
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 31 August 2023
Sec. Parasite and Host
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1219629
This article is part of the Research Topic Trypanosomatids Ecology, Diversity, Phylogeny, and Spatial Syntax: Impacts on the Interaction with Insect Vectors and Vertebrate Hosts View all 5 articles
 Mahmud Usman1
Mahmud Usman1 Audu Joseph Natala2
Audu Joseph Natala2 Isa Danladi Jatau2
Isa Danladi Jatau2 Ndudim Isaac Ogo3
Ndudim Isaac Ogo3 Ghulam Jeelani4
Ghulam Jeelani4 Yasuyuki Goto5
Yasuyuki Goto5 Tomoyoshi Nozaki4
Tomoyoshi Nozaki4 James H. McKerrow6
James H. McKerrow6 Emmanuel Oluwadare Balogun4,6,7,8*
Emmanuel Oluwadare Balogun4,6,7,8*Introduction: Female sand flies are hematophagous, feeding on animals and in the process serve as vectors for Leishmania, the parasites that cause leishmaniasis in humans. Leishmaniasis are a group of parasitic neglected tropical diseases in 98 countries including Nigeria and kills ~60,000 people/year. In Nigeria, Sokoto State is endemic to leishmaniasis but there is a knowledge gap on the identity of the prevalent sand flies and the Leishmania species they transmit. Hence, this cross-sectional study was designed to take inventory of the species of sand flies in Sokoto using genetic methods.
Methods: 1,260 (310 females) sand flies were collected from three Local Government Areas (L.G.A) of Sokoto State- Wamakko, Sokoto South and Kware. Genomic DNA was extracted from each fly and DNA amplification by polymerase chain reaction (PCR) was carried out on the DNA samples using primers targeting the arthropods mitochondrial cytochrome oxidase subunit 1 (mt-coI) gene, and nested PCR with primers targeting the gene for Leishmania internal transcribed spacer-1 (its-1) of ribosomal RNA its-1rRNA. The PCR products were sequenced.
Results: Gene sequence analysis revealed five species of sand flies belonging to the old-world genera namely Phlebotomus and Sergentomyia. The identified species were P. papatasi (6.45%), S. adleri (6.45%), S. affinis (9.7%), S. distincta (9.7%), S. schwetzi (67.7%). Within the sampling period, sand flies were most abundant in the rainy months of August (104/33.5%) and September (116/37.4%) with all the five identified species occurring. Sequence analysis of its-1 gene identified Leishmania infantum in two sand flies (2/310)- P. papatasi (from Sokoto South) and S. affinis (from Wamakko). BLAST search in NCBI and phylogenetic analysis revealed that the sand fly species are related to the species reported in different parts of Africa, while the L. infantum is identical to strain reported in Brazil (KY379083.1).
Discussion: Phlebotomus papatasi and four species belonging to the genus Sergentomyia are the most prevalent sand flies in Sokoto State, Nigeria and they harbor L. infantum solely. The results shed light on why visceral leishmaniasis is the most predominant form of the disease. Therefore, we recommend that adequate care for dogs must be instituted as dogs are the major animal reservoir for L. infantum.
Phlebotomine sand flies are the unique hematophagous insects proven to transmit Leishmania spp. through the bite of infected female that have previously fed on an infected mammal (Cox, 2002). They are vectors of various pathogenic agents that are responsible for leishmaniasis, bartonellosis and various arboviruses, which are potentially fatal diseases of animals and humans (David et al., 2015). Of these diseases, and from global health viewpoint, leishmaniasis are the most important due to their widespread, morbidities and fatalities in humans (Desjeux, 2004; Nihashi et al., 2016).
Leishmaniasis are caused by protozoan parasites belonging to the genus Leishmania, which is further divided into two subgenera- L. (Leishmania) and L. (Viannia). Leishmania protozoa are transmitted through the bite of the female phlebotomine sand fly (Desjeux, 2004). The disease is widely distributed around the world especially in tropical and subtropical areas, affecting at least 12 million people in 98 countries, with an additional 350 million people at risk (Desjeux, 2001). Approximately, twenty (20) Leishmania species are known to be pathogenic to humans, and these species are the major determinants of clinical outcome, which are of cutaneous, mucocutaneous, and visceral in nature (Herwaldt, 1999; Choi and Lerner, 2001; Desjeux, 2004).
The spread of leishmaniasis largely depends on the distribution of the vectors, therefore, the identification of circulating sand fly species in endemic and surrounding areas is important for predictions of the risk and expansion of the diseases. Sand flies are generally identified as adults based on morphologic characteristics (Munstermann, 2004). Unfortunately, morphological classification requires considerable skill as well as taxonomic expertise. In addition, the presence of intraspecific variation and cryptic species frequently complicates classifications based on morphological features (Bauzer et al., 2007). Therefore, other characteristics such as molecular markers have been explored for the development of simpler and more accurate identification of sand flies. Several genetic markers have been used to examine the systematics, relationships, and evolution amongst sand fly species and for population analyses within species (Depaquit et al., 2008; Kato et al., 2008; Kuwahara et al., 2009).
The infection of sand flies with Leishmania promastigotes has been examined by the dissection of individual sand flies under a microscope. For this purpose, specimens need to be fresh, and the dissection of the sand flies require a highly skilled technique due to their minute size. The procedure takes a relatively long time, and additionally, many specimens must be examined to obtain informative data for each area since the infection rate of sand flies with Leishmania is generally very low (0.01–1%) even in endemic areas (Hashiguchi et al., 2003). To improve on conventional methods, several PCR-based techniques which successfully detect the presence of Leishmania species within sand flies have been developed (Kato et al., 2005; Kato et al., 2007; Khalid et al., 2010). However, several improvements were desirable for the analysis of many sand flies with less effort and cost. In addition, it is better to analyze sand flies individually because several species co-exist in most endemic areas and the use of pooled samples may compromise important information on vector epidemiology such as the prevalent sand fly species and the relationships between Leishmania and vector species (Maroli et al., 2013).
Diagnosis of cutaneous leishmaniasis (CL) in Sokoto state was based on patient clinical presentation and microscopic identification (Jiya et al., 2007; Faleke et al., 2008). Improved identification of the causative Leishmania species and their vectors require sophisticated techniques such as molecular biology-based PCR (Schonian et al., 2003; Murray and Cappello, 2008). Molecular-based approaches based on nucleic acids offer greater sensitivity and specificity over the existing diagnostic tests (Cruz et al., 2002; Cruz et al., 2006; Cobo et al., 2007). The techniques permit the detection of infections from very low parasitized samples including those from asymptomatic patients’ samples (Mens et al., 2007). Although DNA-based methods have shown excellent sensitivity and specificity, the introduction of these methods in daily laboratory practice is still uncommon especially in rural endemic regions.
Several molecular markers and polymerase chain reaction (PCR) protocols have been developed for the detection and identification of sand flies and Leishmania (Khalid et al., 2010; Falcão de Oliveira et al., 2016). Many of those molecular tools have already been used in different parts of the world to differentiate species (Anderson et al., 2011; Dawit et al., 2013).
The study area was Sokoto State, located in the semi-arid region of North-Western Nigeria between longitudes 13° 5' E and 5° 15' E. It shares borders with Niger Republic to the North, Kebbi state to the Southwest and Zamfara State to the Southeast (Figure 1). The state has a total land mass of 32,000 km2, and is characterized by two distinct seasons, the short rainy season which runs from May or June to September or October and the long dry season that starts from October till May or June (Victor et al., 1997). The minimum relative humidity is less than 20% for most part of the year and ambient temperature ranges from 22 °C to 43 °C.
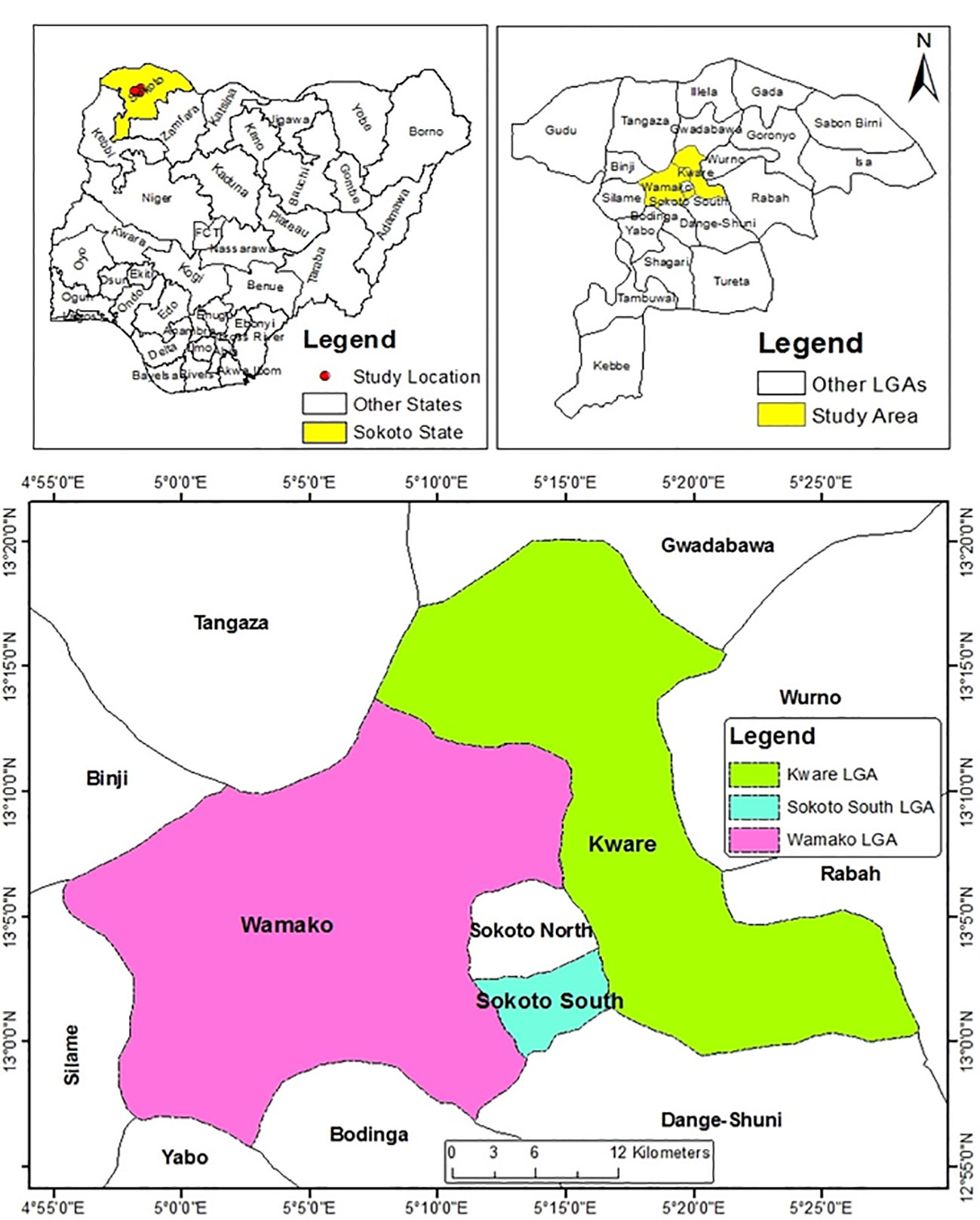
Figure 1 Map of Sokoto State showing the three Local Government Areas Sampled (Source: Department of Geography, A.B.U Zaria).
This cross-sectional study covered three Local Government Areas of Sokoto state.
Sand fly habitats were identified based on reported cases of human cutaneous leishmaniasis within the study area.
Survey was carried out between May and November 2016 (Usman et al., 2020a). Sand flies were collected using improvised sticky traps made of plywood (25 × 40 cm) coated on both sides with engine oil and kept around sand fly breeding sites onto which randomly impinging sand flies adhered. Traps were set thrice weekly. During each trapping session, ten (10) traps were allocated to each site (Refuse dumps and Sewage tanks). Traps were set between 17:30 and 18:30 hr and collected between 06:00 and 07:00 hr of the following day. In the laboratory, sand flies were removed from the sticky traps using dissecting needle, washed in kerosene, soapy water, then clean water (to reduce viscosity), and observed under the microscope for sex identification (Usman et al., 2020a). The female flies were transferred into tubes containing 70% ethanol and stored at 4 °C.
Morphological identification of sand fly specimens was carried out using Kyowa Stereo microscope HWFX (Kyowa, Tokyo, Japan) under the 10× eye pieces and using the established standard keys of Abonnenc (1972) and Lewis (1982). Sand flies were morphologically identified using body size and shape of wings. The presence or absence of external genitalia was used to identify categorize as male or female sand fly (Young and Duncan, 1994).
Genomic DNA was extracted from each of 400 morphologically identified female sand flies using Speedtools Tissue DNA extraction Kit (Biotools B&M Labs, Madrid, Spain). Briefly, each fly was washed with 200 µL distilled water to remove residual ethanol and thoroughly homogenized with a pestle in a laminar air flow. Exactly 130 µL of buffer ATL was added to each sample tube followed by 20 µL of proteinase K and mixed thoroughly by vortexing before incubation at 56 °C overnight in a heating block. The subsequent steps were carried out exactly according to the manufacturer’s guide. The eluted DNA was transferred into 1.5 mL tube with screw cap. Samples were incubated for 15 minutes at 100 °C to inactivate residual proteinase K before storage at -20 °C until required for PCR. The extracted DNA is expected to contain either the gDNA of sand flies alone or a mixture of the gDNA of sand flies and harbored pathogens such as Leishmania spp.
A summary of the PCR conditions and primers used for amplification of the indicated target genes for the Leishmania, and sand flies is provided in “Supplementary Table 1”. Owing to the genus and species specificity of the arthropod mitochondrial cytochrome C oxidase sub-unit 1 gene (mtcoI), it is generally targeted for amplification and sequencing, for the purpose of molecular identification and characterization. Therefore, mtcoI was amplified in a reaction containing 100 ng of the template DNA, 2.5 µM of the forward (5'-GGTCAACAAATCATAAAGATATTGG-3') and reverse (5'-TAAACTTCAGGGTGACCAAAAAATCA-3') primers, 1× of PCR reaction buffer containing 2 mM MgCl2, 250 µM of each dNTPs, and 1.0 U of PfuUltra II Fusion HS DNA Polymerase (Agilent technologies, Santa Clara, CA, USA) all in a total reaction volume of 50 µL. The negative controls were devoid only of DNA samples. Tubes were placed in a thermocycler and the program was run according to protocol by Hebert et al. (2003). The PCR cycling conditions were initial denaturation at 94 °C for 5 mins, 35 cycles of denaturation at 94 °C for 30 seconds, annealing at 55 °C for 1 min, and extension at 72 °C for 1 min, followed by final extension at 72 °C for 10 mins. The PCR products were stored at −20 °C until required.
For diagnosis and species identification, two Leishmania genes- small sub-unit ribosomal RNA (ssu rRNA) and ribosomal internal transcribed spacer-1 (its-1) were targeted for amplification of the species-specific portions by nested PCR (nPCR). The first PCR for amplification of the full length ssurRNA and its-1 were done using the primer pairs (forward: 5'-GGTTCCTTTCCTGATTTACG-3' and reverse: 5'-GGCCGGTAAAGGCCGAATAG-3') and (forward: 5'-CTGGATCATTTTCCGATG-3' and reverse: 5'-TGATACCACTTATCGCACTT-3'), respectively. Twenty five microliter reaction mixtures were prepared containing 50 ng of DNA, 0.5 U of PfuUltra II Fusion HS DNA Polymerase (Agilent technologies, Santa Clara, CA, USA), 1× of reaction buffer containing 2 mM MgCl2, 250 µM of each dNTPs, and 2.5 µM of the respective primers.
The PCR cycling protocol involved initial denaturation at 94 °C for 5 mins, 30 cycles of denaturation at 94 °C for 30 secs, annealing at 60 °C for 30 secs, and extension at 72 °C for 30secs. Final extension was performed at 72 °C for 5 mins. The PCR products were stored at −20 °C until required for the second PCR.
The second PCR of the nested technique to amplify the internal portions both ssu rRNA and its-1 was performed in using the same reaction constituents as the first step except that the DNA template was 5 µL product of the first PCR, and the primers (forward: 5'-TCCCATCGCAACCTCGGTT-3' and reverse: 5'-AAAGCGGGCGCGGTGCTG-3') and (forward: 5'-CATTTTCCGATGATTACACC-3' and reverse: 5'-CGTTCTTCAACGAAATAGG-3'), respectively, were used. The PCR cycling conditions were initial denaturation at 94 °C for 5 mins, 30 cycles of denaturation at 94 °C for 30 secs, annealing at 65 °C for 30 secs, and extension at 72 °C for 10 secs. Final extension was performed at 72 °C for 1min. The PCR products were stored at -20 °C until required for electrophoresis and sequencing.
Agarose (1.5%) was melted in 1× TAE buffer containing SYBR Safe and polymerized in gel tray. Samples were prepared by addition of 10× loading dye, loaded into agarose gel wells alongside 100 bp DNA ladder, and electrophoresed at 80 V for 30 minutes.. The Gels were visualized under UV light and the images were documented. Each DNA band containing the expected specific amplicon was carefully excised using a scalpel blade and purified using QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol.
Sequencing primers were designed based on mtcoI sequences (for sand flies) and its-1 sequences (for Leishmania) that were obtained from the GenBank. The gel-purified PCR products were sequenced using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Carlsbad, CA). Data were analyzed with ABI 3130 genetic analyzer software.
The sequence chromatograms were viewed and edited using ApE genetic analyzer. Each sequence was imported into the NCBI Database and nucleotide Basic Local Alignment Search Tool (BLAST) was used to search for similarity with other sequences in the GenBank using the NCBI database search (https://www.ncbi.nlm.nih.gov/BLAST).
The evolutionary relationship of sand flies and Leishmania specie isolates were determined by the construction of a phylogenetic tree using the Molecular Evolutionary Genetic Analysis (MEGA 7.0) software (Kumar et al., 2016). All species were separated with each one having its own branch. Sequences for all newly collected isolates clustered together with those already published for the respective species of the same or different locality forming groups of the same sub genus. The phylogenetic groupings provided by the tree, coupled with the aforementioned sequencing queries against GenBank, confirmed the molecular and morphological identification of the sampled sand flies and Leishmania species.
Data collected was presented as tables, figures and plates (SPSS Version 20). Values of P < 0.05 were considered significant. Bioclimatic data, including relative humidity, rainfall and average temperature, were obtained from the Nigerian Meteorological Agency as reported by https://www.timeanddate.com/weather/nigeria/sokoto for each month of the study period.
In general, sand flies are small with relatively long legs and erect wings as seen. While for Phlebotomus species, wings are asymmetrical and erect tapering towards the end and close to the body as observed while, Sergentomyia species the wings are symmetrical. While identification of the sand flies and Leishmania genera are possible by microscopic examinations, accurate identification and categorization into their species can only be confirmed using molecular biology tools involving PCR and DNA sequencing. Trap-captured female sand flies were identified microscopically and counted, and then DNA were prepared from the whole individual fly. The extracted DNA, which may also contain DNA of Leishmania, were subjected to two different nPCR for molecular identification of the sand flies and confirmation of the presence or absence of Leishmania parasites. The nPCR assays amplified the 700 bp segment of fly’s mtcoI (Figure 2), and the targeted 400 bp and 280 bp segments of ssurRNA and its-1, respectively, in DNA sample of a Leishmania positive sand fly (Figures 3A–C).
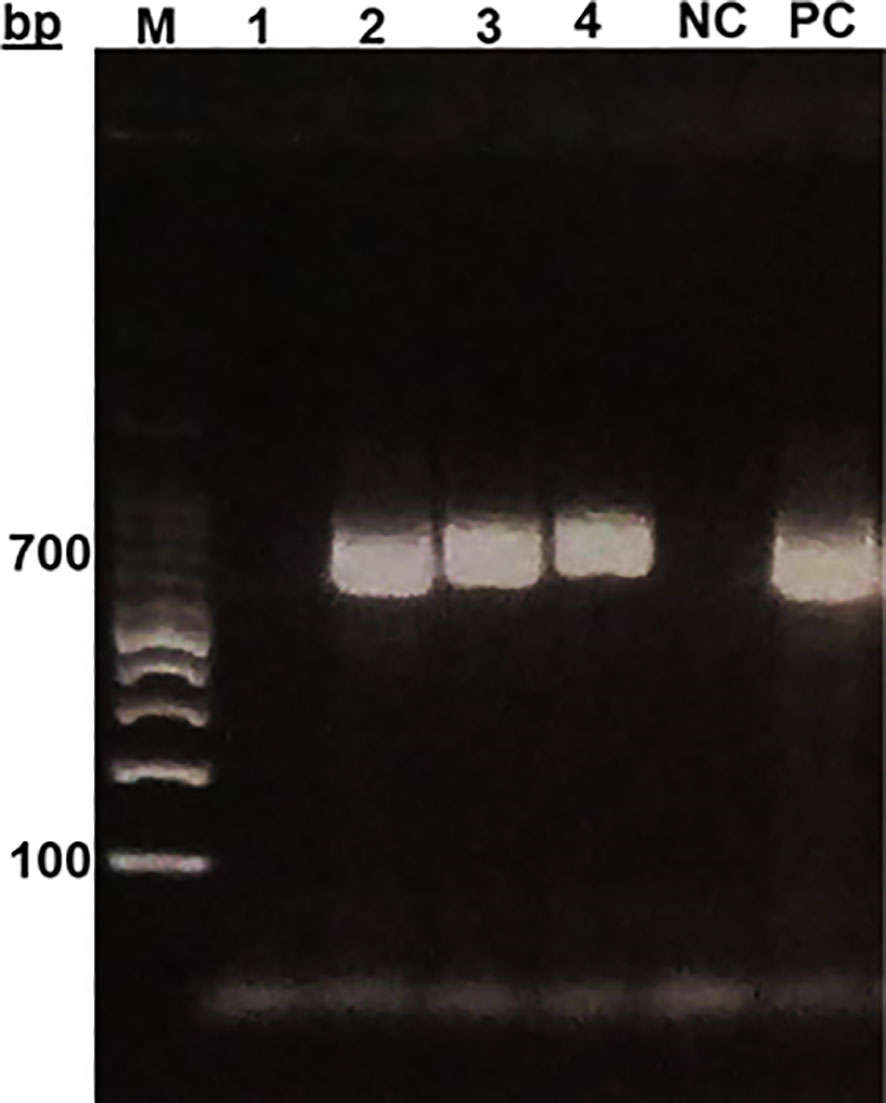
Figure 2 Gel electrophoresis of mt-co1 of sand flies from Sokoto. MM- Molecular marker (100bp DNA ladder) PC (positive control), NC (Negative control).
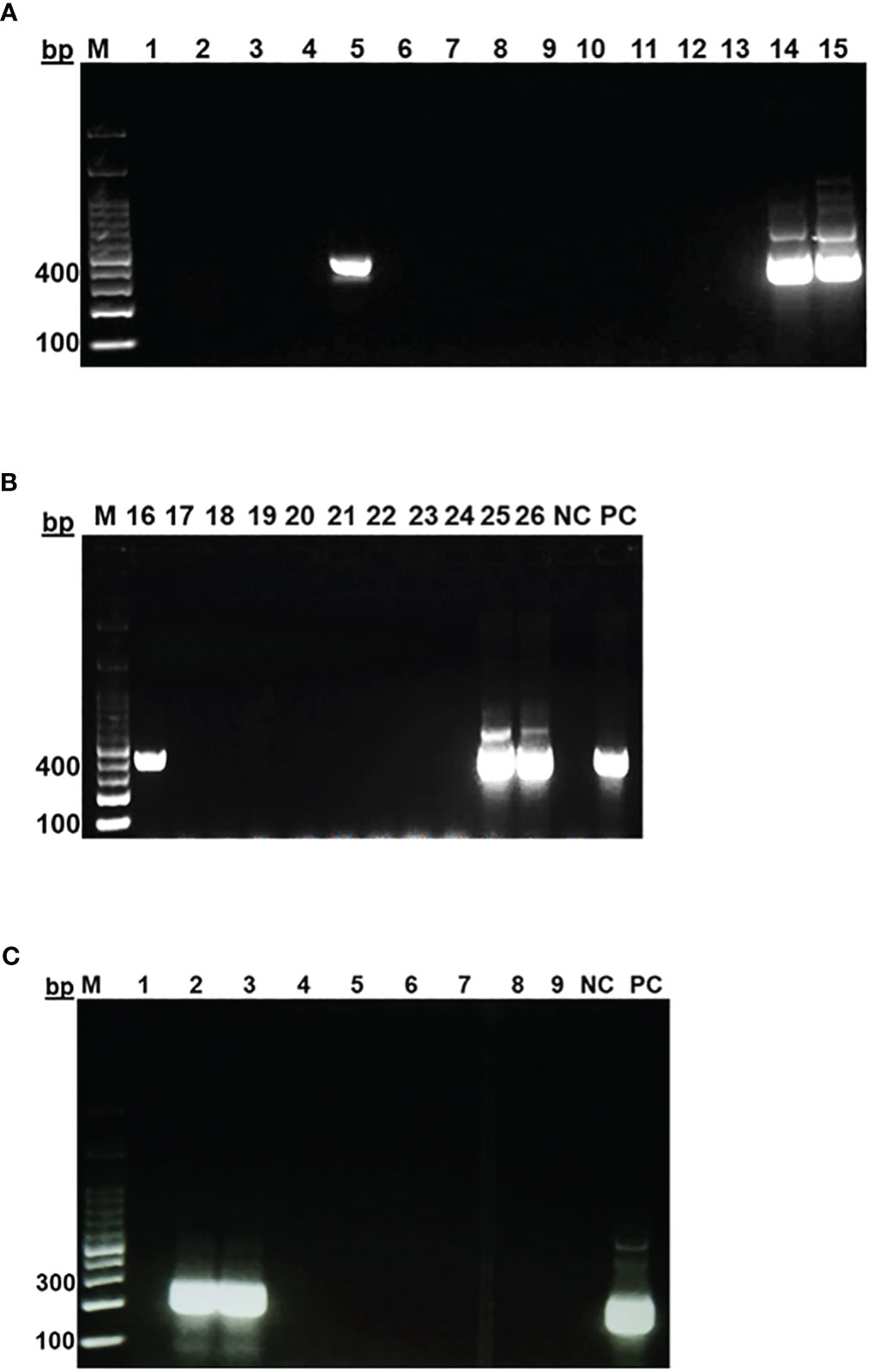
Figure 3 Identification of Leishmania spp. in female sand flies in Sokoto. Gel electrophoresis of PCR products targeting ssu rRNA gene (A, B) and its-1 gene (C). MM- Molecular marker (100bp DNA ladder), NC (Negative control), PC (Positive control).
Sand flies: Amplification of the mtcoI gene was carried out by nPCR on DNA samples prepared from 400 female sand flies. Sequence analysis of the 400 PCR products revealed that they clustered into five different consensus nucleotide sequences- SF-Sokoto 1, SF-Sokoto 2, SF-Sokoto 3, SF-Sokoto 4, and SF-Sokoto 5). BLAST search with each consensus sequence revealed that SF-Sokoto_1 was 99% identical to Phlebotomus papatasi (KR 020560.1), SF-Sokoto 2 was 98% identical to Sergentomyia adleri (KJ746879.1), SF-Sokoto 3 was 99% identical to S. affinis (KJ746893.1), SF-Sokoto 4 was 100% identical to S. schwetzi (KJ481125.1), while SF-Sokoto 5 was 100% identical to S. distincta (KY451790.1). Our results imply that the sand fly population of Sokoto, Nigeria are comprised mainly of P. papatasi, S. adleri, S. affinis, S. distincta, and S. schwetzi. These species of sand flies constitute (20%, 6.5%, 10%, 10%, 67%) of the sampled flies in Sokoto state respectively (Figures 4, 5).
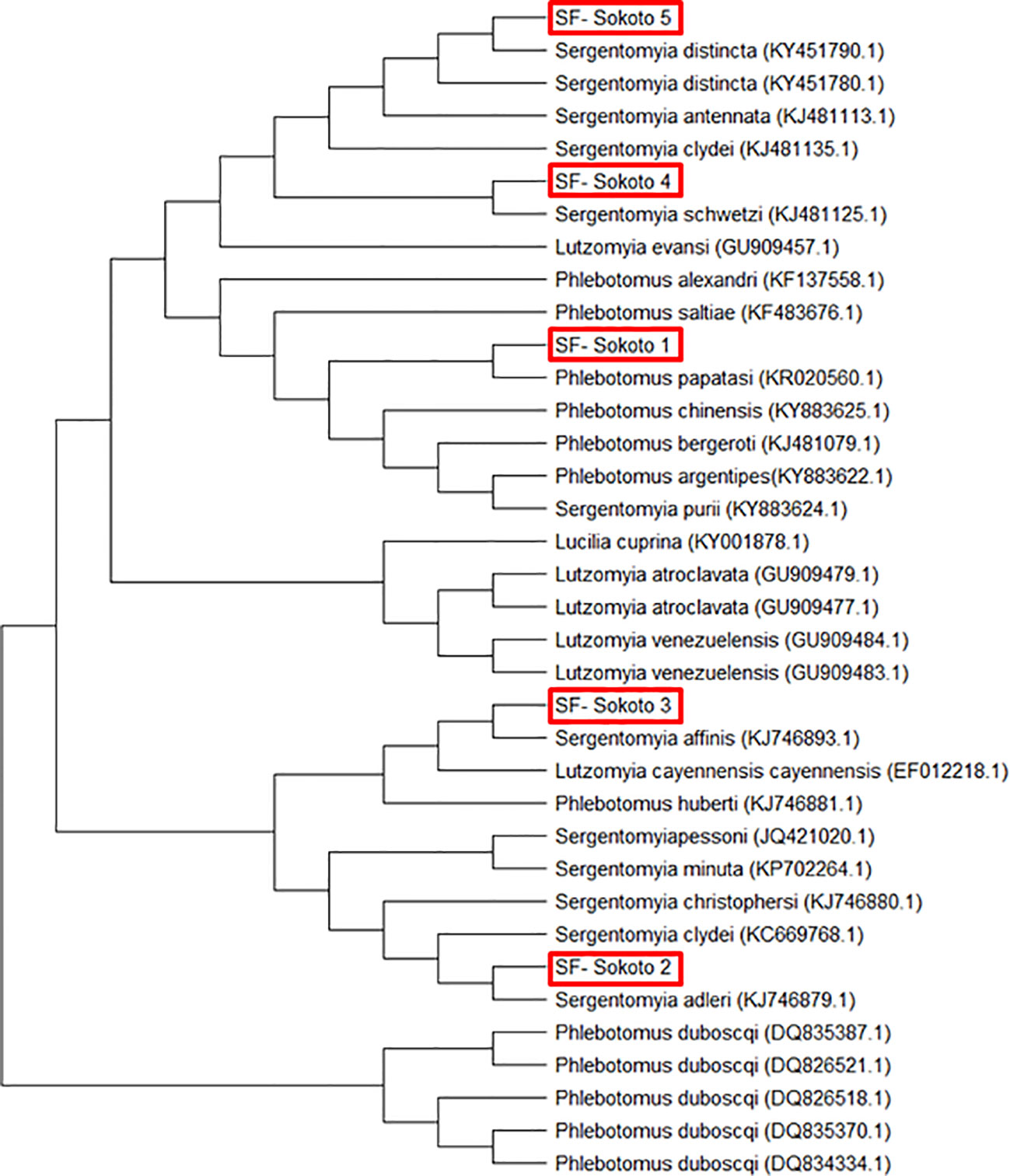
Figure 5 Phylogenetic tree of sand fly species from Sokoto State with other sand fly species sequences obtained from GenBank using MEGA 7.0.
Leishmania species: Sequence analyses of the targeted inner segment of Leishmania its-1 for the 400 DNA samples that were prepared from female sand flies revealed 2 different but related consensus nucleotide sequences– Leish-Sokoto 1 and Leish-Sokoto 2. The sequences have been deposited to the GenBank, with accession codes MN243118.1 and MN243117.1, respectively. These sequences were used to search the GenBank (blastn; http://blast.ncbi.nlm.nih.gov). Both Leish-Sokoto 1 and 2 were identified as Leishmania infantum, with 98% and 100% identities with L infantum isolate from Brazil (KY379083.1) and Greece (KY379081.1), respectively (Figure 6).
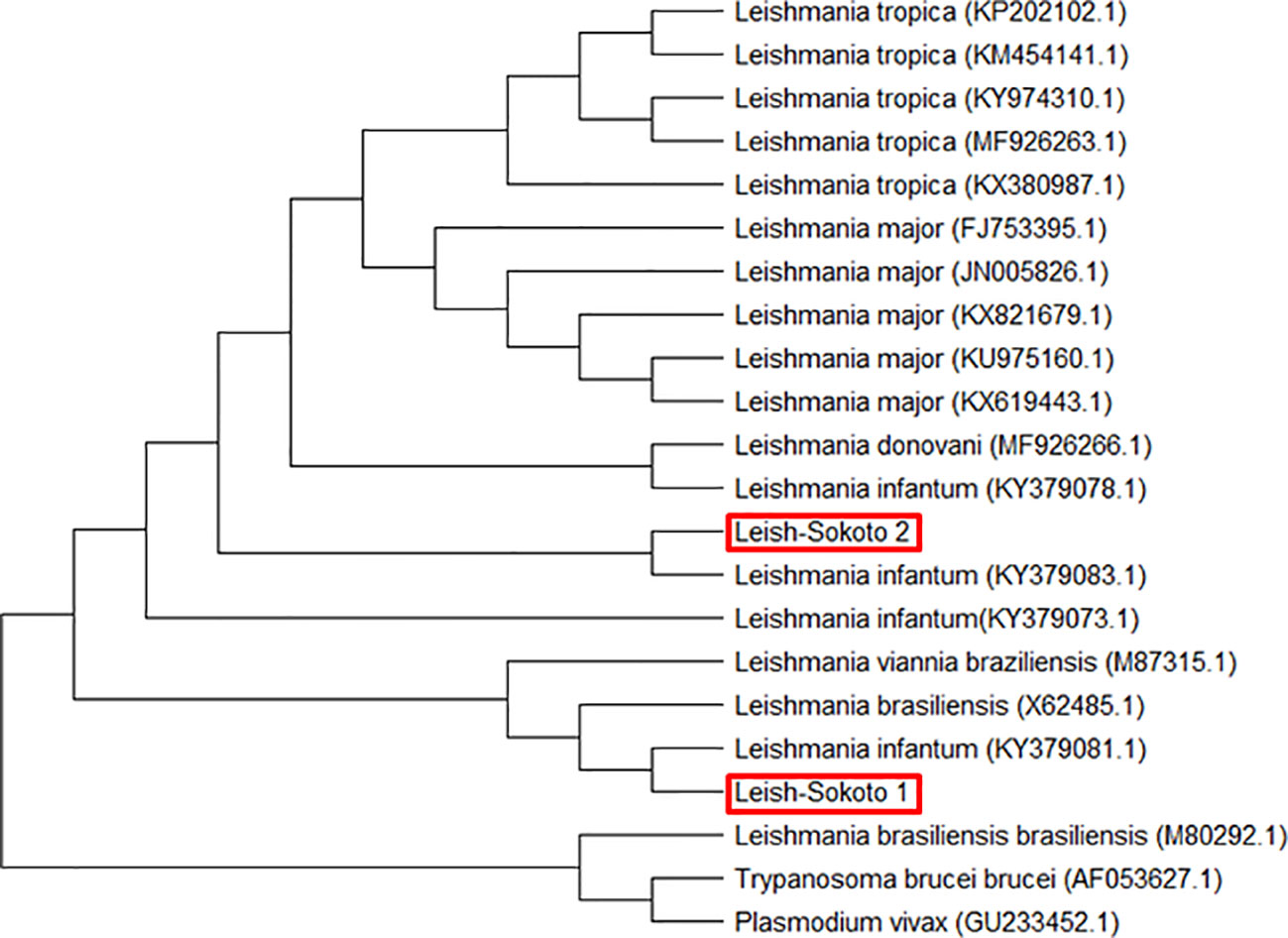
Figure 6 Evolutionary relationship of Leishmania infantum from Sokoto State with other similar isolates from different localities based on ITS-1 gene constructed using MEGA 7.0.
While all the reported 5 sand fly species were found in Wamakko L.G.A., (occurrence rates of 5.3%, P. papatasi; 5.3%, S. adleri; 15.7%, S. affinis; 10.5%, S. distincta; 63.2%, S. schwetzi), only 4 species were captured in Sokoto-South L.G.A., (occurrence rates: 8.3%, P. papatasi; 8.3%, S. adleri; 8.3%, S. distincta; 75.0%, S. schwetzi). S. affinis was absent in Sokoto-South, and no sand fly was captured in Kware L.G.A (Table 1).
The nPCR detected Leishmania DNA in samples prepared from P. papatasi from Sokoto South and S. affinis from Wamakko. DNA sequencing identified the Leishmania as L. infantum (Table 2). Out of the twenty P. papatasi species identified, 5% were positive for L. infantum DNA, and one out of 30 S. affinis species was positive for L. infantum (3%). This is the first time we are recording the presence of Leishmania DNA in Sergentomyia species in Nigeria. Other species (S. distincta, S. adleri and S. schwetzi) were negative for Leishmania DNA. (Table 2). Only 310 out of the 400 PCR products from female sand flies produced readable sequences after direct sequencing, although DNA quantity was not a limiting factor. Total infection rate of Leishmania in sand flies, in both Sokoto South and Wamakko L.G.A of Sokoto State was 0.6% (2/310) and this is epidemiologically significant.
Of the five (5) species of sand flies identified, three (3) were reported in the dry months of May; S. affinis (33.3%), S. distincta (16.7%) and S. schwetzi (50%) and in November only S. schwetzi (100%) was found. All the 5 species were reported in the rainy months of August; P. papatasi (3.8%), S. adleri (1.0%), S. affinis (19.2%), S. distincta (16.4%), S. schwetzi (59.6%) and in September; P. papatasi (9.5%), S. adleri (16.4%), S. affinis (1.7%), S. distincta (6.0%), S. schwetzi (66.4%) (Table 3). These results imply that irrespective of the season, S. schwetzi is the dominant sand fly species, and P. papatasi is the least prevalent and found only during the rainy season. Overall, the sand flies of the Sergentomyia genus account for between 80-100% of the sand fly population in Sokoto State.
Based on the estimated molecular size of the PCR products, five species of sand flies were identified at the study location. The five sand fly (SF) samples were code-named SF-Sokoto 1, SF-Sokoto 2, SF-Sokoto 3, SF-Sokoto 4, and SF-Sokoto 5. The evolutionary relationship of the sand fly species is illustrated as a phylogenetic tree (Figure 5). Phylogenetic analysis revealed that all the five representative flies are of 2 different genera and belong to five different species namely, Phlebotomus papatasi, Sergentomyia adheleri, S. affinis, S. schwetzi, and S, distincta, respectively (Figure 5). Sequences for all newly collected isolates formed five different clusters with those already published for the respective species from Africa. The phylogenetic groupings provided by the tree, coupled with the aforementioned sequencing queries against GenBank confirmed the molecular and morphological identification of the sampled sand flies.
Phylogenetic analysis using the ssu rRNA nucleotide sequences showed two categories of sequences that are pure clusters with each other and with those of L. infantum (KY379073.1, KY379078.1, KY379081.1, KY379083.1) isolates from Europe (Figure 6). The sequences from this study were those from the parasite isolates from Sokoto, code-named Leish-Sokoto 1 and Leish-Sokoto 2 (Figure 6). The evolutionary groupings provided by the tree, coupled with the sequencing queries against GenBank, confirmed the molecular identification of the Leishmania species present in the sampled sand flies as L. infantum.
Leishmaniasis is an important emerging parasitic disease found in 98 countries around the world (World Health Organization, 2010), and domestic dogs are the principal reservoir hosts and while wild canids constitute major sylvatic reservoirs (Ashford, 2000). Intense transmission of leishmaniasis by infected sand flies, from dog to dog or from dog to human, occurs in places where the Leishmania infection rate is very high in dogs (Vercammen et al., 1997). Sokoto State is an endemic focus for leishmaniasis in Nigeria. A 3.5% seroprevalence of canine leishmaniasis was reported in Sokoto State (Usman et al., 2020b). Jiya et al. (2007) reported a 10% prevalence of cutaneous leishmaniasis in school children in the study area, while Faleke et al. (2008) published a case report of cutaneous leishmaniasis in an undergraduate university student in Sokoto State. However, none of the reported cases adopted molecular techniques in the diagnosis of the disease. Visceral leishmaniasis is not routinely diagnosed in west Africa (Coulibaly et al., 2016). This is the first molecular studies on phlebotomine sand flies Leishmania species in the study area. We previously established by morphological observation that male sand fly population in Sokoto is twice the female population (Usman et al., 2020a) but there is no information on the prevalent parasite species as well as the vector species. In this study, we identified five species of phlebotomine sand flies as the potential vectors for leishmaniasis in Sakoto State, Nigeria. Of which sand flies belonging to the old-world genera namely Phlebotomus (Rondani, 1840) and Sergentomyia (França and Parrot, 1920) were identified: One (1) Phlebotomus spp., (P. papatasi) and four Sergentomyia spp. (S. adleri, S. affinis, S. distincta and S. schwetzi). The preponderance of Sergentomyia spp. over Phlebotomus spp. is similar to earlier findings in Cameroon (Rageau, 1951; Rageau and Adam, 1953), some parts of Nigeria (Agwale et al., 1995; Adamu et al., 2012) and in The Gambia (Desjeux et al., 1983).
Sergentomyia schwetzi was the most abundant species and it has been found in all the study areas. This is similar to the findings of Sangare et al. (2009) in Burkina Faso, Dondji et al. (2000) in Cameroon where S. schwetzi was found to be the most abundant species. The other dominant sand flies of the genus Sergentomyia were S. distincta and S. affinis. P. papatasi and S. adleri are the third most abundant species, both were recorded in Sokoto South and Wamakko. In this study, only the areas of suspected transmission were investigated. Moreover, P. papatasi was the only species identified from the genus Phlebotomus. For this reason, this species could be considered as the probable main vector of Leishmania spp. parasites. Until now, P. duboscqi was reported to be the principal vector of Leishmania in Nigeria (Killick-Kendrick, 1990). Our present findings on the skewed occurrence of P. papatasi in Sokoto State, which also coincided with prevalence of Leishmania infections in animals in the same location suggests that P. papatasi is also a major player in the transmission of leishmaniasis in Nigeria. Furthermore, although P. papatasi is the recognized vector of L. major in the old world (Anis et al., 2001), the results herein also implicate P. papatasi may be involved in the transmission of L. infantum corroborating that the multiple sand fly species may be serving as the vectors for L. infantum. We recommend that the vectoral capacity of P. papatasi for transmission of L. infantum should be investigated.
The natural occurrence of Leishmania parasites in sand flies is an important determinant of active transmission in a particular locality (Ready, 2013). Therefore, it is important to conduct routine surveillance of parasites in the midgut of sand flies. Dissection of sand fly gut is the gold-standard method used to study the rate of natural infection in endemic areas. This method is laborious, time consuming and requires a lot of skills and expertise. It also requires a large number of specimens to achieve reasonable epidemiological data (Aransay et al., 2000). Alternatively, molecular techniques allow for DNA detection of a single Leishmania parasite (Pita-Pereira et al., 2005) and probably represent a more sensitive tool than manual dissection and microscopic examination (Nascimento et al., 2007), which may underestimate natural sand fly infection rates in cases of low parasitemia. The Polymerase chain reaction (PCR) is a suitable technique for the detection of Leishmania DNA in sand flies and for identification of Leishmania vectors in different geographical areas (Aransay et al., 2000). Molecular methods are more sensitive and specific, regardless of the number, stage, and location of the parasite in the insect midgut (Perez et al., 1994). The PCR technique was therefore used in this study.
From literature, the role of Sergentomyia spp. in the circulation of Leishmania spp. is becoming more apparent as Leishmania DNA has been identified in several species of the genus. These include the molecular detection of L. major in S. sintoni in Iran (Parvizi and Amirkhani, 2008), S. garnhami in Kenya (Mutinga et al., 1994), S. darlingi in Mali (Berdjane-Brouk et al., 2012), and S. minuta in Portugal (Campino et al., 2013). Furthermore L. donovani has been detected in S. babu in India (Mukherjee et al., 1997), L. infantum in S. dubia, S. magna and S. schewtzi in Senegal (Senghor et al., 2011) and more recently, L. tropica has been found in S. ingrami and S. hamoni in Ghana (Nzelu et al., 2014). Although L. infantum had been detected in S. schwetzi from Senegal (Senghor et al., 2011), the refractoriness of this African species to some Leishmania species infecting humans (including L. donovani, L. infantum and L. major) has also been recently demonstrated (Sadlova et al., 2013). This study is the first to detect the presence of Leishmania sp. (L. infantum) in Sergentomyia species (S. affinis) in Nigeria, which is similar to findings from other parts of the world as previously discussed. This further confirms the possibility of Sergentomyia species becoming a vector of Leishmania parasites, hence should be of great relevance in the epidemiology of leishmaniasis in Nigeria and other parts of Africa. Worthy of mention, according to literature the occurrence of sand flies in the rainy season is uncommon. However, the rainy season in Sokoto State is very short (about 47.17 rainy days in a year) and precipitation is as low as 34.38 mm. In addition, the average daily mean temperature can be as high as 39 °C. These climatic conditions could be the reasons for the presence of sand flies even in the rainy season in Sokoto state.
Leishmania infantum is the causative agent of infantile visceral leishmaniasis in the Mediterranean region of the Old World and in Latin America, where it has been named Leishmania chagasi. (Maurício et al., 2000). It is also an unusual cause of cutaneous leishmaniasis (BenSaid et al., 2006). Leishmania infantum is closely related to L. donovani, and some authors believe that these two species are so close as to be subspecies of each other; (Le Blancq and Peters, 1986) however, phylogenetic analyses can easily distinguish between the two groups, although analysis has shown that some isolates of L. donovani have been classified as L. infantum and that the former includes a number of different genetic groups (Kuhls et al., 2005). P. papatasi supports the development of only L. major (Killick-Kendrick et al., 1994). However, other sand fly species support the development of wider range of Leishmania spp. The detection of phylogenetically and epidemiologically distant species of L. infantum could be due to natural genetic hybridization between L. major and L. infantum as reported by Ravel et al. (2006) and Volf et al. (2007). The reports raise questions about the frequency of such cross species genetic exchanges in nature, modalities of hybrid transmission, and their long-term maintenance as well as consequences of the genetic hybrids. Human infection with L. infantum is zoonotic with dogs serving as the reservoir hosts. Interestingly, Usman et al. (2020b) reported a 3.5% seroprevalence of canine leishmaniasis in the study area. This report may explain the possible existence of L. infantum in Sokoto State, Nigeria.
Visceral leishmaniasis is not routinely diagnosed in West Africa (Coulibaly et al., 2016). For the first time, the occurrence of L. infantum in Sokoto State Nigeria was observed in two species of sand flies (P. papatasi and S. affinis). It is possible that clinicians are misdiagnosing cases of this disease and confusing symptoms with malaria, toxoplasmosis, or another infectious fever. Therefore, the detection of L. infantum in the study area is of great public health significance.
Five species of phlebotomine sand flies belonging to the Old-World genera namely Phlebotomus (Rodani and Berte, in Rodani 1840) and Sergentomyia (França and Parrot, 1920) were identified: One Phlebotomus sp., (P. papatasi) and four Sergentomyia spp. (S. adleri, S. affinis, S. distincta and S. schwetzi) were detected. The LnPCR detected Leishmania DNA in two (2) sand fly species (0.6%) belonging to P. papatasi from Sokoto South and S. affinis from Wamakko, out of the 310 female sand flies analyzed. Bearing in mind the contribution of climate change to thriving of disease vectors (Balogun et al., 2016; Adepoju et al., 2023), it is important to increase surveillance efforts for vectors of parasitic infections. The outcomes may further stimulate efforts towards the discovery of new drugs (Ungogo et al., 2020).
The data presented in the study are deposited in the https://www.ncbi.nlm.nih.gov/nuccore repository, with accession numbers MN243117.1 and MN243118.1.
MU, AN and EB conceptualized the project. AN, IJ, and EB supervised the work. MU, NO, GJ, YG, and EB contributed to the development and writing of the manuscript. YG, TN, JM and EB contributed to validating and reviewing the project. All authors contributed to the article and approved the submitted version.
This work was partly supported with funding from the World Bank through the Africa Center of Excellence for Development (ACE Impacts) project to Africa Center of Excellence for Neglected Tropical Disease and Forensic Biotechnology (ACENTDFB), Ahmadu Bello University, Nigeria, in part by Institution Based Research grant from Tertiary Education Trust Fund (TETF/DR&D/UNI/ZARIA/IBR/2020/VOL.1/46) to EB, in part by Grant-in-Aid for Early-Career Scientists (B) (JP21K15427 to GJ) and with funding from the Japan Society for the Promotion of Science JSPS KAKENHI (21H02722 to YG) and AMED (22wm0225024h0001 to YG). EB is a recipient of the Emerging Global Leader (K43) Award and supported by FIC/NIH under Award Number K43TW012015. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1219629/full#supplementary-material
Abonnenc, E. (1972). Les phlébotomes de la région éthiopienne (Diptera, Psychodidae). Cahiers de l'ORSTOM, série Entomologie médicale et Parasitologie. 55, 1–239.
Adamu, B. S., William, A. I., Balarabe, L. M. (2012). The occurence of phebotomine sand flies in some parts of Southern Bauchi State, Nigeria. Int. J. Adv. Biol. Res. 4, 134–138.
Adepoju, O. A., Afinowi, O. A., Tauheed, A. M., Danazumi, A. U., Dibba, L. B., Balogun, J. B., et al. (2023). Multisectoral perspectives on global warming and vector-borne diseases: a focus on Southern Europe. Curr. Trop. Med. Rep. 10, 47–70. doi: 10.1007/s40475-023-00283-y
Agwale, S. M., Pam, D. D., Dondji, B., Duhlinska, D. D. (1995). Preliminary survey of phlebotomine sand flies (Diptera: Psychodidae) In Northern Nigeria. M e m. Inst. Oswaldo Cruz 90, 557–558. doi: 10.1590/S0074-02761995000500001
Anderson, J. M., Samake, S., Jaramillo-Gutierrez, G., Sissoko, I., Coulibaly, C. A., Traoré, B., et al. (2011). Seasonality and Prevalence of Leishmania major Infection in Phlebotomus duboscqi Neveu-Lemaire from Two Neighboring Villages in Central Mali. PLoS Neglec. Trop. Dis. 10, e1139. 10.1371/journal.pntd.0001139
Anis, E., Leventhal, A., Elkana, Y., Wilamowski, A, Pener, H. (2001). Cutaneous leishmaniasis in Israel in the era of changing environment.Pub. Health Rev. 29, 37–47.
Aransay, A. M., Scoulica, E., Tselentis, Y. (2000). Detection and identification of Leishmania DNA within naturally infected sand flies by semi-nested PCR on minicircle kinetoplastic DNA. Applic. Env. Microbiol. 66, 1933–1938. doi: 10.1128/AEM.66.5.1933-1938.2000
Ashford, R. W. (2000). The leishmaniases as emerging and reemerging zoonoses. Int. J. Parasitol. 30, 1269–1281. doi: 10.1016/S0020-7519(00)00136-3
Balogun, E. O., Nok, A. J., Kita, K. (2016). Global warming and the possible globalization of vector-borne diseases: a call for increased awareness and action. Trop. Med. Health 44, 38. doi: 10.1186/s41182-016-0039-0
Bauzer, L. G., Souza, N. A., Maingon, R. D., Peixoto, A. A. (2007). Lutzomyia longipalpis in Brazil: a complex or a single species? A mini-review. Mem. Instituto Oswaldo Cruz 102, 1–12. doi: 10.1590/S0074-02762007000100001
BenSaid, M., Guerbouj, S., Saghrouni, F. (2006). “Occurrence of Leishmania infantum cutaneous Leishmaniosis in central Tunisia. Trans. R. Soc Trop. Med. Hyg. 100, 521–526. doi: 10.1016/j.trstmh.2005.08.012
Berdjane-Brouk, Z., Koné, A. K., Djimdé, A. A., Charrel, R. N., Ravel, C. (2012). First detection of Leishmania major DNA in Sergentomyia (Spelaeomyia) darlingi from cutaneous Leishmaniosis foci in Mali. PloS One 7, e28266. doi: 10.1371/journal.pone.0028266
Campino, L., Cortes, S., Dionísio, L., Neto, L., Afonso, M. O., Maia, C. (2013). The first detection of Leishmania major in naturally infected Sergentomyia minuta in Portugal. Memórias do Instituto Oswaldo Cruz 108, 516–518. doi: 10.1590/S0074-02762013000400020
Choi, C. M., Lerner, E. A. (2001). Leishmaniosis as an emerging infection. J. Invest. Dermatol. Symp. Proc. 6, 175–182. doi: 10.1046/j.0022-202x.2001.00038.x
Cobo, F., Aliaga, L., Talavera, P., Concha, A. (2007). The histological spectrum of non-granulomatous localized mucosal Leishmaniosis caused by Leishmania infantum.” Ann. Trop. Med. Parasitol. 101, 689–694. doi: 10.1179/136485907X229095
Coulibaly, C. A., Sissoko, I., Traore, B., Diallo, A., Samake, S., Traore, S. F., et al. (2016). Diversity of sand flies (Diptera: Psychodidae: Phlebotominae) in two different eco-climatic and endemic zones of cutaneous Leishmaniosis in Mali, West Africa. J. Med. Entomol. 53, 923–927. doi: 10.1093/jme/tjw060
Cox, F. E. G. (2002). History of human parasitology. Clin. Microbiol. Rev. 15, 595–612. doi: 10.1128/CMR.15.4.595-612.2002
Cruz, I., Canavate, C., Rubio, J. M., Morales, M., Chicharo, C., Laguna, F., et al. (2002). A nested polymerase chain reaction (Ln-PCR) for diagnosing and monitoring Leishmania infantum infection in patients co-infected with human immunodeficiency virus. Trans. R. Soc Trop. Med. Hyg. 96, 185–189. doi: 10.1016/S0035-9203(02)90074-X
Cruz, I., Chicharro, C., Nieto, J. (2006). Comparison of new diagnostic tools for management of pediatric Mediterranean visceral Leishmaniosis. J. Clin. Microbiol. 44, 2343–2347. doi: 10.1128/JCM.02297-05
David, E. B., Seth, C. B., Richard, N. J., Yvonne, M. L., Graham, B. W. (2015). (Diptera: psychodidae: phlebotominae): significance, surveiilance, and control in contingency operations. Armed Forces Pest Manage. Board Tech. Guide 49, 40–50.
Dawit, G., Girma, Z., Simenew, K. (2013). Review on Biology, epidemiology and public health significance of Leishmaniosis. J. Bacteriol. Parasitol. 4, 100166. doi: 10.4172/2155-9597.1000166
Depaquit, J., Lienard, E., Verzeaux-Griffon, A., Ferté, H., Bounamous, A., Gantier, J. C., et al. (2008). Molecular homogeneity in diverse geographical populations of Phlebotomus papatasi (Diptera, Psychodidae) inferred from ND4 mtDNA and ITS2 rDNA epidemiological consequences. Infect. Genet. Evol. 8, 159–170. doi: 10.1016/j.meegid.2007.12.001
Desjeux, P. (2001). The increase of risk factors for Leishmaniosis worldwide. Trans. R. Soc Trop. Med. Hyg. 95, 239–243. doi: 10.1016/S0035-9203(01)90223-8
Desjeux, P. (2004). Leishmaniosis: current situation and new perspectives. Comp. Immunol. Microbiol. Infect. Dis. 27, 305–318. doi: 10.1016/j.cimid.2004.03.004
Desjeux, P., Bryan, J. H., Martin-Saxon, P. (1983). Leishmaniosis in the Gambia. Trans. R. Soc Trop. Med. Hyg. 77, 143–148. doi: 10.1016/0035-9203(83)90052-4
Dondji, B., Duhlinska, D. D., Same-Ekobo, A. (2000). Species composition of the phlebotomine sand fly fauna (Diptera: phlebotominae) in Mokolo Region, Northern Cameroon. Insec. Sci. Applic. 20, 221–226. doi: 10.1017/S1742758400019676
Falcão de Oliveira, E., Casaril, A. E., Fernandes, W. S., Ravanelli, Md. S., Medeiros, M. Jd., Gamarra, R. M. (2016). Monthly distribution of phlebotomine sand flies, and biotic and abiotic factors related to their abundance, in an urban area to which visceral Leishmaniosis is endemic in Corumbá, Brazil. PloS One 11, e0165155. doi: 10.1371/journal.pone.0165155
Faleke, O. O., Lawal, M. D., Magaji, A. A. (2008). Cutaneous Leishmaniosis: Some aspects of epidemiology and a case report. Sokoto J. Veterinary Sci. 7 (1), 13–16.
França, C., Parrot, L. (1920). Introduction ŕ l’étude sys-tématique des Diptčres du genre. Phlebotomus Bul Soc. Path Exot 12, 695708.
Hashiguchi, Y., Otsuru, M., Kamegai, S., Hayashi, S. (2003). “Progress of medical parasitology in Japan,” in Leishmaniosis, vol. 7. (Tokyo, Japan: Megro Parasitological Museum), 537–553.
Hebert, D. N., Cywinska, A., Ball, S. L., deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. R. Soc. London. Ser. B Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Jiya, N. M., Ahmed, H., Jibrin, B., Philips, A. O. (2007). An outbreak of cutaneous Leishmaniosis in a boarding senior secondary school in Sokoto, North – Western Nigeria: clinical presentation and outcome. Nigeria Med. Practice. 5, 86–89.
Kato, H., Gomez, E. A., Yamamoto, Y., Calvopiña, M., Guevara, A. G., Marco, J. D., et al. (2008). Natural infection of Lutzomyia tortura with Leishmania (Viannia) naiffi in an Amazonian area of Ecuador. Am. J. Trop. Med. Hygiene 79, 438–440. doi: 10.4269/ajtmh.2008.79.438
Kato, H., Uezato, H., Gomez, E. A., Terayama, Y., Calvopiña, M., Iwata, H., et al. (2007). Establishment of a mass screening method of sand fly vectors for Leishmania infection by molecular biological methods. Am. J. Trop. Med. Hygiene 77, 324–329. doi: 10.4269/ajtmh.2007.77.324
Kato, H., Uezato, H., Katakura, K., Calvopina, M., Marco, J. D., Barroso, P. A., et al. (2005). Detection and identification of Leishmania species within naturally infected sand flies in the Andean areas of Ecuador by a polymerase chain reaction. Am. J. Trop. Med. Hygiene 72, 87–93. doi: 10.4269/ajtmh.2005.72.87
Khalid, N., Elnaiem, D. E., Aboud, M., Al Rabba, F., Tripet, F. (2010). Morphometric and Molecular differentiation of Phlebotomus (Phlebotomus) sand flies. Med. Veterinary Entomology 24, 352–360. doi: 10.1111/j.1365-2915.2010.00893.x
Killick-Kendrick, R. (1990). Phlebotomine vectors of leishmaniases: a review. Med. Vet. Entomol. 4, 1–24. doi: 10.1111/j.1365-2915.1990.tb00255.x
Killick-Kendrick, R., Killick-Kendrick, M., Tang, Y. (1994). Anthroponotic cutaneous leishmaniasis in Kabul, Afghanistan: the low susceptibility of Phlebotomus papatasi to Leishmania tropica. Trans. R. Soc Trop. Med. Hyg. 88, 252–253. doi: 10.1016/0035-9203(94)90320-4
Kuhls, K., Mauricio, I. L., Pratlong, F., Presber, W., Schönian, G. (2005). “Analysis of ribosomal DNA internal transcribed spacer sequences of the Leishmania donovani complex”. Microbes Infection 7, 1224–1234. doi: 10.1016/j.micinf.2005.04.009
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kuwahara, K., Kato, H., Gomez, E. A., Uezato, H., Mimori, T., Yamamoto, Y. I., et al. (2009). Genetic diversity of ribosomal RNA internal transcribed spacer sequences in Lutzomyia species from areas endemic for New World cutaneous Leishmaniosis. Acta Tropica 112, 131–136. doi: 10.1016/j.actatropica.2009.07.010
Le Blancq, S. M., Peters, W. (1986). Leishmania in the Old World: 4. The distribution of L. donovani sensu lato zymodemes. Trans. R. Soc Trop. Med. Hyg. 80, 367–377. doi: 10.1016/0035-9203(86)90320-2
Lewis, D. J. (1982). A taxonomic review of the genus Phlebotomus (Diptera, Psychodidae). Bull. R. Mus. (Natural History) Entomol. 5, 121–209.
Maroli, M., Feliciangeli, M. D., Bichaud, L., Charrel, R. N., Gradoni, L. (2013). Phlebotomine sand flies and the spreading of leishmaniases and other diseases of public health concern. Med. Vet. Entomol. 27, 123–147. doi: 10.1111/j.1365-2915.2012.01034.x
Maurício, I. L., Stothard, J. R., Miles, M. A. (2000). The strange case of Leishmania chagasi. Parasitol. Today 16, 188–189. doi: 10.1016/S0169-4758(00)01637-9
Mens, P., Spieker, N., Omar, S., Heijnen, M., Schallig, H., Kager, P. A. (2007). Is molecular biology the best alternative for diagnosis of malaria to microscopy? A comparison between microscopy, antigen detection and molecular tests in rural Kenya and urban Tanzania. Trop. Med. Intern. Health 12, 238–244. doi: 10.1111/j.1365-3156.2006.01779.x
Mukherjee, S., Hassan, M. Q., Ghosh, A., Ghosh, K. N., Bhattacharya, A., Adhya, S. (1997). Leishmania DNA in Phlebotomus and Sergentomyia species during a kala-azar epidemic. The Amer. J. Trop. Med. Hyg. 57, 423–425. doi: 10.4269/ajtmh.1997.57.423
Munstermann, L. E. (2004). “Phlebotomine sand flies, the Psychodidae,” in Biology of disease vectors, 2nd ed. Ed. Marquardt, W. C. (CA, USA: Elsevier; San Diego), 141–151.
Murray, T. S., Cappello, M. (2008). The molecular diagnosis of parasitic diseases. Ped. Infect. Dis. J. 27, 163–164. doi: 10.1097/INF.0b013e3181658af0
Mutinga, M. J., Massamba, N. N., Basimike, M., Kamau, C. C., Amimo, F. A., Onyido, A. E. (1994). Cutaneous Leishmaniosis in Kenya: Sergentomyia garnhami (Diptera Psychodidae), a possible vector of Leishmania major in Kitui District: a new focus of the disease. East Afric. Med. J. 71, 424–428.
Nascimento, J. C., Paiva, B. R., Malafronte, R. S., Fernandes, W. D., Galati, E. A. B. (2007). Natural infection of Phlebotomines (Diptera : Psychodidae) in a visceral-Leishmaniosis focus in Mato Grosso do Sul, Brazil. Rev. Institu. Med. Trop. 49, 119–122. doi: 10.1590/S0036-46652007000200011
Nihashi, N., Inaoka, D. K., Tsuge, C., Balogun, E. O., Osada, Y., Goto, Y., et al. (2016). Siccanin is a novel selective inhibitor of trypanosomatid complex II (succinate-ubiquinone reductase) and a potent broad-spectrum anti-trypanosomatid drug candidate. Kala Azar South Asia: Curr. Status Sustain. Challenges, 101–122. doi: 10.1007/978-3-319-47101-3_9
Nzelu, C. O., Kato, H., Puplampu, N., Desewu, K., Odoom, S., Wilson, M. D. (2014). First detection of Leishmania tropica DNA and Trypanosoma species in Sergentomyia sand flies (Diptera: Psychodidae) from an outbreak area of cutaneous Leishmaniosis in Ghana. PLoS Neglec. Trop. Dis. 8, 2630. doi: 10.1371/journal.pntd.0002630
Parvizi, P., Amirkhani, A. (2008)Mitochondrial DNA characterization of Sergentomyia sintoni populations and finding mamMalian Leishmania infections in this sand fly by using ITS-rDNA gene. Available at: .
Perez, J. E., Ogusuku, E., Inga, R., Lopez, M., Monje, J., Paz, L. (1994). Natural Leishmania infection of Leishmania spp. in Peru. Trans. R. Soc Trop. Med. Hyg. 88, 161–164. doi: 10.1016/0035-9203(94)90276-3
Pita-Pereira, D., Alves, C. R., Souza, M. B., Brazil, R. P., Bertho, A. L., Barbosa, A. F. (2005). Identifications of naturally infected Lutzomyia intermedia and Lutzomyia migonei and with Leishmania (Viannia) Braziliensis in Rio de Janeiro (Brazil) revealed by a PCR multiplex non-isotopic hybridization assay. Acta Trop. 99, 905–913. doi: 10.1016/j.trstmh.2005.06.019
Rageau, J., Adam, J. P. (1953). Note sur les phlebotomes d’ Evodoula (Cameroun Francais). Bull. Soc Path. Exot. 46, 587–594.
Ravel, C., Cortes, S., Pratlong, F., Morio, F., Dedet, J. P., Campino, L. (2006). First report of genetic hybrids between two very divergent Leishmania species: Leishmania infantum and Leishmania major. Int. J. Parasitol. 36, 1383–1388. doi: 10.1016/j.ijpara.2006.06.019
Ready, P. D. (2013). Biology of phlebotomine sand flies as vectors of disease agents. Ann. Rev. Entomol. 58, 227–250. doi: 10.1146/annurev-ento-120811-153557
Rondani, C. (1840). “Sopra una Specie di Insetto Dittero,” in Memoria Prima per Servire alla Ditterologia Italiana n°1 (Parma: Donati), 16.
Sadlova, J., Dvorak, V., Seblova, V., Warburg, A., Votypka, J., Volf, P. (2013). Sergentomyia schwetzi is not a competent vector for Leishmania donovani and other Leishmania species pathogenic to humans 6, 186. doi: 10.1186/1756-3305-6-186
Sangare, I., Gantier, J. C., Koalaga, G., Deniau, M., Ouari, A., Giguemdé, R. T. (2009). Sand flies of the South part of Ouagadougou city, Burkina faso. Parasite 16, 231–233. doi: 10.1051/parasite/2009163231
Schonian, G., Nasereddin, A., Dinse, N., Schweynoch, C., Schallig, H. D., Presber, W. (2003). PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn. Microbiol. Infect. Dis. 47, 349 358. doi: 10.1016/S0732-8893(03)00093-2
Senghor, M. W., Faye, M. N., Faye, B., Diana, K., Elguero, E., Gaye, O., et al. (2011). Ecology of phlebotomine sand flies in the rural community of Mont Rolland (Thiès Region, Senegal): area of transmission of canine Leishmaniosis. PloS One 6, e14773. doi: 10.1371/journal.pone.0014773
Ungogo, M. A., Ebiloma, G. U., Ichoron, N., Igoli, J. O., de Koning, H. P., Balogun, E. O. (2020). A review of the antimalarial, antitrypanosomal, and antileishmanial activities of natural compounds isolated from Nigerian flora. Front. Chem. 8, 617448. doi: 10.3389/fchem.2020.617448
Usman, M., Natala, A. J., Jatau, I. D., Ogo, N. I., Balogun, E. O., Alayande, M. O., et al. (2020b). Seroprevalence of canine leishmaniasis in parts of sokoto state, Northwestern Nigeria. Int. J. Curr. Res. 12, 10087–10091. doi: 10.24941/ijcr.37908.02.2020
Usman, M., Natala, A. J., Jatau, I. D., Ogo, N. I., Balogun, E. O., Lawal, M. D., et al. (2020a). Occurrence and monthly dynamics of phlebotomine sand flies in parts of Sokoto State, North-West Nigeria. Niger. J. Parasitol. 41, 109–113. doi: 10.4314/njpar.v41i1.17
Vercammen, F., Berkvens, D., Le Ray, D., Jacquet, D., Vervoort, T. (1997). Development of a slide ELISA for canine Leishmaniosis and comparison with four serological tests. Vet. Rec. 141, 328–330. doi: 10.1136/vr.141.13.328
Volf, P., Benkova, I., Myskova, J., Sadlova, J., Campino, L., Ravel, C. (2007). Increased transmission potential of Leishmania major/Leishmania infantum hybrids. Int. J. Parasitol. 37, 589–593. doi: 10.1016/j.ijpara.2007.02.002
World Health Organization (2010). “Control of the leishmaniosis,” in Report of a meeting of the WHO expert committee on the control of leishmaniases (WHO, Geneva: WHO Technical Report Series).
Young, D. G., Duncan, M. A. (1994). “Guide to the identification and geographic distribution of Lutzomyia sand flies in Mexico, the West Indies, Central and South America (Diptera: Psychodidae),” in Memoirs of the american entomological institute, no. 54 (Gainesville, Florida: Associated Publishers), 881.
Keywords: Phylogenetics, Phlebotomus, Sergentomyia, Leishmania infantum, Sokoto
Citation: Usman M, Natala AJ, Jatau ID, Ogo NI, Jeelani G, Goto Y, Nozaki T, McKerrow JH and Balogun EO (2023) Molecular identification of phlebotomine sand flies and the harbored Leishmania spp. in Sokoto State, Nigeria. Front. Cell. Infect. Microbiol. 13:1219629. doi: 10.3389/fcimb.2023.1219629
Received: 09 May 2023; Accepted: 15 August 2023;
Published: 31 August 2023.
Edited by:
Ana Maria Jansen, Oswaldo Cruz Foundation (Fiocruz), BrazilReviewed by:
Carina Elisei Oliveira, Dom Bosco Catholic University, BrazilCopyright © 2023 Usman, Natala, Jatau, Ogo, Jeelani, Goto, Nozaki, McKerrow and Balogun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emmanuel Oluwadare Balogun, ZW9iYWxvZ3VuQGFidS5lZHUubmc=; b2x1d2FkYXJldXNAeWFob28uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.