- Department of Biochemistry and Molecular Biology, College of Agriculture, Biotechnology & Natural Resources, University of Nevada, Reno, NV, United States
The first step of any immune response is the recognition of foreign molecular structures inside the host organism. An important molecule that is generally foreign to eukaryotic cells is long double-stranded RNA (dsRNA), which can be generated during virus replication. The mechanisms of sensing viral dsRNA are well-studied in mammalian systems but are only poorly understood in insects, including disease vectors such as Culex quinquefasciatus mosquitoes. These mosquitoes are vectors for important arboviruses, such as West Nile virus, and Culex species mosquitoes are distributed across the globe in many temperate and tropical regions. The major antiviral response triggered by dsRNA in mosquitoes is RNA interference – a sequence-specific response which targets complementary viral RNA for degradation. However, here, we aimed to identify whether sequence-independent dsRNA sensing, mimicked by poly(I:C), can elicit an antiviral response. We observed a significant reduction in replication of La Crosse virus (LACV) in Cx. quinquefasciatus mosquito cells following poly(I:C) priming. We identified a number of antimicrobial peptides and Toll receptors that were upregulated at the transcript level by poly(I:C) stimulation. Notably, Toll6 was upregulated and we determined that a knockdown of Toll6 expression resulted also in increased LACV replication. Future efforts require genetic tools to validate whether the observed Toll6 antiviral activity is indeed linked to dsRNA sensing. However, large-scale functional genomic and proteomic approaches are also required to determine which downstream responses are part of the poly(I:C) elicited antiviral response.
1 Introduction
Mosquito-borne diseases have been historically localized to tropical and subtropical regions of the world, however globalization and climate change have led to an expansion of habitat for mosquitoes, increasing the risk of mosquito borne disease transmission. Culex spp. mosquitoes are now ubiquitous worldwide, capable of inhabiting tropical, subtropical, and temperate regions (Ciota and Kramer, 2013). They are principal vectors for a variety of arboviruses such as Japanese encephalitis virus (Sharma et al., 2021; Hernández-Triana et al., 2022), St. Louis encephalitis virus (Reisen, 2003), Usutu virus, and West Nile virus (WNV) (Brugman et al., 2018). Due to the absence of effective vaccine and treatment options for most arboviruses, including Culex-borne viruses, prevention of transmission at the vector level remains the most likely means of reducing disease burden. Developing a greater understanding of mosquito immune pathways may translate into novel vector control methods. However, much of our current understanding of mosquito-virus interactions comes from Aedes aegypti. There is a need to build and expand on these findings in Culex spp. mosquitoes.
The first step of any immune response is sensing the presence of a microbial infection. Innate immune responses are generally activated by pattern recognition receptors (PRRs), which recognize pathogen-associated molecular patterns (PAMPs) (Janeway and Medzhitov, 2002; Lu et al., 2020). Mosquito antiviral responses include several conserved signal transduction pathways resulting in antiviral effector gene expression and restriction of arbovirus replication. These pathways include the Janus kinase/signal transducer and activator of transcription (JAK/STAT), Toll, and immune deficiency (Imd) pathways, all of which can be activated by a variety of PAMPs (Kumar et al., 2018; Tikhe and Dimopoulos, 2021). In mammals, dsRNA is a major PAMP of viruses sensed by a variety of receptors that often result in the activation of antiviral and inflammatory pathways (Chen and Hur, 2022). In mosquitoes, the helicase and endonuclease Dicer-2 (Dcr-2) is responsible for sensing dsRNA during viral infection, but its main known function is to then process dsRNA into siRNAs that target and degrade viral genomes in a sequence-dependent manner as part of the RNA interference (RNAi) response (Bronkhorst and van Rij, 2014; Kumar et al., 2018). Aside from its very defined role in RNAi, Dcr-2 is related to mammalian Rig-I-like receptors, which sense cytosolic dsRNA and activate an interferon-based antiviral response (Deddouche et al., 2008; Baldaccini and Pfeffer, 2021). About a decade ago, researchers identified a signaling function of Dcr-2 to activate the NF-κB-like transcription factor Rel2 following WNV infection of Culex quinquefasciatus-derived Hsu cells (Paradkar et al., 2012; Paradkar et al., 2014). The downstream upregulation of the cytokine-like molecule Vago was shown to activate the JAK/STAT pathway (Paradkar et al., 2012). However, it was not tested whether this signaling was dsRNA mediated or initiated by another aspect of virus infection. This role in immune signaling had also been identified in Drosophila melanogaster Dcr-2 (Deddouche et al., 2008). Another recent study found that the synthetic dsRNA analog, poly(I:C), activated rel2 expression in mosquito cells (Russell et al., 2021). Based on this evidence, mosquito Dcr-2 remains a strong candidate for a role in sensing viral dsRNA and eliciting a sequence-independent immune response to virus infection.
Another mechanism that could be conserved between humans and mosquitoes is dsRNA sensing via Toll receptors. The Toll pathway has been shown to be important for antiviral defenses in mosquitoes (Xi et al., 2008) and Ae. aegypti Toll6 (AaToll6) was recently shown to share residues important for dsRNA binding with human Toll-like receptor 3 (TLR3), a known sensor of dsRNA in humans (Angleró-Rodríguez et al., 2021). The authors also determined that numerous antimicrobial peptides (AMPs) downstream of Toll signaling were upregulated in mosquitoes cells exposed to poly(I:C) (Angleró-Rodríguez et al., 2021), which was previously used to determine that mammalian TLR3 acts as a PRR for dsRNA (Alexopoulou et al., 2001). Angleró-Rodríguez et al. also showed that extracellularly provided poly(I:C) localized to endosomes, where human TLR3 is localized and senses dsRNA. However, it remains unknown whether Toll6 also localizes to endosomes, whether it senses dsRNA and poly(I:C), and if it plays any role in antiviral responses of mosquitoes. Another open question is whether any other Toll receptors are involved in dsRNA sensing. In mammals, other TLRs besides TLR3 are also important for controlling virus infection (Lester and Li, 2014) and multiple TLRs can detect foreign nucleic acids. In Cx. quinquefasciatus, there are 9 Toll genes (Bartholomay et al., 2010), but none have been studied in any depth.
In this study, we aimed to elucidate if and how dsRNA sensing impacts virus replication in Cx. quinquefasciatus-derived Hsu cells. We used poly(I:C) to elicit an immune response and tested whether poly(I:C) priming impacts replication of the orthobunyavirus La Crosse virus (LACV). The advantage of using poly(I:C) over other long dsRNA molecules is that it will not elicit a sequence-specific antiviral response in mosquito cells. We used LACV as our model virus due to its ability to efficiently replicate in Hsu cells (Walsh et al., 2022) and its ability to be transmitted by Culex spp. mosquitoes (Thompson et al., 1972; Harris et al., 2015) despite being traditionally transmitted by Aedes triseriatus (Borucki et al., 2002). We found that LACV was reduced in poly(I:C) primed cells and further tested which AMPs are differentially regulated following poly(I:C) treatment. We used media-based treatment and transfection-based treatment in an attempt to elicit responses through endosomal and cytosolic signaling, respectively. We also found that multiple Toll receptors were upregulated following poly(I:C) treatment and, finally, determined that knockdown of Cx. quinquefasciatus Toll6 (CqToll6) using siRNA increased LACV replication in Hsu cells. While we were not yet able to generate conclusive evidence for any role of Toll6 in dsRNA sensing, we have shown that it is upregulated rapidly upon poly(I:C) stimulation and that Toll6 is involved in antiviral responses in Cx. quinquefasciatus Hsu cells.
2 Materials and methods
2.1 Cell lines
The Cx. quinquefasciatus ovary-derived (Hsu) cell line (Hsu et al., 1970) was grown at 27°C, and 5% CO2 in Dulbecco’s Modified Eagle Medium (DMEM; Corning #10-013-CV; Corning, New York, NY, USA) supplemented with 10% FBS and antibiotics (100 units mL penicillin, 100 μg/mL streptomycin, 5 μg/mL gentamicin).
2.2 Viruses
LACV strain R97841d was initially obtained from a human brain in Tennessee in 2012 and kindly provided by Brandy Russell, CDC Fort Collins, as a Vero p1 stock. Virus was then propagated on Vero cells and concentrated using the Amicon® Ultra-15 Centrifugal Filter (Sigma-Aldrich, #UFC9010 St. Louis, MO, USA) to obtain a high titer stock. Virus titration was performed by standard plaque assay on Vero cells.
2.3 Poly(I:C) stimulation
Hsu cells were seeded into 24-well plates in 0.5 mL of culture medium. Cells were incubated overnight at 27°C to allow for cell adhesion. Poly(I:C) treatment was performed by replacing the culture medium with fresh medium containing 100 μg/mL (50 μg total) of high molecular weight poly(I:C) (Invivogen, tlrl-pic, San Diego, CA, USA) or the equivalent volume of physiological H2O. Transfection of poly(I:C) was performed using Lipofectamine RNAiMAX (Thermo Fisher, #13778075, Waltman, MA, USA) following the manufacturer’s protocol. Briefly, per well, 1.5 µL Lipofectamine RNAiMAX reagent (Thermo Fisher, #13778100, Waltman, MA, USA) was diluted in 25 µL Opti-MEM (Thermo Fisher, #31985062, Waltman, MA, USA). In parallel, 10 μg high molecular weight poly(I:C) were diluted in 25 µL Opti-MEM (Thermo Fisher, #31985062, Waltman, MA, USA). Next, the diluted poly(I:C) was added to the diluted Lipofectamine RNAiMAX reagent (Thermo Fisher, AM1626, Waltman, MA, USA), mixed by pipetting up and down, and the complex was incubated for 5 min at room temperature. An amount of 50 µL of the complex was added to cells with 450 µL fresh complete media. Cells were incubated for the indicated time periods before RNA extraction or virus infection.
2.4 Virus infection
Hsu were seeded into 24-well plates and infected with LACV at an MOI of 1 or 10. Briefly, per well, virus was diluted in 250 µL of DMEM (without additives) and added to each well after removal of complete culture media. For mock infected controls, 250 µL DMEM (without additives) was added to each well. Cells were incubated for 1 h at 27°C to allow for infection. After 1 h, the virus-containing media was removed, and replaced with 0.5 mL of complete culture media. The plate was then incubated at 27°C for 24 or 48 h before RNA extraction.
2.5 RNA extraction
RNA was extracted using the Direct-zol™ RNA miniprep kit (Zymo Research, #D4033; Irvine, CA, USA) following the manufacturer’s protocol. Briefly, culture media was removed from all wells, and 300 µL TRI reagent was added to each well. Plates were incubated with TRI reagent on a rocker for 20 min to ensure complete lysis. Cell lysates were transferred to a microcentrifuge tube and frozen at −80°C prior to RNA extraction following the provided protocol. RNA was quantified using a Qubit Flex Fluorometer (Thermo Fisher, Waltham, MA, USA).
2.6 cDNA synthesis and RT-qPCR
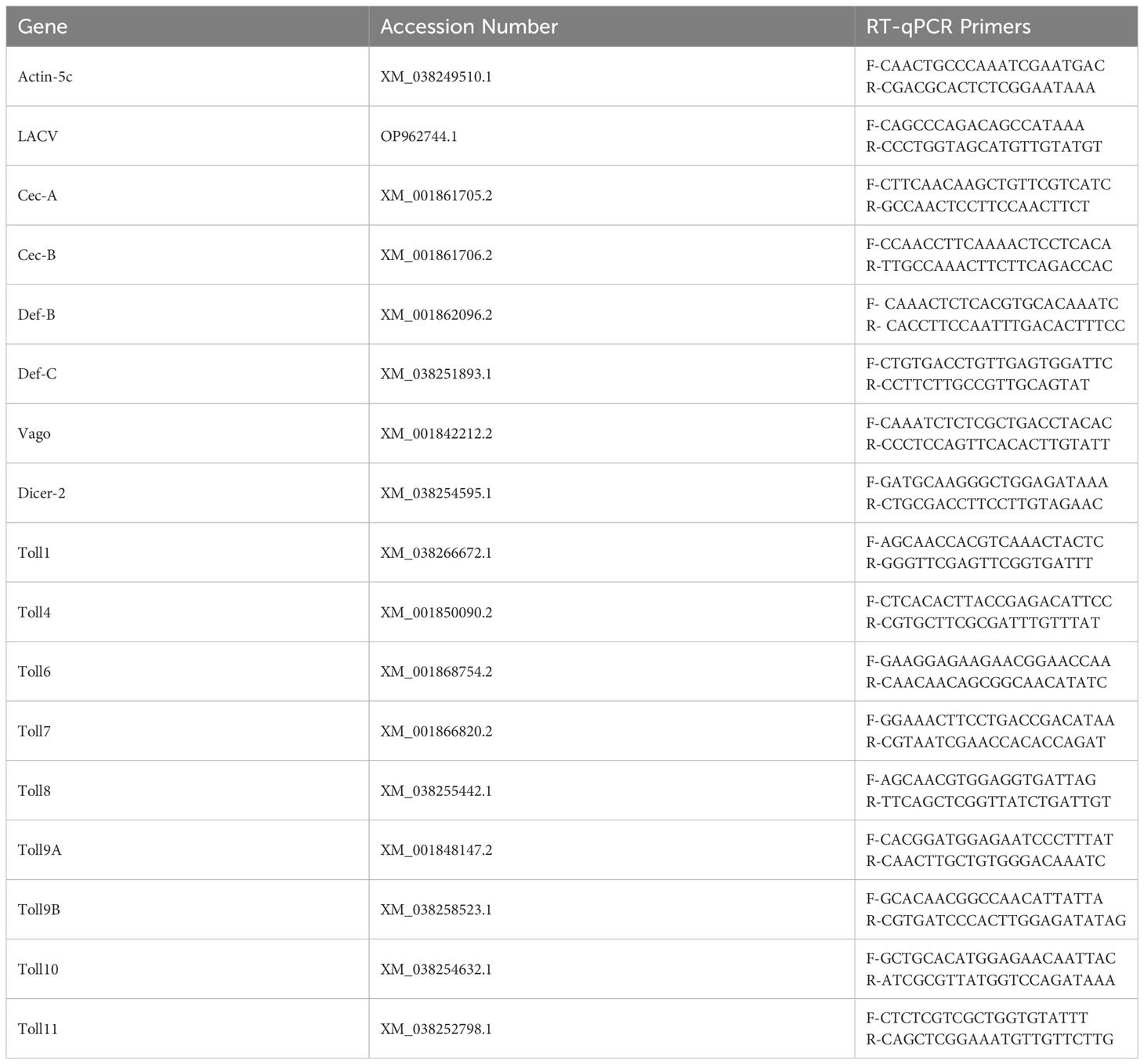
To measure AMP and Toll gene expression, cDNA was first generated from 200 ng of each RNA sample using the High-Capacity cDNA reverse transcription kit (Thermo Fisher, #4368814, Waltman, MA, USA). Quantitative real-time PCR (qRT-PCR) was then performed using iTaq Universal SYBR Green Supermix (Bio-Rad, #1725120, Hercules, CA, USA) and primers (Table 1) on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Each target gene was normalized to a previously tested, reliable housekeeping gene (actin-5c) to quantify gene expression levels.
To determine knockdown efficiency and LACV RNA levels, 100 ng of each RNA sample was used directly in a one-step qRT-PCR reaction using iTaq Universal SYBR Green One-Step Kit (Bio-Rad, #1725150) or iTaq Universal Probes One-Step Kit (Bio-Rad #1725140). Toll6 and LACV RNA levels were again normalized to actin-5c as a housekeeping gene.
2.7 siRNA knockdown
Transfection of gene-specific siRNA and a non-specific siRNA control (RLuc) was done by reverse transfection using Lipofectamine RNAiMAX (Thermo Fisher, #13778075, Waltman, MA, USA) following the manufacturer’s protocol. Briefly, per well, 3 µL of 5 µM siRNA was diluted with 1.5 µL Lipofectamine RNAiMAX reagent (Thermo Fisher, #13778100, Waltman, MA, USA) in Opti-MEM (Thermo Fisher, #31985062, Waltman, MA, USA) to a final volume of 100 μL. The solution was mixed by pipetting up and down, incubated for 20 min at room temperature, and then each complex was added to its respective well in a 24-well plate. Hsu cells in 0.5 mL of culture medium were then added to each well. Cells were incubated for 48 h before RNA extraction or virus infection.
2.8 Phylogenetic analysis
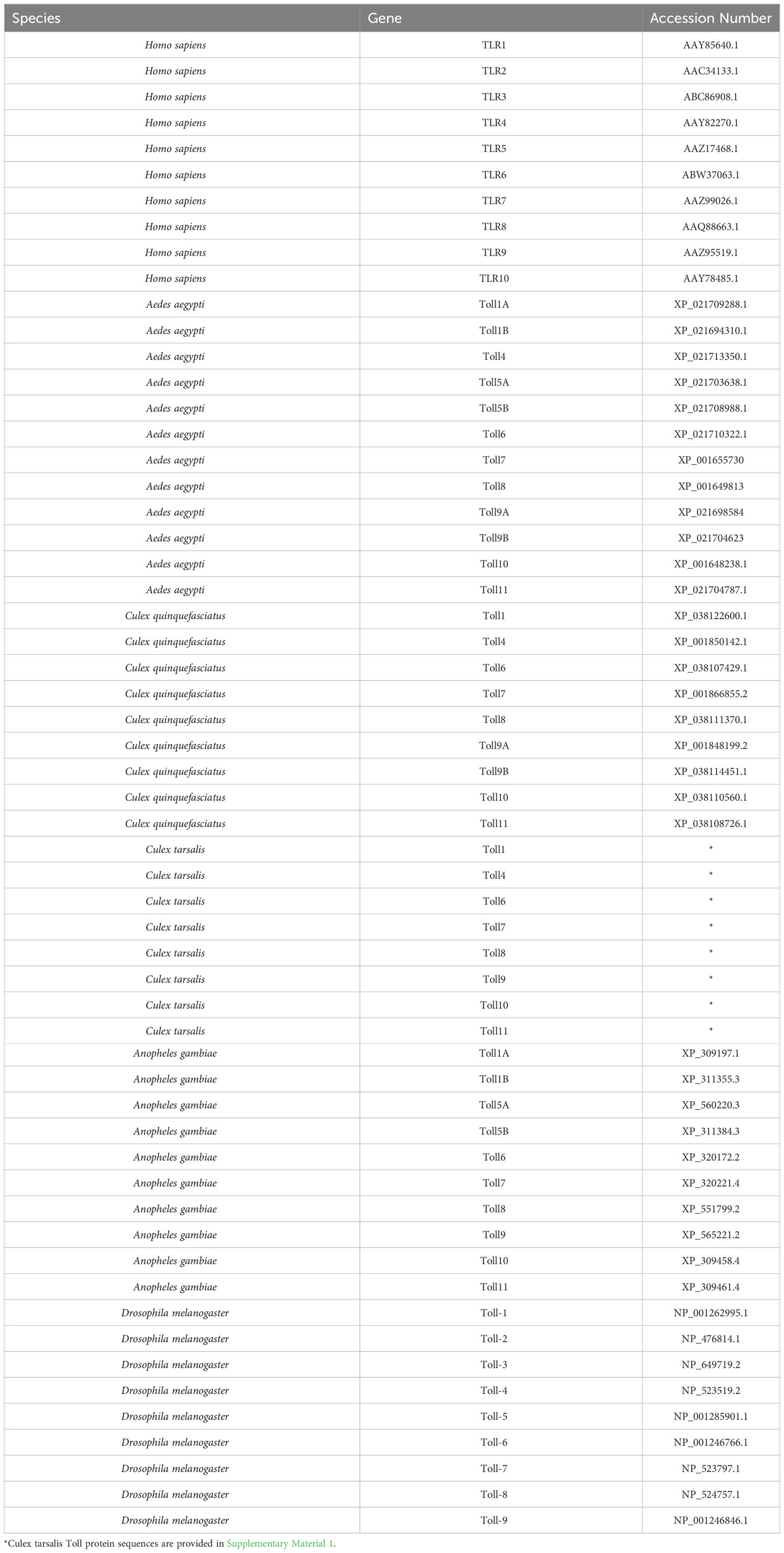
Cx. quinquefasciatus, Ae. aegypti, A. gambiae, D. melanogaster, and H. sapiens Toll receptor protein sequences were collected from the VectorBase and NCBI databases (Table 2). Culex tarsalis protein sequences were identified from the published Cx. tarsalis genome (Main et al., 2021) using blastp in the BLAST command line to search for Toll receptors with the closely related Cx. quinquefasciatus Toll protein sequences. All Cx. tarsalis Toll protein sequences were confirmed to contain a Toll/interleukin-1 receptor (TIR) domain and leucine rich (LRR) motifs using InterPro (Paysan-Lafosse et al., 2023). Sequence alignment and tree generation was performed as done previously (Zhang et al., 2021) with minor variations. Briefly, Toll sequences from all species were aligned using MAFFT (v7.520) with the E-INS-I alignment algorithm to account for sequences with multiple conserved domains and long gaps. The phylogenetic tree was generated using the ExaBayes (version 1.4.1) tool with the optimal amino acid substitution model for the data chosen by default. The analysis was initiated with a random seed and ran for 1,000,000 generations, sampling every 500 generations. Visualization and annotation were done in FigTree (version 1.4.4).
2.9 Statistical analysis
Statistical analysis was performed using ANOVA or unpaired t-test in GraphPad Prism as indicated in figure legends. Outliers from select individual experiments were excluded from the data using the Grubb’s test with a significance level of 0.05 in GraphPad Prism.
3 Results
3.1 Poly(I:C) priming reduces LACV replication in Cx. quinquefasciatus cells
In order to determine if poly(I:C) elicits an antiviral immune response in Cx. quinquefasciatus, we primed Hsu cells with 100 μg mL of poly(I:C) or water prior to infection with LACV. As the duration of poly(I:C) treatment can affect the expression of cytokines in mammals (Kato et al., 2006) and immune effector genes in Ae. aegypti (Angleró-Rodríguez et al., 2021), we chose four different timepoints of poly(I:C) priming time. Hsu cells primed with poly(I:C) for 6, 12, and 24 hours led to a modest but significant (p < 0.05) decrease in LACV RNA levels 24 hours post infection (hpi) at an MOI of 1 (Figure 1A). Poly(I:C) priming for 12 and 24 h also resulted in a significant decrease of LACV RNA at 48 hpi (Figure 1B). To see if a similar effect was maintained at a higher viral load, we primed Hsu cells and infected at an MOI of 10. At 24 hpi, only 24 hours of priming had been able to reduce LACV RNA levels (Figure 1C). However, by 48 hpi LACV RNA levels were again significantly reduced in cells primed with poly(I:C) for 6, 12, and 24 h prior to infection (Figure 1D). Overall, these results showed that poly(I:C) treatment prior to virus infection can reduce LACV RNA levels in Hsu cells across different MOIs and infection periods.
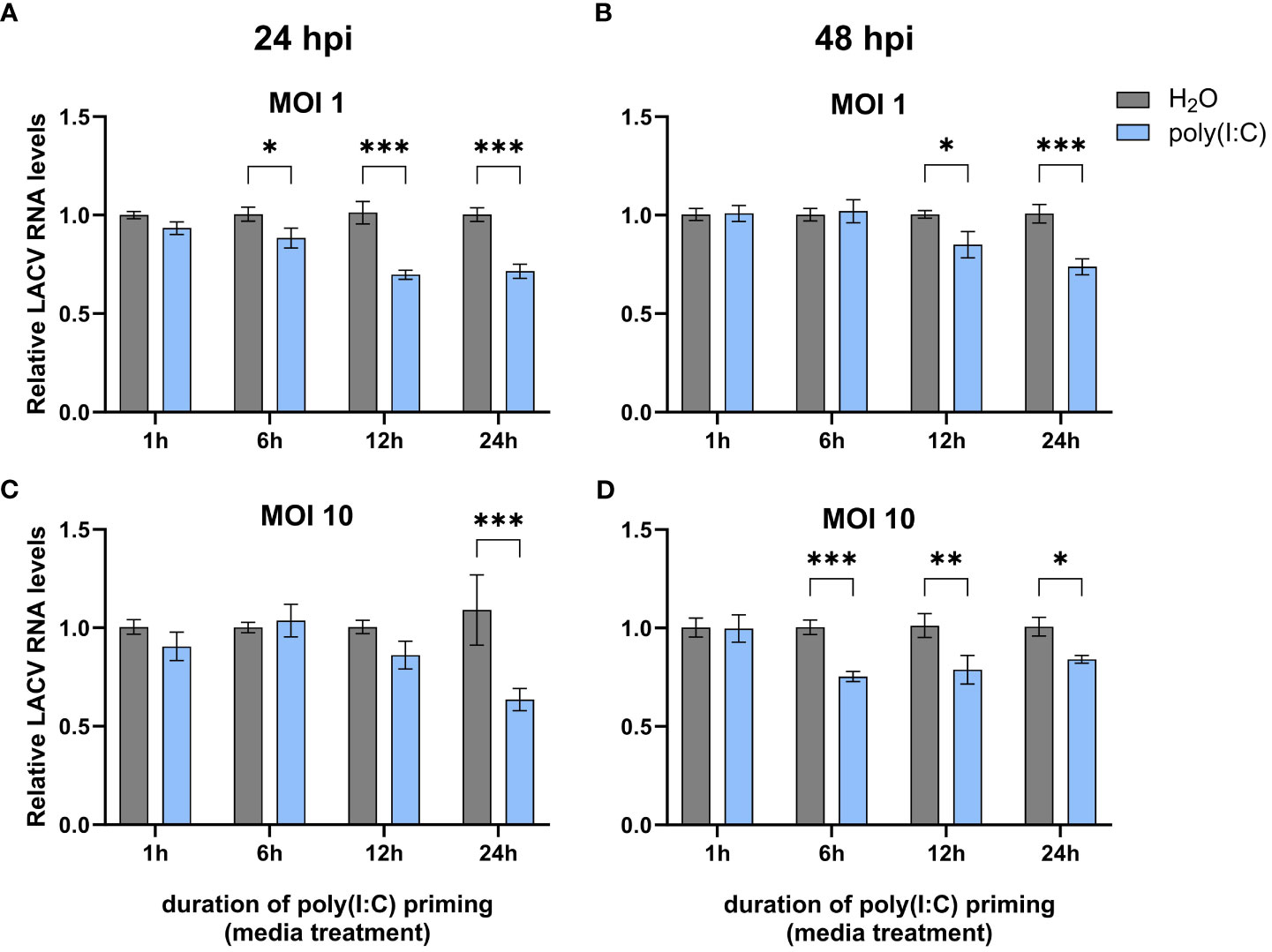
Figure 1 Effect of poly(I:C) priming on LACV RNA levels. Hsu cells were treated with poly(I:C) for 1, 6, 12, or 24 h prior to infection with LACV at MOI 1 (A, B) or 10 (C, D). Cellular RNA was extracted and LACV RNA levels were measured by RT-qPCR at 24 hpi (A, C) or 48 hpi (B, D) and normalized to actin-5c (housekeeping gene). Bars represent the mean +/- SEM of 2 independent experiments with 4 biological replicates each. Significant changes in viral RNA abundance compared to the H2O control are shown as * p < 0.05, ** p < 0.01, *** p < 0.001 as analyzed by two-way ANOVA.
3.2 Poly(I:C) treatment induces modest AMP upregulation in Cx. quinquefasciatus cells
Since we observed a significant decrease in LACV replication, we wanted to determine if any immune genes were differentially regulated following poly(I:C) treatment. We delivered poly(I:C) to Hsu cells with or without a transfection reagent to mimic cytosolic dsRNA sensing and extracellular sensing, respectively, and measured immune effector gene expression at different timepoints post stimulation by RT-qPCR. We measured the mRNA levels of cecropins, defensins, and the cytokine-like molecule Vago, all of which showed increased mRNA levels at varying durations of poly(I:C) treatment with and/or without a transfection reagent (Figures 2A–H). Cecropin-A (Cec-A) mRNA levels were significantly (p < 0.05) increased at 1, 6, and 24 h post treatment (Figure 2A), but there was no increase in Cec-A mRNA following poly(I:C) transfection (Figure 2B). Another cecropin, Cec-B, was upregulated at 1, 6, and 24 h post treatment without a transfection reagent (Figure 2C). Additionally, transfected poly(I:C) upregulated Cec-B’s mRNA levels at 6 and 12 h, returning to basal levels at 24 h (Figure 2D). Transcript abundance of Defensin-B (Def-B) was increased at all timepoints after treatment without a transfection reagent (Figure 2E), with only a slight uptick at 6 h post poly(I:C) transfection (Figure 2F). Def-C mRNA was also upregulated at all timepoints when poly(I:C) was added alone (Figure 2G), but only after 24 h of transfected poly(I:C) treatment (Figure 2H).
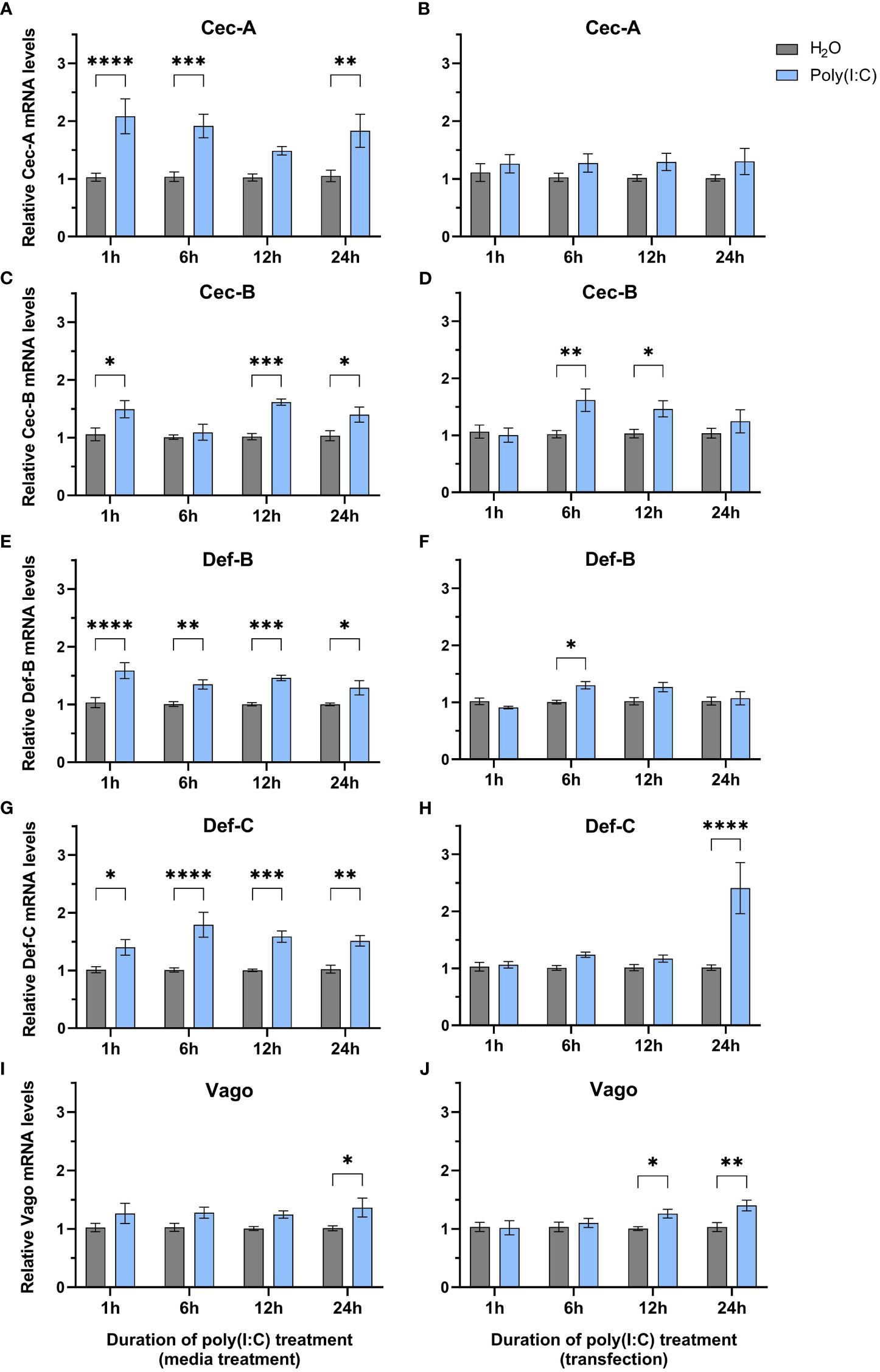
Figure 2 Expression of select immune genes after treatment with poly(I:C). Hsu cells were treated with poly(I:C) alone (A, C, E, G, I) or with poly(I:C) plus a transfection reagent (B, D, F, H, J) for 1, 6, 12, or 24 h. Cellular RNA was extracted and gene expression was measured by RT-qPCR and normalized to actin-5c (housekeeping gene). Bars represent the mean +/- SEM of 3 independent experiments with 4 biological replicates each. Significant changes in mRNA abundance compared to the H2O control are shown as * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, as analyzed by two-way ANOVA.
In contrast to studies in Ae. aegypti Aag2 cells (Angleró-Rodríguez et al., 2021; Russell et al., 2021), we found that vago is upregulated after poly(I:C) treatment without (Figure 2I) and with a transfection reagent (Figure 2J). While statistically significant (p < 0.05), this increase in vago expression was only modest with a 1.4-fold increase at 24 h post treatment (Figure 2I) and a 1.3-fold and 1.4-fold increase at 12 and 24 h post transfection, respectively (Figure 2J).
3.3 CqToll6 is involved in antiviral defenses
In searching for a potential receptor responsible for dsRNA sensing in mosquitoes, Toll6 in A. aegypti was recently shown to share a conserved amino acid motif with human TLR3 that is required for dsRNA recognition (Angleró-Rodríguez et al., 2021). However, little is known about other mosquito toll6 genes. Protein sequence alignment showed that these amino acid residues are also conserved in Cx. quinquefasciatus and Culex tarsalis, but not in D. melanogaster or Anopheles gambiae) (Figure 3A). We thus hypothesize that CqToll6 is involved in dsRNA sensing.
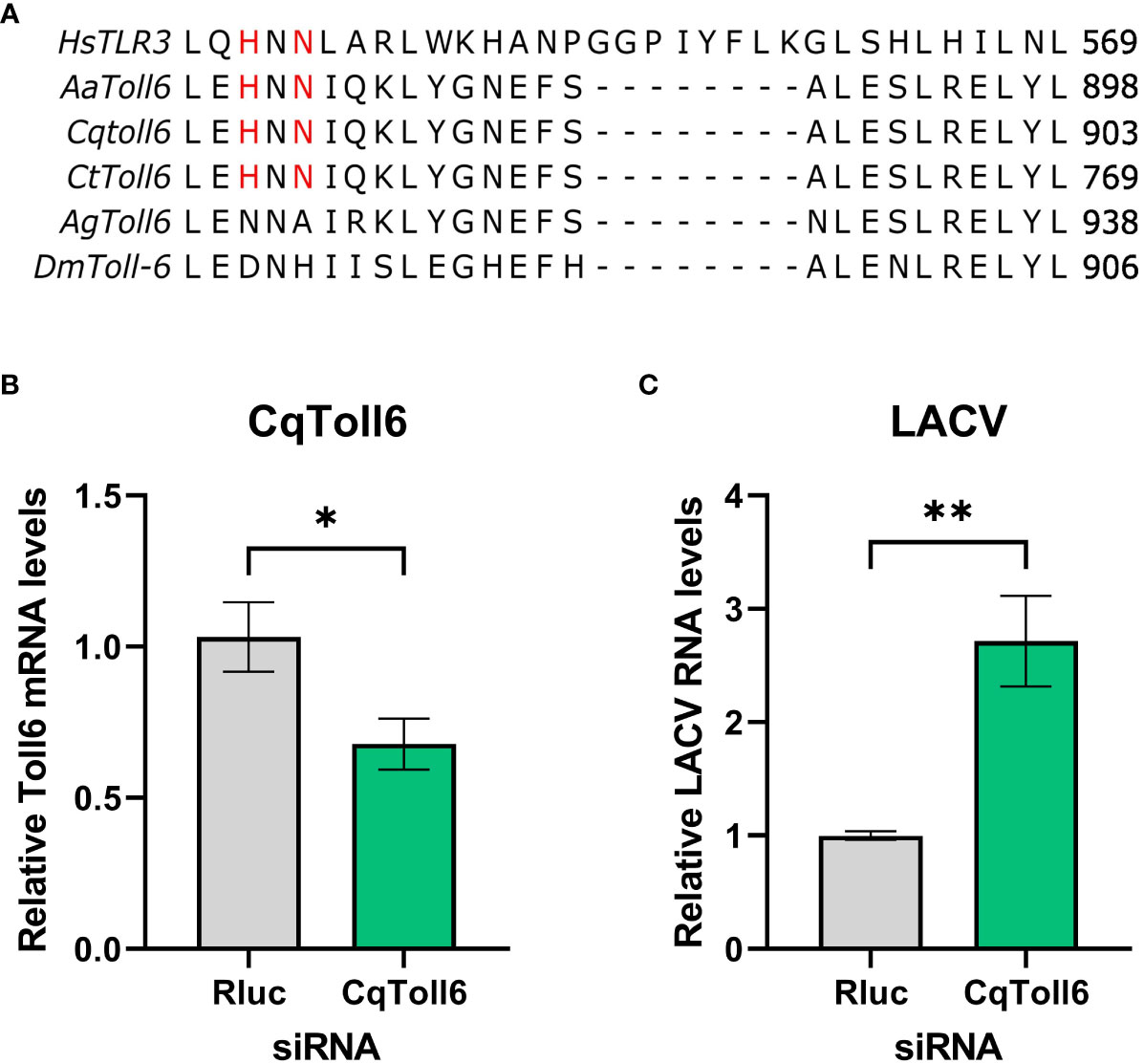
Figure 3 CqToll6 knockdown increases LACV replication. The protein sequences of Toll6 from Culex quinquefasciatus (Cq), Culex tarsalis (Ct), Aedes aegypti (Aa), Anopheles gambiae (Ag), and Drosophila melanogaster (Dm) and TLR3 from Homo sapiens (Hs) were aligned using Clustal Omega (A). The histidine and asparagine residues important for TLR3 dsRNA binding are shown in red. CqToll6 was silenced in Hsu cells using siRNA and validated 48 hours post siRNA transfection (B). Hsu cells were then infected with LACV MOI 1 (C). Cellular RNA was extracted at the time of infection (B) or at 48 hpi (C). CqToll6 knockdown efficiency (B) and viral RNA levels (C) were determined by RT-qPCR and normalized to actin-5c (housekeeping gene). Bars are the mean +/SEM of 2 independent experiments with 4 biological replicates each. Significant changes in RNA abundance compared to the Renilla luciferase targeting control are shown as * p < 0.05, ** p < 0.01 as analyzed by unpaired t-test.
We first wanted to investigate whether CqToll6 is involved in controlling virus replication in Hsu cells. We used siRNA-mediated knockdown of CqToll6 expression in Hsu cells and infected with LACV at an MOI 1. While statistically significant, CqToll6 RNA levels were only reduced modestly (Figure 3B). However, following knockdown, we infected cells with LACV and, at 48 hpi, there was a significant increase in LACV cellular RNA levels (Figure 3C). These results indicate that CqToll6 is antiviral against LACV even after a modest knockdown. Due to the difficulty of obtaining a reliable knockdown in CqToll6 expression, we were unable to directly investigate its role in poly(I:C) sensing.
3.4 Mosquito Toll phylogeny
We also wanted to extend our understanding of the Toll receptor repertoire in Culex species mosquitoes. It is now known that receptors besides TLR3, such as TLR10, can sense poly(I:C) in human cells (Lee et al., 2018), suggesting that multiple mosquito Toll receptors may be responsible for sensing poly(I:C). There are nine Toll genes in Cx. quinquefasciatus mosquitoes, all of which can be detected in the head, thorax, and abdomen of adult Cx. quinquefasciatus mosquitoes (Supplemental Figure 1). To build a comprehensive phylogeny of Toll receptors in vector mosquitoes, we first set out to identify all Toll receptors in the recently assembled genome of Cx. tarsalis (Main et al., 2021). We used the protein sequences of Cx. quinquefasciatus Toll genes to BLAST for homologous genes in the Cx. tarsalis genome. In our final phylogenetic analysis, we included protein sequences of Toll genes from four mosquito species, D. melanogaster and TLRs from humans to generate a phylogenetic tree (Figure 4). Our findings show a clear subcluster of Toll1/5 in mosquitoes that has been described previously (Christophides et al., 2002; Kanzok et al., 2004). However, both Culex spp. appear to have lost the Toll5 gene and do not contain the Toll1 gene duplication seen in Ae. aegypti and An. gambiae. Both Culex spp. also possess a Toll4 that belongs to the larger Toll1/3/4/5 family. We found that Toll9 clustered closely with all human TLRs, suggesting Toll9 may be a conserved PRR considering that mammalian TLRs are essential players in recognizing infections (Leulier and Lemaitre, 2008; Lester and Li, 2014). We could only find one copy of Cx. tarsalis Toll9, despite the presence of two Toll9 copies in Cx. quinquefasciatus and Ae. aegypti. Mosquito Tolls 6-8 tended to cluster with their respective D. melanogaster ortholog in our analysis. The mosquito-specific Toll10 and 11 were conserved between the four mosquito species in our analysis.
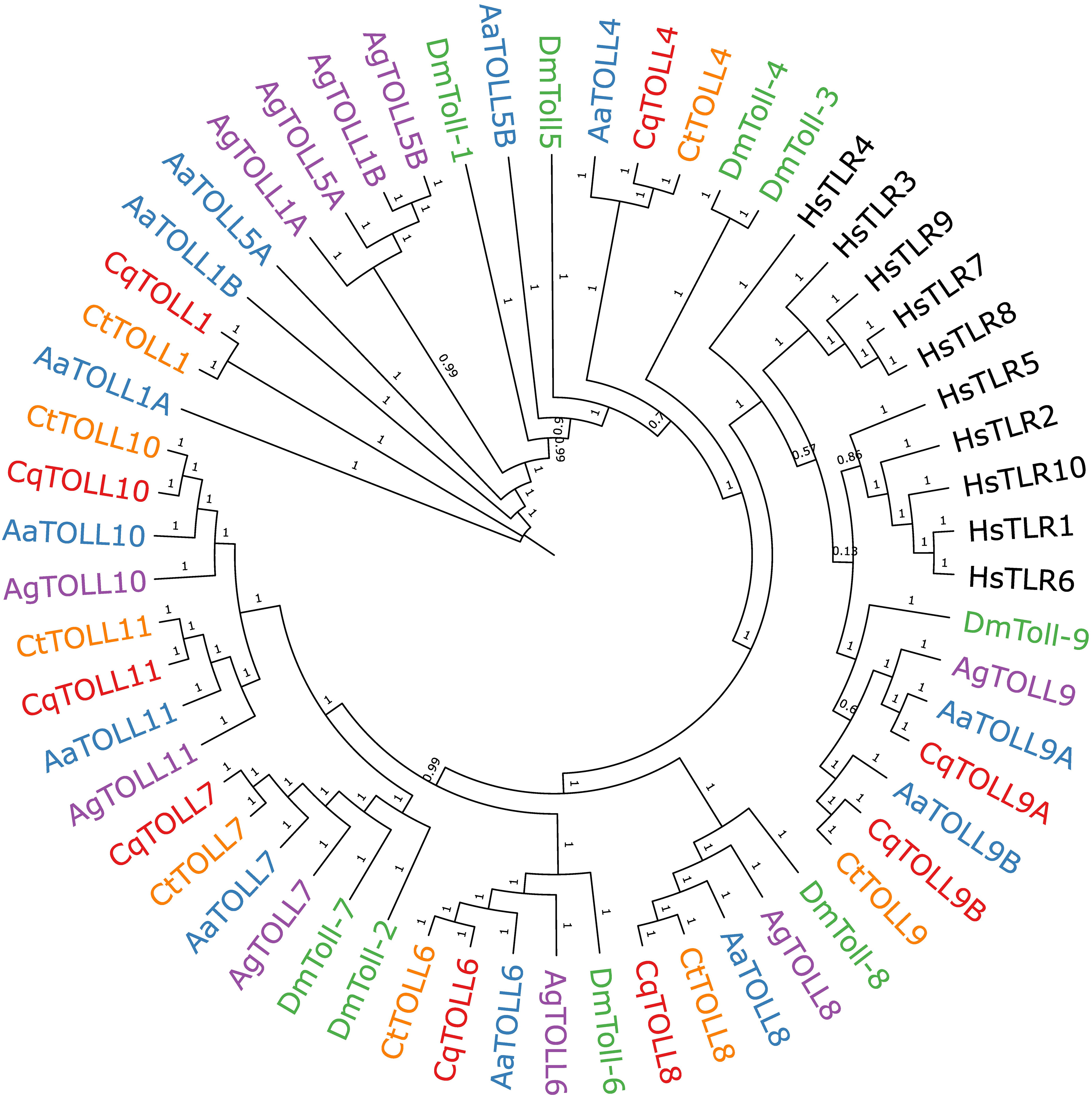
Figure 4 Bayesian inference of phylogeny of Culex quinquefasciatus (Cq), Culex tarsalis (Ct), Aedes aegypti (Aa), Anopheles gambiae (Ag), Drosophila melanogaster (Dm), and Homo sapiens (Hs) full length Toll receptor protein sequences. Branch nodes are labeled with posterior probabilities.
3.5 Select toll genes are differentially regulated following poly(I:C) exposure and LACV infection
To help further our functional understanding of all Culex Toll receptors, we treated Hsu cells with poly(I:C) with or without a transfection reagent and measured mRNA levels of all nine toll genes at 1, 6, 12 and 24 h (Figures 5A–H). In cells treated with poly(I:C) alone, toll6, 8, and 9A were significantly upregulated after 1 h of treatment (Figure 5A) showing the largest increase in toll expression of all treatments and timepoints. In contrast, no toll genes were differentially regulated following poly(I:C) transfection for 1 h (Figure 5B). By 6 h post media treatment, all toll genes were at comparable levels in poly(I:C) and water treated cells (Figure 5C). However, following 6 h of transfection with poly(I:C), toll10 was significantly upregulated (Figure 5D). After 12 h of treatment, media treatment of poly(I:C) resulted in the upregulation of toll9b and toll4 (Figure 5E). However, toll8 was slightly downregulated 12 h post media treatment (Figure 5E), while it was upregulated 12 h post transfection (Figure 5F), possibly suggesting a difference in the timing of signaling effects. By 24 h of treatment, none of the toll genes showed significant changes in expression, despite a small trend for upregulation of multiple toll genes (Figures 5G, H).
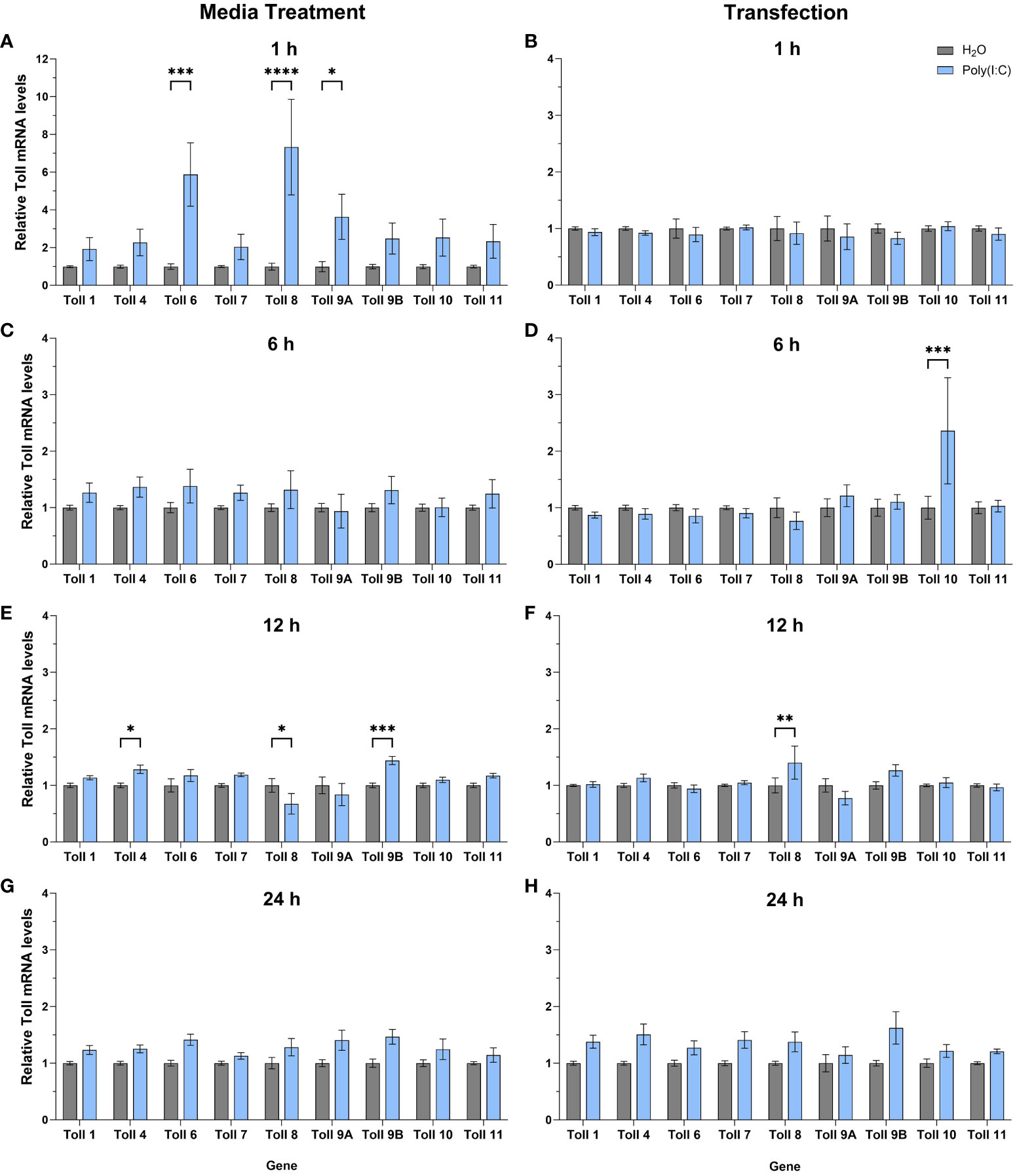
Figure 5 Expression of toll genes after treatment with poly(I:C). Hsu cells were treated with poly(I:C) alone (A, C, E, G) or with poly(I:C) plus a transfection reagent (B, D, F, H), for 1, 6, 12, or 24 hours. Cellular RNA was extracted and gene expression was measured by RT-qPCR and normalized to actin-5c (housekeeping gene). Bars represent the mean +/- SEM of 3 independent experiments with 4 biological replicates each. Significant changes in RNA abundance compared to the H2O control are shown as * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001 as analyzed by two-way ANOVA.
We then sought to determine the expression profile of Cx. quinquefasciatus toll genes in Hsu cells after infection with LACV. At 24 hpi, no toll genes were differentially regulated in Hsu cells compared to mock-infected cells (Figure 6A). However, by 48 hpi, there was a modest, but significant upregulation of toll10 expression in LACV infected cells (Figure 6B). No other toll genes were differentially regulated at 48 hpi with LACV.
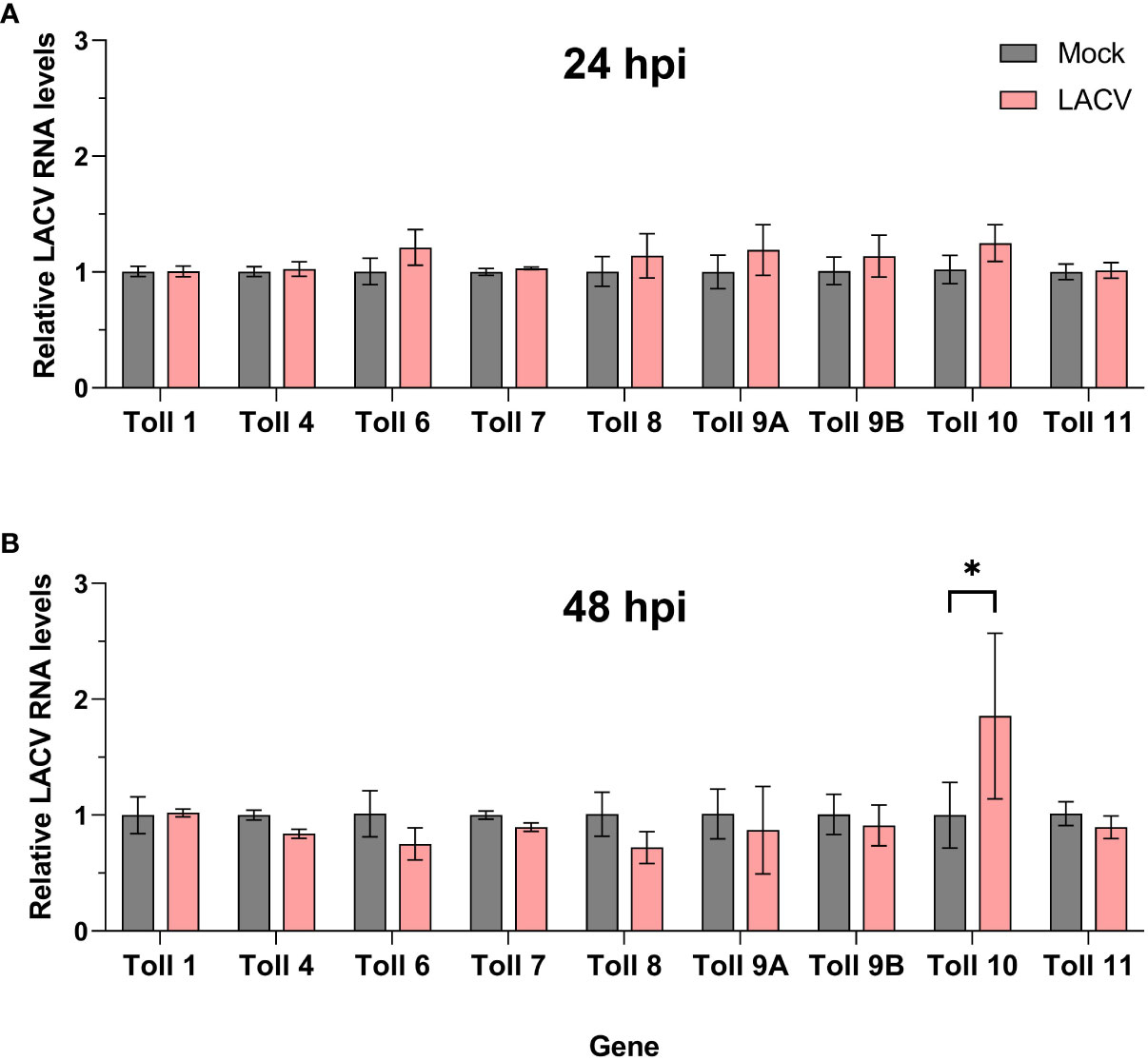
Figure 6 Expression of toll genes after infection with LACV. Hsu cells were infected with LACV MOI 1 for 24 (A) or 48 (B) hours. Cellular RNA was extracted and LACV RNA was measured by RT-qPCR and normalized to actin-5c (housekeeping gene). Bars represent the mean +/- SEM of 2 independent experiments with 4 biological replicates each. Significant changes in RNA abundance compared to the mock infected control are shown as * p < 0.05.
4 Discussion
In this study, we sought to build on the knowledge of how mosquitoes sense virus infection to mount an immune response. In particular, we were interested in the response of Cx. quinquefasciatus to the dsRNA mimic poly(I:C). We found that poly(I:C) treatment elicits an antiviral immune response in Cx. quinquefasciatus Hsu cells that limits LACV replication. This is consistent with studies in multiple cell lines of human origin that have shown varying treatment times with poly(I:C) can limit LACV titers (Luby, 1975; Monteiro et al., 2019). The impact on LACV replication in our study was modest and highly dependent on the duration of poly(I:C) priming prior to infection, with longer treatments more consistently reducing viral RNA levels. This finding differed from studies performed in human cells with CHIKV (Li et al., 2012) and mosquito cells with DENV (Angleró-Rodríguez et al., 2021) where 1 h of poly(I:C) priming was sufficient to reduce virus replication. However, we did observe the upregulation of select immune effector genes after only 1 h of poly(I:C) treatment. It is possible that other genes, ones not measured here, are responsible for controlling LACV replication in Hsu cells and that these are only upregulated after prolonged poly(I:C) incubation. It is worth noting that Hsu cells are slow growing cells compared to, for example, Ae. aegypti Aag2 cells, which might impact the exact timing of transcriptional changes compared to other mosquito cell lines. Transcriptomic profiling of poly(I:C) treated Hsu cells and Cx. quinquefasciatus mosquitoes at varying timepoints would help identify additional factors that may be involved in an antiviral response. It has also been shown that priming Aag2 cells with Rift Valley fever virus results in higher AMP expression after bacterial challenge than virus infection or bacterial challenge alone (Laureti et al., 2023). It may thus be worthwhile to measure AMP expression after poly(I:C) treatment and virus infection to determine if a similar priming effect is observed when a viral PAMP is used prior to a virus infection.
Our finding that certain Cecropins and Defensins are upregulated after poly(I:C) treatment in Cx. quinquefasciatus Hsu cells indicates a conserved response across mosquito species. This result builds on similar findings from two recent studies in Ae. aegypti Aag2 cells (Angleró-Rodríguez et al., 2021; Russell et al., 2021). One AMP, AaCec-E, was upregulated in both studies and expression of its Cx. quinquefasciatus ortholog, Cec-A, was also induced in Hsu cells here. However, Cec-A was not upregulated in Hsu cells when poly(I:C) was added with a transfection reagent, while Russell et al. used transfected poly(I:C) when measuring AaCec-E expression (Russell et al., 2021). This discrepancy could be due to inherent differences in protocols or could point towards a biological difference in the receptors that are responsible for sensing intracellular poly(I:C) when it is transfected. Poly(I:C) delivered to cells without a transfection reagent predominantly localizes to endosomes in many cell types, including mosquito cells (Angleró-Rodríguez et al., 2021), or activates receptors on the cell surface (Palchetti et al., 2015). A transfection reagent allows poly(I:C) to localize to the cytosol (Palchetti et al., 2015), likely initiating cytosolic signaling responses separate from those of the endocytic pathways triggered by simple addition to the media. Yet, in some human cell types, transfected poly(I:C) may be delivered to endosomes (Matsumoto and Seya, 2008) and may thus still be sensed there as well. Given the variability in poly(I:C) localization and the fact that different receptors are likely responsible for detecting poly(I:C) in those compartments, we feel it is important to highlight when a transfection reagent is used in experiments aiming to study potential dsRNA receptors. It will ultimately be crucial to determine which, if any, of the AMPs studied here have a direct antiviral role and what pathways regulate them.
The cytokine Vago was initially shown to be responsible for controlling DCV infection in flies (Deddouche et al., 2008). Later, Vago was found to be upregulated in response to WNV infection and to act as a cytokine-like molecule that activated the JAK-STAT pathway in Cx. quinquefasciatus Hsu cells (Paradkar et al., 2012; Paradkar et al., 2014). Our finding that vago mRNA is modestly but significantly (p < 0.05) upregulated in Hsu cells after treatment with poly(I:C) with and without a transfection reagent provides some evidence that dsRNA can act as a viral stimulus that induces vago expression in Hsu cells. Given that vago upregulation in response to virus infection is dependent on Dcr-2 (Deddouche et al., 2008; Paradkar et al., 2012; Paradkar et al., 2014), which is capable of sensing dsRNA (Blair and Olson, 2014), it follows that poly(I:C) might stimulate vago via Dcr-2 sensing. However, two recent studies have found vago to be unresponsive to poly(I:C) in Ae. aegypti Aag2 cells (Angleró-Rodríguez et al., 2021; Russell et al., 2021). Russell et al. recall that Ae. aegypti encodes two vago genes, and that they could only detect the expression of vago2 without a notable increase, while Angleró-Rodríguez et al. measured vago1 in Aag2 cells and observed no significant increase following poly(I:C) treatment (Asad et al., 2018; Angleró-Rodríguez et al., 2021; Russell et al., 2021). We, along with Paradkar et al., measured the expression of vago1 in Hsu cells (Paradkar et al., 2012). Given that vago is upregulated in vivo in Ae. aegypti in response to YFV (Colpitts et al., 2011), we speculate that different experimental conditions or cell line specific effects may impact vago expression in Aag2 cells. However, there likely remains some variability in the response of vago depending on the specific virus-vector pairing.
As TLR3 is well-known to sense dsRNA (Alexopoulou et al., 2001), and AaToll6 was recently shown to share key residues important for dsRNA binding (Bell et al., 2006; Angleró-Rodríguez et al., 2021), we wanted to determine if other mosquito Toll6 sequences also shared these conserved residues. When we aligned TLR3 with Toll6 of four mosquito species and D. melanogaster, only Culex spp. and Aedes spp., which are important viral vectors, had these key residues conserved in Toll6. This may indicate the importance of Toll6 for sensing viral infection in these species. Given that TLR3 has been shown to recognize dsRNA and induce a type I IFN-mediated response, it is no surprise that it serves an antiviral role against a wide array of viruses in humans (Chen et al., 2021). For example, multiple studies have found TLR3 crucial for controlling dengue virus replication in human cells (Tsai et al., 2009; Nasirudeen et al., 2011). Therefore, we investigated if Toll6 is antiviral in Cx. quinquefasciatus Hsu cells. After infection with LACV for 48 h, there was a significant increase in viral RNA levels, indicating that CqToll6 serves an antiviral role in Hsu cells. Additional studies are warranted to determine whether this antiviral role is mediated by direct dsRNA binding or other mechanisms, and to understand the broader impact it has on arboviruses in mosquitoes. Since siRNA and dsRNA knockdown of Toll6 is difficult to achieve, based on our experience, and introduces a confounding dsRNA molecule to the cells, generating a CRISPR/Cas9 knockout cell line and a CqToll6 expression plasmid would be advantageous for future studies.
We also wanted to build on the phylogenetic understanding of Tolls in mosquito species, given how important TLRs are in human innate immune responses to virus infections (Lester and Li, 2014) and how little is known of mosquito Toll receptors. In addition, in the context of poly(I:C), it is now known that receptors besides TLR3 can sense poly(I:C) in human cells, such as TLR10 (Lee et al., 2018). It is thus plausible to hypothesize that other Toll receptors, besides Toll6, may be responsible for poly(I:C) sensing in mosquitoes. While we used a similar approach to Zhang et al. in our analysis, we observed differences in Toll clustering (Zhang et al., 2021). In that study, the authors only included a subset of mosquito Toll protein sequences in their analysis and found that invertebrate Toll9s clustered with TLR4 (Zhang et al., 2021). Instead, we included the full-length protein sequences for all Tolls in the species we focused on and were able to add to previous studies, finding that mosquito Toll9s, including Culex spp., clustered with another set of human TLRs (Imler and Zheng, 2004; Kanzok et al., 2004; Waterhouse et al., 2007). Mammalian TLRs, in general, are considered bona fide PRRs. The clustering of insect Toll9s with human TLRs has been proposed to be due to the number of cysteine clusters present on the ectodomains. All mammalian TLRs harbor a single cysteine cluster, while most insect Tolls, besides Toll9, possess multiple cysteine clusters (Imler and Zheng, 2004; Leulier and Lemaitre, 2008; Brennan and Gilmore, 2018). Additionally, only the intracellular TIR domain of insect Toll9 clusters with those of human TLRs, and, based on this clustering, it has been proposed that most insect Tolls likely evolved separately, and their function in immunity has been questioned (Imler and Zheng, 2004). While Toll9 has been shown to be involved in Drosophila immunity (Ooi et al., 2002), no Toll9 receptor has been studied in mosquitoes to our knowledge. It is also worth noting that even between Diptera species, the function of any particular Toll receptor may not be conserved (Lima et al., 2021). However, given the demonstrated importance of the Toll pathway in mosquitoes (Xi et al., 2008), the implication of other Toll genes involved in Drosophila antiviral immunity (Nakamoto et al., 2012), and the effect of CqToll6 knockdown seen here, it is reasonable to think that other mosquito Toll receptors also play a role in immune defense against viruses. It will be important to determine the downstream effects any individual Toll receptors may have in response to viral infection, as it is largely unknown which Tolls feed into the canonical mosquito Toll signaling pathway. It was recently shown that the cytokine-like molecule Spaetzle binds AaToll5a and leads to AMP upregulation (Saucereau et al., 2022). Whether transcription such as this results from any Toll receptor sensing a viral PAMP directly remains to be seen.
It has been shown that AaToll6 is upregulated after poly(I:C) treatment in Aag2 cells (Angleró-Rodríguez et al., 2021). In human cells, poly(I:C) induces the upregulation of TLR3 (Bsibsi et al., 2006; Huang et al., 2006), but also other TLRs, such as TLR2 (Melkamu et al., 2009) and TLR4 (Anabel et al., 2014). In addition to poly(I:C) treatment, it has also been shown in human cells and patients that RNA viruses, including Zika virus, can upregulate TLR3 in a positive-feedback fashion (Tanabe et al., 2003; da Silva et al., 2019). Here, we found that the most upregulation of toll genes was in response to non-transfected poly(I:C) after 1 h of treatment. These included toll6, which complements Angleró-Rodríguez et al. studies in Aag2 cells (Angleró-Rodríguez et al., 2021), and toll8 and toll9A as well. However, toll6 was not upregulated after LACV infection, which is not reminiscent of TLR3 in multiple studies in human cells (Tanabe et al., 2003; da Silva et al., 2019). It is possible that toll6 upregulation occurred at other timepoints post infection that we did not investigate, in particular earlier timepoints post infection. The expression of toll-8 in Drosophila was shown to be upregulated after Sindbis virus infection (Xu et al., 2012). In our study, only poly(I:C), and not LACV infection, triggered the upregulation of toll8 in Hsu cells. This may again be a result of timing or indicate that LACV can antagonize dsRNA sensing and signaling. The fact that toll9A, and to some extent toll9B, were upregulated after poly(I:C) treatment encourages future studies into these genes. The Toll receptor that stands out as being the most upregulated after transfected poly(I:C) treatment was toll10, which was also the only upregulated toll gene after LACV infection in our study. It is important to note that the lack of upregulation of any particular toll receptor does not necessarily indicate a lack of potential signaling through that receptor. Likewise, it is still unclear if any observed toll gene upregulation is the result of positive feedback or the result of independent upstream signaling. In addition, the timing of poly(I:C) induced upregulation of toll genes may not directly correlate with the timing of antiviral poly(I:C) priming. Toll protein stability and turnover in Hsu cells is unknown and short increases in mRNA levels may have longer lasting impacts on Toll protein levels and activity.
One limitation of our study is that we only tested one arbovirus. Other viruses, such as WNV or Usutu virus are relevant to the infection of Culex spp. mosquitoes, and we aim to test the impact of poly(I:C) priming on these viruses in the future. Similarly, studies in mosquitoes in vivo will be required to fully understand the relevance of these dsRNA sensing mechanisms. We found that poly(I:C) stimulation results could be variable between experiments and Toll knockdown experiments were difficult to achieve. In the future, we hope to establish Toll6 knockout cell lines to further study its role during virus replication. Key experiments will require large scale transcriptomic analysis to identify other genes that are upregulated following poly(I:C) treatment.
Overall, our data indicates that poly(I:C) induces an antiviral state in Cx. quinquefasciatus Hsu cells, which correlated with an increase in AMP gene expression, similar to what is found in Aag2 cells. We showed that a putative dsRNA receptor in Cx. quinquefasciatus mosquitoes, Toll6, is antiviral against LACV. We were able to expand and update the phylogeny of Toll receptors in multiple mosquito species and analyzed the expression of all toll genes in response to poly(I:C) and virus infection in Cx. quinquefasciatus cells. While many of our results indicate only modest changes in gene expression and virus replication, this may be a result of our slow-growing cell culture system or the redundancy of multiple immune pathways. However, our work presented here will guide future efforts to uncover the exact role of specific mosquito Toll receptors in the sensing of virus infection and guide in vivo studies into the role of Toll receptors in mosquito antiviral immunity.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
BP performed primary data collection, experimental optimization, data analysis, and manuscript drafting. KC performed Toll expression analysis as shown in Figures 5, 6 and helped edit the manuscript. CR conceived and supervised the project, assisted with data visualization, as well as manuscript drafting and editing. All authors have reviewed, edited, and agreed to the final version of this manuscript.
Funding
KC was supported by the Nevada INBRE Undergraduate Research Opportunity Program made possible by a grant from the National Institute of General Medical Sciences (GM103440) from the National Institutes of Health. KC was also supported by the Nevada Undergraduate Research Award program. No other external funding sources were used for this research.
Acknowledgments
We would also like to thank Dr. Won Yim for bioinformatic help extracting coding sequences from the recently published Culex tarsalis genome. We would further like to thank Brandy Russell and the CDC Arbovirus Reference Collection for providing the LACV isolate. We would also like to thank Tran Zen Belila Torres for his efforts to attempt in vivo poly(I:C) treatments via spiked bloodmeal.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1251204/full#supplementary-material
References
Alexopoulou, L., Holt, A. C., Medzhitov, R., Flavell, R. A. (2001). Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413 (6857), 732–738. doi: 10.1038/35099560
Anabel, A. S., Eduardo, P. C., Pedro Antonio, H. C., Carlos, S. M., Juana, N. M., Honorio, T. A., et al. (2014). Human platelets express Toll-like receptor 3 and respond to poly I:C. Hum. Immunol. 75 (12), 1244–1251. doi: 10.1016/j.humimm.2014.09.013
Angleró-Rodríguez, Y. I., Tikhe, C. V., Kang, S., Dimopoulos, G. (2021). Aedes aEgypti Toll pathway is induced through dsRNA sensing in endosomes. Dev. Comp. Immunol. 122, 104138. doi: 10.1016/j.dci.2021.104138
Asad, S., Parry, R., Asgari, S. (2018). Upregulation of Aedes aEgypti Vago1 by Wolbachia and its effect on dengue virus replication. Insect Biochem. Mol. Biol. 92, 45–52. doi: 10.1016/j.ibmb.2017.11.008
Baldaccini, M., Pfeffer, S. (2021). Untangling the roles of RNA helicases in antiviral innate immunity. PloS Pathog. 17 (12), e1010072. doi: 10.1371/journal.ppat.1010072
Bartholomay, L. C., Waterhouse, R. M., Mayhew, G. F., Campbell, C. L., Michel, K., Zou, Z., et al. (2010). Pathogenomics of Culex quinquefasciatus and meta-analysis of infection responses to diverse pathogens. Science 330 (6000), 88–90. doi: 10.1126/science.1193162
Bell, J. K., Askins, J., Hall, P. R., Davies, D. R., Segal, D. M. (2006). The dsRNA binding site of human Toll-like receptor 3. Proc. Natl. Acad. Sci. U.S.A. 103 (23), 8792–8797. doi: 10.1073/pnas.0603245103
Blair, C. D., Olson, K. E. (2014). Mosquito immune responses to arbovirus infections. Curr. Opin. Insect Sci. 3, 22–29. doi: 10.1016/j.cois.2014.07.005
Borucki, M. K., Kempf, B. J., Blitvich, B. J., Blair, C. D., Beaty, B. J. (2002). La Crosse virus: replication in vertebrate and invertebrate hosts. Microbes Infect. 4 (3), 341–350. doi: 10.1016/s1286-4579(02)01547-2
Brennan, J. J., Gilmore, T. D. (2018). Evolutionary origins of toll-like receptor signaling. Mol. Biol. Evol. 35 (7), 1576–1587. doi: 10.1093/molbev/msy050
Bronkhorst, A. W., van Rij, R. P. (2014). The long and short of antiviral defense: small RNA-based immunity in insects. Curr. Opin. Virol. 7, 19–28. doi: 10.1016/j.coviro.2014.03.010
Brugman, V. A., Hernández-Triana, L. M., Medlock, J. M., Fooks, A. R., Carpenter, S., Johnson, N. (2018). The role of culex pipiens L. (Diptera: culicidae) in virus transmission in europe. Int. J. Environ. Res. Public Health 15 (2), 389. doi: 10.3390/ijerph15020389
Bsibsi, M., Persoon-Deen, C., Verwer, R. W., Meeuwsen, S., Ravid, R., Van Noort, J. M. (2006). Toll-like receptor 3 on adult human astrocytes triggers production of neuroprotective mediators. Glia 53 (7), 688–695. doi: 10.1002/glia.20328
Chen, Y. G., Hur, S. (2022). Cellular origins of dsRNA, their recognition and consequences. Nat. Rev. Mol. Cell Biol. 23 (4), 286–301. doi: 10.1038/s41580-021-00430-1
Chen, Y., Lin, J., Zhao, Y., Ma, X., Yi, H. (2021). Toll-like receptor 3 (TLR3) regulation mechanisms and roles in antiviral innate immune responses. J. Zhejiang Univ Sci. B 22 (8), 609–632. doi: 10.1631/jzus.B2000808
Christophides, G. K., Zdobnov, E., Barillas-Mury, C., Birney, E., Blandin, S., Blass, C., et al. (2002). Immunity-related genes and gene families in Anopheles gambiae. Science 298 (5591), 159–165. doi: 10.1126/science.1077136
Ciota, A. T., Kramer, L. D. (2013). Vector-virus interactions and transmission dynamics of West Nile virus. Viruses 5 (12), 3021–3047. doi: 10.3390/v5123021
Colpitts, T. M., Cox, J., Vanlandingham, D. L., Feitosa, F. M., Cheng, G., Kurscheid, S., et al. (2011). Alterations in the Aedes aEgypti transcriptome during infection with West Nile, dengue and yellow fever viruses. PloS Pathog. 7 (9), e1002189. doi: 10.1371/journal.ppat.1002189
da Silva, M. H. M., Moises, R. N. C., Alves, B. E. B., Pereira, H. W. B., de Paiva, A. A. P., Morais, I. C., et al. (2019). Innate immune response in patients with acute Zika virus infection. Med. Microbiol. Immunol. 208 (6), 703–714. doi: 10.1007/s00430-019-00588-8
Deddouche, S., Matt, N., Budd, A., Mueller, S., Kemp, C., Galiana-Arnoux, D., et al. (2008). The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat. Immunol. 9 (12), 1425–1432. doi: 10.1038/ni.1664
Harris, M. C., Yang, F., Jackson, D. M., Dotseth, E. J., Paulson, S. L., Hawley, D. M. (2015). La crosse virus field detection and vector competence of culex mosquitoes. Am. J. Trop. Med. Hyg. 93 (3), 461–467. doi: 10.4269/ajtmh.14-0128
Hernández-Triana, L. M., Folly, A. J., Sewgobind, S., Lean, F. Z. X., Ackroyd, S., Nuñez, A., et al. (2022). Susceptibility of Aedes albopictus and Culex quinquefasciatus to Japanese encephalitis virus. Parasit Vectors 15 (1), 210. doi: 10.1186/s13071-022-05329-0
Hsu, S. H., Mao, W. H., Cross, J. H. (1970). Establishment of a line of cells derived from ovarian tissue of Clex quinquefasciatus Say. J. Med. Entomol 7 (6), 703–707. doi: 10.1093/jmedent/7.6.703
Huang, C. C., Duffy, K. E., San Mateo, L. R., Amegadzie, B. Y., Sarisky, R. T., Mbow, M. L. (2006). A pathway analysis of poly(I:C)-induced global gene expression change in human peripheral blood mononuclear cells. Physiol. Genomics 26 (2), 125–133. doi: 10.1152/physiolgenomics.00002.2006
Imler, J. L., Zheng, L. (2004). Biology of Toll receptors: lessons from insects and mammals. J. Leukoc. Biol. 75 (1), 18–26. doi: 10.1189/jlb.0403160
Janeway, C. A., Jr., Medzhitov, R. (2002). Innate immune recognition. Annu. Rev. Immunol. 20, 197–216. doi: 10.1146/annurev.immunol.20.083001.084359
Kanzok, S. M., Hoa, N. T., Bonizzoni, M., Luna, C., Huang, Y., Malacrida, A. R., et al. (2004). Origin of Toll-like receptor-mediated innate immunity. J. Mol. Evol. 58 (4), 442–448. doi: 10.1007/s00239-003-2565-8
Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., et al. (2006). Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441 (7089), 101–105. doi: 10.1038/nature04734
Kumar, A., Srivastava, P., Sirisena, P., Dubey, S. K., Kumar, R., Shrinet, J., et al. (2018). Mosquito innate immunity. Insects 9 (3), 95. doi: 10.3390/insects9030095
Laureti, M., Lee, R. X., Bennett, A., Wilson, L. A., Sy, V. E., Kohl, A., et al. (2023). Rift valley fever virus primes immune responses in aedes aEgypti cells. Pathogens 12 (4), 563. doi: 10.3390/pathogens12040563
Lee, S. M., Yip, T. F., Yan, S., Jin, D. Y., Wei, H. L., Guo, R. T., et al. (2018). Recognition of double-stranded RNA and regulation of interferon pathway by toll-like receptor 10. Front. Immunol. 9. doi: 10.3389/fimmu.2018.00516
Lester, S. N., Li, K. (2014). Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 426 (6), 1246–1264. doi: 10.1016/j.jmb.2013.11.024
Leulier, F., Lemaitre, B. (2008). Toll-like receptors–taking an evolutionary approach. Nat. Rev. Genet. 9 (3), 165–178. doi: 10.1038/nrg2303
Lima, L. F., Torres, A. Q., Jardim, R., Mesquita, R. D., Schama, R. (2021). Evolution of Toll, Spatzle and MyD88 in insects: the problem of the Diptera bias. BMC Genomics 22 (1), 562. doi: 10.1186/s12864-021-07886-7
Li, Y. G., Siripanyaphinyo, U., Tumkosit, U., Noranate, N., A-Nuegoonpipat, A., Pan, Y., et al. (2012). Poly (I:C), an agonist of toll-like receptor-3, inhibits replication of the Chikungunya virus in BEAS-2B cells. Virol. J. 9, 114. doi: 10.1186/1743-422X-9-114
Lu, Y., Su, F., Li, Q., Zhang, J., Li, Y., Tang, T., et al. (2020). Pattern recognition receptors in Drosophila immune responses. Dev. Comp. Immunol. 102, 103468. doi: 10.1016/j.dci.2019.103468
Luby, J. P. (1975). Sensitivities of neurotropic arboviruses to human interferon. J. Infect. Dis. 132 (4), 361–367. doi: 10.1093/infdis/132.4.361
Main, B. J., Marcantonio, M., Johnston, J. S., Rasgon, J. L., Brown, C. T., Barker, C. M. (2021). Whole-genome assembly of Culex tarsalis. G3 (Bethesda) 11 (2), jkaa063. doi: 10.1093/g3journal/jkaa063
Matsumoto, M., Seya, T. (2008). TLR3: interferon induction by double-stranded RNA including poly(I:C). Adv. Drug Deliv. Rev. 60 (7), 805–812. doi: 10.1016/j.addr.2007.11.005
Melkamu, T., Squillace, D., Kita, H., O'Grady, S. M. (2009). Regulation of TLR2 expression and function in human airway epithelial cells. J. Membr. Biol. 229 (2), 101–113. doi: 10.1007/s00232-009-9175-3
Monteiro, J. T., Schön, K., Ebbecke, T., Goethe, R., Ruland, J., Baumgärtner, W., et al. (2019). The CARD9-Associated C-Type Lectin, Mincle, Recognizes La Crosse Virus (LACV) but Plays a Limited Role in Early Antiviral Responses against LACV. Viruses 11 (3), 303. doi: 10.3390/v11030303
Nakamoto, M., Moy, R. H., Xu, J., Bambina, S., Yasunaga, A., Shelly, S. S., et al. (2012). Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity 36 (4), 658–667. doi: 10.1016/j.immuni.2012.03.003
Nasirudeen, A. M., Wong, H. H., Thien, P., Xu, S., Lam, K. P., Liu, D. X. (2011). RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PloS Negl. Trop. Dis. 5 (1), e926. doi: 10.1371/journal.pntd.0000926
Ooi, J. Y., Yagi, Y., Hu, X., Ip, Y. T. (2002). The Drosophila Toll-9 activates a constitutive antimicrobial defense. EMBO Rep. 3 (1), 82–87. doi: 10.1093/embo-reports/kvf004
Palchetti, S., Starace, D., De Cesaris, P., Filippini, A., Ziparo, E., Riccioli, A. (2015). Transfected poly(I:C) activates different dsRNA receptors, leading to apoptosis or immunoadjuvant response in androgen-independent prostate cancer cells. J. Biol. Chem. 290 (9), 5470–5483. doi: 10.1074/jbc.M114.601625
Paradkar, P. N., Duchemin, J. B., Voysey, R., Walker, P. J. (2014). Dicer-2-dependent activation of Culex Vago occurs via the TRAF-Rel2 signaling pathway. PloS Negl. Trop. Dis. 8 (4), e2823. doi: 10.1371/journal.pntd.0002823
Paradkar, P. N., Trinidad, L., Voysey, R., Duchemin, J. B., Walker, P. J. (2012). Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. U.S.A. 109 (46), 18915–18920. doi: 10.1073/pnas.1205231109
Paysan-Lafosse, T., Blum, M., Chuguransky, S., Grego, T., Pinto, B. L., Salazar, G. A., et al. (2023). InterPro in 2022. Nucleic Acids Res. 51 (D1), D418–d427. doi: 10.1093/nar/gkac993
Reisen, W. K. (2003). Epidemiology of St. Louis encephalitis virus. Adv. Virus Res. 61, 139–183. doi: 10.1016/s0065-3527(03)61004-3
Russell, T. A., Ayaz, A., Davidson, A. D., Fernandez-Sesma, A., Maringer, K. (2021). Imd pathway-specific immune assays reveal NF-κB stimulation by viral RNA PAMPs in Aedes aEgypti Aag2 cells. PloS Negl. Trop. Dis. 15 (2), e0008524. doi: 10.1371/journal.pntd.0008524
Saucereau, Y., Wilson, T. H., Tang, M. C. K., Moncrieffe, M. C., Hardwick, S. W., Chirgadze, D. Y., et al. (2022). Structure and dynamics of Toll immunoreceptor activation in the mosquito Aedes aEgypti. Nat. Commun. 13 (1), 5110. doi: 10.1038/s41467-022-32690-6
Sharma, K. B., Vrati, S., Kalia, M. (2021). Pathobiology of Japanese encephalitis virus infection. Mol. Aspects Med. 81, 100994. doi: 10.1016/j.mam.2021.100994
Tanabe, M., Kurita-Taniguchi, M., Takeuchi, K., Takeda, M., Ayata, M., Ogura, H., et al. (2003). Mechanism of up-regulation of human Toll-like receptor 3 secondary to infection of measles virus-attenuated strains. Biochem. Biophys. Res. Commun. 311 (1), 39–48. doi: 10.1016/j.bbrc.2003.09.159
Thompson, W. H., Anslow, R. O., Hanson, R. P., Defoliart, G. R. (1972). La Crosse virus isolations from mosquitoes in Wisconsin 1964-68. Am. J. Trop. Med. Hyg. 21 (2), 90–96. doi: 10.4269/ajtmh.1972.21.90
Tikhe, C. V., Dimopoulos, G. (2021). Mosquito antiviral immune pathways. Dev. Comp. Immunol. 116, 103964. doi: 10.1016/j.dci.2020.103964
Tsai, Y. T., Chang, S. Y., Lee, C. N., Kao, C. L. (2009). Human TLR3 recognizes dengue virus and modulates viral replication in vitro. Cell Microbiol. 11 (4), 604–615. doi: 10.1111/j.1462-5822.2008.01277.x
Walsh, E., Torres, T. Z. B., Ruckert, C. (2022). Culex mosquito piwi4 is antiviral against two negative-sense RNA viruses. Viruses 14 (12), 2758. doi: 10.3390/v14122758
Waterhouse, R. M., Kriventseva, E. V., Meister, S., Xi, Z., Alvarez, K. S., Bartholomay, L. C., et al. (2007). Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316 (5832), 1738–1743. doi: 10.1126/science.1139862
Xi, Z., Ramirez, J. L., Dimopoulos, G. (2008). The Aedes aEgypti toll pathway controls dengue virus infection. PloS Pathog. 4 (7), e1000098. doi: 10.1371/journal.ppat.1000098
Xu, J., Grant, G., Sabin, L. R., Gordesky-Gold, B., Yasunaga, A., Tudor, M., et al. (2012). Transcriptional pausing controls a rapid antiviral innate immune response in Drosophila. Cell Host Microbe 12 (4), 531–543. doi: 10.1016/j.chom.2012.08.011
Keywords: mosquito, Culex, immune sensing, dsRNA sensing, Toll, arbovirus, La Crosse virus
Citation: Prince BC, Chan K and Rückert C (2023) Elucidating the role of dsRNA sensing and Toll6 in antiviral responses of Culex quinquefasciatus cells. Front. Cell. Infect. Microbiol. 13:1251204. doi: 10.3389/fcimb.2023.1251204
Received: 01 July 2023; Accepted: 14 August 2023;
Published: 30 August 2023.
Edited by:
Ladislav Simo, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Isabelle Dietrich, The Pirbright Institute, United KingdomKevin Maringer, The Pirbright Institute, United Kingdom
Copyright © 2023 Prince, Chan and Rückert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claudia Rückert, Y3J1ZWNrZXJ0QHVuci5lZHU=
 Brian C. Prince
Brian C. Prince Kalvin Chan
Kalvin Chan Claudia Rückert
Claudia Rückert