
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 25 April 2023
Sec. Microbial Vaccines
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1162299
Vibrio alginolyticus is the common pathogen affecting various species of marine organisms. It has been demonstrated that fliR is a necessary virulence factor to adhere and infect their hosts for pathogenic bacteria. Frequent disease outbreaks in aquaculture have highlighted the necessity of developing effective vaccines. In the present study, in order to investigate the function of fliR in V.alginolyticus, the fliR deletion mutant ΔfliR was constructed and its biological properties were evaluated, additionally, the differences in gene expression levels between wild-type and ΔfliR were analyzed by transcriptomics. Finally, ΔfliR was used as a live attenuated vaccine to immunize grouper via the intraperitoneal route to evaluate its protective effect. Results show that fliR gene of V. alginolyticus was identified as being 783 bp in length, encoding 260 amino acids, and showing significant similarity to homologs of other Vibrio species. The fliR-deletion mutant ΔfliR of V. alginolyticus was successfully constructed, and its biological phenotype analysis showed no significant differences in growth capacity and extracellular enzyme activity compared to the wild-type. However, a substantial reduction of motility ability was detected in ΔfliR. Transcriptomic analysis revealed that the absence of fliR gene is responsible for a significantly decreased expression of flagellar genes, including flaA, flaB, fliS, flhB and fliM. The fliR-deletion mainly affects the related pathways involved in cell motility, membrane transport, signal transduction, carbohydrate metabolism, and amino acid metabolism in V. alginolyticus. The efficacy of ΔfliR as a candidate of live attenuated vaccine were evaluated by intraperitoneal injection in grouper. The ΔfliR provided the RPS (Relative protection rate) of 67.2% against V. alginolyticus in groupers. The ΔfliR efficiently stimulated antibody production with specific IgM still detected at 42 d post-vaccination, and significantly elevated the activity of antioxidant enzymes like Catalase (CAT), Superoxide dismutase (SOD), and lactate dehydrogenase (LDH) in the serum. The higher expression levels of immune-related genes were observed in the immune tissues of inoculated grouper compared to the control. In conclusion, ΔfliR effectively improved the immunity of inoculated fish. The results suggest that ΔfliR is an effective live attenuated vaccine against vibriosis in in grouper.
Intensive marine fish aquaculture is developing rapidly in coastal areas of China. With the fast expansion of mariculture, more and more diseases have outbroken and led to the huge economic loss. Vibrio is a kind of ubiquitous pathogens in marine and estuarine environments, and consumption of undercooked seafood or exposure to Vibrio-contaminated seawater may cause Vibrio infection (Adeleye et al., 2010; Mrityunjoy et al., 2013; Baker-Austin et al., 2018; Song et al., 2020). Vibrio alginolyticus is one of the dominant pathogens in the marine environment and associated with a variety of diseases in marine organisms such as fish, crustaceans, and mollusks (Mustapha et al., 2013; Li et al., 2021; Sadat et al., 2021; Yang et al., 2021). Investigations have demonstrated that V. alginolyticus is responsible for ulceration, bleeding, and black spots on the epidermis of various marine organisms, causing extremely bad effect to the aquaculture industry (Abdallah et al., 2011).
The pathogenic processes of Vibrio include adhesion, invasion and in vivo proliferation, acting through the production of pathogenic factors that interfere with the normal metabolism of host cells (Li et al., 2003). Bacterial adhesion to the host is an important step and the initial stage of bacterial infection (Qi et al., 2022). Motility is usually essential for bacterial adhesion. The flagellum is the primary organelle for the motility of many bacteria (Chaban et al., 2015). Bacterial flagella are large macromolecular complexes composed of a hook, filament, and basal body (Zhu et al., 2017). The filament is a helical propeller consisting of a repetitive protein monomer flagellin. The hook is a curved hollow cylinder attached to the basal body and is a flexible gimbal. The basal body is anchored to the cell membrane, houses the secretory apparatus, and provides the power for flagellar rotation (Mukherjee and Kearns, 2014). The flagellar biosynthetic protein fliR is an integral membrane protein thought to be located in the central pore of the MS ring of the bacterial flagellar type III protein exporter along with FlhA, FlhB, FliO, FliP and FliQ (Mukherjee and Kearns, 2014). Recent study have shown that fliR is required for severe damage to the intestinal epithelium in Streptococcus mucilaginous (Sina Rahme et al., 2022). Related study in V. harveyi have shown that FliA, FliR and FlrB are the essential proteins that mediate the bacterial adhesion (Qi et al., 2022). However, the role of fliR in V.alginolyticus has not been demonstrated.
Bacterial diseases in fish are considered a significant problem in aquaculture. Overuse of antibiotics causes the increasing bacterial resistance and drugs residue, and vaccines are emerging as an essential strategy to prevent fish diseases (Minor, 2015). Vaccines include inactivated vaccine, attenuated live vaccine, recombinant vaccine and DNA vaccine. Attenuated vaccines have many noteworthy advantages, including low stress on the fish, the possibility of soaking or oral vaccines, and the ability to induce effective humoral and cell-mediated immune responses (Mohd-Aris et al., 2019). Research on live attenuated vaccines dates back to measles, mumps, rubella, and varicella zoster (Goh et al., 2007; Dhillon and Curran, 2008; Schuster et al., 2008). Attenuated live vaccines for human diseases are among the most successful and cost-effective interventions in the history of medicine (Minor, 2015). Attenuated live vaccines are obtained by removing critical virulent genes from strongly virulent strain and weakening their virulence to form the avirulent strain. Research has indicated that adherence is an essential virulence characteristic of V. alginolyticus (Luo et al., 2016). In the present study, to understand the function of fliR in V. alginolyticus, we firstly constructed fliR-deletion mutant ΔfliR and studied its biological characteristics, then analyzed the differences in gene expression levels between the wild-type and ΔfliR by transcriptomics. Finally, ΔfliR was used as an attenuated live vaccine to immunize grouper via the intraperitoneal route and demonstrate its protective effect.
Healthy pearl gentian grouper (weight: 50.0 ± 5.0 g) was purchased from a fish farm in Donghai Island (Zhanjiang). The experimental fish were cultured in a temporary pond for two weeks before being placed in a tank at 28°C with daily seawater changes. The fish were fed twice daily with a commercial diet. Before sampling, the experiment fish were anesthetized with MS222 (Sigma, USA), and the tissue (liver, spleen and kidney) was surgically sampled and stored in liquid nitrogen. V. alginolyticus HY9901 (wild-type) was isolated from the diseased grouper and conserved in our laboratory. The plasmid used to construct the mutant in this study was the PLP12CM suicide plasmid.
V. alginolyticus HY9901 fliR gene was obtained from the whole bacterial genome assembly (GCA_023206775.1). In this study, in-frame deletion of the fliR gene was performed by overlapping extension PCR. The primers fliR-MF1/fliR-MR1 were used to amplify the upstream homologous arm fragment of fliR with a length of 763 bp. The primers fliR-MF2/fliR-MR2 were used to amplify the downstream homologous arm fragment of fliR with a length of 718 bp. Amplification procedure is following: 98°C for 3 min. 35 cycles of 98°C for 10 sec, 52/58°C for 20 sec and an elongation stage at 72°C for 1 min. The amplified fliR upstream and downstream homologous arm fragments were used as templates for overlapping PCR amplification. Amplification program is following: 98°C for 1 min followed by 35 cycles of 98°C for 10 sec, 68°C for 20 sec and an elongation stage at 72°C for 1 min. The fusion fragment was 1481 bp in length and was ligated to the suicide vector PLP12CM. The recombinant product was transformed into Escherichia coli β2163 receptor cells. The recombinant plasmids PLP12CM-fliR were transferred from E. coli β2163 into V. alginolyticus HY990 by conjugation. The single crossover mutants were screened on LB agar medium containing 20 μg/mL chloramphenicol and 0.3% D-glucose at 28°C. The second crossover mutants were selected on LB agar with 0.4% L-arabinose. The V. alginolyticus fliR-deletion mutants (ΔfliR) were confirmed by PCR with the primers pLP-UF/pLP-UR.
The ΔfliR and wild-type strain were cultured in TSB liquid medium on a rotary shaker (120 rpm) in a 28°C incubator. The OD600 was measured by pipetting the bacterial solution every 2 h.
The ΔfliR was passaged 30 times continuously on the TSB medium, and colonies of each generation were amplified by PCR and sequenced to determine the genetic stability of ΔfliR for subsequent research.
Sterile cellophane was tightly attached to TSA solid medium, and subsequently, wild-type and ΔfliR were coated on the cellophane separately. The solid medium was placed in a constant temperature incubator at 28°C for 24 h, followed by washing the cellophane with sterile phosphate buffered saline (PBS, pH 7.4), aspirating the suspension, and centrifuging at 5000 rpm for 30 min at 4°C. The supernatant obtained was filtered with a membrane (pore size 0.22 μm), 100 µl of the filtered liquid was added to 400 µl of 5 mg/mL azo casein solution, and the mixture was allowed to stand at 37°C for 30 min, then 400 µl of 10% trichloroacetic acid was added, and after standing for 30 min, sodium hydroxide was added. Finally, the mixture was read by spectrophotometer for absorbance at 442 nm.
The ΔfliR and wild-type were inoculated with toothpicks on plates with an agar content of 0.3%. The plates were incubated overnight at 28°C to observe the size of swimming rings and measure the diameter of the swimming rings.
To investigate the pathogenicity of wild-type strains and deletion strains of the strain. The LD50 was determined. The wild-type and ΔfliR were incubated in TSB medium, respectively. After 12 h of incubation, the bacterial solutions were collected, and the concentrations of the two strains were adjusted to 104, 105, 106, 107 and 108 CFU/mL with PBS. Healthy groupers were selected and divided into 2 groups (150 fish/group), each group was further divided into 5 subgroups, and each subset of fish was injected with different concentrations of bacterial solution in turn. The control group of 30 fish was injected with an equal volume of PBS. Mortality and morbidity monitoring was subsequently carried out for 2 weeks.
The wild-type and ΔfliR were incubated in TSB medium with a speed of 100 rpm at 28°C. After 10 h of incubation, the bacterial solution was centrifuged at 6000 g for 15 min, and the sediment was dissolved with Trizol to extract RNA. RNA quality was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, USA) and sequenced using the Illumina Novaseq6000 platform (China Gene Denovo Biotechnology Ltd.) for sequencing. Fastp (version 0.18.0) was used to filter the raw data. Mapping was performed regarding the GCA_001679745.1 genome. Three biological replicates were set for each sample. Transcriptomics data were commissioned for analysis by China Gene Denovo Biotechnology Ltd. Quantitative real-time PCR (qRT-PCR) is used to detect differential gene mRNA expression levels.
Sixty groupers were divided randomly into 2 groups. The injection doses for the experimental and control groups were as described in 2.5. Three fish in each group were randomly selected at 1, 2, 3, 4, 6 and 8 d after injection and anesthetized with an excess of MS-222 (Sigma, USA), and liver, spleen and kidney tissues were collected. Each sample was weighed and grounded in 1 mL of sterile PBS. The suspensions were diluted 10, 100 and 1000 times of the homogenate and applied on TCBS agar plates to determine the number of bacteria. TCBS medium is a common method for isolating pathogenic Vibrio in medical clinics. In healthy fish tissue, there is barely the presence of Vibrio. The experiment was performed three times independently.
Healthy groupers were divided randomly into 2 groups of 300 fish each, and the fish in each group were cultured separately in 10 buckets. The injection doses for the experimental and control groups were as above. After 6 weeks of immunization, each group of fish was divided into 2 subgroups, with subgroup 1 injected with 100 μl of 1 × 109 CFU/mL wild-type and subgroup 2 injected with the same volume of PBS. Clinical signs and mortality of grouper were monitored for 2 weeks after challenge. RPS was calculated by the following formula: RPS (%) = [1-(Mortality of vaccinated fish/Mortality of control fish)] × 100%.
Tail vein blood was taken from the experimental and control groups at 1-7 weeks. Blood samples were left at room temperature for 1 h, centrifuged at 3000 rpm for 10 min at 4°C, and the supernatant (serum) was aspirated and stored at -80°C. For detection of specific IgM against V. alginolyticus, The serum was continuously diluted with 2-fold gradient (from 21 to 212 folds dilution). Specific IgM antibody levels were measured by ELISA at 1-6 weeks after vaccination. After 12 h of incubation, V. alginolyticus HY9901 was resuspended in PBS and adjusted the concentration to 1 × 108 CFU/mL. The 100 µl of bacterial suspension was added to each well of a 96-well microtiter plate and incubated overnight at 4°C. The wells were washed with PBST three times and blocked using 100 μl of 5% non-fat powdered milk (Sangon Biotech, Guangzhou, China) for 2 h. And then, the wells were washed with PBST three times, then added to 2-fold gradient diluted grouper serum and incubated at 37°C for 1 h. The wells were washed with PBST three times, and added 100 μl 1:1000 mouse-anti-pearl gentian grouper IgM polyclonal antibody as first antibody and incubated at 37°C for 1 h. The wells were washed with PBST three times, and incubated 100 μl 1:2000 horseradish peroxidase (HRP)-conjugated goat anti-mouse IgM as the secondary antibody, and incubated for 3 h at room temperature. After washing three times with PBST, color development buffer was added, and the absorbance was read at 450 nm with a spectrophotometer after 20 min of incubation. The experiment was performed three times.
Superoxide dismutase (SOD) is involved in the body’s natural immunity. catalase (CAT) maintains the redox balance of the immune system. lactate dehydrogenase (LDH) rises in the serum when the tissues of an organism are damaged. The activity of these three enzymes was therefore measured after immunization. SOD, LDH, and CAT concentrations were measured using commercial kits from Nanjing Chengjian Institute of Biological Engineering and according to the manufacturer’s instructions. All samples were measured on a microplate reader. The experiment was performed three times.
The liver, spleen and kidney of fish in experimental and control groups were collected for qRT-PCR on days 0, 1, 2, 3, 4, 5 and 6 after inoculation. The qRT-PCR was performed to detect the expression levels of caspase8, CD4, IgM, MHC-IIβ, IL6 and TNF-α. The genes selected are grouper innate and adaptive immunity-related genes. Primers are shown in Table 1, and β-actin was used as an internal reference. The qRT-PCR procedure was 10 min at 95°C. 40 amplification cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 30 s. The expression levels of the related immune genes were analyzed by the 2-ΔΔCT method. The experiment was performed three times.
All statistical analyses were performed using one-way ANOVA, and all data are expressed as mean ± standard error. Statistical significance (P < 0.05) differences between groups were tested by Duncan’s multiple range test.
The fliR gene was identified as being 783 bp in length, encoding 260 amino acids, and multiple sequence comparisons showed significant similarity to homologs of other Vibrio species (Figure 1A). To generate the fliR-deletion mutation ΔfliR, the intermediate region of the fliR gene was deleted by allelic exchange (Figure 1B). The deletion of 717 bp of fliR gene truncation by allele exchange was confirmed by PCR amplification and subsequent sequencing (Figure 1C). The sequence of the fliR has been uploaded to the GenBank database with accession number OQ813764.
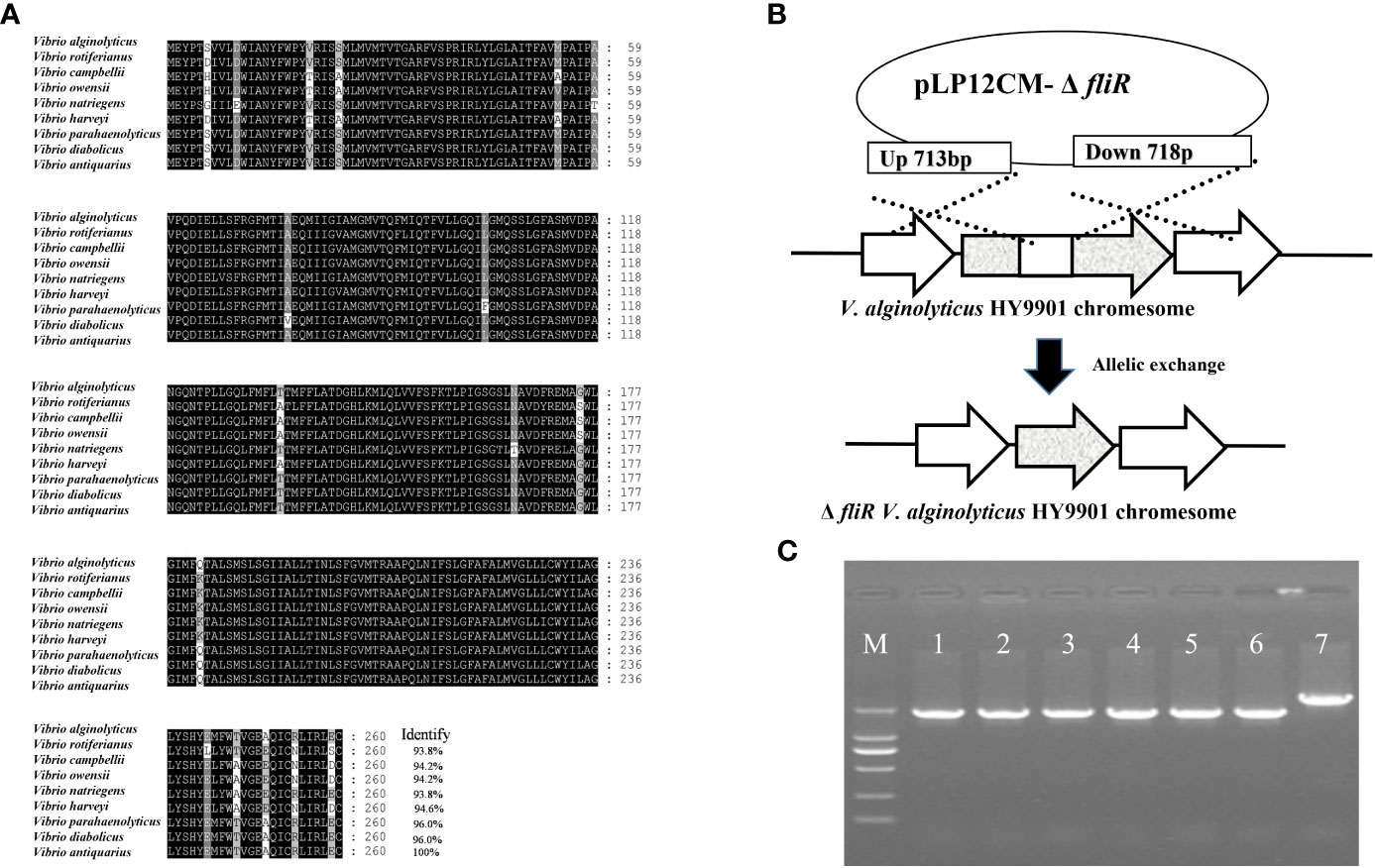
Figure 1 Identification of fliR gene from V. alginolyticus HY9901 strain and construction of fliR mutant strain. Multiple alignments of the fliR amino acid sequences of V. alginolyticus HY9901 with other Vibrio species (A). The fliR gene of V. alginolyticus HY9901 was knocked out by allele exchange (B). PCR amplification of wild-type V. alginolyticus HY9901 (lane 6), fliR mutant strain (lanes 1-5). Lane M is a 1 kb ladder. The PCR product of the mutant strain is smaller than that of wild-type V. alginolyticus HY9901 due to the partial deletion of the corresponding gene. The construction of the deletion strain was confirmed by PCR amplification (C).
The ΔfliR had stable genetics after 30 consecutive passages on TSB medium and its identification by PCR. Characterization of the biological phenotype of ΔfliR demonstrated that the deletion of fliR failed to significantly affect the growth ability and extracellular protease activity of V. alginolyticus (Figure 2A). However, after 12 h of incubation, the motility assay revealed that the swarming diameter of the wild-type was about 35 mm, and the swarming diameter of ΔfliR was only 3 mm, indicating that fliR-deletion had a significant effect on the motility of V. alginolyticus (Figure 2B). The LD50 of the wild-type was found to be 2.37×105 CFU/mL, while the LD50 of ΔfliR was 3.48×107 CFU/mL (Table 2), indicating that the deletion of fliR significantly diminished the pathogenicity of alginolyticus (Table 3).
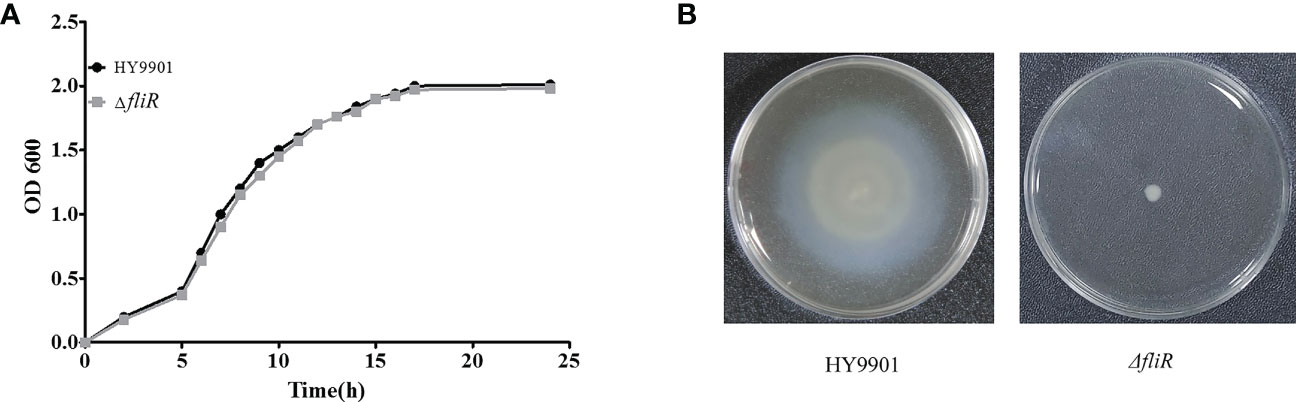
Figure 2 The biological properties of V. alginolyticus HY9901 compared with the fliR mutant strain. HY9901 and mutant strain growth curves (A), values of OD600 of the bacterial suspensions were measured at different time points. Motility measurements. The swarming diameter of the wild-type strain was about 35 mm, and that of the mutant strain was 3 mm (B).
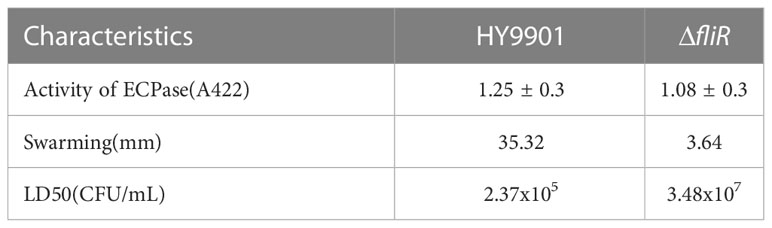
Table 3 The biological properties of Vibrio alginolyticus HY9901 compared with the fliR mutant strain.
To investigate the function of fliR in V. alginolyticus, RNA-seq was used to investigate the related pathways affected because of the absence of fliR gene. The transcriptome of wild-type and ΔfliR was compared to identify differentially expressed genes (DEGs). Gene expression data were normalized in all replicates for differential expression analysis. To analyze the DEG, we chose a false discovery rate of less than 5% and a fold change >1.5 to obtain significant differences (All data are analyzed through the online platform of China Gene Denovo Biotechnology Ltd). Several differentially expressed genes were randomly selected for qPCR to validate transcriptome results (not shown in the article). The results showed that a total of 678 genes were found to be differentially expressed. Among them, 398 genes were downregulated, and 280 genes were upregulated in ΔfliR compared to wild-tpye (Figure 3A). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed that a large number of DEGs were mainly associated with carbohydrate metabolism, amino acid metabolism, cell motility, membrane transport, signal transduction (Figure 3B). Most of these significantly down-regulated genes were associated with flagella (Table 4). Raw transcriptome sequencing data were submitted to GenBank with the accession number PRJNA954769.
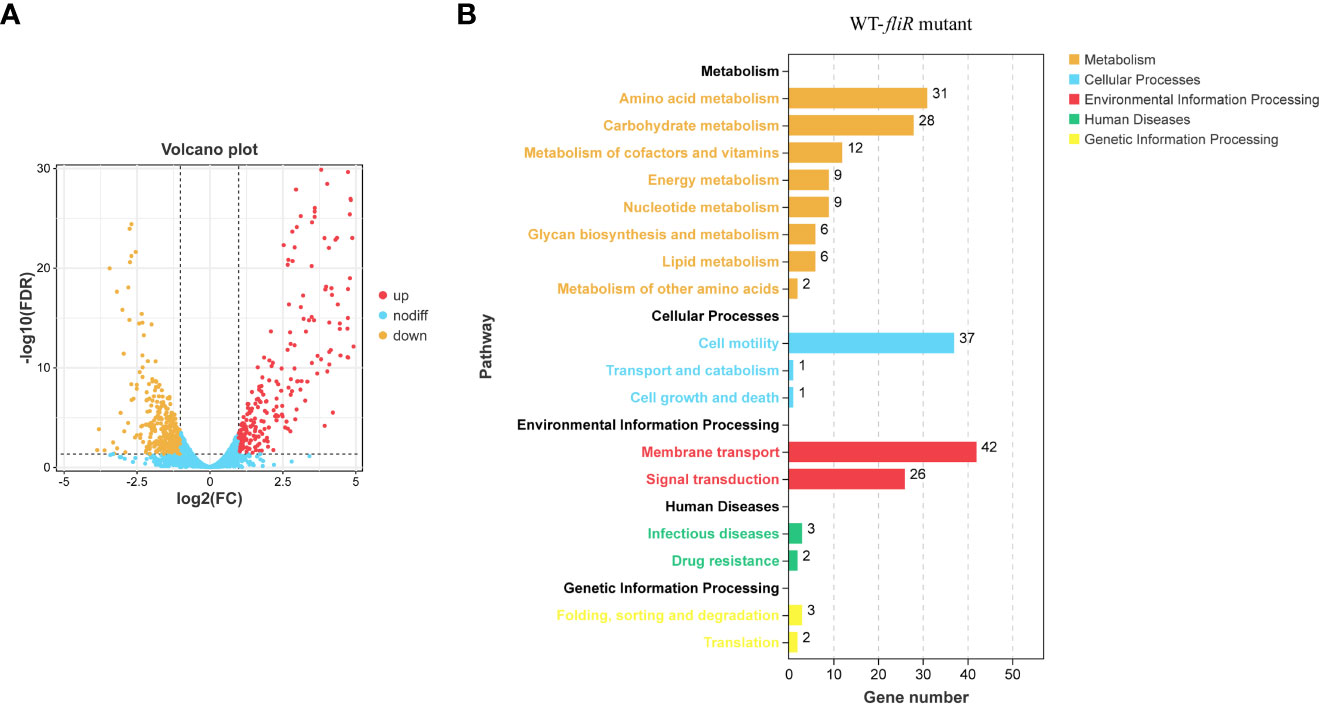
Figure 3 Comparison of the transcriptomes of fliR mutant strains and wild-type strains. The transcriptomes of the wild and mutant strains identified the number of differentially expressed genes, with 280 up-regulated genes and 398 down-regulated genes in the mutant strains (A). The Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis shows that mutations in fliR mainly affect pathways related to carbohydrate metabolism, amino acid metabolism, cell motility, membrane transport, and signal transduction (B).
Following intraperitoneal injection, tissue samples from experimental fish were sampled consecutively for the bacterial load. After 12 h of administration, the maximum numbers of ΔfliR in the liver (Figure 4A), spleen (Figure 4B), and kidney (Figure 4C) were found to be about 106 CFU/g. Bacterial counts were found to diminish in all tissues tested 1 day after inoculation, and a significant decline in counts was detected after 5 days, while only a slight amount of ΔfliR were isolated in the spleen after day 10 after inoculation, while no ΔfliR were detected in the kidney and liver.
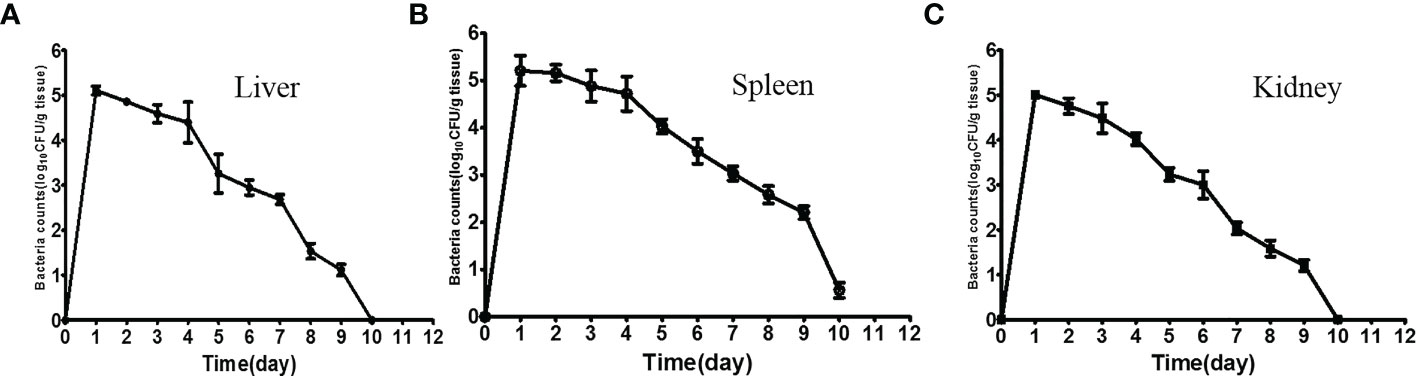
Figure 4 Dynamic distribution of fliR Mutant strain. Bacterial loads in the liver (A), spleen (B), and kidney (C) of experimental fish 12 h after intraperitoneal injection.
After 4 weeks of vaccination, groupers from the immunized and control groups were challenged with wild-type. It was demonstrated that after stimulation with wild-type, mortality was observed in both the control and vaccine groups. In contrast to the immunized group, the control group showed a significantly higher number of mortalities during 3-7 days post-challenge, and the dead grouper exhibited terminal congestion of the caudal, ventral, and dorsal fins and ulcerated lesions with variable sites on the body surface. At 2 weeks post-challenge, the mortality rates were 68.45% and 24.54% in the control and immunized groups, respectively, and the RPS was calculated to be 67.2% (Figure 5).
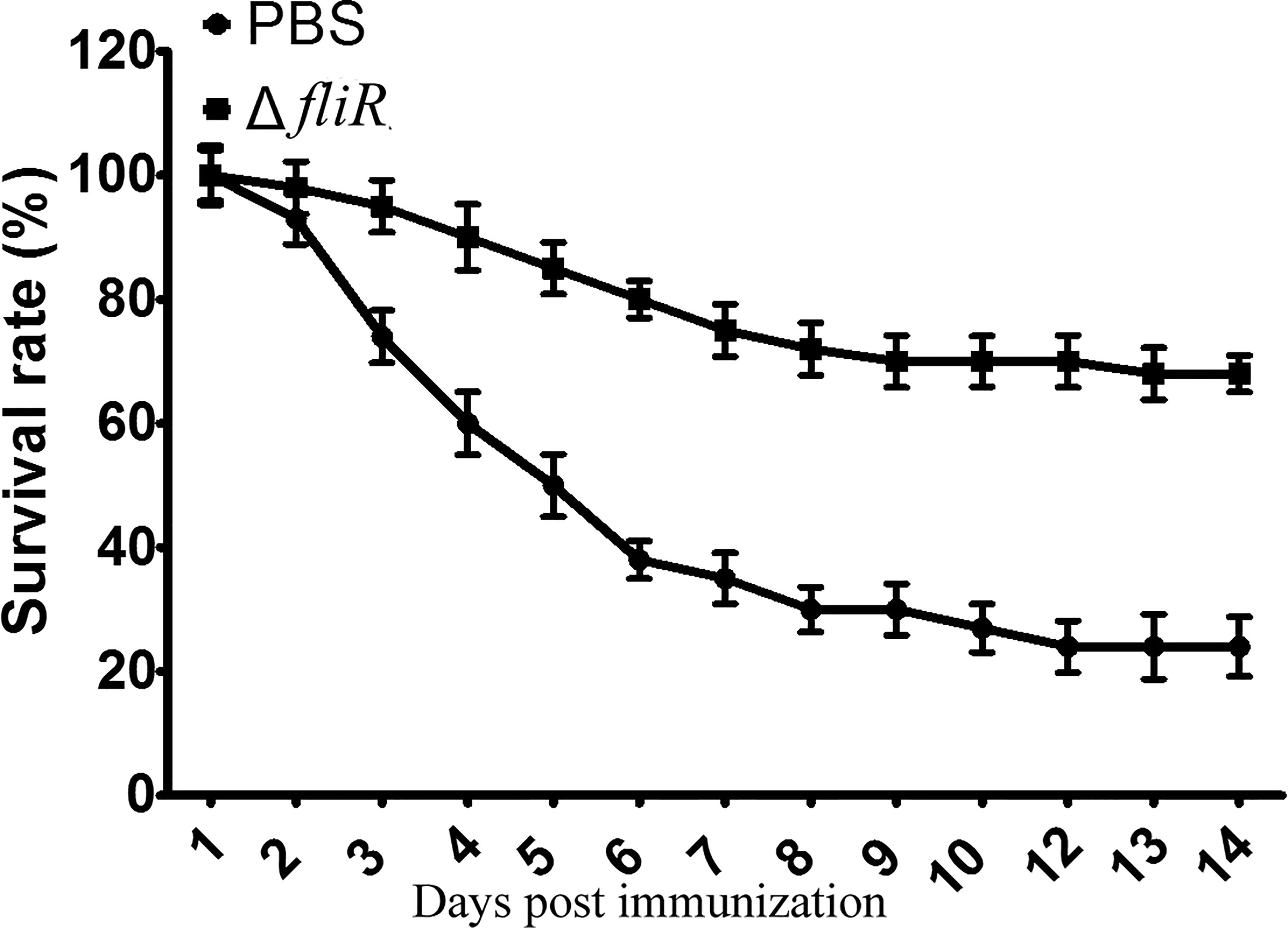
Figure 5 Determination of the immune protection rate of the fliR mutant strain against grouper. After infection with the wild-type strain, the control group had a significantly higher mortality rate three days after injection than the immunized group, with 68.45% and 24.54% mortality in the control and immunized groups, respectively.
The ELISA analyzed the antibody titers of IgM in experimental fish from 1-6 weeks post-vaccination. The results indicated that the specific antibody titers of goupers in experimental group were significantly higher than those in the control group. A trend of continuous increase was observed in the experimental group from 1-4 weeks, with the highest level reached at 4th week, followed by a decrease in titers detected from 5th week (Figure 6).
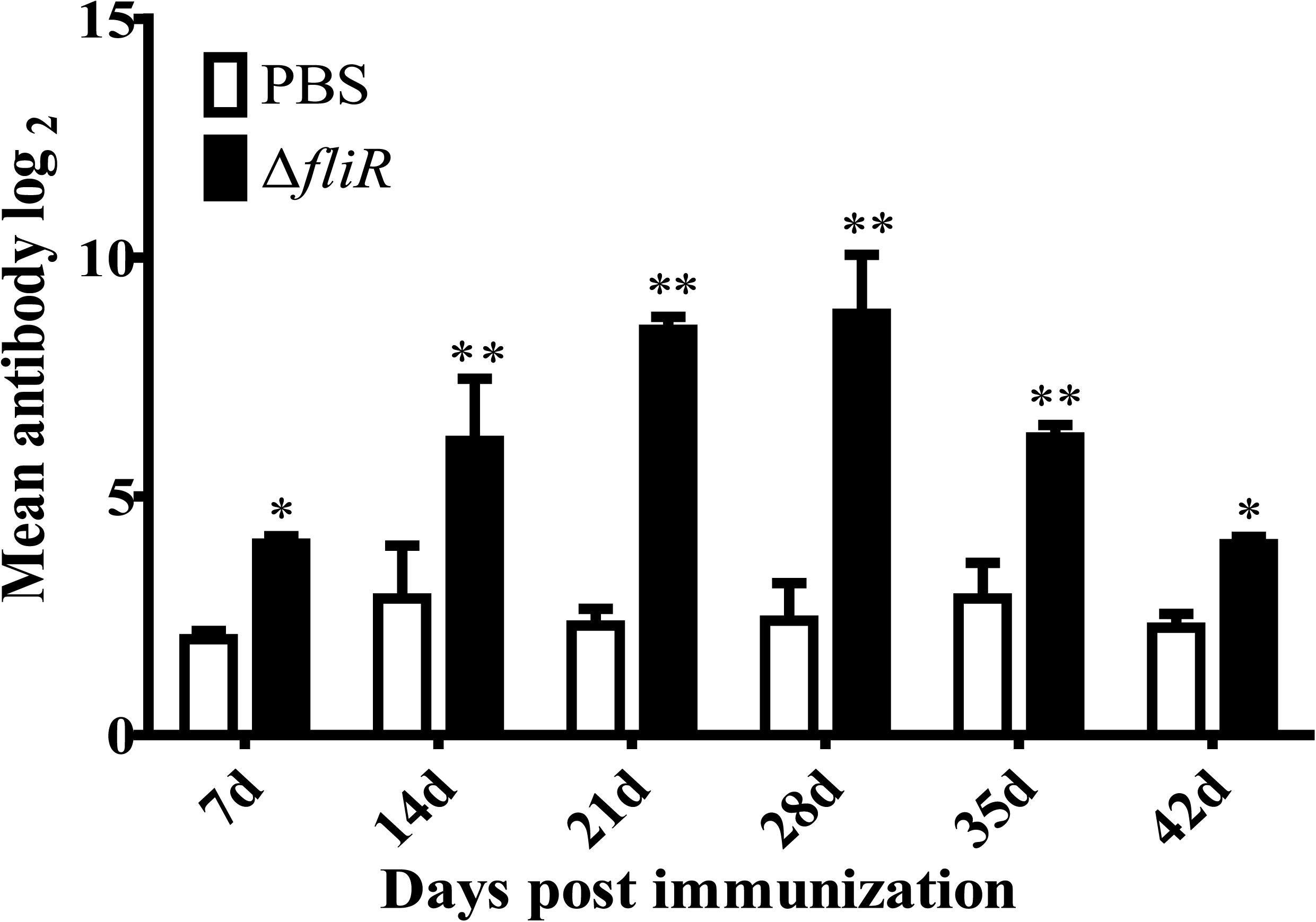
Figure 6 Determination of specific antibody titers following immunization of grouper with the fliR mutant strain. ELISA analyzed the IgM antibody titers in experimental fish 1-6 weeks after administration of the mutant strain. A consistent increase in serum antibody titers was seen in the experimental group from days 1-28, reaching a peak level on day 28, with a subsequent decrease. *indicate significant differences (p<0.05), **indicate extremely significant differences (p<0.01).
The enzyme activities of the groupers were measured in experimental and cotrol groups. Compared to that in the control, SOD activity increased significantly during 1 to 4 weeks post-vaccination, reached its highest value at 4th week, and returned to normal levels from 5th week. CAT activity increased significantly and reached its peak at 2nd week, and then decreased from 3rd week. LDH activity increased significantly and peaked at 2nd week, and returned to normal levels from 3rd week. These results suggest that ΔfliR activates the activity of antioxidant enzymes in the serum of inoculated fish (Figure 7).
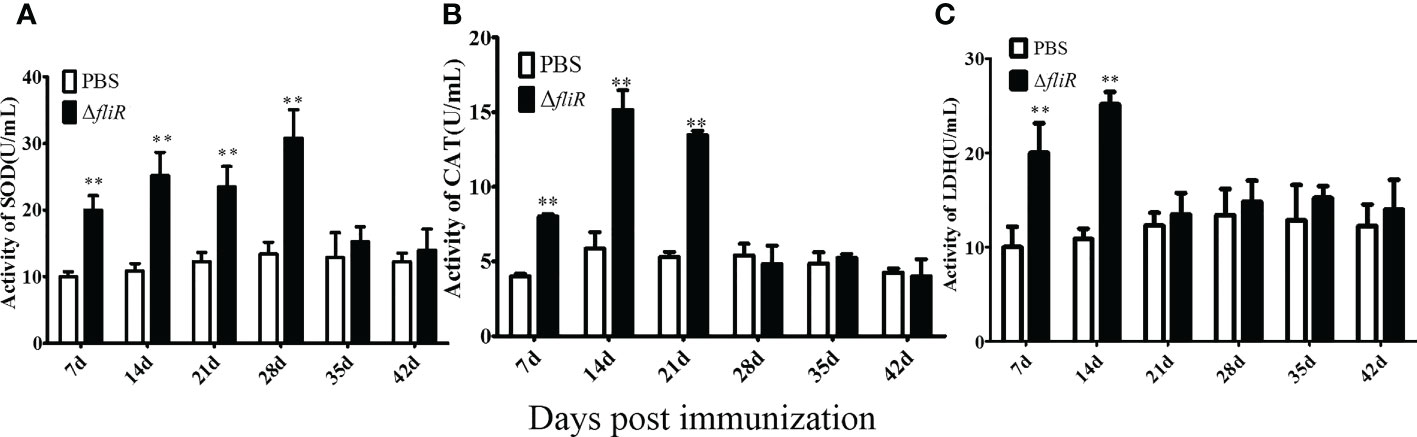
Figure 7 Determination of the enzymatic activities of SOD (A), CAT (B), and LZM (C) in fish serum. SOD activity peaked at week four and returned to normal by week 5. CAT and LDH activity peaked at week two and returned to normal by week 3. *indicate significant differences (p<0.05), **indicate extremely significant differences (p<0.01).
The immune response to the immune-related genes caspase8, CD4, IL6, IgM, MHC-IIβ and TNF-α in the liver (Figure 8), spleen (Figure 9), and kidney (Figure 10) of grouper was assessed by qRT-PCR after immunization. The results demonstrated that caspase8 expression levels were significantly elevated at 3rd and 4th day in the liver, and at 4th day in the spleen. CD4 and IgM expression levels were dramatically elevated and reached peaking value at 2nd day in the liver and kidney, and significantly increased in the spleen at 5th and 6th day. MHC-IIβ expression levels increased considerably in the liver at 3rd, 4th and 6th day, in the spleen at 4th and 5th day, and in the kidney at 5th day. IL6 expression levels notably increased with the highest value appearing at 3rd day in the liver, at 4th day in the spleen, and at 5th day in the kidney. TNF-α expression levels increased remarkably from 2nd day in all 3 tissues, and decreased at 5th day in the liver and spleen. These results suggest that ΔfliR stimulates the expression of innate and adaptive immune-related genes in inoculated fish.
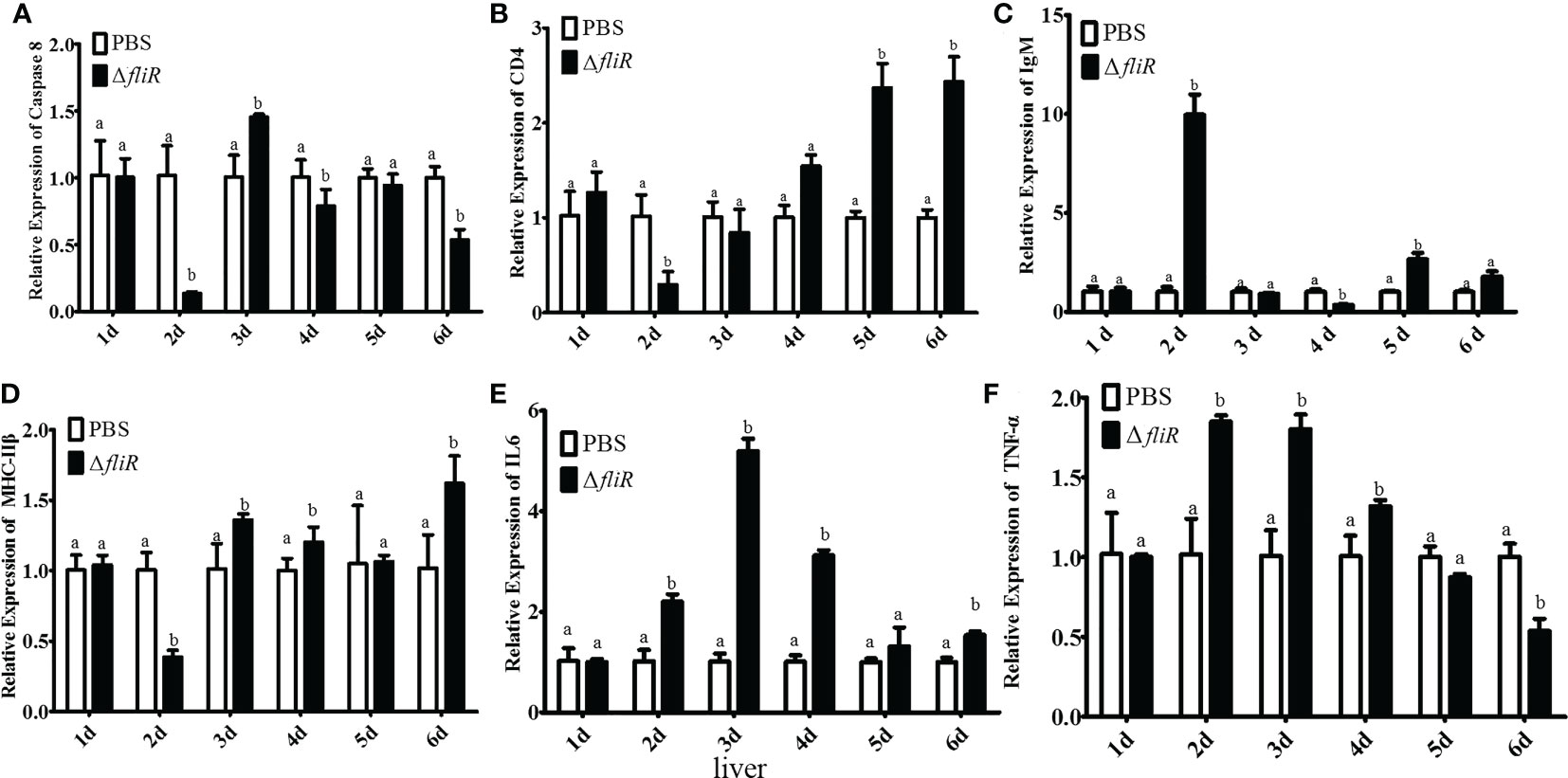
Figure 8 The expression levels of immune-related genes in grouper liver after immunization were detected by qRT-PCR for (A) Caspase8, (B) CD4, (C) IgM, (D) MHC-IIβ, (E) IL6, (F) TNF-α. Groups with notable differences are labeled with different letters above the bars. β-actin was chosen as the reference gene. Data are shown as mean ± SEM (n = 3).
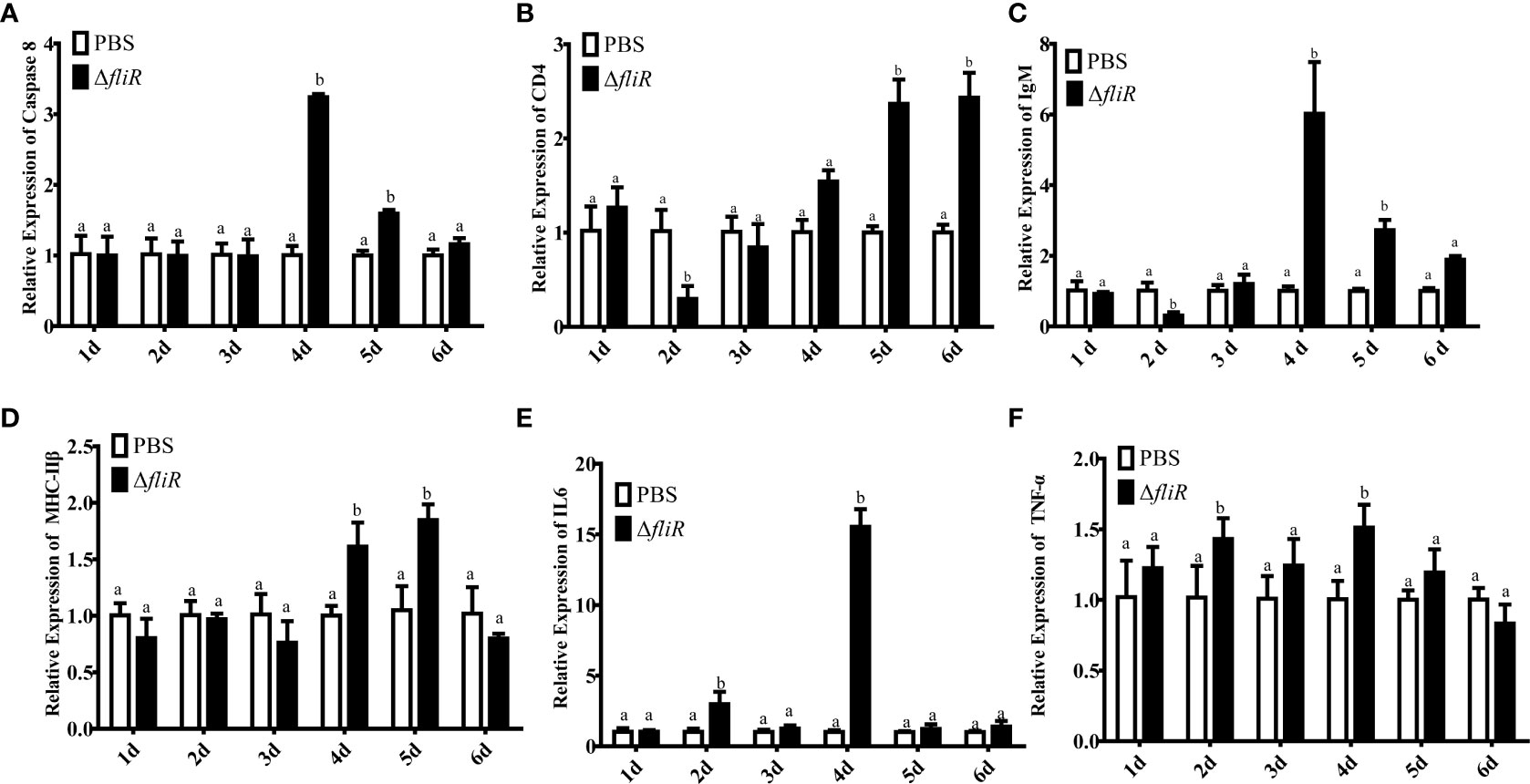
Figure 9 The expression levels of immune-related genes in grouper spleen after immunization were detected by qRT-PCR for (A) Caspase8, (B) CD4, (C) IgM, (D) MHC-IIβ, (E) IL6, (F) TNF-α. Groups with notable differences are labeled with different letters above the bars. β-actin was chosen as the reference gene. Data are shown as mean ± SEM (n = 3).
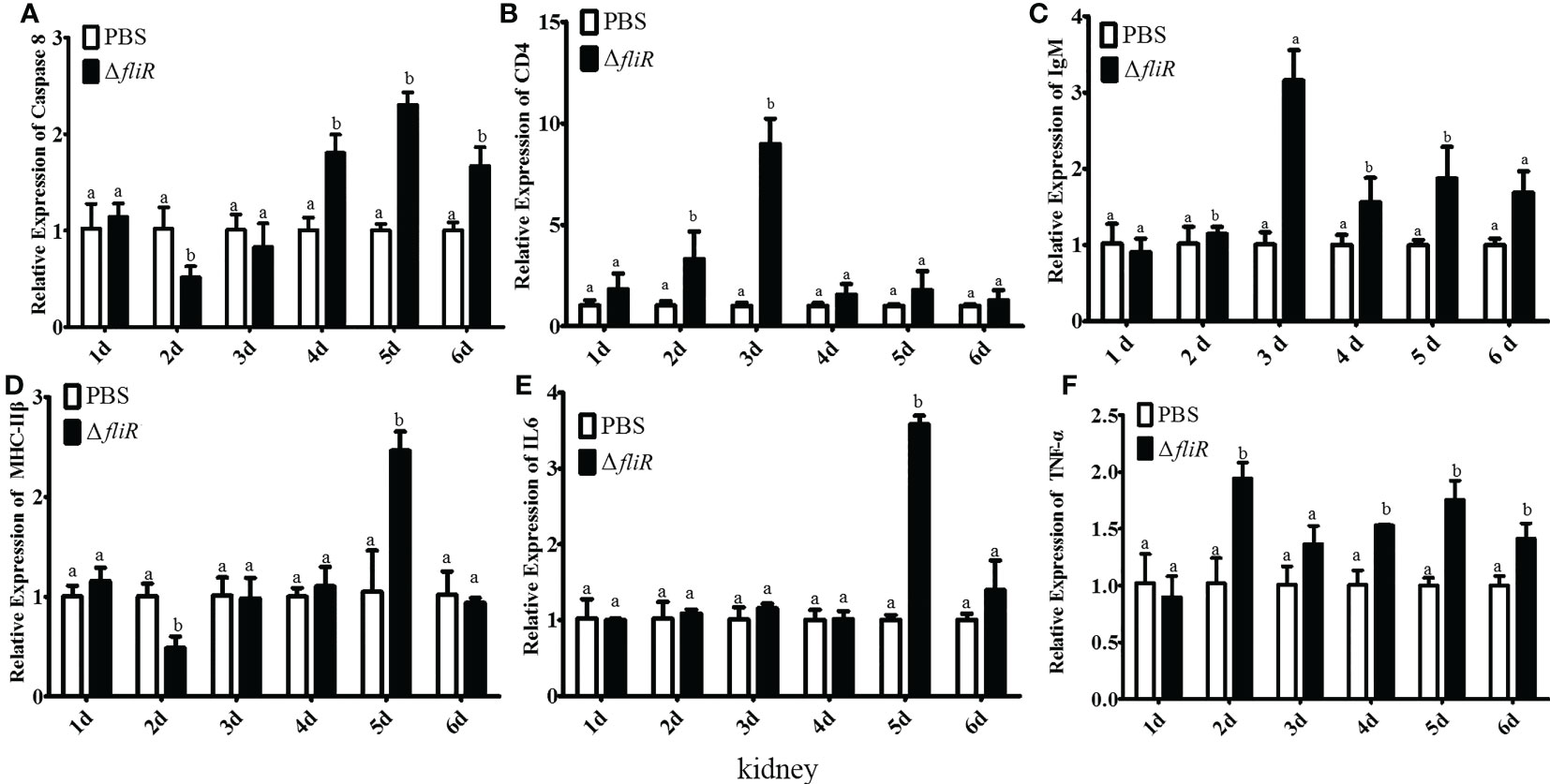
Figure 10 The expression levels of immune-related genes in grouper kidney after immunization were detected by qRT-PCR for (A) Caspase8, (B) CD4, (C) IgM, (D) MHC-IIβ, (E) IL6, (F) TNF-α. Groups with notable differences are labeled with different letters above the bars. β-actin was chosen as the reference gene. Data are shown as mean ± SEM (n = 3).
Bacteria control the flagella for motility, enabling them to swim toward a favorable environment (Xie et al., 2011). Pathobacteria adhere to the surface of the host using flagella during the initial stages of infection, and consequently, flagella contribute essentially to the pathogenicity of bacteria (Li et al., 2003). Assembly of the flagellum is facilitated by its intrinsic protein export apparatus, the type III secretion system (T3SS) in pathogenic bacteria (Minamino et al., 2019). A series of genes regulate bacterial flagella’s number, location, and biosynthesis. V. alginolyticus forms a single flagellum at the polar end of the cell, and FlhFG regulates flagellum formation and number. Overexpression of FlhF in V. alginolyticus contributed to a multipolar flagellar phenotype, while mutation of flhF gene caused a flagellar-free phenotype. The function of FlhG is reversed from that of FlhF (Homma et al., 2022). The absence of FlhA, FlhB, FliI or FliR restrained the twitching motility of Lysobacter enzymogenes, implicating that FlhA, FlhB, FliI and FliR could be involved in regulating the function of twitch motility (Fulano et al., 2020). Silencing of FliA, FliR, and FlrB in V. harveyi significantly reduced bacterial adhesion, biofilm formation, motility, and flagellar synthesis (Qi et al., 2022). Investigation of S. mucilaginous infection of Drosophila melanogaster showed fliR as a virulence factor involved in S. marcescens mediated destruction of intestinal epithelial cells, which ultimately leads to the death of the host (Sina Rahme et al., 2022). Thus, the flagellum is regulated by a range of flagellum-associated genes, and the flagellum genes are involved in pathogenicity to the host.
It has been shown that FliR combined with FliP and FliQ forms the transmembrane output gate complex of the type III flagellin output apparatus and is associated with the injector of T3SS secretion system (Minamino et al., 2019). Scanning under electron cryomicroscopy showed that the core complex contains a single copy of FliR. The FliO ring complex protects FliP against proteolytic degradation and promotes the formation of the stable FliP5FliR1 complex. FlhA and FlhB are associated with the FliR/FliP/FliQ core complex (Minamino et al., 2019). Elimination of csrA gene in H. pylori J99 exhibited the loss of motility and reduced adherence, and transcription of mRNAs of FlaA and FlaB in the mutant strain was reduced to only 40% and 16%, respectively (Kao et al., 2014). After the knockout of FliS in Campylobacter jejuni, mutant strain behaved non-motile, showed reduced levels of FlaA/B and FlaC flagellin, and carried severely truncated flagella (Radomska et al., 2017). In Listeria monocytogenes, after deletion of flhB, fliM or fliY gene, the transcript levels of flagella-related genes flaA, fliM, fliY, lmo0695, lmo0698, fliI and fliS were significantly down-regulated, and bacterial motility and flagellar synthesis was utterly abolished (Cheng et al., 2018). Taken together, fliR has an essential function in the pathogenicity of pathogenic bacteria, however, the role of fliR on V. alginolyticus has not been studied.
The present study demonstrated that the absence of fliR resulted in a significant reduction of mRNAs for flaA, flaB, fliS, flhB and fliM. Mutation of fliR-deletion mainly affected pathways related to cell motility, membrane transport, signal transduction, carbohydrate metabolism, and amino acid metabolism in V. alginolyticus. Hence we observed that the swarming diameter of the fliR mutant strain was significantly smaller compared to the wild strain, suggesting that the absence of fliR resulted in reduced motility of Vibrio alginolyticus.
Aquaculture has become an economically important agribusiness worldwide. The primary obstacle to aquaculture development is the infectious diseases that cause severe economic losses. Live attenuated vaccines offer advantages against diseases caused by some pathogens in mariculture fish. Deletion or mutation of essential virulence-related genes of wild-type to make them weakly virulent strains (Mohd-Aris et al., 2019). The mutant do not cause fish mortality after vaccination and can exist in fish for a long time, effectively stimulating the host to produce antibodies and high expression of immune genes (Minor, 2015).
The experiments of bacterial dynamic distribution and ELISA analysis showed that fliR mutant survived for 10 days in the immune tissues of the inoculated fish, and efficiently stimulated IgM antibody production. SOD is a natural metalloenzyme used to scavenge large amounts of reactive oxygen radicals generated by the body’s redox reactions to maintain the integrity of the cellular structure, genetic material, and to participate in the body’s natural immunity (Garcia et al., 2017). Organisms can eliminate pathogenic bacteria through reactive oxygen species. CAT abolishes excess reactive oxygen species to avoid cell damage and maintain the redox balance of the immune system (Wang et al., 2013). LDH is a glycolytic enzyme and ascends in the serum when the organism’s tissues are damaged (Ding et al., 2017). In this study, the activities of CAT, SOD and LDH in grouper serum were significantly increased after the administration of fliR mutant. Furthermore, ΔfliR significantly increased the innate and adaptive (humoral and cellular) immune-related gene expression levels, and effectively promoted the survival rate of groupers.
In conclusion, we have constructed V. alginolyticus fliR-deletion mutant ΔfliR and analyzed the biological phenotype and transcriptome of the mutant. The protective effect of ΔfliR as a live attenuated vaccine against vibriosis in groupers was evaluated. The ΔfliR effectively stimulated the humoral and cellular immunity of the inoculated fish. The results indicate that ΔfliR is an effective live attenuated vaccine against vibriosis in cultured grouper.
Raw transcriptome sequencing data were submitted to GenBank with the accession number PRJNA954769. The sequence of the fliR has been uploaded to the GenBank database with accession number OQ813764.
The animal study was reviewed and approved by Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals Ethics Committee.
SC and JJ designed the experiments. FD completed the experiments and wrote the manuscript under the guidance of SC. SC and JJ revised FD’s manuscript. XW assisted with the fish rearing and injection experiments and GL assisted with the sampling and blood collection. All authors contributed to the article and approved the submitted version.
This work was funded by the Natural Science Foundation of Shenzhen City (JCYJ20190813114409506 and JCYJ20210324130003009), National Natural Science Foundation of China (No. U20A2065) and Guangdong Natural Science Foundation (No. 2021A1515010532).
The authors of the present study thank the Shenzhen Research Institute of Guangdong Ocean University, the School of Fisheries of Guangdong Ocean University, and the Guangdong Key Laboratory of Aquatic Animal Disease Control and Healthy Farming, for providing the experimental facilities.
The authors declare that the research was conducted without any commercial or financial relationships that could be considered as potential conflicts of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdallah, F. B., Ellafi, A., Lagha, R., Kallel, H., Bakhrouf, A. (2011). Virulence gene expression, proteins secreted and morphological alterations of Vibrio parahaemolyticus and Vibrio alginolyticus in response to long-term starvation in seawater. Afr. J. Microbiol. Res. 5, 792–801. doi: 10.5897/AJMR10.653
Adeleye, I., Daniels, F., Enyinnia, V. (2010). Characterization and pathogenicity of vibrio spp. contaminating seafoods in Lagos, Nigeria. Internet J. Food Safety. 12, 1–9.
Baker-Austin, C., Oliver, J. D., Alam, M., Ali, A., Waldor, M. K., Qadri, F., et al. (2018). Vibrio Spp. infections. Nat. Rev. Dis. Primers. 4, 1–19. doi: 10.1038/s41572-018-0005-8
Chaban, B., Hughes, H. V., Beeby, M. (2015). The flagellum in bacterial pathogens: for motility and a whole lot more. Semin. Cell Dev. Biol. 46, 91–103. doi: 10.1016/j.semcdb.2015.10.032
Cheng, C., Wang, H., Ma, T., Han, X., Yang, Y., Sun, J., et al. (2018). Flagellar basal body structural proteins FlhB, FliM, and FliY are required for flagellar-associated protein expression in Listeria monocytogenes. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00208
Dhillon, S., Curran, M. P. (2008). Live attenuated measles, mumps, rubella, and varicella zoster virus vaccine (Priorix-tetra). Pediatr. Drugs 10, 337–347. doi: 10.2165/00148581-200810050-00007
Ding, J., Karp, J. E., Emadi, A. (2017). Elevated lactate dehydrogenase (LDH) can be a marker of immune suppression in cancer: interplay between hematologic and solid neoplastic clones and their microenvironments. Cancer Biomarkers. 19, 353–363. doi: 10.3233/CBM-160336
Fulano, A. M., Shen, D., Kinoshita, M., Chou, S.-H., Qian, G. (2020). The homologous components of flagellar type III protein apparatus have acquired a novel function to control twitching motility in a non-flagellated biocontrol bacterium. Biomolecules. 10, 733. doi: 10.3390/biom10050733
Garcia, Y. M., Barwinska-Sendra, A., Tarrant, E., Skaar, E. P., Waldron, K. J., Kehl-Fie, T. E. (2017). A superoxide dismutase capable of functioning with iron or manganese promotes the resistance of Staphylococcus aureus to calprotectin and nutritional immunity. PLoS pathogens. 13, e1006125. doi: 10.1371/journal.ppat.1006125
Goh, P., Lim, F., Han, H., Willems, P. (2007). Safety and immunogenicity of early vaccination with two doses of tetravalent measles-mumps-rubella-varicella (MMRV) vaccine in healthy children from 9 months of age. Infection. 35, 326–333. doi: 10.1007/s15010-007-6337-z
Homma, M., Mizuno, A., Hao, Y., Kojima, S. (2022). Functional analysis of the n-terminal region of Vibrio FlhG, a MinD-type ATPase in flagellar number control. J. Biochem. 172, 99–107. doi: 10.1093/jb/mvac047
Kao, C. Y., Sheu, B. S., Wu, J. J. (2014). CsrA regulates Helicobacter pylori J99 motility and adhesion by controlling flagella formation. Helicobacter. 19, 443–454. doi: 10.1111/hel.12148
Li, F., Tian, F., Li, J., Li, L., Qiao, H., Dong, Y., et al. (2021). Isolation and characterization of a podovirus infecting the opportunist pathogen Vibrio alginolyticus and Vibrio parahaemolyticus. Virus Res. 302, 198481. doi: 10.1016/j.virusres.2021.198481
Li, J., Zhou, L., Woo, N. Y. (2003). Invasion route and pathogenic mechanisms of Vibrio alginolyticus to silver sea bream Sparus sarba. J. Aquat. Anim. Health 15, 302–313. doi: 10.1577/H03-034.1
Luo, G., Huang, L., Su, Y., Qin, Y., Xu, X., Zhao, L., et al. (2016). flrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerging Microbes Infections. 5, 1–11. doi: 10.1038/emi.2016.82
Minamino, T., Kawamoto, A., Kinoshita, M., Namba, K. (2019). Molecular organization and assembly of the export apparatus of flagellar type III secretion systems. Bacterial Type III Protein Secretion Systems. 427, 91–107. doi: 10.1007/82_2019_170
Minor, P. D. (2015). Live attenuated vaccines: historical successes and current challenges. Virology. 479, 379–392. doi: 10.1016/j.virol.2015.03.032
Mohd-Aris, A., Muhamad-Sofie, M. H. N., Zamri-Saad, M., Daud, H. M., Ina-Salwany, M. Y. (2019). Live vaccines against bacterial fish diseases: a review. Veterinary world. 12, 1806–1815. doi: 10.14202/vetworld.2019.1806-1815
Mrityunjoy, A., Kaniz, F., Fahmida, J., Shanzida, J., Aftab, U. M., Rashed, N. (2013). Prevalence of Vibrio cholerae in different food samples in the city of Dhaka, Bangladesh. Int. Food Res. J. 20, 1017–1022.
Mukherjee, S., Kearns, D. B. (2014). The structure and regulation of flagella in Bacillus subtilis. Annu. Rev. Genet. 48, 319–340. doi: 10.1146/annurev-genet-120213-092406
Mustapha, S., Mustapha, E. M., Nozha, C. (2013). Vibrio alginolyticus: An emerging pathogen of food borne diseases. Int. J. Sci. Technology. 2, 302–309.
Qi, X., Xu, X., Li, H., Pan, Y., Kraco, E. K., Zheng, J., et al. (2022). fliA, flrB, and fliR regulate adhesion by controlling the expression of critical virulence genes in Vibrio harveyi. Gene. 839, 146726. doi: 10.1016/j.gene.2022.146726
Radomska, K. A., Wösten, M. M., Ordoñez, S. R., Wagenaar, J. A., van Putten, J. P. (2017). Importance of Campylobacter jejuni FliS and FliW in flagella biogenesis and flagellin secretion. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01060
Sadat, A., El-Sherbiny, H., Zakaria, A., Ramadan, H., Awad, A. (2021). Prevalence, antibiogram and virulence characterization of Vibrio isolates from fish and shellfish in Egypt: a possible zoonotic hazard to humans. J. Appl. Microbiol. 131, 485–498. doi: 10.1111/jam.14929
Schuster, V., Otto, W., Maurer, L., Tcherepnine, P., Pfletschinger, U., Kindler, K., et al. (2008). Immunogenicity and safety assessments after one and two doses of a refrigerator-stable tetravalent measles-mumps-rubella-varicella vaccine in healthy children during the second year of life. Pediatr. Infect. Dis. J. 27, 724–730. doi: 10.1097/INF.0b013e318170bb22
Sina Rahme, B., Lestradet, M., Di Venanzio, G., Ayyaz, A., Yamba, M. W., Lazzaro, M., et al. (2022). The fliR gene contributes to the virulence of S. marcescens in a drosophila intestinal infection model. Sci. Rep. 12, 1–10. doi: 10.21203/rs.3.rs-831030/v1
Song, X., Zang, J., Yu, W., Shi, X., Wu, Y. (2020). Occurrence and identification of pathogenic Vibrio contaminants in common seafood available in a chinese traditional market in qingdao, Shandong province. Front. Microbiol. 11, 1488. doi: 10.3389/fmicb.2020.01488
Wang, C., Yue, X., Lu, X., Liu, B. (2013). The role of catalase in the immune response to oxidative stress and pathogen challenge in the clam Meretrix meretrix. Fish Shellfish Immunol. 34, 91–99. doi: 10.1016/j.fsi.2012.10.013
Xie, L., Altindal, T., Chattopadhyay, S., Wu, X.-L. (2011). Bacterial flagellum as a propeller and as a rudder for efficient chemotaxis. Proc. Natl. Acad. Sci. 108, 2246–2251. doi: 10.1073/pnas.1011953108
Yang, B., Zhai, S., Li, X., Tian, J., Li, Q., Shan, H., et al. (2021). Identification of vibrio alginolyticus as a causative pathogen associated with mass summer mortality of the pacific oyster (Crassostrea gigas) in China. Aquaculture. 535, 736363. doi: 10.1016/j.aquaculture.2021.736363
Keywords: Vibrio alginolyticus, fliR gene, live attenuated vaccine, grouper, motility
Citation: Da F, Wan X, Lin G, Jian J and Cai S (2023) Characterization of fliR-deletion mutant ΔfliR from Vibrio alginolyticus and the evaluation as a live attenuated vaccine. Front. Cell. Infect. Microbiol. 13:1162299. doi: 10.3389/fcimb.2023.1162299
Received: 01 March 2023; Accepted: 05 April 2023;
Published: 25 April 2023.
Edited by:
Daniel Yero, Department of Genetics and Microbiology, Autonomous University of Barcelona, SpainReviewed by:
Md Mozammel Hoque, University of Technology Sydney, AustraliaCopyright © 2023 Da, Wan, Lin, Jian and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuanghu Cai, Y2Fpc2hAZ2RvdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.