
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 17 August 2023
Sec. Molecular Bacterial Pathogenesis
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1236866
 Marta Garcia-Lopez1,2
Marta Garcia-Lopez1,2 Celine Lorioux1
Celine Lorioux1 Anais Soares3
Anais Soares3 Sabine Trombert-Paolantoni4
Sabine Trombert-Paolantoni4 Elena Harran2
Elena Harran2 Florence Ayral2
Florence Ayral2 Mathieu Picardeau1
Mathieu Picardeau1 Zouheira Djelouadji2
Zouheira Djelouadji2 Pascale Bourhy1*
Pascale Bourhy1*Leptospirosis is a bacterial zoonotic disease. Humans and dogs are susceptible hosts, with similar clinical manifestations ranging from a febrile phase to multiple organ dysfunction. The incidence of leptospirosis in mainland France is relatively high, at about 1 case per 100,000 inhabitants, but our knowledge of the strains circulating in humans and dogs remains limited. We studied the polymorphism of the lfb1 gene sequences in an exhaustive database, to facilitate the identification of Leptospira strains. We identified 46 species-groups (SG) encompassing the eight pathogenic species of Leptospira. We sequenced the lfb1 gene amplification products from 170 biological samples collected from 2019 to 2021: 110 from humans and 60 from dogs. Epidemiological data, including vaccination status in dogs, were also collected. Three Leptospira species displaying considerable diversity were identified: L. interrogans, with eight lfb1 species-groups (including five new lfb1 species-groups) in humans and dogs; L. kirschneri, with two lfb1 species-groups in humans and dogs; and L. borgpetersenii, with one lfb1 species-group in humans only. The lfb1 species-group L. interrogans SG1, corresponding to serovar Icterohaemorrhagiae or Copenhageni, was frequently retrieved from both humans and dogs (n=67/110; 60.9% and n=59/60; 98.3% respectively). A high proportion of the affected dogs developed the disease despite vaccination (n=30/60; 50%). Genotyping with the polymorphic lfb1 gene is both robust and simple. This approach provided the first global picture of the Leptospira strains responsible for acute infections in mainland France, based on biological samples but without the need for culture. Identification of the Leptospira strains circulating and their changes over time will facilitate more precise epidemiological monitoring of susceptible and reservoir species. It should also facilitate the monitoring of environmental contamination, making it possible to implement preventive measures and to reduce the burden of this disease.
With an estimated one million cases of severe leptospirosis in humans each year, resulting in 60,000 deaths, leptospirosis is considered one of the commonest zoonoses worldwide. It has been recognized as an emerging global public health problem, because its incidence is increasing in both developing and developed countries (World Health Organization, 2003; Costa et al., 2015; Pijnacker et al., 2016; Bourhy et al., 2017). Leptospirosis is essentially considered a tropical disease, as high temperatures and humid climates favour the survival of the causal bacterium (Evangelista and Coburn, 2010; Costa et al., 2015), and it predominantly affects impoverished populations. Nevertheless, it is also widely reported in temperate areas, including Europe (Goarant, 2016; Pijnacker et al., 2016). Leptospirosis also has a major impact on the health of wild and domestic mammals, and can cause major economic losses in the livestock sector (Noguera et al., 2022). Dogs are particularly susceptible to leptospirosis, and vaccines are available to protect them from disease (Azócar-Aedo et al., 2014; Ellis, 2015).
The genus Leptospira is currently subdivided into 68 genomic species, including saprophytic and pathogenic species. Infections in humans and animals are caused by only eight pathogenic species: L. interrogans, L. kirschneri, L. noguchi, L. santarosai, L. mayottensis, L. borgpetersenii, L. alexanderi and L. weilli (Vincent et al., 2019). Serological classification based on polymorphism of the lipopolysaccharide (LPS) O-antigen has made it possible to identify more than 300 serovars (sv) grouped into 26 serogroups (sg) (Nieves C et al., 2023).
Infection with Leptospira spp. occurs predominantly through contact between abraded skin or mucous membranes and water or soil contaminated with the urine of infected animals, such as rodents, which are the main reservoir of human leptospirosis (Ellis, 2015). The rat (Rattus spp.) serves as a host for the most widespread serovars worldwide, Icterohaemorrhagiae and Copenhageni (Boey et al., 2019). Other studies have demonstrated the existence of several other hosts, such as mice, voles, and hedgehogs, harboring other Leptospira serovars (Ayral et al., 2016; Izquierdo-Rodríguez et al., 2020; Jeske et al., 2021). Moreover, livestock can also be a source of environmental contamination (Zarantonelli et al., 2018; Sykes et al., 2022).
Human leptospirosis is not a notifiable disease in France. The French National Reference Center (FNRC) conducts passive surveillance by compiling biologically confirmed cases through a network of partners (hospitals and diagnostic laboratories). The incidence of leptospirosis in mainland France is one of the highest in Europe, with approximately 1 case per 100,000 inhabitants or more than 600 cases per year (Bourhy et al., 2017). This disease is associated with working in contact with animals, environment-related activities and recreational activities linked to water (Nardone et al., 2004; Guillois et al., 2018; Velardo et al., 2022).
Information about the Leptospira strains circulating in susceptible species is scarce in France and, indeed, in Europe generally. There are several reasons for this. The first one is the laborious nature of Leptospira culture methods, which require fresh biological samples and reference laboratories capable of identifying the strains concerned. Moreover, molecular diagnostic methods, such as Polymerase Chain Reactions (PCR), are gradually supplanting serological tests, which remain the only available epidemiological tool for serogroup identification in Leptospira spp. There is, therefore, a crucial need for a new Leptospira genotyping method suitable for direct use on biological samples that is sensitive, discriminant, simple to implement and inexpensive. Identification of the strains in circulation is essential for diagnosis (to ensure that the diagnostic methods used are capable of detecting these strains), epidemiology (to characterise the reservoirs), surveillance (to detect the occurrence of new genotypes) and prevention (to evaluate the efficacy of vaccines and for the development of new vaccines).
Epidemiological studies of circulating strains are difficult to implement because of the lack of clinical isolates. Pathogenic Leptospira strains are slow-growing bacteria that can be grown only on complex culture media. Current knowledge about the epidemiology of leptospirosis is based on serological results obtained with the reference microscopic agglutination test (MAT), which can be used to identify the infecting serogroup (Faine and Stallman, 1982; Picardeau, 2020). However, this technique has known inconsistencies and weaknesses (Kusum et al., 2005; Smythe et al., 2009). Molecular techniques for studying Leptospira epidemiology have been described, including a core-genome MLST scheme (cgMLST) for isolates (Guglielmini et al., 2019). In the absence of isolates, several alternative methods are available for genotyping, including multispacer sequence typing or MST (Zilber et al., 2014), variable number tandem repeat or VNTR methods (Salaün et al., 2006), and multilocus sequence typing or MLST (Ahmed et al., 2006). Various genes, including the 16S rRNA, lipL32, secY, lfb1, and lic12008 genes can also be sequenced for the direct genotyping of strains present in biological samples without the need for isolation in culture (Merien et al., 2005; Marquez et al., 2017; Santos et al., 2018).
Lfb1 is a putative adhesin of the “fibronectin-binding protein” family (Merien et al., 2000) and lfb1 gene sequences have been shown to be congruent with the genomic species classification of pathogenic Leptospira strains (Merien et al., 2005). Perez et al. showed that the lfb1 locus displayed greater phylogenetic polymorphism than the 16S rRNA gene, making it possible to identify strains down to subspecies level (Perez and Goarant, 2010). This identification method has since been used in numerous epidemiological studies in humans and/or animals (Grégoire et al., 2020; Soupé-Gilbert et al., 2022).
The FNRC has an exhaustive database of sequences of strains from many regions of the world and from a wide range of hosts. We used this database to evaluate the lfb1 marker for large-scale use for identification purposes. The objectives of our study were to validate the robustness of the lfb1 method for detecting and genotyping Leptospira strains directly from DNA and to propose a new classification system. We then applied this method to the genetic characterization of strains circulating in humans and dogs with acute leptospirosis in mainland France from 2019 to 2021. This work is completed to describe the incidence and geographical distribution in humans, and also the vaccination status in dogs.
lfb1 sequences were extracted from the genomes of pathogenic species from the Leptospira cgMLST database (https://bigsdb.pasteur.fr/leptospira/). This publicly available web-based database currently contains sequences from 834 pathogenic Leptospira strains from different hosts around the world. A clonal group (CG) is defined as a group of cgMLST allelic profiles differing from at least one other member of the group by no more than 40 allelic mismatches over the 545 genetic loci (Guglielmini et al., 2019).
A phylogenetic tree was generated with BioNumerics V7.6 (Applied-Maths, Saint-Martens-Latem, Belgium). The 46 lfb1 sequence strains for the production of the phylogenetic tree are summarized in Table 1. The lfb1 nucleotide sequences have been deposited in Genbank under accession number OR101259 - OR101474.
PCR-positive samples collected from humans and dogs in mainland France between January 2019 and December 2021 were included in this study. Cases imported from French overseas territories were excluded from the study.
The samples studied were obtained from i) the routine diagnosis of human samples by real-time PCR (RT-PCR) SYBR-Green targeting lfb1 (Merien et al., 2005), which is performed at the FNRC for Leptospirosis at the Pasteur Institute, and ii) dogs testing positive in routine diagnostic RT-PCR targeting the 16S rRNA (rrs) gene of pathogenic Leptospira strains (Waggoner et al., 2014) at the Laboratory of Leptospira and Veterinary Analysis (LAV) at VetAgro Sup (the Veterinary School of Lyon, France).
The human samples and associated data were collected and used in the framework of the surveillance activities of the FNRC for Leptospirosis. These activities are performed in accordance with the mandate awarded to the FNRC by the French Ministry of Health and the French Public Health Code. Dog samples and associated data were collected and sent at by veterinarians from across the country in the context of leptospirosis suspicion. The confirmatory tests were performed by the LAV.
Associated clinical and epidemiological data are entered by clinicians on a document accompanying the samples. These data are often scarce, particularly for human infections. They included the sex and age of the human patient or dog, type of sample, sampling date, geographic information (zip code and region), and vaccination status for dogs. In the absence of geographic data for the patients/dogs, the address of the laboratory or the veterinary clinic was used assuming that the patients/dogs were exposed in the same region.
A case was defined as an individual resident in mainland France at the time of infection with clinical findings suggestive of leptospirosis and either a Microscopic Agglutination Test (MAT) titer ≥100 for at least one pathogenic serovar or other laboratory results indicative of leptospirosis (ELISA for IgM, PCR or culture). The panel of strains used for MAT included the serovars Australis, Autumnalis, Bataviae, Canicola, Castellonis, Copenhageni, Cynopteri, Djasiman, Grippotyphosa, Hardjo, Hebdomadis, Icterohaemorrhagiae, Javanica, Louisiana, Mini, Panama, Pomona, Pyrogenes, Sarmi, Sejroe, Shermani and Tarassovi. The infecting serogroup was determined based on the serovar with the highest titer. If the highest titer was recorded for several different serovars, the serogroup was considered to be undetermined. The incidence of annual leptospirosis in humans was calculated with the French population data for 2019, 2020 and 2021 obtained from the National Institute of Statistics and Economic Studies (INSEE: https://www.insee.fr/).
The map of the estimated mean annual incidence of leptospirosis in humans by region and the distribution of the different infecting species-groups were performed with R Studio version 2022.12.0 software, produced with the GeoJSON and Scatter Pie Plot functions. Background map was extracted from https://www.data.gouv.fr/fr/datasets/contours-des-regions-francaises-sur-openstreetmap/and centroid data was extracted from https://www.ign.fr/reperes/centre-geographique-des-regions-metropolitaines.
The lfb1 genotyping method was applied to 170 clinical samples: 110 DNA samples from humans and 60 from dogs testing positive by RT-PCR for lfb1 and 16S rRNA, respectively (Merien et al., 2005; Waggoner et al., 2014). The human DNA samples were obtained from 60 blood samples, 17 urine samples, 4 cerebrospinal fluid samples and 29 DNA extracts from associated laboratories. The dog DNA samples were extracted from 34 blood samples, 21 urine samples and 5 kidney tissue samples.
For both humans and dogs, DNA was extracted with the QIAamp DNA mini kit (Qiagen, Germany), and PCR was performed as described by Merien et al. (2005). The CFX96 real-time PCR detection system (Bio-Rad) was used for qPCR SYBR green assays. The amplification mixture consisted of 0.4 μM primers (lfb1-F 3’-CATTCATGTTTCGAATCATTTCAAA-5’ and lfb1-R 3’-GGCCCAAGTTCCTTCTAAAAG-5’), 10 μl of SsoFast EvaGreen supermix (Bio-Rad), and 5 μl sample DNA in a total volume of 20 μl. Samples were amplified with the following program: initial denaturation at 98°C for 2 min, followed by 50 cycles of denaturation for 5 s at 98°C and annealing/elongation for 30 s at 57°C. A 331-bp fragment was amplified corresponding to the complete gene lfb1 for L.interrogans serovar Icterohaemorrhagiae.
All positive PCR products were subjected to Sanger sequencing (Eurofins Scientific, Colonia, Germany and Genoscreen, Lille, France).
We selected a total of 834 genomes of pathogenic Leptospira isolates (corresponding to 227 different cgMLST clonal groups or CGs), from which lfb1 sequences were extracted for phylogenetic analysis (Table 1). A single-nucleotide polymorphism (SNP) in the alignment between two sequences was considered significant for the creation of Leptospira species-groups (SGs).
The selected genomes belong to the eight pathogenic Leptospira species and the analysis of a 334 bp fragment from lfb1 identified 46 different Leptospira SGs, distributed as follows: 13 groups for L. interrogans, corresponding to 76 CGs, 5 groups for L. kirschneri (28 CGs), 7 groups for L. noguchii (21 CGs), 6 for L. borgpetersenii (21 CGs), 2 groups for L. alexanderi (2 CGs), 3 groups for L. mayottensis (3 CGs), 4 groups for L. weilli (23 CGs), and 6 groups for L. santarosai (53 CGs) (Figure 1).
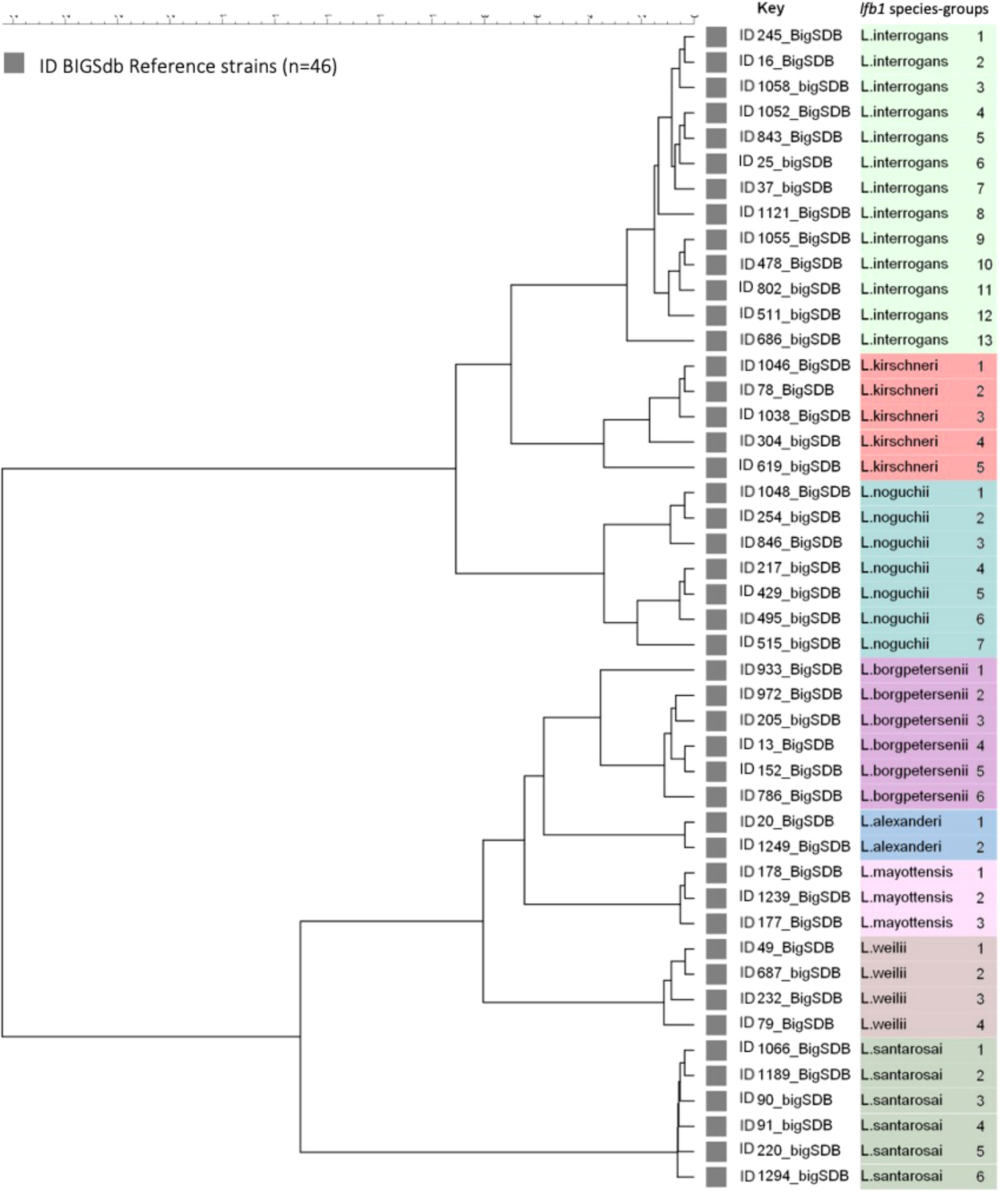
Figure 1 Maximum likelihood tree inferred from a Leptospira spp. Ifb 1 partial gene in 46 reference strains. The ID BIGSdb accession numbers are indicated for the reference strains. Further information about the reference strains is provided in Table 1.
From 2019 to 2021, we collected 110 samples from humans and 60 samples from dogs that had tested positive for leptospirosis by RT-PCR. The lfb1 gene was sequenced from these samples. The sequences obtained and compiled with the classification data proposed above (Figure 1) revealed considerable genetic diversity in samples from humans and dogs, which could be subdivided into three Leptospira species: L. interrogans, L. kirschneri and L. borgpetersenii, corresponding to a total of 11 SGs. We detected L. interrogans in 67/110 (60.9%) of humans and in 59/60 (98.3%) of dog samples, and L. kirschneri in 39/110 (35.5%) of humans and 1/60 (1.7%) in dog samples. L. borgpetersenii was detected only in 4/110 (3.6%) of human samples.
Based on the classification established above, we detected L. interrogans with eight lfb1 SGs and L. kirschneri with two lfb1 SGs in humans and dogs; L. borgpetersenii was represented by a single lfb1 SG found exclusively in humans. More detailed information is provided in Table 2.
The lfb1 phylogenetic analyses of the distribution Leptospira strains from human and dog samples are shown in Figure 2. In human samples, L. interrogans SG1 (sv Icterohaemorrhagiae/Copenhageni) was detected in 53/110 (48.2%), L. interrogans SG5 (sv Bratislava/Lora/Jalna/Muenchen/Bataviae) in 9/110 (8.2%), L. interrogans SG10 (sv Canicola/Pomona) in 2/110 (1.8%), L. kirschneri SG1 (sv Grippotyphosa/Valbuzzi) in 34/110 (30.9%), L. kirschneri SG4 (sv Tsaratsovo) in 5/110 (4.5%), and L. borgpetersenii SG1 (sv Sejroë/Ballum/Castellonis/Mini) in 4/110 (3.6%) samples. Three new L. interrogans SGs were detected in 3/110 (2.7%) samples: SG15, SG17 and SG18. In patients, the diversity SGs were retrieved in all types of biological matrices (blood, urine and CSF) with no particular tropism for an organ, such as the kidney or the brain. For example, the four CSF extracts were identified as L. interrogans SG1 (n=1), L. interrogans SG10 (n=1) and L. kirschneri SG1 (n=2).
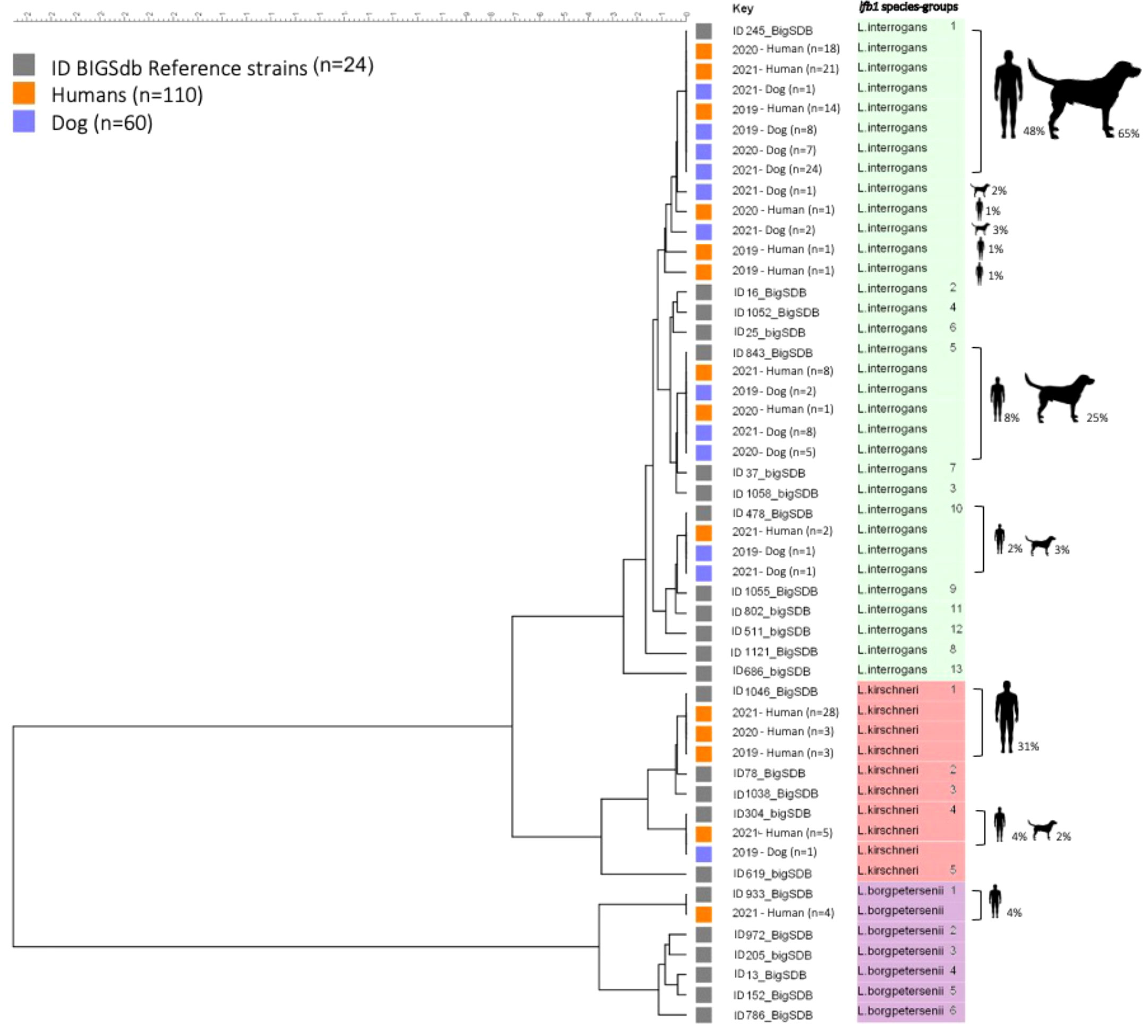
Figure 2 Phylogenetic tree based on fbl 1 sequences from human and dog samples. Maximum likelihood tree inferred from Leptospira spp. lfb 1 partial gene polymorphism in clinical specimens and reference strains. Gray boxes indicate reference strains, orange boxes indicate infected humans and blue boxes indicate infected dogs. The colors on the right (green, red, and purple) correspond to three different species of Leptospira. The ID BIGSdb numbers of the reference strains are indicated. Further information about the reference strains is provided in Table 2.
L. interrogans SG1 (sv Icterohaemorrhagiae/Copenhageni) was the most frequently detected SG in dog samples, being found in 39/60 samples (65.0%), followed by L. interrogans SG5 (sv Bratislava/Lora/Jalna/Muenchen/Bataviae) in 15/60 (25.0%), L. interrogans SG10 (sv Canicola/Pomona) in 2/60 (3.3%) and L. kirschneri SG4 (sv Tsaratsovo) in 1/60 (1.7%). Two new L. interrogans SGs, SG14 and SG16, were identified in 3/60 (5.0%) samples.
676 human cases in 2019, 450 human cases in 2020, and 708 human cases in 2021 of leptospirosis were diagnosed clinically and biologically in mainland France, corresponding to an average incidence per 100,000 inhabitants of 1.04 for 2019, 0.69 for 2020, and 1.08 for 2021 (Supplementary Table S1). The number of cases clearly increased from July to November in 2019 and 2021, and increased to a lesser extent over this period in 2020 (Picardeau, 2020). Detailed information is available from Supplementary Figure S1. For cases with positive MAT results, the most frequently identified serogroup was Icterohaemorrhagiae (34.4%). More detailed information is provided in Supplementary Figure S2.
Over the study period, the number of human specimens analyzed was highest for 2021 (68 cases, 61.8%), followed by 2020 (23 cases) and 2019 (19 cases). In dogs, the number of cases was highest in 2021 (36 cases, 60%), followed by 2020 (12 cases) and 2019 (12 cases). More detailed information is provided in Table 3.
The month with the largest number of notifications of leptospirosis in humans was August with an average number of 30 cases (27.3%) whereas the numbers of notifications for dogs were highest in July and September, with 11 cases in each of these months (18.3%). For human samples, a male preponderance was observed, with 96/110 (87.3%) of cases obtained from male individuals vs. 14/110 (12.7%) from female individuals. More than four fifths (89%) of the patients were adults and the median age was 49 years (5–84 years). A male preponderance was also observed in dogs, with 66.7% of cases (40/60) occurring in male dogs vs. 33.3% (20/60) in females. Just over half the dogs were adults (56.7%) and the median age was four years (0-13 years) (Table 3).
The predominant SG in humans was L. interrogans SG1 - Icterohaemorrhagiae/Copenhageni in the 13 regions of mainland France (Figure 3). The genotypes circulating in human cases were identified in 34 departments (an administrative area equivalent to a county) and 12 regions. The largest number of cases in a region was recorded for Auvergne-Rhône-Alpes in 2021 (Ayral et al., 2016), followed by Pays de la Loire in 2020 (Ellis, 2015), and Bourgogne-Franche-Comté in 2019 (Goarant, 2016). The genotypes in canine cases were identified in 30 departments and 10 regions, with Auvergne-Rhône-Alpes having the largest number of cases in both 2021 (Ayral et al., 2016) and 2020 (Pijnacker et al., 2016). More detailed information is provided in Supplementary Table S2.
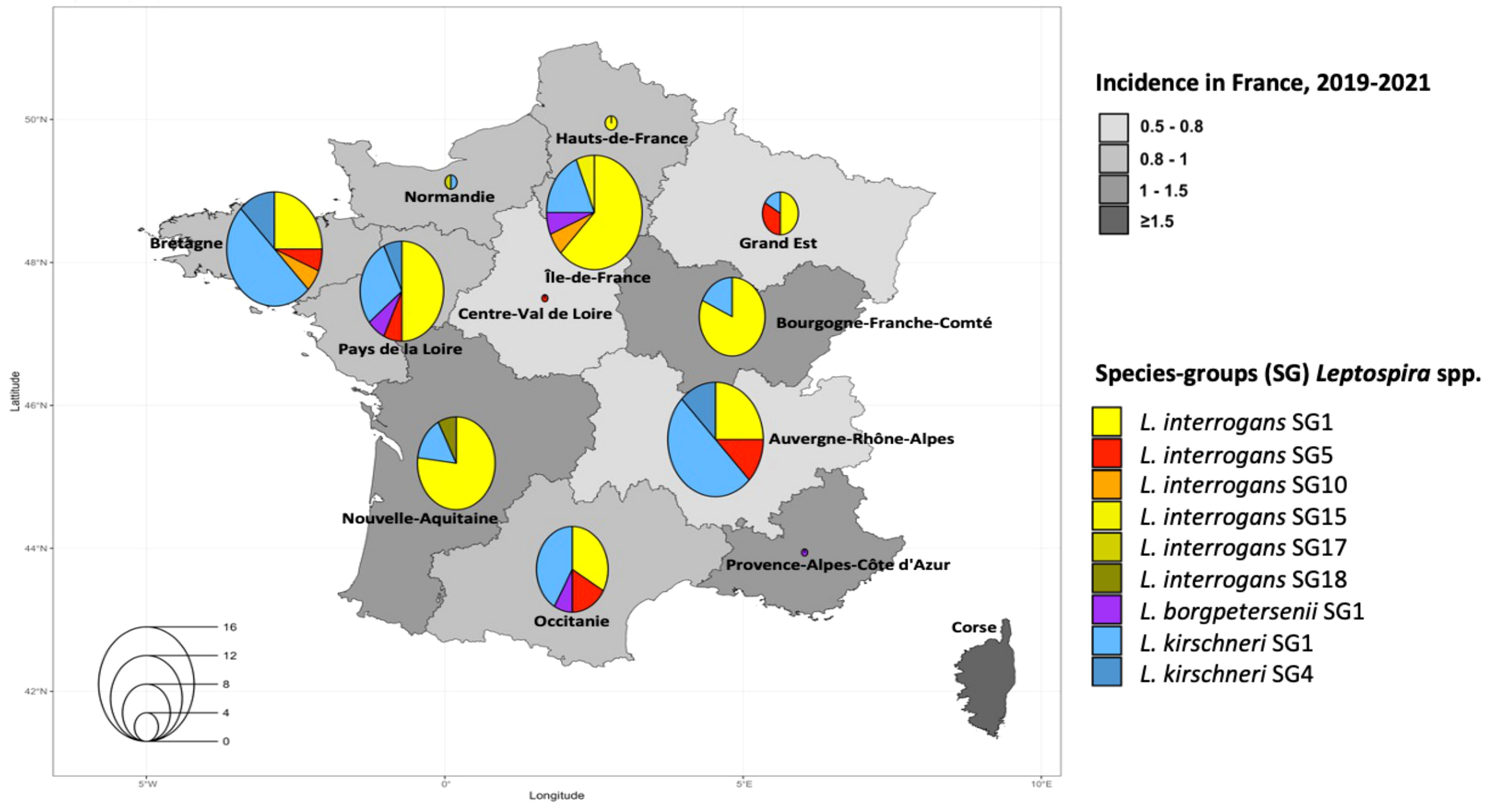
Figure 3 Estimated three-year mean annual human incidence of leptospirosis by region in mainland France (2019-2021). Mean annual incidence is represented as an exponential color gradient from light grey (0.5-0.8) to dark gray (≥1.5), in cases per 100,000 population. Circles indicate the distribution of the different infecting species-groups, identified in 110 humans from our study.
The human population included in this study had not been vaccinated against Leptospira spp. By contrast, 50% (n=30/60) of the infected dogs included in this study had completed the full vaccination protocol for Leptospira. The dogs had mostly been vaccinated with L4 vaccines (63.3% of dogs; vaccine active against serovars Icterohaemorrhagiae, Canicola, Grippotyphosa, and Pomona), L3 vaccines (20%, serovars Icterohaemorrhagiae, Canicola and Grippotyphosa), or L2 vaccines (16.7%, serovars Icterohaemorrhagiae and Canicola). More detailed information is provided in Supplementary Table S3.
The lfb1 gene is found only in pathogenic species of the P1 clade (Vincent et al., 2019). It encodes a putative adhesin of the “fibronectin-binding protein” family (Merien et al., 2000), suggesting that it may be important for virulence and/or colonization. This gene appears to have changed little during evolution, and is a potentially interesting marker for studies of the genetic diversity of pathogenic strains. Comparative analyses of the lfb1 sequences extracted from available Leptospira genomes can be used to define different clusters or species-groups differing by between 1 and 74 nucleotides over a total length of 281 nucleotides. We found that 119 lfb1 sequences from strains belonging to serovars Icterohaemorrhagiae or Copenhageni isolated from all continents and corresponding to L. interrogans SG1 (cgMLST 6) had identical lfb1 sequences. A single SNP may, therefore, be sufficient to define a new species-group. However, some lfb1 species-groups may contain several serovars or clonal groups, precluding precise identification. For example, L. interrogans SG2 is found in 32 genomes corresponding to different CGs (9, 10, 30, 39, 76, 77, 81 and 400) and different serovars. In this case, identification is less precise and may be a limitation of the proposed method.
The diversity of reservoir hosts for leptospirosis is an important parameter to be taken into account when considering the evolution of Leptospira spp. All these arguments suggest that this gene should be a robust phylogenetic marker.
SG diversity was greatest in the species L. interrogans (n=13), for which five new SGs were identified in human or dog samples. L. interrogans is isolated more frequently from both humans and animals than the other pathogenic species. The discovery of new SGs was unexpected and suggests that our knowledge of the strains responsible for disease remains incomplete. The Ifb1 genogroups circulating in humans and dogs were identical for L. interrogans SG1/SG5/SG10, with a strong representation of L. interrogans SG1 (sv Icterohaemorrhagiae or Copenhageni), which is responsible for the most severe forms and for which the predominant reservoir in France is the rat (Boey et al., 2019).
The genogroups of species L. kirschneri were frequent in humans and displayed very little diversity (L. kirschneri SG1 and SG4) probably because the L. kirschneri SG1 group contains many different serovars/cgMLST, rendering high-resolution strain discrimination impossible. Similarly, L. interrogans SG5 and L. borgpetersenii SG1 may correspond to several different clonal groups or serovars. The combination of this approach with more refined tools, such as MLST and VNTR, might facilitate more precise identification in such cases. However, our previous analysis of the core genome of strains isolated from several patients (Grillova et al., 2023) showed that strains belonging to cgMLST CG64 (n=10/12) corresponding to L. kirschneri sv Grippotyphosa sg Grippotyphosa (L. kirschneri SG1) and to cgMLST CG72 corresponding to L. borgpetersenii sg Sejroe sv Sejroe predominated in France. The L. kirschneri SG1 genogroup was not detected in any of the dog samples, possibly due to the low level of exposure of dogs to infected environments or a lower susceptibility to L. kirschneri SG1 strains. Indeed, dogs with few symptoms are not presented to veterinarians and recover spontaneously. The L. kirschneri SG4 group was represented by only two strains isolated from a mouse in Bulgaria (identified as serovar Tsaratsovo sg Pomona) and a strain isolated from a French patient in 1990. Samples corresponding to this group were identified only in 2021, and were obtained from five human patients and one dog. The number of samples analyzed was much larger in 2021 than in 2019 and 2020, and this may have made it possible to identify less frequent genogroups.
L. borgpetersenii SG1 was found exclusively in samples from four patients in 2021. It was not found in dogs. It is possible that the human patients were contaminated by cattle, mice or bats, as previously suggested (Grégoire et al., 2020).
Our data indicate that leptospirosis is common in dogs in mainland France. L. interrogans SG1 (sv Icterohaemorrhagiae/Copenhageni) and L. interrogans SG5 (sv Bratislava/Lora/Jalna/Muenchen/Bataviae) predominate in our study and were clearly linked to clinical leptospirosis, whereas L. kirschneri infections may be associated with mild clinical symptoms or possibly be linked to asymptomatic carriage (Renaud et al., 2013; Goy-Thollot et al., 2018). The genotype distribution in dogs were consistent with the findings of previous studies in France (Renaud et al., 2013; Ayral et al., 2014; André-Fontaine and Triger, 2018; Hidalgo Friaz et al., 2023). Prevention is the best way to protect dogs from leptospirosis. It can be achieved through vaccination, avoiding contact with contaminated water and infected animals. In this study, the human patients were not vaccinated, because vaccination against leptospirosis is reserved for individuals in certain high-risk professions in France. By contrast, the leptospirosis vaccine is one of the recommended the core vaccines for dogs in France. Nevertheless, 50% of dogs included in this study were diagnosed with leptospirosis despite complete vaccination annually. Several studies have reported similar results, demonstrating the relative nature of the efficacy of current vaccines in France (Ayral et al., 2014; André-Fontaine and Triger, 2018; Bertasio et al., 2020). However, quadrivalent vaccines (L4, directed against serogroups Canicola, Icterohaemorrhagiae, Grippotyphosa and Australis) would be a more appropriate vaccine for preventing severe forms and deaths from leptospirosis (Klaasen et al., 2022; Hidalgo Friaz et al., 2023), provided that vaccination protocols are properly respected by owners and veterinarians.
The incidence of leptospirosis in humans over the three years of the study was about 1 per 100,000 inhabitants, but was lower, at only 0.69 per 100,000 inhabitants, in 2020. This decrease is probably related to the COVID-19 pandemic, during which recreational activities were limited and the number of imported cases was decreased by travel restrictions. Similarly, little seasonality was observed in 2020.
Incidence was highest in regions with livestock production, high rainfall, and large rivers. However, some fluctuation was observed from year to year and the data may not necessarily reflect the situation on the ground at a particular time point. For example, people may become infected during their summer vacation but are not diagnosed until they return to their home region.
The French Ministry of Health plans to declare leptospirosis to be a notifiable disease in the near future. This initiative will improve the follow-up of cases and provide associated epidemiological data (number of cases, disease severity, site of exposure, mode of contamination). The FNRC will play an important role in confirming the biological diagnosis and tracking the serogroups/genogroups involved in these infections. The follow-up of serogroups based on MAT shows that Icterohaemorrhagiae is the most frequently encountered serogroup, consistent with the results of Ifb1 genotyping. However, very little information is available for the other serogroups. The regional distribution of origin of samples and of the lfb1 genogroups to identify in mainland France appears to be generally homogeneous for human infection, but not for dogs, for which samples were mostly obtained in the Rhone-Alpes region, in which the LAV laboratory location. More regular surveillance of canine cases in different French regions over a period of several years would be useful as it would provide a more precise idea of the Leptospira genotypes circulating in this species, and would make it possible to establish the epidemiological basis of transmission between humans and dogs. Such surveillance would also make it possible to update preventive measures in humans and animals.
We provide here the first global description of the Leptospira strains responsible for acute infections in humans and dogs in mainland France. We demonstrate that polymorphism of the lfb1 gene is a robust method to provide rapid identification using biological samples. This tool has enabled us to also identify five lfb1 L. interrogans species-groups never before described. The availability of more precise epidemiological data in the future should facilitate the identification of sources of animal and environmental contamination, making it possible to establish public health control interventions.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OR101259 - OR101474.
MG-L: Data curation and analyses. CL, EH: Data curation. AS, ST-P: Samples for diagnostic. FA: Data analysis and review. PB contributed to conception and design of the study and data analyses. MG-L, ZD, MP, PB contributed to the writing and editing of manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This study was supported and financed by the Pasteur Institute of Paris and the Veterinary Analysis Laboratory (LAV) at VetAgro Sup, Lyon, France.
We would like to thank the sampling department of Santé Pulique France for providing the laboratory with samples from hospitals. We thank the staff of the Reference Center for Leptospirosis at the Pasteur Institute, Farida Zinini, Jean-François Mariet and Vallier Sordoillet, for processing the human samples. We also thank the Laboratoire des analyses vétérinaires (LAV) at VetAgro Sup, and Marine Le Guyader, Elisa Boissy and Angeli Kodjo in particular, for providing the samples from dogs.
The authors declare that they have no known competing financial interests or personal relationships that could appear to influence the work reported in this paper.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1236866/full#supplementary-material
Ahmed, N., Devi, S. M., Valverde M de los, A., Vijayachari, P., Machang’u, R. S., Ellis, W. A., et al. (2006). Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrob. 5, 28. doi: 10.1186/1476-0711-5-28
André-Fontaine, G., Triger, L. (2018). MAT cross-reactions or vaccine cross-protection: retrospective study of 863 leptospirosis canine cases. Heliyon [Internet] 4 (11), e00869. doi: 10.1016/j.heliyon.2018.e00869
Ayral, F. C., Bicout, D. J., Pereira, H., Artois, M., Kodjo, A. (2014). Distribution of leptospira serogroups in cattle herds and dogs in France. Am. J. Trop. Med. Hygiene 91 (4), 756. doi: 10.4269/ajtmh.13-0416
Ayral, F., Djelouadji, Z., Raton, V., Zilber, A. L., Gasqui, P., Faure, E., et al. (2016). Hedgehogs and mustelid species: major carriers of pathogenic leptospira, a survey in 28 animal species in France (20122015). PloS One 11 (9), e0162549. doi: 10.1371/journal.pone.0162549
Azócar-Aedo, L., Smits, H. L., Monti, G. (2014). Leptospirosis in dogs and cats: epidemiology, clinical disease, zoonotic implications and prevention. Archivos med. veterinaria 46 (3), 337–348. doi: 10.4067/S0301-732X2014000300002
Bertasio, C., Boniotti, M. B., Lucchese, L., Ceglie, L., Bellinati, L., Mazzucato, M., et al. (2020). Detection of new leptospira genotypes infecting symptomatic dogs: is a new vaccine formulation needed? Pathogens 9 (6), 484. doi: 10.3390/pathogens9060484
Boey, K., Shiokawa, K., Rajeev, S. (2019). Leptospira infection in rats: A literature review of global prevalence and distribution. PloS Negl. Trop. Dis. 13 (8), e0007499. doi: 10.1371/journal.pntd.0007499
Bourhy, P., Septfons, A., Picardeau, M. (2017). Diagnostic, surveillance et épidémiologie de la leptospirose en France. Numéro thématique. La leptospirose dans les régions et départements français d’outre-mer. Bull. Epidemiol. Hebd 8, 131–137. doi: 10.1016/j.medmal.2017.03.364
Costa, F., Hagan, J. E., Calcagno, J., Kane, M., Torgerson, P., Martinez-Silveira, M. S., et al. (2015). Global morbidity and mortality of leptospirosis: A systematic review. PloS Negl. Trop. Dis. 9 (9), e0003898. doi: 10.1371/journal.pntd.0003898
Ellis, W. A. (2015). Animal leptospirosis. Curr. Top. Microbiol. Immunol. 387, 99–137. doi: 10.1007/978-3-662-45059-8_6
Evangelista, K. V., Coburn, J. (2010). Leptospira as an emerging pathogen: a review of its biology, pathogenesis and host immune responses. Future Microbiol. 5 (9), 1413–1425. doi: 10.2217/fmb.10.102
Faine, S., Stallman, N. D. (1982). Amended Descriptions of the Genus Leptospira Noguchi 1917 and the Species L. interrogans (Stimson 1907) Wenyon 1926 and L. biflexa (Wolbach and Binger 1914) Noguchi 1918. Int. J. Sys. Evol. Microbiol. 32 (4), 461–463.
Goarant, C. (2016). Leptospirosis: risk factors and management challenges in developing countries. Res. Rep. Trop. Med. 7, 49–62. doi: 10.2147/RRTM.S102543
Goy-Thollot, I., Djelouadji, Z., Nennig, M., Hazart, G., Hugonnard, M. (2018). Screening for Leptospira DNA in blood and urine from 30 apparently healthy dogs. Rev. Vétérinaire Clinique 53 (3), 79–86. doi: 10.1016/j.anicom.2018.06.003
Grégoire, F., Bakinahe, R., Petitjean, T., Boarbi, S., Delooz, L., Fretin, D., et al. (2020). Laboratory diagnosis of bovine abortions caused by non-maintenance pathogenic leptospira spp.: necropsy, serology and molecular study out of a Belgian experience. Pathogens 9 (6), 413. doi: 10.3390/pathogens9060413
Grillova, L., Cokelaer, T., Mariet, J. F., da Fonseca, J. P., Picardeau, M. (2023). Core genome sequencing and genotyping of Leptospira interrogans in clinical samples by target capture sequencing. BMC Infect. Dis. 23 (1), 157. doi: 10.1186/s12879-023-08126-x
Guglielmini, J., Bourhy, P., Schiettekatte, O., Zinini, F., Brisse, S., Picardeau, M. (2019). Genus-wide Leptospira core genome multilocus sequence typing for strain taxonomy and global surveillance. PloS Negl. Trop. Dis. 13 (4), e0007374. doi: 10.1371/journal.pntd.0007374
Guillois, Y., Bourhy, P., Ayral, F., Pivette, M., Decors, A., Aranda Grau, J. H., et al. (2018). An outbreak of leptospirosis among kayakers in Brittany, North-West France, 2016. Euro Surveill 23 (48), 1700848. doi: 10.2807/1560-7917.ES.2018.23.48.1700848
Hidalgo Friaz, M., Barthélemy, A., Savoie, P., Freyburger, L., Hugonnard, M. (2023). Vaccination contre la leptospirose canine en France : enquête sur les pratiques vétérinaires et leurs motivations. Rev. Vétérinaire Clinique 58 (1), 1–11. doi: 10.1016/j.anicom.2022.12.001
Izquierdo-Rodríguez, E., Fernández-Álvarez, Á, Martín-Carrillo, N., Marchand, B., Feliu, C., Miquel, J., et al. (2020). Pathogenic Leptospira species in rodents from Corsica (France). PloS One 15 (6), e0233776. doi: 10.1371/journal.pone.0233776
Jeske, K., Jacob, J., Drewes, S., Pfeffer, M., Heckel, G., Ulrich, R. G., et al. (2021). Hantavirus–Leptospira coinfections in small mammals from central Germany. Epidemiol. Infect. 149, e97. doi: 10.1017/S0950268821000443
Klaasen, H. L. B. M., van der Veen, M., Dorrestein-Spierenburg, C. M., Cao, Q. (2022). An assessment and comparison of the efficacy of two licensed tetravalent leptospira vaccines for dogs using an improved challenge model. Vaccines (Basel) 10 (9), 1472. doi: 10.3390/vaccines10091472
Kusum, M., Boonsarthorn, N., Biaklang, M., Sina, U., Sawanpanyalert, P., Naigowit, P. (2005). Comparison of leptospiral serovars identification by serology and cultivation in northeastern region, Thailand. J. Med. Assoc. Thai 88 (8), 1098–1102.
Marquez, A., Djelouadji, Z., Lattard, V., Kodjo, A. (2017). Overview of laboratory methods to diagnose Leptospirosis and to identify and to type leptospires. Int. Microbiol. Off. J. Spanish Soc. Microbiol. 20), 184–193. doi: 10.2436/20.1501.01.302
Merien, F., Portnoi, D., Bourhy, P., Charavay, F., Berlioz-Arthaud, A., Baranton, G. (2005). A rapid and quantitative method for the detection of Leptospira species in human leptospirosis. FEMS Microbiol. Lett. 249 (1), 139–147. doi: 10.1016/j.femsle.2005.06.011
Merien, F., Truccolo, J., Baranton, G., Perolat, P. (2000). Identification of a 36-kDa fibronectin-binding protein expressed by a virulent variant of Leptospira interrogans serovar icterohaemorrhagiae. FEMS Microbiol. Lett. 185 (1), 17–22. doi: 10.1111/j.1574-6968.2000.tb09034.x
Nardone, A., Capek, I., Baranton, G., Campèse, C., Postic, D., Vaillant, V., et al. (2004). Risk factors for leptospirosis in metropolitan France: results of a national case-control study, 1999-2000. Clin. Infect. Dis. 39 (5), 751–753. doi: 10.1086/423272
Nieves C, G., Huete, S., Veyrier, F., Picardeau, M. (2023). “Taxonomy and Phylogenomics of Leptospira,” in Phylogenomics: Foundations, Methods, and Pathogen Analysis. Eds. Egor Shitikov, E., Moksrousov, I. (Cambridge, Massachussets: Academic Press).
Noguera, Z. L. P., Charypkhan, D., Hartnack, S., Torgerson, P. R., Rüegg, S. R. (2022). The dual burden of animal and human zoonoses: A systematic review. PloS Negl. Trop. Dis. 16 (10), e0010540. doi: 10.1371/journal.pntd.0010540
Perez, J., Goarant, C. (2010). Rapid Leptospira identification by direct sequencing of the diagnostic PCR products in New Caledonia. BMC Microbiol. 10, 325. doi: 10.1186/1471-2180-10-325
Picardeau, M. (2020) Rapports d’activité du CNR de la Leptospirose. Available at: https://www.pasteur.fr/fr/sante-publique/CNR/les-cnr/leptospirose/rapports-d-activite.
Pijnacker, R., Goris, M. G. A., Te Wierik, M. J. M., Broens, E. M., van der Giessen, J. W. B., de Rosa, M., et al. (2016). Marked increase in leptospirosis infections in humans and dogs in the Netherlands, 2014. Euro Surveill 21 (17). doi: 10.2807/1560-7917.ES.2016.21.17.30211
Renaud, C., Andrews, S., Djelouadji, Z., Lecheval, S., Corrao-Revol, N., Buff, S., et al. (2013). Prevalence of the Leptospira serovars bratislava, grippotyphosa, mozdok and pomona in French dogs. Vet. J. 196 (1), 126–127. doi: 10.1016/j.tvjl.2012.10.002
Salaün, L., Mérien, F., Gurianova, S., Baranton, G., Picardeau, M. (2006). Application of multilocus variable-number tandem-repeat analysis for molecular typing of the agent of leptospirosis. J. Clin. Microbiol. 44 (11), 3954–3962. doi: 10.1128/JCM.00336-06
Santos, L. A., Adhikarla, H., Yan, X., Wang, Z., Fouts, D. E., Vinetz, J. M., et al. (2018). Genomic comparison among global isolates of L. interrogans serovars copenhageni and icterohaemorrhagiae identified natural genetic variation caused by an indel. Front. Cell Infect. Microbiol. 8, 193. doi: 10.3389/fcimb.2018.00193
Smythe, L. D., Wuthiekanun, V., Chierakul, W., Suputtamongkol, Y., Tiengrim, S., Dohnt, M. F., et al. (2009). The microscopic agglutination test (MAT) is an unreliable predictor of infecting Leptospira serovar in Thailand. Am. J. Trop. Med. Hyg 81 (4), 695–697. doi: 10.4269/ajtmh.2009.09-0252
Soupé-Gilbert, M. E., Oedin, M., Kainiu, M., Girault, D., Figuet, O., Brescia, F., et al. (2022). Original Leptospira spp. in island’s native terrestrial mammals: A case study in Pteropus spp. bats of New Caledonia. Transbound Emerg. Dis. 69 (5), e2852–e2862. doi: 10.1111/tbed.14635
Sykes, J. E., Reagan, K. L., Nally, J. E., Galloway, R. L., Haake, D. A. (2022). Role of diagnostics in epidemiology, management, surveillance, and control of leptospirosis. Pathogens 11 (4), 395. doi: 10.3390/pathogens11040395
Velardo, F., Bouziri, H., Adélaïde, L., Oliosi, E., Layan, M., Descamps, A., et al. (2022). A cross-sectional study on infectious health risks regarding freshwater sports practice in Brittany, France. J. Water Health 20 (2), 356–368. doi: 10.2166/wh.2022.232
Vincent, A. T., Schiettekatte, O., Goarant, C., Neela, V. K., Bernet, E., Thibeaux, R., et al. (2019). Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PloS Negl. Trop. Dis. 13 (5), e0007270. doi: 10.1371/journal.pntd.0007270
Waggoner, J. J., Balassiano, I., Abeynayake, J., Sahoo, M. K., Mohamed-Hadley, A., Liu, Y., et al. (2014). Sensitive real-time PCR detection of pathogenic leptospira spp. and a comparison of nucleic acid amplification methods for the diagnosis of leptospirosis. PloS One 9 (11), e112356. doi: 10.1371/journal.pone.0112356
World Health Organization (2003). Human Leptospirosis: Guidance for Diagnosis, Surveillance and Control (Geneva: World Health Organization). Available at: https://apps.who.int/iris/handle/10665/42667. Report No.: WHO/CDS/CSR/EPH 2002.23.
Zarantonelli, L., Suanes, A., Meny, P., Buroni, F., Nieves, C., Salaberry, X., et al. (2018). Isolation of pathogenic Leptospira strains from naturally infected cattle in Uruguay reveals high serovar diversity, and uncovers a relevant risk for human leptospirosis. PloS Negl. Trop. Dis. 12 (9), e0006694. doi: 10.1371/journal.pntd.0006694
Keywords: leptospirosis, zoonotic disease, human, dog, France, lfb1 gene
Citation: Garcia-Lopez M, Lorioux C, Soares A, Trombert-Paolantoni S, Harran E, Ayral F, Picardeau M, Djelouadji Z and Bourhy P (2023) Genetic diversity of Leptospira strains circulating in humans and dogs in France in 2019-2021. Front. Cell. Infect. Microbiol. 13:1236866. doi: 10.3389/fcimb.2023.1236866
Received: 08 June 2023; Accepted: 21 July 2023;
Published: 17 August 2023.
Edited by:
Tao Lin, Baylor College of Medicine, United StatesReviewed by:
Béla Dénes, University of Veterinary Medicine Budapest, HungaryCopyright © 2023 Garcia-Lopez, Lorioux, Soares, Trombert-Paolantoni, Harran, Ayral, Picardeau, Djelouadji and Bourhy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pascale Bourhy, cGJvdXJoeUBwYXN0ZXVyLmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.