- Microbiology Program, Department of Mathematics and Natural Sciences, BRAC University, Dhaka, Bangladesh
“Expanded quantitative urine culture (EQUC)” is an enhanced culture protocol for the detection of viable microbes in urine specimens. Using a large volume of urine and different sets of cultural conditions, EQUC is able to uncover a wide range of bacteria and fungi (yeasts) that were otherwise undetected by the standard urinary culture. In addition to common urinary pathogens, EQUC has been shown to detect emerging and new pathogens, and commensal microbiota. Although the usefulness of EQUC protocol in clinical set up has not yet been fully established, recent studies have demonstrated that EQUC can provide valuable information regarding symptom resolution, treatment responses and diagnosis of major urinary disorders including urinary tract infections, urinary incontinence and other lower urinary tract symptoms. EQUC may also help in evaluating the utility of beneficial microbiota as biotherapeutics. This narrative minireview describes the current research findings regarding the clinical utility of EQUC in characterizing the role of urinary microbiome and uropathogens in health and disease. The literature which are written in English, available on “PubMed” and contain any of the terms- “expanded quantitative urine culture”, “enhanced quantitative urine culture” and “EQUC” in the abstracts were used as the source articles to prepare this minireview.
1 Introduction
It was not long ago when the human bladder was believed to be sterile (Wolfe and Brubaker, 2015; Thomas-White et al., 2016). Using 16s ribosomal RNA (rRNA) sequencing, Wolfe and colleagues for the first time described that the bladder of healthy females contained uncultivable bacteria (Wolfe et al., 2012). However, they were unable to detect whether those uncultivable bacteria were viable. Two years later, using the protocol “expanded quantitative urine culture (EQUC)”, also known as “enhanced quantitative urine culture”, they showed that female urine harbored communities of live bacteria (Hilt et al., 2014). Since then, EQUC, in addition to or independent of standard urine culture (SUC) and next generation sequencing (NGS), have been used to detect urinary microbiota/microbiome (urobiome) from healthy and diseased individuals.
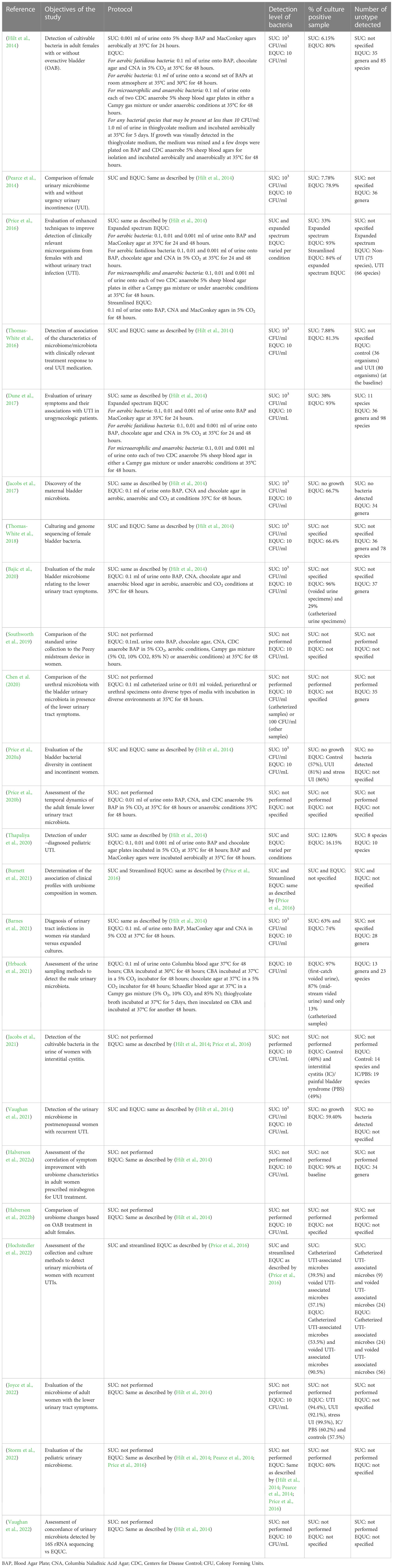
EQUC offers several advantages relative to SUC and NGS. Since the first description of EQUC in 2014, the protocol has been updated in a number of ways to utilize a broad range of growth media, culture conditions and incubation time to detect bacteria and some fungi, particularly yeasts, in urine samples (Table 1). EQUC is able to detect microbes in urine samples that are considered as “no growth” by SUC (Hilt et al., 2014; Brubaker and Wolfe, 2016). One of the strengths of EQUC is its ability to detect slow-growing anaerobic, microaerophilic and fastidious bacteria, in addition to aerobic and facultative anaerobes (Southworth et al., 2019). EQUC is also capable of detecting atypical and subthreshold bacterial species at a detection level of as low as 10 colony-forming unit (CFU)/ml (Xu et al., 2021). Since EQUC detects viable microbes, it is possible to evaluate the antibiotic sensitivity profiles of the bacteria analyzed by EQUC (Thapaliya et al., 2020; Xu et al., 2021). Without whole genome sequencing, NGS alone cannot provide data on antibiotic resistance.
Although EQUC has not been used regularly in a clinical set up due to lack of sufficient data, accumulating evidence have demonstrated the clinical relevance of EQUC in terms of symptoms resolution, treatment response and diagnosis. In the following sections of this review, recent data on the utility of EQUC with respect to several urinary conditions (Table 2) will be discussed.
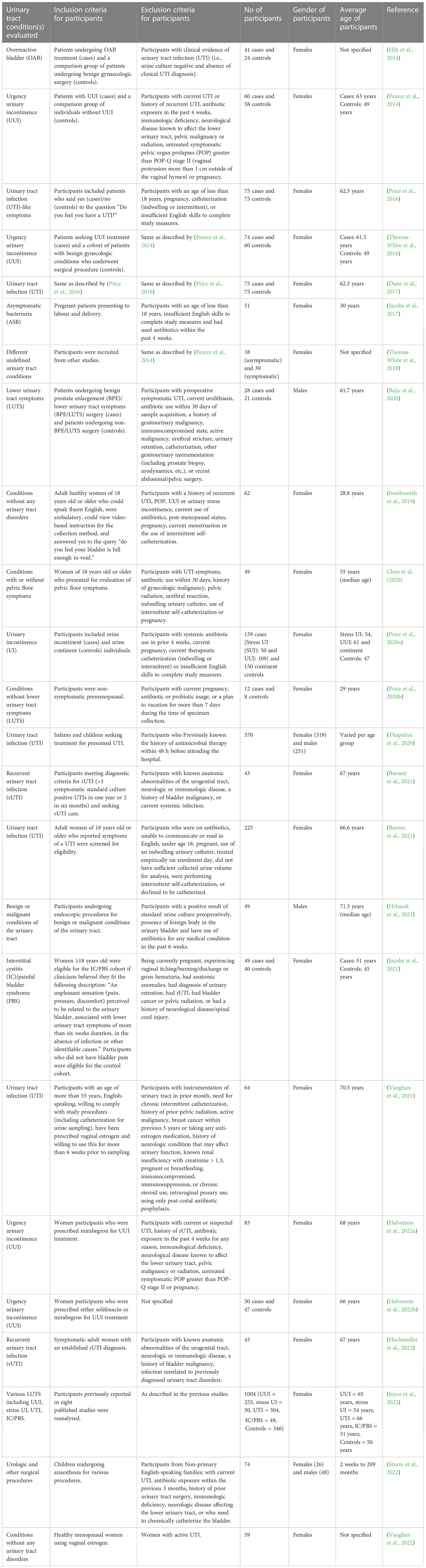
Table 2 Comparisons between participants of different urinary tract conditions reported in the EQUC urinary microbiota studies.
2 Urinary tract infection
A number of studies have used EQUC to detect uropathogens from females suffering from urinary tract infections (UTIs). In 2016, Price et al. evaluated the expanded spectrum EQUC using varying volume of urine as well as different cultural and incubation conditions to identify optimum protocol for detecting uropathogens from UTI and non-UTI cohorts (Price et al., 2016). They proposed a streamlined EQUC protocol that detected 84% of uropathogens compared to 34% uropathogens detected by SUC (Price et al., 2016). EQUC also detected higher number of uropathogens (16.15%) from pediatric patients suffering from UTIs compared to SUC (12.80%) (Thapaliya et al., 2020). EQUC was found to detect multidrug-resistant, extensive drug-resistant, and extended-spectrum β-lactamase producing uropathogens as detected by antibiotic susceptibility testing (Thapaliya et al., 2020). While E. coli was found to be the most common uropathogen detected by both EQUC and SUC, some other uropathogens such as Candida albicans, Provedencia retegerii, and Morganella morganii grew only on EQUC (Thapaliya et al., 2020). The findings of these studies suggest that the EQUC is capable of providing more useful information to clinicians compared to SUC.
Data from a study have shown that 69% of patients with positive EQUC growth responded to antibiotic treatment (Barnes et al., 2021). Between two types of UTIs detected by EQUC, namely Escherichia coli-uropathogen predominant UTIs and non-E. coli-uropathogen predominant UTIs, the latter demonstrated better symptom resolution after antibiotic treatment. These results suggest that EQUC may be better utilized to resolve non-E. coli-uropathogen predominant UTIs. Another study investigated the association of patient-reported symptoms with urobiome composition in female patients experiencing recurrent UTIs (Burnett et al., 2021). They described five distinct clinical profile groups based on EQUC data collected from 49 participants: odor, cloudiness, and current vaginal estrogen use (no culture result association); frequency, low back pain, incomplete emptying, and vaginal estrogen (significantly increased proportion of Lactobacillus-positive cultures); pain/burning, odor, cloudiness, and urgency (high proportions of UTI-associated microbe-positive cultures), frequency, urgency, pain/burning, and current vaginal estrogen use (increased number of no growth cultures) and frequency, urgency, pain/burning, odor, overactive bladder, and sexually active (significantly increased proportion of Klebsiella-positive cultures). However, no association of a single urinary symptom was found with specific uropathogens (Burnett et al., 2021). The findings of this study somewhat contradict with the results of another study, which has reported that the presence of pain, but not frequency and urgency of urination is more effective indicators of UTIs in urogynecologic female patients (Dune et al., 2017). The most common uropathogens found in this patient group were Escherichia coli, Enterococuus faeculis, Aerococcus urinae as detected by EQUC. More such studies with greater sample size are warranted to provide useful insights for diagnostic and therapeutic interventions of UTIs.
When detected by SUC, microbial composition has been observed to vary in the same individual due to different urine collection method. The urine collection method has also been observed as a crucial factor while detecting microbes by EQUC method. In a study of 43 women with recurrent UTIs, culture of voided urine as assessed by EQUC yielded high false positive results and the catheterized urine specimens detected microbes with the highest sensitivity (Hochstedler et al., 2022). Additionally, EQUC detected more unique UTI-associated microbes such as Actinotignum schaalii, Candida species, and Streptococcus anginosus and consistently detected more uropathogens from both catheterized and voided urine specimens compared to SUC. These results therefore indicate that EQUC is capable of providing more clinically relevant data.
A recent study with a large number of samples has detected the urinary microbiome present in the catheterized urine specimens collected from female patients with various lower urinary tract symptoms (LUTS) including UTI, urgency urinary incontinence (UUI), interstitial cystitis/painful bladder syndrome [IC/PBS] and stress urinary incontinence (SUI) by EQUC and observed that Escherichia was the most prevalent bacterium in UTI patients. (Joyce et al., 2022). Additionally, other bacterial genera including Lactobacillus, Streptococcus, Staphylococcus, Corynebacterium, Actinomyces, and Aerococcus were moderately prevalent in UTI cohort. In another study, different species of Lactobacillus were detected in the catheterized urine samples of postmenopausal women with or without UTI as detected by EQUC (Vaughan et al., 2021). It is intriguing that although Lactobacillus is generally considered to be a beneficial bacterium, these studies reported to detect Lactobacillus species from UTI patients.
3 Overreactive bladder and urinary incontinence
The pathophysiology of overreactive bladder (OAB) and urinary incontinence (UI) is not very well-understood. Recent studies using EQUC and NGS have suggested that urobiome may play a role in the pathophysiology of UI. A difference in the urinary microbiota have been observed between OAB/UUI-affected and -unaffected women as detected by EQUC (Hilt et al., 2014; Thomas-White et al., 2016; Thomas-White et al., 2017; Price et al., 2020a). Increased number of microorganisms were detected from the urine samples of the urgency incontinent women compared to the control group (81-85% vs 57-65%) (Thomas-White et al., 2016; Price et al., 2020a). Additionally, relative to control, more types of cultivable bacteria were identified from UUI patients.(Thomas-White et al., 2016) The most frequently isolated bacterial species were found to be Lactobacillus spp. and S. anginonus in the control group, Lactobacillus iners, S. anginonus and S. epidermidis in SUI patients and S. anginosus, L. gasseri, Aerococcus urinae, and Gardnerella vaginalis in UUI patients (Price et al., 2020a). Another study reported that nine genera including Actinomyces, Aerococcus, Arthrobacter, Corynebacterium, Gardnerella, Staphylococcus, Streptococcus, Actinobaculum, Aerococcus, Arthrobacter and Oligella) were more frequently isolated from the UUI cohort than from the control group; the last four genera being found only in UUI patients (Pearce et al., 2014). Similar trend was observed in a study conducted by Hilt and colleagues in which while Lactobacillus, Streptococcus, Corynebacterium, Staphylococcus, Actinomyces, and Bifidobacterium spp. were detected from both individuals with and without OAB, Aerococcus, Actinobaculum and Athrobacter were isolated only from OAB patients (Hilt et al., 2014). The previously mentioned study conducted by Joyce and colleagues isolated S. anginosus, L. gasseri, A. urinae, S. epidermidis, L. iners, Corynebacterium coyleae, Actinomyces neuii, L. jensenii, and Corynebacterium amycolatum in a more significant number in the UUI patients compared to the healthy control (Joyce et al., 2022). Interestingly, members of the UUI cohort were found to have richer and more abundant urobiome compared to UTI and other cohorts in this study suggesting possible urobiome dysbiosis resulting from overgrowth of bacteria in the UUI patients.
Evaluation of the urobiome profile of patients with UUI before and after treatment have demonstrated longituidinal changes in microbial composition that is associated with symptoms resolution (Thomas-White et al., 2017; Halverson et al., 2022a; Halverson et al., 2022b). In a UUI cohort, the clinical responders to solifenacin, an anticholinergic medication, had a less diverse bacterial community at the baseline, whereas non-responders had a more diverse urobiome (Thomas-White et al., 2016). Additionally, following solifenacin administration, certain microbiota profiles were found to predict the clinically significant response to treatment (Thomas-White et al., 2016). Interestingly, in another UUI cohort treated with mirabegron, a beta-3 agonist approved for treatment of UUI, no difference in the urobiome diversity was observed between the responders and the non-responders at the baseline. However, after 12 weeks of treatment, the urobiome of the responder was found to be significantly richer (Halverson et al., 2022a). In order to evaluate why the microbial characteristics of two cohorts treated by two UUI medication differ, a follow up study was performed to analyze the data obtained from those two studies by using a uniform approach (Halverson et al., 2022b). The re-analysis revealed that alterations in the pre-treatment urobiome occurred in the solifenacin-treated participants only, but not in the mirabegron-treated participants. Additionally, an increased diversity of post-treatment urobiome were found to be associated with treatment response irrespective of medication (Halverson et al., 2022b). Taken together, findings of these studies have indicated that UUI medications affect patients’ microbial profiles differently and further studies are warranted to determine the mechanisms of action of these medications in regards to urobiome.
4 Other lower urinary tract symptoms
Apart from UTI and OAB/UI, many patients experience other urinary symptoms involving the bladder, urethra, prostate (in men) and other parts of the lower urinary tract. These symptoms include, but are not limited to, urine hesitancy, intermittent stream, straining, prolonged micturition, feeling of incomplete bladder emptying, nocturia etc (Lepor, 2005). A study examined the association of lower urinary tract microbiota (LUTM) with lower urinary tract symptoms (LUTS) in male patients with and without benign prostate enlargement (BPE) using EQUC and 16S rRNA sequencing and found a positive relationship between LUTM and LUTS (Bajic et al., 2020). LUTM was detected in catheterized urine of 57.1% of men with severe LUTS, 30.0% of men with moderate LUTS, and 22.2% of men with mild LUTS (Bajic et al., 2020). It was revealed that EQUC detected bacteria in 96% of voided urine specimens and 29% of catheterized urine specimens. The bacterial genus Streptococcus (mostly S. anginosus) and the fungal genus Candida were more abundant in catheterized urine of patients with severe LUTS as detected by EQUC (Bajic et al., 2020). These findings might have clinical implications as S. anginosus are found to be associated with UUI symptoms in women (Pearce et al., 2014; Joyce et al., 2022) and Candida species are associated with patients with symptom flare of urological chronic pelvic pain syndrome (Nickel et al., 2016). All patients with more than 50% relative abundance of Escherichia and Klebsiella species, two of the most common uropathogens, in their voided urine had moderate-to-severe LUTS (Bajic et al., 2020). When the microbial composition in the samples collected from voided urine, catheterized urine, periurethral swab and transurethral swab of women suffering from pelvic floor symptoms was evaluated, different microbiota was observed in bladder relative to urethral, periurethral and voided urine microbiota (Chen et al., 2020). It has been suggested that this difference in microbiota may be influenced by clinical features such as menopausal status and sexual activity (Chen et al., 2020). Using EQUC protocol, other studies have also demonstrated that the lower urinary tract microbiota has been related to menstruation and vaginal intercourse (Price et al., 2020b).
In a cohort of women with interstitial cystitis/painful bladder syndrome (IC/PBS), Lactobacillus and Streptococcus were found to be the most common bacteria, detected in 49.0% EQUC positive samples (Jacobs et al., 2021). While Lactobacillus was not demonstrated to affect IC/PBS symptom response, Streptococcus was found to be associated with less severe symptoms. Additionally, samples collected from most participants with active IC/PBS symptoms did not contain bacteria (Jacobs et al., 2021). Altogether, findings of this study have suggested that bacteria may not impact the symptoms of IC/PBS in the observed cohort.
5 Discussion
Ninety percent of urine samples that yielded growth by EQUC protocol have been reported as “no growth” by the SUC protocol (Hilt et al., 2014; Pearce et al., 2014). EQUC is particularly useful in assessing the urine specimens from patients with recurrent UTIs and other urinary symptoms in which the bladder microbes might remain undetected by SUC. Longituidinal studies of microbiota evaluating the symptoms and treatment response of UUI revealed that EQUC is capable of catching microbes that may be used as a predictive biomarker. Therefore, EQUC could potentially be used to detect microbial biomarkers to monitor disease progression, and to develop diagnostic, therapeutic, and prognostic tools (Antunes-Lopes et al., 2020).
With the advent of EQUC, a number of studies have reported that while beneficial commensal bacteria, including Lactobacillus, are present in the bladder of healthy females, these bacteria have also been detected in females experiencing urinary symptoms (Hilt et al., 2014; Pearce et al., 2014; Jacobs et al., 2017; Thomas-White et al., 2018; Joyce et al., 2022). It is suggested that the positive and negative contributions of Lactobacillus to the bladder health may be determined at the species level and under certain clinical conditions, they can become opportunistic uropathogens (Joyce et al., 2022). It is also possible that the presence of these beneficial bacteria in urinary patients is transient and is linked to symptom responses. A study by Thomas-White and colleagues demonstrated that health-associated commensal bacteria including L. iners and L. crispatus, found in both the bladder and vagina of the same individual are functionally highly similar and the authors suggested that these bacteria could provide protection against urinary infections (Thomas-White et al., 2018). Contrarily, a harmful effect of L. iners has been suggested in a study by Annelis and colleagues. They proposed that L. iners may outcompete Bacillus Calmette-Guerin (BCG), that has been used to manipulate the bladder microenvironment for the treatment of non-muscle invasive bladder cancer, by competing for binding to urothelial fibronectin (Annels et al., 2020). It is intriguing that the same species of Lactobacillus may play opposite role under different physiological conditions. Further studies are necessary to investigate the beneficial role of these commensals to evaluate their possible use as biotherapeutics, which will in turn assist to reduce antibiotic-resistant urinary infections. Indeed, the use of this kind of microbiota-mediated treatment must be evaluated considering the health status of the individual patient.
Cumulative studies have suggested that dysbiosis of urobiome influences the microenvironments of the urinary tract and thereby contributing to the onset and progression of the bladder cancer and prostate cancer (Alfano et al., 2016; Annels et al., 2020; Chipollini et al., 2020; Cai et al., 2021; James et al., 2023). Urobiome dysbiosis has also been found to affect the treatment response to the bladder cancer (Annels et al., 2020). Most of these studies have used DNA sequencing methods to detect and characterize the urobiome in cancer patients. A number of anaerobic and other bacteria detected by these methods are not culturable by SUC. EQUC may be utilized to examine whether viable bacteria are required to stimulate the urobiome-mediated tumour-inducing microenvironments. Modulation of the urobiome via probiotics, fecal microbiota transplantation and other microbiome-based therapeutics might be useful in preventing and predicting the urinary tract cancers as well as improving treatment response to these cancers.
It is likely that detection of diverse microbial community by EQUC protocol would capture contamination, which is also apparent from the studies that used both voided urine and catharized urine, and found that the latter contained lower load of microbes compared to the former when analyzed by EQUC due to the presence of urethral microbiota in the voided urine (Bajic et al., 2020; Hrbacek et al., 2021; Hochstedler et al., 2022). Vulvovaginal contamination was also observed in clean-catch voided urine (Wolfe et al., 2012). Taken together, these results suggest that catheterized urine specimens detected via EQUC would provide clinically relevant information. It is therefore suggested that when analyzed by EQUC, urine samples must be collected by catheter to reduce the chance of contamination (Price et al., 2016).
One limitation of EQUC is its inability to detect fungal species except yeasts. Our understanding of the urinary microbiome is incomplete unless the resident fungal species is considered (Ackerman and Underhill, 2017). The role of the resident fungi and their relationship with the other urinary bacteria warrants future research.
The ability of EQUC to detect a vast array of viable and clinically significant microbes makes it an excellent diagnostic, therapeutic and prognostic tool to study urinary microbiota in health and disease. Besides epidemiological studies, more basic research involving experimental and interventional studies with a larger sample size is warranted to acquire more information regarding the clinical utility of EQUC.
Author contributions
ND: conceptualization, writing (original draft preparation) and editing. AA: conceptualization, writing and editing. NT: writing. NK: writing. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ackerman, A. L., Underhill, D. M. (2017). The mycobiome of the human urinary tract: potential roles for fungi in urology. Ann. Transl. Med. 5 (2), 31. doi: 10.21037/atm.2016.12.69
Alfano, M., Canducci, F., Nebuloni, M., Clementi, M., Montorsi, F., Salonia, A. (2016). The interplay of extracellular matrix and microbiome in urothelial bladder cancer. Nat. Rev. Urol. 13 (2), 77–90. doi: 10.1038/nrurol.2015.292
Annels, N. E., Simpson, G. R., Pandha, H. (2020). Modifying the non-muscle invasive bladder cancer immune microenvironment for optimal therapeutic response. Front. Oncol. 10. doi: 10.3389/fonc.2020.00175
Antunes-Lopes, T., Vale, L., Coelho, A. M., Silva, C., Rieken, M., Geavlete, B., et al. (2020). The role of urinary microbiota in lower urinary tract dysfunction: A systematic review. Eur. Urol. Focus 6 (2), 361–369. doi: 10.1016/j.euf.2018.09.011
Bajic, P., Van Kuiken, M. E., Burge, B. K., Kirshenbaum, E. J., Joyce, C. J., Wolfe, A. J., et al. (2020). Male bladder microbiome relates to lower urinary tract symptoms. Eur. Urol. Focus 6 (2), 376–382. doi: 10.1016/j.euf.2018.08.001
Barnes, H. C., Wolff, B., Abdul-Rahim, O., Harrington, A., Hilt, E. E., Price, T. K., et al. (2021). A randomized clinical trial of standard versus expanded cultures to diagnose urinary tract infections in women. J. Urol. 206 (5), 1212–1221. doi: 10.1097/JU.0000000000001949
Brubaker, L., Wolfe, A. (2016). The urinary microbiota: a paradigm shift for bladder disorders? Curr. Opin. Obstet. Gynecol. 28 (5), 407–412. doi: 10.1097/GCO.0000000000000298
Burnett, L. A., Hochstedler, B. R., Weldon, K., Wolfe, A. J., Brubaker, L. (2021). Recurrent urinary tract infection: Association of clinical profiles with urobiome composition in women. Neurourol. Urodyn. 40 (6), 1479–1489. doi: 10.1002/nau.24707
Cai, Y., Ji, W., Sun, C., Xu, R., Chen, X., Deng, Y., et al. (2021). Interferon-induced transmembrane protein 3 shapes an inflamed tumor microenvironment and identifies immuno-hot tumors. Front. Immunol. 12. doi: 10.3389/fimmu.2021.704965
Chen, Y. B., Hochstedler, B., Pham, T. T., Acevedo-Alvarez, M., Mueller, E. R., Wolfe, A. J. (2020). The urethral microbiota: A missing link in the female urinary microbiota. J. Urol. 204 (2), 303–309. doi: 10.1097/JU.0000000000000910
Chipollini, J., Wright, J. R., Nwanosike, H., Kepler, C. Y., Batai, K., Lee, B. R., et al. (2020). Characterization of urinary microbiome in patients with bladder cancer: Results from a single-institution, feasibility study. Urol. Oncol. 38 (7), 615–621. doi: 10.1016/j.urolonc.2020.04.014
Dune, T. J., Price, T. K., Hilt, E. E., Thomas-White, K. J., Kliethermes, S., Brincat, C., et al. (2017). Urinary symptoms and their associations with urinary tract infections in urogynecologic patients. Obstet. Gynecol. 130 (4), 718–725. doi: 10.1097/AOG.0000000000002239
Halverson, T., Mueller, E. R., Brubaker, L., Wolfe, A. J. (2022a). Symptom improvement with mirabegron treatment is associated with urobiome changes in adult women. Int. Urogynecol. J. 33 (5), 1319–1328. doi: 10.1007/s00192-022-05190-w
Halverson, T., Mueller, E. R., Brubaker, L., Wolfe, A. J. (2022b). Urobiome changes differ based on OAB treatment in adult females. Int. Urogynecol. J. 34 (6), 1271–1277. doi: 10.1007/s00192-022-05416-x
Hilt, E. E., McKinley, K., Pearce, M. M., Rosenfeld, A. B., Zilliox, M. J., Mueller, E. R., et al. (2014). Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J. Clin. Microbiol. 52 (3), 871–876. doi: 10.1128/JCM.02876-13
Hochstedler, B. R., Burnett, L., Price, T. K., Jung, C., Wolfe, A. J., Brubaker, L. (2022). Urinary microbiota of women with recurrent urinary tract infection: collection and culture methods. Int. Urogynecol. J. 33 (3), 563–570. doi: 10.1007/s00192-021-04780-4
Hrbacek, J., Morais, D., Cermak, P., Hanacek, V., Zachoval, R. (2021). Alpha-diversity and microbial community structure of the male urinary microbiota depend on urine sampling method. Sci. Rep. 11 (1), 23758. doi: 10.1038/s41598-021-03292-x
Jacobs, K. M., Price, T. K., Thomas-White, K., Halverson, T., Davies, A., Myers, D. L., et al. (2021). Cultivable bacteria in urine of women with interstitial cystitis: (Not) what we expected. Female Pelvic Med. Reconstr. Surg. 27 (5), 322–327. doi: 10.1097/SPV.0000000000000854
Jacobs, K. M., Thomas-White, K. J., Hilt, E. E., Wolfe, A. J., Waters, T. P. (2017). Microorganisms identified in the maternal bladder: discovery of the maternal bladder microbiota. AJP Rep. 7 (3), e188–e196. doi: 10.1055/s-0037-1606860
James, C., Gomez, K., Desai, S., Patel, H. D., Rac, G., Doshi, C. P., et al. (2023). Impact of intravesical Bacillus Calmette-Guerin and chemotherapy on the bladder microbiome in patients with non-muscle invasive bladder cancer. Front. Cell Infect. Microbiol. 13. doi: 10.3389/fcimb.2023.1125809
Joyce, C., Halverson, T., Gonzalez, C., Brubaker, L., Wolfe, A. J. (2022). The urobiomes of adult women with various lower urinary tract symptoms status differ: A re-analysis. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.860408
Lepor, H. (2005). Pathophysiology of lower urinary tract symptoms in the aging male population. Rev. Urol. 7 (Suppl 7), S3–S11.
Nickel, J. C., Stephens, A., Landis, J. R., Mullins, C., van Bokhoven, A., Lucia, M. S., et al. (2016). Assessment of the lower urinary tract microbiota during symptom flare in women with urologic chronic pelvic pain syndrome: A MAPP network study. J. Urol. 195 (2), 356–362. doi: 10.1016/j.juro.2015.09.075
Pearce, M. M., Hilt, E. E., Rosenfeld, A. B., Zilliox, M. J., Thomas-White, K., Fok, C., et al. (2014). The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. mBio 5 (4), e01283–e01214. doi: 10.1128/mBio.01283-14
Price, T. K., Dune, T., Hilt, E. E., Thomas-White, K. J., Kliethermes, S., Brincat, C., et al. (2016). The clinical urine culture: enhanced techniques improve detection of clinically relevant microorganisms. J. Clin. Microbiol. 54 (5), 1216–1222. doi: 10.1128/JCM.00044-16
Price, T. K., Lin, H., Gao, X., Thomas-White, K. J., Hilt, E. E., Mueller, E. R., et al. (2020a). Bladder bacterial diversity differs in continent and incontinent women: a cross-sectional study. Am. J. Obstet Gynecol 223 (5), 729.E1–729.E10. doi: 10.1016/j.ajog.2020.04.033
Price, T. K., Wolff, B., Halverson, T., Limeira, R., Brubaker, L., Dong, Q., et al. (2020b). Temporal dynamics of the adult female lower urinary tract microbiota. mBio 11 (2), e00475–20. doi: 10.1128/mBio.00475-20
Southworth, E., Hochstedler, B., Price, T. K., Joyce, C., Wolfe, A. J., Mueller, E. R. (2019). A cross-sectional pilot cohort study comparing standard urine collection to the peezy midstream device for research studies involving women. Female Pelvic Med. Reconstr. Surg. 25 (2), e28–e33. doi: 10.1097/SPV.0000000000000693
Storm, D. W., Copp, H. L., Halverson, T. M., Du, J., Juhr, D., Wolfe, A. J. (2022). A Child's urine is not sterile: A pilot study evaluating the Pediatric Urinary Microbiome. J. Pediatr. Urol. 18 (3), 383–392. doi: 10.1016/j.jpurol.2022.02.025
Thapaliya, J., Khadka, P., Thapa, S., Gongal, C. (2020). Enhanced quantitative urine culture technique, a slight modification, in detecting under-diagnosed pediatric urinary tract infection. BMC Res. Notes 13 (1), 5. doi: 10.1186/s13104-019-4875-y
Thomas-White, K., Brady, M., Wolfe, A. J., Mueller, E. R. (2016). The bladder is not sterile: History and current discoveries on the urinary microbiome. Curr. Bladder Dysfunct. Rep. 11 (1), 18–24. doi: 10.1007/s11884-016-0345-8
Thomas-White, K., Forster, S. C., Kumar, N., Van Kuiken, M., Putonti, C., Stares, M. D., et al. (2018). Culturing of female bladder bacteria reveals an interconnected urogenital microbiota. Nat. Commun. 9 (1), 1557. doi: 10.1038/s41467-018-03968-5
Thomas-White, K. J., Hilt, E. E., Fok, C., Pearce, M. M., Mueller, E. R., Kliethermes, S., et al. (2016). Incontinence medication response relates to the female urinary microbiota. Int. Urogynecol. J. 27 (5), 723–733. doi: 10.1007/s00192-015-2847-x
Thomas-White, K. J., Kliethermes, S., Rickey, L., Lukacz, E. S., Richter, H. E., Moalli, P., et al. (2017). Evaluation of the urinary microbiota of women with uncomplicated stress urinary incontinence. Am. J. Obstet. Gynecol. 216 (1), 55 e1–55 e16. doi: 10.1016/j.ajog.2016.07.049
Vaughan, M. H., Mao, J., Karstens, L. A., Ma, L., Amundsen, C. L., Schmader, K. E., et al. (2021). The urinary microbiome in postmenopausal women with recurrent urinary tract infections. J. Urol. 206 (5), 1222–1231. doi: 10.1097/JU.0000000000001940
Vaughan, M. H., Zemtsov, G. E., Dahl, E. M., Karstens, L., Ma, L., Siddiqui, N. Y. (2022). Concordance of urinary microbiota detected by 16S ribosomal RNA amplicon sequencing vs expanded quantitative urine culture. Am. J. Obstet. Gynecol. 227 (5), 773–775. doi: 10.1016/j.ajog.2022.06.031
Wolfe, A. J., Brubaker, L. (2015). "Sterile urine" and the presence of bacteria. Eur. Urol. 68 (2), 173–174. doi: 10.1016/j.eururo.2015.02.041
Wolfe, A. J., Toh, E., Shibata, N., Rong, R., Kenton, K., Fitzgerald, M., et al. (2012). Evidence of uncultivated bacteria in the adult female bladder. J. Clin. Microbiol. 50 (4), 1376–1383. doi: 10.1128/JCM.05852-11
Keywords: expanded quantitative urine culture (EQUC), enhanced urine culture, urinary tract infection (UTI), urgency urinary incontinence (UUI), microbiome, urobiome, urine, bladder
Citation: Deen NS, Ahmed A, Tasnim NT and Khan N (2023) Clinical relevance of expanded quantitative urine culture in health and disease. Front. Cell. Infect. Microbiol. 13:1210161. doi: 10.3389/fcimb.2023.1210161
Received: 21 April 2023; Accepted: 14 July 2023;
Published: 01 August 2023.
Edited by:
Soumya Panigrahi, Case Western Reserve University, United StatesReviewed by:
Fengping Liu, Jiangnan University, ChinaSalequl Islam, Jahangirnagar University, Bangladesh
Copyright © 2023 Deen, Ahmed, Tasnim and Khan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nadia S. Deen, bmFkaWEuc3VsdGFuYUBicmFjdS5hYy5iZA==
 Nadia S. Deen
Nadia S. Deen Akash Ahmed
Akash Ahmed Nazifa Tabassum Tasnim
Nazifa Tabassum Tasnim Nabila Khan
Nabila Khan