- 1Department of Veterinary Pathobiology, School of Veterinary Medicine and Biomedical Sciences, Texas A&M University, College Station, TX, United States
- 2Department of Computer Science, Georgia State University, Atlanta, GA, United States
- 3Department of Biological Sciences, and School of Biomedical Sciences, Kent State University, Kent, OH, United States
- 4Brain Health Research Institute, Kent State University, Kent, OH, United States
- 5Texas A&M Institute for Genomics Sciences and Society, Texas A&M University, College Station, TX, United States
- 6Department of Molecular and Cellular Medicine, Texas A&M University Health Science Center, Texas A&M University, College Station, TX, United States
Lyme disease (LD), the most prevalent tick-borne disease of humans in the Northern Hemisphere, is caused by the spirochetal bacterium of Borreliella burgdorferi (Bb) sensu lato complex. In nature, Bb spirochetes are continuously transmitted between Ixodes ticks and mammalian or avian reservoir hosts. Peromyscus leucopus mice are considered the primary mammalian reservoir of Bb in the United States. Earlier studies demonstrated that experimentally infected P. leucopus mice do not develop disease. In contrast, C3H mice, a widely used laboratory strain of Mus musculus in the LD field, develop severe Lyme arthritis. To date, the exact tolerance mechanism of P. leucopus mice to Bb-induced infection remains unknown. To address this knowledge gap, the present study has compared spleen transcriptomes of P. leucopus and C3H/HeJ mice infected with Bb strain 297 with those of their respective uninfected controls. Overall, the data showed that the spleen transcriptome of Bb-infected P. leucopus mice was much more quiescent compared to that of the infected C3H mice. To date, the current investigation is one of the few that have examined the transcriptome response of natural reservoir hosts to Borreliella infection. Although the experimental design of this study significantly differed from those of two previous investigations, the collective results of the current and published studies have consistently demonstrated very limited transcriptomic responses of different reservoir hosts to the persistent infection of LD pathogens.
Importance: The bacterium Borreliella burgdorferi (Bb) causes Lyme disease, which is one of the emerging and highly debilitating human diseases in countries of the Northern Hemisphere. In nature, Bb spirochetes are maintained between hard ticks of Ixodes spp. and mammals or birds. In the United States, the white-footed mouse, Peromyscus leucopus, is one of the main Bb reservoirs. In contrast to humans and laboratory mice (e.g., C3H mice), white-footed mice rarely develop clinical signs (disease) despite being (persistently) infected with Bb. How the white-footed mouse tolerates Bb infection is the question that the present study has attempted to address. Comparisons of genetic responses between Bb-infected and uninfected mice demonstrated that, during a long-term Bb infection, C3H mice reacted much stronger, whereas P. leucopus mice were relatively unresponsive.
Introduction
Lyme disease (LD) is the most prevalent tick-borne disease in North America and Europe with an estimate of ~476,000 annual cases in the United States (U.S.) alone (Mead, 2015; Kugeler et al., 2021). LD is caused by some members of Borreliella burgdorferi (Bb) sensu lato complex, which is divided into more than 20 genospecies. At least three genospecies (Borreliella afzelii, Bb sensu stricto, Borreliella garinii) have the capacity to cause LD in humans (Baranton et al., 1992; Canica et al., 1993; Kurtenbach et al., 2006). The horizontal transmission of LD is mediated by hard ticks of Ixodes spp., which acquire spirochetes from Bb-infected vertebrate carriers such as rodents and birds (Piesman and Gern, 2004; Barbour and Gupta, 2021). In the U.S., the white-footed mouse, Peromyscus leucopus, is considered the main mammalian reservoir of Bb spirochetes (Levine et al., 1985; Donahue et al., 1987). Clinical signs of LD patients are often flu-like symptoms that can be followed by arthritis and neurological or cardiac abnormalities (Smith et al., 2011; Radolf et al., 2021).
When Bb infects mammals, innate immune effectors such as toll-like receptor (TLR) 2 and myeloid differentiation antigen 88 (MyD88) become fully engaged (Hirschfeld et al., 1999; Wooten et al., 2002a; Wooten et al., 2002b; Bolz et al., 2004). MyD88 subsequently activates a toll/IL-1 receptor-domain-containing adapter-inducing interferon-β (TRIF) signaling (Petnicki-Ocwieja et al., 2013). Moreover, Bb outer surface lipoproteins lead to activation of nuclear factor kappa-light-chain-enhancer (NF-κB) (Wooten et al., 1996; Ebnet et al., 1997). This cascade leads to secretion of pro-inflammatory cytokines and chemokines for neutrophils, monocytes, and lymphocytes during LD infection in mice (Wooten and Weis, 2001; Verhaegh et al., 2017; Bockenstedt et al., 2021). The mounting immune response is also accompanied by production of interferons (IFN), IFNα and IFNβ (type I INF), and IFNγ in response to Bb antigens (Ganapamo et al., 2000; Crandall et al., 2006; Miller et al., 2008). As a result of the activated innate immune system, the acquired humoral immune response is developed, and this leads to lowering the spirochetal burden in various tissues (Barthold et al., 1997; McKisic and Barthold, 2000). In LD patients, Th1 lymphocytes and mononuclear cells mediate B lymphocyte production of neutralizing anti-Bb antibodies such as IgG1 and IgG3, which induce opsonization and activate complement defense mechanisms (Hechemy et al., 1988; Bockenstedt et al., 2021).
Similar to LD human patients, some laboratory mouse strains develop arthritis and carditis upon LD infection (Barthold et al., 1990; Armstrong et al., 1992; Brown and Reiner, 1999). The degree of susceptibility to Bb-induced arthritis in mice is predetermined by a number of factors including inter-strain genetic variability and the fitness variation of Bb strains (Barthold et al., 1992; Ma et al., 1998; Hanincová et al., 2008; Lin et al., 2014). The different capacity of mouse strains to regulate the localized inflammatory response is a major factor for the arthritis development (Ma et al., 1998). By using C3H/He mice, which develop severe Lyme arthritis (Barthold et al., 1990; Barthold et al., 1992), it was demonstrated that IFNα and IFNβ have an important role during an early LD infection. The upregulation of the interferon responsive genes contributes to the pathogenesis of LD arthritis in C3H/He mice (Crandall et al., 2006; Miller et al., 2008). Type I INF response can also be associated with differential regulation of genes that are involved in tissue repair processes (Crandall et al., 2006; Lochhead et al., 2021). In contrast to C3H strains, C57BL/6 mice, which are deficient in an intense interferon response to Bb infection (Crandall et al., 2006), develop only mild Lyme arthritis (Barthold et al., 1990; Crandall et al., 2006). In addition to type I IFN, interleukin 10 (IL-10), an anti-inflammatory cytokine, has a crucial role in the regulation of arthritis severity (Sonderegger et al., 2012). C57BL/6 mice lacking IL-10 exhibit more severe arthritis upon LD infection as opposed to their Bb-infected wild-type C57BL/6 controls (Brown et al., 1999).
In contrast to C3H mice, previous studies demonstrated that experimentally Bb-infected P. leucopus mice do not develop disease (Schwanz et al., 2011). Both Bb-infected and uninfected control P. leucopus mice were shown to have similar white blood cell counts and wheel-running activity (Schwanz et al., 2011). The resistance of persistently Bb-infected P. leucopus mice to the disease was also shown to be age-dependent since only infant mice could develop carditis and multifocal arthritis upon Bb infection (Moody et al., 1994). Of note, however, the tolerance of white-footed mice to Bb-induced disease is not absolute. A previous study demonstrated that a fraction (e.g., ~7-10%) of wild-caught P. leucopus population had clinical signs of LD (Burgess et al., 1990). The lack of detectable LD in the majority of Bb-infected P. leucopus population can potentially be explained by a long-term co-existence between the host and the pathogen. The outcome of this host-pathogen adaptation is likely to have created a balance between the strong but non-sterilizing immune response of reservoir hosts (Schwan et al., 1989; Baum et al., 2012; Cook and Barbour, 2015; Rogovskyy et al., 2015) and the persistent presence of Bb spirochetes. Unfortunately, to date, the exact factors and mechanism of tolerance or susceptibility of P. leucopus mice to Bb-induced disease are unknown. In the present study, we have explored infection-induced changes in gene expression to understand how P. leucopus mice are able to tolerate Bb infection. For that, we compared responses to Bb infection between P. leucopus and Mus musculus (C3H/HeJ, referred to here as C3H) mice at the transcriptomic level. The overall data showed that the spleen transcriptome of P. leucopus mice was much more quiescent compared to that of the C3H mice.
Materials and methods
Bacterial strain
Borreliella burgdorferi strain 297 was a generous gift of Troy Bankhead. Spirochetes were grown in liquid Barbour-Stoenner-Kelly medium supplemented with 6% rabbit serum (referred to here as BSK-II; Gemini Bio-Products, CA, USA) and incubated at 35°C under 2% CO2.
Mouse experiment
Six males of P. leucopus mice acquired from The Peromyscus Genetic Stock Center (the University of South Carolina, SC, USA) and 6 males of C3H/HeJ (C3H) mice purchased from The Jackson Laboratory (Bar Harbor, ME, USA) were split into 4 groups of three mice each. After an adaptation period, each of 3 P. leucopus (5-8 weeks of age) and 3 C3H (4-6 weeks of age) mice was subcutaneously (s.c.) inoculated in the shoulder region with 100-μl inoculum containing ~1.0 x105 cells of B. burgdorferi 297. The remaining 6 animals (uninfected controls) that were within the respective age range were s.c. inoculated with 100 μl of sterile saline.
To confirm the infection, ~50 μl of blood samples taken via maxillary bleed from the 6 Bb-inoculated mice at day 7 postinoculation (pi) were cultured in BSK-II at 35°C under 2% CO2. At day 70 pi, ear pinnae, bladder, tibiotarsal joint, and heart tissues were harvested from both infected and uninfected groups of mice and cultured in BSK-II as described (Rogovskyy et al., 2016). Dark-field microscopy was utilized on a weekly basis over a four-week period to examine tissue cultures for the presence or absence of viable spirochetes (Table S1). At day 70 pi, spleens were also harvested from the 12 mice. All spleens were individually preserved in Invitrogen RNAlater stabilization solution (Thermo Fisher Scientific, MA, USA) and stored at - 80°C until RNA extraction.
RNA extraction, library preparation, and sequencing
Total RNA from spleen tissues was isolated by using QIAGEN RNeasy Mini Kit (QIAGEN, CA, USA) according to manufacturer’s instruction. The quality and concentration of the RNA samples were determined by using Agilent Tapestation 2200 system on the RNA screen tape (Agilent Technologies, CA, USA) and Qubit Broad Range flurometric assay (Thermo Fisher Scientific, MA, USA), respectively. All RNA samples were normalized to 80 ng/μL for input into the Illumina TruSeq Stranded mRNA LS library preparation kit (Illumina, CA, USA). Individual libraries were constructed and barcoded according to the manufacturer’s protocol (Illumina). Sequencing library quality was assessed via Agilent TapeStation 2200 D1000 DNA screen tape (Agilent Technologies) showing a final library size of approximately 270 bp and were quantified with the Qubit High Sensitivity dsDNA assay (Thermo Fisher Scientific). All libraries were normalized and pooled in equimolar concentration for sequencing on an Illumina NextSeq 500 75 cycle High Output kit to generate approximately 400 million 75 base-pair, single-end sequencing reads (approximately 33 million reads per sample). All raw data was uploaded to Illumina BaseSpace (basespace.illumina.com) for FASTQ generation and demultiplexing of sequencing reads for their respective samples. All sequences were deposited to the GenBank Sequence Read Archive (SRA) and their SRA accession numbers are as follows: SAMN32740077, SAMN32740078, SAMN32740079, SAMN32740080, SAMN32740081, SAMN32740082, SAMN32740083, SAMN32740084, SAMN32740085, SAMN32740086, SAMN32740087, SAMN32740088.
Reads mapping
Single reads were trimmed and mapped by STAR (version 020201) (Dobin et al., 2013) to Peromyscus leucopus (white-footed mouse) draft genome assembly (UCI_PerLeu_2.1; BioProject PRJNA533285 https://www.ncbi.nlm.nih.gov/assembly/GCF_004664715.2) (Long et al., 2019), and to Mus musculus C3H/HeJ strain (C3H_HeJ_v1; BioProject PRJNA310854; https://www.ncbi.nlm.nih.gov/assembly/GCA_001632575.1/). To facilitate functional annotation and pathways analyses, once Peromyscus reads were mapped to the UCI_PerLeu_2.1 assembly, the respective gene IDs were converted to mouse mm38 assembly IDs (https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/) using TBLASTN because the latter had more extensive functional and gene ontology annotations as the most frequently used mouse reference genome. The identified genes were also cross-referenced with the human genome assembly GRCh38.p13 using Ensembl BioMart tools (http://ensembl.org/biomart/martview). BAM files were combined for each mouse (in other words, each sample was represented by a single BAM file), and expression of each gene was inferred by HTSeq-count (Putri et al., 2022) followed by DESeq2 analysis (Love et al., 2014b). Transcript counts and R code for differential gene expression analysis are available at https://github.com/RNAdetective/Bb_Pleu-C3H_study.
Identification and pathway analyses of differentially expressed genes
Differential gene expression analysis was performed using DESeq2 (Love et al., 2014b). The P values were adjusted by the Benjamini–Hochberg method (Benjamini and Hochberg, 1995). A corrected P value (padj) of 0.05 and 0.1 and log2 fold change (FC) of 2 and 1.5 were set as the strict and relaxed thresholds for differentially expressed genes (DEGs), respectively. To better understand the biological significance of identified DEGs, functional annotation of DEGs, and their interaction relationships were examined using STRING database analysis (string-db.org) (von Mering et al., 2005), which considered both direct physical interactions as well as indirect interactions through participation in the same pathways. The functional enrichment analyses of DEGs were carried out using the relaxed cutoffs, where a medium confidence score of 0.4 was used. Their connection degree and correlation were shown in the interaction network. Additional functional annotation and pathway enrichment analyses were conducted using the Reactome and InnateDB (Sidiropoulos et al., 2017; Fabregat et al., 2018; Jassal et al., 2020). The latter is a specialized database specifically tailored toward capturing interactions linked with the innate immune response (Breuer et al., 2013).
Results
Differentially expressed genes identified in the spleen transcriptome of Bb-infected P. leucopus and C3H mice
When only the strict cut-off values were considered (padj 0.05/2 FC), 34 DEGs were identified in the spleen transcriptome of Bb-infected P. leucopus mice relative to their uninfected control group. Out of the 34 DEGs, 12 and 22 genes were found to be up- and down-regulated, respectively (Tables S2, S3). Additional 47 DEGs were detected using relaxed cut-off values (padj 0.1/1.5 FC). Thus, a total of 35 (Table S2) and 46 (Table S3) genes were up- and down-regulated in Bb-infected P. leucopus mice, respectively. For Bb-infected C3H mice, 421 DEGs, which included 159 up-regulated and 262 down-regulated genes, were identified when relaxed cut-off values were considered. Of the 421 DEGs, 92 genes had strict cut-off values with 39 up-regulated and 53 down-regulated DEGs (Tables S4, S5).
Pathway analyses of differentially expressed genes identified in the spleen transcriptome of Bb-infected P. leucopus mice by the Reactome and InnateDB
To better understand functional changes associated with the Bb infection in P. leucopus mice, the genes that were homologous between P. leucopus and the reference mouse mm38 genome were identified using BLASTN, because the latter had more comprehensive functional annotations than what was available for P. leucopus. Thus, a total of 53 (15 up-regulated and 38 down-regulated) DEGs identified in Bb-infected P. leucopus mice that also had homologs in the mouse genome were subjected to the Reactome and InnateDB pathway analyses in order to identify whether these genes shared common functional annotations and/or pathways (Breuer et al., 2013; Sidiropoulos et al., 2017; Fabregat et al., 2018; Jassal et al., 2020).
The data showed that the top 5 significant pathways detected through the Reactome were associated with the following: erythrocyte uptake of oxygen and release of carbon dioxide, erythrocyte uptake of carbon dioxide and release of oxygen, O2/CO2 exchange in erythrocytes, hydroxycarboxylic acid-binding receptors and auto-degradation of Cdh1 by Cdh1:APC/C (ST3). Moreover, the pathway overrepresentation analysis in the InnateDB (Breuer et al., 2013) identified the following top 5 overrepresented pathways (a corrected P value of < 0.05): hepatitis C, protein processing in endoplasmic reticulum, adipocytokine signaling pathway, peroxisome proliferator-activated receptor (PPAR) signaling pathway, and TLR signaling pathway.
Pathway analyses of differentially expressed genes identified in the spleen transcriptome of Bb-infected C3H mice by the Reactome and InnateDB
A total of 159 up-regulated and 262 down-regulated DEGs detected in Bb-infected C3H mice were also subjected to the Reactome pathway and InnateDB network analyses. The top 5 significant pathways identified for the up-regulated genes via the Reactome were associated with cell cycle, cell cycle check points, activation of ATM- and Rad3-related (ATR) kinase in response to replication stress, cell cycle miotic, and activation of the pre-replicative complex. The InnateDB analysis of the up-regulated genes resulted in identification of the top 5 significant pathways associated with cell cycle, cell cycle mitotics, G1/S-specific transcription, G2/M checkpoints, and activation of ATR kinase in response to replication stress.
Pathways associated with platelet activation, signaling and aggregation, nitric oxide stimulation of guanylate cyclase, hemostasis, platelet adhesion to exposed collagen, and formation of fibrin clot (clotting cascade) were detected by the Reactome analysis for the down-regulated DEGs. Lastly, the top 5 significant pathways identified for the down-regulated genes via the InnateDB network were related to hemostasis, nitric oxide stimulation of guanylate cyclase, platelet homeostasis, platelet activation, signaling and aggregation, and platelet adhesion to exposed collagen.
Pathway analyses of differentially expressed genes shared by both Bb-infected mouse species
Only 3 DEGs were commonly identified for both Bb-infected mouse species. The HMGB3 gene encoding High Mobility Group Protein B3 was up- and down-regulated in infected C3H and P. leucopus mice, respectively. The top 10 interactors of the network identified by STRING for HMGB3 are shown in Figure 1. The Slc16a2 gene, which encodes solute carrier family 16 member 2 (monocarboxylate transporter 8) was down- and up-regulated in Bb-infected C3H and P. leucopus mice, respectively. The top 10 interactors of the Slc16a2 network are indicated in Figure 2. Lastly, the pathway analysis of the trypsin 5 (Try5)-encoding gene, which was up-regulated in both Bb-infected P. leucopus and C3H mice, identified the top 10 interactors of Try5 (Figure 3).
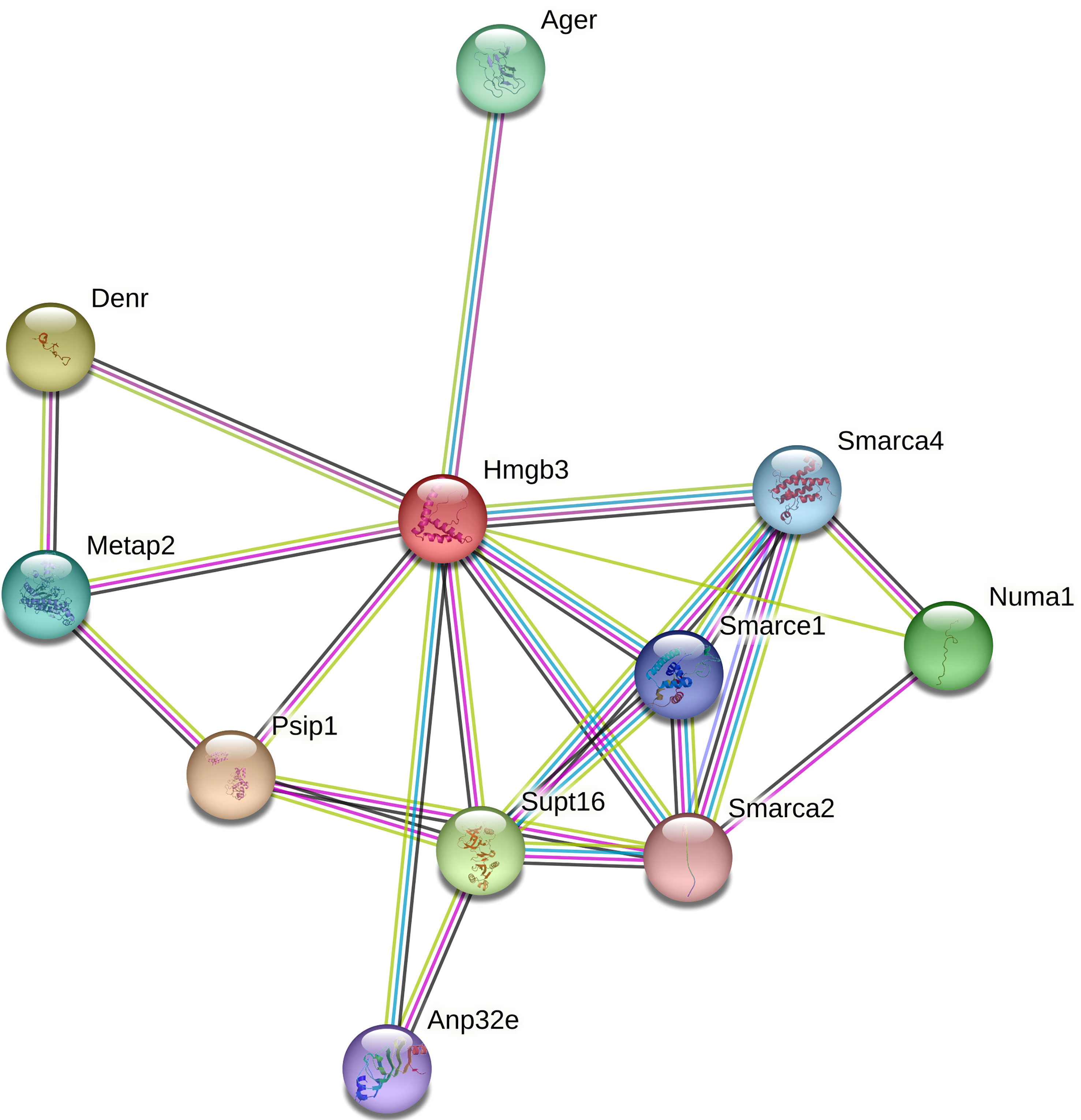
Figure 1 The protein-protein interaction network of HMGB3. The protein-protein interaction analyses were performed using the STRING database. Shown are HMGB3 (High Mobility Group Protein B3) and its top 10 protein interactors (listed clockwise): Ager (Advanced glycosylation end product-specific receptor), Smarca1, Smarca2, and Smarca4 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily A members 1, 2 and 4, respectively), Numa1 (Nuclear mitotic apparatus protein 1), Supt16 (Component of the FACT complex), Anp32e (Acidic leucine-rich nuclear phosphoprotein 32 family member E protein), PsiP1 (PC4 and SFRS1-interacting protein), Metap2 (Methionine aminopeptidase), and Denr (Density-regulated protein). Different line colors represent different types of interaction evidence between the proteins.
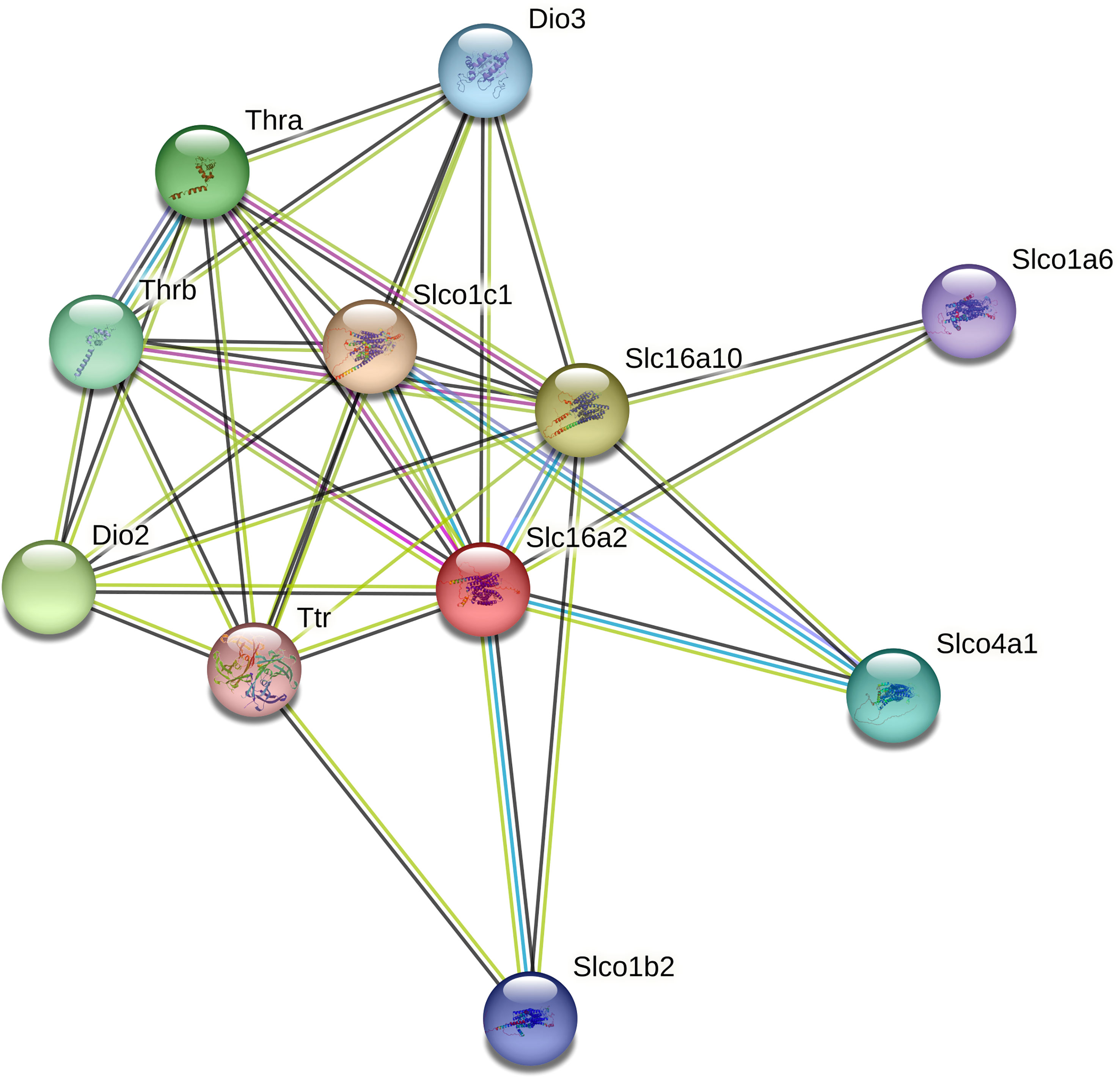
Figure 2 The protein-protein interaction network of Slc16a2. The protein-protein interaction analyses were performed using the STRING database. Shown are Slc16a2 (Solute carrier family 16 member 2 (Monocarboxylate transporter 8)) and its top 10 protein interactors (listed clockwise): Slco1c1 (Solute carrier organic anion transporter family member 1C1), Slc16a10 (Solute carrier family 16 member 10), Slco1a6 (Solute carrier organic anion transporter family member 1A6), Slco4a1 (Solute carrier organic anion transporter family member 4A1), Slco1b2 (Solute carrier organic anion transporter family member 1B2), Ttr (Transthyretin), Dio2 (Thyroxine 5-deiodinase type II), Thrb (Thyroid hormone receptors beta), Thra (Thyroid hormone receptors alpha), and Dio3 (Deiodinase, iodothyronine type III). Different line colors represent different types of interaction evidence between the proteins.
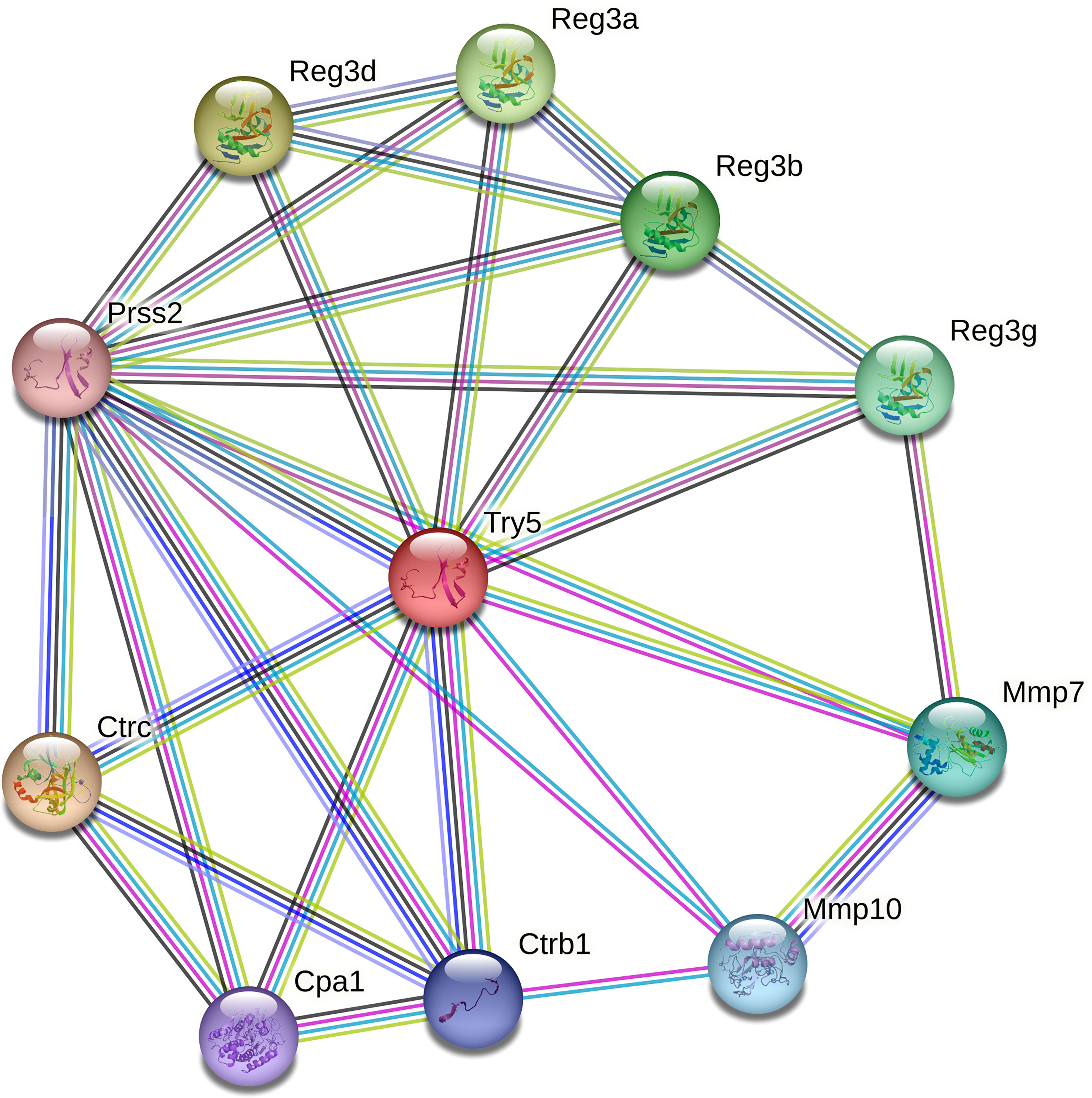
Figure 3 The protein-protein interaction network of Try5. The protein-protein interaction analyses were performed using the STRING database. Shown are Try5 (Trypsin 5) and its top 10 protein interactors (listed clockwise): Reg3a (Regenerating islet-derived 3 alpha), Reg3b (Regenerating islet-derived 3 beta), Reg3g (Regenerating islet-derived 3 gamma), Mmp7 (Matrix metallopeptidase 7), Mmp10 (Matrix metallopeptidase 10), Ctrb1 (Chymotrypsinogen B1), Cpa1 (Carboxypeptidase A1), Ctrc (Chymotrypsin C), Prss2 (Protease, serine 2), and Reg3d (Regenerating islet-derived 3 delta). Different line colors represent different types of interaction evidence between the proteins.
Protein-protein interaction analyses by STRING
In order to better understand the biological functions of DEGs, protein-protein (PPI) interaction analyses were performed using the STRING database (Szklarczyk et al., 2019; Szklarczyk et al., 2021a; Szklarczyk et al., 2021b). The STRING PPI analysis was utilized to detect functionally associated proteins and their interconnections (Szklarczyk et al., 2019; Szklarczyk et al., 2021a; Szklarczyk et al., 2021b). Consequently, protein networks were constructed to show potential relationships of DEGs with different pathways.
When DEG categories were considered separately, the functional enrichment analysis of the 35 genes up-regulated in Bb-infected P. leucopus mice demonstrated only a few protein-protein interactions (15 nodes and 3 edges). In contrast, when the 46 down-regulated genes were analyzed, the network included a total of 32 nodes and 76 edges with 14 proteins related to immune system process pathways. Of the 14 proteins, 11, 8, and 3 proteins were found to be specifically associated with innate immune response, viral defense response, and positive regulation of type I INF production, respectively (Figure 4). The latter pathway also included genes encoding TLR7, signal transducer and activator of transcription 1 (STAT1), and ATP-dependent RNA helicase (DHX58).
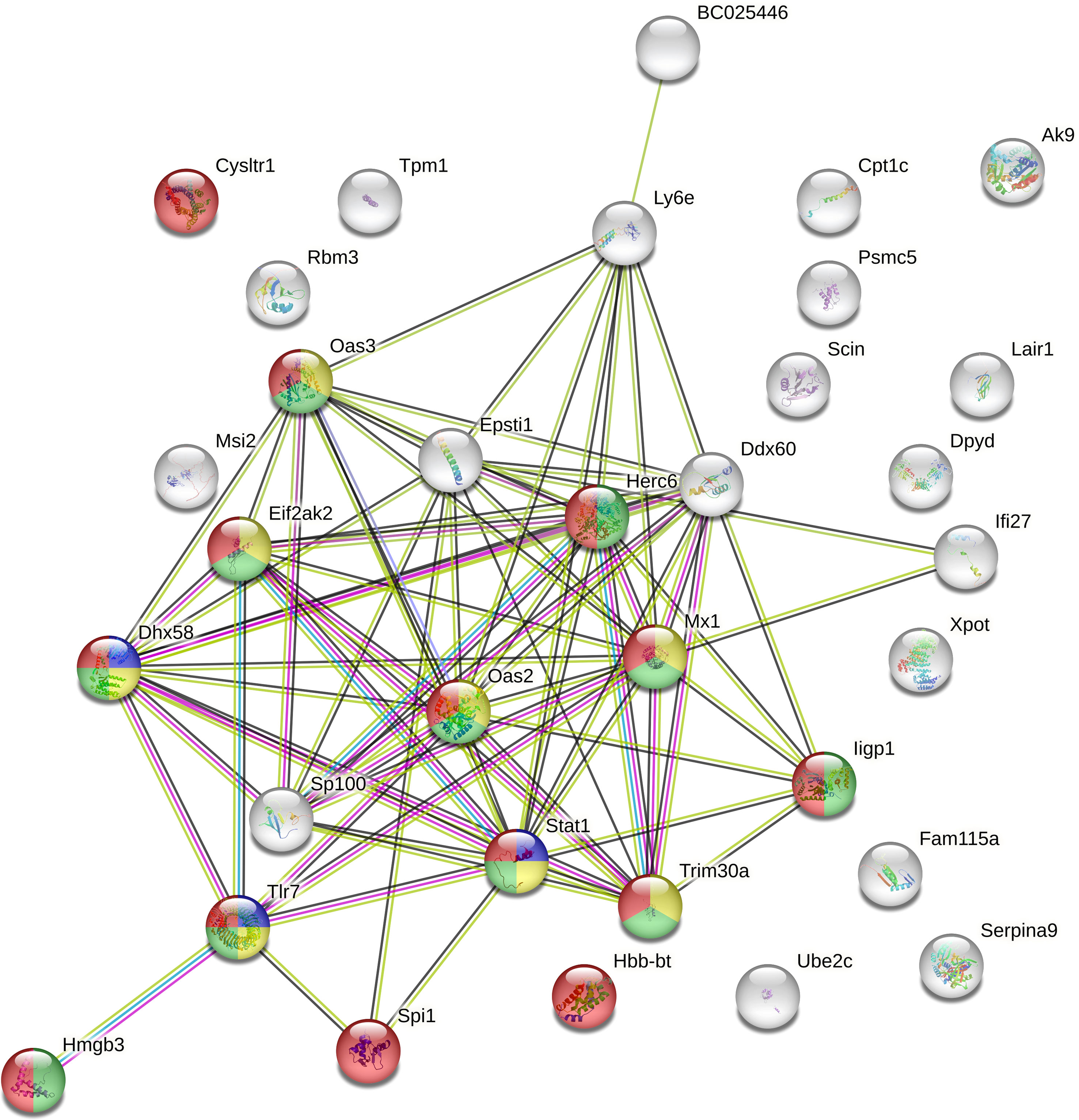
Figure 4 The protein-protein interaction network of down-regulated genes detected in Borreliella burgdorferi-infected Peromyscus leucopus mice. The protein-protein interaction analyses of 46 down-regulated genes identified in the spleen transcriptome of B burgdorferi-infected P. leucopus mice compared to their uninfected controls were performed using the STRING database. Colored are 14 proteins that are related to immune system process pathways (red), of which 11, 8, and 3 proteins are specifically associated with innate immune response (green), viral defense response (yellow), and positive regulation of type I INF production (blue), respectively.
When the entire set of 81 DEGs identified in Bb-infected P. leucopus mice was considered, the STRING analysis demonstrated significant enrichment of functional interactions among these genes (P value of < 0.1.0e-16), and overrepresentation of the following Gene Ontology (GO) molecular function terms: heterocyclic compound binding (25 genes), organic cyclic compound binding (25 genes), RNA binding (12 genes), double-stranded RNA binding (5 genes), and 2-5-oligoadenylate synthetase activity (2 genes) (Figure S1).
The STRING analysis of the 159 genes up-regulated in Bb-infected C3H mice identified 148 nodes and 1,551 edges in the network. In tissue expression and biological process terms, a total of 44 and 4 proteins were associated with immune system and B cell activation, respectively. Furthermore, in biological process term, 50 and 60 proteins were found to be related to pathways of cell cycle and cellular response to stimulus, respectively.
A total of 391 nodes and 2,095 edges were constructed by STRING for the 421 DEGs identified in Bb-infected C3H mice. In biological process term, some DEGs were enriched in the blood coagulation pathway (n=18). Out of 10 genes, whose proteins were associated with the regulation of blood coagulation, 9 and 1 genes were down- and up-regulated, respectively. The up-regulated gene encodes chymotrypsin-like elastase family member 2A (Cela2a). A total of 9 genes were associated with the platelet activation pathway, of which 8 and 1 genes were down- and up-regulated, respectively. The up-regulated gene encodes CD40 ligand (Cd40lg). When down-regulated genes were only considered (n=262), a total of 245 nodes and 407 edges were constructed by STRING with identification of 38 homeostasis-associated proteins via the Reactome (Figure S2). In biological process term, the STRING analysis showed that down-regulated genes were also enriched in wound healing (n=22), regulation of wound healing (n=11), response to stimulus (n=136), and response to stress (n=64).
Discussion
In the present investigation, comparative transcriptomic analyses of spleens harvested from Bb-infected and uninfected P. leucopus mice have been performed. The spleen was selected because it is one of the major lymphoid organs that together with the bone marrow contains most immune cells (Zhao et al., 2012; Bronte and Pittet, 2013). The experimental design also included the laboratory “control” mouse strain – Bb-infected and uninfected C3H mice. To understand how P. leucopus mice are able to tolerate Bb infection, the transcriptome data generated from spleens of infected and uninfected mice were analyzed within each animal species by three complementing bio-molecular tools. The overall data demonstrated that Bb-infected P. leucopus mice had five times lower number of DEGs (n=81) than infected C3H mice (n=421). Out of the 81 DEGs, approximately 57% (n=46) and 43% (n=35) of genes were found to be down- and up-regulated, respectively. As opposed to the 46 down-regulated genes, many of whom were associated with immune response, the 35 up-regulated genes were not related to any immune response pathways. The overall data demonstrated that the Bb-induced response of the spleen transcriptome was more quiescent in P. leucopus mice compared to that of C3H mice.
The present results are similar to the findings of a previous study, where the spleen transcriptome analyses of B. afzelii-infected and uninfected bank voles (Myodes glareolus) and yellow-necked mice (Apodemus flavicollis) were performed (Zhong et al., 2020). Although both animal species are considered natural reservoir hosts of B. afzelii in Europe (Zhong et al., 2019), Borreliella-infected bank voles have on average a magnitude higher spirochetal loads in their tissues (Raberg, 2012; Zhong et al., 2019). The comparative analyses between infected and uninfected animals within each species identified only 8 and 5 DEGs in B. afzelii-infected bank voles and yellow-necked mice, respectively (Zhong et al., 2020). Furthermore, similar to the current data, other study had also shown no significant involvement of inflammation or innate immunity genes, when transcriptomes of skin and blood samples taken at 5 weeks postinfection were compared between Bb strain Sh-2-82-infected and uninfected P. leucopus mice (Long et al., 2019). Overall, the current and previous results could collectively suggest that reservoir hosts of LD pathogens moderate the immune system to avoid development of exaggerated (immune) responses.
A few examples of immune related genes that, during the persistent Bb infection, were down-regulated in P. leucopus mice and remained unresponsive in C3H mice include TLR7-encoding gene, type I IFN-associated genes, and members of the lymphocyte antigen-6 (Ly6) gene family. TLR7 recognizes bacterial RNA and peptidoglycan, and becomes activated in response to bacterial exposure by dendritic cells (DCs) (Mancuso et al., 2009). It was shown that TLR7 mRNA became up-regulated in Bb-exposed human oligodendrocyte cell line (Parthasarathy and Philipp, 2018). Overall, activation of TLRs by Bb lipoproteins leads to the induction of type I IFN (Petzke et al., 2009; Love et al., 2014a; Lochhead et al., 2021), which in turn primes macrophages for producing proinflammatory cytokines (Bockenstedt et al., 2021).
Through the use of laboratory mouse strains of Mus musculus, it has been consistently demonstrated that the severity of LD arthritis is driven by type I IFN (Crandall et al., 2006; Miller et al., 2008). Early type I IFN induction also positively correlates with the Bb capacity to disseminate in C3H/HeJ mice (Petzke et al., 2016). Type I IFN signaling may also lead to the accumulation of naïve B lymphocytes in Bb-infected mice (Hastey et al., 2014). Surprisingly, the present study did not identify any DEGs associated with the interferon signaling pathways in Bb-infected C3H mice. The latter cannot be solely explained by the lack of a functional TLR4 pathway in this particular mouse strain (Poltorak et al., 1998), partly because the classic enterobacterium-type lipopolysaccharide (LPS) (Brubaker et al., 2015), one of TLR4 ligands, is not produced by Bb spirochetes (Takayama et al., 1987). In contrast, a number of types I IFN-associated genes (n=6) were down-regulated in Bb-infected P. leucopus mice. These findings are consistent with the previous study, which demonstrated that a set of genes involved in IFNα signaling was down-regulated in B. afzelii-infected bank voles and yellow-necked mice compared to their uninfected controls (Zhong et al., 2020). In addition to type I IFN-associated genes, two members of Ly6 gene family were also found to be down-regulated in Bb-infected P. leucopus mice. The 6E and 6A-2/6E-1-like proteins are known to be expressed by different types of mouse and human immune cells (e.g., B and T lymphocytes, natural killer (NK) cells, monocytes, and DCs) and involved in regulation of activation, proliferation, and differentiation of T lymphocytes (Ortega et al., 1986; Toulon et al., 1988; Lee et al., 2013).
Notably, of the 421 DEGs identified in Bb-infected C3H mice, a number of genes (n=38) were associated with different aspects of hemostasis. In the blood coagulation pathway alone, there were a total of 19 down-regulated genes with some involved in the regulation of blood coagulation (n=10) and others associated with platelet activation (n=9). Similar results were obtained by an earlier study, where blood, spleen and liver transcriptomes of LPS-treated-treated Mus musculus (BALB/cAnNCrl, referred here as BALB/c) and P. leucopus mice were compared to their respective non-treated controls (Balderrama-Gutierrez et al., 2021). The previous data showed that it was only in the blood of LPS-treated BALB/c mice where DEGs associated with negative regulation of blood coagulation pathways were detected (Balderrama-Gutierrez et al., 2021). The present study also demonstrated that the Cela2a and Cd40lg genes were up-regulated only in Bb-infected C3H mice. Cela2a hydrolyzes elastin and reduces platelet hyperactivation (Esteghamat et al., 2019), and Cd40lg is involved in immunoglobulin class switching, activation of B cell and T cell proliferation, enhancement of IL10 production, and induction of NF-kappa-B production (Armitage et al., 1992; Borrow et al., 1996; Van Kooten and Banchereau, 1996). As opposed to P. leucopus, Bb-infected C3H mice had also an overabundance of up-regulated cell cycle genes (n=50), whose products are known to play an important role in the host immune response (e.g., macrophage expansion and antibody class switching) (Laphanuwat and Jirawatnotai, 2019).
Overall, there were only 3 DEGs that were commonly identified in both animal species when infected with Bb (Table 1). The HMGB3 gene was up- and down-regulated in Bb-infected C3H and P. leucopus mice, respectively. As multifunctional molecules, HMGB proteins are proposed to play a role in the nucleic-acid-mediated activation of innate immune responses (Yanai et al., 2009). Being part of the cytosolic receptor–IRF3/NF-kB signaling pathway (Yanai et al., 2009), HMGB proteins can also stimulate TLR2, which, through its engagement with Bb lipoproteins, initiates expression of NF-κB-dependent cytokines (Shin et al., 2008). Another example of “shared” DEGs includes the Slc16a2 gene, which encodes solute carrier family 16 member 2 (monocarboxylate transporter 8) in P. leucopus and C3H mice. This active and specific thyroid hormone transporter mediates cellular uptake and transmembrane transporter activity of thyroxine (T4), triiodothyronine (T3), reverse triiodothyronine (rT3) and diidothyronine (Friesema et al., 2003; Friesema et al., 2005). Thyroid hormones have an important role in activation of neutrophils, NK cells, DCs, and B and T lymphocytes during infection (Rubingh et al., 2020). The corresponding genes were up- and down-regulated in Bb-infected P. leucopus and C3H mice, respectively. Lastly, both mouse species had only one “shared” gene up-regulated in response to Bb infection. The Try5 gene encodes anionic trypsin-2-like protein and trypsin 5 in P. leucopus and C3H mice, respectively.
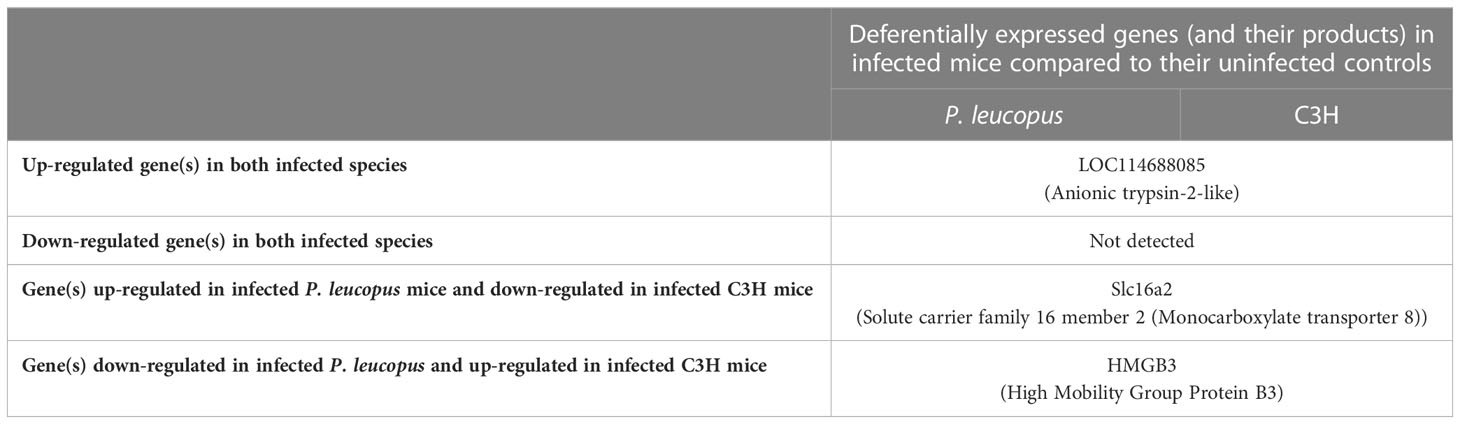
Table 1 A list of genes that are deferentially expressed in both Borreliella burgdorferi-infected Peromyscus leucopus and C3H mice.
Some limitations of this study include small sample sizes and the use of male mice only. Another caveat is that spirochetal burdens were not determined in spleens of the infected mice to see if potentially higher bacterial loads in C3H mice would explain overall higher numbers of DEGs identified in the laboratory mice compared to P. leucopus mice. In addition to addressing the above limitations, future studies are also warranted to confirm the identified DEGs through other approaches (e.g., qRT-PCR, proteomics).
In summary, the present investigation is one of the few that have examined the transcriptome response of mammalian reservoir hosts to Borreliella infection. Although the experimental design of this study is different from the above-mentioned earlier investigations (e.g., reservoir host, Borreliella strain, time point of harvest, tissue type analyzed), the collective results of all studies have consistently demonstrated a very limited response of natural reservoir hosts to the persistent infection of LD pathogens. In order to decipher tolerance mechanism of P. leucopus and other reservoir hosts to Bb infection, future investigations that would pursue some of the data leads identified in the present and earlier studies are highly warranted.
Data availability statement
The data presented in the study are deposited in the GenBank Sequence Read Archive (SRA) depository and the SRA accession numbers are SAMN32740077, SAMN32740078, SAMN32740079, SAMN32740080, SAMN32740081, SAMN32740082, SAMN32740083, SAMN32740084, SAMN32740085, SAMN32740086, SAMN32740087, SAMN32740088.
Ethics statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Texas A&M University.
Author contributions
AG analyzed the data and produced the manuscript draft. AH performed sequencing and raw data assembly. IM and AZ were involved in the data analysis. CN and HP analyzed the data, contributed to the data visualization and writing of the manuscript. DT provided resources and contributed to the data analysis. AR concepted and developed the study, provided resources, oversaw the project, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The work at Texas A&M University was partially supported by National Institutes of Health (NIH) grant R03AI135159-02, Department of Veterinary Pathobiology, Texas A&M School of Veterinary Medicine and Biomedical Sciences, and Texas A&M AgriLife. The analysis at Kent State University was partially supported by NIH National Institute on Aging award R21AG064479-01, and Brain Health Research Institute Pilot Award and Healthy Communities Research Initiative Launch Pad Award from Kent State University. The work at Georgia State University was partially supported by NIH grant 1R21CA241044-01A1, National Science Foundation grant IIS-2212508, and by Georgia State University Molecular Basis of Disease Fellowship.
Acknowledgments
The authors would like to thank David C. Gillis for his assistance with the mouse experiment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1115350/full#supplementary-material
References
Armitage, R. J., Fanslow, W. C., Strockbine, L., Sato, T. A., Clifford, K. N., Macduff, B. M., et al. (1992). Molecular and biological characterization of a murine ligand for CD40. Nature 357 (6373), 80–82. doi: 10.1038/357080a0
Armstrong, A. L., Barthold, S. W., Persing, D. H., Beck, D. S. (1992). Carditis in Lyme disease susceptible and resistant strains of laboratory mice infected with Borrelia burgdorferi. Am. J. Trop. Med. Hyg. 47 (2), 249–258. doi: 10.4269/ajtmh.1992.47.249
Balderrama-Gutierrez, G., Milovic, A., Cook, V. J., Islam, M. N., Zhang, Y., Kiaris, H., et al. (2021). An infection-tolerant mammalian reservoir for several zoonotic agents broadly counters the inflammatory effects of endotoxin. mBio 12 (2). doi: 10.1128/mBio.00588-21
Baranton, G., Postic, D., Saint Girons, I., Boerlin, P., Piffaretti, J. C., Assous, M., et al. (1992). Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int. J. Syst. Bacteriol. 42 (3), 378–383. doi: 10.1099/00207713-42-3-378
Barbour, A. G., Gupta, R. S. (2021). The family Borreliaceae (Spirochaetales), a diverse group in two genera of tick-borne spirochetes of mammals, birds, and reptiles. J. Med. Entomol. 58 (4), 1513–1524. doi: 10.1093/jme/tjab055
Barthold, S. W., Beck, D. S., Hansen, G. M., Terwilliger, G. A., Moody, K. D. (1990). Lyme borreliosis in selected strains and ages of laboratory mice. J. Infect. Dis. 162 (1), 133–138. doi: 10.1093/infdis/162.1.133
Barthold, S. W., Feng, S., Bockenstedt, L. K., Fikrig, E., Feen, K. (1997). Protective and arthritis-resolving activity in sera of mice infected with Borrelia burgdorferi. Clin. Infect. Dis. 25 Suppl 1, S9–17. doi: 10.1086/516166
Barthold, S. W., Sidman, C. L., Smith, A. L. (1992). Lyme borreliosis in genetically resistant and susceptible mice with severe combined immunodeficiency. Am. J. Trop. Med. Hyg. 47 (5), 605–613. doi: 10.4269/ajtmh.1992.47.605
Baum, E., Hue, F., Barbour, A. G. (2012). Experimental infections of the reservoir species Peromyscus leucopus with diverse strains of Borrelia burgdorferi, a Lyme disease agent. MBio 3 (6), e00434–e00412. doi: 10.1128/mBio.00434-12
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. Ser. B 57, 289–300. doi: 10.2307/2346101
Bockenstedt, L. K., Wooten, R. M., Baumgarth, N. (2021). Immune response to Borrelia: Lessons from Lyme disease spirochetes. Curr. Issues Mol. Biol. 42, 145–190. doi: 10.21775/cimb.042.145
Bolz, D. D., Sundsbak, R. S., Ma, Y., Akira, S., Kirschning, C. J., Zachary, J. F., et al. (2004). MyD88 plays a unique role in host defense but not arthritis development in Lyme disease. J. Immunol. 173 (3), 2003–2010. doi: 10.4049/jimmunol.173.3.2003
Borrow, P., Tishon, A., Lee, S., Xu, J., Grewal, I. S., Oldstone, M. B., et al. (1996). CD40L-deficient mice show deficits in antiviral immunity and have an impaired memory CD8+ CTL response. J. Exp. Med. 183 (5), 2129–2142. doi: 10.1084/jem.183.5.2129
Breuer, K., Foroushani, A. K., Laird, M. R., Chen, C., Sribnaia, A., Lo, R., et al. (2013). InnateDB: Systems biology of innate immunity and beyond-recent updates and continuing curation. Nucleic Acids Res. 41 (Database issue), D1228–D1233. doi: 10.1093/nar/gks1147
Bronte, V., Pittet, M. J. (2013). The spleen in local and systemic regulation of immunity. Immunity 39 (5), 806–818. doi: 10.1016/j.immuni.2013.10.010
Brown, C. R., Reiner, S. L. (1999). Experimental Lyme arthritis in the absence of interleukin-4 or gamma interferon. Infect. Immun. 67 (7), 3329–3333. doi: 10.1128/iai.67.7.3329-3333.1999
Brown, J. P., Zachary, J. F., Teuscher, C., Weis, J. J., Wooten, R. M. (1999). Dual role of interleukin-10 in murine Lyme disease: Regulation of arthritis severity and host defense. Infect. Immun. 67 (10), 5142–5150. doi: 10.1128/iai.67.10.5142-5150.1999
Brubaker, S. W., Bonham, K. S., Zanoni, I., Kagan, J. C. (2015). Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 33, 257–290. doi: 10.1146/annurev-immunol-032414-112240
Burgess, E. C., French, J. B., Jr., Gendron-Fitzpatrick, A. (1990). Systemic disease in Peromyscus leucopus associated with Borrelia burgdorferi infection. Am. J. Trop. Med. Hyg. 42 (3), 254–259. doi: 10.4269/ajtmh.1990.42.254
Canica, M. M., Nato, F., du Merle, L., Mazie, J. C., Baranton, G., Postic, D. (1993). Monoclonal antibodies for identification of Borrelia afzelii sp. nov. associated with late cutaneous manifestations of Lyme borreliosis. Scand. J. Infect. Dis. 25 (4), 441–448. doi: 10.3109/00365549309008525
Cook, V., Barbour, A. G. (2015). Broad diversity of host responses of the white-footed mouse Peromyscus leucopus to Borrelia infection and antigens. Ticks Tick. Borne Dis. 6 (5), 549–558. doi: 10.1016/j.ttbdis.2015.04.009
Crandall, H., Dunn, D. M., Ma, Y., Wooten, R. M., Zachary, J. F., Weis, J. H., et al. (2006). Gene expression profiling reveals unique pathways associated with differential severity of Lyme arthritis. J. Immunol. 177 (11), 7930–7942. doi: 10.4049/jimmunol.177.11.7930
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29 (1), 15–21. doi: 10.1093/bioinformatics/bts635
Donahue, J. G., Piesman, J., Spielman, A. (1987). Reservoir competence of white-footed mice for Lyme disease spirochetes. Am. J. Trop. Med. Hyg. 36 (1), 92–96. doi: 10.4269/ajtmh.1987.36.92
Ebnet, K., Brown, K. D., Siebenlist, U. K., Simon, M. M., Shaw, S. (1997). Borrelia burgdorferi activates nuclear factor-kappa B and is a potent inducer of chemokine and adhesion molecule gene expression in endothelial cells and fibroblasts. J. Immunol. 158 (7), 3285–3292. doi: 10.4049/jimmunol.158.7.3285
Esteghamat, F., Broughton, J. S., Smith, E., Cardone, R., Tyagi, T., Guerra, M., et al. (2019). CELA2A mutations predispose to early-onset atherosclerosis and metabolic syndrome and affect plasma insulin and platelet activation. Nat. Genet. 51 (8), 1233–1243. doi: 10.1038/s41588-019-0470-3
Fabregat, A., Jupe, S., Matthews, L., Sidiropoulos, K., Gillespie, M., Garapati, P., et al. (2018). The reactome pathway knowledgebase. Nucleic Acids Res. 46 (D1), D649–d655. doi: 10.1093/nar/gkx1132
Friesema, E. C., Ganguly, S., Abdalla, A., Manning Fox, J. E., Halestrap, A. P., Visser, T. J. (2003). Identification of monocarboxylate transporter 8 as a specific thyroid hormone transporter. J. Biol. Chem. 278 (41), 40128–40135. doi: 10.1074/jbc.M300909200
Friesema, E. C., Jansen, J., Milici, C., Visser, T. J. (2005). Thyroid hormone transporters. Vitam. Horm. 70, 137–167. doi: 10.1016/s0083-6729(05)70005-4
Ganapamo, F., Dennis, V. A., Philipp, M. T. (2000). Early induction of gamma interferon and interleukin-10 production in draining lymph nodes from mice infected with Borrelia burgdorferi. Infect. Immun. 68 (12), 7162–7165. doi: 10.1128/iai.68.12.7162-7165.2000
Hanincová, K., Ogden, N. H., Diuk-Wasser, M., Pappas, C. J., Iyer, R., Fish, D., et al. (2008). Fitness variation of Borrelia burgdorferi sensu stricto strains in mice. Appl. Environ. Microbiol. 74 (1), 153–157. doi: 10.1128/aem.01567-07
Hastey, C. J., Ochoa, J., Olsen, K. J., Barthold, S. W., Baumgarth, N. (2014). MyD88- and TRIF-independent induction of type I interferon drives naive B cell accumulation but not loss of lymph node architecture in Lyme disease. Infect. Immun. 82 (4), 1548–1558. doi: 10.1128/iai.00969-13
Hechemy, K. E., Harris, H. L., Duerr, M. J., Benach, J. L., Reimer, C. B. (1988). Immunoglobulin G subclasses specific to Borrelia burgdorferi in patients with Lyme disease. Ann. N. Y. Acad. Sci. 539, 162–169. doi: 10.1111/j.1749-6632.1988.tb31849.x
Hirschfeld, M., Kirschning, C. J., Schwandner, R., Wesche, H., Weis, J. H., Wooten, R. M., et al. (1999). Cutting edge: Inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by toll-like receptor 2. J. Immunol. 163 (5), 2382–2386. doi: 10.4049/jimmunol.163.5.2382
Jassal, B., Matthews, L., Viteri, G., Gong, C., Lorente, P., Fabregat, A., et al. (2020). The reactome pathway knowledgebase. Nucleic Acids Res. 48 (D1), D498–d503. doi: 10.1093/nar/gkz1031
Kugeler, K. J., Schwartz, A. M., Delorey, M. J., Mead, P. S., Hinckley, A. F. (2021). Estimating the frequency of Lyme disease diagnoses, United States 2010-2018. Emerg. Infect. Dis. 27 (2), 616–619. doi: 10.3201/eid2702.202731
Kurtenbach, K., Hanincová, K., Tsao, J. I., Margos, G., Fish, D., Ogden, N. H. (2006). Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat. Rev. Microbiol. 4 (9), 660–669. doi: 10.1038/nrmicro1475
Laphanuwat, P., Jirawatnotai, S. (2019). Immunomodulatory roles of cell cycle regulators. Front. Cell Dev. Biol. 7. doi: 10.3389/fcell.2019.00023
Lee, P. Y., Wang, J. X., Parisini, E., Dascher, C. C., Nigrovic, P. A. (2013). Ly6 family proteins in neutrophil biology. J. Leukoc. Biol. 94 (4), 585–594. doi: 10.1189/jlb.0113014
Levine, J. F., Wilson, M. L., Spielman, A. (1985). Mice as reservoirs of the Lyme disease spirochete. Am. J. Trop. Med. Hyg. 34 (2), 355–360. doi: 10.4269/ajtmh.1985.34.355
Lin, Y. P., Benoit, V., Yang, X., Martinez-Herranz, R., Pal, U., Leong, J. M. (2014). Strain-specific variation of the decorin-binding adhesin DbpA influences the tissue tropism of the Lyme disease spirochete. PloS Pathog. 10 (7), e1004238. doi: 10.1371/journal.ppat.1004238
Lochhead, R. B., Strle, K., Arvikar, S. L., Weis, J. J., Steere, A. C. (2021). Lyme arthritis: Linking infection, inflammation and autoimmunity. Nat. Rev. Rheumatol. 17 (8), 449–461. doi: 10.1038/s41584-021-00648-5
Long, A. D., Baldwin-Brown, J., Tao, Y., Cook, V. J., Balderrama-Gutierrez, G., Corbett-Detig, R., et al. (2019). The genome of Peromyscus leucopus, natural host for Lyme disease and other emerging infections. Sci. Adv. 5 (7), eaaw6441. doi: 10.1126/sciadv.aaw6441
Love, M. I., Huber, W., Anders, S. (2014b). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Love, A. C., Schwartz, I., Petzke, M. M. (2014a). Borrelia burgdorferi RNA induces type I and III interferons via toll-like receptor 7 and contributes to production of NF-κB-dependent cytokines. Infect. Immun. 82 (6), 2405–2416. doi: 10.1128/iai.01617-14
Ma, Y., Seiler, K. P., Eichwald, E. J., Weis, J. H., Teuscher, C., Weis, J. J. (1998). Distinct characteristics of resistance to Borrelia burgdorferi-induced arthritis in C57BL/6N mice. Infect. Immun. 66 (1), 161–168. doi: 10.1128/iai.66.1.161-168.1998
Mancuso, G., Gambuzza, M., Midiri, A., Biondo, C., Papasergi, S., Akira, S., et al. (2009). Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat. Immunol. 10 (6), 587–594. doi: 10.1038/ni.1733
McKisic, M. D., Barthold, S. W. (2000). T-cell-independent responses to Borrelia burgdorferi are critical for protective immunity and resolution of Lyme disease. Infect. Immun. 68 (9), 5190–5197. doi: 10.1128/iai.68.9.5190-5197.2000
Mead, P. S. (2015). Epidemiology of Lyme disease. Infect. Dis. Clin. North Am. 29 (2), 187–210. doi: 10.1016/j.idc.2015.02.010
Miller, J. C., Ma, Y., Bian, J., Sheehan, K. C., Zachary, J. F., Weis, J. H., et al. (2008). A critical role for type I IFN in arthritis development following Borrelia burgdorferi infection of mice. J. Immunol. 181 (12), 8492–8503. doi: 10.4049/jimmunol.181.12.8492
Moody, K. D., Terwilliger, G. A., Hansen, G. M., Barthold, S. W. (1994). Experimental Borrelia burgdorferi infection in Peromyscus leucopus. J. Wildl. Dis. 30 (2), 155–161. doi: 10.7589/0090-3558-30.2.155
Ortega, G., Korty, P. E., Shevach, E. M., Malek, T. R. (1986). Role of Ly-6 in lymphocyte activation. I. Characterization of a monoclonal of a monoclonal antibody to a nonpolymorphic Ly-6 specificity. J. Immunol. 137 (10), 3240–3246. doi: 10.4049/jimmunol.137.10.3240
Parthasarathy, G., Philipp, M. T. (2018). Intracellular TLR7 is activated in human oligodendrocytes in response to Borrelia burgdorferi exposure. Neurosci. Lett. 671, 38–42. doi: 10.1016/j.neulet.2018.01.058
Petnicki-Ocwieja, T., Chung, E., Acosta, D. I., Ramos, L. T., Shin, O. S., Ghosh, S., et al. (2013). TRIF mediates toll-like receptor 2-dependent inflammatory responses to Borrelia burgdorferi. Infect. Immun. 81 (2), 402–410. doi: 10.1128/iai.00890-12
Petzke, M. M., Brooks, A., Krupna, M. A., Mordue, D., Schwartz, I. (2009). Recognition of Borrelia burgdorferi, the Lyme disease spirochete, by TLR7 and TLR9 induces a type I IFN response by human immune cells. J. Immunol. 183 (8), 5279–5292. doi: 10.4049/jimmunol.0901390
Petzke, M. M., Iyer, R., Love, A. C., Spieler, Z., Brooks, A., Schwartz, I. (2016). Borrelia burgdorferi induces a type I interferon response during early stages of disseminated infection in mice. BMC Microbio.l 16, 29. doi: 10.1186/s12866-016-0644-4
Piesman, J., Gern, L. (2004). Lyme borreliosis in Europe and north America. Parasitology 129 Suppl, S191–S220. doi: 10.1017/s0031182003004694
Poltorak, A., He, X., Smirnova, I., Liu, M. Y., Van Huffel, C., Du, X., et al. (1998). Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science 282 (5396), 2085–2088. doi: 10.1126/science.282.5396.2085
Putri, G. H., Anders, S., Pyl, P. T., Pimanda, J. E., Zanini, F. (2022). Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 38 (10), 2943–2945. doi: 10.1093/bioinformatics/btac166
Raberg, L. (2012). Infection intensity and infectivity of the tick-borne pathogen Borrelia afzelii. J. Evol. Biol. 25 (7), 1448–1453. doi: 10.1111/j.1420-9101.2012.02515.x
Radolf, J. D., Strle, K., Lemieux, J. E., Strle, F. (2021). Lyme disease in humans. Curr. Issues Mol. Biol. 42, 333–384. doi: 10.21775/cimb.042.333
Rogovskyy, A. S., Casselli, T., Tourand, Y., Jones, C. R., Owen, J. P., Mason, K. L., et al. (2015). Evaluation of the importance of VlsE antigenic variation for the enzootic cycle of Borrelia burgdorferi. PloS One 10 (4), e0124268. doi: 10.1371/journal.pone.0124268
Rogovskyy, A. S., Gillis David, C., Ionov, Y., Gerasimov, E., Zelikovsky, A., Bäumler Andreas, J. (2016). Antibody response to Lyme disease spirochetes in the context of VlsE-mediated immune evasion. Infect. Immun. 85 (1), e00890–e00816. doi: 10.1128/IAI.00890-16
Rubingh, J., van der Spek, A., Fliers, E., Boelen, A. (2020). The role of thyroid hormone in the innate and adaptive immune response during infection. Compr. Physiol. 10 (4), 1277–1287. doi: 10.1002/cphy.c200003
Schwan, T. G., Kime, K. K., Schrumpf, M. E., Coe, J. E., Simpson, W. J. (1989). Antibody response in white-footed mice (Peromyscus leucopus) experimentally infected with the Lyme disease spirochete (Borrelia burgdorferi). Infect. Immun. 57 (11), 3445–3451. doi: 10.1128/iai.57.11.3445-3451.1989
Schwanz, L. E., Voordouw, M. J., Brisson, D., Ostfeld, R. S. (2011). Borrelia burgdorferi has minimal impact on the Lyme disease reservoir host Peromyscus leucopus. Vector Borne Zoonotic Dis. 11 (2), 117–124. doi: 10.1089/vbz.2009.0215
Shin, O. S., Isberg, R. R., Akira, S., Uematsu, S., Behera, A. K., Hu, L. T. (2008). Distinct roles for MyD88 and toll-like receptors 2, 5, and 9 in phagocytosis of Borrelia burgdorferi and cytokine induction. Infect. Immun. 76 (6), 2341–2351. doi: 10.1128/iai.01600-07
Sidiropoulos, K., Viteri, G., Sevilla, C., Jupe, S., Webber, M., Orlic-Milacic, M., et al. (2017). Reactome enhanced pathway visualization. Bioinformatics 33 (21), 3461–3467. doi: 10.1093/bioinformatics/btx441
Smith, B. G., Cruz, A. I., Jr., Milewski, M. D., Shapiro, E. D. (2011). Lyme disease and the orthopaedic implications of Lyme arthritis. J. Am. Acad. Orthop. Surg. 19 (2), 91–100. doi: 10.5435/00124635-201102000-00004
Sonderegger, F. L., Ma, Y., Maylor-Hagan, H., Brewster, J., Huang, X., Spangrude, G. J., et al. (2012). Localized production of IL-10 suppresses early inflammatory cell infiltration and subsequent development of IFN-γ-mediated Lyme arthritis. J. Immunol. 188 (3), 1381–1393. doi: 10.4049/jimmunol.1102359
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47 (D1), D607–d613. doi: 10.1093/nar/gky1131
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021a). Correction to ‘The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets’. Nucleic Acids Res. 49 (18), 10800. doi: 10.1093/nar/gkab835
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021b). The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49 (D1), D605–d612. doi: 10.1093/nar/gkaa1074
Takayama, K., Rothenberg, R. J., Barbour, A. G. (1987). Absence of lipopolysaccharide in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 55 (9), 2311–2313. doi: 10.1128/iai.55.9.2311-2313.1987
Toulon, M., Palfree, R. G., Palfree, S., Dumont, F. J., Hämmerling, U. (1988). Ly-6 A/E antigen of murine T cells is associated with a distinct pathway of activation. requirements for interferon and exogenous interleukin 2. Eur. J. Immunol. 18 (6), 937–942. doi: 10.1002/eji.1830180616
Van Kooten, C., Banchereau, J. (1996). CD40-CD40 ligand: A multifunctional receptor-ligand pair. Adv. Immunol. 61, 1–77. doi: 10.1016/s0065-2776(08)60865-2
Verhaegh, D., Joosten, L. A. B., Oosting, M. (2017). The role of host immune cells and Borrelia burgdorferi antigens in the etiology of Lyme disease. Eur. Cytokine Netw. 28 (2), 70–84. doi: 10.1684/ecn.2017.0396
von Mering, C., Jensen, L. J., Snel, B., Hooper, S. D., Krupp, M., Foglierini, M., et al. (2005). STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 33 (Database issue), D433–D437. doi: 10.1093/nar/gki005
Wooten, R. M., Ma, Y., Yoder, R. A., Brown, J. P., Weis, J. H., Zachary, J. F., et al. (2002a). Toll-like receptor 2 is required for innate, but not acquired, host defense to Borrelia burgdorferi. J. Immunol. 168 (1), 348–355. doi: 10.4049/jimmunol.168.1.348
Wooten, R. M., Ma, Y., Yoder, R. A., Brown, J. P., Weis, J. H., Zachary, J. F., et al. (2002b). Toll-like receptor 2 plays a pivotal role in host defense and inflammatory response to Borrelia burgdorferi. Vector Borne Zoonotic Dis. 2 (4), 275–278. doi: 10.1089/153036602321653860
Wooten, R. M., Modur, V. R., McIntyre, T. M., Weis, J. J. (1996). Borrelia burgdorferi outer membrane protein a induces nuclear translocation of nuclear factor-kappa b and inflammatory activation in human endothelial cells. J. Immunol. 157 (10), 4584–4590. doi: 10.4049/jimmunol.157.10.4584
Wooten, R. M., Weis, J. J. (2001). Host-pathogen interactions promoting inflammatory Lyme arthritis: Use of mouse models for dissection of disease processes. Curr. Opin. Microbiol. 4 (3), 274–279. doi: 10.1016/s1369-5274(00)00202-2
Yanai, H., Ban, T., Wang, Z., Choi, M. K., Kawamura, T., Negishi, H., et al. (2009). HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 462 (7269), 99–103. doi: 10.1038/nature08512
Zhao, E., Xu, H., Wang, L., Kryczek, I., Wu, K., Hu, Y., et al. (2012). Bone marrow and the control of immunity. Cell Mol. Immunol. 9 (1), 11–19. doi: 10.1038/cmi.2011.47
Zhong, X., Lundberg, M., Råberg, L. (2020). Comparison of spleen transcriptomes of two wild rodent species reveals differences in the immune response against Borrelia afzelii. Eco.l Evol. 10 (13), 6421–6434. doi: 10.1002/ece3.6377
Keywords: Lyme borreliosis, Borreliella burgdorferi, Borrelia, Peromyscus leucopus, C3H mice, differentially expressed genes
Citation: Gaber AM, Mandric I, Nitirahardjo C, Piontkivska H, Hillhouse AE, Threadgill DW, Zelikovsky A and Rogovskyy AS (2023) Comparative transcriptome analysis of Peromyscus leucopus and C3H mice infected with the Lyme disease pathogen. Front. Cell. Infect. Microbiol. 13:1115350. doi: 10.3389/fcimb.2023.1115350
Received: 03 December 2022; Accepted: 23 March 2023;
Published: 11 April 2023.
Edited by:
Armin Alaedini, Columbia University, United StatesReviewed by:
Maria Kazimirova, Slovak Academy of Sciences, SlovakiaAlan Barbour, University of California, Irvine, United States
Yi-Pin Lin, Wadsworth Center, United States
Copyright © 2023 Gaber, Mandric, Nitirahardjo, Piontkivska, Hillhouse, Threadgill, Zelikovsky and Rogovskyy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Artem S. Rogovskyy, YXJvZ292c2t5eUB0YW11LmVkdQ==
 Alhussien M. Gaber
Alhussien M. Gaber Igor Mandric
Igor Mandric Caroline Nitirahardjo3
Caroline Nitirahardjo3 Helen Piontkivska
Helen Piontkivska Andrew E. Hillhouse
Andrew E. Hillhouse David W. Threadgill
David W. Threadgill Alex Zelikovsky
Alex Zelikovsky Artem S. Rogovskyy
Artem S. Rogovskyy