
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 03 August 2023
Sec. Clinical Microbiology
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1112229
This article is part of the Research Topic Clinical metagenomics-based diagnostics for infectious diseases View all 25 articles
Background: Infectious disease is a large burden on public health globally. Metagenomic next-generation sequencing (mNGS) has become popular as a new tool for pathogen diagnosis with numerous advantages compared to conventional methods. Recently, research on mNGS increases yearly. However, no bibliometric analysis has systematically presented the full spectrum of this research field. Therefore, we reviewed all the publications associated with this topic and performed this study to analyze the comprehensive status and future hotspots of mNGS for infectious disease diagnosis.
Methods: The literature was searched in the Web of Science Core Collection and screened without year or language restrictions, and the characteristics of the studies were also identified. The outcomes included publication years, study types, journals, countries, authorship, institutions, frontiers, and hotspots with trends. Statistical analysis and visualization were conducted using VOSviewer (version 1.6.16) and CiteSpace (version 6.1. R3).
Results: In total, 325 studies were included in the analysis after screening. Studies were published between 2009 and 2022 with a significantly increasing number from 1 to 118. Most of the studies were original articles and case reports. Frontiers in Cellular and Infection Microbiology and Clinical Infectious Disease were the most commonly cited and co-cited journals. Institutions and researchers from China contributed the most to this field, followed by those from the USA. The hotspots and frontiers of these studies are pneumonia, tuberculosis, and central nervous system infections.
Conclusion: This study determined that mNGS is a hot topic in the diagnosis of infectious diseases with development trends and provides insights into researchers, institutions, hotspots and frontiers in mNGS, which can offer references to related researchers and future research.
As a series of common and frequently occurring diseases, infectious diseases have always been a heavy burden to global health due to their high morbidity and mortality rate, especially in developing countries (McArthur, 2019). First, some ancient infectious diseases, such as tuberculosis (TB) caused by mycobacterium tuberculosis (MTB), have plagued mankind for thousands of years, and a quarter of the global population is infected with MTB (Kherabi et al., 2022). Additionally, some explosive epidemics, including seasonal influenza and COVID-19, have caused great losses in health, finances, and emotions (Calabrò et al., 2022; Schäfer et al., 2022; Xie et al., 2022). Precise pathogen diagnosis is a key part of infectious disease management. However, rare pathogens, emerging infectious diseases, and the prevalence of drug resistance present huge challenges to the diagnosis and further management (Mancuso et al., 2021; Vitiello, 2022).
Etiology is the gold standard for infectious disease diagnosis (Esposito, 2016). The detection technology of the traditional etiological method is culture with high positive predictive value. However, the limitations, including narrow pathogen spectrum, low positive rate, and long detection cycle, are nonnegligible. These drawbacks may lead to delays in diagnosis, the irrational use of antibiotics, etc., which can result in poor prognosis (Carbo et al., 2021; Chen et al., 2022). Metagenomic next-generation sequencing (mNGS), based on high-throughput sequencing technology, is a burgeoning unbiased pathogen detection method (Chiu and Miller, 2019; Gu et al., 2019). mNGS mainly consists of nucleic acid extraction, library preparation, host sequence exclusion and pathogen sequence enrichment (Jia et al., 2021). It can characterize all DNA or RNA in samples and analyze the entire microbiome, human host genome, and transcriptome in clinical samples (Chiu and Miller, 2019). The advantages, such as high throughput, wide coverage, high accuracy, and efficiency have brought new light to clinical use. According to the study by Duan et al., mNGS presented significantly higher sensitivity than the traditional culture method (67.4% vs 23.6%, p < 0.001) and similar specificity (68.8% vs 81.3%; p = 0.41) (Duan et al., 2021). Furthermore, mNGS is more effective than the conventional method that it required 3 days to identify 67.23% of cases of tuberculosis (TB), whereas 49.58% detected using conventional methods required over 90 days (Shi et al., 2020). Additionally, it can markedly increase the detection rate for some rare pathogens with a low positive rate of culture or insufficient clinical precedents (Simner et al., 2018). Therefore, it has been successfully applied in the diagnosis and treatment of difficult and critical infectious diseases, the identification of unknown pathogens, drug resistance gene monitoring, and epidemiological tracking investigations (Chiu and Miller, 2019; Han et al., 2019).
Bibliometrics is a new cross-science approach that can analyze all knowledge carriers using mathematical and statistical methods (Blakeman, 2018). It has been widely used to guide researchers in specific fields via the quantitative research assessment of academic output to improve research efficiency (Tang et al., 2018; Yeung et al., 2018). Citation analysis is the core part of bibliometric analysis because the number of citations can reflect the impact of an article to a certain extent (Feijoo et al., 2014; Xiong et al., 2022). Many medical articles have explored certain research fields via bibliometric analysis have been published, including diabetes (Zhao et al., 2016), tuberculosis (Xiong et al., 2022), and vaccines(Zhang et al., 2019). With the widespread use of mNGS, the number of studies about this topic has increased in recent years. However, there is no study concerning the overview of mNGS in pathogen diagnosis based on bibliometric analysis. Thus, we performed this study to identify the current status, future hotspots, and frontiers of mNGS by analyzing the publications from core collection in Web of Science (WoSCC).
We extracted literature from the Science Citation Index database in the WoSCC via the Sichuan University Library website. The search strategy was “(TI= (metagenomic next-generation sequencing)) OR TI= (mNGS),” and we downloaded all data within one day on October 1, 2022. There were no restrictions on language or publication time to ensure comprehensive search results. Two researchers (SKH and JMF) screened the literature by excluding meeting abstracts, letters, book chapters, corrections, and articles not related to this topic. If two investigators have a discrepancy, the third author (LYC) will be consulted.
Two researchers (SKH and DL) extracted data from all the included studies. The extracted data included the title, abstract, keywords, references, source journal, publication date, total citations of all databases, authors with affiliation, country, and journal impact factor (IF). The institutions and countries were counted based on the corresponding authors. Journal IFs were obtained from Journal Citation Reports (JCR) 2022 to reflect the academic influence. Two researchers (NW and LYC) from the Center of Infectious Diseases in West China Hospital analyzed and screened the keywords, excluding some words without significant relevance, and merged synonyms (e.g., pcr and PCR). The third investigator (LYC) will be consulted when divergence occurs.
Microsoft (version 2022) was used to organize the publications and analyze their basic characteristics. The number of publications and citations annually were plotted by Stata (version 16.0). Bibliometric visualization was completed by VOSviewer (version 1.6.16) and CiteSpace (version 6.1. R3), including the journals, authors, countries, affiliations, and keywords in this research field regarding the use of mNGS in pathogen detection. VOSviewer was applied in co-citation and co-occurrence analysis. In the collaborative network map, each node represented one element (i.e., journal, country, author), and the size of each circle was weighted using document numbers and the line in the visualization reflected the relatedness of links. In the density map of keywords com-occurrence analysis, the colors ranged from blue to green to yellow. Yellow areas represented research hotspots and directions in this field. The “author keywords” was set in co-occurrence analysis. CiteSpace was applied to track the trend of keywords. The significance test was not used for statistical analysis because no control was involved.
In total, 378 studies were conducted. After excluding meeting abstracts (26), letters (17), book chapters (4), corrections (4), and articles unrelated to the topic (2), we finally included 325 studies for further analysis (Figure 1). The study types (i.e., article, review, case report) were counted and the distribution is shown in Figure 2. Original article (n=214) and case report (n=96) accounted for the majority (65.8% and 29.5%). All the studies were published over 13 years from 2009 to 2022, and the number of publications increased during the past years, with an upwards trend (Figure 3A). The annual citation account was also conducted. From 2013 to 2020, the total citations increased significantly, which is consistent with the annual publications (Figure 3B). The top 20 most cited studies with details are displayed in Table 1.
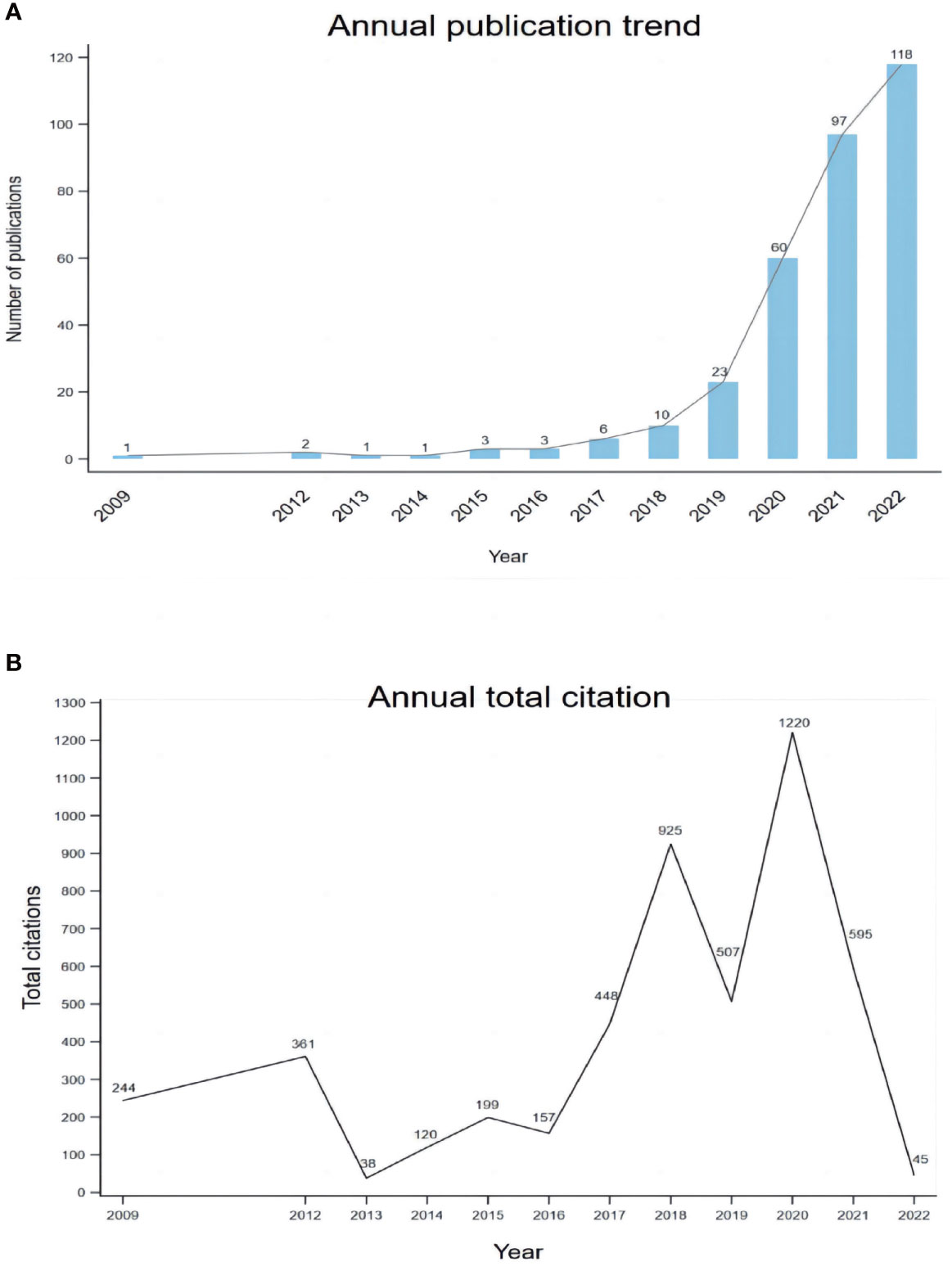
Figure 3 The trend of publications on mNGS in infectious diseases diagnosis from 2009 to 2022 (A) and the annual number of total citations (B).
A total of 325 publications were published in 114 journals, and the top 10 cited journals are listed in Table 2. Most were published in Frontiers in Cellular and Infection Microbiology (n=28), followed by Frontiers in Medicine (n=22), Infection and Drug Resistance (n=21), BMC Infectious Diseases (n=20), Frontiers in Microbiology (n=20), International Journal of Infectious Diseases (n=15), Journal of Clinical Microbiology (n=9), Frontiers in Public Health (n=8), Annals of Translational Medicine (n=7), BMC Pulmonary Medicine (n=7), and PLoS One (n=7). The journals’ IFs ranged from 11.677 to 3.320. The collaborative network of journals is shown in Figure 4A.
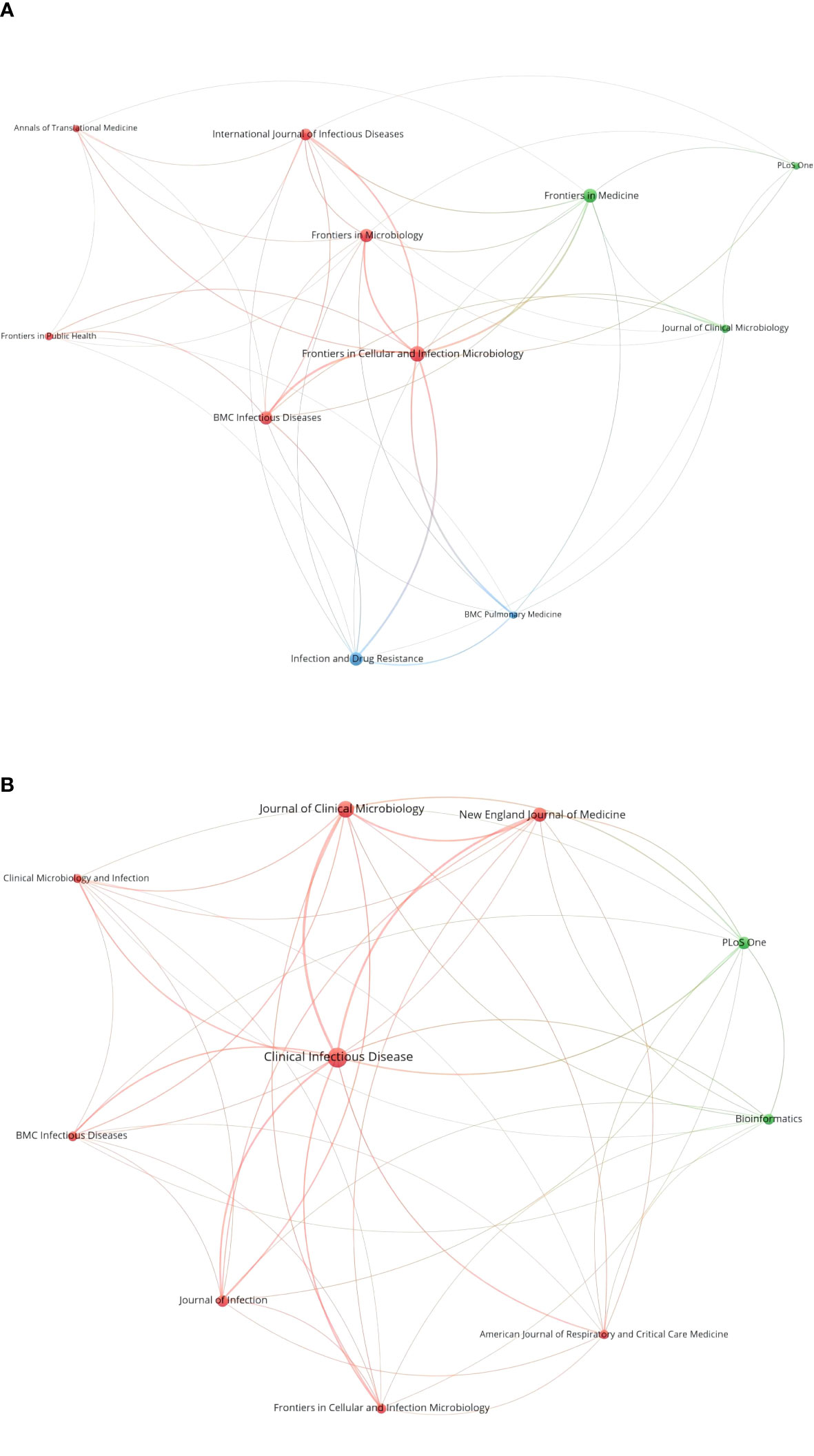
Figure 4 Collaborative network of the top 10 cited journals (A) and the top 10 co-cited journals (B).
The top 10 co-cited journals are listed in Table 2. Clinical Infectious Diseases ranked first with 604 co-citations, followed by Journal of Clinical Microbiology (390), New England Journal of Medicine (288), PLoS One (241), and Journal of Infection (203). The network map of co-cited journals is shown in Figure 4B.
We list the top 11 countries based on the corresponding authors who published at least two studies (Table 3). China occupies the majority of the contributing countries (70%), followed by the USA and France, respectively. In addition, India, the Netherlands, Switzerland, Japan, Brazil, England, Vietnam, and Germany also contributed significantly to this field. According to the total citations, England ranked the first, followed by the USA, France, and China. The collaborative network of contributing countries is shown in Figure 5. The strength of the lines shows that the USA connected with most countries and had a great impact on their research. People R China and England follow the USA with similar strengths.
In Table 4, the institutions with a large number of publications are listed. Eight institutions (based on the corresponding authors) contributed more than 10 studies, from People R China (7) and the USA (1). The top five institutions are Zhejiang University (20), Fudan University (11), Fujian Medical University (10), Tianjin Medical University (10), and Zhengzhou University (10). The collaborative network is displayed in Figure 6
Figure 7A presents the collaborative network of authors (authors who have published more than five studies) in the publications. Most of them were from China. These authors are divided into groups with quite a few collaborative works. The top 22 authors with large numbers of publications (≥5) are listed in Table 5. Han Xia from Hugobiotech in China ranked first with 14 publications, followed by Jing Feng (9), Charles Y Chiu (8), Bin Yang (8), Qing Miao (7), Hui Wang (7), and so on. In Figure 7B, the co-citation network map of authors is presented.
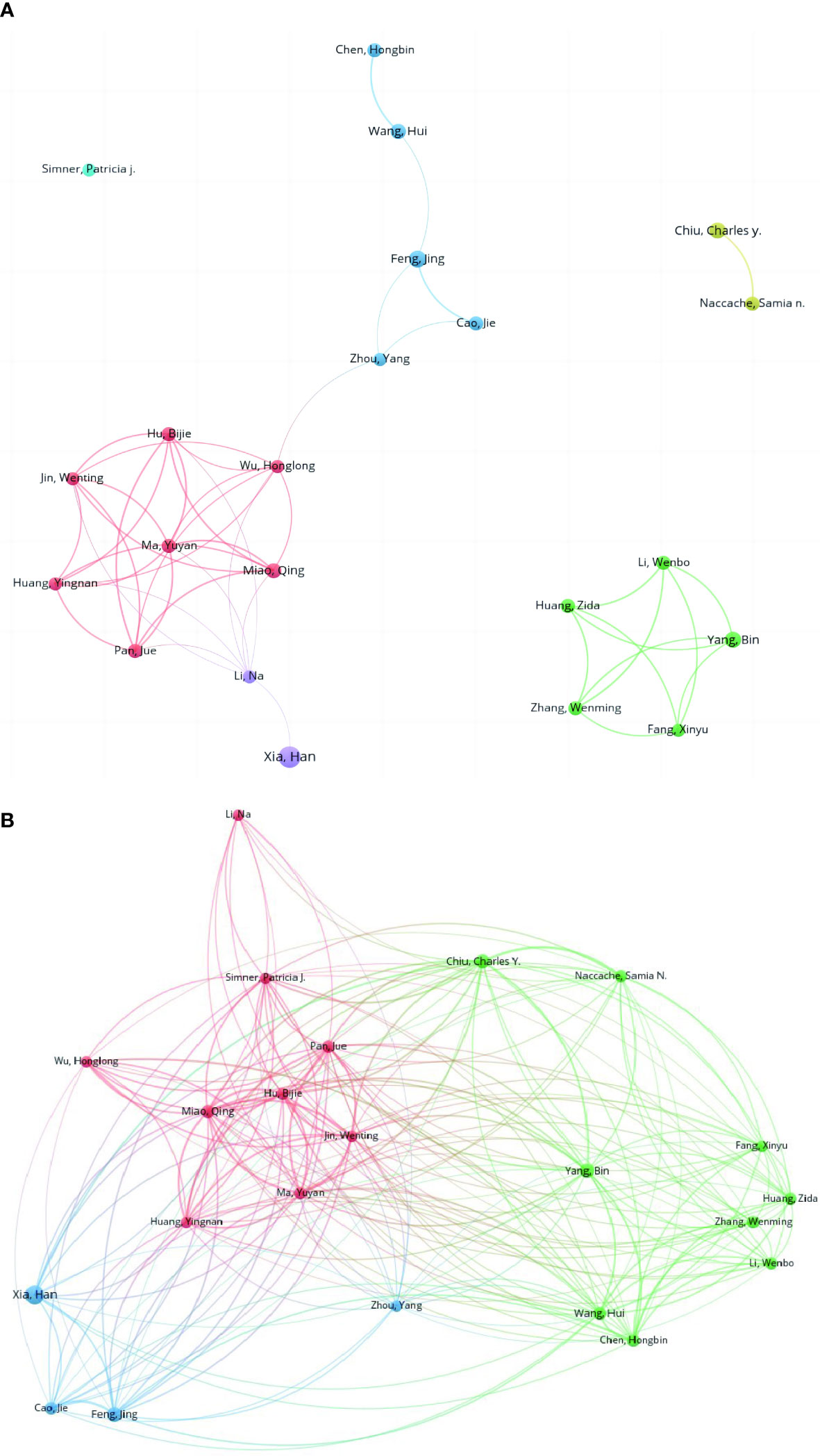
Figure 7 Collaborative network of the co-authorship (A) and co-citation relationships of authors (B).
Twenty-four keywords were retained and their co-occurrence frequencies were calculated. The keyword co-occurrence network map is shown in Figure 8A. From the density map of keyword co-occurrence in Figure 8B, the keywords mainly focused on diagnosis, infection, encephalitis, community-acquired pneumonia, viruses, pathogens, pneumonia, children, cerebrospinal fluid, MTB, and meningitis.
Figure 9 presents the top 14 keywords with the strongest citation burst. “Genome”, “Encephalitis”, “Next-generation sequencing” and “Virus” were listed as the top 4 keywords with citation bursts of more than 3.50. The trend of some screened keywords and shows that some of them have been constantly heated thus far, such as genome and virus, while some of them have been focused on in recent years and can be considered as frontiers in the field, such as “children” (2017-2020), “meningoencephalitis” (2019-2020), “Mycobacterium tuberculosis (2019-2020)” and “cerebrospinal fluid (2019-2020)”. There are similarities between hotspots and frontiers.
Pathogenic infection is a leading cause of disease and remains a severe problem for public health, which is involved in more than 20% of deaths per year globally (GBD 2019 Antimicrobial Resistance Collaborators, 2022). To accurately identify pathogens and precisely treat the disease, mNGS was initiated at the right moment. The utility of mNGS in providing clinically actionable information was first approved in a 14-year-old boy with neuroleptospirosis in 2014 (Wilson et al., 2014). At present, an increasing number of clinical cohort studies and case reports have confirmed the value of mNGS in pathogen diagnosis (Chiu and Miller, 2019; Li et al., 2021). However, the studies on mNGS are mostly comprised of case reports and cohort studies. To the best of our knowledge, this is the first bibliometric analysis focused on mNGS in pathogen detection.
The number of publications significantly increased since 2014. After the first clinical use, a large number of cases have been diagnosed using the new tool, and as a result, more studies were published. In addition to pathogen diagnosis, the use of mNGS in other fields also contributed to the increasing number of publications, such as the surveillance of antimicrobial resistance in the food supply (Oniciuc et al., 2018), prediction of the cause of infection, and evaluation of risk (Gliddon et al., 2018; Langelier et al., 2018). The total citations had a similar trend to the publications. However, there are some fluctuations that may be caused by some extremely highly cited studies in certain years. The classification of the study types revealed that researchers mainly focus on the utility of mNGS in some notable cases and assess the diagnostic performance in some infectious diseases via cohort studies, such as TB and meningitis. With the large number of original articles, some reviews and meta-analysis will be published to summarize the development or efficacy of mNGS in the future.
The studies were mostly published in journals related to general and integrative medicine (Frontiers in Medicine, Annals of Translational Medicine) and infectious diseases (Frontiers in Cellular and Infection Microbiology, Infection and Drug Resistance, BMC Infectious Diseases). This illustrates that the application of mNGS is currently limited in infectious diseases now. Frontiers in Cellular and Infection Microbiology was the most popular cited journal, and also ranked 6th in the co-cited journals, which means it is vital in this field. Clinical Infectious Diseases ranked first in co-cited journals with high IF (20.999). The average IF of the top 10 popular journals was 5.948 and that of the top 10 co-cited journals was 31.165. More articles can be published in more fields in the future based on these high-quality references.
Our study showed that the publications are from many countries. However, People R China and the USA showed such a powerful influence that publications from People R China accounted for approximately 70% of the total. This may be related to the epidemiology in China. Although China has achieved great economic, public health, and healthcare development after the Severe Acute Respiratory Syndrome outbreak in 2003, there are still various infectious diseases (Lu et al., 2016; Liu et al., 2018). Second, owing to the complex ecological environment, the spectrum of pathogens is broad and includes many rare pathogens. Since mNGS is effective for pathogen identification and can aid doctors in rapid and accurate diagnosis, it has attracted a great deal of attention with plenty of literature published in China. The USA is the birthplace of mNGS for pathogen detection and has lead to extensive cohort studies to examine the efficacy. Furthermore, the USA and People R China have a strong relationship in cooperation with other countries. It can be speculated that mNGS is widely clinically applied in China, and the basic molecular mechanism has been studied extensively in the USA. Additionally, according to the average citations, England ranked the top. The study published in 2009 by Adams et al. from England was the first study to detect pathogens using mNGS in the world with 244 citations, which lead to the highest average citations of England (Adams et al., 2009). With the effort of many countries, mNGS can be improved for better application.
Considering the research institutions, Zhejiang University, Fudan University, and BGI in China contributed significantly to this field, while the University of California San Francisco in the USA also played a key role. It can be concluded that collaboration among authors is relatively fixed and independent. This is possibly because mNGS is used for detecting rare pathogens, which are distributed regionally, such as leishmaniasis in the central and western regions of China (Tang et al., 2022), hepatic echinococcosis in cattle-producing areas, and Dengue fever in tropical areas (Ren et al., 2022). Furthermore, mNGS is still in the development stage and there is currently no deep cross-team collaboration at the moment, and different teams have different research focuses. However, the lack of communication and cooperation among institutions certainly limited the development in this field. Therefore, the exchanges of research findings and more cooperation are essential and the academic barriers should be reduced to promote the development of mNGS.
Based on the results of author analysis, authors from China occupy the vast majority, which is similar to the distribution of countries and institutions. Xia Han from Hugobiotech is the most productive with 14 publications, followed by Feng Jing from Beijing Chaoyang Hospital with 9. Chiu Charles from the University of California San Francisco is one the most influential researches in this field. He is the corresponding author of the first case report of mNGS by Wilson et al. which was published in New England Journal of Medicine as a milestone. Then, a clinical trial investigating the detection efficacy of mNGS in body fluids was published in Nature Medicine with a high IF (87.241) and citation (Gu et al., 2021).
Keywords can reflect the core contents and topics of the studies. Through the keyword analysis, the results showed that pneumonia, tuberculosis, central nervous system (CNS) infection, and children had a high frequency or upwards trend.
Pneumonia is one of the most common disease manifestations in respiratory infection and the leading cause of death, especially community acquired pneumonia (Biscevic-Tokic et al., 2013; Wunderink and Waterer, 2017; Wang et al., 2022). Since a variety of pathogens can cause pneumonia and some of the symptoms are nonspecific, the detection rate of traditional culture combined with clinical experience is still low and ineffective (Schlaberg et al., 2017; Zhu et al., 2018). mNGS has shown better diagnostic efficiency for pneumonia and other respiratory system infections (Hogan et al., 2021; Zhan et al., 2021). Chen et al. demonstrated that mNGS had high diagnostic accuracy in bronchoalveolar lavage fluid (BALF) with 78% sensitivity and 77% specificity (Chen et al., 2022). In a retrospective study focused on mixed respiratory infection cases, compared with the conventional method, the sensitivity was significantly higher (97.2% vs 13.9%, p < 0.001). However, the specificity was the opposite (63.2% vs 94.7%; p = 0.07) (Wang et al., 2019). Besides, the prevalence of coronavirus disease 2019 (COVID-19) has also contributed to making pneumonia a hotspot in recent years. This novel coronavirus was identified by RNA-based mNGS and the genome was revealed to provide insights for future research (Chen et al., 2020a). Additionally, acute respiratory distress syndrome (ARDS) in severe pneumonia, such as COVID-19, is critical and rapidly progresses, resulting in high morality and poor prognosis for survivors (Griffiths et al., 2019). mNGS is confirmed to be a valuable tool to improve the management of ARDS and provide better quality of life for survivors (Zhang et al., 2020). In summary, mNGS is a promising technique to detect pathogens in pneumonia and other respiratory infections.
Similar to pneumonia, TB caused by MTB is another hot topic in mNGS. MTB diagnosis has been a challenge for decades. Because of the specific biological properties of MTB, the positive rate of culture is low and takes a long time (Wang et al., 2020; Floyd et al., 2022). Thus, the diagnosis of TB is a problem that usually leads to TB-related death. mNGS was tested in pulmonary TB and showed outstanding performance in diagnosis with high sensitivity and specificity (Chen et al., 2020b). In addition, the multiorgan lesions also contribute to diagnosis difficulty, especially extrapulmonary TB (EPTB) (Suárez et al., 2019). Pang et al. revealed that the positive rate for EPTB by traditional tests was only 12.8% (Pang et al., 2019). A retrospective study in China assessed the clinical efficacy of mNGS in EPTB diagnosis and found that the positive rate was significantly higher than other routine methods (p < 0.001). Another meta-analysis also proved that mNGS is more effective than traditional tests in TB meningitis (Yu et al., 2020). However, the detection threshold and the efficacy in TB-related coinfection are still controversial and deserve further research (Shi et al., 2020). All of the evidence indicated that mNGS is a potential resolution for TB diagnosis and more research is necessary in the future.
CNS infections are life-threatening, usually resulting in poor prognosis (Yu et al., 2022). Because of the lack of specificity in clinical presentation and cerebrospinal fluid (CSF) parameters, as well as the low pathogen content in CSF, diagnosis remains a challenge (Leonard, 2017; Yu et al., 2022). Although the utility of mNGS matures in some samples such as blood and BALF, it remains unclear whether mNGS has better diagnostic efficiency in CSF. A meta-analysis focused on mNGS for bacterial meningoencephalitis elucidated that the diagnostic efficacy is satisfactory with an estimated AUC (area under curve) of the summary receiver operating characteristic curve of 0.91 (Kanaujia et al., 2022). However, since available evidence is divided and limited, more cohort studies or meta-analysis are expected to eliminate the discrepancy and present more comprehensive and reliable conclusions (Chen et al., 2021).
For children or pediatrics, it is considered another frontier. Infectious disease is the leading cause of death in children, especially those under 5 years old. Because of the various pathogens, occult onset, atypical clinical symptoms, and rapid progression, pediatric infection is difficult to diagnose. Timely and accurate tools are necessary for the selection of effective medical interventions to improve the prognosis (Tao et al., 2022). Many studies elucidated that mNGS can provide useful information for suspected cases and identify infection or noninfection (Yan et al., 2021; Tao et al., 2022). Nowadays, large number of case report have demonstrated the successful use of mNGS in pediatric infection. However, the overall diagnostic performance has not been validated, which is worth illustrating in the future studies.
Summarily, all the hotspots and frontiers indicate that mNGS is a potential test and can be widely used.
This bibliometric analysis presents the status of mNGS in pathogen detection, including journals, countries, contributors, institutions, research hotspots, and frontiers via data visualization, and the studies in this field can be rapidly accessed from the Supplementary Material. Scholars in the field can quickly understand the status by reading this study and contribute more to this field.
Our study has some limitations. First, the citation analysis was based on the Web of Science Core Collection, which might have missed some important literature indexed by other databases, such as Google Scholar and Scopus, resulting in biased results. Second, we used an accurate title search which means that a small number of publications that did not mention metagenomic next-generation sequencing may not have been included. Third, since bibliometrics includes several secondary studies (such as reviews), the keywords and research focus of the secondary research may be different from those of the original studies, which may also lead to bias. In addition, due to the purpose of the study, we can only describe the overview of this research field without a detailed mechanism.
In conclusion, this study determined that mNGS has become a current research hotspot and related research has grown exponentially. This study shows the research hotspots in mNGS, research institutions, and researchers of most related research that can provide useful references for workers in infectious diseases in the future.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Conceptualization, YX. Methodology, SH, JW, JF. Software, SH, JW. Formal Analysis, YX, DL. Writing – Original Draft Preparation, SH. Writing – Review and Editing, YX, LC. All authors contributed to the article and approved the submitted version.
We thank all the authors who contributed to this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1112229/full#supplementary-material
Supplementary Table 1 | The 325 studies from the Web of Science Core Collection.
Adams, I. P., Glover, R. H., Monger, W. A., Mumford, R., Jackeviciene, E., Navalinskiene, M., et al. (2009). Next-generation sequencing and metagenomic analysis: a universal diagnostic tool in plant virology. Mol. Plant Pathol. 10, 537–545. doi: 10.1111/j.1364-3703.2009.00545.x
Biscevic-Tokic, J., Tokic, N., Musanovic, A. (2013). Pneumonia as the most common lower respiratory tract infection. Med. Arch. 67, 442–445. doi: 10.5455/medarh.2013.67.442-445
Blakeman, K. (2018). Bibliometrics in a digital age: help or hindrance. Sci. Prog. 101, 293–310. doi: 10.3184/003685018X15337564592469
Calabrò, G. E., D’Ambrosio, F., Fallani, E., Ricciardi, W. (2022). Influenza vaccination assessment according to a value-based health care approach. Vaccines (Basel) 10, 1675. doi: 10.3390/vaccines10101675
Carbo, E. C., Blankenspoor, I., Goeman, J. J., Kroes, A. C. M., Claas, E. C. J., De Vries, J. J. C. (2021). Viral metagenomic sequencing in the diagnosis of meningoencephalitis: a review of technical advances and diagnostic yield. Expert Rev. Mol. Diagn. 21, 1139–1146. doi: 10.1080/14737159.2021.1985467
Chen, S., Kang, Y., Li, D., Li, Z. (2022). Diagnostic performance of metagenomic next-generation sequencing for the detection of pathogens in bronchoalveolar lavage fluid in patients with pulmonary infections: Systematic review and meta-analysis. Int. J. Infect. Dis. 122, 867–873. doi: 10.1016/j.ijid.2022.07.054
Chen, L., Liu, W., Zhang, Q., Xu, K., Ye, G., Wu, W., et al. (2020a). RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 9, 313–319. doi: 10.1080/22221751.2020.1725399
Chen, P., Sun, W., He, Y. (2020b). Comparison of metagenomic next-generation sequencing technology, culture and GeneXpert MTB/RIF assay in the diagnosis of tuberculosis. J. Thorac. Dis. 12, 4014–4024. doi: 10.21037/jtd-20-1232
Chen, J., Zhang, R., Liu, L., Qi, T., Wang, Z., Song, W., et al. (2021). Clinical usefulness of metagenomic next-generation sequencing for the diagnosis of central nervous system infection in people living with HIV. Int. J. Infect. Dis. 107, 139–144. doi: 10.1016/j.ijid.2021.04.057
Chiu, C. Y., Miller, S. A. (2019). Clinical metagenomics. Nat. Rev. Genet. 20, 341–355. doi: 10.1038/s41576-019-0113-7
Duan, H., Li, X., Mei, A., Li, P., Liu, Y., Li, X., et al. (2021). The diagnostic value of metagenomic next⁃generation sequencing in infectious diseases. BMC Infect. Dis. 21, 62. doi: 10.1186/s12879-020-05746-5
Esposito, S. (2016). Infectious diseases: pathophysiology, diagnostics and prevention. Int. J. Mol. Sci. 17, E1464. doi: 10.3390/ijms17091464
Feijoo, J. F., Limeres, J., Fernández-Varela, M., Ramos, I., Diz, P. (2014). The 100 most cited articles in dentistry. Clin. Oral. Investig. 18, 699–706. doi: 10.1007/s00784-013-1017-0
Floyd, S., Klinkenberg, E., de Haas, P., Kosloff, B., Gachie, T., Dodd, P. J., et al. (2022). Optimising Xpert-Ultra and culture testing to reliably measure tuberculosis prevalence in the community: findings from surveys in Zambia and South Africa. BMJ Open 12, e058195. doi: 10.1136/bmjopen-2021-058195
GBD 2019 Antimicrobial Resistance Collaborators (2022). Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 400, 2221–2248. doi: 10.1016/S0140-6736(22)02185-7
Gliddon, H. D., Herberg, J. A., Levin, M., Kaforou, M. (2018). Genome-wide host RNA signatures of infectious diseases: discovery and clinical translation. Immunology 153, 171–178. doi: 10.1111/imm.12841
Griffiths, M. J. D., McAuley, D. F., Perkins, G. D., Barrett, N., Blackwood, B., Boyle, A., et al. (2019). Guidelines on the management of acute respiratory distress syndrome. BMJ Open Respir. Res. 6, e000420. doi: 10.1136/bmjresp-2019-000420
Gu, W., Deng, X., Lee, M., Sucu, Y. D., Arevalo, S., Stryke, D., et al. (2021). Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat. Med. 27, 115–124. doi: 10.1038/s41591-020-1105-z
Gu, W., Miller, S., Chiu, C. Y. (2019). Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. 14, 319–338. doi: 10.1146/annurev-pathmechdis-012418-012751
Han, D., Li, Z., Li, R., Tan, P., Zhang, R., Li, J. (2019). mNGS in clinical microbiology laboratories: on the road to maturity. Crit. Rev. Microbiol. 45, 668–685. doi: 10.1080/1040841X.2019.1681933
Hogan, C. A., Yang, S., Garner, O. B., Green, D. A., Gomez, C. A., Dien Bard, J., et al. (2021). Clinical impact of metagenomic next-generation sequencing of plasma cell-free DNA for the diagnosis of infectious diseases: A multicenter retrospective cohort study. Clin. Infect. Dis. 72, 239–245. doi: 10.1093/cid/ciaa035
Jia, X., Hu, L., Wu, M., Ling, Y., Wang, W., Lu, H., et al. (2021). A streamlined clinical metagenomic sequencing protocol for rapid pathogen identification. Sci. Rep. 11, 4405. doi: 10.1038/s41598-021-83812-x
Kanaujia, R., Biswal, M., Angrup, A., Ray, P. (2022). Diagnostic accuracy of the metagenomic next-generation sequencing (mNGS) for detection of bacterial meningoencephalitis: a systematic review and meta-analysis. Eur. J. Clin. Microbiol. Infect. Dis. 41, 881–891. doi: 10.1007/s10096-022-04445-0
Kherabi, Y., Tunesi, S., Kay, A., Guglielmetti, L. (2022). Preventive therapy for contacts of drug-resistant tuberculosis. Pathogens 11, 1189. doi: 10.3390/pathogens11101189
Langelier, C., Kalantar, K. L., Moazed, F., Wilson, M. R., Crawford, E. D., Deiss, T., et al. (2018). Integrating host response and unbiased microbe detection for lower respiratory tract infection diagnosis in critically ill adults. Proc. Natl. Acad. Sci. U.S.A. 115, E12353–E12362. doi: 10.1073/pnas.1809700115
Leonard, J. M. (2017). Central nervous system tuberculosis. Microbiol. Spectr. 5. doi: 10.1128/microbiolspec.TNMI7-0044-2017
Li, N., Cai, Q., Miao, Q., Song, Z., Fang, Y., Hu, B. (2021). High-throughput metagenomics for identification of pathogens in the clinical settings. Small Methods 5, 2000792. doi: 10.1002/smtd.202000792
Liu, Q., Xu, W., Lu, S., Jiang, J., Zhou, J., Shao, Z., et al. (2018). Landscape of emerging and re-emerging infectious diseases in China: impact of ecology, climate, and behavior. Front. Med. 12, 3–22. doi: 10.1007/s11684-017-0605-9
Lu, J., Milinovich, G. J., Hu, W. (2016). A brief historical overview of emerging infectious disease response in China and the need for a One Health approach in future responses. One Health 2, 99–102. doi: 10.1016/j.onehlt.2016.07.001
Mancuso, G., Midiri, A., Gerace, E., Biondo, C. (2021). Bacterial antibiotic resistance: the most critical pathogens. Pathogens 10, 1310. doi: 10.3390/pathogens10101310
McArthur, D. B. (2019). Emerging infectious diseases. Nurs. Clin. North Am. 54, 297–311. doi: 10.1016/j.cnur.2019.02.006
Oniciuc, E. A., Likotrafiti, E., Alvarez-Molina, A., Prieto, M., Santos, J. A., Alvarez-Ordóñez, A. (2018). The present and future of whole genome sequencing (WGS) and whole metagenome sequencing (WMS) for surveillance of antimicrobial resistant microorganisms and antimicrobial resistance genes across the food chain. Genes (Basel) 9, E268. doi: 10.3390/genes9050268
Pang, Y., An, J., Shu, W., Huo, F., Chu, N., Gao, M., et al. (2019). Epidemiology of extrapulmonary tuberculosis among inpatients, China 2008-2017. Emerg. Infect. Dis. 25, 457–464. doi: 10.3201/eid2503.180572
Ren, Z.-Z., Zheng, Y., Sun, T., Wang, G.-Y., Chen, X.-M., Zhou, Y.-M. (2022). A survey of clinical and laboratory characteristics of the dengue fever epidemic from 2017 to 2019 in Zhejiang, China. Med. (Baltimore) 101, e31143. doi: 10.1097/MD.0000000000031143
Schäfer, S. K., Kunzler, A. M., Kalisch, R., Tüscher, O., Lieb, K. (2022). Trajectories of resilience and mental distress to global major disruptions. Trends Cognit. Sci. 1171–1189. doi: 10.1016/j.tics.2022.09.017.
Schlaberg, R., Chiu, C. Y., Miller, S., Procop, G. W., Weinstock, G., Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology, et al. (2017). Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch. Pathol. Lab. Med. 141, 776–786. doi: 10.5858/arpa.2016-0539-RA
Shi, C.-L., Han, P., Tang, P.-J., Chen, M.-M., Ye, Z.-J., Wu, M.-Y., et al. (2020). Clinical metagenomic sequencing for diagnosis of pulmonary tuberculosis. J. Infect. 81, 567–574. doi: 10.1016/j.jinf.2020.08.004
Simner, P. J., Miller, S., Carroll, K. C. (2018). Understanding the promises and hurdles of metagenomic next-generation sequencing as a diagnostic tool for infectious diseases. Clin. Infect. Dis. 66, 778–788. doi: 10.1093/cid/cix881
Suárez, I., Fünger, S. M., Kröger, S., Rademacher, J., Fätkenheuer, G., Rybniker, J. (2019). The diagnosis and treatment of tuberculosis. Dtsch Arztebl Int. 116, 729–735. doi: 10.3238/arztebl.2019.0729
Tang, C., He, S., Wang, N., Chen, L. (2022). A case report and literature review: diagnosis and treatment of human immunodeficiency virus coinfected with visceral leishmania by metagenomic next-generation sequencing in China. Ann. Transl. Med. 10, 497. doi: 10.21037/atm-22-1351
Tang, H., Huang, W., Ma, J., Liu, L. (2018). SWOT analysis and revelation in traditional Chinese medicine internationalization. Chin. Med. 13, 5. doi: 10.1186/s13020-018-0165-1
Tao, Y., Yan, H., Liu, Y., Zhang, F., Luo, L., Zhou, Y., et al. (2022). Diagnostic performance of metagenomic next-generation sequencing in pediatric patients: A retrospective study in a large children’s medical center. Clin. Chem. 68, 1031–1041. doi: 10.1093/clinchem/hvac067
Vitiello, A. (2022). Sars-Cov-2 and risk of antiviral drug resistance. Ir J. Med. Sci. 191, 2367–2368. doi: 10.1007/s11845-021-02820-y
Wang, J., Han, Y., Feng, J. (2019). Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm Med. 19, 252. doi: 10.1186/s12890-019-1022-4
Wang, W.-H., Takeuchi, R., Jain, S.-H., Jiang, Y.-H., Watanuki, S., Ohtaki, Y., et al. (2020). A novel, rapid (within hours) culture-free diagnostic method for detecting live Mycobacterium tuberculosis with high sensitivity. EBioMedicine 60, 103007. doi: 10.1016/j.ebiom.2020.103007
Wang, D., Willis, D. R., Yih, Y. (2022). The pneumonia severity index: Assessment and comparison to popular machine learning classifiers. Int. J. Med. Inf. 163, 104778. doi: 10.1016/j.ijmedinf.2022.104778
Wilson, M. R., Naccache, S. N., Samayoa, E., Biagtan, M., Bashir, H., Yu, G., et al. (2014). Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N Engl. J. Med. 370, 2408–2417. doi: 10.1056/NEJMoa1401268
Wunderink, R. G., Waterer, G. (2017). Advances in the causes and management of community acquired pneumonia in adults. BMJ 358, j2471. doi: 10.1136/bmj.j2471
Xie, X., Zhang, N., Fu, J., Wang, Z., Ye, Z., Liu, Z. (2022). The potential for traditional Chinese therapy in treating sleep disorders caused by COVID-19 through the cholinergic anti-inflammatory pathway. Front. Pharmacol. 13. doi: 10.3389/fphar.2022.1009527
Xiong, Y., Wei, J., Cai, Y., Zhang, Y., Feng, L., Zhang, Y. (2022). Analysis of the research hotspot of drug treatment of tuberculosis: A bibliometric based on the top 50 cited literatures. BioMed. Res. Int. 2022, 9542756. doi: 10.1155/2022/9542756
Yan, G., Liu, J., Chen, W., Chen, Y., Cheng, Y., Tao, J., et al. (2021). Metagenomic next-generation sequencing of bloodstream microbial cell-free nucleic acid in children with suspected sepsis in pediatric intensive care unit. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.665226
Yeung, A. W. K., Heinrich, M., Atanasov, A. G. (2018). Ethnopharmacology-A bibliometric analysis of a field of research meandering between medicine and food science? Front. Pharmacol. 9. doi: 10.3389/fphar.2018.00215
Yu, L., Zhang, Y., Zhou, J., Zhang, Y., Qi, X., Bai, K., et al. (2022). Metagenomic next-generation sequencing of cell-free and whole-cell DNA in diagnosing central nervous system infections. Front. Cell Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.951703
Yu, G., Zhao, W., Shen, Y., Zhu, P., Zheng, H. (2020). Metagenomic next generation sequencing for the diagnosis of tuberculosis meningitis: A systematic review and meta-analysis. PloS One 15, e0243161. doi: 10.1371/journal.pone.0243161
Zhan, Y., Xu, T., He, F., Guan, W.-J., Li, Z., Li, S., et al. (2021). Clinical evaluation of a metagenomics-based assay for pneumonia management. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.751073
Zhang, P., Chen, Y., Li, S., Li, C., Zhang, S., Zheng, W., et al. (2020). Metagenomic next-generation sequencing for the clinical diagnosis and prognosis of acute respiratory distress syndrome caused by severe pneumonia: a retrospective study. PeerJ 8, e9623. doi: 10.7717/peerj.9623
Zhang, Y., Quan, L., Xiao, B., Du, L. (2019). The 100 top-cited studies on vaccine: a bibliometric analysis. Hum. Vaccin Immunother. 15, 3024–3031. doi: 10.1080/21645515.2019.1614398
Zhao, X., Guo, L., Lin, Y., Wang, H., Gu, C., Zhao, L., et al. (2016). The top 100 most cited scientific reports focused on diabetes research. Acta Diabetol. 53, 13–26. doi: 10.1007/s00592-015-0813-1
Keywords: metagenomic next-generation sequencing, pathogen diagnosis, bibliometric analysis, Web of Science, VOSviewer, CiteSpace
Citation: He S, Wei J, Feng J, Liu D, Wang N, Chen L and Xiong Y (2023) The application of metagenomic next-generation sequencing in pathogen diagnosis: a bibliometric analysis based on Web of Science. Front. Cell. Infect. Microbiol. 13:1112229. doi: 10.3389/fcimb.2023.1112229
Received: 30 November 2022; Accepted: 18 July 2023;
Published: 03 August 2023.
Edited by:
Jeyaprakash Rajendhran, Madurai Kamaraj University, IndiaReviewed by:
Jiaxu Liang, The Fifth Clinical Medical College of Henan University of Chinese Medicine, ChinaCopyright © 2023 He, Wei, Feng, Liu, Wang, Chen and Xiong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liyu Chen, MTQ0OTEyOTFAcXEuY29t; Ying Xiong, NjE3MTE0NDVAcXEuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.