristin-dalfopristin; E, erythromycin; NIT, nitrofurantoin; LZD, linezolid; STR, streptomycin; GM, gentamicin.
The MIC values of different EFAs against VRE-fm isolates in the planktonic form are listed in Table 2. Within the set range of EFA concentrations, the results showed that LOA and AA did not have antibacterial activity. By comparing MIC50 and MIC90, it was found that DHA exhibited the best antimicrobial activity, while EPA and GLA showed slightly lower than DHA, and ALA had a weak inhibitory effect on VRE-fm isolates.
Compared to control group, a large number of DHA molecules were adsorbed to the cell wall of strain V27 in the experimental group, and the surface became rough (Figure 2). When observing a large number of fields, bacterial cells with significantly damaged cell membranes were not observed.
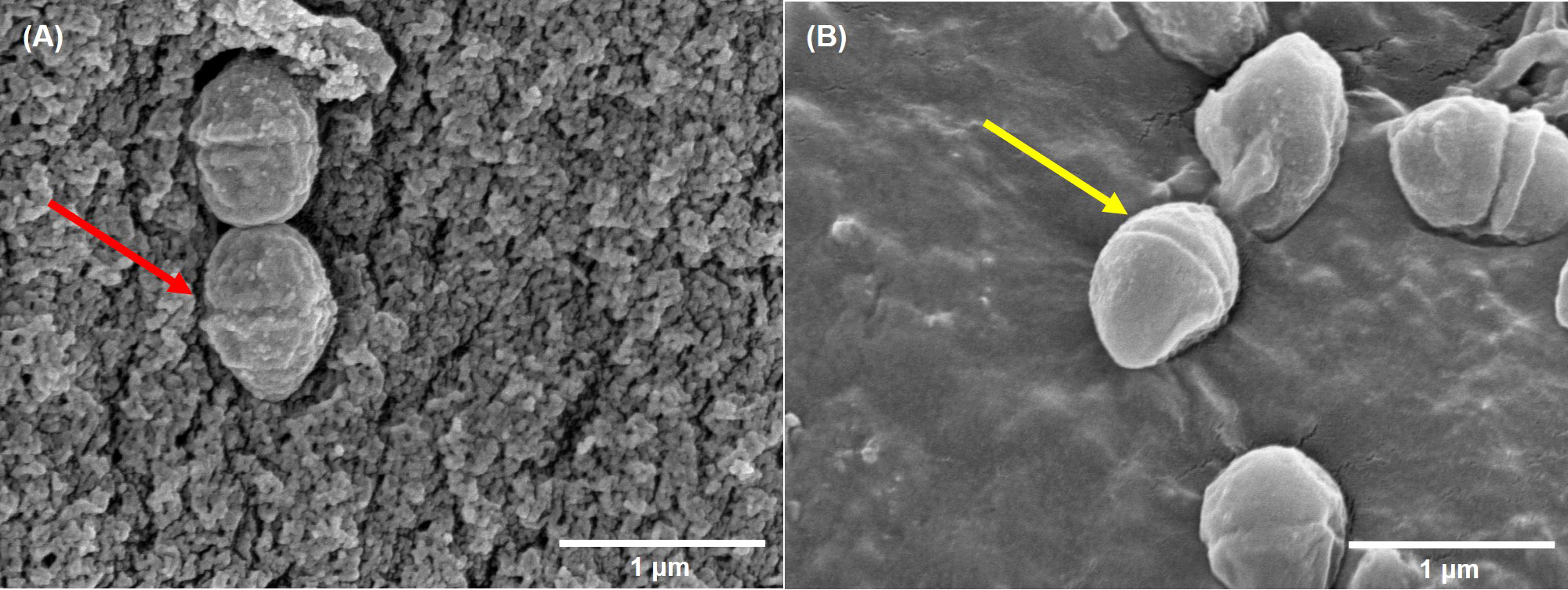
Figure 2 The scanning electron microscope imaging. (A) incubated with DHA; (B) incubated without DHA. The red arrow indicates rough cells; The yellow arrow indicates smooth cells.
To investigate the antibiofilm effect of EFAs, we explored the optimal condition for biofilm formation in vitro. Different concentrations of glucose were added to trypticase-soy broth (TSB) to test the biofilm formation of VRE-fm isolates. The results are shown in Figure 3. Although there was no statistically significant difference in biofilm biomass when different concentrations of glucose were added, the mean value of biofilm biomass in the TSB+2% glucose group was the highest. Therefore, TSB containing 2% glucose (TSBG) medium was chosen as the final condition in the study.
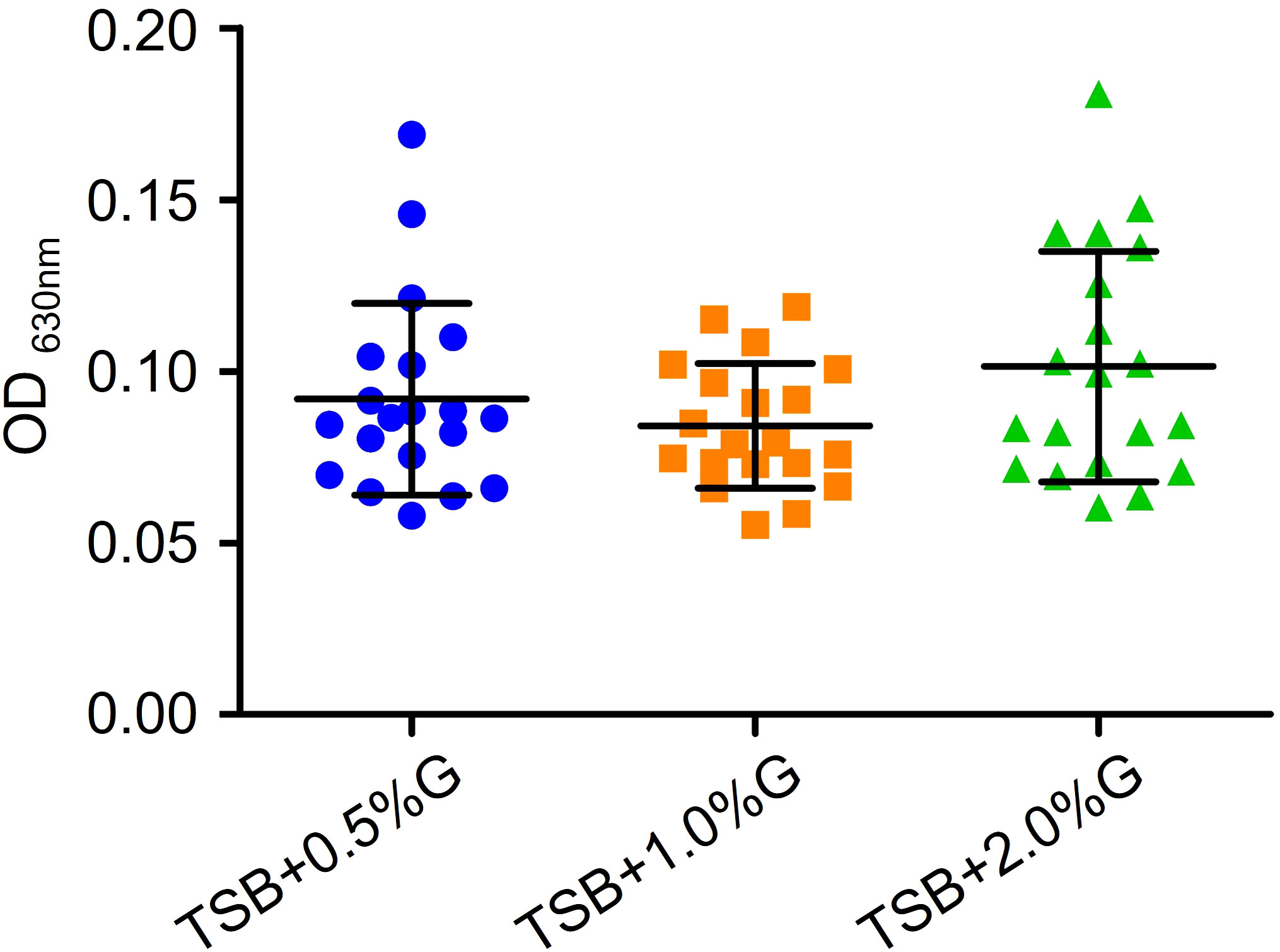
Figure 3 Comparison of the effects of different concentrations of glucose in TSB broth on the biofilm formation of VRE-fm isolates. G, glucose.
In the TSBG medium, 6 out of 20 isolates had moderate biofilm formation ability, while 14 isolates had weak ability (Figure 4A). There were no statistically significant differences in biofilm formation capacity between VRE-fm isolates from drain fluid and urine sources (Figure 4B).
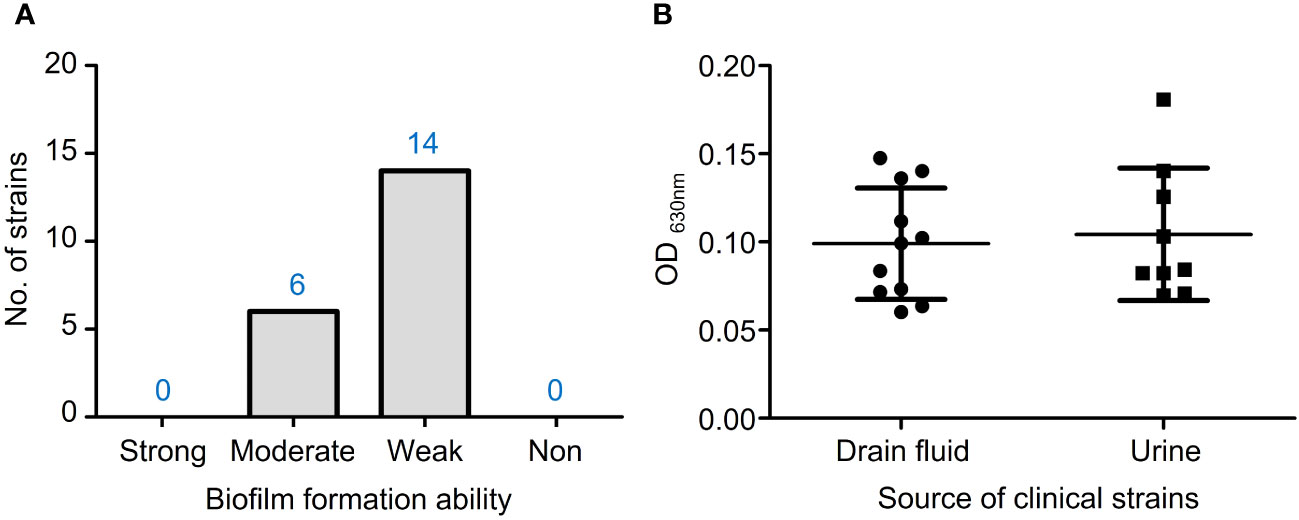
Figure 4 (A) The biofilm formation ability of VRE-fm isolates in TSB+2% glucose; (B) Comparison of the biofilm biomass of VRE-fm isolates from different clinical sources.
To study the antibiofilm activity of these EFAs, 6 isolates of VRE-fm (V05, V06, V08, V09, V22, and V27) with moderate biofilm formation ability were selected for the experiments. The antibiofilm activities of the EFAs against the VRE-fm isolates were different from those of the antimicrobial activities. All EFAs at 1 mM exhibited excellent activity in inhibiting and eradicating biofilms (Figure 5). The biofilm inhibition rates for EPA, DHA, ALA, LOA, AA, and GLA against these VRE-fm isolates were 45.3%, 48.9%, 50.0%, 52.4%, 57.8%, and 58.0%, respectively (Figure 5A). While the biofilm eradication rates for EPA, DHA, ALA, LOA, GLA, and AA were 54.1%, 54.1%, 56.6%, 60.8%, 63.2%, and 63.4%, respectively (Figure 5B). GLA showed the best antibiofilm activity against VRE-fm among the EFAs with antibacterial activity.
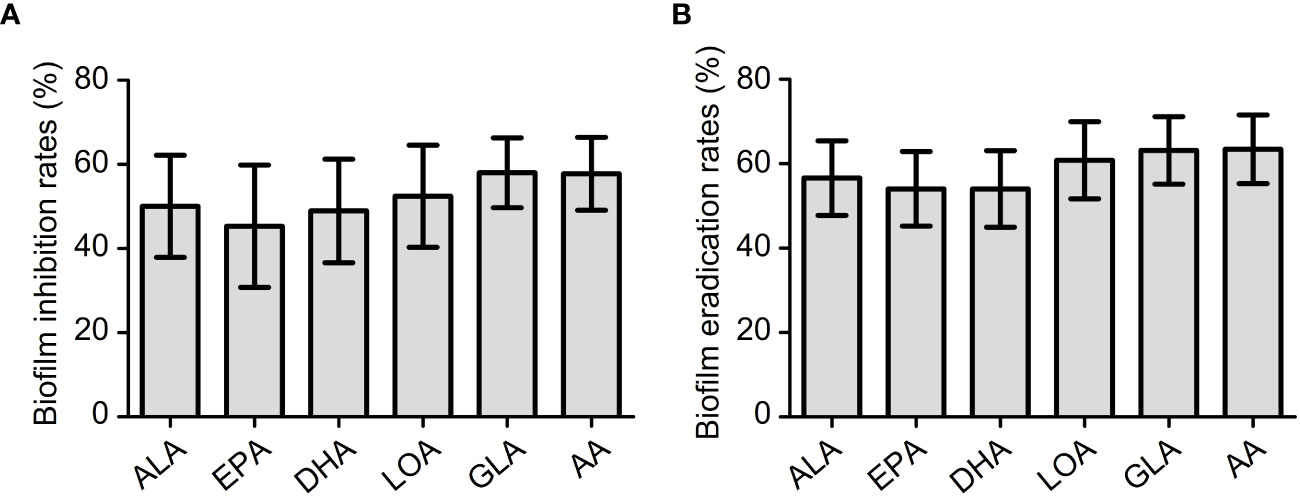
Figure 5 (A) The biofilm inhibition rates (%) of EFAs against VRE-fm isolates; (B) The biofilm eradication rates (%) of EFAs against VRE-fm isolates. ALA, α-linolenic acid; EPA, eicosapentaenoic acid; DHA, docosahexaenoic acid; LOA, linoleic acid; GLA, γ-linolenic acid; AA, arachidonic acid.
In all VRE-fm isolates, the genes acm, esp, and sagA were detected in 20/20 (100%). Only one isolate with low biofilm forming capacity did not detect the atlA gene (95.0%, 19/20). However, agg and gelE genes were not detected in any isolates (Table 3).
To illustrate the antibiofilm mechanism of GLA, the expression levels of the biofilm-related gene (acm, atlA, esp, and sagA) in six VRE-fm isolates were analyzed. Compared to the control group, GLA down-regulated the expression of the VRE-fm atlA gene (0.232-fold) and the difference was statistically significant (P < 0.01), while the gene expression levels of acm (1.013-fold), esp (0.373-fold), and sagA (0.931-fold) compared to the control had no statistical difference (P > 0.05) (Figure 6).
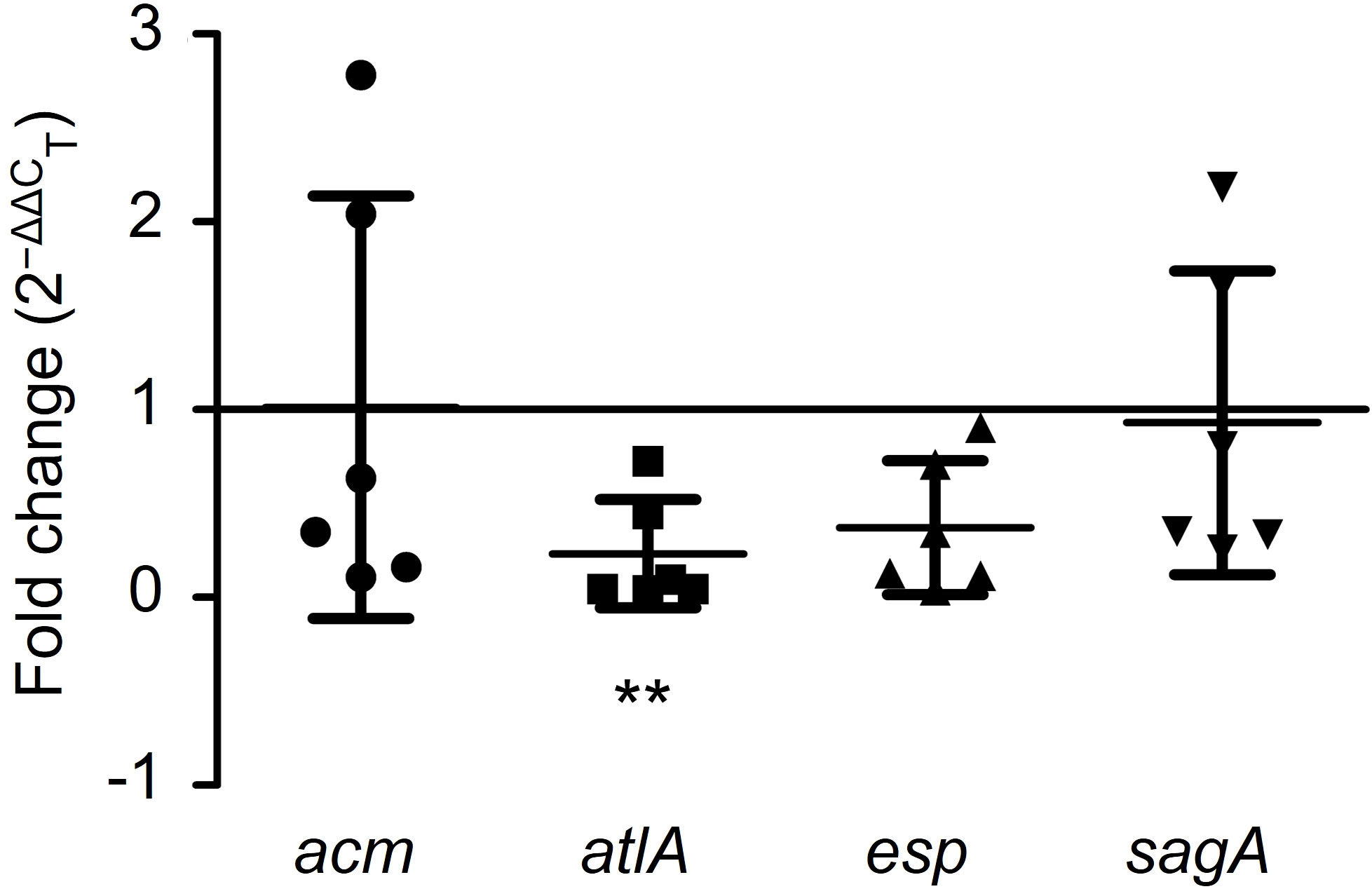
Figure 6 The expression levels (fold change) of biofilm-related genes in mature biofilms of the six VRE-fm isolates treated with γ-linolenic acid (GLA). **P < 0.01 compared to control by Dunnett’s t test.
Figure 7 further demonstrated the regulatory effects of GLA on the expression levels of biofilm-related genes in different VRE-fm isolates. Among six tested VRE-fm isolates, the expression levels of the acm, atlA, esp, and sagA genes were down-regulated in four isolates (V05, V08, V22 and V27). Compared to the control group, there was a significant difference in the GLA-treated groups of V05, V22 and V27 but there was no significant difference for V08. Meanwhile, compared to other isolates, the expression levels of acm and sagA genes were different in the treatment groups of V06 and V09 isolates, which were up-regulated in GLA-treated groups.
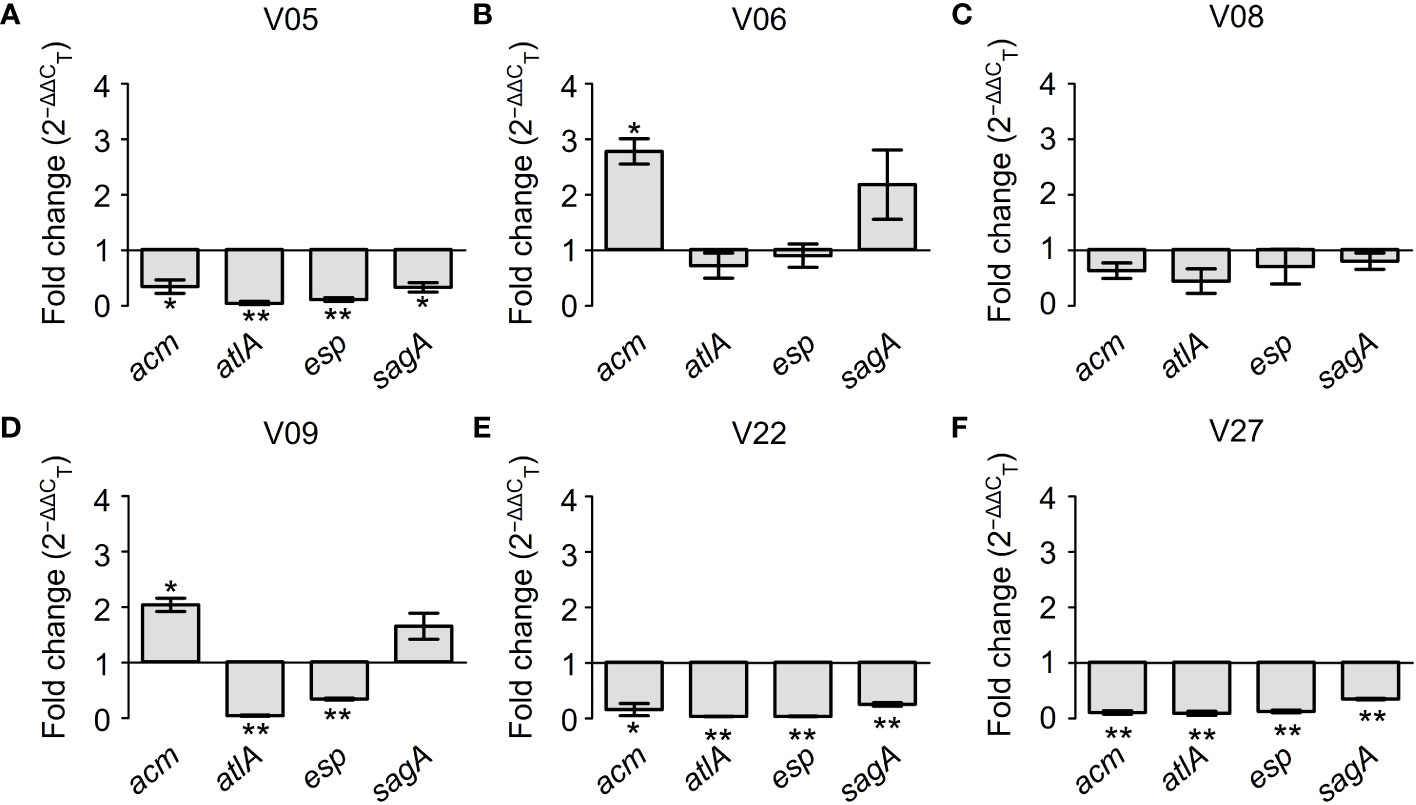
Figure 7 The expression levels (fold change) of biofilm-related genes in mature biofilms of each VRE-fm isolate treated with γ-linolenic acid (GLA). *P < 0.05 and **P < 0.01 compared to the control by the Dunnett’s t test. (A) V05; (B) V06; (C) V08; (D) V09; (E) V22; (F) V27.
The emergence of VRE-fm poses a significant threat to public health. Additionally, the ability of E. faecium to form biofilms further enhances its resilience and makes it challenging to eradicate infections caused by this pathogen. In recent years, there has been growing interest in the potential antimicrobial and antibiofilm effects of EFAs against clinically isolated VRE-fm. In this study, we not only evaluated the antimicrobial and antibiofilm abilities of EFAs against VRE-fm but also first analyzed the possible molecular mechanisms of GLA eradication of VRE-fm biofilms.
In this study, the VRE-fm isolates were highly resistant to most traditional antibiotics, only highly susceptible to quinupristin-dalfopristin (90.0%), tigecycline (100%), and linezolid (100%). Therefore, infections caused by VRE-fm are difficult to treat due to the limited choice of antibiotics. The intrinsic resistance to β-lactams (penicillin and ampicillin) of VRE-fm strains is due to the high expression of penicillin-binding proteins that have low affinity to β-lactams, allowing peptidoglycan synthesis and bacterial growth (Montealegre et al., 2017), which makes enterococci about 100 times more resistant than streptococci (Murray, 1997). resistance to fluoroquinolones (ciprofloxacin and levofloxacin) is mediated by alteration of target enzymes, including DNA gyrase and topoisomerase IV, as well as by decreasing intracellular accumulation (Piddock, 1999). Resistance to gentamicin is due to the low permeability of the E. faecium cell wall to the large gentamicin molecules, but resistance to high-level gentamicin is conferred by alteration of the binding site in the 30S ribosomal subunit or production of modifying enzymes that lead to inactivation of gentamicin (Osuka et al., 2016). Although both vancomycin and teicoplanin belong to glycopeptide antibiotics, teicoplanin still maintains activity against some VRE-fm isolates in the study. It may be caused by the carrying vanB gene, making the isolate lower level resistant to vancomycin and susceptible to teicoplanin (Kuzma et al., 2022).
Because E. faecium is becoming increasingly resistant to traditional antibiotics, it is urgent to find new antimicrobial agents as a supplement to traditional antibiotics. One promising agent with antimicrobial activity is unsaturated fatty acids for the prevention and treatment of microbial infections (Chanda et al., 2018). Several studies have shown that unsaturated fatty acids can fight against a wide range of bacteria species (Sun et al., 2016; Churchward et al., 2017; Pushparaj Selvadoss et al., 2018). In the study, six EFAs are used to test antimicrobial activity against VRE-fm. In terms of the chemical structure of EFAs (for example, the number of double bonds, the relative position of the double bonds and the carboxyl group) which was shown in our previous work (Wang et al., 2022), it is difficult to find a pattern to explain the different antibacterial activities displayed by them. The antibacterial effect of EFAs may depend mainly on the interaction between EFAs and the cell membrane of VRE-fm strains, so it is not possible to find a pattern solely from the chemical structure of EFAs. The cell wall of strain V27 adsorbed a large number of DHA molecules was observed by SEM. It suggests that DHA may play an inhibitory role in interfering with the growth by preventing the nutrients transportation into the cell, which is also discovered by Dasa et al. (Dasa et al., 2016) No bacteria with significantly damaged cell membranes were observed, which indicates that direct destruction of the cell membrane of VRE-fm is not the primary antibacterial mechanism of DHA. This result is different from that observed by Sun et al. (Sun et al., 2016) Besides, Cartron et al. found that cis-6-hexadecanoic acid could disrupt the proton motive force and the electron transfer pathways of Staphylococcus aureus (Cartron et al., 2014), while Won et al. reported that oleic acid could inhibit the membrane enzymes of Streptococcus mutans (Won et al., 2007), which may be also involved in the antibacterial effects of DHA in the current study.
Biofilms play a crucial role in the pathogenesis of E. faecium infections, as they protect bacteria from the host’s immune response and improve their resistance to antibiotics. According to the reports by Maurya et al. (Maurya et al., 2021), the maximum biofilm formation of E. faecium was achieved in the TSB medium, followed by brain heart infusion broth and Luria Bertani broth. Thus, TSB was selected as the basis in the current study. Since glucose has a significant impact on E. faecalis biofilm formation (Tendolkar et al., 2004), different concentrations of glucose were added to TSB to test the biofilm formation of VRE-fm isolates. Finally, TSB+2% glucose medium was selected for the subsequent biofilm study. It is well known that biofilm cells are generally more resistant to antibiotics than their planktonic counterparts, due to the thick mature biofilm, slower metabolic rate, and higher rates of horizontal gene transfer of antibiotic resistance genes within the biofilm community (Komiyama et al., 2016; Lim et al., 2017). Therefore, biofilm-associated E. faecium infections are often difficult to eradicate (Ch’ng et al., 2019). Multidrug resistance and biofilm make the treatment of VRE-fm strain infection even more challenging. Numerous studies indicate that unsaturated fatty acids have the activity of inhibiting or eradicating of biofilms formed by a wide range of bacteria, including even fungi (Kim et al., 2018; Ramanathan et al., 2018; Cui et al., 2019; Wang et al., 2022). These findings are similar to those in the current study. It suggests that EFAs may play a role in the clinical treatment of infections in the future.
The initial attachment is the first step in the formation of a biofilm. Various surface adhesins are involved in this stage. The absence of surface adhesins, including enterococcal surface protein (Esp), aggregation substance (Agg), and adhesion to collagen of E. faecium (Acm), will reduce adherence to human cells and weaken biofilm formation (Nallapareddy et al., 2007; Soares et al., 2014). Hydrolases, including secreted antigen A (SagA), autolysin (AtlA), and gelatinase (GelE), also play a crucial role in biofilm formation. SagA, which is secreted during biofilm formation, is an important component of the biofilm matrix. It binds to a broad spectrum of extracellular matrix proteins. When SagA is hydrolyzed by proteinase K, the thickness of the biofilm will decrease (Paganelli et al., 2015). autolysin (AtlA) plays a role in the stage of biofilm growth and maturation and is mainly responsible for facilitating the release of eDNA by hydrolyzing E. faecium cells (Xie et al., 2022). GelE mainly hydrolyzes gelatin, collagen, casein, and hemoglobin, which assists AtlA in the release of eDNA (Dunny et al., 2014; Kiruthiga et al., 2020). In the study, the esp, sagA, and acm genes were detected in 20/20 (100%), while the atlA gene was detected in 19/20 (95.0%) VRE-fm isolates. Only one isolate with low biofilm-forming ability did not detect the atlA gene. The results suggest that the atlA gene alone cannot determine whether an isolate can form a biofilm but may affect the ability of biofilm formation. However, agg and gelE genes were not detected in any isolates. This may be because the two genes are present mainly in E. faecalis (Kiruthiga et al., 2020), they are not crucial genes for E. faecium to form a biofilm.
In terms of eradicating biofilms, in addition to directly killing VRE-fm, further study is needed on how EFAs affect biofilm-related genes which include acm, atlA, esp, and sagA. GLA with the highest biofilm eradication rate, and six VRE-fm isolates (V05, V06, V08, V09, V22, and V27) with moderate biofilm formation ability were used to illustrate the mechanism. In this study, GLA could significantly down-regulate the expression of the atlA gene in VRE-fm strains, which inhibit the release of eDNA, may result in the eradication of biofilm. The finding suggests that GLA may offer a potential alternative to traditional antibiotics to combat E. faecium infections. The acm gene was up-regulated in two isolates under the action of GLA, which suggests that these isolates may resist EFAs eradicating or repairing biofilm through high expression of the acm gene. These inferences require further work to confirm.
In summary, DHA and GLA showed excellent antibacterial and antibiofilm activity against VRE-fm isolates in the current study, and they are promising next generation antimicrobial agents. Through analysis of relative expression levels of biofilm-related genes, we found that the expression level of atlA gene in VRE-fm strains was significantly down-regulated, which may be an important molecular mechanism for EFAs to eradicate biofilms.
A total of 20 clinical VRE-fm isolates (non-replicative) were collected from the Department of Infectious Diseases and Clinical Microbiology, Beijing Chao-Yang Hospital, Capital Medical University (Beijing, China). These strains were identified using matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) (VITEK-MS; bioMérieux, France; IVD version 3.0). The verified strains were routinely refreshed from frozen stocks at -20°C and inoculated at least twice on Columbia blood agar at 35°C for 24 h before all experiments.
The antimicrobial susceptibility of E. faecium to penicillin (P), ampicillin (AM), vancomycin (VA), tigecycline (TGC), ciprofloxacin (CIP), levofloxacin (LEV), tetracycline (TE), quinupristin-dalfopristin (QDA), erythromycin (E), nitrofurantoin (NIT), linezolid (LZD), high-level streptomycin (STR), and high-level gentamicin (GM), were tested by broth microdilution using the Vitek 2 compact system (bioMerieux, Marcy l’Etoile, France). The minimum inhibitory concentration (MIC) of teicoplanin (TEC) was determined by Etest (bioMerieux, Marcy l’Etoile, France). The interpretation of sensitive (S), intermediate sensitive (I), and resistant (R) was carried out according to the guideline of Clinical and Laboratory Standards Institute (CLSI, 2023). Because the CLSI guideline does not offer recommended MIC susceptibility breakpoints for TGC against E. faecium, the MICs of TGC were analyzed using the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2023). E. faecalis ATCC 29212 was used as a quality control strain for the antimicrobial susceptibility testing.
The antimicrobial activity of EFAs (ALA, EPA, DHA, LOA, GLA, and AA) was evaluated using a microdilution method as previously reported (Wang et al., 2022). Briefly, VRE-fm isolates (1.0-2.0 × 108 CFU/mL) inoculated 1:200 into each well of 96-well round bottom microtiter plates with 200 μL of Mueller Hinton II broth containing different concentrations of EFAs. The EFAs were two-fold diluted from 0.125 mM to 1 mM. The MIC values were read and recorded as the lowest concentration of compounds that inhibited visible growth of bacteria after a 24 h incubation period at 35°C. All experiments were performed at least three times.
To investigate the effect of EFAs on the morphology of VRE-fm cells, the strain V27 incubated with DHA was selected for SEM imaging. Strain V27 at a final concentration about 7.5×105 CFU/mL was incubated with and without 1 mM DHA for 1 h with shaking at 35°C. The samples were fixed in 2.5% glutaraldehyde for 5 h at 4°C, then were dehydrated in a sequential-graded ethanol (30%, 50%, 70%, 80%, 90%, and 100%), and then two times with 100% ethanol for 15 min. Finally, the samples were sputter coated with gold, followed by microscopic examinations by using a SU8020 SEM (Hitachi, Japan) (Rani et al., 2022).
For biofilm formation, the method described by Cruz et al. was followed with some modifications (Cruz et al., 2018). Briefly, VRE-fm cultures were diluted 1:100 in fresh trypticase-soy broth (TSB) containing 0.5%, 1% or 2% glucose. Then, 200 µL of suspension (1×106 CFU/mL) was added to each well of the microtiter plate and incubated at 35°C for 24 h. Wells with sterile medium were used as negative controls.
To determine the biofilm mass, the wells were washed three times with phosphate buffered saline (PBS; pH 7.2) to remove planktonic cells. The remaining attached cells were fixed with 200 μL methanol for 15 min, stained with 200 μL crystal violet (0.2%) for 20 min, and washed with distilled water to remove excess crystal violet. Then, the bound crystal violet was extracted with 200 μL of 33% acetic acid. The OD values were measured at 630 nm using a Multiskan EX microplate photometer (Thermo Fisher Scientific, Waltham, MA, United States). The experiments were independently repeated three times. The OD cut-off value (ODc) for biofilm formation was defined as three standard deviations above the mean OD value of the negative control.
The isolates were classified as strong biofilm producers (OD 630 nm > 4×ODc), moderate biofilm producers (2×ODc < OD 630 nm ≤ 4×ODc), weak biofilm producers (ODc < OD 630 nm ≤ 2×ODc), and non-biofilm producers (OD 630 nm ≤ ODc) (El-Houssaini et al., 2019).
For the biofilm inhibition assay, E. faecium isolates were inoculated into microtiter plates with TSB + 2% glucose (TSBG) containing 1 mM EFAs. TSBG medium without EFAs was used as control. After incubation of microplates at 35°C for 24 h, the biofilm formed was measured using the crystal violet method as described above. The biofilm inhibition rate (%) was calculated according to the formula: Inhibition (%) = (OD control - OD sample)/OD sample × 100% (Wang et al., 2021).
For the biofilm eradication assay, mature biofilms of E. faecium were pre-formed in TSBG medium culturing for 24 h. After incubation, the non-adherent cells were removed by washing with PBS twice. Then, TSBG containing 1 mM EFAs was added to the wells. The mature biofilms treated with fresh TSBG medium were used as a control. The microtiter plates were incubated at 35°C for an additional 24 h, and the biofilm mass was detected using the crystal violet method described above. The biofilm eradication rate (%) was calculated according to the formula: Eradication (%) = (OD control - OD sample)/OD sample × 100% (Wang et al., 2021).
To detect the presence of biofilm-associated genes from E. faecium isolates, a qPCR assay was used. Genomic DNA was extracted from strains inoculated on Columbia blood agar using a TIANamp Bacteria DNA kit (Tiangen Biotech (Beijing) Co., Ltd., Beijing, China) according to the manufacturer’s protocol. Specific primers were used for the PCR testing of all E. faecium isolates to detect biofilm-related genes encoding Acm, Agg, AtlA, Esp, GelE, and SagA. The primers used for the detection are shown in Table 4. The PCR procedure was 95°C for 3 min, 40 cycles (95°C for 15 s, and 60°C for 1 min).
To determine the effects of GLA on the transcription of E. faecium genes related to biofilms, the gene expression levels of acm, atlA, esp, and sagA were evaluated by a qRT-PCR method. Sequences of gene-specific primers and the reference gene encoding 16S rRNA (16S), which serves as an internal control for the determination of gene expression level, are presented in Table 4. Some primers used in this study were designed by Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast), according to selected sequences obtained from the GenBank database.
The mature biofilm of E. faecium was established according to the previous method. After the biofilm was treated with or without GLA for 24 h, the cells were collected by centrifugation (5000 × g for 10 min). Total RNA from E. faecium was extracted using the RNA extraction reagent kit (Tiangen Biotech, Beijing, China) according to the manufacturer′s instructions. The concentration, purity, and quality of the isolated RNA samples were determined using a Nano Drop One Spectrophotometer (Thermo Scientific, Waltham, MA, USA). RNA (1 μg) from each sample was immediately reverse transcribed into cDNA using SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen, Carlsbad, CA, USA). qRT-PCR was analyzed using the PowerUp SYBR Green Master Mix (Applied Biosystems, Life Technologies, Austin, USA) in an Applied Biosystems 7500 real-time PCR system (Applied Biosystems, Waltham, MA, USA). The amplification reactions were carried out in a 20 µL reaction volume involving 10 μL of 2× PowerUp SYBR Green Master Mix, 2 μL of cDNA, 1 μL of Forward Primer, 1 μL of Reverse Primer, and 6 μL of H2O. The reaction conditions were 95°C for 3 min, 40 cycles (95°C for 15 s, and 60°C for 1 min). Gene expression was normalized with the reference gene 16S using 2−ΔΔCT method (Livak and Schmittgen, 2001).
Statistical analysis was performed using the IBM SPSS Statistics 20.0 software program (IBM, Armonk, NY, USA). Data were expressed as means ± standard deviation (SD) of at least three independent experiments. Statistical comparison between two groups was performed using Student’s t-test. Multiple comparisons between the control and different treatment groups were analyzed using one-way analysis of variance (ANOVA) followed by Dunnett’s t test. A P-value < 0.05 was considered statistically significant.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
The studies involving humans were approved by Ethics Committee of Beijing Chao-Yang Hospital, Capital Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
MW: Conceptualization, Methodology, Project administration, Writing – original draft. PW: Methodology, Writing – original draft. TL: Investigation, Writing – original draft. QW: Validation, Writing – original draft. MS: Formal Analysis, Writing – original draft. LG: Supervision, Writing – review & editing. SW: Funding acquisition, Visualization, Writing – review & editing.
The authors declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by Beijing Natural Science Foundation (Grant No. 7234369) and Beijing Chao-Yang Hospital Science and Technology Innovation Fund (Grant No. 22kcjjyb-19).
The authors appreciate Prof. Dan Luo and Dr. Shiwang Xie, Beijing Institute of Nanoenergy and Nanosystems, Chinese Academy of Sciences, for their help in scanning electron microscope imaging. In addition, the authors would like to thank all members of the Department of Infectious Diseases and Clinical Microbiology staff at Beijing Chao-Yang Hospital (Beijing, China) for contributing to this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Cairns, K. A., Udy, A. A., Peel, T. N., Abbott, I. J., Dooley, M. J., Peleg, A. Y. (2023). Therapeutics for vancomycin-resistant enterococcal bloodstream infections. Clin. Microbiol. Rev. 36, e0005922. doi: 10.1128/cmr.00059-22
Cartron, M. L., England, S. R., Chiriac, A. I., Josten, M., Turner, R., Rauter, Y., et al. (2014). Bactericidal activity of the human skin fatty acid cis-6-hexadecanoic acid on Staphylococcus aureus. Antimicrob. Agents Chemother. 58, 3599–3609. doi: 10.1128/aac.01043-13
Chanda, W., Joseph, T. P., Guo, X. F., Wang, W. D., Liu, M., Vuai, M. S., et al. (2018). Effectiveness of omega-3 polyunsaturated fatty acids against microbial pathogens. J. Zhejiang Univ. Sci. B. 19, 253–262. doi: 10.1631/jzus.B1700063
Ch’ng, J. H., Chong, K. K. L., Lam, L. N., Wong, J. J., Kline, K. A. (2019). Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 17, 82–94. doi: 10.1038/s41579-018-0107-z
Churchward, C. P., Alany, R. G., Kirk, R. S., Walker, A. J., Snyder, L. A. S. (2017). Prevention of ophthalmia neonatorum caused by Neisseria gonorrhoeae using a fatty acid-based formulation. mBio 8, e00534-17. doi: 10.1128/mBio.00534-17
CLSI (2023). “Performance standards for antimicrobial susceptibility testing,” in CLSI supplement M100, 33rd ed (Wayne, Pennsylvania, United States: Clinical and Laboratory Standards Institute).
Cruz, C. D., Shah, S., Tammela, P. (2018). Defining conditions for biofilm inhibition and eradication assays for Gram-positive clinical reference strains. BMC Microbiol. 18, 173. doi: 10.1186/s12866-018-1321-6
Cui, C., Song, S., Yang, C., Sun, X., Huang, Y., Li, K., et al. (2019). Disruption of quorum sensing and virulence in Burkholderia cenocepacia by a structural analogue of the cis-2-dodecenoic acid Signal. Appl. Environ. Microbiol. 85, e00105–e00119. doi: 10.1128/aem.00105-19
Dasa, K. T., Westman, S. Y., Millati, R., Cahyanto, M. N., Taherzadeh, M. J., Niklasson, C. (2016). Inhibitory effect of long-chain fatty acids on biogas production and the protective effect of membrane bioreactor. Biomed. Res. Int. 2016, 7263974. doi: 10.1155/2016/7263974
Desbois, A. P. (2012). Potential applications of antimicrobial fatty acids in medicine, agriculture and other industries. Recent Pat. Antiinfect. Drug Discovery 7, 111–122. doi: 10.2174/157489112801619728
Dunny, G. M., Hancock, L. E., Shankar, N. (2014). “Enterococcal biofilm structure and role in colonization and disease,” in Enterococci: from commensals to leading causes of drug resistant infection (Boston: Massachusetts Eye and Ear Infirmary), 409–434.
El-Houssaini, H. H., Elnabawy, O. M., Nasser, H. A., Elkhatib, W. F. (2019). Correlation between antifungal resistance and virulence factors in Candida albicans recovered from vaginal specimens. Microb. Pathog. 128, 13–19. doi: 10.1016/j.micpath.2018.12.028
EUCAST (2023). Breakpoint tables for interpretation of MICs and zone diameters, version 13.0 (Växjö, Sweden: European Committee on Antimicrobial Susceptibility Testing).
Kaur, N., Chugh, V., Gupta, A. K. (2014). Essential fatty acids as functional components of foods- a review. J. Food Sci. Technol. 51, 2289–2303. doi: 10.1007/s13197-012-0677-0
Kim, Y. G., Lee, J. H., Raorane, C. J., Oh, S. T., Park, J. G., Lee, J. (2018). Herring oil and omega fatty acids inhibit Staphylococcus aureus biofilm formation and virulence. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01241
Kiruthiga, A., Padmavathy, K., Shabana, P., Naveenkumar, V., Gnanadesikan, S., Malaiyan, J. (2020). Improved detection of esp, hyl, asa1, gelE, cylA virulence genes among clinical isolates of enterococci. BMC Res. Notes. 13, 170. doi: 10.1186/s13104-020-05018-0
Komiyama, E. Y., Lepesqueur, L. S., Yassuda, C. G., Samaranayake, L. P., Parahitiyawa, N. B., Balducci, I., et al. (2016). Enterococcus species in the oral cavity: prevalence, virulence factors and antimicrobial susceptibility. PloS One 11, e0163001. doi: 10.1371/journal.pone.0163001
Kuzma, J., Palcová, L., Timko, J., Bastová, V., Janošcová, V., Chmelař, D. (2022). Detection and molecular characterization of VRE isolates in Slovakia from stool samples positive for Clostridioides difficile toxins. Folia. Microbiol. (Praha). 67, 975–984. doi: 10.1007/s12223-022-01002-2
Lagatolla, C., Mehat, J. W., La Ragione, R. M., Luzzati, R., Di Bella, S. (2022). In vitro and in vivo studies of oritavancin and fosfomycin synergism against vancomycin-resistant Enterococcus faecium. Antibiotics (Basel). 11, 1334. doi: 10.3390/antibiotics11101334
Lim, S. Y., Teh, C. S. J., Thong, K. L. (2017). Biofilm-related diseases and omics: global transcriptional profiling of Enterococcus faecium reveals different gene expression patterns in the biofilm and planktonic cells. Omics. 21, 592–602. doi: 10.1089/omi.2017.0119
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Maurya, A., Kumar, R., Singh, A., Raj, A. (2021). Investigation on biofilm formation activity of Enterococcus faecium under various physiological conditions and possible application in bioremediation of tannery effluent. Bioresour. Technol. 339, 125586. doi: 10.1016/j.biortech.2021.125586
Montealegre, M. C., Roh, J. H., Rae, M., Davlieva, M. G., Singh, K. V., Shamoo, Y., et al. (2017). Differential penicillin-binding protein 5 (PBP5) levels in the Enterococcus faecium clades with different levels of ampicillin resistance. Antimicrob. Agents. Chemother. 61, e02034-16. doi: 10.1128/aac.02034-16
Murray, B. E. (1997). Vancomycin-resistant enterococci. Am. J. Med. 102, 284–293. doi: 10.1016/s0002-9343(99)80270-8
Nallapareddy, S. R., Sillanpää, J., Ganesh, V. K., Höök, M., Murray, B. E. (2007). Inhibition of Enterococcus faecium adherence to collagen by antibodies against high-affinity binding subdomains of Acm. Infect. Immun. 75, 3192–3196. doi: 10.1128/iai.02016-06
Osuka, H., Nakajima, J., Oishi, T., Funayama, Y., Ebihara, T., Ishikawa, H., et al. (2016). High-level aminoglycoside resistance in Enterococcus faecalis and Enterococcus faecium causing invasive infection: twelve-year surveillance in the Minami Ibaraki Area. J. Infect. Chemother. 22, 61–63. doi: 10.1016/j.jiac.2015.09.003
O’Toole, R. F., Leong, K. W. C., Cumming, V., Van Hal, S. J. (2023). Vancomycin-resistant Enterococcus faecium and the emergence of new sequence types associated with hospital infection. Res. Microbiol. 174, 104046. doi: 10.1016/j.resmic.2023.104046
Paganelli, F. L., de Been, M., Braat, J. C., Hoogenboezem, T., Vink, C., Bayjanov, J., et al. (2015). Distinct sagA from hospital-associated clade A1 Enterococcus faecium strains contributes to biofilm formation. Appl. Environ. Microbiol. 81, 6873–6882. doi: 10.1128/aem.01716-15
Panda, S. K., Buroni, S., Swain, S. S., Bonacorsi, A., da Fonseca Amorim, E. A., Kulshrestha, M., et al. (2022). Recent advances to combat ESKAPE pathogens with special reference to essential oils. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1029098
Patil, A., Banerji, R., Kanojiya, P., Saroj, S. D. (2021). Foodborne ESKAPE biofilms and antimicrobial resistance: lessons learned from clinical isolates. Pathog. Glob. Health 115, 339–356. doi: 10.1080/20477724.2021.1916158
Piddock, L. J. (1999). Mechanisms of fluoroquinolone resistance: an update 1994-1998. Drugs. 58 Suppl 2, 11–18. doi: 10.2165/00003495-199958002-00003
Pushparaj Selvadoss, P., Nellore, J., Balaraman Ravindrran, M., Sekar, U., Tippabathani, J. (2018). Enhancement of antimicrobial activity by liposomal oleic acid-loaded antibiotics for the treatment of multidrug-resistant Pseudomonas aeruginosa. Artif. Cells Nanomed. Biotechnol. 46, 268–273. doi: 10.1080/21691401.2017.1307209
Ramanathan, S., Ravindran, D., Arunachalam, K., Arumugam, V. R. (2018). Inhibition of quorum sensing-dependent biofilm and virulence genes expression in environmental pathogen Serratia marcescens by petroselinic acid. Anton. Leeuw. Int. J. G. 111, 501–515. doi: 10.1007/s10482-017-0971-y
Rani, S., Verma, S., Singh, H., Ram, C. (2022). Antibacterial activity and mechanism of essential oils in combination with medium-chain fatty acids against predominant bovine mastitis pathogens. Lett. Appl. Microbiol. 74, 959–969. doi: 10.1111/lam.13675
Rathnayake, I. U., Hargreaves, M., Huygens, F. (2012). Antibiotic resistance and virulence traits in clinical and environmental Enterococcus faecalis and Enterococcus faecium isolates. Syst. Appl. Microbiol. 35, 326–333. doi: 10.1016/j.syapm.2012.05.004
Santajit, S., Indrawattana, N. (2016). Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int. 2016, 2475067. doi: 10.1155/2016/2475067
Soares, R. O., Fedi, A. C., Reiter, K. C., Caierão, J., d’Azevedo, P. A. (2014). Correlation between biofilm formation and gelE, esp, and agg genes in Enterococcus spp. clinical isolates. Virulence. 5, 634–637. doi: 10.4161/viru.28998
Sun, M., Zhou, Z., Dong, J., Zhang, J., Xia, Y., Shu, R. (2016). Antibacterial and antibiofilm activities of docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) against periodontopathic bacteria. Microb. Pathog. 99, 196–203. doi: 10.1016/j.micpath.2016.08.025
Tendolkar, P. M., Baghdayan, A. S., Gilmore, M. S., Shankar, N. (2004). Enterococcal surface protein, Esp, enhances biofilm formation by Enterococcus faecalis. Infect. Immun. 72, 6032–6039. doi: 10.1128/iai.72.10.6032-6039.2004
Wang, Y., Pei, Z., Lou, Z., Wang, H. (2021). Evaluation of anti-biofilm capability of cordycepin against Candida albicans. Infect. Drug Resist. 14, 435–448. doi: 10.2147/idr.s285690
Wang, S., Wang, P., Liu, J., Yang, C., Wang, Q., Su, M., et al. (2022). Antibiofilm activity of essential fatty acids against Candida albicans from vulvovaginal candidiasis and bloodstream infections. Infect. Drug Resist. 15, 4181–4193. doi: 10.2147/idr.s373991
Won, S. R., Hong, M. J., Kim, Y. M., Li, C. Y., Kim, J. W., Rhee, H. I. (2007). Oleic acid: an efficient inhibitor of glucosyltransferase. FEBS Lett. 581, 4999–5002. doi: 10.1016/j.febslet.2007.09.045
Xie, Y., Wang, L., Yang, Y., Zha, L., Zhang, J., Rong, K., et al. (2022). Antibacterial and anti-biofilm activity of diarylureas against Enterococcus faecium by suppressing the gene expression of peptidoglycan hydrolases and adherence. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1071255
Keywords: enterococcus faecium, multidrug resistance, biofilm, VRE, fatty acid
Citation: Wei M, Wang P, Li T, Wang Q, Su M, Gu L and Wang S (2023) Antimicrobial and antibiofilm effects of essential fatty acids against clinically isolated vancomycin-resistant Enterococcus faecium. Front. Cell. Infect. Microbiol. 13:1266674. doi: 10.3389/fcimb.2023.1266674
Received: 25 July 2023; Accepted: 13 September 2023;
Published: 29 September 2023.
Edited by:
Adline Princy Solomon, SASTRA University, IndiaReviewed by:
Poonam Mudgil, Western Sydney University, AustraliaCopyright © 2023 Wei, Wang, Li, Wang, Su, Gu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Gu, ZGlkY20yMDA2QG1haWwuY2NtdS5lZHUuY24=; Shuai Wang, d2FuZ3BrcUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.