- 1Key Laboratory of Animal Genetic and Breeding of Sichuan Province, Sichuan Animal Science Academy, Chengdu, China
- 2College of Animal and Veterinary Sciences, Southwest Minzu University, Chengdu, China
Introduction: Despite long-term integrated control programs for Eimeria stiedai infection in China, hepatic coccidiosis in rabbits persists. Th1, Th2, Th17, Treg, Th9, and Th21 cells are involved in immune responses during pathogen infection. It is unclear whether Th cell subsets are also involved in E. stiedai infection. Their roles in the immunopathology of this infection remain unknown. Therefore, monitoring these T-cell subsets’ immune responses during primary infection of E. stiedai at both transcriptional (mRNA) and protein (cytokines) levels is essential.
Methods: In experimentally infected New Zealand white rabbits, mRNA expression levels of their transcript—TBX2 (Th1), GATA3 (Th2), RORC (Th17), Foxp3 (Treg), SPI1 (Th9), and BCL6 (Th21)—were evaluated using quantitative real-time polymerase chain reaction (qRT-PCR), whereas Th1 (IFN-g and TNF-a), Th2 (IL4), Th17 (IL17A and IL6), Treg (IL10 and TGF-b1), Th9 (IL9), and Th21 (IL21) cytokines were measured using enzyme-linked immunosorbent assays (ELISAs).
Results: We found that levels of TBX2, GATA3, RORC, SPI1, and BCL6 in the livers of infected rabbits were elevated on days 5 and 15 post-infection (PI). The concentrations of their distinctive cytokines IFN-g and TNF-a for Th1, IL4 for Th2, IL17A for Th17, IL9 for Th9, IL21 for Th21, and IL10 for Treg IL10 were also significantly increased on days 5 and 15 PI, respectively (p < 0.05). On day 23 PI, GATA3 with its cytokine IL4, RORC with IL17A, Foxp3 with IL10 and TGF-b1, and SPI1 with IL9 were significantly decreased, but TBX2 with IFN-g and IL6 remained elevated.
Discussion: Our findings are the first evidence of Th1/Th2/Treg/Th17/Th9/Th21 changes in E. stiedai-infected rabbits and provide insights into immune regulation mechanisms and possible vaccine development.
Introduction
Hepatic coccidiosis in rabbits is caused by Eimeria stiedai. First discovered more than 100 years ago, it has a high host specificity, infectivity, and lethality. Post-weaning rabbits are particularly susceptible, with mortality as high as 80% (Gomez-Bautista et al., 1987; Pakandl, 2009). Unlike the other 10 species of rabbit intestinal coccidia, E. stiedai lives in the rabbit liver to complete its entire development (Duszynski and Couch, 2013). The life cycle of E. stiedai occurs both in the external environment and within the host rabbit, and generally experiences three stages, Sporogony, Schizogony or Merogony, and Gametogony. Firstly, sporulated oocysts are consumed by rabbits and rupture, then released sporozoites invade the liver and bile duct epithelial cells where merogony occurs. Merozoites multiply within and are released from epithelial cells and continue to increase until they complete a repeat of at least four generations. Lastly, the life cycle enters the gametogony stage, macrogametes and microgametes combine into zygotes and eventually develop into oocysts passed out in the feces. E. stiedai causes severe damage to the hepatobiliary epithelial structure, affects the normal hepatobiliary function, leads to slow growth and high mortality in diseased rabbits (Oliveira et al., 2011; Jing et al., 2016), and causes severe harm to the rabbit industry. It is listed as a second-class animal disease by the Ministry of Agriculture of the People’s Republic of China.
T helper cells with specific cytokines (IFN-γ, IL4, IL10, IL17A, IL21, IL9, TNF-α, and TGF-β1) play essential roles in parasitic diseases (Pesce et al., 2006; Seki et al., 2012; Pang et al., 2014; Li et al., 2017; Kalantari et al., 2019; Bai et al., 2021; Al-Masoudi et al., 2021; Parhizgar et al., 2021; Li et al., 2022). Usually, interferon-γ (IFN-γ) is induced by TBX2, but in New Zealand white rabbits infected with Schistosoma japonicum (S. japonicum), it was found that the transcription factor TBX2 was positively correlated to TNF-α, but not to IFN-γ, which was correlated to another Th1 transcriptional factor, IRF8 (Farwa et al., 2018); thus, our study aimed to evaluate the correlation between T helper cells, transcription factors (TBX2, GATA3, Foxp3, RORC, SPI1, and BCL6), and cytokines (IFN-γ, IL4, IL10, IL17A, IL21, IL9, TNF-α, and TGF-β1) in rabbits infected with E. stiedai.
The Th1 cytokine IFN-γ sometimes protects against damage, while the Th2 cytokine IL4 enhances fibrogenesis (McManus et al., 2010; Du et al., 2011). Treg cells that produce IL10 cytokines play a crucial role in the immune balance; it acts as an immunosuppressant or a fibrotic inducer (Layland et al., 2010; Adalid-Peralta et al., 2011; Zhou et al., 2015). For example, in Schistosoma mansoni-infected mice, higher expression levels of hepatic IL17A were found in CBA mice, suggesting that its lesions may be associated with IL17A produced by Th17 cells (Kalantari et al., 2019). However, lacking the IL21/IL21R axis in S. mansoni-infected mice attenuated granulomatous inflammation and liver fibrosis formation in mice (Pesce et al., 2006). There were significantly increased levels of Th9 (IL9) in the liver of C57BL/6 mice infected by S. japonica in the early stage, suggesting that IL9 may be involved in liver injury (Li et al., 2017). However, whether these Th subsets and their cytokines also play an immuno-protective or damaging role in E. stiedai infection is unknown.
TBX2 induces the production of interferon-γ (IFN-γ) and orchestrates the Th1 cell migration by regulating the expression of chemokines and chemokine receptors. TBX2 and GATA3 regulate Th1/Th2 cell fate using a synchronous mechanism, by directly binding to the genetic locus of cytokines: TBX2 binds IFN-γ whereas GATA3 binds IL4 and other Th2 cytokines such as IL5 and IL13, leading to the expression of these proteins (Kanhere et al., 2012).
Transcriptional factor Foxp3 serves as a lineage specification factor of Treg cells, and is induced either during native Treg (nTreg) development in the thymus or in the presence of TGF-β1 and retinoic acid. High levels of IL17 is expressed by Th17 under the transcriptional regulation of RORC. In some studies, it was suggested that the signature transcription factor RORC of Th17 cells is also induced by TGF-β1, thus leading to differentiation of Treg and Th17 lineages. In the absence of a second signal derived from proinflammatory cytokines, Foxp3 can inhibit RORC function and promote Treg differentiation. However, when cells also receive signals from proinflammatory cytokines (e.g., IL6), the function of Foxp3 is inhibited and Th17 differentiation is induced. Thus, it is the balance between Foxp3 and RORC function that determines the fate of CD4+T cells and the type of immune response that would be generated (Ziegler and Buckner, 2009; Ren and Li, 2017).
PU.1 (SPI1) is a transcription factor that belongs to the E-twenty six (ETS) family of proteins and is a key factor for the differentiation of the IL9-producing Th9 subset, in humans and mice (Chang et al., 2010; Ramming et al., 2012). BCL6 served as a master regulator of Th21 differentiation, and Th21 differentiation does not occur in vivo in the absence of BCL6 (BCL6 −/−) (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009). While BCL6 is vital in both GC B cells and Th21 cells, it can control Th21 differentiation mainly by regulating genes of GC B cells (Crotty et al., 2010).
In E. stiedai infection, it is unclear whether different mechanisms are involved. For these reasons, we measured dynamic changes in Th cells, including Th1, Th2, Th17, Treg, Th9, and Th21, during different stages of E. stiedai infection. Our findings provide a theoretical basis to control for protective and therapeutic vaccinations.
Materials and methods
Ethics statement
This study was carried out in full compliance with the framework of noncommercial scientific research, authorized by the Animal Ethics Committee of Sichuan Animal Science Academy (approval no. AECSASA2023-18).
Parasites
The E. stiedai strain in this study was provided by the Department of Parasitology at Sichuan Agricultural University. Three coccidia-free New Zealand white rabbits (5 weeks old) were orally infected with E. stiedai to obtain more oocysts. Then high numbers of unsporulated oocysts were harvested from liver with grinding, filtration, and centrifugation. Finally, the oocysts were sporulated in 2.5% potassium dichromate solution at 28°C and stored at 4°C.
Experimental rabbits
We divided 36 female New Zealand white rabbits aged 50 days into a control group (n = 18, six rabbits for each time point) and an experiment group (n = 18, six rabbits for each time point). They received 1.0 × 105 E. stiedai orally and were euthanized on days 5, 15, and 23 post-infection (PI). All rabbits were monitored for 5 days by testing the daily collected fecal samples to confirm the infection. Blood samples were collected for quantitative real-time polymerase chain reaction (qRT-PCR) and enzyme-linked immunosorbent assays (ELISAs).
Blood sample collection
We collected 3 ml of blood from the rabbits on days 5, 15, and 23 PI. Serum was also collected and stored at −80°C to evaluate Th1/Th2/Treg/Th17/Th9/Th21-related cellular cytokines.
RNA extraction and cDNA preparation
RNA was extracted from the isolated blood using the TRIzol method (TRIzol reagent provided by Invitrogen, Carlsbad, CA, USA), following the manufacturer’s protocols. cDNA preparation was performed in a 20-μl mixture condition using Hifar® II 1st Strand cDNA Synthesis SuperMix for qRT-PCR (Yeasen Biotechnology Co., Ltd) as follows: 4 μl of RNA, 2 μl of 5× gDNA digester Buffer, 1 μl of gDNA, 10 μl of 2× Hifar® II SuperMix plus, and 3 μl of sterile RNase-free water. The amplification conditions were 25°C for 5 min, 42°C for 30 min, and 85°C for 5 min.
Primer design and qRT-PCR amplification
CDS sequences of TBX2, GATA3, Foxp3, RORC, BCL6, and SPI1 genes and housekeeping GAPDH as a reference gene were retrieved from the GenBank database with their accession numbers as illustrated in C 1. For qRT-PCR expression profiles, primer pairs of each target gene were designed by Premier 6.0 software, where their length ranged from 18 to 25 bp with 55%–65% of GC content and different annealing temperatures (Table 1). All primers were synthesized by Tsingke Biotechnology (Chengdu) Co., Ltd. qRT-PCR was performed in a 20-μl mixture condition using Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (Yeasen Biotechnology Co., Ltd) as follows: 1 μl of cDNA, 10 μl of Master Mix, 0.4 μl of forward primer, 0.4 μl of reverse primer, and 8.2 μl of sterile RNase-free water. The amplification conditions started with initial denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 10 s, 57°C for 30 s at annealing temperature, and the melting curve stage at 95°C for 15 s, 55°C for 15 s, followed by 95°C for 15 s.
ELISA
According to the manufacturer’s instructions (Shanghai MLBIO Biotechnology Co. Ltd), an ELISA test was performed to measure nine cytokine levels—IFN-γ, TNF-α, IL4, IL10, TGF-β1 IL17A, IL6, IL9, and IL21—for the control and experiment group. Concentrations of plasma cytokines were calculated by using ELISA Calc v 0.1 software.
Hepatic histopathology
Rabbits were euthanized on days 5, 15, and 23 PI. Partial left lobes of the livers from all rabbits were fixed in 10% neutral buffered formalin and embedded in paraffin for histological examination.
Total hepatic oocysts
The total liver weight was determined by electronic weighing, then 10 g of the left liver lobe was ground, dissolved in 50 ml of physiological saline, and filtered with an 80-mesh filter screen. Counts were performed with a hemocytometer.
Statistical analysis
The general linear model procedure in SPSS 22.0 version was used to analyze mRNA expression levels of TBX2, GATA3, Foxp3, RORC, BCL6, and SPI1, as well as plasma cytokine levels of IFN-γ, TNF-α, IL4, IL6, IL10, IL9, IL17, IL21, and TGF-β1. Differences were considered significantly different when p < 0.05. Regression and correlation analyses were performed to detect the relationships between each mRNA transcriptional factor and every cytokine individually. Another set of regression and correlation was performed to determine the relationships between different mRNA transcriptional factors and the relationships between transcriptional factors and oocyst data, all at three time points (on days 5, 15, and 23 PI). |r| values ≥ 0.8 were considered statistically significant.
Results
Hepatic histopathological changes
No rabbit died during the infection period, and the rabbits were euthanized on days 5, 15, and 23 PI. The effect of E. stiedai infection on rabbit livers was investigated using stained paraffin-embedded tissues. In control group, as shown in Figures 1A–C and in the experiment group, as shown in Figures 1D–F, in the experiment group, substantial numbers of inflammatory cells had infiltrated around the portal area, collagen depositions were slightly stained, and intrahepatic bile duct epithelia were proliferated in the liver tissue. The intrahepatic bile ducts continued to be exaggerated extensively on days 15 and 23 PI (Figures 1E, F). Several merozoites were observed (black arrow) (Figure 1E). On day 23 PI, the proliferation of bile duct epithelial and numerous eggs of E. stiedai cells was observed (Figure 1F).
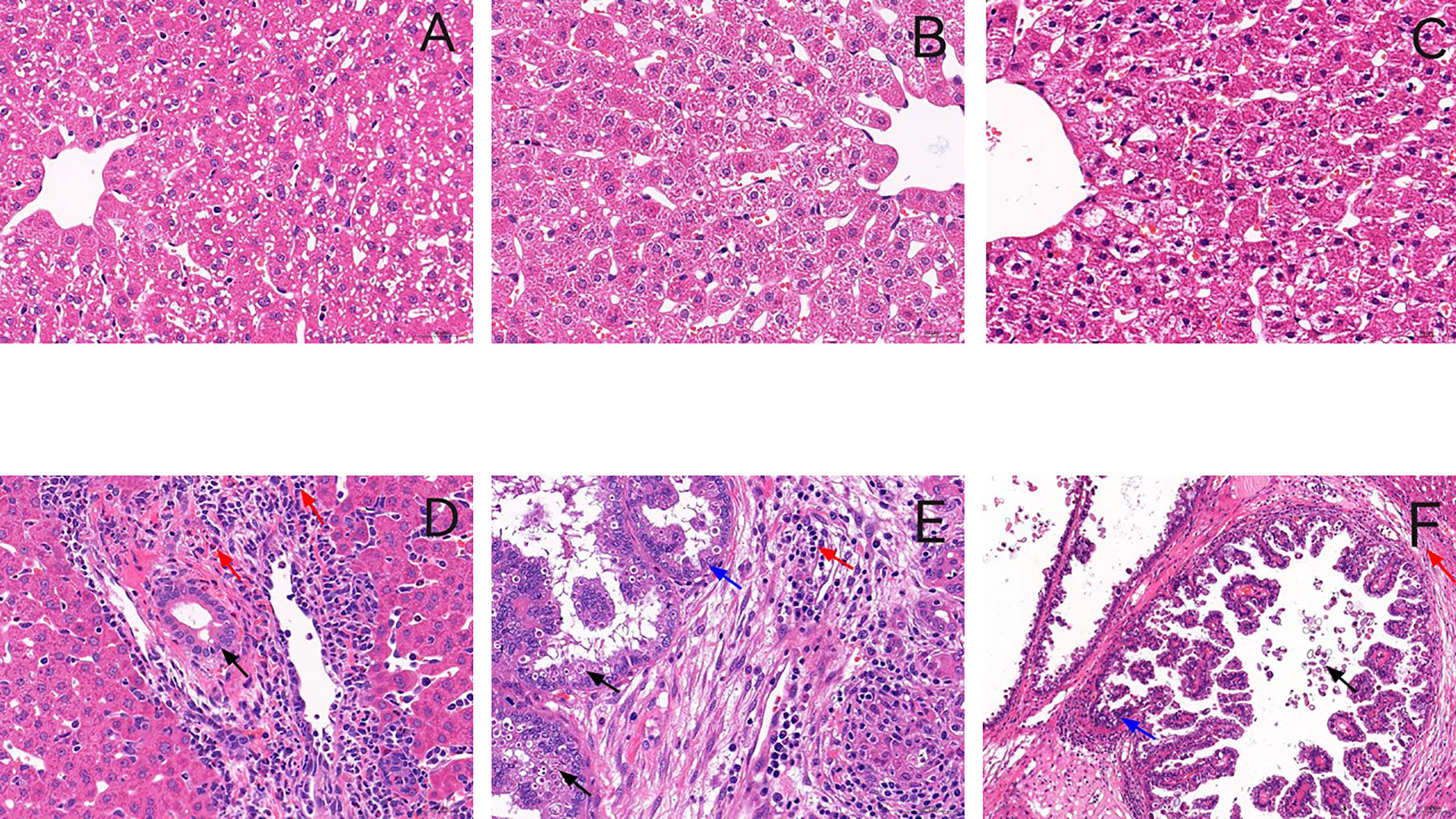
Figure 1 Representative histology of liver sections stained with hematoxylin and eosin. (A–C) show liver sections from the control group in three time points, and (D–F) show liver sections from the experiment group in three time points (on days 5, 15, and 23 PI) (×400). In the experiment group, massive inflammatory cells were infiltrated around the portal areas accompanied by cholangiocyte proliferation on day 5 PI (D). The number of inflammatory cells increased, and bile duct hyperplasia was exaggerated on day 15 PI (E). Many schizonts were observed (black arrow). On day 23 PI (post-infection) bile duct expansion, a proliferation of bile duct epithelial cells and fibrous tissue hyperplasia, and numerous Eimeria stiedai eggs appeared (F).
T helper-1 response: TBX2 mRNA expressions and IFN-γ and TNF-α serum levels
On days 5 and 15 PI, TBX2 mRNA expressions and the levels of IFN-γ and TNF-α in serum were significantly increased after the infection compared to the control group. However, on day 23 PI, the infected rabbits showed a significant decrease in TNF-α (p < 0.05), whereas TBX2 mRNA (p < 0.05) expression and IFN-γ (p > 0.05) were elevated on day 23 PI (Figures 2A, 3A, B).
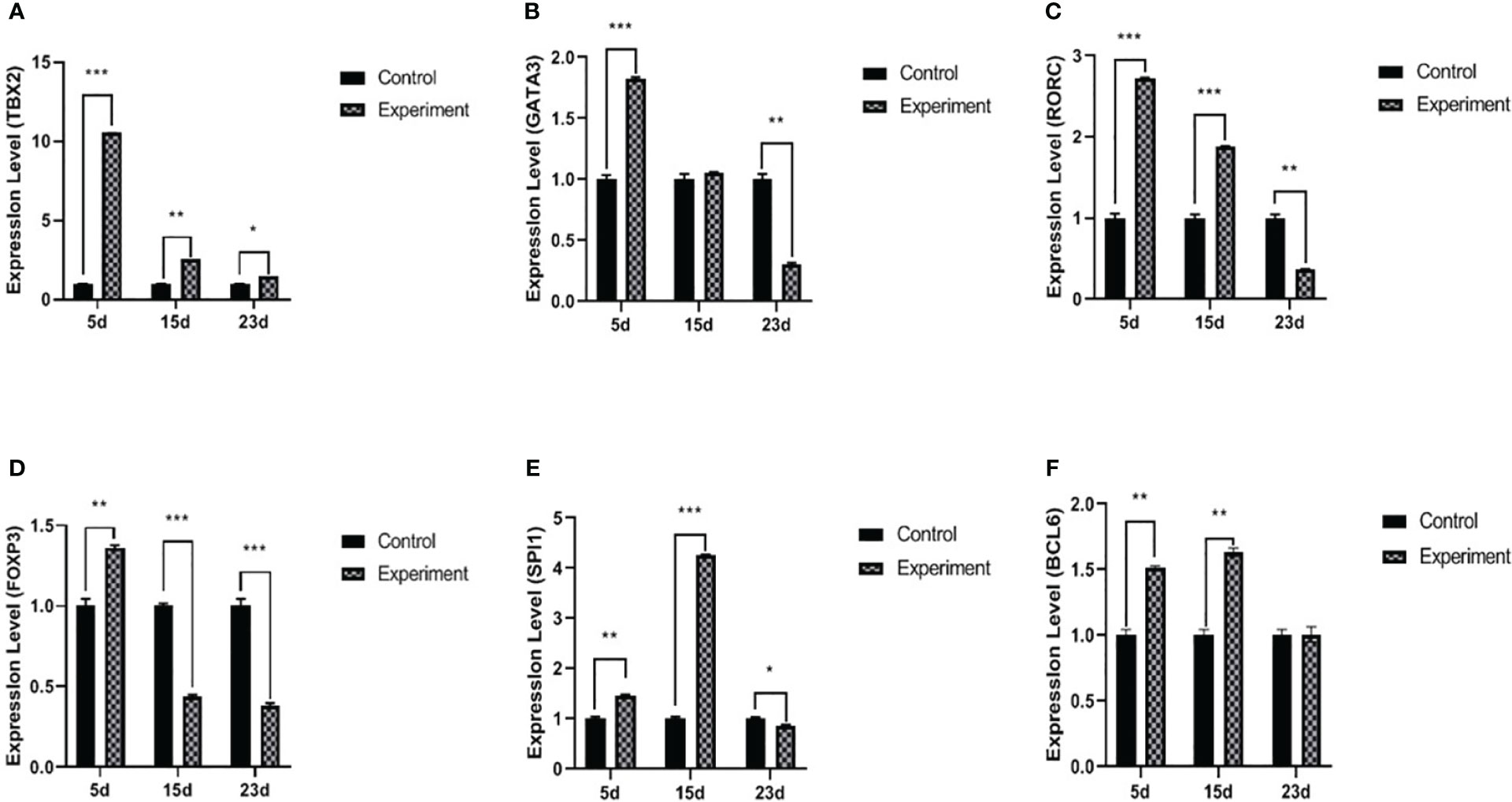
Figure 2 mRNA expression levels of TBX2 (A), GATA3 (B), RORC (C), Foxp3 (D), SPI1 (E), and BCL6 (F) transcriptional factors. The experiment group on days 5, 15, and 23 PI, compared with the control group, *p < 0.05, **p < 0.01, ***p < 0.001.
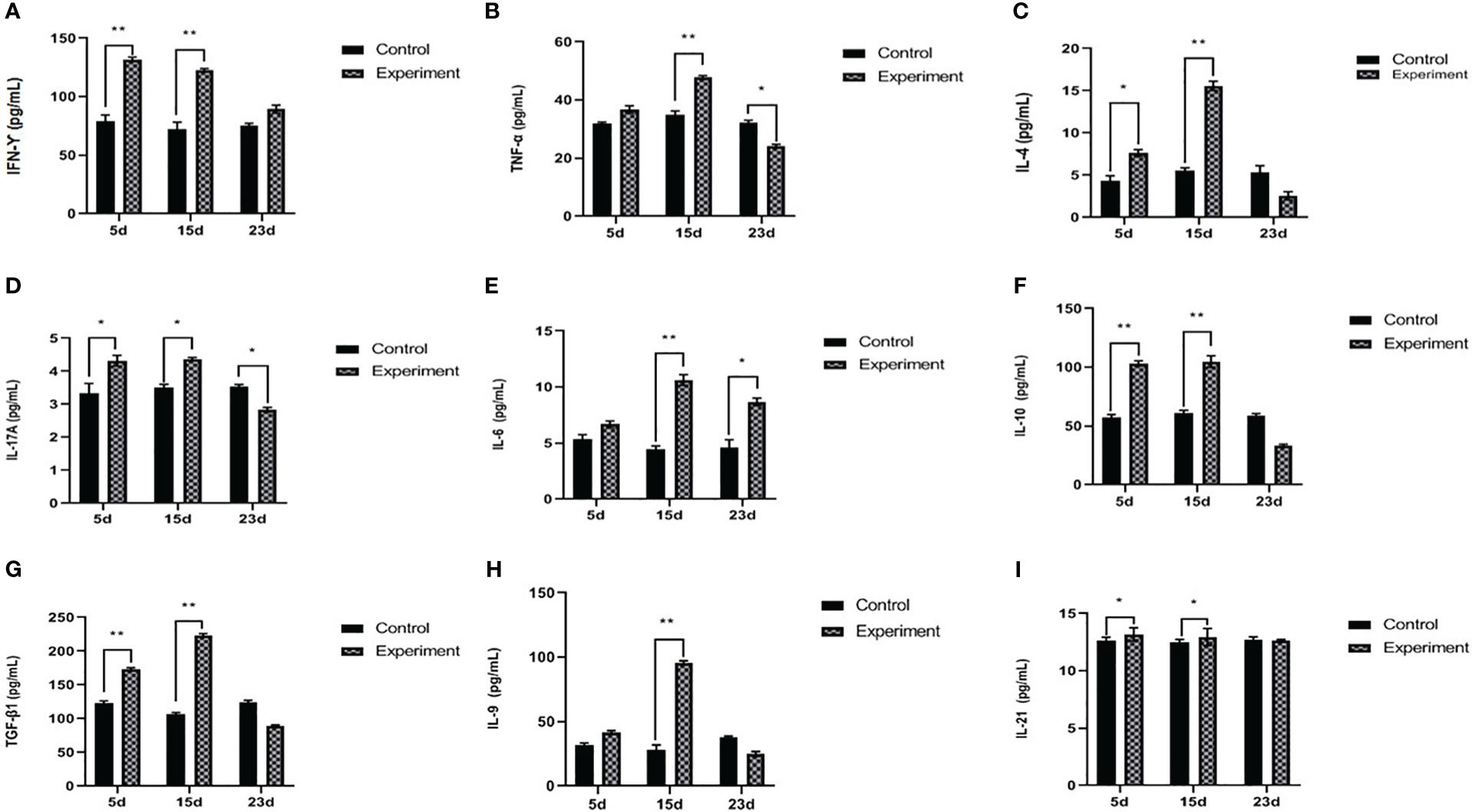
Figure 3 Serum concentration of IFN-γ (A), TNF-α (B), IL4 (C), IL17A (D), IL6 (E), IL10 (F), TGF-β1 (G), IL9 (H), and IL21 (I). The experiment group on days 5, 15, and 23 PI, compared with the control group, *p < 0.05, **p < 0.01.
T helper-2 response: GATA3 expressions and IL4 serum levels
On day 5 PI, GATA3 mRNA and IL4 significantly increased after the infection compared to the control. Furthermore, on day 15 PI, IL4 was significantly increased (p < 0.01) without any effect on GATA3 mRNA. On day 23 PI, the infected group showed a decrease in GATA3 (p < 0.01), and IL4 was not significantly different (p > 0.05) (Figures 2B, 3C).
T helper-17 response: RORC mRNA expressions and IL17A and IL6 serum levels
The levels of RORC mRNA (p < 0.001) and IL17A (p < 0.01) in serum were significantly increased on day 5 PI, but there was no statistical difference in the IL6 serum levels. On day 15 PI, RORC mRNA (p < 0.001), IL17A (p < 0.05), and IL6 (p < 0.01) were significantly upregulated. On day 23 PI, RORC mRNA (p < 0.01) and IL17A (p < 0.05) oppositely decreased. Interestingly, IL6 serum levels were upregulated during the three periods in the experiment groups (Figures 2C, 3D, E).
T regulatory response: Foxp3 mRNA expressions and IL10 and TGF-β1 serum levels
The level of Foxp3 was significantly increased (p < 0.001) on day 5 PI, and IL10 and TGF-β1 were also significantly increased (p < 0.01), compared with normal control. Interestingly, on day 15 PI, the level of Foxp3 was significantly decreased (p < 0.001); however, IL10 and TGF-β1 were still significantly increased (p < 0.01) compared with normal control. On day 23 PI, the levels of Foxp3 and relative cytokines (IL10 and TGF-β1) in serum were significantly decreased (p < 0.05) (Figures 2D, 3F, G).
T helper-9 response: SPI1 mRNA expressions and IL9 serum levels
On day 5 PI, the level of SPI1 mRNA was significantly increased (p < 0.01); however, there was no statistical difference in IL9 (p > 0.05). On day 15 PI, SPI1 mRNA (p < 0.001) and IL9 (p < 0.01) were significantly elevated. On day 23 PI, the level of SPI1 mRNA was significantly decreased (p < 0.05), but there was no statistical difference in the concentration of IL9 between the normal control and infected groups (Figures 2E, 3H).
T helper-21 response: BCL6 mRNA expressions and IL21 serum levels
BCL6 mRNA and IL21 were increased with the infection (p < 0.01 and p < 0.05, respectively) on day 5 PI. This influence was relatively stable on day 15 PI (BCL6, p < 0.01 and IL21, p < 0.05). On day 23 PI, the levels of BCL6 mRNA and IL21 showed no statistical difference between the normal control and infected group (p > 0.05) (Figures 2F, 3I).
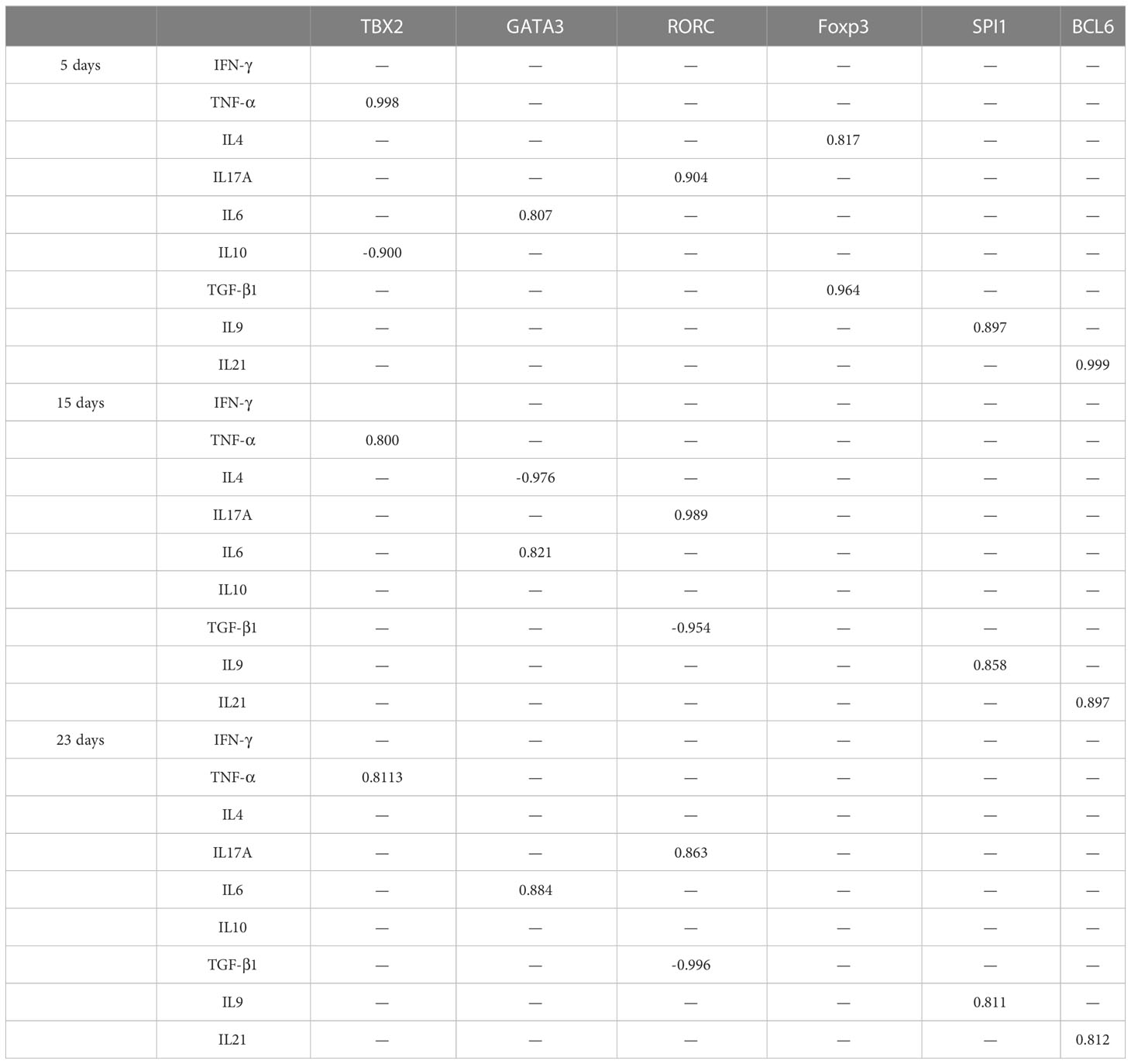
Relationships between different transcriptional factors and their cytokines
There are relationships between Th1/Th2/Th17/Treg/Th9/Th21 transcriptional factors with their cytokine lineages, where positive correlations (r ≥ 0.8) were found between RORC and IL17A, SPI1 and IL9, and BCL6 and IL21 in the experiment groups throughout the whole period of infection as shown in Table 2. TBX2 was positively correlated to TNF-α on days 5 and 15 PI. Cross-correlations were detected between transcriptional factors and cytokines of other T helper subsets, where TBX2 was negatively correlated to IL10 on day 5 PI. GATA3 was positively correlated to IL6 during the whole period of infection. GATA3 was negatively correlated to IL4 on day 5 PI. Foxp3 reported a positive correlation with IL4 and TGF-β1 (r ≥ 0.8) on day 5 PI, and RORC negatively correlated to TGF-β1 on days 15 and 23 PI.
The relationships between different transcriptional factors are illustrated in Figure 4.
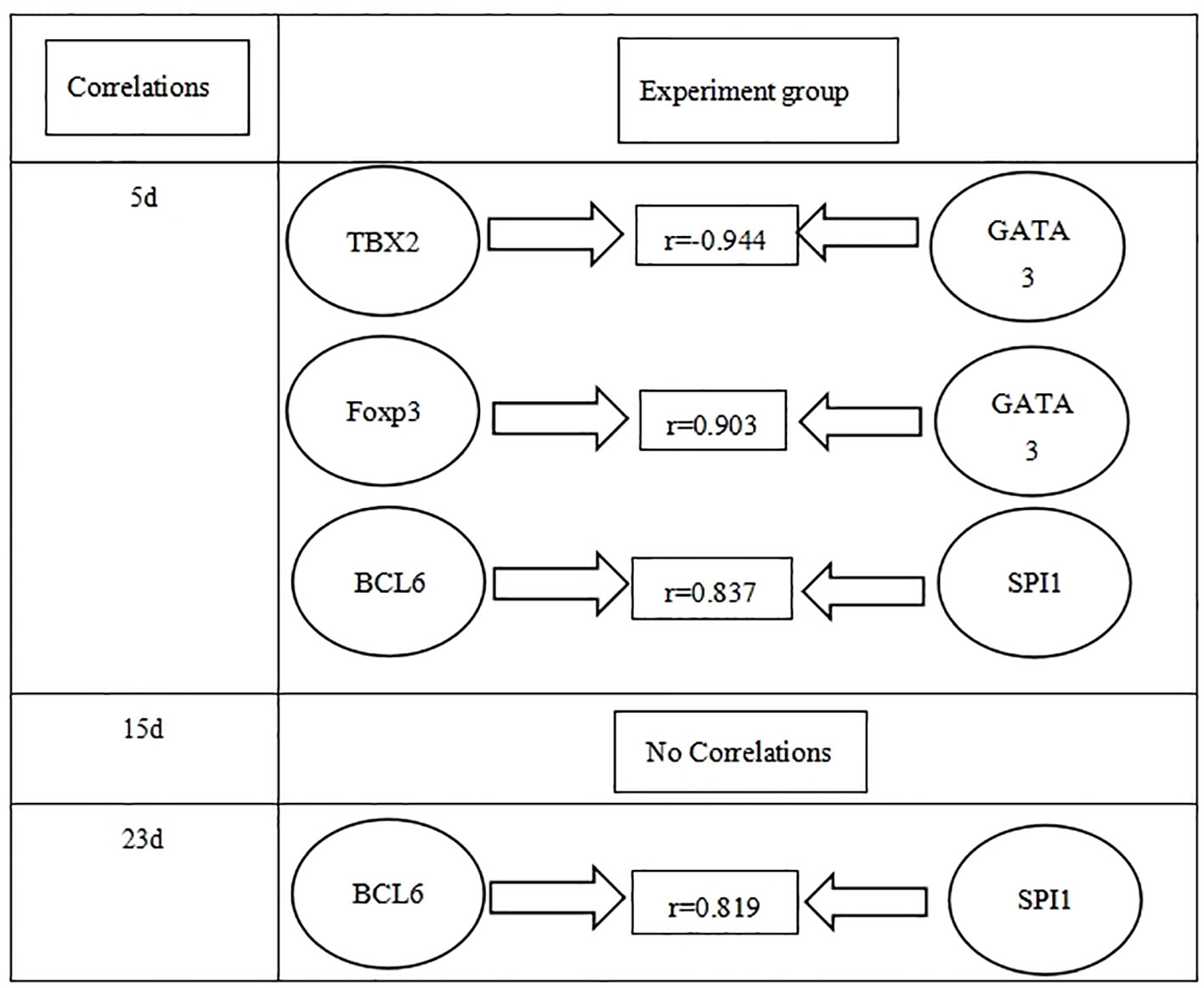
Figure 4 Correlations between different transcriptional factors in the experiment group in three time points (days 5, 15, and 23 PI).
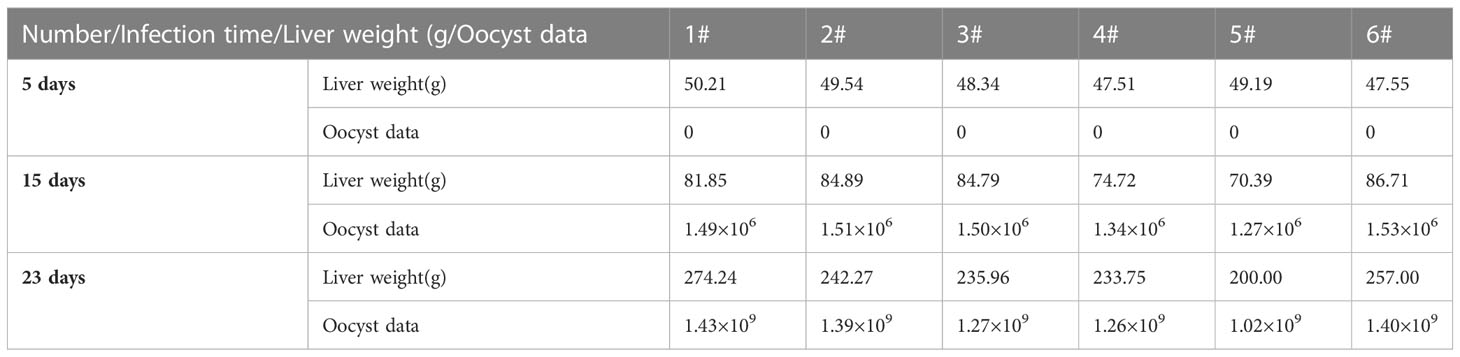
Hepatic weight and oocyst data are illustrated in Table 3.
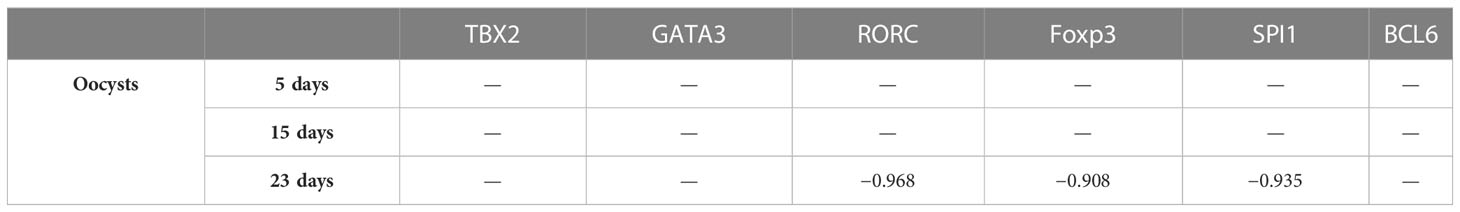
Correlation between transcriptional factors and oocyst data are illustrated in Table 4.
Discussion
We studied dynamic changes in Th mRNA expression levels of their transcript—TBX2 (Th1), GATA3 (Th2), RORC (Th17), Foxp3 (Treg), SPI1 (Th9), and BCL6 (Th21)—and analyzed the potential effects of their imbalance on disease outcomes in the rabbit. Moreover, E. stiedai infections provide an excellent model to study the roles of Th cell transcriptional factors. In the early stage of the infection with E. stiedai (on day 5 PI), Th1 dominated at both transcriptional and protein levels. We also found a Treg response in the late stage of infection. Furthermore, our study also showed that Th17, Th9, and Th21 were involved in E. stiedai infection. However, helminth infection is characterized by Th2-dominated immune responses, which may contribute to chronic infection and the host’s inability to clear the parasites (Kim et al., 2012; Uddin et al., 2012). For Th1, the transcription factor TBX2 was positively correlated to TNF-α, but not IFN-γ, which Farwa found was correlated to another Th1 transcriptional factor IRF8 (Farwa et al., 2018). Mechanistically, TBX2-dependent innate IFN-γ is essential in T. gondii infection T-bet to produce IFN-γ, which is essential for host resistance to intracellular infection by maintaining IRF8 inflammatory dendritic cells at the infection site (Lopez-Yglesias et al., 2021). A report showed that loss of TBX2 had some effect on IFN-γ production in T cells but not parasite-specific T cells (Harms Pritchard et al., 2015).
For Th2, the expression levels of GATA3 and IL4 appeared to be elevated on days 5 and 15 PI, and GATA3 mRNA expression was uncorrelated to IL4, similar to GATA3 mRNA expression, which was essential for IL13 production but not IL4 in the experiment group, whereas IL4 appeared to be a STAT6-dependent stimulation (Zhu, 2010). Regarding Treg, the expression levels of Foxp3 also appeared to be elevated. TGF-β1 was also induced simultaneously, similar to an observation by Tang et al. (2015) on day 5 PI. Foxp3 appeared to have elevated expression levels, and the same result appeared in S. japonicum infection, which was a potent inducer for Foxp3 (Finlay et al., 2014). For Th17, RORC expression was correlated to IL17A. On days 5 and 15 PI, Th17 mRNA and IL17A expression levels were elevated, similarly in S. mansoni-infected mice, in which the liver developed significant hepatic granulomatous inflammation, with higher expression levels of IL17 and RORC, suggesting that its lesions may be associated with IL17 produced by Th 17 cells (Kalantari et al., 2019). For Th9, the expression levels of SPI1 and IL9 appeared to be elevated on days 5 and 15 PI, and SPI1 expression was correlated to IL9. The expression of IL9 and its transcription factor SPI1 in hydatid patients was significantly higher in focal tissue, suggesting the formation of chronic granuloma in hydatid by upregulating the expression of Th9 cells, IL9, and transcription factor SPI1 during infection (Tuxun et al., 2015). The expression levels of Th21, BCL6, and IL21 appeared to be elevated on days 5 and 15 PI, and BCL6 expression was correlated to IL21. A correlation study of IL21 and transcription factor BCL6 showed that IL21 was positively correlated to BCL6, which was upregulated on Th21 cells, and IL21 is the primary effector molecule of Tfh cells. (Geng et al., 2018).
In addition, many cross-modulations were observed; TBX2 suppressed GATA3 expression on day 5 PI (Zhu, 2010). Cross-modulations between GATA3 and Foxp3 were observed in Treg cells, where GATA3 expression enhanced the accumulation of Treg in the inflammatory site (Turner et al., 2011). On days 5 and 23 PI, we also observed the cross-modulations between SPI1 and BCL6. The expression of B-cell lymphoma protein 6 (BCL6) depends on IRF8 and SPI1 in vivo, which is essential for developing germinal center B cells, and SPI1 acts as a regulator of BCL6 expression during GC B-cell development (Wang et al., 2019). GATA3 promotes Th2 differentiation while inhibiting Th1, and TBX2 has been shown to promote the development of Th1 and IFN-γ production (Szabo et al., 2000), but a study contradicted this finding (Cobb et al., 2010). A study showed that the IL21/IL21R signaling axis regulated the effector functions of T-cell subsets (e.g., production of cytokines) by enhancing TBX2 and STAT4 expression in human T cells, thereby enhancing IFN-γ production (Solaymani-Mohammadi et al., 2019). Treg only regulated TGF-β1 and was independent of type 2 cytokines (Mishra et al., 2014). In general, IL6 combined with TGF-β induced differentiation of Th17 cells from initial CD4+T cells and was also polarized into Treg cells after co-stimulation with IL2 and TGF-β (Kimura and Kishimoto, 2010). Our study found interesting findings: GATA3, Foxp3, RORC, and SPI1 mRNA and its relative cytokines (such as IL4, IL10, IL17A, and IL9 in the serum) were significantly decreased in the late stage, which suggests that they were suppressed.
In the experiment group, liver weight increased, and the significant proliferation of eggs of E. stiedai made liver lesions severe (Table 3). In the liver sections, massive inflammatory cells were infiltrated around the portal areas accompanied by cholangiocyte proliferation on day 5 PI (Figure 1D). TBX2, GATA3, Foxp3, RORC, BCL6, SPI1, and relative cytokine expression levels were elevated, which resisted E. stiedai infection. Several merozoites were observed on day 15 PI (Figure 1E) (black arrow). TBX2, RORC, BCL6, SPI1, and relative cytokine expression levels were elevated, and Foxp3 expression levels were decreased, likely due to dynamic Treg/Th17 changes. On day 23 PI, the appearance of numerous eggs of E. stiedai was consistent with previous reports (Figure 1F), except TBX2 expression levels were elevated; GATA3, RORC, BCL6, and SPI1 expression levels were decreased; and Th1 dominated at both transcriptional and protein levels, but this contradicted a study carried out by Uddin et al. (2012). In addition, RORC, Foxp3, and SPI1 were negatively correlated to oocyst numbers on day 23 PI, and expression levels of cytokines including IL17A, IL10, IL9, and TGF-β1 decreased. No statistically significant difference of relationships between TBX2, GATA3, BCL6, and oocyst output was observed in the experiment groups at three time points (Table 4). In the experiment group, the severe liver damage was observed during the proliferation of E. stiedai, likely through suppressing the expression of RORC, Foxp3, SPI1, cytokines (IL17A, IL10, and IL9), and TGF-β1.
Conclusion
Our findings provide the first evidence of dynamic changes in Th1/Th2/Th17/Treg/Th9/Th21 and confirm an imbalance of these cell populations in E. stiedai-infected rabbits. Additional studies are needed to elucidate the detailed roles of these Th cells in the pathogenesis of E. stiedai.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was reviewed and approved by the Institutional Animal Care Committee of Sichuan Animal Science Academy.
Author contributions
Conceptualization: X-DC, R-MK, and Y-GY. Methodology: X-DC. Software: LX, C-JM, and X-JW. Validation: J-FY, YW, and JX. Formal analysis: X-DC and YC. Resources: Y-GY. Data curation: R-MK and Y-GY. Writing—original draft preparation: X-DC. Writing—review and editing: Y-GY. Visualization: X-DC. Supervision: Y-GY. Project administration: R-MK and Y-GY. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Sichuan Provincial Financial Operation Special Project (SASA2023CZYX006), the Sichuan Province Regional Innovation Cooperation Project (2023YFQ0037), and the Sichuan Provincial Natural Science Foundation (2022NSFSC0081).
Acknowledgments
We thank the Sichuan Agricultural University for providing us with E. stiedai.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adalid-Peralta, L., Fragoso, G., Fleury, A., Sciutto, E. (2011). Mechanisms underlying the induction of regulatory T cells and its relevance in the adaptive immune response in parasitic infections. Int. J. Biol. Sci. 7 (9), 1412–1426. doi: 10.7150/ijbs.7.1412
Al-Masoudi, H. K., Al-Hamadani, K. C., Khiarull, I. A. (2021). Interleukin 17 cytokine profiles in patients with cystic Echinococcosis in babylon province, Iraq. Arch. Razi Inst. 76 (5), 1493–1500. doi: 10.22092/ari.2021.355855.1730
Bai, Y., Guan, F., Zhu, F., Jiang, C., Xu, X., Zheng, F., et al. (2021). IL-33/ST2 Axis deficiency exacerbates hepatic pathology by regulating Treg and Th17 cells in murine Schistosomiasis Japonica. J. Inflammation Res. 14, 5981–5998. doi: 10.2147/JIR.S336404
Chang, H. C., Sehra, S., Goswami, R., Yao, W., Yu, Q., Stritesky, G. L., et al. (2010). The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat. Immunol. 11 (6), 527–534. doi: 10.1038/ni.1867
Cobb, D., Hambright, D., Smeltz, R. B. (2010). T-bet-independent effects of IL-12 family cytokines on regulation of Th17 responses to experimental T. cruzi infection. J. Leukoc. Biol. 88 (5), 965–971. doi: 10.1189/jlb.0410238
Crotty, S., Johnston, R. J., Schoenberger, S. P. (2010). Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 11 (2), 114–120. doi: 10.1038/ni.1837
Du, X., Wu, J., Zhang, M., Gao, Y., Zhang, D., Hou, M., et al. (2011). Upregulated expression of cytotoxicity-related genes in IFN-γ knockout mice with Schistosoma Japonicum infection. J. BioMed. Biotechnol. 2011, 864945. doi: 10.1155/2011/864945
Duszynski, D. W., Couch, L. (2013). The biology and identification of the Coccidia (Apicomplexa) of rabbits of the world. Newnes 23, 56–23. doi: 10.34/0347-356(67)87-6
Farwa, A., He, C., Xia, L., Zhou, H. (2018). Immune modulation of Th1, Th2, and Treg transcriptional factors differing from cytokine levels in Schistosoma Japonicum infection. Parasitol. Res. 117 (1), 115–126. doi: 10.1007/s00436-017-5678-5
Finlay, C. M., Walsh, K. P., Mills, K. H. (2014). Induction of regulatory cells by helminth parasites: exploitation for the treatment of inflammatory diseases. Immunol. Rev. 259 (1), 206–230. doi: 10.1111/imr.12164
Geng, X. H., Zhang, F. B., Jing, M., Zhao, H., An, M. T., Pang, N. N., et al. (2018). Expression of IL-9, PU.1 and IRF-4 mRNA in liver tissues in patients with alveolar echinococcosis. Chin. J. Immunol. 34 (3), 412–416.
Gomez-Bautista, M., Rojo-Vazquez, F. A., Alunda, J. M. (1987). The effect of the host’s age on the pathology of Eimeria stiedai infection in rabbits. Vet. Parasitol. 24 (2), 47–57. doi: 10.1016/0304-4017(87)90129-4
Harms Pritchard, G., Hall, A. O., Christian, D. A., Wagage, S., Fang, Q., Muallem, G., et al. (2015). Diverse roles for T-bet in the effector responses required for resistance to infection. J. Immunol. 194 (3), 1131–1140. doi: 10.4049/jimmunol.1401617
Jing, J., Liu, C., Zhu, S. X., Jiang, Y. M., Wu, L. C., Song, H. Y., et al. (2016). Pathological and ultrastructural observations and liver function analysis of Eimeria stiedai-infected rabbits. Vet. Parasitol. 223, 165–172. doi: 10.1016/j.vetpar.2016.04.020
Johnston, R. J., Poholek, A. C., DiToro, D., Yusuf, I., Eto, D., Barnett, B., et al. (2009). Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Sci. (New York N.Y.). 325 (5943), 1006–1010. doi: 10.1126/science.1175870
Kalantari, P., Bunnell, S. C., Stadecker, M. J. (2019). The C-type lectin receptor-driven, Th17 cell-mediated severe pathology in schistosomiasis: Not all immune responses to helminth parasites are Th2 dominated. Front. Immunol. 10. doi: 10.3389/fimmu.2019.00026
Kanhere, A., Hertweck, A., Bhatia, U., Gokmen, M. R., Perucha, E., Jackson, I., et al. (2012). T-bet and GATA3 orchestrate Th1 and Th2 differentiation through lineage-specific targeting of distal regulatory elements. Nat. Commun. 3, 1268. doi: 10.1038/ncomms2260
Kim, E. M., Bae, Y. M., Choi, M. H., Hong, S. T. (2012). Cyst formation, increased anti-inflammatory cytokines and expression of chemokines support for Clonorchis sinensis infection in FVB mice. Parasitol. Int. 61 (1), 124–129. doi: 10.1016/j.parint.2012.07.001
Kimura, A., Kishimoto, T. (2010). IL6: regulator of Treg/Th17 balance. Eur. J. Immunol. 40 (7), 1830–1835. doi: 10.1002/eji.201040391
Layland, L. E., Mages, J., Loddenkemper, C., Hoerauf, A., Wagner, H., Lang, R., et al. (2010). Pronounced phenotype in activated regulatory T cells during a chronic helminth infection. J. Immunol. 184 (2), 713–724. doi: 10.4049/jimmunol.0901435.2010.4.9
Li, B., Qi, X., Liu, Y., Yan, Y., Shan, J., Cai, X., et al. (2022). Monocyte-derived macrophages: The supplements of hepatic macrophage in Echinococcus multilocularis infected mice. Immun. Inflammation Dis. 10 (10), e699. doi: 10.1002/iid3.699
Li, L., Xie, H., Wang, M., Qu, J., Cha, H., Yang, Q., et al. (2017). Characteristics of IL9 induced by Schistosoma japonicum infection in C57BL/6 mouse liver. Sci. Rep. 7, 2343. doi: 10.1038/s41598-017-02422-8
Lopez-Yglesias, A. H., Burger, E., Camanzo, E., Martin, A. T., Araujo, A. M., Kwok, S. F., et al. (2021). T-bet-dependent ILC1- and NK cell-derived IFN-γ mediates cDC1-dependent host resistance against Toxoplasma gondii. PloS Pathog. 17, e1008299. doi: 10.1371/journal.ppat.1008299
McManus, D. P., Gray, D. J., Li, Y., Feng, Z., Williams, G. M., Stewart, D., et al. (2010). Schistosomiasis in the people’s republic of China: the era of the three gorges dam. Clin. Microbiol. Rev. 23 (2), 442–466. doi: 10.1128/CMR.00044-09
Mishra, P. K., Palma, M., Bleich, D., Loke, P., Gause, W. C. (2014). Systemic impact of intestinal helminth infections. Mucosal Immunol. 7 (4), 753–762. doi: 10.1038/mi.2014.23
Nurieva, R. I., Chung, Y., Martinez, G. J., Yang, X. O., Tanaka, S., Matskevitch, T. D., et al. (2009). Bcl6 mediates the development of T follicular helper cells. Sci. (New York N.Y.). 325 (5943), 1001–1005. doi: 10.1126/science.1176676
Oliveira, U. C., Fraga, J. S., Licois, D., Pakandl, M., Gruber, A. (2011). Development of molecular assays for the identification of the 11 Eimeria species of the domestic rabbit (Oryctolagus cuniculus). Vet. Parasitol. 176 (2), 275–280. doi: 10.1016/j.vetpar.2011.10.054
Pakandl, M. (2009). Coccidia of rabbit: a review. Folia parasitologica. 56 (3), 153–166. doi: 10.14411/fp.2009.019
Pang, N., Zhang, F., Ma, X., Zhang, Z., Zhao, H., Xin, Y., et al. (2014). Th9/IL9 profile in human echinococcosis: their involvement in immune response during infection by Echinococcus granulosus. Mediators Inflamm. 2014, 781649. doi: 10.1155/2014/781649
Parhizgar, N., Borji, H., Moazeni, M., Saboor Darbandi, M., Heidarpour, M. (2021). The effects of Zataria multiflora aromatic water and nano emulsion of Z. multiflora essential oil on the serum level of IFN-γ and IL4 in mice with hydatidosis. J. Parasit Dis. 45 (3), 733–737. doi: 10.1007/s12639-021-01353-6
Pesce, J., Kaviratne, M., Ramalingam, T. R., Thompson, R. W., Urban J.R., J. F., Cheever, A. W., et al. (2006). The IL21 receptor augments Th2 effector function and alternative macrophage activation. J. Clin. Invest. 116 (7), 2044–2055. doi: 10.1172/JCI27727
Ramming, A., Druzd, D., Leipe, J., Schulze-Koops, H., Skapenko, A. (2012). Maturation-related histone modifications in the PU.1 promoter regulate Th9-cell development. Blood 119 (20), 4665–4674. doi: 10.1182/blood-2011-11-392589
Ren, J., Li, B. (2017). The functional stability of Foxp3 and RORγt in Treg and Th17 and their therapeutic applications. Adv. Protein Chem. Struct. Biol. 107, 155–189. doi: 10.1016/bs.apcsb.2017.10.002
Seki, T., Kumagai, T., Kwansa-Bentum, B., Furushima-Shimogawara, R., Anyan, W. K., Miyazawa, Y., et al. (2012). Interleukin-4 (IL4) and IL-13 suppress excessive neutrophil infiltration and hepatocyte damage during acute murine Schistosomiasis Japonica. Infect. Immun. 80 (1), 159–168. doi: 10.1128/IAI.05581-11
Solaymani-Mohammadi, S., Eckmann, L., Singer, S. M. (2019). Interleukin (IL)-21 in inflammation and immunity during parasitic diseases. Front. Cell Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00401
Szabo, S. J., Kim, S. T., Costa, G. L., Zhang, X., Fathman, C. G., Glimcher, L. H. (2000). A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100 (6), 655–669. doi: 10.1016/s0092-8674(00)80702-3
Tang, J., Zhu, X., Zhao, J., Fung, M., Li, Y., Gao, Z., et al. (2015). Tissue transglutaminase-regulated transformed growth factor-β1 in the parasite links Schistosoma japonicum infection with liver fibrosis. Mediators Inflamm. 2015, 659378. doi: 10.1155/2015/659378
Turner, J. D., Jenkins, G. R., Hogg, K. G., Aynsley, S. A., Paveley, R. A., Cook, P. C., et al. (2011). CD4+CD25+ regulatory cells contribute to the regulation of colonic Th2 granulomatous pathology caused by schistosome infection. PloS Negl. Trop. Dis. 5 (8), e1269. doi: 10.1371/journal.pntd.0001269
Tuxun, T., Apaer, S., Ma, H-Z., Zhang, H., Aierken, A., Lin, R-Y., et al. (2015). The potential role of th9 cell related cytokine and transcription factors in patients with hepatic alveolar echinococcosis. J. Immunol. Res. 895416.
Uddin, M. H., Li, S., Bae, Y. M., Choi, M. H., Hong, S. T. (2012). Strain variation in the susceptibility and immune response to Clonorchis sinensis infection in mice. Parasitol. Int. 61 (1), 118–123. doi: 10.1016/j.parint.2012.07.002
Wang, H., Jain, S., Li, P., Lin, J. X., Oh, J., Qi, C., et al. (2019). Transcription factors IRF8 and PU.1 are required for follicular B cell development and BCL6-driven germinal center responses. Proc. Natl. Acad. Sci. U.S.A. 116 (19), 9511–9520. doi: 10.1073/pnas.1901258116
Yu, D., Rao, S., Tsai, L. M., Lee, S. K., He, Y., Sutcliffe, E. L., et al. (2009). The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity 31 (3), 457–468. doi: 10.1016/j.immuni.2009.07.002
Zhou, S., Jin, X., Chen, X., Zhu, J., Xu, Z., Wang, X., et al. (2015). Heat shock protein 60 in eggs specifically induces Tregs and reduces liver immunopathology in mice with Schistosomiasis Japonica. PLoS One 10 (9), e0139133. doi: 10.1371/journal.pone.0139133
Zhu, J. (2010). Transcriptional regulation of Th2 cell differentiation. Immunol. Cell Biol. 88, 244–249. doi: 10.1038/icb.2009.114
Keywords: E. stiedai, TBX2, GATA3, RORC, Foxp3, SPI1, BCL6
Citation: Chen X-D, Xie J, Wei Y, Yu J-F, Cao Y, Xiao L, Wu X-J, Mao C-J, Kang R-M and Ye Y-G (2023) Immune modulation of Th1/Th2/Treg/Th17/Th9/Th21 cells in rabbits infected with Eimeria stiedai. Front. Cell. Infect. Microbiol. 13:1230689. doi: 10.3389/fcimb.2023.1230689
Received: 29 May 2023; Accepted: 11 July 2023;
Published: 01 August 2023.
Edited by:
Chao Yan, Xuzhou Medical University, ChinaReviewed by:
Yue Xie, Sichuan Agricultural University, ChinaXiao-Xuan Zhang, Qingdao Agricultural University, China
Copyright © 2023 Chen, Xie, Wei, Yu, Cao, Xiao, Wu, Mao, Kang and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Run-Min Kang, YW5nZWxpbmFfMDcwOEBob3RtYWlsLmNvbQ==; Yong-Gang Ye, eWV5ZzAyMDJAc2luYS5jb20=
 Xiao-Di Chen
Xiao-Di Chen Jing Xie1
Jing Xie1