- 1Department of Clinical Laboratory Medicine, Taizhou Municipal Hospital, Taizhou, China
- 2State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
- 3Nanxiang Branch of Ruijin Hospital, Shanghai Jiaotong University, Shanghai, China
- 4Institutes of Biomedical Sciences, Shanxi University, Taiyuan, China
Introduction: The objective of this study is to thoroughly analyze the detailed genomic characteristics of clinical strain 211703 of Aeromonas caviae, which co-carrying blaRSA-1 and blaNDM-1 genes. 211703 was isolated from the patient’s cerebrospinal fluid drainage sample in a Chinese tertiary hospital.
Methods: Carbapenemase NDM was detected by the immunocolloidal gold technique. The MIC values were determined by VITEK2. The whole genome sequence of 211703 was analyzed using phylogenetics, genomic comparison, and extensive dissection.
Results: This study revealed that 211703 only contained a single 4.78 Mb chromosome (61.8% GC content), and no plasmids were discovered in 211703. 15 different types of resistant genes were detected in the genome of 211703, including blaRSA-1 harbored on integrative and mobilizable element (IME) Tn7413a, and blaNDM-1 harbored on integrative and conjugative element (ICE). The ICE and IME were all carried on the chromosome of 211703 (c211703). Detailed comparison of related IMEs/ICEs showed that they shared similar conserved backbone regions, respectively. Comprehensive annotation revealed that blaRSA-1 was carried by the gene cassette of a novel integron In2148 on Tn7413a, and blaNDM-1 was captured by an insertion sequence ISCR14-like on the ICE of 211703. We speculated that mobile genetic elements (MGEs) such as ICE and IME facilitated the spread of resistance genes such as blaRSA-1 and blaNDM-1.
Discussion: In conclusion, this study provides an overall understanding of the genomic characterization of clinically isolated A. caviae 211703, and an in-depth discussion of multiple acquisition methods of drug resistance genes in Aeromonas. To the best of our knowledge, this is the first report of A. caviae carrying blaRSA-1 even both blaRSA-1 and blaNDM-1, and this is the first bacterium carrying blaRSA-1 isolated from the clinical setting.
Introduction
The genus Aeromonas comprises a group of Gram-negative bacterium worldwide distributed in aquatic and soil environments (Fernández-Bravo and Figueras, 2020). Among 42 different species included in the genus Aeromonas, three species (Aeromonas hydrophila, Aeromonas caviae, and Aeromonas veronii) account for the vast majority of human infections and clinical isolates (Pessoa et al., 2022). In recent years, Aeromonas have gradually increased, and it has become a common opportunistic pathogen that causes human infection (Bhowmick and Bhattacharjee, 2018; Pessoa et al., 2022). The main reason of carbapenem resistance in Aeromonas is carrying cphA gene encoding a CphA metallo-β-lactamase (Wu et al., 2012). In addition, Aeromonas has the ability to acquire different types of carbapenemase genes. In 2007, French scholars Neuwirth. C et al. reported a carbapenem-resistant clinical isolate of A. caviae for the first time, which produced a class B carbapenemase IMP-19 (Neuwirth et al., 2007). Subsequently, in 2008, Swedish scholars Bala´zs Libisch et al. reported the first clinical isolate of A. hydrophila producing VIM-4 (Libisch et al., 2008). Following this, environmental and clinical isolates of Aeromonas spp. producing class A carbapenemase KPC were reported in Brazil, the USA, Japan, and China, respectively (Montezzi et al., 2015; Hughes et al., 2016; Hu et al., 2019; Sekizuka et al., 2019; Yang et al., 2022). In 2017, Indian researchers Shalini Anandan and colleagues made the initial discovery of A. caviae strain carrying the blaOXA-181 gene, located within the composite transposon Tn2013. This was confirmed through whole genome sequencing. (Anandan et al., 2017). In 2018, Japanese scholar Kohei Uechi et al. reported a clinical isolate of A. hydrophila encoding GES-24 carbapenemase, and blaGES-24 was located in the integron (Uechi et al., 2018b). In 2022, Shuguang Xu et al. discovered an A. caviae strain carrying the blaNDM gene located on its chromosome. (Xu et al., 2022). In the same year, our research team reported blaNDM-1 harboring in plasmid pK433-NDM carried in A. caviae K433 isolated from patient with community-acquired pneumonia (Luo et al., 2022). Overall, Aeromonas is becoming more prevalent in the clinic, and multidrug resistant A. caviae is emerging as a potential threat to human society and public health in recent years.
Ambler class B β-lactamase New Delhi metallo-β-lactamase (NDM) is one of the most common carbapenemases in Enterobacterales (Nordmann et al., 2011; Porreca et al., 2018; Wu et al., 2019), which can hydrolyze nearly all classes of β-lactam antibiotics. blaNDM-1, the first identified gene of 41 known variants encoding NDM, was initially detected in a Klebsiella pneumoniae strain in New Delhi in 2008 (Yong et al., 2009). Since then, blaNDM-1 is prevalent in various species and has rapidly spread all over the world (Dortet et al., 2014). In addition to blaNDM-1, blaRSA-1, a noval gene encoding class A β-lactamase, was first discovered and identified in Indian river sediments in 2018 (Marathe et al., 2018). Open reading frames (ORFs) of blaRSA-1 were synthesized and cloned in E. coli in further experiments. The clone expressing class A enzyme encoded by blaRSA-1 was verified to efficiently hydrolyze benzylpenicillin, cephalothin (first generation cephalosporin), and cefotaxime (third generation cephalosporin). The clone exhibited a β-lactam resistance phenotype typical for Extended Spectrum Beta-Lactamase (ESBL) production and showed a great resistance challenge (Marathe et al., 2018). Although blaNDM-1 and blaRSA-1 are significant drug resistance genes, strains that produce both NDM and RSA have not been reported.
Integrative and conjugative element (ICE) (Delavat et al., 2017; Botelho and Schulenburg, 2021) and integrative and mobilizable element (IME) (Bellanger et al., 2014; Guedon et al., 2017) are two types of mobile genetic elements (MGEs) primarily resided in the bacterial cell’s chromosome, and both of them have the ability to carry resistance genes. ICEs can be transferred between cells by conjugation, which function is self-encoded. The core components of an ICE typically include attL (attachment site at the left end of the ICE), int (integrase), xis (excisionase), rlx (relaxase), nic/oriT (nick site, origin of conjugative replication), cpl (coupling protein), a P (TivB)- or F (TivF)-type T4SS machinery (mating pair formation), and attR (attachment site at the right end of the ICE). IMEs are autonomous in integration but nonautonomous in conjugation. IMEs contained no conjugal transfer genes and typically have attL, int, rlx, oriT, and attR.
In this work, a carbapenem-resistant A. caviae strain 211703 co-carrying blaNDM-1 and blaRSA-1 was identified in clinical setting. According to the sequencing results, the whole genome of A. caviae strain 211703 contained only the chromosome (c211703), and no plasmid was found. All resistance genes including blaNDM-1 and blaRSA-1, and antimicrobial susceptibility profiles of A. caviae 211703 were obtained. The genetic dissection of strain 211703 of A. caviae revealed the presence of ICE carrying the blaNDM-1 gene and IME carrying the blaRSA-1 gene, both of which had integrated into the chromosome.
Comprehensive genomic comparisons of above ICE and IME with their closely related elements were performed, respectively. According to our understanding, this is the first report of A. caviae strain co-carrying blaNDM-1 and blaRSA-1. At the same time, this is the first report of strains carrying blaRSA-1 gene found in a clinical setting. This research will provide a new understanding for the genetic context of novel antibiotic resistance genes in A. caviae, and in-depth insights for the integration, transformation, conjugation, and mobilization of MGEs related to resistance genes under the selection pressure.
Materials and methods
Sample collection and species identification
On 10 October 2021, a patient (52-year-old male) was admitted to the neurosurgery department, due to sudden unconsciousness and vomiting for three and a half hours. The diagnosis was spontaneous subarachnoid hemorrhage, hemorrhage in the third, fourth and lateral ventricles. After admission, right ventricle drainage was performed. Fluid replacement, brain protection, prevention of bleeding infection, and neurological rehabilitation were given. On the eighth day of admission, the patient was considered intracranial infection with the body temperature 37.9°C and persistent unconsciousness, and a carbapenem-resistant A. caviae strain 211703 was isolated from the patient’s cerebrospinal fluid drainage sample initially identified by Vitek 2. Later, bacterial species identification using average nucleotide identity (ANI) analysis (http://www.ezbiocloud.net/tools/ani) based on genome sequences finally proved that 211703 belongs to A. caviae (Richter and Rossello-Mora, 2009).
Antibiotic susceptibility test and carbapenemases phenotype detection
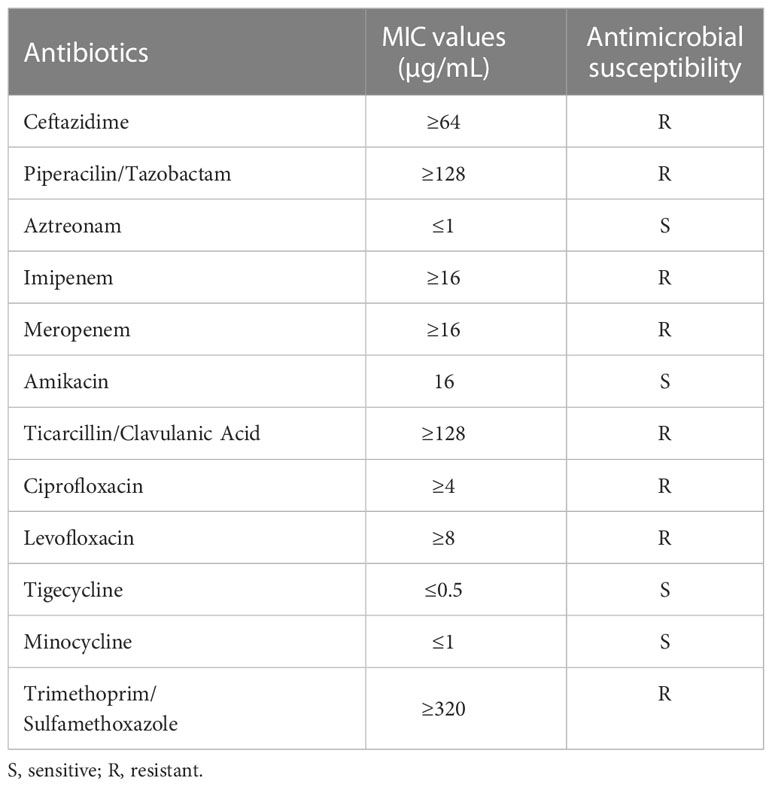
The drug minimum inhibitory concentrations (MICs) of 211703 were determined by BioMérieux VITEK2 (Table 1). The antibiotic susceptibility test results were determined by the Clinical and Laboratory Standards Institute (CLSI) guidelines (2021). The production of NDM carbapenemase in 211703 was detected on an NG-Test CARBA 5 (NG Biotech, Guipry, France), a rapid diagnostic test based on the immunocolloidal gold technique (Figure S1).
Conjugal transfer
Conjugal transfer experiments were carried out with rifampicin-resistant Escherichia coli EC600 being used as a recipient, and the strain 211703 as a donor. The donor and recipient strains were grown in three milliliters (mL) brain heart infusion (BHI) broth overnight at 37°C. And then, 50 μL of donor strain culture was mixed with 500 μL of recipient strain culture (v:v = 1:10) and 4.5 mL of fresh BHI broth. In addition, 100 μL of the mixture was applied onto a cellulose filter membrane (pore size, 0.22 μm) already placed on a BHI agar plate. After incubation at 37°C for 16h-18h, the filter membrane was taken out and vortexed in 1 mL of BHI broth. The vortex mixtures were plated on BHI agar plates containing 2 mg/L imipenem and 1,500 mg/L rifampicin for the selection of the transconjugants. However, repeated conjugation experiments failed to acquire transconjugants.
Sequencing and sequence assembly
Bacterial genomic DNA of A. caviae 211703 was isolated using the Gentra Puregene Yeast/Bact. Kit (Qiagen, Valencia, CA). The TruePrepTM DNA Library Prep Kit V2 and the SQU-LSK109 Ligation Sequencing kit were used for libraries preparation separately. After the preparation of the library was completed, it was separately sequenced on an Illumina HiSeq X Ten platform (Illumina Inc., San Diego, CA, USA) and GridION X5 platform (Oxford Nanopore, UK). Raw data from the HiSeq X Ten platform and the GridION X5 platform were trimmed to obtain the high-quality clean reads (clean data) by Canu v1.8 (https://canu.readthedocs.io/en/latest/index.html). The paired-end short Illumina reads and the long Nanopore reads were assembled de novo utilizing Unicycler v0.4.8.0 (https://github.com/rrwick/Unicycler).
Whole-genome phylogeny and genetic background analysis
A total of 62 sequences of A. caviae sequenced at three levels (scaffold, chromosome, and complete) were downloaded from NCBI (last accessed on 15 Aug 2022), which were isolated from various sources from 2004 to 2021 (Table S1). Assembly method of 211703 genome that were used in the phylogenetic analysis was Unicycler v0.4.8.0. MUMmer v3.1 was used for alignments against the reference genome to create a core genome alignment (Delcher et al., 2003). The core-genome length for phylogenetic analysis was 4.5-Mb. A total of 394,778 single nucleotide polymorphisms (SNPs) in the backbone regions were identified and extracted, and a maximum-likelihood phylogenetic tree was constructed based on the SNPs’ dataset (Figure 1). Matrix of pairwise SNP distances among studied genomes was shown in Table S2. The phylogenetic tree and related background information (collection date, location, isolation source, and host) were shown using the Interactive Tree of Life (iTOL) programs (Letunic and Bork, 2021).
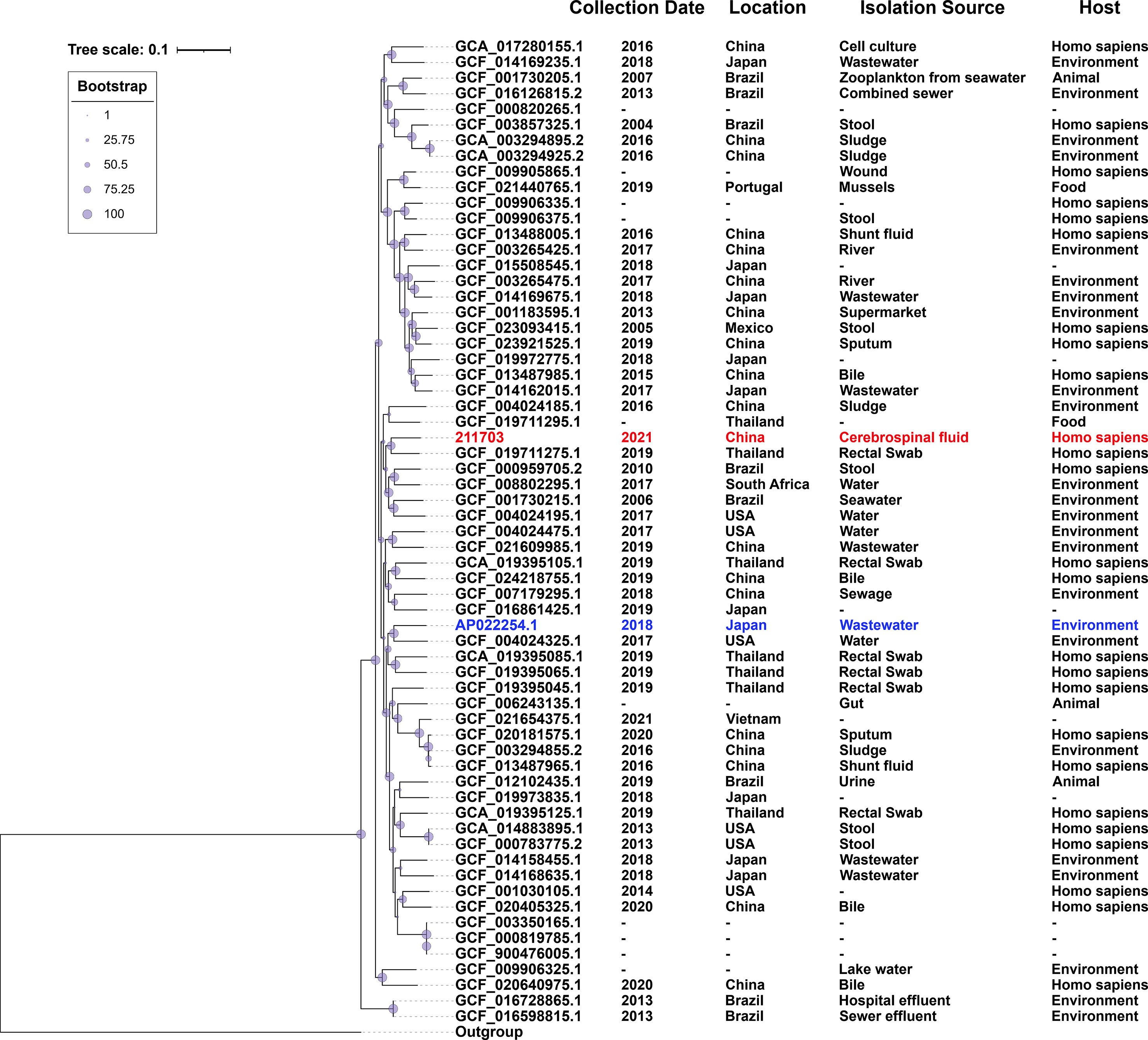
Figure 1 Population distribution of A. caviae 211703 with 62 A. caviae genomes. The phylogenetic tree was constructed by the Maximum-likelihood method. The degree of support (percentage) for each cluster of associated taxa, as determined by bootstrap analysis, was shown with blue dots next to each branch. The bar corresponded to the scale of sequence divergence. 211703 was indicated in red and AP022254.1 was indicated in blue.
Sequence annotation and comparison
ORFs and pseudogenes of 211703 genome were predicted using RAST 2.0 (Brettin et al., 2015). Further detailed dissection and manual annotation were performed using BLASTP/BLASTN (Boratyn et al., 2013). Annotation of resistance genes, MGEs, and other features was carried out using online databases such as CARD (Alcock et al., 2020), ResFinder (Zankari et al., 2012), ISfinder (Siguier et al., 2006), and INTEGRALL (Moura et al., 2009), and the Tn Number Registry (Roberts et al., 2008). Alignments with homologous sequences of ICE harboring on 211703 were performed by using the BRIG tool (Alikhan et al., 2011). Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.31 and BLASTN (Edgar, 2004). Comparison diagrams of ICEs/IMEs regions were drawn using Inkscape 1.1 (https://inkscape.org/en).
Nucleotide sequence accession number
The complete sequence of A. caviae 211703 (chromosome c211703) was submitted to the GenBank database, under accession number CP092181. The Illumina reads of A. caviae 211703 was deposited as a SRA in the GenBank database (accession number PRJNA934671).
Results
Antimicrobial susceptibility test
Antimicrobial susceptibility tests were performed on A. caviae 211703. The results showed that 211703 exhibited resistance to a broad range of tested antimicrobials except aztreonam, amikacin, tigecycline, and minocycline (Table 1). The production of NDM carbapenemase was confirmed by immunocolloidal gold technique, which was consistent with the result of antibiotic susceptibility tests (Figure S1). Corresponding to the results of multidrug resistance profile, 15 different types of resistant genes were detected from A. caviae 211703 (Table S3), which confer resistance to aminoglycosides (arr2 and aphA6), β-lactams (blaMOX-6, blaNDM-1, and blaRSA-1), macrolides (mph(A), mph(E), and msr(E)), quinolones (qnrVC1), sulphonamides (sul1), and other antibiotics.
Phylogenetic Analysis of Aeromonas caviae 211703
Based on the genome of 211703 and all A. caviae strains available from GenBank of NCBI whose assembly levels are scaffold, chromosome, or complete, phylogenetic analysis was performed to explore the evolutionary relationships and potential associations (Sayers et al., 2021). A total of 62 A. caviae strains from GenBank were included, and the related information of these strains were collected and verified manually (Table S1). From the background information, the main collection sites of these strains were China (18/62), Japan (10/62), Brazil (9/62), Thailand (7/62), and the USA (6/62). Among these 62 strains, the chromosome sequence of A. caviae WP8-S18-ESBL-04 (accession number AP022254, GCF_014169735.1, the standard strain of A. caviae) was used as the reference. Strain OnP3.1 (accession number CP050851, GCF_017310215.1, the standard strain of A. hydrophila) was used as the outgroup. After multiple sequence alignment, a total of 394,778 SNPs located in the core genome regions were identified and extracted. Using these SNPs’ dataset, a maximum likelihood (ML) phylogenetic tree was constructed. Figure 1 showed the phylogenetic tree and marked basic background information of each node/strain such as the collection date, location, isolation source, and host. The most closely related strain to 211703 was GCF_019711275.1, which was isolated from a rectal swab in Thailand in 2019. They both belonged to the same branch on the evolutionary tree.
Overview of the Aeromonas caviae 211703
The whole genome sequence of A. caviae 211703 was obtained by high-throughput sequencing. Genome sequencing result revealed that A. caviae 211703 contained only chromosome (c211703, accession number CP092181) with a length of 4,783,384 bp, and no plasmid was found in its genome after verification (Table 2). The mean G+C content of c211703 was 61.8% and 4,400 ORFs were predicted on c211703. Based on the six key housekeeping genes (gltA, groL, gyrB, metG, ppsA, and recA) identified on c211703, MLST typing method proved that 211703 belongs to ST928.
Among the 16 resistance locus existing on c211703, 15 of them were concentrated on one ICE (6/16, including blaNDM-1) and one IME (9/16, including blaRSA-1) (Table S3; Figure S2). Further, detailed genome annotation identified the location and genetic context of ICE and IME (designated as Tn7413a). In order to explore the similarities and differences in the acquisition of resistance genes and the genetic environment, precise comparisons were performed with related ICEs and IMEs, respectively.
Genetic characterization of the IME Tn7413a
The IME Tn7413a carrying blaRSA-1 was 35,097 bp in length, located on A. caviae 211703 from 2,715,927 bp to 2,751,023 bp (Figure S2). Accurate annotation and genomic dissection revealed that Tn7413a was bordered by a pair of 18-bp attL/attR (attachment site at the left/right end) (Figure 2). According to genes function, Tn7413a can be divided into backbone regions and accessory regions. Backbone regions with a length of 16.2 kb were consisted by genes such as int, repC, hipA, and hipB. Integron In2148 inserted into the backbone regions, as the accessory region of Tn7413a.

Figure 2 Comparison of Tn7413a with related IMEs. Genes were denoted by arrows. Genes, mobile genetic elements, and other features were colored based on their functional classification. Shading denoted regions of homology (nucleotide identity > 95%). A single quotation mark in front of the gene name indicated pseudogene.
In2148 was a novel concise class 1 integron containing primary components 5’-conserved region (5’-CS), 3’-conserved region (3’-CS), and a gene cassette array (GCA) qnrVC1–arr2–blaRSA-1–dfrA1–gcuC. The blaRSA-1 gene was found to be located in the GCA of In2148, most likely due to the integration of an integron, which facilitated its spread. In the downstream of 3’-CS, there was a reverse putative resistance unit IS26–mph(A)–IS6100 unit carrying mph(A), encoding proteins conferring resistance to macrolides (Partridge et al., 2018). In the upstream of IS26–mph(A)–IS6100 unit, IS26 was shared by IS26–mph(E)–IS26 unit followed by a truncated tniA encoding DD(35)E transposase TniA. A pair of 25-bp IRi (inverted repeat at the integrase end) and IRt (inverted repeat at the tni end) flanked In2148. On the outside of IRi and IRt, there were 5-bp DRs (direct repeats, target site duplication signals for transposition), respectively.
Eight Tn7413a-related IMEs Tn7413b-h and Tn7412
To investigate the structural variations among these IMEs, a BLAST analysis was performed using the backbone region sequences of Tn7413a as a reference. BLAST analysis revealed that a total of seven IMEs in the GenBank database were similar to Tn7413a, sharing 100% coverage and >99% identity for gene int from Tn7413a. These seven IMEs were included in this analysis, and the gene structures of these IMEs were detailed annotated manually. Due to their structural similarity to Tn7413a, these seven IMEs were designated as Tn7413b-h as listed in Table S4.
Tn7413b-h ranged in length from 29,639 bp to 59,408 bp, and were all distributed in Aeromonas. Similar to Tn7413a, the boundaries of these seven IMEs were a pair of 18-bp attL/attR. The backbone regions of Tn7413b-h were almost identical to that of Tn7413a, but differed in the accessory regions (Figure 2). In all the seven IMEs except Tn7413d, there was only one accessory region with integron as the primary content. Two insertion sequences insertions in Tn7413d resulted in its backbone gene hsdS being interrupted into three parts. In Tn7413b-d and Tn7413f-h, In2148 in Tn7413a was replaced by In27. In27 was a concise class 1 integron, carrying GCA dfrA12–gcuF–aadA2 (Partridge et al., 2009; Partridge, 2011). In Tn7413e, In2148 in Tn7413a was replaced by In384, a concise class 1 integron carrying GCA dfrA12–gcuF. In Tn7413c-e, the insertion sequence ISAve3 inserted upstream of the integron. For all these seven IMEs Tn7413b-h, there was the putative resistance unit chrA–orf98 unit downstream of the 3’-CS of the integron they carried. Further, the chrA–orf98 unit was all followed by an IS26–mph(A)–IS6100 unit, which was same as in Tn7413a. For Tn7413c-e and Tn7413h, all contained single or multiple Tn4352, a composite transposon involved in kanamycin resistance (Wrighton and Strike, 1987). For Tn7413b, the most identical IME related to Tn7413a, its integron In27 contained the IS26–mph(E)–IS26 unit, which was identical to Tn7413a. Tn7413g contained a putative resistance unit ISCR2–floR unit. ISCR elements were bounded by an origin (oriIS) downstream and a terminus (terIS) upstream, responsible for capturing resistance genes (Partridge, 2011; Partridge et al., 2018). In Tn7413h, there was a novel composite transposon newly designated Tn7419, flanked by 8-bp DRs both sides. Both ends of Tn7419 were insertion sequence ISAhy4, and the middle part were cmeA, cmeB, and cmeC, encoding membrane fusion proteins related to RND efflux system. Meanwhile, downstream of Tn7419, there were IS26–catA2–IS26 unit and IS26–tetA(D)–IS26 unit sharing IS26, carrying resistance genes catA2 and tetA(D), respectively.
Furthermore, in order to explore the genetic environment differences of blaRSA-1, the genetic environment of the previously reported blaRSA-1, which belonged to Aeromonas spp. strain 1402 was also included in this study (Marathe et al., 2018). The results showed that the gene fragment carrying blaRSA-1 in former report was an IME with a length of 50,823 bp, which was also bounded by a pair of 18-bp length attL/attR (Figure 2). Backbone regions of Tn7412 also contained genes such as int, hipA, and hipB, but other genes in the backbone region were quite different from Tn7413a and Tn7413b-h. The accessory region of Tn7412 was mainly composed of concise class 1 integron In1357, containing GCA qacG–blaRSA-1–dfrA1–gcuC. Coincidentally, blaRSA-1 was also located in the gene cassette of the integron, consistent with Tn7413a. After the 3’-CS of In1357, there were chrA–orf98 unit and IS26–mph(A)–IS6100 unit.
Genetic characterization of ICE of Aeromonas caviae 211703
ICE of A. caviae 211703 was located on chromosome from 3,757,651 bp to 3,865,845 bp with a length of 108,195 bp, bounded by a pair of 19-bp attL/attR (Figure S2). Genomic annotation revealed that ICE of A. caviae 211703 can also be divided into backbone regions and accessory regions according to their functions (Figure 3). The backbone regions consisted of two parts, including backbone of maintenance harboring gene int and backbone of conjugal transfer. Accurate annotation demonstrated that there were two accessory regions on A. caviae 211703, ars locus and blaNDM-1 region. ars locus were a series of resistance genes encoding proteins related to arsenic resistance. blaNDM-1 region from ICE of 211703 was formed by multiple recombination mediated by ISCR14-like (Figure 4). Unlike common structures of genetic contexts harboring blaNDM-1 previously reported (ΔTn125 harboring blaNDM-1 captured by ISCR1) (Wu et al., 2019; Luo et al., 2021), the blaNDM-1 genetic context was formed by the insertion sequence ISCR14-like mediated capture of ΔTn125 harboring blaNDM-1 and bleMBL. Downstream of ΔISAba125 were another ISCR14-like with floR, tetR(G), and tetA(G), which together constituted the end of the blaNDM-1 region from ICE of 211703. There were four ISCR14-like elements upstream of the ISCR14-like captured ΔTn125, which indicated that multiple insertions or recombination occurred.
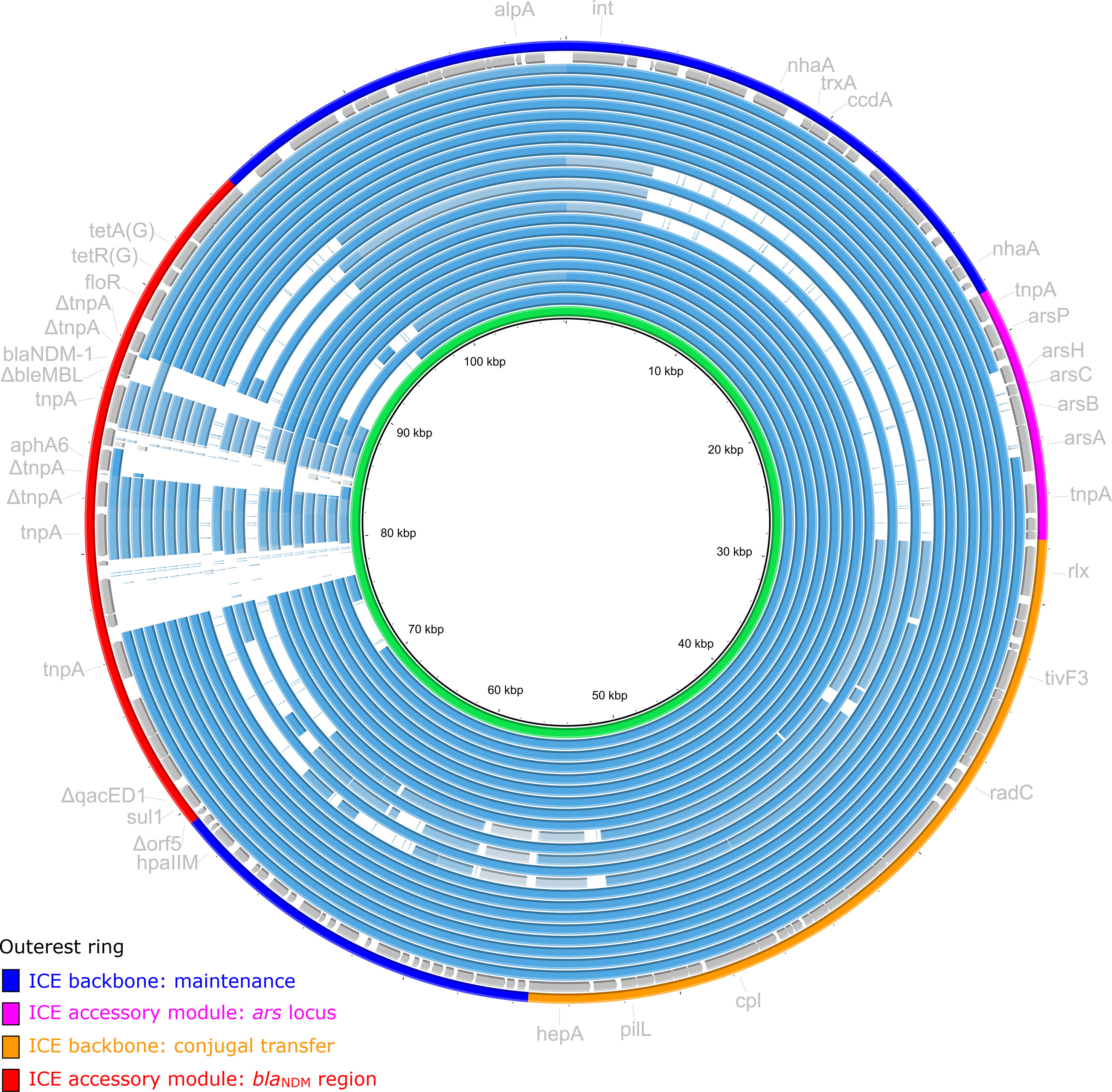
Figure 3 Alignment of ICE of A. caviae 211703 with related ICEs. ICE of A. caviae 211703 with 21 other similar ICEs deposited in the GenBank database were included. The rings of plasmids were arranged in the order (from inner to outer) as described in Table S5. The second outer ring colored in gray were annotations of ICE of A. caviae 211703. Highlighted on the outer ring represented functional regions (backbone regions and accessory regions).
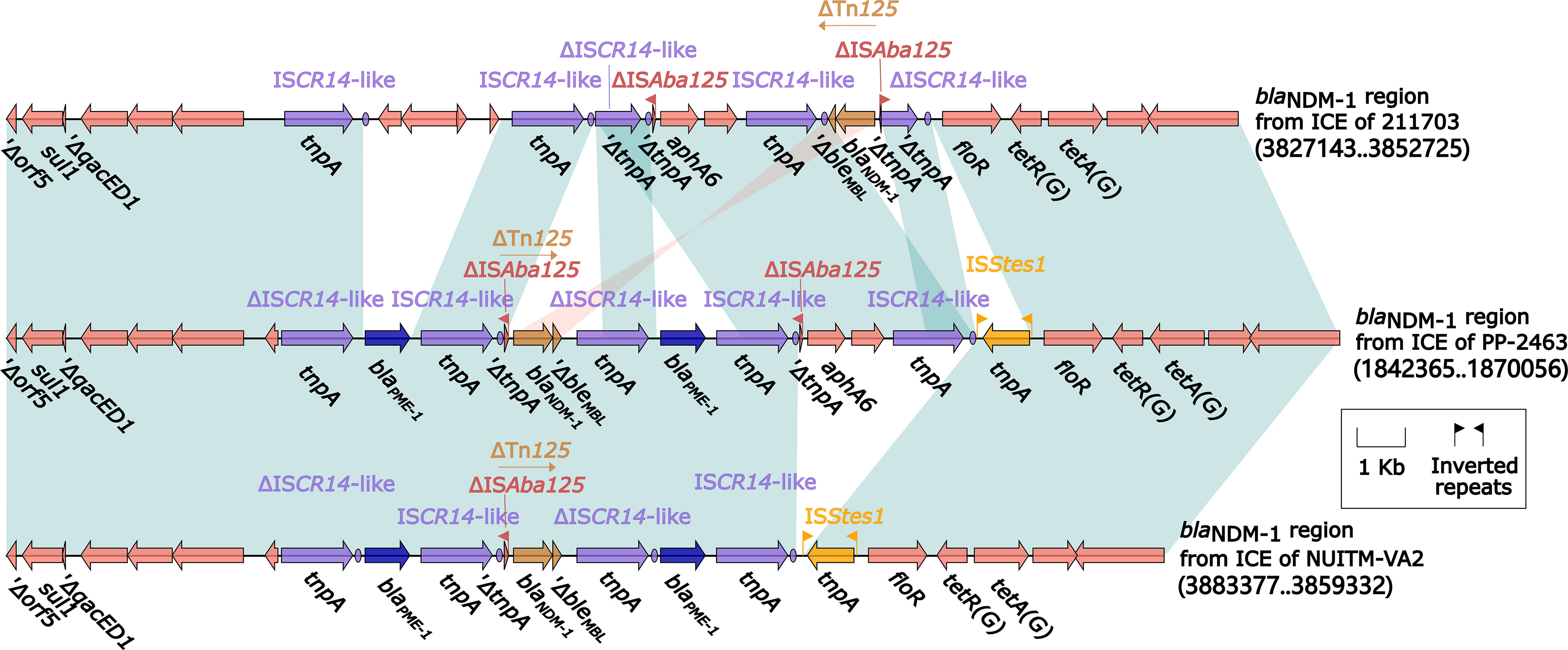
Figure 4 Comparison of ICEs harboring blaNDM-1 region from 211703, PP-2463, and NUITM-VA2. Genes were denoted by arrows. Genes, mobile genetic elements, and other features were colored based on their functional classification. Shading denoted regions of homology (nucleotide identity > 95%). A single quotation mark in front of the gene name indicated pseudogene.
BLAST analysis revealed that a total of 21 ICEs in the GenBank database were almost identical to ICE of A. caviae 211703 (100% coverage and >95% identity for gene int from ICE of A. caviae 211703) (Table S5). The 21 ICEs ranged in length from 88,228 bp to 112,147 bp. Most of these ICEs (14/21) were distributed in Pseudomonas, where ICE of Pseudomonas juntendi PP-2463 was highly identical to ICE of A. caviae 211703. P. juntendi PP-2463 also isolated from the same city (Taizhou, China) with A. caviae 211703, which indicated that ICE of A. caviae 211703 may be transferred from Pseudomonas to Aeromonas. Only one of the 21 ICEs originated from Aeromonas (ICE of NUITM-VA2). As shown in Figure 3, most of these ICEs (17/21) maintained high concordance with the ICE of 211703 in the backbone region of maintenance, ars locus, and the backbone region of conjugal transfer, and the differences mainly occurred in the accessory region where the blaNDM-1 region was located.
Among the 21 ICEs, the ICE of P. juntendi PP_2463 (sharing the most similar accessory regions with the ICE of A. caviae 211703) and the ICE of A. caviae NUITM-VA2 (the only one ICE from A. caviae), were specially annotated and analyzed. Detailed annotation of their blaNDM-1 region and comparison with blaNDM-1 region from ICE of 211703 were performed (Figure 4). Results showed that the genetic contexts of blaNDM-1 region from ICE of PP-2463/NUITM-VA2 shared a common structure in upstream(Δorf5–sul1–ΔqacED1–ISCR14-like) with blaNDM-1 region from ICE of 211703, following by a replacement of gene blaPME-1. The genetic structures identified included an ISCR14-like ΔTn125 carrying the blaNDM-1 gene, as well as a ΔISCR14-like–blaPME-1–ISCR14-like structure, which was not present in the ICE carrying blaNDM-1 in strain 211703. The only difference found between the blaNDM-1 regions from ICE of PP-2463 and NUITM-VA2 was an insertion of aphA6–ISCR14-like, which was also present in the blaNDM-1 region from ICE of 211703. Despite this variation, all three blaNDM-1 regions shared a common structure downstream, consisting of floR–tetR(G)–tetA(G). However, there was an additional insertion of the ISStes1 sequence downstream of the blaNDM-1 regions in both ICE of PP-2463 and NUITM-VA2.
Discussion
Carbapenem-resistant Aeromonas has received more attention since the first isolate from France harboring an acquired blaIMP gene was reported in 2007 (Neuwirth et al., 2007; Pessoa et al., 2022). So far, carbapenem resistance genes such as blaKPC (Montezzi et al., 2015), blaNDM (Luo et al., 2022), blaOXA (Anandan et al., 2017), and blaGES (Uechi et al., 2018a) have been reported many times in Aeromonas. From a geographic perspective, carbapenem-resistant Aeromonas were distributed almost all over the world, such as the USA (Hughes et al., 2016), China (Luo et al., 2022), Japan (Sekizuka et al., 2019), Brazil (Conte et al., 2022), and Europe (Neuwirth et al., 2007). From the perspective of host distribution, Aeromonas were reported in different hosts such as human (Luo et al., 2022) and environment (river sediment, wastewater treatment plant effluent, and hospital effluent) (Hu et al., 2019; Sekizuka et al., 2019; Conte et al., 2022). In the “One Health” approach, the emergence of Aeromonas carrying antibiotic resistance genes indicates that the drug-resistant Aeromonas is becoming an important threat to human public health. To the best of our knowledge, the newly discovered strain 211703 of A. caviae in this study, which carrying the novel blaRSA-1 gene, is the first such bacterium to be isolated from a clinical setting worldwide. It is also the first report of Aeromonas harboring a new drug resistance gene combination of blaRSA-1 and blaNDM-1. At the same time, the blaRSA-1 and blaNDM-1 gene in this study were acquired by A. caviae in two different ways, indicating that the drug resistance of A. caviae changes rapidly under selection pressure, marking a further increase in the threat of A. caviae to human society.
The blaRSA-1 gene was first discovered in Indian river sediments in 2018 (Marathe et al., 2018). Through the verification of metagenomics and drug susceptibility experiments, as well as phylogenetic analysis, the blaRSA-1 gene was determined to be a new beta-lactam antibiotic resistance gene. In previous study, the blaRSA-1 gene was located in the integron gene cassette, but the gene environment was not analyzed by detailed annotation. After 2018, there has been no related report of blaRSA-1 gene. In this study, we identified strain A. caviae 211703 from clinical setting carrying the blaRSA-1 gene. Compared with previous report, we speculated the blaRSA-1 gene was transferred from India to China geographically, and from the river sediments to the clinic in terms of infection environment. The only similarity is that both of the blaRSA-1 genes are located in the gene cassette of the integron. We carried out detailed annotation and comparative genomics analysis of the gene environment where the blaRSA-1 is located, and found that there are many IMEs (Tn7413b-h) that are similar in structure to the IME (Tn7413a) where the integron In2148 carrying blaRSA-1 is located (Figure 2; Table S4). The IMEs Tn7413a-h backbone regions are relatively conserved and intact, although their accessory regions have undergone recombination events such as replacement or insertion. The IMEs Tn7413a-h are all derived from Aeromonas and have highly conserved and homologous backbone regions, indicating a widespread distribution of this category of IMEs within Aeromonas. At the same time, the blaRSA-1 gene is located in the gene cassette of In2148, which can be speculated that the gene blaRSA-1 carrying by the IME Tn7413a was derived by the capture from integron. Therefore, it is foreseeable that the blaRSA-1 gene cassette has gained a stronger capability to spread. In addition, the IME Tn7413a harboring blaRSA-1 is likely to spread in Aeromonas through conjugal transfer and other methods, although the IME itself does not have the ability to transfer. Its threat cannot be underestimated.
blaNDM-1 is a drug resistance gene encoding carbapenemase, which has strong resistance to carbapenems, and has been widely reported in Enterobacterales all over the world (Nordmann et al., 2011; Porreca et al., 2018; Wu et al., 2019). In Aeromonas, blaNDM-1 was discovered in the last year or two (Wang et al., 2021; Luo et al., 2022). In the strain A. caviae 211703 of this study, the genetic environment in which blaNDM-1 is located is slightly different from the previous reports (Figure 4). The ΔTn125 containing blaNDM-1 in the strain A. caviae 211703 was carried by the ICE of the 211703 chromosome (c211703), and was captured by ISCR14-like rather than the common ISCR1, and was not further integrated into the integron. In addition, more than one insertion and recombination of ISCR14-like related elements occurred in the blaNDM-1 region from ICE of 211703, and similar phenomenon also appeared in the blaNDM-1 regions of Pseudomonas PP-2463 and Aeromonas NUITM-VA2. The blaNDM-1 regions from 211703, PP-2463, and NUITM-VA2 showed high homology, indicating that such blaNDM-1 environments have existed across species. This new blaNDM-1 genetic environment may herald a new way of recombination, which needs to be focused on. Further, the similar related ICE included in this study are mainly derived from Pseudomonas (Figure 3; Table S5). The ICE with the highest homology to ICE in A. caviae 211703 was also isolated in Pseudomona (PP-2463), and a high degree of structural similarity was shared by blaNDM-1 regions from both A. caviae 211703 and Pseudomonas PP-2463. This may indicate that the ICE in A. caviae 211703 was transferred from Pseudomonas.
Conclusion
In this work, we characterized and deciphered the genomic features and population distribution of 211703, a newly identified extensively drug resistant A. caviae strain harboring both blaRSA-1 and blaNDM-1 isolated from the clinical patient. To the best of our knowledge, this is the first report of A. caviae harboring blaRSA-1 even both blaRSA-1 and blaNDM-1 from clinical setting in the world. For the first time, we detailed annotated the integron where the blaRSA-1 gene is located and deeply analyzed its related genetic environment. Further, the uncommon genetic environment of blaNDM-1 was discovered in this study and comparative genomics analysis was performed. The coexistence of blaRSA-1 and blaNDM-1 can result in more extensive antibiotic resistance in the host bacteria, and is a significant threat to public health. This study would not only provide an overall understanding of the genomic characterization of clinically isolated carbapenem-resistant A. caviae, but also provide an in-depth discussion of the multiple acquisition methods of drug resistance genes in Aeromonas. We hope that this study will contribute to the understanding of the drug resistance of Aeromonas, and provide a richer perspective on how drug resistance genes are carried in Aeromonas.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The specimens were obtained with the patient’s consent. The use of human specimens and all related experimental protocols were reviewed and approved by the Ethics Committee of Taizhou Municipal Hospital, Zhejiang, China, in accordance with the medical research regulations of the Ministry of Health, China. Research and all related procedures involving biohazardous materials were approved by the Biosafety Committee of Taizhou Municipal Hospital. This research was conducted in China.
Author contributions
Conceptualization, XL, JF, ZY; and FW; methodology, XL, JZ and MX; data analysis, JF, XL, JZ, LY; resources, LY, DH, and PW; writing—original draft, XL and ZY; writing—review and editing, FW and JF. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Foundation of Taizhou Science and Technology Bureau (22ywa43), the Medical and Health Science and Technology Project of Zhejiang Province (2023KY1323), the Key Laboratory of Microbiol Technology and Bioinformatics of Zhejiang Province Foundation (2021E10010KFJJ09), and the National Natural Science Foundation of China (82002207).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1131059/full#supplementary-material
Supplementary Figure 1 | Confirmation of production of NDM.
Supplementary Figure 2 | Location of ICE and IME on A. caviae 211703.
Supplementary Table 1 | Background of isolates included in phylogenetic analysis.
Supplementary Table 2 | Pairwise.
Supplementary Table 3 | Resistance genes in A. caviae 211703.
Supplementary Table 4 | Major features of IMEs characterized in this work.
Supplementary Table 5 | Major features of ICEs characterized in this work.
References
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48 (D1), D517–D525. doi: 10.1093/nar/gkz935
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., Beatson, S. A. (2011). BLAST ring image generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics 12, 402. doi: 10.1186/1471-2164-12-402
Anandan, S., Gopi, R., Devanga Ragupathi, N. K., Muthuirulandi Sethuvel, D. P., Gunasekaran, P., Walia, K., et al. (2017). First report of bla(OXA-181)-mediated carbapenem resistance in aeromonas caviae in association with pKP3-a: Threat for rapid dissemination. J. Glob. Antimicrob. Resist. 10, 310–314. doi: 10.1016/j.jgar.2017.07.006
Bellanger, X., Payot, S., Leblond-Bourget, N., Guedon, G. (2014). Conjugative and mobilizable genomic islands in bacteria: Evolution and diversity. FEMS Microbiol. Rev. 38 (4), 720–760. doi: 10.1111/1574-6976.12058
Bhowmick, U. D., Bhattacharjee, S. (2018). Bacteriological, clinical and virulence aspects of aeromonas-associated diseases in humans. Pol. J. Microbiol. 67 (2), 137–149. doi: 10.21307/pjm-2018-020
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41 (Web Server issue), W29–W33. doi: 10.1093/nar/gkt282
Botelho, J., Schulenburg, H. (2021). The role of integrative and conjugative elements in antibiotic resistance evolution. Trends Microbiol. 29 (1), 8–18. doi: 10.1016/j.tim.2020.05.011
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365. doi: 10.1038/srep08365
Conte, D., Mesa, D., Jové, T., Zamparette, C. P., Sincero, T. C. M., Palmeiro, J. K., et al. (2022). Novel insights into bla(GES) mobilome reveal extensive genetic variation in hospital effluents. Microbiol. Spectr. 10 (4), e0246921. doi: 10.1128/spectrum.02469-21
Delavat, F., Miyazaki, R., Carraro, N., Pradervand, N., van der Meer, J. R. (2017). The hidden life of integrative and conjugative elements. FEMS Microbiol. Rev. 41 (4), 512–537. doi: 10.1093/femsre/fux008
Delcher, A. L., Salzberg, S. L., Phillippy, A. M. (2003). Using MUMmer to identify similar regions in large sequence sets. Curr. Protoc. Bioinf. Chapter 10, Unit 10 13. doi: 10.1002/0471250953.bi1003s00
Dortet, L., Poirel, L., Nordmann, P. (2014). Worldwide dissemination of the NDM-type carbapenemases in gram-negative bacteria. BioMed. Res. Int. 2014, 249856. doi: 10.1155/2014/249856
Edgar, R. C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 5, 113. doi: 10.1186/1471-2105-5-113
Fernández-Bravo, A., Figueras, M. J. (2020). An update on the genus aeromonas: Taxonomy, epidemiology, and pathogenicity. Microorganisms 8(1), 129. doi: 10.3390/microorganisms8010129
Guedon, G., Libante, V., Coluzzi, C., Payot, S., Leblond-Bourget, N. (2017). The obscure world of integrative and mobilizable elements, highly widespread elements that pirate bacterial conjugative systems. Genes (Basel) 8(11), 337. doi: 10.3390/genes8110337
Hu, X., Yu, X., Shang, Y., Xu, H., Guo, L., Liang, Y., et al. (2019). Emergence and characterization of a novel IncP-6 plasmid harboring bla (KPC-2) and qnrS2 genes in aeromonas taiwanensis isolates. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02132
Hughes, H. Y., Conlan, S. P., Lau, A. F., Dekker, J. P., Michelin, A. V., Youn, J. H., et al. (2016). Detection and whole-genome sequencing of carbapenemase-producing aeromonas hydrophila isolates from routine perirectal surveillance culture. J. Clin. Microbiol. 54 (4), 1167–1170. doi: 10.1128/jcm.03229-15
Letunic, I., Bork, P. (2021). Interactive tree of life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49 (W1), W293–W296. doi: 10.1093/nar/gkab301
Libisch, B., Giske, C. G., Kovács, B., Tóth, T. G., Füzi, M. (2008). Identification of the first VIM metallo-beta-lactamase-producing multiresistant aeromonas hydrophila strain. J. Clin. Microbiol. 46 (5), 1878–1880. doi: 10.1128/jcm.00047-08
Luo, X., Mu, K., Zhao, Y., Zhang, J., Qu, Y., Hu, D., et al. (2022). Emergence of bla NDM- 1-carrying aeromonas caviae K433 isolated from patient with community-acquired pneumonia. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.825389
Luo, X., Yin, Z., Zeng, L., Hu, L., Jiang, X., Jing, Y., et al. (2021). Chromosomal integration of huge and complex bla NDM-carrying genetic elements in enterobacteriaceae. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.690799
Marathe, N. P., Janzon, A., Kotsakis, S. D., Flach, C. F., Razavi, M., Berglund, F., et al. (2018). Functional metagenomics reveals a novel carbapenem-hydrolyzing mobile beta-lactamase from Indian river sediments contaminated with antibiotic production waste. Environ. Int. 112, 279–286. doi: 10.1016/j.envint.2017.12.036
Montezzi, L. F., Campana, E. H., Corrêa, L. L., Justo, L. H., Paschoal, R. P., da Silva, I. L., et al. (2015). Occurrence of carbapenemase-producing bacteria in coastal recreational waters. Int. J. Antimicrob. Agents 45 (2), 174–177. doi: 10.1016/j.ijantimicag.2014.10.016
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., Correia, A. (2009). INTEGRALL: A database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25 (8), 1096–1098. doi: 10.1093/bioinformatics/btp105
Neuwirth, C., Siebor, E., Robin, F., Bonnet, R. (2007). First occurrence of an IMP metallo-beta-lactamase in aeromonas caviae: IMP-19 in an isolate from France. Antimicrob. Agents Chemother. 51 (12), 4486–4488. doi: 10.1128/aac.01462-06
Nordmann, P., Poirel, L., Walsh, T. R., Livermore, D. M. (2011). The emerging NDM carbapenemases. Trends Microbiol. 19 (12), 588–595. doi: 10.1016/j.tim.2011.09.005
Partridge, S. R. (2011). Analysis of antibiotic resistance regions in gram-negative bacteria. FEMS Microbiol. Rev. 35 (5), 820–855. doi: 10.1111/j.1574-6976.2011.00277.x
Partridge, S. R., Kwong, S. M., Firth, N., Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31 (4): e00088–17. doi: 10.1128/cmr.00088-17
Partridge, S. R., Tsafnat, G., Coiera, E., Iredell, J. R. (2009). Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33 (4), 757–784. doi: 10.1111/j.1574-6976.2009.00175.x
Pessoa, R. B. G., de Oliveira, W. F., Correia, M., Fontes, A., Coelho, L. (2022). Aeromonas and human health disorders: Clinical approaches. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.868890
Porreca, A. M., Sullivan, K. V., Gallagher, J. C. (2018). The epidemiology, evolution, and treatment of KPC-producing organisms. Curr. Infect. Dis. Rep. 20 (6), 13. doi: 10.1007/s11908-018-0617-x
Richter, M., Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106 (45), 19126–19131. doi: 10.1073/pnas.0906412106
Roberts, A. P., Chandler, M., Courvalin, P., Guédon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60 (3), 167–173. doi: 10.1016/j.plasmid.2008.08.001
Sayers, E. W., Cavanaugh, M., Clark, K., Pruitt, K. D., Schoch, C. L., Sherry, S. T., et al. (2021). GenBank. Nucleic Acids Res. 49 (D1), D92–D96. doi: 10.1093/nar/gkaa1023
Sekizuka, T., Inamine, Y., Segawa, T., Hashino, M., Yatsu, K., Kuroda, M. (2019). Potential KPC-2 carbapenemase reservoir of environmental aeromonas hydrophila and aeromonas caviae isolates from the effluent of an urban wastewater treatment plant in Japan. Environ. Microbiol. Rep. 11 (4), 589–597. doi: 10.1111/1758-2229.12772
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., Chandler, M. (2006). ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 34 (Database issue), D32–D36. doi: 10.1093/nar/gkj014
Uechi, K., Tada, T., Sawachi, Y., Hishinuma, T., Takaesu, R., Nakama, M., et al. (2018a). A carbapenem-resistant clinical isolate of aeromonas hydrophila in Japan harbouring an acquired gene encoding GES-24 beta-lactamase. J. Med. Microbiol. 67 (11), 1535–1537. doi: 10.1099/jmm.0.000842
Uechi, K., Tada, T., Sawachi, Y., Hishinuma, T., Takaesu, R., Nakama, M., et al. (2018b). A carbapenem-resistant clinical isolate of aeromonas hydrophila in Japan harbouring an acquired gene encoding GES-24 β-lactamase. J. Med. Microbiol. 67 (11), 1535–1537. doi: 10.1099/jmm.0.000842
Wang, Y., Liu, H., Zhang, L., Sun, B. (2021). Application of modified carbapenem inactivation method and its derivative tests for the detection of carbapenemase-producing aeromonas. Infect. Drug Resist. 14, 3949–3960. doi: 10.2147/IDR.S330115
Wrighton, C. J., Strike, P. (1987). A pathway for the evolution of the plasmid NTP16 involving the novel kanamycin resistance transposon Tn4352. Plasmid 17 (1), 37–45. doi: 10.1016/0147-619x(87)90006-0
Wu, C. J., Chen, P. L., Wu, J. J., Yan, J. J., Lee, C. C., Lee, H. C., et al. (2012). Distribution and phenotypic and genotypic detection of a metallo-β-lactamase, CphA, among bacteraemic aeromonas isolates. J. Med. Microbiol. 61 (Pt 5), 712–719. doi: 10.1099/jmm.0.038323-0
Wu, W., Feng, Y., Tang, G., Qiao, F., McNally, A., Zong, Z. (2019). NDM metallo-beta-Lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 32 (2), e00115–18. doi: 10.1128/CMR.00115-18
Xu, S., Tu, J., Zhang, L., Chen, Y., Dong, X., Chi, X., et al. (2022). Detection of NDM-1-Positive aeromonas caviae from bacteremia by using whole-genome sequencing. Infect. Drug Resist. 15, 2835–2841. doi: 10.2147/idr.S360353
Yang, C., Guo, M., Yang, H., Wen, Y., Zhu, Z., Wang, T., et al. (2022). bla(KPC-24)-Harboring aeromonas veronii from the hospital sewage samples in China. Microbiol. Spectr. 10 (3), e0055522. doi: 10.1128/spectrum.00555-22
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53 (12), 5046–5054. doi: 10.1128/AAC.00774-09
Keywords: Aeromonas caviae, blaNDM-1, blaRSA-1, ICE, IME
Citation: Luo X, Yin Z, Yu L, Zhang J, Hu D, Xu M, Wang P, Wang F and Feng J (2023) Genomic analysis of chromosomal cointegrated blaNDM-1-carrying ICE and blaRSA-1-carrying IME from clinical multidrug resistant Aeromonas caviae. Front. Cell. Infect. Microbiol. 13:1131059. doi: 10.3389/fcimb.2023.1131059
Received: 24 December 2022; Accepted: 14 March 2023;
Published: 23 March 2023.
Edited by:
Parveen Kumar, University of Alabama at Birmingham, United StatesReviewed by:
Matej Medvecky, University of Hradec Kralove, CzechiaRitesh Sevalkar, University of Alabama at Birmingham, United States
Surbhi Gupta, Department of Microbiology and Immunology, University of Michigan, United States
Copyright © 2023 Luo, Yin, Yu, Zhang, Hu, Xu, Wang, Wang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiao Feng, ZmVuZ2ppYW8tMTIxOUAxNjMuY29t; Fengling Wang, d2FuZ2ZlbmdsaW5nXzIwMDVAMTYzLmNvbQ==
†These authors have contributed equally to this work
 Xinhua Luo
Xinhua Luo Zhe Yin
Zhe Yin Lianhua Yu1
Lianhua Yu1 Fengling Wang
Fengling Wang Jiao Feng
Jiao Feng