
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol., 09 March 2023
Sec. Bacteria and Host
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1144210
This article is part of the Research TopicMycobacterial Dormancy, Culturability, and Resuscitation: State-of-the-art, Challenges, and Future ProspectsView all 10 articles
Mycobacterium abscessus (Mab), an emerging opportunistic pathogen, predominantly infects individuals with underlying pulmonary diseases such as cystic fibrosis (CF). Current treatment outcomes for Mab infections are poor due to Mab’s inherent antibiotic resistance and unique host interactions that promote phenotypic tolerance and hinder drug access. The hypoxic, mucus-laden airways in the CF lung and antimicrobial phagosome within macrophages represent hostile niches Mab must overcome via alterations in gene expression for survival. Regulatory mechanisms important for the adaptation and long-term persistence of Mab within the host are poorly understood, warranting further genetic and transcriptomics study of this emerging pathogen. DosRSMab, a two-component signaling system (TCS), is one proposed mechanism utilized to subvert host defenses and counteract environmental stress such as hypoxia. The homologous TCS of Mycobacterium tuberculosis (Mtb), DosRSMtb, is known to induce a ~50 gene regulon in response to hypoxia, carbon monoxide (CO) and nitric oxide (NO) in vitro and in vivo. Previously, a small DosRMab regulon was predicted using bioinformatics based on DosRMtb motifs however, the role and regulon of DosRSMab in Mab pathogenesis have yet to be characterized in depth. To address this knowledge gap, our lab generated a Mab dosRS knockout strain (MabΔdosRS) to investigate differential gene expression, and phenotype in an in vitro hypoxia model of dormancy. qRT-PCR and lux reporter assays demonstrate Mab_dosR and 6 predicted downstream genes are induced in hypoxia. In addition, RNAseq revealed induction of a much larger hypoxia response comprised of >1000 genes, including 127 differentially expressed genes in a dosRS mutant strain. Deletion of DosRSMab led to attenuated growth under low oxygen conditions, a shift in morphotype from smooth to rough, and down-regulation of 216 genes. This study provides the first look at the global transcriptomic response of Mab to low oxygen conditions encountered in the airways of CF patients and within macrophage phagosomes. Our data also demonstrate the importance of DosRSMab for adaptation of Mab to hypoxia, highlighting a distinct regulon (compared to Mtb) that is significantly larger than previously described, including both genes conserved across mycobacteria as well as Mab-specific genes.
Mycobacterium abscessus (Mab) is an opportunistic pathogen capable of causing skin, soft tissue and pulmonary infections in immunocompromised individuals and individuals with pre-existing lung disease such as cystic fibrosis (CF) and bronchiectasis (Brown-Elliott and Wallace, 2002; Olivier et al., 2003; Harris and Kenna, 2014; Lee et al., 2015; Lopeman et al., 2019). Impaired innate immune defenses and viscous mucus within the CF lung contribute to reduced clearance of bacterial pathogens leading to increased rates of infections and morbidity (Lyczak et al., 2002; Chmiel and Davis, 2003). Mab is the most common rapidly-growing mycobacterial (RGM) species recovered from the lungs of CF patients (Esther et al., 2010). Within the CF population and patients with underlying lung dysfunction, infections caused by Mab are associated with lung function decline, increased hospital visits, prolonged hospital stays and in some cases exclusion from lung transplants (Olivier et al., 2003; Esther et al., 2010; Lopeman et al., 2019). Due to Mab’s inherent antibiotic resistance, treatment options are limited, resulting in extremely low cure rates of less than 50% (Greendyke and Byrd, 2008; Hurst-Hess et al., 2017; Molina-Torres et al., 2018; Story-Roller et al., 2018; Lopeman et al., 2019).
The development of effective treatment strategies for Mab is hindered by discrepancies between in vitro and in vivo susceptibilities associated with Mab’s unique lifestyle (Greendyke and Byrd, 2008; Philley et al., 2016; Molina-Torres et al., 2018). Akin to Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis, Mab resides within pulmonary macrophages and within granulomas which limit antibiotic accessibility and promote drug tolerance, making treatment of an inherently antibiotic resistant pathogen even more difficult (Bernut et al., 2016; Peddireddy et al., 2017). The viscous mucus in the CF lung represents an additional hostile niche within the host to which Mab must adapt (Lyczak et al., 2002; Chmiel and Davis, 2003; Miranda-CasoLuengo et al., 2016; Dubois et al., 2019). One key host-derived stress encountered by Mab in all three of these microenvironments is decreased oxygen tension, with oxygen tension estimated to be ~1% O2 within these niches (Worlitzsch et al., 2002; Cunningham-Bussel et al., 2013; Hudock et al., 2017). Thus, to successfully cause an infection, Mab must encode mechanisms to adapt and persist under hypoxic conditions. Transcriptional responses to host-derived cues/stresses are not well-defined in this NTM pathogen, requiring further studies to understand how Mab adapts its physiology and virulence factor expression to cause insidious, persistent infections.
Two-component signaling (TCS) is a mechanism commonly utilized by prokaryotes to regulate virulence gene expression in response to host-derived cues (Yarwood et al., 2001; Walters et al., 2006; Gonzalo-Asensio et al., 2008; Gooderham and Hancock, 2009). A typical TCS consists of a sensor histidine kinase (HK) responsible for signal recognition and subsequent phosphorylation of a cognate response regulator (RR) which binds DNA motifs within promoter regions to drive alterations in gene expression (Stock et al., 1989; West and Stock, 2001; Mascher et al., 2006; Salazar and Laub, 2015). Mab encodes 11 TCS, 5 orphan RRs and 1 orphan HK each with a corresponding ortholog in Mtb; however, in-depth studies of Mab TCS have not been performed (Bretl et al., 2011). The well-documented atypical TCS DosRS/TMtb, is known to control a ~50 gene regulon to counteract hypoxic and nitrosative stress encountered within macrophages and granulomas (Sherman et al., 2001; Park et al., 2003; Voskuil et al., 2003; Leistikow et al., 2010; Rohde et al., 2012; Cunningham-Bussel et al., 2013; Peterson et al., 2020; Kundu and Basu, 2021). DosRS/TMtb,contains 2 HKs (DosS and DosT) rather than one which are responsible for phosphorylating the RR DosR at different stages of hypoxia (Roberts et al., 2004). Although Mtb dosT contributes to signaling in early stages of hypoxia, it is constitutively expressed and is not part of the DosRMtb regulon (Honaker et al., 2009). The DosRMtb regulon includes autoregulation of dosRS itself, as well as heat shock proteins, triacylglycerol synthases, ferrodoxins, universal stress proteins, diacylglycerol acyltransferases, and nitroreductase which are implicated in dormancy, resuscitation, phenotypic drug tolerance and increased lipid metabolism (Park et al., 2003; Voskuil et al., 2003; Leistikow et al., 2010; Galagan et al., 2013; Aguilar-Ayala et al., 2017). Induction of Mtb dosR within animal models capable of forming hypoxic granulomas (rhesus macaques, guinea pigs, and C3HeB/FeJ mice) and attenuation of mutants lacking DosRSMtb highlight the importance of this TCS for Mtb pathogenesis (Converse et al., 2009; Gautam et al., 2015a, Gautam et al., 2015b; Mehra et al., 2015).
According to whole genome sequence data, Mab encodes a DosR ortholog (Mab_3891c) with a high level of homology with DosRMtb (~72% identity) adjacent to and upstream of DosSMab (Mab_3890c) with lower similarity (~51% identify) to its counterpart in Mtb. Mab does not appear to encode a secondary orphan HK analogous to DosT, with the closest homolog to dosT being dosSMab/Mab_3890C (53% identity). At the time this study was initiated, only two reports made mention of DosRSMab. A bioinformatics study by Gerasimova et al. used Mtb DosR promoter motifs to predict a small DosRMab regulon consisting of only 6 genes (Gerasimova et al., 2011). A subsequent transcriptomics study by Miranda Caso-Luengo et al. demonstrated induction of the 6 predicted DosRMab regulated genes plus 56 other genes upon exposure to nitric oxide (NO) (Miranda-CasoLuengo et al., 2016). It remained unclear whether induction of these genes occurs through signaling of DosRSMab or whether hypoxia is an induction cue.
Although there is considerable overlap in the repertoires of TCS encoded by different mycobacteria, few cross-species transcriptomic studies are available comparing TCS regulons and regulatory networks between e.g. Mtb and NTM pathogens. Given the diversity of conditions encountered by Mab as an environmental, opportunistic pathogen, and larger genome (compared to Mtb) comprised of ~800 species-specific genes, the potential for unique gene sets in Mab TCS regulons is high (Malhotra et al., 2017; Wee et al., 2017). A hypoxia model mimicking the physiologic conditions in the mucus of CF airways, within granulomas and intramacrophage compartments was used to evaluate the role of DosRSMab and assess transcriptional regulation mediated by this TCS. Our work demonstrates DosRSMab is important for maximal growth and survival in hypoxia and regulates a potentially larger set of genes than previously predicted. RNAseq revealed upregulation of >1000 genes in hypoxia including 127 putative DosRSMab regulated genes. Information gained from this study identifies the importance of the DosRSMab TCS in adaptation to hypoxia for survival and provides valuable knowledge of a novel set of hypoxia-induced genes in this species for future studies.
MabΔdosRS was generated via recombineering as described by van Kessell and Hatfull in the strain Mab 390S obtained from the Thomas Byrd lab (Byrd and Lyons, 1999; van Kessel and Hatfull, 2007). In brief, an allelic exchange substrate (AES) was engineered containing an apramycin resistance cassette flanked by ~1000 nucleotides upstream and downstream of the dosRS operon via round the horn PCR and fast cloning (Moore and Prevelige, 2002; Li et al., 2011). Mab::pJV53 competent cells induced with .02% acetamide for 4 hours were electroporated with 100ng AES, recovered in 7H9 OADC media for 24 hours, and plated on 7H10 agar supplemented with apramycin 50 µg/ml. Complement strain, MabΔdosRS+C, containing dosRS with its native promoter was engineered using round the horn (Moore and Prevelige, 2002) and fast cloning (Li et al., 2011) in the integrating vector pUAB400 followed by electroporation into MabΔdosRS (Singh et al., 2006). Mab 390S and MabΔdosRS were transformed with pMV306hspG13lux (Addgene #26161) to generate the constitutive lux strains, Mab 390S Phsp60-lux and MabΔdosRS Phsp60-lux. Lux reporters under the control of PdosR and P2489 were constructed in the background plasmid pMV306hspG13lux to generate Mab 390S PdosR-lux, MabΔdosRS PdosR-lux, Mab 390S P2489-lux, and MabΔdosRS P2489-lux, via replacement of Phsp60 (hsp60 promoter) using round-the horn cloning (Moore and Prevelige, 2002; Andreu et al., 2010; Li et al., 2011). Refer to Table S1 for strains, plasmids and primers used for cloning.
Cultures were grown in 7H9-OADC+.05% tyloxapol from glycerol stocks at 37°C while shaking unless otherwise noted. For growth kinetics assays, strains were inoculated from mid-log phase starter cultures into 13ml of media in filter-capped T-25 flasks to an OD600 = 0.02. Hypoxic cultures were grown standing in a hypoxic incubator set at 1% O2 while aerated controls were cultured at 20% O2 while shaking at 100 rpm. Re-aeration studies were conducted after cultures were subjected to hypoxia or grown at 20% O2 for 30 days. Optical density readings (OD600nm) were taken on days 2, 3, 5, 8 and 10.
RNA was extracted as previously described (Rohde et al., 2007) in triplicate from hypoxic cultures (1% O2) and normoxic cultures (20% O2) for qRT-PCR and RNAseq. At designated time points, cultures were pelleted at 4300 rpm for 5 minutes, resuspended in guanidine thiocyanate buffer, pelleted again at 12,000 rpm for 5 min, and stored at -80°C until processing. Thawed pellets were resuspended in 65°C Trizol then lysed using 0.1mM silicon beads in a BeadBeater at max speed for 1 minute 2x followed by cooling on ice for 1 minute between bead beating. Isolation of total RNA from Trizol lysates was performed using chloroform extraction and Qiagen RNeasy column purification. Total RNA was treated with Turbo DNase (Invitrogen) to eliminate DNA contamination. 50 ng/µl of total RNA was used to generate cDNA using iScript™ cDNA synthesis kit (Bio-Rad) for qRT-PCR reactions carried out in a QuantStudio7 thermocycler. Primers used for qRT-PCR are listed in Table S1. RNA samples for RNAseq analysis were pooled at equal RNA concentrations from three biological experiments as previously described (Tomlinson et al., 2021; Tomlinson et al., 2022) prior to library preparation at a concentration of 50 ng/µl in 20 µl. Only RNA samples with RIN>6 as determined by Tapestation analysis were utilized. RNA samples were sequenced by Microbial Genome Sequencing Center (MiGs) using Illumina sequencing protocol aligning reads to the Mab ATCC19977 genome (accession #CU458896). RNAseq data reflects a minimum of 12M paired end reads per sample. Due to incompatibility of Ribo-zero rRNA removal kit (Illumina) with Mab which resulted in high levels of rRNA, MiGS designed custom depletion probes (Table S2) for rRNA depletion. Raw data was received from MiGs as fastq files followed by analysis using CLC Genomics Workbench 12 (Qiagen Bioinformatics). Illumina paired importer tool was used to eliminate failed reads using the quality score parameter option set to Illumina Pipelines 1.8. Expression browser tool (v1.1) was used to calculate gene expression with an output of transcript per million (TPM). Differential gene expression is expressed as a log2FC of ≤-1 or ≥1 and visualized as scatter plots created in GraphPad Prism 9.
Bio-luminescent reporter strains were grown to mid-log phase, diluted to .02 OD in 13ml in T25 flasks and grown in either 20% O2 or 1% O2 for 1, 5 and 20 days. 200 ul of each culture was aliquoted in triplicate into 96 well white bottomed plate to measure luminescence via Synergy H4 reader (Biotek). Fold change of luminescence was analyzed comparing individual strains in 1% O2 to 20% O2 using Mab 390S Phsp60-lux or MabΔdosRS Phsp60-lux as internal controls (1%O2(PdosR or P2489/Phsp60))/(20% O2(PdosR or P2489/Phsp60)).
Our first goal was to develop a tractable in vitro model to investigate the mechanisms employed by Mab to persist under physiologically relevant oxygen-limited conditions. To do this, Mab 390S was grown under standing conditions in a 1% O2 atmosphere to mimic the pO2 observed within the CF lung, macrophages and granulomas (Worlitzsch et al., 2002; Cunningham-Bussel et al., 2013; Hudock et al., 2017). Mab 390S grew steadily at 1% O2, reaching OD600 ~0.7 by day 5 with continued increase to ~0.9 by day 10 (Figure S1). Somewhat unexpectedly, the intended aerobic control culture (20% O2 atmosphere, standing) had a very similar growth profile, reaching only a slightly higher OD ~1.2 by day 10 (Figure S1). In contrast, Mab 390S grown in 20% O2 with shaking (100rpm) reached a maximum density by day 5 (OD600~1.6) and subsequently plateaued up to day 10 (Figure S1). This difference is likely due to the microaerobic conditions experienced by bacilli growing below the media surface, as previously seen with Mtb and BCG strains grown in standing conditions (Cunningham and Spreadbury, 1998; Purkayastha et al., 2002). To maximize the contrast between aerobic and hypoxic conditions, Mab cultures shaking in 20% O2 served as references for all hypoxia experiments. Thus, Mab is not only able to survive but to actively replicate under in-vivo like hypoxia conditions. It is worth noting that exposure of Mab to sudden hypoxia using BD Gaspack anaerobic pouches led to rapid sterilization of the cultures (data not shown). This may indicate a lower threshold of O2 levels needed for Mab viability was exceeded or that slower, adaptive responses are necessary to survive.
Exploiting this model to profile Mab differential gene expression (DGE) in response to hypoxia, RNAseq transcriptomic analysis was conducted on wild-type Mab 390S cultured at 1% O2 and 20% O2 on day 5. This time point reflects the maximal difference in OD600nm between hypoxic and aerated cultures and precedes the plateau in growth curves (Figure S1). The scatter plot in Figure 1A graphically depicts the dramatic genome-wide alterations in Mab 390S gene expression induced by in vivo-like low oxygen condition. The Mab hypoxia response included induction of 1,190 genes (≥ 1 log2 fold change compared to 20% O2) and downregulation of 1,062 genes (≤ 1 log2 fold change compared to 20% O2). This represented a larger set of hypoxia-induced genes than observed in Mtb in two studies under similar 1% O2 hypoxia conditions (induction of ~400 genes detected via microarray and ~682 via RNAseq) (Rustad et al., 2009; Vilcheze et al., 2022). These data highlight the functional genomic differences between Mab and Mtb, in particular their distinct patterns of gene regulation in response to hypoxia which are discussed in detail below.
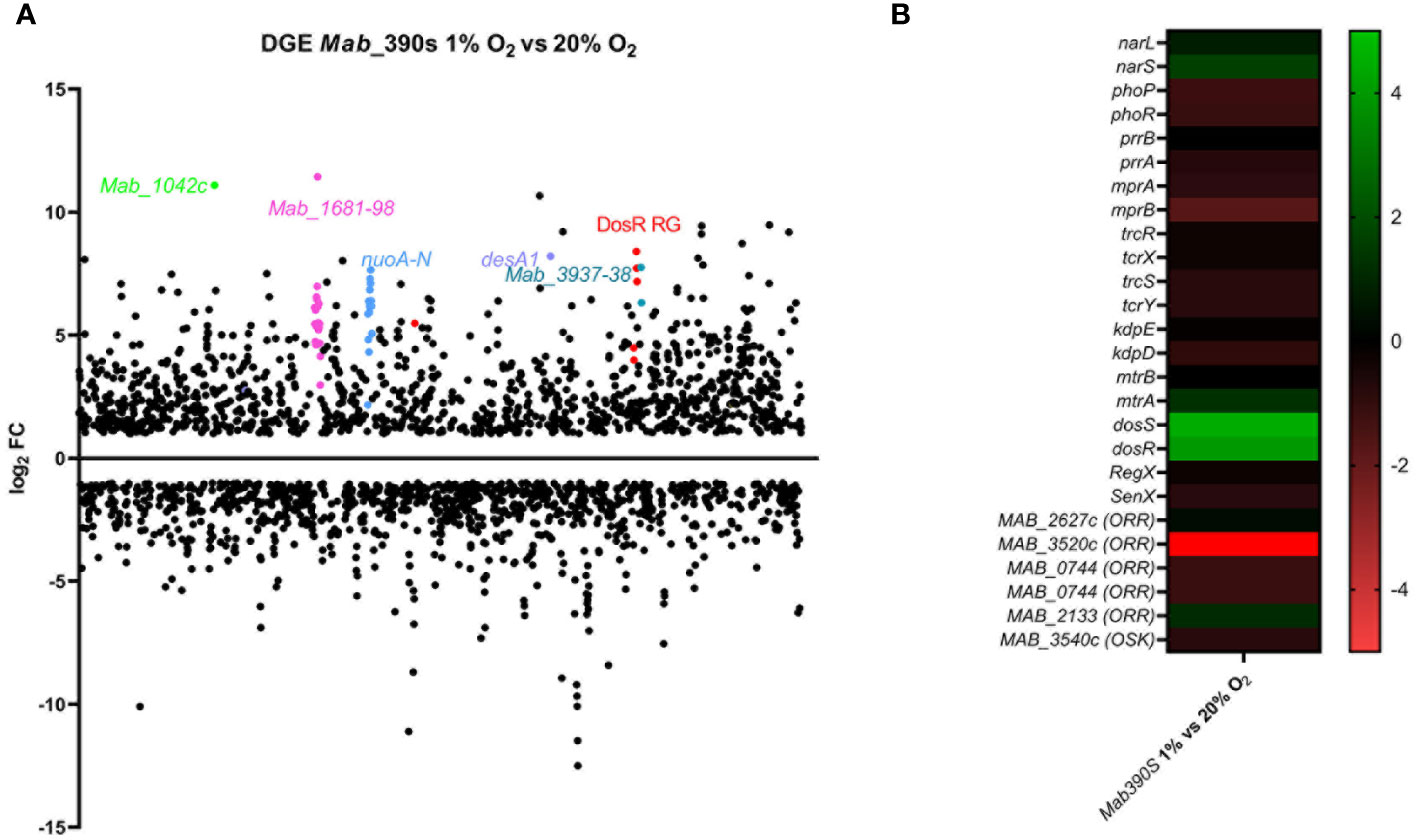
Figure 1 Transcriptome analysis of Mab390S in defined hypoxia model. DGE is visualized as (A) scatter plot depicting changes in gene expression reported as TPM with ≥ or ≤ log2FC on Day 5 for Mab 390S 1% O2 vs Mab 390S 20% O2. Dots represent individual genes (neon green=Mab_1042C, pink=Mab_1681-1698, blue=nuo operon, purple= desA1, turquoise=Mab_3937 & 3938, red= predicted DosR regulated genes (RG). (B) Heat map showing DGE of TCS of Mab 390S in 1% O2 vs 20% O2. In gene names, orphan response regulators and orphan sensor kinases are denoted as ORR and OSK, respectively.
To identify putative regulators of hypoxia adaptation in Mab, we assessed the differential expression of TCSs and other annotated transcription factors in the 1% hypoxia model. Of the 11 TCS orthologous to Mtb, only dosRS, mtrA, narS and the orphan RRs Mab_2133 and Mab_3520c displayed DGE (Figure 1B). Mab_dosRS exhibited the largest magnitude of gene induction with log2FC of 3.9 and 4.5, respectively, pointing to DosRSMab as an important TCS facilitating adaptation to hypoxic stress. The other TCS components were minimally induced with a log2FC of 1.6 for narS, 1.2 for mtrA and 1.1 for the orphan RR Mab_2133. The roles of narS, mtrA and Mab_2133 have not been determined in Mab. However, in the context of Mtb, the regulons of DosRS and NarLS TCS display partial overlap and protein-protein interactions between the RR from these two TCSs, DosR and NarL, have been detected (Malhotra et al., 2015). mtrA is essential in Mtb due its role in replication, whereas in Mab mtrA was reported to be non-essential, pointing to potential differences in the role of this TCS between the two species (Fol et al., 2006; Akusobi et al., 2022). The Mtb ortholog (Rv3143) of orphan RR Mab_2133 is implicated in nitrate metabolism in the absence of oxygen, binds to nuo subunits required for electron transport and is within nuo operon (Plocinska et al., 2022). Mab’s nuo operon displays synteny with the Mtb nuo operon including the orphan RR Mab_2133. Whereas Rv3143 was moderately induced by hypoxia in a DosR-dependent manner (Kendall et al., 2004), the upregulation of Mab_2133 in our hypoxia model was not altered in the absence of DosRS (Table S3). The only TCS gene displaying substantial downregulation was Mab_3520c (log2FC = -9.7). The Mtb ortholog of Mab_3520c, Rv0260c, is known to be upregulated in hypoxia and to interact with DosS via protein-protein interaction independent of DosR; however, the function of Rv0260c has not been identified (Gautam et al., 2019; Vilcheze et al., 2022). The opposite pattern of regulation of Rv0260c (induced) and Mab_3520c (repressed) in hypoxia implies they are utilized differently for adaptation to hypoxia. Thus, our transcriptomic analyses of Mab under physiologic hypoxia conditions point to Mab DosRS as an important TCS aiding in adaptation to hypoxia.
In addition to induction of the Mab_dosRS TCS, we also observed upregulation of 80 single component transcription factors (TF) in hypoxia (Table S3). Due to the high number of TF, only the genes with a log2FC ≥ 3 are included in Table S4 with the most highly induced genes being Mab_4180 (lclR), Mab_2606c (TetR family), Mab_4332 (TetR family) and Mab_3018 (GntR family). The Mtb orthologs for Mab_4332 (Rv0273c) and Mab_3018 (Rv0586) have 64.14% and 45.53% identity, respectively. Rv0273c has been identified as a regulator of inhA, an enoyl-ACP reductase involved in mycolic acid synthesis, and Rv0586 is known to mediate lipid metabolism in Mtb (Santangelo Mde et al., 2009; Yousuf et al., 2018; Zhu et al., 2018). Both Mab_4180 and Mab_2606c share less than 30% sequence homology with the Mtb orthologs and no known function has been identified. Although few Mab transcriptional regulators have been characterized, the large number of regulators with altered expression under hypoxic stress are likely key nodes in the regulatory networks needed to adapt in vivo.
Due to the number of TFs and their magnitude of modulation in response to hypoxia, including upregulation of Mab_dosRS, the broad scope of transcriptional changes was not surprising. The list of hypoxia-induced genes included loci involved in fatty acid and cholesterol metabolism, components of the NADH-quinone oxidoreductase subunits (nuoA-N), 6 epoxide hydrolases (ephD), ATP synthase subunits, 5 mammalian cell entry operons (MCE), members of the glycopeptidolipid locus (GPL), and 520 hypothetical genes (Tables S3, 4). Several pathways critical for pathogenesis of Mtb, such as fatty acid and cholesterol metabolism, are also induced in various hypoxia models (Wayne model, 1% O2) and within granulomas (Wilburn et al., 2018; Yang et al., 2021; Vilcheze et al., 2022). All four MCE loci in Mtb are differentially expressed in response to hypoxia, suggesting an important role for this family of lipid/cholesterol transporters in adaptation to this stress. While mce2 and mce3 were induced in a hypoxia model similar to ours (1% O2, 5 days), mce1 was repressed, and mce4 expression remained unchanged (Vilcheze et al., 2022). A separate study by Rathor et al. found that mce4 was upregulated after much longer durations of hypoxia stress (Rathor et al., 2016). Mce1 and Mce4 are known to play a role in the transport of fatty acids and cholesterol (Wilburn et al., 2018; Klepp et al., 2022), whereas the function of Mce2 and Mce3 have not been determined yet. In contrast, five of the seven MCE systems encoded by Mab were upregulated in 1% O2 after 5 days (Tables S3, 4). Although the biological role of MCE complexes in Mab have not been studied, their distinct expression profile suggest they may be important for in vivo survival. The Mab hypoxia-induced gene set also included a large number of genes implicated in β-oxidation pathways - 14 fadE genes, 8 fadD genes, and one of each fadA and fadH (Tables S3, 4). Upregulation of 6 ephD genes (an epoxide hydrolase predicted to alter the amount of epoxymycolates in the cell wall), members of the GPL locus accounting for smooth morphology (Tables S3, 4), and arabinosyltransferases A and B (Supplemental Table 1) involved with arabinogalactan synthesis indicate Mab may undergo cell wall remodeling in response to hypoxia (Amin et al., 2008; Gutierrez et al., 2018; Madacki et al., 2018).
In addition to β-oxidation, metabolic pathway induction included nuo subunits A-N, ATPase subunits, and cytochrome P450 genes (Tables S3, 4). NuoA-N are subunits of the proton pumping NADH dehydrogenase type 1 responsible for transferring electrons to menaquinone in the electron transport chain (ETC) in an energy conserving manner to generate a PMF (Weinstein et al., 2005). Although these genes are upregulated in the RGM Mycobacterium smegmatis (Msmeg) during slowed growth and in E.coli in anaerobic conditions, this is not a feature observed in the hypoxic response of slow-growing mycobacteria (SGM) like Mtb (Unden and Bongaerts, 1997; Berney and Cook, 2010). In contrast to Mab, the nuo operon and ATPase subunits are downregulated in hypoxic Mtb, further highlighting the distinct stress responses and energy metabolism of these two species in response to hypoxia (Cook et al., 2014; Vilcheze et al., 2022). Of the 25 Mab cytochrome P450s, 14 were induced in hypoxia (Supplemental Table 1). This data is consistent with hypoxic induction a large number of cytochrome P450s in the RGM Msmeg but not in the SGM Mtb with the exception of only 2 cytochrome P450s (Sherman et al., 2001; Berney and Cook, 2010; Ortega Ugalde et al., 2019). The roles of individual Mab cytochrome P450s remain unknown but the functions of the Mtb orthologs are dependent on their ferredoxin redox partners and include cholesterol degradation, redox balance, and virulence (Capyk et al., 2009). Our data supports hypoxic induction of 2 ferredoxins (Mab_0914c & Mab_2049c) and 3 ferredoxin reductases (Mab_0930, Mab_2047c and Mab_4356c) (Table S3). Induction of nuoA-N, ATPAse subunits, the large number of cytochrome P450s and ferredoxins implies Mab may employ different sets of genes for anaerobic respiration in its response to hypoxia and adapation.
Mab’s transcriptional adaptation to hypoxia also comprised a large set of downregulated genes including but not limited to multiple TF, tRNAs, 30S and 50S ribosomal proteins, alternative sigma factors, and 431 hypothetical proteins (Table S3). Downregulation of genes involved in essential processes such as protein synthesis (e.g. tRNAs, ribosomal proteins and sigma factors) are consistent with the slowed growth observed in hypoxic Mab cultures. Included among the most downregulated genes in hypoxia (Table S3) is the orphan response regulator Mab_3520c (Figure 1B) and three adjacent upstream genes (nirD/MAB_3521c, nirB/MAB_3522c), and nark3/MAB_3523c) predicted to be involved in nitrite reduction and extrusion (Malm et al., 2009). Mtb nirB and nirD orthologs are reportedly induced in nutrient starvation but minimal to no DGE occurred in hypoxia at 1% O2 (Vilcheze et al., 2022). However, in the Wayne model of hypoxia Mtb nirB displayed induction and functional nirBD genes were required for growth in hypoxia when nitrite was used as the sole nitrogen source (Akhtar et al., 2013).
Elucidation of the global transcriptional responses of Mab to hypoxia for the first time revealed the DosRS TCS is employed during hypoxic adaptation, yet much remains unknown about the role of DosRS in gene regulation and Mab pathogenesis. The DosRMab regulon was previously predicted to consist of only 6 genes - Mab_3890 (dosS), Mab_3891 (dosR), Mab_2489 (universal stress protein, USP), Mab_3902c (ortholog of Rv2004c), Mab_3903 (nitroreductase) and Mab_3904 (USP) – based solely on bioinformatic analysis (Gerasimova et al., 2011). However, at the time this study was initiated, the role of DosRSMab signaling in gene regulation, including induction of this gene set, remained to be experimentally demonstrated. To enable determination of the DosRSMab regulon and role of this TCS in Mab adaptation to hypoxia, we generated MabΔdosRS(knockout mutant) using recombineering and the corresponding complemented strain (MabΔdosRS+C) expressing a single, integrated dosRS allele driven by its native promoter (van Kessel and Hatfull, 2007). In addition to confirming strain genotypes by PCR and DNA sequencing (data not shown), the absence of dosRS transcripts in MabΔdosRS and restoration to wild-type levels in MabΔdosRS+C was validated by qRT-PCR (Figure 2A). We next assessed transcript levels of 4 genes (in addition to dosRS operon itself) previously predicted to be DosR-dependent. Loss of a functional DosRS system resulted in down-regulation of all predicted DosRMab-regulated genes by >2 log (MAB_2489, MAB_3902c, MAB_3903) or > 1 log in the case of MAB_3904 with restoration to wild-type levels in the complemented strain (Figure 2B), consistent with DosR-mediated induction of these genes.
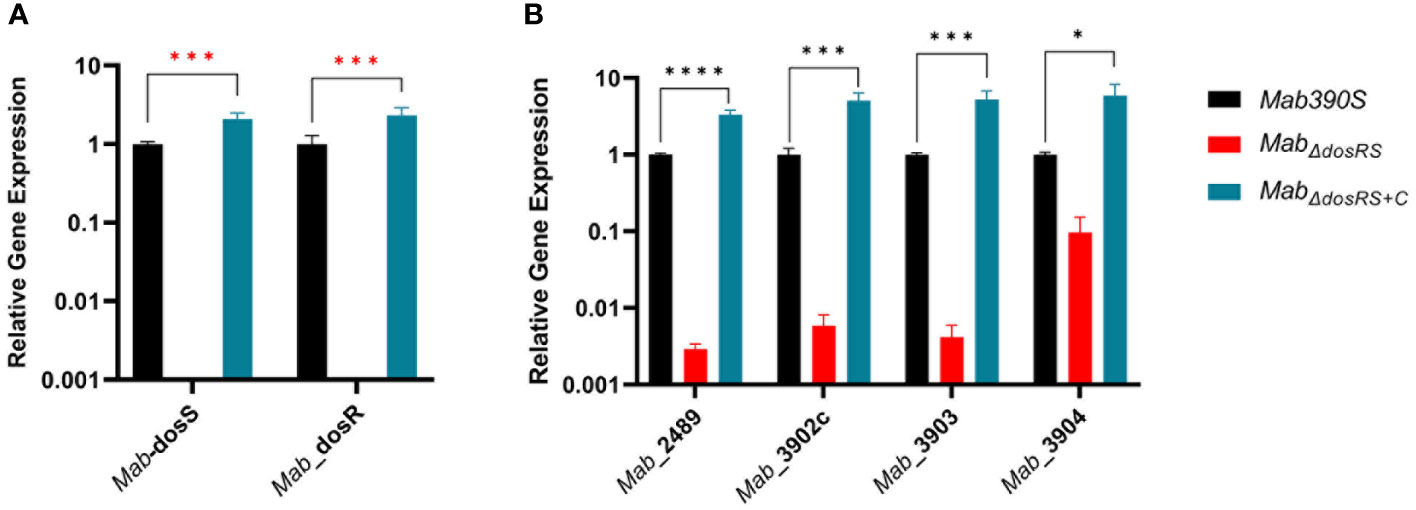
Figure 2 Validation of MabΔdosRS and MabΔdosRS+C. qRT-PCR was performed to (A) confirm the deletion and restoration of dosS and dosR in MabΔdosRS and MabΔdosRS+C, respectively, and (B) assess the effect on predicted downstream genes. Mab 390S (black), MabΔdosRS (red) and MabΔdosRS+C (blue). Data is representative of 3 experiments performed in triplicate. P values were calculated via one -way ANOVA using GraphPad. Red stars indicate Ct values were not detected for Mab dosR nor dosS in the mutant strain. *P-value <.05, ***P-value <.001 ****P-value <.0001.
As detailed above, to verify the role of DosRS under in vivo relevant conditions, we compared the growth kinetics assays of Mab 390S, MabΔdosRS, and MabΔdosRS+C in hypoxic (1% O2, standing) versus aerobic (20% O2, shaking) conditions. Strains were monitored over a 30-day period using CFU/ml as the readout at day 5, 20, and 30 and grown in normoxic conditions after plating. Cultures grown in 20% O2 reached maximum growth on day 5 with no difference in growth between strains at any time point confirming fully aerated cultures are not dependent on DosRS for replication (Figure 3A). At 1% O2 maximum growth was achieved by day 20 in Mab 390S with a ~2-log decrease in MabΔdosRSand ~log decrease in MabΔdosRS+C (Figure 3B) suggesting a functional DosRS is required for maximal growth and survival during hypoxic stress. By day 30 in 1% O2, Mab 390S displayed a slight decrease in CFU compared to day 20 however, this decline was also observed in fully aerated cultures indicating that hypoxic stress was not the cause (Figures 3A, B). These data support the conclusion that DosRSMab is necessary for growth in hypoxic environments that mimic the physiologic environments of the CF lung, within macrophages and granulomas (Worlitzsch et al., 2002; Cunningham-Bussel et al., 2013; Hudock et al., 2017). Unexpectedly, a morphotype transition from smooth to rough occurred in MabΔdosRS after pro-longed exposure to 1% O2. On day 20 and 30 an observable change in morphology occurred only in the mutant strain resulting in a heterogenous population of smooth and rough colonies (Figures 4A–D), indicating a DosR-dependent inducible alteration in cell wall composition for MabΔdosRS. This is corroborated by the fact that in hypoxic liquid culture assays, MabΔdosRS alone adopted a biofilm-like pellicle layer that was resistant to disruption, whereas other strains maintained a homogeneous composition (data not shown).
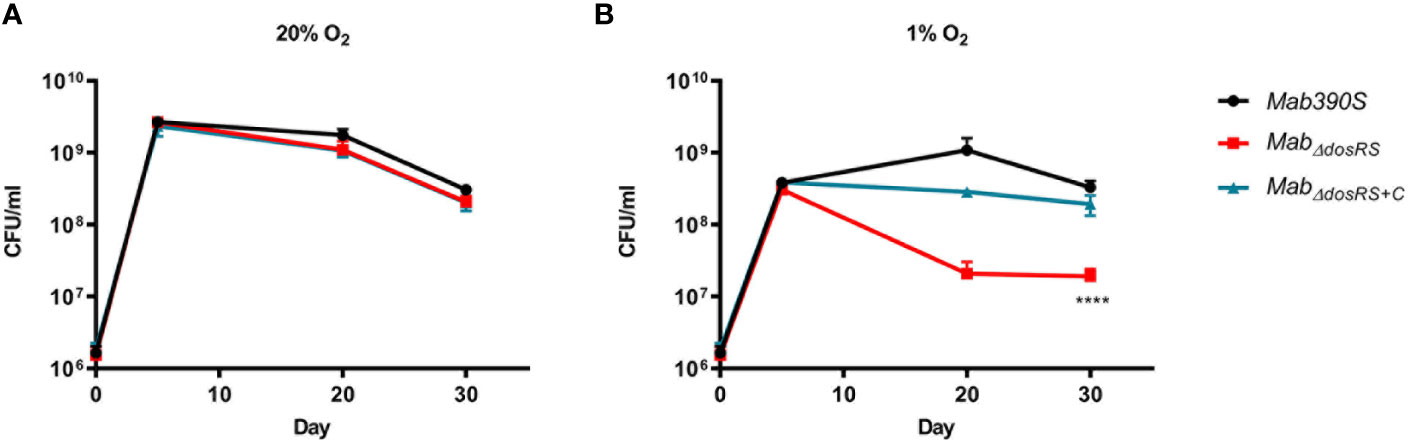
Figure 3 MabΔdosRS is attenuated in hypoxia. Growth kinetics in hypoxia were assessed via serial dilutions, spot plating and enumeration of CFU/ml on day 5, 20 and 30. (A) Growth kinetics at 20% O2 (B) Growth kinetics at 1% O2. Mab 390S (black), MabΔdosRS (red) and MabΔdosRS+C (blue). Data reflects 3 independent experiments performed in triplicate. P-values were calculated via one-way ANOVA using GraphPad, ****P-value <.0001.
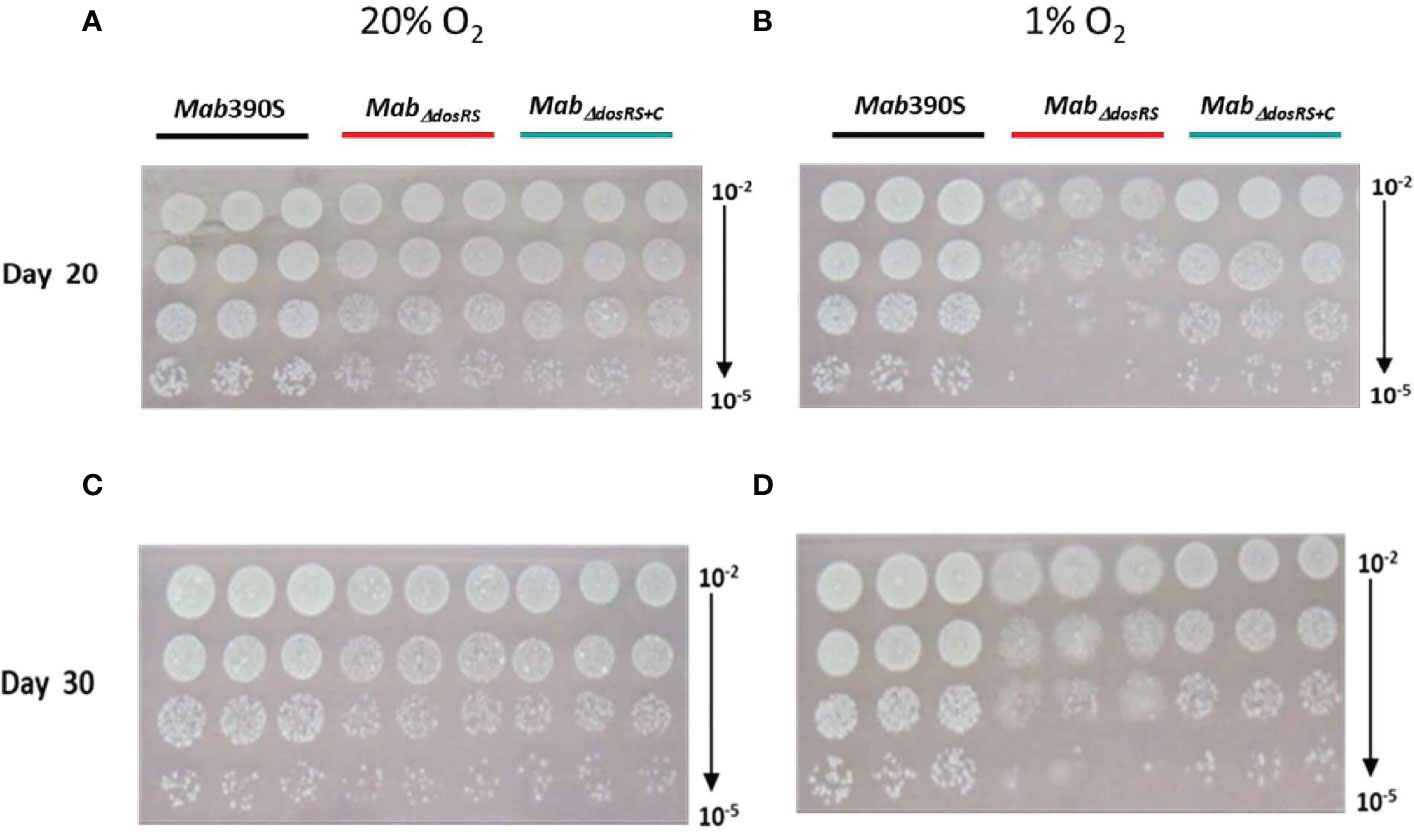
Figure 4 Hypoxia-induced morphological changes in MabΔdosRS. Cultures grown at 20% or 1% O2 were spot plated on Day 20 and Day 30 and incubated under normoxic conditions. (A) Day 20 at 20% O2 (B) Day 20 at 1% O2 (C) Day 30 at 20% O2 (D) Day 30 at 1% O2.
Prompted by the inability of MtbΔdosR to resuscitate after re-aeration from hypoxia (Leistikow et al., 2010; Veatch and Kaushal, 2018), we investigated the ability of MabΔdosRS to resuscitate after 30 days in hypoxia. Day 30 cultures taken from 20% O2 and 1% O2 were diluted to an OD600nm of 0.02 and grown in 20% O2 while shaking to evaluate the recovery of MabΔdosRS after prolonged exposure to hypoxia. OD600nm was taken over a 10-day period (Day 0, 2, 3, 5, 8, and 10) to monitor growth kinetics after re-introduction of O2. All strains originating from aerobic conditions displayed similar growth curves after re-culturing and reached maximal OD by day 5 (Figure 5A). In contrast, after being subjected to hypoxia for 30 days, MabΔdosRS displayed reduced growth compared to Mab 390S and MabΔdosRS+C taken from 1% O2 (Figure 5B). Attenuated growth in 1% O2 and the inability to resuscitate after re-aeration for MabΔdosRS supports a critical role for DosRSMab in mediating adaptation to changing oxygen levels encountered within the host during both dormancy and reactivation.
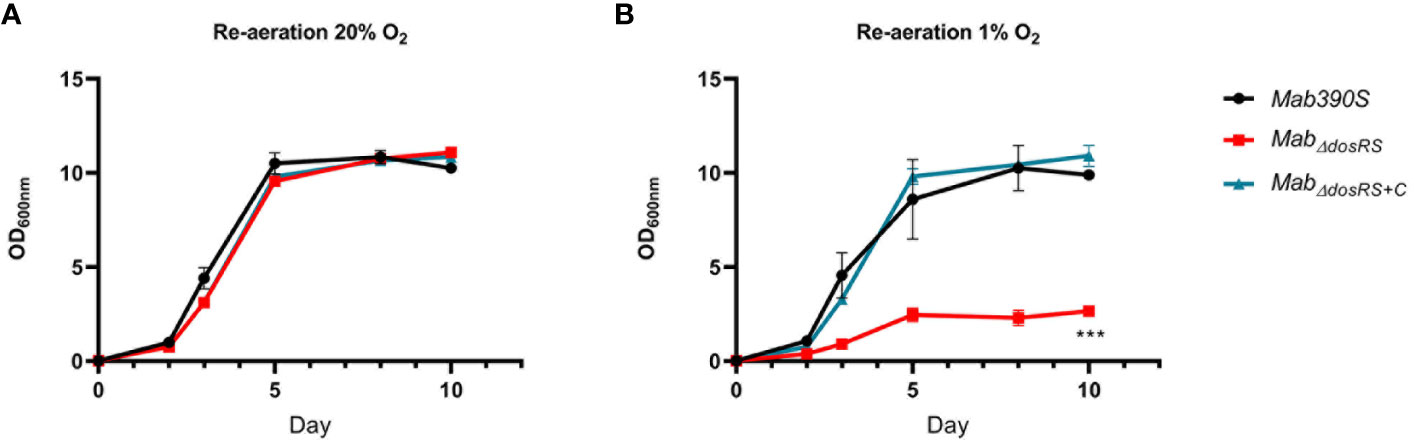
Figure 5 DosRS is required for resuscitation after hypoxia. OD600nm was taken over a 10-day period of re-aerated cultures after 30 days of growth in either (A) 20% O2 or (B) 1% O2. Mab390S (black), MabΔdosRS (red) and MabΔdosRS+C (blue). Data reflects 2 independent experiments performed in triplicate. P-values were calculated via one-way ANOVA using GraphPad, ***P-value <.001.
We next analyzed DGE between Mab390S and MabΔdosRS in hypoxia via RNAseq to experimentally identify DosRS regulated genes (Figure 6A). Cultures of Mab 390S and MabΔdosRS were grown at 1% O2 for 5 days at which point RNA was extracted from three independent experiments for analysis. In the absence of DosRS, 216 genes were expressed at lower levels relative to Mab 390S after exposure to 1% O2 (Figure 6A), of which 127 genes were also induced in Mab 390S by hypoxia (Table S3). This pattern is consistent with DosRS-dependent hypoxia induction, suggesting that Mab DosR may control a much larger regulon than previously predicted. In subsequent analyses, we defined putative DosRS-dependent hypoxia induced genes as those whose transcript levels were decreased by log2FC ≥ 1 in the MabΔdosRS in 1% O2 and were induced by log2FC ≥ 1 in Mab 390S 1% O2 (Table S3). The top 20 putative DosRS-dependent genes induced most highly by hypoxia (Table S5) include 4 of the genes previously predicted in silico to be members of the DosRSMab regulon. Notably, two of the most highly upregulated genes, Mab_3937 and Mab_3354 (desA1), appear to be Mab-specific members of this regulon. Mab_3937, a hypothetical protein with no known ortholog, is predicted to be in an operon with Mab_3938 and Mab_3939, encoding a clp protease subunit (ClpC2) with orthologous counterparts in Mtb and Msmeg which are essential genes. (Sassetti et al., 2003; Miranda-CasoLuengo et al., 2016; Kester et al., 2021). Both desA1 (Mab_3354) and desA2 (Mab_1237), desaturase enzymes with predicted roles in the biosynthesis of the mycolic acid component of mycobacterial cell walls, exhibited hypoxic DGE in the dosRS mutant (Yeruva et al., 2016; Bailo et al., 2022). Recently desA2 but not desA1 was predicted to be an essential gene in Mab, Mtb and Msmeg suggesting even slight downregulation could lead to detrimental alterations in the cell wall (Akusobi et al., 2022; Bailo et al., 2022). Although Mtb has orthologs of these genes (ClpC2, desA1, and desA2), there is no evidence of regulation by DosRMtb, illustrating the potential for conserved TCS to interact with conserved target genes in distinct ways. Additionally, no known DosRMtb-regulated genes have been deemed essential, further highlighting the unique nature of DosRSMab-mediated hypoxia response. Among the 127 hypoxia-induced putative DosRMab-dependent genes is a large gene cluster (Mab_1681-1698) containing hypothetical proteins, daunorubicin resistance efflux pump subunit (drrA), and a putative mce operon (Table S5). Whereas some putative Mab MCE gene clusters have clear orthologs in Mtb (e.g. MAB_4146c-4155c with Mce4, Rv3492c-3501c), the predicted Mab MCE proteins encoded by MAB_1681-1698 do not directly correspond to a specific MCE loci in Mtb. Rather, they share low homology with components of different Mtb MCE complexes, necessitating further research to fully assess the role of this Mab-specific MCE during hypoxia.
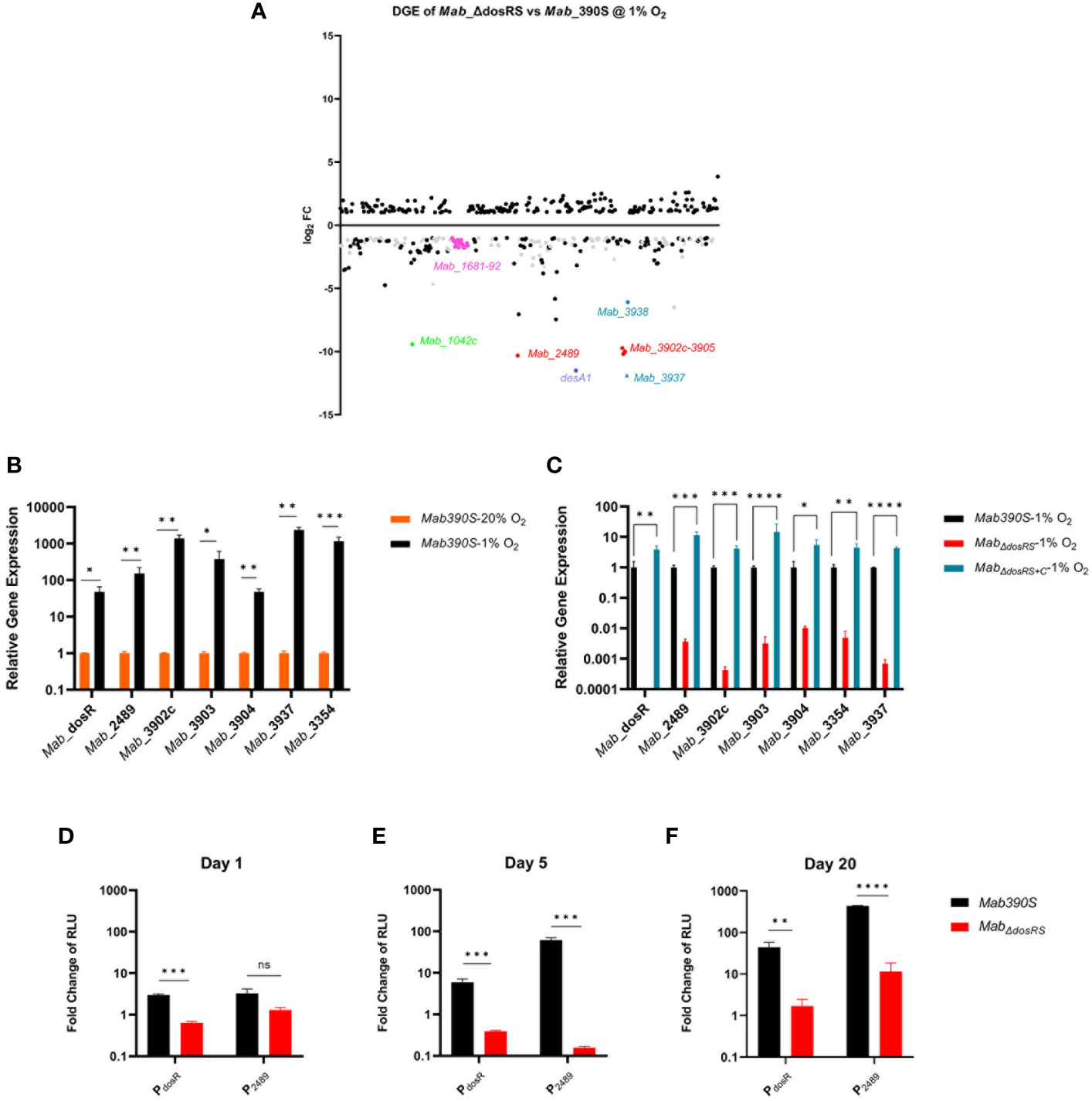
Figure 6 Mab DosR dependent DGE in 1% O2. (A) Scatter plot of RNAseq analysis of MabΔdosRS vs Mab 390S after 5 days in 1% O2. Figure shows genes with log2FC ≤ -1 and ≥ 1. Gray circles are conserved hypothetical genes and gray triangles are Mab genes with no ortholog in Mtb. Names of select genes of interest are labeled (neon green=Mab_1042C, pink=Mab_1681-1698, purple= desA1, turquoise (triangle denotes no known ortholog in Mtb) =Mab_3937 & 3938, red= predicted DosR regulated genes (RG). (B, C) qRT-PCR assessment of select DosR-dependent genes. Gene expression was calculated as relative quantitation via the 2-ΔΔCt method using sigA as the reference gene. (B) Hypoxia induced gene expression. Transcript levels of select genes in Mab 390S 20% O2 (orange bars) and Mab 390S 1% O2 (black bars). (C) DosR dependent gene expression. Transcript levels of select genes in MabΔdosRS (red bars) and MabΔdosRS+C(blue bars) were compared to Mab 390S (black bars) in 1% O2 on Day 5. (D-F) Kinetics of DosR-dependent gene induction measured luciferase reporter assays. Promoter activity in 1% O2 was quantified by measuring luminescence compared to 20% O2 cultures and normalized to lux-hsp60 constitutive promoter as a reference signal, (1%O2(PdosR or P2489/Phsp60))/(20% O2(PdosR or P2489/Phsp60)) and expressed as fold change of relative light units (RLU) (D) 24hours, (E) Day 5, and (F) Day 20. Mab 390S (black bars) and MabΔdosRS (red bars). qRT-PCR and luciferase assay data are representative of 3 independent assays performed in triplicate. P-values were calculated using t-test (B, D–F) and one-way ANOVA (C). P-values were calculated via GraphPad, *P-value <.05, ** P-value <.01, ***P-value <.001, ****P-value. Not significant is denoted as ns.
Comparing the ~ 50 genes of the DosRMtb regulon with the putative DosRMab regulated genes identified in this study (Supplemental Table 3), we only discovered 6 shared orthologs including 4 conserved hypothetical proteins (CHPs) plus DosR (Mab_3891c) and DosS (Mab_3890c). In addition to the transcriptional regulator Mab_3891c, we found nine other transcriptional regulators to be induced by hypoxia in a putative DosR-dependent manner (Tables S3–5) with none of their Mtb orthologs regulated by DosRMtb. The transcriptional response of Mab to hypoxia is further differentiated from Mtb by the lack of regulation of any of the 7 putative triacylglycerol synthases (Tgs) by hypoxia or DosR, a characteristic of in vitro dormancy and hypoxia for Mtb (Table S3) (Voskuil et al., 2004; Deb et al., 2009). Hypoxic Mtb positively regulates tgs1 via DosR for synthesis of triacylglycerol (TAG) for energy storage and utilization. The absence of DosRMab mediated induction of any Tgs enzymes under hypoxic stress points to a different mechanism for energy storage and utilization in Mab vs Mtb (Park et al., 2003; Daniel et al., 2004). The scope of the Mab DosR regulon precludes a comprehensive discussion of every downstream gene, many of which encode uncharacterized conserved hypothetical proteins. However, this RNAseq dataset is evidence for a species-specific regulon larger than predicted bioinformatically that likely contains novel mechanisms of hypoxia adaptation and pathogenesis.
Comparative transcriptomics also revealed upregulation of 200 genes in MabΔdosRS compared to Mab 390S (Figure 6A and Table S3), however the magnitude of induction was low ranging between log2FC of 1-3.6. Among the upregulated genes are 81 hypothetical proteins, 5 transcription factors, 30S and 50S ribosomal proteins. Included in the top 20 most highly induced genes in MabΔdosRS are Mab_3521c (nitrite reductase) and Mab_3523c (nitrite extrusion protein) which were observed to be highly downregulated in hypoxic Mab 390S (Table S3). These data suggests that DosRS may act as a repressor of a subset of genes in hypoxic conditions, a hypothesis that remains to be experimentally validated. The low-level induction of other genes in the knockout strain may also result from indirect effects of DosRS on mycobacterial physiology under hypoxic stress.
To validate RNAseq results, two of the putative Mab DosR-dependent genes most differentially expressed in the mutant strain (Mab_3354 and Mab_3937) were assessed via qRT-PCR along with 5 of the predicted Mab DosR genes (Figures 6B, C). Similar to RNAseq studies, the effect of hypoxia on gene expression was assessed in Mab 390S (1% vs 20% O2) and the requirement for DosRS for DGE in response to hypoxia by comparing Mab 390S, MabΔdosRS and MabΔdosRS+C. All genes assessed via qRT-PCR were induced by ≥ 10-fold in 1% O2 compared to 20% O2 for Mab 390S with Mab_3937, Mab_3902c, and Mab_3354 having the highest gene induction consistent with RNAseq results (Figure 6B). These same genes displayed loss of induction in MabΔdosRS by ≥ 10-fold similar to RNAseq results with restoration by MabΔdosRS+C in hypoxia (Figure 6C). The most dramatic change in gene expression in MabΔdosRS was Mab_3902c and Mab_3937 with > 100-fold reduction compared to Mab 390S. Results from qRT-PCR corroborate RNAseq results and accentuate the magnitude of differential gene expression of predicted and newly discovered hypoxia-induced DosRSMab regulated genes.
In addition to qRT-PCR, bacterial luciferase (Lux) reporter strains were used to evaluate the kinetics of hypoxia-dependent changes in gene expression across a broader time course. The integrating shuttle plasmid pMV306 luxG13 optimized for mycobacteria consists of the constitutive Phsp60 and PG13 promoters driving expression of luxAB and luxCDE, respectively (Andreu et al., 2010). Lux reporter constructs in which the constitutive Phsp60 promoter was replaced with promoters from two hypoxia inducible genes, DosR and Mab_2489 (PDosR and P2489) were introduced into Mab 390S and MabΔdosRS. Reporter assays performed on Days 1, 5, 20 identified temporal changes in gene expression with both promoters for Mab 390S but not in MabΔdosRS (Figures 6D–F). The lux reporter under the control of PdosR shows sustained induction over time, indicating that dosR is expressed throughout early and late stages of hypoxia, a trait not observed in Mtb (Rustad et al., 2008). Although the Mab 390S P2489-lux reporter displays modest induction of ~ 3-fold change on day 1, there is not a significant difference compared to MabΔdosRSP2489 (Figure 6D). However, on days 5 and 20 Mab 390S P2489-lux was highly induced compared to MabΔdosRS P2489-lux with fold changes of 61 and 433, respectively (Figures 6E, F). It should be noted that MabΔdosRS P2489-lux did exhibit low-level induction on Day 20, which may be attributable to the activity of other transcription factors. However, as noted previously, there was a significant difference compared to the expression of P2489 in wild-type Mab 390S. Lux reporters facilitated dynamic monitoring of DosRS activation by hypoxia and provide a valuable tool to explore DosR-mediated gene regulation in vitro and in vivo in response to various stresses (e.g. NO, CO, antibiotics) or host microenvironments (e.g intramacrophage, granuloma, airway mucus).
Infections caused by Mab, particularly within the CF population, are a major cause of concern due to the lack of efficacious antibiotics and the resulting inability to clear the infections from the airways. The poor correlation between in vitro drug susceptibility profiles and in vivo efficacy when treating Mab infections suggest that host-driven adaptations of Mab may contribute to treatment failures (Nessar et al., 2012). Host-derived cues encountered by Mab within the viscous mucus layer of CF airways, phagosomal compartments of macrophages, and during residence within granulomas may trigger upregulation of antimicrobial resistance mechanisms (Lyczak et al., 2002; Worlitzsch et al., 2002; Cunningham-Bussel et al., 2013; Hudock et al., 2017). Extrapolating from studies on Mtb (Gold and Nathan, 2017; Boldrin et al., 2020; Joshi et al., 2021), in vivo stresses such as hypoxia may also promote the development of phenotypically drug-tolerant persisters. Thus, a better understanding of Mab’s physiological states and stress responses required for long-term persistence within the human host may lead to more effective treatment strategies.
Successful bacterial pathogens like Mtb and Mab employ extensive repertoires of transcription factors, including TCS, for coordinating gene expression to counteract host antimicrobial factors and immune pressure. The transcriptional regulatory networks and role of TCS of Mtb have been extensively characterized in multiple in vitro and in vivo models of infection (Bacon and Marsh, 2007; Rohde et al., 2012; Li et al., 2019; Stupar et al., 2022; Vilcheze et al., 2022). In contrast, few transcriptomic studies defining Mab stress responses, or the role of specific transcription factors have been reported (Miranda-CasoLuengo et al., 2016; Dubois et al., 2019). Given the well-documented relevance of hypoxia during Mab-host interactions (e.g. mucus of CF airway, macrophage phagosome, granuloma) (Lyczak et al., 2002; Worlitzsch et al., 2002; Cunningham-Bussel et al., 2013; Hudock et al., 2017), we sought to identify molecular mechanisms that enable Mab to adapt to these low-oxygen niches. In the better characterized pathogen Mtb, the master regulator of hypoxia adaptation is the atypical TCS DosRS/T, which regulates a ~50 gene regulon upon induction by hypoxia and NO stress (Park et al., 2003; Voskuil et al., 2003). Mab encodes orthologs of eleven of the twelve TCS found in Mtb, including DosRS (dosT homolog missing). However, as detailed in this study, the scope and content of Mab regulons controlled by these TCS may be less conserved. Prior to initiation of our study, only two reports had mentioned Mab DosRS: i) a bioinformatics study predicting a 6 gene regulon based on previously known Mtb DosR binding motifs (Gerasimova et al., 2011) and ii) transcriptomic study assessing the affect of NO exposure on the predicted genes (Miranda-CasoLuengo et al., 2016). These studies, however, did not directly demonstrate DosRS-mediated regulation of the predicted genes, define the transcriptional response of Mab to hypoxia nor the full extent of the DosR regulon, or identify a DosRS phenotype. To begin to address these knowledge gaps we developed a hypoxic model of 1% O2 to mimic physiologically relevant oxygen tensions Mab encounters in vivo (Worlitzsch et al., 2002; Cunningham-Bussel et al., 2013; Hudock et al., 2017) to assess transcriptomics and growth kinetics of Mab in the presence or absence of DosRSMab.
Genome wide transcriptomics identified DosRS as the main TCS activated during hypoxia and analysis of a defined mutant lacking DosRS revealed a potentially larger regulon than previously predicted. We identified 216 genes to be downregulated in MabΔdosRSversus Mab 390S, 127 of which were upregulated in hypoxia. This gene set was deemed the putative hypoxia-induced DosR regulon-Mab_3902c, Mab_3903 and Mab_3904), 2 novel genes displaying the highest DGE (MAB_3937 and desA1), 9 transcription factors, and 57 hypothetical genes among others. 22 of the 57 hypothetical genes are species-specific, further illustrating the unique nature of the regulons controlled by orthologous TCS. Not only is the Mab DosRS regulon likely larger than previously predicted, but it also appears to be notably larger than the well-studied Mtb DosR regulon. Surprisingly, the only orthologs in common between the Mtb DosR and Mab DosR regulons were the 6 genes originally predicted from the bioinformatics study and Mab_1040, an ortholog of the hypothetical protein Rv3129 which is documented as an antigen in tuberculosis patients with latent infections (Park et al., 2003; Black et al., 2009; Lin et al., 2009). One of the hallmarks of Mtb hypoxia adaptations in vivo and in vitro is the marked upregulation of tgs1 for energy storage and utilization (Garton et al., 2008; Deb et al., 2009). The induction of Mab_3551c, the primary TAG synthase gene in Mab (Viljoen et al., 2016), is not observed in our Mab in vitro hypoxia model, suggesting Mab mechanisms of hypoxia adaptation or cues for regulation of lipid storage are distinctive from Mtb. Studies are underway to discriminate between genes whose expression is altered directly by DosR (DosR binding to promoter) versus indirectly (promoter regulated by secondary TF).
During the course of our study, Belardinelli et al. also reported on characterization of the Mab DosR regulon as part of efforts to repurpose antimalarial drugs that inhibit Mtb DosR as therapeutics for Mab (Belardinelli et al., 2022). Their transcriptomic comparison of Mab ATCC 19977 and MabΔdosRS in microaerobic conditions identified 180 genes downregulated in a DosRS-dependent manner. Of these 180 genes, only 45 overlapped with our list of 216 genes downregulated in the dosRS mutant at 1% O2. Both studies included the 6 previously predicted genes and the 2 genes most highly differentially expressed on our list (Mab_3937, desA1) plus 37 other genes. Of the 45 genes in common between these 2 studies, Belardinelli et al., reported 38 DosR binding motifs supporting the assertion that Mab DosR regulon is larger than previously predicted. Discrepant findings between this report and our study could be attributable to differences in strains (ATCC 19977 vs 390S), hypoxic models (20% O2 standing vs 1% O2 standing), time points (24hr vs 5 days), or media (Dubos-Tween albumin vs 7H9-OADC+.05% tyloxapol). Regardless, both clearly highlight the broad scope and unique nature of the DosRSMab regulon and provide a framework for future studies to fully elucidate the role of this important two-component system.
The importance of DosR-regulated genes for hypoxia adaptation was evident from growth deficits seen on day 20 and 30 and impaired resuscitation after reaeration in MabΔdosRS compared to Mab 390S (Figure 3). Mab_ΔdosRS differentially expressed genes in hypoxia contain 7 genes predicted to be essential in a recent TnSeq study under aerobic conditions (Akusobi et al., 2022), possibly accounting for these phenotypes. Included in this list is desA2, a desaturase enzyme that is responsible for mycolic acid biosynthesis and is essential for growth in the RGM Msmeg (Bailo et al., 2022). Strong induction of desA1 in hypoxia suggests that it, along with desA2, may play role in cell wall modification in response to this stress. It is worth noting that, despite exclusion from the list of Mab predicted essential genes, the orthologous desaturase in Msmeg was deemed essential (Singh et al., 2016). Additionally, the MCE operon Mab_1693-Mab_1698 was differentially expressed in the mutant strain and may contribute to decreased importation of mycolic acids further disrupting cell wall integrity. The 6 other predicted essential genes possibly contributing to the MabΔdosRS growth phenotype are 2 conserved hypothetical proteins (Mab_3268c-Mab_3269c), a DNA helicase (Mab_3511c), a protoporphyrinogen oxidase, a prephenate dehydratase (Mab_0132), and phosphoribosylformylglycinamidine synthase (Mab_0698).
In addition to growth/survival deficits and the inability to resuscitate, we also observed DosRS-dependent hypoxia-induced morphological changes. After 20 days in hypoxia, MabΔdosRS displayed heterogeneous morphology consisting of smooth and rough colonies. This phenotype was not present in strains expressing DosRS or in fully aerated cultures, evidence that this TCS mediates dramatic remodeling of the cell wall in response hypoxia. The smooth and rough morphotypes of Mab, reflective of different compositions of the outer cell wall, have been shown to impact interactions with macrophages, immune stimulation and inflammation, antibiotic susceptibility, and virulence (Jonsson et al., 2007; Catherinot et al., 2009; Ruger et al., 2014; Roux et al., 2016; Li et al., 2020). Rough strains are able to trigger apoptosis of macrophages and grow extracellularly as aggregates known as cords, and are associated with worse clinical outcomes (Li et al., 2020) Mechanisms involved in smooth to rough transitions have not been fully elucidated. However, genomic comparisons between the two morphotypes revealed SNPs and indels in the gpl locus and in mmpl4b and mps1 genes (Pawlik et al., 2013). In addition to total or partial loss of GPL due to mutations affecting its biosynthesis or transport, recent studies including identification of GPL+ rough clinical isolates suggest other mechanisms may also govern S➔R morphotype switching (Gutierrez et al., 2021). For example, Daher et al. reported that glycosylation patterns of GPL can alter Mab surface properties (Daher et al., 2022). An inducible transition from smooth to rough was also observed following exposure to subinhibitory doses of aminoglycoside antibiotics providing evidence for transcriptional modulation of morphotype in response to stress (Tsai et al., 2015). Belardinelli et al. reported no differences in GPL content between Mab ATCC 19977 and isogenic ΔdosRS mutant after microaerobic culture for 24 hours (Belardinelli et al., 2022). Our observation of a switch to rough morphotype in a ΔdosRS mutant after extended culture at 1% O2 may reflect either adaptive cell wall remodeling triggered by lower O2 levels or longer duration of stress in our model. Alternatively, rather than affecting GPL levels per se, inactivation of the DosRS regulon may impact GPL modifications or biosynthesis of unique cell wall constituents. Intriguingly, Pawlik et al. reported that expression of dosR was elevated in an R versus S strain (Pawlik et al., 2013). This seems to contrast with our data suggesting that DosRS positively regulates GPLs, or at least the smooth phenotype (e.g. loss of DosRS➔rough phenotype in hypoxia). Whether this is a direct correlation or stress induced side-effect stemming from the loss of GPL remains to be determined. It is clear that much remains to be learned regarding how Mab regulates the composition of its complex cell wall during infection and the roles of TCS like DosRS in host-pathogen interactions.
In addition to determination of the DosRS-dependent component of Mab hypoxia adaptation, this is the first transcriptomics study designed to identify genome-wide changes in Mab gene expression in a defined, physiologically relevant model of hypoxia. RNAseq analysis of wild-type Mab390S in 1% O2 versus 20% O2 identified an additional 1,063 DosRS-independent hypoxia-induced genes with putative roles in Mab adaptation in hypoxia. Differential gene expression of such a large group of genes in hypoxia outside of the DosR regulon points to a sophisticated mechanism of regulation for adaptation beyond TCS. This gene set included 80 TF, lipid metabolism and transport, energetics, secondary metabolism, cell wall synthesis plus induction of 540 hypothetical proteins, (Table S3). Our data highlights the necessity of adaptation to hypoxia via a large repertoire of genes including but not limited to the TCS DosRS. Further investigation of unique Mab DosR regulated genes and species-specific Mab genes employed for hypoxic adaptation will provide beneficial insights into Mab pathogenesis.
The datasets presented in this study are deposited in the NCBI database, accession number PRJNA932814 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA932814).
BS and KR: Conception and design of experiments. BS and BT: Analysis and interpretation of data for RNAseq experiments. BS and KR: Preparation and revision of manuscript. KR and LS: Approval of final manuscript. All authors contributed to the article and approved the submitted version.
LS acknowledges funding from the National Health Institute grant AI124458.
We would like to thank George Walters-Marrah for assistance with construction of luciferase reporter plasmids. We also thank Dr. Rushmi Gupta and Dr. Mollie Jewett for technical support and guidance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1144210/full#supplementary-material
Aguilar-Ayala, D. A., Tilleman, L., Van Nieuwerburgh, F., Deforce, D., Palomino, J. C., Vandamme, P., et al. (2017). The transcriptome of mycobacterium tuberculosis in a lipid-rich dormancy model through RNAseq analysis. Sci. Rep. 7 (1), 17665. doi: 10.1038/s41598-017-17751-x
Akhtar, S., Khan, A., Sohaskey, C. D., Jagannath, C., Sarkar, D. (2013). Nitrite reductase NirBD is induced and plays an important role during in vitro dormancy of mycobacterium tuberculosis. J. Bacteriol 195 (20), 4592–4599. doi: 10.1128/JB.00698-13
Akusobi, C., Benghomari, B. S., Zhu, J., Wolf, I. D., Singhvi, S., Dulberger, C. L., et al. (2022). Transposon mutagenesis in mycobacterium abscessus identifies an essential penicillin-binding protein involved in septal peptidoglycan synthesis and antibiotic sensitivity. Elife 11. doi: 10.7554/eLife.71947.sa2
Amin, A. G., Goude, R., Shi, L., Zhang, J., Chatterjee, D., Parish, T. (2008). EmbA is an essential arabinosyltransferase in mycobacterium tuberculosis. Microbiol. (Reading) 154 (Pt 1), 240–248. doi: 10.1099/mic.0.2007/012153-0
Andreu, N., Zelmer, A., Fletcher, T., Elkington, P. T., Ward, T. H., Ripoll, J., et al. (2010). Optimisation of bioluminescent reporters for use with mycobacteria. PloS One 5 (5), e10777. doi: 10.1371/journal.pone.0010777
Bacon, J., Marsh, P. D. (2007). Transcriptional responses of mycobacterium tuberculosis exposed to adverse conditions in vitro. Curr. Mol. Med. 7 (3), 277–286. doi: 10.2174/156652407780598566
Bailo, R., Radhakrishnan, A., Singh, A., Nakaya, M., Fujiwara, N., Bhatt, A. (2022). The mycobacterial desaturase DesA2 is associated with mycolic acid biosynthesis. Sci. Rep. 12 (1), 6943. doi: 10.1038/s41598-022-10589-y
Belardinelli, J. M., Verma, D., Li, W., Avanzi, C., Wiersma, C. J., Williams, J. T., et al. (2022). Therapeutic efficacy of antimalarial drugs targeting DosRS signaling in mycobacterium abscessus. Sci. Transl. Med. 14 (633), eabj3860. doi: 10.1126/scitranslmed.abj3860
Berney, M., Cook, G. M. (2010). Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PloS One 5 (1), e8614. doi: 10.1371/journal.pone.0008614
Bernut, A., Nguyen-Chi, M., Halloum, I., Herrmann, J. L., Lutfalla, G., Kremer, L. (2016). Mycobacterium abscessus-induced granuloma formation is strictly dependent on TNF signaling and neutrophil trafficking. PloS Pathog. 12 (11), e1005986. doi: 10.1371/journal.ppat.1005986
Black, G. F., Thiel, B. A., Ota, M. O., Parida, S. K., Adegbola, R., Boom, W. H., et al. (2009). Immunogenicity of novel DosR regulon-encoded candidate antigens of mycobacterium tuberculosis in three high-burden populations in Africa. Clin. Vaccine Immunol. 16 (8), 1203–1212. doi: 10.1128/CVI.00111-09
Boldrin, F., Provvedi, R., Cioetto Mazzabo, L., Segafreddo, G., Manganelli, R. (2020). Tolerance and persistence to drugs: A main challenge in the fight against mycobacterium tuberculosis. Front. Microbiol. 11, 1924. doi: 10.3389/fmicb.2020.01924
Bretl, D. J., Demetriadou, C., Zahrt, T. C. (2011). Adaptation to environmental stimuli within the host: Two-component signal transduction systems of mycobacterium tuberculosis. Microbiol. Mol. Biol. Rev. 75 (4), 566–582. doi: 10.1128/MMBR.05004-11
Brown-Elliott, B. A., Wallace, R. J., Jr. (2002). Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15 (4), 716–746. doi: 10.1128/CMR.15.4.716-746.2002
Byrd, T. F., Lyons, C. R. (1999). Preliminary characterization of a mycobacterium abscessus mutant in human and murine models of infection. Infect. Immun. 67 (9), 4700–4707. doi: 10.1128/IAI.67.9.4700-4707.1999
Capyk, J. K., Kalscheuer, R., Stewart, G. R., Liu, J., Kwon, H., Zhao, R., et al. (2009). Mycobacterial cytochrome p450 125 (cyp125) catalyzes the terminal hydroxylation of c27 steroids. J. Biol. Chem. 284 (51), 35534–35542. doi: 10.1074/jbc.M109.072132
Catherinot, E., Roux, A. L., Macheras, E., Hubert, D., Matmar, M., Dannhoffer, L., et al. (2009). Acute respiratory failure involving an r variant of mycobacterium abscessus. J. Clin. Microbiol. 47 (1), 271–274. doi: 10.1128/JCM.01478-08
Chmiel, J. F., Davis, P. B. (2003). State of the art: Why do the lungs of patients with cystic fibrosis become infected and why can't they clear the infection? Respir. Res. 4, 8. doi: 10.1186/1465-9921-4-8
Converse, P. J., Karakousis, P. C., Klinkenberg, L. G., Kesavan, A. K., Ly, L. H., Allen, S. S., et al. (2009). Role of the dosR-dosS two-component regulatory system in mycobacterium tuberculosis virulence in three animal models. Infect. Immun. 77 (3), 1230–1237. doi: 10.1128/IAI.01117-08
Cook, G. M., Hards, K., Vilcheze, C., Hartman, T., Berney, M. (2014). Energetics of respiration and oxidative phosphorylation in mycobacteria. Microbiol. Spectr. 2 (3). doi: 10.1128/9781555818845.ch20
Cunningham, A. F., Spreadbury, C. L. (1998). Mycobacterial stationary phase induced by low oxygen tension: Cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J. Bacteriol 180 (4), 801–808. doi: 10.1128/JB.180.4.801-808.1998
Cunningham-Bussel, A., Zhang, T., Nathan, C. F. (2013). Nitrite produced by mycobacterium tuberculosis in human macrophages in physiologic oxygen impacts bacterial ATP consumption and gene expression. Proc. Natl. Acad. Sci. U.S.A. 110 (45), E4256–E4265. doi: 10.1073/pnas.1316894110
Daher, W., Leclercq, L. D., Johansen, M. D., Hamela, C., Karam, J., Trivelli, X., et al. (2022). Glycopeptidolipid glycosylation controls surface properties and pathogenicity in mycobacterium abscessus. Cell Chem. Biol. 29 (5), 910–924 e917. doi: 10.1016/j.chembiol.2022.03.008
Daniel, J., Deb, C., Dubey, V. S., Sirakova, T. D., Abomoelak, B., Morbidoni, H. R., et al. (2004). Induction of a novel class of diacylglycerol acyltransferases and triacylglycerol accumulation in mycobacterium tuberculosis as it goes into a dormancy-like state in culture. J. Bacteriol 186 (15), 5017–5030. doi: 10.1128/JB.186.15.5017-5030.2004
Deb, C., Lee, C. M., Dubey, V. S., Daniel, J., Abomoelak, B., Sirakova, T. D., et al. (2009). A novel in vitro multiple-stress dormancy model for mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PloS One 4 (6), e6077. doi: 10.1371/journal.pone.0006077
Dubois, V., Pawlik, A., Bories, A., Le Moigne, V., Sismeiro, O., Legendre, R., et al. (2019). Mycobacterium abscessus virulence traits unraveled by transcriptomic profiling in amoeba and macrophages. PloS Pathog. 15 (11), e1008069. doi: 10.1371/journal.ppat.1008069
Esther, C. R., Jr., Esserman, D. A., Gilligan, P., Kerr, A., Noone, P. G. (2010). Chronic mycobacterium abscessus infection and lung function decline in cystic fibrosis. J. Cyst Fibros 9 (2), 117–123. doi: 10.1016/j.jcf.2009.12.001
Fol, M., Chauhan, A., Nair, N. K., Maloney, E., Moomey, M., Jagannath, C., et al. (2006). Modulation of mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol. Microbiol. 60 (3), 643–657. doi: 10.1111/j.1365-2958.2006.05137.x
Galagan, J. E., Minch, K., Peterson, M., Lyubetskaya, A., Azizi, E., Sweet, L., et al. (2013). The mycobacterium tuberculosis regulatory network and hypoxia. Nature 499 (7457), 178–183. doi: 10.1038/nature12337
Garton, N. J., Waddell, S. J., Sherratt, A. L., Lee, S. M., Smith, R. J., Senner, C., et al. (2008). Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PloS Med. 5 (4), e75. doi: 10.1371/journal.pmed.0050075
Gautam, U. S., McGillivray, A., Mehra, S., Didier, P. J., Midkiff, C. C., Kissee, R. S., et al. (2015a). DosS is required for the complete virulence of mycobacterium tuberculosis in mice with classical granulomatous lesions. Am. J. Respir. Cell Mol. Biol. 52 (6), 708–716. doi: 10.1165/rcmb.2014-0230OC
Gautam, U. S., Mehra, S., Kaushal, D. (2015b). In-vivo gene signatures of mycobacterium tuberculosis in C3HeB/FeJ mice. PloS One 10 (8), e0135208. doi: 10.1371/journal.pone.0135208
Gautam, U. S., Mehra, S., Kumari, P., Alvarez, X., Niu, T., Tyagi, J. S., et al. (2019). Mycobacterium tuberculosis sensor kinase DosS modulates the autophagosome in a DosR-independent manner. Commun. Biol. 2, 349. doi: 10.1038/s42003-019-0594-0
Gerasimova, A., Kazakov, A. E., Arkin, A. P., Dubchak, I., Gelfand, M. S. (2011). Comparative genomics of the dormancy regulons in mycobacteria. J. Bacteriol 193 (14), 3446–3452. doi: 10.1128/JB.00179-11
Gold, B., Nathan, C. (2017). Targeting phenotypically tolerant mycobacterium tuberculosis. Microbiol. Spectr. 5 (1). doi: 10.1128/microbiolspec.TBTB2-0031-2016
Gonzalo-Asensio, J., Mostowy, S., Harders-Westerveen, J., Huygen, K., Hernandez-Pando, R., Thole, J., et al. (2008). PhoP: A missing piece in the intricate puzzle of mycobacterium tuberculosis virulence. PloS One 3 (10), e3496. doi: 10.1371/journal.pone.0003496
Gooderham, W. J., Hancock, R. E. (2009). Regulation of virulence and antibiotic resistance by two-component regulatory systems in pseudomonas aeruginosa. FEMS Microbiol. Rev. 33 (2), 279–294. doi: 10.1111/j.1574-6976.2008.00135.x
Greendyke, R., Byrd, T. F. (2008). Differential antibiotic susceptibility of mycobacterium abscessus variants in biofilms and macrophages compared to that of planktonic bacteria. Antimicrob. Agents Chemother. 52 (6), 2019–2026. doi: 10.1128/AAC.00986-07
Gutierrez, A. V., Baron, S. A., Sardi, F. S., Saad, J., Coltey, B., Reynaud-Gaubert, M., et al. (2021). Beyond phenotype: The genomic heterogeneity of co-infecting mycobacterium abscessus smooth and rough colony variants in cystic fibrosis patients. J. Cyst Fibros 20 (3), 421–423. doi: 10.1016/j.jcf.2021.02.002
Gutierrez, A. V., Viljoen, A., Ghigo, E., Herrmann, J. L., Kremer, L. (2018). Glycopeptidolipids, a double-edged sword of the mycobacterium abscessus complex. Front. Microbiol. 9, 1145. doi: 10.3389/fmicb.2018.01145
Harris, K. A., Kenna, D. T. (2014). Mycobacterium abscessus infection in cystic fibrosis: molecular typing and clinical outcomes. J. Med. Microbiol. 63 (Pt 10), 1241–1246. doi: 10.1099/jmm.0.077164-0
Honaker, R. W., Leistikow, R. L., Bartek, I. L., Voskuil, M. I. (2009). Unique roles of DosT and DosS in DosR regulon induction and mycobacterium tuberculosis dormancy. Infect. Immun. 77 (8), 3258–3263. doi: 10.1128/IAI.01449-08
Hudock, T. A., Foreman, T. W., Bandyopadhyay, N., Gautam, U. S., Veatch, A. V., LoBato, D. N., et al. (2017). Hypoxia sensing and persistence genes are expressed during the intragranulomatous survival of mycobacterium tuberculosis. Am. J. Respir. Cell Mol. Biol. 56 (5), 637–647. doi: 10.1165/rcmb.2016-0239OC
Hurst-Hess, K., Rudra, P., Ghosh, P. (2017). Mycobacterium abscessus WhiB7 regulates a species-specific repertoire of genes to confer extreme antibiotic resistance. Antimicrob. Agents Chemother. 61 (11). doi: 10.1128/AAC.01347-17
Jonsson, B. E., Gilljam, M., Lindblad, A., Ridell, M., Wold, A. E., Welinder-Olsson, C. (2007). Molecular epidemiology of mycobacterium abscessus, with focus on cystic fibrosis. J. Clin. Microbiol. 45 (5), 1497–1504. doi: 10.1128/JCM.02592-06
Joshi, H., Kandari, D., Bhatnagar, R. (2021). Insights into the molecular determinants involved in mycobacterium tuberculosis persistence and their therapeutic implications. Virulence 12 (1), 2721–2749. doi: 10.1080/21505594.2021.1990660
Kendall, S. L., Movahedzadeh, F., Rison, S. C., Wernisch, L., Parish, T., Duncan, K., et al. (2004). The mycobacterium tuberculosis dosRS two-component system is induced by multiple stresses. Tuberculosis (Edinb) 84 (3-4), 247–255. doi: 10.1016/j.tube.2003.12.007
Kester, J. C., Kandror, O., Akopian, T., Chase, M. R., Zhu, J., Rubin, E. J., et al. (2021). ClpX is essential and activated by single-strand DNA binding protein in mycobacteria. J. Bacteriol 203 (4). doi: 10.1128/JB.00608-20
Klepp, L. I., Sabio, Y. G. J., FabianaBigi (2022). Mycobacterial MCE proteins as transporters that control lipid homeostasis of the cell wall. Tuberculosis (Edinb) 132, 102162. doi: 10.1016/j.tube.2021.102162
Kundu, M., Basu, J. (2021). Applications of transcriptomics and proteomics for understanding dormancy and resuscitation in mycobacterium tuberculosis. Front. Microbiol. 12, 642487. doi: 10.3389/fmicb.2021.642487
Lee, M. R., Sheng, W. H., Hung, C. C., Yu, C. J., Lee, L. N., Hsueh, P. R. (2015). Mycobacterium abscessus complex infections in humans. Emerg. Infect. Dis. 21 (9), 1638–1646. doi: 10.1126/scitranslmed.abj3860
Leistikow, R. L., Morton, R. A., Bartek, I. L., Frimpong, I., Wagner, K., Voskuil, M. I. (2010). The mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J. Bacteriol 192 (6), 1662–1670. doi: 10.1128/JB.00926-09
Li, X., Lv, X., Lin, Y., Zhen, J., Ruan, C., Duan, W., et al. (2019). Role of two-component regulatory systems in intracellular survival of mycobacterium tuberculosis. J. Cell Biochem. 120 (8), 12197–12207. doi: 10.1002/jcb.28792
Li, C., Wen, A., Shen, B., Lu, J., Huang, Y., Chang, Y. (2011). FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 11, 92. doi: 10.1186/1472-6750-11-92
Li, B., Ye, M., Zhao, L., Guo, Q., Chen, J., Xu, B., et al. (2020). Glycopeptidolipid genotype correlates with the severity of mycobacterium abscessus lung disease. J. Infect. Dis. 221 (Suppl 2), S257–S262. doi: 10.1093/infdis/jiz475
Lin, M. Y., Reddy, T. B., Arend, S. M., Friggen, A. H., Franken, K. L., van Meijgaarden, K. E., et al. (2009). Cross-reactive immunity to mycobacterium tuberculosis DosR regulon-encoded antigens in individuals infected with environmental, nontuberculous mycobacteria. Infect. Immun. 77 (11), 5071–5079. doi: 10.1128/IAI.00457-09
Lopeman, R. C., Harrison, J., Desai, M., Cox, J. A. G. (2019). Mycobacterium abscessus: Environmental bacterium turned clinical nightmare. Microorganisms 7 (3). doi: 10.3390/microorganisms7030090
Lyczak, J. B., Cannon, C. L., Pier, G. B. (2002). Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15 (2), 194–222. doi: 10.1128/CMR.15.2.194-222.2002
Madacki, J., Laval, F., Grzegorzewicz, A., Lemassu, A., Zahorszka, M., Arand, M., et al. (2018). Impact of the epoxide hydrolase EphD on the metabolism of mycolic acids in mycobacteria. J. Biol. Chem. 293 (14), 5172–5184. doi: 10.1074/jbc.RA117.000246
Malhotra, V., Agrawal, R., Duncan, T. R., Saini, D. K., Clark-Curtiss, J. E. (2015). Mycobacterium tuberculosis response regulators, DevR and NarL, interact in vivo and co-regulate gene expression during aerobic nitrate metabolism. J. Biol. Chem. 290 (13), 8294–8309. doi: 10.1074/jbc.M114.591800
Malhotra, S., Vedithi, S. C., Blundell, T. L. (2017). Decoding the similarities and differences among mycobacterial species. PloS Negl. Trop. Dis. 11 (8), e0005883. doi: 10.1371/journal.pntd.0005883
Malm, S., Tiffert, Y., Micklinghoff, J., Schultze, S., Joost, I., Weber, I., et al. (2009). The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of mycobacterium tuberculosis. Microbiol. (Reading) 155 (Pt 4), 1332–1339. doi: 10.1099/mic.0.023275-0
Mascher, T., Helmann, J. D., Unden, G. (2006). Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70 (4), 910–938. doi: 10.1128/MMBR.00020-06
Mehra, S., Foreman, T. W., Didier, P. J., Ahsan, M. H., Hudock, T. A., Kissee, R., et al. (2015). The DosR regulon modulates adaptive immunity and is essential for mycobacterium tuberculosis persistence. Am. J. Respir. Crit. Care Med. 191 (10), 1185–1196. doi: 10.1164/rccm.201408-1502OC
Miranda-CasoLuengo, A. A., Staunton, P. M., Dinan, A. M., Lohan, A. J., Loftus, B. J. (2016). Functional characterization of the mycobacterium abscessus genome coupled with condition specific transcriptomics reveals conserved molecular strategies for host adaptation and persistence. BMC Genomics 17, 553. doi: 10.1186/s12864-016-2868-y
Molina-Torres, C. A., Tamez-Pena, L., Castro-Garza, J., Ocampo-Candiani, J., Vera-Cabrera, L. (2018). Evaluation of the intracellular activity of drugs against mycobacterium abscessus using a THP-1 macrophage model. J. Microbiol. Methods 148, 29–32. doi: 10.1016/j.mimet.2018.03.020
Moore, S. D., Prevelige, P. E., Jr. (2002). A P22 scaffold protein mutation increases the robustness of head assembly in the presence of excess portal protein. J. Virol. 76 (20), 10245–10255. doi: 10.1128/JVI.76.20.10245-10255.2002
Nessar, R., Cambau, E., Reyrat, J. M., Murray, A., Gicquel, B. (2012). Mycobacterium abscessus: A new antibiotic nightmare. J. Antimicrob. Chemother. 67 (4), 810–818. doi: 10.1093/jac/dkr578
Olivier, K. N., Weber, D. J., Wallace, R. J., Jr., Faiz, A. R., Lee, J. H., Zhang, Y., et al. (2003). Nontuberculous mycobacteria. I: Multicenter prevalence study in cystic fibrosis. Am. J. Respir. Crit. Care Med. 167 (6), 828–834. doi: 10.1164/rccm.200207-678OC
Ortega Ugalde, S., Boot, M., Commandeur, J. N. M., Jennings, P., Bitter, W., Vos, J. C. (2019). Function, essentiality, and expression of cytochrome P450 enzymes and their cognate redox partners in mycobacterium tuberculosis: Are they drug targets? Appl. Microbiol. Biotechnol. 103 (9), 3597–3614. doi: 10.1007/s00253-019-09697-z
Park, H. D., Guinn, K. M., Harrell, M. I., Liao, R., Voskuil, M. I., Tompa, M., et al. (2003). Rv3133c/dosR is a transcription factor that mediates the hypoxic response of mycobacterium tuberculosis. Mol. Microbiol. 48 (3), 833–843. doi: 10.1046/j.1365-2958.2003.03474.x
Pawlik, A., Garnier, G., Orgeur, M., Tong, P., Lohan, A., Le Chevalier, F., et al. (2013). Identification and characterization of the genetic changes responsible for the characteristic smooth-to-rough morphotype alterations of clinically persistent mycobacterium abscessus. Mol. Microbiol. 90 (3), 612–629. doi: 10.1111/mmi.12387
Peddireddy, V., Doddam, S. N., Ahmed, N. (2017). Mycobacterial dormancy systems and host responses in tuberculosis. Front. Immunol. 8, 84. doi: 10.3389/fimmu.2017.00084
Peterson, E. J. R., Abidi, A. A., Arrieta-Ortiz, M. L., Aguilar, B., Yurkovich, J. T., Kaur, A., et al. (2020). Intricate genetic programs controlling dormancy in mycobacterium tuberculosis. Cell Rep. 31 (4), 107577. doi: 10.1016/j.celrep.2020.107577
Philley, J. V., DeGroote, M. A., Honda, J. R., Chan, M. M., Kasperbauer, S., Walter, N. D., et al. (2016). Treatment of non-tuberculous mycobacterial lung disease. Curr. Treat Options Infect. Dis. 8 (4), 275–296. doi: 10.1007/s40506-016-0086-4
Plocinska, R., Wasik, K., Plocinski, P., Lechowicz, E., Antczak, M., Blaszczyk, E., et al. (2022). The orphan response regulator Rv3143 modulates the activity of the NADH dehydrogenase complex (Nuo) in mycobacterium tuberculosis via protein-protein interactions. Front. Cell Infect. Microbiol. 12, 909507. doi: 10.3389/fcimb.2022.909507
Purkayastha, A., McCue, L. A., McDonough, K. A. (2002). Identification of a mycobacterium tuberculosis putative classical nitroreductase gene whose expression is coregulated with that of the acr aene within macrophages, in standing versus shaking cultures, and under low oxygen conditions. Infect. Immun. 70 (3), 1518–1529. doi: 10.1128/IAI.70.3.1518-1529.2002
Rathor, N., Garima, K., Sharma, N. K., Narang, A., Varma-Basil, M., Bose, M. (2016). Expression profile of mce4 operon of mycobacterium tuberculosis following environmental stress. Int. J. Mycobacteriol 5 (3), 328–332. doi: 10.1016/j.ijmyco.2016.08.004
Roberts, D. M., Liao, R. P., Wisedchaisri, G., Hol, W. G., Sherman, D. R. (2004). Two sensor kinases contribute to the hypoxic response of mycobacterium tuberculosis. J. Biol. Chem. 279 (22), 23082–23087. doi: 10.1074/jbc.M401230200
Rohde, K. H., Abramovitch, R. B., Russell, D. G. (2007). Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2 (5), 352–364. doi: 10.1016/j.chom.2007.09.006
Rohde, K. H., Veiga, D. F., Caldwell, S., Balazsi, G., Russell, D. G. (2012). Linking the transcriptional profiles and the physiological states of mycobacterium tuberculosis during an extended intracellular infection. PloS Pathog. 8 (6), e1002769. doi: 10.1371/journal.ppat.1002769
Roux, A. L., Viljoen, A., Bah, A., Simeone, R., Bernut, A., Laencina, L., et al. (2016). The distinct fate of smooth and rough mycobacterium abscessus variants inside macrophages. Open Biol. 6 (11). doi: 10.1098/rsob.160185
Ruger, K., Hampel, A., Billig, S., Rucker, N., Suerbaum, S., Bange, F. C. (2014). Characterization of rough and smooth morphotypes of mycobacterium abscessus isolates from clinical specimens. J. Clin. Microbiol. 52 (1), 244–250. doi: 10.1128/JCM.01249-13
Rustad, T. R., Harrell, M. I., Liao, R., Sherman, D. R. (2008). The enduring hypoxic response of mycobacterium tuberculosis. PloS One 3 (1), e1502. doi: 10.1371/journal.pone.0001502
Rustad, T. R., Sherrid, A. M., Minch, K. J., Sherman, D. R. (2009). Hypoxia: a window into mycobacterium tuberculosis latency. Cell Microbiol. 11 (8), 1151–1159. doi: 10.1111/j.1462-5822.2009.01325.x
Salazar, M. E., Laub, M. T. (2015). Temporal and evolutionary dynamics of two-component signaling pathways. Curr. Opin. Microbiol. 24, 7–14. doi: 10.1016/j.mib.2014.12.003
Santangelo Mde, L., Blanco, F., Campos, E., Soria, M., Bianco, M. V., Klepp, L., et al. (2009). Mce2R from mycobacterium tuberculosis represses the expression of the mce2 operon. Tuberculosis (Edinb) 89 (1), 22–28. doi: 10.1016/j.tube.2008.09.002
Sassetti, C. M., Boyd, D. H., Rubin, E. J. (2003). Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48 (1), 77–84. doi: 10.1046/j.1365-2958.2003.03425.x
Sherman, D. R., Voskuil, M., Schnappinger, D., Liao, R., Harrell, M. I., Schoolnik, G. K. (2001). Regulation of the mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin 98, 13, 7534–7539. doi: 10.1073/pnas.121172498
Singh, A., Mai, D., Kumar, A., Steyn, A. J. (2006). Dissecting virulence pathways of mycobacterium tuberculosis through protein-protein association. Proc. Natl. Acad. Sci. U.S.A. 103 (30), 11346–11351. doi: 10.1073/pnas.0602817103
Singh, A., Varela, C., Bhatt, K., Veerapen, N., Lee, O. Y., Wu, H. H., et al. (2016). Identification of a desaturase involved in mycolic acid biosynthesis in mycobacterium smegmatis. PloS One 11 (10), e0164253. doi: 10.1371/journal.pone.0164253
Stock, J. B., Ninfa, A. J., Stock, A. M. (1989). Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53 (4), 450–490. doi: 10.1128/mr.53.4.450-490.1989
Story-Roller, E., Maggioncalda, E. C., Cohen, K. A., Lamichhane, G. (2018). Mycobacterium abscessus and beta-lactams: Emerging insights and potential opportunities. Front. Microbiol. 9, 2273. doi: 10.3389/fmicb.2018.02273
Stupar, M., Furness, J., De Voss, C. J., Tan, L., West, N. P. (2022). Two-component sensor histidine kinases of mycobacterium tuberculosis: Beacons for niche navigation. Mol. Microbiol. 117 (5), 973–985. doi: 10.1111/mmi.14899
Tomlinson, B. R., Denham, G. A., Torres, N. J., Brzozowski, R. S., Allen, J. L., Jackson, J. K., et al. (2022). Assessing the role of cold-shock protein c: a novel regulator of acinetobacter baumannii biofilm formation and virulence. Infect. Immun. 90 (10), e0037622. doi: 10.1128/iai.00376-22
Tomlinson, B. R., Malof, M. E., Shaw, L. N. (2021). A global transcriptomic analysis of staphylococcus aureus biofilm formation across diverse clonal lineages. Microb. Genom 7 (7). doi: 10.1099/mgen.0.000598
Tsai, S. H., Lai, H. C., Hu, S. T. (2015). Subinhibitory doses of aminoglycoside antibiotics induce changes in the phenotype of mycobacterium abscessus. Antimicrob. Agents Chemother. 59 (10), 6161–6169. doi: 10.1128/AAC.01132-15
Unden, G., Bongaerts, J. (1997). Alternative respiratory pathways of escherichia coli: Energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320 (3), 217–234. doi: 10.1016/S0005-2728(97)00034-0
van Kessel, J. C., Hatfull, G. F. (2007). Recombineering in mycobacterium tuberculosis. Nat. Methods 4 (2), 147–152. doi: 10.1038/nmeth996
Veatch, A. V., Kaushal, D. (2018). Opening pandora's box: Mechanisms of mycobacterium tuberculosis resuscitation. Trends Microbiol. 26 (2), 145–157. doi: 10.1016/j.tim.2017.08.001
Vilcheze, C., Yan, B., Casey, R., Hingley-Wilson, S., Ettwiller, L., Jacobs, W. R., Jr. (2022). Commonalities of mycobacterium tuberculosis transcriptomes in response to defined persisting macrophage stresses. Front. Immunol. 13, 909904. doi: 10.3389/fimmu.2022.909904
Viljoen, A., Blaise, M., de Chastellier, C., Kremer, L. (2016). MAB_3551c encodes the primary triacylglycerol synthase involved in lipid accumulation in mycobacterium abscessus. Mol. Microbiol. 102 (4), 611–627. doi: 10.1111/mmi.13482
Voskuil, M. I., Schnappinger, D., Visconti, K. C., Harrell, M. I., Dolganov, G. M., Sherman, D. R., et al. (2003). Inhibition of respiration by nitric oxide induces a mycobacterium tuberculosis dormancy program. J. Exp. Med. 198 (5), 705–713. doi: 10.1084/jem.20030205
Voskuil, M. I., Visconti, K. C., Schoolnik, G. K. (2004). Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb) 84 (3-4), 218–227. doi: 10.1016/j.tube.2004.02.003
Walters, S. B., Dubnau, E., Kolesnikova, I., Laval, F., Daffe, M., Smith, I. (2006). The mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol. Microbiol. 60 (2), 312–330. doi: 10.1111/j.1365-2958.2006.05102.x
Wee, W. Y., Dutta, A., Choo, S. W. (2017). Comparative genome analyses of mycobacteria give better insights into their evolution. PloS One 12 (3), e0172831. doi: 10.1371/journal.pone.0172831
Weinstein, E. A., Yano, T., Li, L. S., Avarbock, D., Avarbock, A., Helm, D., et al. (2005). Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc. Natl. Acad. Sci. U.S.A. 102 (12), 4548–4553. doi: 10.1073/pnas.0500469102
West, A. H., Stock, A. M. (2001). Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26 (6), 369–376. doi: 10.1016/S0968-0004(01)01852-7
Wilburn, K. M., Fieweger, R. A., VanderVen, B. C. (2018). Cholesterol and fatty acids grease the wheels of mycobacterium tuberculosis pathogenesis. Pathog. Dis. 76 (2). doi: 10.1093/femspd/fty021
Worlitzsch, D., Tarran, R., Ulrich, M., Schwab, U., Cekici, A., Meyer, K. C., et al. (2002). Effects of reduced mucus oxygen concentration in airway pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109 (3), 317–325. doi: 10.1172/JCI0213870
Yang, H., Wang, F., Guo, X., Liu, F., Liu, Z., Wu, X., et al. (2021). Interception of host fatty acid metabolism by mycobacteria under hypoxia to suppress anti-TB immunity. Cell Discovery 7 (1), 90. doi: 10.1038/s41421-021-00301-1
Yarwood, J. M., McCormick, J. K., Schlievert, P. M. (2001). Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of staphylococcus aureus. J. Bacteriol 183 (4), 1113–1123. doi: 10.1128/JB.183.4.1113-1123.2001
Yeruva, V. C., Savanagouder, M., Khandelwal, R., Kulkarni, A., Sharma, Y., Raghunand, T. R. (2016). The mycobacterium tuberculosis desaturase DesA1 (Rv0824c) is a Ca(2+) binding protein. Biochem. Biophys. Res. Commun. 480 (1), 29–35. doi: 10.1016/j.bbrc.2016.10.014
Yousuf, S., Angara, R. K., Roy, A., Gupta, S. K., Misra, R., Ranjan, A. (2018). Mce2R/Rv0586 of mycobacterium tuberculosis is the functional homologue of FadR(E. coli). Microbiol. (Reading) 164 (9), 1133–1145. doi: 10.1099/mic.0.000686
Keywords: nontuberculous mycobacteria (NTM), Mycobacterium abcessus, hypoxia, two-component system (TCS), RNAseq, DosR
Citation: Simcox BS, Tomlinson BR, Shaw LN and Rohde KH (2023) Mycobacterium abscessus DosRS two-component system controls a species-specific regulon required for adaptation to hypoxia. Front. Cell. Infect. Microbiol. 13:1144210. doi: 10.3389/fcimb.2023.1144210
Received: 13 January 2023; Accepted: 15 February 2023;
Published: 09 March 2023.
Edited by:
Elena G. Salina, Research Center of Biotechnology of the Russian Academy of Sciences, RussiaReviewed by:
Martin I. Voskuil, University of Colorado Denver, United StatesCopyright © 2023 Simcox, Tomlinson, Shaw and Rohde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyle H. Rohde, S3lsZS5yb2hkZUB1Y2YuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.