- Department of Microbial Pathogenesis and Immunology, School of Medicine, Texas A&M University, Bryan, TX, United States
The long non-coding RNAs (lncRNAs) are evolutionarily conserved classes of non-coding regulatory transcripts of > 200 nucleotides in length. They modulate several transcriptional and post-transcriptional events in the organism. Depending on their cellular localization and interactions, they regulate chromatin function and assembly; and alter the stability and translation of cytoplasmic mRNAs. Although their proposed range of functionality remains controversial, there is increasing research evidence that lncRNAs play a regulatory role in the activation, differentiation and development of immune signaling cascades; microbiome development; and in diseases such as neuronal and cardiovascular disorders; cancer; and pathogenic infections. This review discusses the functional roles of different lncRNAs in regulation of host immune responses, signaling pathways during host-microbe interaction and infection caused by obligate intracellular bacterial pathogens. The study of lncRNAs is assuming significance as it could be exploited for development of alternative therapeutic strategies for the treatment of severe and chronic pathogenic infections caused by Mycobacterium, Chlamydia and Rickettsia infections, as well as commensal colonization. Finally, this review summarizes the translational potential of lncRNA research in development of diagnostic and prognostic tools for human diseases.
1 Introduction
The genomes of simplest micro-organisms to complex genome of humans encode a vast repertoire of biological information that needs to be unraveled at the molecular level. Next-generation sequencing and recent developments in computational programming and annotation methods have expanded our understanding of DNA beyond the protein-coding regions of the genome (Liu et al., 2013; Uszczynska-Ratajczak et al., 2018). A comparison between the genomes of different species showed functional, evolutionary, and genomic similarities in their protein-coding regions. This prompted a paradigm shift from mRNA-centric view of host gene regulation to the non-coding regions of the genome. Recent studies and consortiums like ENCODE (The Encyclopedia of DNA Elements) and FANTOM (The Functional Annotation of the Mammalian Genome) have characterized a section of the non-coding regions of the genome and identifies their regulatory role in host transcriptome (Chen et al., 2017). Among such loci are the long non-coding RNAs (lncRNAs) (Uszczynska-Ratajczak et al., 2018).
LncRNAs are classically defined as transcripts of at least > 200 nucleotides in length (Uszczynska-Ratajczak et al., 2018). The recent GENCODE v. 42 release shows out of 252,416 annotated transcripts, 89,305 are mRNA, while 57,936 are annotated lncRNAs in the human genome (Frankish et al., 2021). New tools like single cell sequencing, rapid amplification of cDNA ends sequencing (RACE-seq), capture long sequencing (CLS) method, photoactivatable ribonucleotide-enhanced cross-linking and immunoprecipitation (PAR-CLIP), capture hybridization analysis of RNA targets (CHART) and in vitro RNA antisense purification (RAP) have expanded our understanding of lncRNA transcriptome and interactome (Agliano et al., 2019). The tools have validated these transcripts and annotated their genomic location and structure (Chen et al., 2017). Studies have shown lncRNAs modulate diverse biological pathways at transcriptional, post-transcriptional and epigenetic levels by acting as decoys, scaffold, guides, sponges or enhancers (Wang and Chang, 2011; St.Laurent et al., 2015). As a result, they play a critical role in diverse cellular processes like embryonic development, stem cell pluripotency, neuronal and cardiac differentiation (DiStefano, 2018). They have also been linked to several cancer, immune response regulation, host-pathogen interaction and microbiota interaction (DiStefano, 2018; Agliano et al., 2019).
The current review has two main objectives. The first objective is to understand the organization of lncRNAs in human genome and how they are functionally classified and regulated. Often, the lncRNAs are spatio-temporally regulated and their genome location is biologically linked with the cellular pathways they are likely to influence in the host (Herriges et al., 2014; Dempsey et al., 2018). The second objective is to shed light on the role of several annotated and discovered lncRNAs that regulate the immune responses and pathogenesis during host-pathogen interaction with a special focus on the obligate intracellular pathogens. The review also identifies gap in lncRNA research and discusses lncRNA based therapeutic approaches in diagnosis and treatment of infectious diseases.
2 Classification of LncRNA
The classification of lncRNAs is mostly empirical and so far, there are no standard parameters to classify them. However, lncRNAs can be classified according to different features as follows: (A) genome location; (B) mode of action on DNA sequence. The most widely used classification of lncRNAs is based on their positioning in the genome which could be sub-divided into five classes of lncRNA (Ma et al., 2013; St.Laurent et al., 2015).
2.1 Based on genome location
2.1.1 Intergenic and intronic lncRNAs
LncRNAs that are located and transcribed from different intergenic regions are designated intergenic lncRNAs (Figure 1A). LncRNAs transcribed from introns of protein-coding genes are designated intronic lncRNAs (Figure 1B). Long-intergenic non-coding RNAs (lincRNAs) are the most widely studied type of lncRNAs. They are transcriptionally activated in a mechanism similar to mRNA and are more conserved across species than intronic lncRNA (Khalil et al., 2009; Guttman et al., 2010; Ma et al., 2013) and antisense transcripts (Guttman et al., 2010; Ma et al., 2013). Their expression is more tissue-specific and stable as compared to other types of lncRNAs (Guttman et al., 2010; Cabili et al., 2011).
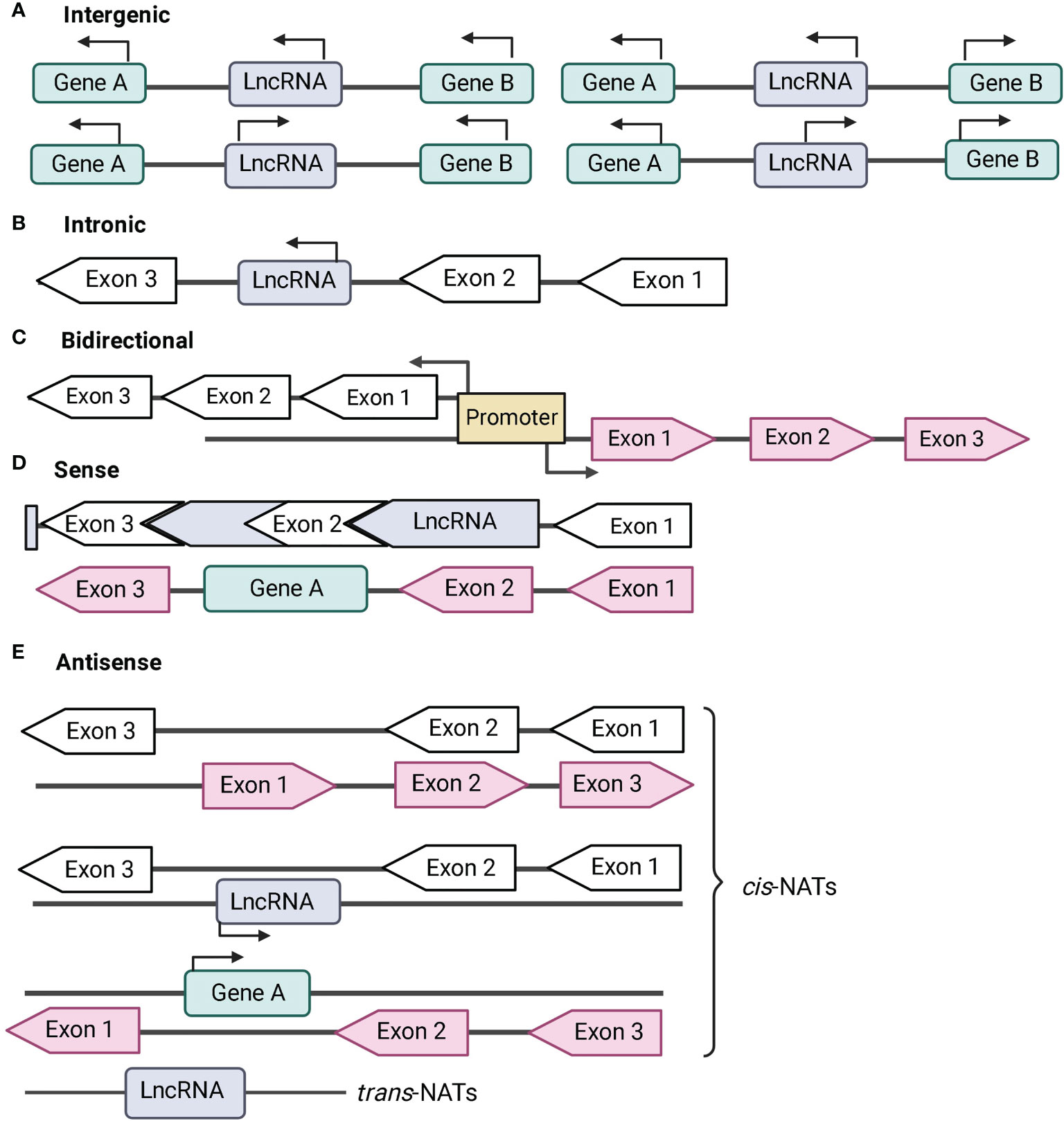
Figure 1 Classification of long non-coding RNA (lncRNA) based on genomic location. Protein-coding genes are represented in green and their exons are represented in black-bordered, arrow-shaped box. Introns are denoted by solid black lines. LncRNAs are represented in blue and their exons are represented in pink-arrow-shaped box. Direction of transcription is noted by the direction of the arrows. (A) Intergenic lncRNAs are transcribed from the intergenic regions of both the strands. (B) Intronic lncRNAs are transcribed from introns of protein-coding genes. (C) Bidirectional lncRNAs are located head-to-head with a protein-coding gene within 1 kb under a common bidirectional promoter. (D) Sense lncRNAs are transcribed from the sense strand of the protein-coding genes and contain exons from protein-coding genes. The lncRNA may partially overlap with the protein coding genes or may entirely overlap with the protein-coding gene through introns. (E) Antisense lncRNAs or naturally antisense transcripts (NATs) are transcribed from the antisense strand of protein-coding genes. NATs comprises of cis-NATs and trans-NATs. cis-NATs can overlap with exons or introns or may entirely overlap with the protein-coding sequence. trans-NATs is complementary to a target protein-coding gene and it is located in distal part of the genome.
2.1.2 Bidirectional lncRNAs
Bidirectional lncRNAs are located head-to-head within 1kb of protein-coding genes, are transcribed under a bidirectional promoter (Figure 1C). This results in similar expression pattern between the lncRNA transcripts and its protein-coding gene (Carpenter et al., 2013; Atianand et al., 2017). LincRNAs and bidirectional lncRNAs do not overlap with other genes, and are therefore called as non-overlapping lncRNAs (Statello et al., 2021). Bi-directional or divergent promoters of lncRNAs are predicted as sites for evolutionary divergence to new genes or for cis-regulation of protein-coding genes.
2.1.3 Sense lncRNAs
Sense lncRNAs are a type of overlapping lncRNAs that are transcribed from the sense strand of protein-coding genes containing exons (Figure 1D). Interestingly, some sense lncRNAs can function as both RNA and protein-coding gene. For instance, steroid receptor RNA activator (SRA) gene can translate into protein, and the transcript can also function as scaffold for co-activators and repressors to form complexes regulating transcription (Hubé et al., 2011).
2.1.4 Antisense lncRNAs or Natural antisense lncRNAs (NATs)
Antisense or NAT lncRNAs are transcribed from the antisense strand of protein-coding genes. They are the most abundant type of lncRNAs found in the mouse and human genome (Agliano et al., 2019). NATs are further classified into cis-NATs and trans-NATs (Figure 1E). cis-NATs are complementary to its overlapping protein-coding gene, while trans-NATs are located in different positions of the genome and are complementary to a protein-coding gene (Ohhata et al., 2022).
2.2 Based on mode of action on DNA sequences
2.2.1 cis-lncRNA
cis-lncRNAs are lncRNAs that can regulate expression of genes located in the neighboring region (Ohhata et al., 2022). Most of the lncRNA from the global transcriptome exert cis-regulatory effects on neighboring protein-coding gene (Engreitz et al., 2016). For instance, AS-IL1α (AS- interleukin-1 α-subunit) (Chan et al., 2015), lnc-IL7R (lnc-interleukin-7 receptor α-subunit) (Hadjicharalambous et al., 2018), and IL-1β-enhancer RNA are examples of cis-acting lncRNA (Engreitz et al., 2016; Robinson et al., 2020). LincRNA-Cox2 act as an enhancer RNA (eRNA) in cis to regulate neighboring gene Ptgs2 (Elling et al., 2018). Cis-lncRNAs because of their co-occurrence with neighboring gene are ideal candidates for investigation of their biological role in host. This is usually based on the hypothesis that, cis-lncRNAs could be regulating the same biological pathways as their neighboring protein-coding gene (Robinson et al., 2020).
cis-lncRNA regulates neighboring gene either at transcriptional or epigenetic level. It represses transcription either by binding to the promoter of target gene and blocking the formation of pre-initiation complex, or by interacting with the transcription factors (TFs) (St.Laurent et al., 2015). For instance, lncRNA of DHFR (di-hydrofolate reductase) can form a stable triplex structure with the promoter region (0.8-7.3Kb region upstream) of DHFR and TFIIβ to displace the pre-initiation complex for transcriptional inhibition (Martianov et al., 2007; Ma et al., 2013). SRG1 is ~0.4-1.9Kb of size transcript that spans the promoter region of SER3 (phosphoglycerate dehydrogenase gene in Saccharomyces cervisiae). The resultant SRG1-RNA prevents the TF binding to SER3 promoter to inhibit transcription (Martens et al., 2004; Ma et al., 2013). Interaction of cis-lncRNA with chromatin modifying enzyme (CME) complex to regulate protein-coding gene expression can be negative or positive regulatory event. Xist is a 19 Kb lncRNA which interacts with polycomb repressive complex 2 (PRC2) to trimethylate H3K27 that transcriptionally represses genes encoded on X-chromosome (Froberg et al., 2013; Maclary et al., 2017). GAL10-ncRNA (Gal10p-noncoding RNA) is ~4Kb lncRNA that recruits Rpd3S HDAC (Rpd3 small histone deacetylase complex) to reduce acetylation of H3 histone and transcriptionally represses GAL1 expression (Houseley et al., 2008; Leng et al., 2021). HOTTIP (HOXA transcript at the distal tip) is ~3.8 kb lncRNA in human that recruits MLL chromatin modifying complex to maintain a domain of active chromatin over the 5’ end of HOXA (homeobox A cluster) (Wang et al., 2011; Ghafouri-Fard et al., 2020)
2.2.2 trans-lncRNA
LncRNAs that can regulate expression of genes located in different region are called trans-lncRNAs. For instance, lincRNA-Cox2 is upregulated (Carpenter et al., 2013) and lincRNA-erythroid prosurvival (lincRNA-EPS) is downregulated (Robinson et al., 2020) in response to Toll-like receptor (TLR)-4 and TLR-2 stimulation in murine dendritic cells (DCs) and bone marrow derived macrophages (BMDM). These lncRNAs regulate large number of distally located interferon stimulated genes (ISGs) and NF-κB regulated genes (Carpenter et al., 2013). HOTAIR (HOX antisense intergenic RNA) is a lncRNA transcribed from the HOXC (homeobox C cluster) gene locus in chromosome 12. It is exported by Suz-Twelve protein to regulate the homologous target sites at HOXD (homeobox D cluster) gene located in chromosome 2 (Rinn et al., 2007).
3 Biogenesis and regulation of LncRNA
The biogenesis of LncRNAs is similar to that of mRNAs and regulated by several transcriptional and epigenetic regulators. The study of the differences in biogenesis, processing and turnover of lncRNAs is important as it influences their cellular fates and functions.
3.1 Transcriptional processing of LncRNA
LncRNAs in comparison to mRNA contain fewer but longer exons. Their cellular expression levels are low and exhibit low primary sequence conservation. They are mainly transcribed by RNA polymerase II, but also can be transcribed by other RNA polymerases. Following transcription, they undergo splicing in a mechanism similar to that required for mRNA maturation. The matured lncRNA transcripts often comprise of 7-methyl guanosine (m7G) cap at their 5’ ends and polyadenylation (poly-A tail) at 3’ ends.
At the chromatin level, lncRNA and mRNA show H3K4me3 (trimethylation of histone 3 at Lys4) at the promoters. However, lncRNA that shows chromatin enrichment of H3K27ac (acetylation of histone 3 Lys27) are repressed by CME complexes; Swr1, Rsc, Ino80 and Isw2 (van Attikum et al., 2007). The initiation sites of lncRNA for transcription are divergent like most of the eukaryotic promoters. Most of the cis-lncRNA are transcribed antisense to the protein-coding gene by RNA polymerase II. Usually, these mRNA-lncRNA pair has coordinated expression, however transcription elongation is productive only in sense direction (Core et al., 2008). Asymmetric distribution of polyA- and splicing signals also determine elongation and stability of mRNA-lncRNA pairs. For instance, polyadenylation signals in U1 snRNP are enriched in the cis antisense direction, while the splicing signals are enriched in the sense direction (Almada et al., 2013).
3.2 Post-transcriptional processing of lncRNA
Besides, 5’-capping, 3’-polyadenylation, splicing and chemical base modification, lncRNAs undergo alternative types of processing to form mature lncRNAs. For instance, RNaseP-mediated cleavage is an alternative method of canonical cleavage and polyadenylation to process 3’ ends of most lncRNAs. MALAT1 (metastasis associated lung adenocarcinoma) and NEAT1 (nuclear enriched abundant transcript 1) are highly abundant types of nuclear lncRNA, that localizes to the nuclear speckles and paraspeckles respectively (Quinn and Chang, 2016). They have a tRNA-like-cloverleaf structure in the 3’end and A/U-rich sequence which form a stable triple helix. RNaseP cleaves tRNA-like-cloverleaf structure from the immature MALAT1 and NEAT1 to produce mature transcripts. The mature MALAT1 transcript is known as MALAT1−associated small cytoplasmic RNAs (mascRNAs) which structurally mimics tRNA and is exported to the cytoplasm (Wilusz et al., 2008). mascRNA subunit is cleaved, the U–A•U RNA triple helix at the 3′ end of mature MALAT1 increases the stability of the transcript that enables it to regulate alternative splicing (Tripathi et al., 2010; Brown et al., 2012; Zong et al., 2016). A NAT of MALAT1 was recently discovered and annotated as TALAM1. It contributes to the stability of MALAT1 by promoting the 3′ end cleavage and maturation of MALAT1. MALAT1 regulates the transcription and stability of TALAM1 through a feed-forward positive regulatory loop at the MALAT1 locus, that leads to high cellular levels of MALAT1 (Zong et al., 2016). On the other hand, NEAT1 is transcribed into two isoforms: a short form with a canonical poly (A)-tail (NEAT1_1) and a long unspliced form that lacks polyadenylation (NEAT1_2). NEAT1_2 is 22.7 kilobases (kb) and is processed by RNase P (Wilusz et al., 2012; Cornelis et al., 2016). Post cleavage by RNAseP, NEAT1_2 is processed into an RNA-stabilizing triple helix at its 3′ end and an unstable tRNA-like by-product (Statello et al., 2021).
4 Localization of LncRNA
Cellular localization of lncRNA can determine molecular contacts and hence its biological functions such as modulation of mRNA stability, signaling pathways, regulatory site of action and transcriptional expression of target gens. It can therefore be argued as a fundamental feature for understanding of lncRNA molecular mechanisms. Based on their localization, lncRNAs can be classified into the classes described below.
4.1 Nuclear lncRNAs
Nuclear localization of lncRNAs is governed by several factors such as splicing signals (Melé et al., 2017), polyadenylation signal at 3’ end (Rosenberg et al., 2015), nuclear retention elements and repeats (Zuckerman and Ulitsky, 2019; Guo et al., 2020). For example, in mouse embryonic stem cells (mESCs), splicing inhibitor peptidylprolyl isomerase E suppresses splicing of a subset of lncRNAs, leading to significant nuclear retention of many lncRNAs in mESCs (Guo et al., 2020). LncRNA CCAT1 (colon cancer-associated transcript 1) produces two isoforms: the long isoform (CCAT1-L) is nuclear and contains an internal polyadenylation site corresponding with the 3′ end of the short isoform (CCAT1-S), which is cytoplasmic (Xiang et al., 2014). In addition, lncRNAs can often contain nuclear retention signals or motifs that recruit nuclear factors, which promote the nuclear localization of the lncRNA. For example, the lncRNA maternally expressed gene 3 (MEG3) contains a 356-nucleotide nuclear retention element that associates with U1 snRNP (Azam et al., 2019). Repeat elements also drive lncRNA nuclear retention (Lubelsky and Ulitsky, 2018; Shukla et al., 2018). For instance, the lncRNA functional intergenic repeating RNA element (FIRRE) contains many unique repeats which promote FIRRE interaction with heterogeneous nuclear ribonucleoproteins (hnRNP) and chromatin localization (Hacisuleyman et al., 2016). In summary, nuclear lncRNAs regulate transcriptional expression of target genes in cis or in trans or at epigenetic level by being tethered to the chromatin in membrane-less nuclear domains.
4.2 Cytoplasmic lncRNAs
Cytoplasmic lncRNAs are expected to perform regulatory roles in the cytoplasm and may share the same processing and export pathways with mRNAs. Cytoplasmic lncRNAs are sorted into either organelles, or distributed in the cytoplasm, or interact with different RNA-binding proteins (RBPs) or microRNAs (Rackham et al., 2011). Linc-MD1 regulates muscle differentiation timing by sponging two miRNAs. LncRNA H19, Linc-MD1 and PTENP1 act as microRNA decoy of let-7, miR-133b and PTEN respectively to inhibit their transcription. LincRNA-p21 is both cytoplasmic and ribosomal lncRNA that negatively regulates JUNB and CTNNB1 translation in HeLa cells (Noh et al., 2018). LncRNA co-localization with ribosomes or endoplasmic reticulum (ER) regulates translation such as for ZFAS1 regulates mRNAs encoding proteins from the ribosomal complex. LncRNA UCA1 act as decoy of hnRNP1 to prevent it’s binding to p27-mRNA and restrict translation. Cytoplasmic lncRNAs like LINK-A, lnc-DC, NKILA and Lethe regulates different cellular signaling response. LINK-A activates BRK kinase and promote HIF1α stabilization. Lnc-DC promotes STAT3 phosphorylation, while NKILA inhibit NF-κB signaling and Lethe reduces RelA transcriptional turnover (Noh et al., 2018).
4.3 Mitochondria associated lncRNAs
Mitochondrial associated lncRNAs often function in regulation of mitochondrial metabolism, apoptosis and the crosstalk of mitochondria with nuclei (Zhao et al., 2018). For instance, survival associated mitochondrial melanoma specific oncogenic non-coding RNA (SAMMSON) is a lncRNA that maintains mitochondrial homeostasis, mitochondrial 16S rRNA maturation and expression of mitochondria-encoded polypeptides (Leucci et al., 2016). The mitochondria-encoded lncRNAs; lncND5, lncND6 and lncCytB form intermolecular duplexes with mRNAs and regulate their stability and expression (Statello et al., 2021). The RNA component of mitochondrial RNA-processing endoribonuclease (RMRP) is exported to the cytoplasm through its association with RBP HuR by exportin 1. RMRP is then exported to mitochondria, where it stabilized by G-rich RNA sequence-binding factor 1 (GRSF1), which allows its enrichment in the mitochondrial matrix (Quinn and Chang, 2016).
4.4 Exosome associated lncRNAs
Transcriptome analyses of human blood samples have identified many lncRNAs which localize into the exosomes through an unknown mechanism. Exosomal lncRNA like MALAT1 (Liu J. et al., 2021), growth arrest-specific 5 (GAS5) (Patel et al., 2022), H19 (Wang et al., 2020), HOTAIR (Yang et al., 2019), small ubiquitin-like modifier 1 SUMO1 pseudogene 3 (SUMO1P3) (Li et al., 2021b) and X-inactive specific transcript (XIST) (Lan et al., 2021) were previously shown upregulated in the blood, lung and breast tissues of cancer patients (Li et al., 2021b). It is noteworthy that the gut microbiota (Díaz-Garrido et al., 2021; Park et al., 2021), intracellular pathogens like Salmonella Typhimurium (Imamura et al., 2018), Mycobacterium tuberculosis (Liu M. et al., 2021) and obligate intracellular pathogens like Chlamydia psittaci (Radomski et al., 2019), Rickettsia spp. (Liu Y. et al., 2021) and Anaplasma phagocytophilum are internalized into host cells via endocytosis (Zhou et al., 2020). They communicate with DCs and phagocytes’ host-derived factors through the exosome for activation of antibacterial immunity. A transcriptome analysis of serum derived exosomes during Orientia tsutsugamushi infection showed downregulation of Let7 family of microRNAs (a type of non-coding RNA). Let7 regulates immune response and cytokine activation by modulating TLR-4 signaling pathway (Jiang et al., 2020). These findings suggest that exosomal lncRNAs may modulate host immune response to intracellular bacterial infections.
In summary, lncRNAs have complex spatio-temporal regulation within the cell that is linked to sequence encoded localization signals. All these findings may help in prediction of their mechanism of functioning in the cell.
5 Molecular mechanisms of functioning of lncRNA
As discussed previously, lncRNAs regulate different cellular functions at transcriptional, post-transcriptional, translational or epigenetic level depending on their sub-cellular localization (Statello et al., 2021). Briefly, they primarily function as:
5.1 Signals
LncRNAs which are expressed at a specific time and cellular location in response to stimuli are signal lncRNAs (Figure 2A). They can interact with CME such as histone methyltransferases to silence their target genes either by inhibition of transcription or through heterochromatin formation (Wang and Chang, 2011).
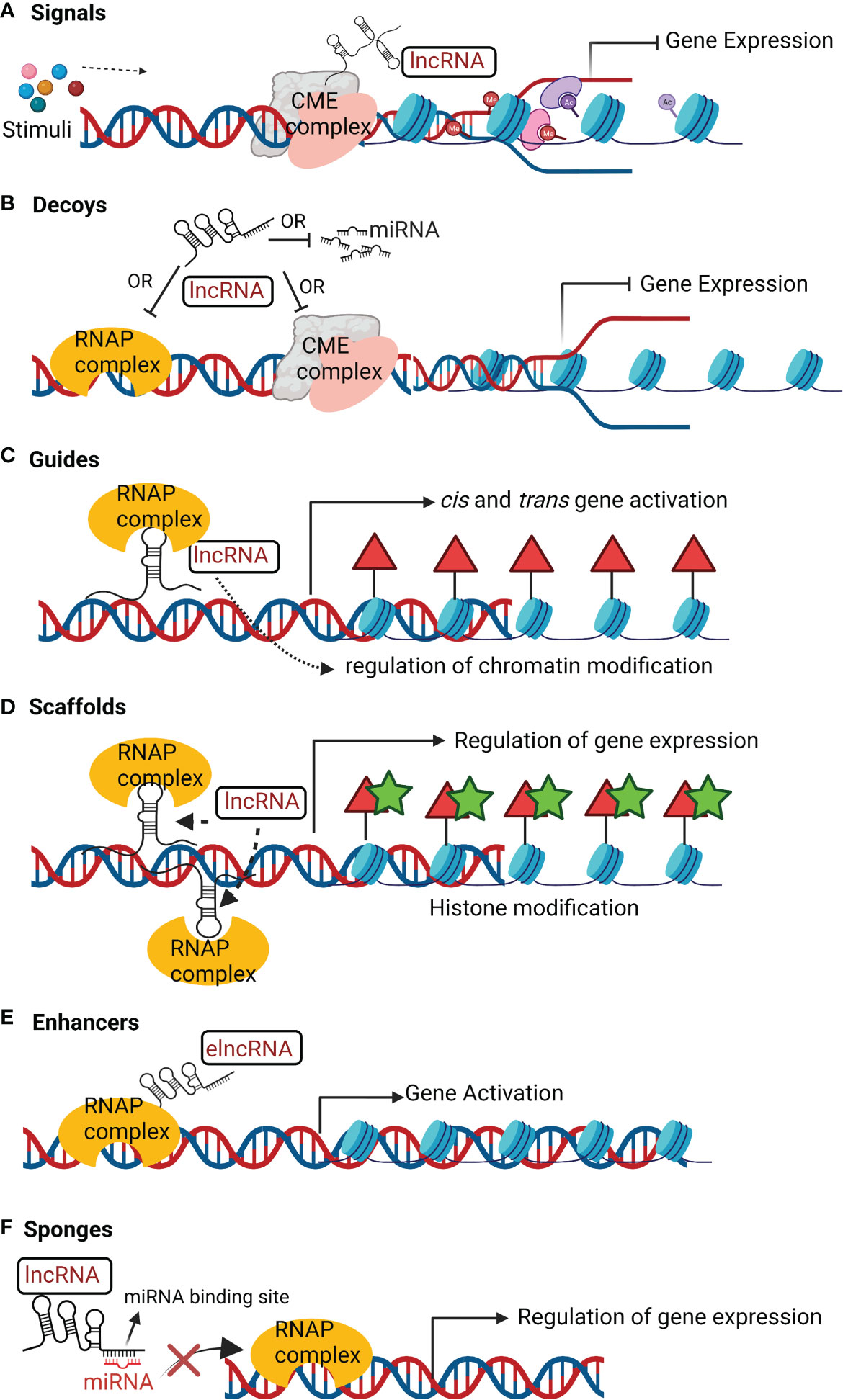
Figure 2 Mechanisms of action of lncRNAs. (A) Signals; lncRNAs receive a signal in response to stimuli to interact with chromatin-modifying enzyme (CME) complex to inhibit transcription. (B) Decoys; lncRNAs can interact with ribonucleoprotein (RNAP) complex or CME complex or miRNA to inhibit these regulatory factors from binding to the target gene and inhibiting its transcription. (C) Guides; lncRNAs can recruit RNAP complex and transcription factors to a specific genomic location and help to regulate chromatin arrangement for gene activation. (D) Scaffolds; lncRNAs can assemble several RNAP complex to inhibit or activate gene transcription. (E) Enhancers; enhancer lncRNA (elncRNA) enhance interaction of RNAP complex and chromatin to promote gene activation. (F) Sponges; LncRNA can act as sponges of miRNA. They have sequence complementary to miRNA and therefore can bind to miRNA, limiting its availability to transcription complex for regulation of target gene transcription.
5.2 Decoys
Decoy lncRNAs act as transcriptional repressors in an indirect way by reducing the availability of the regulatory factors (Figure 2B) that includes: microRNA (miRNA), CME complexes, catalytic proteins, and TFs (Ahmad et al., 2021). For instance, the decoy lncRNA PANDA (P21 associated ncRNA DNA damage activated) regulates p53-dependent apoptosis. NFYA (Nuclear Transcription Factor Y Subunit Alpha) is a TF that activates transcription of apoptosis-related genes. PANDA binds to NFYA which prevents its binding to the promoter of apoptosis related genes (Liu J. et al., 2018). Taurine upregulated 1 (TUG1) and MEG3 are other instances of lncRNAs decoys miRNA from target mRNA (Balas and Johnson, 2018; Da et al., 2021). LncPRESS1 (lncRNA p53 regulated and ESC associated 1) is a pluripotency-associated lncRNA. lncPRESS act a decoy of deacetylase Sirtuin-6 and sequesters it from the chromatin, which promotes transcriptional activation by H3K56ac and H3K9ac of pluripotency-related genes (Jain et al., 2016).
5.3 Guides
LncRNA as guides (Figure 2C) regulate the expression of target genes by reprogramming CME complex or regulating the recruitment of epigenetic modifiers to the regulatory loci of target genes (Statello et al., 2021). For instance, KCNQ1 overlapping transcript 1 (KCNQ1OT1) and HOTAIR binds to PRC2, to regulate its target gene expression (Ahmad et al., 2021). LncRNA TCF7(transcription factor 7) recruits the SWItch/Sucrose Non-Fermentable (SWI/SNF) complex to the promoter of TCF7 that activates the Wnt signaling pathway to promote self-renewal of liver cancer stem cells (Gao et al., 2020).
5.4 Scaffolds
LncRNAs act as scaffold (Figure 2D) by providing structural support in assembly of regulatory protein complexes like ribo-nucleoprotein (RNAP) complex (Statello et al., 2021). They facilitate the assembly of RNAP complex to either transcriptionally inhibit or activate target protein-coding genes (Statello et al., 2021). For example, lncRNA TERC scaffolds the assembly of telomerase complex and associated accessory proteins with reverse transcriptase in a single RNAP (Wang and Chang, 2011). Under certain events, lncRNAs like MALAT1, ANRIL, and TUG1 act as molecular scaffolds to PRC1 and PRC2 to transcriptionally repress or activate target genes (Balas and Johnson, 2018).
Several lncRNAs act as scaffold for the assembly of nuclear condensates. Nuclear condensates are membrane-less compartments encompassing RNA-protein complex involved in regulation of cellular activities (Statello et al., 2021). NEAT1 act as scaffold for organization and function of paraspeckles (Jiang et al., 2017; Yamazaki et al., 2018). NEAT1_2 is essential for assembly of paraspeckles (Yamazaki et al., 2018). The middle region (8–16.6 kb) of NEAT1_2 comprises of two sub-domains of (~12–13 kb) and (~15.4–16.6 kb) length that act as scaffold to the paraspeckle core proteins NONO and SFPQ (Yamazaki et al., 2018). The mechanism behind the structural assembly of NEAT1_2 into the paraspeckles is not clear. LncRNA can also act as scaffold to bring distal chromatin regions into proximity. For example, FIRRE is transcribed from X chromosome. It functions in trans as scaffold with the nuclear matrix factor hnRNPU to maintain a nuclear domain (Lewandowski et al., 2019).
5.5 Enhancers
LncRNAs that act as enhancers can be broadly categorized into enhancer-RNAs (eRNAs) and enhancer-associated lncRNAs (elncRNAs). eRNAs are unstable, short, bidirectional capped and unspliced transcripts that lacks a 3’ poly-A tail (Statello et al., 2021). ElncRNAs are mostly unidirectional, 3’-polyadenylated and spliced. eRNAs can drive directly the expression of target genes through chromatin looping and interaction with scaffold proteins (Figure 2E). This interaction leads to regulatory contacts between enhancers and promoters of the distally located target genes (Statello et al., 2021). For instance, eRNA P53BER (p53-bound enhancer region) (Melo et al., 2013) and elncRNA SWINGN (SWI/SNF interacting GAS6 eRNA) (Grossi et al., 2020) recruit chromatin-activating complexes to the promoters of the target protein-coding genes. In response to oestrogen receptor (ER) transcription activation, eNRIP is another bi-directionally transcribed eRNA, that recruits cohesin to form chromatin loops. This interaction promotes contact between the eNRIP1 and the promoters of nuclear receptor interacting protein 1 (NRIP1) and trefoil factor 1 (TFF1) (Li et al., 2013).
5.6 Sponges
LncRNAs as sponges contain sites complementary to miRNA for interaction (Figure 2F). This results in reduced miRNA availability to target mRNAs (Statello et al., 2021). For instance, lncRNA PNUTS contains seven complementary sequences to miRNA-205, which reduces the availability of miRNA-205 to bind and inhibit mRNAs of the zinc finger E-box-binding homeobox 1 (ZEB1) and ZEB2 (Grelet et al., 2017). The sponging of miRNA-205 by lncRNA PNUTS leads to increased expression of ZEB1 and ZEB2 which promotes epithelial to mesenchymal transition, and breast cancer cell migration and invasion (Grelet et al., 2017). LncRNA CCAT1 is sponge for miRNA-7 which upregulates the expression of the TF homeobox gene B1, thereby promoting the proliferation and metastasis of esophageal squamous cell carcinoma (Gao et al., 2020).
In conclusion, the review discusses different classes of lncRNA, their subcellular localization, and mechanism of action and how these characteristics determine the functionality of lncRNAs, their target genes expression and downstream immune signaling events in host cells.
6 Role of lncRNAs in regulation of immune response against microbial components
Macrophages and DCs are the central players of host innate immune defense against pathogens. They recognize specific pathogen-associated molecular patterns (PAMPs) on invading pathogens through their pattern recognition receptors (PRRs). PRRs like lipopolysaccharide (LPS), bacterial nucleic acids, lipoproteins and peptidoglycan upon host recognition trigger specific immune signaling pathways mediated either by TF, NF-κB or the Interferon regulatory factor (IRF) family of proteins (Li and Wu, 2021). IRF mediates selective expression of interferons (IFN) and AIM2 inflammasomes which leads to activation of apoptotic and cell autonomous immunity against intracellular pathogen (Li and Wu, 2021; Pant et al., 2022) like Listeria monocytogenes, Legionella pneumophila (Omotade and Roy, 2020), Salmonella Typhimurium, Francisella spp., Rickettsia spp., Anaplasma phagocytophilum (Rikihisa, 2011) and Orientia tsutsugamushi (Deretic, 2011; Ko et al., 2013). Type-I IFN can also dampen inflammasome signaling and promote bacterial infection like Mycobacterium tuberculosis (Novikov et al., 2011). NF-κB activation usually results in acute activation of pro-inflammatory pathways. Infact, NF-κB mediated induction of host responses is known to be modulated by other pathogens like Coxiella burnetii to trigger anti-apoptotic pathways; dampen inflammatory cell damage; and promote selective survival in bacterial replicative niche (Lührmann and Roy, 2007).
Many aspects of signaling response are regulated both at transcriptional and post-transcriptional level which allows fine-tuning of immune response against infections (Yoshinaga and Takeuchi, 2019). Recent studies have shown lncRNAs involvement in regulation of differentiation and activation of macrophages, DCs and adaptive immune cells like T- and B- lymphocytes (Walther and Schulte, 2021). Several high-throughput sequencing studies have deciphered lncRNAs regulatory effect on immune response upon infection or PRR based stimulation in vitro and in vivo (Walther and Schulte, 2021). In addition, functional conservation of these immune signaling pathways in different infection models makes it essential to discuss the role of these lncRNAs in regulation of immune response, and further investigate their role in obligate intracellular bacterial infections.
6.1 LncRNAs in TLR- NF-κB dependent immunity
eLncRNA IL-1β and lncRNA IL-1β-RBT46 act as enhancers of IL1β after TLR4-MyD88-NFκB activation by LPS (Walther and Schulte, 2021). Nuclear-lncRNA PACER is a positive feedback regulator that acts as decoy of NF-κB p50 subunit to promote NF-κB p50/p65 heterodimer formation. This leads to immune gene expression in human macrophages in TLR-4 dependent manner (Krawczyk and Emerson, 2014). lncRNA CARLR is induced upon LPS-treatment which binds to p65 subunit of NF-κB to promote pro-inflammatory gene expression (Castellanos-Rubio et al., 2017). IL7-AS as described previously acts as chromatin modifier by association with p300 and SWI/SNF complex to drive expression of CCL2 and IL-6 in TLR-MAPK/NF-κB pathway (Walther and Schulte, 2021). LincRNA- tumor necrosis factor alpha-induced protein 3 (TNFAIP3) interacts with chromatin regulator HMGB1 to form HMGB1/NF-κB complex. This interaction facilitates NFκB binding to promoter region of IL-6 (Ma et al., 2017). LncRNA EPAV act as decoy of SFPQ protein from the p65 promoter of NF-κB to regulate NF-κB activation in mouse (Walther and Schulte, 2021).
LincRNA-Cox2 can act as positive and negative regulator of NF-κB through different mechanisms. It promotes IκBα degradation which causes translocation of NF-κB into the nucleus for pro-inflammatory cytokine expression (Xue et al., 2019). It negatively regulates TLR-induced ISG CCL5 expression by binding to hnRNPA/B and A2/B1 in the nucleus and inhibiting RNA polymerase II recruitment to the CCL5 promoter in Pam3CSK4-stimulated cells (Figure 3A). In another study, in vitro assays revealed a physical association between NF-kB subunits (RelA and p50) and the SWI/SNF complex in LPS-stimulated murine macrophages. siRNA knockdown of lincRNA-Cox2 showed decreased association between NF-kB subunits and the SWI/SNF complex (Hu et al., 2016). These findings indicated lincRNA-Cox2 can bind the SWI/SNF complex to modulate chromatin remodeling, and NF-kB dependent transcription of CCL5 (Figure 3A). LncRNA IL1α-AS upon TLR activation with LPS, Pam3CSK4 or poly (I:C) in cells recruits RNA polymerase II to induce expression of IL1α (Chan et al., 2015). LncRNA FIRRE is induced upon macrophage treatment with LPS, which promotes hnRNPU dependent transcriptional stabilization of IL-1β, IL-12β and VCAM1(Lu et al., 2017). TMC3-AS1 negatively regulates IL-10 by interacting with p65 in the nucleus, which prevents its binding to the NF-κB in the promoter region of IL-10 in macrophages and intestinal epithelial cell lines (Ye et al., 2020).
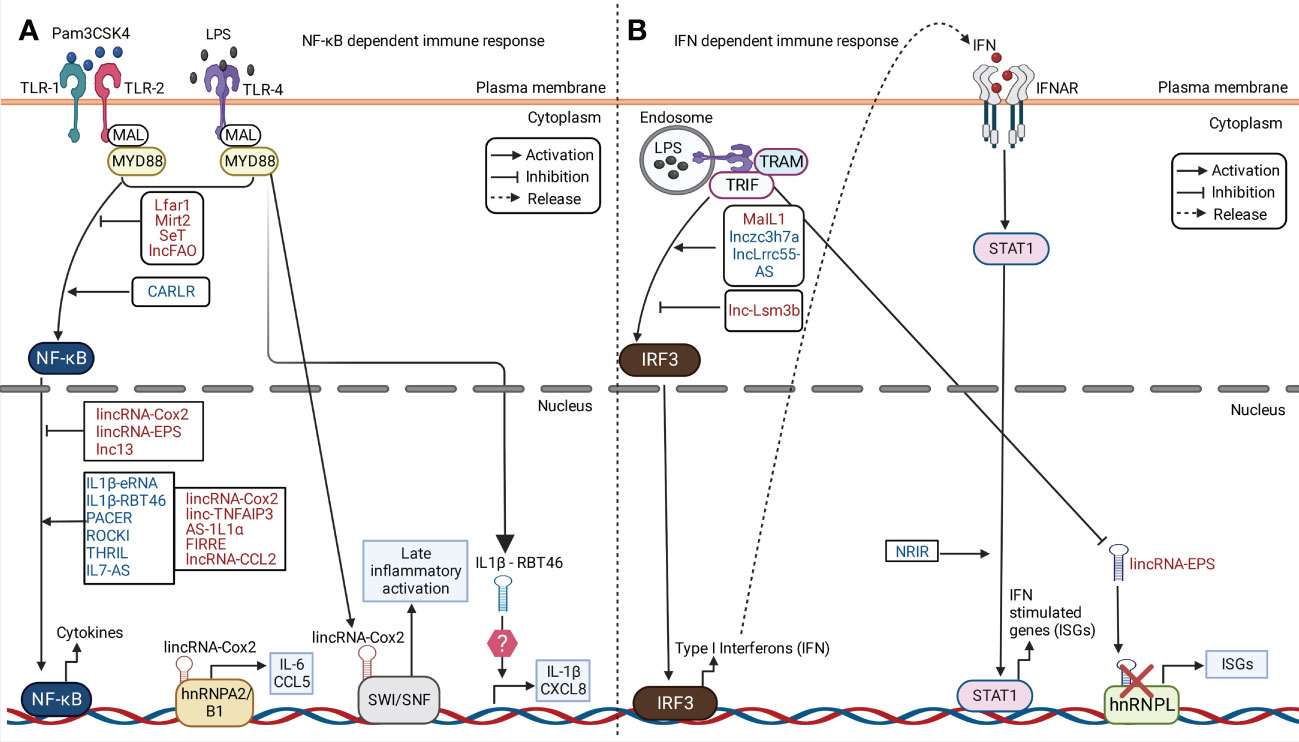
Figure 3 LncRNA in regulation of immune response against microbial components. Examples of cytoplasmic and nuclear lncRNAs acting as positive and negative regulators of NF-κB and IFN signaling pathway in PRR dependent pathway is illustrated in the figure. Examples of murine and human lncRNAs are represented in red and blue fonts respectively. (A) In response to Pam3CSK4 or LPS stimulation, lncRNAs positively or negatively regulate the activation of NF-κB signaling via TLR-1, 2 or 4. This leads to downstream regulation of cytokines expression. For instance, lincRNA-Cox2 is upregulated in murine phagocytes. It binds to heterogeneous nuclear ribonucleoprotein (hnRNP)A/B and hnRNPA2/B1 leading to both the activation and repression of different genes. LincRNA-Cox2 can bind the SWItch/Sucrose non-fermentable (SWI/SNF) complex, leading to activation of late inflammatory genes. Human IL-1β-RBT46 is upregulated following LPS stimulation, which leads to enhanced expression of IL-1β and CXCL8. (B) Upon LPS stimulation, lncRNAs like MaIL1, Lnczc3h7a, and lnc-Lsm3b positively and negatively regulate the activation of Interferon (IFN) regulatory factor 3 (IFN3) which leads to subsequent regulation of IFN production. lincRNA- EPS is downregulated following LPS stimulation leading to the upregulation of IFN stimulatory genes (ISGs).
LncRNAs can also dampen the activation of PRR induced NF-κB gene expression. lincRNA-EPS and GAPLINC are downregulated in vitro upon TLR-MyD88-NFκB activation (Walther and Schulte, 2021). LncRNA THRIL was previously shown to be downregulated upon Pam3CSK4 stimulation (Figure 3A). This study suggested THRIL forms a transcriptional complex with hnRNPL at the promoter of TNF-α to drive its expression (Li et al., 2014). LncRNA Mirt2 is a negative feedback regulator of TLR-MyD88 pathway that interacts with TRAF6 and inhibits its auto-ubiquitination and oligomerization in the cytoplasm (Du et al., 2017). LncRNA lncFAO is another negative regulator which binds to the mitochondrial enzyme HADHB that promotes fatty acid oxidation and downregulates IL-1β to reduce inflammation (Nakayama et al., 2020).
6.2 LncRNAs in IFN dependent immunity
A study showed macrophage interferon-regulatory lncRNA 1 (MaIL1) positively regulates TRIF-TBK1-IRF3 induced type-I IFN expression in human macrophages after treatment with LPS. MaIL1 activates type-I IFN production (Figure 3B) through formation of complex with optineurin in a TLR4-MyD88-NF-κB dependent manner (Aznaourova et al., 2020). This results in ubiquitin-dependent aggregation of the optineurin signaling platform and phosphorylation of TF IRF3 by TBK1(Aznaourova et al., 2020). In clinical settings, MaIL1 expression correlated with IFNβ levels in broncho-alveolar lavage from pneumonia patients (Aznaourova et al., 2020). LncRNA NRIR is highly expressed in LPS treated human macrophages in TLR-4 dependent manner to promote ISG (CXCL10) expression through an unknown mechanism (Walther and Schulte, 2021). Murine macrophage systems have identified an array of lncRNA in IRF activation such as lncRNA Lnczc3h7a. It acts as a molecular scaffold to TRIM25 ubiquitin ligase and RIG-I (Lin et al., 2019). LncRNA LRRC55-AS upon STAT-dependent activation promotes type-I IFN production (Figure 3B) by supporting de-methylation and inactivation of PP2A (IRF3 inhibitor) by the PME1 protein (Zhou et al., 2019). LncRNA LSM3B negatively regulates RIG-I signaling through binding with RIG-I monomers to limit IFN production (Jiang et al., 2018). LincRNA-EPS interacts with chromatin repressor hnRNPL at target promoters to inhibit cytokine, chemokine and ISG expression (Figure 3B). Upon TLR-MyD88-NF-κB activation, lincRNA-EPS is repressed which leads to immune gene activation (Atianand et al., 2016).
6.3 LncRNAs in phagocyte differentiation and polarization
Myeloid specific human transcript HOTAIRM1 was previously reported as upregulated during granulocytic differentiation and regulated expression of the myeloid differentiation markers CD11b and CD18 (Walther and Schulte, 2021). Morrbid negatively regulates expression of the pro-apoptotic factor Bim in cis, by promoting PRC2 binding to the Bim promoter (Kotzin et al., 2016). Morrbid deletion in vivo showed reduced counts of eosinophil, neutrophil, and monocytes suggesting its role in life span of myeloid cells. lnc-DC is a lncRNA which interacts with TF STAT3 to promote STAT3 phosphorylation by preventing its de-phosphorylation by SHP1 (Walther and Schulte, 2021). A study showed NEAT1 is required for LPS-dependent maturation of DCs (Dai et al., 2021). Lnc-M2 was previously reported to be upregulated in differentiated macrophages (activated with M-CSF, Th2 cytokines IL-4 and IL-13) compared to M0 macrophages in a STAT3-dependent manner. It forms a complex with protein kinase A (PKA) to promote its phosphorylation and M2 polarization (Chen Y. et al., 2020). These findings suggest roles of lncRNAs beyond PRR dependent immunity to developmental role in host myeloid cell differentiation, polarization and repair.
Besides the role of lncRNAs in immune cell response and differentiation, they are also engaged in regulation of epithelial cells, endothelial cells and fibroblast’s PAMP induced inflammatory responses (Agliano et al., 2019). The lncRNAs involvement in PRR induced phagocytic and DCs immune signaling pathways may also contribute to regulatory functions in the other cell types and adaptive immunity. However, these questions are unexplored and have not been discussed in this review.
Collectively, this section discusses several examples of lncRNA playing a central role in the regulation of immune responses to PAMPs and bacterial nucleic acids. However, a better understanding of the role of these lncRNAs during acute pathogenic infections across different species may provide critical insights into designing innovative therapeutic strategies to target these infections.
7 Role of lncRNAs during infection by intracellular pathogens
LncRNAs are either host-derived or expressed by pathogens and their response may play a crucial role in modulating bacterial infections. The general outcome of intracellular pathogenic infection may depend upon how these lncRNAs modulate target genes to either promote or restrict the pathogens’ replication inside host (Agliano et al., 2019).
In this review, we discuss the functions of lncRNAs by categorizing them in response to infections caused by (a) facultative intracellular pathogens, (b) obligate intracellular pathogens, and (c) commensals. It highlights the functions of discovered lncRNAs in infection models like M. tuberculosis, L. monocytogenes and S. Typhimurium. These discovered lncRNAs could also be investigated for their role in regulation of host immune response against different obligate intracellular bacterial infections. A list of these lncRNA in regulation of immune response to several bacterial infections is summarized in Table 1 of the review.
7.1 LncRNAs during infection by facultative intracellular pathogens
7.1.1 Mycobacterium tuberculosis
Previous studies in TB patients and in vitro infected samples showed differential expression of several lncRNA and mRNA. Functional analysis of these transcripts showed majority of the lncRNA regulate immune signaling pathways such as TGF-β, IFN-γ, JAK-STAT; T- and B- cells differentiation and adaptive immune responses (Fathizadeh et al., 2020). PCED1B-AS1 is a lncRNA that act as sponge of miRNA-155 in macrophages to relieve its effect on Forkhead Box O3 (FOXO3) and Ras Homolog MTORC1 binding (Rheb). This results in inhibition of apoptosis and increase in autophagy to clear Mtb. Previously reported study on CD14+ monocytes isolated from TB patients showed downregulation of PCED1B-AS1 which led to enhanced autophagy and reduced caspase-3 expression to attenuate apoptosis (Yao et al., 2014). These findings proposed PCED1B-AS1 as an early diagnostic biomarker for Mtb infection (Fathizadeh et al., 2020). LincRNA-EPS was previously reported as downregulated in monocytes isolated from Mtb patients. Further a knockdown of lincRNA-EPS in RAW264.7 macrophages followed by infection with M. bovis (Mbv) BCG (Bacillus Calmette–Guérin) showed inhibition of apoptosis and increase in autophagy due to enhanced signaling of JNK/MAPK pathway and IFN-γ production (Ke et al., 2020). On the contrary, lincRNA-Cox2 silencing in RAW264.7 cells showed increased intracellular growth of Mbv BCG. LincRNA-Cox2 upregulation was linked to enhanced binding of NF-κB to the promoter of inducible Nitric Oxide Synthase (iNOS) and increases nitric oxide (NO) production and M1 macrophage polarization. This resulted in bactericidal activity against Mtb or Mbv infection (Fathizadeh et al., 2020). Previous studies have also shown pathogen-specific regulation by MEG3. MEG3 was downregulated in IFN-γ-treated THP1-derived-macrophages after infection with Mbv BCG, but it was upregulated in M. smegmatis (Msm) infected cells (Agliano et al., 2019). In vitro silencing of MEG3 showed increased conversion of LC3-I to LC3-II during Mbv BCG infection in human macrophages to induce autophagy (Figure 4A). The study also showed that MEG3 knockdown in cells resulted in reduced mTOR-mediated phosphorylation of ribosomal protein S6 kinase (p70-S6K) at Thr389 and increased accumulation of p62 compared to uninfected macrophages which is necessary for induction of selective autophagy (Pawar et al., 2016). In summary, the evidences suggest regulatory control by these lncRNAs in production of IFN-γ and autophagic clearance of Mtb or Mbv in the host.

Figure 4 LncRNA in modulation of host immune response to facultative and obligate intracellular bacterial infection. (A) (left panel) During Mycobacterium tuberculosis infection, lncRNA-CD244 in human CD8+ T lymphocytes is upregulated in a CD244-dependent manner. LncRNA-CD244 interacts with the chromatin modification enzyme enhancer of zeste homolog 2 (EZH2), which leads to trimethylation of H3K27 at the tumor necrosis factor alpha (TNF-α) and interferon gamma (IFN-γ) loci. This represses the expression TNF-α and IFN-γ and promotes Mycobacterium replication. (right panel) Maternally expressed gene 3 (MEG3) is downregulated in THP-1 cells during M. bovis Bacillus Calmette–Guerin (BCG) infection. MEG3 increases LC3A/B conversion. LC3s decorate the Mycobacterium containing phagosomes and activate autophagy for bacterial clearance. (B) During Brucella abortus infection, PAMPSs on B. abortus activate the TLRs and TGF-β signaling. The TLRs via unknown mechanism inhibit expression of Gm28309 (LncRNA in mouse)/P33714 (human ortholog of Gm28309). Gm28309 sponges miR-3068-5p in steady state. During infection, downregulation of Gm28309 leads to release of miR-3068-5p in cytoplasm that degrades κB-Ras2 (an inhibitor of NF-κB) inducing p65 phosphorylation. In parallel, TGF-β activates TAK1 and IKK kinase mediates phosphorylation of IKB-α and p65, leading to translocation and activation of NF-κB signaling pathway. This leads to assembly of NLRP3 inflammasome, and secretion of IL-1β and IL-18. (C) Chlamydia trachomatis infection leads to upregulation of lncRNAs; MIAT, ZFAS1 and ZEB-AS1 via unknown mechanism. MIAT and ZFAS1 activate p38/MAPK/ERK signaling pathway through unclear mechanisms. ZEB1-AS1 activates p38/MAPK/ERK axis by acting as sponge of miR-1224-5p, which in turn cannot bind to MAP4K4-mRNA to inhibit its translation. FGD5-AS1 is also upregulated in response to infection. It activates the Wnt/β-Catenin pathway via unknown mechanism. The upregulation of these lncRNAs is linked to inhibition of apoptosis and increased C. trachomatis replication. (D) Rickettsia conorii infection in mouse phagocytic cells upregulates enhancer-lncRNA (elncRNA); NONMMUT013718 and NONMMUT024103 via unknown mechanism. They have been associated with an increase in the expression of Id2 (inhibitor of DNA binding 2) and Apol10b (apolipoprotein 10b). These genes contribute to activation of Type I-IFN signaling through unknown mechanisms.
Therapies to improve CD8+ T cell functioning is one of the promising strategy against Mtb infection (Fathizadeh et al., 2020). LncRNA-CD244 is found exclusively in humans and it overlaps with 5’-UTR of glutathione S-transferase theta-1 (GSTθ1) in CD8+ T cells (Wang et al., 2015). LncRNA-CD244 has been suggested as a potential target in anti-TB therapy. CD244+ CD8+ T cells isolated from peripheral blood mononuclear cells (PBMCs) of TB patients expressed higher levels of co-stimulatory molecule CD244 and lncRNA-CD244 compared to healthy controls through an unknown mechanism (Fathizadeh et al., 2020). Silencing of this lncRNA resulted in restricted Mtb growth. Moreover, immunoprecipitation studies in CD8+ T cells isolated from TB patients showed lncRNA-CD244 interacted with chromatin modification enzyme enhancer of zeste homolog 2 (EZH2) (Figure 4A), which catalyzed H3K27me3 at promoters of IFN-γ and TNF-α to inhibit their production (Wang et al., 2015). Previous studies have also shown that both the isoforms of NEAT1 were upregulated in T-helper (Th) cells derived from TB patients compared to healthy controls and its deletion resulted in reduced survival of Mtb in macrophages (Agliano et al., 2019). LncRNA XLOC_012582 positively regulates SOCS3 which led to increase in cytokine expression in B cells from TB patients (Fathizadeh et al., 2020).
RNA-sequencing studies from TB patients showed reduced expression of LncRNA-TGS1-1 and increased expression of miRNA-143 which represses downstream immune gene activation (Fathizadeh et al., 2020). Microarrays studies from plasma of TB patients have also identified NR_038221, ENST00000427151 and ENST00000354432 as potential biomarkers for early detection of TB (He et al., 2017).
7.1.2 Salmonella Typhimurium
Nettoie Salmonella pas Theiler’s (NeST) or Tmevpg1 is an antisense lncRNA located within IFN-γ and is expressed in CD4+ Th1 cells, CD8+ T cells, and natural killer (NK) cells in humans and mice (Agliano et al., 2019). NeST expression is enhanced during STm infection and this promotes bacterial clearance in vivo (Gomez et al., 2013). Transgenic B10.S mice expressing either SJL/J- or B10.S-derived NeST RNA demonstrated decreased STm pathogenesis compared to WT B10.S without NeST. This study further showed that co-transfection of NeST with WD repeat domain 5 (WDR5) cDNA in transgenic B10.S resulted in interaction of NeST with WDR5. These mice during STm infection showed increased H3K4me3 marks at IFN-γ locus in activated CD8+ T cells. Further, polymorphisms in NeST resulted in difference in T-cell mediated responses which modulated the inflammatory activities of cells to confer resistance against STm infection (Gomez et al., 2013). RNA-sequencing studies on STm-infected HeLa cells showed significant upregulation of NEAT1_2 compared to uninfected cells. NEAT1_2 was also induced in response to heat-killed STm which suggested that NEAT1_2 upregulation was as a result of PRR response of HeLa cells to STm PAMPs (Imamura et al., 2018). Knockout and knockdown of NEAT1_2 in HeLa cells resulted in reduced expression of TNF superfamily member 9 (TNFSF9) and CCL2 that led to increased susceptibility to STm infection. In normal HeLa cells, NEAT1_2 is readily degraded into unstable transcripts by MTR4 and RRP6 (components of nuclear exosome targeting (NEXT)-RNA exosome pathway). During STm infection, MTR4 and RRP6 levels is reduced leading to decreased degradation of NEAT1_2 (Imamura et al., 2018). Intracellular pathogens hijack metabolic pathways to foster replication in host cells. Recent research identified glucose transporters were encoded by genes SLC2A3 and SLC2A14. They are located near NEAT1_2 loci and were significantly upregulated in STm-infected HeLa cells and were essential for STm intracellular replication (Agliano et al., 2019).
7.1.3 Listeria monocytogenes
Lm infection leads to increased expression of AS-IL1α which transcriptionally activates IL-1α by recruiting RNA polymerase II to IL-1α promoter (Chan et al., 2015). LincRNA-EPS was previously reported as downregulated during Lm infection. Further, lincRNA-EPS deficient BMDM and DCs when infected with Lm displayed reduced bacterial replication and increased expression of TNF-α, IL-6, and iNOS (Agliano et al., 2020). This concluded that lincRNA-EPS downregulation is essential for activation of NF- κB dependent pro-inflammatory response to limit Lm infection (Atianand et al., 2016). LincRNA-Cox2 is another well characterized lncRNA which is upregulated during Lm infection in vitro and in vivo. It also promotes Lm clearance via activation of NF- κB dependent pro-inflammatory gene expression through increased association with SWI/SNF complex and interactions with hnRNA-A/B and A2/B1 (Carpenter et al., 2013). Sros1 is a lncRNA that was reported as downregulated in IFN-γ-stimulated and Lm-infected macrophages. In uninfected cells, Sros1 regulates JAK-STAT and MAPK pathways (Li et al., 2021a). However, Lm infection activates miRNA-1 that degrades Sros1, which in turn relieves RNA binding protein Cell Cycle Associated Protein 1 (CAPRIN1) from Stat1. This stabilizes Stat1 transcription and increases STAT1 translation leading to enhanced IFN-γ signaling and bacterial clearance (Li et al., 2021a).
7.1.4 Legionella pneumophila
Lp is an intracellular pathogen that employs a Dot/IcM Type IV secretion system (T4SS) apparatus to infect and replicate inside human phagocytes. Recent studies have discovered lncRNAs MaIL1 and Linc01215 are induced during Lp infection in a TLR4-NF-κB dependent manner to activate type-I IFN production and limit Lp replication inside macrophages. Concurrently, MaIL1 knockdown in macrophages resulted in an increased Lp intracellular replication. The growth could be restricted following treatment of Lp-infected macrophages with type-I IFN (Aznaourova et al., 2020). C. burnetii (Cb) is phylogenetically related to Lp and infects phagocytes in a similar mechanism using the T4SS. We hypothesize the signaling mechanisms involved in Lp pathogenesis would be similar in Cb infection. Although the type-I IFN response could counter or dampen Cb infection in a tissue-specific manner unlike Lp infection. It would be interesting to explore type-I IFN dependent lncRNA MaIL1 and other discovered lncRNAs in regulation of Cb pathogenesis.
7.1.5 Pseudomonas aeruginosa
Transcriptome analysis of Pa-infected bronchial epithelial cells from patients with cystic fibrosis (CF) compared to non-CF epithelial cells revealed 108 differentially expressed lncRNAs. Of them, MEG9 and BLACAT1, and MEG3 were significantly downregulated (Balloy et al., 2017) MEG3 was also reported as downregulated in lung tissues from Pa-infected mice in TLR4-NF-κB dependent manner. Pa infection downregulates MEG3 isoform-4 which relieves miRNA-138. The miRNA-138 binds to IL-1β to reduce its expression, and thereby control inflammation and promote bacterial colonization in vivo (Li et al., 2018).
7.1.6 Brucella abortus
Trancriptome analysis of THP-1 cells infected with B. abortus S2308 strain showed downregulation of lncRNA Gm28309 in a TLR4 dependent manner. The reduced expression of Gm28309 resulted in the activation of the NF-κB pathway and increased expression of TGF-β. TGF-β signaling activates TAK1 and IKK kinase mediated phosphorylation of p65. Also, Gm28309 act as sponge to miR-3068-5p in steady state. However, during infection miR-3068-5p is relesaed in the cytoplasm that binds and degrades κB-Ras2, promoting p65 phosphorylation (Figure 4B). The activation of the NF-κB pathway recruits the NLRP3 inflammasome, and secretion of IL-1β and IL-18 to modulate Ba infection in vitro (Deng et al., 2020). However, the exact role of Gm28309 in Ba pathogenesis is unclear. Another study reported increased expression of NeST and IFN-γ in PBMCs isolated from patients with brucellosis (Gheitasi et al., 2019).
7.2 LncRNAs during colonization by commensals
Commensals are obligate anaerobic opportunistic pathogens that constitute the human microbiota. They regulate many host cellular processes like metabolism, immune response, physiology and dysbiosis can lead to autoimmune disorders and microbiota associated cancers (Hou et al., 2022). LncRNAs are also emerging as a factor to regulate microbiota composition and host-microbiota interaction (Díaz-Garrido et al., 2021). Of them, A recent study has proposed lncRNA can act as molecular signatures in gut tissues and their spatio-temporal expression patterns could serve as biomarker to differentiate between a healthy and dysbiotic gut (Liang et al., 2015). It is noteworthy; these commensals triggered lncRNAs could also be explored in pathogenesis by obligate intracellular pathogens and therefore are discussed in this review.
One of the attributing factors to colorectal cancer (CRC) is overgrowth of Fusobacterium nucleatum (Fn). Recent trancriptome analysis of Fn-infected CRC cells revealed 43 upregulated lncRNAs that includes Keratin 7 antisense RNA (KRT7-AS) and endogenous retroviral-associated adenocarcinoma lncRNA (EVADR) (Chen S. et al., 2020). KRT7-AS promotes metastasis by CRC cells in nude mice by upregulating KRT7 in NF-κB dependent manner. CRC infection with Fn leads to upregulation of EVADR leading to increased metastasis of CRC cells both in vivo and in vitro. EVADR acts as scaffold to Y-box binding protein 1 (YBX1) leading to increased translation of SNAIL, SLUG, and ZEB1, which are required for epithelial to mesenchymal transition (EMT) (Lu et al., 2022). Another RNA sequencing analysis found 75 lncRNAs and 49 lncRNAs in probiotic and pathogen mediated carcinogenesis. Four lncRNAs: FRMD6-AS2, DIRC3, LIFR-AS1, and MRPL23-AS1 were proposed as unique signatures to CRC prognosis. LINC00355, KCNQ1OT1, LINC00491, and HOTAIR were associated with poor survival of CRC cells (Wen et al., 2020b). Fn is also an etiological agent for oral squamous cell carcinoma (OSCC). lncRNA MIR4435-2HG-5p was previously reported to be upregulated in Fn-infected OSCC cells. It act as sponge to miRNA-296-5p, leading to activation of serine/threonine kinase Akt2 and Akt2 induced tumorigenesis (Zhang et al., 2020).
Enterotoxigenic Bacteroides fragilis (ETBF) can induce tumor in dysbiotic gut. LncRNA Bacteroides fragilis-associated lncRNA1 (BFAL1) was previously reported to be upregulated in ETBF-infected CRC cells. BFAL1 over-expression promoted tumorigenesis by acting as sponge to microRNAs; miR-155-5p and miR-200a-3p, which in turn activated RHEB/mTOR pathway (Bao et al., 2019).
Lnc-GNAT1 was reported to be downregulated in Helicobacter pylori (Hp) induced gastric cancer. It negatively regulates the Wnt/β-Catenin signaling to regulate cancer cell proliferation (Liu L. et al., 2018). Lnc-SGK1 induces expression of glucocorticoid-inducible kinase 1 (SGK1) to induce Th2 and Th17 differentiation and inhibit Th1 differentiation in Hp-infected gastric cancer (Yao et al., 2016). Previous serological studies have also identified potential lncRNA signatures in Hp-infected cancer cells like H19, LINC00152, lncRNA AF147447, RP11-169F17.1, and RP11-669 N7.2 (Wen et al., 2020b).
7.3 LncRNAs during infection by obligate intracellular pathogens
Obligate intracellular pathogens include members of Anaplasma, Rickettsia, Orientia, Wolbachia, Chlamydia, and C. burnetii. Although lncRNA research in obligate intracellular pathogenesis is at nascent stage, recent studies have identified some lncRNAs role during infection and are discussed in the current section.
7.3.1 Chlamydia trachomatis
Ct is an obligate intracellular pathogen which is a leading cause of sexually transmitted infection and reproductive complications. It infects host cells by modulating host cellular responses at transcriptional and translational level (Dzakah et al., 2021). Previous transcriptome analyses of Ct-infected HeLa cells have identified lncRNAs in inhibition of host cell apoptosis. For instance, lncRNA zinc finger antisense 1 (ZFAS1) was previously reported as upregulated in Ct infection. It was predicted to regulate MAPK pathways and its silencing led to decrease in Chlamydia plasmid protein pORF5-mediated-apoptosis and Ct virulence (Figure 4C). The study concluded pORF5 crucial role in activation of lncRNA ZFAS1, and ZFAS1 mediated induction of MAPK/p38 pathway to inhibit host cell apoptosis (Wen et al., 2020a).
A microarray analysis of IFN-γ-treated HeLa cells infected with Ct revealed 1718 lncRNAs as differentially expressed. Gene enrichment analysis showed these lncRNAs regulated host pathways involved in apoptosis, NOD-like receptor, TNF and RIG-I-like receptor signaling (Luo et al., 2022b). LncRNAs MIAT, ZEB1-AS1, and IRF1 were silenced in HeLa which resulted in increased apoptosis. MIAT-silenced cell lines showed reduced expression of Bcl-2, and increased activation of caspase-3. This indicated MIAT was involved in inhibition of mitochondria-mediated cellular apoptosis to promote Ct intracellular growth (Luo et al., 2022b). A follow-up study by Luo et al. studied the molecular mechanisms of ZEB-AS1 in Ct infection. siRNA mediated silencing of ZEB1-AS1 increased host cell apoptosis and impeded Ct growth. This was marked by downregulation of Bcl-2/Bax ratio that led to reduced mitochondrial membrane potential, release of cytochrome c, and caspase-3 activation. Further analysis revealed ZEB1-AS1 acted as a sponge for miRNA-1224-5p which transcriptionally activated MAP4K4 (Figure 4C). In conclusion, ZEB1-AS1 regulated miR-1224-5p/MAP4K4 axis to inhibit cellular apoptosis and promoted intracellular Ct infection (Luo et al., 2022a).
A previous microarray study also showed lncRNA FYVE, RhoGEF, and PH domain containing five antisense RNA1 (FGD5-AS1) was upregulated during Ct infection. Gene ontology analysis showed they enriched for DNA replication and Wnt signaling pathway. FGD5-AS1 positively regulated the expression of the Wnt/β-Catenin pathway (Figure 4C), inhibiting apoptosis and promoting intracellular replication of Ct in host (Wen et al., 2021).
7.3.2 Rickettsia conorii
RNA-sequencing analysis to identify host lncRNA during Rickettsia infection was previously conducted by Chowdhury et al. C3H/HeN mice were infected with R. conorii (Rc) and transcriptome analysis was conducted from the infected lung tissues. This analysis identified 74,964 non-coding RNAs, out of which 206 lncRNAs were upregulated, and 277 were downregulated. The study identified two highly upregulated elncRNAs NONMMUT013718 and NONMMUT024103. NONMMUT013718 and NONMMUT024103 interacted with the promoter regions of Id2 (inhibitor of DNA binding 2) and Apol10b (apolipoprotein 10b) respectively to enhance their transcription (Figure 4D). Further, in vivo and in vitro infection with Rc showed increased expression of both lncRNAs and their associated genes in the macrophages. These findings were the first experimental evidence demonstrating regulatory role of lncRNAs in modulation of protective immunity against Rc infection (Chowdhury et al., 2019).
7.3.3 Wolbachia
Wolbachia is an obligate intracellular pathogen that infects Aedes aegypti. Genome wide analysis of A. aegypti Aag2 cells infected with Wolbachia identified 3035 differentially expressed lncRNAs. Gene enrichment and network analysis revealed that the identified lncRNAs acted in trans to target genes and belongs to oxidative phosphorylation pathway. Further, competitive endogenous RNA (ceRNA) network analysis showed these lncRNA enriched for cellular oxidant detoxification pathway. Of them, lncRNA aae-lnc-7598 had the highest upregulation (Mao et al., 2022). Silencing of aae-lnc-7598 resulted in significant downregulation of antioxidant catalase 1B (CAT1B) gene and increase in mitochondria-mediated reactive oxygen species (ROS) in host cells. Another lncRNA, aae-lnc-0165 was downregulated after Wolbachia infection in host cells, which increased expression of REL1 through binding of aae-miRNA-980-5p, leading to Toll activation (Mao et al., 2022). Notably, these lncRNAs activate the anti-Dengue Toll pathway during Wolbachia infection. Wolbachia doesn’t infect human, but it can stay as an endosymbiont in vectors like A. aegypti. A. aegypti is a major vector responsible for transmission of Dengue virus. Taken together, we propose that these Wolbachia regulated lncRNAs could be explored as potential therapeutic markers to limit Dengue virus infection (Bensaoud et al., 2021; Mao et al., 2022).
7.3.4 Anaplasma phagocytophilum
Ap infects the host via tick bite. It replicates within neutrophils and modulate host signaling responses mediated by secreted effectors such as AnkA. AnkA recruits HDAC1 to deacetylate H3 leading to transcriptional repression of several host immune response genes (Rennoll-Bankert et al., 2015). A previously reported AnkA-binding-CHIP sequencing analysis showed AnkA binding sites in the intergenic regions of target genes and transcriptionally regulated their expression in cis. NONCODE databases alignment showed AnkA binding sites in the promoter region of TNF/LT. The locus overlapped with a lncRNA named as lincRNA-TUCP. Further studies are required to dissect role of lincRNA-TUCP in regulation of TNF/LT locus during Ap infection (Dumler et al., 2016). There is also a limited understanding on how intracellular pathogens may regulate the expression of lncRNAs. AnkA effector of Ap was found to regulate the TNF/LT locus and the locus overlaps with linc-TUCP (Dumler et al., 2016). Future research could probe the prospective role of AnkA as a nucleo-modulin in regulating the linc-TUCP/TNF/LT locus during Ap infection. More studies should be conducted to investigate and identify bacterial effectors in regulation of host lncRNAs and their impact on downstream signaling pathways during infection.
8 LncRNAs as therapeutic targets
LncRNAs linked to host-pathogen interaction are promising targets for development of diagnostic and prognostic biomarkers. This possibility is supported by the clinical advantages represented by several of their molecular characteristics such as high tissue-specificity and regulation of specific biological targets in the host which circumvents undesired effects that arises from using protein-based drugs. Additionally, the non-translational nature, low endogenous expression and fast transcriptional turnover of lncRNA can result in greater biological effects with lower doses.
Some obligate intracellular pathogens and parasites infect via exosomes that are internalized by cells to form endosomes. Exosomes from gut microbes have been shown to improve metabolic functions (Li et al., 2021b). Conversely, fecal exosomes from mice infected with Pseudomonas panacis promotes glucose intolerance and insulin resistance (Li et al., 2021b). There is growing evidence on the regulation of cellular responses by exosomal lncRNAs. For instance, exosomal lncRNAs such as LNMAT2 (lymph node metastasis associated transcript 2), MALAT1, Gm26809 and NEAT1 etc. have been involved in metabolic reprogramming of tumor microenvironment (Sun et al., 2018), macrophage polarization and inflammatory diseases (Liu R. et al., 2018). Of note, MALAT1 over-expression suppresses macrophage activation, IL-10 response during Leishmannia and Plasmodium infection (Hewitson et al., 2020). Depletion of NEAT1_2 in HeLa cells promotes increased STm replication (Imamura et al., 2018). Furthermore, NEAT1_1 and NEAT1_2 knockout in macrophages have previously shown decreased IL-6 levels and delayed Mtb clearance (Huang et al., 2018). MALAT1 and NEAT1 are well characterized regulators of immune response in different pathogenic infection models. They may also be studied in regulation of inflammatory response, metabolic reprogramming and macrophage polarization during obligate bacterial infections. One advantage of targeting exosomal lncRNA is their stability inside exosomes, tissue-specificity and often their response is disease-specific. The criteria are ideal for development of lncRNA based biomarkers (Mosquera-Heredia et al., 2021). This may also lead to the development of highly specific disease markers leading to other research questions including- could these discovered lncRNAs be used to develop infection specific biomarkers for bioterror agents like C. burnetii?
The most recent technique of targeting lncRNA for therapeutics is based on the use of antisense oligonucleotides (ASOs). ASOs specifically binds lncRNA based on their sequence complementarities that induce RNase H mediated cleavage of lncRNA (Lai et al., 2020; Lee and Mendell, 2020). However, ASO mediated targeting of lncRNA in clinical settings hasn’t been successful due to in vivo toxicity and lack of proper delivery systems. Current researches have attempted to improve the pharmacological properties of ASOs by chemically modifying them to enhance the binding affinity to their target lncRNA, thereby increasing resistance to degradation by nucleases and reducing immunostimulatory activity (Seth et al., 2008). Fused aptamers may also be used for enhanced specificity in the intracellular delivery of oligo-based drugs (Dassie and Giangrande, 2013). Another strategy is the use of small molecules for targeting lncRNAs. This strategy of targeting lncRNA has been less successful because of the complexity associated with development of a high affinity small molecule that can bind lncRNA. Additionally, it requires identification of RNA motifs which can be only achieved from complete understanding of the lncRNA structure (Warner et al., 2018). Often, these lncRNAs fold into several molecular domains and can engage in different molecular interactions making it difficult for designing a small molecule targeting the lncRNA (Statello et al., 2021).
9 Conclusion
Advancements in RNA biology techniques, single cell sequencing and CHIP-seq assays have shown that lncRNAs are central regulators for innate immune activation, inflammasome assembly, and host-pathogen interactions. However, the field of study is challenging due to the complexities arising from the unstable nature of RNAs as well as lack of technological advancements in studying the 3-D conformation and the molecular interaction of RNA in vitro and in vivo. It is also becoming increasingly evident that their sequence, expression levels, processing, cellular localization, organization and interaction with other molecules can define their functionality. Another bottleneck in lncRNA research is the poor sequence conservation across species. Therefore, it becomes difficult to correlate and extrapolate results between human and mouse experimental systems. This is partially mitigated by initial screening, synteny and motif conservation analysis among the lncRNAs from different species. Additionally, the existing databases and computational tools require improvement in the current algorithms to not only precisely assess orthologs of lncRNAs but also predict their functional conservation and interactome across species (Zheng et al., 2021). One promising strategy to overcome this limitation could be the use of humanized mice models to study lncRNAs in response to infections. For instance, the role of lncRNA-CD244 in Mtb infection was studied in severe combined immuno-deficiency mice (SCID) mice engrafted with lncRNA-CD244-depressed human CD8+ T cells. Comparable studies can vastly expand the knowledge in the field.
The review of current literature suggests that lncRNAs regulate cell specification and disease. These functions require deeper understanding of physio-pathological processes so that they can be therapeutically targeted with high specificity. Cellular RNA may be a promising target for drug discovery. Synthesizing functionally relevant motifs of lncRNAs or the whole RNA complementary to a target host molecule for drug development may prove to be an effective strategy for lncRNA-directed therapeutics. For example, (a) lncRNAs as sponges of miRNAs or affecting the splicing mechanisms of a protein coding gene, or (b) compounds that could regulate biological processes in host for clinical development. The advantage of such lncRNA-directed therapeutics would be that these oligonucleotides may target complex and undruggable cellular targets. Overall, development of better tools for comprehensive molecular understanding of lncRNAs in regulation of immune responses and host-pathogen interaction might help in development of lncRNA-based therapeutics for intervention of infectious and inflammatory diseases.
Author contributions
AA wrote the review. All authors contributed to the article and approved the submitted version.
Funding
This review was supported by the Defense Threat Reduction Agency, contract no. HDTRA1210002. The views expressed in this article are those of the authors and do not reflect the official policy or position of the US Department of Defense or the US Army.
Acknowledgment
All the figures in this review were created using Biorender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agliano, F., Fitzgerald, K. A., Vella, A. T., Rathinam, V. A., Medvedev, A. E. (2020). Long non-coding RNA LincRNA-EPS inhibits host defense against Listeria monocytogenes infection. Front. Cell. Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00481
Agliano, F., Rathinam, V. A., Medvedev, A. E., Vanaja, S. K., Vella, A. T. (2019). Long noncoding RNAs in host–pathogen interactions. Trends Immunol. 40, 492–510. doi: 10.1016/j.it.2019.04.001
Ahmad, P., Bensaoud, C., Mekki, I., Rehman, M. U., Kotsyfakis, M. (2021). Long non-coding RNAs and their potential roles in the vector–Host–Pathogen triad. Life 11, 1–12. doi: 10.3390/life11010056
Almada, A. E., Wu, X., Kriz, A. J., Burge, C. B., Sharp, P. A. (2013). Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature 499, 360–363. doi: 10.1038/nature12349
Atianand, M. K., Caffrey, D. R., Fitzgerald, K. A. (2017). Immunobiology of long noncoding RNAs. Annu. Rev. Immunol. 35, 177–198. doi: 10.1146/annurev-immunol-041015-055459
Atianand, M. K., Hu, W., Satpathy, A. T., Shen, Y., Ricci, E. P., Alvarez-Dominguez, J. R., et al. (2016). A long noncoding RNA LincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165, 1672–1685. doi: 10.1016/j.cell.2016.05.075
Azam, S., Hou, S., Zhu, B., Wang, W., Hao, T., Bu, X., et al. (2019). Nuclear retention element recruits U1 snRNP components to restrain spliced lncRNAs in the nucleus. RNA Biol. 16, 1001–1009. doi: 10.1080/15476286.2019.1620061
Aznaourova, M., Janga, H., Sefried, S., Kaufmann, A., Dorna, J., Volkers, S. M., et al. (2020). Noncoding RNA MaIL1 is an integral component of the TLR4-TRIF pathway. Proc. Natl. Acad. Sci. U. S. A. 117, 9042–9053. doi: 10.1073/pnas.1920393117
Balas, M. M., Johnson, A. M. (2018). Exploring the mechanisms behind long noncoding RNAs and cancer. Non-coding RNA Res. 3, 108–117. doi: 10.1016/j.ncrna.2018.03.001
Balloy, V., Koshy, R., Perra, L., Corvol, H., Chignard, M., Guillot, L., et al. (2017). Bronchial epithelial cells from cystic fibrosis patients express a specific long non-coding RNA signature upon Pseudomonas aeruginosa infection. Front. Cell. Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00218
Bao, Y., Tang, J., Qian, Y., Sun, T., Chen, H., Chen, Z., et al. (2019). Long noncoding RNA BFAL1 mediates enterotoxigenic Bacteroides fragilis-related carcinogenesis in colorectal cancer via the RHEB/mTOR pathway. Cell Death Dis. 10, 675. doi: 10.1038/s41419-019-1925-2
Bensaoud, C., Martins, L. A., Aounallah, H., Hackenberg, M., Kotsyfakis, M. (2021). Emerging roles of non-coding RNAs in vector-borne infections. J. Cell Sci. 134, 5. doi: 10.1242/jcs.246744
Brown, J. A., Valenstein, M. L., Yario, T. A., Tycowski, K. T., Steitz, J. A. (2012). Formation of triple-helical structures by the 3’-end sequences of MALAT1 and MENβ noncoding RNAs. Proc. Natl. Acad. Sci. U. S. A. 109, 19202–19207. doi: 10.1073/pnas.1217338109
Cabili, M. N., Trapnell, C., Goff, L., Koziol, M., Tazon-Vega, B., Regev, A., et al. (2011). Integrative annotation of human Large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927. doi: 10.1101/gad.17446611
Carpenter, S., Aiello, D., Atianand, M. K., Ricci, E. P., Gandhi, P., Hall, L. L., et al. (2013). A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792. doi: 10.1126/science.1240925
Castellanos-Rubio, A., Kratchmarov, R., Sebastian, M., Garcia-Etxebarria, K., Garcia, L., Irastorza, I., et al. (2017). Cytoplasmic form of carlr lncRNA facilitates inflammatory gene expression upon NF-κB activation. J. Immunol. 199, 581–588. doi: 10.4049/jimmunol.1700023
Chan, J., Atianand, M., Jiang, Z., Carpenter, S., Aiello, D., Elling, R., et al. (2015). A natural antisense transcript, AS-IL1α, controls inducible transcription of the proinflammatory cytokine IL-1α. J. Immunol. 195, 1359–1363. doi: 10.4049/jimmunol.1500264
Chen, Y., Li, H., Ding, T., Li, J., Zhang, Y., Wang, J., et al. (2020). Lnc-M2 controls M2 macrophage differentiation via the PKA/CREB pathway. Mol. Immunol. 124, 142–152. doi: 10.1016/j.molimm.2020.06.006
Chen, S., Su, T., Zhang, Y., Lee, A., He, J., Ge, Q., et al. (2020). Fusobacterium nucleatum Promotes colorectal cancer metastasis by modulating KRT7-AS/KRT7. Gut Microbes 11, 511–525. doi: 10.1080/19490976.2019.1695494
Chen, X., Yan, C. C., Zhang, X., You, Z. H. (2017). Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief. Bioinform. 18, 558–576. doi: 10.1093/bib/bbw060
Chowdhury, I. H., Narra, H. P., Sahni, A., Khanipov, K., Fofanov, Y., Sahni, S. K. (2019). Enhancer associated long non-coding RNA transcription and gene regulation in experimental models of rickettsial infection. Front. Immunol. 10. doi: 10.3389/fimmu.2018.03014
Core, L. J., Waterfall, J. J., Lis, J. T. (2008). Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Sci. (80-.) 322, 1845–1848. doi: 10.1126/science.1162228
Cornelis, G., Souquere, S., Vernochet, C., Heidmann, T., Pierron, G. (2016). Functional conservation of the lncRNA NEAT1 in the ancestrally diverged marsupial lineage: evidence for NEAT1 expression and associated paraspeckle assembly during late gestation in the opossum Monodelphis domestica. RNA Biol. 13, 826–836. doi: 10.1080/15476286.2016.1197482
Da, M., Zhuang, J., Zhou, Y., Qi, Q., Han, S. (2021). Role of long noncoding RNA taurine-upregulated gene 1 in cancers. Mol. Med. 27, 51. doi: 10.1186/s10020-021-00312-4
Dai, W., Wang, M., Wang, P., Wen, J., Wang, J., Cha, S., et al. (2021). LncRNA NEAT1 ameliorates LPS−Induced inflammation in MG63 cells by activating autophagy and suppressing the NLRP3 inflammasome. Int. J. Mol. Med. 47, 607–620. doi: 10.3892/ijmm.2020.4827
Dassie, J. P., Giangrande, P. H. (2013). Current progress on aptamer-targeted oligonucleotide therapeutics. Ther. Deliv. 4, 1527–1546. doi: 10.4155/tde.13.118
Dempsey, J., Zhang, A., Cui, J. Y. (2018). Coordinate regulation of long non-coding RNAs and protein-coding genes in germ-free mice. BMC Genomics 19, 1–24. doi: 10.1186/s12864-018-5235-3
Deng, X., Guo, J., Sun, Z., Liu, L., Zhao, T., Li, J., et al. (2020). Brucella-induced downregulation of lncRNA Gm28309 triggers macrophages inflammatory response through the miR-3068-5p/NF-κB pathway. Front. Immunol. 11, 1–14. doi: 10.3389/fimmu.2020.581517
Deretic, V. (2011). Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 240, 92–104. doi: 10.1111/j.1600-065X.2010.00995.x
Díaz-Garrido, N., Badia, J., Baldomà, L. (2021). Microbiota-derived extracellular vesicles in interkingdom communication in the gut. J. Extracell. Vesicles 10, e12161. doi: 10.1002/jev2.12161
DiStefano, J. K. (2018). The emerging role of long noncoding RNAs in human disease. Methods Mol. Biol. 1706, 91–110. doi: 10.1007/978-1-4939-7471-9_6
Du, M., Yuan, L., Tan, X., Huang, D., Wang, X., Zheng, Z., et al. (2017). The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat. Commun. 8, 2049. doi: 10.1038/s41467-017-02229-1
Dumler, J. S., Sinclair, S. H., Pappas-Brown, V., Shetty, A. C. (2016). Genome-wide Anaplasma phagocytophilum AnkA-DNA interactions are enriched in intergenic regions and gene promoters and correlate with infection-induced differential gene expression. Front. Cell. Infect. Microbiol. 6, 1–15. doi: 10.3389/fcimb.2016.00097
Dzakah, E. E., Huang, L., Xue, Y., Wei, S., Wang, X., Chen, H., et al. (2021). Host cell response and distinct gene expression profiles at different stages of Chlamydia trachomatis infection reveals stage-specific biomarkers of infection. BMC Microbiol. 21, 3. doi: 10.1186/s12866-020-02061-6
Elling, R., Robinson, E. K., Shapleigh, B., Liapis, S. C., Covarrubias, S., Katzman, S., et al. (2018). Genetic models reveal cis and trans immune-regulatory activities for lincRNA-Cox2. Cell Rep. 25, 1511–1524.e6. doi: 10.1016/j.celrep.2018.10.027
Engreitz, J. M., Haines, J. E., Perez, E. M., Munson, G., Chen, J., Kane, M., et al. (2016). Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455. doi: 10.1038/nature20149
Fathizadeh, H., Hayat, S. M. G., Dao, S., Ganbarov, K., Tanomand, A., Asgharzadeh, M., et al. (2020). Long non-coding RNA molecules in tuberculosis. Int. J. Biol. Macromol. 156, 340–346. doi: 10.1016/j.ijbiomac.2020.04.030
Frankish, A., Diekhans, M., Jungreis, I., Lagarde, J., Loveland, J. E., Mudge, J. M., et al. (2021). GENCODE 2021. Nucleic Acids Res. 49, D916–D923. doi: 10.1093/nar/gkaa1087
Froberg, J. E., Yang, L., Lee, J. T. (2013). Guided by RNAs: X-inactivation as a model for lncRNA function. J. Mol. Biol. 425, 3698–3706. doi: 10.1016/j.jmb.2013.06.031
Gao, N., Li, Y., Li, J., Gao, Z., Yang, Z., Li, Y., et al. (2020). Long non-coding RNAs: the regulatory mechanisms, research strategies, and future directions in cancers. Front. Oncol. 10. doi: 10.3389/fonc.2020.598817
Ghafouri-Fard, S., Dashti, S., Taheri, M. (2020). The HOTTIP (HOXA transcript at the distal tip) lncRNA: review of oncogenic roles in human. Biomed. Pharmacother. 127, 110158. doi: 10.1016/j.biopha.2020.110158
Gheitasi, R., Jourghasemi, S., Pakzad, I., Hosseinpour Sarmadi, V., Samieipour, Y., Sekawi, Z., et al. (2019). A potential marker in brucellosis, long non coding RNA IFNG-AS1. Mol. Biol. Rep. 46, 6495–6500. doi: 10.1007/s11033-019-05095-w
Gomez, J. A., Wapinski, O. L., Yang, Y. W., Bureau, J.-F., Gopinath, S., Monack, D. M., et al. (2013). The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell 152, 743–754. doi: 10.1016/j.cell.2013.01.015
Grelet, S., Link, L. A., Howley, B., Obellianne, C., Palanisamy, V., Gangaraju, V. K., et al. (2017). A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 19, 1105–1115. doi: 10.1038/ncb3595
Grossi, E., Raimondi, I., Goñi, E., González, J., Marchese, F. P., Chapaprieta, V., et al. (2020). A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat. Commun. 11, 936. doi: 10.1038/s41467-020-14623-3
Guo, C.-J., Ma, X.-K., Xing, Y.-H., Zheng, C.-C., Xu, Y.-F., Shan, L., et al. (2020). Distinct processing of lncRNAs contributes to non-conserved functions in stem cells. Cell 181, 621–636.e22. doi: 10.1016/j.cell.2020.03.006
Guttman, M., Garber, M., Levin, J. Z., Donaghey, J., Robinson, J., Adiconis, X., et al. (2010). Ab Initio Reconstruction of cell type–specific transcriptomes in mouse reveals the conserved multi-exonic structure of LincRNAs. Nat. Biotechnol. 28, 503–510. doi: 10.1038/nbt.1633
Hacisuleyman, E., Shukla, C. J., Weiner, C. L., Rinn, J. L. (2016). Function and evolution of local repeats in the Firre locus. Nat. Commun. 7, 11021. doi: 10.1038/ncomms11021
Hadjicharalambous, M. R., Roux, B. T., Feghali-Bostwick, C. A., Murray, L. A., Clarke, D. L., Lindsay, M. A. (2018). Long non-coding RNAs are central regulators of the IL-1β-Induced inflammatory response in normal and idiopathic pulmonary lung fibroblasts. Front. Immunol. 9. doi: 10.3389/fimmu.2018.02906
He, J., Ou, Q., Liu, C., Shi, L., Zhao, C., Xu, Y., et al. (2017). Differential expression of long non-coding RNAs in patients with tuberculosis infection. Tuberculosis 107, 73–79. doi: 10.1016/j.tube.2017.08.007
Herriges, M. J., Swarr, D. T., Morley, M. P., Rathi, K. S., Peng, T., Stewart, K. M., et al. (2014). Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev. 28, 1363–1379. doi: 10.1101/gad.238782.114
Hewitson, J. P., West, K. A., James, K. R., Rani, G. F., Dey, N., Romano, A., et al. (2020). MALAT1 suppresses immunity to infection through promoting expression of maf and IL-10 in Th cells. J. Immunol. 204, 2949–2960. doi: 10.4049/jimmunol.1900940
Hou, K., Wu, Z.-X., Chen, X.-Y., Wang, J.-Q., Zhang, D., Xiao, C., et al. (2022). Microbiota in health and diseases. Signal Transduction Targeting Ther. 7, 135. doi: 10.1038/s41392-022-00974-4
Houseley, J., Rubbi, L., Grunstein, M., Tollervey, D., Vogelauer, M. (2008). A ncRNA modulates histone modification and mRNA induction in the yeast gal gene cluster. Mol. Cell 32, 685–695. doi: 10.1016/j.molcel.2008.09.027
Hu, G., Gong, A.-Y., Wang, Y., Ma, S., Chen, X., Chen, J., et al. (2016). LincRNA-Cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF-mediated chromatin remodeling. J. Immunol. 196, 2799–2808. doi: 10.4049/jimmunol.1502146
Huang, S., Huang, Z., Luo, Q., Qing, C. (2018). The expression of lncRNA NEAT1 in human tuberculosis and its antituberculosis effect. BioMed. Res. Int. 2018, 9529072. doi: 10.1155/2018/9529072
Hubé, F., Velasco, G., Rollin, J., Furling, D., Francastel, C. (2011). Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 39, 513–525. doi: 10.1093/nar/gkq833
Imamura, K., Takaya, A., Ishida, Y.-I., Fukuoka, Y., Taya, T., Nakaki, R., et al. (2018). Diminished nuclear RNA decay upon Salmonella infection upregulates antibacterial noncoding RNAs. EMBO J. 37, e97723. doi: 10.15252/embj.201797723
Jain, A. K., Xi, Y., McCarthy, R., Allton, K., Akdemir, K. C., Patel, L. R., et al. (2016). LncPRESS1 is a p53-regulated LncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol. Cell 64, 967–981. doi: 10.1016/j.molcel.2016.10.039
Jiang, L., Belinskaya, T., Zhang, Z., Chan, T.-C., Ching, W.-M., Chao, C.-C. (2020). Regulation of serum exosomal MicroRNAs in mice infected with Orientia tsutsugamushi. Microorganisms 9, 80. doi: 10.3390/microorganisms9010080
Jiang, L., Shao, C., Wu, Q.-J., Chen, G., Zhou, J., Yang, B., et al. (2017). NEAT1 scaffolds RNA-binding proteins and the microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol. 24, 816–824. doi: 10.1038/nsmb.3455
Jiang, M., Zhang, S., Yang, Z., Lin, H., Zhu, J., Liu, L., et al. (2018). Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell 173, 906–919.e13. doi: 10.1016/j.cell.2018.03.064
Ke, Z., Lu, J., Zhu, J., Yang, Z., Jin, Z., Yuan, L. (2020). Down-regulation of LincRNA-EPS regulates apoptosis and autophagy in BCG-infected RAW264.7 macrophages via JNK/MAPK signaling pathway. Infect. Genet. Evol. 77, 104077. doi: 10.1016/j.meegid.2019.104077
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D., et al. (2009). Many human Large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. 106, 11667–11672. doi: 10.1073/pnas.0904715106
Ko, Y., Choi, J.-H., Ha, N.-Y., Kim, I.-S., Cho, N.-H., Choi, M.-S. (2013). Active escape of Orientia tsutsugamushi from cellular autophagy. Infect. Immun. 81, 552–559. doi: 10.1128/IAI.00861-12
Kotzin, J. J., Spencer, S. P., McCright, S. J., Kumar, D. B. U., Collet, M. A., Mowel, W. K., et al. (2016). The long non-coding RNA morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 537, 239–243. doi: 10.1038/nature19346
Krawczyk, M., Emerson, B. M. (2014). P50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. Elife 3, e01776. doi: 10.7554/eLife.01776
Lai, F., Damle, S. S., Ling, K. K., Rigo, F. (2020). Directed RNase h cleavage of nascent transcripts causes transcription termination. Mol. Cell 77, 1032–1043.e4. doi: 10.1016/j.molcel.2019.12.029
Lan, F., Zhang, X., Li, H., Yue, X., Sun, Q. (2021). Serum exosomal lncRNA XIST is a potential non-invasive biomarker to diagnose recurrence of triple-negative breast cancer. J. Cell. Mol. Med. 25, 7602–7607. doi: 10.1111/jcmm.16009
Lee, J.-S., Mendell, J. T. (2020). Antisense-mediated transcript knockdown triggers premature transcription termination. Mol. Cell 77, 1044–1054.e3. doi: 10.1016/j.molcel.2019.12.011
Leng, H., Liu, S., Lei, Y., Tang, Y., Gu, S., Hu, J., et al. (2021). FACT interacts with Set3 HDAC and fine-tunes GAL1 transcription in response to environmental stimulation. Nucleic Acids Res. 49, 5502–5519. doi: 10.1093/nar/gkab312
Leucci, E., Vendramin, R., Spinazzi, M., Laurette, P., Fiers, M., Wouters, J., et al. (2016). Melanoma addiction to the long non-coding RNA SAMMSON. Nature 531, 518–522. doi: 10.1038/nature17161
Lewandowski, J. P., Lee, J. C., Hwang, T., Sunwoo, H., Goldstein, J. M., Groff, A. F., et al. (2019). The Firre locus produces a trans-acting RNA molecule that functions in hematopoiesis. Nat. Commun. 10, 5137. doi: 10.1038/s41467-019-12970-4
Li, Z., Chao, T.-C., Chang, K.-Y., Lin, N., Patil, V. S., Shimizu, C., et al. (2014). The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. 111, 1002–1007. doi: 10.1073/pnas.1313768111
Li, R., Fang, L., Pu, Q., Bu, H., Zhu, P., Chen, Z., et al. (2018). MEG3-4 is a miRNA decoy that regulates il-1β abundance to initiate and then limit inflammation to prevent sepsis during lung infection. Sci. Signal. 11, eaao2387. doi: 10.1126/scisignal.aao2387
Li, C., Han, H., Li, X., Wu, J., Li, X., Niu, H., et al. (2021a). Analysis of lncRNA, miRNA, and mRNA expression profiling in type I IFN and type II IFN overexpressed in porcine alveolar macrophages. Int. J. Genomics 2021, 6666160. doi: 10.1155/2021/6666160
Li, C., Ni, Y. Q., Xu, H., Xiang, Q. Y., Zhao, Y., Zhan, J. K., et al. (2021b). Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduction Targeting Ther. 6, 1–31. doi: 10.1038/s41392-021-00779-x
Li, W., Notani, D., Ma, Q., Tanasa, B., Nunez, E., Chen, A. Y., et al. (2013). Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520. doi: 10.1038/nature12210
Li, D., Wu, M. (2021). Pattern recognition receptors in health and diseases. Signal Transduction Targeting Ther. 6, 291. doi: 10.1038/s41392-021-00687-0
Liang, L., Ai, L., Qian, J., Fang, J. Y., Xu, J. (2015). Long noncoding RNA expression profiles in gut tissues constitute molecular signatures that reflect the types of microbes. Sci. Rep. 5, 1–8. doi: 10.1038/srep11763
Lin, H., Jiang, M., Liu, L., Yang, Z., Ma, Z., Liu, S., et al. (2019). The long noncoding RNA Lnczc3h7a promotes a TRIM25-mediated RIG-I antiviral innate immune response. Nat. Immunol. 20, 812–823. doi: 10.1038/s41590-019-0379-0
Liu, J., Ben, Q., Lu, E., He, X., Yang, X., Ma, J., et al. (2018). Long noncoding RNA PANDAR blocks CDKN1A gene transcription by competitive interaction with p53 protein in gastric cancer. Cell Death Dis. 9, 168. doi: 10.1038/s41419-017-0246-6
Liu, G., Mattick, J. S., Taft, R. J. (2013). A meta-analysis of the genomic and transcriptomic composition of complex life. Cell Cycle 12, 2061–2072. doi: 10.4161/cc.25134
Liu, J., Niu, Z., Zhang, R., Peng, Z., Wang, L., Liu, Z., et al. (2021). MALAT1 shuttled by extracellular vesicles promotes M1 polarization of macrophages to induce acute pancreatitis via miR-181a-5p/HMGB1 axis. J. Cell. Mol. Med. 25, 9241–9254. doi: 10.1111/jcmm.16844
Liu, L., Shuai, T., Li, B., Zhu, L., Li, X. (2018). Long Non−Coding RNA lnc−GNAT1−1 inhibits gastric cancer cell proliferation and invasion through the Wnt/β−Catenin pathway in Helicobacter pylori infection. Mol. Med. Rep. 18, 4009–4015. doi: 10.3892/mmr.2018.9405
Liu, R., Tang, A., Wang, X., Chen, X., Zhao, L., Xiao, Z., et al. (2018). Inhibition of LncRNA NEAT1 suppresses the inflammatory response in IBD by modulating the intestinal epithelial barrier and by exosome-mediated polarization of macrophages. Int. J. Mol. Med. 42, 2903–2913. doi: 10.3892/ijmm.2018.3829
Liu, M., Wang, Z., Ren, S., Zhao, H. (2021). Exosomes derived from Mycobacterium tuberculosis-infected MSCs induce a pro-inflammatory response of macrophages. Aging (Albany NY) 13, 11595–11609. doi: 10.18632/aging.202854
Liu, Y., Zhou, C., Su, Z., Chang, Q., Qiu, Y., Bei, J., et al. (2021). Endothelial exosome plays a functional role during rickettsial infection. MBio 12, e00769–21. doi: 10.1128/mBio.00769-21
Lu, Y., Liu, X., Xie, M., Liu, M., Ye, M., Li, M., et al. (2017). The NF-κB–responsive long noncoding RNA FIRRE regulates posttranscriptional regulation of inflammatory gene expression through interacting with hnRNPU. J. Immunol. 199, 3571–3582. doi: 10.4049/jimmunol.1700091
Lu, X., Xu, Q., Tong, Y., Zhang, Z., Dun, G., Feng, Y., et al. (2022). Long non-coding RNA EVADR induced by Fusobacterium nucleatum infection promotes colorectal cancer metastasis. Cell Rep. 40, 111127. doi: 10.1016/j.celrep.2022.111127
Lubelsky, Y., Ulitsky, I. (2018). Sequences enriched in alu repeats drive nuclear localization of long RNAs in human cells. Nature 555, 107–111. doi: 10.1038/nature25757
Lührmann, A., Roy, C. R. (2007). Coxiella burnetii Inhibits activation of host cell apoptosis through a mechanism that involves preventing cytochrome c release from mitochondria. Infect. Immun. 75, 5282–5289. doi: 10.1128/IAI.00863-07
Luo, F., Wen, Y., Zhao, L., Su, S., Lei, W., Chen, L., et al. (2022a). LncRNA ZEB1-AS1/miR-1224-5p/MAP4K4 axis regulates mitochondria-mediated hela cell apoptosis in persistent Chlamydia trachomatis infection. Virulence 13, 444–457. doi: 10.1080/21505594.2022.2044666
Luo, F., Wen, Y., Zhao, L., Su, S., Zhao, Y., Lei, W., et al. (2022b). Chlamydia trachomatis Induces lncRNA MIAT upregulation to regulate mitochondria-mediated host cell apoptosis and chlamydial development. J. Cell. Mol. Med. 26, 163–177. doi: 10.1111/jcmm.17069
Ma, L., Bajic, V. B., Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA Biol. 10, 924–933. doi: 10.4161/rna.24604
Ma, S., Ming, Z., Gong, A.-Y., Wang, Y., Chen, X., Hu, G., et al. (2017). A long noncoding RNA, lincrna-TNFAIP3, acts as a coregulator of NF-κB to modulate inflammatory gene transcription in mouse macrophages. FASEB J. 31, 1215–1225. doi: 10.1096/fj.201601056R
Maclary, E., Hinten, M., Harris, C., Sethuraman, S., Gayen, S., Kalantry, S. (2017). PRC2 represses transcribed genes on the imprinted inactive X chromosome in mice. Genome Biol. 18, 82. doi: 10.1186/s13059-017-1211-5
Mao, W., Zeng, Q., She, L., Yuan, H., Luo, Y., Wang, R., et al. (2022). Wolbachia utilizes lncRNAs to activate the anti-dengue toll pathway and balance reactive oxygen species stress in Aedes aegypti through a competitive endogenous RNA network. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.823403
Martens, J. A., Laprade, L., Winston, F. (2004). Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429, 571–574. doi: 10.1038/nature02538
Martianov, I., Ramadass, A., Serra Barros, A., Chow, N., Akoulitchev, A. (2007). Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445, 666–670. doi: 10.1038/nature05519
Melé, M., Mattioli, K., Mallard, W., Shechner, D. M., Gerhardinger, C., Rinn, J. L. (2017). Chromatin environment, transcriptional regulation, and splicing distinguish LincRNAs and mRNAs. Genome Res. 27, 27–37. doi: 10.1101/gr.214205.116
Melo, C. A., Drost, J., Wijchers, P. J., van de Werken, H., de Wit, E., Vrielink, J. A. F. O., et al. (2013). eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell 49, 524–535. doi: 10.1016/j.molcel.2012.11.021
Mosquera-Heredia, M. I., Morales, L. C., Vidal, O. M., Barceló, E., Silvera-Redondo, C., Vélez, J. I., et al. (2021). Exosomes: potential disease biomarkers and new therapeutic targets. Biomedicines 9, 1061. doi: 10.3390/biomedicines9081061
Nakayama, Y., Fujiu, K., Yuki, R., Oishi, Y., Morioka, M. S., Isagawa, T., et al. (2020). A long noncoding RNA regulates inflammation resolution by mouse macrophages through fatty acid oxidation activation. Proc. Natl. Acad. Sci. U. S. A. 117, 14365–14375. doi: 10.1073/pnas.2005924117
Noh, J. H., Kim, K. M., McClusky, W. G., Abdelmohsen, K., Gorospe, M. (2018). Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip. Rev. RNA 9, e1471–e1471. doi: 10.1002/wrna.1471
Novikov, A., Cardone, M., Thompson, R., Shenderov, K., Kirschman, K. D., Mayer-Barber, K. D., et al. (2011). Mycobacterium tuberculosis Triggers host type I IFN signaling to regulate il-1β production in human macrophages. J. Immunol. 187, 2540–2547. doi: 10.4049/jimmunol.1100926
Ohhata, T., Suzuki, M., Sakai, S., Ota, K., Yokota, H., Uchida, C., et al. (2022). CCIVR facilitates comprehensive identification of cis-natural antisense transcripts with their structural characteristics and expression profiles. Sci. Rep. 12, 15525. doi: 10.1038/s41598-022-19782-5
Omotade, T. O., Roy, C. R. (2020). Legionella pneumophila Excludes autophagy adaptors from the ubiquitin-labeled vacuole in which it resides. Infect. Immun. 88, e00793–e00719. doi: 10.1128/IAI.00793-19
Pant, A., Yao, X., Lavedrine, A., Viret, C., Dockterman, J., Chauhan, S., et al. (2022). Interactions of autophagy and the immune system in health and diseases. Autophagy Rep. 1, 438–515. doi: 10.1080/27694127.2022.2119743
Park, J., Kim, N.-E., Yoon, H., Shin, C. M., Kim, N., Lee, D. H., et al. (2021). Fecal microbiota and gut microbe-derived extracellular vesicles in colorectal cancer. Front. Oncol. 11. doi: 10.3389/fonc.2021.650026
Patel, R. S., Impreso, S., Lui, A., Vidyarthi, G., Albear, P., Patel, N. A. (2022). Long noncoding RNA GAS5 contained in exosomes derived from human adipose stem cells promotes repair and modulates inflammation in a chronic dermal wound healing model. Biol. (Basel). 11, 426. doi: 10.3390/biology11030426
Pawar, K., Hanisch, C., Palma Vera, S. E., Einspanier, R., Sharbati, S. (2016). Down regulated lncRNA MEG3 eliminates Mycobacteria in macrophages via autophagy. Sci. Rep. 6, 19416. doi: 10.1038/srep19416
Quinn, J. J., Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10
Rackham, O., Shearwood, A.-M. J., Mercer, T. R., Davies, S. M. K., Mattick, J. S., Filipovska, A. (2011). Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 17, 2085–2093. doi: 10.1261/rna.029405.111
Radomski, N., Karger, A., Franzke, K., Liebler-Tenorio, E., Jahnke, R., Matthiesen, S., et al. (2019). Chlamydia Psittaci-infected dendritic cells communicate with NK cells via exosomes to activate antibacterial immunity. Infect. Immun. 88, e00541–e00519. doi: 10.1128/IAI.00541-19
Rennoll-Bankert, K. E., Garcia-Garcia, J. C., Sinclair, S. H., Dumler, J. S. (2015). Chromatin-bound bacterial effector ankyrin a recruits histone deacetylase 1 and modifies host gene expression. Cell. Microbiol. 17, 1640–1652. doi: 10.1111/cmi.12461
Rikihisa, Y. (2011). Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin. Microbiol. Rev. 24, 469–489. doi: 10.1128/CMR.00064-10
Rinn, J. L., Kertesz, M., Wang, J. K., Squazzo, S. L., Xu, X., Brugmann, S. A., et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. doi: 10.1016/j.cell.2007.05.022
Robinson, E. K., Covarrubias, S., Carpenter, S. (2020). The how and why of LncRNA function: an innate immune perspective. Biochim. Biophys. Acta - Gene Regul. Mech. 1863, 194419. doi: 10.1016/j.bbagrm.2019.194419
Rosenberg, A. B., Patwardhan, R. P., Shendure, J., Seelig, G. (2015). Learning the sequence determinants of alternative splicing from millions of random sequences. Cell 163, 698–711. doi: 10.1016/j.cell.2015.09.054
Seth, P. P., Siwkowski, A., Allerson, C. R., Vasquez, G., Lee, S., Prakash, T. P., et al. (2008). Design, synthesis and evaluation of constrained methoxyethyl (cMOE) and constrained ethyl (cEt) nucleoside analogs. Nucleic Acids Symp. Ser. 52, 553–554. doi: 10.1093/nass/nrn280
Shukla, C. J., McCorkindale, A. L., Gerhardinger, C., Korthauer, K. D., Cabili, M. N., Shechner, D. M., et al. (2018). High-throughput identification of RNA nuclear enrichment sequences. EMBO J. 37, e98452. doi: 10.15252/embj.201798452
Statello, L., Guo, C. J., Chen, L. L., Huarte, M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118. doi: 10.1038/s41580-020-00315-9
St.Laurent, G., Wahlestedt, C., Kapranov, P. (2015). The landscape of long noncoding RNA classification. Trends Genet. 31, 239–251. doi: 10.1016/j.tig.2015.03.007
Sun, Z., Yang, S., Zhou, Q., Wang, G., Song, J., Li, Z., et al. (2018). Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol. Cancer 17, 82. doi: 10.1186/s12943-018-0831-z
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938. doi: 10.1016/j.molcel.2010.08.011
Uszczynska-Ratajczak, B., Lagarde, J., Frankish, A., Guigó, R., Johnson, R. (2018). Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 19, 535–548. doi: 10.1038/s41576-018-0017-y
van Attikum, H., Fritsch, O., Gasser, S. M. (2007). Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 26, 4113–4125. doi: 10.1038/sj.emboj.7601835
Walther, K., Schulte, L. N. (2021). The role of LncRNAs in innate immunity and inflammation. RNA Biol. 18, 587–603. doi: 10.1080/15476286.2020.1845505
Wang, K. C., Chang, H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914. doi: 10.1016/j.molcel.2011.08.018
Wang, X., Pei, X., Guo, G., Qian, X., Dou, D., Zhang, Z., et al. (2020). Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J. Cell. Physiol. 235, 6896–6904. doi: 10.1002/jcp.29585
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. doi: 10.1038/nature09819
Wang, Y., Zhong, H., Xie, X., Chen, C. Y., Huang, D., Shen, L., et al. (2015). Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc. Natl. Acad. Sci. 112, E3883–E3892. doi: 10.1073/pnas.1501662112
Warner, K. D., Hajdin, C. E., Weeks, K. M. (2018). Principles for targeting RNA with drug-like small molecules. Nat. Rev. Drug Discovery 17, 547–558. doi: 10.1038/nrd.2018.93
Wen, Y., Chen, H., Luo, F., Zhao, L., Shu, M., Su, S., et al. (2020a). Chlamydia trachomatis Plasmid protein pORF5 up-regulates ZFAS1 to promote host cell survival via MAPK/p38 pathway. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.593295
Wen, Y., Chen, H., Luo, F., Zhou, H., Li, Z. (2020b). Roles of long noncoding RNAs in bacterial infection. Life Sci. 263, 118579. doi: 10.1016/j.lfs.2020.118579
Wen, Y., Luo, F., Zhao, L., Su, S., Lei, W., Liu, Y., et al. (2021). Long non-coding RNA FGD5-AS1 induced by Chlamydia trachomatis infection inhibits apoptosis via wnt/β-catenin signaling pathway. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.701352
Wilusz, J. E., Freier, S. M., Spector, D. L. (2008). 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135, 919–932. doi: 10.1016/j.cell.2008.10.012
Wilusz, J. E., JnBaptiste, C. K., Lu, L. Y., Kuhn, C.-D., Joshua-Tor, L., Sharp, P. A. (2012). A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack Poly(A) tails. Genes Dev. 26, 2392–2407. doi: 10.1101/gad.204438.112
Xiang, J.-F., Yin, Q.-F., Chen, T., Zhang, Y., Zhang, X.-O., Wu, Z., et al. (2014). Human colorectal cancer-specific CCAT1-l LncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 24, 513–531. doi: 10.1038/cr.2014.35
Xue, Z., Zhang, Z., Liu, H., Li, W., Guo, X., Zhang, Z., et al. (2019). LincRNA-Cox2 regulates NLRP3 inflammasome and autophagy mediated neuroinflammation. Cell Death Differ. 26, 130–145. doi: 10.1038/s41418-018-0105-8
Yamazaki, T., Souquere, S., Chujo, T., Kobelke, S., Chong, Y. S., Fox, A. H., et al. (2018). Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038–1053.e7. doi: 10.1016/j.molcel.2018.05.019
Yang, L., Peng, X., Li, Y., Zhang, X., Ma, Y., Wu, C., et al. (2019). Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol. Cancer 18, 78. doi: 10.1186/s12943-019-0990-6
Yao, S., Huang, D., Chen, C. Y., Halliday, L., Wang, R. C., Chen, Z. W. (2014). CD4+T cells contain early extrapulmonary tuberculosis (TB) dissemination and rapid TB progression and sustain multieffector functions of CD8+T and CD3– lymphocytes: mechanisms of CD4+T cell immunity. J. Immunol. 192, 2120–2132. doi: 10.4049/jimmunol.1301373
Yao, Y., Jiang, Q., Jiang, L., Wu, J., Zhang, Q., Wang, J., et al. (2016). Lnc-SGK1 induced by helicobacter pylori infection and highsalt diet promote TH2 and TH17 differentiation in human gastric cancer by SGK1/Jun b signaling. Oncotarget 7, 20549–20560. doi: 10.18632/oncotarget.7823
Ye, M., Xie, M., Zhu, J., Wang, C., Zhou, R., Li, X. (2020). LPS-inducible lncRNA TMC3-AS1 negatively regulates the expression of IL-10. Front. Immunol. 11. doi: 10.3389/fimmu.2020.01418
Yoshinaga, M., Takeuchi, O. (2019). Post-transcriptional control of immune responses and its potential application. Clin. Transl. Immunol. 8, e1063–e1063. doi: 10.1002/cti2.1063
Zhang, S., Li, C., Liu, J., Geng, F., Shi, X., Li, Q., et al. (2020). Fusobacterium nucleatum Promotes epithelial-mesenchymal transiton through regulation of the lncrna MIR4435-2HG/miR-296-5P/Akt2/SNAI1 signaling pathway. FEBS J. 287, 4032–4047. doi: 10.1111/febs.15233
Zhao, Y., Sun, L., Wang, R. R., Hu, J.-F., Cui, J. (2018). The effects of mitochondria-associated long noncoding RNAs in cancer mitochondria: new players in an old arena. Crit. Rev. Oncol. Hematol. 131, 76–82. doi: 10.1016/j.critrevonc.2018.08.005
Zheng, H., Talukder, A., Li, X., Hu, H. (2021). A systematic evaluation of the computational tools for LncRNA identification. Brief. Bioinform. 22, bbab285. doi: 10.1093/bib/bbab285
Zhou, Y., Li, M., Xue, Y., Li, Z., Wen, W., Liu, X., et al. (2019). Interferon-inducible cytoplasmic lncLrrc55-AS promotes antiviral innate responses by strengthening IRF3 phosphorylation. Cell Res. 29, 641–654. doi: 10.1038/s41422-019-0193-0
Zhou, W., Tahir, F., Wang, J. C. Y., Woodson, M., Sherman, M. B., Karim, S., et al. (2020). Discovery of exosomes from tick saliva and salivary glands reveals therapeutic roles for CXCL12 and IL-8 in wound healing at the tick–human skin interface. Front. Cell Dev. Biol. 8. doi: 10.3389/fcell.2020.00554
Zong, X., Nakagawa, S., Freier, S. M., Fei, J., Ha, T., Prasanth, S. G., et al. (2016). Natural antisense RNA promotes 3’ end processing and maturation of MALAT1 lncRNA. Nucleic Acids Res. 44, 2898–2908. doi: 10.1093/nar/gkw047
Keywords: long non-coding RNAs (lncRNAs), host-pathogen interaction, obligate intracellular pathogens, microbiota, inflammation, phagocytes, biomarker
Citation: Arunima A, van Schaik EJ and Samuel JE (2023) The emerging roles of long non-coding RNA in host immune response and intracellular bacterial infections. Front. Cell. Infect. Microbiol. 13:1160198. doi: 10.3389/fcimb.2023.1160198
Received: 06 February 2023; Accepted: 07 April 2023;
Published: 21 April 2023.
Edited by:
Jianfeng Wang, Jilin University, ChinaReviewed by:
Duc-Cuong Bui, University of Texas Medical Branch at Galveston, United StatesLi Xing, Shanxi University, China
Copyright © 2023 Arunima, van Schaik and Samuel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James E. Samuel, anNhbXVlbEB0YW11LmVkdQ==
 Aryashree Arunima
Aryashree Arunima Erin J. van Schaik
Erin J. van Schaik James E. Samuel
James E. Samuel