- 1Department of Medical Research Center, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Department of Gastroenterology, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 3Department of Cardiology, Peking Union Medical College Hospital, Chinese Academy of Medical Science and Peking Union Medical College, Beijing, China
- 4Department of Endocrinology, Peking Union Medical College Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing, China
- 5Department of Emergency Medicine, Peking Union Medical College Hospital, Peking Union Medical College, Chinese Academy of Medical Sciences, Beijing, China
Background: Acute respiratory distress syndrome (ARDS) is the most common cause of organ failure in acute pancreatitis (AP) patients, which associated with high mortality. Specific changes in the gut microbiota have been shown to influence progression of acute pancreatitis. We aimed to determine whether early alterations in the gut microbiota is related to and could predict ARDS occurrence in AP patients.
Methods: In this study, we performed 16S rRNA sequencing analysis in 65 AP patients and 20 healthy volunteers. The AP patients were further divided into two groups: 26 AP-ARDS patients and 39 AP-nonARDS patients based on ARDS occurrence during hospitalization.
Results: Our results showed that the AP-ARDS patients exhibited specific changes in gut microbiota composition and function as compared to subjects of AP-nonARDS group. Higher abundances of Proteobacteria phylum, Enterobacteriaceae family, Escherichia-Shigella genus, and Klebsiella pneumoniae, but lower abundances of Bifidobacterium genus were found in AP-ARDS group compared with AP-nonARDS groups. Random forest modelling analysis revealed that the Escherichia-shigella genus was effective to distinguish AP-ARDS from AP-nonARDS, which could predict ARDS occurrence in AP patients.
Conclusions: Our study revealed that alterations of gut microbiota in AP patients on admission were associated with ARDS occurrence after hospitalization, indicating a potential predictive and pathogenic role of gut microbiota in the development of ARDS in AP patients.
Introduction
Acute pancreatitis (AP), one of the most common gastrointestinal diseases, is an acute inflammatory disease with an increasing incidence worldwide (Tenner et al., 2013; Greenberg et al., 2016). In patients with AP, persistent organ failure (OF) can reach 35% and is a key determinant of mortality (Johnson and Abu-Hilal, 2004; Shah and Rana, 2020). The most common cause of OF in AP is acute respiratory distress syndrome (ARDS) (Garg et al., 2005). ARDS is a type of acute, diffuse inflammatory lung injury that can lead to a high mortality rate of up to 48% (Schmandt et al., 2021). Even after five years of rehabilitation, surviving ARDS patients still suffer from poor long-term quality of life; including exercise limitation, difficulty in returning to work, and high medical costs (Herridge et al., 2011). However, missed or delayed diagnosis of ARDS remains a common and challenging problem worldwide. Nearly two-thirds of patients had a delayed or missed diagnosis of ARDS. The miss rate was approximately 40%, and the diagnosis of half of mild ARDS patients was delayed (Bellani et al., 2020). Early recognition of ARDS ensures that patients receive appropriate treatment which relieves lung injury and improves prognosis, therefore, effective prediction methods for ARDS are urgently required (Fan et al., 2018; Pan et al., 2018; Bellani et al., 2020).
AP is strongly associated with gut microbiota imbalance and an impaired epithelial barrier (Besselink et al., 2009). Compared with the healthy group, the diversity of the gut microbiota decreased; with a greater abundance of pathogenic bacteria in AP patients and lower numbers of commensal beneficial genera (Zhang et al., 2018; Zhu et al., 2019). According to the revised Atlanta classification 2012, AP can be divided into three grades: mild AP (MAP), moderately severe AP (MSAP) with transient OF, and severe AP (SAP) with persistent OF (Banks et al., 2013). In AP patients with different degrees of severity, the dominant gut microbiota also varied; with Bacteroides in MAP, Escherichia-Shigella in MSAP, and Enterococcus in SAP (Yu et al., 2020). Similar results have been reported in animal models; gut microbiota-depleted AP rats were found to have lower levels of inflammatory factors (Zheng et al., 2019; Li et al., 2020). The degree of gut barrier injury and bacterial translocation are important prognostic factors for AP (Besselink et al., 2009).
Disorganized microbiota and damaged intestinal epithelium in AP patients make it easier for the endotoxin diffusion, immune cell migration, and bacteria translocation. The lung environment may be more susceptible to the gut microbiota in patients with AP. Owing to the increased gut permeability, inflammatory factors and activated trypsin could function as the gut-lung axis, thus triggering and promoting lung disease in patients with AP (Shah and Rana, 2020). In addition, gram-negative infections promote release of endotoxins and these can translocate through high-permeability gut mucosa and contribute to the development of ARDS in AP patients (AP-ARDS; “ARDS” mean for ARDS in general, “AP-ARDS” mean for ARDS in acute pancreatitis.) (Gray et al., 2003). Bacteria can also translocate from the gut to the lung (Mukherjee and Hanidziar, 2018). Previous studies also found evidence of bacteria translocation in ARDS patients (Dickson et al., 2016; Dickson et al., 2020). The composition of gut-associated bacteria, especially Bacteroidetes and Enterobacteriaceae, increased in the lower respiratory tract of ARDS patients (Dickson et al., 2016; Siwicka-Gieroba and Czarko-Wicha, 2020). Further study found that the increase of Escherichia coli in lung was related to higher mortality of ARDS patients (Zhang et al., 2021). Studies have confirmed that the lung microbiota is associated with alveolar inflammation in ARDS (Dickson et al., 2016). Michihito et al. revealed that alterations in lung microbiota are correlated with serum IL-6 levels and hospital mortality in patients with ARDS (Kyo et al., 2019). Therefore, the gut microbiota may be involved in the pathogenesis of AP-ARDS, however, the relationship between the gut microbiota and AP-ARDS remains unknown. If early changes in gut microbiota in AP-ARDS patients can be found, they may help in the early recognition of AP-ARDS, promote early intervention, and even improve patient outcomes.
Therefore, we wanted to investigate the relationship between gut microbiota and AP-ARDS by comparing the microbiota among three groups: healthy controls, AP patients without ARDS (AP-nonARDS), and AP-ARDS patients. By collecting the gut microbiota at the early stage of AP, we investigated whether gut microbiota was related to and could help predict and recognize AP-ARDS. Our study explored the potential effect of the gut-lung axis in AP-ARDS and provide identify biomarkers for prediction and early recognition of AP-ARDS.
Methods
Study population
This prospective and observational cohort study was conducted at Peking Union Medical College Hospital, Beijing, China. Twenty healthy volunteers and 75 patients were enrolled between June 2018 and July 2021. Ten AP patients were excluded due to history of comorbidities and medicine intake. All patients fulfilled the AP diagnostic criteria according to the 2012 revised Atlanta criteria and were admitted within 24 h of onset (Banks et al., 2013).
The exclusion criteria were as follows: patients with chronic pancreatitis, immunosuppressive disease, inflammatory bowel disease, cancer, irritable bowel syndrome, gastroenteritis, or necrotizing enterocolitis; and use of antibiotics, probiotics, laxatives, or Chinese herbs within two months before symptom onset. Informed consent was obtained from all participants. This study was approved by the Ethics Committee of PUMCH (Identifier: JS1826; date of approval:20th February 2018. Period of validity: February 2018 to August 2020)
For all patients, no ARDS was diagnosed during the first fecal sampling; however, some patients developed ARDS during hospitalization. Patients were diagnosed with ARDS according to the Berlin definition (Ranieri et al., 2012). According to PaO2/FiO2 levels, patients with ARDS were divided into three groups: mild ARDS (MARDS), moderate ARDS, and severe ARDS (Ranieri et al., 2012). Considering the higher rate of mechanical ventilation in moderate ARDS and severe ARDS patients (Fan et al., 2018) as well as the small sample size of severe ARDS group (n=5), we combined moderate ARDS and severe ARDS as non-MARDS group for subgroup analysis.
Collection and analysis of clinical characteristics
Demographic and clinical data were collected from medical record libraries, including age, sex, body mass index (BMI), smoking history, drinking history, combined diseases, disease severity-related scores, local complications, systematic complications, and clinical outcomes. Definitions of local and systematic complications can be found in previous studies (Yu et al., 2020; Hu et al., 2021b; Yu et al., 2021).
Statistical analysis of clinical characteristics was performed using SPSS Statistics 26.0 (IBM Corp., Armonk, NY, USA). The mean ± standard deviation (SD) was used to represent the data distribution. However, when the data did not fit a normal distribution, the median (interquartile range [IQR]) was used. For categorical variables, we performed the χ2 test or Fisher’s exact test; while for continuous variables, we performed the nonparametric Mann-Whitney test. A difference was considered significant when the two-sided p value was less than 0.05.
Sample collection, DNA extraction, and 16S rRNA gene sequencing
Patients with AP have difficulty defecating owing to fasting and water deprivation. Therefore, we used rectal swabs for fecal sampling, as previous studies have described (Yu et al., 2020; Yu et al., 2021). The fecal samples were immediately collected after admission, and all samples were collected within 24 h of AP onset. Then, these samples were stored at − 80°C, and microbial DNA was extracted as soon as possible. We then performed PCR amplification, library construction, Illumina (San Diego, CA, USA) MiSeq sequencing, and sequence quality control, using previously reported methods (Yu et al., 2020; Yu et al., 2021).
Bioinformatics analysis
Amplicon sequence variant (ASV) analysis was performed using EasyAmplicon (Version 1.10). We use the -derep_fullength command in VSEARCH (version 2.15) to create dereplication, denoised these unique sequences into ASVs by the -unoise3 algorithm in USEARCH (Version 10.0), created an ASVs table using the -usearch_global command, and then completed the ASVs classification using the Sintax algorithm command.
Microbiota composition
Alpha diversity analysis, including the Chao and Simpson indices, was performed using Mothur software (1.30.2). The dilution curve was plotted using R software to calculate the microbial diversity at different numbers of sequences. In the beta diversity analysis, principal coordinate analysis (PCoA) was performed using the R package vegan (v2.5-6).
Based on taxonomic information, community structure analysis can be performed at various taxonomic levels. The composition of microbiota at the phylum, family, genus, and species levels was determined using the stat package in R software. Relevant analytical methods were used to detect variation in microbes between the different groups and pairwise comparisons were calculated using the Wilcoxon rank-sum test.
Functional annotation
Linear discriminant analysis (LDA) effect size (LefSe; http://huttenhower.sph.harvard.edu/galaxy) was performed to identify potential biomarkers in the different groups (LDA score>2, p<0.05). Microbiota phenotypes were predicted using BugBase, based on normalized ASVs. The significance of the functional difference was evaluated using the Wilcoxon rank-sum test in the BugBase prediction analysis. The Random Forest R package was used to build a random forest regression model. We randomly divided the 65 samples into training sets (70%) and testing sets (30%) according to the 16S amplicon sequence and clinical characteristics. Bioinformatics analysis and visualization were performed using the R software. Detailed analysis methods can be found in our previous studies (Hu et al., 2021a; Hu et al., 2021b).
Results
Clinical characteristics of AP-ARDS patients
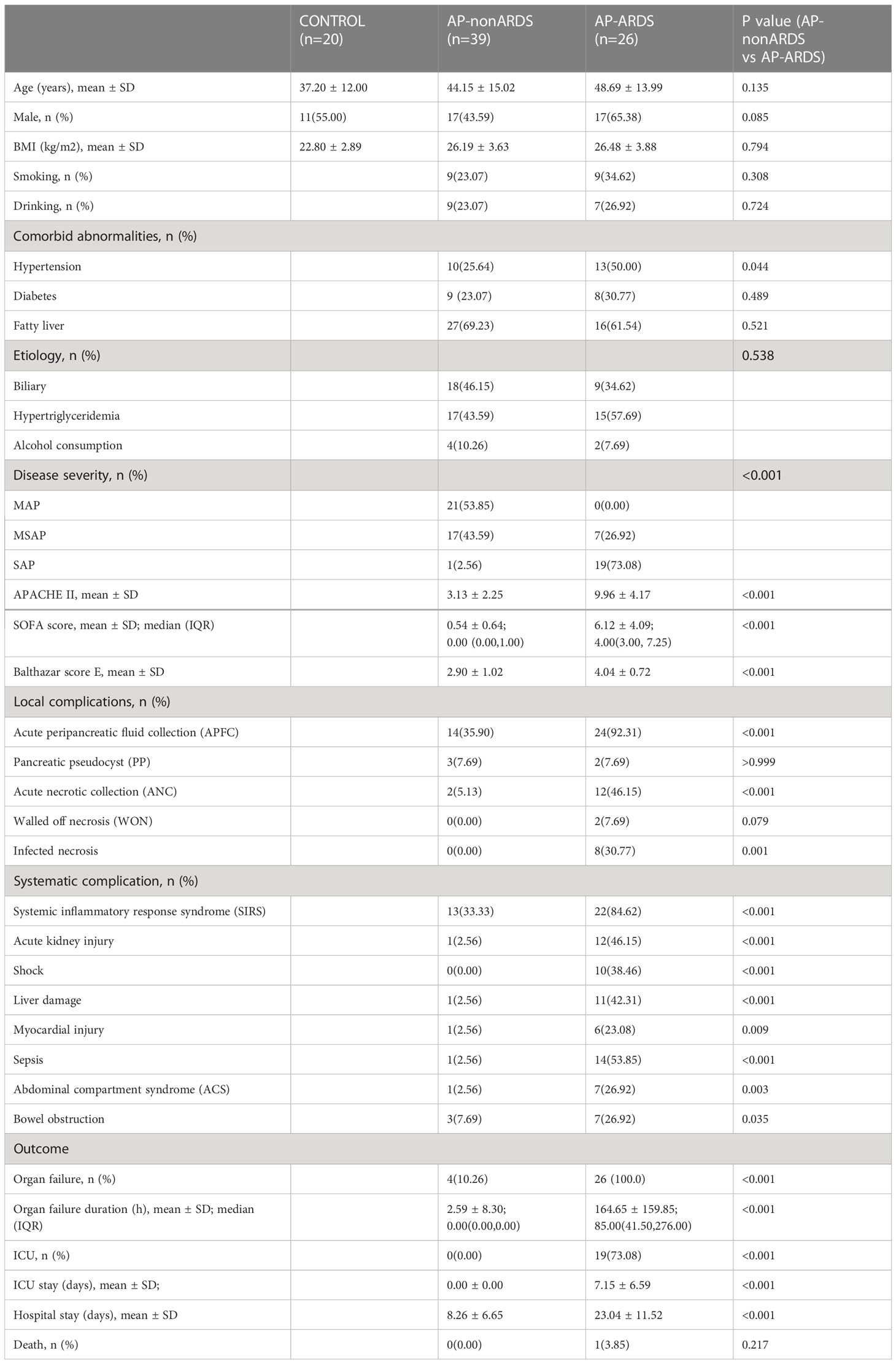
Sixty-five AP patients and 20 healthy individuals were included in the study. Rectal swabs were collected before the occurrence of ARDS. Twenty-six patients with AP developed ARDS (AP-ARDS; mild ARDS: n=12; moderate ARDS: n=9; severe ARDS: n=5) and 39 patients did not (AP-nonARDS). The average diagnosis time of ARDS was 3.46 ± 1.92 d after AP onset. Table 1 shows the demographic and clinical characteristics of the two groups. Demographic characteristics were generally balanced, however, patients with ARDS had more severe symptoms than those without ARDS (Table 1). The AP-ARDS group had a higher proportion of SAP (2.56% vs. 73.08%; p<0.001) and higher disease severity-related scores compared to the AP-nonARDS group. The occurrence of acute peripancreatic fluid collection (35.90% vs. 92.31%; p<0.001), systematic complications, organ failure (10.26% vs. 100.00%; p<0.001), and ICU admission (0.00% vs. 73.08%; p<0.001) was also significantly increased in the AP-ARDS group, except for bowel obstruction and mental status. Furthermore, the total duration of organ failure (median 0.00, IQR 0.00–0.00; vs median 85.00, IQR 41.50–276.00; p<0.001); ICU stay (0.00 ± 0.00 vs. 7.15 ± 6.59; p<0.001) and hospital stay (8.26 ± 6.65 vs. 23.04 ± 11.52; p<0.001) were both longer in AP-ARDS group.
Taxonomic features of gut microbiota in AP-ARDS patients
We analyzed 745,895 reads that were clustered into 1910 ASVs. No statistically significant differences in the richness and diversity of the gut microbiota were noted between the AP-nonARDS and AP-ARDS groups. In the alpha diversity analysis, there were no significant differences in the Chao index (p>0.05 between any two groups; Figure 1A). Compared with healthy controls, the Simpson index decreased in both the AP-nonARDS and AP-ARDS groups, but no differences were found between the AP-nonARDS and AP-ARDS groups (Figure 1B). In the rarefaction curve analysis, the curve tended to plateau as the number of reads increased, demonstrating that microbiota in the healthy control, AP-nonARDS, and AP-ARDS groups were abundant and evenly distributed (Figure 1C). PCoA for the beta diversity results clearly distinguished the three groups, but overlap did occur between the AP-nonARDS group and AP-ARDS group. This indicated a significant difference in the microbiota structure between healthy controls and patients with AP, possible similarities between the AP-nonARDS group and AP-ARDS group (Figure 1D).
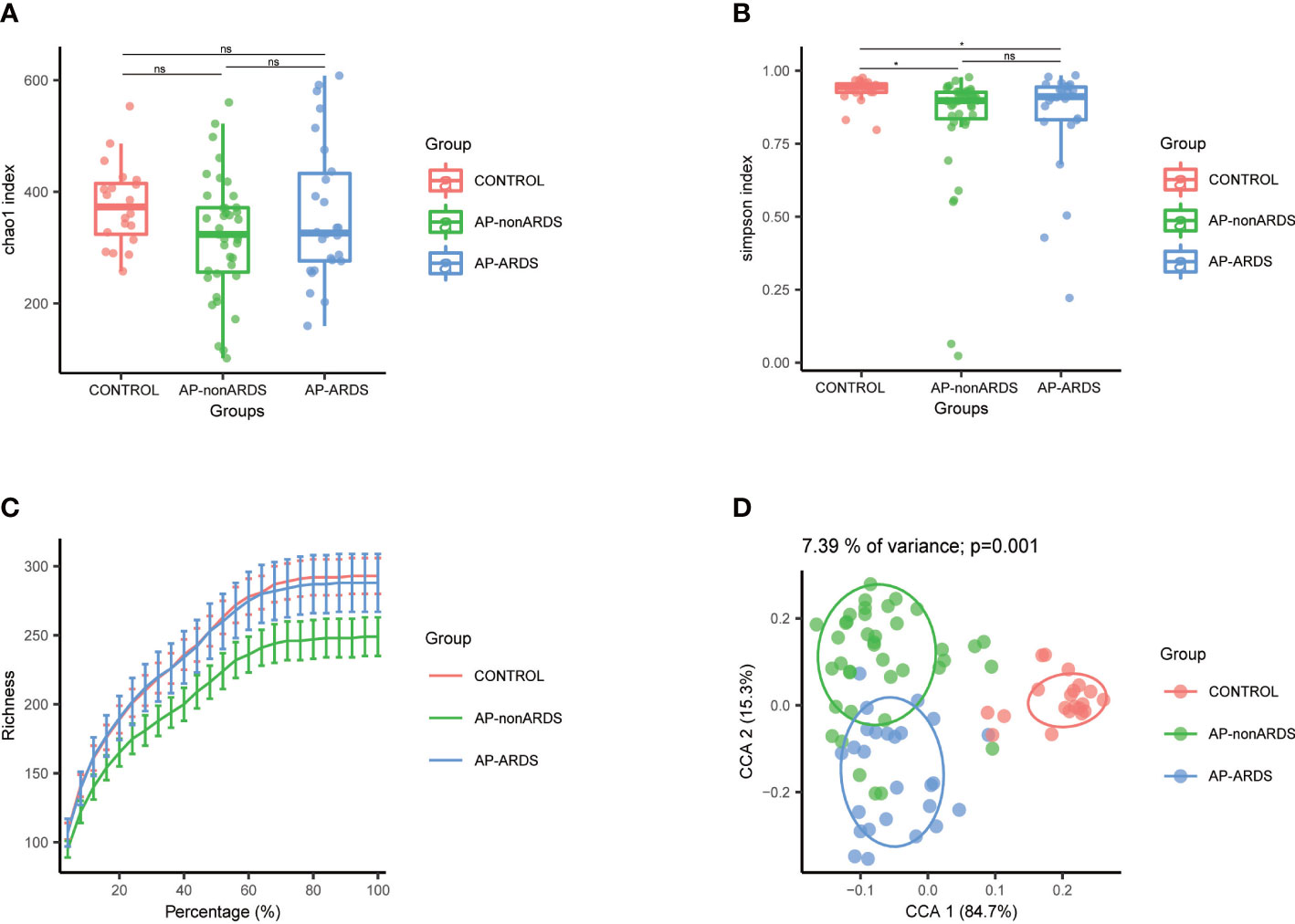
Figure 1 Diversity analysis of Control, AP-nonARDS and AP-ARDS group. (A) Chao index of α analysis; (B) Simpson index of α analysis. There are significant differences between the Control group and AP patients, but no significant difference between AP-nonARDS and AP-ARDS group. (C) Rarefaction curves analysis. (D). Principal coordinate analysis (PCoA). CONTROL, healthy population; AP-nonARDS, AP patients without ARDS; AP-ARDS, AP patients with ARDS. * P < 0.05; ns: not significant.
The composition of the gut microbiota was significantly different among the three groups. At the phylum level, Proteobacteria and Bacteroidetes were both increased in patients with AP compared to healthy controls. Proteobacteria showed a gradually increase with disease progression (Figure 2A). At the family level, Enterobacteriaceae, Enterococcaceae, Bacteroidaceae, Clostridiales Incertae Sedis XI, and Prevotellaceae increased, while Ruminococcaceae decreased in patients with AP compared to healthy controls. In particular, the abundance of Enterobacteriaceae and Enterococcaceae increased with disease progression (Figure 2B). At the genus level, Escherichia-Shigella, Bacteroides, and Enterococcus were more abundant in patients with AP, while Bifidobacterium and Blautia were more abundant in healthy controls. Escherichia-Shigella and Enterococcus gradually increased while Bifidobacterium decreased with disease progression (Figure 2C). Compared to the AP-nonARDS group, 25 ASVs were enriched and 22 ASVs were depleted in the AP-ARDS group (Figure 2D). Figure 2E shows the top 11 different bacteria between the AP-ARDS and AP-nonARDS groups at the species level. Klebsiella pneumoniae (ASV_101, p<0.001; ASV_71, p<0.001), Prevotella copri (ASV_30, p<0.001; ASV_111, p=0.002), and Clostridium ramosum (ASV_150, p=0.002) showed a significant increase; and Bifidobacterium longum (ASV_14, p=0.003) decreased in the AP-ARDS group compared to the AP-nonARDS group. Among these microbiota, Clostridium ramosum (ASV_150) showed a gradual increase in the healthy to AP-nonARDS to AP-ARDS groups, whereas Bifidobacterium longum (ASV_40) showed a gradual decrease (Figure 2E).
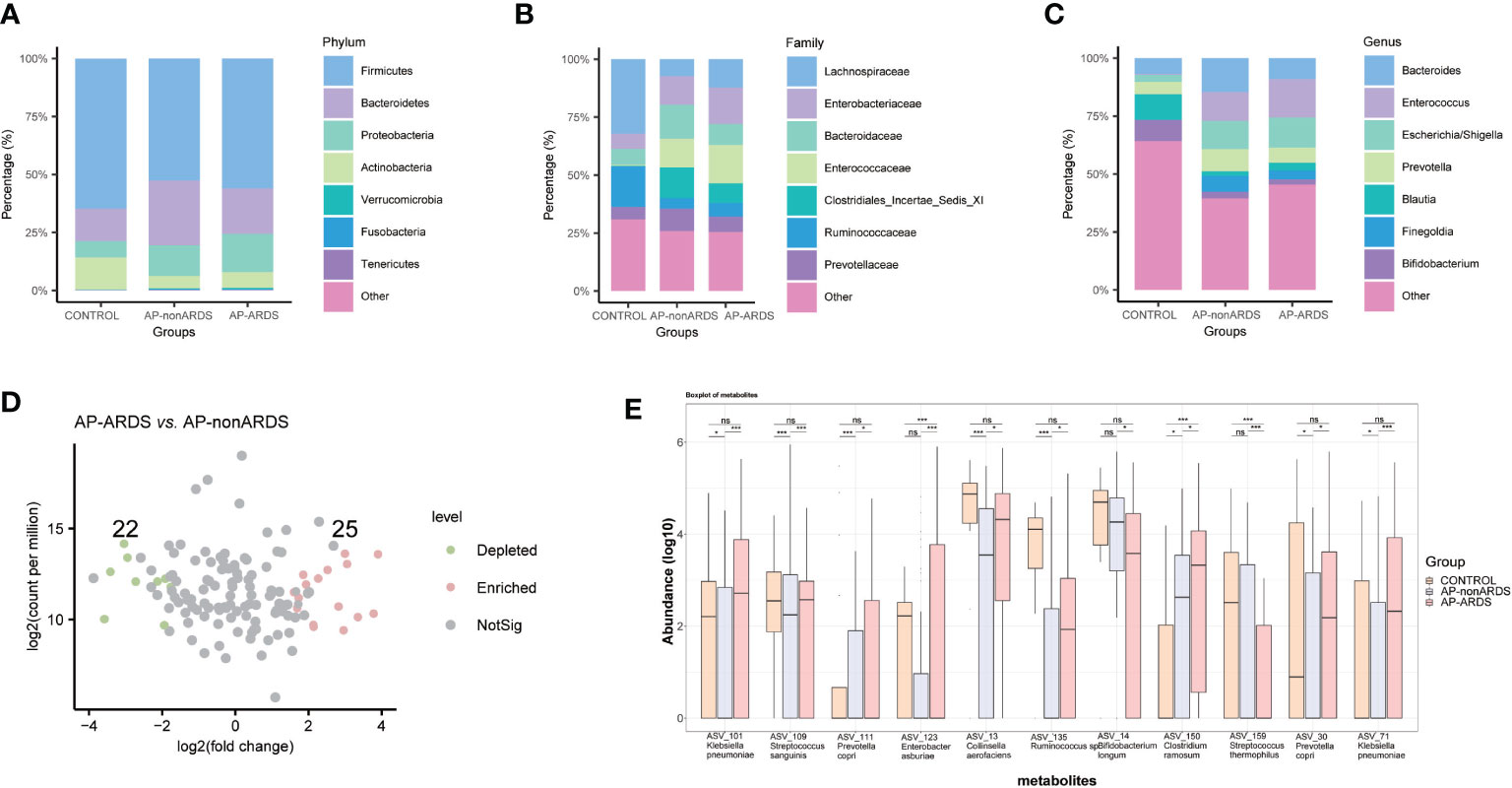
Figure 2 Gut microbiota composition at (A) phylum, (B) family, (C) genus levels. (D) Different amplicon sequence variants (ASVs) between AP-ARDS and AP-nonARDS group. (green= depleted in AP-ARDS group; red = enriched in AP-ARDS group; gray = no significantly difference). (E) Relative abundances of different species between AP-nonARDS and AP-ARDS groups. * P < 0.05; *** P < 0.001; ns: not significant.
We performed a subgroup analysis according to ARDS severity, and identified some microbiota showing similar trends. At the phylum level, Proteobacteria increased in the non-MARDS group compared to that in the MARDS group (Figure 3A). At the family level, Enterobacteriaceae increased in the non-MARDS group (Figure 3B). At the genus level, Escherichia-Shigella was more abundant in the non-MARDS group (Figure 3C).
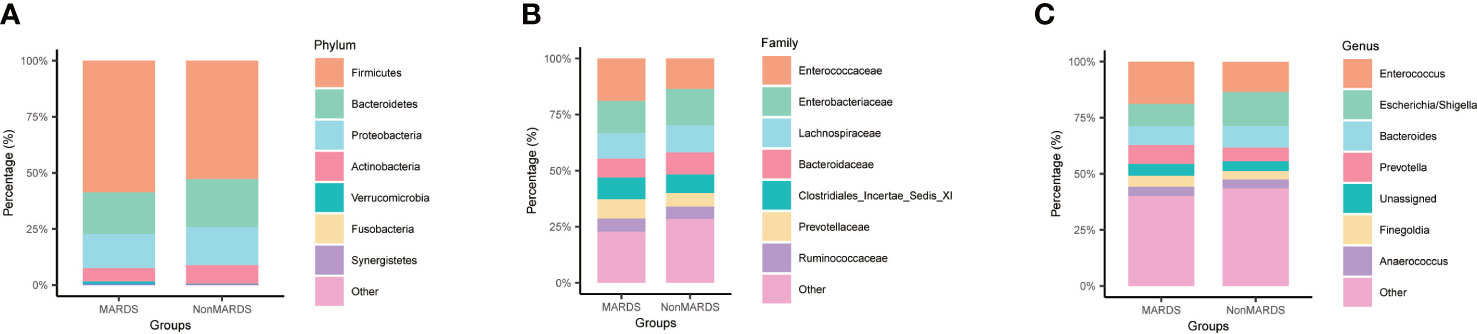
Figure 3 Subgroup analysis of gut microbiota composition at phylum (A), family (B), and genus (C) levels. MARDS, mild ARDS; NonMARDS, moderate ARDS and severe ARDS.
Alterations of gut microbiota in AP-ARDS patients are associated with more severe manifestations
LEFSe analysis also revealed that Enterobacteriaceae and Escherichia-Shigella were dominant in AP-ARDS group while Enterococcaceae and Enterococcus were dominant in AP-nonARDS group (Figure 4A).
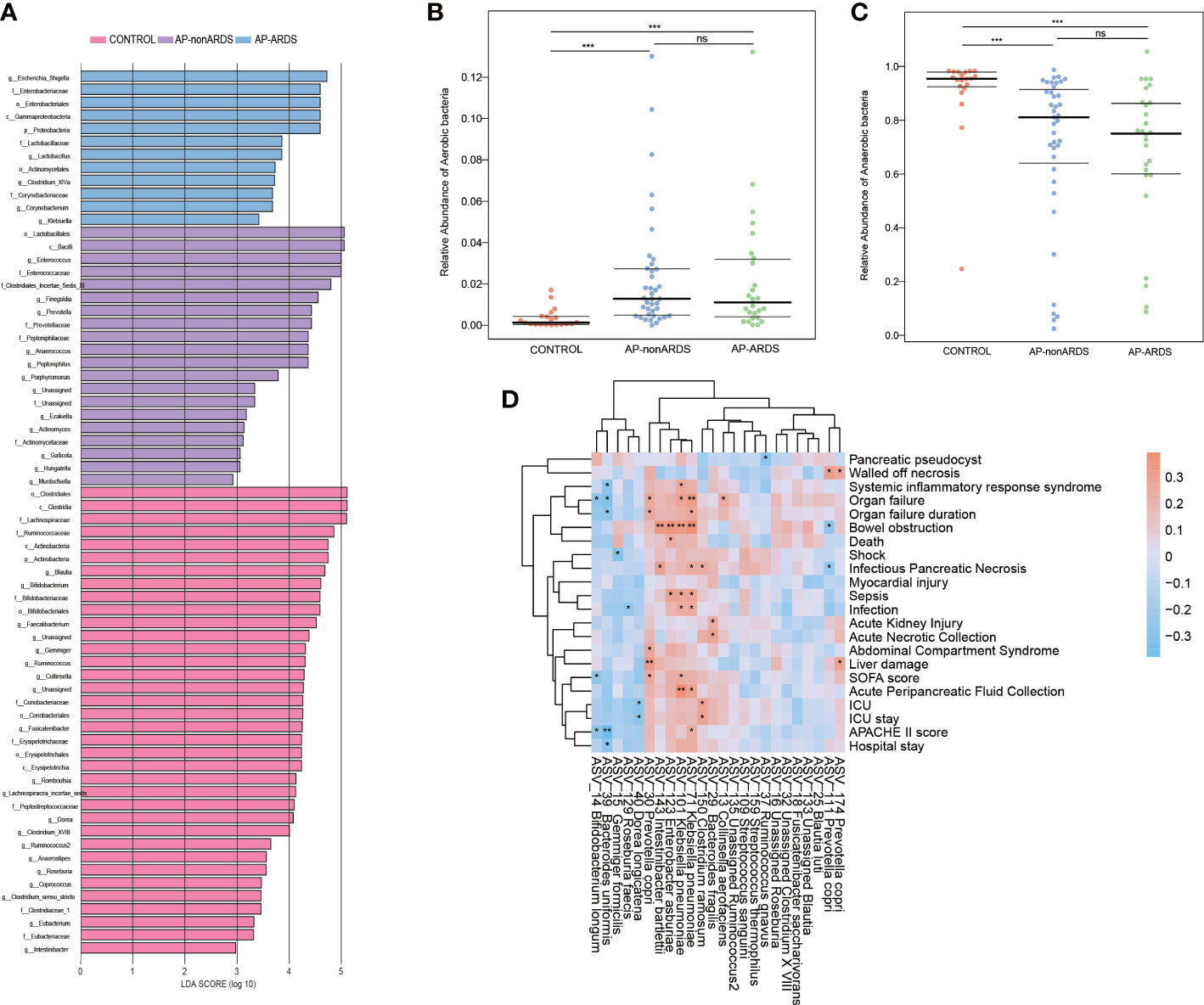
Figure 4 Microbial Function Analysis and Clinical Correlation Analysis. (A). Linear discriminant analysis (LDA) Effect Size (LEfSe) analysis. (B). Relative abundance of aerobic bacteria in BugBase analysis. (C). Relative abundance of anaerobic bacteria in BugBase analysis. (D). Spearman correlation of clinical characteristics and different species between AP-ARDS and AP-nonARDS group. * P < 0.05; *** P < 0.001, ns: not significant.
BugBase functional analysis predicted oxygen utilizing, gram staining, oxidative stress tolerance, biofilm forming, pathogenic potential, mobile element containing, and oxygen tolerance. Compared with healthy controls, anaerobic bacteria decreased in the AP-nonARDS and AP-ARDS groups (CONTROL vs. AP-nonARDS, CONTROL vs. AP-ARDS, both p<0.001). Although there was no significant difference, anaerobic bacteria showed a decreasing trend in AP-ARDS compared with AP-nonARDS (Figure 4B). In contrast, aerobic bacteria increased in the AP-nonARDS and AP-ARDS groups (Figure 4C).
Spearman correlation analysis was performed to investigate the relationship between microbiota and clinical outcomes. Two subspecies of Klebsiella pneumoniae, ASV_101 and ASV_71, were positively correlated with multiple clinical characteristics, including organ failure, bowel obstruction, sepsis, infection, and acute peripancreatic fluid collection. Prevotella copri (ASV_30) was positively correlated with the occurrence and duration of organ failure. Clostridium ramosum (ASV_150) was associated with ICU admission and length of hospital stay. As a probiotic, Bifidobacterium longum (ASV_14) negatively correlated with organ failure, Sequential Organ Failure Assessment score (SOFA score), and Acute Physiology And Chronic Health II score (APACHII score) (Figure 4D).
The progression of AP-ARDS is closely associated with Enterobacteriaceae
Considering the significant increase in Enterobacteriaceae and its potential pathogenicity, we performed further analyses of Enterobacteriaceae. Figure 5A shows the relative abundance of Enterobacteriaceae increased with disease progression. Further analysis revealed that almost all genera of the Enterobacteriaceae family were increased in the AP-ARDS group (Figure 5B). Random forest identified Escherichia-shigella as the most significant feature for distinguishing AP-ARDS from AP-nonARDS (Figure 5C).
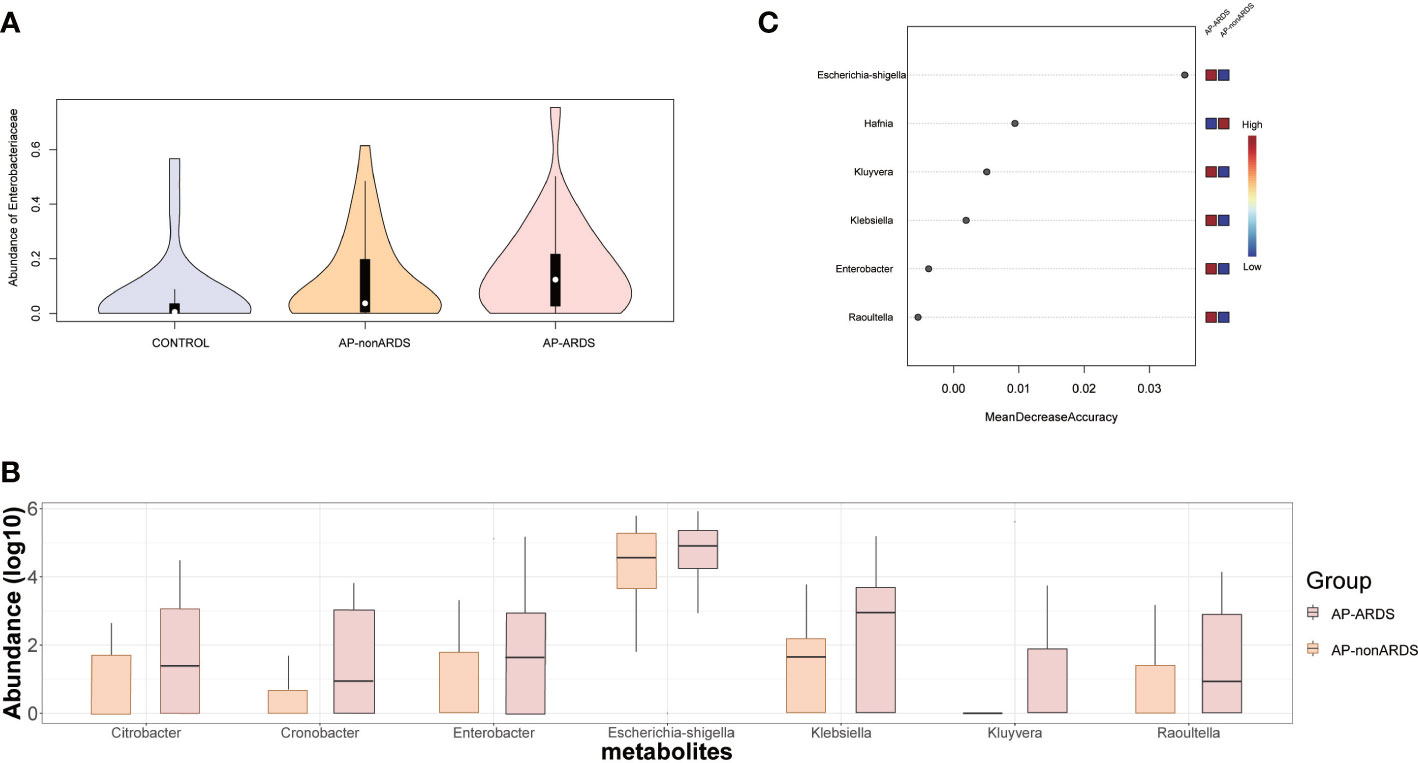
Figure 5 Enterobacteriaceae Analysis and Model Predicting. (A) The relative abundance of Enterobacteriaceae in Control, AP-nonARDS, and AP-ARDS group. (B) Major genus in Enterobacteriaceae family between AP-nonARDS, and AP-ARDS group. (C) Random forest model predicting. It screened out the Escherichia-shigella genus as the most significant feature for predicting ARDS.
Discussion
To our knowledge, this is the first study to explore the relationship between gut microbiota and AP-ARDS and reveals gut microbiota as a predictive biomarker for ARDS. The 16S rRNA sequencing analysis revealed differences of microbiota composition and function between the AP-ARDS and AP-nonARDS groups. Subgroup analysis suggested that gut microbiota composition was also related to the severity of ARDS. Before patients were diagnosed with AP-ARDS, the gut microbiota already had the characteristics of ARDS in AP patients. This indicates that the gut microbiota can be a potential biomarker for prediction and early recognition of AP-ARDS, thereby improving AP-ARDS diagnosis and treatment.
In our characteristics analysis, AP patients with ARDS were more serious than that in the non-ARDS group. AP-severity-associated changes in the gut microbiota were also observed in the AP-ARDS group compared with the AP-nonARDS group. However, we also observed some microbiota changes that might be related to the occurrence and development of AP-ARDS. The enrichment of Enterobacteriaceae and Escherichia-Shigella, and the reduction of Bifidobacterium were associated with AP-ARDS. Previous studies have focused on the lung microbiota of ARDS patients and found that the composition was affected by the gut microbiota (Dickson et al., 2016; Dickson, 2018; Dickson et al., 2020). In our study, similar changes in composition were also observed in the lung microbiota.
In the normal population, the most dominant phylum in the gut microbiota is Firmicutes, followed by Bacteroidetes, Actinobacteria, and Proteobacteria (Bozzi Cionci et al., 2018). In our study, the composition of healthy controls was consistent with the normal population; but in AP patients, Proteobacteria significantly increased with disease severity. Previous studies have found Proteobacteria overgrowth in patients with AP, particularly SAP (Zhu et al., 2019; Yu et al., 2020; Zhu et al., 2021). In addition, Proteobacteria in the lung microbiota are closely associated with inflammatory lung disease and positively related to alveolar TNF-α (Dickson et al., 2016). A higher abundance of Proteobacteria was a distinguishing feature of ventilator-associated pneumonia (Fromentin et al., 2021). The enrichment of Proteobacteria might be a biomarker of inflammatory status in patients.
In our study, Enterobacteriaceae and Escherichia-Shigella were dominant in AP-ARDS patients. The overall levels of Enterobacteriaceae, and the individual Enterobacteriaceae genera, increased significantly in patients with ARDS. Escherichia-Shigella, a genus of Enterobacteriaceae family, is an opportunistic pathogen and more abundant in the sicker group. Random forest analysis identified Escherichia-Shigella as the most significant feature for distinguishing ARDS from non-ARDS. Multiple studies have shown that gut-associated bacteria in the lung microbiota, especially Enterobacteriaceae, are more abundant in ARDS patients. The abundance of Enterobacteriaceae in the lung microbiota was strongly associated with serum IL-6 level and the development of ARDS (Dickson et al., 2016; Dickson, 2018; Mukherjee and Hanidziar, 2018; Kyo et al., 2019). The composition of Enterobacteriaceae in the lung can help to identify ARDS patients (Dickson et al., 2020). The enrichment of gut-associated bacteria could also be a biomarker for ARDS patients (Fromentin et al., 2021). Suppression of the gut microbiota could improve the prognosis of critically ill patients (Silvestri et al., 2012).
At the species level, potentially pathogenic bacteria, including Klebsiella pneumoniae, Prevotella copri, and Clostridium ramosum, increased significantly in AP-ARDS patients. Klebsiella pneumoniae, a common pathogen of the Enterobacteriaceae family, normally colonizes respiratory tract and gut (Chen et al., 2021; Wolff et al., 2021). Dickson et al. revealed that Klebsiella pneumoniae overgrowth in the lung was strongly associated with ARDS (Dickson et al., 2020). In addition, Klebsiella pneumoniae infection can influence both the gut microbiome and lung metabolome (Wu et al., 2020; Jiang et al., 2022). After inoculation of mice with Klebsiella pneumoniae, the diversity and composition of the gut microbiota changed and contributed to lung microbiota dysbiosis within several hours (Jiang et al., 2022). Therefore, Klebsiella pneumoniae in the gut may influence lung inflammation through bacterial translocation. Prevotella could increase the host sensitivity to intestinal inflammation (Iljazovic et al., 2021). Prevotella copri is the most well-known of the Prevotella genus and is positively correlated with many inflammatory diseases, such as rheumatoid arthritis and ankylosing spondylitis (Tett et al., 2021). Transplantation of Prevotella copri induces dysbiosis of inflammatory and immune functions and can induce arthritis in mice (Maeda et al., 2016; Qian et al., 2022). Although less well studied, Clostridium ramosum has been proven to be positively correlated with Covid-19 disease severity, as well as infection and bacteremia (Zuo et al., 2020).
In our study, the levels of probiotics, such as Bifidobacterium and Bifidobacterium longum, decreased in patients with ARDS. Bifidobacterium longum was negatively correlated with organ failure and disease severity scores. As a beneficial bacterium, Bifidobacterium can help maintain gut barrier function, inhibit bacterial translocation, reduce lung inflammation, and therefore improve prognosis (Akshintala et al., 2019; Zhu et al., 2019; Zhu et al., 2021). In the current study, Bifidobacterium decreased in both AP patients and mice (Chen et al., 2017; Huang et al., 2017; Zhu et al., 2019; Li et al., 2020). Bifidobacterium longum can inhibit viral-induced lung inflammation and injury in mice (Groeger et al., 2020). Supplementation with Bifidobacterium longum has shown promising benefits for many diseases, such as irritable bowel syndrome, atopic dermatitis, and obesity (Schellekens et al., 2021; Fang et al., 2022; Sabaté and Iglicki, 2022). Therefore, probiotics have been used in the treatment of AP despite controversy.
Previous studies found the enrichment of gut-associated bacteria in the lung is closely associated with ARDS. However, whether the changes in the gut and lung microbiota are consistent has never been studied. In our study, the variation in gut microbiota in ARDS patients is similar to those seen in lung microbiota in previous studies, which suggests that changes in the lung microbiota might be due to the translocation of the gut microbiota in ARDS patients.
The gut-lung axis is a potential mechanism by which the gut microbiota influences lung inflammation. Gut microbiota can influence local immunity, systemic inflammation, and host immune suppression (Budden et al., 2017; Mukherjee and Hanidziar, 2018; Siwicka-Gieroba and Czarko-Wicha, 2020). Gut microbiota activate immune cells, which can migrate from the gut to the lung and assist in resisting systemic inflammatory disease (He et al., 2017; Mjösberg and Rao, 2018), and release metabolites and endotoxins to influence host immune response (Segain et al., 2000; Artis, 2008; Lin and Zhang, 2017). Additionally, gut microbiota dysbiosis damages the integrity of the intestinal barrier and enables bacterial translocation (Wang et al., 2022). Bacteria in the gut can translocate to the lung through the lymphatic or blood circulation systems and thus mediate lung inflammation (Mukherjee and Hanidziar, 2018). Enterobacteriaceae, Escherichia-Shigella, and several gut-associated bacteria have been detected in pancreatic fluid which suggested bacteria translocation could occur in AP patients and lead to infected pancreatic necrosis (Li et al., 2013; Hanna et al., 2014; Schmidt et al., 2014). Further studies have revealed that the composition of the lung microbiota can be easily changed, even if the immigration of gut-associated bacteria is transient (Dickson et al., 2016).
Gut microbiota could be transferred to the lung by several possible mechanisms. First, intestinal mucosal permeability may be impaired owing to dysbiosis of the microbiota. In our functional analysis, there was a difference in anaerobic bacterial composition between the AP-ARDS and AP-nonARDS groups. Dysbiosis of anaerobic bacteria is correlated with intestinal epithelial integrity and promotes overgrowth of pathogenic bacteria (Hong et al., 2018; Zhou and Liao, 2021). The proliferation of pathogenic bacteria can consume fatty acids, change intestinal pH, inhibit the growth of probiotics, and damage the gut chemical barrier (Wang et al., 2022). In addition, the overgrowth of pathogens restricts the function of immune cells, such as Tregs, Th2, and B cells; promotes the production of inflammatory factors, such as IL-1β, IL-6, and TNF-α; and thus damages the gut immune barrier (Zhou and Liao, 2021; Wang et al., 2022). As a normal pathogen, Escherichis-Shigella is associated with epithelial cell injury and is strongly correlated with AP and ARDS severity (Zhu et al., 2019; Pan et al., 2021). Through reduced butyrate production and increased oxidative stress, Escherichis-Shigella could penetrate the intestinal barrier, reach the basolateral layer, and spread rapidly to adjacent cells (Fokam Tagne et al., 2018; Dong et al., 2020). A second possible mechanism may involve the lung microenvironment which is important for bacterial colonization; normally, the alveolar ecosystem is not appropriate for bacterial reproduction (Dickson et al., 2017), however, the lung barrier could be damaged in AP patients. Previous studies have shown that inflammatory factors can migrate from the gut to the lung, recruit neutrophils in the blood, and cause lung inflammation (Mjösberg and Rao, 2018; Zhou and Liao, 2021). The inflammatory cascade amplified the inflammatory response and provided a more favorable inflammatory lung microenvironment for bacterial colonization. In ARDS patients, the influx of nutrient-rich edema and establishment of stark oxygen gradients will damage the local host defenses of the lung and make it easier for bacteria to translocate from the gut to the lung (Dickson, 2018). Therefore, patients may be more sensitive to disruption of the gut microbiota. However, these hypotheses have not been fully confirmed.
The translocation of bacteria from the gut to the lung has important clinical implications. Previous studies have proposed that the lung microbiota from bronchoalveolar lavage fluid (BAL) could help distinguish ARDS. However, rectal swabs are simpler to collect than BAL, therefore, gut microbiota data are easier to acquire than the lung; making gut microbiota a better prospect. In addition, our study found that the gut microbiota changed before the ARDS diagnosis. Prior to the occurrence of AP-ARDS, the gut microbiota already had characteristics relating to ARDS. Therefore, gut microbiota can be an important predictor of ARDS. Among them, Proteobacteria, Enterobacteriaceae, and Escherichia-Shigella were also found increased in lung microbiota in previous studies. These bacteria may help build prediction models for AP-ARDS that could assist clinicians in decision-making and prevent the occurrence and development of AP-ARDS.
Considering the potential function of microbiota dysbiosis, restoring immune competence and disturbing microbiota is a promising therapy for AP-ARDS (Mukherjee and Hanidziar, 2018). However, the effects of probiotics on patients with AP remain controversial. Some trials have revealed that probiotic supplements may have no benefit in the clinical outcomes of AP patients (Isenmann et al., 2004; Mazaki et al., 2006; Dellinger et al., 2007; de Vries et al., 2007) and probiotic treatment may even worsen the prognosis of patients with AP. Probiotics could cause bacteremia despite the rarity and transfer of antibiotic resistance from probiotics to pathogenic bacteria may worsen infection (Salminen et al., 2002; Cannon et al., 2005; Connolly et al., 2005; Feld et al., 2008). Besselink et al. illustrated that probiotic supplementation increases the occurrence of organ failure and mortality in patients with SAP (Besselink et al., 2008).
According to our study results, this poor response might be related to the overgrowth of pathogens and a disrupted intestinal mucosal barrier. For example, Klebsiella pneumoniae infection can inhibit Bifidobacterium production (Jiang et al., 2022) and the abundance of Prevotella is negatively associated with Bacteroides (Tett et al., 2021). Therefore, reducing pathogenic bacteria may promote the growth of probiotics, reduce barrier damage, and thus improve the efficacy of probiotic supplements. Targeted antibiotics are an effective strategy. Germ-free or antibiotic-treated animals are consistently protected from ARDS, and prophylactic administration of antibiotics decreases both mortality and multiple organ dysfunction syndromes, including ARDS (Dickson, 2016; Dickson, 2018). Supplementation with short-chain fatty acids (SCFAs) is another effective treatment option. Studies have found that oral supplementation with SCFAs could decrease susceptibility to bacterial infection, indicating that adjusting the gut microbiota could prevent bacterial pneumonia (Seki et al., 2021). These treatment concepts can be applied for AP patients to prevent ARDS (Siwicka-Gieroba and Czarko-Wicha, 2020). However, considering the potential harm caused by probiotics and antibiotics, targeted therapy should be provided to high-risk ARDS patients. Therefore, the prediction or early recognition of ARDS is essential. Collecting gut microbiota in the early stages of AP could help recognize and diagnose ARDS, and thus guide clinical management.
Gut microbiota could help identify high risk population for developing ARDS. Early identification gives time for appropriate intervention which could help improve prognosis. However, our study has some limitations. First, the specific role of microbiota changes in the disease is unclear. Our study can only provide correlations and suggest that the microbiota might help predict ARDS. However, the mechanism by which microbiota causes pathological conditions remains unknown. Zhang et al. found that different initial sites of infection could influence lung microbiota in patients with septic ARDS. ARDS patients with initial intrapulmonary infection tend to have higher abundance of gut-associated in lung (Zhang et al., 2022). To determine the specific role of gut-lung axis, it is better to make a more nuanced classification in the future. Second, the 16S rRNA sequence analysis could not predict the real composition and function of the microbiota community because it is based on the 16S rRNA sequence library. 16S rRNA analysis cannot completely replace metagenomic analysis but can help guide further studies. Third, the detection time 16S rRNA is long now. For clinical application, quick PCR kit target to specific bacteria is still needed.
In conclusion, this is the first study to report the relationship between gut microbiota and AP-ARDS. Gut microbiota showed a potential predicting ability for AP-ARDS. Dysbiosis of gut microbiota is strongly correlated with AP-ARDS. Enterobacteriaceae and Escherichia-Shigella are important prediction biomarkers for AP-ARDS. In the future, gut microbiota in early stage of patients with AP may help predict and allow early recognition of AP-ARDS, aid therapy planning, and thus improve patients’ quality of life and reduce morbidity of ARDS in AP patients. Further studies will improve our understanding of the role of microbiota in ARDS.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA893348.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of PUMCH (JS1826). The patients/participants provided their written informed consent to participate in this study.
Author contributions
XH and ZH conceived this study and drafted the manuscript. RZ and WS performed data analysis and reviewed the manuscript. ZH, RZ, WS, LG, ZY, and XS collected rectal swabs and clinical data. SZ revised the manuscript. HS and DW contributed to the study design, managed this study and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from Chinese Natural Science Foundation, grant number 82270405 and 32170788, National Key Clinical Specialty Construction Project, grant number ZK108000, National High Level Hospital Clinical Research Funding, grant number 2022-PUMCH-A-026, 2022-PUMCH-B-016, 2022-PUMCH-B-023, and Beijing Nova Program.
Acknowledgments
This work was supported by Public Laboratory Platform, National Science and Technology Key Infrastructure on Translational Medicine in Peking Union Medical College Hospital.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AP, acute pancreatitis; AP-ARDS, acute pancreatitis with acute respiratory distress syndrome; AP-nonARDS, acute pancreatitis without acute respiratory distress syndrome; ARDS, acute respiratory distress syndrome; ASV, amplicon sequence variant; BAL, bronchoalveolar lavage fluid; BMI, body mass index; ICU, intensive care unit; IQR, interquartile range; LDA, linear discriminant analysis; MAP, mild acute pancreatitis; MARDS, mild acute respiratory distress syndrome; MSAP, moderate severe pancreatitis; OF, organ failure; PCoA, principal coordinate analysis; SAP, severe acute pancreatitis; SCFAs, short-chain fatty acids; SD, standard deviation.
References
Akshintala, V. S., Talukdar, R., Singh, V. K., Goggins, M. (2019). The gut microbiome in pancreatic disease. Clin. Gastroenterol. Hepatol. 17 (2), 290–295. doi: 10.1016/j.cgh.2018.08.045
Artis, D. (2008). Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 8 (6), 411–420. doi: 10.1038/nri2316
Banks, P. A., Bollen, T. L., Dervenis, C., Gooszen, H. G., Johnson, C. D., Sarr, M. G., et al. (2013). Classification of acute pancreatitis–2012: revision of the Atlanta classification and definitions by international consensus. Gut 62 (1), 102–111. doi: 10.1136/gutjnl-2012-302779
Bellani, G., Pham, T., Laffey, J. G. (2020). Missed or delayed diagnosis of ARDS: a common and serious problem. Intensive Care Med. 46 (6), 1180–1183. doi: 10.1007/s00134-020-06035-0
Besselink, M. G., van Santvoort, H. C., Boermeester, M. A., Nieuwenhuijs, V. B., van Goor, H., Dejong, C. H., et al. (2009). Timing and impact of infections in acute pancreatitis. Br. J. Surg. 96 (3), 267–273. doi: 10.1002/bjs.6447
Besselink, M. G., van Santvoort, H. C., Buskens, E., Boermeester, M. A., van Goor, H., Timmerman, H. M., et al. (2008). Probiotic prophylaxis in predicted severe acute pancreatitis: A randomised, double-blind, placebo-controlled trial. Lancet 371 (9613), 651–659. doi: 10.1016/s0140-6736(08)60207-x
Bozzi Cionci, N., Baffoni, L., Gaggìa, F., Di Gioia, D. (2018). Therapeutic microbiology: The role of bifidobacterium breve as food supplement for the Prevention/Treatment of paediatric diseases. Nutrients 10 (11), 1–27. doi: 10.3390/nu10111723
Budden, K. F., Gellatly, S. L., Wood, D. L., Cooper, M. A., Morrison, M., Hugenholtz, P., et al. (2017). Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 15 (1), 55–63. doi: 10.1038/nrmicro.2016.142
Cannon, J. P., Lee, T. A., Bolanos, J. T., Danziger, L. H. (2005). Pathogenic relevance of lactobacillus: A retrospective review of over 200 cases. Eur. J. Clin. Microbiol. Infect. Dis. 24 (1), 31–40. doi: 10.1007/s10096-004-1253-y
Chen, J., Huang, C., Wang, J., Zhou, H., Lu, Y., Lou, L., et al. (2017). Dysbiosis of intestinal microbiota and decrease in paneth cell antimicrobial peptide level during acute necrotizing pancreatitis in rats. PloS One 12 (4), e0176583. doi: 10.1371/journal.pone.0176583
Chen, C. M., Wang, M., Li, X. P., Li, P. L., Tian, J. J., Zhang, K., et al. (2021). Homology analysis between clinically isolated extraintestinal and enteral klebsiella pneumoniae among neonates. BMC Microbiol. 21 (1), 25. doi: 10.1186/s12866-020-02073-2
Connolly, E., Abrahamsson, T., Björkstén, B. (2005). Safety of d(-)-lactic acid producing bacteria in the human infant. J. Pediatr. Gastroenterol. Nutr. 41 (4), 489–492. doi: 10.1097/01.mpg.0000176179.81638.45
Dellinger, E. P., Tellado, J. M., Soto, N. E., Ashley, S. W., Barie, P. S., Dugernier, T., et al. (2007). Early antibiotic treatment for severe acute necrotizing pancreatitis: a randomized, double-blind, placebo-controlled study. Ann. Surg. 245 (5), 674–683. doi: 10.1097/01.sla.0000250414.09255.84
de Vries, A. C., Besselink, M. G., Buskens, E., Ridwan, B. U., Schipper, M., van Erpecum, K. J., et al. (2007). Randomized controlled trials of antibiotic prophylaxis in severe acute pancreatitis: relationship between methodological quality and outcome. Pancreatology 7 (5-6), 531–538. doi: 10.1159/000108971
Dickson, R. P. (2016). The microbiome and critical illness. Lancet Respir. Med. 4 (1), 59–72. doi: 10.1016/s2213-2600(15)00427-0
Dickson, R. P. (2018). The lung microbiome and ARDS. it is time to broaden the model. Am. J. Respir. Crit. Care Med. 197 (5), 549–551. doi: 10.1164/rccm.201710-2096ED
Dickson, R. P., Erb-Downward, J. R., Freeman, C. M., McCloskey, L., Falkowski, N. R., Huffnagle, G. B., et al. (2017). Bacterial topography of the healthy human lower respiratory tract. mBio 8 (1), 1–12. doi: 10.1128/mBio.02287-16
Dickson, R. P., Schultz, M. J., van der Poll, T., Schouten, L. R., Falkowski, N. R., Luth, J. E., et al. (2020). Lung microbiota predict clinical outcomes in critically ill patients. Am. J. Respir. Crit. Care Med. 201 (5), 555–563. doi: 10.1164/rccm.201907-1487OC
Dickson, R. P., Singer, B. H., Newstead, M. W., Falkowski, N. R., Erb-Downward, J. R., Standiford, T. J., et al. (2016). Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 1 (10), 16113. doi: 10.1038/nmicrobiol.2016.113
Dong, R., Bai, M., Zhao, J., Wang, D., Ning, X., Sun, S. (2020). A comparative study of the gut microbiota associated with immunoglobulin a nephropathy and membranous nephropathy. Front. Cell Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.557368
Fan, E., Brodie, D., Slutsky, A. S. (2018). Acute respiratory distress syndrome: Advances in diagnosis and treatment. Jama 319 (7), 698–710. doi: 10.1001/jama.2017.21907
Fang, Z., Pan, T., Li, L., Wang, H., Zhu, J., Zhang, H., et al. (2022). Bifidobacterium longum mediated tryptophan metabolism to improve atopic dermatitis via the gut-skin axis. Gut Microbes 14 (1), 2044723. doi: 10.1080/19490976.2022.2044723
Feld, L., Schjørring, S., Hammer, K., Licht, T. R., Danielsen, M., Krogfelt, K., et al. (2008). Selective pressure affects transfer and establishment of a lactobacillus plantarum resistance plasmid in the gastrointestinal environment. J. Antimicrob. Chemother. 61 (4), 845–852. doi: 10.1093/jac/dkn033
Fokam Tagne, M. A., Noubissi, P. A., Fankem, G. O., Kamgang, R. (2018). Effects of oxalis barrelieri l. (Oxalidaceae) aqueous extract on diarrhea induced by shigella dysenteriae type 1 in rats. Health Sci. Rep. 1 (2), e20. doi: 10.1002/hsr2.20
Fromentin, M., Ricard, J. D., Roux, D. (2021). Respiratory microbiome in mechanically ventilated patients: A narrative review. Intensive Care Med. 47 (3), 292–306. doi: 10.1007/s00134-020-06338-2
Garg, P. K., Madan, K., Pande, G. K., Khanna, S., Sathyanarayan, G., Bohidar, N. P., et al. (2005). Association of extent and infection of pancreatic necrosis with organ failure and death in acute necrotizing pancreatitis. Clin. Gastroenterol. Hepatol. 3 (2), 159–166. doi: 10.1016/s1542-3565(04)00665-2
Gray, K. D., Simovic, M. O., Chapman, W. C., Blackwell, T. S., Christman, J. W., May, A. K., et al. (2003). Endotoxin potentiates lung injury in cerulein-induced pancreatitis. Am. J. Surg. 186 (5), 526–530. doi: 10.1016/j.amjsurg.2003.07.010
Greenberg, J. A., Hsu, J., Bawazeer, M., Marshall, J., Friedrich, J. O., Nathens, A., et al. (2016). Clinical practice guideline: Management of acute pancreatitis. Can. J. Surg. 59 (2), 128–140. doi: 10.1503/cjs.015015
Groeger, D., Schiavi, E., Grant, R., Kurnik-Łucka, M., Michalovich, D., Williamson, R., et al. (2020). Intranasal bifidobacterium longum protects against viral-induced lung inflammation and injury in a murine model of lethal influenza infection. EBioMedicine 60, 102981. doi: 10.1016/j.ebiom.2020.102981
Hanna, E. M., Hamp, T. J., McKillop, I. H., Bahrani-Mougeot, F., Martinie, J. B., Horton, J. M., et al. (2014). Comparison of culture and molecular techniques for microbial community characterization in infected necrotizing pancreatitis. J. Surg. Res. 191 (2), 362–369. doi: 10.1016/j.jss.2014.05.003
He, Y., Wen, Q., Yao, F., Xu, D., Huang, Y., Wang, J. (2017). Gut-lung axis: The microbial contributions and clinical implications. Crit. Rev. Microbiol. 43 (1), 81–95. doi: 10.1080/1040841x.2016.1176988
Herridge, M. S., Tansey, C. M., Matté, A., Tomlinson, G., Diaz-Granados, N., Cooper, A., et al. (2011). Functional disability 5 years after acute respiratory distress syndrome. N Engl. J. Med. 364 (14), 1293–1304. doi: 10.1056/NEJMoa1011802
Hong, G., Zheng, D., Zhang, L., Ni, R., Wang, G., Fan, G. C., et al. (2018). Administration of nicotinamide riboside prevents oxidative stress and organ injury in sepsis. Free Radic. Biol. Med. 123, 125–137. doi: 10.1016/j.freeradbiomed.2018.05.073
Hu, X., Fan, Y., Li, H., Zhou, R., Zhao, X., Sun, Y., et al. (2021a). Impacts of cigarette smoking status on metabolomic and gut microbiota profile in Male patients with coronary artery disease: A multi-omics study. Front. Cardiovasc. Med. 8. doi: 10.3389/fcvm.2021.766739
Hu, X., Gong, L., Zhou, R., Han, Z., Ji, L., Zhang, Y., et al. (2021b). Variations in gut microbiome are associated with prognosis of hypertriglyceridemia-associated acute pancreatitis. Biomolecules 11 (5), 1–16. doi: 10.3390/biom11050695
Huang, C., Chen, J., Wang, J., Zhou, H., Lu, Y., Lou, L., et al. (2017). Dysbiosis of intestinal microbiota and decreased antimicrobial peptide level in paneth cells during hypertriglyceridemia-related acute necrotizing pancreatitis in rats. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00776
Iljazovic, A., Roy, U., Gálvez, E. J. C., Lesker, T. R., Zhao, B., Gronow, A., et al. (2021). Perturbation of the gut microbiome by prevotella spp. enhances host susceptibility to mucosal inflammation. Mucosal Immunol. 14 (1), 113–124. doi: 10.1038/s41385-020-0296-4
Isenmann, R., Rünzi, M., Kron, M., Kahl, S., Kraus, D., Jung, N., et al. (2004). Prophylactic antibiotic treatment in patients with predicted severe acute pancreatitis: a placebo-controlled, double-blind trial. Gastroenterology 126 (4), 997–1004. doi: 10.1053/j.gastro.2003.12.050
Jiang, Q., Xu, Q., Kenéz, Á., Chen, S., Yang, G. (2022). Klebsiella pneumoniae infection is associated with alterations in the gut microbiome and lung metabolome. Microbiol. Res. 263, 127139. doi: 10.1016/j.micres.2022.127139
Johnson, C. D., Abu-Hilal, M. (2004). Persistent organ failure during the first week as a marker of fatal outcome in acute pancreatitis. Gut 53 (9), 1340–1344. doi: 10.1136/gut.2004.039883
Kyo, M., Nishioka, K., Nakaya, T., Kida, Y., Tanabe, Y., Ohshimo, S., et al. (2019). Unique patterns of lower respiratory tract microbiota are associated with inflammation and hospital mortality in acute respiratory distress syndrome. Respir. Res. 20 (1), 246. doi: 10.1186/s12931-019-1203-y
Li, X. Y., He, C., Zhu, Y., Lu, N. H. (2020). Role of gut microbiota on intestinal barrier function in acute pancreatitis. World J. Gastroenterol. 26 (18), 2187–2193. doi: 10.3748/wjg.v26.i18.2187
Li, Q., Wang, C., Tang, C., He, Q., Li, N., Li, J. (2013). Bacteremia in patients with acute pancreatitis as revealed by 16S ribosomal RNA gene-based techniques*. Crit. Care Med. 41 (8), 1938–1950. doi: 10.1097/CCM.0b013e31828a3dba
Lin, L., Zhang, J. (2017). Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 18 (1), 2. doi: 10.1186/s12865-016-0187-3
Maeda, Y., Kurakawa, T., Umemoto, E., Motooka, D., Ito, Y., Gotoh, K., et al. (2016). Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol 68 (11), 2646–2661. doi: 10.1002/art.39783
Mazaki, T., Ishii, Y., Takayama, T. (2006). Meta-analysis of prophylactic antibiotic use in acute necrotizing pancreatitis. Br. J. Surg. 93 (6), 674–684. doi: 10.1002/bjs.5389
Mjösberg, J., Rao, A. (2018). Lung inflammation originating in the gut. Science 359 (6371), 36–37. doi: 10.1126/science.aar4301
Mukherjee, S., Hanidziar, D. (2018). More of the gut in the lung: How two microbiomes meet in ARDS. Yale J. Biol. Med. 91 (2), 143–149.
Pan, L. L., Li, B. B., Pan, X. H., Sun, J. (2021). Gut microbiota in pancreatic diseases: possible new therapeutic strategies. Acta Pharmacol. Sin. 42 (7), 1027–1039. doi: 10.1038/s41401-020-00532-0
Pan, C., Liu, L., Xie, J. F., Qiu, H. B. (2018). Acute respiratory distress syndrome: Challenge for diagnosis and therapy. Chin. Med. J. (Engl) 131 (10), 1220–1224. doi: 10.4103/0366-6999.228765
Qian, X., Zhang, H. Y., Li, Q. L., Ma, G. J., Chen, Z., Ji, X. M., et al. (2022). Integrated microbiome, metabolome, and proteome analysis identifies a novel interplay among commensal bacteria, metabolites and candidate targets in non-small cell lung cancer. Clin. Transl. Med. 12 (6), e947. doi: 10.1002/ctm2.947
Ranieri, V. M., Rubenfeld, G. D., Thompson, B. T., Ferguson, N. D., Caldwell, E., Fan, E., et al. (2012). Acute respiratory distress syndrome: The Berlin definition. Jama 307 (23), 2526–2533. doi: 10.1001/jama.2012.5669
Sabaté, J. M., Iglicki, F. (2022). Effect of bifidobacterium longum 35624 on disease severity and quality of life in patients with irritable bowel syndrome. World J. Gastroenterol. 28 (7), 732–744. doi: 10.3748/wjg.v28.i7.732
Salminen, M. K., Tynkkynen, S., Rautelin, H., Saxelin, M., Vaara, M., Ruutu, P., et al. (2002). Lactobacillus bacteremia during a rapid increase in probiotic use of lactobacillus rhamnosus GG in Finland. Clin. Infect. Dis. 35 (10), 1155–1160. doi: 10.1086/342912
Schellekens, H., Torres-Fuentes, C., van de Wouw, M., Long-Smith, C. M., Mitchell, A., Strain, C., et al. (2021). Bifidobacterium longum counters the effects of obesity: Partial successful translation from rodent to human. EBioMedicine 63, 103176. doi: 10.1016/j.ebiom.2020.103176
Schmandt, M., Glowka, T. R., Kreyer, S., Muders, T., Muenster, S., Theuerkauf, N. U., et al. (2021). Secondary ARDS following acute pancreatitis: Is extracorporeal membrane oxygenation feasible or futile? J. Clin. Med. 10 (5), 1–11. doi: 10.3390/jcm10051000
Schmidt, P. N., Roug, S., Hansen, E. F., Knudsen, J. D., Novovic, S. (2014). Spectrum of microorganisms in infected walled-off pancreatic necrosis - impact on organ failure and mortality. Pancreatology 14 (6), 444–449. doi: 10.1016/j.pan.2014.09.001
Segain, J. P., Raingeard de la Blétière, D., Bourreille, A., Leray, V., Gervois, N., Rosales, C., et al. (2000). Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for crohn's disease. Gut 47 (3), 397–403. doi: 10.1136/gut.47.3.397
Seki, D., Mayer, M., Hausmann, B., Pjevac, P., Giordano, V., Goeral, K., et al. (2021). Aberrant gut-microbiota-immune-brain axis development in premature neonates with brain damage. Cell Host Microbe 29 (10), 1558–1572.e1556. doi: 10.1016/j.chom.2021.08.004
Shah, J., Rana, S. S. (2020). Acute respiratory distress syndrome in acute pancreatitis. Indian J. Gastroenterol. 39 (2), 123–132. doi: 10.1007/s12664-020-01016-z
Silvestri, L., de la Cal, M. A., van Saene, H. K. (2012). Selective decontamination of the digestive tract: the mechanism of action is control of gut overgrowth. Intensive Care Med. 38 (11), 1738–1750. doi: 10.1007/s00134-012-2690-1
Siwicka-Gieroba, D., Czarko-Wicha, K. (2020). Lung microbiome - a modern knowledge. Cent Eur. J. Immunol. 45 (3), 342–345. doi: 10.5114/ceji.2020.101266
Tenner, S., Baillie, J., DeWitt, J., Vege, S. S. (2013). American College of gastroenterology guideline: management of acute pancreatitis. Am. J. Gastroenterol. 108 (9), 1400–1415; 1416. doi: 10.1038/ajg.2013.218
Tett, A., Pasolli, E., Masetti, G., Ercolini, D., Segata, N. (2021). Prevotella diversity, niches and interactions with the human host. Nat. Rev. Microbiol. 19 (9), 585–599. doi: 10.1038/s41579-021-00559-y
Wang, Z., Li, F., Liu, J., Luo, Y., Guo, H., Yang, Q., et al. (2022). Intestinal microbiota - an unmissable bridge to severe acute pancreatitis-associated acute lung injury. Front. Immunol. 13. doi: 10.3389/fimmu.2022.913178
Wolff, N. S., Jacobs, M. C., Wiersinga, W. J., Hugenholtz, F. (2021). Pulmonary and intestinal microbiota dynamics during gram-negative pneumonia-derived sepsis. Intensive Care Med. Exp. 9 (1), 35. doi: 10.1186/s40635-021-00398-4
Wu, T., Xu, F., Su, C., Li, H., Lv, N., Liu, Y., et al. (2020). Alterations in the gut microbiome and cecal metabolome during klebsiella pneumoniae-induced pneumosepsis. Front. Immunol. 11. doi: 10.3389/fimmu.2020.01331
Yu, S., Xiong, Y., Fu, Y., Chen, G., Zhu, H., Mo, X., et al. (2021). Shotgun metagenomics reveals significant gut microbiome features in different grades of acute pancreatitis. Microb. Pathog. 154, 104849. doi: 10.1016/j.micpath.2021.104849
Yu, S., Xiong, Y., Xu, J., Liang, X., Fu, Y., Liu, D., et al. (2020). Identification of dysfunctional gut microbiota through rectal swab in patients with different severity of acute pancreatitis. Dig Dis. Sci. 65 (11), 3223–3237. doi: 10.1007/s10620-020-06061-4
Zhang, P., Chen, Y., Zheng, W., Wu, M., Wu, Z., Lu, Y., et al. (2021). Changes of lung microbiome of acute respiratory distress syndrome before and after treatment under open airway. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 33 (9), 1063–1068. doi: 10.3760/cma.j.cn121430-20210414-00558
Zhang, P., Liu, B., Zheng, W., Chen, Y., Wu, Z., Lu, Y., et al. (2022). Pulmonary microbial composition in sepsis-induced acute respiratory distress syndrome. Front. Mol. Biosci. 9. doi: 10.3389/fmolb.2022.862570
Zhang, X. M., Zhang, Z. Y., Zhang, C. H., Wu, J., Wang, Y. X., Zhang, G. X. (2018). Intestinal microbial community differs between acute pancreatitis patients and healthy volunteers. BioMed. Environ. Sci. 31 (1), 81–86. doi: 10.3967/bes2018.010
Zheng, J., Lou, L., Fan, J., Huang, C., Mei, Q., Wu, J., et al. (2019). Commensal escherichia coli aggravates acute necrotizing pancreatitis through targeting of intestinal epithelial cells. Appl. Environ. Microbiol. 85 (12), 1–15. doi: 10.1128/aem.00059-19
Zhou, X., Liao, Y. (2021). Gut-lung crosstalk in sepsis-induced acute lung injury. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.779620
Zhu, Y., He, C., Li, X., Cai, Y., Hu, J., Liao, Y., et al. (2019). Gut microbiota dysbiosis worsens the severity of acute pancreatitis in patients and mice. J. Gastroenterol. 54 (4), 347–358. doi: 10.1007/s00535-018-1529-0
Zhu, Y., Mei, Q., Fu, Y., Zeng, Y. (2021). Alteration of gut microbiota in acute pancreatitis and associated therapeutic strategies. BioMed. Pharmacother. 141, 111850. doi: 10.1016/j.biopha.2021.111850
Keywords: acute pancreatitis, acute respiratory distress syndrome, gut microbiota, disease prediction, biomarker
Citation: Hu X, Han Z, Zhou R, Su W, Gong L, Yang Z, Song X, Zhang S, Shu H and Wu D (2023) Altered gut microbiota in the early stage of acute pancreatitis were related to the occurrence of acute respiratory distress syndrome. Front. Cell. Infect. Microbiol. 13:1127369. doi: 10.3389/fcimb.2023.1127369
Received: 19 December 2022; Accepted: 20 February 2023;
Published: 06 March 2023.
Edited by:
Leticia Reyes, University of Wisconsin-Madison, United StatesReviewed by:
Fons Van Den Berg, Amsterdam University Medical Center, NetherlandsPeng Ge, Dalian Medical University, China
Copyright © 2023 Hu, Han, Zhou, Su, Gong, Yang, Song, Zhang, Shu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huijun Shu, c2hqcHVtY2gyMDAyQGFsaXl1bi5jb20=; Dong Wu, d3Vkb25nQHB1bWNoLmNu
†These authors have contributed equally to this work and share first authorship
 Xiaomin Hu1,2†
Xiaomin Hu1,2† Ziying Han
Ziying Han Wan Su
Wan Su Liang Gong
Liang Gong Zihan Yang
Zihan Yang Shuyang Zhang
Shuyang Zhang Dong Wu
Dong Wu