- 1AI Hospital/Macro Signal Dynamics Research and Development Center (ai@ku), Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa, Japan
- 2Department of Bio-engineering Nursing, Graduate School of Nursing, Ishikawa Prefectural Nursing University, Kahoku, Japan
- 3Faculty of Health Sciences, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa, Japan
- 4Biotechnology Center, University of Yaoundé I, Yaoundé, Cameroon
- 5Department of Animal Biology and Physiology of the Faculty of Science, University of Yaoundé I, Yaoundé, Cameroon
- 6Department of Tropical Medicine, Medical Microbiology and Pharmacology, John A. Burns School of Medicine, University of Hawaii at Manoa, Honolulu, HI, United States
- 7Institute of Medical Research and Medicinal Plant Studies, University of Yaoundé I, Yaoundé, Cameroon
- 8Faculty of Pharmacy, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa, Japan
- 9Department of Pharmacy, Faculty of Pharmacy, Hasanuddin University, Makassar, Indonesia
- 10Advanced Health Care Science Research Unit, Institute for Frontier Science Initiative, Kanazawa University, Kanazawa, Japan
- 11Division of Health Sciences, Graduate School of Medicine, Osaka University, Suita, Japan
The presence of pathogens and the state of diseases, particularly skin diseases, may alter the composition of human skin microbiome. HIV infection has been reported to impair gut microbiome that leads to severe consequences. However, with cutaneous manifestations, that can be life-threatening, due to the opportunistic pathogens, little is known whether HIV infection might influence the skin microbiome and affect the skin homeostasis. This study catalogued the profile of skin microbiome of healthy Cameroonians, at three different skin sites, and compared them to the HIV-infected individuals. Taking advantage on the use of molecular assay coupled with next-generation sequencing, this study revealed that alpha-diversity of the skin microbiome was higher and beta-diversity was altered significantly in the HIV-infected Cameroonians than in the healthy ones. The relative abundance of skin microbes such as Micrococcus and Kocuria species was higher and Cutibacterium species was significantly lower in HIV-infected people, indicating an early change in the human skin microbiome in response to the HIV infection. This phenotypical shift was not related to the number of CD4 T cell count thus the cause remains to be identified. Overall, these data may offer an important lead on the role of skin microbiome in the determination of cutaneous disease state and the discovery of safe pharmacological preparations to treat microbial-related skin disorders.
1 Introduction
Skin is one of the most important innate immunological barriers available in humans. Together with the skin microbiome, this outer layer of protective elements have been known to play a vital role in a human’s life (Segre, 2006). In fact, impairment of the skin microbiome has been suggested to be closely associated with the occurrence of several dermatological conditions (Fredricks, 2001; Dethlefsen et al., 2007; Lunjani et al., 2019). Dysbiosis of the skin microbiome is involved in the occurrence of several dermatological disorders (Prescott et al., 2017) such as acne, atopic dermatitis, psoriasis, rosacea, and seborrheic dermatitis (Byrd et al., 2018). Recently, it has been reported that skin microbiota composition is subject to alteration in response to pharmacotherapeutic interventions e.g., the application of systemic antimicrobial drugs (Bayal et al., 2019). Thus, identifying factors that affect skin microbiota is crucial as they are important for maintaining skin homeostasis, thereby controlling human health.
Human immunodeficiency virus (HIV) infection can cause serious immune dysfunctions in the affected patients, which might modify the dynamic state of the human microbiome (Williams et al., 2016; D’Angelo et al., 2017). Over the years, HIV infection can progressively develop into acquired immune deficiency syndrome (AIDS) which leads to a more severe condition or even the death of the infected patients (Deeks et al., 2015). Almost a million people succumbed from AIDS each year, especially in developing countries, such as the ones located in Africa (Deeks et al., 2015; World Health Organization, 2022). In Cameroon, for example, HIV infection has been listed as the leading cause of all deaths and remains a threatening public health issue (Mbuagbaw et al., 2016). Of all HIV-related signatures, opportunistic dermatological infections such as prurigo, Kaposi’s sarcoma (KS), and other skin lesions are commonly observed in the AIDS patients (Deeks et al., 2015), implicating potential disturbances in the composition of skin microbiome. Although much focus has been given to study the skin-related disorders, research investigating the tripartite relationship between the immune-deficient status of the HIV-infected patient, the occurrence of dermatological conditions, and the composition of the skin microbiome remains underrepresented.
With the development of modern and advanced tools, human microbiome research has progressed exponentially (NIH Human Microbiome Portfolio Analysis Team, 2019). In relation to the alteration of microbiome under HIV-infected and/or AIDS-affected situations, several studies have been reported on gut (Dillon et al., 2016; Monaco et al., 2016), oral (Zhang et al., 2015; Ali et al., 2021; Li et al., 2021a), serum (Ali et al., 2021), and lung microbiome (Zhu et al., 2022). However, taking the topic of skin microbiome into account, to the best of our knowledge, no study has been reported on the skin microbiome in AIDS patients nor in HIV infected individuals without AIDS and how this may be associated to the presence of dermatological infections. In this study, we are seeking to characterize the composition of skin microbiota in HIV-infected patients, who may or may not have developed AIDS, to assess the presence of a dysbiosis due to immune suppression and how this dysbiosis is associated to the presence of skin conditions like prurigo or KS. Our findings shall provide important insights for further research to decipher the relationship between the patient’s skin microbiota and the development of opportunistic infections of the skin.
2 Materials and methods
2.1 Ethical considerations
This study was approved by the National Ethics Committee of Cameroon (approval no. 2018/06/1045/CE/CNERSH/SP), the hospital where the research was conducted, and the Medical Ethics Committee of Kanazawa University, where the next generation sequencing (NGS) was performed (approval no. 894). All research was performed in accordance with the Declaration of Helsinki. All participants were informed by a written document about the research, and written informed consent was obtained from all participants.
2.2 Study design and settings
This was a part of a cross-sectional study conducted in an HIV-clinic of the Efoulan District Hospital in Yaoundé, Cameroon, and in a single university in Japan. The inclusion criterion was being aged 21–65 years, because (1) the age of adulthood in Cameroon is 21 years old and thus it is deemed necessary to be 21 years old or older to understand and consent to this research, and (2) the skin microbiome of older people (older than ~65 years old) is reported to be affected by aging itself (Nagase et al., 2020) and thus several studies limit the age of participants being younger than or equal to 65 years old (Carrieri et al., 2021; Li et al., 2021b). The exclusion criteria were those who (1) had oral/topical antibiotics 1 week prior to the study, (2) were pregnant, or (3) were in a critical situation such as severe pneumonia, sepsis, pulmonary tuberculosis, toxoplasmosis, cryptococcosis, and meningoencephalitis. The participants were requested not to (1) take a bath or shower or (2) use emollients/creams on the sampling site, after midnight of the day of sampling (Nakatsuji et al., 2017), as these may alter the bacterial composition and interfere DNA extraction. Those who did not follow the instruction requested by the researchers were excluded.
2.3 Sample collection
Skin swabs were collected as described previously (Benderli et al., 2019; Ogai et al., 2022). In brief, Puritan HydraFlock Sterile Flocked Swab (25-3306-H; Puritan Medical Products Co., ME, USA) was presoaked in normal saline (S5815; Teknova, CA, USA) with 0.1% Tween 20 (28353-14; Nacalai Tesque, Inc., Kyoto, Japan) solution. Swabbing was then performed in a 5 × 5 cm2 on designated positions or in the entire area of KS. After swabbing, each swab head was cut and placed in a 1.5 mL microcentrifuge tube, carried in a cooler box with ice, and stored at -30°C until DNA extraction. To minimize sample degradation, DNA extraction was done in the same day of sampling.
2.4 DNA extraction
DNA from the swab head was performed as described previously (Ogai et al., 2018; Benderli et al., 2019; Ogai et al., 2022). Briefly, the swab heads were processed with the Kaneka Easy DNA Extraction Kit version 2 (KN-T110005; Kaneka Corp., Tokyo, Japan), followed by the enzymatic DNA extraction process with QIAamp DNA Mini Kit (51304; Qiagen N.V., Venlo, The Netherlands). The extracted DNA samples were stored at -30°C until NGS preparation.
2.5 NGS
The extracted DNA samples, along with the negative control (DNA extracted with swab head only) and positive control (DNA extracted from the ZymoBIOMICS Microbial Community Standard [D6300; Zymo Research Corp., Irvine, CA, USA]), were dedicated to the NGS analysis as described previously (Ogai et al., 2022). The same amount of the 16S rRNA gene from each sample was used to amplify the V3-V4 hypervariable region (Castelino et al., 2017; Zeeuwen et al., 2017; Ogai et al., 2018). After indexing with the Nextera XT Index Kit version 2 (FC-131-2001 to 2004; Illumina, Inc., San Diego, CA, USA), the library solution was loaded onto the MiSeq System (SY-410-1003; Illumina) with MiSeq Reagent Kit (version 3, 600 cycles; MS-102-3003; Illumina) and 15% PhiX Control (version 3; FC-110-3001; Illumina). The data from negative and positive controls are shown in Supplementary Figure S1.
2.6 Sequence analysis
The sequence analysis was done according to the Qiime2 instruction (Bolyen et al., 2019). Briefly, the raw fastq sequences were first quality-filtered and chimera-eliminated by DADA2 plugin (Callahan et al., 2016). Prior to analysis, the samples whose sequencing depth were <5,000 were discarded. For taxonomic classification of amplicon sequence variants (ASVs), q2-feature-classifier plugin (Bokulich et al., 2018) was used to construct the naïve Bayes classifier with the Silva database (version 138) (Quast et al., 2013). Beta diversity was calculated based on the Bray–Curtis distance followed by principal coordinate analysis by using q2-diversity plugin. Alpha diversity indices [observed ASVs, Faith’s phylogenetic diversity (PD), and Shannon index] were calculated by the q2-diversity plugin with the rarefaction at 5,000 depth of sequences.
2.7 Statistical analysis
All statistical analyses were performed using R statistical software (R Core Team, 2020) version 4.2.2. The data were expressed as means ± standard deviation or n (%) where appropriate. The boxplot denotes the 25th, 50th, and 75th percentile boxes with 25th percentile - 1.5 × IQR to 75th percentile + 1.5 × IQR whiskers. The relative abundance between the HIV-positive and HIV-negative population was compared by using the linear models for differential abundance analysis method (LinDA) (Zhou et al., 2022) with incorporating age and sex as covariates. For multiple comparisons of bacterial abundance, Benjamini-Hochberg’s false discovery rate control was employed. The significant differences in beta diversity between the two groups was assessed by a permutational analysis of variance with 9,999 permutations by adonis2 function of car package (Fox and Weisberg, 2018) of R software. Alpha diversity between the two groups were compared by using Mann–Whitney U test. For correlation analysis, Spearman’s correlation coefficient was used. P-values < 0.05 was considered statistically significant.
3 Results
3.1 16S rRNA-Based metagenomic analysis of skin microbes between the healthy and HIV-infected Cameroonians
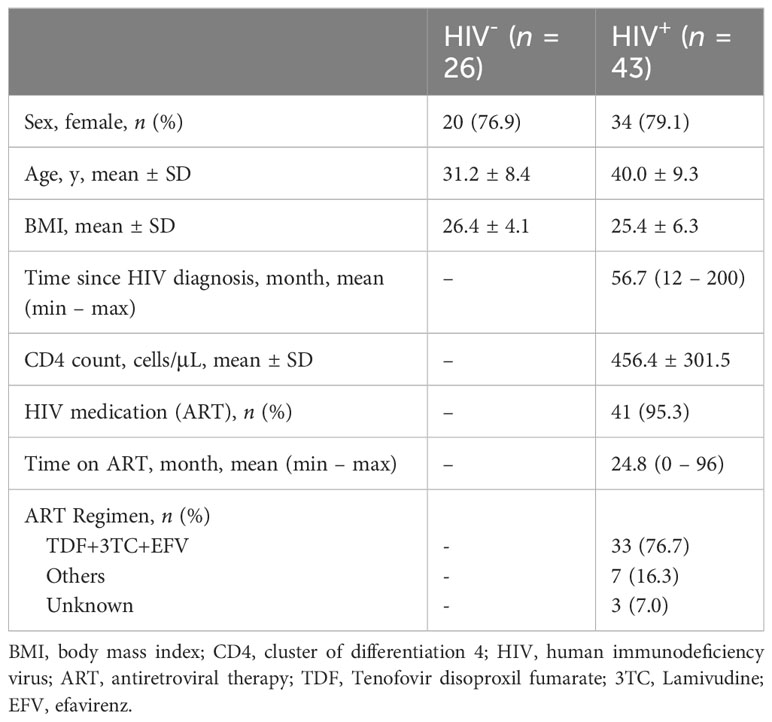
This study first analyzed the relative abundance and prevalence of skin-resident microbes collected from healthy Cameroonians and the ones infected with HIV: Healthy (n = 26) and HIV-infected (n = 43). The demographic information is described in Table 1. Data of 16S rRNA gene sequencing analysis of skin swabs from healthy Cameroonians were overlapped with our previous study (Ogai et al., 2022). However, we applied ASV-based analysis to compare microbiome difference in higher resolution.
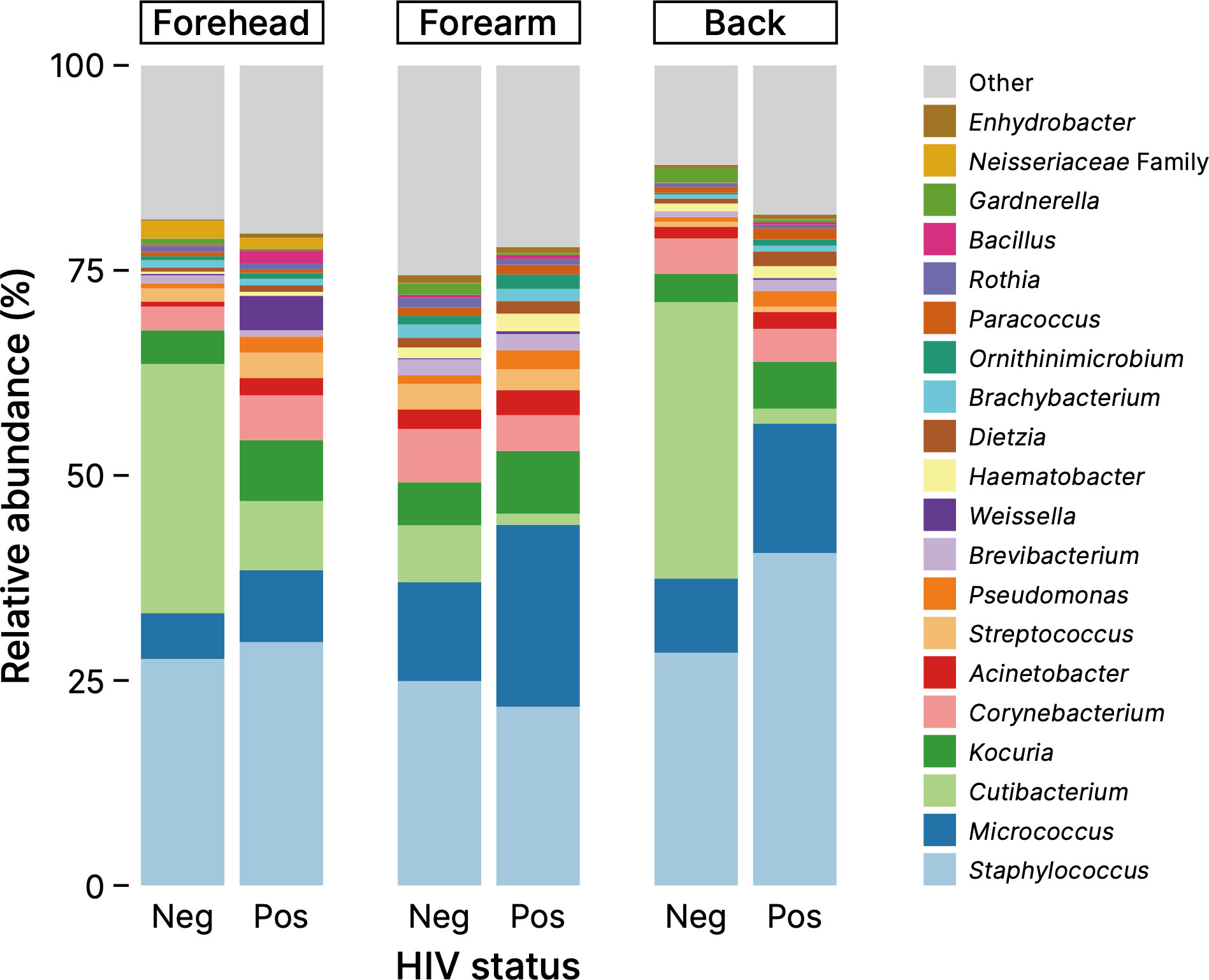
Figure 1 shows a comparison of the skin microbiome compositions between healthy Cameroonians and their HIV-infected counterparts in the three skin sites: forehead, right forearm, and the mid-upper back. The composition of skin microbiome of all samples are shown in Supplementary Figure S2. The microbiome of the Cameroonian skin samples, in the presence or absence of HIV infection, can be classified into more than 20 genera that primarily belong to three phyla: Actinobacteria, Firmicutes, and Proteobacteria. At the genus level, most obtained sequences were Staphylococcus, Cutibacterium, Micrococcus, Corynebacterium, Kocuria, and, to a lesser extent, Acinetobacter and Streptococcus (Figure 1). In the samples collected from the surface of KS, the main bacterial genera were common, but their proportions were more diverse (Supplementary Figure S3).
The microbial differences between the HIV-positive and HIV-negative participants were further confirmed by the beta diversity analysis using the Bray–Curtis distance. The principal component analysis of beta diversity showed that the skin samples (forehead, forearm, and back skin) obtained from HIV-infected Cameroonian constituted significantly different clusters from healthy Cameroonian participants (Figure 2). This result suggests that there is a difference of microbiome profiles between healthy and HIV-infected people.
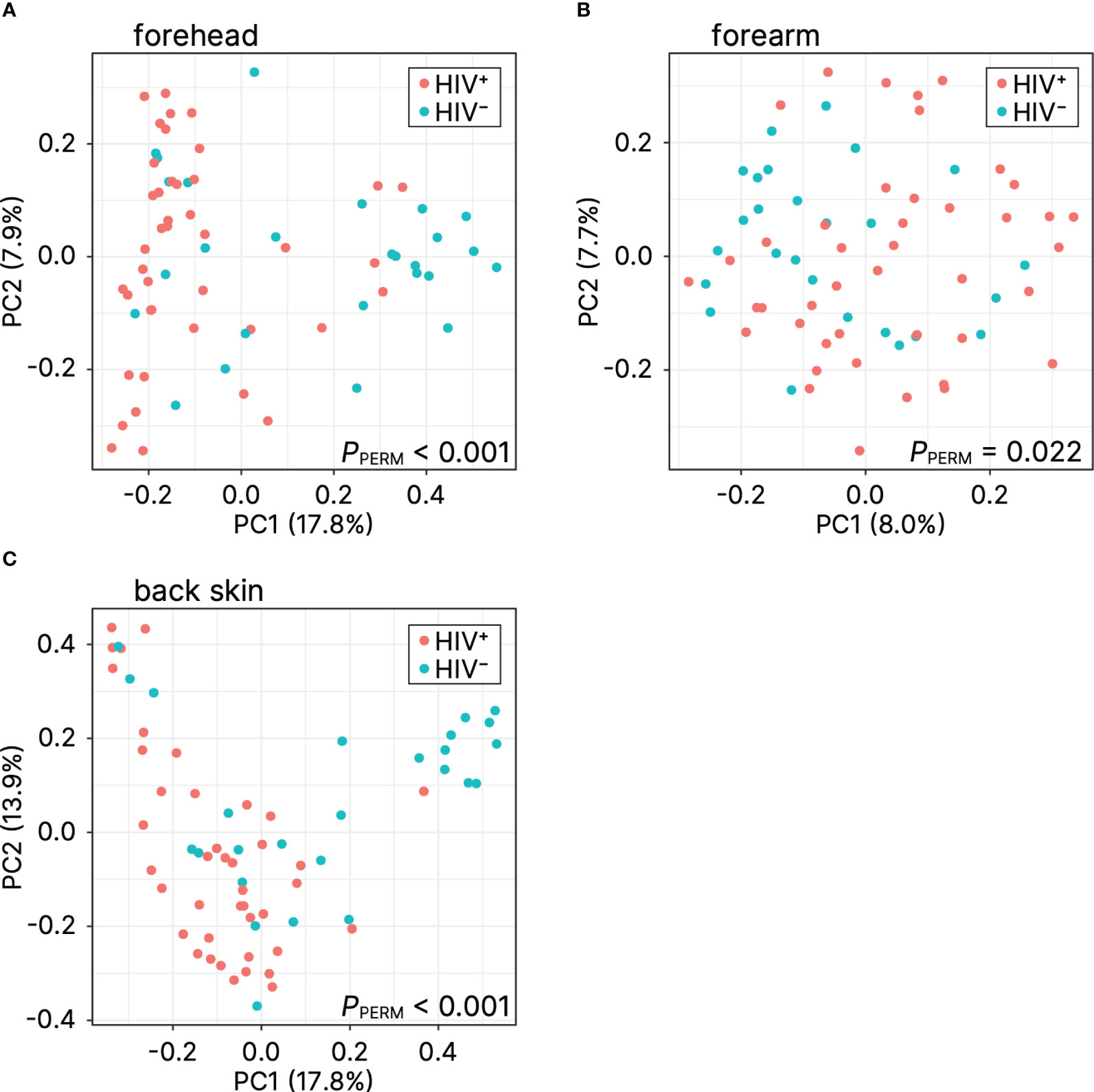
Figure 2 Principal coordinate analysis of beta diversity (Bray–Curtis distance) for (A) forehead, (B) forearm, and (C) back skin. PC, principal coordinate; PPERM, P-value of the permutational analysis of variance.
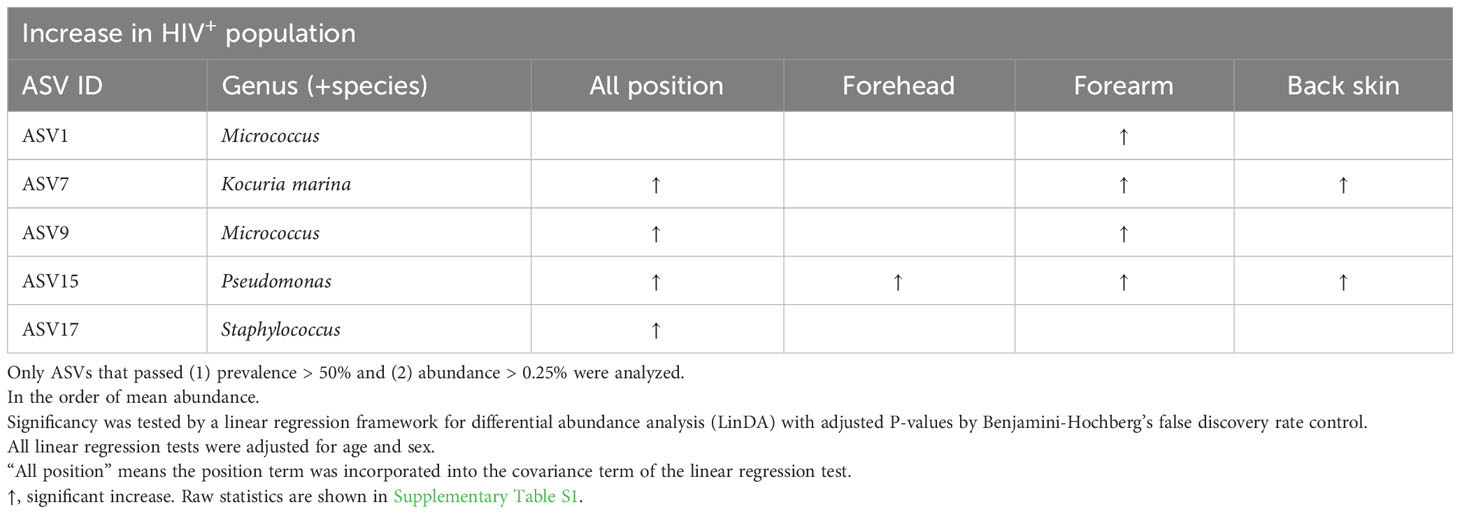
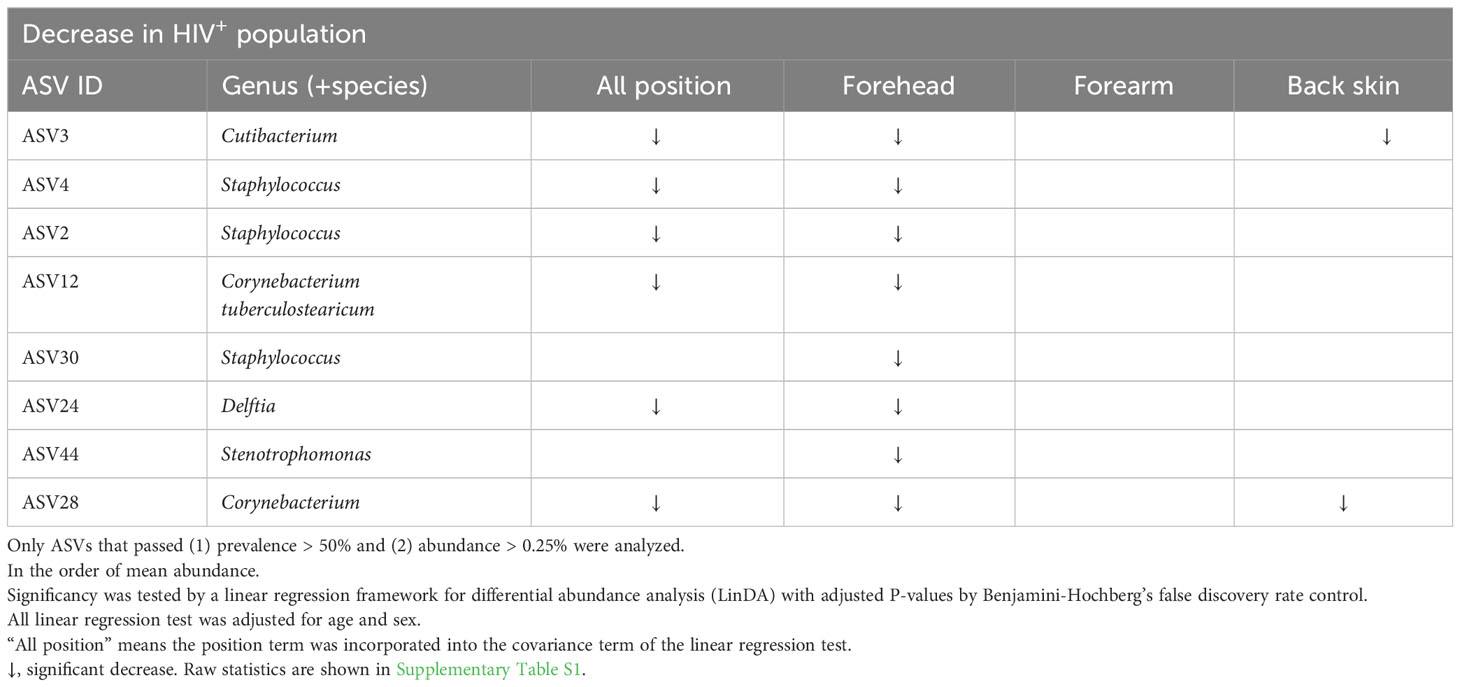
ASV-based analysis showed that the genus or species of Micrococcus (ASV1, ASV9), Kocuria marina (ASV7), Pseudomonas (ASV15), Staphylococcus (ASV17) were significantly higher in all or some parts of skin sites of the HIV-infected Cameroonians in comparison to their healthy counterparts (Figure 3; Table 2). In contrast, the genus or species of Cutibacterium (ASV3), Staphylococcus (ASV2, ASV4, ASV30), Corynebacterium tuberculostearicum (ASV12), Corynebacterium (ASV28), Delftia (ASV24), Stenotrophomonas (ASV44) were significantly lower in the HIV-infected Cameroonians (Figure 3; Table 3).
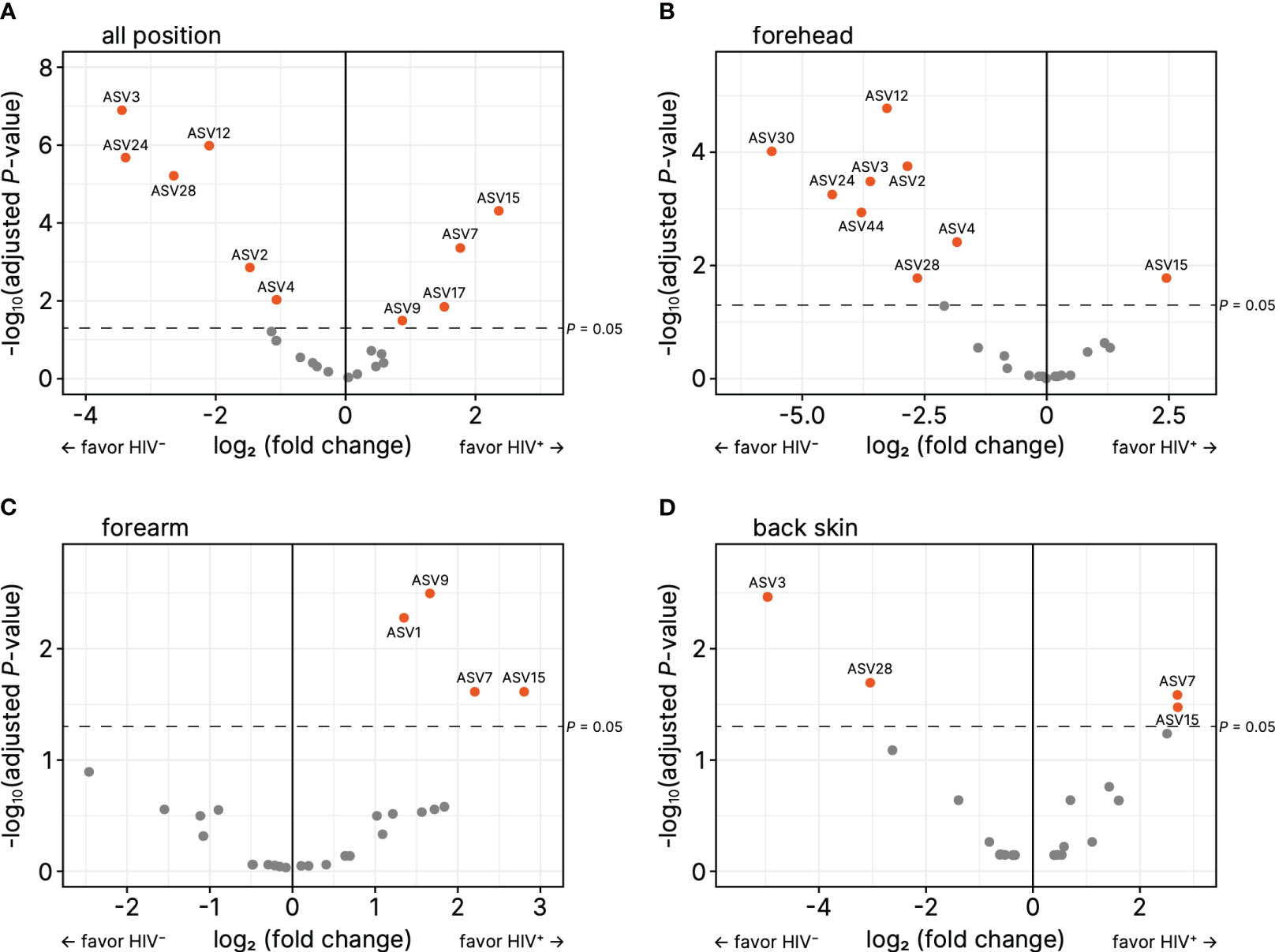
Figure 3 Volcano plots for relative abundance of amplicon sequence variances (ASVs) on (A) forehead, (B) forearm, and (C) back skin. The horizontal axes denote the logarithm of relative fold change (based on HIV-negative) to base 2, and the vertical axes denote the negative of logarithm of adjusted P-values to base 10. The ASVs that have P < 0.05 were orange colored.
Overall, these findings indicated that HIV infection does affect the composition of skin microbiome in the Cameroonians.
3.2 Higher alpha diversity in HIV-infected Cameroonians
We calculated the alpha diversity metrics of the skin microbiome between healthy Cameroonians and the HIV-infected individuals in the forehead, forearm and the back skin, namely the number of observed ASVs, Faith’s PD, and Shannon index (Figure 4). HIV-infected Cameroonians have a much diverse skin-resident flora at the forehead and the back, with a significantly higher alpha diversity metrices. However, no significant difference in the skin microbiome diversity of the forearm skin between both groups. These findings suggest that HIV-infected people have more diverse bacterial species than their healthy counterparts in some parts of their skin.
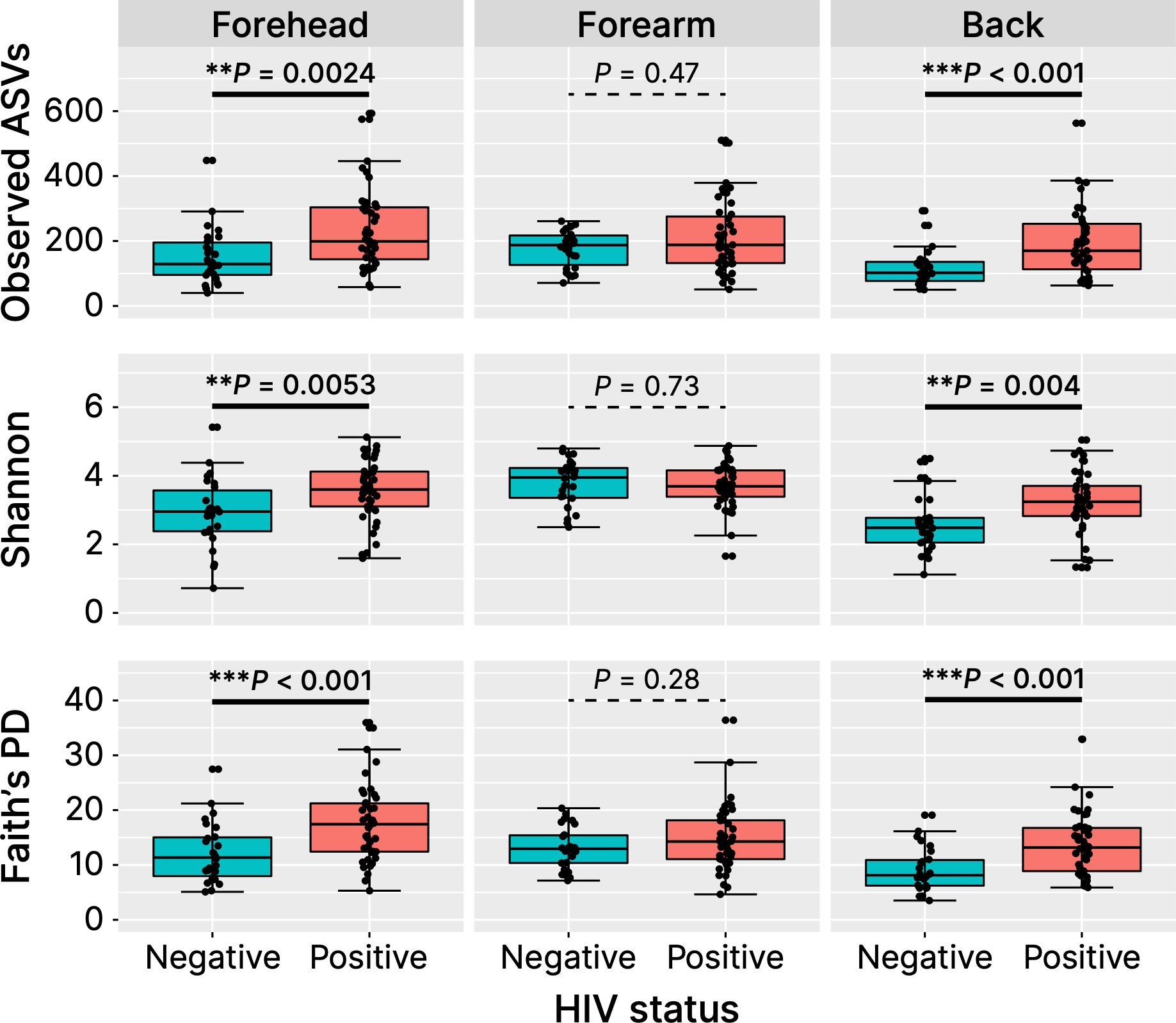
Figure 4 Alpha diversity metrices. ASVs, amplicon sequence variances; PD, phylogenetic diversity. **P < 0.01, ***P < 0.001.
3.3 Irrelevant correlation of CD4+ T cells count with bacterial species abundance
The close correlation of CD4 T cells count with the progressiveness of HIV status has been widely suggested (Deeks et al., 2015; Kestens and Mandy, 2017). To investigate whether the number of CD4 T cells is likely correlated with the discrepancies of skin microbe composition observed in this study, an experiment to determine the CD4 T cell count was carried out. As revealed in Table 4, we observed that there is no correlation between the number of CD4 T cell count with the abundance of bacterial species recovered from the skin samples of HIV-infected Cameroonians.
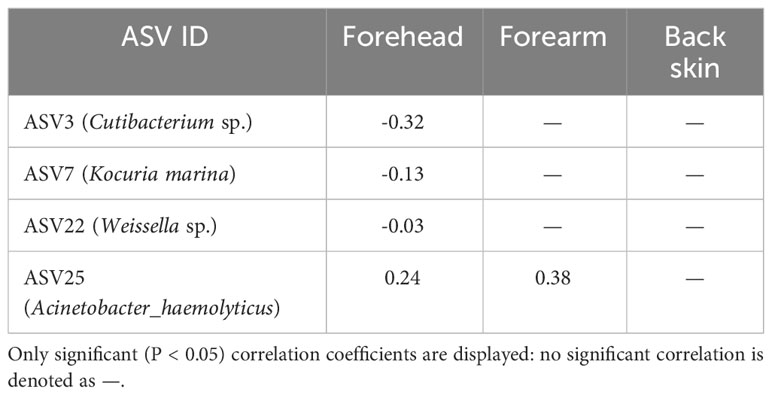
Table 4 Spearman’s correlation coefficients between the CD4+ cell number and the relative abundance of each amplicon sequence variance (ASV).
4 Discussion
In this study, we carried out comprehensive analysis of the skin microbiome profile of healthy Cameroonians, at three different skin sites: forehead, right forearm, and the mid-upper back, and compared them to the ones infected with HIV. Taking advantage of the NGS method and ASV-based bioinformatic analysis, we assessed the dynamic variations of Cameroonians skin microbiome and ascertain whether topological elements might play distinctive roles in the skin microbiome diversity in the event of HIV infection. Our ASV-based analysis indicated that, in HIV-infected Cameroonians, the relative number of some bacteria increased and some decreased. Furthermore, our results clearly showed that the alpha-diversity of the skin microbiome was greater, and beta-diversity was distinct from healthy counterparts. These findings collectively suggest that HIV infection affects human skin microbiome.
To inspect the compositional difference in detail, we observed that the relative abundance of Cutibacterium species (spp.) was significantly lower in the HIV-infected Cameroonians than their healthy counterparts. In our previous study, we confirmed that Cutibacterium spp. was less observed in the skin of healthy Cameroonians than in that of healthy Japanese people (Ogai et al., 2022), probably due to the less sebum-rich ecological nature of Cameroonians skin that might support Cutibacterium spp. growth. It is important to note that the role of Cutibacterium spp. in the cutaneous homeostasis and skin health has been reported (Bolla et al., 2020; Claesen et al., 2020; Rozas et al., 2021). Hence, although the cause of Cutibacterium spp. decline remains to be identified, this finding is particularly interesting because HIV infection may further reduce the already low levels of Cutibacterium spp., which may adversely affect to the skin homeostasis and lead to undesirable phenotypes observed during the state of disease.
It is widely known that the skin microbiome can be modified by the skin-related diseases such as atopic dermatitis and psoriasis. For example, higher Staphylococcus aureus and Corynebacterium is reported in atopic dermatitis (Kobayashi et al., 2015), and colonization of Staphylococcus, Streptococcus, Finegoldia, and Corynebacterium is reported to be involved in psoriasis (Hsu et al., 2020). On the other hand, changes in skin microbiome of HIV+ patients (Figure 1; Tables 2, 3) are different from those found in major skin diseases, highlighting the uniqueness of skin microbiome under HIV infection. However, there are emerging interest in skin “mycobiome” in recent years, and it is reported that Malassezia and Candida fungi are involved in psoriasis (Gupta et al., 2022). Analysing fungal composition, or mycobiome, of HIV patients’ skin might also give new insights about HIV-related changes in skin microorganisms.
The composition of skin microbiome can be affected by several intrinsic factors, such as aging, sex, and racial differences (Wilantho et al., 2017; Khmaladze et al., 2020; Boxberger et al., 2021; Kim et al., 2021). Additionally, extrinsic factors such as hygiene, lifestyle, climate and/or geographical differences are also stated to be accountable for variations in the skin microbiome profile (Lehtimaki et al., 2017; Khmaladze et al., 2020; Boxberger et al., 2021). We previously showed that Cameroonian people have significantly higher alpha diversity in skin microbiome than Japanese people do, which has been attributed to both intrinsic and extrinsic factors (Ogai et al., 2022). In this study, we observed that the alpha diversity was significantly higher in the HIV-positive individuals. Given that the participants in this study resided in the close region (Yaoundé city) of the country, and sex and age are not so different (young- to middle- aged), it is speculated that HIV infection itself, other than environmental and innate factors, influences the diversity of the skin microbiota. Further studies would be desired whether increased alpha-diversity indicates the likelihood of opportunistic skin infection. Moreover, the cause for such differences shall be an interesting venue for future research. Is it the result of HIV infection? How might HIV infection alter the number of bacterial species? Since we found that there is no correlation between the number of CD4 T cell count with the abundance of bacterial species recovered from the skin samples of HIV-infected Cameroonians (Table 4), we consider that difference in the cellular adaptive immune status, at least the CD4 T cells, might not the primary cause for the different skin microbiome.
Another important question is whether the observed skin microbiome distinction alters the landscape of skin homeostasis in HIV-infected people, such as the integrity of skin structure and skin opportunistic infections. Such information might provide insights on how skin microbiome modulates its physiology and how those interactions may influence the patients state during the course of HIV infection. This, in the end, may provide important leads in the discovery of effective pharmacological preparations to manage microbial-related disorders in the skin.
Through this study, we could obtain the changes of skin microbiome in HIV patients. Although we need further studies to look into the relationship between the microbial changes and skin condition and immunological status, information of skin microbiome could be used for predicting and preventing the opportunistic infection of the skin in HIV patients.
5 Conclusion
In this study, we have shown the distinct skin microbiome between HIV-positive and HIV-negative Cameroonian people. This study may provide a brief but fundamental information to better understand skin symptoms and crosstalk between skin and body via the skin microbiome under the influence of HIV infection.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ddbj.nig.ac.jp/, DRA011596 and DRA016187.
Ethics statement
The studies involving humans were approved by National Ethics Committee of Cameroon (approval no. 2018/06/1045/CE/CNERSH/SP) Medical Ethics Committee of Kanazawa University (approval no. 894). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
Conceptualization: GE, SO, and TK. Methodology: KO, YL, AH, AM, SO, and TK. Software: KO. Validation: KO, BN, AM, FN, and TK. Formal analysis: KO, BN, and AM. Investigation: KO, BN, YL, JA, BJ, HA, LE, AH, AM, GL, and TK. Resources: BN, HA, LE, RM, RL, and GL. Data curation: KO, AM, FN, and TK. Writing—original draft preparation: KO, FN, and TK. Writing—review and editing: KO, FN, and TK. Visualization: KO and AM. Supervision: RM, RL, GL, SO, and TK. Project administration: GL, SO, and TK. Funding acquisition: KO, YL, GL, SO, and TK. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Japan Society for the Promotion of Science KAKENHI (17H04428, 19KK0243, and 21H03221 to SO), Kanazawa University Sakigake Project (to SO), Kanazawa University Overseas Research Support for Young Scientists (to KO), and Takeda Science Foundation Research Grant (to TK).
Acknowledgments
We thank the staff of the Efoulan district hospital for their collaboration and support, especially Dr. Akame, the HIV clinic coordinator. We also thank the researchers and laboratory staff at the Biotechnology Centre, University of Yaoundé I, Cameroon, for processing and archiving the samples. We appreciate all the participants of this study for their time and contribution.
Conflict of interest
KO belongs to the Department of Bio-engineering Nursing, which was funded by Saraya Co., Ltd. Osaka, Japan, from April 1, 2023. The funder was not involved in any stage of this study, and thus does not constitute any conflict of interest on this study.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1211899/full#supplementary-material
References
Ali, Z., Shahzadi, I., Majeed, A., Malik, H. M. T., Waseem, S., Ahmed, I., et al. (2021). Comparative analysis of the serum microbiome of HIV infected individuals. Genomics 113, 4015–4021. doi: 10.1016/j.ygeno.2021.10.005
Bayal, N., Nagpal, S., Haque, M. M., Patole, M. S., Valluri, V., Suryavanshi, R., et al. (2019). 16S rDNA based skin microbiome data of healthy individuals and leprosy patients from India. Sci. Data 6, 225. doi: 10.1038/s41597-019-0232-1
Benderli, N. C., Ogai, K., Lloyd, Y. M., Arios, J. P., Jiyarom, B., Awanakam, A. H., et al. (2019). Feasibility of microbial sample collection on the skin from people in Yaounde, Cameroon. Drug Discovery Ther. 13, 360–364. doi: 10.5582/ddt.2019.01075
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome 6, 90. doi: 10.1186/s40168-018-0470-z
Bolla, B. S., Erdei, L., Urban, E., Burian, K., Kemeny, L., Szabo, K. (2020). Cutibacterium acnes regulates the epidermal barrier properties of HPV-KER human immortalized keratinocyte cultures. Sci. Rep. 10, 12815. doi: 10.1038/s41598-020-69677-6
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Boxberger, M., Cenizo, V., Cassir, N., La Scola, B. (2021). Challenges in exploring and manipulating the human skin microbiome. Microbiome 9, 125. doi: 10.1186/s40168-021-01062-5
Byrd, A. L., Belkaid, Y., Segre, J. A. (2018). The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155. doi: 10.1038/nrmicro.2017.157
Callahan, B. J., Mcmurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Carrieri, A. P., Haiminen, N., Maudsley-Barton, S., Gardiner, L. J., Murphy, B., Mayes, A. E., et al. (2021). Explainable AI reveals changes in skin microbiome composition linked to phenotypic differences. Sci. Rep. 11, 4565. doi: 10.1038/s41598-021-83922-6
Castelino, M., Eyre, S., Moat, J., Fox, G., Martin, P., Ho, P., et al. (2017). Optimisation of methods for bacterial skin microbiome investigation: primer selection and comparison of the 454 versus MiSeq platform. BMC Microbiol. 17, 23. doi: 10.1186/s12866-017-0927-4
Claesen, J., Spagnolo, J. B., Ramos, S. F., Kurita, K. L., Byrd, A. L., Aksenov, A. A., et al. (2020). A Cutibacterium acnes antibiotic modulates human skin microbiota composition in hair follicles. Sci. Transl. Med. 12, eaay5445. doi: 10.1126/scitranslmed.aay5445
D’Angelo, C., Reale, M., Costantini, E. (2017). Microbiota and probiotics in health and HIV infection. Nutrients 9, 615. doi: 10.3390/nu9060615
Deeks, S. G., Overbaugh, J., Phillips, A., Buchbinder, S. (2015). HIV infection. Nat. Rev. Dis. Primers 1, 15035. doi: 10.1038/nrdp.2015.35
Dethlefsen, L., Mcfall-Ngai, M., Relman, D. A. (2007). An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449, 811–818. doi: 10.1038/nature06245
Dillon, S. M., Frank, D. N., Wilson, C. C. (2016). The gut microbiome and HIV-1 pathogenesis: a two-way street. AIDS 30, 2737–2751. doi: 10.1097/QAD.0000000000001289
Fox, J., Weisberg, S. (2018). An R Companion to Applied Regression. (Thousand Oaks, CA, USA:SAGE Publications).
Fredricks, D. N. (2001). Microbial ecology of human skin in health and disease. J. Investig. Dermatol. Symp Proc. 6, 167–169. doi: 10.1046/j.0022-202x.2001.00039.x
Gupta, M., Weinberg, J. M., Yamauchi, P. S., Patil, A., Grabbe, S., Goldust, M. (2022). Psoriasis: Embarking a dynamic shift in the skin microbiota. J. Cosmet Dermatol. 21, 1402–1406. doi: 10.1111/jocd.14273
Hsu, D. K., Fung, M. A., Chen, H.-L. (2020). Role of skin and gut microbiota in the pathogenesis of psoriasis, an inflammatory skin disease. Med. Microecol 4, 100016. doi: 10.1016/j.medmic.2020.100016
Kestens, L., Mandy, F. (2017). Thirty-five years of CD4 T-cell counting in HIV infection: From flow cytometry in the lab to point-of-care testing in the field. Cytometry B Clin. Cytom 92, 437–444. doi: 10.1002/cyto.b.21400
Khmaladze, I., Leonardi, M., Fabre, S., Messaraa, C., Mavon, A. (2020). The skin interactome: A holistic "Genome-microbiome-exposome" Approach to understand and modulate skin health and aging. Clin. Cosmet Investig. Dermatol. 13, 1021–1040. doi: 10.2147/CCID.S239367
Kim, J. H., Son, S. M., Park, H., Kim, B. K., Choi, I. S., Kim, H., et al. (2021). Taxonomic profiling of skin microbiome and correlation with clinical skin parameters in healthy Koreans. Sci. Rep. 11, 16269. doi: 10.1038/s41598-021-95734-9
Kobayashi, T., Glatz, M., Horiuchi, K., Kawasaki, H., Akiyama, H., Kaplan, D. H., et al. (2015). Dysbiosis and staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity 42, 756–766. doi: 10.1016/j.immuni.2015.03.014
Lehtimaki, J., Karkman, A., Laatikainen, T., Paalanen, L., Von Hertzen, L., Haahtela, T., et al. (2017). Patterns in the skin microbiota differ in children and teenagers between rural and urban environments. Sci. Rep. 7, 45651. doi: 10.1038/srep45651
Li, S., Su, B., He, Q. S., Wu, H., Zhang, T. (2021a). Alterations in the oral microbiome in HIV infection: causes, effects and potential interventions. Chin. Med. J. (Engl) 134, 2788–2798. doi: 10.1097/CM9.0000000000001825
Li, Z., Xia, J., Jiang, L., Tan, Y., An, Y., Zhu, X., et al. (2021b). Characterization of the human skin resistome and identification of two microbiota cutotypes. Microbiome 9, 47. doi: 10.1186/s40168-020-00995-7
Lunjani, N., Hlela, C., O'mahony, L. (2019). Microbiome and skin biology. Curr. Opin. Allergy Clin. Immunol. 19, 328–333. doi: 10.1097/ACI.0000000000000542
Mbuagbaw, J., Jingi, A. M., Noubiap, J. J., Kaze, A. D., Nansseu, J. R., Bigna, J. J., et al. (2016). Patterns and trends in mortality among HIV-infected and HIV-uninfected patients in a major Internal Medicine Unit in Yaounde, Cameroon: a retrospective cohort study. JRSM Open 7, 2054270416654859. doi: 10.1177/2054270416654859
Monaco, C. L., Gootenberg, D. B., Zhao, G., Handley, S. A., Ghebremichael, M. S., Lim, E. S., et al. (2016). Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe 19, 311–322. doi: 10.1016/j.chom.2016.02.011
Nagase, S., Ogai, K., Urai, T., Shibata, K., Matsubara, E., Mukai, K., et al. (2020). Distinct skin microbiome and skin physiological functions between bedridden older patients and healthy people: A single-center study in Japan. Front. Med. (Lausanne) 7, 101. doi: 10.3389/fmed.2020.00101
Nakatsuji, T., Chen, T. H., Narala, S., Chun, K. A., Two, A. M., Yun, T., et al. (2017). Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 9, eaah4680. doi: 10.1126/scitranslmed.aah4680
NIH Human Microbiome Portfolio Analysis Team (2019). A review of 10 years of human microbiome research activities at the US National Institutes of Health, Fiscal Years 2007-2016. Microbiome 7, 31. doi: 10.1186/s40168-019-0620-y
Ogai, K., Nagase, S., Mukai, K., Iuchi, T., Mori, Y., Matsue, M., et al. (2018). A comparison of techniques for collecting skin microbiome samples: swabbing versus tape-stripping. Front. Microbiol. 9, 2362. doi: 10.3389/fmicb.2018.02362
Ogai, K., Nana, B. C., Lloyd, Y. M., Arios, J. P., Jiyarom, B., Awanakam, H., et al. (2022). Skin microbiome profile of healthy Cameroonians and Japanese. Sci. Rep. 12, 1364. doi: 10.1038/s41598-022-05244-5
Prescott, S. L., Larcombe, D. L., Logan, A. C., West, C., Burks, W., Caraballo, L., et al. (2017). The skin microbiome: impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ J. 10, 29. doi: 10.1186/s40413-017-0160-5
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team (2020) R: A Language and Environment for Statistical Computing. Available at: https://www.r-project.org/ (Accessed 5 January 2020).
Rozas, M., Hart De Ruijter, A., Fabrega, M. J., Zorgani, A., Guell, M., Paetzold, B., et al. (2021). From dysbiosis to healthy skin: major contributions of cutibacterium acnes to skin homeostasis. Microorganisms 9, 628. doi: 10.3390/microorganisms9030628
Segre, J. A. (2006). Epidermal barrier formation and recovery in skin disorders. J. Clin. Invest. 116, 1150–1158. doi: 10.1172/JCI28521
Wilantho, A., Deekaew, P., Srisuttiyakorn, C., Tongsima, S., Somboonna, N. (2017). Diversity of bacterial communities on the facial skin of different age-group Thai males. PeerJ 5, e4084. doi: 10.7717/peerj.4084
Williams, B., Landay, A., Presti, R. M. (2016). Microbiome alterations in HIV infection a review. Cell Microbiol. 18, 645–651. doi: 10.1111/cmi.12588
World Health Organization (2022) Fact sheets HIV-AIDS. Available at: https://www.who.int/news-room/fact-sheets/detail/hiv-aids (Accessed 13 August 2022).
Zeeuwen, P., Boekhorst, J., Ederveen, T. H. A., Kleerebezem, M., Schalkwijk, J., Van Hijum, S., et al. (2017). Reply to Meisel et al. J. Invest. Dermatol. 137, 961–962. doi: 10.1016/j.jid.2016.11.013
Zhang, F., He, S., Jin, J., Dong, G., Wu, H. (2015). Exploring salivary microbiota in AIDS patients with different periodontal statuses using 454 GS-FLX Titanium pyrosequencing. Front. Cell Infect. Microbiol. 5, 55. doi: 10.3389/fcimb.2015.00055
Zhou, H., He, K., Chen, J., Zhang, X. (2022). LinDA: linear models for differential abundance analysis of microbiome compositional data. Genome Biol. 23, 95. doi: 10.1186/s13059-022-02655-5
Keywords: skin microbiome, AIDS, HIV, Cameroon, Cutibacterium
Citation: Ogai K, Nana BC, Lloyd YM, Arios JP, Jiyarom B, Awanakam H, Esemu LF, Hori A, Matsuoka A, Nainu F, Megnekou R, Leke RGF, Ekali GL, Okamoto S and Kuraishi T (2023) Skin microbiome profile in people living with HIV/AIDS in Cameroon. Front. Cell. Infect. Microbiol. 13:1211899. doi: 10.3389/fcimb.2023.1211899
Received: 25 April 2023; Accepted: 10 October 2023;
Published: 31 October 2023.
Edited by:
Carlo Contini, University of Ferrara, ItalyReviewed by:
Fengping Liu, Jiangnan University, ChinaIndrashis Podder, College of Medicine & Sagore Dutta Hospital, India
Copyright © 2023 Ogai, Nana, Lloyd, Arios, Jiyarom, Awanakam, Esemu, Hori, Matsuoka, Nainu, Megnekou, Leke, Ekali, Okamoto and Kuraishi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriel Loni Ekali, ZWxvbmkyMDAwQHlhaG9vLmNvbQ==; Shigefumi Okamoto, c29rYW1vdG9Ac2Focy5tZWQub3Nha2EtdS5hYy5qcA==; Takayuki Kuraishi, dGt1cmFpc2hpQHN0YWZmLmthbmF6YXdhLXUuYWMuanA=
 Kazuhiro Ogai
Kazuhiro Ogai Benderli Christine Nana
Benderli Christine Nana Yukie Michelle Lloyd6
Yukie Michelle Lloyd6 John Paul Arios
John Paul Arios Boonyanudh Jiyarom
Boonyanudh Jiyarom Livo Forgu Esemu
Livo Forgu Esemu Firzan Nainu
Firzan Nainu Rosette Megnekou
Rosette Megnekou Gabriel Loni Ekali
Gabriel Loni Ekali Shigefumi Okamoto
Shigefumi Okamoto Takayuki Kuraishi
Takayuki Kuraishi