
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell. Infect. Microbiol., 05 May 2023
Sec. Bacteria and Host
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1180708
This article is part of the Research TopicNew Advances in Obligate Intracellular Bacteria: Pathogenesis and Host InteractionsView all 9 articles
As a highly conserved, multifunctional protein with multiple domains, p62/SQSTM1 plays a crucial role in several essential cellular activities, particularly selective autophagy. Recent research has shown that p62 is crucial in eradicating intracellular bacteria by xenophagy, a selective autophagic process that identifies and eliminates such microorganisms. This review highlights the various roles of p62 in intracellular bacterial infections, including both direct and indirect, antibacterial and infection-promoting aspects, and xenophagy-dependent and independent functions, as documented in published literature. Additionally, the potential applications of synthetic drugs targeting the p62-mediated xenophagy mechanism and unresolved questions about p62’s roles in bacterial infections are also discussed.
Despite the progress achieved with the development and use of antibiotics and vaccines, the mortality rates associated with infectious diseases remains high. Moreover, antibiotic resistance amongst pathogenic microorganisms has emerged as an alarming global challenge (Rex et al., 2017; Abbara et al., 2022). Intracellular bacterial infections have long been a severe threat to public health. Unlike extracellular microbes that often result in acute infections, intracellular bacteria have the tendency to cause chronic or persistent infections. This type of bacterial infection can remain latent in the host for life, presenting an ongoing threat to the health of the individual (Grant and Hung, 2013). Consequently, research into the mechanisms of intracellular bacterial infection and the host’s defense against such invaders has become a crucial and trending topic in basic biomedical research.
The p62/SQSTM1 protein, a multifunctional protein with a molecular weight of 62-kDa, is found in the cytoplasm in scattered dots or aggregates and can be transported between the cytoplasm and nucleus (Zhang and Costa, 2021). Initially discovered in 1995 for its ability to specifically bind with the src homology 2 domain of p56lck, regulated by phosphorylation of Ser-59 in a phosphotyrosine-independent way, p62 has since been implicated in the pathogenesis of cardiometabolic disease, neurodegenerative disease, malignant tumors, and infectious diseases (Park et al., 1995; Seto et al., 2013; Jeong et al., 2019; Ma et al., 2019; Tang et al., 2021). Additionally, studies have shown that p62 plays a crucial role in xenophagy, a type of selective autophagy that targets intracellular pathogenic microbes and enables their elimination (Knodler and Celli, 2011). This article delves into the role of p62 protein, specifically its involvement in p62-mediated xenophagy, in the infection of various intracellular bacteria like Salmonella enterica serovar Typhimurium and Legionella pneumophila. In this review, we highlight the current understanding of the mechanisms underlying intracellular bacterial infections and how our innate immune system functions in response to such pathogens (Table 1).
Comprised of 440 amino acids, p62 is composed of six domains from the N-terminal to C-terminal, including the Phox and Bem1 (PB1) domain (104 amino acids), ZZ-type zinc finger (ZZ) domain (36 amino acids), TRAF6-binding (TB) domain (27 amino acids), LC3-interacting region (LIR) (22 amino acids), Keap1-interacting region (KIR) (6 amino acids), and ubiquitin-associated (UBA) domain (49 amino acids) (Pankiv et al., 2007; Gal et al., 2009; Jiang et al., 2009; Jain et al., 2010; Zhang and Costa, 2021). Notably, p62 has been shown to play a significantly increasing role in multiple intracellular bacterial infections through its involvement in xenophagy against bacteria. The mechanisms behind microbial pathogenesis involve the active invasion of pathogens into host cells or their passive uptake via phagocytosis. Once inside, pathogen-associated molecular patterns such as bacterial surface lipopolysaccharide (LPS) or viral nucleic acid products can alter signaling pathways like nuclear factor kappa B (NF-κB) and mitogen-activated protein kinase, eventually leading to the formation of pre-autophagosomal structures (PAS) facilitated by the endoplasmic reticulum and the Golgi apparatus (Jounai et al., 2007; Xu et al., 2007). Host cells then mark the microbes for destruction by ubiquitination, which generates specific “eat me” signals and enables selective autophagy (Boyle and Randow, 2013). Subsequently, Ser 403 of the UBA domain (aa 391-436) in p62 undergoes phosphorylation by Tank Binding Kinase 1(TBK1), enabling the recruitment of p62 to ubiquitin-coated microbes and initiating its self-oligomerization (Matsumoto et al., 2011; Pilli et al., 2012). During selective autophagy, self-oligomerization of p62 plays a crucial role in delivering ubiquitinated cargos to the autophagy pathway, a process driven by the PB1 domain (Nakamura et al., 2010). The UBA domain in p62 then recognizes and binds to the ubiquitin coat of microbes, while the LIR of p62 interacts with LC3 tagged on the phagophore (PG) originating from PAS. The process is facilitated by the direct binding of N-terminal degrons to the ZZ domain of p62 (Cha-Molstad et al., 2017). Therefore, the microbe is specifically targeted and gradually enveloped by PG via bridging of p62, which leads to the formation of autophagosome. Ultimately, the autophagosome is fused with the lysosome to form the autolysosome in which the microbes are degraded and destroyed by acid hydrolase (Shen et al., 2021; Tripathi-Giesgen et al., 2021). In addition, the TB domain of p62 interacts with TRAF6, leading to the activation of NF-κB signaling pathway which induces further autophagy activation (Min et al., 2018). The domain architecture of p62, functions of each domain, and simplified mechanisms of p62-mediated xenophagy targeting bacteria are summarized in Figure 1.
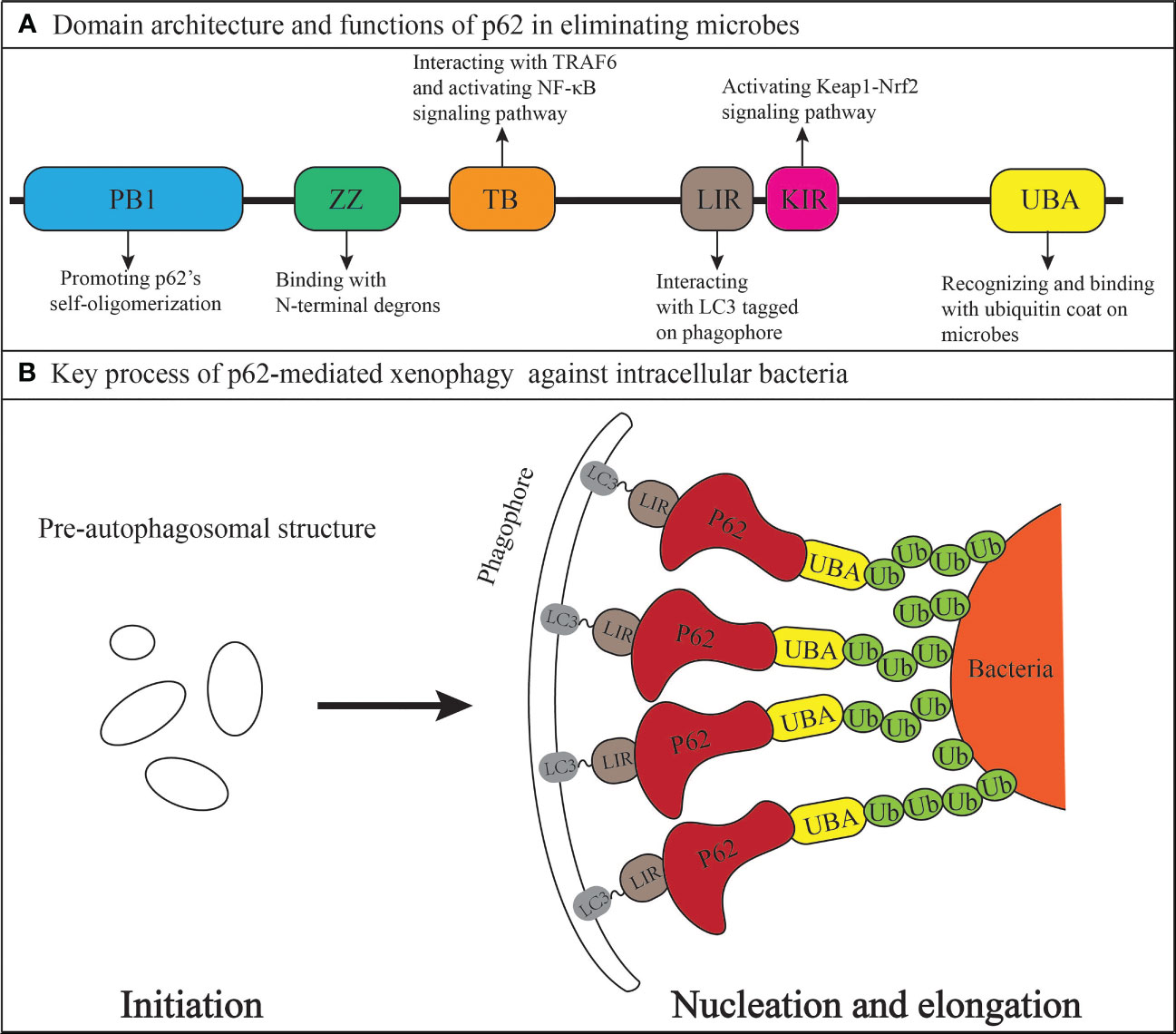
Figure 1 The xenophagy adaptor p62/SQSTM1 (A) Domain architecture and anti-bacterial functions. (B) Mechanisms in linking the bacteria to a growing phagophore (domains other than the LIR and UBA in the p62 protein are contained within the red region where the word p62 is located).
The posttranslational modification of p62 is critical in the process of xenophagy targeting invaders. A study demonstrated that during Mycobacterium tuberculosis (Mtb) infection, human protein phosphatase Mg2+/Mn2+-dependent 1A (PPM1A) inhibit xenophagy via directly dephosphorylating p62 on S403, resulting in impairment of antibacterial effect of host cells. And treatment with a selective PPM1A inhibitor can inhibit the intracellular survival of Mtb in macrophages and in the lungs of infected mice (Berton et al., 2022).
In addition to phosphorylation, ubiquitination of p62 has recently been confirmed to participate in xenophagy against intracellular bacteria. RNF166, a family member of E3 ligases, was found indispensable for recruitment of p62 to Salmonella upon infection via directly ubiquitinating p62 at K91 and K189 in a K29- and K33-linked manner, which inhibits bacterial replication through xenophagy but without altering host cell’s autophagic flux (Heath et al., 2016). Notably, studies have revealed that during Salmonella infection, the E3 ligase RNF213 may contribute to the formation of a bacterial ubiquitin coat by directly ubiquitinating bacterial LPS. Moreover, RNF213 has been shown to facilitate the recruitment of ubiquitin-dependent autophagy adaptors such as p62 to Salmonella and promote subsequent antibacterial xenophagy. These findings suggest that non-proteinaceous substances may also undergo ubiquitylation and recognition in p62-mediated antibacterial xenophagy (Otten et al., 2021).
There are some typical bacterial proteins involved in host cell’s p62-mediated xenophagy (Table 2). As cell surface protein of Mtb, Rv1468c was found to recruit ubiquitin and bind with polyubiquitin via its UBA domain upon Mtb’s infection in macrophages, which promotes p62-mediated xenophagy targeting Mtb (Chai et al., 2019). Meanwhile, researchers also found that deletion of Rv1468c in Mtb reduces colocalizations of Mtb with LC3 and autophagy targeting to Mtb and promotes bacterial intracellular survival.
It is reported that Acinetobacter baumannii could utilize its virulent protein to trigger incomplete autophagy by interfering the fusion of autophagosomes with lysosomes to enhance its survival in infected host cells (An et al., 2019). Meanwhile, Isochorismatase encoded by Acinetobacter baumannii was testified to mediate the xenophagy-induced clearance of Acinetobacter baumannii via facilitating recruitment of autophagy adaptor p62 and NDP52 to bacteria both in vitro and in vivo (Wang et al., 2016). However, the specific molecular target of Isochorismatase in the physiological process is unclear.
Although p62-mediated xenophagy plays an essential role in host’s defense against invasive microbes, bacteria have evolved mechanisms to avoid xenophagy through secreting a large cohort of virulence factors called effectors. SidE, an effector protein belonging to the SidE family, help L. pneumophila evade xenophagy by disrupting recruitment of p62 to L. pneumophila-containing vacuole (LCV) (Omotade and Roy, 2020). Moreover, the study indicates that SidE’s interference with recruitment of p62 to LCV is due to SidE’s phosphoribosyl-ubiquitination of proteins on LCV, and this non-canonical ubiquitination disenables p62 to efficiently recognize bacteria and initiate subsequent xenophagy.
ATG14 is an essential protein in promoting autophagosome-endolysosome fusion in autophagy process (Diao et al., 2015). Sayaka Shizukuishi et al. reported that during streptococcus pneumoniae infection, CbpC, which belongs to pneumococcal cell surface proteins, could bind to p62 and act as a decoy for autophagic degradation of Atg14, thus suppressing host’s xenophagy targeting intracellular pneumococci and promoting bacterial survival within host cells (Shizukuishi et al., 2020).
SseL, a virulent protein originating from Salmonella’s type 3 secretion systems(T3SS), was demonstrated to inhibit host cell’s xenophagy and promote Salmonella’s replication (Mesquita et al., 2012). Mechanistically, SseL deubiquitinates autophagic substrates targeted by p62 and induced by Salmonella, which inhibits the recruitment of p62 and LC3 to SCV-associated aggregates.
Belonging to obligate intracellular bacteria, Rickettsia parkeri could evade host cell’s xenophagy and easily spread from cell to cell (Helminiak et al., 2022). Encoded by all sequenced Rickettsia species, patatin-like phospholipase A2 enzyme (Pat1) was found to enable Rickettsia parkeri’s evasion of recognition by p62-mediated xenophagy through avoidance of polyubiquitin recruitment (Borgo et al., 2022).
The antimicrobial function of p62 is not limited to xenophagy, but may also involve the Nrf2 (nuclear factor erythroid 2-related factor 2)-Keap1 (kelch-like ECH-associated protein 1) pathway. Previous studies have demonstrated that upon sequestration of p62 to Salmonella and its subsequent oligomerization, Ser351 of the KIR domain (amino acids 346-359) is phosphorylated, leading to recruitment of Keap1 onto p62-positive microbes. This results in the activation of Nrf2, which translocate into the nucleus and induces the transcription of cytoprotective genes, including scavenger receptors and enzymes involved in the pentose phosphate pathway to generate nicotinamide adenine dinucleotide phosphate. The scavenger receptors increase the number of bacteria in phagosomes, whereas the enzymes elevate the level of reactive oxygen species in phagosomes through activation of NADPH oxidase, both of which aid in eliminating intracellular bacteria (Harvey et al., 2011; Bonilla et al., 2013; Ishimura et al., 2014).
In turn, certain bacteria can exploit host Nrf2-Keap1 pathway via modifying p62. A study showed that during Coxiella burnetiid infection, instead of binding with pathogen-containing vacuole directly, p62 is recruited to the vicinity of the parasitophorous vacuole in which Coxiella burnetiid resides, which is independent of UBA domain or LIR of p62. In addition, the levels of phosphorylated p62(S349) were remarkably increased throughout infection, coincident with activation of Nrf2-Keap1 pathway (Ichimura et al., 2013; Winchell et al., 2018). However, downregulating p62 seems to have no significant impact on bacterial growth in THP-1 cells, which indicates that p62 is not absolutely required for certain bacterial intracellular replication and is utilized by bacteria during their intracellular growth (Winchell et al., 2018).
p62-mediated xenophagy in intracellular bacterial infection may be affected by another autophagy adapter NDP52. Septins, a kind of conserved GTP-binding proteins, form cages around intracytosolic Shigella but not Listeria to restrict bacterial proliferation (Mostowy et al., 2010). During Shigella’s infection, p62 and NDP52 are recruited interdependently to the Shigella-containing septin cages and regulate each other’s xenophagic activity (Mostowy et al., 2011).
However, when host cells are under Listeria’s invasion, p62 and NDP52 can be recruited independently of each other. Likewise, another research suggested that though sharing similar kinetics of recruitment to Salmonella, p62 and NDP52 are recruited independently of one another to target different microdomains surrounding bacteria to facilitate xenophagy (Cemma et al., 2011).
In fact, p62 does not always play an antimicrobial role in all cases of intracellular bacterial infection. Usually accompanied by Burkholderia cenocepacia infection, cystic fibrosis (CF) is caused by mutations in the cftr gene encoding the cystic fibrosis transmembrane conductance regulator (CFTR), which usually results in a deletion of phenylalanine at position 508 (△F508) (Luciani et al., 2011). The △F508 cell’s normal processing of the CFTR protein to the epithelial cell surface is hindered, resulting in an aggresome-prone protein that forms intracellular aggregates which sequester autophagy molecule beclin1 (BECN1) and further inhibit the early stages of autophagosome formation (Deretic, 2010; Luciani et al., 2010; Luciani et al., 2011). Surprisingly, the researchers found that knockdown of p62 in △F508 macrophages increased B. cepacia’s colocalization with LC3 and inhibited the growth of bacteria, and vice versa (Abdulrahman et al., 2013). Mechanistically, downregulation of p62 liberated BECN1 from aggregates, allowing its redistribution in the cytosol and recruitment by the B. cepacia-containing vacuole and recuperating autophagy in △F508 macrophages. Furthermore, depletion of p62 enables autophagy adapter NDP52 and NBR1 to facilitate the delivery of B. cepacia to autophagosomes and inhibit the bacteria’s growth.
Moreover, while p62 has been found to target intracellular S. flexneri that are trapped in septin cages to initiate xenophagy and curb bacterial replication, recent studies also suggest that p62 can potentially enhance the proliferation of free S. flexneri that are not entrapped in septin cages (Mostowy et al., 2011; Lobato-Marquez et al., 2019). Additionally, p62 has been shown to boost the metabolic activity of intracellular Shigella not trapped in septin cages. However, the exact mechanism by which p62 promotes the metabolism of free Shigella requires more systematic exploration.
p62 may facilitate clearance of invaders through producing substances with antibacterial activity. For example, upon Mtb invaded host cells, p62 could capture certain cytosolic proteins, such as ribosomal protein rpS30 precursor FAU and ubiquitin, and then deliver them from the cytosol into conventional autophagic organelles, leading to their degradation by proteolysis into smaller peptides which possess antimicrobial properties (Ponpuak and Deretic, 2011). Subsequently, these vesicles containing small antimicrobial peptides are transported into autophagosomes in which Mtb resides and kill them, which indirectly contributes to xenophagy.
To explore a new effective pharmaceutical means to eliminate various drug-resistant intracellular bacteria, Yoon Jee Lee et al. developed and synthesized chemical p62 agonists targeting the N-degron pathway and facilitating p62-mediated xenophagy under microbes’ invasion (Lee et al., 2022). They found that these p62 agonists rescue Hela cell’s autophagic activities from suppression by S. Typhimurium. Mechanistically, to promote xenophagy targeting S. Typhimurium, the p62 agonists bind directly to the ZZ domain of p62, facilitating transportation of the pathogens to the autophagosome. Furthermore, the p62 agonists was confirmed to suppress the growth of Mtb, Gram-negative Escherichia coli and the Gram-positive Streptococcus pyogenes through xenophagy, suggesting its potential as a drug in killing a broad range of pathogenic bacteria. More encouragingly, in addition to its antimicrobial effect, the anti-inflammatory effect of p62 agonists were also observed both in vitro and in vivo. Interestingly, the antimicrobial efficacy is independent of rapamycin-modulated core autophagic pathways and is synergistic with the reduced production of inflammatory cytokines. Collectively, manual chemical activation of universally conserved p62-dependent xenophagy could be a promising treatment for infectious diseases caused by various bacterial strains. Previously, cytokine TNF-α was confirmed to stimulate the activity of p62-mediated xenophagy targeting intracytosolic Shigella and Listeria (Mostowy et al., 2011). However, whether TNF-α will affect overall autophagy flux which is required during normal metabolic activity of cell needs further investigation.
Apart from its role in eliminating bacteria through xenophagy, p62 is also pivotal in reducing inflammasome activity and mitigating acute pulmonary inflammation resulting from L. pneumophila. This is achieved through the direct binding of p62 to NLR proteins of the inflammasome, further underscoring the potential of p62 as a therapeutic target in managing the severity of Legionnaires’ disease (Ohtsuka et al., 2014).
Although much progress has been made regarding roles of p62 in host’s antibacterial mechanisms, some mysteries therein still need to be unveiled. For example, it is demonstrated that Chlamydia trachomatis infection of HeLa cells could induce p62-dependent xenophagy 24h post-infection. However, a decrease of xenophagy levels at 48h post-infection was observed in study, indicating a defect in sustained autophagy initiation with limited p62 protein after lysosomal degradation of autophagosomes (Wang et al., 2021). Moreover, downregulation of p62 did not influence the morphology and number of C. trachomatis infecting HeLa cells, suggesting that there must be other antibacterial mechanisms other than xenophagy which play significant roles in host cell clearance of C. trachomatis.
As a cytoplasmic protein, the role of p62 outside the cells is rarely investigated. A study focusing on sepsis indicated that treatment of macrophages and monocytes with lipopolysaccharide phosphorylated intracellular p62 at Ser403 and induced pyroptosis which enable passive release of p62. Furthermore, extracellular p62 binds directly with insulin receptor (INSR) on cell surface to activate downstream NF-κB-dependent metabolic programming, which mediates bacterial septic death in mice (Zhou et al., 2020). However, whether extracellular p62 could facilitate or inhibit intracellular bacterial growth still needs to be further considered and explored.
To sum up, functioning as a cytoprotective protein in most cases, p62 defends against pathogen’s invasion mainly in a xenophagy-dependent way during intracellular bacterial infection. However, some mysteries regarding roles of p62 in bacterial infection still wait to be explored, and uncovering these mysteries will help us further understand the mechanisms of bacterial infection and develop targeted and effective antibacterial drugs.
YZ, SH, and LS conceived and pursued the design of the review. YZ and LS wrote the manuscript. YZ, SH, and LS critically revised the manuscript. All authors read the manuscript. All authors substantially contributed to the discussions of content and reviewed or edited the manuscript before final submission.
The study was funded by National Natural Science Foundation of China grant 32270185, the State Key Laboratory of Pathogen and Biosecurity, Academy of Military Medical Science grant SKLPBS2132 and Science Development Project of Jilin province (20220303001SF).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbara, S., Guillemot, D., El Oualydy, S., Kos, M., Poret, C., Breant, S., et al. (2022). Antimicrobial resistance and mortality in hospitalized patients with bacteremia in the greater Paris area from 2016 to 2019. Clin. Epidemiol. 14, 1547–1560. doi: 10.2147/CLEP.S385555
Abdulrahman, B. A., Khweek, A. A., Akhter, A., Caution, K., Tazi, M., Hassan, H., et al. (2013). Depletion of the ubiquitin-binding adaptor molecule SQSTM1/p62 from macrophages harboring cftr DeltaF508 mutation improves the delivery of burkholderia cenocepacia to the autophagic machinery. J. Biol. Chem. 288, 2049–2058. doi: 10.1074/jbc.M112.411728
An, Z., Huang, X., Zheng, C., Ding, W. (2019). Acinetobacter baumannii outer membrane protein a induces HeLa cell autophagy via MAPK/JNK signaling pathway. Int. J. Med. Microbiol. 309, 97–107. doi: 10.1016/j.ijmm.2018.12.004
Berton, S., Chen, L., Liang, Y. C., Xu, Z., Afriyie-Asante, A., Rajabalee, N., et al. (2022). A selective PPM1A inhibitor activates autophagy to restrict the survival of mycobacterium tuberculosis. Cell Chem. Biol. 29, 1126–1139 e1112. doi: 10.1016/j.chembiol.2022.03.006
Bonilla, D. L., Bhattacharya, A., Sha, Y., Xu, Y., Xiang, Q., Kan, A., et al. (2013). Autophagy regulates phagocytosis by modulating the expression of scavenger receptors. Immunity 39, 537–547. doi: 10.1016/j.immuni.2013.08.026
Borgo, G. M., Burke, T. P., Tran, C. J., Lo, N. T. N., Engstrom, P., Welch, M. D. (2022). A patatin-like phospholipase mediates rickettsia parkeri escape from host membranes. Nat. Commun. 13, 3656. doi: 10.1038/s41467-022-31351-y
Boyle, K. B., Randow, F. (2013). The role of ‘eat-me’ signals and autophagy cargo receptors in innate immunity. Curr. Opin. Microbiol. 16, 339–348. doi: 10.1016/j.mib.2013.03.010
Cemma, M., Kim, P. K., Brumell, J. H. (2011). The ubiquitin-binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria-associated microdomains to target salmonella to the autophagy pathway. Autophagy 7, 341–345. doi: 10.4161/auto.7.3.14046
Chai, Q., Wang, X., Qiang, L., Zhang, Y., Ge, P., Lu, Z., et al. (2019). A mycobacterium tuberculosis surface protein recruits ubiquitin to trigger host xenophagy. Nat. Commun. 10, 1973. doi: 10.1038/s41467-019-09955-8
Cha-Molstad, H., Yu, J. E., Feng, Z., Lee, S. H., Kim, J. G., Yang, P., et al. (2017). p62/SQSTM1/Sequestosome-1 is an n-recognin of the n-end rule pathway which modulates autophagosome biogenesis. Nat. Commun. 8, 102. doi: 10.1038/s41467-017-00085-7
Deretic, V. (2010). Autophagy in infection. Curr. Opin. Cell Biol. 22, 252–262. doi: 10.1016/j.ceb.2009.12.009
Diao, J., Liu, R., Rong, Y., Zhao, M., Zhang, J., Lai, Y., et al. (2015). ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520, 563–566. doi: 10.1038/nature14147
Gal, J., Strom, A. L., Kwinter, D. M., Kilty, R., Zhang, J., Shi, P., et al. (2009). Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J. Neurochem. 111, 1062–1073. doi: 10.1111/j.1471-4159.2009.06388.x
Grant, S. S., Hung, D. T. (2013). Persistent bacterial infections, antibiotic tolerance, and the oxidative stress response. Virulence 4, 273–283. doi: 10.4161/viru.23987
Harvey, C. J., Thimmulappa, R. K., Sethi, S., Kong, X., Yarmus, L., Brown, R. H., et al. (2011). Targeting Nrf2 signaling improves bacterial clearance by alveolar macrophages in patients with COPD and in a mouse model. Sci. Transl. Med. 3, 78ra32. doi: 10.1126/scitranslmed.3002042
Heath, R. J., Goel, G., Baxt, L. A., Rush, J. S., Mohanan, V., Paulus, G. L. C., et al. (2016). RNF166 determines recruitment of adaptor proteins during antibacterial autophagy. Cell Rep. 17, 2183–2194. doi: 10.1016/j.celrep.2016.11.005
Helminiak, L., Mishra, S., Kim, H. K. (2022). Pathogenicity and virulence of rickettsia. Virulence 13, 1752–1771. doi: 10.1080/21505594.2022.2132047
Ichimura, Y., Waguri, S., Sou, Y. S., Kageyama, S., Hasegawa, J., Ishimura, R., et al. (2013). Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell 51, 618–631. doi: 10.1016/j.molcel.2013.08.003
Ishimura, R., Tanaka, K., Komatsu, M. (2014). Dissection of the role of p62/Sqstm1 in activation of Nrf2 during xenophagy. FEBS Lett. 588, 822–828. doi: 10.1016/j.febslet.2014.01.045
Jain, A., Lamark, T., Sjottem, E., Larsen, K. B., Awuh, J. A., Overvatn, A., et al. (2010). p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 285, 22576–22591. doi: 10.1074/jbc.M110.118976
Jeong, S. J., Zhang, X., Rodriguez-Velez, A., Evans, T. D., Razani, B. (2019). p62/SQSTM1 and selective autophagy in cardiometabolic diseases. Antioxid Redox Signal 31, 458–471. doi: 10.1089/ars.2018.7649
Jiang, J., Parameshwaran, K., Seibenhener, M. L., Kang, M. G., Suppiramaniam, V., Huganir, R. L., et al. (2009). AMPA receptor trafficking and synaptic plasticity require SQSTM1/p62. Hippocampus 19, 392–406. doi: 10.1002/hipo.20528
Jounai, N., Takeshita, F., Kobiyama, K., Sawano, A., Miyawaki, A., Xin, K. Q., et al. (2007). The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. U.S.A. 104, 14050–14055. doi: 10.1073/pnas.0704014104
Knodler, L. A., Celli, J. (2011). Eating the strangers within: host control of intracellular bacteria via xenophagy. Cell Microbiol. 13, 1319–1327. doi: 10.1111/j.1462-5822.2011.01632.x
Lee, Y. J., Kim, J. K., Jung, C. H., Kim, Y. J., Jung, E. J., Lee, S. H., et al. (2022). Chemical modulation of SQSTM1/p62-mediated xenophagy that targets a broad range of pathogenic bacteria. Autophagy 18 (12), 2926–2945. doi: 10.1080/15548627.2022.2054240
Lobato-Marquez, D., Krokowski, S., Sirianni, A., Larrouy-Maumus, G., Mostowy, S. (2019). A requirement for septins and the autophagy receptor p62 in the proliferation of intracellular shigella. Cytoskeleton (Hoboken) 76, 163–172. doi: 10.1002/cm.21453
Luciani, A., Villella, V. R., Esposito, S., Brunetti-Pierri, N., Medina, D., Settembre, C., et al. (2010). Defective CFTR induces aggresome formation and lung inflammation in cystic fibrosis through ROS-mediated autophagy inhibition. Nat. Cell Biol. 12, 863–875. doi: 10.1038/ncb2090
Luciani, A., Villella, V. R., Esposito, S., Brunetti-Pierri, N., Medina, D. L., Settembre, C., et al. (2011). Cystic fibrosis: a disorder with defective autophagy. Autophagy 7, 104–106. doi: 10.4161/auto.7.1.13987
Ma, S., Attarwala, I. Y., Xie, X. Q. (2019). SQSTM1/p62: a potential target for neurodegenerative disease. ACS Chem. Neurosci. 10, 2094–2114. doi: 10.1021/acschemneuro.8b00516
Matsumoto, G., Wada, K., Okuno, M., Kurosawa, M., Nukina, N. (2011). Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol. Cell 44, 279–289. doi: 10.1016/j.molcel.2011.07.039
Mesquita, F. S., Thomas, M., Sachse, M., Santos, A. J., Figueira, R., Holden, D. W. (2012). The salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PloS Pathog. 8, e1002743. doi: 10.1371/journal.ppat.1002743
Min, Y., Kim, M. J., Lee, S., Chun, E., Lee, K. Y. (2018). Inhibition of TRAF6 ubiquitin-ligase activity by PRDX1 leads to inhibition of NFKB activation and autophagy activation. Autophagy 14, 1347–1358. doi: 10.1080/15548627.2018.1474995
Mostowy, S., Bonazzi, M., Hamon, M. A., Tham, T. N., Mallet, A., Lelek, M., et al. (2010). Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe 8, 433–444. doi: 10.1016/j.chom.2010.10.009
Mostowy, S., Sancho-Shimizu, V., Hamon, M. A., Simeone, R., Brosch, R., Johansen, T., et al. (2011). p62 and NDP52 proteins target intracytosolic shigella and listeria to different autophagy pathways. J. Biol. Chem. 286, 26987–26995. doi: 10.1074/jbc.M111.223610
Nakamura, K., Kimple, A. J., Siderovski, D. P., Johnson, G. L. (2010). PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NF-kappaB activation. J. Biol. Chem. 285, 2077–2089. doi: 10.1074/jbc.M109.065102
Ohtsuka, S., Ishii, Y., Matsuyama, M., Ano, S., Morishima, Y., Yanagawa, T., et al. (2014). SQSTM1/p62/A170 regulates the severity of legionella pneumophila pneumonia by modulating inflammasome activity. Eur. J. Immunol. 44, 1084–1092. doi: 10.1002/eji.201344091
Omotade, T. O., Roy, C. R. (2020). Legionella pneumophila excludes autophagy adaptors from the ubiquitin-labeled vacuole in which it resides. Infect. Immun. 88 (8), e00793–19. doi: 10.1128/IAI.00793-19
Otten, E. G., Werner, E., Crespillo-Casado, A., Boyle, K. B., Dharamdasani, V., Pathe, C., et al. (2021). Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. Nature 594, 111–116. doi: 10.1038/s41586-021-03566-4
Pankiv, S., Clausen, T. H., Lamark, T., Brech, A., Bruun, J. A., Outzen, H., et al. (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145. doi: 10.1074/jbc.M702824200
Park, I., Chung, J., Walsh, C. T., Yun, Y., Strominger, J. L., Shin, J. (1995). Phosphotyrosine-independent binding of a 62-kDa protein to the src homology 2 (SH2) domain of p56lck and its regulation by phosphorylation of ser-59 in the lck unique n-terminal region. Proc. Natl. Acad. Sci. U.S.A. 92, 12338–12342. doi: 10.1073/pnas.92.26.12338
Pilli, M., Arko-Mensah, J., Ponpuak, M., Roberts, E., Master, S., Mandell, M. A., et al. (2012). TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity 37, 223–234. doi: 10.1016/j.immuni.2012.04.015
Ponpuak, M., Deretic, V. (2011). Autophagy and p62/sequestosome 1 generate neo-antimicrobial peptides (cryptides) from cytosolic proteins. Autophagy 7, 336–337. doi: 10.4161/auto.7.3.14500
Rex, J. H., Talbot, G. H., Goldberger, M. J., Eisenstein, B. I., Echols, R. M., Tomayko, J. F., et al. (2017). Progress in the fight against multidrug-resistant bacteria 2005-2016: modern noninferiority trial designs enable antibiotic development in advance of epidemic bacterial resistance. Clin. Infect. Dis. 65, 141–146. doi: 10.1093/cid/cix246
Seto, S., Tsujimura, K., Horii, T., Koide, Y. (2013). Autophagy adaptor protein p62/SQSTM1 and autophagy-related gene Atg5 mediate autophagosome formation in response to mycobacterium tuberculosis infection in dendritic cells. PloS One 8, e86017. doi: 10.1371/journal.pone.0086017
Shen, Q., Shi, Y., Liu, J., Su, H., Huang, J., Zhang, Y., et al. (2021). Acetylation of STX17 (syntaxin 17) controls autophagosome maturation. Autophagy 17, 1157–1169. doi: 10.1080/15548627.2020.1752471
Shizukuishi, S., Ogawa, M., Matsunaga, S., Tomokiyo, M., Ikebe, T., Fushinobu, S., et al. (2020). Streptococcus pneumoniae hijacks host autophagy by deploying CbpC as a decoy for Atg14 depletion. EMBO Rep. 21, e49232. doi: 10.15252/embr.201949232
Tang, J., Li, Y., Xia, S., Li, J., Yang, Q., Ding, K., et al. (2021). Sequestosome 1/p62: a multitasker in the regulation of malignant tumor aggression (Review). Int. J. Oncol. 59 (4), 77. doi: 10.3892/ijo.2021.5257
Tripathi-Giesgen, I., Behrends, C., Alpi, A. F. (2021). The ubiquitin ligation machinery in the defense against bacterial pathogens. EMBO Rep. 22, e52864. doi: 10.15252/embr.202152864
Wang, F., Zhang, H., Lu, X., Zhu, Q., Shi, T., Lu, R., et al. (2021). Chlamydia trachomatis induces autophagy by p62 in HeLa cell. World J. Microbiol. Biotechnol. 37, 50. doi: 10.1007/s11274-021-03014-5
Wang, Y., Zhang, K., Shi, X., Wang, C., Wang, F., Fan, J., et al. (2016). Critical role of bacterial isochorismatase in the autophagic process induced by acinetobacter baumannii in mammalian cells. FASEB J. 30, 3563–3577. doi: 10.1096/fj.201500019R
Winchell, C. G., Dragan, A. L., Brann, K. R., Onyilagha, F. I., Kurten, R. C., Voth, D. E. (2018). Coxiella burnetii subverts p62/Sequestosome 1 and activates Nrf2 signaling in human macrophages. Infect. Immun. 86 (5), e00608–17. doi: 10.1128/IAI.00608-17
Xu, Y., Jagannath, C., Liu, X. D., Sharafkhaneh, A., Kolodziejska, K. E., Eissa, N. T. (2007). Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 27, 135–144. doi: 10.1016/j.immuni.2007.05.022
Zhang, Z., Costa, M. (2021). p62 functions as a signal hub in metal carcinogenesis. Semin. Cancer Biol. 76, 267–278. doi: 10.1016/j.semcancer.2021.04.014
Keywords: intracellular bacterial infection, p62 (sequestosome 1(SQSTM1), xenophagy, bacterial protein, microdomain
Citation: Zhou Y, Hua S and Song L (2023) The versatile defender: exploring the multifaceted role of p62 in intracellular bacterial infection. Front. Cell. Infect. Microbiol. 13:1180708. doi: 10.3389/fcimb.2023.1180708
Received: 06 March 2023; Accepted: 25 April 2023;
Published: 05 May 2023.
Edited by:
Jun Jiao, Beijing Institute of Microbiology and Epidemiology, ChinaReviewed by:
Erin J. Van Schaik, Texas A&M University, United StatesCopyright © 2023 Zhou, Hua and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Song, bHNvbmdAamx1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.