- 1National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention, Shanghai, China
- 2National Center for International Research on Tropical Diseases, Shanghai, China
- 3National Health Commission (NHC) Key Laboratory of Parasite and Vector Biology (National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention), Shanghai, China
- 4World Health Organization (WHO) Collaborating Centre for Tropical Diseases, National Center for International Research on Tropical Diseases, Shanghai, China
- 5Shanghai Municipal Center for Disease Control and Prevention, Shanghai, China
China has now achieved the elimination of malaria, but it still faces severe challenges in the post-elimination stage. China continues to be plagued by imported malaria cases, and preventing re-transmission of imported malaria is critical. The effectiveness of antimalarial drugs for malaria control largely depends on the study of drug resistance markers in vitro. Monitoring molecular markers of parasite-associated drug resistance can help predict and manage drug resistance. There is currently a lack of systematic reviews of molecular markers for indigenous and imported malaria in China. Therefore, this review summarizes the published articles related to molecular marker polymorphism of indigenous and imported malaria cases in China in the past two decades, to study the mutation frequency and distribution of crt, mdr1, dhps, dhfr and K13 gene resistance-related loci. This can provide a whole picture of molecular markers and the resistance mutations of imported cases in China, which has certain significance for drug resistance surveillance planning, safe and effective treatment, and preventing the recurrence of local transmission by imported malaria in China in the future.
Introduction
Malaria is one of the most devastating infectious diseases. It imposes a severe burden on developing countries, especially in Southeast Asia and Africa. Of the five species of Plasmodium that infect humans, Plasmodium falciparum and Plasmodium vivax have the highest infection and morbidity rates. According to the 2022 World Health Organization (WHO) World Malaria Report, there are an estimated 247 million malaria cases globally in 2021, with an increase from 245 million in 2020 (WHO, 2022). The increase in malaria cases has been influenced by the disruption of services during the COVID-19 pandemic. Malaria deaths are estimated at 619,000 in 2021, a slight decrease compared to 2020 (WHO, 2022).
The widespread resistance of parasites to antimalarial drugs has attracted great attention in recent years. The emergence of parasite resistance has severely hindered the progress of malaria elimination. Resistance to chloroquine (CQ) in P. falciparum had emerged in Thailand as early as the late 1950s, and over time the drug was no longer effective (Payne, 1987). In the 1960s and 1970s, these resistant strains spread steadily through South America, Southeast Asia and India (Liu et al., 1995; Wellems and Plowe, 2001). Resistance also generally emerged in the late 1970s following the entry of sulfadoxine-pyrimethamine (SP) as an alternative treatment for falciparum malaria (Suebsaeng et al., 1986). Malaria treatment has been plagued by recurring resistance in parasites until the advent of artemisinin. Artemisinin-based combination therapies (ACTs) were introduced in the 1990s and are now used to treat uncomplicated malaria (Fairhurst and Dondorp, 2016). With the widespread use of ACTs in malaria-endemic countries and the increase in insecticide-treated bed nets, malaria morbidity and mortality have decreased significantly globally (Bhattarai et al., 2007). However, artemisinin resistance was first reported in western Cambodia in 2008, and the emergence of resistance resulted in lower parasite clearance rates (Noedl et al., 2008; Dondorp et al., 2009; Ashley et al., 2014). Since then, artemisinin-resistant parasites have spread rapidly across Southeast Asia (Phyo et al., 2012; Kyaw et al., 2013). Recent studies have confirmed the emergence of artemisinin partial resistance in several areas of Africa, notably in Rwanda, Guyana and Papua New Guinea (Maïga-Ascofaré and May, 2016; Uwimana et al., 2021). In the absence of effective antimalarial drugs, the emergence of artemisinin resistance in Africa is likely to lead to a repeat of high mortality rates and this needs to be addressed through resistance surveillance. At present, P. falciparum is resistant to almost all antimalarial drugs. Meanwhile, P. vivax also developed resistance to chloroquine and primaquine in Southeast Asia during the 1990s (Murphy et al., 1993; Looareesuwan et al., 1997).
China has also suffered from malaria since ancient times. After long-term unremitting struggle and efforts, China reported zero indigenous cases in 2017 (Feng et al., 2018). Although WHO certified China as a malaria-free country in 2021, China is still plagued by imported malaria cases (Liu, 2014; WHO, 2022). In the context of current antimalarial drug resistance globally, the threat of imported drug-resistant parasites must be monitored and addressed timely. The effectiveness of antimalarial drugs for malaria control relies heavily on the study of resistance markers in vitro, and monitoring parasite-associated molecular markers of resistance can help predict and manage drug resistance (Fairhurst and Dondorp, 2016; Haldar et al., 2018). This review summarizes the published articles related to molecular marker polymorphism of indigenous and imported malaria cases in China in the past two decades, to study the mutation frequency and distribution of resistance-related loci. This can provide better understanding of antimalarial drug resistance marker surveillance in China, which has certain significance for the surveillance and management of imported cases and preventing re-transmission in the future.
Study selection method
PubMed and CNKI databases were searched for peer-reviewed articles from China published between 1 January 2001 and 1 May 2022, that had genotyped the crt, mdr1, dhfr, dhps and pfK13 genes of P. falciparum or P. vivax (Figure 1). The following search terms were used: “((malaria OR falciparum OR vivax) AND (molecular marker OR crt OR mdr1 OR dhfr OR dhps OR K13)) AND (China OR China-Myanmar border) AND ((“2001/01/01” [Date - Publication]: “2022/05/01” [Date - Publication]))”. Studies related to parasites in the last two decades were screened and those based on transgenic parasites, books and modelling studies were excluded. In addition, the following information was extracted from the studies: the year samples were collected, study area, source of imported cases, sample size, and gene polymorphism.
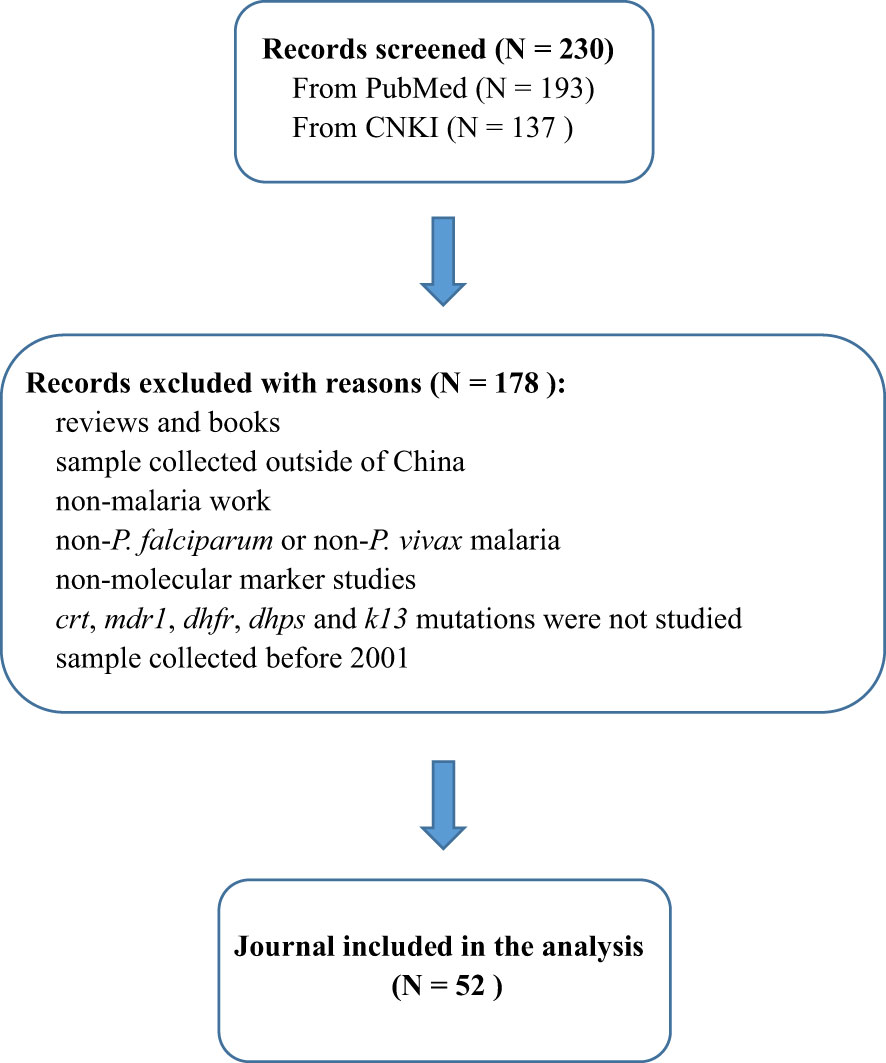
Figure 1 Flow diagram of study selection. The figure shows the number of manually screened and excluded publications identified in PubMed and CNKI, and finally screened the 52 publications counted in this study.
Results
Literature screening results
According to the search conditions, a total of 230 literatures were obtained from PubMed and CNKI databases. Reviews and books, non-malaria studies and molecular marker studies were excluded. According to the abstract and results of the literature, studies containing non-P. falciparum or non-P. vivax, non-crt gene, non-mdr1 gene, non-dhfr gene, non-dhps gene, non-K13 gene and studies with samples collected before 2001 were all excluded. Finally, 52 literatures were included in this study (Figure 1). According to statistics, the total number of samples included in the study was 17,754, and its time span was from 2001 to 2019.
The distribution of samples
Among all samples, indigenous samples (76.7%) were collected from 2001 to 2014 and imported samples (23.3%) were collected from 2004 to 2019. The collection time of these samples was not continuous, and there were gaps in several years, which may be due to the time lag of sample research or the lack of research in this area. The proportion of imported samples was significantly higher than that of indigenous samples among different gene groups (P < 0.05) (Figure 2). For more than 63.5% of the 52 studies, the years of sample collection were from 2012 to 2019 (Supplementary Tables 1–8). More than 90% of the imported P. falciparum malaria samples in China were mainly imported from Africa, whereas all imported P. vivax cases in China came from Southeast Asia, especially Myanmar.
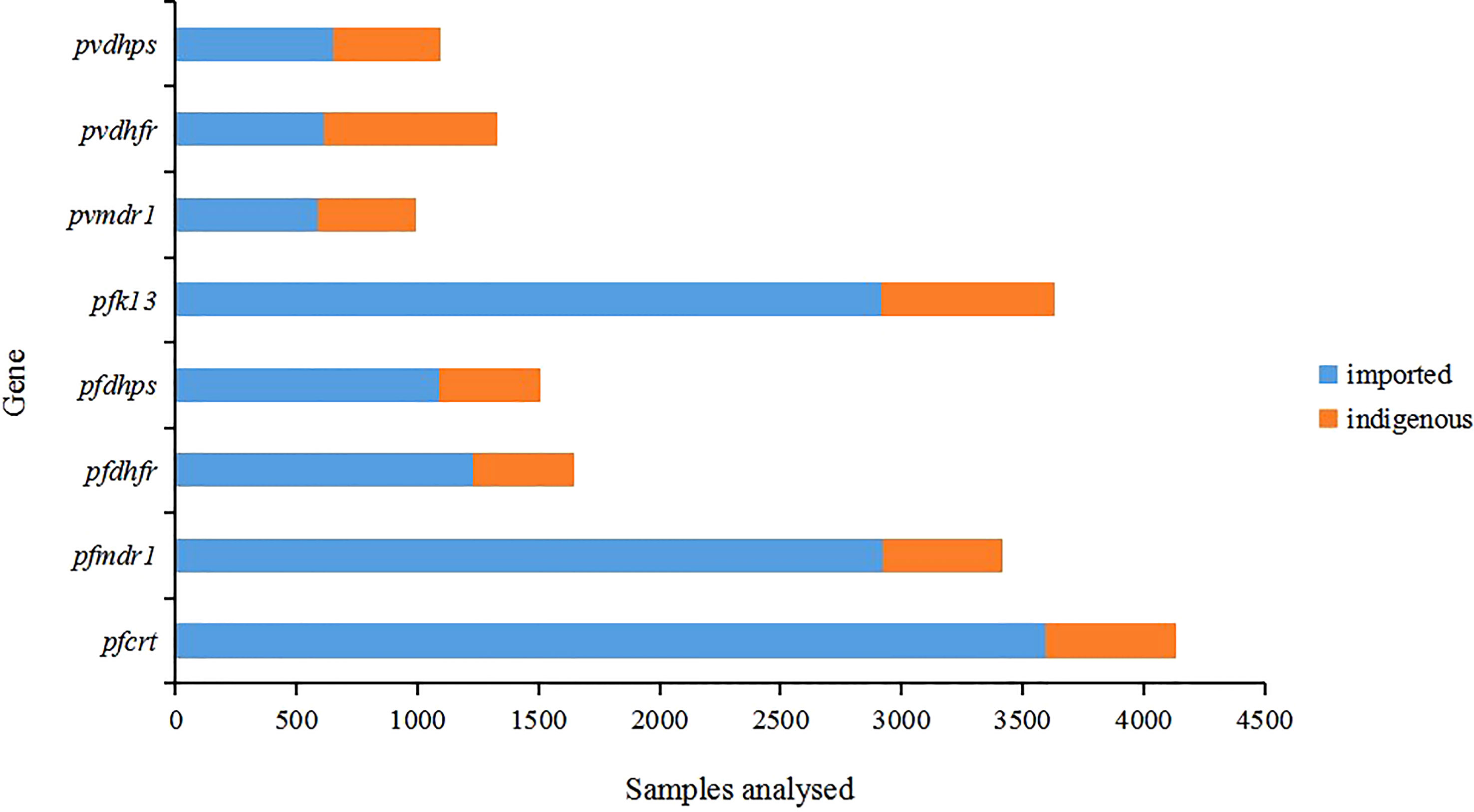
Figure 2 Distribution of samples by gene. All samples (n=17754) were from published studies (n=52). The indigenous cases were collected from 2001 to 2014 and the imported cases were collected from 2004 to 2019.
Molecular marker of resistance
In P. falciparum related resistance genes, the pfcrt gene of indigenous samples had the highest mutation frequency of K76T (87.1%), while M74I/T, N75E/D/K and A220S mutations were more common (37.5%, 37.3% and 35%, respectively) (Figure 3). Only K76T mutation was detected in samples from 2001 to 2007. In imported samples, mutations frequency at sites 74, 75 and 76 were similar (32.6%, 32.2% and 34.8%, respectively) and usually mutated simultaneously. In indigenous samples, the most common mutations of pfmdr1 gene were N86Y and Y184F, accounting for 14% and 12.6%, respectively. The N1042D mutation of pfmdr1 was detected in some samples, but the proportion was very low (only 0.8%) (Supplementary Table 2). Similar to the indigenous samples, the most common mutation of pfmdr1 in the imported samples were N86Y and Y184F (22.7% and 34.7%, respectively) (Figure 3). The difference was that the mutation of N1042D was not detected in the imported samples, but the D1246Y mutation of pfmdr1 was detected in the imported African samples, which was not detected in the indigenous samples. In indigenous samples, the mutations in N51I, C59R, S108N and I164L of pfdhfr were all over 50%, and C59R had the highest mutation frequency (92.4%). In imported samples, the mutation frequency of N51I, C59R and S108N in pfdhfr gene were high, accounting for 92.3%, 84.9% and 97.1%, respectively (Figure 3). The difference was that the mutation frequency of I164L in indigenous samples (69.5%) was significantly higher than that in imported samples (0.24%) (P < 0.05). For pfdhps, the highest mutation frequency was K540E/N (78.4%) in indigenous samples and A437G (75.7%) in imported samples. Mutations of I431V and A613S were detected in a small part of the imported samples. For pfK13, F446I had the highest mutation frequency, accounting for 31.4% of indigenous samples and 3.7% of imported samples (Figure 3).
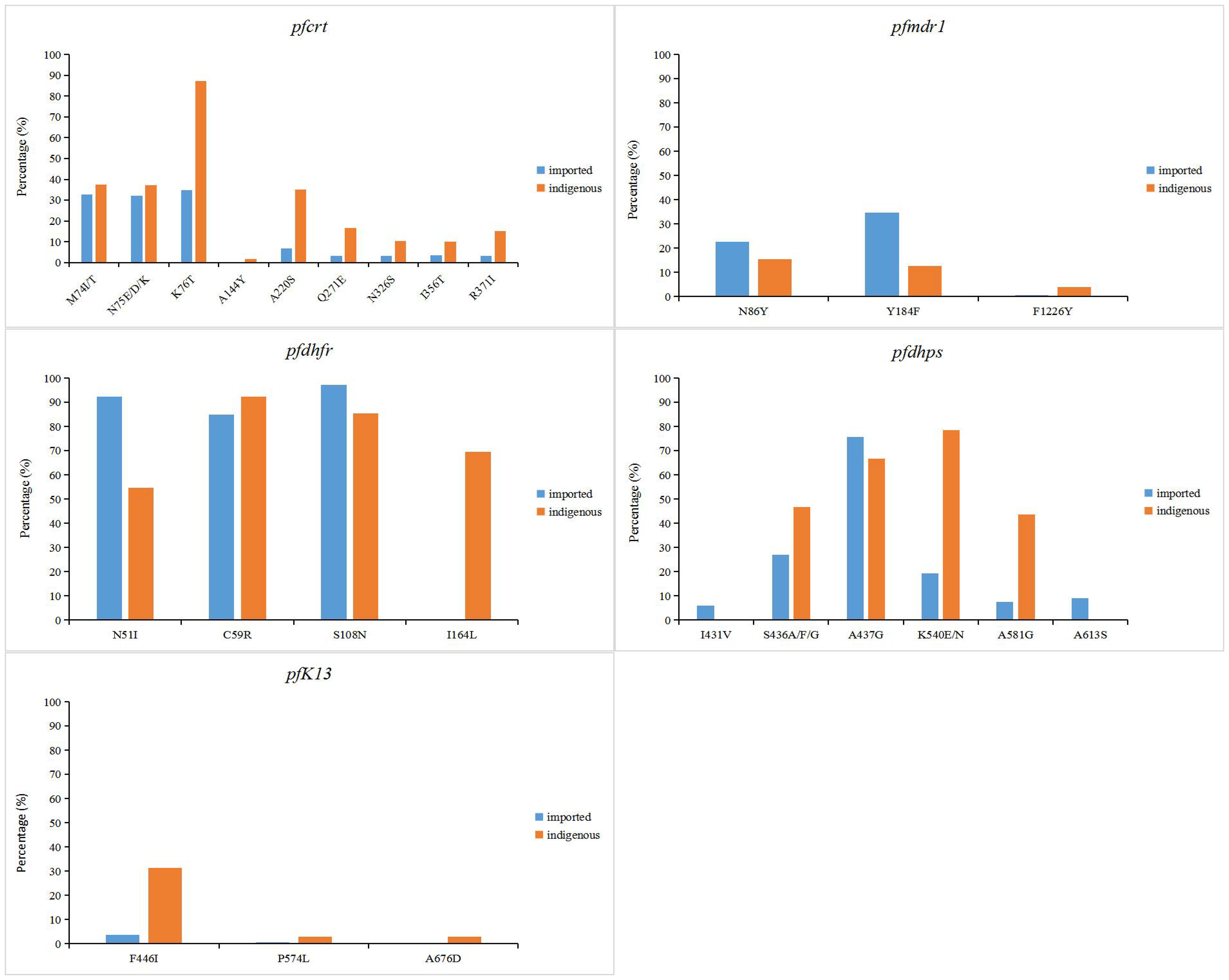
Figure 3 Point mutation frequencies of pfcrt, pfmdr1, pfdhfr, pfdhps and pfK13 genes. The mutation sites with frequencies greater than 1% were shown in the figure. The indigenous cases of P. falciparum were collected from 2001 to 2014, while the imported cases were collected from 2007 to 2019.
In P. vivax related resistance genes, the mutation frequency of T958M and F1076L of pvmdr1 gene was the highest in indigenous samples, which were 71.6% and 75.6%, respectively (Figure 4). Different from the indigenous samples, in addition to T958M (74.1%) and F1076L (64.9%), the mutation of pvmdr1 gene was also higher at G698S (62%) and M908L (60.8%) in imported samples. In indigenous samples, the mutation frequency of S58R, T61M and S117N/T were high in pvdhfr gene, which were 44.4%, 40.1% and 67.3%, respectively, and the mutation frequency of A383G site in pvdhps gene was the highest (61.5%) (Figure 4). The highest mutation frequency of pvdhfr gene in imported samples was S58R (58.6%) and the mutation rate of A383G site in pvdhps gene was the highest (71.3%).
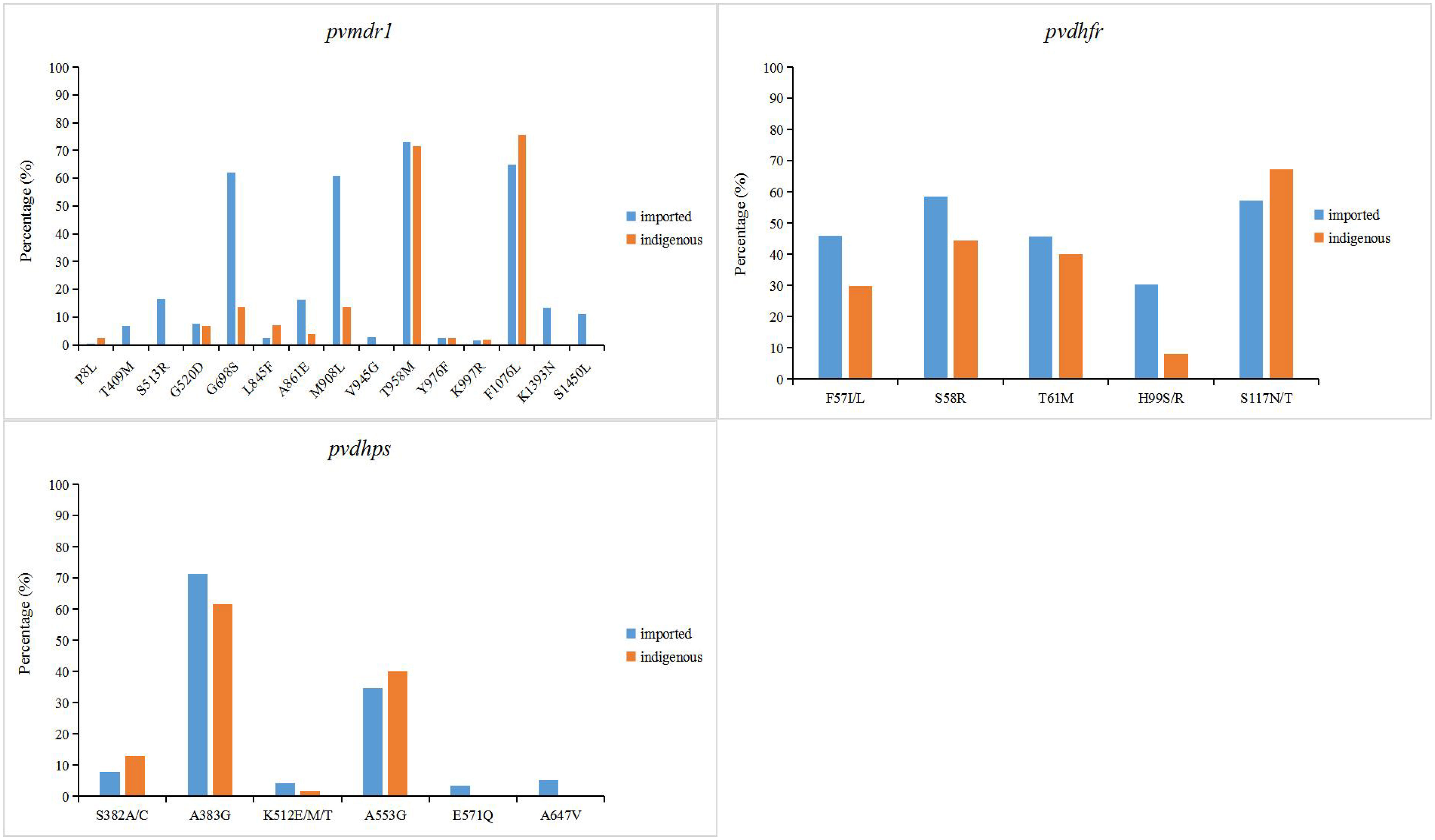
Figure 4 Point mutation frequencies of pvmdr1, pvdhfr and pvdhps genes. The mutation sites with frequencies greater than 1% were shown in the figure. The indigenous cases of P. vivax were collected from 2006 to 2012, while the imported cases were collected from 2004 to 2019.
Based on the collected results, the period from 2001 to 2012 was defined as the early stage of the study, and the period from 2012 to 2019 as the late stage. In indigenous samples, pfcrt, pfmdr1, pfdhfr, pfdhps, pvdhfr, and pvdhps genes in the early stage were not significantly different from those in the late stage (P >0.05). Different from the late stage, mutations at N11Y, K189T, R225K, E252Q, I352T and P441L were newly found in less than five samples, but F446I was still the main mutation in pfk13. In the pvmdr1 gene, mutations at P8L, G520D, G698S, L845F, A861E and M908L were newly discovered in the early stage, which were not found in the late stage. In the imported malaria samples, the mutation of each gene in the early stage showed no significant trend than the later stage (P >0.05).
Discussion and conclusion
The study of genetic molecular markers of drug resistance can provide a better understanding of drug action and drug resistance mechanisms that are critical to achieving malaria treatment and control of transmission. The mutation of P. falciparum chloroquine resistance transporter gene (pfcrt) K76T has been confirmed to be closely related to CQ resistance, and those P. falciparum carrying the K76T mutation are generally accompanied by M74I, N75E and A220S mutations (Babiker et al., 2001; Djimdé et al., 2001; Sa and Twu, 2010). This phenomenon was also shown in the collection of indigenous and imported malaria samples in China, with a trend toward higher mutation frequency at sites 74-76 of pfcrt gene (Figure 3). In addition, mutations at loci Q271E, N326S, I356T, and R371I were found in which reported in the border area of Yingjiang County in Yunnan Province and Kachin area in Myanmar, which may be due to the frequent migration of populations in border areas to spread parasite drug resistance (Yang et al., 2007; Carrara et al., 2013; Wang et al., 2020; Si et al., 2021). P. falciparum multidrug resistance 1 (pfmdr1) genes are associated with resistance to CQ, mefloquine, quinine and artemisinin in P. falciparum, especially the N86Y allele (Sidhu et al., 2006; Feng et al., 2015; Gil and Krishna, 2017; Muiruri et al., 2018). From the data collected, mutations at the N86Y and Y184F sites of pfmdr1 were common, mutations in Y184F and N86Y occurred simultaneously in most samples. (Figure 3). The F1226Y mutation was detected in only a few samples, studies confirmed that parasites with the F1226Y mutation increases parasite resistance to quinine and lumefantrine (Wang et al., 2020). Notably, the N1042D mutation was detected in the indigenous samples and studies have found that parasites with the N1042D mutation have increased susceptibility to pyronaridine (Bai et al., 2018), but the D1246Y mutation was detected in the imported African samples, which indicated the geographical differences in the production of malaria resistance (Xu et al., 2018; Zhou et al., 2019; She et al., 2020; Huang et al., 2021; Zhao et al., 2021).Mutations in dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) are associated with parasite resistance to SP drugs (Triglia et al., 1997). The study found that mutations at N51I, C59R, S108N, and I164L of pfdhfr gene were associated with pyrimethamine resistance (Triglia et al., 1997; Happi et al., 2005). And mutations at S436A, A437G, K540E, A581G and A613S of pfdhp gene were associated with sulfadoxine resistance (Kublin et al., 2002; Berglez et al., 2004; Pearce et al., 2009). The mutation frequency of these sites of pfdhfr and pfdhp, which we collected, was high. This phenomenon is similar to studies in Myanmar, Thailand, and Cambodia in Southeast Asia (Anderson et al., 2005; Khim et al., 2005; Huang et al., 2012). It is estimated that P. vivax, which is endemic in Southeast Asia, has shown resistance to SP (Figure 3). Different from the indigenous samples imported samples of falciparum malaria from Africa have high mutation frequency (>80%) in N51I, C59R, and S108N of pfdhfr, and have lower mutation frequency at I164L (Figure 3). It can indicate that indigenous parasites may have developed high pyrimidine resistance. Therefore, close monitoring of the associated resistance to imported malaria in Africa is critical. The kelch13 (K13) propeller domain of P. falciparum has been shown to be associated with artemisinin resistance (Cheeseman et al., 2012; Miotto et al., 2013). It has been confirmed that the F446I, N458Y, M476I, Y493H, R539T, I543T, P553L, R561H and C580Y site mutations of the pfK13 gene are closely related to artemisinin resistance (Huang et al., 2015; WHO, 2018; Feng et al., 2019). The F446I mutation of the pfK13 gene was found to be predominant in the border areas of Myanmar and Yunnan Province of China (Supplementary Table 8).And the mutations of A578S and P574L loci of pfK13 were more common in African imported samples (Supplementary Table 8). Although zero indigenous cases have been reported in China since 2017, cross-border malaria transmission due to Anopheles mosquitoes in the China-Myanmar border area and human importation from malaria-endemic areas in Southeast Asia and Africa make malaria management and drug resistance monitoring difficult (Feng et al., 2019).
.The proportion of imported vivax malaria cases has increased since the absence of indigenous cases in China, and there is still a risk of transmission (Zhang et al., 2022). P. vivax is the geographically most widespread malaria parasite and imposes a severe burden on global public health (Mueller et al., 2009; Howes et al., 2016). From the data collected, imported samples of vivax malaria in recent years came from Southeast Asia, especially Myanmar, where the malaria burden is the heaviest (Supplementary Tables 5–7). China’s Yunnan Province borders Myanmar, so there is a high risk of cross-border transmission of mosquito vectors, which still requires high attention. Due to the limitation of in vitro culture of P. vivax, the research of drug resistance molecular markers is full of challenges, so the research mainly focuses on the homologous molecular markers with P. falciparum (Wang et al., 2022). P. vivax multidrug resistance gene (pvmdr1), homologous to pfmdr1, has been identified as a possible genetic molecular marker of CQ resistance (Brega et al., 2005; Suwanarusk et al., 2007). The frequency of M908L (41.8%) or T958M (72.5%) mutations in the pvmdr1 gene was higher in imported vivax samples of China (Figure 4). It is worth noting that mutations at the M908L, T958M, Y976F and F1076L sites have been confirmed to be closely related to CQ resistance (Brega et al., 2005; Chehuan et al., 2013; Li et al., 2020). Interestingly, the mutation frequencies of Y976F (2.4%) and F1076L (69.2%) in the China-Myanmar border area of imported P. vivax samples were significantly different (P<0.05) (Figure 4). This finding is consistent with studies in the Myanmar region, where Y976F is also rare (Nyunt et al., 2017), but these results are quite different from studies in the Thai-Cambodian border region, where the incidence of Y976F is high (Tantiamornkul et al., 2018). The resistance of P. vivax to SP has been shown to be caused by mutations in two genes, dihydrofolate reductase (pvdhfr) and dihydropteroate synthase (pvdhps) (Tjitra et al., 2002; Korsinczky et al., 2004). Among them, the mutations of pvdhfr codons 57, 58, 117, 173 and pvdhps codons 382, 383 and 553 were found to be associated with pyrimethamine and sulfadoxine resistance (Imwong et al., 2001; Imwong et al., 2003; Korsinczky et al., 2004; Huang et al., 2022). The pvdhfr gene with 57 site (36.3%) mutation mainly occurred in the border area between China and Myanmar, which was an imported case from Myanmar. The mutation at locus 117 of pvdhfr gene mainly occurred in the Myanmar, and the point mutation frequency of loci 58 and 117 were high, which were 51.0% and 62.7%, respectively (Figure 4). Almost all of the collected data carried at least one of the two mutations, 383 and 553, indicating that almost all of these parasites had developed resistance to sulfadoxine (Figure 4).
This study found that there was a lack of sample collection and testing in some years, which may be due to fewer studies in this period or delayed reporting. Most of the study period was from 2012 to 2019. Therefore, no significant trend in mutation sites of each gene has been observed over time. In addition, it was found that some mutation sites (especially pfK13 gene) were detected in a small number of cases of these genes studied in this study. Due to the small number of detected mutations, it is necessary to continue to detect whether they have an impact on drug resistance in the future. At present, China has been eliminated malaria, but the risk of imported malaria continues to exist globally, especially resistant malaria in Southeast Asia and Africa. Therefore, surveillance for susceptibility of imported malaria to commonly used antimalarial drugs in China (including increased monitoring of the piperaquine resistance gene related gene pfpm2-3) should be integrated into the routine case surveillance in all provinces, which can provide complete evidence and address the risk of resistance to antimalarial drugs in time to ensure that treatment effects to prevent malaria retransmission.
Author contributions
SW collected and analyzed the data. ZX revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Bill & Melinda Gates Foundation (No. INV-018913).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1167220/full#supplementary-material
References
Anderson, T. J., Nair, S., Sudimack, D., Williams, J. T., Mayxay, M., Newton, P. N., et al. (2005). Geographical distribution of selected and putatively neutral SNPs in southeast Asian malaria parasites. Mol. Biol. Evol. 22, 2362–2374. doi: 10.1093/molbev/msi235
Ashley, E. A., Dhorda, M., Fairhurst, R. M., Amaratunga, C., Lim, P., Suon, S., et al. (2014). Spread of artemisinin resistance in plasmodium falciparum malaria. N Engl. J. Med. 371, 411–423. doi: 10.1056/NEJMoa1314981
Babiker, H. A., Pringle, S. J., Abdel-Muhsin, A., Mackinnon, M., Hunt, P., Walliker, D. (2001). High-level chloroquine resistance in Sudanese isolates of plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene pfcrt and the multidrug resistance gene pfmdr1. J. Infect. Dis. 183, 1535–1538. doi: 10.1086/320195
Bai, Y., Zhang, J., Geng, J., Xu, S., Deng, S., Zeng, W., et al. (2018). Longitudinal surveillance of drug resistance in plasmodium falciparum isolates from the China-Myanmar border reveals persistent circulation of multidrug resistant parasites. Int. J. Parasitol. Drugs Drug Resist. 8, 320–328. doi: 10.1016/j.ijpddr.2018.05.003
Berglez, J., Iliades, P., Sirawaraporn, W., Coloe, P., Macreadie, I. (2004). Analysis in escherichia coli of plasmodium falciparum dihydropteroate synthase (DHPS) alleles implicated in resistance to sulfadoxine. Int. J. Parasitol. 34, 95–100. doi: 10.1016/j.ijpara.2003.09.009
Bhattarai, A., Ali, A. S., Kachur, S. P., Mårtensson, A., Abbas, A. K., Khatib, R., et al. (2007). Impact of artemisinin-based combination therapy and insecticide-treated nets on malaria burden in Zanzibar. PloS Med. 4, e309. doi: 10.1371/journal.pmed.0040309
Brega, S., Meslin, B., De Monbrison, F., Severini, C., Gradoni, L., Udomsangpetch, R., et al. (2005). Identification of the plasmodium vivax mdr-like gene (pvmdr1) and analysis of single-nucleotide polymorphisms among isolates from different areas of endemicity. J. Infect. Dis. 191, 272–277. doi: 10.1086/426830
Carrara, V. I., Lwin, K. M., Phyo, A. P., Ashley, E., Wiladphaingern, J., Sriprawat, K., et al. (2013). Malaria burden and artemisinin resistance in the mobile and migrant population on the Thai-Myanmar border 1999-2011: an observational study. PloS Med. 10, e1001398. doi: 10.1371/journal.pmed.1001398
Cheeseman, I. H., Miller, B. A., Nair, S., Nkhoma, S., Tan, A., Tan, J. C., et al. (2012). A major genome region underlying artemisinin resistance in malaria. Science 336, 79–82. doi: 10.1126/science.1215966
Chehuan, Y. F., Costa, M. R., Costa, J. S., Alecrim, M. G., Nogueira, F., Silveira, H., et al. (2013). In vitro chloroquine resistance for plasmodium vivax isolates from the Western Brazilian Amazon. Malar J. 12, 226. doi: 10.1186/1475-2875-12-226
Djimdé, A., Doumbo, O. K., Cortese, J. F., Kayentao, K., Doumbo, S., Diourté, Y., et al. (2001). A molecular marker for chloroquine-resistant falciparum malaria. N Engl. J. Med. 344, 257–263. doi: 10.1056/NEJM200101253440403
Dondorp, A. M., Nosten, F., Yi, P., Das, D., Phyo, A. P., Tarning, J., et al. (2009). Artemisinin resistance in plasmodium falciparum malaria. N Engl. J. Med. 361, 455–467. doi: 10.1056/NEJMoa0808859
Fairhurst, R. M., Dondorp, A. M. (2016). Artemisinin-resistant plasmodium falciparum malaria. Microbiol. Spectr. 4 (3), 10.1128. doi: 10.1128/9781555819453.ch22
Feng, J., Kong, X., Xu, D., Yan, H., Zhou, H., Tu, H., et al. (2019). Investigation and evaluation of genetic diversity of plasmodium falciparum kelch 13 polymorphisms imported from southeast Asia and Africa in southern China. Front. Public Health 7, 95. doi: 10.3389/fpubh.2019.00095
Feng, J., Zhang, L., Huang, F., Yin, J. H., Tu, H., Xia, Z. G., et al. (2018). Ready for malaria elimination: zero indigenous case reported in the people’s republic of China. Malar J. 17, 315. doi: 10.1186/s12936-018-2444-9
Feng, J., Zhou, D., Lin, Y., Xiao, H., Yan, H., Xia, Z. (2015). Amplification of pfmdr1, pfcrt, pvmdr1, and K13 propeller polymorphisms associated with plasmodium falciparum and plasmodium vivax isolates from the China-Myanmar border. Antimicrob. Agents Chemother. 59, 2554–2559. doi: 10.1128/AAC.04843-14
Gil, J. P., Krishna, S. (2017). pfmdr1 (Plasmodium falciparum multidrug drug resistance gene 1): a pivotal factor in malaria resistance to artemisinin combination therapies. Expert Rev. Anti Infect. Ther. 15, 527–543. doi: 10.1080/14787210.2017.1313703
Haldar, K., Bhattacharjee, S., Safeukui, I. (2018). Drug resistance in plasmodium. Nat. Rev. Microbiol. 16, 156–170. doi: 10.1038/nrmicro.2017.161
Happi, C. T., Gbotosho, G. O., Folarin, O. A., Akinboye, D. O., Yusuf, B. O., Ebong, O. O., et al. (2005). Polymorphisms in plasmodium falciparum dhfr and dhps genes and age related in vivo sulfadoxine-pyrimethamine resistance in malaria-infected patients from Nigeria. Acta Trop. 95, 183–193. doi: 10.1016/j.actatropica.2005.06.015
Howes, R. E., Battle, K. E., Mendis, K. N., Smith, D. L., Cibulskis, R. E., Baird, J. K., et al. (2016). Global epidemiology of plasmodium vivax. Am. J. Trop. Med. Hyg 95, 15–34. doi: 10.4269/ajtmh.16-0141
Huang, F., Cui, Y., Yan, H., Liu, H., Guo, X., Wang, G., et al. (2022). Prevalence of antifolate drug resistance markers in plasmodium vivax in China. Front. Med. 16, 83–92. doi: 10.1007/s11684-021-0894-x
Huang, F., Takala-Harrison, S., Jacob, C. G., Liu, H., Sun, X., Yang, H., et al. (2015). A single mutation in K13 predominates in southern China and is associated with delayed clearance of plasmodium falciparum following artemisinin treatment. J. Infect. Dis. 212, 1629–1635. doi: 10.1093/infdis/jiv249
Huang, F., Tang, L., Yang, H., Zhou, S., Liu, H., Li, J., et al. (2012). Molecular epidemiology of drug resistance markers of plasmodium falciparum in yunnan province, China. Malar J. 11, 243. doi: 10.1186/1475-2875-11-243
Huang, F., Yan, H., Xue, J. B., Cui, Y. W., Zhou, S. S., Xia, Z. G., et al. (2021). Molecular surveillance of pfcrt, pfmdr1 and pfk13-propeller mutations in plasmodium falciparum isolates imported from Africa to China. Malar J. 20, 73. doi: 10.1186/s12936-021-03613-5
Imwong, M., Pukrittakayamee, S., Looareesuwan, S., Pasvol, G., Poirreiz, J., White, N. J., et al. (2001). Association of genetic mutations in plasmodium vivax dhfr with resistance to sulfadoxine-pyrimethamine: geographical and clinical correlates. Antimicrob. Agents Chemother. 45, 3122–3127. doi: 10.1128/AAC.45.11.3122-3127.2001
Imwong, M., Pukrittayakamee, S., Rénia, L., Letourneur, F., Charlieu, J. P., Leartsakulpanich, U., et al. (2003). Novel point mutations in the dihydrofolate reductase gene of plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob. Agents Chemother. 47, 1514–1521. doi: 10.1128/AAC.47.5.1514-1521.2003
Khim, N., Bouchier, C., Ekala, M. T., Incardona, S., Lim, P., Legrand, E., et al. (2005). Countrywide survey shows very high prevalence of plasmodium falciparum multilocus resistance genotypes in Cambodia. Antimicrob. Agents Chemother. 49, 3147–3152. doi: 10.1128/AAC.49.8.3147-3152.2005
Korsinczky, M., Fischer, K., Chen, N., Baker, J., Rieckmann, K., Cheng, Q. (2004). Sulfadoxine resistance in plasmodium vivax is associated with a specific amino acid in dihydropteroate synthase at the putative sulfadoxine-binding site. Antimicrob. Agents Chemother. 48, 2214–2222. doi: 10.1128/AAC.48.6.2214-2222.2004
Kublin, J. G., Dzinjalamala, F. K., Kamwendo, D. D., Malkin, E. M., Cortese, J. F., Martino, L. M., et al. (2002). Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of plasmodium falciparum malaria. J. Infect. Dis. 185, 380–388. doi: 10.1086/338566
Kyaw, M. P., Nyunt, M. H., Chit, K., Aye, M. M., Aye, K. H., Aye, M. M., et al. (2013). Reduced susceptibility of plasmodium falciparum to artesunate in southern Myanmar. PloS One 8, e57689. doi: 10.1371/journal.pone.0057689
Li, J., Zhang, J., Li, Q., Hu, Y., Ruan, Y., Tao, Z., et al. (2020). Ex vivo susceptibilities of plasmodium vivax isolates from the China-Myanmar border to antimalarial drugs and association with polymorphisms in Pvmdr1 and pvcrt-o genes. PloS Negl. Trop. Dis. 14, e0008255. doi: 10.1371/journal.pntd.0008255
Liu, D. Q. (2014). Surveillance of antimalarial drug resistance in China in the 1980s-1990s. Infect. Dis. Poverty 3, 8. doi: 10.1186/2049-9957-3-8
Liu, D. Q., Liu, R. J., Ren, D. X., Gao, D. Q., Zhang, C. Y., Qui, C. P., et al. (1995). Changes in the resistance of plasmodium falciparum to chloroquine in hainan, China. Bull. World Health Organ 73, 483–486.
Looareesuwan, S., Buchachart, K., Wilairatana, P., Chalermrut, K., Rattanapong, Y., Amradee, S., et al. (1997). Primaquine-tolerant vivax malaria in Thailand. Ann. Trop. Med. Parasitol. 91, 939–943. doi: 10.1080/00034983.1997.11813222
Maïga-Ascofaré, O., May, J. (2016). Is the A578S single-nucleotide polymorphism in K13-propeller a marker of emerging resistance to artemisinin among plasmodium falciparum in Africa? J. Infect. Dis. 213, 165–166. doi: 10.1093/infdis/jiv414
Miotto, O., Almagro-Garcia, J., Manske, M., Macinnis, B., Campino, S., Rockett, K. A., et al. (2013). Multiple populations of artemisinin-resistant plasmodium falciparum in Cambodia. Nat. Genet. 45, 648–655. doi: 10.1038/ng.2624
Mueller, I., Galinski, M. R., Baird, J. K., Carlton, J. M., Kochar, D. K., Alonso, P. L., et al. (2009). Key gaps in the knowledge of plasmodium vivax, a neglected human malaria parasite. Lancet Infect. Dis. 9, 555–566. doi: 10.1016/S1473-3099(09)70177-X
Muiruri, P., Juma, D. W., Ingasia, L. A., Chebon, L. J., Opot, B., Ngalah, B. S., et al. (2018). Selective sweeps and genetic lineages of plasmodium falciparum multi-drug resistance (pfmdr1) gene in Kenya. Malar J. 17, 398. doi: 10.1186/s12936-018-2534-8
Murphy, G. S., Basri, H., Andersen, E. M., Bangs, M. J., Mount, D. L., Gorden, J., et al. (1993). Vivax malaria resistant to treatment and prophylaxis with chloroquine. Lancet 341, 96–100. doi: 10.1016/0140-6736(93)92568-E
Noedl, H., Se, Y., Schaecher, K., Smith, B. L., Socheat, D., Fukuda, M. M. (2008). Evidence of artemisinin-resistant malaria in western Cambodia. N Engl. J. Med. 359, 2619–2620. doi: 10.1056/NEJMc0805011
Nyunt, M. H., Han, J. H., Wang, B., Aye, K. M., Aye, K. H., Lee, S. K., et al. (2017). Clinical and molecular surveillance of drug resistant vivax malaria in myanmar, (2009-2016). Malar J. 16, 117. doi: 10.1186/s12936-017-1770-7
Payne, D. (1987). Spread of chloroquine resistance in plasmodium falciparum. Parasitol. Today 3, 241–246. doi: 10.1016/0169-4758(87)90147-5
Pearce, R. J., Pota, H., Evehe, M. S., El-H, B., Mombo-Ngoma, G., Malisa, A. L., et al. (2009). Multiple origins and regional dispersal of resistant dhps in African plasmodium falciparum malaria. PloS Med. 6, e1000055. doi: 10.1371/journal.pmed.1000055
Phyo, A. P., Nkhoma, S., Stepniewska, K., Ashley, E. A., Nair, S., Mcgready, R., et al. (2012). Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet 379, 1960–1966. doi: 10.1016/S0140-6736(12)60484-X
Sa, J. M., Twu, O. (2010). Protecting the malaria drug arsenal: halting the rise and spread of amodiaquine resistance by monitoring the PfCRT SVMNT type. Malar J. 9, 374. doi: 10.1186/1475-2875-9-374
She, D., Wang, Z., Liang, Q., Lu, L., Huang, Y., Zhang, K., et al. (2020). Polymorphisms of pfcrt, pfmdr1, and K13-propeller genes in imported falciparum malaria isolates from Africa in guizhou province, China. BMC Infect. Dis. 20, 513. doi: 10.1186/s12879-020-05228-8
Si, Y., Zeng, W., Li, N., Wang, C., Siddiqui, F., Zhang, J., et al. (2021). In vitro susceptibility of plasmodium falciparum isolates from the China-Myanmar border area to piperaquine and association with candidate markers. Antimicrob. Agents Chemother. 65 (10), e02305-20. doi: 10.1128/AAC.02305-20
Sidhu, A. B., Uhlemann, A. C., Valderramos, S. G., Valderramos, J. C., Krishna, S., Fidock, D. A. (2006). Decreasing pfmdr1 copy number in plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J. Infect. Dis. 194, 528–535. doi: 10.1086/507115
Suebsaeng, L., Wernsdorfer, W. H., Rooney, W. (1986). Sensitivity to quinine and mefloquine of plasmodium falciparum in Thailand. Bull. World Health Organ 64, 759–765.
Suwanarusk, R., Russell, B., Chavchich, M., Chalfein, F., Kenangalem, E., Kosaisavee, V., et al. (2007). Chloroquine resistant plasmodium vivax: in vitro characterisation and association with molecular polymorphisms. PloS One 2, e1089. doi: 10.1371/journal.pone.0001089
Tantiamornkul, K., Pumpaibool, T., Piriyapongsa, J., Culleton, R., Lek-Uthai, U. (2018). The prevalence of molecular markers of drug resistance in plasmodium vivax from the border regions of Thailand in 2008 and 2014. Int. J. Parasitol. Drugs Drug Resist. 8, 229–237. doi: 10.1016/j.ijpddr.2018.04.003
Tjitra, E., Baker, J., Suprianto, S., Cheng, Q., Anstey, N. M. (2002). Therapeutic efficacies of artesunate-sulfadoxine-pyrimethamine and chloroquine-sulfadoxine-pyrimethamine in vivax malaria pilot studies: relationship to plasmodium vivax dhfr mutations. Antimicrob. Agents Chemother. 46, 3947–3953. doi: 10.1128/AAC.46.12.3947-3953.2002
Triglia, T., Menting, J. G., Wilson, C., Cowman, A. F. (1997). Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 94, 13944–13949. doi: 10.1073/pnas.94.25.13944
Uwimana, A., Legrand, E., Stokes, B. H., Ndikumana, J. M., Warsame, M., Umulisa, N., et al. (2021). Author correction: emergence and clonal expansion of in vitro artemisinin-resistant plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 27, 1113–1115. doi: 10.1038/s41591-021-01365-y
Wang, Z., Wei, C., Pan, Y., Wang, Z., Ji, X., Chen, Q., et al. (2022). Polymorphisms of potential drug resistant molecular markers in plasmodium vivax from China-Myanmar border during 2008-2017. Infect. Dis. Poverty 11, 43. doi: 10.1186/s40249-022-00964-2
Wang, S., Xu, S., Geng, J., Si, Y., Zhao, H., Li, X., et al. (2020). Molecular surveillance and in vitro drug sensitivity study of plasmodium falciparum isolates from the China-Myanmar border. Am. J. Trop. Med. Hyg 103, 1100–1106. doi: 10.4269/ajtmh.20-0235
Wellems, T. E., Plowe, C. V. (2001). Chloroquine-resistant malaria. J. Infect. Dis. 184, 770–776. doi: 10.1086/322858
WHO (2018). Artemisinin resistance and artemisinin-based combination therapy efficacy. World Health Organization. Available at: https://www.who.int/malaria/areas/drug_resistance/en/.
WHO (2022). World malaria report 2022. World Health Organization. Available at: https:///www.who.int/publications/i/item/9789240064898.
Xu, C., Wei, Q., Yin, K., Sun, H., Li, J., Xiao, T., et al. (2018). Surveillance of antimalarial resistance pfcrt, Pfmdr1, and Pfkelch13 polymorphisms in African plasmodium falciparum imported to Shandong province, China. Sci. Rep. 8, 12951. doi: 10.1038/s41598-018-31207-w
Yang, Z., Zhang, Z., Sun, X., Wan, W., Cui, L., Zhang, X., et al. (2007). Molecular analysis of chloroquine resistance in plasmodium falciparum in yunnan province, China. Trop. Med. Int. Health 12, 1051–1060. doi: 10.1111/j.1365-3156.2007.01882.x
Zhang, L., Yi, B., Xia, Z., Huang, F. (2022). Plasmodium vivax in the elimination phase - china 2013-2020. China CDC Wkly 4, 609–613. doi: 10.46234/ccdcw2022.133
Zhao, D., Zhang, H., Ji, P., Li, S., Yang, C., Liu, Y., et al. (2021). Surveillance of antimalarial drug-resistance genes in imported plasmodium falciparum isolates from Nigeria in henan, China 2012-2019. Front. Cell Infect. Microbiol. 11, 644576. doi: 10.3389/fcimb.2021.644576
Keywords: Plasmodium falciparum, Plasmodium vivax, polymorphism, gene, China
Citation: Wang S, Huang F, Yan H, Yin J and Xia Z (2023) A review of malaria molecular markers for drug resistance in Plasmodium falciparum and Plasmodium vivax in China. Front. Cell. Infect. Microbiol. 13:1167220. doi: 10.3389/fcimb.2023.1167220
Received: 16 February 2023; Accepted: 26 April 2023;
Published: 09 May 2023.
Edited by:
Jude Marek Przyborski, Justus-Liebig-University Gießen, GermanyReviewed by:
Amadou Niangaly, Université des Sciences, des Techniques et des Technologies de Bamako, MaliAnurag Shukla, Drexel University, United States
Copyright © 2023 Wang, Huang, Yan, Yin and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhigui Xia, eGlhemdAbmlwZC5jaGluYWNkYy5jbg==
 Siqi Wang
Siqi Wang Fang Huang
Fang Huang He Yan
He Yan Jianhai Yin
Jianhai Yin Zhigui Xia1,2,3,4*
Zhigui Xia1,2,3,4*