
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 17 May 2023
Sec. Microbiome in Health and Disease
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1119992
This article is part of the Research TopicGut Microbe and Colorectal CancerView all 7 articles
 Ming Gu†
Ming Gu† Weixiang Yin†
Weixiang Yin† Jiaming Zhang†
Jiaming Zhang† Junfeng Yin
Junfeng Yin Xiaofei Tang
Xiaofei Tang Jie Ling
Jie Ling Zhijie Tang
Zhijie Tang Weijuan Yin
Weijuan Yin Xiangjun Wang*
Xiangjun Wang* Qing Ni*
Qing Ni* Yunxiang Zhu*
Yunxiang Zhu* Tuo Chen*
Tuo Chen*Colorectal cancer (CRC) is a major health burden, accounting for approximately 10% of all new cancer cases worldwide. Accumulating evidence suggests that the crosstalk between the host mucins and gut microbiota is associated with the occurrence and development of CRC. Mucins secreted by goblet cells not only protect the intestinal epithelium from microorganisms and invading pathogens but also provide a habitat for commensal bacteria. Conversely, gut dysbiosis results in the dysfunction of mucins, allowing other commensals and their metabolites to pass through the intestinal epithelium, potentially triggering host responses and the subsequent progression of CRC. In this review, we summarize how gut microbiota and bacterial metabolites regulate the function and expression of mucin in CRC and novel treatment strategies for CRC.
Globally, colorectal cancer (CRC) has become a major health burden due to its higher incidence and mortality. Epidemiological data indicate that CRC ranks third in incidence with 1.9 million new cases and is the second most common cause of cancer mortality (Sung et al., 2021). In China, over 300,000 new cases and 191,000 deaths are reported annually (Chen et al., 2016; Chen et al., 2018b). As with many diseases, the etiology of CRC is multi-factors involving genetic and environmental factors (Song and Chan, 2019). While genetic susceptibility implicated in CRC is well-described, the incidence of CRC in genetic predisposition syndromes, including familial adenomatous polyposis, Peutz–Jeghers syndrome, and Lynch syndrome, only accounts for a minority of CRC cases (Boland et al., 2018; Hryhorowicz et al., 2022). Thus, it is suggested that environmental factors play a major role in the initiation and progression of CRC (Keum and Giovannucci, 2019). Among environmental factors, the gut microbiome has been increasingly considered a modulator of CRC (Zou et al., 2018).
The community of bacteria, fungi, archaea, phages, and protists is referred to as the microbiota. These microorganisms within the gastrointestinal tract are named “gut microbiota.” There are approximately 1013 to 1014 bacteria living in the gut, which contain 10 times more than human cells and outnumber human genes by a factor of 100 (Sears, 2005; Wardman et al., 2022). These microorganisms play an important role in maintaining the intestinal epithelium (Hill et al., 2017), harvesting energy (Vandeputte, 2020), and maturing immunity (Shi et al., 2017). Meanwhile, the shift in their composition has been associated with cardiovascular diseases (Brown and Hazen, 2018), metabolic diseases (Maruvada et al., 2017), and digestive diseases (such as inflammatory bowel disease and CRC) (Kostic et al., 2014; O'Keefe, 2016). Accumulating evidence shows that the initiation of CRC is triggered by the dysfunction of colonic mucosal barrier colonization by specific gut microbiota (Sheng et al., 2012; Wong and Yu, 2019). These bacteria cause changes in the tumor microenvironment, allowing for colonization by opportunistic bacteria that facilitate disease progression.
The mucus layer acts as the first gatekeeper against environmental and microbial insults. Among the components of the mucus layer, mucins, mainly secreted by goblet cells, are found throughout the gastrointestinal epithelium (Johansson and Hansson, 2016). The mucus layer not only creates a physical barrier between the host and commensals but also provides an energy source for bacterial growth (Johansson et al., 2015; Desai et al., 2016). In healthy individuals, the gut microbiota is accompanied by a thicker mucus layer. In contrast, thinner mucus and gut dysbiosis have been implicated in the development of CRC through the underlying mechanism of gut microbiota and its metabolites stimulating mucus secretion (Petersson et al., 2011; Earle et al., 2015). In this review, we summarize the recent studies that focus on the role of microbiota and bacterial metabolites in mucins in CRC and offer different bacteria-targeted therapies for mucin regulation (Figure 1).
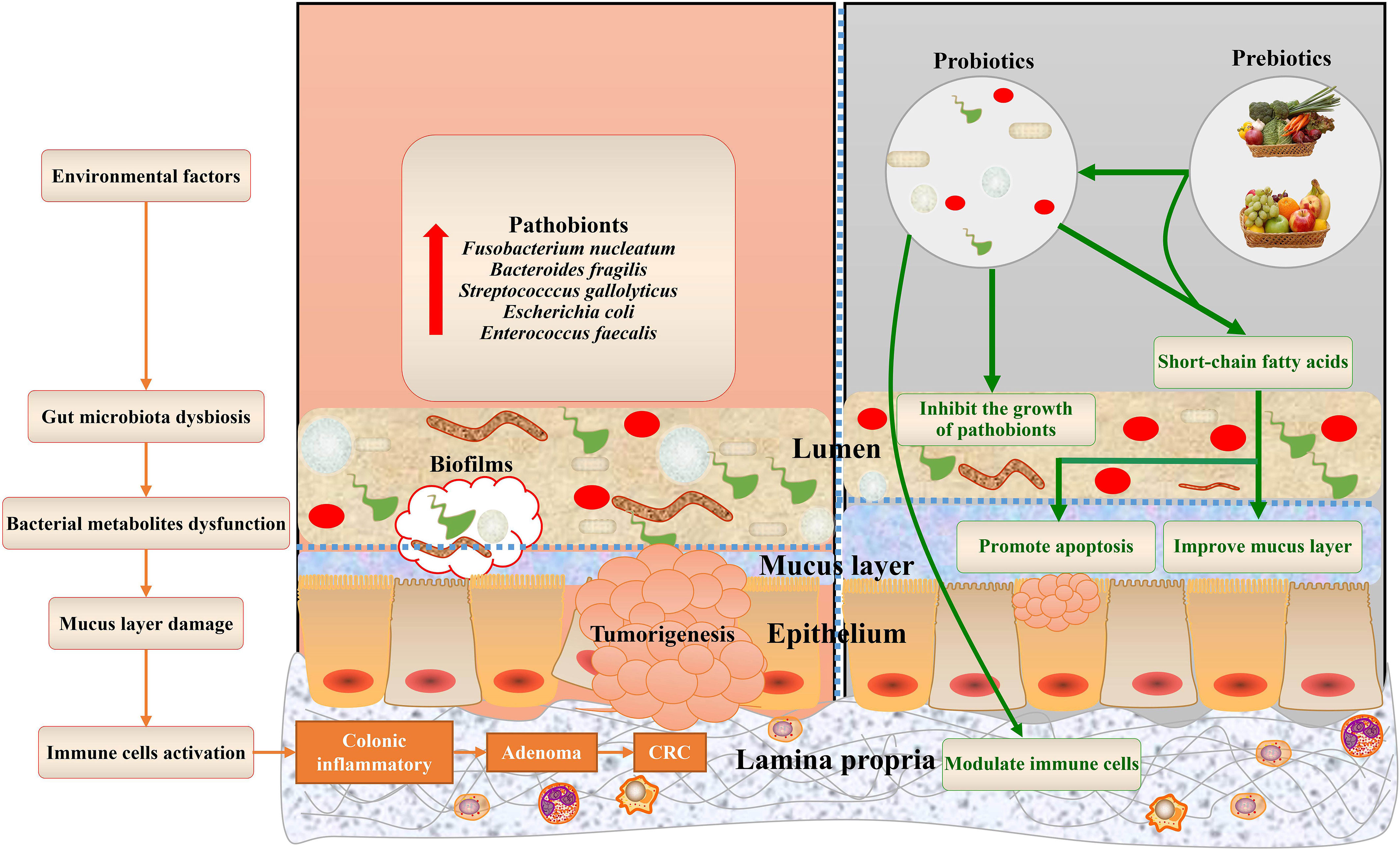
Figure 1 Gut microbiota bacterial metabolites associated with mucins of colorectal cancer, and potential bacteria-related therapies. The dysbiosis of the gut microbiota and dysfunctions of bacterial metabolites, aggravated by environmental factors, contribute to mucus layer damage during colorectal cancer. This schematic also summarizes the health benefits of prebiotics and probiotics that cause alterations to mucins of CRC.
In contrast to the small intestine (which comprises a single mucus layer), two distinct mucus layers are involved in the colon. It is composed of an outer layer exposed to commensal microbiota and an inner layer that is firmly and densely attached to the epithelium (Pothuraju et al., 2020). The inner mucus layer is rich in mucin-2 (MUC2), produced by specialized cells of the host called goblet cells, and permits less bacterial penetration into the intestinal epithelium (Desai et al., 2016). Moreover, the numerous O-linked glycans in the outer layer cannot only provide bacterial habitats but also serve as an energy source for bacteria (Zhang et al., 2021). The mucus layer is constantly renewed and can be rapidly adjusted to alternations in the intestinal microenvironment against bacterial invasion and activation of inflammatory responses. However, the dysfunction of the mucus layer allows the microbiota to come into contact with the intestinal epithelium, affecting the initiation and progression of CRC via initiating modifications of epithelial cells and triggering intestinal inflammation responses (Coleman et al., 2018; Yu, 2018). Hence, we focus on the mucins, including MUC2, mucin-5AC (MUC5AC), mucin-5B (MUC5B), and mucin-6 (MUC6), and systematically review their composition and function in CRC.
MUC2 is the most abundant colonic mucin and forms the basis of the mucus layer. It covers the surface of intestinal mucosa in the form of gelatin (Yamashita and Melo, 2018). It is accepted that MUC2 protein plays an important role in keeping the intestinal tract healthy, while abnormal levels of MUC2 can be found in CRC patients. Numerous studies have indicated that MUC2 mucin production was reduced in patients with CRC (Bu et al., 2010; Al-Khayal et al., 2016), and higher MUC2 expression was negatively correlated with TNM stage, lymphatic metastasis, and prognosis of CRC (Elzagheid et al., 2013; Li et al., 2018). Moreover, murine models have demonstrated that MUC2−/− mice allowed bacteria to contact with the intestinal epithelium, resulting in inflammation and colon cancer (Wenzel et al., 2014). An absence of MUC2 mucin expression was closely related to high methylation modification of the MUC2 promoter and a glycosylation defect of the MUC2 gene in CRC cells (Yonezawa and Sato, 1997; Biemer-Hüttmann et al., 2000).
MUC5AC is mainly secreted by gastric goblet cells, which belong to gastric mucins. Concurrent with these studies, the expression of MUC5AC was not observed in normal colorectal epithelial cells, but its expression was significantly increasing in CRC tissues (Krishn et al., 2016). Abnormal expression of MUC5AC was related to microsatellite instability (MSI) status and poor differentiation. Moreover, MSI status was determined by MUC5AC demethylation, indicating that MUC5AC hypomethylation was a promising marker for MSI in CRC (Renaud et al., 2015). Higher MUC5 expression in CRC patients was positively associated with a high lymph node metastasis rate, poor cell differentiation, and late-stage CRC (Wang et al., 2017). In vitro, blocking the expression of MUC5AC in SW620 cells by siRNA technology significantly induced cell apoptosis and G1-phase cell cycle arrest and inhibited tumor cell invasion and migration (Zhu et al., 2016). The above results suggest that MUC5AC acts as a potential target for the treatment of CRC.
MUC5B is mainly expressed in the bronchus, gland, cervix, gallbladder, and pancreas and less expressed in a subset of goblet cells at the bottom of the colonic crypts in humans (Vandenhaute et al., 1997; van Klinken et al., 1998). Consistent with the mechanism of MUC5AC, overexpression of MUC5B is associated with poor outcomes in different types of gastrointestinal cancers. In HT-29 MTX cells and LS174T cells, respectively, belonging to gastric and intestinal cancer cell lines, the expression of MUC5B was significantly increased (Lesuffleur et al., 1995). To better understand the abnormal expression of the MUC5B on the pathogenesis of cancer cells, silencing the MUC5B gene in the colon cancer cell line LS174T and gastric cancer cell line KATO-III efficiently restrained cell proliferation and migration by regulating the Wnt/β-catenin pathway (Lahdaoui et al., 2017).
MUC6, which is highly similar to MUC5AC, belongs to gastric mucins, but it is mainly rich in glandular epithelial cells. Lower expression of MUC6 has been reported to be associated with increased tumor cell mobility in CRC (Tsai et al., 2015). Moreover, overexpression of the MUC6 in patients with CRC had long PFS and cancer-specific survival (Betge et al., 2016). Although current studies indicate that MUC6 plays a protective in the occurrence and development of CRC, the specific mechanism needs to be verified in future experiments.
Despite several studies emphasizing the role of mucins in CRC, the modulating effects of gut microbiota on mucins are often ignored. Some species of pathogenic and commensal bacteria could degrade mucins or use them as attachment sites, promoting their colonization and replication. These invasive strains blinding the intestinal epithelium drive the transition to a pro-inflammatory microenvironment that accelerates colorectal tumorigenesis. Several pathogenic and commensal bacteria have been associated with CRC, including Fusobacterium nucleatum, Bacteroides fragilis, Streptococcus gallolyticus, Escherichia coli, and Enterococcus faecalis. Moreover, it has been shown that the influence of gut microbiota on the mucus layer of CRC requires the formation of bacterial biofilms. Here, we highlight the potential role of gut microbiota and their biofilms in regulating the mucins of CRC.
The obligate anaerobic, Gram-negative bacterial species Fusobacterium nucleatum (F. nucleatum) is a normal inhabitant of the human gut and mouth. It has been recognized as an opportunistic pathogen implicated in CRC (Tahara et al., 2014; Chen et al., 2017). Recent studies have demonstrated that an abundance of F. nucleatum is enriched in CRC tissue in comparison to normal tissue (Kostic et al., 2012; Warren et al., 2013; Ye et al., 2017; King and Hurley, 2020). The increasing number of F. nucleatum in CRC patients is associated with poor survival (Mima et al., 2016). Pro-tumorigenic effects of F. nucleatum on CRC were associated with dysfunction of the intestinal mucosal barrier and the secretion of pro-inflammatory factors. Invasive strains of F. nucleatum accelerated mucin secretion, which resulted in the rapid depletion of mucin stores from goblet cells and subsequently breached the mucus layer (Dharmani et al., 2011). Meanwhile, the FadA adhesion protein secreted by F. nucleatum provoked the β-catenin signaling pathway in intestinal epithelial cells by interacting with E-cadherin, leading to upregulation of pro-inflammatory responses and pro-oncogenic pathways in colorectal cancer cases (Rubinstein et al., 2013; Rubinstein et al., 2019). Additionally, supplementation with F. nucleatum isolated from a patient with inflammatory bowel disease (IBD) in ApcMin/+ mice promoted tumor progression (Hooper and Macpherson, 2010; Kostic et al., 2013). Therefore, F. nucleatum could serve as a potential bacterial marker for the diagnosis and treatment of CRC.
Bacteroides fragilis (B. fragilis), belonging to the Bacteroidetes phylum, is a common obligate anaerobic, Gram-negative gut bacterium. Although B. fragilis acted as a common colonic symbiote with an affinity for mucosal colonization, enterotoxigenic B. fragilis (ETBF), a subset of B. fragilis secreting a specific enterotoxin, had been shown to promote the development of CRC (Wu et al., 2009). ETBF exhibited more stable colonization in the colonic epithelial crypts of CRC and rapidly damaged the structure and function of colonic epithelial cells, such as by cleaving the tumor suppressor protein E-cadherin (Dai et al., 2019). The accumulation of ETBF strains in crypts has been shown to be essential for tumor formation via activator of transcription 3 (STAT-3) and an IL-17-dependent pro-carcinogenic inflammatory response (Wu et al., 2009; DeStefano Shields et al., 2016). Thus, ETBF has a role in triggering mucosal inflammation and promoting the carcinogenesis of colorectal cancer. Further research is needed to ascertain how the production of toxic metabolites from B. fragilis influences carcinogenesis by regulating the mucosal barrier.
Streptococcus gallolyticus (S. gallolyticus), formerly known as Streptococcus bovis (S. bovis) biotype I, is a Gram-positive bacterium of humans belonging to the Firmicutes family. It acted as one of the few opportunistic pathogens that was a reported risk factor for CRC (Corredoira-Sánchez et al., 2012; Boleij and Tjalsma, 2013). Previous studies showed that an abundance of S. gallolyticus was enriched in CRC-mucosal tissues as compared to healthy tissue (Abdulamir et al., 2010). Another study published in 2018 found that tumor-bearing mice had an increased level (up to 1,000-fold) of S. gallolyticus in the gut (Aymeric et al., 2018). S. gallolyticus was mainly found entrapped in the mucus layer through the Pil3 pilus. Overexpression of MUC5AC in CRC could favor the adhesion of S. gallolyticus through Pil3 pili and thereby promotes colonization of S. gallolyticus (Martins et al., 2015; Martins et al., 2016). However, the role of S. gallolyticus in the occurrence and development of CRC is still controversial. One study within CRC patients showed that S. gallolyticus was more prevalent in pre-malignant tissue and drove carcinogenesis (Pasquereau-Kotula et al., 2018). On the contrary, another study revealed that S. gallolyticus probably only promoted tumor development after CRC had already begun (Butt et al., 2018). It should be noted that S. gallolyticus contributing to mucins before or after initiation of CRC certainly needs further experimental exploration.
Escherichia. coli (E. coli) is a Gram-negative, facultative anaerobic bacteria of the Enterobacteriaceae family. While E. coli is a gut commensal bacterium, more studies have shown that higher levels of E. coli were colonized in the colonic mucosa of CRC patients compared with that in healthy people (Denizot et al., 2015; Veziant et al., 2016). E. coli binding to the host intestinal epithelium damages the mucus layer and promotes colitis, which eventually leads to dysplasia and CRC (Martin et al., 2004; Elliott et al., 2013). E. coli can reduce the mucus layer and promote tumor growth due to the production of enterotoxins. In vitro, incubation of E. coli with HT-29 colon carcinoma cells resulted in reduced MUC2 glycoprotein levels via the secretion of Shiga toxins (Xue et al., 2014). Moreover, polyketide synthase (pks) island harbored by E. coli codes for the production of colibactin, which had been found in CRC patients and promoted colonic carcinogenesis (Cougnoux et al., 2014). Colonization with pks+ E. coli induced carcinogenesis via mucus damage and thereby promoted more pks+ E. coli binding to the intestinal epithelium, which increased colonic epithelial cell double-strand DNA breaks (Lasry et al., 2016; Dziubańska-Kusibab and Berger, 2020). Thus, genotoxic compounds from E. coli play a major role in promoting colorectal tumorigenesis.
Enterococcus faecalis (E. faecalis) belongs to the Firmicutes and is a Gram-positive, facultatively anaerobic bacteria in humans of the gut commensal bacterium. Despite E. faecalis being part of normal gut flora, accumulating evidence suggests that systemic infection and CRC are closely related to the colonization of E. faecalis. Some studies have shown that higher E. faecalis levels were detected in patients with CRC compared with healthy controls (Balamurugan et al., 2008; Zhou et al., 2016). Supplementation of E. faecalis in the IL-10 knockout mice promoted colitis and resulted in CRC (Lucas et al., 2017). E. faecalis contributed to CRC pathogenesis due to its reactive oxygen species (ROS) production, which induces DNA damage and chromosomal instability in the colonic epithelium (Huycke et al., 2002). Moreover, E. faecalis binding mucin layers via biofilm or pilus promoted intestinal colonization and translocated through the intestine, causing systemic infection (Khan et al., 2018; Banla et al., 2019). According to evidence, E. faecalis may serve as biomarkers for the diagnosis and treatment of CRC with infection.
Biofilms are formed on the surface of gastric or intestinal epithelia and interact with the secreted or membrane-bound mucin, which affects mucin production. It has been reported that mucus-invasive biofilms are present in the colon of over 50% of CRC patients, whereas they are found in only 13% of healthy individuals (Dejea and Sears, 2016). Biofilms tend to invade the colonic mucus layer and present an important factor in CRC.
E. coli formed biofilms and used mucus as a source of energy through its digestion, which harbored its virulence genes associated with CRC (Sicard et al., 2018). F. nucleatum is considered to be a central player in the formation of biofilms. A clinical study has found that F. nucleatum and its biofilms were enriched in CRC tissues, which indicated that these bacterial species had a propensity for biofilm formation (Nakatsu et al., 2015). The increased presence of B. fragilis and Enterobacteriaceae and their ability to form biofilms could play a role in the development of CRC. Once these biofilm-positive bacteria invaded the colonic mucosal layer and came into direct contact with mucosal epithelial cells, they could cause CRC development in this population (Dejea et al., 2018). Hence, the mechanism driving the presence of tumor-associated biofilms in the mucus layer of CRC requires further investigation.
Dietary components in the large intestine are fermented by the microbial community to produce a wide range of metabolites. The major fermentation products are short-chain fatty acids, bile acids, and tryptophan, which are crucial for gut homeostasis (Feng et al., 2018). More evidence has become increasingly clear that the microbiota’s metabolic products strongly influence the intestinal mucus layer formation and development of CRC (Gill and Rowland, 2002; Schwabe and Jobin, 2013). Below, we describe the role of bacterial metabolites in regulating the mucin of CRC.
In the metabolites of the gut microbiota, short-chain fatty acids (SCFAs) are considered the most important bacterial products. The nonabsorbable dietary fibers and resistant starches are selectively fermented by microorganisms, resulting in the production of SCFAs (butyrate, propionate, and acetate) (Koh et al., 2016). SCFAs could create a barrier between the lumen and the near-gut epithelium, leading to the activation of the MUC2 expression in the intestinal barrier and showing anti-inflammatory effects by regulating G protein-coupled receptors (Maslowski et al., 2009). Various studies have demonstrated that SCFAs also aid in improving epithelial barrier function by maintaining a good balance between intestinal immunity and inflammation (Schulthess et al., 2019; Markowiak-Kopeć and Śliżewska, 2020). In addition, SCFAs inhibited the colonization of F. nucleatum in patients with CRC due to shortening intestinal transit time and a change in the PH of the gut (Mehta et al., 2017). Among SCFAs, butyrate played an important role in colonic inflammation and was mainly produced by Firmicutes, Eubacterium, Ruminococcaceae, and Clostridia (Ohira et al., 2017). In clinical trials, fecal butyrate levels and butyrate-producing bacterial species were significantly decreased in patients with advanced colorectal adenoma (Chen et al., 2013). Furthermore, butyrate is thought to have a preventative impact on CRC by regulating mucin expression. A study of the effect of butyrate on mucin secretion in LS174T CRC cells indicated that butyrate could increase MUC2 levels by acetylation and methylation of histones of the MUC2 promoter (Burger-van Paassen et al., 2009). Also, treatment with butyrate in LS174T cells significantly increased mucin protein content and improved probiotic strains, thereby inhibiting the attachment of pathogenic E. coli (Jung et al., 2015). Above all, SCFAs, especially butyrate, are important to maintain intestinal mucus layer homeostasis and prevent CRC.
In the context of lipid metabolism, bile acids (BAs) and their derivative molecules play an important role in human metabolism. BAs are synthesized in the host liver and subsequently translated by the gut microbiota to secondary BAs (lithocholic acids and deoxycholic) in the colon (Sayin et al., 2013). A diet containing saturated fats increased the production of BAs and risk of CRC by inducing gut dysbiosis (Liu et al., 2020). In Apcmin/+ mice, supplementation with BAs could enhance the relative abundance of Akkermansia and Bacteroides and decrease SCFAs and MUC2 expression, leading to cancer progression via activating STAT3 signaling (Wang et al., 2019). Among secondary BAs, deoxycholic acid (DCA) was considered a tumor promoter in CRC. Fecal concentrations of DCA increased the risk of CRC (Ocvirk et al., 2020), and enhanced DCAs were also found in patients with intra-mucosal carcinomas and numerous polypoid adenomas (Yachida and Mizutani, 2019). Interestingly, treatment of HM3 colon cancer cells with DCA resulted in abnormal expression of MUC2 by positive multiple pathways (Lee et al., 2010). Furthermore, pseudo-germ-free Apcmin/+ mice induced by antibiotic streptomycin received fecal microbiota from DCA-fed animals, leading to low-grade inflammation and promoting intestinal carcinogenesis (Cao et al., 2017). Thus, BAs are positively correlated with the incidence of CRC, and understanding interactions between BAs and mucins is helpful for CRC therapy.
Among the metabolism of amino acids supplied through food high in protein, tryptophan (Trp), which acts as a vital amino acid, plays an important role in the maintenance of inflammatory response and intestinal permeability. Some of Trp is catalyzed by host tryptophanase into kynurenine, while others are catabolized by bacteria (Lactobacillus, Clostridium sporogenes, etc.) to serotonin, tryptamine, and indole derivatives (indole-3-ethanol-IEt, indole-3-pyruvate-IPyA, and indole-3-aldehyde-I3A and 3-indole-propionic acid (IPA)) (Devlin et al., 2016). These indole derivatives could strengthen the mucosal layer and enhance MUC2 expression by regulating aryl hydrocarbon receptor (AhR) and pregnane X receptor (PXR) (Venkatesh et al., 2014; Gheorghe et al., 2019; Scott et al., 2020). Indole derivatives activating AhR facilitate the proliferation of epithelial cells and the expression of antimicrobial peptide and mucin production while reducing LPS-mediated inflammation (Venkatesh et al., 2014; Lanis et al., 2017; Taleb, 2019). Trp metabolites also enhanced intestinal integrity through the activation of PXR (Venkatesh et al., 2014). Moreover, supplementation with IPA in rats with a high-fat diet could repair the intestinal mucosal barrier via increased MUC2 expression (Li et al., 2021). Therefore, Trp is considered a potential target for CRC treatment, and more research is needed to fully comprehend its role in modulating mucus layer synthesis during carcinogenesis.
As described previously, the gut microbiota and its metabolites play a crucial role in the intestinal mucus layer of CRC. Therefore, obtaining favorable modulations of the gut microbiome and metabolic activities to protect gut barrier function is a promising strategy for CRC prevention and treatment. The various strategies, such as probiotics, prebiotics, and fecal microbiota transplantation, are considered below.
Probiotics are live microorganisms that, when administered in adequate amounts, confer a health benefit on the host. The probiotics exert a protective effect against CRC by competing with pro-carcinogenic microbiota, modulating host immunity, and enhancing the intestinal barrier (Fong et al., 2020). On an ecological level, some probiotics could suppress the proliferation of pathogenic bacteria by secreting antimicrobial peptides. The consumption of probiotics like Lactobacillus and Bifidobacterium could reduce the abundance of Clostridium, Bifidobacterium, Roseburia, and Faecalibacterium bacteria enriched in CRC patients and inhibit the colonization of commensal bacteria such as E. coli, E. faecalis, F. nucleatum, and S. gallolyticus (Chen et al., 2019). Other probiotics may function in CRC prevention by modifying the immune response. A chemical-induced animal model study revealed that orally administered VSL#3 probiotic cocktail meliorated colitis-associated tumor development through the reduction of STAT-3 expression (Do et al., 2016). Moreover, probiotic administration could strengthen the mucosal barrier in CRC treatment. One clinical trial indicated that a combination of prebiotic inulin and two probiotic strains, B. lactis Bb12 and L. rhamnosus GG, improved epithelial barrier function and reduced colorectal proliferation in patients with adenomatous or cancerous lesions (Rafter et al., 2007). Additionally, several studies have reported that probiotics reduced the frequency of severe diarrhea and abdominal discomfort in CRC patients induced by immunotherapy and chemotherapy by repairing the gut barrier (Liu et al., 2011; Demers et al., 2014). Therefore, probiotics confer health benefits to the gut barrier function of CRC.
Prebiotics are derived from nondigestible carbohydrates in the diet, which are defined as “composition selectively fermented by microorganisms conferring host health benefits” (Gibson et al., 2017). Prebiotics are selectively utilized by host microorganisms, prompting the production of beneficial metabolites to restore intestinal homeostasis and barrier integrity (Nagpal and Yadav, 2017). Thus, consuming prebiotic-rich dietary foods that are high in fiber and low in fat and processed meat have been suggested to protect against CRC. A high-fiber diet showed a better response to prebiotics, which resulted in longer transit time in the intestine and greater immune surveillance to inhibit the mucosal colonization of invasive-adherent bacteria (Mehta et al., 2017). A recent meta-analysis study demonstrated that a high-fiber intake, particularly of whole grains and dairy products, was associated with a decreased risk of CRC (Aune et al., 2011). By contrast, western diets that were rich in red and processed meat influenced the integrity of the intestinal mucus layer, altered gut microbiota, and increased the risk of CRC (Tan and Chen, 2016).
In general, the main prebiotics included fructose-oligosaccharides (FOS) and galacto-oligosaccharides (GOS). Several studies have shown the protective effects of FOS and GOS against CRC progression via modulating gut microbiota and mucus layer function (Valcheva and Dieleman, 2016; Davani-Davari et al., 2019). FOS from nondigestible carbohydrates is absorbed by the small intestine and transferred to the colon, where they contribute to the specific stimulation of endogenous probiotics (lactobacilli species and bifidobacteria) (Tuohy et al., 2001). A study of the effects of FOS on the gut microbiotas of healthy humans revealed that FOS supplementation could increase the concentration of bifidobacteria in the feces, along with stabilizing neutral sterols and host bile acid content, which were involved in CRC progression (Bouhnik et al., 1996). Furthermore, feeding with FOS showed a promising increase in the relative abundance of Lactobacillus and Bifidobacterium and the intestinal mucosal barrier in rats (Lima et al., 2018). In the ApcMin/+ mouse model, feeding of FOS effectively inhibited the development of tumors in the colon by activating the antitumor immunity (Pierre et al., 1997). Besides influencing the microbiota, another prebiotic, GOS, is selectively degraded by the gut microbiota, leading to the production of SCFAs, which can, in turn, reduce the risk of CRC development via regulating mucus barrier functions (Ohtsuka et al., 1991; Rowland et al., 1998). In vitro, GOS induced increased expression of MUC2 at the transcript levels and its co-secreted molecule trefoil factor-3 in LS174T cells (Figueroa-Lozano et al., 2020). In vivo, GOS supplementation for 4 days resulted in higher expression of MUC2 at the transcript level in BALB/c mice (Leforestier et al., 2009). Moreover, oral administration of GOS (derived from lactulose) for 20 weeks inhibited colon tumors in the CRC rat model (Fernández et al., 2018). Overall, although the diet containing FOS and GOS provides beneficial effects on gut homeostasis, the mechanism of prebiotics on the mucus layers of CRC needs further exploration.
Based on the crucial role of the intestinal microbiome in the pathogenesis of CRC, fecal microbiota transplantation (FMT) involved in bacteria-related therapies is gaining more attention. FMT refers to fecal stools from healthy donors transferred to patients via a nasoenteric tube or endoscope (Brandt and Aroniadis, 2013). The aim of FMT is to normalize gut dysbiosis and treat various gastrointestinal diseases, including IBD, Clostridium difficile infection (CDI), and irritable bowel syndrome (Smits et al., 2013; Choi and Cho, 2016). Currently, FMT is an established therapy for recurrent and refractory CDI with an over 90% success rate in clinical studies (Quraishi et al., 2017; Chen et al., 2018a). Although its application in the treatment of CRC patients is unexplored, some studies have been conducted on the use of FMT in murine models with CRC. Wild-type and germ-free mice fed with fecal samples from CRC patients prompted tumor cell proliferation compared to healthy stool-fed mice under dextran sulfate sodium salt/azoxymethane-induced colorectal tumorigenesis (Wong et al., 2017). Furthermore, fecal transplants from wild mice to laboratory mice also resisted chemically induced CRC (Rosshart et al., 2017). Thus, FMT may be a novel macrobiotic therapy for CRC, and further clinical studies are required to explore the safety and mechanisms of FMT in mucins of CRC.
The gut microbiota, bacterial metabolites, and host mucus layer are key players in protecting and maintaining the colon. In this review, we have outlined the profound effects of colonic microbiota and their ability to produce metabolites on the intestinal mucus layer that support colonic health and prevent CRC development (Table 1). Thus, studies on bacteria-targeted therapies for mucin provided many new ideas for CRC prevention and treatment. The interventions involved in prebiotics, probiotics, and FMT improve the mucus layer as a strategy for the prevention or treatment of CRC. In conclusion, a better understanding of the interplay between gut microbiota, bacterial metabolites, and the mucus barrier will shed light on novel therapeutic approaches to intestinal diseases, especially CRC.
TC read the literature related to the topic and drafted the manuscript for publication. MG, WXY and JZ participated in searching the literature and preparing figures. JY, XT, JL, ZT and WJY participated in revising the manuscript. XW, QN and YZ participated in designing and revising the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the “Sail Plan” Talent Training Project of the Affiliated Hospital of Yangzhou University and the Excellent Doctoral Program of Yangzhou “Green Poplar Golden Phoenix” (Grant No. YZLYJFJH2021YXBS041).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
CRC, colorectal cancer; IBD, inflammatory bowel disease; MUC2, mucin-2; MUC5AC, mucin-5 subtype AC; MUC5B, mucin-5B; MUC6, mucin-6; F. nucleatum, Fusobacterium nucleatum; B. fragilis, Bacteroides fragilis; ETBF, enterotoxigenic B. fragilis; S. gallolyticus, Streptococccus gallolyticus; S. bovis, Streptococcus bovis; E. coli, Escherichia coli; pks, polyketide synthase; E. faecalis, Enterococcus faecalis; SCFAs, short-chain fatty acids; BAs, bile acids; DCA, deoxycholic acid; Trp, tryptophan; AhR, aryl hydrocarbon receptor; PXR, pregnane X receptor; IPA, 3-indole-propionic acid; FMT, fecal microbiota transplantation; FOS, fructose-oligosaccharides; GOS, galacto-oligosaccharides; CDI, clostridium difficile infection.
Abdulamir, A. S., Hafidh, R. R., Bakar, F. A. (2010). Molecular detection, quantification, and isolation of streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol. Cancer 9, 249. doi: 10.1186/1476-4598-9-249
Al-Khayal, K., Abdulla, M., Al-Obaid, O., Zubaidi, A., Vaali-Mohammed, M. A., Alsheikh, A., et al. (2016). Differential expression of mucins in middle Eastern patients with colorectal cancer. Oncol. Lett. 12 (1), 393–400. doi: 10.3892/ol.2016.4672
Aune, D., Chan, D. S., Lau, R., Vieira, R., Greenwood, D. C., Kampman, E., et al. (2011). Dietary fibre, whole grains, and risk of colorectal cancer: systematic review and dose-response meta-analysis of prospective studies. Bmj 343, d6617. doi: 10.1136/bmj.d6617
Aymeric, L., Donnadieu, F., Mulet, C., du Merle, L., Nigro, G., Saffarian, A., et al. (2018). Colorectal cancer specific conditions promote streptococcus gallolyticus gut colonization. Proc. Natl. Acad. Sci. U.S.A. 115 (2), E283–e291. doi: 10.1073/pnas.1715112115
Balamurugan, R., Rajendiran, E., George, S., Samuel, G. V., Ramakrishna, B. S. (2008). Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, desulfovibrio and enterococcus faecalis in the feces of patients with colorectal cancer. J. Gastroenterol. Hepatol. 23 (8 Pt 1), 1298–1303. doi: 10.1111/j.1440-1746.2008.05490.x
Banla, L. I., Pickrum, A. M., Hayward, M., Kristich, C. J., Salzman, N. H. (2019). Sortase-dependent proteins promote gastrointestinal colonization by enterococci. Infect. Immun. 87 (5), e00853–18. doi: 10.1128/iai.00853-18
Betge, J., Schneider, N. I., Harbaum, L., Pollheimer, M. J., Lindtner, R. A., Kornprat, P., et al. (2016). MUC1, MUC2, MUC5AC, and MUC6 in colorectal cancer: expression profiles and clinical significance. Virchows Arch. 469 (3), 255–265. doi: 10.1007/s00428-016-1970-5
Biemer-Hüttmann, A. E., Walsh, M. D., McGuckin, M. A., Simms, L. A., Young, J., Leggett, B. A., et al. (2000). Mucin core protein expression in colorectal cancers with high levels of microsatellite instability indicates a novel pathway of morphogenesis. Clin. Cancer Res. 6 (5), 1909–1916.
Boland, P. M., Yurgelun, M. B., Boland, C. R. (2018). Recent progress in lynch syndrome and other familial colorectal cancer syndromes. CA Cancer J. Clin. 68 (3), 217–231. doi: 10.3322/caac.21448
Boleij, A., Tjalsma, H. (2013). The itinerary of streptococcus gallolyticus infection in patients with colonic malignant disease. Lancet Infect. Dis. 13 (8), 719–724. doi: 10.1016/s1473-3099(13)70107-5
Bouhnik, Y., Flourié, B., Riottot, M., Bisetti, N., Gailing, M. F., Guibert, A., et al. (1996). Effects of fructo-oligosaccharides ingestion on fecal bifidobacteria and selected metabolic indexes of colon carcinogenesis in healthy humans. Nutr. Cancer 26 (1), 21–29. doi: 10.1080/01635589609514459
Brandt, L. J., Aroniadis, O. C. (2013). An overview of fecal microbiota transplantation: techniques, indications, and outcomes. Gastrointest Endosc 78 (2), 240–249. doi: 10.1016/j.gie.2013.03.1329
Brown, J. M., Hazen, S. L. (2018). Microbial modulation of cardiovascular disease. Nat. Rev. Microbiol. 16 (3), 171–181. doi: 10.1038/nrmicro.2017.149
Bu, X. D., Li, N., Tian, X. Q., Li, L., Wang, J. S., Yu, X. J., et al. (2010). Altered expression of MUC2 and MUC5AC in progression of colorectal carcinoma. World J. Gastroenterol. 16 (32), 4089–4094. doi: 10.3748/wjg.v16.i32.4089
Burger-van Paassen, N., Vincent, A., Puiman, P. J., van der Sluis, M., Bouma, J., Boehm, G., et al. (2009). The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem. J. 420 (2), 211–219. doi: 10.1042/bj20082222
Butt, J., Blot, W. J., Teras, L. R., Visvanathan, K., Le Marchand, L., Haiman, C. A., et al. (2018). Antibody responses to streptococcus gallolyticus subspecies gallolyticus proteins in a Large prospective colorectal cancer cohort consortium. Cancer Epidemiol. Biomarkers Prev. 27 (10), 1186–1194. doi: 10.1158/1055-9965.Epi-18-0249
Cao, H., Xu, M., Dong, W., Deng, B., Wang, S., Zhang, Y., et al. (2017). Secondary bile acid-induced dysbiosis promotes intestinal carcinogenesis. Int. J. Cancer 140 (11), 2545–2556. doi: 10.1002/ijc.30643
Chen, J., Pitmon, E., Wang, K. (2017). Microbiome, inflammation and colorectal cancer. Semin. Immunol. 32, 43–53. doi: 10.1016/j.smim.2017.09.006
Chen, W., Sun, K., Zheng, R., Zeng, H., Zhang, S., Xia, C., et al. (2018b). Cancer incidence and mortality in chin. Chin. J. Cancer Res. 30 (1), 1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01
Chen, M. L., Takeda, K., Sundrud, M. S. (2019). Emerging roles of bile acids in mucosal immunity and inflammation. Mucosal Immunol. 12 (4), 851–861. doi: 10.1038/s41385-019-0162-4
Chen, H. M., Yu, Y. N., Wang, J. L., Lin, Y. W., Kong, X., Yang, C. Q., et al. (2013). Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am. J. Clin. Nutr. 97 (5), 1044–1052. doi: 10.3945/ajcn.112.046607
Chen, W., Zheng, R., Baade, P. D., Zhang, S., Zeng, H., Bray, F., et al. (2016). Cancer statistics in chin. CA Cancer J. Clin. 66 (2), 115–132. doi: 10.3322/caac.21338
Chen, T., Zhou, Q., Zhang, D., Jiang, F., Wu, J., Zhou, J. Y., et al. (2018a). Effect of faecal microbiota transplantation for treatment of clostridium difficile infection in patients with inflammatory bowel disease: a systematic review and meta-analysis of cohort studies. J. Crohns Colitis 12 (6), 710–717. doi: 10.1093/ecco-jcc/jjy031
Choi, H. H., Cho, Y. S. (2016). Fecal microbiota transplantation: current applications, effectiveness, and future perspectives. Clin. Endosc 49 (3), 257–265. doi: 10.5946/ce.2015.117
Coleman, O. I., Lobner, E. M., Bierwirth, S., Sorbie, A., Waldschmitt, N., Rath, E., et al. (2018). Activated ATF6 induces intestinal dysbiosis and innate immune response to promote colorectal tumorigenesis. Gastroenterology 155 (5), 1539–1552.e1512. doi: 10.1053/j.gastro.2018.07.028
Corredoira-Sánchez, J., García-Garrote, F., Rabuñal, R., López-Roses, L., García-País, M. J., Castro, E., et al. (2012). Association between bacteremia due to streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis I) and colorectal neoplasia: a case-control study. Clin. Infect. Dis. 55 (4), 491–496. doi: 10.1093/cid/cis434
Cougnoux, A., Dalmasso, G., Martinez, R., Buc, E., Delmas, J., Gibold, L., et al. (2014). Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 63 (12), 1932–1942. doi: 10.1136/gutjnl-2013-305257
Dai, Z., Zhang, J., Wu, Q., Chen, J., Liu, J., Wang, L., et al. (2019). The role of microbiota in the development of colorectal cancer. Int. J. Cancer 145 (8), 2032–2041. doi: 10.1002/ijc.32017
Davani-Davari, D., Negahdaripour, M., Karimzadeh, I., Seifan, M., Mohkam, M., Masoumi, S. J., et al. (2019). Prebiotics: definition, types, sources, mechanisms, and clinical applications. Foods 8 (3), 92. doi: 10.3390/foods8030092
Dejea, C. M., Fathi, P., Craig, J. M., Boleij, A., Taddese, R., Geis, A. L., et al. (2018). Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359 (6375), 592–597. doi: 10.1126/science.aah3648
Dejea, C. M., Sears, C. L. (2016). Do biofilms confer a pro-carcinogenic state? Gut Microbes 7 (1), 54–57. doi: 10.1080/19490976.2015.1121363
Demers, M., Dagnault, A., Desjardins, J. (2014). A randomized double-blind controlled trial: impact of probiotics on diarrhea in patients treated with pelvic radiation. Clin. Nutr. 33 (5), 761–767. doi: 10.1016/j.clnu.2013.10.015
Denizot, J., Desrichard, A., Agus, A., Uhrhammer, N., Dreux, N., Vouret-Craviari, V., et al. (2015). Diet-induced hypoxia responsive element demethylation increases CEACAM6 expression, favouring crohn's disease-associated escherichia coli colonisation. Gut 64 (3), 428–437. doi: 10.1136/gutjnl-2014-306944
Desai, M. S., Seekatz, A. M., Koropatkin, N. M., Kamada, N., Hickey, C. A., Wolter, M., et al. (2016). A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167 (5), 1339–1353.e1321. doi: 10.1016/j.cell.2016.10.043
DeStefano Shields, C. E., Van Meerbeke, S. W., Housseau, F., Wang, H., Huso, D. L., Casero, R. A., Jr., et al. (2016). Reduction of murine colon tumorigenesis driven by enterotoxigenic bacteroides fragilis using cefoxitin treatment. J. Infect. Dis. 214 (1), 122–129. doi: 10.1093/infdis/jiw069
Devlin, A. S., Marcobal, A., Dodd, D., Nayfach, S., Plummer, N., Meyer, T., et al. (2016). Modulation of a circulating uremic solute via rational genetic manipulation of the gut microbiota. Cell Host Microbe 20 (6), 709–715. doi: 10.1016/j.chom.2016.10.021
Dharmani, P., Strauss, J., Ambrose, C., Allen-Vercoe, E., Chadee, K. (2011). Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect. Immun. 79 (7), 2597–2607. doi: 10.1128/iai.05118-11
Do, E. J., Hwang, S. W., Kim, S. Y., Ryu, Y. M., Cho, E. A., Chung, E. J., et al. (2016). Suppression of colitis-associated carcinogenesis through modulation of IL-6/STAT3 pathway by balsalazide and VSL3. J. Gastroenterol. Hepatol. 31 (8), 1453–1461. doi: 10.1111/jgh.13280
Dziubańska-Kusibab, P. J., Berger, H. (2020). Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 26 (7), 1063–1069. doi: 10.1038/s41591-020-0908-2
Earle, K. A., Billings, G., Sigal, M., Lichtman, J. S., Hansson, G. C., Elias, J. E., et al. (2015). Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18 (4), 478–488. doi: 10.1016/j.chom.2015.09.002
Elliott, T. R., Hudspith, B. N., Wu, G., Cooley, M., Parkes, G., Quiñones, B., et al. (2013). Quantification and characterization of mucosa-associated and intracellular escherichia coli in inflammatory bowel disease. Inflammation Bowel Dis. 19 (11), 2326–2338. doi: 10.1097/MIB.0b013e3182a38a92
Elzagheid, A., Emaetig, F., Buhmeida, A., Laato, M., El-Faitori, O., Syrjänen, K., et al. (2013). Loss of MUC2 expression predicts disease recurrence and poor outcome in colorectal carcinoma. Tumour Biol. 34 (2), 621–628. doi: 10.1007/s13277-012-0588-8
Feng, Q., Chen, W. D., Wang, Y. D. (2018). Gut microbiota: an integral moderator in health and disease. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00151
Fernández, J., Moreno, F. J., Olano, A., Clemente, A., Villar, C. J., Lombó, F. (2018). A galacto-oligosaccharides preparation derived from lactulose protects against colorectal cancer development in an animal model. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02004
Figueroa-Lozano, S., Ren, C., Yin, H., Pham, H., van Leeuwen, S., Dijkhuizen, L., et al. (2020). The impact of oligosaccharide content, glycosidic linkages and lactose content of galacto-oligosaccharides (GOS) on the expression of mucus-related genes in goblet cells. Food Funct. 11 (4), 3506–3515. doi: 10.1039/d0fo00064g
Fong, W., Li, Q., Yu, J. (2020). Gut microbiota modulation: a novel strategy for prevention and treatment of colorectal cancer. Oncogene 39 (26), 4925–4943. doi: 10.1038/s41388-020-1341-1
Gheorghe, C. E., Martin, J. A., Manriquez, F. V., Dinan, T. G., Cryan, J. F., Clarke, G. (2019). Focus on the essentials: tryptophan metabolism and the microbiome-gut-brain axis. Curr. Opin. Pharmacol. 48, 137–145. doi: 10.1016/j.coph.2019.08.004
Gibson, G. R., Hutkins, R., Sanders, M. E., Prescott, S. L., Reimer, R. A., Salminen, S. J., et al. (2017). Expert consensus document: the international scientific association for probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14 (8), 491–502. doi: 10.1038/nrgastro.2017.75
Gill, C. I., Rowland, I. R. (2002). Diet and cancer: assessing the risk. Br. J. Nutr. 88 Suppl 1, S73–S87. doi: 10.1079/bjn2002632
Hill, D. R., Huang, S., Nagy, M. S., Yadagiri, V. K., Fields, C., Mukherjee, D., et al. (2017). Bacterial colonization stimulates a complex physiological response in the immature human intestinal epithelium. Elife 6, e29132. doi: 10.7554/eLife.29132
Hooper, L. V., Macpherson, A. J. (2010). Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 10 (3), 159–169. doi: 10.1038/nri2710
Hryhorowicz, S., Kaczmarek-Ryś, M., Lis-Tanaś, E., Porowski, J., Szuman, M., Grot, N., et al. (2022). Strong hereditary predispositions to colorectal cancer. Genes (Basel) 13 (12), 2326. doi: 10.3390/genes13122326
Huycke, M. M., Abrams, V., Moore, D. R. (2002). Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 23 (3), 529–536. doi: 10.1093/carcin/23.3.529
Johansson, M. E., Hansson, G. C. (2016). Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 16 (10), 639–649. doi: 10.1038/nri.2016.88
Johansson, M. E., Jakobsson, H. E., Holmén-Larsson, J., Schütte, A., Ermund, A., Rodríguez-Piñeiro, A. M., et al. (2015). Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe 18 (5), 582–592. doi: 10.1016/j.chom.2015.10.007
Jung, T. H., Park, J. H., Jeon, W. M., Han, K. S. (2015). Butyrate modulates bacterial adherence on LS174T human colorectal cells by stimulating mucin secretion and MAPK signaling pathway. Nutr. Res. Pract. 9 (4), 343–349. doi: 10.4162/nrp.2015.9.4.343
Keum, N., Giovannucci, E. (2019). Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 16 (12), 713–732. doi: 10.1038/s41575-019-0189-8
Khan, Z., Siddiqui, N., Saif, M. W. (2018). Enterococcus faecalis infective endocarditis and colorectal carcinoma: case of new association gaining ground. Gastroenterol. Res. 11 (3), 238–240. doi: 10.14740/gr996w
King, M., Hurley, H. (2020). The link between fusobacteria and colon cancer: a fulminant example and review of the evidence. Immune Netw. 20 (4), e30. doi: 10.4110/in.2020.20.e30
Koh, A., De Vadder, F., Kovatcheva-Datchary, P., Bäckhed, F. (2016). From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165 (6), 1332–1345. doi: 10.1016/j.cell.2016.05.041
Kostic, A. D., Chun, E., Robertson, L., Glickman, J. N., Gallini, C. A., Michaud, M., et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14 (2), 207–215. doi: 10.1016/j.chom.2013.07.007
Kostic, A. D., Gevers, D., Pedamallu, C. S., Michaud, M., Duke, F., Earl, A. M., et al. (2012). Genomic analysis identifies association of fusobacterium with colorectal carcinoma. Genome Res. 22 (2), 292–298. doi: 10.1101/gr.126573.111
Kostic, A. D., Xavier, R. J., Gevers, D. (2014). The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146 (6), 1489–1499. doi: 10.1053/j.gastro.2014.02.009
Krishn, S. R., Kaur, S., Smith, L. M., Johansson, S. L., Jain, M., Patel, A., et al. (2016). Mucins and associated glycan signatures in colon adenoma-carcinoma sequence: prospective pathological implication(s) for early diagnosis of colon cancer. Cancer Lett. 374 (2), 304–314. doi: 10.1016/j.canlet.2016.02.016
Lahdaoui, F., Messager, M., Vincent, A., Hec, F., Gandon, A., Warlaumont, M., et al. (2017). Depletion of MUC5B mucin in gastrointestinal cancer cells alters their tumorigenic properties: implication of the wnt/β-catenin pathway. Biochem. J. 474 (22), 3733–3746. doi: 10.1042/bcj20170348
Lanis, J. M., Alexeev, E. E., Curtis, V. F., Kitzenberg, D. A., Kao, D. J., Battista, K. D., et al. (2017). Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol. 10 (5), 1133–1144. doi: 10.1038/mi.2016.133
Lasry, A., Zinger, A., Ben-Neriah, Y. (2016). Inflammatory networks underlying colorectal cancer. Nat. Immunol. 17 (3), 230–240. doi: 10.1038/ni.3384
Lee, H. Y., Crawley, S., Hokari, R., Kwon, S., Kim, Y. S. (2010). Bile acid regulates MUC2 transcription in colon cancer cells via positive EGFR/PKC/Ras/ERK/CREB, PI3K/Akt/IkappaB/NF-kappaB and p38/MSK1/CREB pathways and negative JNK/c-Jun/AP-1 pathway. Int. J. Oncol. 36 (4), 941–953. doi: 10.3892/ijo_00000573
Leforestier, G., Blais, A., Blachier, F., Marsset-Baglieri, A., Davila-Gay, A. M., Perrin, E., et al. (2009). Effects of galacto-oligosaccharide ingestion on the mucosa-associated mucins and sucrase activity in the small intestine of mice. Eur. J. Nutr. 48 (8), 457–464. doi: 10.1007/s00394-009-0036-8
Lesuffleur, T., Roche, F., Hill, A. S., Lacasa, M., Fox, M., Swallow, D. M., et al. (1995). Characterization of a mucin cDNA clone isolated from HT-29 mucus-secreting cells. the 3' end of MUC5AC? J. Biol. Chem. 270 (23), 13665–13673. doi: 10.1074/jbc.270.23.13665
Li, J., Zhang, L., Wu, T., Li, Y., Zhou, X., Ruan, Z. (2021). Indole-3-propionic acid improved the intestinal barrier by enhancing epithelial barrier and mucus barrier. J. Agric. Food Chem. 69 (5), 1487–1495. doi: 10.1021/acs.jafc.0c05205
Li, C., Zuo, D., Yin, L., Lin, Y., Li, C., Liu, T., et al. (2018). Prognostic value of MUC2 expression in colorectal cancer: a systematic review and meta-analysis. Gastroenterol. Res. Pract. 2018, 6986870. doi: 10.1155/2018/6986870
Lima, G. C., Vieira, V. C. C., Cazarin, C. B. B., Ribeiro, R. D. R., Junior, S. B., de Albuquerque, C. L., et al. (2018). Fructooligosaccharide intake promotes epigenetic changes in the intestinal mucosa in growing and ageing rats. Eur. J. Nutr. 57 (4), 1499–1510. doi: 10.1007/s00394-017-1435-x
Liu, Z., Qin, H., Yang, Z., Xia, Y., Liu, W., Yang, J., et al. (2011). Randomised clinical trial: the effects of perioperative probiotic treatment on barrier function and post-operative infectious complications in colorectal cancer surgery - a double-blind study. Aliment Pharmacol. Ther. 33 (1), 50–63. doi: 10.1111/j.1365-2036.2010.04492.x
Liu, T., Song, X., Khan, S., Li, Y., Guo, Z., Li, C., et al. (2020). The gut microbiota at the intersection of bile acids and intestinal carcinogenesis: an old story, yet mesmerizing. Int. J. Cancer 146 (7), 1780–1790. doi: 10.1002/ijc.32563
Lucas, C., Barnich, N., Nguyen, H. T. T. (2017). Microbiota, inflammation and colorectal cancer. Int. J. Mol. Sci. 18 (6), 1310. doi: 10.3390/ijms18061310
Markowiak-Kopeć, P., Śliżewska, K. (2020). The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 12 (4), 1107. doi: 10.3390/nu12041107
Martin, H. M., Campbell, B. J., Hart, C. A., Mpofu, C., Nayar, M., Singh, R., et al. (2004). Enhanced escherichia coli adherence and invasion in crohn's disease and colon cancer. Gastroenterology 127 (1), 80–93. doi: 10.1053/j.gastro.2004.03.054
Martins, M., Aymeric, L., du Merle, L., Danne, C., Robbe-Masselot, C., Trieu-Cuot, P., et al. (2015). Streptococcus gallolyticus Pil3 pilus is required for adhesion to colonic mucus and for colonization of mouse distal colon. J. Infect. Dis. 212 (10), 1646–1655. doi: 10.1093/infdis/jiv307
Martins, M., Porrini, C., du Merle, L., Danne, C., Robbe-Masselot, C., Trieu-Cuot, P., et al. (2016). The Pil3 pilus of streptococcus gallolyticus binds to intestinal mucins and to fibrinogen. Gut Microbes 7 (6), 526–532. doi: 10.1080/19490976.2016.1239677
Maruvada, P., Leone, V., Kaplan, L. M., Chang, E. B. (2017). The human microbiome and obesity: moving beyond associations. Cell Host Microbe 22 (5), 589–599. doi: 10.1016/j.chom.2017.10.005
Maslowski, K. M., Vieira, A. T., Ng, A., Kranich, J., Sierro, F., Yu, D., et al. (2009). Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461 (7268), 1282–1286. doi: 10.1038/nature08530
Mehta, R. S., Nishihara, R., Cao, Y., Song, M., Mima, K., Qian, Z. R., et al. (2017). Association of dietary patterns with risk of colorectal cancer subtypes classified by fusobacterium nucleatum in tumor tissue. JAMA Oncol. 3 (7), 921–927. doi: 10.1001/jamaoncol.2016.6374
Mima, K., Nishihara, R., Qian, Z. R., Cao, Y., Sukawa, Y., Nowak, J. A., et al. (2016). Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 65 (12), 1973–1980. doi: 10.1136/gutjnl-2015-310101
Nagpal, R., Yadav, H. (2017). Bacterial translocation from the gut to the distant organs: an overview. Ann. Nutr. Metab. 71 Suppl 1, 11–16. doi: 10.1159/000479918
Nakatsu, G., Li, X., Zhou, H., Sheng, J., Wong, S. H., Wu, W. K., et al. (2015). Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat. Commun. 6, 8727 doi: 10.1038/ncomms9727
O'Keefe, S. J. (2016). Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 13 (12), 691–706. doi: 10.1038/nrgastro.2016.165
Ocvirk, S., Wilson, A. S., Posma, J. M., Li, J. V., Koller, K. R., Day, G. M., et al. (2020). A prospective cohort analysis of gut microbial co-metabolism in Alaska native and rural African people at high and low risk of colorectal cancer. Am. J. Clin. Nutr. 111 (2), 406–419. doi: 10.1093/ajcn/nqz301
Ohira, H., Tsutsui, W., Fujioka, Y. (2017). Are short chain fatty acids in gut microbiota defensive players for inflammation and atherosclerosis? J. Atheroscler Thromb. 24 (7), 660–672. doi: 10.5551/jat.RV17006
Ohtsuka, K., Tsuji, K., Nakagawa, Y., Ueda, H., Ozawa, O., Uchida, T., et al. (1991). Utilization and metabolism of [U-14C]4' galactosyllactose (O-beta-D-galactopyranosyl-(1–-4)-O-beta-D-galactopyranosyl-(1—-4)- d-glucopyranose) in rats. J. Nutr. Sci. Vitaminol (Tokyo) 37 (2), 173–184. doi: 10.3177/jnsv.37.173
Pasquereau-Kotula, E., Martins, M., Aymeric, L., Dramsi, S. (2018). Significance of streptococcus gallolyticus subsp. gallolyticus association with colorectal cancer. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00614
Petersson, J., Schreiber, O., Hansson, G. C., Gendler, S. J., Velcich, A., Lundberg, J. O., et al. (2011). Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am. J. Physiol. Gastrointest Liver Physiol. 300 (2), G327–G333. doi: 10.1152/ajpgi.00422.2010
Pierre, F., Perrin, P., Champ, M., Bornet, F., Meflah, K., Menanteau, J. (1997). Short-chain fructo-oligosaccharides reduce the occurrence of colon tumors and develop gut-associated lymphoid tissue in Min mice. Cancer Res. 57 (2), 225–228.
Pothuraju, R., Krishn, S. R., Gautam, S. K., Pai, P., Ganguly, K., Chaudhary, S., et al. (2020). Mechanistic and functional shades of mucins and associated glycans in colon cancer. Cancers (Basel) 12 (3), 649. doi: 10.3390/cancers12030649
Quraishi, M. N., Widlak, M., Bhala, N., Moore, D., Price, M., Sharma, N., et al. (2017). Systematic review with meta-analysis: the efficacy of faecal microbiota transplantation for the treatment of recurrent and refractory clostridium difficile infection. Aliment Pharmacol. Ther. 46 (5), 479–493. doi: 10.1111/apt.14201
Rafter, J., Bennett, M., Caderni, G., Clune, Y., Hughes, R., Karlsson, P. C., et al. (2007). Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am. J. Clin. Nutr. 85 (2), 488–496. doi: 10.1093/ajcn/85.2.488
Renaud, F., Vincent, A., Mariette, C., Crépin, M., Stechly, L., Truant, S., et al. (2015). MUC5AC hypomethylation is a predictor of microsatellite instability independently of clinical factors associated with colorectal cancer. Int. J. Cancer 136 (12), 2811–2821. doi: 10.1002/ijc.29342
Rosshart, S. P., Vassallo, B. G., Angeletti, D., Hutchinson, D. S., Morgan, A. P., Takeda, K., et al. (2017). Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171 (5), 1015–1028.e1013. doi: 10.1016/j.cell.2017.09.016
Rowland, I. R., Rumney, C. J., Coutts, J. T., Lievense, L. C. (1998). Effect of bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis 19 (2), 281–285. doi: 10.1093/carcin/19.2.281
Rubinstein, M. R., Baik, J. E., Lagana, S. M., Han, R. P., Raab, W. J., Sahoo, D., et al. (2019). Fusobacterium nucleatum promotes colorectal cancer by inducing wnt/β-catenin modulator annexin A1. EMBO Rep. 20 (4), e47638. doi: 10.15252/embr.201847638
Rubinstein, M. R., Wang, X., Liu, W., Hao, Y., Cai, G., Han, Y. W. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating e-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14 (2), 195–206. doi: 10.1016/j.chom.2013.07.012
Sayin, S. I., Wahlström, A., Felin, J., Jäntti, S., Marschall, H. U., Bamberg, K., et al. (2013). Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17 (2), 225–235. doi: 10.1016/j.cmet.2013.01.003
Schulthess, J., Pandey, S., Capitani, M., Rue-Albrecht, K. C., Arnold, I., Franchini, F., et al. (2019). The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50 (2), 432–445.e437. doi: 10.1016/j.immuni.2018.12.018
Schwabe, R. F., Jobin, C. (2013). The microbiome and cancer. Nat. Rev. Cancer 13 (11), 800–812. doi: 10.1038/nrc3610
Scott, S. A., Fu, J., Chang, P. V. (2020). Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. U.S.A. 117 (32), 19376–19387. doi: 10.1073/pnas.2000047117
Sears, C. L. (2005). A dynamic partnership: celebrating our gut flora. Anaerobe 11 (5), 247–251. doi: 10.1016/j.anaerobe.2005.05.001
Sheng, Y. H., Hasnain, S. Z., Florin, T. H., McGuckin, M. A. (2012). Mucins in inflammatory bowel diseases and colorectal cancer. J. Gastroenterol. Hepatol. 27 (1), 28–38. doi: 10.1111/j.1440-1746.2011.06909.x
Shi, N., Li, N., Duan, X., Niu, H. (2017). Interaction between the gut microbiome and mucosal immune system. Mil Med. Res. 4, 14. doi: 10.1186/s40779-017-0122-9
Sicard, J. F., Vogeleer, P., Le Bihan, G., Rodriguez Olivera, Y., Beaudry, F., Jacques, M., et al. (2018). N-acetyl-glucosamine influences the biofilm formation of escherichia coli. Gut Pathog. 10, 26. doi: 10.1186/s13099-018-0252-y
Smits, L. P., Bouter, K. E., de Vos, W. M., Borody, T. J., Nieuwdorp, M. (2013). Therapeutic potential of fecal microbiota transplantation. Gastroenterology 145 (5), 946–953. doi: 10.1053/j.gastro.2013.08.058
Song, M., Chan, A. T. (2019). Environmental factors, gut microbiota, and colorectal cancer prevention. Clin. Gastroenterol. Hepatol. 17 (2), 275–289. doi: 10.1016/j.cgh.2018.07.012
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249. doi: 10.3322/caac.21660
Tahara, T., Yamamoto, E., Suzuki, H., Maruyama, R., Chung, W., Garriga, J., et al. (2014). Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 74 (5), 1311–1318. doi: 10.1158/0008-5472.can-13-1865
Taleb, S. (2019). Tryptophan dietary impacts gut barrier and metabolic diseases. Front. Immunol. 10. 2113 doi: 10.3389/fimmu.2019.02113
Tan, J., Chen, Y. X. (2016). Dietary and lifestyle factors associated with colorectal cancer risk and interactions with microbiota: fiber, red or processed meat and alcoholic drinks. Gastrointest Tumors 3 (1), 17–24. doi: 10.1159/000442831
Tsai, J. H., Lin, Y. L., Cheng, Y. C., Chen, C. C., Lin, L. I., Tseng, L. H., et al. (2015). Aberrant expression of annexin A10 is closely related to gastric phenotype in serrated pathway to colorectal carcinoma. Mod Pathol. 28 (2), 268–278. doi: 10.1038/modpathol.2014.96
Tuohy, K. M., Kolida, S., Lustenberger, A. M., Gibson, G. R. (2001). The prebiotic effects of biscuits containing partially hydrolysed guar gum and fructo-oligosaccharides–a human volunteer study. Br. J. Nutr. 86 (3), 341–348. doi: 10.1079/bjn2001394
Valcheva, R., Dieleman, L. A. (2016). Prebiotics: definition and protective mechanisms. Best Pract. Res. Clin. Gastroenterol. 30 (1), 27–37. doi: 10.1016/j.bpg.2016.02.008
Vandenhaute, B., Buisine, M. P., Debailleul, V., Clément, B., Moniaux, N., Dieu, M. C., et al. (1997). Mucin gene expression in biliary epithelial cells. J. Hepatol. 27 (6), 1057–1066. doi: 10.1016/s0168-8278(97)80150-x
Vandeputte, D. (2020). Personalized nutrition through the gut microbiota: current insights and future perspectives. Nutr. Rev. 78 (12 Suppl 2), 66–74. doi: 10.1093/nutrit/nuaa098
van Klinken, B. J., Dekker, J., van Gool, S. A., van Marle, J., Büller, H. A., Einerhand, A. W. (1998). MUC5B is the prominent mucin in human gallbladder and is also expressed in a subset of colonic goblet cells. Am. J. Physiol. 274 (5), G871–G878. doi: 10.1152/ajpgi.1998.274.5.G871
Venkatesh, M., Mukherjee, S., Wang, H., Li, H., Sun, K., Benechet, A. P., et al. (2014). Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and toll-like receptor 4. Immunity 41 (2), 296–310. doi: 10.1016/j.immuni.2014.06.014
Veziant, J., Gagnière, J., Jouberton, E., Bonnin, V., Sauvanet, P., Pezet, D., et al. (2016). Association of colorectal cancer with pathogenic escherichia coli: focus on mechanisms using optical imaging. World J. Clin. Oncol. 7 (3), 293–301. doi: 10.5306/wjco.v7.i3.293
Wang, S., Dong, W., Liu, L., Xu, M., Wang, Y., Liu, T., et al. (2019). Interplay between bile acids and the gut microbiota promotes intestinal carcinogenesis. Mol. Carcinog 58 (7), 1155–1167. doi: 10.1002/mc.22999
Wang, H., Jin, S., Lu, H., Mi, S., Shao, W., Zuo, X., et al. (2017). Expression of survivin, MUC2 and MUC5 in colorectal cancer and their association with clinicopathological characteristics. Oncol. Lett. 14 (1), 1011–1016. doi: 10.3892/ol.2017.6218
Wardman, J. F., Bains, R. K., Rahfeld, P., Withers, S. G. (2022). Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 20 (9), 542–556. doi: 10.1038/s41579-022-00712-1
Warren, R. L., Freeman, D. J., Pleasance, S., Watson, P., Moore, R. A., Cochrane, K., et al. (2013). Co-Occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome 1 (1), 16. doi: 10.1186/2049-2618-1-16
Wenzel, U. A., Magnusson, M. K., Rydström, A., Jonstrand, C., Hengst, J., Johansson, M. E., et al. (2014). Spontaneous colitis in Muc2-deficient mice reflects clinical and cellular features of active ulcerative colitis. PloS One 9 (6), e100217. doi: 10.1371/journal.pone.0100217
Wong, S. H., Yu, J. (2019). Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 16 (11), 690–704. doi: 10.1038/s41575-019-0209-8
Wong, S. H., Zhao, L., Zhang, X., Nakatsu, G., Han, J., Xu, W., et al. (2017). Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology 153 (6), 1621–1633.e1626. doi: 10.1053/j.gastro.2017.08.022
Wu, S., Rhee, K. J., Albesiano, E., Rabizadeh, S., Wu, X., Yen, H. R., et al. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15 (9), 1016–1022. doi: 10.1038/nm.2015
Xue, Y., Zhang, H., Wang, H., Hu, J., Du, M., Zhu, M. J. (2014). Host inflammatory response inhibits escherichia coli O157:H7 adhesion to gut epithelium through augmentation of mucin expression. Infect. Immun. 82 (5), 1921–1930. doi: 10.1128/iai.01589-13
Yachida, S., Mizutani, S. (2019). Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 25 (6), 968–976. doi: 10.1038/s41591-019-0458-7
Yamashita, M. S. A., Melo, E. O. (2018). Mucin 2 (MUC2) promoter characterization: an overview. Cell Tissue Res. 374 (3), 455–463. doi: 10.1007/s00441-018-2916-9
Ye, X., Wang, R., Bhattacharya, R., Boulbes, D. R., Fan, F., Xia, L., et al. (2017). Fusobacterium nucleatum subspecies animalis influences proinflammatory cytokine expression and monocyte activation in human colorectal tumors. Cancer Prev. Res. (Phila) 10 (7), 398–409. doi: 10.1158/1940-6207.capr-16-0178
Yonezawa, S., Sato, E. (1997). Expression of mucin antigens in human cancers and its relationship with malignancy potential. Pathol. Int. 47 (12), 813–830. doi: 10.1111/j.1440-1827.1997.tb03713.x
Yu, L. C. (2018). Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis. J. BioMed. Sci. 25 (1), 79. doi: 10.1186/s12929-018-0483-8
Zhang, Y., Wang, L., Ocansey, D. K. W., Wang, B., Wang, L., Xu, Z. (2021). Mucin-type O-glycans: barrier, microbiota, and immune anchors in inflammatory bowel disease. J. Inflammation Res. 14, 5939–5953. doi: 10.2147/jir.S327609
Zhou, Y., He, H., Xu, H., Li, Y., Li, Z., Du, Y., et al. (2016). Association of oncogenic bacteria with colorectal cancer in south China. Oncotarget 7 (49), 80794–80802. doi: 10.18632/oncotarget.13094
Zhu, X., Long, X., Luo, X., Song, Z., Li, S., Wang, H. (2016). Abrogation of MUC5AC expression contributes to the apoptosis and cell cycle arrest of colon cancer cells. Cancer Biother Radiopharm 31 (7), 261–267. doi: 10.1089/cbr.2016.2054
Keywords: mucins, gut microbiota, bacterial metabolites, bacteria-related therapies, colorectal cancer
Citation: Gu M, Yin W, Zhang J, Yin J, Tang X, Ling J, Tang Z, Yin W, Wang X, Ni Q, Zhu Y and Chen T (2023) Role of gut microbiota and bacterial metabolites in mucins of colorectal cancer. Front. Cell. Infect. Microbiol. 13:1119992. doi: 10.3389/fcimb.2023.1119992
Received: 09 December 2022; Accepted: 03 May 2023;
Published: 17 May 2023.
Edited by:
Xingmin Sun, University of South Florida, United StatesReviewed by:
Cuncong Zhong, University of Kansas, United StatesCopyright © 2023 Gu, Yin, Zhang, Yin, Tang, Ling, Tang, Yin, Wang, Ni, Zhu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangjun Wang, eXp5c3d4akAxNjMuY29t; Qing Ni, WXpuaXFpbmdAMTYzLmNvbQ==; Yunxiang Zhu, eXh6aHVAeXp1LmVkdS5jbg==; Tuo Chen, Y2hlbnR1b3lzaEAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.