
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 21 April 2023
Sec. Bacteria and Host
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1141115
This article is part of the Research Topic Proceedings of the Legionella 2022 View all 10 articles
A correction has been applied to this article in:
Corrigendum: Antibiotic susceptibility pattern of Portuguese environmental Legionella isolates
Introduction: Legionnaires’ Disease is a pneumonia caused by Legionella spp., currently treated empirically with fluoroquinolones and macrolides. In this study, we aim to describe the antibiotic susceptibility pattern of environmental Legionella recovered in the south of Portugal.
Methods: Minimal inhibitory concentration (MIC) determination of 57 Legionella isolates (10 Lp sg 1, 32, Lp sg 2-14 15 L. spp) was achieved by broth microdilution, as described by EUCAST, for azithromycin, clarithromycin, ciprofloxacin, levofloxacin, and doxycycline.
Results: Fluoroquinolones were the most active antibiotic, displaying the lowest MIC values in contrast to doxycycline which had the highest. MIC90 and epidemiological cut-off (ECOFF) values were, respectively, 0.5/1 mg/L for azithromycin, 0.125/0.25 mg/L for clarithromycin, 0.064/0.125 mg/L for ciprofloxacin, 0.125/0.125 mg/L for levofloxacin and 16/32 mg/L for doxycycline.
Discussion: MIC distributions were higher than reported by EUCAST for all antibiotics. Interestingly, two phenotypically resistant isolates with high-level quinolone resistance were identified. This is the first time that MIC distributions, lpeAB and tet56 genes have been investigated in Portuguese environmental isolates of Legionella.
Legionnaires’ Disease (LD) is a pneumonia, often severe, caused by waterborne pathogens of the Legionella genera. Despite more than 20 of the 65 known Legionella species being able to cause human infection (Chambers et al., 2021), Legionella pneumophila (Lp) alone is responsible for more than 90% of reported cases (European Centre for Disease Prevention and Control, 2022). Transmission to human hosts occurs primarily through aerosolized water particles, with only one description of person-to-person transmission of LD (Correia et al., 2016). Legionella is ubiquitous in water, colonizing natural and artificial environments, such as cooling systems, air conditioners and water supply networks.
The antibiotics used in LD treatment include fluoroquinolones, macrolides, and tetracyclines, given their high cellular penetration, azithromycin, and levofloxacin being considered the first line (Mandell et al., 2007; Woodhead et al., 2011). However, cases of unresolved LD or with slow treatment have already been described. Bruin et al., described one case of susceptibility loss to ciprofloxacin treatment, derived from a mutation in position 83 of the gyrA gene which encodes a subunit of the DNA gyrase (Bruin et al. ). Additionally, resistance by target mutation has been comproved in vitro for macrolides and rifampicin (Nielsen et al., 2000; Descours et al., 2017).
Recently, other resistance mechanisms were reported in this bacterium associated with the LpeAB operon, an analog of the AcrAB efflux pump of Escherichia coli (Massip et al., 2017). Mutations upstream of lpeAB gene, have been described as responsible for a decreased susceptibility to azithromycin and erythromycin in Lp Paris strain. This LpeAB efflux pump has been found in clinical and environmental isolates (Vandewalle-Capo et al., 2017; Nata and Løva, 2019; Cocuzza et al., 2021). Tet56, a tetracycline destructase of the TetX family has been identified in L. longbeacheae, L. nautarum, and L. jordanis isolates (Forsberg et al., 2015; Joseph et al., 2016). Although the presence of Tet56 may not be clinically impactful, as tetracyclines are regarded as an alternative in LD treatment, tigecycline, a synthetic tetracycline, has been successfully used in cases of LD, when treatment with first-line antibiotics has failed (Valve et al., 2010; Slawek et al., 2017).
Antibiotic pollution is a worrying problem. The presence of antibiotics in different aquatic environments, usually in low concentrations (Hughes et al., 2013; Danner et al., 2019; Rodriguez-Mozaz et al., 2020), is known to influence resistance mechanisms (Andersson and Hughes, 2014; Murray et al., 2018; Chow et al., 2021), promote virulence factors (Andersson and Hughes, 2014), and decrease antibiotic susceptibility (Sulyok et al., 2017). However, the full extent of the impact caused by chronic exposure remains unknown (Janecko et al., 2016). As the source of LD infection is from water and soils, special concern should be given to environmental Legionella populations.
The high efficacy of LD antibiotic treatment, over reliance on urinary antigen tests and reduction in culture, and the lack of consensus in antimicrobial susceptibility testing (AST) methodology, are contributing factors to the absence of standardization, and clinically defined breakpoint (Portal et al., 2021a). In this study, we aim to characterize Portuguese environmental Legionella susceptibility patterns, define ECOFF, and contribute more data on the subject.
Isolates were collected (n = 57) from November 2021 to June 2022. They were obtained from water samples screened for Legionella presence in LAIST (Laboratório de Análises de Água, Técnico Lisboa, Universidade de Lisboa), in accordance with the standardized procedures described in the ISO 11731 (International Organization for Standardization, 2017). All isolates were obtained from aerosol producing equipment. Isolates were discriminated into three groups (Lp sg 1, Lp sg 2-14, L. spp) based on serological identification with latex agglutination test (OXOID, UK). Bacterial isolates were then preserved at -80°C in a thioglycolate medium until tested.
In this study, the control strains Lp sg1 ATCC 33152 and Paris was used.
Five antibiotics were selected to be studied: azithromycin (AZT); clarithromycin (CLA); ciprofloxacin (CIP); levofloxacin (LEV) and doxycycline (DOX) [Sigma Aldrich, USA]. Antibiotics were resuspended in the advised solvents, respectively: methanol, DMSO, HCl 1% solution, and water for the last two. The range of concentrations tested were respectively: 0.016 – 4 mg/L; 0.004 – 2 mg/L; 0.016 – 1 mg/L; 0.008 – 0.5 mg/L; and 0.5 – 128 mg/L.
Tests were performed in accordance with European Committee on Antimicrobial Susceptibility Testing (EUCAST) and Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute, 2018; European Committee on Antimicrobial Susceptibility Testing, 2021). Briefly, bacterial suspensions of 0.5 McFarland were prepared using buffered yeast extract medium (BYE). Antibiotics were diluted in ultrapure water in ranges equal to the minimal inhibitory concentration (MIC) distribution of EUCAST (European Committee on Antimicrobial Susceptibility Testing, 2021). Forty microliters of each antibiotic dilution were added to the test wells of the microplate before adding the 160 µL of the bacterial suspension. For positive and negative controls bacterial suspension and BYE medium were used, respectively. All isolates were tested in duplicate.
Microplates were incubated at 37°C for 48 hours in a humid chamber. After incubation, the microplates were read manually according to by EUCAST and CLSI guidelines. Additionally, absorbances were read in a spectrophotometer at a wavelength of 600 nm and the inhibition rate was calculated using the formula:
Where AbsX represents the absorbance of the sample; AbsC- and AbsC+ the absorbance of the negative and positive control, respectively; IR the inhibition rate. To determine MIC a cut-off value of 90% inhibition was used.
ECOFFs were defined through the ECOFFinder program available at the EUCAST website, that follows the methodologies described by Turnidge et al. (Turnidge et al., 2006; European Committee on Antimicrobial Susceptibility Testing, 2023).
Molecular amplification of lpeAB and tet56 genes was performed through conventional PCR to determine the presence of possible resistance mechanisms. Briefly, for the gene lpeAB the specific primers lpp2879_detec_F2 (5′-GTGATGATTGTCTTATTGG TGCGA-3′) and lpp2879_detec_R3 (5′-ATGGCGTTTAAGATGATGGT GATT-3′) were used (Vandewalle-Capo et al., 2017) and, for the gene tet56 the primers Fw (5’- ATGTCTAAAAATATCAAAATTCTCGTC-3’) and Rv (5’- CTATGATGATTCATATTGAGGTAAGG-3’) (Forsberg et al., 2015). The presence of the lpeAB gene was investigated in all isolates tested for azithromycin. The presence of the tet56 gene was investigated in Lp sg1 and Lp sg 2-14 isolates when MIC for doxycycline was raised to EUCAST tentative highest MICs, 2 mg/L and 32 mg/L respectively, and in all L. spp isolates.
To compare the two reading methods, the MIC distributions between serological groups from the present study to the EUCAST values and with the MICs values described in selected studies (Wilson et al., 2018; Assaidi et al., 2020; Cocuzza et al., 2021), non-parametric Mann-Whitney and Kruskal-Wallis test were performed using XLSTAT Software (Addinsoft, France).
Thirty eight of the collected isolates were from water distribution systems of hospitals, hotels, and other public spaces, two isolates were from industrial cooling towers, one came from residual waters and, the remaining were of unknown origin. Water samples were collected from the southern region of Portugal, mainly from the Lisbon metropolitan area and Alentejo. In the studied population the three groups are represented: Lp sg 1 (n = 10), Lp sg 2-14 (n = 32), and L. spp (n = 15).
MIC distributions and ECOFFs are shown in Table 1. Overall, both quinolones achieved the lowest ECOFF values: 0.125 mg/L for both CIP and LEV; 0.25 mg/L for CLA; 1 mg/L for AZT; and 32 mg/L for DOX, which is the highest ECOFF.
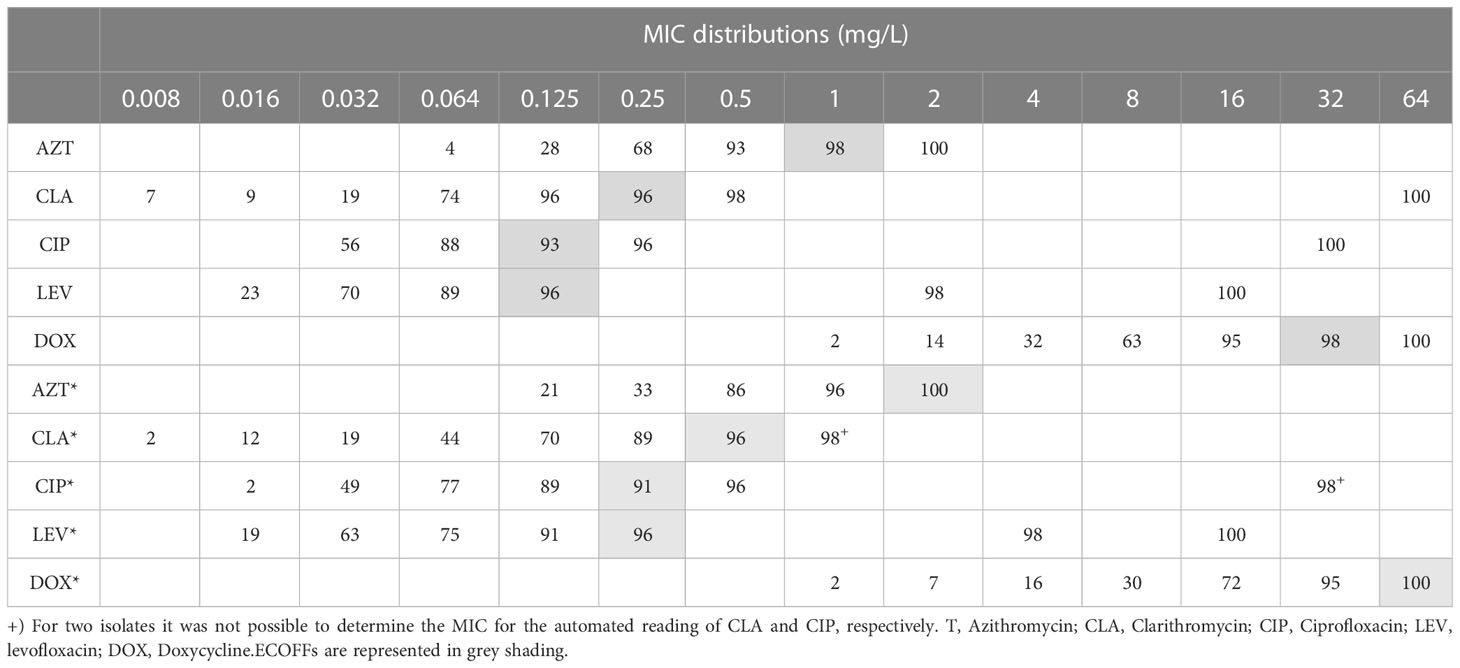
Table 1 Cumulative percentages of MIC distribution of environmental Legionella isolates (n = 57) obtained by manual and automated reading (signalized with *).
Comparison of MIC distributions between serological groups (Table 2) showed that Lp sg 1 had the highest MIC values. The ranges of the Lp sg 2-14 and L. spp groups were identical for LEV. For CIP the Lp sg 2-14 group achieved the lowest MIC range and for the other antibiotics, the L. spp group had the lowest range. Generally, the groups followed the above-described trend in MICs, except for CIP and CLA in the Lp sg 1 and L. spp groups, as in these, CLA presented a lower MIC 90 and range than CIP.
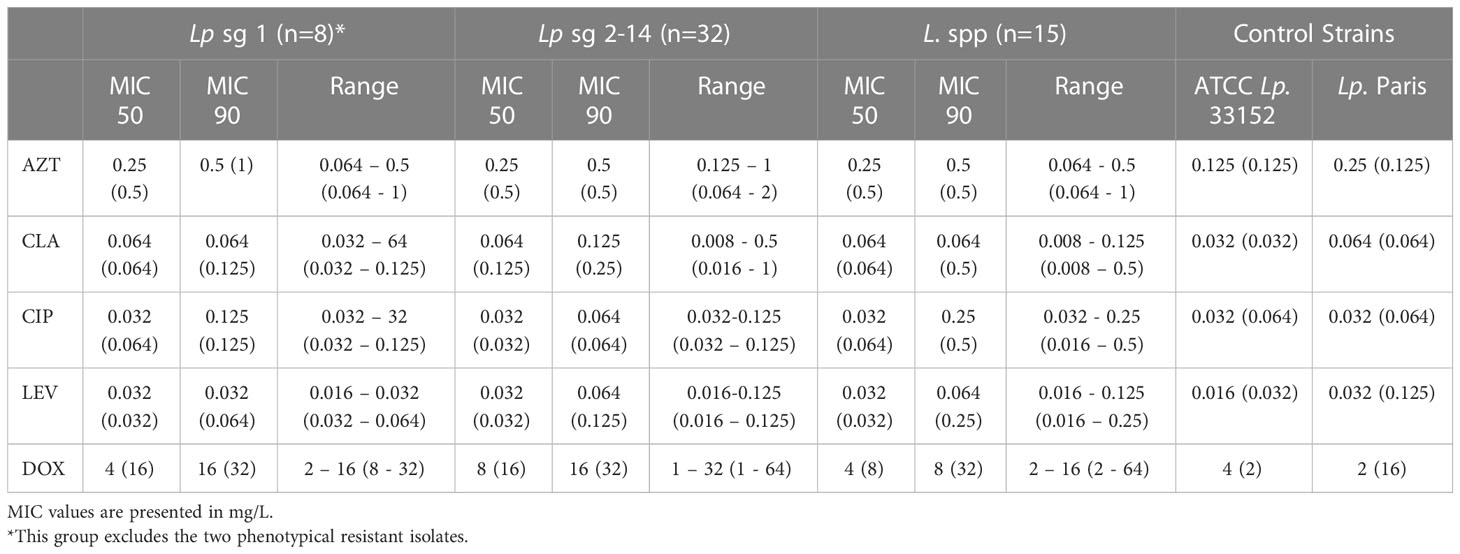
Table 2 MIC50, MIC90, and range values of five antibiotics for segregated serological groups of 57 environmental isolates obtained by manual and automated reading (within parenthesis).
Regarding MIC 50, values were similar in all groups for all antibiotics except for DOX in the Lp sg 2-14. Contrarily for MIC 90, values do not follow any trend in the three groups (Table 2).
A closer analysis of discriminatory MIC 50 and MIC 90 values demonstrated that the presence in two isolates showed a disequilibrium in these parameters. These discrepancies were caused by two phenotypically resistant strains (Table 1, supplementary material S1). Both isolates were collected from a hotel water system. The results of these two isolates have been retested in triplicate. These two isolates were also investigated for mutations in the QRDR of gyrA and no mutations were found in positions 83 and 87 (E. coli numbering, Table S3 and Figure S1), suggesting a resistance mechanism different from one starting at GyrA position 83 (Almahmoud et al., 2009).
MIC distributions obtained through manual and absorbance readings were revealed to have statistically significant differences (p-value< 0.05). For all serological groups and antibiotics tested, the absorbance reading method produced higher MICs. Isolates Lp sg 1 and Lp sg 2-14 showed similar MIC in both methods, except for DOX and CLA. The MICs of L. spp were at least twice as high, except for AZT. DOX was the antibiotic that revealed the greatest discrepancy between the reading methods (Table 3).
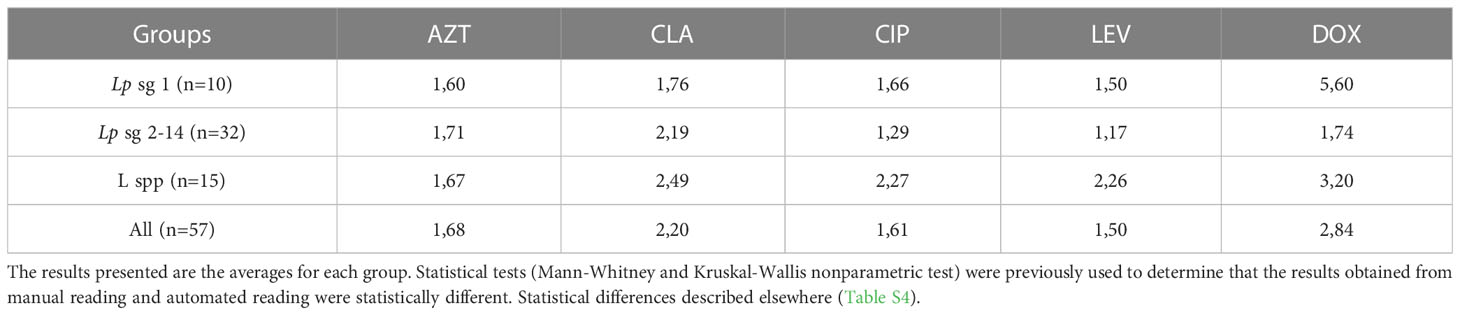
Table 3 Ratios comparing automatic to the manual reading of MICs discriminated by serological group of isolates and antibiotic.
The evaluation results of the efflux pump gene presence showed that 67% (6/9) of the Lp sg 1 isolates and 60% (12/20) of the Lp sg 2-14, with MIC values, for AZT, greater than 0.125 mg/L, revealed the presence of the efflux pump gene, as well as 53% (8/15) of the studied L. spp. Overall, the lpeAB gene was present in 62% (18/29) of Lp sg 1 and Lp sg 2-14 isolates analyzed. The results showed that the number of isolates without the lpeAB gene was more prevalent for MICs equal to or less than 0.250 mg/L. Conversely, for higher MICs, isolates with this gene predominate (Figure 1, supplementary material S2).
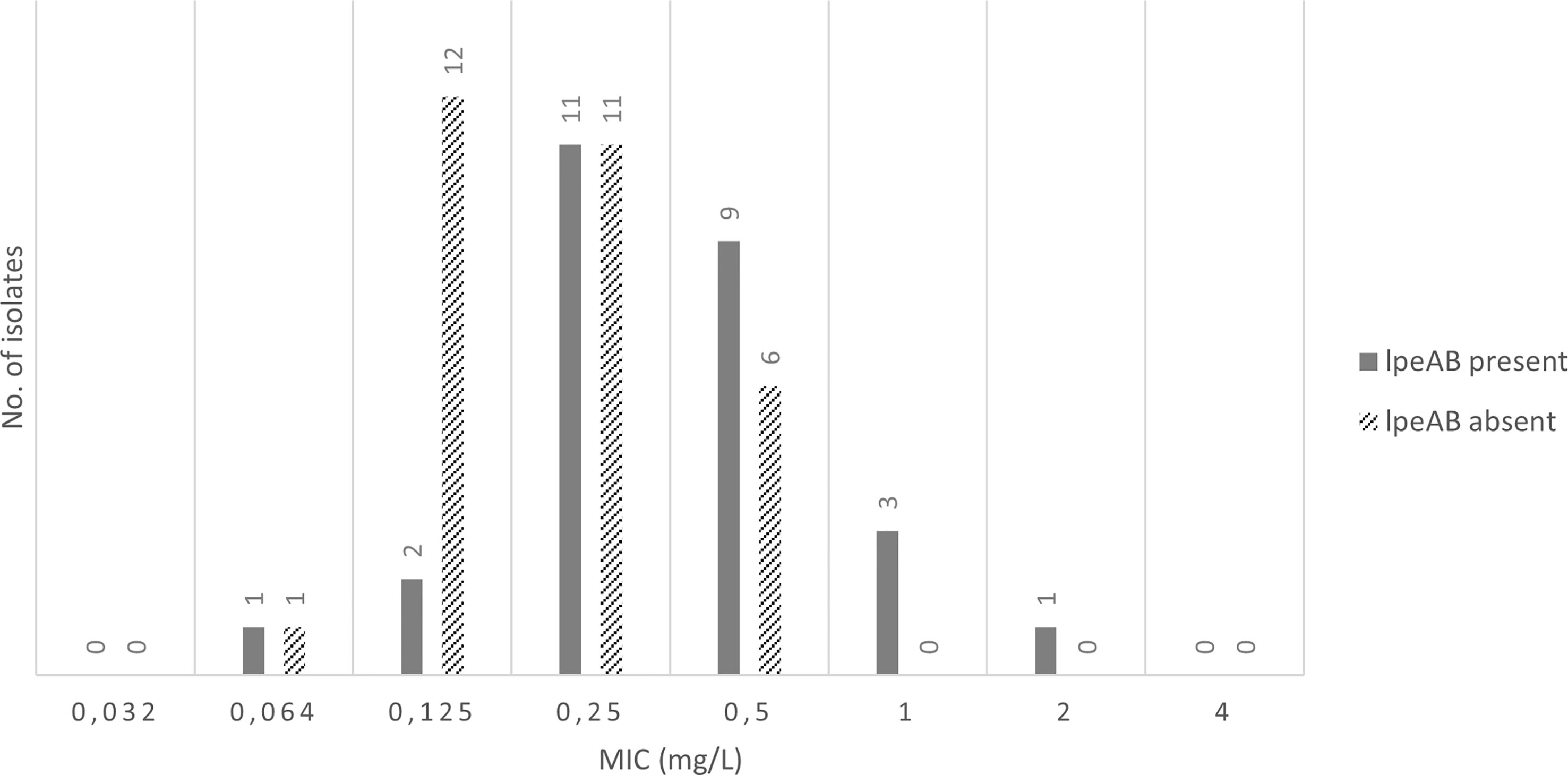
Figure 1 Presence of IpeAB gene and its distribution according to the AZT MIC of the 57 environmental isolates under study.
Converse to the presence of the lpeAB gene, the tet56 gene was found in only one L. spp isolate of those analysed (1/21) and identified by MALDI-ToF as Legionella anisa.
In summary, the achieved results did not show significant differences comparing the MIC distributions in the three groups, except the MIC distributions of CLA and DOX in Lp sg 2-14 and L. spp. MIC distributions obtained manually and automatedly were found to be significantly different (p-value< 0.05). By comparison, our MIC distributions is statistically different from EUCAST and selected studies, Cocuzza et al. (Cocuzza et al., 2021); Assaidi et al. (Assaidi et al., 2020); Wilson et al. (Wilson et al., 2018), except for AZT where similar distributions are found (Table S4).
In 1999 LD was included for mandatory clinical and laboratory notification as part of the Portuguese infectious disease surveillance scheme. The disease is potentially life-threatening if not treated with the proper antibiotics.
Environmental monitoring of Legionella, in our country, became mandatory after the publication of Law n. 52/2018, of August 20 (Law n.o 52/2018, of Aug 20, 2023). The Law proposed periodic sampling of water from heat transfer equipment associated with heating, ventilation, air systems, air conditioning units, or air treatment, which produce water aerosols. However, in our study, most of the isolates were recovered from water distribution systems in hospitals and hotels. Growth inhibition, due to a high concentration of disinfectant or another factor, could be a plausible explanation for this lower number of isolates from heat transfer equipment (García et al., 2007; Sanli, 2019).
Regarding the serological distribution, the predominant group was Lp sg 2-14, which matches with previous studies (Xiong et al., 2016; Graells et al., 2018; Torre et al., 2018; Assaidi et al., 2020; Cocuzza et al., 2021; Zhan et al., 2022).
Legionella susceptibility analysis is important to predict the evolution of antibiotic resistance and assess its impact on the treatment of LD. This topic has been the subject of several studies, and the results obtained show there is a need for standardization of methodologies, and the establishment of guidelines for all groups of Legionella isolates to optimize the detection of resistances. In this study, susceptibility against five antibiotics was evaluated in 57 environmental isolates, and our results corroborate the trend in the effectiveness already reported, with floroquinolones being the most effective antibiotic and tetracyclines the least effective (Xiong et al., 2016; Vandewalle-Capo et al., 2017; Wilson et al., 2018; Assaidi et al., 2020; Cocuzza et al., 2021).
In contrast, MIC 50 and MIC 90 values are not concordant with the literature, being higher for quinolones and macrolides (Vandewalle-Capo et al., 2017; Wilson et al., 2018; Cocuzza et al., 2021). Nonetheless, Xiong et al. (2016) obtained similarly high values for DOX in the Lp sg 1 group, and Assaidi et al. (2020), reported elevated MIC 50 and MIC 90 for all antibiotics compared to current guidelines, except for AZT (Xiong et al., 2016; Assaidi et al., 2020). In comparison with EUCAST’s values, our MIC 50 and MIC 90 are bigger in Lp sg 1 and Lp sg 2-14 for AZT and CIP and in LEV and DOX for Lp sg 1, CLA for Lp sg 1, and LEV and DOX for Lp sg 2-14 agreed with the EUCAST guidelines (European Committee on Antimicrobial Susceptibility Testing, 2021).
EUCAST guidelines do not describe any MIC distributions for L. spp, although two other studies and these studies were in agreement with our results (Xiong et al., 2016; Assaidi et al., 2020).
The distribution of our MICs proved to be statistically different from those presented by EUCAST and in selected studies for Lp isolates (Wilson et al., 2018; Assaidi et al., 2020; Cocuzza et al., 2021; European Committee on Antimicrobial Susceptibility Testing, 2021). The comparison was performed using only studies that employed EUCAST methodologies, but the differences showed how important it is to carry out this type of investigation to provide additional data on Lp susceptibility so that an ECOFF can be defined. The global trend of increasing resistance levels implies a reduction of antibacterial treatment effectiveness that could have worrying consequences.
MIC values obtained through automated reading are higher than manual, making a defined cut-off difficult when comparing both methods. Operator variability and subjectiveness can contribute to a lack of standardization and as most results are read manually, so the comparison of MICs values can be controversial in studies.
To the best of our knowledge, this is the first time two phenotypically resistant isolates have been reported with such elevated MIC values (Table S1). These isolates possess an elevated risk, given their diminished susceptibilities to several of the first line antibiotics. While the LD mortality rate typically does not surpass 10% (European Centre for Disease Prevention and Control, 2022), cases have achieved rates of nearly 30%, in hospital-related LD cases (Beauté, 2017). The effect on increased morbidity caused by strains with this high MIC is unknown. Curiously, presence of the efflux pump gene, lpeAB was found in both isolates as expected given their MIC values for AZT.
Presence of lpeAB, in our isolates, was high. Previous studies reported a 50% prevalence (Portal et al., 2021b), but no studies have reported the lpeAB gene outside of L. pneumophila. This is the first time resistance has been reported in non pneumophila Legionella. Given the similarities between the MIC distributions for L. spp, with and without lpeAB, it is difficult to determine a clear cut-off value to distinguish the wild-type and lpAB positive isolates. This is mainly due to the small population analyzed.
Overall, these results do not diverge from EUCAST. Apart from a single Lp sg 2-14 all remaining isolates from this group and Lp sg 1 did not change the EUCAST distribution for LpeAB-positive isolates. Previous studies have also reported the presence of the efflux pump gene in isolates whose MIC values for AZT are equal to or exceed 0.125 mg/L (Vandewalle-Capo et al., 2017; Nata and Løva, 2019; Portal et al., 2021b). However, the presence of lpeAB-negative isolates with elevated MIC values as high as 0.5 mg/L was also observed in this study, and in previous works (Cocuzza et al., 2021; Yang et al., 2022). In this situation defining an ECOFF is a complex question given that the purpose is to discriminate between wild-type isolates and isolates containing acquired resistance mechanisms, and the efflux pump LpeAB has been described as an acquired resistance mechanism (Turnidge et al., 2006). Therefore, additional studies are required to provide robust ECOFF values.
The tet56 gene was detected in one isolate with a MIC of 2 mg/L. This gene codes for a tetracycline destructase, and was first described in L. longbeacheae (Forsberg et al., 2015).
This study reports the susceptibility patterns of environmental Portuguese isolates, whose MIC distributions surpass EUCAST reference values. We also describe two phenotypically resistant isolates with high-level floroquinolone resistance. It would be interesting to pursue further genotypical characterization of these isolates, in particular the parC, gyrB, parE, rrl, rplD and rplV genes.
The study highlights the presence of the efflux pump gene lpeAB in Portuguese isolates, and the presence of the tet56 gene in a single L. anisa isolate which, to the best of our knowledge, is the first reported instance regarding presence of tetracycline destructase in this Legionella species.
The current epidemiologic trends of growing incidence of LD cases in several countries, allied with the global antibiotic resistance accentuates the task of establishing robust ECOFF values, clinical breakpoints, and a standardized methodology. The present research points to the current need to increase knowledge about environmental populations of Legionella to predict the potential emergence of antibiotic resistance in clinical isolates.
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
CC carried out the experiments, analyzed the data, and wrote the original draft manuscript. LR made data interpretations and revised and edited the manuscript. FF isolated Legionella and made data interpretations. RS collected the environmental samples and revised the manuscript. PP verified the data and revised the manuscript. MC designed the study, verified the data, and revised and edited the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1141115/full#supplementary-material
Almahmoud, I., Kay, E., Schneider, D., Maurin, M. (2009). Mutational paths towards increased fluoroquinolone resistance in legionella pneumophila. J. Antimicrob. Chemother. 64 (2), 284–293. doi: 10.1093/jac/dkp173
Andersson, D. I., Hughes, D. (2014). Microbiological effects of sublethal levels of antibiotics. Nat. Publ. Gr. 12, 1–14. doi: 10.1038/nrmicro3270
Assaidi, A., Ellouali, M., Latrache, H., Timinouni, M., Zahir, H., Karoumi, A., et al. (2020). Antimicrobial susceptibility patterns of Legionella spp. strains isolated from water systems in Morocco. Microb. Drug Resist. 26 (8), 991–996. doi: 10.1089/mdr.2019.0218
Law n.o 52/2018, of Aug 20 (Portugal). Available at: https://dre.pt/application/conteudo/116108098 (Accessed Jan 6 2023).
Beauté, J. (2017). Legionnaires’ disease in Europe, 2015 Vol. 35 (Stockholm: European Centre for Disease Prevention and Control).
Bruin, J. P., Koshkolda, T., Lu, C., Diederen, B. M. W., Boer, J. W. D., Mouton, J. W. (2014). Isolation of ciprofloxacin-resistant legionella pneumophila in a patient with severe pneumonia. J. Antimicrob. Chemother. 1 (Ld), 2869–2871. doi: 10.1093/jac/dku196
Chambers, S. T., Slow, S., Scott-thomas, A., Murdoch, D. R. (2021). Legionellosis caused by non-Legionella pneumophila species, with a focus on legionella longbeachae. Microorganisms 9 (2), 1–18. doi: 10.3390/microorganisms9020291
Chow, L. K. M., Ghaly, T. M., Gillings, M. R. (2021). A survey of sub-inhibitory concentrations of antibiotics in the environment. J. Environ. Sci. 99, 21–27. doi: 10.1016/j.jes.2020.05.030
Clinical and Laboratory Standards Institute (2018). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, approved standard, 11th ed. CLSI document M07-A11 Vol. 112 (Pensilvania: Clinical and Laboratory Standards Institute).
Cocuzza, C. E., Martinelli, M., Perdoni, F., Giubbi, C., Vinetti, M. E. A., Calaresu, E., et al. (2021). Antibiotic susceptibility of environmental Legionella pneumophila strains isolated in northern Italy. Int. J. Environ. Res. Public Health 18 (17), 0–9. doi: 10.3390/ijerph18179352
Correia, A. M., Ferreira, J. S., Borges, V., Nunes, A., Gomes, B., Capucho, R., et al. (2016). Probable person-to-Person transmission of legionnaires’ disease. N. Engl. J. Med. 374 (5), 497–498. doi: 10.1056/NEJMc1505356
Danner, M. C., Robertson, A., Behrends, V., Reiss, J. (2019). Antibiotic pollution in surface fresh waters: Occurrence and effects. Sci. Total Environ. 664, 793–804. doi: 10.1016/j.scitotenv.2019.01.406
Descours, G., Ginevra, C., Jacotin, N., Forey, F., Chastang, J., Kay, E., et al. (2017). Ribosomal mutations conferring macrolide resistance in legionella pneumophila. Antimicrob. Agents Chemother. 61 (3), 1–12. doi: 10.1093/jac/dkp173
European Centre for Disease Prevention and Control (2022). Legionnaires’ disease annual epidemiological report for 2020 Vol. 13 (Stockholm: European Centre for Disease Prevention and Control).
European Committee on Antimicrobial Susceptibility Testing (2021). Guidance document on antimicrobial susceptibility testing of legionella pneumophila Vol. 5 (Växjö: European Committee on Antimicrobial Susceptibility Testing).
European Committee on Antimicrobial Susceptibility Testing (2023) MIC and zone diameter distributions and ECOFFs. Available at: https://www.eucast.org/mic_and_zone_distributions_and_ecoffs (Accessed Jan 6 2023).
Forsberg, K. J., Patel, S., Wencewicz, T. A., Dantas, G. (2015). The tetracycline destructases: A novel family of tetracycline-inactivating enzymes. Chem. Biol. 22 (7), 888–897. doi: 10.1016/j.chembiol.2015.05.017
García, M. T., Jones, S., Pelaz, C., Millar, R. D., Abu Kwaik, Y. (2007). Acanthamoeba polyphaga resuscitates viable non-culturable Legionella pneumophila after disinfection. Environ. Microbiol. 9 (5), 1267–1277. doi: 10.1111/j.1462-2920.2007.01245.x
Graells, T., Hernández-García, M., Pérez-Jové, J., Guy, L., Padilla, E. (2018). Legionella pneumophila recurrently isolated in a Spanish hospital: Two years of antimicrobial resistance surveillance. Environ. Res. 166, 638–646. doi: 10.1016/j.envres.2018.06.045
Hughes, S. R., Kay, P., Brown, L. E. (2013). Global synthesis and critical evaluation of pharmaceutical data sets collected from river systems. Environ. Sci. Technol. 47 (2), 661–677. doi: 10.1021/es3030148
International Organization for Standardization (2017). Water quality — enumeration of legionella Vol. 46 (Geneva: International Organization for Standardization).
Janecko, N., Pokludova, L., Blahova, J., Svobodova, Z., Literak, I. (2016). Implications of fluoroquinolone contamination for the aquatic environment–a review. Environ. Toxicol. Chem. 35 (11), 2647–2656. doi: 10.1002/etc.3552
Joseph, S. J., Cox, D., Wolff, B., Morrison, S. S., Kozak-Muiznieks, N. A., Frace, M., et al. (2016). Dynamics of genome change among legionella species. Sci. Rep. 6, 1–14. doi: 10.1038/srep33442
Mandell, L. A., Wunderink, R. G., Anzueto, A., Bartlett, J. G., Campbell, G. D., Dean, N. C., et al. (2007). Infectious diseases society of America/American thoracic society consensus guidelines on the management of community-acquired pneumonia in adults. Clin. Infect. Dis. 44 (SUPPL. 2), S27–S72. doi: 10.1086/511159
Massip, C., Descours, G., Ginevra, C., Doublet, P., Jarraud, S., Gilbert, C. (2017). Macrolide resistance in Legionella pneumophila: The role of LpeAB efflux pump. J. Antimicrob. Chemother. 72 (5), 1327–1333. doi: 10.1093/jac/dkw594
Murray, A. K., Zhang, L., Yin, X., Zhang, T., Buckling, A., Snape, J., et al. (2018). Novel insights into selection for antibiotic resistance in complex microbial communities. MBio 9 (4), e00969–18. doi: 10.1128/mBio.00969-18
Nata, O. B., Løva, A. (2019). Susceptibility of legionella pneumophila to antimicrobial agents and the presence of the efflux pump LpeAB. J Antimicrob Chemother 2879, 1545–1550. doi: 10.1093/jac/dkz081
Nielsen, K., Hindersson, P., Hoiby, N., Bangsborg, J. M. (2000). Sequencing of the rpoB gene in Legionella pneumophila and characterization of mutations associated with rifampin resistance in the legionellaceae. Antimicrob. Agents Chemother. 44 (10), 2679–2683. doi: 10.1128/AAC.44.10.2679-2683.2000
Portal, E., Descours, G., Ginevra, C., Mentasti, M., Afshar, B., Chand, M., et al. (2021a). Legionella antibiotic susceptibility testing: Is it time for international standardization and evidence-based guidance? J. Antimicrob. Chemother. 76 (5), 1113–1116. doi: 10.1093/jac/dkab027
Portal, E., Sands, K., Portnojs, A., Chalker, V. J., Spiller, O. B. (2021b). Legionella antimicrobial sensitivity testing: comparison of microbroth dilution with BCYE and LASARUS solid media. J Antimicrob Chemother 14 (8), 1197–1204. doi: 10.2217/fmb-2019-0049
Rodriguez-Mozaz, S., Vaz-Moreira, I., Varela Della Giustina, S., Llorca, M., Barceló, D., Schubert, S., et al. (2020). Antibiotic residues in final effluents of European wastewater treatment plants and their impact on the aquatic environment. Environ. Int. 140, 105733. doi: 10.1016/j.envint.2020.105733
Sanli, N. O. (2019). Evaluation of biocidal efficacy of chloramine T trihydrate on planktonic and sessile bacteria in a model cooling tower water system. Water Sci. Technol. 79 (3), 526–536. doi: 10.2166/wst.2019.076
Slawek, D., Altshuler, D., Dubrovskaya, Y., Louie, E. (2017). Tigecycline as a second-line agent for legionnaires’ disease in severely ill patients. Open Forum Infect. Dis. 4 (4), 9–11. doi: 10.1093/ofid/ofx184
Sulyok, K., Kreizinger, Z., Wehmann, E., Lysnyansky, I., Bányai, K., Marton, S., et al. (2017). Mutations associated with decreased susceptibility to seven antimicrobial families in field and laboratory-derived Mycoplasma bovis strains. Antimicrob. Agents Chemother. 61 (2), 1–14. doi: 10.1128/AAC.01983-16
Torre, I., Alfano, R., Borriello, T., De Giglio, O., Iervolino, C., Montagna, M. T., et al. (2018). Environmental surveillance and in vitro activity of antimicrobial agents against Legionella pneumophila isolated from hospital water systems in campania, south Italy: a 5-year study. Environ. Res. 164, 574–579. doi: 10.1016/j.envres.2018.02.030
Turnidge, J., Kahlmeter, G., Kronvall, G. (2006). Statistical characterization of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin. Microbiol. Infect. 12 (5), 418–425. doi: 10.1111/j.1469-0691.2006.01377.x
Valve, K., Vaalasti, A., Anttila, V. J., Vuento, R. (2010). Disseminated Legionella pneumophila infection in an immunocompromised patient treated with tigecycline. Scand. J. Infect. Dis. 42 (2), 152–155. doi: 10.3109/00365540903359895
Vandewalle-Capo, M., Massip, C., Descours, G., Charavit, J., Chastang, J., Billy, P. A., et al. (2017). Minimum inhibitory concentration (MIC) distribution among wild-type strains of Legionella pneumophila identifies a subpopulation with reduced susceptibility to macrolides owing to efflux pump genes. Int. J. Antimicrob. Agents 50 (5), 684–689. doi: 10.1016/j.ijantimicag.2017.08.001
Wilson, R. E., Hill, R. L. R., Chalker, V. J., Mentasti, M., Ready, D. (2018). Antibiotic susceptibility of Legionella pneumophila strains isolated in England and Wales 2007–17. J. Antimicrob. Chemother. 73 (10), 2757–2761. doi: 10.1093/jac/dky253
Woodhead, M., Blasi, F., Ewig, S., Garau, J., Huchon, G., Ieven, M., et al. (2011). New guidelines for the management of adult lower respiratory tract infections. Eur. Respir. J. 38 (6), 1250–1251. doi: 10.1111/j.1469-0691.2011.03672.x
Xiong, L., Yan, H., Shi, L., Mo, Z. (2016). Antibiotic susceptibility of Legionella strains isolated from public water sources in Macau and guangzhou. J. Water Health 14 (6), 1041–1046. doi: 10.2166/wh.2016.056
Yang, J. L., Sun, H., Zhou, X., Yang, M., Zhan, X. Y. (2022). Antimicrobial susceptibility profiles and tentative epidemiological cutoff values of legionella pneumophila from environmental water and soil sources in China. Front. Microbiol. 13 (924706). doi: 10.3389/fmicb.2022.924709
Keywords: Legionella, broth microdilution, MIC, fluoroquinolones, macrolides, environmental, antibiotic susceptibility
Citation: Cruz C, Rodrigues L, Fernandes F, Santos R, Paixão P and Chasqueira MJ (2023) Antibiotic susceptibility pattern of Portuguese environmental Legionella isolates. Front. Cell. Infect. Microbiol. 13:1141115. doi: 10.3389/fcimb.2023.1141115
Received: 10 January 2023; Accepted: 24 March 2023;
Published: 21 April 2023.
Edited by:
Diane S J. Lindsays, NHS Greater Glasgow and Clyde, United KingdomReviewed by:
Jianzhong Huang, GlaxoSmithKline, United StatesCopyright © 2023 Cruz, Rodrigues, Fernandes, Santos, Paixão and Chasqueira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Jesus Chasqueira, bWpjaGFzcXVlaXJhQG5tcy51bmwucHQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.