- Department of Microbiology and Molecular Genetics, University of Pittsburgh School of Medicine, Pittsburgh, PA, United States
Resistance Nodulation Division (RND) efflux systems are ubiquitous transporters in gram-negative bacteria that provide protection against antimicrobial agents and thereby enhance survival in virtually all environments these prokaryotes inhabit. Vibrio cholerae is a dual lifestyle enteric pathogen that spends much of its existence in aquatic environments. An unwitting encounter with a human host can lead to V. cholerae intestinal colonization by strains that encode cholera toxin and toxin co-regulated pilus virulence factors leading to potentially fatal cholera diarrhea and dissemination in the environment. Adaptive response mechanisms to host factors encountered by these pathogens are therefore critical both to engage survival mechanisms such as RND-mediated transporters and to induce timely expression of virulence factors. Sensing of cues encountered in the host may therefore activate more than protective responses such as efflux systems, but also be coordinated to initiate expression of virulence factors. This review summarizes recent advances that contribute towards the understanding of RND efflux physiological functions and how the transport systems interface with the regulation of virulence factor production in V. cholerae.
Vibrio cholerae is an enteric pathogen and the etiological agent of pandemic cholera
Vibrio cholerae is a comma-shaped, gram-negative bacterium that causes the acute diarrheal disease cholera. Cholera is an enteric infection characterized by a rapid onset of profuse watery diarrhea that can quickly lead to dehydration and death if left untreated (Phillips, 1968). Seven cholera pandemics have been documented since 1817 with the seventh and ongoing pandemic beginning in 1961 [reviewed by (Kaper et al., 1995)]. Currently, ~1.3 billion people are at risk for cholera with a global burden of ~2.9 million cholera cases and 95,000 deaths annually (Ali et al., 2015).
V. cholerae inhabits aquatic ecosystems including fresh (Bina et al., 2020; Daboul et al., 2020) and brackish water (Chakraborty et al., 1997) where these bacteria associate with aquatic invertebrates and hydrolyze chitin (Huq et al., 1983; Meibom et al., 2004). Humans naturally acquire cholera from consumption of contaminated food or water. Following ingestion, V. cholerae traverses the stomach and colonizes the crypts of the small intestine where cholera toxin (CT) production and activity leads to the majority of diarrhea induced in the host (Herrington et al., 1988). V. cholerae adaptive responses to cues encountered in the human gastrointestinal tract elicit transcriptome reprogramming by activating environmental sensing systems including two-component regulatory systems (TCS), the key virulence regulator ToxR, and other regulatory proteins [reviewed by (Childers and Klose, 2007)]. Numerous TCS have been documented to contribute towards V. cholerae survival in the host. For example, the Cad system mediates acid tolerance (Merrell and Camilli, 1999; Merrell and Camilli, 2000), CarRS contributes to biofilm production and antimicrobial peptide resistance (Bilecen and Yildiz, 2009; Herrera et al., 2014; Bilecen et al., 2015), the PhoBR system regulates phosphate uptake and metabolism (Pratt et al., 2010), LuxO acts as a quorum-sensing regulator (Zhu et al., 2002), the VarSA system responds to environmental pH and salt (Jang et al., 2011), the VieSA system responds to the second messenger cyclic diguanylate (Tischler and Camilli, 2005; Martinez-Wilson et al., 2008), FlrBC is a flagellar regulatory protein (Correa et al., 2000), the VxrAB system senses cell-wall stress and regulates type VI secretion (Cheng et al., 2015; Teschler et al., 2017; Peschek et al., 2020), and the osmoregulator OmpR-EnvZ system modulates virulence, adaptation to alkaline pH and responses to membrane intercalating agents (Kunkle et al., 2019; Kunkle et al., 2020a; Kunkle et al., 2020b). In many of these systems the specific environmental cues that stimulate their activity are poorly understood.
V. cholerae must negotiate a range of contrasting physiological environments to reach the site of colonization in the small intestine; the ability to coordinate the expression of virulence genes in response to environmental cues is therefore critical to cholera pathogenesis. This process is orchestrated by a hierarchical regulatory pathway called the ToxR virulence regulon which consists of five major regulatory proteins: AphA, AphB, TcpP, ToxR, and ToxT (Figure 1) (Childers and Klose). The regulon is induced by AphA and AphB binding to the promoter and initiating transcription of tcpPH (Kovacikova and Skorupski, 1999; Skorupski and Taylor, 1999). ToxR’s ancestral role is to modulate expression of outer membrane porins OmpU and OmpT (Provenzano and Klose, 2000; Provenzano et al., 2001), but together with TcpP, ToxR induces transcription of toxT (Hase and Mekalanos, 1998). ToxT then directly activates the expression of the genes encoding the two primary V. cholerae virulence factors, CT and the toxin coregulated pilus (TCP) (Figure 1) (DiRita et al., 1991). TCP is a type IV pilus required for colonization, and CT is the enterotoxin that causes the overwhelming majority of secretory diarrhea that is the hallmark of cholera and essential for V. cholerae epidemic spread [7]. Late during infection when V. cholerae reaches high cell densities in the intestinal lumen, a phenotypic shift occurs in the pathogen’s transcriptome that results in the repression of virulence and upregulation of genes involved in dissemination and transmission (Merrell et al., 2002; Bina et al., 2003; Xu et al., 2003; Larocque et al., 2005; Schild et al., 2007). Dissemination genes enhance V. cholerae survival in aquatic ecosystems whereas transmission genes induce a transient hyperinfectious phenotype that promotes epidemic spread (Merrell et al., 2002). Signals received by the bacteria that lead to expression of late infection genes are unknown.
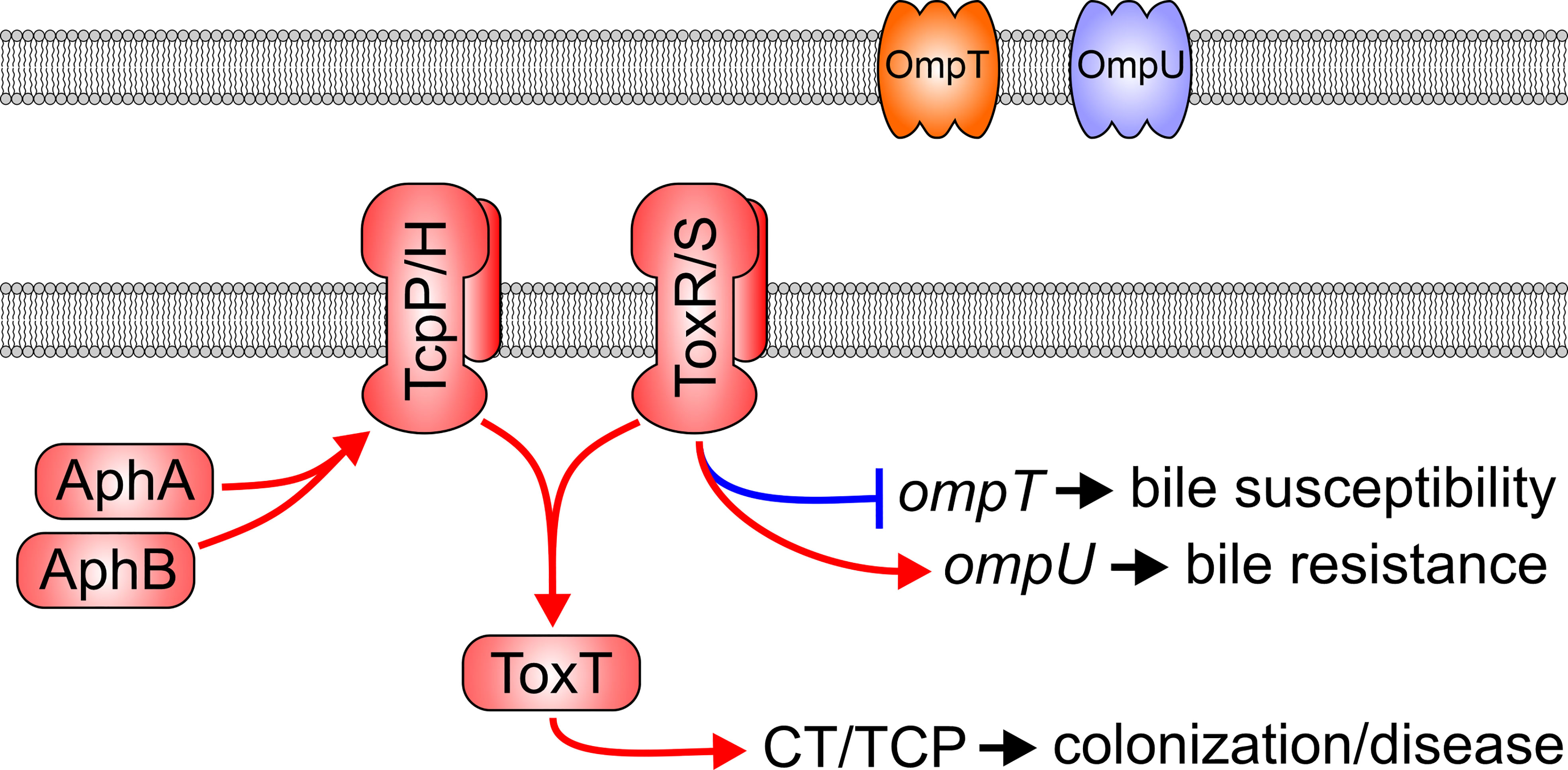
Figure 1 The Vibrio cholerae ToxR virulence regulon. In response to low cell density and low oxygen tension, the AphA and AphB transcription factors bind to the promoter and activate transcription of the tcpPH operon. TcpP/H and ToxR/S then bind to the toxT promoter to induce its transcription. ToxT then activates the transcription of multiple virulence genes including the genes encoding for production of cholera toxin (CT) and the toxin co-regulated pilus (TCP). ToxR/S, independently of TcpP/H, also modulates transcription of outer membrane porins OmpU and OmpT.
Similar to TcpP and TcpH where TcpH stabilizes TcpP (Hase and Mekalanos, 1998), the virulence regulator ToxR, together with its cognate accessory protein ToxS, forms a transmembrane one-component transcription factor that responds to environmental cues and is critical to V. cholerae pathogenesis (Miller et al., 1987; Miller et al., 1989). The ToxR/S heterodimer consists of two proteins; the ToxR moiety spans the membrane with a DNA-binding N-terminal cytoplasmic and a C-terminal periplasmic sensing domain (PPD). The toxR gene is encoded upstream of its accessory toxS which gene product is also anchored in the membrane and is required for full ToxR activity (Miller et al., 1989). Agonists have been shown to promote interactions between ToxR and ToxS, and/or affect the oxidation state of the PPD’s cysteine residues to activate ToxR (Fengler et al., 2012; Midgett et al., 2017). Interestingly, all ToxR agonists described to date [bile salts (Provenzano et al., 2000; Ante et al., 2015b), indole (Howard et al., 2019) and cyclo(Phe-Pro) (Bina and Bina, 2010)] are substrates of the Resistance Nodulation Division (RND) efflux systems suggesting the possibility of a small molecule feedback mechanism between ToxR and RND-mediated efflux.
V. cholerae must overcome several barriers in transit across the gastrointestinal tract including toxic metabolites produced by the resident microbiota, bile salts and products of the innate immune system [reviewed by (Peterson and Gellings, 2018)]. V. cholerae transcriptome modulation enhances resistance against these harmful products by restricting outer membrane permeability and by increasing efflux system activity (Wibbenmeyer et al., 2002; Bina et al., 2008). Multiple drug efflux systems belonging to the RND superfamily are essential for intrinsic antimicrobial resistance against a wide range of compounds in most gram-negative bacteria [reviewed by (Li et al., 2015; Colclough et al., 2020)]. While RND systems contribute to antimicrobial resistance, numerous studies indicate that these transporters fulfill critical physiological functions independent of their role in antibiotic resistance [reviewed by (Piddock, 2006; Alvarez-Ortega et al., 2013)]. This review provides an update on the contributions of the RND efflux systems to V. cholerae host-pathogen interactions.
RND efflux systems are ubiquitous transporters among gammaproteobacteria
RND efflux pumps were first discovered in the early 1990’s because of their contribution to antibiotic resistance (Poole et al., 1993). Subsequent studies revealed that RND efflux transporters were ubiquitous in gram-negative bacteria and that most bacteria encoded multiple and distinct RND transporters in their chromosome [reviewed by (Li et al., 2015)]. More recently, RND system gene clusters encoded on plasmids were identified in multiple antibiotic resistant pathogens enhancing the potential for their horizontal spread (Lv et al., 2020). RND transporters efflux a wide variety of structurally and chemically diverse compounds including various classes of antibiotics, antimicrobial peptides, detergents and dyes (Li et al., 2015). This sets RND systems apart from other transporters and provides intrinsic resistance to diverse antimicrobials in their respective host bacteria. Broad substrate specificity has been recognized as an important factor in the evolution of multiple drug resistance in gram-negative bacteria including pathogens identified by the Centers for Disease Control as priority health threats due to emergent antibiotic resistance (Solomon and Oliver, 2014).
RND efflux systems comprise remarkable tripartite transporters consisting of an outer membrane pore protein ortholog of Escherichia coli TolC, a periplasmic adapter protein belonging to the membrane fusion protein family, and the integral inner membrane RND superfamily pump protein (Colclough et al., 2020). The transporter’s structure exhibits a 3:6:3 stoichiometry (pump to membrane fusion to pore protein peptide ratio) that forms a quaternary complex with an interior channel that spans the entire width of the cell envelope (Figure 2) (Daury et al., 2016). The RND pump protein features an entry portal domain at the periplasm-cytoplasmic interface thought to facilitate the passage of substrate molecules from the periplasm and the cytoplasmic membrane for efflux into the extracellular milieu. The transporter’s pump antiport activity is provided by the proton motive force that powers the efflux activity (Venter et al., 2015). Interestingly, the V. cholerae VexEF RND transporter requires sodium for activity; however, the underlying mechanism governing this requirement remains to be investigated (Rahman et al., 2007).
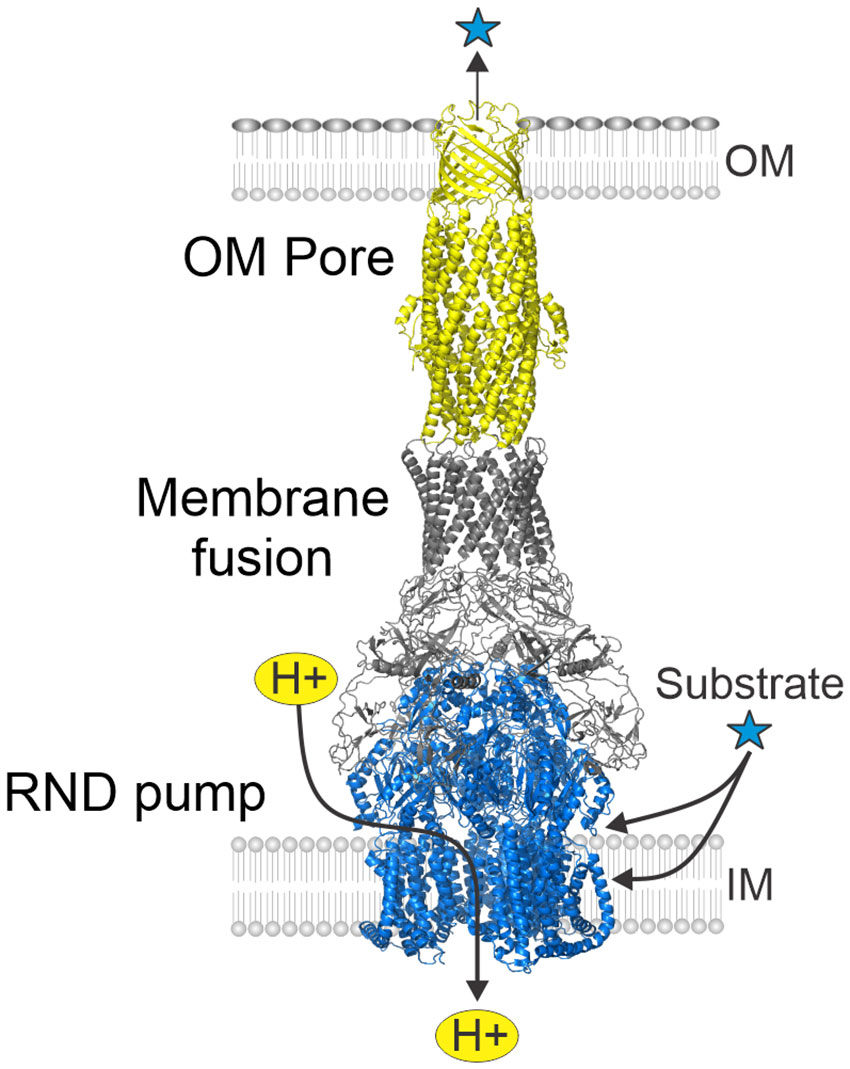
Figure 2 Structure of tripartite RND efflux systems. RND efflux systems consists of an outer membrane pore protein (yellow) orthologous to E. coli TolC, a membrane fusion protein (gray) and an inner membrane RND-family pump protein (blue). The RND systems function as substrate-proton antiporters and collect substrates from the periplasm and cytoplasmic membrane for export into the extremal milieu. Figure derived from PDB ID: 5V5S (Wang et al., 2017).
Crystallized protein domains of RND efflux systems in the presence and absence of substrates or inhibitors provided a wealth of information on assembly and function [see (Alav et al., 2021; Athar et al., 2023)]. Analyses of the crystal structures of multiple RND transporter homologues revealed that RND pump proteins are structurally conserved; they all include multiple substrate entry portals and several substrate binding sites which mediate uptake of substrates from the periplasm and from within the cytoplasmic membrane. Large substrate-binding pockets along the periplasm-cytoplasmic membrane interface appear to facilitate selection of substrates with defined physicochemical properties with specific amino acid residues determining the selectivity and specificity of the binding pocket. Collectively, multiple binding sites and substrate entry portals explain the broad substrate specificity of RND transporters. Substrate molecules are transported across the transporter to the external environment by a peristaltic pump mechanism that results from the rotation of the three RND pump components. TolC’s export channel is composed of an outer membrane-spanning β-barrel domain joined to a periplasm-spanning α-helical domain that directly interact with the membrane-fusion proteins which function as adaptors between the pore and the pump (Koronakis et al., 2000).
RND efflux systems in V. cholerae are structurally related to those in other gram-negative bacteria
V. cholerae, like most gram-negative bacteria harbors multiple RND transporter operons. There are six RND systems encoded in its genome: VexAB, VexCD, VexEF, VexGH, VexIJK, and VexLM (Bina et al., 2006; Bina et al., 2008); five are located on the large chromosome and vexLM on the small chromosome. These operons display an arrangement typical of RND genes in most bacteria with the membrane fusion coding sequence located directly upstream of the RND pump ORF (Figure 3). The vexIJK operon is an exception to this rule because it includes two membrane fusion genes (i.e., vexI and vexJ); the biological significance of the two membrane fusion proteins is unknown. None of the six V. cholerae RND efflux operons include a gene coding an outer membrane pore. The TolC homologue appears to be a shared outer membrane pore among the six RND systems in V. cholerae. The tolC (VC2436) coding sequence is encoded as a single, self-standing gene on the large chromosome (Bina and Mekalanos, 2001). TolC’s RND pore function was inferred from experiments showing that a tolC deletion mutant exhibited the same antimicrobial resistance and virulence factor production phenotypes as an otherwise isogenic RND efflux pump negative strain that retains tolC (Bina and Mekalanos, 2001; Weng et al., 2021). VexR, a transcriptional regulator belonging to the TetR family has been reported to positively regulate the expression of the vexRAB operon (Bina et al., 2008; Taylor et al., 2015), while BreR, another member of the TetR family is a transcriptional regulator that represses vexCD expression (Cerda-Maira et al., 2008; Cerda-Maira et al., 2013).
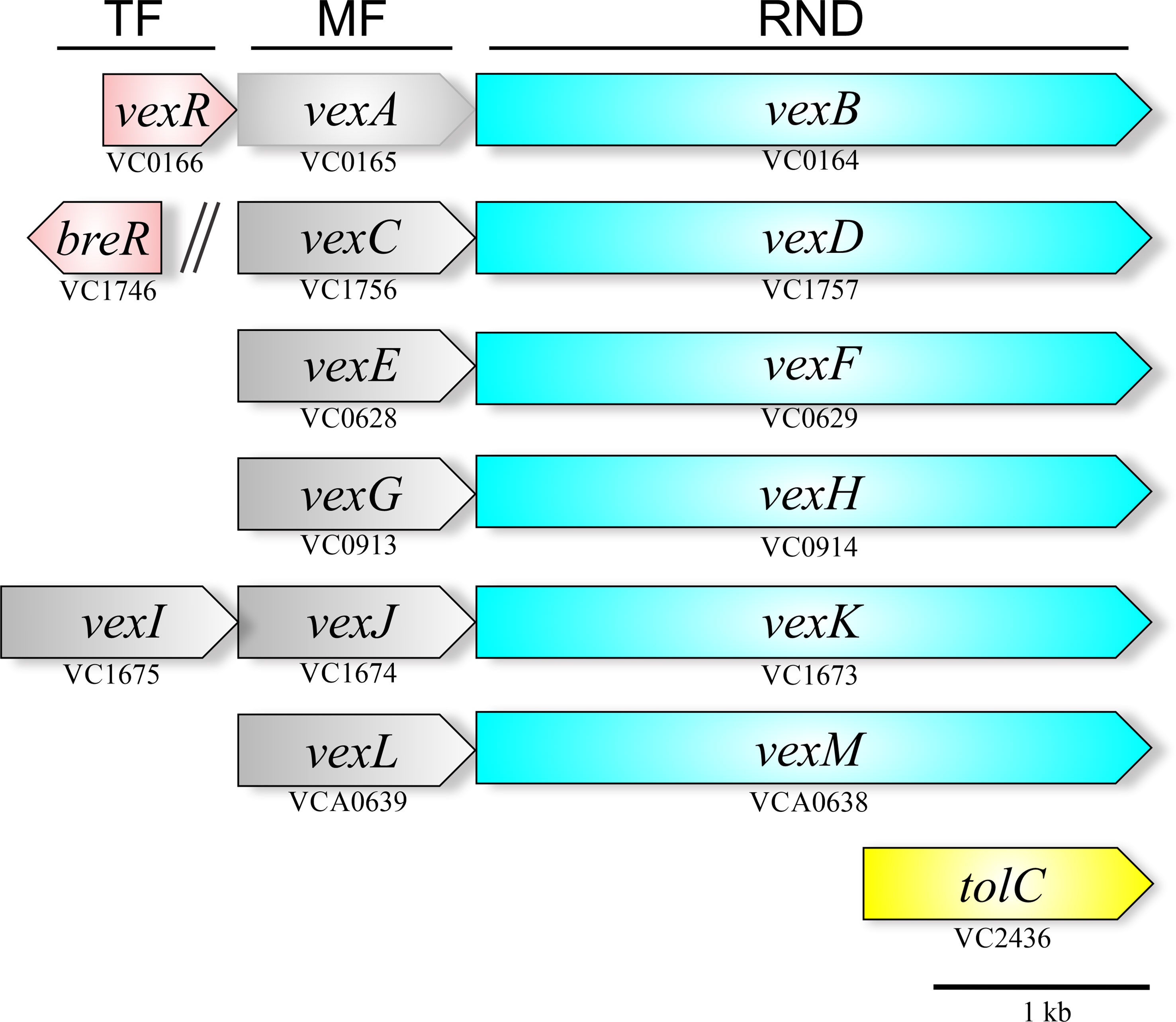
Figure 3 Genetic arrangement of the V. cholerae RND efflux operons. The six operons encode a membrane fusion protein (gray) upstream of the RND-family pump protein (blue). Five of the operons are encoded on the large chromosome with one on the small chromosome (vexLM). Two TetR-family regulators (red) have been linked to the RND systems, VexR, which regulates the vexRAB operon, and BreR, which regulates the vexCD operon. The outer membrane pore protein is provided by tolC (yellow) is encoded separately in the genome.
RND systems mediate V. cholerae antimicrobial resistance
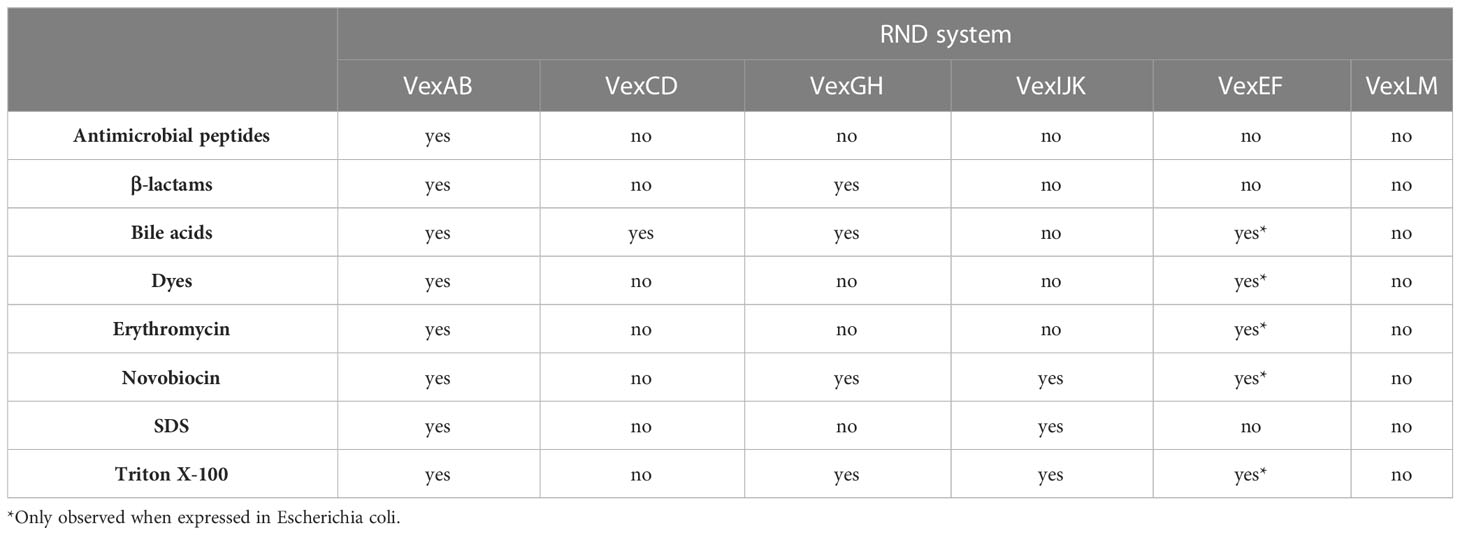
RND efflux systems afford gram-negative bacteria intrinsic antimicrobial resistance while contributing to the evolution of multiple antibiotic resistant pathogens. V. cholerae RND systems have been linked to xenobiotic resistance, but their contributions to the evolution of multiple antibiotic resistant strains have not been investigated. RND transporters provide resistance in vivo against multiple toxic molecules including bile salts, detergents, and antimicrobial peptides to which V. cholerae is exposed to while colonizing the human small intestine (Alav et al., 2021). The contribution of RND efflux system towards V. cholerae antimicrobial resistance was tested experimentally by comparing a mutant lacking all six RND pump protein operons to otherwise isogenic strains encoding single or select RND pumps (Bina et al., 2006; Bina et al., 2008). VexEF was also assessed by expression of the operon in E. coli (Rahman et al., 2007). Deletion of all six RND transporters resulted in hypersensitivity to multiple antimicrobial compounds including antibiotics, antimicrobial peptides, bile salts, dyes and detergents (Table 1) (Bina et al., 2008). VexAB conferred resistance to bile acids, detergents, defensins, dyes and several antibiotics, including erythromycin, polymyxin B, penicillin, ampicillin and novobiocin demonstrating a broad-spectrum multiple drug efflux phenotype indicative of VexAB being the main contributor to V. cholerae intrinsic antimicrobial resistance. Furthermore, the vexB single deletion mutant and the RND-null strain displayed identical minimal inhibitory concentrations (MIC) of erythromycin, polymyxin B, and penicillin, attributing resistance to these antibiotics solely to the VexAB RND system (Bina et al., 2006). The remaining efflux pumps revealed comparatively limited substrate specificities that were redundant with those of VexAB. No compounds tested in these experiments were substrates for the vexLM operon gene products suggesting a higher extent of specificity and/or efflux of compounds not evaluated; for example, those encountered in the aquatic environment. The vexCD operon functioned exclusively in bile acids efflux (Bina et al., 2006; Bina et al., 2008) while VexGH was implicated in resistance to ampicillin, bile acids, novobiocin and the nonionic detergent Triton X-100 (Taylor et al., 2012). VexIJK efflux mediated resistance to novobiocin, SDS and Triton X-100 (Bina et al., 2008; Taylor et al., 2012) and a combination vexBDHK deletion mutant exhibited an antimicrobial susceptibility profile identical to the RND deficient mutant (Bina et al., 2008; Taylor et al., 2012). VexB, VexD, VexK, and VexH are likely the only RND pumps that contribute to in vitro antimicrobial resistance. Taken together, these experiments indicate that V. cholerae expresses multiple, functionally redundant pump systems to efflux bile acids and detergents supporting an important role for RND proteins towards adaptation during colonization of the small intestine. Redundant bile efflux mechanisms provide an adaptation that allows pathogens to overcome an intrinsic colonization barrier in the small intestine where bile acts as a natural host defense against microorganisms.
Although VexB, VexD, VexH, and VexK efflux pumps exhibited redundant specificity for some substrates, they did not contribute equally to antimicrobial resistance. For example, all four pumps enhanced resistance to bile acids, but in the presence of VexB the loss of the other three transporters did not have an impact (Bina et al., 2008). Further, the contribution of VexH and VexK to bile resistance was only observed in a vexBD negative background (Bina et al., 2008; Taylor et al., 2012). These observations suggest that VexB and VexD are the major contributors to bile acid resistance in vitro, while the contribution of VexH and VexK are minor under laboratory experimental conditions. Transcriptomic analyses of cholera patient stool and a human volunteer study indicated that vexAB, vexGH and vexIJK are expressed in the human intestine (Bina et al., 2003; Lombardo et al., 2007). Taken together, these results support the notion that VexAB is the principal RND efflux system in V. cholerae and that VexIJK and VexGH may play a greater role in vivo.
Importantly, these observations may not reflect the contributions of individual RND transporters to resistance under alternate experimental conditions, including in vivo because RND transporter expression is regulated in response to environmental cues. Moreover, individual V. cholerae RND transport systems expressed episomally in E. coli revealed that recombinant vexEF led to decreased susceptibility against multiple antimicrobials (e.g. bile acids, dyes, erythromycin, novobiocin and Triton X-100), indicating that VexEF is capable of acting as a broad spectrum multiple drug efflux system (Rahman et al., 2007). In contrast, deletion of vexF in V. cholerae did not affect antimicrobial resistance for reasons that remain unclear but may related to intrinsic low expression levels (Bina et al., 2008).
RND efflux contributes to V. cholerae host colonization
V. cholerae lacking RND efflux systems exhibits severe attenuation in infant mouse colonization which was hypothesized to result from an abundance of antimicrobial compounds in the gastrointestinal tract (Bina et al., 2008). RND-mediated efflux together with outer membrane selective permeability is hypothesized to synergistically allow bacteria to overcome the cytotoxic effect of bile acids, detergents and antimicrobial peptides commonly found in the intestine (Bina and Mekalanos, 2001; Bina et al., 2008; Weng et al., 2021). Competitive index experiments in which strains are compared pairwise for infant mouse colonization revealed that the vexBDH, vexBDK and RND null mutants could not be recovered from infant mice when administered at a 1:1 ratio with WT. All the other single and combination mutants were colonization competent comparably to WT including the vexDHKFM mutant with an intact vexB. These results confirmed that VexB can complement the loss of the other five RND efflux systems, and that the other RND transporters were redundant with VexB in this animal model (Bina et al., 2008). In contrast, vexBDH, vexBDK and vexBDHK mutants inoculated into mice at a 100:1 mutant to WT ratio exhibited 3.7, 4.4 and 4.1 log reductions in recovery relative to wild type (WT), respectively. Furthermore, the RND null mutant remained unrecoverable at the same inoculation ratio with WT as well (Taylor et al., 2012). This finding further supports the notion that all six RND systems contribute towards colonization.
Separate from the strictly protective physiological function which impact colonization of the small intestine by enhancing antimicrobial resistance, RND efflux evolved to tie into virulence gene expression. CT and TCP transcription in RND-efflux deficient V. cholerae is decreased by ~70% when the bacteria are grown in laboratory conditions that induce virulence factor expression, suggesting a regulatory link between RND-mediated efflux and expression of the ToxR regulon (Bina and Mekalanos, 2001; Bina et al., 2008; Weng et al., 2021). Interestingly, the contribution of individual RND efflux systems towards antimicrobial resistance mirrored their impact on virulence factor production. For example, a vexBDHK mutant expressed lower amounts of CT and TCP but the RND null mutant even less, suggesting that the VexLM and VexEF RND systems, which do not contribute towards antimicrobial resistance, do play a role in virulence factor production (Bina et al., 2008; Taylor et al., 2012). In contrast, levels of CT and TCP production of vexBDH, vexBDK, vexBHK, and vexDHK mutants did not differ significantly from WT, suggesting that the VexAB, VexCD, VexGH, and VexIJK efflux systems are redundant for CT and TCP production (Bina et al., 2008; Taylor et al., 2012). The presence of VexAB alone complemented the loss of the other RND systems with regards to virulence factor production. The observation that vexBDH and vexBDK mutants were attenuated for colonization, but not for CT and TCP production, led to the conclusion that the RND systems played a dual role in V. cholerae pathogenesis with the RND efflux systems being critical both for resistance to antimicrobial compounds present in the gastrointestinal tract and for virulence factor production thereby impacting colonization multifold. VexLM and VexEF RND contribution towards V. cholerae colonization remains subject of speculation as it may result from its impact on virulence factor production, enhancement of antimicrobial resistance, through some other, yet to be determined activity or any combination thereof.
RND efflux activity influences V. cholerae two-component system activity
In vitro virulence inducing conditions (i.e., AKI conditions) consist of an artificial set of in vitro parameters that activate the ToxR regulon and have been shown to mimic specific aspects of the events that induce expression of virulence factors in vivo (Iwanaga and Yamamoto, 1985). The effects of the RND efflux systems on the V. cholerae transcriptome were investigated by whole genome RNA sequencing of the O1 El Tor strain N16961 grown under AKI conditions. Loss of RND-mediated efflux resulted in reprogramming of the V. cholerae transcriptome with >350 genes being identified as differentially expressed between the efflux null mutant and WT (Bina et al., 2018). Functional classification of differentially expressed genes revealed that most had predicted functions in cell metabolism, transport and binding, virulence and regulation. Most of the genes in the ToxR virulence regulon were downregulated, suggesting that RND-mediated efflux is required for virulence factor production and may have contributed to the in vivo colonization defect observed in RND efflux impaired strains. The RND efflux-dependent transcriptome was also enriched for TCS, environmentally responsive regulatory systems that consist of a membrane-linked sensor kinase that relays environmental cues to a response regulator by a phosphorelay mechanism (Mitrophanov and Groisman, 2008). The phosphorylated response regulator typically functions as a transcription factor to transduce extracellular cues into transcriptional responses. Genes associated with TCS represented 57% of the differentially expressed regulatory genes in the RND mutant, whereas TCS represent just 17% of the regulatory genes in the V. cholerae genome (Heidelberg et al., 2000). Given that the RND systems transport substrates from the periplasm and cytoplasmic membrane, this observation suggested a novel feedback mechanism between environmental adaptation and RND-mediated efflux where metabolites normally removed from the cell by the RND systems accumulate intracellularly and initiate feedback mechanisms by environmental sensors (e.g., one- and two-component regulatory systems) leading to the expression of adaptive responses. Further investigation between the link between efflux and adaptation are described below.
RND efflux activity modulates adaptive responses via ToxR
LeuO is a recently described LysR-type regulator in V. cholerae that regulates biofilm production and acid tolerance response (Moorthy and Watnick, 2005; Ante et al., 2015a). ToxR activates leuO transcription by binding to a consensus promoter site; an event that appears to occur constitutively in the absence of RND-mediated efflux, leading to elevated leuO transcription (Bina et al., 2018). Interestingly, ToxR’s periplasmic sensing domain is required for activation of leuO transcription suggestive of an efflux-dependent small molecule feedback mechanism tying ToxR to leuO expression. Consistent with these observations, ToxR induced leuO expression in the presence of multiple RND efflux substrates including bile acids, indole, and diketopiperazines, the former two being RND efflux substrates that are abundant in the human gastrointestinal tract (Bina et al., 2009; Bina and Bina, 2010; Bina et al., 2013; Ante et al., 2015b; Howard et al., 2019; Bina et al., 2021). Furthermore, the RND efflux inhibitor phenylalanine-arginine β-naphthylamide induced leuO expression by a mechanism dependent on the presence of the ToxR periplasmic sensing domain suggestive of a regulatory feedback circuit that inhibits efflux of the transporter’s native substrates to modulate the ToxR regulon. These findings led to the formulation of a model whereby RND systems influence adaptive responses as a direct result of endogenous and exogenous metabolite efflux from the periplasmic space where they are otherwise sensed by ToxR (Figure 4). In the absence of RND efflux activity, through mutation or inhibition by chemicals, metabolites normally expelled accumulate in the periplasmic space and activate ToxR by interacting with its periplasmic sensing domain. ToxR, independent of TcpP then induces expression of leuO, which leads to virulence repression and altered cell physiology (Bina et al., 2018). Metabolite accumulation may also result from high cell density, consistent with the observation that LeuO is expressed in stationary phase of growth. Overexpression of leuO negatively regulates aphA thereby repressing production of CT and TCP. Given LeuO’s natural expression during stationary phase of growth when cell densities are high, it is tempting to speculate that this novel ToxR regulon branch is involved in V. cholerae’s late infection phenotype.
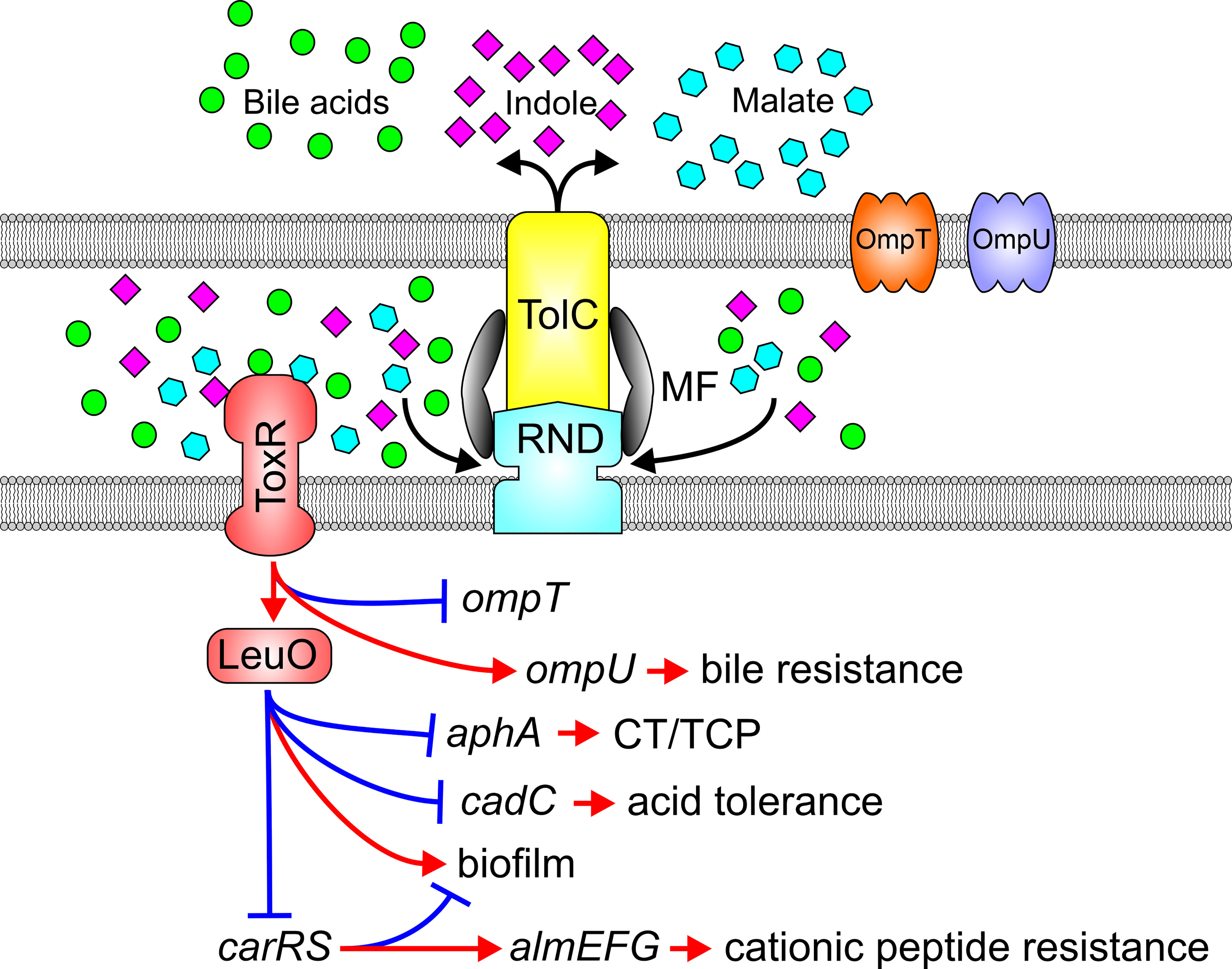
Figure 4 Substrates of the RND efflux systems mediate adaptive responses via ToxR. Impaired RND-mediated efflux results in the periplasmic accumulation of endogenous and exogenous RND substrates such as bile acids (green balls), indole (violet diamonds) or malate (blue hexagons) which engage the ToxR periplasmic domain to activate ToxR and downstream adaptive responses.
The effects of efflux on ToxR and LeuO signaling pathways extend beyond virulence to additional adaptive responses that are likely important to the V. cholerae pathogenic lifecycle including host adaptation or dissemination or transmission. ToxR and LeuO play additional roles in bile resistance independently of RND transporters thereby further protecting V. cholerae from the toxic effect of bile in the intestine. ToxR and LeuO also mediate acid tolerance via LeuO-mediated repression of the tightly regulated cadC-cadBA system critical for maintaining pH homeostasis and cell survival in acidic milieus (Ante et al., 2015a). Furthermore, ToxR regulates antimicrobial peptide resistance via LeuO-dependent repression of the carRS TCS (Bina et al., 2016). CarRS positively regulates the almEFG operon which modifies lipopolysaccharides with glycine and diglycine to mediate high-level cationic antimicrobial resistance in V. cholerae (Bilecen and Yildiz, 2009; Herrera et al., 2014; Bilecen et al., 2015). Lastly, ToxR and LeuO also contribute to biofilm production, which is critical for hyperinfectivity and environmental survival, but the mechanism by which this occurs has not been fully resolved (Moorthy and Watnick, 2005; Bilecen et al., 2015; Kazi et al., 2016).
Reciprocal relationship between RND-mediated efflux and the Cpx response
The Cpx system is widespread among Gammaproteobacteria where it functions to mitigate envelope stress resulting from environmental perturbations such as high salinity, magnesium ion concentration, oxidative stress, low iron, and proteins containing aberrant disulfide bonds (Hews et al., 2019). CpxA is a membrane-associated histidine kinase sensor; when stimulated this sensor phosphorylates the CpxR response regulator which then binds to a conserved consensus sequence in the promoter of target genes to modulate adaptive responses (Taylor et al., 2014). CpxR also regulates expression of cpxP, which is divergently expressed from the cpxAB operon. CpxP is a periplasmic protein thought to interact with the periplasmic domain of CpxA to modulate its activity (Hews et al., 2019). Initial studies revealed that tolC deletion in V. cholerae activated the Cpx system, suggesting a possible link between RND-mediated efflux and the Cpx system (Slamti and Waldor, 2009). Consistent with this finding the Cpx system was upregulated in RND-deficient V. cholerae (Bina et al., 2018). Targeted studies revealed that inactivation of the VexB or VexH RND transporters activated the Cpx system (Kunkle et al., 2017). The link between the RND efflux and the Cpx system extended beyond efflux-dependent activation of the Cpx system, as vexRAB, vexGH and tolC were identified as members of the Cpx regulon and positively regulated by CpxR, suggesting that the VexAB and VexGH RND transporters are components of the V. cholerae Cpx response intended to mitigate extracytoplasmic stress (Taylor et al., 2014; Acosta et al., 2015b; Kunkle et al., 2017).
Constitutive activation of the Cpx system in RND-impaired cells suggests that periplasmic accumulation of metabolites normally expelled from V. cholerae periplasm by the VexB and VexH RND transporters are likely responsible for Cpx activation. A Cpx-inducing metabolite was identified from a transposon library screened for suppressors of the Cpx system in RND efflux negative V. cholerae (Kunkle et al., 2017). Transposon insertions into succinate dehydrogenase coding sequence or genes involved in synthesis of vibriobactin, an iron siderophore, suppressed expression of the Cpx system. This suggested that the RND transporters are likely involved in vibriobactin efflux, a finding confirmed with directed studies showing that VexAB and VexGH mediate siderophore’s secretion (Figure 5) (Kunkle et al., 2017). This led to the formulation of a model where in the absence of VexAB or VexGH, vibriobactin accumulates in the periplasm, titrated iron from iron-containing members of the electron transport chain such as succinate dehydrogenase either leads to formation of aberrant disulfide bonds or misfolded membrane proteins, which in turn stimulate activation of the Cpx system. Consistent with these observations, the Cpx system functions as an adaptation to iron-limiting conditions (Acosta et al., 2015b) V. cholerae may encounter in vivo. Interestingly, ectopic overexpression of CpxR was shown to repress V. cholerae virulence factor production (Acosta et al., 2015a), suggesting another potential link between the Cpx system, RND-mediated efflux and virulence.
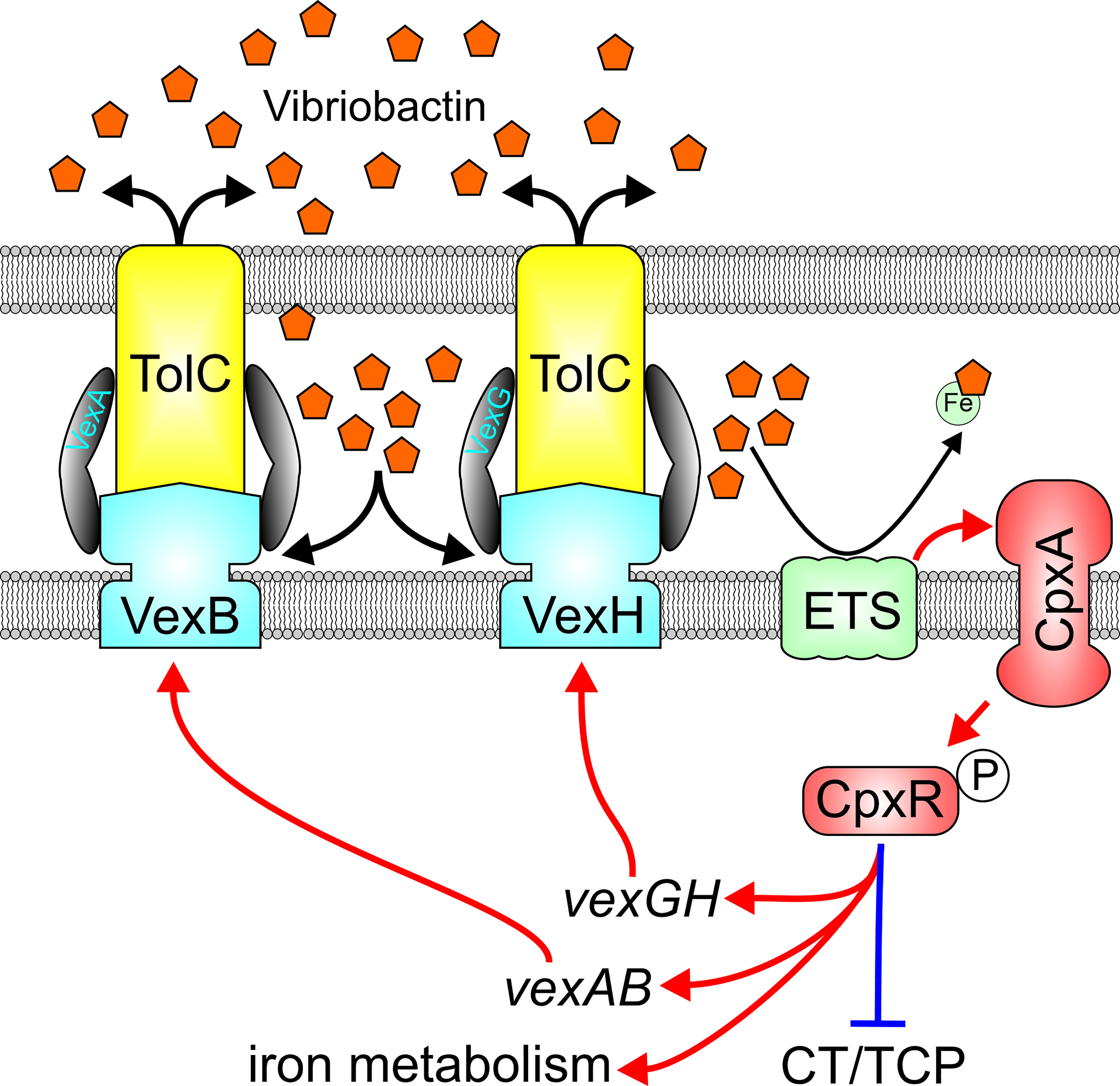
Figure 5 Reciprocal relationship between the VexAB and VexGH RND systems and the Cpx system. VexAB and VexGH contribute to efflux of the siderophore vibriobactin, and impaired efflux results in vibriobactin accumulation in the periplasm which titrates iron from iron-containing components of the electron transport system (ETS) resulting in protein misfolding and activation of the cpx system. Cpx system activation results in induction of vexAB, vexGH and iron homeostasis genes while repressing virulence gene expression.
The EnvZ-OmpR regulon is integrated into the ToxR virulence regulon through RND efflux activity
The EnvZ-OmpR TCS is activated in response to high osmolarity or low pH among Enterobacteriaceae where EnvZ is a membrane associated histidine kinase sensor and OmpR is the DNA binding response regulator (Sarma and Reeves, 1977; Hall and Silhavy, 1981; Slauch et al., 1988). OmpR has also been linked to virulence (Bernardini et al., 1990; Lee et al., 2000; Schwan, 2009; Reboul et al., 2014; Tipton and Rather, 2017; Lin et al., 2018; Silva et al., 2018) and acid tolerance (Heyde and Portalier, 1987; Bang et al., 2000; Hu et al., 2009; Gao et al., 2011; Stincone et al., 2011; Quinn et al., 2012) in gram-negative bacteria. The V. cholerae envZ-ompR operon was found to be upregulated in the absence of RND mediated efflux, suggesting that it may contribute to the virulence repression phenotype observed in this background (Bina et al., 2008; Bina et al., 2018). Deletion of ompR in an RND-negative V. cholerae background raised virulence factor production (Kunkle et al., 2019). Conversely, ompR overexpression in WT cells repressed CT and TCP biosynthesis confirming that OmpR acts as a virulence repressor (Kunkle et al., 2020a). Transcriptomic studies revealed that OmpR controlled expression of a large regulon that included outer membrane proteins, virulence factors and genes involved in acid tolerance (Kunkle et al., 2020b). Interestingly, the V. cholerae EnvZ-OmpR system did not respond to canonical inductive stimuli (i.e. high salt or low pH) typical of Enterobacteriaceae, but instead was induced in response to membrane intercalating agents including bile salts, detergent-like molecules, and alkaline pH (Slamti and Waldor, 2009; Kunkle et al., 2019). Interestingly, AKI growth media passed through a C18 reverse phase chromatography column revealed the presence of multiple bile acids acting as activating factors responsible for ompR induction in RND negative V. cholerae (Kunkle et al., 2019). Induction of ompR in RND deficient V. cholerae likely resulted from intracellular accumulation of bile salts in the RND-null mutant (Figure 6). The link between RND efflux and OmpR-dependent virulence repression was linked to OmpR directly repressing aphB transcription, thus, revealing a new regulatory arm of the ToxR regulon (Kunkle et al., 2019). Integration of OmpR into the ToxR regulon represents a third mechanism by which RND-mediated efflux contributes virulence. Subsequent experiments revealed that the EnvZ-OmpR system is also activated in response to alkaline pH, and shown to regulate multiple acid-tolerance genes through a phenotype that appeared to be independent of the RND efflux but could have contributed to decreased colonization fitness of an ompR mutant in infant mice (Kunkle et al., 2020b).
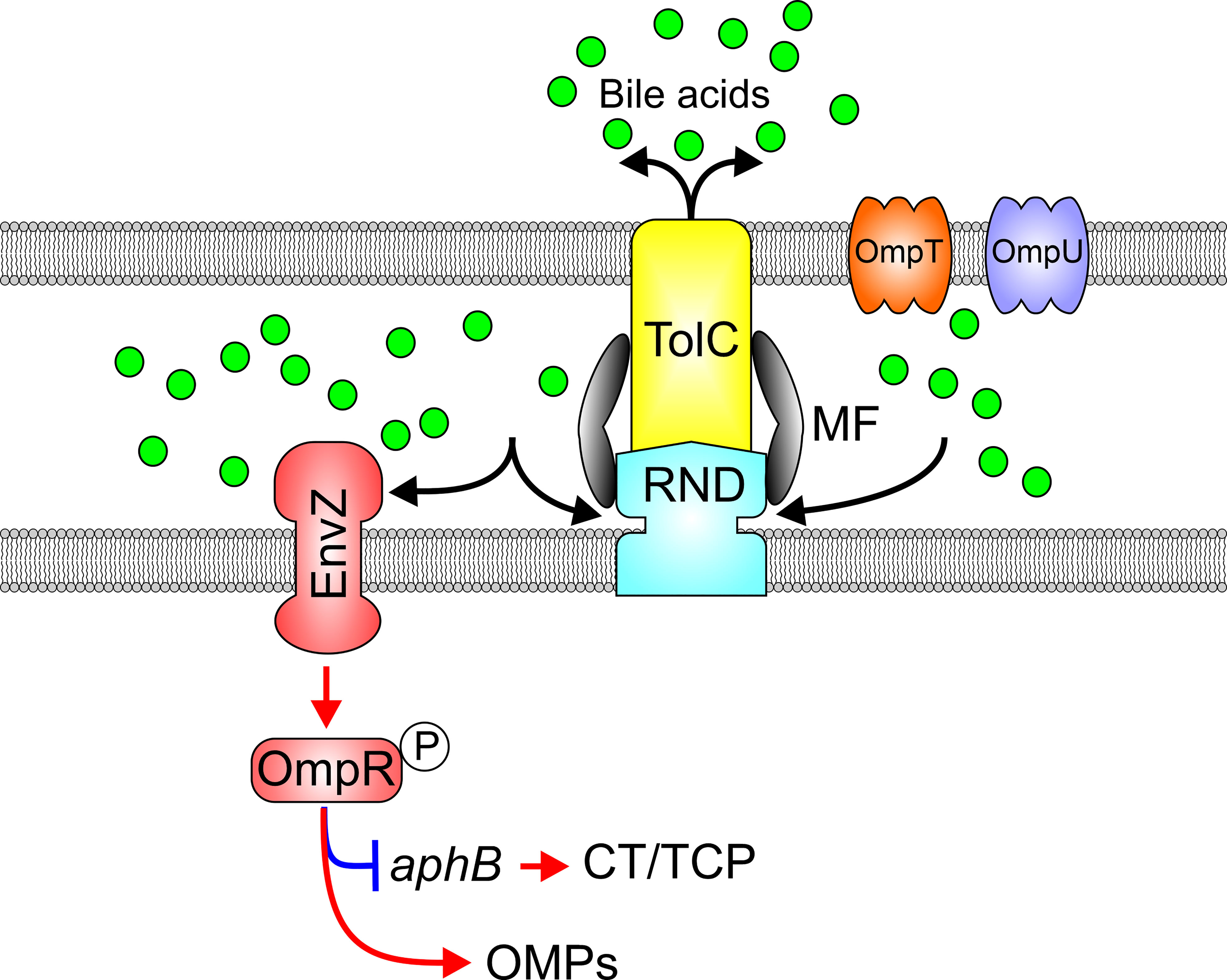
Figure 6 RND efflux influences the activation state of the EnvZ-OmpR TCS via efflux of bile acids. Bile acids (green balls) and other membrane intercalating agents induce EnvZ kinase activity and phosphorylation of OmpR which in turn modulates outer membrane porin expression and represses virulence factor production by inhibiting aphB transcription.
Conclusions
The lifecycle of pathogenic V. cholerae relies on a continuum of adaptations required to transition between disparate environments within the aquatic ecosystem and survival through the human gastrointestinal tract, from traversing the gastric acid barrier of the stomach to colonization in the duodenum, jejunum and ileum [reviewed in (Kaper et al., 1995)]. Each of these anatomical sites presents distinct environmental cues produced from the host and resident flora, and within each site local environmental differences between the lumen and the epithelium are dramatic. The ability of V. cholerae to colonize and replicate in each of these respective sites requires rapid transcriptional adaptation in response to environmental cues including exogenous and endogenous cues that are substrates of RND efflux systems such as organic acids, antimicrobial peptides, bile salts, fatty acids, and indole (Bina et al., 2013; Taylor et al., 2014; Ante et al., 2015b; Bina et al., 2018; Howard et al., 2019; Kunkle et al., 2019; Kunkle et al., 2020b; Bina et al., 2021). These compounds are concentration-dependent effectors and their influence on adaptive responses is directly linked to RND-mediated efflux. The intracellular accumulation of each of these effectors is therefore linked to cell metabolism and the maintenance of the proton motive force which is the energy source for RND-mediated efflux. Thus as V. cholerae enters stationary phase, a time when a stringent response is induced and the proton motive force begins to dissipate, these molecules may accumulate intracellularly and induce adaptive responses that are necessary for transmission and dissemination (Bina et al., 2018). Further, as endogenous metabolites are expelled from V. cholerae by the RND efflux systems, competitive inhibition of efflux by exogenous compounds (e.g., bile acids in the small intestine) may further drive adaptive responses contributing to colonization and persistence. Finally, concentration gradients of molecules in the host GI tract may have a profound impact on the pathogenic success of V. cholerae in vivo (Duane et al., 1986). For example, bile acids are important signaling molecules that affect numerous virulence-associated phenotypes (Provenzano and Klose, 2000; Provenzano et al., 2000; Wibbenmeyer et al., 2002; Cerda-Maira et al., 2008; Yang et al., 2013; Ante et al., 2015b; Midgett et al., 2017; Bina et al., 2021) and the bile-acid gradient across the mucous layer covering the intestinal epithelium may drive differential phenotypic evolution of V. cholerae localized to the epithelial surface compared to the lumen.
The broad specificity of RND transporters is critical for their roles in the evolution of multiple drug resistant pathogens and studies to develop efflux inhibitors to reverse efflux-mediated resistance are ongoing (Compagne et al., 2023). However, the RND systems likely evolved to fulfil physiological functions in bacteria, suggesting a paradigm where the contribution RND efflux systems to antimicrobial resistance is secondary to their native functions in maintaining cell homeostasis. This conclusion is supported by V. cholerae studies described herein showing that the loss of RND transport systems have widespread effects on multiple physiological functions including iron homeostasis, acid tolerance, biofilm production, and membrane remodeling. Similar findings have been reported in multiple other pathogens suggesting that the function of the RND systems in bacterial physiology is conserved across gram-negative pathogens [reviewed by (Alvarez-Ortega et al., 2013; Colclough et al., 2020)]. As many of these RND-dependent phenotypes are important for pathogenesis, the use of RND inhibitors represents a novel avenue for the development of antivirulence therapeutics that specifically impact physiology and adaptation rather than just antimicrobial resistance. Targeting bacterial physiology provides an alternative therapeutic strategy to treat infections that has the potential to circumvent primary and secondary drug resistance mechanisms. While there have been limited efforts in this approach, experimental results suggest it is feasible and merits further exploration.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by the National Institutes of Health under Award Numbers: AI132460 and AI166889. The content is solely the responsibility of the authors.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acosta, N., Pukatzki, S., Raivio, T. L. (2015a). The cpx system regulates virulence gene expression in vibrio cholerae. Infect. Immun. 83, 2396–2408. doi: 10.1128/IAI.03056-14
Acosta, N., Pukatzki, S., Raivio, T. L. (2015b). The vibrio cholerae cpx envelope stress response senses and mediates adaptation to low iron. J. Bacteriol. 197, 262–276. doi: 10.1128/JB.01957-14
Alav, I., Kobylka, J., Kuth, M. S., Pos, K. M., Picard, M., Blair, J. M. A., et al. (2021). Structure, assembly, and function of tripartite efflux and type 1 secretion systems in gram-negative bacteria. Chem. Rev. 121, 5479–5596. doi: 10.1021/acs.chemrev.1c00055
Ali, M., Nelson, A. R., Lopez, A. L., Sack, D. A. (2015). Updated global burden of cholera in endemic countries. PloS Negl. Trop. Dis. 9, e0003832. doi: 10.1371/journal.pntd.0003832
Alvarez-Ortega, C., Olivares, J., Martinez, J. L. (2013). RND multidrug efflux pumps: what are they good for? Front. Microbiol. 4, 7. doi: 10.3389/fmicb.2013.00007
Ante, V. M., Bina, X. R., Bina, J. E. (2015a). The LysR-type regulator LeuO regulates the acid tolerance response in vibrio cholerae. Microbiol. (Reading) 161, 2434–2443. doi: 10.1099/mic.0.000194
Ante, V. M., Bina, X. R., Howard, M. F., Sayeed, S., Taylor, D. L., Bina, J. E. (2015b). Vibrio cholerae leuO transcription is positively regulated by ToxR and contributes to bile resistance. J. Bacteriol. 197, 3499–3510. doi: 10.1128/JB.00419-15
Athar, M., Gervasoni, S., Catte, A., Basciu, A., Malloci, G., Ruggerone, P., et al. (2023). Tripartite efflux pumps of the RND superfamily: what did we learn from computational studies? Microbiol. (Reading) 169. doi: 10.1099/mic.0.001307
Bang, I. S., Kim, B. H., Foster, J. W., Park, Y. K. (2000). OmpR regulates the stationary-phase acid tolerance response of salmonella enterica serovar typhimurium. J. Bacteriol. 182, 2245–2252. doi: 10.1128/JB.182.8.2245-2252.2000
Bernardini, M. L., Fontaine, A., Sansonetti, P. J. (1990). The two-component regulatory system ompR-envZ controls the virulence of shigella flexneri. J. Bacteriol. 172, 6274–6281. doi: 10.1128/jb.172.11.6274-6281.1990
Bilecen, K., Fong, J. C., Cheng, A., Jones, C. J., Zamorano-Sanchez, D., Yildiz, F. H. (2015). Polymyxin b resistance and biofilm formation in vibrio cholerae are controlled by the response regulator CarR. Infect. Immun. 83, 1199–1209. doi: 10.1128/IAI.02700-14
Bilecen, K., Yildiz, F. H. (2009). Identification of a calcium-controlled negative regulatory system affecting vibrio cholerae biofilm formation. Environ. Microbiol. 11, 2015–2029. doi: 10.1111/j.1462-2920.2009.01923.x
Bina, X. R., Bina, J. E. (2010). The cyclic dipeptide Cyclo(Phe-pro) inhibits cholera toxin and toxin-coregulated pilus production in O1 El tor vibrio cholerae. J. Bacteriol. 192, 3829–3832. doi: 10.1128/JB.00191-10
Bina, R. F., Bina, J. E., Weng, Y. (2020). Genome sequence of vibrio cholerae strain RFB16, isolated from north park lake in Allegheny county, Pennsylvania. Microbiol. Resour Announc 9. doi: 10.1128/MRA.00111-20
Bina, X. R., Howard, M. F., Ante, V. M., Bina, J. E. (2016). Vibrio cholerae LeuO links the ToxR regulon to expression of lipid a remodeling genes. Infect. Immun. 84, 3161–3171. doi: 10.1128/IAI.00445-16
Bina, X. R., Howard, M. F., Taylor-Mulneix, D. L., Ante, V. M., Kunkle, D. E., Bina, J. E. (2018). The vibrio cholerae RND efflux systems impact virulence factor production and adaptive responses via periplasmic sensor proteins. PLoS Pathog. 14, e1006804. doi: 10.1371/journal.ppat.1006804
Bina, T. F., Kunkle, D. E., Bina, X. R., Mullett, S. J., Wendell, S. G., Bina, J. E. (2021). Bile salts promote ToxR regulon activation during growth under virulence-inducing conditions. Infect. Immun. 89, e0044121. doi: 10.1128/IAI.00441-21
Bina, J. E., Mekalanos, J. J. (2001). Vibrio cholerae tolC is required for bile resistance and colonization. Infect. Immun. 69, 4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001
Bina, X. R., Philippart, J. A., Bina, J. E. (2009). Effect of the efflux inhibitors 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta-naphthylamide on antimicrobial susceptibility and virulence factor production in vibrio cholerae. J. Antimicrob. Chemother. 63, 103–108. doi: 10.1093/jac/dkn466
Bina, X. R., Provenzano, D., Nguyen, N., Bina, J. E. (2008). Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect. Immun. 76, 3595–3605. doi: 10.1128/IAI.01620-07
Bina, J. E., Provenzano, D., Wang, C., Bina, X. R., Mekalanos, J. J. (2006). Characterization of the vibrio cholerae vexAB and vexCD efflux systems. Arch. Microbiol. 186, 171–181. doi: 10.1007/s00203-006-0133-5
Bina, X. R., Taylor, D. L., Vikram, A., Ante, V. M., Bina, J. E. (2013). Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo(Phe-pro). mBio 4, e00366-13. doi: 10.1128/mBio.00366-13
Bina, J., Zhu, J., Dziejman, M., Faruque, S., Calderwood, S., Mekalanos, J. (2003). ToxR regulon of vibrio cholerae and its expression in vibrios shed by cholera patients. Proc. Natl. Acad. Sci. U.S.A. 100, 2801–2806. doi: 10.1073/pnas.2628026100
Cerda-Maira, F. A., Kovacikova, G., Jude, B. A., Skorupski, K., Taylor, R. K. (2013). Characterization of BreR interaction with the bile response promoters breAB and breR in vibrio cholerae. J. Bacteriol. 195, 307–317. doi: 10.1128/JB.02008-12
Cerda-Maira, F. A., Ringelberg, C. S., Taylor, R. K. (2008). The bile response repressor BreR regulates expression of the vibrio cholerae breAB efflux system operon. J. Bacteriol. 190, 7441–7452. doi: 10.1128/JB.00584-08
Chakraborty, S., Nair, G. B., Shinoda, S. (1997). Pathogenic vibrios in the natural aquatic environment. Rev. Environ. Health 12, 63–80. doi: 10.1515/REVEH.1997.12.2.63
Cheng, A. T., Ottemann, K. M., Yildiz, F. H. (2015). Vibrio cholerae response regulator VxrB controls colonization and regulates the type VI secretion system. PLoS Pathog. 11, e1004933. doi: 10.1371/journal.ppat.1004933
Childers, B. M., Klose, K. E. (2007). Regulation of virulence in vibrio cholerae: the ToxR regulon. Future Microbiol. 2, 335–344. doi: 10.2217/17460913.2.3.335
Colclough, A. L., Alav, I., Whittle, E. E., Pugh, H. L., Darby, E. M., Legood, S. W., et al. (2020). RND efflux pumps in gram-negative bacteria; regulation, structure and role in antibiotic resistance. Future Microbiol. 15, 143–157. doi: 10.2217/fmb-2019-0235
Compagne, N., Vieira da Cruz, A., Muller, R. T., Hartkoorn, R. C., Flipo, M., Pos, K. M. (2023). Update on the discovery of efflux pump inhibitors against critical priority gram-negative bacteria. Antibiotics (Basel) 12. doi: 10.3390/antibiotics12010180
Correa, N. E., Lauriano, C. M., Mcgee, R., Klose, K. E. (2000). Phosphorylation of the flagellar regulatory protein FlrC is necessary for vibrio cholerae motility and enhanced colonization. Mol. Microbiol. 35, 743–755. doi: 10.1046/j.1365-2958.2000.01745.x
Daboul, J., Weghorst, L., Deangelis, C., Plecha, S. C., Saul-Mcbeth, J., Matson, J. S. (2020). Characterization of vibrio cholerae isolates from freshwater sources in northwest Ohio. PLoS One 15, e0238438. doi: 10.1371/journal.pone.0238438
Daury, L., Orange, F., Taveau, J. C., Verchere, A., Monlezun, L., Gounou, C., et al. (2016). Tripartite assembly of RND multidrug efflux pumps. Nat. Commun. 7, 10731. doi: 10.1038/ncomms10731
DiRita, V. J., Parsot, C., Jander, G., Mekalanos, J. J. (1991). Regulatory cascade controls virulence in vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 88, 5403–5407. doi: 10.1073/pnas.88.12.5403
Duane, W. C., Levitt, M. D., Staley, N. A., Mchale, A. P., Wiegand, D. M., Fetzer, C. A. (1986). Role of the unstirred layer in protecting the murine gastric mucosa from bile salt. Gastroenterology 91, 913–918. doi: 10.1016/0016-5085(86)90694-3
Fengler, V. H., Boritsch, E. C., Tutz, S., Seper, A., Ebner, H., Roier, S., et al. (2012). Disulfide bond formation and ToxR activity in vibrio cholerae. PLoS One 7, e47756. doi: 10.1371/journal.pone.0047756
Gao, H., Zhang, Y., Han, Y., Yang, L., Liu, X., Guo, Z., et al. (2011). Phenotypic and transcriptional analysis of the osmotic regulator OmpR in yersinia pestis. BMC Microbiol. 11, 39. doi: 10.1186/1471-2180-11-39
Hall, M. N., Silhavy, T. J. (1981). Genetic analysis of the major outer membrane proteins of escherichia coli. Annu. Rev. Genet. 15, 91–142. doi: 10.1146/annurev.ge.15.120181.000515
Hase, C. C., Mekalanos, J. J. (1998). TcpP protein is a positive regulator of virulence gene expression in vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 95, 730–734. doi: 10.1073/pnas.95.2.730
Heidelberg, J. F., Eisen, J. A., Nelson, W. C., Clayton, R. A., Gwinn, M. L., Dodson, R. J., et al. (2000). DNA Sequence of both chromosomes of the cholera pathogen vibrio cholerae. Nature 406, 477–483. doi: 10.1038/35020000
Herrera, C. M., Crofts, A. A., Henderson, J. C., Pingali, S. C., Davies, B. W., Trent, M. S. (2014). The vibrio cholerae VprA-VprB two-component system controls virulence through endotoxin modification. MBio 5. doi: 10.1128/mBio.02283-14
Herrington, D. A., Hall, R. H., Losonsky, G., Mekalanos, J. J., Taylor, R. K., Levine, M. M. (1988). Toxin, toxin-coregulated pili, and the toxR regulon are essential for vibrio cholerae pathogenesis in humans. J. Exp. Med. 168, 1487–1492. doi: 10.1084/jem.168.4.1487
Hews, C. L., Cho, T., Rowley, G., Raivio, T. L. (2019). Maintaining integrity under stress: envelope stress response regulation of pathogenesis in gram-negative bacteria. Front. Cell Infect. Microbiol. 9, 313. doi: 10.3389/fcimb.2019.00313
Heyde, M., Portalier, R. (1987). Regulation of major outer membrane porin proteins of escherichia coli K 12 by pH. Mol. Gen. Genet. 208, 511–517. doi: 10.1007/BF00328148
Howard, M. F., Bina, X. R., Bina, J. E. (2019). Indole inhibits ToxR regulon expression in vibrio cholerae. Infect. Immun. 87. doi: 10.1128/IAI.00776-18
Hu, Y., Lu, P., Wang, Y., Ding, L., Atkinson, S., Chen, S. (2009). OmpR positively regulates urease expression to enhance acid survival of yersinia pseudotuberculosis. Microbiology 155, 2522–2531. doi: 10.1099/mic.0.028381-0
Huq, A., Small, E. B., West, P. A., Huq, M. I., Rahman, R., Colwell, R. R. (1983). Ecological relationships between vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45, 275–283. doi: 10.1128/aem.45.1.275-283.1983
Iwanaga, M., Yamamoto, K. (1985). New medium for the production of cholera toxin by vibrio cholerae O1 biotype El tor. J. Clin. Microbiol. 22, 405–408. doi: 10.1128/jcm.22.3.405-408.1985
Jang, J., Jung, K. T., Park, J., Yoo, C. K., Rhie, G. E. (2011). The vibrio cholerae VarS/VarA two-component system controls the expression of virulence proteins through ToxT regulation. Microbiol. (Reading) 157, 1466–1473. doi: 10.1099/mic.0.043737-0
Kaper, J. B., Morris, J. G., Jr., Levine, M. M. (1995). Cholera. Clin. Microbiol. Rev. 8, 48–86. doi: 10.1128/CMR.8.1.48
Kazi, M. I., Conrado, A. R., Mey, A. R., Payne, S. M., Davies, B. W. (2016). ToxR antagonizes h-NS regulation of horizontally acquired genes to drive host colonization. PLoS Pathog. 12, e1005570. doi: 10.1371/journal.ppat.1005570
Koronakis, V., Sharff, A., Koronakis, E., Luisi, B., Hughes, C. (2000). Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405, 914–919. doi: 10.1038/35016007
Kovacikova, G., Skorupski, K. (1999). A vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J. Bacteriol. 181, 4250–4256. doi: 10.1128/JB.181.14.4250-4256.1999
Kunkle, D. E., Bina, X. R., Bina, J. E. (2017). The vibrio cholerae VexGH RND efflux system maintains cellular homeostasis by effluxing vibriobactin. mBio 8. doi: 10.1128/mBio.00126-17
Kunkle, D. E., Bina, X. R., Bina, J. E. (2020b). Vibrio cholerae OmpR contributes to virulence repression and fitness at alkaline pH. Infect. Immun. 88. doi: 10.1128/IAI.00141-20
Kunkle, D. E., Bina, T. F., Bina, X. R., Bina, J. E. (2019). Vibrio cholerae OmpR represses the ToxR regulon in response to membrane intercalating agents that are prevalent in the human gastrointestinal tract. Infect. Immun. 88 (3), e00912–19. doi: 10.1128/IAI.00912-19
Kunkle, D. E., Bina, T. F., Bina, X. R., Bina, J. E. (2020a). Vibrio cholerae OmpR represses the ToxR regulon in response to membrane intercalating agents that are prevalent in the human gastrointestinal tract. Infect. Immun. 88. doi: 10.1128/IAI.00912-19
Larocque, R. C., Harris, J. B., Dziejman, M., Li, X., Khan, A. I., Faruque, A. S., et al. (2005). Transcriptional profiling of vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect. Immun. 73, 4488–4493. doi: 10.1128/IAI.73.8.4488-4493.2005
Lee, A. K., Detweiler, C. S., Falkow, S. (2000). OmpR regulates the two-component system SsrA-ssrB in salmonella pathogenicity island 2. J. Bacteriol. 182, 771–781. doi: 10.1128/JB.182.3.771-781.2000
Li, X. Z., Plesiat, P., Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/CMR.00117-14
Lin, T. H., Chen, Y., Kuo, J. T., Lai, Y. C., Wu, C. C., Huang, C. F., et al. (2018). Phosphorylated OmpR is required for type 3 fimbriae expression in klebsiella pneumoniae under hypertonic conditions. Front. Microbiol. 9, 2405. doi: 10.3389/fmicb.2018.02405
Lombardo, M. J., Michalski, J., Martinez-Wilson, H., Morin, C., Hilton, T., Osorio, C. G., et al. (2007). An in vivo expression technology screen for vibrio cholerae genes expressed in human volunteers. Proc. Natl. Acad. Sci. U.S.A. 104, 18229–18234. doi: 10.1073/pnas.0705636104
Lv, L., Wan, M., Wang, C., Gao, X., Yang, Q., Partridge, S. R., et al. (2020). Emergence of a plasmid-encoded resistance-Nodulation-Division efflux pump conferring resistance to multiple drugs, including tigecycline, in klebsiella pneumoniae. mBio 11. doi: 10.1128/mBio.02930-19
Martinez-Wilson, H. F., Tamayo, R., Tischler, A. D., Lazinski, D. W., Camilli, A. (2008). The vibrio cholerae hybrid sensor kinase VieS contributes to motility and biofilm regulation by altering the cyclic diguanylate level. J. Bacteriol. 190, 6439–6447. doi: 10.1128/JB.00541-08
Meibom, K. L., Li, X. B., Nielsen, A. T., Wu, C. Y., Roseman, S., Schoolnik, G. K. (2004). The vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U.S.A. 101, 2524–2529. doi: 10.1073/pnas.0308707101
Merrell, D. S., Butler, S. M., Qadri, F., Dolganov, N. A., Alam, A., Cohen, M. B., et al. (2002). Host-induced epidemic spread of the cholera bacterium. Nature 417, 642–645. doi: 10.1038/nature00778
Merrell, D. S., Camilli, A. (1999). The cadA gene of vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol. Microbiol. 34, 836–849. doi: 10.1046/j.1365-2958.1999.01650.x
Merrell, D. S., Camilli, A. (2000). Regulation of vibrio cholerae genes required for acid tolerance by a member of the "ToxR-like" family of transcriptional regulators. J. Bacteriol. 182, 5342–5350. doi: 10.1128/JB.182.19.5342-5350.2000
Midgett, C. R., Almagro-Moreno, S., Pellegrini, M., Taylor, R. K., Skorupski, K., Kull, F. J. (2017). Bile salts and alkaline pH reciprocally modulate the interaction between the periplasmic domains of vibrio cholerae ToxR and ToxS. Mol. Microbiol. 105, 258–272. doi: 10.1111/mmi.13699
Miller, V. L., Dirita, V. J., Mekalanos, J. J. (1989). Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J. Bacteriol. 171, 1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989
Miller, V. L., Taylor, R. K., Mekalanos, J. J. (1987). Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell 48, 271–279. doi: 10.1016/0092-8674(87)90430-2
Mitrophanov, A. Y., Groisman, E. A. (2008). Signal integration in bacterial two-component regulatory systems. Genes Dev. 22, 2601–2611. doi: 10.1101/gad.1700308
Moorthy, S., Watnick, P. I. (2005). Identification of novel stage-specific genetic requirements through whole genome transcription profiling of vibrio cholerae biofilm development. Mol. Microbiol. 57, 1623–1635. doi: 10.1111/j.1365-2958.2005.04797.x
Peschek, N., Herzog, R., Singh, P. K., Sprenger, M., Meyer, F., Frohlich, K. S., et al. (2020). RNA-Mediated control of cell shape modulates antibiotic resistance in vibrio cholerae. Nat. Commun. 11, 6067. doi: 10.1038/s41467-020-19890-8
Peterson, K. M., Gellings, P. S. (2018). Multiple intraintestinal signals coordinate the regulation of vibrio cholerae virulence determinants. Pathog. Dis. 76. doi: 10.1093/femspd/ftx126
Phillips, R. A. (1968). Asiatic Cholera (with emphasis on pathophysiological effects of the disease). Annu. Rev. Med. 19, 69–80. doi: 10.1146/annurev.me.19.020168.000441
Piddock, L. J. (2006). Multidrug-resistance efflux pumps - not just for resistance. Nat. Rev. Microbiol. 4, 629–636. doi: 10.1038/nrmicro1464
Poole, K., Krebes, K., Mcnally, C., Neshat, S. (1993). Multiple antibiotic resistance in pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175, 7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993
Pratt, J. T., Ismail, A. M., Camilli, A. (2010). PhoB regulates both environmental and virulence gene expression in vibrio cholerae. Mol. Microbiol. 77, 1595–1605. doi: 10.1111/j.1365-2958.2010.07310.x
Provenzano, D., Klose, K. E. (2000). Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. U.S.A. 97, 10220–10224. doi: 10.1073/pnas.170219997
Provenzano, D., Lauriano, C. M., Klose, K. E. (2001). Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in vibrio cholerae virulence. J. Bacteriol. 183, 3652–3662. doi: 10.1128/JB.183.12.3652-3662.2001
Provenzano, D., Schuhmacher, D. A., Barker, J. L., Klose, K. E. (2000). The virulence regulatory protein ToxR mediates enhanced bile resistance in vibrio cholerae and other pathogenic vibrio species. Infect. Immun. 68, 1491–1497. doi: 10.1128/IAI.68.3.1491-1497.2000
Quinn, M. J., Resch, C. T., Sun, J., Lind, E. J., Dibrov, P., Hase, C. C. (2012). NhaP1 is a K+(Na+)/H+ antiporter required for growth and internal pH homeostasis of vibrio cholerae at low extracellular pH. Microbiology 158, 1094–1105. doi: 10.1099/mic.0.056119-0
Rahman, M. M., Matsuo, T., Ogawa, W., Koterasawa, M., Kuroda, T., Tsuchiya, T. (2007). Molecular cloning and characterization of all RND-type efflux transporters in vibrio cholerae non-O1. Microbiol. Immunol. 51, 1061–1070. doi: 10.1111/j.1348-0421.2007.tb04001.x
Reboul, A., Lemaitre, N., Titecat, M., Merchez, M., Deloison, G., Ricard, I., et al. (2014). Yersinia pestis requires the 2-component regulatory system OmpR-EnvZ to resist innate immunity during the early and late stages of plague. J. Infect. Dis. 210, 1367–1375. doi: 10.1093/infdis/jiu274
Sarma, V., Reeves, P. (1977). Genetic locus (ompB) affecting a major outer-membrane protein in escherichia coli K-12. J. Bacteriol. 132, 23–27. doi: 10.1128/jb.132.1.23-27.1977
Schild, S., Tamayo, R., Nelson, E. J., Qadri, F., Calderwood, S. B., Camilli, A. (2007). Genes induced late in infection increase fitness of vibrio cholerae after release into the environment. Cell Host Microbe 2, 264–277. doi: 10.1016/j.chom.2007.09.004
Schwan, W. R. (2009). Survival of uropathogenic escherichia coli in the murine urinary tract is dependent on OmpR. Microbiology 155, 1832–1839. doi: 10.1099/mic.0.026187-0
Silva, I. N., Pessoa, F. D., Ramires, M. J., Santos, M. R., Becker, J. D., Cooper, V. S., et al. (2018). The OmpR regulator of burkholderia multivorans controls mucoid-to-Nonmucoid transition and other cell envelope properties associated with persistence in the cystic fibrosis lung. J. Bacteriol. 200. doi: 10.1128/JB.00216-18
Skorupski, K., Taylor, R. K. (1999). A new level in the vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 31, 763–771. doi: 10.1046/j.1365-2958.1999.01215.x
Slamti, L., Waldor, M. K. (2009). Genetic analysis of activation of the vibrio cholerae cpx pathway. J. Bacteriol. 191, 5044–5056. doi: 10.1128/JB.00406-09
Slauch, J. M., Garrett, S., Jackson, D. E., Silhavy, T. J. (1988). EnvZ functions through OmpR to control porin gene expression in escherichia coli K-12. J. Bacteriol. 170, 439–441. doi: 10.1128/jb.170.1.439-441.1988
Solomon, S. L., Oliver, K. B. (2014). Antibiotic resistance threats in the united states: stepping back from the brink. Am. Fam Physician 89, 938–941.
Stincone, A., Daudi, N., Rahman, A. S., Antczak, P., Henderson, I., Cole, J., et al. (2011). A systems biology approach sheds new light on escherichia coli acid resistance. Nucleic Acids Res. 39, 7512–7528. doi: 10.1093/nar/gkr338
Taylor, D. L., Ante, V. M., Bina, X. R., Howard, M. F., Bina, J. E. (2015). Substrate-dependent activation of the vibrio cholerae vexAB RND efflux system requires vexR. PLoS One 10, e0117890. doi: 10.1371/journal.pone.0117890
Taylor, D. L., Bina, X. R., Bina, J. E. (2012). Vibrio cholerae VexH encodes a multiple drug efflux pump that contributes to the production of cholera toxin and the toxin co-regulated pilus. PLoS One 7, e38208. doi: 10.1371/journal.pone.0038208
Taylor, D. L., Bina, X. R., Slamti, L., Waldor, M. K., Bina, J. E. (2014). Reciprocal regulation of resistance-nodulation-division efflux systems and the cpx two-component system in vibrio cholerae. Infect. Immun. 82, 2980–2991. doi: 10.1128/IAI.00025-14
Teschler, J. K., Cheng, A. T., Yildiz, F. H. (2017). The two-component signal transduction system VxrAB positively regulates vibrio cholerae biofilm formation. J. Bacteriol. 199. doi: 10.1128/JB.00139-17
Tipton, K. A., Rather, P. N. (2017). An ompR-envZ two-component system ortholog regulates phase variation, osmotic tolerance, motility, and virulence in acinetobacter baumannii strain AB5075. J. Bacteriol. 199. doi: 10.1128/JB.00705-16
Tischler, A. D., Camilli, A. (2005). Cyclic diguanylate regulates vibrio cholerae virulence gene expression. Infect. Immun. 73, 5873–5882. doi: 10.1128/IAI.73.9.5873-5882.2005
Venter, H., Mowla, R., Ohene-Agyei, T., Ma, S. (2015). RND-type drug e ffl ux pumps from gram-negative bacteria: molecular mechanism and inhibition. Front. Microbiol. 6, 377. doi: 10.3389/fmicb.2015.00377
Wang, Z., Fan, G., Hryc, C. F., Blaza, J. N., Serysheva, I. I., Schmid, M. F., et al. (2017). An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. Elife 6. doi: 10.7554/eLife.24905
Weng, Y., Fields, E. G., Bina, T. F., Budnick, J. A., Kunkle, D. E., Bina, X. R., et al. (2021). Vibrio cholerae TolC is required for expression of the ToxR regulon. Infect. Immun. 89, e0024221. doi: 10.1128/IAI.00242-21
Wibbenmeyer, J. A., Provenzano, D., Landry, C. F., Klose, K. E., Delcour, A. H. (2002). Vibrio cholerae OmpU and OmpT porins are differentially affected by bile. Infect. Immun. 70, 121–126. doi: 10.1128/IAI.70.1.121-126.2002
Xu, Q., Dziejman, M., Mekalanos, J. J. (2003). Determination of the transcriptome of vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc. Natl. Acad. Sci. U.S.A. 100, 1286–1291. doi: 10.1073/pnas.0337479100
Yang, M., Liu, Z., Hughes, C., Stern, A. M., Wang, H., Zhong, Z., et al. (2013). Bile salt-induced intermolecular disulfide bond formation activates vibrio cholerae virulence. Proc. Natl. Acad. Sci. U.S.A. 110, 2348–2353. doi: 10.1073/pnas.1218039110
Keywords: Vibrio cholerae, pathogenesis, efflux, adaptation, virulence
Citation: Bina XR and Bina JE (2023) Vibrio cholerae RND efflux systems: mediators of stress responses, colonization and pathogenesis. Front. Cell. Infect. Microbiol. 13:1203487. doi: 10.3389/fcimb.2023.1203487
Received: 10 April 2023; Accepted: 25 April 2023;
Published: 15 May 2023.
Edited by:
Jun (Jay) Zhu, University of Pennsylvania, United StatesReviewed by:
Daniele Provenzano, The University of Texas Rio Grande Valley, United StatesMary Brockett, University of Pennsylvania, United States
Copyright © 2023 Bina and Bina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James E. Bina, amJpbmFAcGl0dC5lZHU=
 X. Renee Bina
X. Renee Bina James E. Bina
James E. Bina