
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 26 July 2023
Sec. Virus and Host
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1222805
This article is part of the Research Topic Genomic variation and immune escape of viral pathogens View all 5 articles
Live-attenuated influenza A viruses (LAIV) may be superior to inactivated or subunit vaccines since they can be administered via mucosal routes to induce local immunity in the respiratory tract. In addition, LAIV are expected to trigger stronger T-cell responses that may protect against a broader range of antigen-drifted viruses. However, the development of LAIV is challenging since a proper balance between immunogenicity and safety has to be reached. In this study, we took advantage of reverse genetics to generate three LAIV based on the pandemic H1N1 2009 (pH1N1/09) virus strain: ΔPA-X, which is defective in the synthesis of the accessory PA-X protein, NS1(1-126) lacking 93 amino acids at the C-terminus of the NS1 protein, and a combination of both. Characterization of these recombinant viruses using a novel porcine bronchiolar epithelial cell line (T3) revealed that the ΔPA-X mutant replicated similar to wild type (WT) virus. However, in contrast to the parental virus the ΔPA-X mutant allowed transcription of genes involved in cell cycle progression and limits apoptosis. The NS1(1-126) mutant also replicated comparable to WT virus, but triggered the release of type I and III IFN and several chemokines and cytokines. Surprisingly, only the NS1(1-126)/ΔPA-X double mutant was significantly attenuated on T3 cells, and this was associated with enhanced transcription of genes of the innate immune system and complete absence of apoptosis induction. In conclusion, these findings indicate that NS1 and PA-X act in a concerted manner to manipulate the host cell response, which may help to develop swine LAIV vaccine with a more favorable balance of safety and immunogenicity.
Swine influenza A viruses (SIAV) are frequently circulating in pig farms worldwide leading to respiratory disease and significant economic losses. Pigs are also susceptible to both human and avian influenza viruses and have been regarded as a “mixing vessel” where reassortment of gene segments may occur (Scholtissek, 1995). These reassortant viruses shall have new antigenic properties and – following transmission to humans – have the potential to cause a pandemic in the naive human population. In fact, the 2009 H1N1 pandemic virus (pH1N1/09), a triple reassortant virus with gene segments derived from human, porcine and avian IAV (Hajjar and McIntosh, 2010; Mena et al., 2016), is an example for such an event. It is believed that vaccination of pigs may not only reduce the burden of disease in the animals but may also lower the risk of emergence of potentially pandemic viruses.
Inactivated whole influenza virus vaccines (WIV) are already in use for the immunization of SIAV (Gasparini et al., 2011). Although these vaccines can reduce the morbidity of influenza disease in pigs, they also have limitations. Inactivated vaccines do not efficiently stimulate the cellular arm of the immune system and mostly rely on the induction of antibodies that are specifically directed to the variable envelope antigens HA and NA. Consequently, WIV-induced immune responses are specifically directed against the antigens of the virus strain from which the vaccine is derived from, but may not protect against antigen-drifted IAV strains. Moreover, WIV are usually administered via the parenteral route, and trigger the synthesis of serum antibodies which are only partially secreted into the mucosal tissues of the respiratory tract. Therefore, WIV fail to induce sterilizing immunity and do not completely abrogate shedding of influenza viruses from the upper respiratory tract (Heinen et al., 2001; Vincent et al., 2008; Vincent et al., 2010). In contrast, vaccines based on live-attenuated influenza viruses (LAIV) are administered via the intranasal route, which triggers mucosal immune responses that are able to neutralize virus directly at the entry site (Laurie et al., 2010; Dutta et al., 2016). Moreover, LAIV activate the cellular arm of the immune system and therefore act in a more broadly protective manner (Cheng et al., 2013; Janssens et al., 2022). However, it has always been challenging in the past to find the right balance between sufficient attenuation and the remaining immunogenicity of LAIV.
Influenza A viruses expressing a modified nonstructural protein 1 (NS1) have attracted a lot of interest as LAIVs for veterinary use (Nogales et al., 2022). The NS1 protein is a multifunctional protein that has been shown to antagonize type I interferon (IFN-I) production by inhibiting activation of the RIG-I/TRIM25 complex (Kochs et al., 2007), blocking activation of the NFκB transcription factor (Geiss et al., 2002), and by interacting with the host factor CPSF30 (Nogales et al., 2018; Ji et al., 2021). The binding to the cellular 30 kDa subunit of CPSF leads to inhibition of 3’ end formation of cellular pre-mRNAs leading to inhibition and global deregulation of host transcription (host shut-off) (Nemeroff et al., 1998; Nacken et al., 2021). Interestingly, some IAV strains such as pH1N1/09 bind less efficiently to CPSF30 (Clark et al., 2017) and show reduced host shut-off activity, which might explain the relatively mild clinical disease caused by pH1N1/09 (Swedish, 2011). NS1 is also known to block the antiviral effects of IFN-induced antiviral proteins, such as dsRNA-dependent protein kinase R (PKR) and 2’5’-oligoadenylate synthetize (OAS)/RNase L (Min and Krug, 2006; Min et al., 2007; Nogales et al., 2018).
Three functionally distinct domains have been deciphered in the NS1 protein: The N-terminal RNA binding domain (RBD, amino acid 1-73), the effector domain (amino acid 74-207), and the “disordered tail” (amino acid 208-230) (Kim et al., 2021). Analysis of A/Swine/Texas/4199-2/98 (H3N2) (TX/98) with C-terminal truncated NS1 protein (1-73, 1-99, or 1-126) revealed a decreased ability of the recombinant viruses to block IFN-I and IFN-III synthesis, consequently showing an attenuated phenotype in pigs (Solórzano et al., 2005). Subsequent studies demonstrated that intranasal administration of TX/98-NS1(1-126) triggered a mucosal immune response that lead to complete protection against homologous challenge and nearly complete protection against an antigenic H3N2 variant (Richt et al., 2006; Vincent et al., 2007). In 2017, the Tx/98-NS1(1-126) LAIV was licensed in the US for the prevention of influenza in pigs from one day of age and was shown to efficiently reduce nasal shedding after challenge with heterologous IAV strains (Genzow et al., 2018; Kaiser et al., 2019). However, reassortant strains containing LAIV genes in combination with genes from endemic field strains were detected, indicating a substantial degree of LAIV replication and shedding (Mancera Gracia et al., 2020; Sharma et al., 2020).
Apart from NS1, the PA-X protein has also been found to mediate host shut-off activity (Firth et al., 2012). PA-X is translated from an overlapping protein-coding region in IAV segment 3. It is expressed at low levels due to the low efficiency of the ribosomal frameshift (Muramoto et al., 2013). PA-X is composed of the 191 N-terminal amino acids of the PA protein but has a distinct C-terminal sequence of 61 amino acids (Muramoto et al., 2013; Chaimayo et al., 2018). Some IAV, including pH1N1/09, have a truncated form of PA-X lacking 20 amino acids at the C-terminus (Lee et al., 2017). PA-X specifically degrades host mRNA, in particular the mRNA of cellular RNA polymerase II, without affecting viral mRNA (Gaucherand et al., 2019). Recent findings suggest that NS1 and PA-X act in a concerted manner in order to manipulate the host innate immune response (Nogales et al., 2017; Nogales et al., 2018; Gaucherand et al., 2019; Dunagan et al., 2021).
In view of the safety issues associated with swine LAIV vaccines, the present study aimed to identify a strategy to further attenuate SIAV replication but retaining its capacity to stimulate the immune system. To this end, we generated different recombinant porcine originated pH1N1/09 viruses which include a truncated NS1 protein, a knock-out of the PA-X protein, or a combination of both. Using the recently established porcine bronchiolar epithelial porAEC (“T3”) cell line (López-Gálvez et al., 2021; Fleurot et al., 2022), we compared the recombinant viruses with respect to attenuated replication and the induction of chemokines and cytokines such as IFN-I and IFN-III. Our data support the idea that both NS1 and PA-X contribute to the viral host shut-off activity and may explain why the previously licensed NS1-truncated LAIV were not sufficiently attenuated. We propose that the NS1(1-126)/ΔPA-X mutant virus might be a promising swine LAIV vaccine candidate which deserves further evaluation in vivo.
The T3 porcine bronchiolar epithelial cell line was generated by InSCREENeX GmbH (Braunschweig, Germany) and kindly provided by Sasha Trapp (Institut national de la recherche agronomique, INRA, France). The cells were maintained at 37°C and 5% CO2 in porAEC medium with supplement (InSCREENeX, INS-ME-1024) in collagen-coated (InSCREENeX, INS-SU-1017) flasks. Madin-Darby canine kidney type II cells (MDCK-II) were kindly provided by Georg Herrler (University of Veterinary Medicine, Hannover, Germany) and maintained at 37°C and 5% CO2 with minimum essential medium (MEM, Gibco) supplemented with 5% of fetal bovine serum (FBS). Human embryonic kidney (HEK) 293T cells (ATCC) were maintained at 37°C and 5% CO2 with Dulbecco’s Modified Eagle Medium (DMEM, Gibco) supplemented with 10% of fetal bovine serum (FBS). Human type II pneumocyte lung tumor cells (A549) (Sigma-Aldrich) were maintained at 37°C and 5% CO2 in Ham’s F12 medium (Gibco) supplemented with 5% of fetal bovine serum (FBS).
The pHW2000 plasmids encoding the 8 segments of A/Hamburg/4/2009 (H1N1) were kindly provided by Martin Schwemmle (Institute of Virology, University of Freiburg, Germany). The pHW2000-NS plasmid encoding the genomic segment 8 was modified by introducing 4 stop codons into the NS1 open reading frame as previously described (Solórzano et al., 2005), resulting in the C-terminally truncated NS1(1-126) protein. The pHW2000-ΔPA-X plasmid was generated by modifying the nucleotide sequence 592-UCCUUUCGU-600 in the PA gene to 592-UCGUUCAGA-600, following a published strategy (Nogales et al., 2017; Chaimayo et al., 2018). For generation of recombinant virus, a co-culture of MDCK-II and HEK 293T cells was transfected with Lipofectamine 2000 (Life Technologies) and a mixture of the pHW2000 plasmids encoding the 8 viral genomic segments (2 µg of each plasmid). Following incubation of the cells for 24 hours, the medium was replaced by Opti-MEM medium (Life Technologies) containing 1% penicillin/streptomycin and 1 µg/ml of N-tosyl-L-phenylalanine chloromethyl ketone-treated trypsin (TPCK-trypsin, Merck KGgA). The following day, half of the medium was replaced by DMEM containing 0.2% (w/v) of bovine albumin serum (BSA) and 1 µg/ml of TPCK-trypsin. Following an incubation of the cells for 24 hours, the cell culture supernatant was collected, and cell debris removed by low-speed centrifugation. For production of stocks, the viruses were passaged twice on MDCK-II cells in the presence of TPCK-trypsin. Two days post infection (p.i.), the cell culture supernatant was collected, FBS was added (5% final concentration), and cell debris removed by centrifugation. Viruses were aliquoted and stored at -70°C.
The recombinant viruses were titrated on MDCK-II cells grown in 96-well cell culture plates. The viruses were serially diluted (10-fold dilution steps) and 40 µl of each dilution was added to the wells in quadruplicates. Following an incubation for 90 minutes at 37°C, 160 µl of MEM medium containing 1% (w/v) of methylcellulose were added to each well and the cells incubated for 24 hours at 37°C in a 5% CO2 atmosphere. The cells were fixed with 3.7% formalin in PBS (w/v), permeabilized with PBS containing 0.25% of Triton X-100 (v/v), and infected cells detected by indirect immunofluorescence using a monoclonal antibody directed to the influenza virus nucleoprotein (Ocaña-Macchi et al., 2009). Tissue culture infectious dose 50% (TCID50)/ml was calculated according to the Reed-Muench formula (Lei et al., 2020).
To evaluate the replication kinetics of the viruses, MDCK-II, T3 and A549 cells were grown in 6-well cell culture plates and inoculated (3 replicates) with virus for 90 minutes using a multiplicity of infection (MOI) of 0.0001 TCID50/cell. Thereafter, the inoculum was removed, the cells washed once, and maintained in 2.5 ml/well of FBS-deficient MEM medium supplemented with 1% (v/v) penicillin/streptomycin, 1 µg/ml of TPCK-trypsin, either in the presence or absence of 1 µM of ruxolitinib (Selleckchem). At the indicated time points p.i., 250 µl of cell culture supernatant were collected and titrated on MDCK-II cells as described in section 2.2.
To evaluate the plaques characteristics induced by the viruses, MDCK-II and T3 cells were grown in 24-well cell culture plates and inoculated (3 replicates) with virus for 90 minutes using serial diluted virus from 105 to 102 TCID50/ml. Thereafter, the inoculum was removed, the cells washed once, and maintained for 72 hours in 1ml/well of FBS-deficient MEM medium containing 1.2% Avicel (RC-581NF, FMC Biopolymer) supplemented with 1 µg/ml of TPCK-trypsin. Plaques were stained as described in section 2.2. Immunofluorescence images were acquired using a Lionheart FX automated microscope (BioTek) and area was determined using Image J software (Version 1.53t, NIH).
Cells grown in 24-well cell culture plates were inoculated with virus using an MOI of 1 TCID50/cell. Following an incubation for 90 minutes at 37°C, the inoculum was removed, the cells washed three times with PBS, and fresh MEM medium added. The cells were lysed using RAI lysis buffer (Macherey-Nagel). RNA extraction from cell lysates was performed using the NucleoMag Vet kit (Macherey-Nagel) according to the manufacturer’s protocol. Reverse transcription from RNA to cDNA and real time quantitative PCR (qPCR) was performed with the AgPath-ID™ One-Step RT-PCR kit (Life Technologies) using the 7500 Real-Time PCR System (ThermoFisher). Data were acquired and analyzed using the 7500 Fast System SDS software v1.4 (Fisher Scientific) according to the double delta Ct method (2-ΔΔCT) (Livak and Schmittgen, 2001). The endogenous reference gene 18S was used for normalization. Oligonucleotide and probes used in this study are listed in Supplementary Table 1.
MDCK-II cells were grown in 6-well cell culture plates and infected at an MOI of 3 TCDI50/cell and cultured for 20 hours at 37°C. Infected cells were lysed using NP40 lysis buffer (ThermoFisher, cat. No. J60766.AP), containing protease inhibitor cocktail (cOmplete, Merck cat. N° 04693132001). The cell lysate proteins were separated by SDS-PAGE containing a gradient from 4-12% acrylamide (Gene script, cat. No. M00653), in a reducing condition, and then transferred to nitrocellulose membranes. The blots were incubated overnight at 4°C with blocking buffer (LI-COR Biosciences) and subsequently 60 minutes incubation with mouse anti-influenza NS1 N-terminus (1:200, Santa Cruz Biotechnology, cat. no. sc-130568). The blots were washed 4 times with PBS containing 0.1% (v/v) Tween 20 and thereafter incubated in the dark for 60 minutes with goat anti-mouse IgG IRDye hb800CW (LI-COR, cat. no. 926-32210). Following several wash steps with PBS containing 0.1% (v/v) Tween 20, the blots were scanned with the Odyssey Infrared Imaging system (LI-COR Biosciences) and the scans analyzed by Image Studio version 5.2 software (LI-COR Biosciences).
T3 cells grown in 24-well plates were inoculated for 90 minutes at 37°C with the recombinant influenza viruses using an MOI of 1 TCID50/cell. Thereafter, the inoculum was removed and the cells were washed twice with warm PBS, fresh medium was added, then cells were returned to the CO2 incubator at 37°C. At 24 or 48 hours p.i., the cells were washed twice and RNA was extracted using TRIzol Reagent (Life Technologies). The RNA quality was assessed by fragment analysis (5200 Fragment Analyzer CE instrument, Agilent) and sequencing was performed using an Illumina® NovaSeq6000 sequencer (Illumina). Data analysis was performed with R software version 4.2.1 (2022-06-23) (R Core Team 2022). Differential gene expression (DGE) analysis of the experimental groups was performed using the Bioconductor software package DESeq2 v1.36.0. Venn diagrams were created using the list of differentially expressed genes uploaded on an online Venn diagram tool (https://bioinformatics.psb.ugent.be). ClusterProfiler v.4.4.4 (Wu et al., 2021) was used to identify gene ontology terms that were significantly modulated. Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) was performed with ClusterProfiler v4.4.4 (Wu et al., 2021) using MSigDb (Liberzon et al., 2015).
As described above, T3 cells were seeded in 96-well plates and inoculated for 90 minutes at 37°C with recombinant influenza viruses using an MOI of 1 TCID50/cell. The inoculum was removed, the cells washed three times with PBS and then further maintained at 37°C in a 5% CO2 atmosphere. At 24, 48, and 72 hours p.i., caspase 3/7 activity was monitored in cell lysates using the Caspase-Glo-3/7 kit (Promega; cat. no. G8090) according to the manufacturer`s instructions. LDH level was monitored in the supernatant using the LDG-Glo-Cytotoxicity assay kit (Promega; cat. no. J2380) according to the manufacturer’s instructions.
Data analysis and figures were done using GraphPad Prism 8 Software (GraphPad Software). Multiple comparison one-way and two-way ANOVA test were used to determine statistical significance in terms of gene expression, viral titers, and protein activity. P value lower than 0.05 was considered as statistically significant.
Using reverse genetics, we produced three mutant viruses based on the pandemic virus A/Hamburg/4/2009(H1N1) which lacked either the PA-X protein (ΔPA-X), the last 93 amino acids of the NS1 protein [NS1(1-126)], or a combination of NS1(1-126)/ΔPA-X mutations (Figures 1A, B). The mutations were confirmed by Sanger sequencing of the targeted genome segments. Western blot analysis of Madin-Darby canine kidney cell type II (MDCK-II) infected cell lysates indicated a molecular weight shift for the NS1(1-126) protein (Figure 1C). We speculate that the stability of this truncated protein is highly compromised and subject to faster degradation than full-length NS1 protein which explain the weaker expression compared to WT NS1. Unfortunately, we couldn’t detect the PA-X protein in wild-type virus infected cells, most likely because of low expression levels, high turnover rates (Levene et al., 2021) and no specific available antibody against pH1N1/09 PA-X.
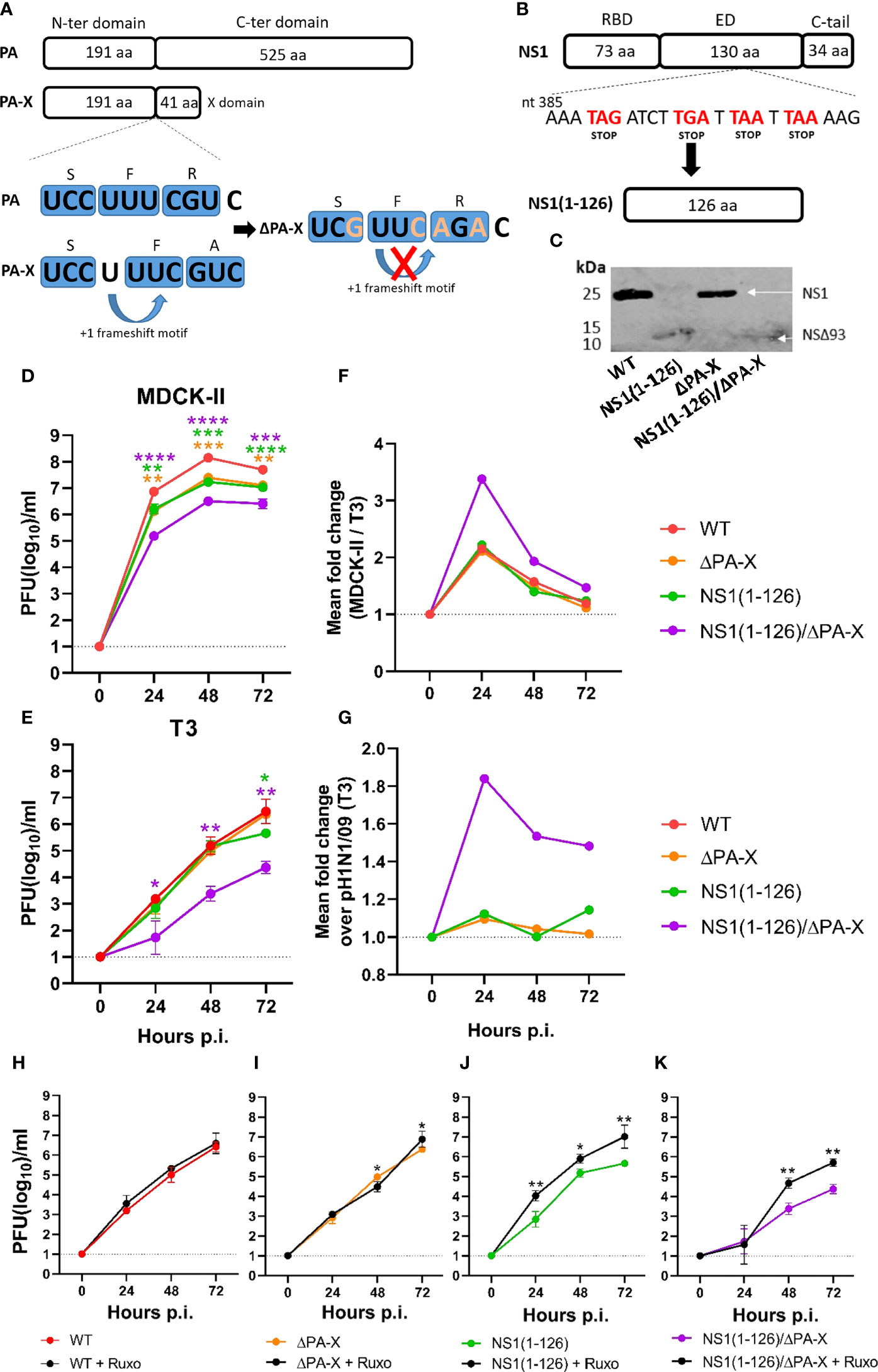
Figure 1 Replication of NS1 and PA-X mutants in MDCK-II and T3 cells. (A) Schematic representation of the PA and PA-X proteins and the mutations (orange letters) introduced into the frameshift motif to inhibit PA-X synthesis. (B) Schematic representation of NS1 protein and the inclusion of four stop codons (red letters) resulting in the truncated NS1(1-126) protein. (C) MDCK-II cells were infected with the indicated viruses using an MOI of 3 TCID50/cell and lysed 20 hours p.i. The NS1 and NS1(1-126) proteins were detected by Western blot using a monospecific antibody directed to the NS1 N-terminus. (D) MDCK-II and (E, H–K) T3 cells were infected with the indicated viruses using an MOI of 0.0001 TCID50/cell and maintained at 37°C in the presence or absence of ruxolitinib. At the indicated times, cell culture supernatant was collected and infectious virus titers determined. (F) Fold change in MDCK-II over T3 cells. (G) Fold change in T3 cells infected with the mutant viruses compared to WT virus. Significant differences were determined comparing WT to NS1(1-126), ΔPA-X, or NS1(1-126)/ΔPA-X (D–E) and each virus with ruxolitinib compared to the same virus without ruxolitinib using the two-way ANOVA test (H–K). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 indicate significant difference.
Virus replication kinetics were analyzed using two different cell lines, MDCK-II cells, a commonly used cell line for influenza A virus replication (Zhai et al., 2012), and the more relevant porcine bronchiolar epithelial cell-derived T3 cells (López-Gálvez et al., 2021; Fleurot et al., 2022). In fact, transcriptomic profiling of T3 cells demonstrated their similarity to primary upper respiratory tract epithelial cells (Supplementary Figure 1).
Both cell types were infected with the recombinant pH1N1/09 mutants using an MOI of 0.0001 TCID50/cell. Virus released into the cell culture supernatant was collected at 0, 24, 48, and 72 hours post infection. (p.i.), and the infectious titers determined. In MDCK-II cells, infectious titers of both NS1(1-126) and ΔPA-X were reduced by one log10 compared to WT virus (Figure 1D). For the double mutant NS1(1-126)/ΔPA-X, titers dropped by two log10 when compared to WT pH1N1/09 (Figure 1D). In T3 cells, all recombinant viruses replicated slower and to generally lower titers than in MDCK-II cells (Figures 1E, F). The ΔPA-X mutant did not show any signs of attenuation and NS1(1-126) showed a drop of infectious titer only at 72 hours p.i. (Figures 1E, G). Similar to what has been observed in MDCK-II cells, the NS1(1-126)/ΔPA-X mutant was the most attenuated virus reaching only 104 pfu/ml at 72 hours p.i. (Figures 1E, G). Replication of WT virus and ΔPA-X mutant in T3 cells was not affected by ruxolitinib (Figures 1H, I), an inhibitor of the JAK1 and 2 signaling pathway (Verstovsek, 2009). However, replication of NS1(1-126) and NS1(1-126)/ΔPA-X was partially rescued in the presence of the inhibitor, reaching similar titers as WT virus (Figures 1J, K). These findings indicate that both NS1 and PA-X protein need to be modified in order to suppress virus replication in T3 cells.
The attenuation of the NS1(1-126) and NS1(1-126)/ΔPA-X mutants was also evident from the smaller size of plaques that were formed and/or lower stained antigen expressed by these viruses in MDCK-II cells (Supplementary Figures 2A, B), as well as from replication kinetics performed in A549 cells (Supplementary Figure 2C), a human type II pneumocyte-derived tumor cell line, which has been frequently used to study influenza A virus infection in human (Giard et al., 1973; Ujie et al., 2019). Cell infection but no plaques formation in T3 cells highlight the more resistant phenotype of these cells to influenza infection
To study the cellular response, T3 cells were infected with WT and mutant viruses at an MOI of 1 TCID50/cell and the transcriptome of the cells was determined at 24 and 48 hours p.i. by RNA sequencing. These time points were chosen since RT-qPCR analysis did not indicate earlier induction of the IFNL3 and MX1 gene transcription (Supplementary Figure 3A). At 24 hours p.i., principal component analysis (PCA) showed clustering of WT pH1N1/09 and ΔPA-X mutant virus with mock-infected cells, but clear separation from NS1(1-126) and NS1(1-126)/ΔPA-X with respect to principal component 1 (PC1) (Figure 2A). At 48 hours p.i., the pronounced effect by the NS1(1-126) and NS1(1-126)/ΔPA-X mutants on PC1 were maintained, but now the WT and ΔPA-X mutant viruses also clearly separated from mock-treated cells on PC2 (Figure 2B).
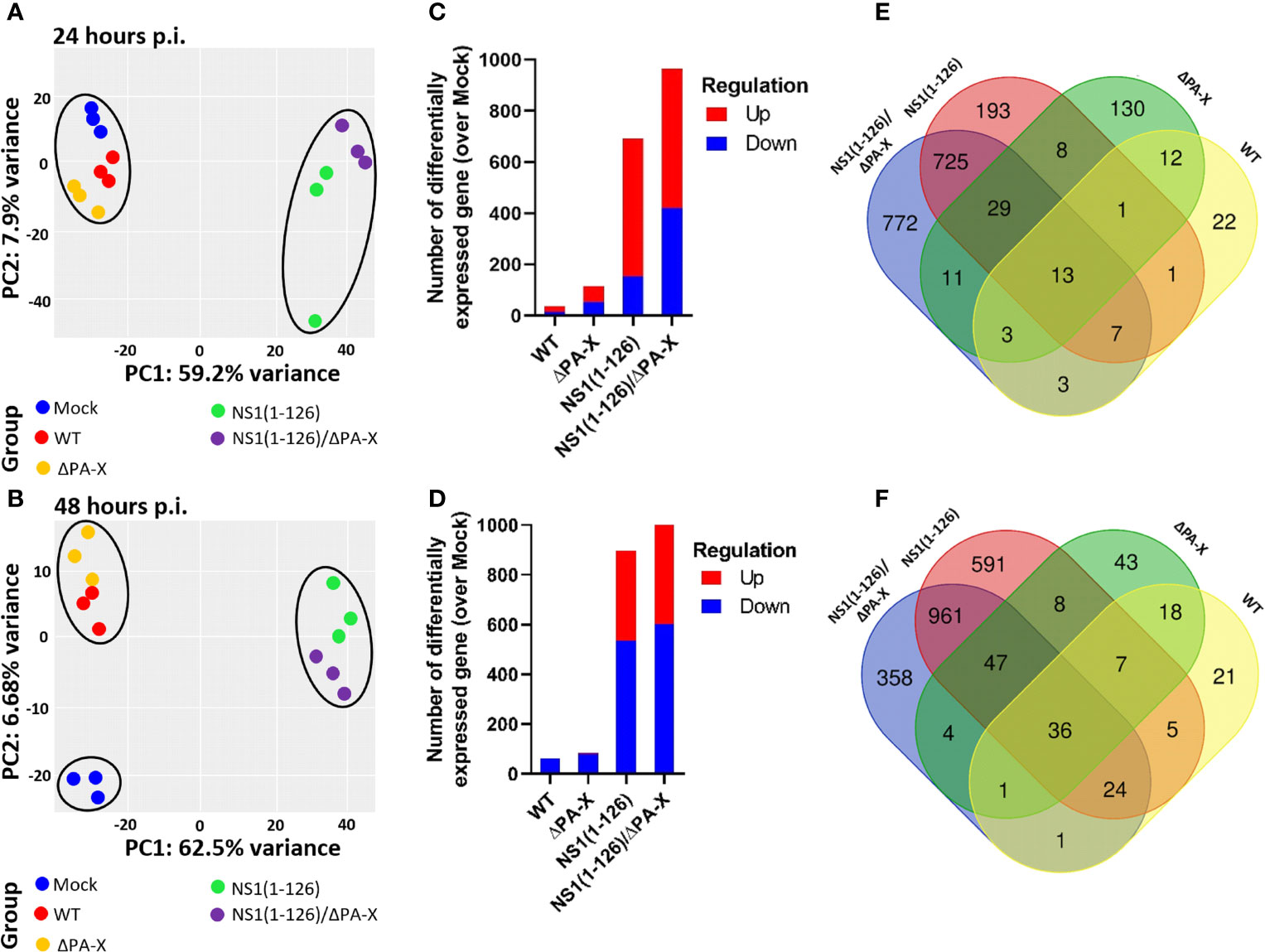
Figure 2 General effects of NS1 and PA-X on T3 cell gene expression. (A, B) PC1 and PC2 analysis of T3 cells at 24 hours (A) and 48 hours (B) p.i. with either WT, ΔPA-X, NS1(1-126) or NS1(1-126)/ΔPA-X virus. (C, D) Number of differentially expressed genes (DEGs) in infected cells over Mock infected cells at 24h p.i. (C) and 48 hours p.i. (D). (E, F) Venn diagrams showing the overlap of DEGs in T3 cells at 24 hours p.i. (E) and 48 hours p.i. (F) with the indicated viruses.
In addition to PCA, differentially expressed genes (DEGs) were compared in infected over mock-treated cells. While WT virus and ΔPA-X mutant induced only a few DEGs (Figures 2C, D), the NS1(1-126) mutant and in particular the NS1(1-126)/ΔPA-X mutant triggered the induction of a high number of DEGs in T3 cells (Figures 2C, D). At 48 hours p.i., the number of downregulated genes increased, especially in NS1(1-126) and NS1(1-126)/ΔPA-X infected cells (Figure 2D). A heatmap of the 500 most significantly differentially expressed genes showed clustering between mock, WT and ΔPA-X on the one hand and between NS1(1-126) and NS1(1-126)/ΔPA-X on the other hand (Supplementary Figure 4). Venn diagrams demonstrated the pivotal role of the NS1(1-126) mutant in triggering transcriptional changes at 24 and 48 hours p.i., and also highlighted the distinct effect of the ΔPA-X mutant on T3 gene expression (Figures 2E, F). At 24 hours p.i. in particular, the majority of the ΔPA-X induced DEGs was found to be unique to this mutant (Figure 2E). More than 50% of the differentially expressed genes in NS1(1-126)/ΔPA-X infected cells were not shared by cells infected with the NS1(1-126) mutant (Figure 2E).
A volcano plot pointed out that most of the transcripts that were induced in T3 cells by the NS1(1-126) and NS1(1-126)/ΔPA-X mutants belong to the IFN responsive gene cluster, while this response appeared to be absent in the WT and ΔPA-X infected T3 cells (Supplementary Figures 5, 6). Analysis of genes involved in pathogen sensing, IFN responses, and inflammation revealed that the transcription of genes involved in RNA sensing was upregulated in T3 cells only if the cells were infected with NS1(1-126) or NS1(1-126)/ΔPA-X, whereas infection with either WT pH1N1/09 or ΔPA-X mutant did not enhance transcription of these genes (Figures 3A, B, “PRR sensing and receptor”). We found that compared to NS1(1-126) the NS1(1-126)/ΔPA-X mutant triggered a more pronounced upregulation of TLR3, IRF7, RIG-I, and STAT1 transcription early during the infection (24 hours p.i.), although this difference was not apparent at 48 hours p.i. (Figures 3A, B, “PRR sensing and receptor”).
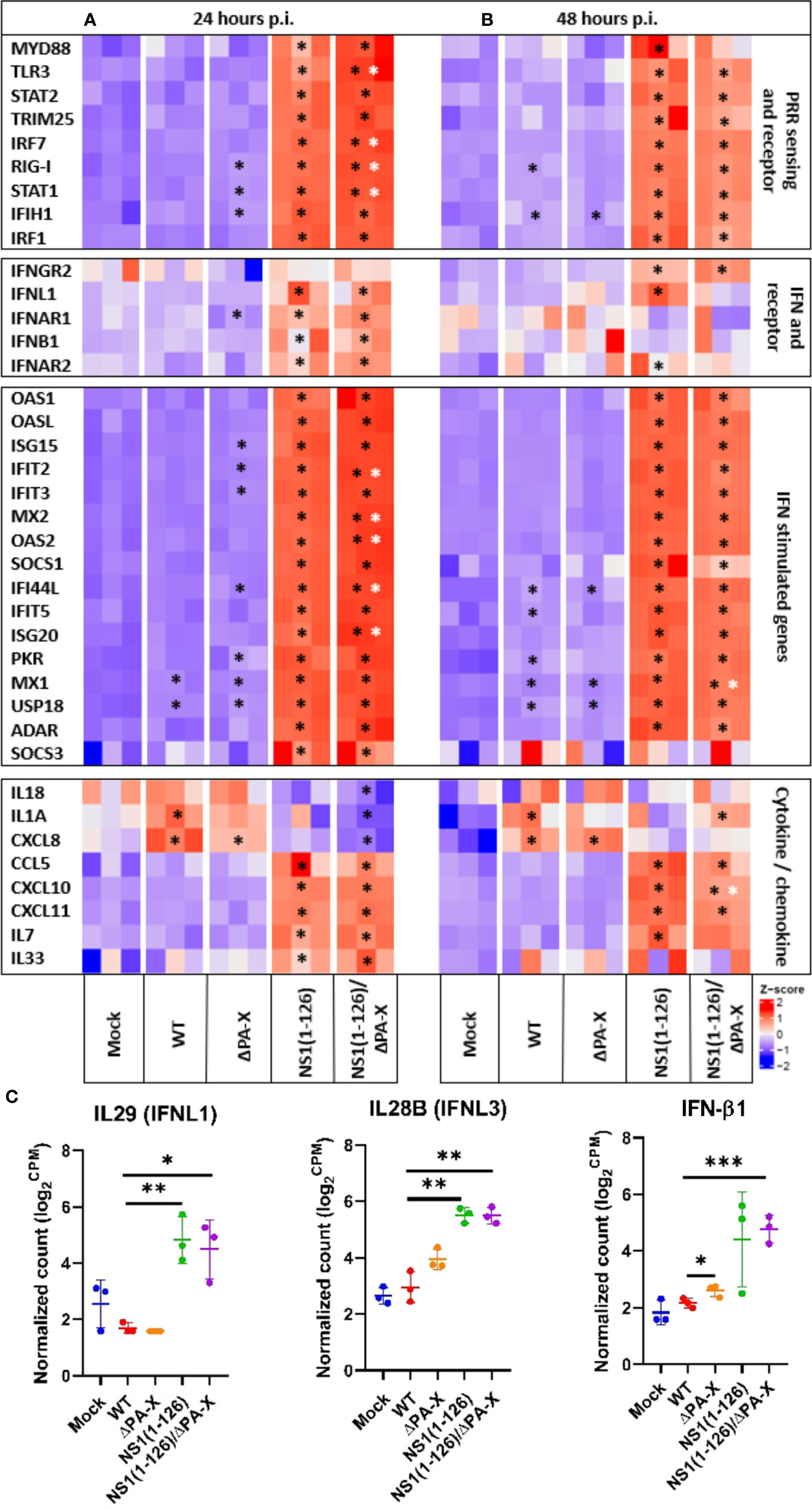
Figure 3 Relative expression levels of genes of the innate immune system in infected T3 cells. Heatmap illustrating the relative expression levels of genes of the innate immune system including PRR sensing and adapter genes, IFN and IFN receptor genes, IFN-stimulated genes and various cytokine and chemokine genes of T3 cells 24 hours (A) and 48 hours (B) p.i. Genes from infected cells showing significant differences to mock-infected cells are indicated by black stars, while genes of NS1(1-126)/ΔPA-X infected cells showing significant differences compared to NS1(1-126) infected cells are indicated by white stars. The Z score of each heat map is represented in the lower right corner. The adjusted p-value of each comparison is shown in Supplementary Tables 2, 3. (C) Normalized count in log2 count per million (CPM) obtained from the transcriptomic reads count at 24 hours p.i. *p<0.05, **p<0.01, ***p<0.001, indicate significant difference.
Transcriptomic analysis showed that the IFN response was exclusively triggered by the NS1(1-126) and NS1(1-126)/ΔPA-X mutants (Figures 3A, B, “IFN and IFN receptor”, and C); moreover, IFN-III appeared to be particularly affected, in accordance with the epithelial characteristics of the T3 cells. Indeed, the expression of IFN-β1 was only transiently upregulated in cells infected with either the NS1(1-126) or the NS1(1-126/ΔPA-X mutant (Figures 3A, B, “IFN and IFN receptor”, and C). RT-qPCR analysis confirmed the upregulation of IFN-β1 by the NS1(1-126)/ΔPA-X mutant at 24 hours p.i. (Figure 4), while transcription of IFNL1 and IFNL3 genes was triggered by both the NS1(1-126) and the NS1(1-126)/ΔPA-X mutant as evident from the transcriptome and RT-qPCR analysis (Figures 3A, B, “IFN and IFN receptor”, C, 4). IFN-α was not detected neither by RT-qPCR (Supplementary Figure 3B) nor by transcriptomic analysis.
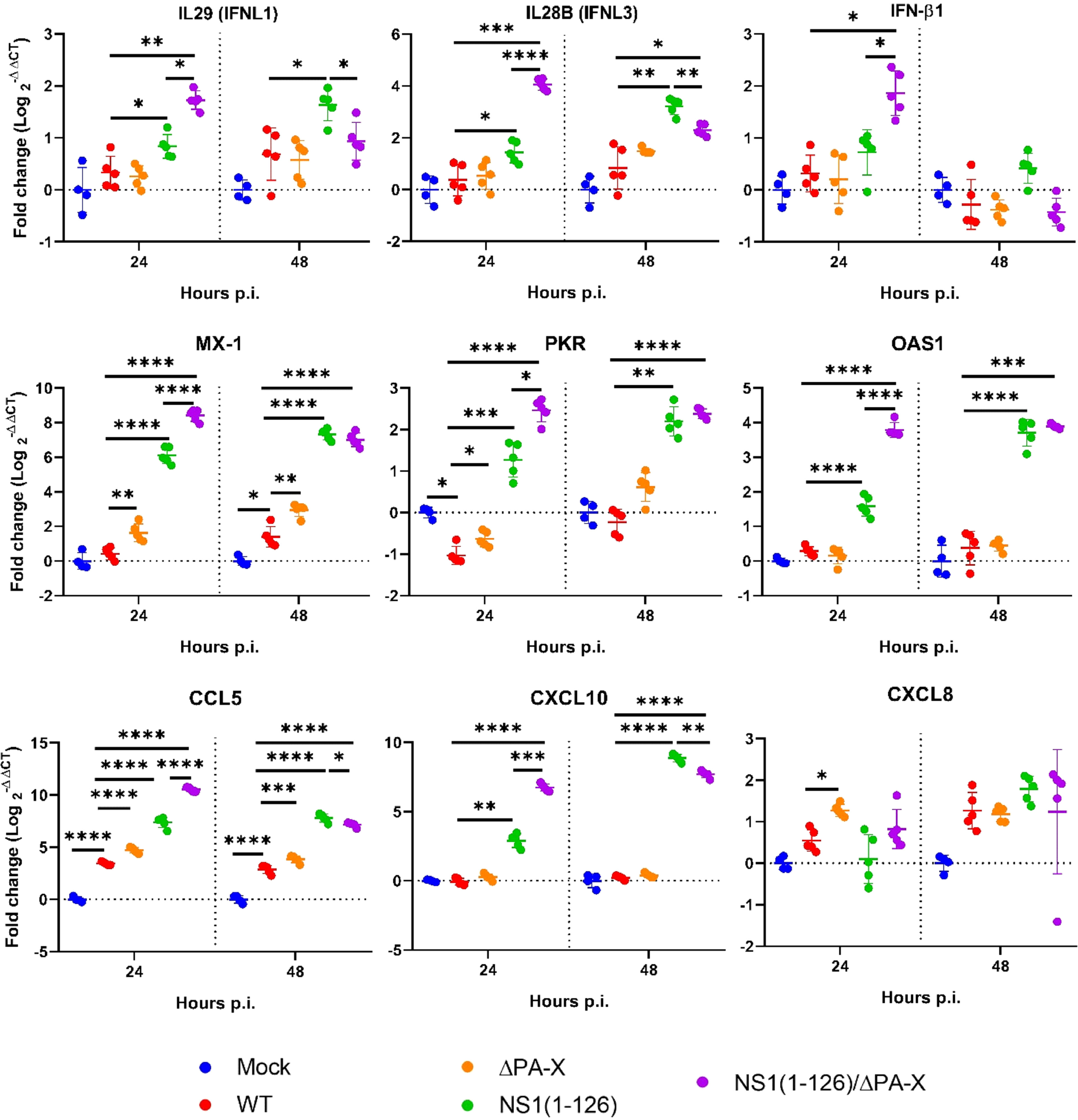
Figure 4 Relative expression levels of selected genes as assessed by RT-qPCR. T3 cells were infected with the indicated viruses using an MOI of 1 TCID50/cell. At 24 and 48 hours p.i., real-time RT-qPCR was performed to quantify mRNA levels relative to their expression in mock-infected cells. Significant differences were determined comparing Mock to WT, WT to NS1(1-126), ΔPA-X, or NS1(1-126)/ΔPA-X and NS1(1-126) to NS1(1-126)/ΔPA-X using multiple comparison one-way ANOVA test. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001 indicate significant difference.
IFNs trigger the expression of interferon-stimulated genes (ISGs), of which several have antiviral functions (e.g., MX1, OAS and PKR), but they also activate the transcription of genes that encode for factors that terminate the IFN response (e.g. SOCS family, USP18). In accordance with the induction of PRR and IFN, infection with NS1(1-126) or NS1(1-126)/ΔPA-X mutants resulted in dramatically high ISG levels while the transcriptome of cells infected with WT pH1N1/09 was quite similar to the transcriptome of mock-infected cells (Figures 3A, B, “IFN stimulated genes” and 4, Supplementary Tables 2, 3). Interestingly, the ΔPA-X mutant also increased expression levels of some ISGs including MX1, PKR and USP18 as evident from both transcriptome analysis and RT-qPCR even though enhanced IFN expression was not observed (Figure 4; Supplementary Figure 3B). Moreover, the NS1(1-126)/ΔPA-X mutant induced significantly higher levels of some ISGs including IFIT2, MX2, OAS2, IFI44L, and IFIT5, compared to NS1(1-126) at 24 hours p.i. (Figures 3A, B, “IFN stimulated genes”). Interestingly, the NS1(1-126)/ΔPA-X mutant induced a faster IFN response in T3 cells compared to the NS1(1-126) mutant with higher levels of mRNA detected by RT-qPCR at 24 hours p.i. However, the IFN response observed for the NS1(1-126)/ΔPA-X mutant at 48 hours p.i. dropped below the response observed for the NS1(1-126) mutant (Figure 4).
IAV infection usually results in the expression of inflammatory and homeostatic cytokines and chemokines that recruit immune cells to the site of infection (Gu et al., 2019). We therefore evaluated the capacity of WT and mutant pH1N1/09 to trigger the expression of cytokines and chemokines in T3 cells. At both 24 and 48 hours p.i., a significantly upregulated transcription of inflammatory cytokines including IL-1α, IL-18 and CXCL8 was detected for the WT virus and the ΔPA-X mutant by transcriptome analysis (Figures 3A, B, “cytokine/chemokine”) and by RT-qPCR (Figure 4). The NS1(1-126) mutant showed no significant changes compared to mock-treated cells. Interestingly, the NS1(1-126)/ΔPA-X mutant significantly downregulated transcription of these pro-inflammatory cytokines compared to mock-infected cells at 24 hours p.i., but significantly upregulated IL-1α at 48 hours p.i. (Figures 3A, B, “cytokine/chemokine”). Moreover, homeostatic cytokines such as IL-33, an alarmin involved in tissue homeostasis and repair, or the hematopoietic growth factor IL-7, were transcriptionally upregulated by the NS1(1-126)/ΔPA-X and NS1(1-126) mutants at 24 hours p.i., while the WT pH1N1/09 virus and the ΔPA-X mutant did not trigger such a response (Figures 3A, B “Cytokine/chemokine”).
In addition to cytokines, changes in chemokine levels were also noticed, in particular if the cells were infected with the NS1(1-126) or the NS1(1-126)/ΔPA-X mutant. Indeed, infection with these virus mutants induced enhanced transcription of chemokine genes that are involved in the recruitment of innate immune cells such as CCL5, CXCL8, and CXCL10, both at 24 and 48 hours p.i. (Figures 3A, B, “cytokine/chemokine” and 4). Moreover, the NS1(1-126) and NS1(1-126)/ΔPA-X mutants, but not WT pH1N1/09 and the ΔPA-X mutant, induced higher levels of chemokines involved in the recruitment of lymphocytes such as CXCL10 and CXCL11.
To better understand how T3 porcine alveolar epithelial cells respond to infection by WT and mutant pH1N1/09, a gene ontology analysis using the “biological process” gene sets (GO-BP) was performed (Figure 5). We observed that infection with either WT virus or ΔPA-X mutant resulted in strong enrichment of gene transcripts encoding proteins involved in cell cycle regulation, such as E2F targets and G2/M checkpoint hallmarks (Figures 5A, B). Both E2F targets and G2/M checkpoint gene products ensure and promote correct cell cycle progression and induce apoptosis in damaged cells (Stark and Taylor, 2004; Iaquinta and Lees, 2007). E2F targets and G2M checkpoint remained upregulated for the WT virus and the ΔPA-X at both 24 and 48 hours p.i. (Figures 5A, B, E, F). The NS1(1-126) mutant had no impact on E2F targets and G2M checkpoint at both 24 and 48 hours p.i. (Figures 5C, G). However, infection with the NS1(1-126)/ΔPA-X mutant led to a significant suppression of E2F targets at 24 hours p.i. (Figure 5D). In line with our previous observations (see Figure 3), IFN transcripts were only slightly upregulated in cells infected with either the WT pH1N1/09 or the ΔPA-X mutant. In contrast, the strongest IFN transcripts upregulation was found in NS1(1-126) and NS1(1-126)/ΔPA-X infected cells, at both time points (Figure 5). Interestingly, the GO-BP “inflammatory response” was activated by all four viruses. In addition to that, the gene transcripts involved in oxidative phosphorylation were highly downregulated in T3 cells at 48 hours p.i. with either the NS1(1-126)/ΔPA-X or the NS1(1-126) mutant (Figures 5G, H). The ability to suppress oxidative phosphorylation is intriguing since an upregulation was reported to be beneficial for influenza replication (Foo et al., 2022). Overall, these data indicate that infection of T3 cells by WT pH1N1/09 virus or the ΔPA-X mutant resulted in enhanced transcription of genes involved in cell cycle progression while NS1(1-126)/ΔPA-X mutants lost this capacity.
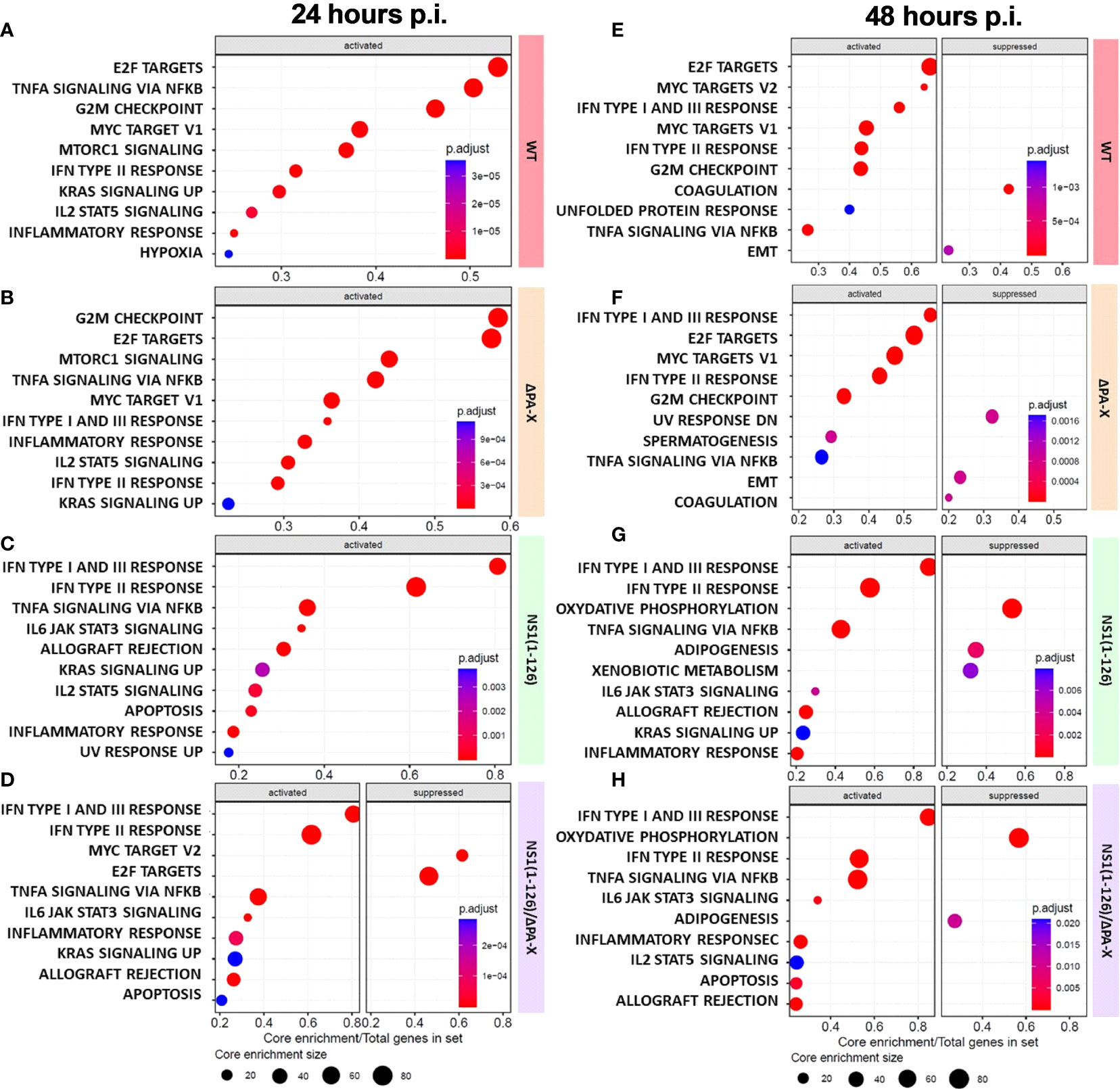
Figure 5 Gene set enrichment analysis (GSEA) of infected compared to mock-treated T3 cells. (A–H) GSEA of the 10 most enriched gene sets (activated or suppressed) at 24 hours (A–D) and 48 hours (E–H) p.i. of T3 cells with the indicated viruses. The enriched gene sets are displayed on the left side and the core enrichment sizes are depicted at the bottom of each panel. The adjusted P-values for each GSEA are represented by the relative size of the dots shown. The viruses used are displayed on the right-hand side of each panel.
To better understand how ΔPA-X mutant modulates the host response compared to WT virus, and how the response induced by the NS1(1-126) mutant was modulated in the absence of the PA-X protein, we analyzed the top 10 gene set enrichments of cells infected with either the ΔPA-X or the NS1(1-126)/ΔPA-X mutant compared to cells infected by either WT virus or NS1(1-126) mutant respectively (Figures 6A–C). Strikingly, even though both WT virus and ΔPAX mutant induced the upregulation of cell cycle gene transcripts. (Figures 5A, B, E, F), the ΔPA-X mutant additionally enhanced the transcription of E2F targets and G2/M checkpoints at 24 hours p.i. (Figure 6A), which also induced a strong upregulation of cyclin-dependent kinase (CDK), G1/S and G2/mitotic-specific cyclin (CCN), and cell cycle gene (CDC) (Figure 6D). Notably, top 10 genes set enrichment revealed that the inflammatory response (TNF-α signaling via NFκB) was also suppressed in the absence of PA-X. Interestingly, cholesterol homeostasis gene sets were upregulated (Figure 6A), suggesting that PA-X plays a role in regulating steroid biogenesis, a phenomenon previously observed for influenza A virus infection (Bai et al., 2022) but not yet linked to PA-X. Of note, no changes were observed for the ΔPA-X mutant compared to WT pH1N1/09 at 48 hours p.i. (Supplementary Figure 7).
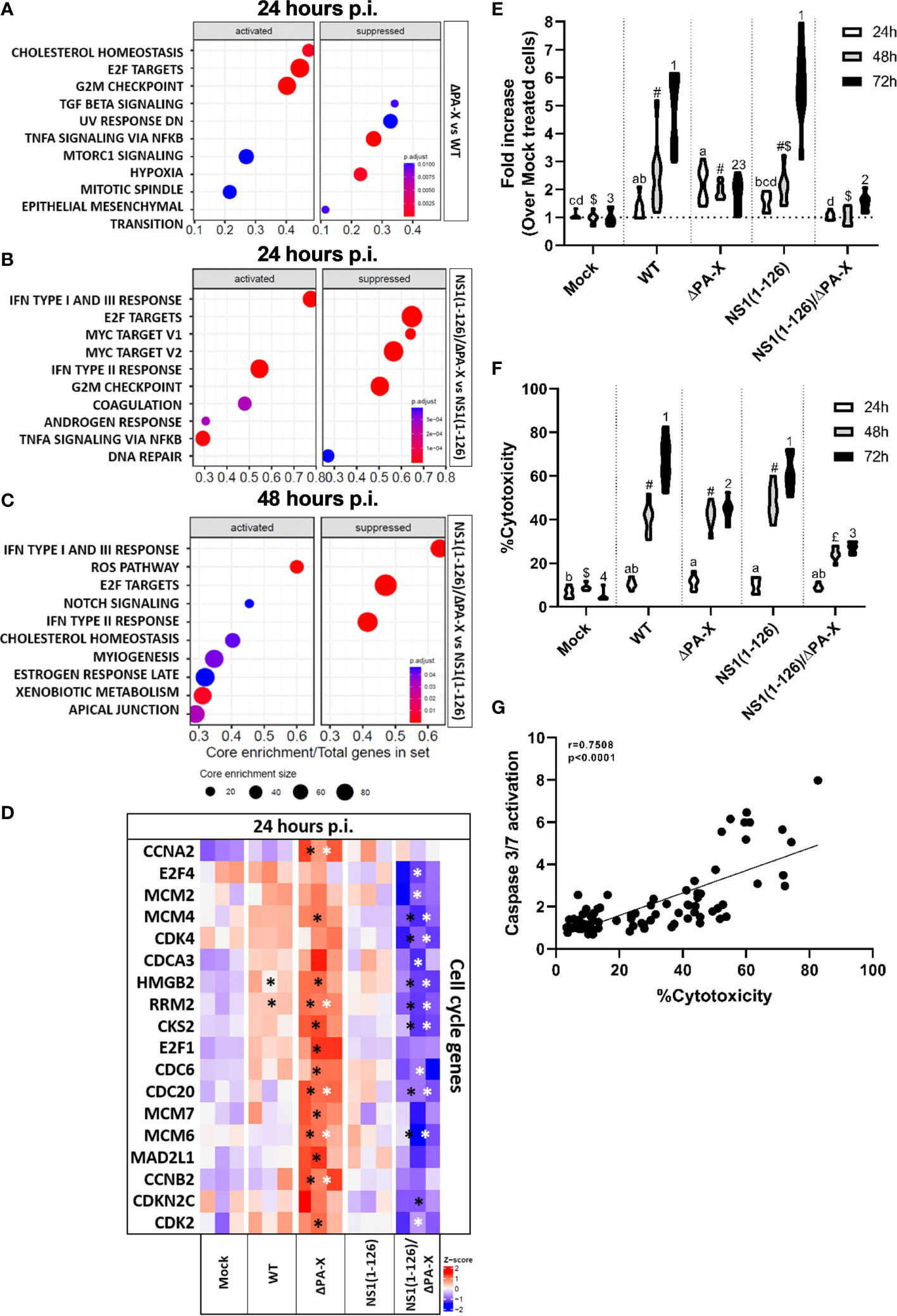
Figure 6 Comparison of ΔPA-X to WT and NS1(1-126)/ΔPA-X to NS1(1-126) virus-infected T3 cells. (A) GSEA of the 10 most enriched gene sets in T3 cells infected with ΔPA-X compared to WT 24 hours p.i. (B) GSEA of the 10 most enriched pathways in T3 cells infected with NS1(1-126)/ΔPA-X compared to NS1(1-126) at 24 hours p.i. (C) GSEA of the 10 most enriched pathways in T3 cells infected with NS1(1-126)/ΔPA-X compared to NS1(1-126) at 48 hours p.i. (D) Heatmaps illustrating the relative expression levels of important differentially expressed cell cycle genes in T3 cells 24 hours p.i. with the indicated viruses. Genes from infected cells showing significant differences to mock-infected cells are indicated by black stars while genes of ΔPA-X and NS1(1-126)/ΔPA-X virus-infected cells showing significant differences compared to WT and NS1(1-126) virus-infected cells, respectively, are indicated by white stars. The Z-score indicates upregulated (red color) or downregulated gene expression (blue color). The adjusted p-value of each comparison is presented in Supplementary Table 4. (E) Detection of caspase-3/7 activity in T3 cells at 24, 48, and 72 hours p.i. with the indicated viruses. (F) Detection of LDH release in the supernatant in T3 cells at 24, 48, and 72 hours p.i. with the indicated virus. For (E, F), conditions showing no significant difference at 24 hours p.i. are marked by the same letter, no significant difference at 48 hours p.i. are marked by the same symbol, and no difference at 72 hours p.i. are marked by the same number. Activation of caspase 3/7 and LDH release was correlated (G). Significant differences were assessed using a multiple comparison two-way ANOVA test (E, F) and Spearman’s correlation (G). Genes from infected cells showing significant differences to mock-infected cells are indicated by *.
Interestingly, avoiding PA-X expression in the NS1(1-126) background caused different gene set enrichments when compared to virus with fully functional NS1 (Figures 6B, C). Infection with the NS1(1-126)/ΔPA-X mutant caused a pronounced downregulation of cell cycle gene transcripts compared to the NS1(1-126) mutant, and this response was still present at 48 hours p.i. (Figures 6B, C). CDK, CDC and CNN gene family transcripts were significantly more downregulated by the NS1(1-126)/ΔPA-X mutant compared to the NS1(1-126) mutant (Figure 6D; Supplementary Table 4).
Cell cycle disturbance is often associated with activation of apoptosis (Pucci et al., 2000). To figure out whether PA-X triggers apoptosis or not, caspase 3/7 activity was measured at 24, 48, and 72 hours p.i. with the WT and mutant viruses (Figure 6E). Infection of T3 cells with WT pH1N1/09 resulted in increasing levels of active caspase 3/7 from 24 to 72 hours p.i. Interestingly, the ΔPA-X mutant induced a weak caspase 3/7 activation along the experiment, which might be due to a downstream effect of disturbance of cell cycle regulation. Moreover, NS1(1-126) did not show any difference compared to WT pH1N1/09 with increasing caspase 3/7 activity over time. Even though infection with the NS1(1-126)/ΔPA-X mutant resulted in reduced transcription of cell cycle genes, this mutant induced a very weak and late apoptosis activation, that it was lower to this induced by the ΔPA-X mutant. To confirm that the activation of caspase 3/7 induce apoptosis, release of lactate dehydrogenase (LDH) in the supernatant was measured at 24, 48 and 72 hours p.i. with the WT and mutant viruses (Figure 6F). Increasing level of LDH was measured for WT and NS1(1-126) from 24 to 72 hours p.i., while LDH release increase between 24 et 48 hours p.i. but remained at the same level between 48 and 72 hours p.i. for ΔPA-X and NS1(1-126)/ΔPA-X. Activation of caspase 3/7 and LDH release upon infection with WT and mutant viruses show strong correlation over time (Figure 6G).
IAV with modified NS1 genes has long been considered as potential LAIV candidates (Nogales et al., 2022). However, it turned out that it is extremely difficult to achieve an appropriate balance between attenuation and safety on one hand and immunogenicity and vaccine efficacy on the other hand (McLean and Belongia, 2021). In addition to the NS1 protein, the recently discovered accessory PA-X protein was also shown to exhibit host shut-off activity, and therefore represents an attractive target for the generation of LAIV (Hayashi et al., 2015; Nogales et al., 2018; Hilimire et al., 2020; Dunagan et al., 2021).
In the present work, we took advantage of reverse genetics to generate viruses with a truncated NS1(1-126) protein and/or a knockout of the PA-X protein that were based on a swine-origin human pandemic influenza virus isolate, A/Hamburg/4/2009 (H1N1), and investigated the response of a recently established T3 porcine bronchiolar epithelial cell line to infection with these viruses. We found that infection of T3 cells with pH1N1/09 WT virus resulted in a moderate suppression of host gene expression including type I and type III IFNs. In line with the absence of IFN, the Janus kinase inhibitor ruxolitinib had no effect on WT virus replication.
The ΔPA-X virus showed a very similar replication kinetics as WT virus, and like WT virus, was not affected by ruxolitinib. In line with this phenotype, infection with ΔPA-X virus did not result in the upregulation of IFNs and ISGs expression. However, in contrast to WT virus a significant increase in the transcription of cell cycle-related genes was observed at 24 hours p.i. Moreover, ΔPA-X virus did not trigger apoptosis to the same extent as WT virus, although both viruses replicated to similar titers. This suggests that the PA-X protein of pH1N1/09 does not cause a global host shut-off but may specifically target only certain genes. Our findings seem to contradict previous works showing that PA-X is a key factor that mediates a global host shut-off (Hayashi et al., 2015; Gaucherand et al., 2019; Ma et al., 2019). However, this discrepancy might be explained by the fact that pH1N1/09 expresses a C-terminally truncated PA-X protein of 232 amino acids (Shi et al., 2012), whereas most IAV encode PA-X proteins of 252 amino acids. The missing C-terminal region of 20 amino acids is known to contribute to PA-X host shutoff activity (Bavagnoli et al., 2015; Gao et al., 2015; Hu et al., 2018). Indeed, apart from pandemic 2009 H1N1 viruses, equine H7N7, canine H3N8, canine H3N2 and bat influenza viruses, show a truncation of the PA-X C-terminal domain, which most likely represents an adaptation to their hosts. The truncated C-terminus of the PA-X protein might explain the little impact the ΔPA-X virus had on the transcription of most genes in infected T3 cells. However, loss of PA-X in the related A/California/04/2009 (H1N1) has been reported to cause a reduction in viral replication and pathogenicity as well as increased innate immune responses in mice (Gao et al., 2015; Hayashi et al., 2015; Lee et al., 2017). Nevertheless, it should also be considered that the PA-X protein may mediate host shut-off activity in a species- and/or cell type-specific manner.
In contrast to WT and ΔPA-X virus, infection of T3 cells with NS1(1-126) virus triggered the transcription of various host factors including IFN genes and ISGs. Consequently, this virus replicated to lower titers on these cells and the attenuation was overcome by the Janus kinase inhibitor ruxolitinib, indicating that the attenuation of the virus was at least in part due to the induction of IFN. Interestingly, the corresponding NS1(1-126) mutant of TX/98 was found to replicate on porcine PK15 cells in a much more attenuated manner than our pH1N1/09 based NS1(1-126) mutant (Solórzano et al., 2005). This discrepancy could be a result of cell-type specific differences between PK15 and T3 cells, but might also be a consequence of the different IAV strains used. It should be noted that the NS1 protein of the pH1N1/09 WT virus is characterized by an 11 amino acid truncation, which has been linked to inefficient inhibition of host gene expression without affecting virus replication (Tu et al., 2011). In addition, the 2009 pandemic H1N1 IAV NS1 protein has been shown to be unable to block general host gene expression in human or porcine cells due to low binding to cellular pre-mRNA processing protein CPSF30 (Hale et al., 2010).
In contrast to the NS1(1-126) mutant, the NS1(1-126)/ΔPA-X double mutant was clearly attenuated in T3 cells. The attenuated replication of NS1(1-126)/ΔPA-X was partially restored by ruxolitinib, indicating that the attenuation was only partially mediated by IFN. In line with this observation, the NS1(1-126)/ΔPA-X double mutant induced expression of IFNs and ISGs at higher levels than the NS1(1-126) mutant early in infection (24 hours p.i., see Figure 4). However, in contrast to NS1(1-126) virus, the NS1(1-126)/ΔPA-X double mutant strongly suppressed the transcription of genes that are involved in cell cycle regulation. The reasons for this synergistic effect are not known but it might be speculated that there is a cross-talk between cell cycle regulators (suppressed by PA-X) and components of the innate immune system (suppressed by NS1). It seemed that upregulation of cell cycle gene expression was accompanied by downregulation of proapoptotic factors and vice versa, suppression of cell cycle genes resulted in the upregulated of proapoptotic genes (see Figure 6). However, caspase-3/7 activity, a key initiator caspase of the apoptotic pathway, remained low in NS1(1-126)/ΔPA-X infected T3 cells for the whole 72 hours of infection time. Either the virus did not trigger apoptosis (in contrast to WT virus) or it actively suppressed it. Caspase-3 activation has been shown to be essential for efficient IAV propagation (Wurzer et al., 2003) as it results in enlargement of the nuclear pores which promotes nuclear export of viral ribonucleoprotein complexes late in the infection cycle (Mühlbauer et al., 2015). However, no correlation between caspase 3/7 activity and virus replication efficacy was observed as the ΔPA-X virus replicated as efficiently as WT virus although caspase-3/7 activity was reduced. It is not excluded that apoptosis inhibition together with elevated IFN- I and -III response might be necessary to reduce virus replication as seen for NS1(1-126)/ΔPA-X in T3 cells. As cells infected with either the ΔPA-X or the NS1(1-126)/ΔPA-X mutant showed low caspase-3/7 activity, it is likely that the absence of PA-X-mediated host shut-off activity is responsible for this phenomenon. Future studies should focus on the mechanisms of how PA-X interferes with apoptosis regulation.
In conclusion, our findings demonstrate that NS1 and PA-X act in a concerted manner to modulate the host response thereby promoting viral replication. The NS1(1-126)/ΔPA-X double mutant might represent an attractive LAIV candidate. It not only showed attenuated virus replication in a bronchiolar epithelial cell line and a human type II pneumocyte-derived tumor cell line but also induced a cocktail of cytokines and chemokines that may stimulate the immune response. In particular, type III IFN was identified as a potent mucosal adjuvant that was demonstrated to strongly promote mucosal immunity against influenza virus infection in a mouse model (Klinkhammer et al., 2018; Ye et al., 2019). Moreover, the induction of IFN will not only restrict LAIV infection but will also prevent co-infection with IAV field strains thereby reducing the risk that reassortant viruses will emerge. The lack of apoptosis in NS1(1-126)/ΔPA-X virus-infected cells is another plus, since this will reduce LAIV-induced cell death and tissue damage in the respiratory tract. In vivo studies are currently performed in order to primarily evaluate the efficacy and safety of these potential swine LAIV vaccine in the pig model but also to assess if potential translation of such LAIV vaccine to human would be conceivable.
The data presented in the study are deposited in the ENA arrayexpress repository, accession number E-MTAB-13128.
RA acquired and analyzed data, and wrote the manuscript. GZ and AS conceived the idea, designed the study and reviewed the manuscript. OG-N helped with interpretation of the data and reviewed the manuscript. All authors gave their final approval for submission.
SNF Grant number IZCOZO_189903. Harnessing interferon-lambda as a mucosal adjuvant of the respiratory tract in pigs. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
We would like to thank Sascha Trapp (INRAE, France) for providing the T3 cells, and Martin Schwemmle (University of Freiburg, Germany) for providing plasmids. We are grateful to Sylvie Python, Noelle Donze, and Sushila Gordon-Lennox for their technical support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1222805/full#supplementary-material
Bai, T., Chen, Y., Beck, S., Stanelle-Bertram, S., Mounogou, N. K., Chen, T., et al. (2022). H7N9 avian influenza virus infection in men is associated with testosterone depletion. Nat. Commun. 13, 6936. doi: 10.1038/s41467-022-34500-5
Bavagnoli, L., Cucuzza, S., Campanini, G., Rovida, F., Paolucci, S., Baldanti, F., et al. (2015). The novel influenza A virus protein PA-X and its naturally deleted variant show different enzymatic properties in comparison to the viral endonuclease PA. Nucleic Acids Res. 43, 9405–9417. doi: 10.1093/nar/gkv926
Chaimayo, C., Dunagan, M., Hayashi, T., Santoso, N., Takimoto, T. (2018). Specificity and functional interplay between influenza virus PA-X and NS1 shutoff activity. PloS Pathog. 14, e1007465. doi: 10.1371/journal.ppat.1007465
Cheng, X., Zengel, J. R., Suguitan, A. L., Xu, Q., Wang, W., Lin, J., et al. (2013). Evaluation of the humoral and cellular immune responses elicited by the live attenuated and inactivated influenza vaccines and their roles in heterologous protection in ferrets. J. Infect. Dis. 208, 594–602. doi: 10.1093/infdis/jit207
Clark, A. M., Nogales, A., Martinez-Sobrido, L., Topham, D. J., DeDiego, M. L. (2017). Functional evolution of influenza virus NS1 protein in currently circulating human 2009 pandemic H1N1 viruses. J. Virol. 91, e00721-17. doi: 10.1128/JVI.00721-17
Dunagan, M. M., Hardy, K., Takimoto, T. (2021). Impact of influenza A virus shutoff proteins on host immune responses. Vaccines 9, 629. doi: 10.3390/vaccines9060629
Dutta, A., Huang, C.-T., Lin, C.-Y., Chen, T.-C., Lin, Y.-C., Chang, C.-S., et al. (2016). Sterilizing immunity to influenza virus infection requires local antigen-specific T cell response in the lungs. Sci. Rep. 6, 32973. doi: 10.1038/srep32973
Firth, A. E., Jagger, B. W., Wise, H. M., Nelson, C. C., Parsawar, K., Wills, N. M., et al. (2012). Ribosomal frameshifting used in influenza A virus expression occurs within the sequence UCC_UUU_CGU and is in the +1 direction. Open Biol. 2, 120109. doi: 10.1098/rsob.120109
Fleurot, I., López-Gálvez, R., Barbry, P., Guillon, A., Si-Tahar, M., Bähr, A., et al. (2022). TLR5 signalling is hyper-responsive in porcine cystic fibrosis airways epithelium. J. Cystic Fibrosis 21, e117–e121. doi: 10.1016/j.jcf.2021.08.002
Foo, J., Bellot, G., Pervaiz, S., Alonso, S. (2022). Mitochondria-mediated oxidative stress during viral infection. Trends Microbiol. 30, 679–692. doi: 10.1016/j.tim.2021.12.011
Gao, H., Xu, G., Sun, Y., Qi, L., Wang, J., Kong, W., et al. (2015). PA-X is a virulence factor in avian H9N2 influenza virus. J. Gen. Virol. 96, 2587–2594. doi: 10.1099/jgv.0.000232
Gasparini, R., Amicizia, D., Lai, P. L., Panatto, D. (2011). Live attenuated influenza vaccine - A review.
Gaucherand, L., Porter, B. K., Levene, R. E., Price, E. L., Schmaling, S. K., Rycroft, C. H., et al. (2019). The influenza A virus endoribonuclease PA-X usurps host mRNA processing machinery to limit host gene expression. Cell Rep. 27, 776–792.e7. doi: 10.1016/j.celrep.2019.03.063
Geiss, G. K., Salvatore, M., Tumpey, T. M., Carter, V. S., Wang, X., Basler, C. F., et al. (2002). Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: The role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. U.S.A. 99, 10736–10741. doi: 10.1073/pnas.112338099
Genzow, M., Goodell, C., Kaiser, T. J., Johnson, W., Eichmeyer, M. (2018). Live attenuated influenza virus vaccine reduces virus shedding of newborn piglets in the presence of maternal antibody. Influenza Other Respi Viruses 12, 353–359. doi: 10.1111/irv.12531
Giard, D. J., Aaronson, S. A., Todaro, G. J., Arnstein, P., Kersey, J. H., Dosik, H., et al. (1973). In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors2. JNCI: J. Natl. Cancer Institute 51, 1417–1423. doi: 10.1093/jnci/51.5.1417
Gu, Y., Hsu, A. C.-Y., Pang, Z., Pan, H., Zuo, X., Wang, G., et al. (2019). Role of the innate cytokine storm induced by the influenza A virus. Viral Immunol. 32, 244–251. doi: 10.1089/vim.2019.0032
Hajjar, S. A., McIntosh, K. (2010). The first influenza pandemic of the 21st century. Ann. Saudi Med. 30, 1–10. doi: 10.5144/0256-4947.59365
Hale, B. G., Steel, J., Medina, R. A., Manicassamy, B., Ye, J., Hickman, D., et al. (2010). Inefficient control of host gene expression by the 2009 pandemic H1N1 influenza A virus NS1 protein. J. Virol. 84, 6909–6922. doi: 10.1128/JVI.00081-10
Hayashi, T., MacDonald, L. A., Takimoto, T. (2015). Influenza A virus protein PA-X contributes to viral growth and suppression of the host antiviral and immune responses. J. Virol. 89, 6442–6452. doi: 10.1128/JVI.00319-15
Heinen, P. P., van Nieuwstadt, A. P., de Boer-Luijtze, E. A., Bianchi, A. T. J. (2001). Analysis of the quality of protection induced by a porcine influenza A vaccine to challenge with an H3N2 virus. Vet. Immunol. Immunopathol. 82, 39–56 doi: 10.1016/S0165-2427(01)00342-7
Hilimire, T. A., Nogales, A., Chiem, K., Ortego, J., Martinez-Sobrido, L. (2020). Increasing the safety profile of the master donor live attenuated influenza vaccine. Pathogens 9, 86. doi: 10.3390/pathogens9020086
Hu, J., Ma, C., Liu, X. (2018). PA-X: a key regulator of influenza A virus pathogenicity and host immune responses. Med. Microbiol. Immunol. 207, 255–269. doi: 10.1007/s00430-018-0548-z
Iaquinta, P. J., Lees, J. A. (2007). Life and death decisions by the E2F transcription factors. Curr. Opin. Cell Biol. 19, 649–657. doi: 10.1016/j.ceb.2007.10.006
Janssens, Y., Joye, J., Waerlop, G., Clement, F., Leroux-Roels, G., Leroux-Roels, I. (2022). The role of cell-mediated immunity against influenza and its implications for vaccine evaluation. Front. Immunol. 13. doi: 10.3389/fimmu.2022.959379
Ji, Z., Wang, X., Liu, X. (2021). NS1: A key protein in the “Game” Between influenza A virus and host in innate immunity. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.670177
Kaiser, T. J., Smiley, R. A., Fergen, B., Eichmeyer, M., Genzow, M. (2019). Influenza A virus shedding reduction observed at 12 weeks post-vaccination when newborn pigs are administered live-attenuated influenza virus vaccine. Influenza Other Respi Viruses 13, 274–278. doi: 10.1111/irv.12630
Kim, H. J., Jeong, M. S., Jang, S. B. (2021). Structure and activities of the NS1 influenza protein and progress in the development of small-molecule drugs. IJMS 22, 4242. doi: 10.3390/ijms22084242
Klinkhammer, J., Schnepf, D., Ye, L., Schwaderlapp, M., Gad, H. H., Hartmann, R., et al. (2018). IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. eLife 7, e33354. doi: 10.7554/eLife.33354
Kochs, G., García-Sastre, A., Martínez-Sobrido, L. (2007). Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 81, 7011–7021. doi: 10.1128/JVI.02581-06
Laurie, K. L., Carolan, L. A., Middleton, D., Lowther, S., Kelso, A., Barr, I. G. (2010). Multiple infections with seasonal influenza A virus induce cross-protective immunity against A(H1N1) pandemic influenza virus in a ferret model. J. Infect. Dis. 202, 1011–1020. doi: 10.1086/656188
Lee, J., Yu, H., Li, Y., Ma, J., Lang, Y., Duff, M., et al. (2017). Impacts of different expressions of PA-X protein on 2009 pandemic H1N1 virus replication, pathogenicity and host immune responses. Virology 504, 25–35. doi: 10.1016/j.virol.2017.01.015
Lei, C., Yang, J., Hu, J., Sun, X. (2020). On the calculation of TCID50 for quantitation of virus infectivity. Virol. Sin. 36, 141–144. doi: 10.1007/s12250-020-00230-5
Levene, R. E., Shrestha, S. D., Gaglia, M. M. (2021). The influenza A virus host shutoff factor PA-X is rapidly turned over in a strain-specific manner. J. Virol. 95, e02312-20. doi: 10.1128/JVI.02312-20
Liberzon, A., Birger, C., Thorvaldsdóttir, H., Ghandi, M., Mesirov, J. P., Tamayo, P. (2015). The molecular signatures database hallmark gene set collection. Cell Syst. 1, 417–425. doi: 10.1016/j.cels.2015.12.004
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
López-Gálvez, R., Fleurot, I., Chamero, P., Trapp, S., Olivier, M., Chevaleyre, C., et al. (2021). Airway administration of flagellin regulates the inflammatory response to Pseudomonas aeruginosa. Am. J. Respir. Cell Mol. Biol. 65, 378–389. doi: 10.1165/rcmb.2021-0125OC
Ma, J., Li, S., Li, K., Wang, X., Li, S. (2019). Effects of the PA-X and PB1-F2 proteins on the virulence of the 2009 pandemic H1N1 influenza A virus in mice. Front. Cell. Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00315
Mancera Gracia, J. C., Pearce, D. S., Masic, A., Balasch, M. (2020). Influenza A virus in swine: epidemiology, challenges and vaccination strategies. Front. Vet. Sci. 7. doi: 10.3389/fvets.2020.00647
McLean, H. Q., Belongia, E. A. (2021). Influenza vaccine effectiveness: new insights and challenges. Cold Spring Harb. Perspect. Med. 11, a038315. doi: 10.1101/cshperspect.a038315
Mena, I., Nelson, M. I., Quezada-Monroy, F., Dutta, J., Cortes-Fernández, R., Lara-Puente, J. H., et al. (2016). Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. eLife 5, e16777. doi: 10.7554/eLife.16777
Min, J.-Y., Krug, R. M. (2006). The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. U.S.A. 103, 7100–7105. doi: 10.1073/pnas.0602184103
Min, J.-Y., Li, S., Sen, G. C., Krug, R. M. (2007). A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363, 236–243. doi: 10.1016/j.virol.2007.01.038
Mühlbauer, D., Dzieciolowski, J., Hardt, M., Hocke, A., Schierhorn, K. L., Mostafa, A., et al. (2015). Influenza virus-induced caspase-dependent enlargement of nuclear pores promotes nuclear export of viral ribonucleoprotein complexes. J. Virol. 89, 6009–6021. doi: 10.1128/JVI.03531-14
Muramoto, Y., Noda, T., Kawakami, E., Akkina, R., Kawaoka, Y. (2013). Identification of novel influenza A virus proteins translated from PA mRNA. J. Virol. 87, 2455–2462. doi: 10.1128/JVI.02656-12
Nacken, W., Schreiber, A., Masemann, D., Ludwig, S. (2021). The effector domain of the influenza A virus nonstructural protein NS1 triggers host shutoff by mediating inhibition and global deregulation of host transcription when associated with specific structures in the nucleus. mBio 12, e02196-21. doi: 10.1128/mBio.02196-21
Nemeroff, M. E., Barabino, S. M. L., Li, Y., Keller, W., Krug, R. M. (1998). Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ End formation of cellular pre-mRNAs. Mol. Cell 1, 991–1000. doi: 10.1016/S1097-2765(00)80099-4
Nogales, A., DeDiego, M. L., Martínez-Sobrido, L. (2022). Live attenuated influenza A virus vaccines with modified NS1 proteins for veterinary use. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.954811
Nogales, A., Martinez-Sobrido, L., Topham, D., DeDiego, M. (2018). Modulation of innate immune responses by the influenza A NS1 and PA-X proteins. Viruses 10, 708. doi: 10.3390/v10120708
Nogales, A., Rodriguez, L., DeDiego, M. L., Topham, D. J., Martínez-Sobrido, L. (2017). Interplay of PA-X and NS1 proteins in replication and pathogenesis of a temperature-sensitive 2009 pandemic H1N1 influenza A virus. J. Virol. 91, e00720-17. doi: 10.1128/JVI.00720-17
Ocaña-Macchi, M., Bel, M., Guzylack-Piriou, L., Ruggli, N., Liniger, M., McCullough, K. C., et al. (2009). Hemagglutinin-dependent tropism of H5N1 avian influenza virus for human endothelial cells. J. Virol. 83, 12947–12955. doi: 10.1128/JVI.00468-09
Pucci, B., Kasten, M., Giordano, A. (2000). Cell cycle and apoptosis. Neoplasia 2, 291–299. doi: 10.1038/sj.neo.7900101
Richt, J. A., Lekcharoensuk, P., Lager, K. M., Vincent, A. L., Loiacono, C. M., Janke, B. H., et al. (2006). Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J. Virol. 80, 11009–11018. doi: 10.1128/JVI.00787-06
Scholtissek, C. (1995). Molecular evolution of influenza viruses 137–143. doi: 10.1007/978-1-4613-1407-3_12
Sharma, A., Zeller, M. A., Li, G., Harmon, K. M., Zhang, J., Hoang, H., et al. (2020). Detection of live attenuated influenza vaccine virus and evidence of reassortment in the U.S. swine population. J. Vet. Diagn. Invest. 32, 301–311. doi: 10.1177/1040638720907918
Shi, M., Jagger, B. W., Wise, H. M., Digard, P., Holmes, E. C., Taubenberger, J. K. (2012). Evolutionary conservation of the PA-X open reading frame in segment 3 of influenza A virus. J. Virol. 86, 12411–12413. doi: 10.1128/JVI.01677-12
Solórzano, A., Webby, R. J., Lager, K. M., Janke, B. H., García-Sastre, A., Richt, J. A. (2005). Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. JVI 79, 7535–7543. doi: 10.1128/JVI.79.12.7535-7543.2005
Stark, G. R., Taylor, W. R. (2004). Analyzing the G2/M checkpoint, in: checkpoint controls and cancer. Humana Press New Jersey 280, 051–082. doi: 10.1385/1-59259-788-2:051
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550. doi: 10.1073/pnas.0506580102
Swedish, K. A. (2011). 2009 pandemic influenza A (H1N1): diagnosis, management, and prevention— Lessons learned. Curr. Infect. Dis. Rep. 13, 169–174. doi: 10.1007/s11908-010-0157-5
Tu, J., Guo, J., Zhang, A., Zhang, W., Zhao, Z., Zhou, H., et al. (2011). Effects of the C-terminal truncation in NS1 protein of the 2009 pandemic H1N1 influenza virus on host gene expression. PloS One 6, e26175. doi: 10.1371/journal.pone.0026175
Ujie, M., Takada, K., Kiso, M., Sakai-Tagawa, Y., Ito, M., Nakamura, K., et al. (2019). Long-term culture of human lung adenocarcinoma A549 cells enhances the replication of human influenza A viruses. J. Gen. Virol. 100, 1345–1349. doi: 10.1099/jgv.0.001314
Verstovsek, S. (2009). Therapeutic potential of JAK2 inhibitors. Long-term culture of human lung adenocarcinoma A549 cells enhances the replication of human influenza A viruses, 100, 10. doi: 10.1182/asheducation-2009.1.636
Vincent, A. L., Ciacci-Zanella, J. R., Lorusso, A., Gauger, P. C., Zanella, E. L., Kehrli, M. E., et al. (2010). Efficacy of inactivated swine influenza virus vaccines against the 2009 A/H1N1 influenza virus in pigs. Vaccine 28, 2782–2787. doi: 10.1016/j.vaccine.2010.01.049
Vincent, A. L., Lager, K. M., Janke, B. H., Gramer, M. R., Richt, J. A. (2008). Failure of protection and enhanced pneumonia with a US H1N2 swine influenza virus in pigs vaccinated with an inactivated classical swine H1N1 vaccine. Vet. Microbiol. 126, 310–323. doi: 10.1016/j.vetmic.2007.07.011
Vincent, A. L., Ma, W., Lager, K. M., Janke, B. H., Webby, R. J., García-Sastre, A., et al. (2007). Efficacy of intranasal administration of a truncated NS1 modified live influenza virus vaccine in swine. Vaccine 25, 7999–8009. doi: 10.1016/j.vaccine.2007.09.019
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141. doi: 10.1016/j.xinn.2021.100141
Wurzer, W. J., Planz, O., Ehrhardt, C., Giner, M., Silberzahn, T., Pleschka, S., et al. (2003). Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J. 22, 2717–2728. doi: 10.1093/emboj/cdg279
Ye, L., Schnepf, D., Becker, J., Ebert, K., Tanriver, Y., Bernasconi, V., et al. (2019). Interferon-λ enhances adaptive mucosal immunity by boosting release of thymic stromal lymphopoietin. Nat. Immunol. 20, 593–601. doi: 10.1038/s41590-019-0345-x
Keywords: influenza A virus, NS1 protein, PA-X protein, host shut-off, cytokine, innate immune response, transcriptomic profiling, virus attenuation
Citation: Avanthay R, Garcia-Nicolas O, Zimmer G and Summerfield A (2023) NS1 and PA-X of H1N1/09 influenza virus act in a concerted manner to manipulate the innate immune response of porcine respiratory epithelial cells. Front. Cell. Infect. Microbiol. 13:1222805. doi: 10.3389/fcimb.2023.1222805
Received: 16 May 2023; Accepted: 05 July 2023;
Published: 26 July 2023.
Edited by:
Slobodan Paessler, University of Texas Medical Branch at Galveston, United StatesReviewed by:
Anan Jongkaewwattana, National Center for Genetic Engineering and Biotechnology (BIOTEC), ThailandCopyright © 2023 Avanthay, Garcia-Nicolas, Zimmer and Summerfield. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Artur Summerfield, YXJ0dXIuc3VtbWVyZmllbGRAdW5pYmUuY2g=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.