
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 30 May 2023
Sec. Bacteria and Host
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1158888
This article is part of the Research TopicThe role of DAMPs in infectionView all 4 articles
Salmonella Enteritidis is an important intracellular pathogen, which can cause gastroenteritis in humans and animals and threaten life and health. S. Enteritidis proliferates in host macrophages to establish systemic infection. In this study, we evaluated the effects of Salmonella pathogenicity island-1 (SPI-1) and SPI-2 to S. Enteritidis virulence in vitro and in vivo, as well as the host inflammatory pathways affected by SPI-1 and SPI-2. Our results show that S. Enteritidis SPI-1 and SPI-2 contributed to bacterial invasion and proliferation in RAW264.7 macrophages, and induced cytotoxicity and cellular apoptosis of these cells. S. Enteritidis infection induced multiple inflammatory responses, including mitogen-activated protein kinase (ERK-mediated) and Janus kinase-signal transducer and activator of transcript (STAT) (STAT2-mediated) pathways. Both SPI-1 and SPI-2 were necessary to induce robust inflammatory responses and ERK/STAT2 phosphorylation in macrophages. In a mouse infection model, both SPIs, especially SPI-2, resulted in significant production of inflammatory cytokines and various interferon-stimulated genes in the liver and spleen. Activation of the ERK- and STAT2-mediated cytokine storm was largely affected by SPI-2. S. Enteritidis ΔSPI-1-infected mice displayed moderate histopathological damage and drastically reduced bacterial loads in tissues, whereas only slight damage and no bacteria were observed in ΔSPI-2- and ΔSPI-1/SPI-2-infected mice. A survival assay showed that ΔSPI-1 mutant mice maintained a medium level of virulence, while SPI-2 plays a decisive role in bacterial virulence. Collectively, our findings indicate that both SPIs, especially SPI-2, profoundly contributed to S. Enteritidis intracellular localization and virulence by activating multiple inflammatory pathways.
Salmonella enterica is a Gram-negative and facultative intracellular pathogen that can lead to various diseases, such as gastroenteritis and even lethal systemic infections, in human and animal hosts (Kurtz et al., 2017; Grenz et al., 2018). Salmonellosis is usually caused by contaminated food and water, and is the most common foodborne disease in humans, especially in developing countries with substandard water supplies and poor sanitation (Parry et al., 2002; Wain et al., 2015). In March 2022, a multi-country outbreak of Salmonella was linked to chocolate products. In total, 151 genetically related cases of Salmonella suspected to be associated with the consumption of the implicated chocolate products were reported in 11 countries (WHO, 2022). The Salmonella genus contains six subspecies and more than 2600 serovars, most of which are pathogenic to humans and animals (LeLièvre et al., 2019). Two major salmonellosis-causing pathogens, S. Typhimurium and S. Enteritidis, are responsible for more than 1 million infections in the U.S. every year (CDC, 2023).
Salmonella pathogenicity islands (SPIs) are large regions of bacterial DNA encoding Salmonella virulence factors. The type III secretion systems (T3SSs) encoded by SPI-1 and SPI-2 are complicated machineries that play crucial roles for Salmonella pathogenesis (Que et al., 2013). Salmonella SPIs allow the transportation of effector proteins into the host cytoplasm, playing particularly important functions during Salmonella infection (Srikanth et al., 2011; Jennings et al., 2017). The SPI-1 gene cluster is a 40-kb region, and its encoded 39 genes include T3SS-1 and effector proteins, as well as the transcription regulatory factors that modulate the expression of numerous virulence genes inside and outside SPI-1 (Hansen-Wester and Hensel, 2001; Zhang et al., 2018). Comparative genomic analysis showed that the SPI-2 T3SS effector repertoire was very different among different Salmonella serovars, with subtle variations infrequently occurring between strains of the same serovar (Nuccio and Bäumler, 2014).
Previous studies of individual Salmonella effectors revealed a series of complex and diverse new biochemical activities. These effectors frequently display exquisite specificity and potency in subverting, avoiding, or reprogramming host cellular processes (Keszei et al., 2014; Rolhion et al., 2016; Paul et al., 2022). Different effector proteins of SPI-1 and SPI-2 could stimulate or inhibit innate immune responses (Jennings et al., 2017). Macrophages usually represent the first line of host defenses against bacterial pathogens and are vital niches in the pathogenic process of Salmonella (Jiang et al., 2021). Cytokines play an important role in host-pathogen intervention by inducing the release of either pro- or anti-inflammatory signaling molecules to regulate immune responses (Paul et al., 2022). However, a prolonged or excessive cytokine storm underlies many chronic inflammatory diseases and can be lethal to the host (O'Neill et al., 2009; Kawai and Akira, 2010; Brandes et al., 2013).
Previous studies investigating the transmission and colonization of Salmonella have focused on S. Typhimurium (Lee et al., 2005; Rana and Kulshreshtha, 2006; Jones et al., 2007). Molecular analysis revealed how S. Typhimurium manipulates host cell signaling through effector proteins. Recent studies show that S. Enteritidis is the most prevalent serovar among Salmonella isolates from children with diarrhea (Zhao et al., 2022). However, the contribution and related signaling pathways of SPI-1 and SPI-2 in inflammation caused by S. Enteritidis during the course of infection remain unclear. Previous studies investigated the functions of SPI-1 and SPI-2 mainly through use of genetic mutant strains; for example, ssaRmut and invAmut representing mutations of SPI-1 and SPI-2, respectively (Sekirov et al., 2010). However, the effects of S. Enteritidis in the context of complete SPI-1 and SPI-2 deficiency have rarely been studied. Moreover, although SPI participates in Salmonella virulence, whether it is directly involved in activation of other signaling pathways, such as Janus kinase-signal transducer and activator of transcript (JAK-STAT), remains to be understood.
In this study, to evaluate the contribution of S. Enteritidis SPIs on the inflammatory pathways, bacterial colonization and virulence in systemic infection, we evaluated the effects of S. Enteritidis SPI-1 and SPI-2 to bacterial invasion of the host cellular cytosol, intracellular survival, and induction of cellular apoptosis. The host inflammatory pathways and bacterial virulence affected by SPI-1 and SPI-2 were also determined in macrophages and in a mouse infection model. This study will improve understanding of the molecular mechanisms employed by different Salmonella SPIs to promote the bacterial survival during infection of the host.
Specific pathogen-free C57BL/6J mice (female, 6–8 weeks) were purchased from Beijing Vital River Laboratory Animal Technology (Beijing, China). Mice were raised in an environment with a light/dark cycle of 12 h, a constant temperature of 25°C and a humidity of 50%. A standard chow diet was provided for mice ad libitum throughout the experiments. All animal studies were conducted according to protocols approved by the Committee on the Ethics of Animal Experiments of Yangzhou University [Approval ID: SYXK (Su) 2017-0044].
Bacterial strains and plasmids used in the present study are shown in Supplementary Table 1. The S. Enteritidis strain CMCC50041 (GenBank Accession No. CP013097.1) was used as the wild-type (WT) strain and for the construction of SPI-1 and SPI-2 mutants. SPI-1 and SPI-2 mutants of S. Enteritidis were previously constructed (Xiong et al., 2019). The ΔSPI-1/SPI-2 double-deficient strain in this study was constructed following the λ-Red recombinase gene replacement method (Datsenko and Wanner, 2000; Jiang et al., 2017). Primer sequences to generate and confirm the mutant strains are listed in Supplementary Table 2. All bacterial strains were cultured on Luria-Bertani (LB) agar plates or in LB broth with necessary antibiotics at appropriate concentrations (e.g., 100 μg/mL ampicillin and 20 μg/mL chloramphenicol).
The murine RAW264.7 macrophage cell line was provided by the Shanghai Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Gibco, Waltham, MA, USA) at 37°C in a 5% CO2 incubator.
The λ-Red system, an effective and widely used approach to inactivate the chromosomal genes in Escherichia coli and S. Typhimurium (Tischer et al., 2006), was used to produce SPI-1 and SPI-2 deficient mutants. ΔSPI-1/SPI-2 double mutant strain was constructed using ΔSPI-2 forward and reverse primers based on the ΔSPI-1 deficient strain. All gene deletions were verified by PCR analysis and sequencing. All primers used for amplification of certain regions in SPI-1 and SPI-2 of S. Enteritidis are shown in Supplementary Table 2. Growth curves of S. Enteritidis C50041 WT, ΔSPI-1, ΔSPI-2, and ΔSPI-1/SPI-2 deletion mutants were determined to evaluate the influence of SPI deficiency on Salmonella growth. Biochemical tests were performed using an API 20E identification kit (bioMerieux SA, Lyon, France) in accordance with the manufacturer’s protocol.
S. Enteritidis colonies were cultured overnight at 37°C with agitation (180 rpm) in LB liquid broth. Log-phase bacteria were obtained by diluting the stationary-phase bacteria to fresh LB at a ratio of 1: 40 and subculturing to the mid-log-phase (3 h). Bacteria were pelleted, washed three times with cold phosphate-buffered saline (PBS), and resuspended with DMEM. The bacterial concentration was estimated based on the optical density at 600 nm, and the CFUs were calculated by serial dilution on agar plates. Before infection, RAW264.7 macrophages were seeded in 24‐well plates at a density of 2 × 105 cells/well overnight in DMEM containing 10% FBS without antibiotics to obtain 80% confluence. Next, Salmonella was added at a multiplicity of infection of 100:1. Plates were centrifuged at 1,000 × g for 8 min to synchronize infection and allowed to invade for 1 h. After washing cells twice with DPBS, DMEM supplemented with 100 μg/mL gentamicin was used to kill the extracellular bacteria for 1 h.
For invasion assays, RAW264.7 cells continued to be infected for 0.5 h, washed twice with DPBS, and lysed with 1% Triton X-100. Subsequently CFU numbers of intracellular bacteria were calculated to determine the bacterial invasion by plating on LB agar. For the proliferation assay, cells continued to be infected for 0.5 h, washed twice with DPBS, and continued to be maintained in culture medium containing 10 μg/mL gentamicin for 2, 4, 6, or 8 h to quantify bacterial proliferation.
Murine RAW264.7 macrophages were seeded in 24-well plates at a density of 2 × 105 cells per well and infected with Salmonella WT and mutants as described above. Cells cultured in DMEM were set as the negative control group, which was not infected with bacteria. After Salmonella infection for 2 h, culture supernatants were harvested for the quantification of lactate dehydrogenase (LDH) with an LDH Cytotoxicity Assay Kit (Beyotime, Shanghai, China). The absorbance value of the DMEM control group was subtracted from all treatment groups according to the manufacturer’s instructions. In addition, cells were collected and centrifuged at 1,000 × g for 10 min and resuspended in 400 μL of binding buffer. Early and late apoptosis among different groups were conducted using an Annexin V-FITC/PI Apoptosis Detection Kit (Vazyme, Nanjing, China). Percentages of apoptotic cells were analyzed using a FACS Aria flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) with FlowJo_v10 software (FowJo, Ashland, OR, USA).
Female C57BL/6J mice were randomly divided into 5 groups (n = 13 per group, n = 5 for tissue collection and n = 8 for survival monitoring). For oral infections, water and food were withdrawn for 4 h before C57BL/6J mice were infected intragastrically with 106 CFU of S. Enteritidis C50041 WT or the indicated ΔSPI-1, ΔSPI-2, or ΔSPI-1/SPI-2-deficient strains in a 200-μL volume. Bacteria were prepared and CFU numbers were determined as described above. Drinking water and food were offered at 2 h post infection. To evaluate the cytokine levels in livers and spleens of mice, tissues were homogenized using a Precellys 24 homogenizer (Rockville, MD, USA). Cytokine expression levels in organs were measured using qRT-PCR. To evaluate the activation of MAPK and JAK-STAT signaling pathways in the tissues, homogenized specimens were centrifuged for 10 min at 12,000 × g at 4°C. p-ERK and p-STAT2 levels in collected supernatants were measured by Western blotting. For analysis of the bacterial burden in tissues, infected mice were sacrificed 4 days post infection. Next, mouse cecums, livers and spleens were aseptically collected, homogenized in sterile ice-cold PBS, serially ten‐fold diluted, and plated on LB plates for CFU determination. Before homogenization, the cecums were aseptically removed and rinsed three times with sterile PBS. Meanwhile, tissue samples from the spleen and liver were removed for pathological analysis. The mortality of mice in different infection groups was recorded daily and monitored for 2 weeks.
Cells were collected and lysed in Cell Lysis Buffer for Western (Beyotime) followed by centrifugation at 12,000 × g at 4°C for 5 min. The supernatant was harvested and the protein concentration was determined by BCA Protein Assay Kit (Beyotime). Protein samples were subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and the polyvinylidene fluoride membrane was used for protein transfer. The membrane was blocked with 1% bovine serum albumin (BSA) in Tris-buffered saline containing 0.05% Tween 20 (TBST) for 2 h at room temperature, followed by primary antibodies and horseradish peroxidase (HRP)-conjugated IgG secondary antibodies (1:5000 dilution). Antibodies used in this study were listed below: rabbit anti-p-STAT2 (Y690) (Abcam, Cambridge, UK), rabbit monoclonal anti-p-ERK1&2 (Thr202/Tyr204) (Cell Signaling Technology, Danvers, MA, USA), mouse monoclonal anti-β-actin (C4) (Santa Cruz Biotechnology, Dallas, TX, USA), HRP-conjugated goat anti-rabbit IgG H&L (Abcam), and goat anti-mouse IgG H&L-HRP (Calbiochem, San Diego, CA, USA). The immunoblotting was conducted with the SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, Waltham, MA, USA). Images were captured with the Amersham™ Imager 600 System (GE Healthcare, Chicago, IL, USA). Band densities were analyzed using ImageJ software (https://imagej.nih.gov/ij/) and normalized to the corresponding internal marker β-actin.
Total mRNA preparations of RAW264.7 macrophage and mouse tissues were extracted with a Total RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). For preparation of tissue mRNA, mouse livers and spleens were harvested and stored using liquid nitrogen. The frozen tissues were homogenized in RLT buffer before extraction. RNA concentrations were measured using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). cDNA was synthesized using a PrimeScript RT Reagent Kit (Takara, Dalian, China). qRT-PCR was performed with a QuantStudio 6 Real-Time PCR System (Applied Biosystems, Foster, CA, USA). The total volume of PCR reaction was 20 μL, and it was conducted in triplicate using the Fast Start Universal SYBR Green Master (Roche, Basel, Switzerland) according to the manufacturer’s protocols. The real-time PCR efficiencies (E) for the reference gene and target genes were determined using the diluted cDNA templates, and calculated based on the equation: E = 10[–1/slope] (Rasmussen, 2001). Normalized levels of gene expression were calculated following the Pfaffl method based on E and the CT deviation of the treatment sample versus the control, and expressed in comparison to the reference gene (Pfaffl, 2001). β-actin gene was used as the reference control. Primer sequences for the qRT-PCR are shown in Supplementary Table 3.
Mouse livers and spleens were collected, formalin-fixed, and embedded in paraffin. The sections were deparaffinized, antigen-retrieved with sodium citrate buffer, permeabilized in Triton X-100 (0.2%) for 0.5 h, and then blocked with PBS containing 5% BSA for 1 h. Mouse p-ERK1&2 and p-STAT2 were stained using rabbit monoclonal anti-p-ERK1&2 and rabbit anti-p-STAT2 (Y690) antibodies with the blocking buffer overnight at 4°C. Cy3-conjugated goat anti-rabbit secondary antibody (Abcam) was diluted in blocking buffer and incubated for 1 h at room temperature according to the manufacturer’s instructions. The nuclear DNA was stained with 6-Diamidino-2-phenylindole (DAPI). The sections were mounted with the coverslips and Prolong Diamond Antifade Mountant (Thermo Fisher Scientific). Images were captured using the fluorescence microscope (Leica, Wetzlar, Germany).
Mouse tissues were fixed with 10% neutral formalin for 24 h, then embedded with paraffin, cut into sections (5 μm), and stained using hematoxylin and eosin (H&E) by standard techniques. Histology sections were observed using an Eclipse Ci-L microscope (Nikon, Tokyo, Japan). Pathological assessment of livers and spleens were performed by two pathologists in a blinded manner using the criteria described in Supplementary Table 4. The sum of the individual scores for each tissue sample was determined as the combined pathological score.
Data were comparatively analyzed with the unpaired Student’s t test and were shown as mean ± SEM. All statistical analysis was performed with GraphPad Prism (version 8.0.2, GraphPad Software, San Diego, CA, USA). The significant differences were determined when the P-value was ≤ 0.05 (*), 0.01 (**), and 0.001 (***).
Salmonella ΔSPI-1, ΔSPI-2, and ΔSPI-1/SPI-2-deficient strains were constructed to identify roles for each SPI in bacterial virulence of macrophages and a mouse model. Using the λ-Red system, SPI-1 (39 kb) and SPI-2 (40 kb) were deleted to generate ΔSPI-1 and ΔSPI-2, respectively, while both gene cassettes were deleted to generate the ΔSPI-1/SPI-2-deficient strain (Figure 1A). Each strain was confirmed by PCR analysis and sequencing, which showed that mutants were constructed correctly (Figure 1B). Our assessment of growth rates confirmed that bacterial growth was unaffected by deletion of SPI-1 or SPI-2 (Figure 1C). In addition, mutant strains were biochemically characterized using an API 20E identification kit, which revealed no differences between mutant strains relative to the WT strain (Figure 1D).
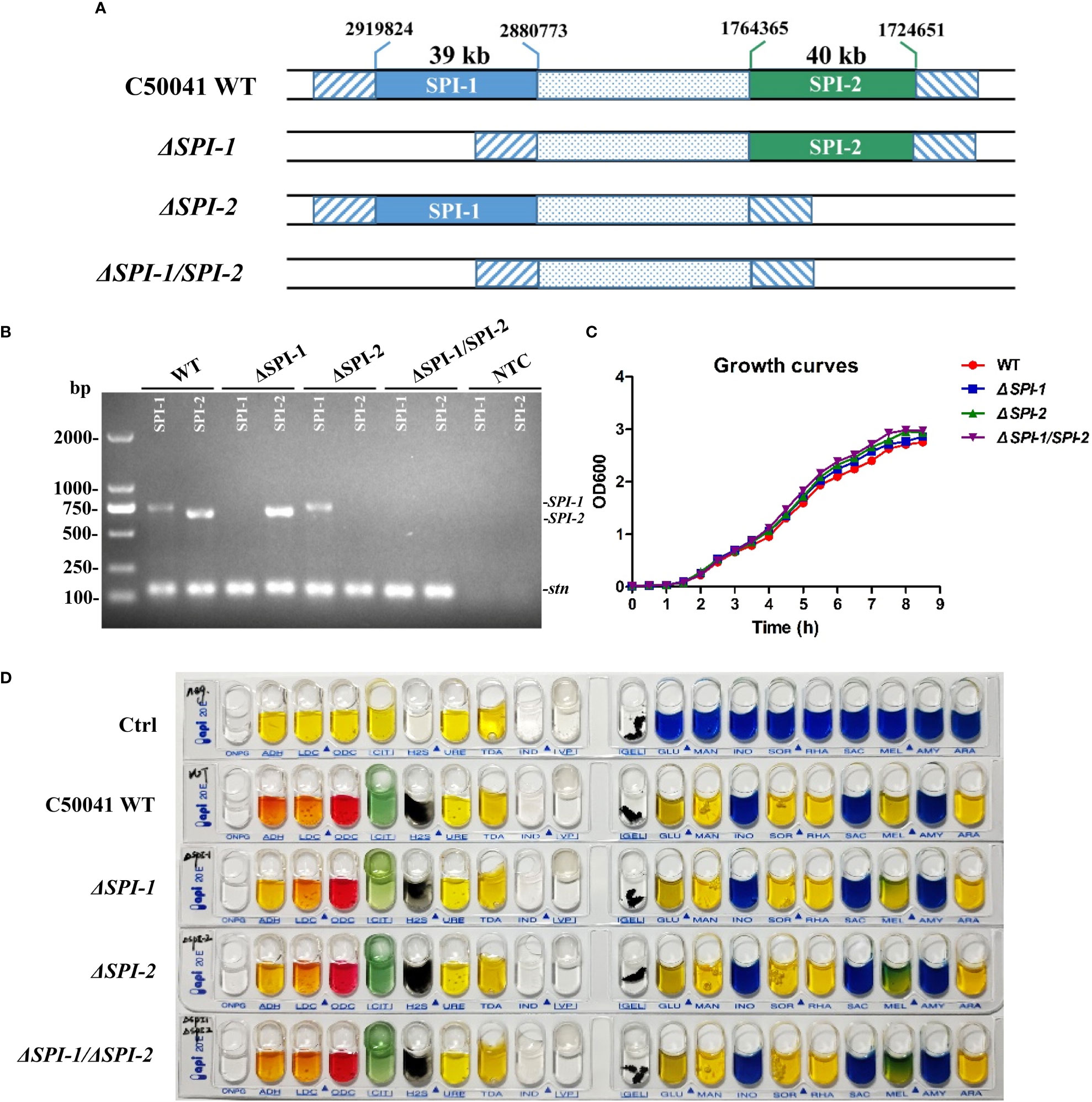
Figure 1 Construction of S. Enteritidis ΔSPI-1, ΔSPI-2 and ΔSPI-1/SPI-2 deficient mutants and biochemical identification of the deficient strains. (A) Schematic representation of the construction for S. Enteritidis ΔSPI-1, ΔSPI-2 and ΔSPI-1/SPI-2 mutants. The Salmonella deficient mutants were produced by using the λ-Red recombinase gene replacement method. (B) All gene deletions were verified by PCR analysis and by sequencing. The stn gene was used as the reference control. (C) Growth curves of S. Enteritidis C50041 wild-type, ΔSPI-1, ΔSPI-2 and ΔSPI-1/SPI-2 deletion strains. Bacteria were cultured in LB medium at 37°C with 180 rpm, and the OD600 values of bacterial cultures were determined in 0.5 h intervals. (D) Biochemical tests of S. Enteritidis WT, ΔSPI-1, ΔSPI-2 and ΔSPI-1/SPI-2 strains using the API 20E identification kit. Sterile water was used as control.
To study the effects of SPI-1 and SPI-2 on bacterial invasion and proliferation in vitro, the RAW264.7 macrophage cells were infected with C50041 WT, ΔSPI-1, ΔSPI-2, or ΔSPI-1/SPI-2 strains. Mutants showed significantly decreased abilities to invade macrophages relative to WT (Figure 2A). In addition, intracellular proliferation of WT S. Enteritidis was substantially increased 2 h following infection compared with the three mutants (Figure 2B). Compared with WT S. Enteritidis, LDH release caused by mutant strains in macrophages was significantly decreased (Figure 2C). In addition, both SPIs independently resulted in the apoptosis of murine macrophages following infection (Figure 2D).
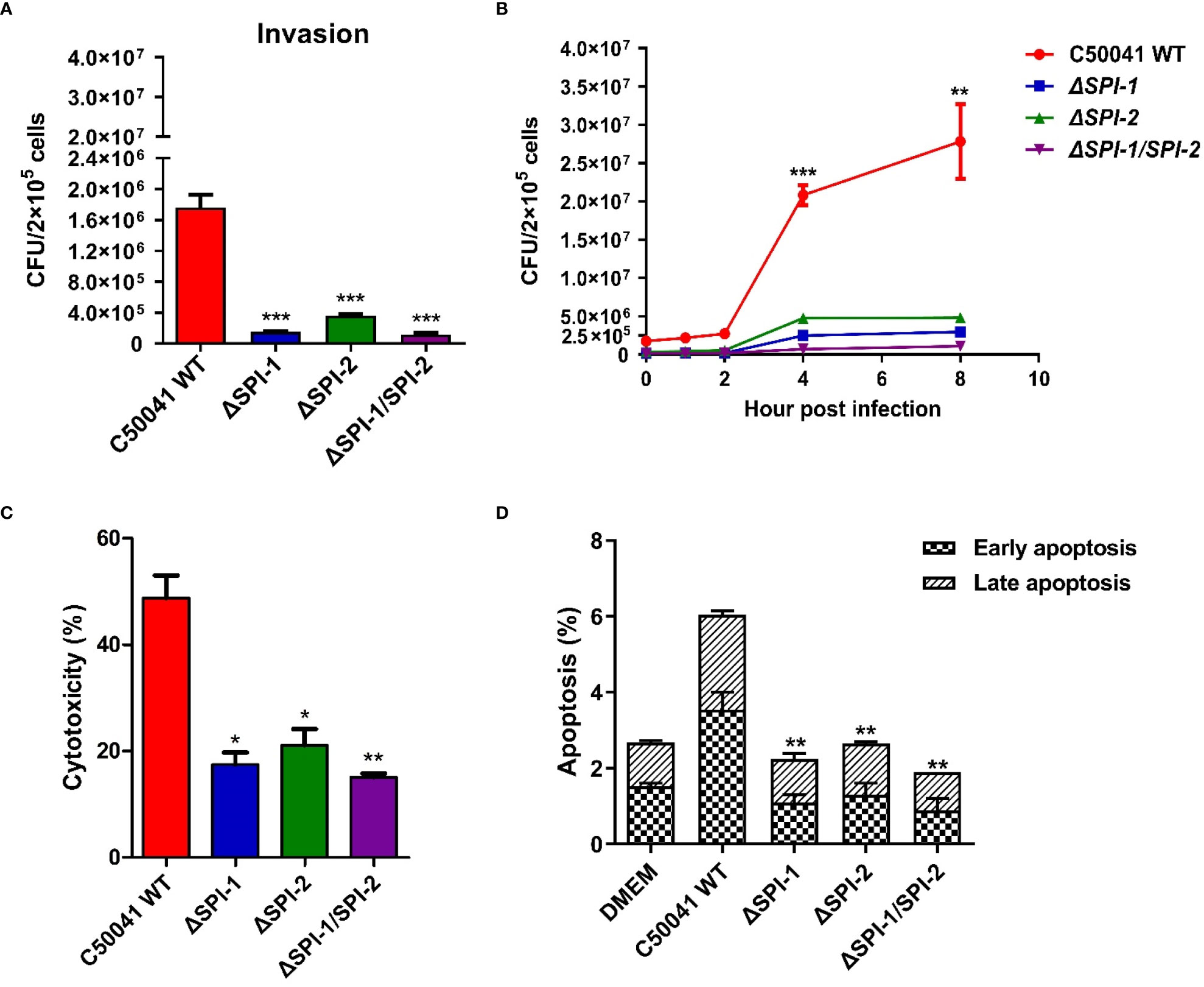
Figure 2 Intracellular localization and virulence evaluation of S. Enteritidis ΔSPI-1, ΔSPI-2 and ΔSPI-1/SPI-2 deficient strains in macrophages. (A) The invasion was determined in RAW264.7 following the infection of S. Enteritidis WT, ΔSPI-1, ΔSPI-2 and ΔSPI-1/SPI-2 deficient strains. (B) The proliferation was determined at different time points in RAW264.7 macrophages following the infection of S. Enteritidis WT, ΔSPI-1, ΔSPI-2 and ΔSPI-1/SPI-2 deficient strains. (C) After Salmonella infection for 2 h, the supernatants were harvested for the quantification of LDH levels. LDH levels were determined with the LDH Cytotoxicity Assay Kit. (D) Apoptosis analysis with the Annexin V-FITC/PI staining and flow cytometry. The cell apoptosis among different groups was determined with the Annexin V-FITC/PI Apoptosis Detection Kit. P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***) were considered statistically significant.
The effects of SPI-1 and SPI-2 on activation of inflammatory pathways were evaluated in murine macrophages following Salmonella infection. The real-time PCR efficiencies for the reference gene and target genes were 2.04 for β-actin, 2.07 for IL-1β, 2.02 for IL-6, 2.15 for IFN-β, 2.08 for ISG15, 2.16 for IFIT1, 1.96 for IFIT2, 1.98 for IFIT3b and 2.05 for CXCL2 (Supplementary Figure 1). The results show that SPI-1 and SPI-2 of S. Enteritidis induced multiple inflammatory responses, including MAPK (ERK-mediated) and JAK-STAT (STAT2-mediated) pathways (Figures 3A, B). Moreover, S. Enteritidis SPI-1 and SPI-2 contributed to robust production of inflammatory cytokines such as interleukin (IL)-1β and IL-6, and interferon-stimulated genes (ISGs) such as IFN-β, ISG15, IFIT1, and IFIT3b (Figure 3C). Inflammatory responses 3 h after infection were stronger compared with those at 1 h. Collectively, these results demonstrate that both SPI-1 and SPI-2 of S. Enteritidis are necessary to induce inflammation in macrophages.
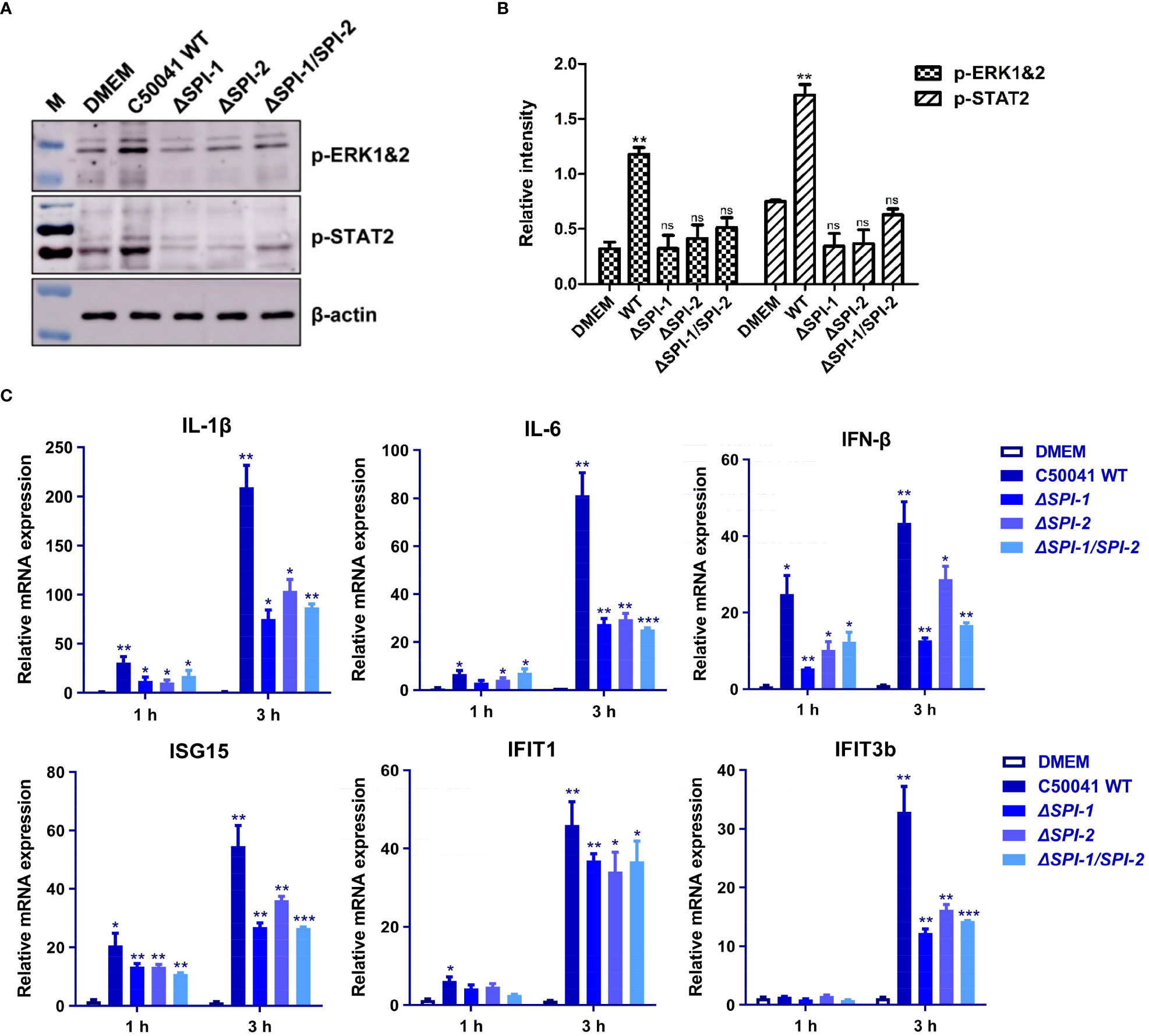
Figure 3 The SPI-1 and SPI-2 promoted S. Enteritidis to trigger inflammation storm via two different inflammatory pathways in macrophages. (A) Salmonella SPI-1 and SPI-2 induced the robust activation of MAPK and STAT signaling pathways. Macrophages were infected with S. Enteritidis WT, ΔSPI-1, ΔSPI-2 and ΔSPI-1/SPI-2 deficient strains, and the phosphorylation of ERK and STAT2 was determined. (B) Relative intensity of p-ERK and p-STAT2 were semi-quantified using ImageJ and presented as bar graphs. (C) Salmonella SPI-1 and SPI-2 contributed to significant increase of inflammatory cytokines and ISGs. The RAW264.7 macrophage was infected with S. Enteritidis WT, ΔSPI-1, ΔSPI-2 and ΔSPI-1/SPI-2 deficient strains, and total mRNA was extracted. The expression levels of inflammatory cytokines and various ISGs were determined by using qRT-PCR. Cells cultured in DMEM alone were served as the control group. P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***) were considered statistically significant. ns, not significant.
To evaluate the contribution of SPI-1 and SPI-2 to the virulence of S. Enteritidis in vivo, immune responses following Salmonella infection were determined in a mouse model. The results show that deficiency of the two SPIs, especially SPI-2, significantly abolished the production of inflammatory cytokines and various ISGs in liver and spleen (Figures 4A, B). In addition, infection with WT Salmonella resulted in robust phosphorylation of ERK and STAT2. However, the SPI-1 mutant displayed reduced p-ERK and p-STAT2 levels. Notably, ΔSPI-2 and ΔSPI-1/SPI-2-deficient strains completely abolished the activation of the two inflammatory pathways (Figures 4C, D).
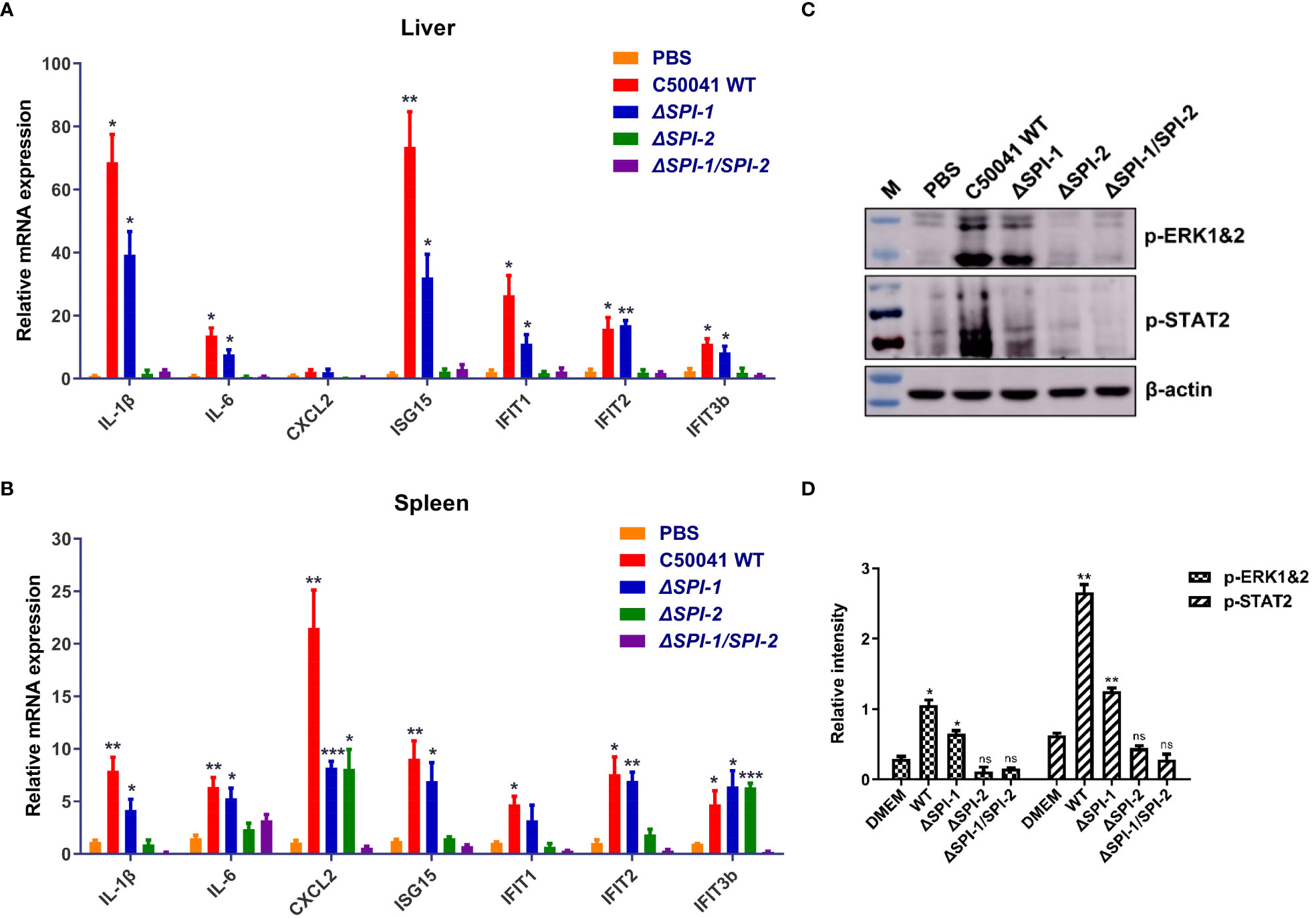
Figure 4 The Salmonella SPI-1 and SPI-2 resulted in the cytokine storm in livers and spleens following infection in mice. Inflammatory cytokines and ISGs were determined in livers (A) and spleens (B) of mice using qRT-PCR. On 4 day following infection, the tissues were harvested and homogenized in RLT buffer for the mRNA extraction. The cytokine expression levels in the organs were measured using qRT-PCR. (C) Western blotting analysis of the phosphorylation of ERK- and STAT2-mediated inflammatory pathways in the spleen following Salmonella infection. On 4 day following infection, the spleens were homogenized and centrifuged to harvest the supernatant for the immunoblotting analysis. (D) Relative intensity of p-ERK and p-STAT2 were semi-quantified using ImageJ and presented as bar graphs. P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***) were considered statistically significant. ns, not significant.
To further characterize the evoked immune response, we performed immunofluorescence assays to examine activation of inflammation-related signaling pathways in the liver and spleen of mice following Salmonella infection. Expression of p-ERK1&2 and p-STAT2 was evaluated as markers of the activation of MAPK and STAT signaling pathways. The results show that WT S. Enteritidis induced the most pronounced activation of p-ERK1&2 and p-STAT2 in liver and spleen tissues of infected mice, while the SPI-1 mutant induced moderate activation of inflammatory signaling. However, neither ΔSPI-2- or ΔSPI-1/SPI-2-infected mice displayed obvious inflammation activation, showing similar results to the PBS control group (Figure 5).
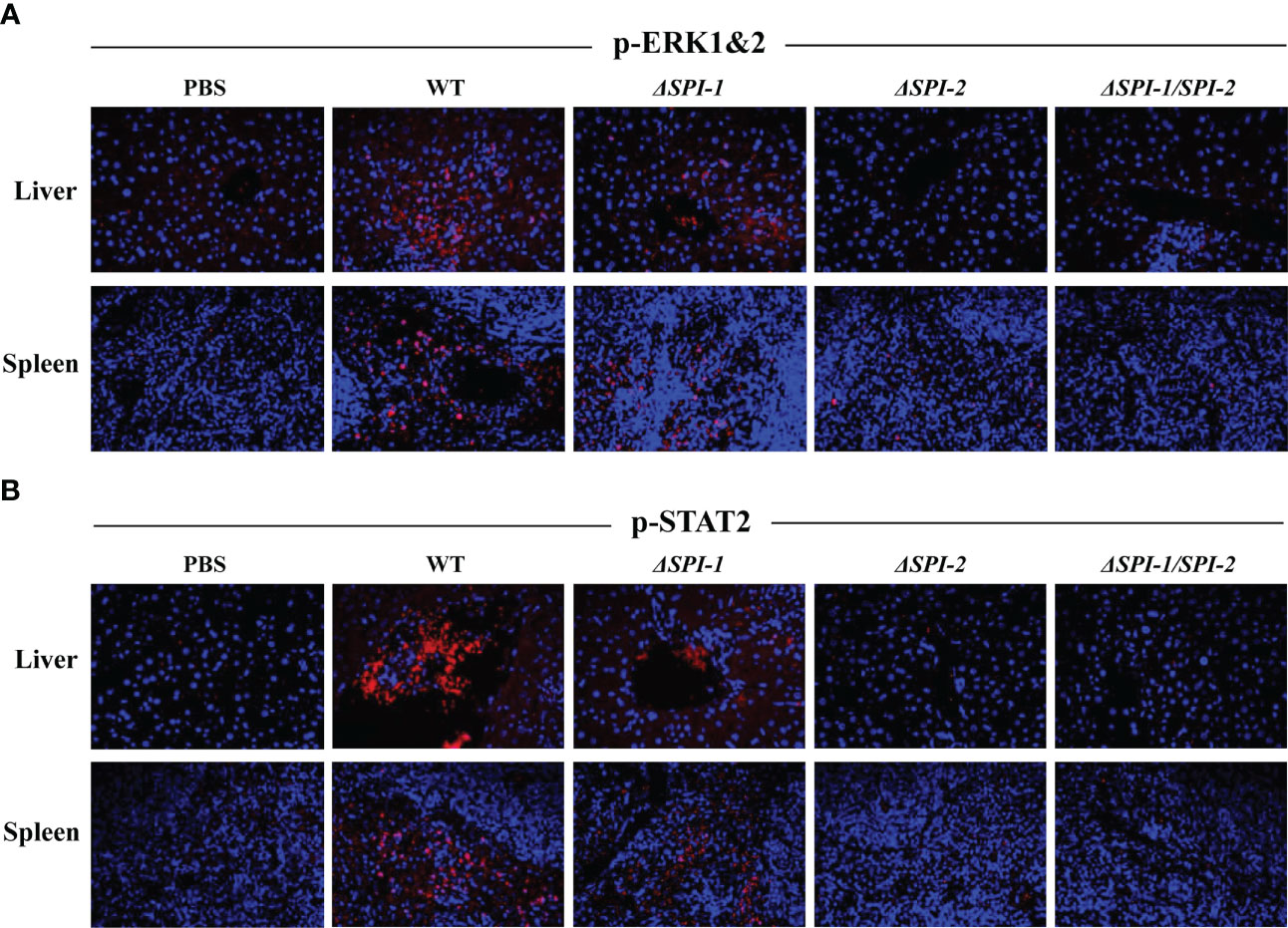
Figure 5 S. Enteritidis WT induced the pronounced activation of p-ERK1&2 and p-STAT2 in the livers and spleens of the infected mice. The activation of p-ERK1&2 (A) and p-STAT2 (B) in mouse liver and spleen were stained using the corresponding antibodies. The nuclear DNA was stained with DAPI. The sections were mounted with the coverslips and Prolong Diamond Antifade Mountant. Images were captured using the Leica confocal microscope at 400 × magnification.
Pro-inflammatory cytokines activate inflammatory processes that result in macrophage recruitment and polymorphonuclear neutrophil infiltration, which lead to cellular injury in the host. Characteristic signs of this process include strong multifocal neutrophilic infiltration, multifocal necrosis, severe diffusion, and cell swelling. To investigate the impact of SPI on pathological manifestations of liver and spleen injury, we performed histopathologic analyses of S. Enteritidis‐infected mice. As shown in Figure 6, liver and spleen tissues from mice infected with WT S. Enteritidis C50041 exhibited severe inflammation and damage on day 4 post infection. The average pathological scores for liver and spleen of WT-infected mice were 5 and 3.7 with severe neutrophil infiltration and necrosis. S. Enteritidis ΔSPI-1-infected mice demonstrated moderate histopathological changes in liver and spleen tissues with the average pathological scores of 2.3 and 1.7 respectively as a result of infection. However, tissues from mice infected with ΔSPI-2 and ΔSPI-1/SPI-2 mutants displayed slight damage and inflammation similar to that observed in control mice. The average pathological scores for liver and spleen of ΔSPI-2-infected mice were 0.7 and 0.3 with mild neutrophil infiltration. Slight inflammation and even no lesions were observed in ΔSPI-1/SPI-2-infected mice with the average pathological score of 0.3 for the spleen.
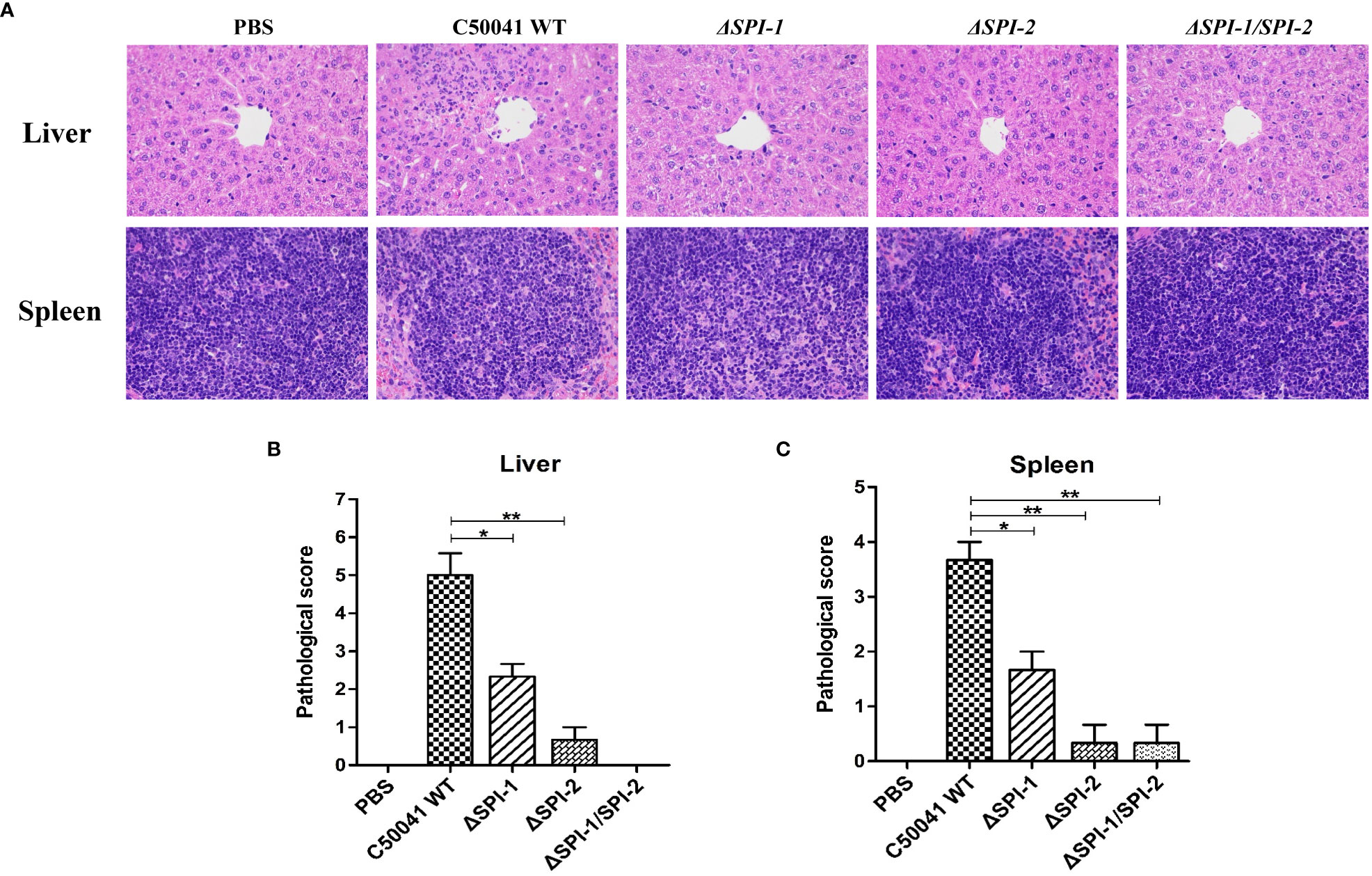
Figure 6 S. Enteritidis SPI-1 and SPI-2 contributed to inflammation storm and tissue damages in the mouse infection model. (A) Histopathological assessment of the livers and spleens from mice infected with S. Enteritidis WT, ΔSPI-1, ΔSPI-2, and ΔSPI-1/SPI-2. Mice were infected with different bacteria, and the livers and spleens were stained with H&E after paraformaldehyde fixation at 4 dpi. Images were captured using a microscope at 400 × magnification. Pathological scores of livers (B) and spleens (C) were evaluated from mice infected with Salmonella C50041, ΔSPI-1, ΔSPI-2, and ΔSPI-1/SPI-2. Pathological evaluations were performed using criteria described in Supplementary Table 4. P < 0.05 (*) and P < 0.01 (**) were considered statistically significant.
To determine the effects of SPI-1 and SPI-2 on the virulence of S. Enteritidis, bacterial localization and mouse survival were monitored following Salmonella infection. The results show that numbers of bacteria were significantly decreased in liver, spleen and cecum tissues 4 days after challenge with the SPI-1-deficient strain, suggesting that the SPI-1 mutant maintained a moderate level of virulence. However, bacteria could not be detected in ΔSPI-2- or ΔSPI-1/SPI-2-infected mice, suggesting that strains lacking SPI-2 or SPI-1/SPI-2 were avirulent (Figures 7A–C). Furthermore, no deaths were observed when mice were challenged with ΔSPI-2 or ΔSPI-1/SPI-2-deficient strains. In contrast, 20% of animals infected with the same dose of S. Enteritidis ΔSPI-1 died on the sixth day post‐infection and 60% of mice succumbed to infection by the ninth day (Figure 7D). These results showed that SPI-1 and SPI-2 enhanced S. Enteritidis virulence by causing cytokine storm and promoting bacterial colonization (Figure 8).
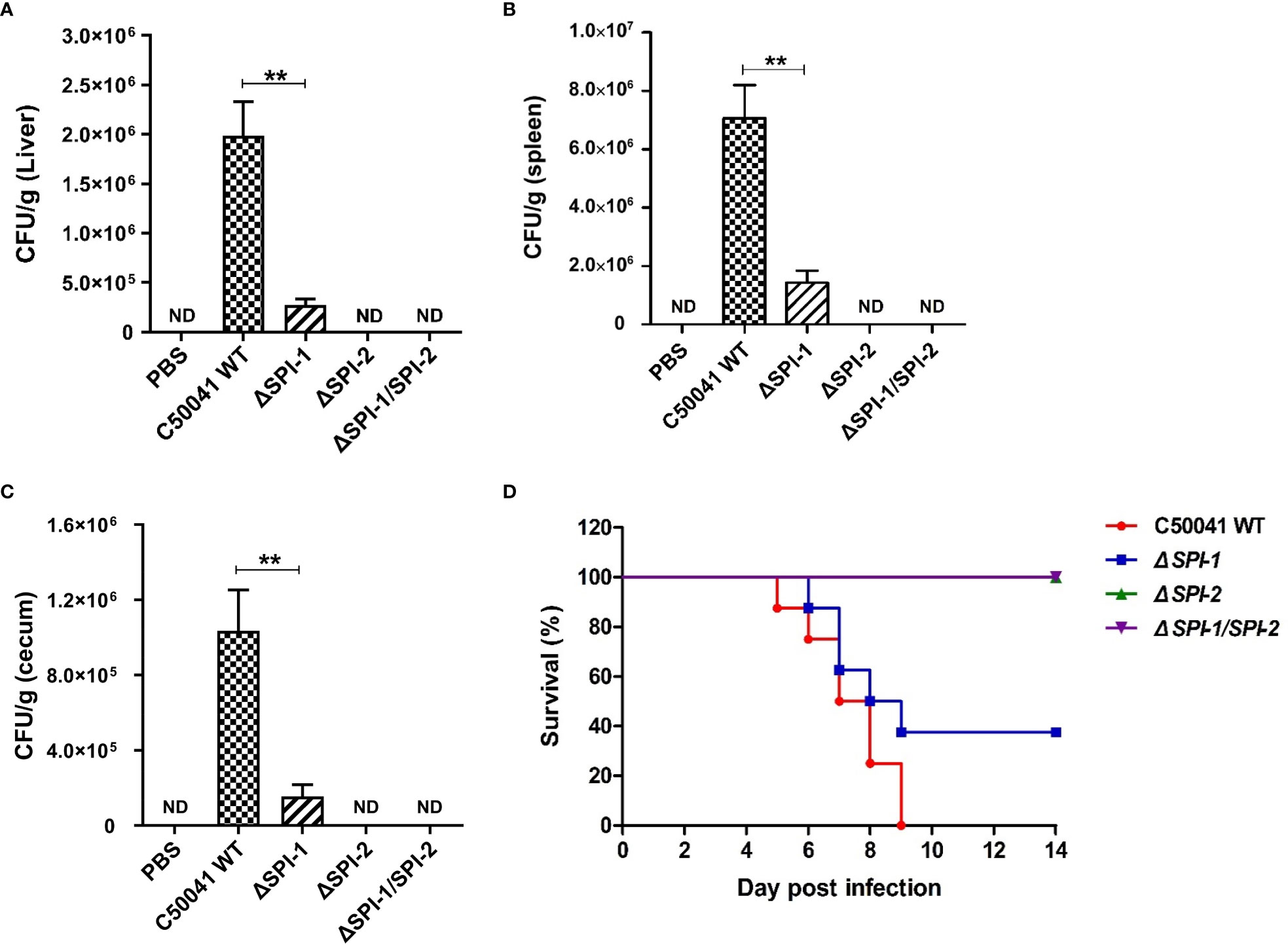
Figure 7 SPI-1 and SPI-2 promoted S. Enteritidis localization in tissues and the bacterial virulence in the mouse infection model. SPI-1 and SPI-2 promoted Salmonella localization in mouse liver (A), spleen (B) and cecum (C). The bacterial burden in tissues were determined on day 4 post infection. Mice were sacrificed, and the livers, spleens and cecums were aseptically collected, homogenized with sterile ice-cold PBS, serially 10‐fold diluted, and plated on LB plates for the determination of CFU. (D) Percentage survival of mice after infection with C50041, ΔSPI-1, ΔSPI-2 and ΔSPI-1/SPI-2. Mice were fasted for 4 h and given 106 CFU of bacteria in 0.1 mL PBS intragastrically. The mortality of mice in different infection groups was recorded daily and monitored for two weeks. P < 0.01 (**) was considered statistically significant. ND, not detected.
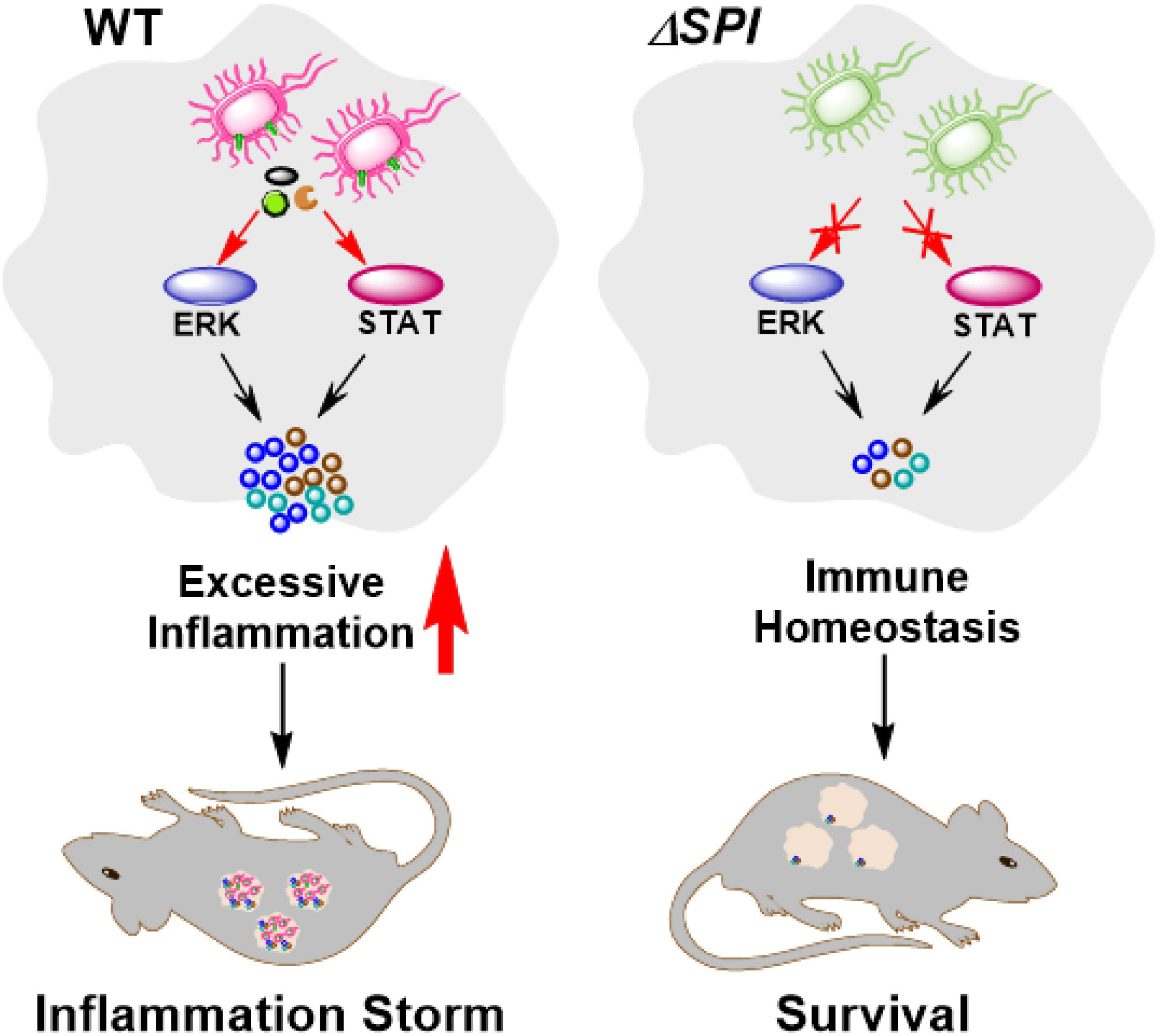
Figure 8 Schematic of S. Enteritidis pathopoiesis by SPI-1 and SPI-2 through inducing cytokine storm via multi-inflammatory signaling pathways. SPI-1 and SPI-2 activate the ERK- and STAT-mediated inflammatory responses and contribute to severe tissue damages, thus promoting the transmission, intracellular localization and bacterial virulence of S. Enteritidis.
S. Enteritidis causes both gastrointestinal and systemic diseases in a wide range of mammalian hosts, including humans (Jennings et al., 2017). It is estimated that more than 1 billion cases of Salmonella gastroenteritis occur every year, resulting in about 3.5 million deaths worldwide (De Masi et al., 2017). Salmonella uses two kinds of specialized T3SSs, which facilitate the bacterial colonization and survival within the host. The two T3SSs, encoded by SPI-1 and SPI-2, secrete effectors into the host cell to cause numerous events that result in symptoms of human enteritis (Brown et al., 2005; Chakravortty et al., 2005). The SPI-1 effector SipA activated caspase-3-mediated inflammation in macrophages and epithelial cells (Srikanth et al., 2010; McIntosh et al., 2017). However, Salmonella SPI-1 effectors, such as SipB and SipD, reduced the level of nuclear factor κB nuclear translocation to reduce inflammation (Lin et al., 2021). Thus, different effectors in SPIs exhibit various functions with respect to inflammation.
Salmonella replication inside macrophages is the key step to induce systemic infection (Gutiérrez et al., 2021). Salmonella can induce apoptosis of many types of cells, such as macrophages and dendritic cells (Kiama et al., 2006). Our results show that both SPI-1 and SPI-2 were required for S. Enteritidis-induced apoptosis of murine macrophages. Previous studies demonstrated that Salmonella-induced apoptosis is SPI-1-dependent (Hueffer and Galán, 2004). However, other studies claimed that apoptosis induced by S. Typhimurium could be SPI-1-independent (van der Velden et al., 2000), suggesting various roles of SPI-1 in different serovars. The present study revealed that S. Enteritidis SPI-1 and SPI-2 caused elevated LDH release, suggesting that both SPIs enhanced bacterial cytotoxicity, and induced apoptosis following infection of macrophages. These features may help Salmonella colonize and spread rapidly in the host.
Salmonella infection induces strong inflammatory responses and subsequently makes use of the inflammatory environment generated during the infection process to overcome the resident microbiota and proliferate in the host (Winter et al., 2010; Faber et al., 2017). Inflammation caused by Salmonella infection can also disturb the host immune system to enhance bacterial replication in infected cells. Here, we found that inflammatory cytokines and various ISGs mediated by MAPK and STAT signaling pathways were associated with S. Enteritidis SPI-1 and SPI-2, suggesting a critical role for Salmonella SPI-1 and SPI-2 in cytokine storm activation. Moreover, both SPI-1 and SPI-2 are required for complete induction of inflammation in vitro.
We investigated the contribution of SPI-1 and SPI-2 to induction of mouse tissue inflammation and bacterial burden following S. Enteritidis infection. Histopathological results revealed focal neutrophil infiltration and necrosis in tissues of mice infected with the ΔSPI-1 mutant at 4 days post infection. In ΔSPI-2-infected mice, however, only steatosis and a few neutrophils were observed. These results indicate that S. Enteritidis SPI-2 had a higher capacity to induce inflammatory responses in mice than SPI-1. Moreover, efficient dissemination and colonization of the cecum, liver and spleen requires SPI-2 but not SPI-1 functions. A previous study reported that SPI-1 alone is not enough to cause cecal inflammation in the presence of a sufficient number of intestinal microbiota (Sekirov et al., 2010). We detected the bacterial colonization in the tissues of mice at 4 days following infection, which was the late stage of infection as mice died at 5 days. At this time, mice underwent a systematic infection, and thus high bacterial loads were detected in the livers and spleens. As the Salmonella counts in the whole cecum contents could be much higher than that of the cecum itself (Elder et al., 2016), the bacterial load in the washed cecum was relatively lower. In this study, the bacterial load was determined in the washed cecum tissues, not the whole cecum contents. Thus, the bacterial counts from the liver and spleen reached numbers higher than those obtained from the cecum. Similar results have also been reported in other studies. The bacterial loads of livers and spleens were higher than that of colons following challenge with S. Enteritidis (van Ampting et al., 2012), ileums following challenge with S. Typhimurium (Xu et al., 2018), and cecums following challenge with S. Enteritidis (Wang et al., 2022). Collectively, these results indicate that S. Enteritidis SPIs, especially SPI-2, could induce inflammatory damage of murine liver and spleen by activating multiple inflammatory signaling pathways.
A previous study showed that a S. Typhimurium SPI‐1 mutant strain was unable to damage eukaryotic cells (Riquelme et al., 2016). Another study reported that a S. Typhimurium strain with disrupted SPI-1 was not completely attenuated, causing delayed colitis that became obvious at days 3 and 4 following infection (Hapfelmeier et al., 2005). In this study, we found that S. Enteritidis SPI-1 deficiency could not induce inflammatory responses or apoptosis in vitro. However, the SPI-1-deficient strain contributed to moderate inflammation, tissue damage, and virulence in a mouse model. These results indicate different roles for SPI-1 during Salmonella infection in vitro and in vivo.
Inflammation might be primarily induced by the translocation of various Salmonella effector proteins, which trigger the activation of pro-inflammatory signaling. Although both SPI-1 and SPI-2 contain pro-inflammatory and anti-inflammatory effectors, we found that the entire SPI-1 or SPI-2 exhibited a pro-inflammatory phenotype during infection of macrophages and mice. WT S. Enteritidis caused significant inflammation of mouse liver and spleen on day 4 following infection, which was attributed to excessive production of inflammatory cytokines. Other studies reported that SPI-1 was insufficient to induce the mouse inflammation according to assessments of intestinal pathology (Sekirov et al., 2010). However, the presence of SPI-2 alone was shown to be sufficient to induce tissue inflammation in a mouse infection model, similar to that induced by ΔSPI-1/SPI-2. These results suggest that the virulence of S. Enteritidis requires functional SPIs, especially SPI-2, in vivo. Notably, ΔSPI-1 and ΔSPI-2 mutants were highly attenuated in the macrophage infection assay (by 80% and 70%, respectively). However, SPI-2 played a more important role in colonization and virulence of S. Enteritidis in the mouse model. Overall, these results indicate that the complex immune environment in vivo may impact the function of different SPIs. Taken together, S. Enteritidis promoted its intracellular colonization by activating multiple inflammatory pathways via SPI-1 and SPI-2 to contribute to cytokine storm, tissue damage and bacterial virulence.
In conclusion, our findings reveal that SPI-1 and SPI-2 are important virulence factors during S. Enteritidis infection in vitro and in vivo. SPI-1 plays an important role in the invasion of the host cellular cytosol, intracellular survival, and induction of cellular apoptosis in vitro, but it does not seem to be required for cytokine storm induction in an in vivo infection model. SPI-2 has a more important role than SPI-1 in systemic infection by activating MAPK- and STAT2-mediated inflammation. S. Enteritidis contributed to SPI-2-induced cytokine storm and tissue damage to enhance intracellular bacterial replication in the mouse liver and spleen. We identified that both SPIs, especially SPI-2, were required for S. Enteritidis colonization and virulence in a mouse model. Our findings increase understanding of the relative roles of SPI-1 and SPI-2 in generation of systemic inflammation and bacterial virulence during S. Enteritidis infection.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by the Committee on the Ethics of Animal Experiments of Yangzhou University.
All authors have contributed to the manuscript. DX, XJ and ZP conceived the study. DX designed the experiments, performed the assays and analysed the results. LS and YC performed the experiments and analysed the results. DX, XJ and ZP wrote the paper. All authors read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the National Natural Science Foundation of China (32102669, 31972685, 31920103015 and 32102679), the Natural Science Foundation of Jiangsu Province (BK20210802), the China Postdoctoral Science Foundation (2020M681748), the Postdoctoral Science Foundation of Jiangsu Province (2021K095A), the 111 Project (D18007), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1158888/full#supplementary-material
Brandes, M., Klauschen, F., Kuchen, S., Germain, R. N. (2013). A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell 154, 197–212. doi: 10.1016/j.cell.2013.06.013
Brown, N. F., Vallance, B. A., Coombes, B. K., Valdez, Y., Coburn, B. A., Finlay, B. B. (2005). Salmonella pathogenicity island 2 is expressed prior to penetrating the intestine. PloS Pathog. 1, e32. doi: 10.1371/journal.ppat.0010032
Centers for Disease Control and Prevention (2023) Salmonella. Available at: https://www.cdc.gov/salmonella/index.html (Accessed 2, 2023).
Chakravortty, D., Rohde, M., Jäger, L., Deiwick, J., Hensel, M. (2005). Formation of a novel surface structure encoded by Salmonella pathogenicity island 2. EMBO J. 24, 2043–2052. doi: 10.1038/sj.emboj.7600676
Datsenko, K. A., Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97, 6640–6645. doi: 10.1073/pnas.120163297
De Masi, L., Yue, M., Hu, C., Rakov, A. V., Rankin, S. C., Schifferli, D. M. (2017). Cooperation of adhesin alleles in Salmonella-host tropism. mSphere 2, e00066–e00017. doi: 10.1128/mSphere.00066-17
Elder, J. R., Chiok, K. L., Paul, N. C., Haldorson, G., Guard, J., Shah, D. H. (2016). The Salmonella pathogenicity island 13 contributes to pathogenesis in streptomycin pre-treated mice but not in day-old chickens. Gut Pathog. 8, 16. doi: 10.1186/s13099-016-0098-0
Faber, F., Thiennimitr, P., Spiga, L., Byndloss, M. X., Litvak, Y., Lawhon, S., et al. (2017). Respiration of microbiota-derived 1, 2-propanediol drives Salmonella expansion during colitis. PloS Pathog. 13, e1006129. doi: 10.1371/journal.ppat.1006129
Grenz, J. R., Cott Chubiz, J. E., Thaprawat, P., Slauch, J. M. (2018). HilE regulates HilD by blocking DNA binding in Salmonella enterica serovar typhimurium. J. Bacteriol. 200, e00750–e00717. doi: 10.1128/JB.00750-17
Gutiérrez, S., Fischer, J., Ganesan, R., Hos, N. J., Cildir, G., Wolke, M., et al. (2021). Salmonella typhimurium impairs glycolysis-mediated acidification of phagosomes to evade macrophage defense. PloS Pathog. 17, e1009943. doi: 10.1371/journal.ppat.1009943
Hansen-Wester, I., Hensel, M. (2001). Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3, 549–559. doi: 10.1016/s1286-4579(01)01411-3
Hapfelmeier, S., Stecher, B., Barthel, M., Kremer, M., Müller, A. J., Heikenwalder, M., et al. (2005). The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J. Immunol. 174, 1675–1685. doi: 10.4049/jimmunol.174.3.1675
Hueffer, K., Galán, J. E. (2004). Salmonella-induced macrophage death: multiple mechanisms, different outcomes. Cell. Microbiol. 6, 1019–1025. doi: 10.1111/j.1462-5822.2004.00451.x
Jennings, E., Thurston, T. L. M., Holden, D. W. (2017). Salmonella SPI-2 type III secretion system effectors: molecular mechanisms and physiological consequences. Cell Host Microbe 22, 217–231. doi: 10.1016/j.chom.2017.07.009
Jiang, L., Feng, L., Yang, B., Zhang, W., Wang, P., Jiang, X., et al. (2017). Signal transduction pathway mediated by the novel regulator LoiA for low oxygen tension induced Salmonella typhimurium invasion. PloS Pathog. 13, e1006429. doi: 10.1371/journal.ppat.1006429
Jiang, L., Wang, P., Song, X., Zhang, H., Ma, S., Wang, J., et al. (2021). Salmonella typhimurium reprograms macrophage metabolism via T3SS effector SopE2 to promote intracellular replication and virulence. Nat. Commun. 12, 879. doi: 10.1038/s41467-021-21186-4
Jones, M. A., Hulme, S. D., Barrow, P. A., Wigley, P. (2007). The Salmonella pathogenicity island 1 and Salmonella pathogenicity island 2 type III secretion systems play a major role in pathogenesis of systemic disease and gastrointestinal tract colonization of Salmonella enterica serovar typhimurium in the chicken. Avian Pathol. 36, 199–203. doi: 10.1080/03079450701264118
Kawai, T., Akira, S. (2010). The role of pattern-recognition receptors in innate immunity: update on toll-like receptors. Nat. Immunol. 11, 373–384. doi: 10.1038/ni.1863
Keszei, A. F., Tang, X., McCormick, C., Zeqiraj, E., Rohde, J. R., Tyers, M., et al. (2014). Structure of an SspH1-PKN1 complex reveals the basis for host substrate recognition and mechanism of activation for a bacterial E3 ubiquitin ligase. Mol. Cell. Biol. 34, 362–373. doi: 10.1128/MCB.01360-13
Kiama, S. G., Dreher, D., Cochand, L., Kok, M., Obregon, C., Nicod, L., et al. (2006). Host cell responses of Salmonella typhimurium infected human dendritic cells. Immunol. Cell Biol. 84, 475–481. doi: 10.1111/j.1440-1711.2006.01461.x
Kurtz, J. R., Goggins, J. A., McLachlan, J. B. (2017). Salmonella infection: interplay between the bacteria and host immune system. Immunol. Lett. 190, 42–50. doi: 10.1016/j.imlet.2017.07.006
Lee, Y. J., Mo, I. P., Kang, M. S. (2005). Safety and efficacy of Salmonella gallinarum 9R vaccine in young laying chickens. Avian Pathol. 34, 362–366. doi: 10.1080/03079450500180895
LeLièvre, V., Besnard, A., Schlusselhuber, M., Desmasures, N., Dalmasso, M. (2019). Phages for biocontrol in foods: what opportunities for Salmonella sp. control along the dairy food chain? Food Microbiol. 78, 89–98. doi: 10.1016/j.fm.2018.10.009
Lin, H. H., Chen, H. L., Weng, C. C., Janapatla, R. P., Chen, C. L., Chiu, C. H. (2021). Activation of apoptosis by Salmonella pathogenicity island-1 effectors through both intrinsic and extrinsic pathways in Salmonella-infected macrophages. J. Microbiol. Immunol. Infect. 54, 616–626. doi: 10.1016/j.jmii.2020.02.008
McIntosh, A., Meikle, L. M., Ormsby, M. J., McCormick, B. A., Christie, J. M., Brewer, J. M., et al. (2017). SipA activation of caspase-3 is a decisive mediator of host cell survival at early stages of Salmonella enterica serovar typhimurium infection. Infect. Immun. 85, e00393–e00317. doi: 10.1128/IAI.00393-17
Nuccio, S. P., Bäumler, A. J. (2014). Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. mBio 5, e00929–e00914. doi: 10.1128/mBio.00929-14
O'Neill, L. A., Bryant, C. E., Doyle, S. L. (2009). Therapeutic targeting of toll-like receptors for infectious and inflammatory diseases and cancer. Pharmacol. Rev. 61, 177–197. doi: 10.1124/pr.109.001073
Parry, C. M., Hien, T. T., Dougan, G., White, N. J., Farrar, J. J. (2002). Typhoid fever. N. Engl. J. Med. 347, 1770–1782. doi: 10.1056/NEJMra020201
Paul, P., Patel, P., Verma, S. K., Mishra, P., Sahu, B. R., Panda, P. K., et al. (2022). The hha-TomB toxin-antitoxin module in Salmonella enterica serovar typhimurium limits its intracellular survival profile and regulates host immune response. Cell Biol. Toxicol. 38, 111–127. doi: 10.1007/s10565-021-09587-z
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. doi: 10.1093/nar/29.9.e45
Que, F., Wu, S., Huang, R. (2013). Salmonella pathogenicity island 1 (SPI-1) at work. Curr. Microbiol. 66, 582–587. doi: 10.1007/s00284-013-0307-8
Rana, N., Kulshreshtha, R. C. (2006). Cell-mediated and humoral immune responses to a virulent plasmid-cured mutant strain of Salmonella enterica serotype gallinarum in broiler chickens. Vet. Microbiol. 115, 156–162. doi: 10.1016/j.vetmic.2006.01.011
Rasmussen, R. (2001). “Quantification on the LightCycler,” in Rapid cycle real-time PCR, methods and applications. Eds. Meuer, S., Wittwer, C., Nakagawara, K. (Heidelberg: Springer Press), 21–34.
Riquelme, S., Varas, M., Valenzuela, C., Velozo, P., Chahin, N., Aguilera, P., et al. (2016). Relevant genes linked to virulence are required for Salmonella typhimurium to survive intracellularly in the social amoeba dictyostelium discoideum. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01305
Rolhion, N., Furniss, R. C., Grabe, G., Ryan, A., Liu, M., Matthews, S. A., et al. (2016). Inhibition of nuclear transport of NF-κB p65 by the Salmonella type III secretion system effector SpvD. PloS Pathog. 12, e1005653. doi: 10.1371/journal.ppat.1005653
Sekirov, I., Gill, N., Jogova, M., Tam, N., Robertson, M., de Llanos, R., et al. (2010). Salmonella SPI-1-mediated neutrophil recruitment during enteric colitis is associated with reduction and alteration in intestinal microbiota. Gut Microbes 1, 30–41. doi: 10.4161/gmic.1.1.10950
Srikanth, C. V., Mercado-Lubo, R., Hallstrom, K., McCormick, B. A. (2011). Salmonella effector proteins and host-cell responses. Cell. Mol. Life Sci. 68, 3687–3697. doi: 10.1007/s00018-011-0841-0
Srikanth, C. V., Wall, D. M., Maldonado-Contreras, A., Shi, H., Zhou, D., Demma, Z., et al. (2010). Salmonella pathogenesis and processing of secreted effectors by caspase-3. Science 330, 390–393. doi: 10.1126/science.1194598
Tischer, B. K., von Einem, J., Kaufer, B., Osterrieder, N. (2006). Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40, 191–197. doi: 10.2144/000112096
van Ampting, M. T., Loonen, L. M., Schonewille, A. J., Konings, I., Vink, C., Iovanna, J., et al. (2012). Intestinally secreted c-type lectin Reg3b attenuates salmonellosis but not listeriosis in mice. Infect. Immun. 80, 1115–1120. doi: 10.1128/IAI.06165-11
van der Velden, A. W., Lindgren, S. W., Worley, M. J., Heffron, F. (2000). Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype typhimurium. Infect. Immun. 68, 5702–5709. doi: 10.1128/IAI.68.10.5702-5709.2000
Wain, J., Hendriksen, R. S., Mikoleit, M. L., Keddy, K. H., Ochiai, R. L. (2015). Typhoid fever. Lancet 385, 1136–1145. doi: 10.1016/S0140-6736(13)62708-7
Wang, X., Kang, X., Pan, M., Wang, M., Zhang, J., Song, H. (2022). Evaluation of the protective immune response induced by an rfbG-deficient Salmonella enterica serovar enteritidis strain as a live attenuated DIVA (differentiation of infected and vaccinated animals) vaccine in chickens. Microbiol. Spectr. 10, e0157422. doi: 10.1128/spectrum.01574-22
Winter, S. E., Thiennimitr, P., Winter, M. G., Butler, B. P., Huseby, D. L., Crawford, R. W., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. doi: 10.1038/nature09415
World Health Organization (2022)Multi-country outbreak of Salmonella typhimurium linked to chocolate products–Europe and the united states of America. Available at: https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON369.
Xiong, D., Song, L., Geng, S., Jiao, Y., Zhou, X., Song, H., et al. (2019). Salmonella coiled-coil- and TIR-containing TcpS evades the innate immune system and subdues inflammation. Cell Rep. 28, 804–818. doi: 10.1016/j.celrep.2019.06.048
Xu, X., Gong, L., Wang, B., Wu, Y., Wang, Y., Mei, X., et al. (2018). Glycyrrhizin attenuates Salmonella enterica serovar typhimurium infection: new insights into its protective mechanism. Front. Immunol. 9. doi: 10.3389/fimmu.2018.02321
Zhang, K., Riba, A., Nietschke, M., Torow, N., Repnik, U., Pütz, A., et al. (2018). Minimal SPI1-T3SS effector requirement for Salmonella enterocyte invasion and intracellular proliferation in vivo. PloS Pathog. 14, e1006925. doi: 10.1371/journal.ppat.1006925
Keywords: Salmonella Pathogenicity Island, Inflammatory pathway, bacterial colonization, macrophage, mouse infection model
Citation: Xiong D, Song L, Chen Y, Jiao X and Pan Z (2023) Salmonella Enteritidis activates inflammatory storm via SPI-1 and SPI-2 to promote intracellular proliferation and bacterial virulence. Front. Cell. Infect. Microbiol. 13:1158888. doi: 10.3389/fcimb.2023.1158888
Received: 04 February 2023; Accepted: 18 May 2023;
Published: 30 May 2023.
Edited by:
Mathieu Coureuil, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Abhinav Upadhyay, University of Connecticut, United StatesCopyright © 2023 Xiong, Song, Chen, Jiao and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinan Jiao, amlhb0B5enUuZWR1LmNu; Zhiming Pan, em1wYW5AeXp1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.