- 1Division of Medical Zoology, Department of Infection and Immunity, Jichi Medical University, Tochigi, Japan
- 2Department of International Medical Zoology, Faculty of Medicine, Kagawa University, Kita-gun, Kagawa, Japan
A rare sugar, allose, was reported to inhibit the development of Plasmodium parasites in Anopheles mosquitoes; however, the mechanism remains unknown. The present study addressed the inhibitory mechanism of allose on the development of the Plasmodium parasite by connecting it with bacteria involvement in the midgut. In addition, further inhibitory sugars against Plasmodium infection in mosquitoes were explored. Antibiotic-treated and antibiotic-untreated Anopheles stephensi were fed fructose with or without allose. The mosquitoes were infected with luciferase-expressing Plasmodium berghei, and parasite development was evaluated by luciferase activity. Bacterial composition analysis in gut of their mosquitoes was performed with comprehensive 16S ribosomal RNA sequencing. As the result, allose inhibited the development of oocysts in mosquitoes regardless of prior antibiotic treatment. Microbiome analysis showed that the midgut bacterial composition in mosquitoes before and after blood feeding was not affected by allose. Although allose inhibited transient growth of the midgut microbiota of mosquitoes after blood feeding, neither toxic nor inhibitory effects of allose on the dominant midgut bacteria were observed. Ookinete development in the mosquito midgut was also not affected by allose feeding. Additional 15 sugars including six monosaccharides, four polyols, and five polysaccharides were tested; however, no inhibitory effect against Plasmodium development in mosquitoes was observed. These results indicated that allose inhibits parasite development in midgut stage of the mosquito independently of midgut microbiota. Although further studies are needed, our results suggest that allose may be a useful material for the vector control of malaria as a “transmission-blocking sugar.”
1 Introduction
Malaria is a life-threatening disease caused by Plasmodium parasites, with the estimated number of deaths reaching 627,000 per year in 2020 (World Health Organization (WHO), 2021). Plasmodium parasites are transmitted by Anopheles mosquitoes, ubiquitously distributing in endemic areas (Moffett et al., 2007; Autino et al., 2012). Vector control is a cornerstone in malaria control, owing to the lack of reliable vaccines, the emergence of drug resistance, and unaffordable potent antimalarials. Studies on transmission-blocking strategies, which aim to interrupt the life cycle of the Plasmodium species within mosquitoes, are progressing, and elucidation of more detailed transmission mechanisms including interaction between Anopheles mosquitoes and Plasmodium is expected.
The midgut of the mosquito is important not only for digestion and uptake of nutrients but also as the entry site of pathogens including Plasmodium parasites. When Anopheles mosquitoes ingest Plasmodium-infected blood from the host, parasites mate and develop into motile ookinetes within 16–20 h (Blandin et al., 2004). Ookinetes traverse across the midgut epithelium within 24 h after the feeding and transform into oocysts on the basal side of the midgut (Baton and Ranford-Cartwright, 2004; Blandin et al., 2004). Sporozoites develop and proliferate within oocysts over about the next 10 days. The sporozoites released from oocysts invade the salivary glands, in which they wait for transmission to the next host via a mosquito bite (Blandin et al., 2004; Frischknecht et al., 2004). The early phase of the parasite infection, from ingestion to invasion into midgut epithelium, is the bottleneck for the parasite in the mosquito cycle and, therefore, is considered to be a suitable target for a transmission-blocking strategy.
Mosquitoes develop protective mechanisms against infectious agents in the midgut, which include the immune system and biological barriers. Parasite invasion and development in the midgut are affected by immune-related factors such as thioester-containing protein 1 (TEP1) (Blandin et al., 2004; Osta, 2004; Dong et al., 2006; Baxter et al., 2010; Povelones et al., 2011) and antimicrobial peptides such as gambicin regulated by the immune deficiency (IMD) pathway through transcription factor Rel2 (Vizioli et al., 2001; Dong et al., 2006), as well as nitric oxide and reactive oxygen species (ROS) induced by invasion of gut epithelium (Luckhart et al., 1998; Molina-Cruz et al., 2008). In addition, malaria parasites are continuously exposed to microbes in the mosquito gut. This naturally acquired microbial flora can modulate the mosquito’s vectorial capacity. Comprehensive functional approaches have been undertaken to elucidate the interplay between commensal bacteria and the development of the malaria parasite in its natural vector. Mosquitoes treated with antibiotics, which diminish bacteria quantity, are known to be more susceptible to Plasmodium parasite infection (Dong et al., 2009). Administration of some characterized bacteria, such as Enterobacter sp. and Serratia marcescens, reduced the development of Plasmodium oocysts by eliciting anti-Plasmodium immunity in mosquitoes (Cirimotich et al., 2011; Bando et al., 2013; Bahia et al., 2014).
In nature, both male and female Anopheles mosquitoes ordinarily ingest sugars from floral nectar, but only females suck blood at a time of reproduction. Sugars function not only as an energy source but also as prebiotics for gut microbiota. As examples, oligosaccharides, which are composed of two to 10 monosaccharides, improve the host gastrointestinal condition by promoting beneficial microbiota (Mano et al., 2018), and polyols and hydrogenated carbohydrates are known to affect the ecology of oral microflora and exhibit a cariostatic effect in mammals (Msomi et al., 2021). Sucrose, an oligosaccharide, maintains antiviral immunity in the gut of Aedes aegypti (Almire et al., 2021). A recent study reported that glucose induces the expansion of Asaia bogorensis, a commensal bacterium in the mosquito gut, that remodels glucose metabolism, resulting in the promotion of Plasmodium gametogenesis and parasite infection by increasing the pH of the mosquito midgut (Wang et al., 2021). However, a rare sugar, allose, which has been shown to influence ROS generation in mammalian and plant cells (Murata et al., 2003; Kimura et al., 2005; Kano et al., 2013), was shown to inhibit Plasmodium infection in mosquitoes (Tokuda et al., 2015). Currently, it remains unknown how allose inhibits malarial parasite development and whether it affects commensal bacterium in the mosquito gut.
The present study addressed the inhibitory mechanism of allose on the development of the Plasmodium parasite and the involvement of the microbiota in midgut fed allose in its mechanism. In addition, other inhibitory sugars potentially affecting the development of Plasmodium were explored.
2 Materials and methods
2.1 Mosquito and malaria parasite maintenance
Anopheles stephensi SDA-500 were maintained at 26°C with 60%–80% relative humidity under the condition of 13-h light/11-h dark at the Jichi Medical University. Larvae were fed with fish food, Hikari (KYORIN Co., Ltd., Hyogo, Japan). Adults were fed on filter paper soaked with 8% fructose (FUJIFILM Wako, Osaka, Japan). Luciferase-expressing P. berghei ANKA234 under the control of elongation factor 1-α promoter (Pb-luc) was injected into an Institute of Cancer Research (ICR) mouse intraperitoneally (Matsuoka et al., 2015). The mouse was intraperitoneally administered 100 µL of pyrimethamine (1 mg/mL) daily. Infected blood was stained with Giemsa’s stain solution (Merck KGaA, Darmstadt, Germany) to determine parasitemia. All procedures were carried out in accordance with the ethical guidelines, and this study was approved by the Institutional Animal Experiment Committee of the Jichi Medical University (approval numbers: 20003-01).
2.2 Sugar feeding, antibiotic treatment, and infection of An stephensi with P berghei
Sixteen sugars—seven monosaccharides (D-glucose, D-fructose, D-galactose, L-sorbose, D-mannose, D-xylose, and D-allose), four polyols (D-sorbitol, xylitol, erythritol, and maltitol), and five oligosaccharides (sucrose, D-lactose, lactulose, D-maltose, and D-raffinose) (Tokyo Chemical Industry Co., Ltd., Tokyo, Japan)—were used in this study to evaluate their inhibitory effects on the development of Plasmodium. Of these, D-allose, erythritol, L-sorbose, and xylitol are categorized as rare sugars as they are rarely present in nature. Mosquitoes were fed 8% fructose with 100 mM of each abovementioned sugar for 24 h ad libitum and fasted for the following 24 h. The mosquitoes were allowed to feed on Pb-luc–infected mice showing 1.5%–2.0% parasitemia for 30 min, and unfed and partially blood-fed mosquitoes were removed from cages. Fully engorged mosquitoes were maintained on 8% (444 mM) fructose containing 100 mM of each sugar for an additional 10 days at 21°C. The concentration of additional sugar was matched with the effective concentration of allose (100 mM) (Tokuda et al., 2015). For the antibiotic treatment, mosquitoes were fed 8% fructose with 100 mM allose containing penicillin (200 U/mL) and streptomycin (200 µg/mL; Thermo Fisher Scientific Inc., Waltham, MA, USA) for 24 h ad libitum. After starvation for the following 24 h, the mosquitoes were allowed to feed on Pb-luc–infected mice showing 10% parasitemia for 30 min. The mosquitoes were maintained as mentioned above. Infection of Pb-luc in mosquitoes fed with the infected blood after 10 days was determined by measuring luciferase activity and by counting oocysts in the midgut (Matsuoka et al., 2015).
2.3 Luciferase assay
Luciferase activity was measured using a Luciferase assay kit according to the manufacturer’s protocol (Promega Inc., Madison, WI, USA). Briefly, the whole body of mosquitoes was individually homogenized with disposable homogenizer in a 1.5-mL tube containing cell lysis buffer. The homogenized samples were centrifuged at 15,000 × g for 1 min, and 10 µL of each supernatant was mixed with 50 µL of substrate in a 96-well black microplate. Luminescence was measured by a SpectraMax® M5 microplate reader (Molecular Devices, LLC., San Jose, CA, USA).
2.4 Effect of allose on Plasmodium parasite ex vivo
To evaluate the effect of allose on ookinete development, mosquitoes were dissected at 24 h after infection, and midguts were collected. The midgut containing ookinetes was suspended individually in Roswell Park Memorial Institute (RPMI) 1640 medium (Nakarai Tesque Inc., Kyoto, Japan) supplemented with 20% heat-inactivated fetal calf serum, penicillin (100 U/mL), streptomycin (100 µg/mL; Thermo Fisher Scientific Inc.), and 25 mM Hydroxyethylpiperazine ethane sulfonic acid (HEPES) (pH 7.4; Tokyo Chemical Industry Co., Ltd.). The suspension was fixed and stained with Giemsa’s stain solution, and, then, ookinetes were counted under a microscope. The number of red blood cells (RBCs) in each sample was also counted to compensate the amount of ingested blood in each mosquito, and the ookinete count per 106 RBCs was determined.
2.5 Midgut sample collection and DNA extraction for microbiota analysis
Mosquitoes were collected on days 1, 3, and 7 post-infection with Pb-luc at 21°C and prior to the infection as a control. They were anesthetized on ice, sterilized with 70% ethanol containing 1% hypochlorite, and then rinsed with phosphate-buffered saline (PBS). Midguts were taken out and washed with sterilized PBS (−), and 10 midguts were pooled in a 2-mL tube containing 0.5 mL of PBS with 0.1-mm Zirconia/Silica beads (Bio Spec Products Inc., Bartlesville, OK, USA). The midgut samples containing microbiota in the gut were mechanically disrupted by a bead beater (TAITEC Co., Saitama, Japan) at 3,500 rpm for 1 min, and DNA was purified using the ReliaPre DNA Clean-Up and Concentration System (Promega Inc.).
2.6 16S ribosomal RNA–based metagenomic analysis of mosquito midgut microbiota
Bacterial 16S ribosomal RNA (16S rRNA) fragments containing hypervariable V3–V4 region were amplified, and index sequences were added by PCR with Tks Gflex DNA polymerase (Takara Bio Inc., Shiga, Japan) using primer sets according to the manufacturer’s protocol (https://jp.support.illumina.com/downloads/16s_metagenomic_sequencing_library_preparation.html ; accessed on 1 February, 2022). The amplified fragments were purified using a FastGene PCR/Gel extraction kit (Nippon Genetics Co., Ltd., Tokyo, Japan), and the concentration was quantified using a Qbit3 Fluorometer with Qbit dsDNA HS assay kit (Thermo Fisher Scientific Inc.). Paired-end sequencing analysis was conducted on the MiSeq® platform with MiSeq Reagent Kit v3 for 600 cycles (Illumina, Inc., San Diego, CA, USA). Sequence data were trimmed and merged by DADA2 plugin in QIIME2 program version 2020.6 (Callahan et al., 2016; Bolyen et al., 2019). The sequences were clustered in amplicon sequence variants (ASVs) by feature-classifier plugin with the dataset of SILVA version 138 with ≥ 99% sequence identity (Quast et al., 2013; Yilmaz et al., 2014; Bokulich et al., 2018). To compare the diversities between groups, beta-diversity analyses with the Bray–Curtis dissimilarity index were performed with the q2-diversity plugin in QIIME2 at a sampling depth of 1,500 (Anderson, 2001).
2.7 Quantification of midgut bacteria in mosquitoes
Midgut bacteria in mosquitoes were quantified by quantitative PCR (qPCR) with KOD -SYBR- (TOYOBO Co., Ltd., Osaka, Japan) using Thermal Cycler Dice Real Time System Lite (Takara Bio Inc.). The bacterial 16S rRNA was used as the target, and the An. stephensi glyceraldehyde 3-phosphate dehydrogenase (Asgapdh) was used as the reference. The primer sequences were 5′-ACHCCTACGGGDGGCWGCAG-3′ (16S-q-337F) and 5′-GTDTYACCGCGGYTGCTGGCAC-3′ (16S-q-514R) for amplification of the bacterial 16S rRNA gene and 5′-GCCGTCGGCAAGGTCATCCC-3′ (Asgapdh-q-F) and 5′-TTCATCGGTCCGTTGGCGGC-3′ (Asgapdh-q-R) for that of the Asgapdh (Yamamoto et al., 2016). Relative quantities of 16S rRNA were determined by the comparative cycle threshold (ΔΔCt) method using the Asgapdh as the reference.
2.8 Isolation and identification of mosquito midgut bacteria
Mosquitoes were anesthetized on ice and washed with 70% ethanol containing 1% hypochlorite followed by sterilized PBS (−). The midgut was removed, rinsed with sterilized PBS (−), and homogenized in 20 µL of sterilized PBS (−). Ten microliters of the homogenate was spread on brain-heart infusion agar plate (Sigma Aldrich Inc., St. Louis, MO, USA), and the plate was incubated at 28°C for 3 days under aerobic and anaerobic conditions. For the identification of bacteria, the bacterial 16S rRNA was amplified with KOD -One- (TOYOBO Co., Ltd.) using a pair of primers: 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Delong, 1992). The amplicon was purified using a FastGene PCR/Gel extraction Kit (Nippon Genetics Co., Ltd.), and the sequence was determined by the dideoxy chain termination method with a specific primer, 519F (5′-CAGCMGCCGCGGTAAT-3′), using a BigDye Terminator v.3.1 Cycle Sequencing Kit (Thermo Fisher Scientific Inc.). Homology was performed with the nucleotide BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and a search was conducted of the taxonomic database (https://www.ncbi.nlm.nih.gov/taxonomy ) of the National Center for Biotechnology Information (NCBI) database.
2.9 Effect of allose on bacterial growth in vitro
A single colony of bacteria isolated from mosquito midgut was cultured in Luria-Bertani (LB) liquid medium at 28°C overnight. A part of the bacterial culture was transferred into LB liquid medium containing 8% fructose with 100 mM glucose or allose, and the optical density at a wavelength of 600 nm (OD600) of the suspensions was adjusted to 0.02 by spectrophotometer. The bacteria were further cultured at 28°C, and the growth was monitored for 36 h.
2.10 Sequence data availability
The microbiome sequencing data obtained in this study were registered in the NCBI/GenBank/ the DNA data bank of Japan (DDBJ) database under the accession numbers DRR337101-DRR337141. The 16S rRNA sequences of Leucobacter sp. and Phyllobacterium sp. isolated from mosquito midgut are available under the accession numbers LC669551 and LC669552, respectively.
2.11 Statistical analysis
Statistical analyses were performed using R version 4.0.3. Welch’s t-test with Bonferroni’s modification was applied to ookinete counting, quantification of bacteria by qPCR, and evaluation of bacterial growth. Wilcoxon rank sum test with Bonferroni’s modification was conducted for the development of Plasmodium parasites within mosquitoes given different sugars. One-way analysis of variance with Holm’s modification was applied to assess the quantification of midgut bacterial amount the time-dependent changes of the relative abundance of the six dominant midgut bacteria genera.
3 Results
3.1 Allose inhibits development of Plasmodium parasite in the antibiotic-treated mosquito
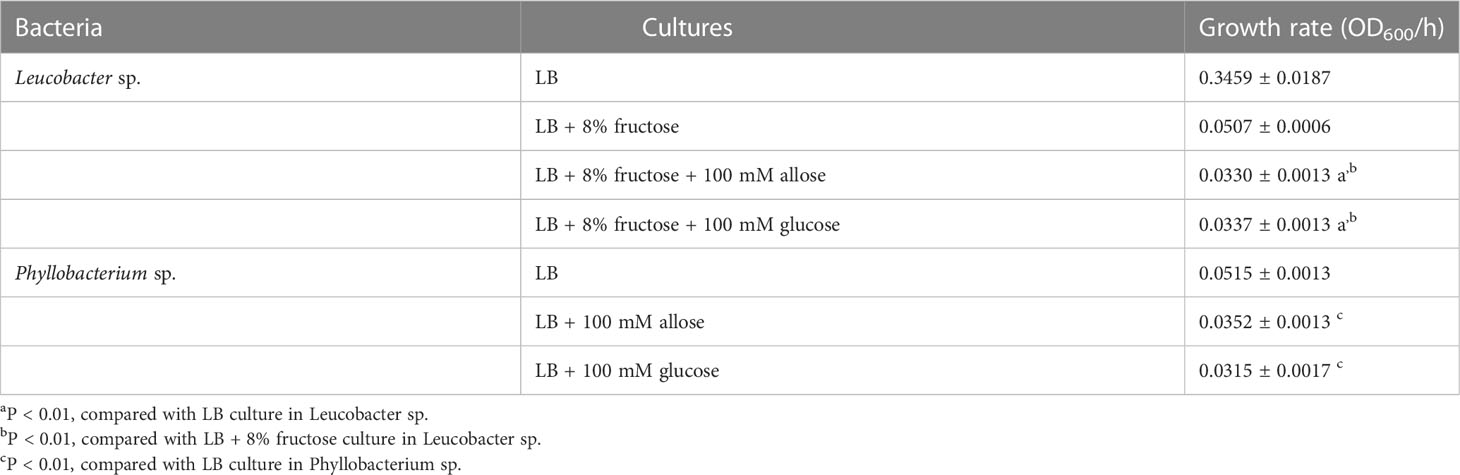
Allose inhibits the development of P. berghei within An. stephensi by assessing the development of oocysts (Tokuda et al., 2015). To reconfirm this finding, luciferase activity was investigated in allose-fed mosquitoes infected with Pb-luc. As shown in Figure 1; Supplementary Figure 4, luciferase activities in the infected mosquitoes were consistent with oocyst counts at 10 days after Pb-luc infection. The inhibitory effect of allose was evaluated in mosquitoes treated with antibiotics, which have diminished microbiota in the midgut. Similar to untreated mosquitoes, the development of Plasmodium was inhibited by allose feeding in the antibiotic-treated mosquitoes (Figure 2). This result suggested that allose inhibits development of the Plasmodium parasite in the mosquito through a midgut microbiota-independent mechanism.
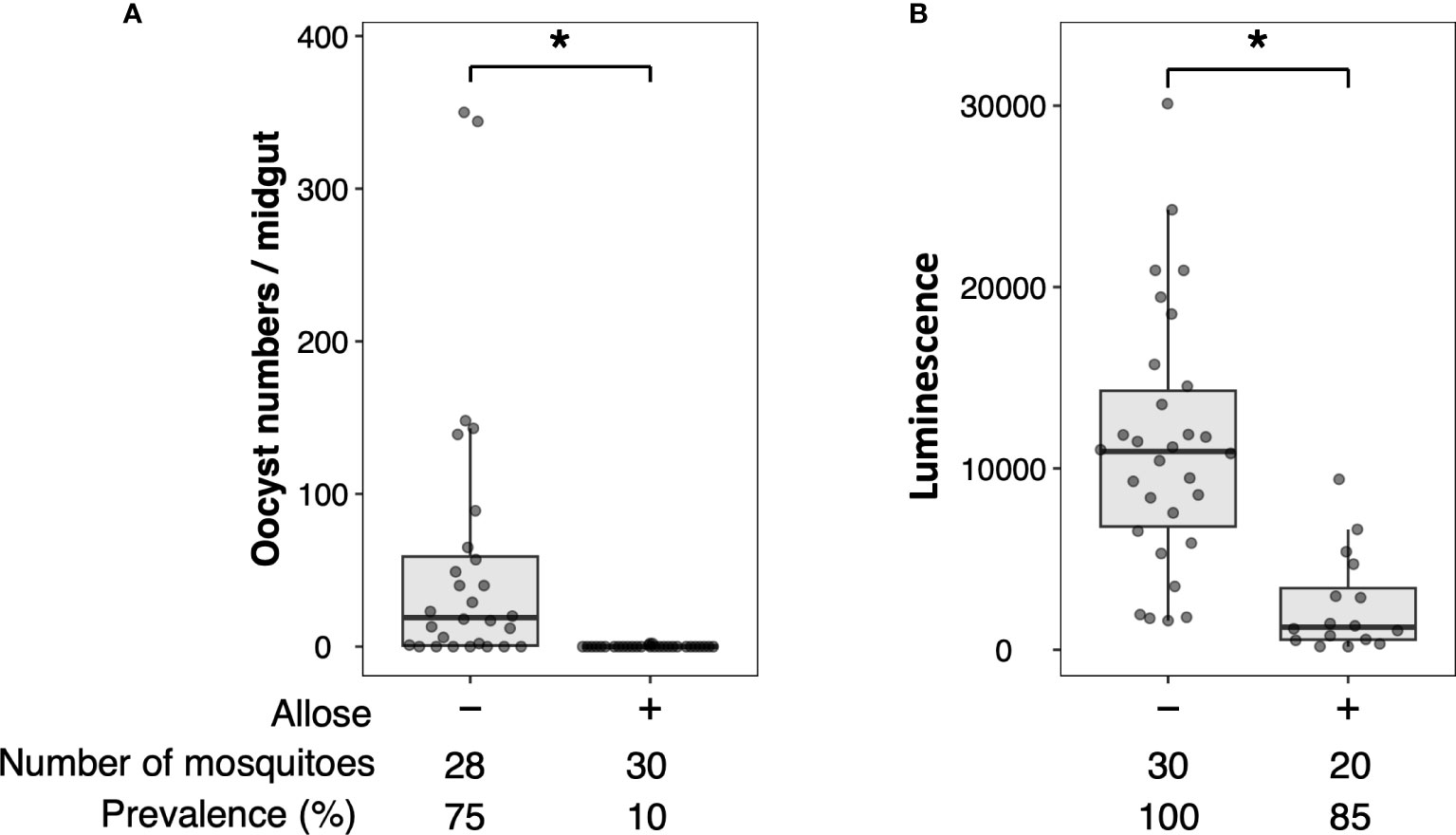
Figure 1 Allose inhibits oocyst development in the mosquito midgut. Allose-fed and allose-unfed mosquitoes were infected by luciferase-expressing Plasmodium berghei, and infectivity was determined 10 days post-infection by counting of the number of oocysts (A) and by measuring the luciferase activity (B). The number of mosquitoes is indicated under each group. The prevalence of infection was calculated number of detected oocyst or luminescence sample divided by number of total sample. The results represent the standard deviation of the mean (SD) of biological replicates. Statistical significance was determined by Wilcoxon’s rank sum test with Bonferroni’s modification. *P < 0.01.
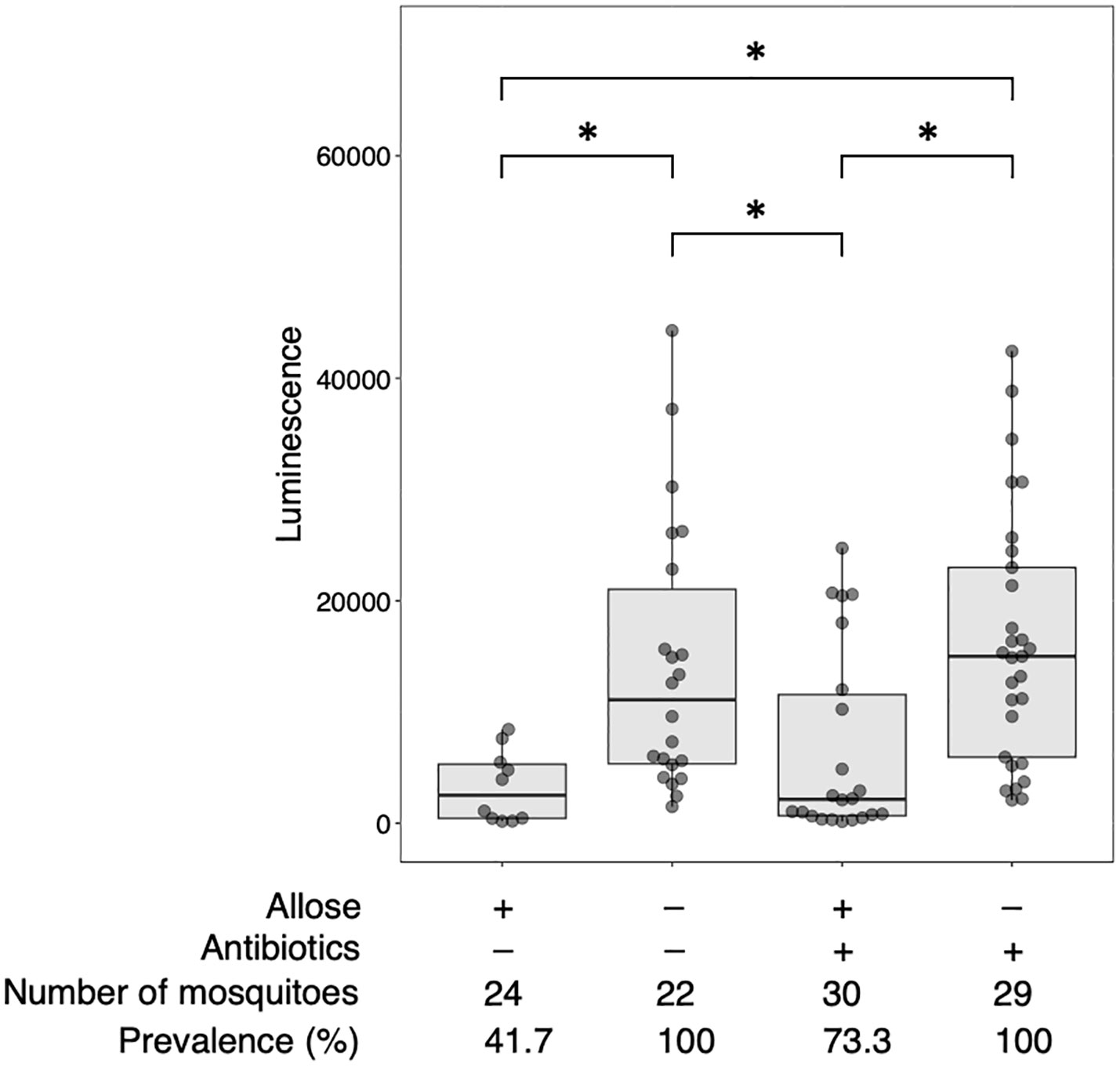
Figure 2 Allose inhibits the development of Plasmodium parasite in the mosquito by a midgut microbiota-independent mechanism. Antibiotic-treated and antibiotic-untreated mosquitoes were fed fructose with or without allose and then infected by luciferase-expressing Plasmodium berghei. Infectivity was determined by a luciferase assay. The number of mosquitoes is indicated under each group. The prevalence of infection was calculated number of detected oocyst or luminescence sample divided by number of total sample. The results represent the standard deviation of the mean (SD) of biological replicates. Statistical significance was determined by Wilcoxon’s rank sum test with Bonferroni’s modification. *P < 0.01.
3.2 Allose affects the proliferation of midgut microbiota but not bacterial composition
As ingested sugars can affect midgut microbiota that may impact the development of parasites directly or indirectly, quantitative and qualitative changes of bacterial communities in the midgut of allose-fed mosquitoes were assessed. Midgut microbiota was quantified by qPCR. The relative quantity tended to be suppressed in allose-fed mosquitoes at 3 days after infection with no significant difference compared to allose-unfed mosquitoes and was comparable between groups on day 7 (Figure 3A). The decrease of bacterial quantities in the allose-fed group seemed to be attributed to inhibition of the rapid proliferation of gut bacteria initiated by blood feeding (Figure 3A). The composition of midgut bacteria was identified by 16S rRNA–based metagenomic analysis and compared between allose-fed and allose-unfed mosquitoes. In total, 2,706,788 reads were obtained from 40 pooled samples, and de novo assembly of the reads generated 427 ASV. Of these, 113 ASVs consisting of 1,146,318 reads (42.4% of total reads) were not categorized in bacteria at the taxonomic level in the SILVA classifier, and, finally, 314 ASVs consisting of 1,497,066 reads (57.6%) were classified into families and genera of bacteria (Supplementary Table S1). The most abundant bacteria belong to the genus Phyllobacterium in the family Rhizobiaceae (51.4% of bacterial reads, 769,144 reads, 25 ASVs), followed by the genus Leucobacter in the family Microbacteriaceae (19.9% of bacterial reads, 297,535 reads, 12 ASVs), the genus Staphylococcus in the family Staphylococcaceae (3.5% of bacterial reads, 51,813 reads, 34 ASVs), the family Peptococcaceae (2.7% of bacterial reads, 40,358 reads, 23 ASVs), the genus Proteus in the family Morganellaceae (2.4% of bacterial reads, 35,621 reads, 7 ASVs), and the genus Pseudomonas in the family Pseudomonadaceae (2.3% of bacterial reads, 33,916 reads, 8 ASVs) (Supplementary Table S1). Other bacteria families consisting of less than 2% of reads were categorized as “others” in this study (17.9% of bacterial reads, 268,679 reads, 205 ASVs). More detailed information obtained by the metagenomic analysis is provided in Supplementary Table S1; Figure 3B shows the time-dependent changes of the relative abundance of the six dominant midgut bacteria genera. The most abundant genus, Phyllobacterium, consisted of 35.6%–57.0%, on average, in the allose-unfed group, and the abundance did not change dramatically before (day 0) or after the infection (days 1–7). In the allose-fed group, Phyllobacterium consisted of 19.3%–73.1% on average during the course before (day 0) and after post infection (days 0–7), and a slight decrease of the composition ratio (19.3%) was observed on day 1 post-infection with no significant difference compared to the allose-unfed group. The composition ratio of the second most abundant genus, Leucobacter, was slightly increased in the allose-unfed group during the course before (day 0) and after post infection (days 0–3), whereas the ratio of the allose-fed group was slightly decreased on day 1 post-infection with no significant difference compared to the allose-unfed group, similar to that of Phyllobacterium. Similarly, although the composition ratio of the other four genera—Staphylococcus, Peptococcaceae, Proteus, and Pseudomonas—were slightly changed during the course before (day 0) and after infection (days 0–7), the differences between two groups were not significant (Figure 3B). In addition, heatmap clustering analysis based on the relative abundance of each genus and beta-diversity analysis showed an allose-independent cluster formation (Supplementary Figure S1) and no significant diversity between the allose-fed and allose-unfed groups during the course (Supplementary Table S1; PERMANOVA, pseudo-F = 1.03, P = 0.371), respectively. These results strongly suggested that allose affects the transient growth of midgut microbiota initiated by blood feeding but not their diversity.
Figure 3 Allose affects the midgut quantity of bacteria but not their composition. Quantitative and qualitative changes of bacterial communities in the midgut were determined on days 0, 1, 3, and 7 after allose feeding. (A) Midgut bacteria of allose-fed (black circle) and allose-unfed (gray triangle) mosquitoes were quantified by qPCR. Relative quantities of the bacterial 16S rRNA were normalized to the An. stephensi glyceraldehyde 3-phosphate dehydrogenase as the reference. Error bars indicate standard deviation (n = 5). (B) Midgut bacterial composition of allose-fed (open circle) and allose-unfed (open triangle) mosquitoes was determined by 16S rRNA–based metagenomic analysis. The relative abundance of the six dominant genera—Phyllobacterium, Leucobacter, Staphylococcus, Peptococcaceae, Proteus, and Pseudomonas—are shown. The results are expressed as the mean titer and standard deviation for five biological experiments in each group.
3.3 Allose did not inhibit growth of bacteria isolated from mosquito midgut
A bacterial growth assay in vitro was performed to assess whether allose inhibits the growth of bacteria in the midgut. Two bacteria in the mosquito midgut were successfully isolated and identified as Leucobacter sp. and Phyllobacterium sp., both of which are dominant in the gut, based on the sequencing analysis of 16S rRNA fragments with 96.8%–99.7% and 96.4%–99.2% identities, respectively, in the BLASTn analysis. The effect of allose on the growth of these bacteria was assessed in vitro. The growth of Leucobacter sp. in LB medium containing 100 mM allose and 8% fructose was comparable with that containing 100 mM glucose and 8% fructose as a control (Table 1). In addition to the growth rate, the maximum of the growth (stationary phase) in LB medium containing 100 mM allose and 8% fructose was comparable with that containing 8% fructose with 100 mM glucose as a control (Supplementary Figure S3A). Similarly, the growth of Phyllobacterium sp. in LB medium containing 100 mM allose was comparable with that containing 100 mM glucose as a control (Table 1; Supplementary Figure S3B) although the growth was inhibited by the presence of 8% fructose in the medium. These results indicated that allose does not have a bactericidal or growth inhibitory effect on these bacteria.
3.4 Effect of allose on ookinete development in the mosquito gut
Allose might impact the development of Plasmodium parasites rather than midgut microbiota. The effect of allose on ookinete development within infected mosquitoes was evaluated ex vivo. The number of ookinetes in the midgut of allose-fed mosquitoes was comparable to that observed in mosquitoes fed fructose alone at 24 h after infection (Figure 4). In addition, ookinetes showing abnormal morphology were not observed in the midgut of allose-fed mosquitoes compared with that in the midgut of allose-unfed mosquitoes (Supplementary Figure S2). These results indicated that allose feeding did not inhibit ookinete development of Plasmodium parasites at the midgut stage of the infected mosquito.
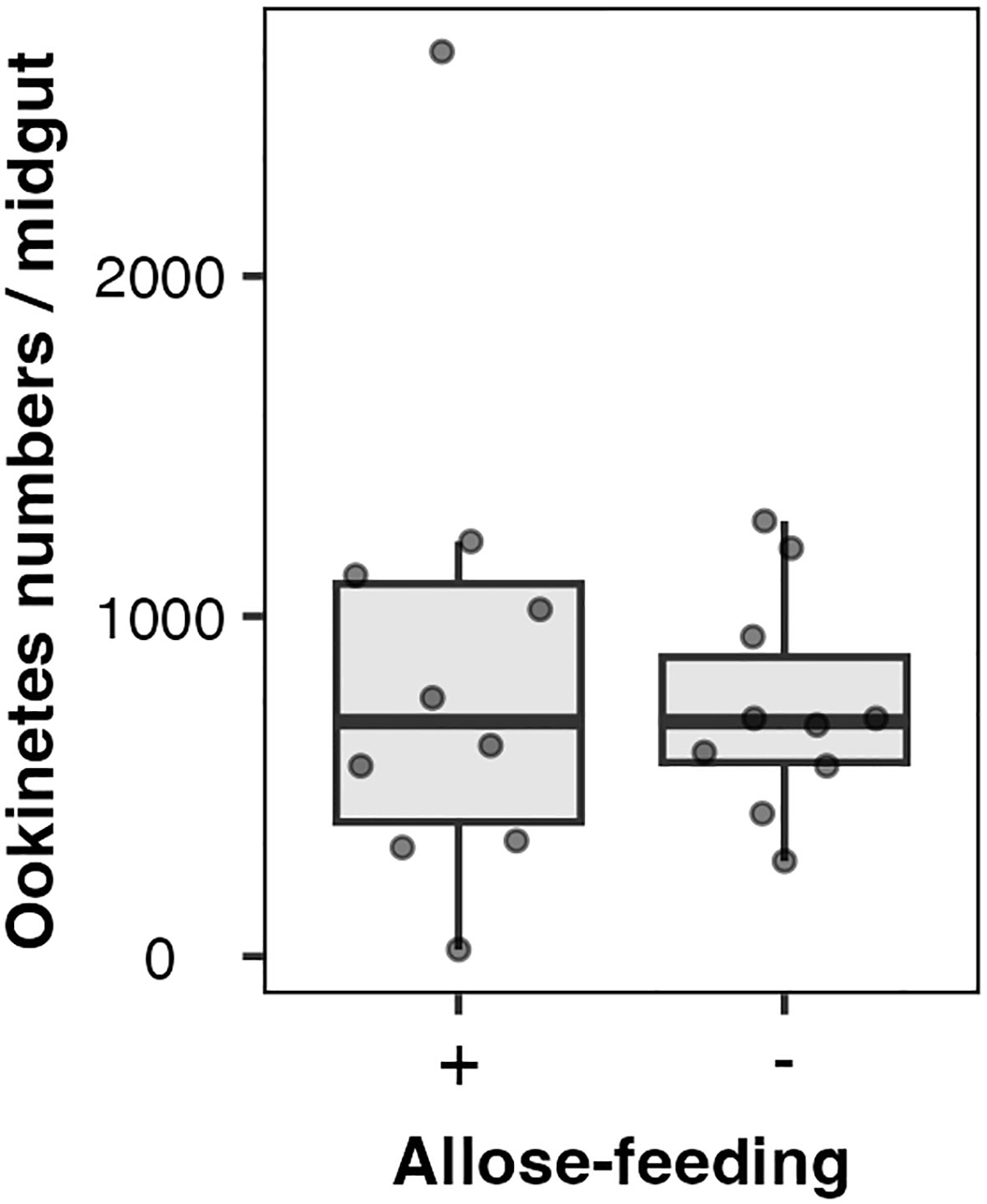
Figure 4 Allose does not inhibit ookinete development in the mosquito midgut. Ten mosquitoes were dissected 24 h after infection, and ookinete count per a midgut was determined. The experiment was repeated three times, and the results are represented as the standard deviation of the mean (SD) of biological replicates.
3.5 Exploration of sugars that impact Plasmodium development
Additional sugars that impact the development of Plasmodium in mosquitoes were explored by targeting the 16 sugars listed above in the Materials and Methods section. Their sugars about the ability to block Plasmodium parasites have not been evaluated, except for glucose (Wang et al., 2021). As shown in Figure 5, no apparent inhibitory effects on the development of Plasmodium in the mosquito were observed with most of the sugars tested. Although fructose, sorbitol, and xylitol slightly inhibited the development of the Plasmodium parasite, the inhibition was not significant. These results indicated that these sugars do not appear to have inhibitory effects on the development of Plasmodium in mosquitoes.
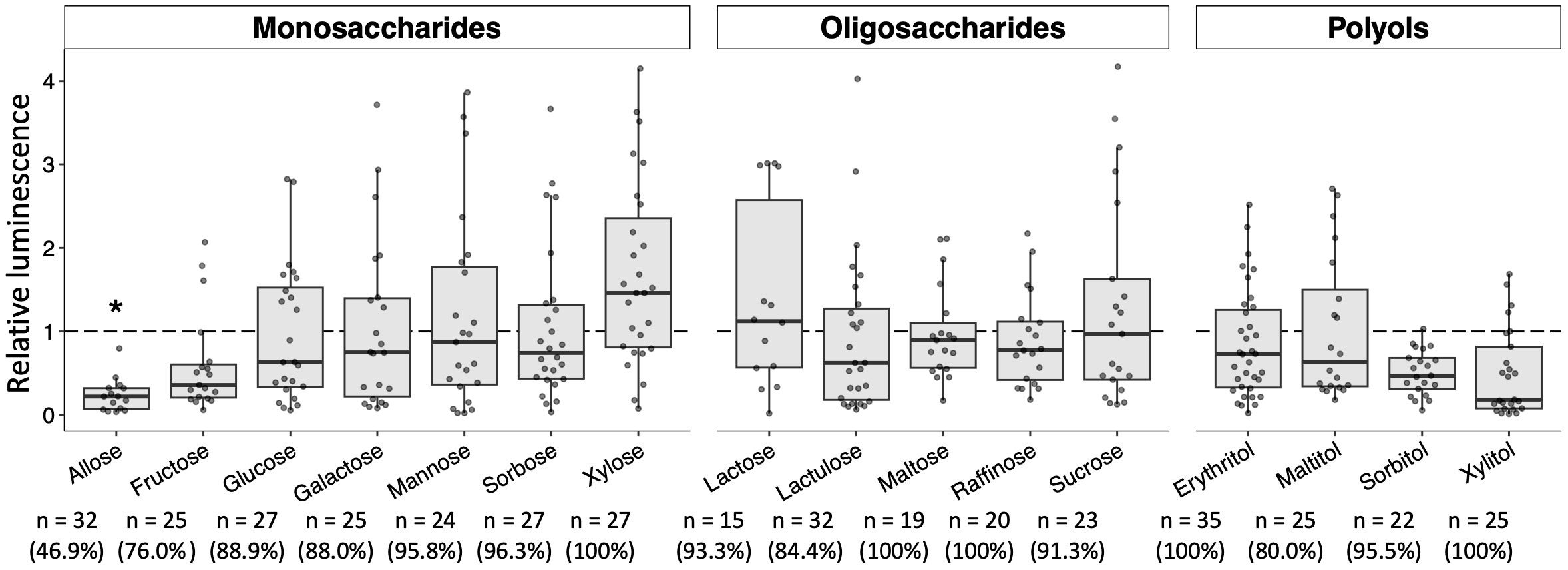
Figure 5 Effect of various sugars on oocyst development in the mosquito midgut. Mosquitoes were fed 8% fructose with 100 mM of each sugar and infected by luciferase-expressing Plasmodium berghei. Infectivity was determined by a luciferase assay. Infection of Pb-luc in mosquitoes fed the infected blood after 10 days was determined by measuring luciferase assay. Relative luminescence indicates the luminescence of each explored sugar-fed group normalized by mean luminescence of control group not added their explored sugar in 8% fructose. The number of mosquitoes is indicated under each group. The prevalence of infection was calculated number of detected oocyst or luminescence sample divided by number of total sample. The results represent the standard deviation of the mean (SD) of biological replicates. Statistical significance was determined by Wilcoxon’s rank sum test with Bonferroni’s modification. *P < 0.01.
4 Discussion
The midgut stage in the mosquito is one of the bottlenecks in the life cycle of Plasmodium parasites, and it can be a target of a transmission-blocking strategy. A previous study showed that allose inhibits oocyst development of Plasmodium parasites in the mosquito. However, the underlying mechanism remains unknown (Tokuda et al., 2015). The present study showed that inhibition by allose is effective even when mosquitoes were treated with antibiotics, strongly suggesting that the primary inhibitory mechanism is independent of the midgut microbiota of the mosquito. In addition, ookinete development in the mosquito was not inhibited by allose feeding. These results suggested that allose mainly inhibits development of the parasite after or possibly during its traverse across the midgut epithelium, e.g., ookinete migration across the midgut epithelium, ookinete-to-oocyst developmental transition, and the early phase of oocyst formation.
The interaction of ookinetes with the midgut surface via specific molecules is crucial for the invasion into midgut epithelium (Osta, 2004). Multiple pathways associated with protein–protein interactions between ookinete proteins [enolase, von Willebrand factor A domain–related protein (WARP), membrane-attack ookinete protein (MAOP), Plasmodium perforin-like proteins (PPLP5), subtilisin-like protease (SUB2), cell-traversal protein for ookinetes and sporozoites (CelTOS), secreted ookinete adhesive protein (SOAP), and ookinete surface P28 and P25 proteins] and mosquito midgut surface molecules [enolase-binding protein (EBP), aminopeptidase 1 (APN1), annexin-like proteins, carboxypeptidase B, croquemort scavenger receptor homolog, and calreticulin] were suggested to be involved in the invasion process (Siden-Kiamos et al., 2000; Soo Han et al., 2001; Tomas et al., 2001; Yuda et al., 2001; Dessens et al., 2003; Kadota et al., 2004; Kotsyfakis et al., 2005; Kariu et al., 2006; Dinglasan et al., 2007; Ecker et al., 2007; Lavazec et al., 2007; Rodríguez M del et al., 2007; González-Lázaro et al., 2009; Ghosh et al., 2011). In addition, several carbohydrate-binding proteins, i.e., lectins, bind specifically to the midgut epithelial cells of mosquitoes, and carbohydrate moieties are suggested to play a part in the binding of ookinetes to midgut epithelium of the mosquito (Rudin and Hecker, 1989; Zieler et al., 1999; Osta, 2004). Interestingly, Plasmodium gallinaceum ookinetes were reported to specifically interact with a carbohydrate ligand on the midgut surface of Aedes aegypti, and the specific interaction was competitively blocked by the monosaccharide N-acetylneuraminic acid (Zieler et al., 1999). Therefore, allose may inhibit specific binding of P. berghei to midgut epithelium of Anopheles mosquitoes. Another possible inhibitory mechanism of allose is related to mosquito immunity against Plasmodium parasites. Parasite invasion and development in the midgut is affected by host immunity, including immune-related factors (Blandin et al., 2004; Osta, 2004; Dong et al., 2006; Baxter et al., 2010; Povelones et al., 2011), antimicrobial peptides (Vizioli et al., 2001; Dong et al., 2006), and nitric oxide and ROS (Luckhart et al., 1998; Molina-Cruz et al., 2008). ROS have been shown to have a role in insect innate immune responses as a potent pathogen-killing agent, and allose is reported to induce ROS generation by activating nicotinamide adenine dinucleotide phosphate (NADPH) oxidase through the activity of glucose 6-phosphate dehydrogenase (Kano et al., 2013). RNA interference (RNAi)-mediated silencing of catalase, a ROS detoxification enzyme, was reported to result in decreased P. berghei infection (Molina-Cruz et al., 2008) and decreased bacterial load in the mosquito midgut (Bahia et al., 2013). These findings suggest that allose might inhibit the development of Plasmodium parasites and affect midgut bacteria via ROS generation in the mosquito midgut.
Several gut symbiotic bacteria, such as Serratia spp., Enterobacter sp., Chromobacterium sp., and Asaia sp., are reported to confer resistance to Plasmodium infection through activation of mosquito immunity (Cirimotich et al., 2011; Bando et al., 2013; Bahia et al., 2014; Ramirez et al., 2014; Bai et al., 2019; Wang et al., 2021). Initially, we expected that allose may work as a prebiotic for such symbiotic bacteria, resulting in the inhibition of Plasmodium development in the mosquito. However, the composition of midgut microbiota in the mosquito was not affected by allose. It seems that allose does not promote the growth of specific bacteria in the gut. In addition, none of the abovementioned bacteria were detected as a major midgut microbiota in our colonized mosquitoes. The microbial community of mosquitoes varies among populations and is associated with locality, climate, and their nutrition (Boissière et al., 2012; Muturi et al., 2018; Sandeu et al., 2022). Because the principal inhibitory mechanism of allose to Plasmodium is independent from commensal microbes, allose is expected to inhibit Plasmodium development in any population of mosquitoes.
Allose inhibited transient growth of midgut microbiota of mosquitoes initiated by blood feeding without toxic and inhibitory effects on the dominant bacteria. Several probable reasons may explain this observation: 1) allose is not suitable nutrition for midgut microbiota, 2) allose inhibits the bacterial metabolic system necessary for growth, and 3) allose enhances innate immune responses, resulting in the inhibition of bacterial growth. In this study, an equal amount of fructose was given to allose-fed mosquitoes as the control group, and allose was shown to be catabolized in several bacteria such as Aerobacter aerogenes, Escherichia coli, and Listeria monocytogenes (Gibbins and Simpson, 1964; Poulsen et al., 1999; Zhang et al., 2018). In addition, allose did not inhibit the in vitro growth of Leucobacter sp. and Phyllobacterium sp., the dominant bacteria isolated from the mosquito midgut. Recently, modulation of the pH of the mosquito midgut by changing its bacterial composition and metabolites was reported to influence Plasmodium gametogenesis (Wang et al., 2021). Our preliminary study showed that allose did not change the pH of the mosquito gut (data not shown). These findings suggested that allose does not directly affect the growth of microbiota in the mosquito gut; rather, the event might be caused by host immunity such as ROS generation.
This study explored 15 additional sugars that might impact the development of Plasmodium in mosquitoes. These sugars having various properties were screened, but none of the sugars tested inhibited parasite development, suggesting that allose has unique characteristics. Although pure allose is costly, an affordable and commercially available sweetener, Rare Sugar Sweet (https://www.matsutani.co.jp/english/products/raresugar.html ), which contains D-allose in addition to D-glucose, D-fructose, D-mannose, and another rare sugar, D-psicose, inhibits the development of Plasmodium in mosquitoes (Tokuda et al., 2015). Recently, toxic sugar baits were reported to be used as a mosquito control strategy (Fiorenzano et al., 2017); the principle is using fruits and plant as an attractant to attract mosquitoes, but it has some risks for other organisms. Because allose is an eco-friendly natural product, there are fewer limitations to using the attractive toxic sugar bait applications: mixing this sugar instead of an insecticide in a field study to control transmission of Plasmodium parasites by mosquitoes.
The present study showed that allose inhibited the development of Plasmodium parasites in the mosquito independently of midgut microbiota. Such unique activity appears to be specific to allose. Further study will be needed to disclose the underlying mechanisms, especially focusing on the mosquito immunity elicited by allose. This sugar may be a useful material for vector control of malaria as a “transmission-blocking sugar” in the future (Ferreira et al., 2018).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Experiment Committee of Jichi Medical University (approval numbers: 22002-01).
Author contributions
Conceptualization: DM, MA, and HK. Methodology: DM, DY, AT, MA, and HK. Software: DM. Validation: DM, AT, MA, and HK. Formal analysis: DM. Investigation: DM and MA. Resources: DM, DY, AT, MA, and HK. Writing—original draft preparation, DM and HK. Writing—review and editing: DM, DSY, AT, MA, and HK; visualization: DM; supervision: AM and HK; project administration: DM and HK. Funding acquisition: DM and HK. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by JSPS KAKENHI grant numbers 19K23849, 20K16237, and 20H03155.
Acknowledgments
We would like to thank Dr. Shinya Watanabe (Division of Bacteriology, Department of Infection and Immunity, Jichi Medical University) for the technical support of Miseq analysis. We also would like to thank Chisato Seki and Keiko Watano (Division of Medical Zoology, Jichi Medical University) for maintenance of the mosquito colony.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1162918/full#supplementary-material
References
Almire, F., Terhzaz, S., Terry, S., McFarlane, M., Gestuveo, R. J., Szemiel, A. M., et al. (2021). Sugar feeding protects against arboviral infection by enhancing gut immunity in the mosquito vector aedes aegypti. PloS Pathog. 17, e1009870. doi: 10.1371/journal.ppat.1009870
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecology. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Autino, B., Noris, A., Russo, R., Castelli, F. (2012). Epidemiology of malaria in endemic areas. Mediterr J. Hematol. Infect. Dis. 4, e2012060. doi: 10.4084/MJHID.2012.060
Bahia, A. C., Dong, Y., Blumberg, B. J., Mlambo, G., Tripathi, A., BenMarzouk-Hidalgo, O. J., et al. (2014). Exploring Anopheles gut bacteria for Plasmodium blocking activity. Environ. Microbiol. 16, 2980–2994. doi: 10.1111/1462-2920.12381
Bahia, A. C., Oliveira, J. H. M., Kubota, M. S., Araújo, H. R. C., Lima, J. B. P., Ríos-Velásquez, C. M., et al. (2013). The role of reactive oxygen species in Anopheles aquasalis response to Plasmodium vivax infection. PloS One 8, e57014. doi: 10.1371/journal.pone.0057014
Bai, L., Wang, L., Vega-Rodríguez, J., Wang, G., Wang, S. (2019). A gut symbiotic bacterium Serratia marcescens renders mosquito resistance to Plasmodium infection through activation of mosquito immune responses. Front. Microbiol. 10. Available: https://www.frontiersin.org/article/doi: 10.3389/fmicb.2019.01580
Bando, H., Okado, K., Guelbeogo, W. M., Badolo, A., Aonuma, H., Nelson, B., et al. (2013). Intra-specific diversity of Serratia marcescens in Anopheles mosquito midgut defines Plasmodium transmission capacity. Sci. Rep. 3, 1641. doi: 10.1038/srep01641
Baton, L. A., Ranford-Cartwright, L. C. (2004). Plasmodium falciparum ookinete invasion of the midgut epithelium of Anopheles stephensi is consistent with the time bomb model. Parasitology. 129, 663–676. doi: 10.1017/S0031182004005979
Baxter, R. H. G., Steinert, S., Chelliah, Y., Volohonsky, G., Levashina, E. A., Deisenhofer, J. (2010). A heterodimeric complex of the LRR proteins LRIM1 and APL1C regulates complement-like immunity in anopheles gambiae. PNAS 107, 16817–16822. doi: 10.1073/pnas.1010575107
Blandin, S., Shiao, S.-H., Moita, L. F., Janse, C. J., Waters, A. P., Kafatos, F. C., et al. (2004). Complement-like protein TEP1 is a determinant of vectorial capacity in the malaria vector anopheles gambiae. Cell 116, 661–670. doi: 10.1016/S0092-8674(04)00173-4
Boissière, A., Tchioffo, M. T., Bachar, D., Abate, L., Marie, A., Nsango, S. E., et al. (2012). Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PloS Pathogens. 8, e1002742. doi: 10.1371/journal.ppat.1002742
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 6, 90. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., Holmes, S. P. (2016). DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cirimotich, C. M., Dong, Y., Clayton, A. M., Sandiford, S. L., Souza-Neto, J. A., Mulenga, M., et al. (2011). Natural microbe-mediated refractoriness to Plasmodium infection in anopheles gambiae. Science 332, 855–858. doi: 10.1126/science.1201618
Delong, E. F. (1992). Archaea in coastal marine environments. Proc. Natl. Acad. Sci. U.S.A. 89, 5685–5689. doi: 10.1073/pnas.89.12.5685
Dessens, J. T., Sidén-Kiamos, I., Mendoza, J., Mahairaki, V., Khater, E., Vlachou, D., et al. (2003). SOAP, a novel malaria ookinete protein involved in mosquito midgut invasion and oocyst development. Mol. Microbiol. 49, 319–329. doi: 10.1046/j.1365-2958.2003.03566.x
Dinglasan, R. R., Kalume, D. E., Kanzok, S. M., Ghosh, A. K., Muratova, O., Pandey, A., et al. (2007). Disruption of Plasmodium falciparum development by antibodies against a conserved mosquito midgut antigen. Proc. Natl. Acad. Sci. 104, 13461–13466. doi: 10.1073/pnas.0702239104
Dong, Y., Aguilar, R., Xi, Z., Warr, E., Mongin, E., Dimopoulos, G. (2006). Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PloS Pathogens. 2, e52. doi: 10.1371/journal.ppat.0020052
Dong, Y., Manfredini, F., Dimopoulos, G. (2009). Implication of the mosquito midgut microbiota in the defense against malaria parasites. PloS Pathogens. 5, e1000423. doi: 10.1371/journal.ppat.1000423
Ecker, A., Pinto, S. B., Baker, K. W., Kafatos, F. C., Sinden, R. E. (2007). Plasmodium berghei: plasmodium perforin-like protein 5 is required for mosquito midgut invasion in anopheles stephensi. Exp. Parasitol. 116, 504–508. doi: 10.1016/j.exppara.2007.01.015
Ferreira, T. N., Pita-Pereira, D., Costa, S. G., Brazil, R. P., Moraes, C. S., Díaz-Albiter, H. M., et al. (2018). Transmission blocking sugar baits for the control of Leishmania development inside sand flies using environmentally friendly beta-glycosides and their aglycones. Parasites Vectors. 11, 614. doi: 10.1186/s13071-018-3122-z
Fiorenzano, J. M., Koehler, P. G., Xue, R.-D. (2017). Attractive toxic sugar bait (ATSB) for control of mosquitoes and its impact on non-target organisms: a review. IJERPH. 14, 398. doi: 10.3390/ijerph14040398
Frischknecht, F., Baldacci, P., Martin, B., Zimmer, C., Thiberge, S., Olivo-Marin, J.-C., et al. (2004). Imaging movement of malaria parasites during transmission by Anopheles mosquitoes. Cell. Microbiol. 6, 687–694. doi: 10.1111/j.1462-5822.2004.00395.x
Ghosh, A. K., Coppens, I., Gårdsvoll, H., Ploug, M., Jacobs-Lorena, M. (2011). Plasmodium ookinetes coopt mammalian plasminogen to invade the mosquito midgut. Proc. Natl. Acad. Sci. 108, 17153–17158. doi: 10.1073/pnas.1103657108
Gibbins, L. N., Simpson, F. J. (1964). The incorporation of d-allose into the glycolytic pathway by aerobacter aerogenes. Can. J. Microbiol. 10, 829–836. doi: 10.1139/m64-108
González-Lázaro, M., Dinglasan, R. R., Hernández-Hernández F de la, C., Rodríguez, M. H., Laclaustra, M., Jacobs-Lorena, M., et al. (2009). Anopheles gambiae croquemort SCRBQ2, expression profile in the mosquito and its potential interaction with the malaria parasite plasmodium berghei. Insect Biochem. Mol. Biol. 39, 395–402. doi: 10.1016/j.ibmb.2009.03.008
Kadota, K., Ishino, T., Matsuyama, T., Chinzei, Y., Yuda, M. (2004). Essential role of membrane-attack protein in malarial transmission to mosquito host. Proc. Natl. Acad. Sci. 101, 16310–16315. doi: 10.1073/pnas.0406187101
Kano, A., Fukumoto, T., Ohtani, K., Yoshihara, A., Ohara, T., Tajima, S., et al. (2013). The rare sugar d-allose acts as a triggering molecule of rice defence via ROS generation. J. Exp. Botany. 64, 4939–4951. doi: 10.1093/jxb/ert282
Kariu, T., Ishino, T., Yano, K., Chinzei, Y., Yuda, M. (2006). CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Mol. Microbiol. 59, 1369–1379. doi: 10.1111/j.1365-2958.2005.05024.x
Kimura, S., Zhang, G.-X., Nishiyama, A., Nagai, Y., Nakagawa, T., Miyanaka, H., et al. (2005). D-allose, an all-cis aldo-hexose, suppresses development of salt-induced hypertension in Dahl rats. J. Hypertension. 23, 1887–1894. doi: 10.1097/01.hjh.0000182523.29193.e3
Kotsyfakis, M., Ehret-Sabatier, L., Siden-Kiamos, I., Mendoza, J., Sinden, R. E., Louis, C. (2005). Plasmodium berghei ookinetes bind to Anopheles gambiae and Drosophila melanogaster annexins. Mol. Microbiol. 57, 171–179. doi: 10.1111/j.1365-2958.2005.04664.x
Lavazec, C., Boudin, C., Lacroix, R., Bonnet, S., Diop, A., Thiberge, S., et al. (2007). Carboxypeptidases b of anopheles gambiae as targets for a Plasmodium falciparum transmission-blocking vaccine. Infection Immunity. 75, 1635–1642. doi: 10.1128/IAI.00864-06
Luckhart, S., Vodovotz, Y., Cui, L., Rosenberg, R. (1998). The mosquito anopheles stephensi limits malaria parasite development with inducible synthesis of nitric oxide. Proc. Natl. Acad. Sci. U.S.A. 95, 5700–5705. doi: 10.1073/pnas.95.10.5700
Mano, M. C. R., Neri-Numa, I. A., da Silva, J. B., Paulino, B. N., Pessoa, M. G., Pastore, G. M. (2018). Oligosaccharide biotechnology: an approach of prebiotic revolution on the industry. Appl. Microbiol. Biotechnol. 102, 17–37. doi: 10.1007/s00253-017-8564-2
Matsuoka, H., Tomita, H., Hattori, R., Arai, M., Hirai, M. (2015). Visualization of malaria parasites in the skin using the luciferase transgenic parasite, plasmodium berghei. Trop. Med. Health 43, 53–61. doi: 10.2149/tmh.2014-18
Moffett, A., Shackelford, N., Sarkar, S. (2007). Malaria in Africa: vector species’ niche models and relative risk maps. PloS One 2, e824. doi: 10.1371/journal.pone.0000824
Molina-Cruz, A., DeJong, R. J., Charles, B., Gupta, L., Kumar, S., Jaramillo-Gutierrez, G., et al. (2008). Reactive oxygen species modulate Anopheles gambiae immunity against bacteria and Plasmodium*. J. Biol. Chem. 283, 3217–3223. doi: 10.1074/jbc.M705873200
Msomi, N., Erukainure, O., Islam, M. (2021). Suitability of sugar alcohols as antidiabetic supplements: a review. J. Food Drug Analysis. 29, 1–14. doi: 10.38212/2224-6614.3107
Murata, A., Sekiya, K., Watanabe, Y., Yamaguchi, F., Hatano, N., Izumori, K., et al. (2003). A novel inhibitory effect of d-allose on production of reactive oxygen species from neutrophils. J. Bioscience Bioengineering. 96, 89–91. doi: 10.1016/S1389-1723(03)90104-6
Muturi, E. J., Lagos-Kutz, D., Dunlap, C., Ramirez, J. L., Rooney, A. P., Hartman, G. L., et al. (2018). Mosquito microbiota cluster by host sampling location. Parasites Vectors. 11, 468. doi: 10.1186/s13071-018-3036-9
Osta, M. A. (2004). Effects of mosquito genes on Plasmodium development. Science. 303, 2030–2032. doi: 10.1126/science.1091789
Poulsen, T. S., Chang, Y.-Y., Hove-Jensen, B. (1999). D-allose catabolism of Escherichia coli: involvement of alsI and regulation of als regulon expression by allose and ribose. J. Bacteriol. 181, 7126–7130.
Povelones, M., Upton, L. M., Sala, K. A., Christophides, G. K. (2011). Structure-function analysis of the Anopheles gambiae LRIM1/APL1C complex and its interaction with complement C3-like protein TEP1. PloS Pathogens. 7, e1002023. doi: 10.1371/journal.ppat.1002023
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramirez, J. L., Short, S. M., Bahia, A. C., Saraiva, R. G., Dong, Y., Kang, S., et al. (2014). Chromobacterium Csp_P reduces malaria and dengue infection in vector mosquitoes and has entomopathogenic and in vitro anti-pathogen activities. PloS Pathogens. 10, e1004398. doi: 10.1371/journal.ppat.1004398
Rodríguez M del, C., Martínez-Barnetche, J., Alvarado-Delgado, A., Batista, C., Argotte-Ramos, R. S., Hernández-Martínez, S., et al. (2007). The surface protein Pvs25 of Plasmodium vivax ookinetes interacts with calreticulin on the midgut apical surface of the malaria vector anopheles albimanus. Mol. Biochem. Parasitol. 153, 167–177. doi: 10.1016/j.molbiopara.2007.03.002
Rudin, W., Hecker, H. (1989). Lectin-binding sites in the midgut of the mosquitoes Anopheles stephensi liston and Aedes aegypti l. (Diptera: Culicidae). Parasitol. Res. 75, 268–279. doi: 10.1007/BF00931811
Sandeu, M. M., Maffo, C. G. T., Dada, N., Njiokou, F., Hughes, G. L., Wondji, C. S. (2022). Seasonal variation of microbiota composition in Anopheles gambiae and Anopheles coluzzii in two different eco-geographical localities in Cameroon. Med. Vet. Entomol. 36, 3. doi: 10.1111/mve.12583
Siden-Kiamos, I., Vlachou, D., Margos, G., Beetsma, A., Waters, A. P., Sinden, R. E., et al. (2000). Distinct roles for pbs21 and pbs25 in the in vitro ookinete to oocyst transformation of plasmodium berghei. J. Cell Sci. 113, 3419–3426. doi: 10.1242/jcs.113.19.3419
Soo Han, Y., Thompson, J., Kafatos, F. C., Barillas-Mury, C. (2001). Molecular interactions between Anopheles stephensi midgut cells and Plasmodium berghei: the time bomb theory of ookinete invasion of mosquitoes. EMBO J. 20, 1483–1483. doi: 10.1038/sj.emboj.7593651b
Tokuda, M., Arai, M., Okuma, K. (2015)Malaria transmission prevention agent having rare sugar as effective component thereof and malarial parasite growth regulating agent. In: Japanese Patent JP 201406207. Available at: https://patentimages.storage.googleapis.com/16/8e/a6/35ab0e2cf2f05b/EP3124028B1.pdf (Accessed 13 July 2022).
Tomas, A. M., Margos, G., Dimopoulos, G., van Lin, L. H. M., de Koning-Ward, T. F., Sinha, R., et al. (2001). P25 and P28 proteins of the malaria ookinete surface have multiple and partially redundant functions. EMBO J. 20, 3975–3983. doi: 10.1093/emboj/20.15.3975
Vizioli, J., Bulet, P., Hoffmann, J. A., Kafatos, F. C., Muller, H.-M., Dimopoulos, G. (2001). Gambicin: a novel immune responsive antimicrobial peptide from the malaria vector anopheles gambiae. Proc. Natl. Acad. Sci. 98, 12630–12635. doi: 10.1073/pnas.221466798
Wang, M., An, Y., Gao, L., Dong, S., Zhou, X., Feng, Y., et al. (2021). Glucose-mediated proliferation of a gut commensal bacterium promotes Plasmodium infection by increasing mosquito midgut pH. Cell Rep. 35, 108992. doi: 10.1016/j.celrep.2021.108992
World Health Organization (WHO) (2021) World malaria report. Available at: https://www.who.int/publications-detail-redirect/9789240040496 (Accessed 10 Feb).
Yamamoto, D. S., Sumitani, M., Kasashima, K., Sezutsu, H., Matsuoka, H. (2016). Inhibition of malaria infection in transgenic anopheline mosquitoes lacking salivary gland cells. PloS Pathogens. 12, e1005872. doi: 10.1371/journal.ppat.1005872
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “All-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Yuda, M., Yano, K., Tsuboi, T., Torii, M., Chinzei, Y. (2001). Von willebrand factor a domain-related protein, a novel microneme protein of the malaria ookinete highly conserved throughout Plasmodium parasites. Mol. Biochem. Parasitology. 116, 65–72. doi: 10.1016/S0166-6851(01)00304-8
Zhang, L., Wang, Y., Liu, D., Luo, L., Wang, Y., Ye, C. (2018). Identification and characterization of als genes involved in d-allose metabolism in lineage II strain of listeria monocytogenes. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00621
Keywords: allose, microbiota, midgut, Anopheles, Plasmodium, luciferase assay, next-generation sequencing
Citation: Mizushima D, Yamamoto DS, Tabbabi A, Arai M and Kato H (2023) A rare sugar, allose, inhibits the development of Plasmodium parasites in the Anopheles mosquito independently of midgut microbiota. Front. Cell. Infect. Microbiol. 13:1162918. doi: 10.3389/fcimb.2023.1162918
Received: 21 February 2023; Accepted: 28 June 2023;
Published: 20 July 2023.
Edited by:
Shahid Karim, University of Southern Mississippi, United StatesReviewed by:
Yuemei Dong, Johns Hopkins University, United StatesJessica Molina-Franky, Beckman Research Institute, City of Hope, United States
Copyright © 2023 Mizushima, Yamamoto, Tabbabi, Arai and Kato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daiki Mizushima, ZG1penVzaGltYUBqaWNoaS5hYy5qcA==
 Daiki Mizushima
Daiki Mizushima Daisuke S. Yamamoto
Daisuke S. Yamamoto Ahmed Tabbabi
Ahmed Tabbabi Meiji Arai
Meiji Arai Hirotomo Kato
Hirotomo Kato