- 1Department of Clinical Laboratory, Medical School of Chinese PLA, Beijing, China
- 2Department of Clinical Laboratory, First Medical Center of Chinese PLA General Hospital, Beijing, China
- 3Department of Clinical Laboratory, Xuanhan People’s Hospital, Xuanhan, Sichuan, China
- 4Department of Clinical Laboratory, Hainan Modern Women and Children’s Hospital, Haikou, China
- 5Department of Clinical Microbiology and Immunology, College of Pharmacy and Medical Laboratory, Army Medical University, Chongqing, China
- 6Department of Basic Medical Laboratory, The 980th Hospital of the PLA Joint Logistical Support Force, Bethune International Peace Hospital, Shijiazhuang, China
Objective: Aeromonas caviae (A. caviae) is one of the major etiological agents in human intestinal infections reported to be associated with a broad spectrum of extra-intestinal infections with increasing incidence over recent years. Although previous studies have established its significance as a causative agent of both bloodstream and gastrointestinal infections, the characteristics of A. caviae that cause extra-intestinal infections remain unilluminated.In this single-center retrospective study, we investigated epidemiological characteristics, antimicrobial resistance genes and phenotypes, virulence genes, and phyloevolution of 47 clinical A. caviae isolated from patients with extra-intestinal infections from 2017 to 2020.
Methods: A. caviae strains were identified by biochemical tests and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF/MS), ultimately confirmed to species level by whole-genome sequencing (WGS). Antimicrobial resistance and virulence genes were identified using the Comprehensive Antibiotic Resistance Database (CARD) and the virulence factor database (VFDB), respectively. Phylogenetic analysis of 47 clinical strains was performed by combining with 521 A. caviae strains from NCBI database.
Results: A. caviae was an opportunistic pathogen in immunocompromised patients, especially those with underlying hepatobiliary diseases and malignancies. 19 out of 47 isolates were identified as multidrug resistance (MDR) strains. Piperacillin-tazobactam, levofloxacin, gentamicin, amikacin with a resistance rate of less than 10% remained as options to treat extra-intestinal infections. 24 out of 47 isolates exhibited non-susceptibility to cephalosporins and cephamycins, all of which carried β-lactamase gene, including blaMOX, blaPER-3, blaOXA, blaNDM, and blaCphA. Most stains (98%, 46/47) carried at least one of the virulence genes, but extra-intestinal infections had a low mortality rate. Phylogenetic analysis indicated the risk of nosocomial transmission but revealed no outbreak. However, the emergence of MDR and β-lactamase resistance genes in extra-intestinal isolates of A. caviae is becoming an increasing risk to public health and requires attention.
Conclusions: This study strengthen our understanding of A.caviae isolated from extra-intestinal infections. It may contribute to the management of extra-intestinal infections as well as the prevention and control of drug resistance.
Introduction
Aeromonas spp. belongs to Aeromonadaceae, a kind of Gram-negative, rod-shaped, facultative anaerobic bacteria, which are ubiquitous in the aquatic environment, soil, fish, animals and foodstuffs (Janda & Abbott, 2010; Fernandez-Bravo & Figueras, 2020). More than 30 different Aeromonas species that threaten various environmental niches and human health have been identified (Hoel et al., 2019; Fernandez-Bravo & Figueras, 2020). Aeromonas veronii bv.sobria, Aeromonas hydrophila, and Aeromonas caviae (A. caviae) are pathogenic to humans and are responsible for a broad spectrum of human intestinal and extra-intestinal infections (Janda & Abbott, 2010). Gastroenteritis associated with Aeromonas infection is primarily caused by the consumption of contaminated water or foods. Extra-intestinal infections in humans include biliary tract infection, urinary tract infection, soft-tissue infection, peritonitis, pneumonia, and bacteremia. Previous studies have indicated that Aeromonas causes infections in both immunosuppressed and immunocompetent persons (Dwivedi et al., 2008; Chuang et al., 2011). Individuals with underlying diseases, such as liver cirrhosis, malignancies, and biliary tract diseases, are at a higher risk of infection when exposed to the pathogens (Janda & Abbott, 2010; Chao et al., 2013).
A. caviae has been proved to reside in the human gastrointestinal tract and is considered a significant etiological agent of gastrointestinal infections. Although intestinal infections caused by A. caviae may be self-limited, invasive extra-intestinal infections in immunocompromised individuals or patients with severe underlying diseases may be life-threatening or even fatal. The increasing number of extra-intestinal infections worldwide poses severe threats to public health (Luo et al., 2022; Xu et al., 2022). A study in Japan found A. caviae to be the most frequent pathogen causing Aeromonas bacteremia (Kimura et al., 2013). According to Lamy et al., A. caviae and A.veronii were the most common Aeromonas species causing bacteremia and gastroenteritis in France (Lamy et al., 2009). Chen et al. reported that A. caviae predominated in primary bacteremia and biliary tract infections in southern Taiwan (Chen et al., 2021). However, epidemiological characteristics, antimicrobial resistance profiles, virulence genes and phyloevolution of A. caviae causing extra-intestinal infections remain unclear.
The present study aims to investigate the epidemiological characteristics of extra-intestinal infections caused by A. caviae and to assess the antimicrobial resistance profiles and virulence genes to provide relevant microbiological data as the basis for effective strategies in the prevention and control of drug resistance. Phyloevolution analysis was subsequently conducted to understand the evolutionary path of A. caviae and the relationship between species. 47 clinical A. caviae causing extra-intestinal infections within a 4-year period were collected at the First Medical Center of Chinese PLA General Hospital. Aeromonas spp. were identified, and virulence and resistance genes were detected through whole-genome sequencing (WGS). Phylogenetic characteristics of 568 strains, including 521 strains obtained from the National Center for Biotechnology Information (NCBI) database and 47 clinical strains in our study, were analyzed.
Materials and methods
Data collection
A retrospective study of 47 clinical A. caviae isolated from 46 patients with extra-intestinal infections from 2017 to 2020 at the First Medical Center of Chinese PLA General Hospital, a 3000-bed teaching hospital, was performed. Clinical data of 46 patients, including demographics, source of infection, underlying diseases, microbiological data, and respective outcomes, were retrieved from medical records. Diagnosis of bacteremia, biliary tract infection, urinary tract infection, pneumonia, and peritonitis was based on clinical, bacteriological, and radiological investigations.
Bacterial strains and species identification
Clinical isolates were obtained from patients with A. caviae extra-intestinal infections. The clinical samples of sputum and urine were plated on the blood agar, and then cultured at 37°C for 24 h to obtain the single colony. Blood, ascites, and bile were inoculated into BACTEC culture bottles using the BACT/ALERT 3D system (BioMerieux, Lyon, France). The suspicious Gram-negative bacteria were characterized by positive oxidase test, D-glucose fermentation, motility test, absence of growth in 6.5% sodium chloride, resistance to the vibriostatic agent O/129 (150 ug), and then identified using matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF/MS) (Bruker, Bremen, Germany). Single colonies were mixed with matrix solution, dried completely, and then MALDI-TOF/MS was tested according to the manufacturer’s protocols. Results were evaluated using an identification database and exported for local preservation and statistical analysis. An appraisal credibility score of > 95% was considered reliable (Jamal et al., 2014). Final species identification was confirmed by WGS. A. caviae isolates were stored in 20% glycerol at -70°C for subsequent studies.
Antimicrobial susceptibility testing
AST was performed by the broth microdilution method using the VITEK 2 Compact System (BioMerieux, Lyon, France). The antimicrobial agents tested included piperacillin/tazobactam (TZP), cefoxitin (FOX), cefuroxime (CXM), ceftazidime (CAZ), cefotaxime (CTX), cefepime (FEP), imipenem (IPM), meropenem (MPN), amikacin (AMK), gentamicin (GEN), ciprofloxacin (CIP), levofloxacin (LEV), tetracycline (TE), trimethoprim/sulfamethoxazole (STX), chloramphenicol (C), and aztreonam (ATM), The minimum inhibitory concentrations (MICs) were interpreted according to the recommendations of the Clinical and Laboratory Standards Institute (CLSI) for Aeromonas spp. (Institute, 2015) E.coli ATCC25922 was used as the quality-control strain. Multidrug resistance (MDR) was defined as non-susceptibility to at least one agent in three or more antimicrobial categories (Magiorakos et al., 2012).
Whole-genome sequencing and analysis
Genomic DNA of A. caviae was extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Purified DNA of all 47 A. caviae strains underwent WGS using the Illumina HiSeq (Illumina, San Diego, California) platform. Sequence reads were assembled using the SOAPdenovo software (version 2.04) (Li et al., 2010). Antimicrobial resistance genes were identified using the Comprehensive Antibiotic Resistance Database (CARD) (Alcock et al., 2020). We used the basic local alignment search tool (BLAST) to detect antimicrobial resistance genes with more than 70% coverage and 80% identity in each genome sequence. BLAST was used to identify virulence genes on the virulence factor database (VFDB) (https://www.mgc.ac.cn/VFs/main.htm).
Phylogenetic analysis
Based on the global distribution of A. caviae, we retrieved the assembled data of 521 A. caviae strains from the NCBI database (44 from patients, 181 from animals, 224 from environment, and 72 strains with no detailed source informations). A. caviae strain AP022254.1 was used as the reference for comparison. Reads were mapped using Burrow-Wheeler Alignerg (BWA, v0.7.12), and single nucleotide polymorphisms (SNPs) were identified using SAMtools (v1.3) to obtain mutation site information. A General Feature Format (Gff) file of each genome was generated using the genome annotation program Prokka (v1.11), then used to obtain the alignment of core genes on Roary (v3.11). Chromosomal SNP alleles were concatenated for each strain to generate a multiple alignment of all SNPs, and RAxML (v8.2.4) was run using the general time-reversible (GTR) model with a gamma distribution to construct a maximum likelihood phylogenetic tree. Average nucleotide identities (ANIs) among strains found in China and those found outside of China were calculated using JSpeciesWS to evaluate genome similarity.
Statistical analysis
All data were analyzed using SPSS 20.0 (SPSS Inc., Chicago, IL, USA). Continuous variables were presented as mean ± standard deviation (SD).
Results
Epidemiological data of patients infected with A. caviae
A total of 47 clinical isolates were obtained from 46 patients with extra-intestinal A. caviae infections at the First Medical Center of Chinese PLA General Hospital from 2017 to 2020. Two strains, S122 and S128, were isolated from different specimens of the same patient with a separation interval of 48 days. The patients had a mean age ± standard deviation of 68.9 ± 13.7 years and were predominantly male (male to female ratio, 3.2). More than 60% were elderly patients aged ≥65 years (63%, 29/46). Hepatobiliary diseases (57%, 26/46) and malignancies (30%, 14/46) were two common underlying diseases. The most common source of infection was biliary tract infection (41%, 19/46), followed by bacteremia (26%, 12/46), pneumonia (17%, 8/46), urinary tract infection (15%, 7/46) and peritonitis (2%, 1/46). Among the 46 patients, two patients died in hospitalization, and five who were given poor prognoses died after discharge, resulting in an in-hospital mortality rate of 4%. 21 patients were co-infected with other bacteria, including Enterococcus spp. (8 cases), Escherichia coli (8 cases), Klebsiella spp. (4 cases), Pseudomonas aeruginosa (4 cases), Citrobacter spp. (2 cases), Acinetobacter spp. (1 case), and 5 patients were infected with two or three pathogens (Table 1 and Table S1).
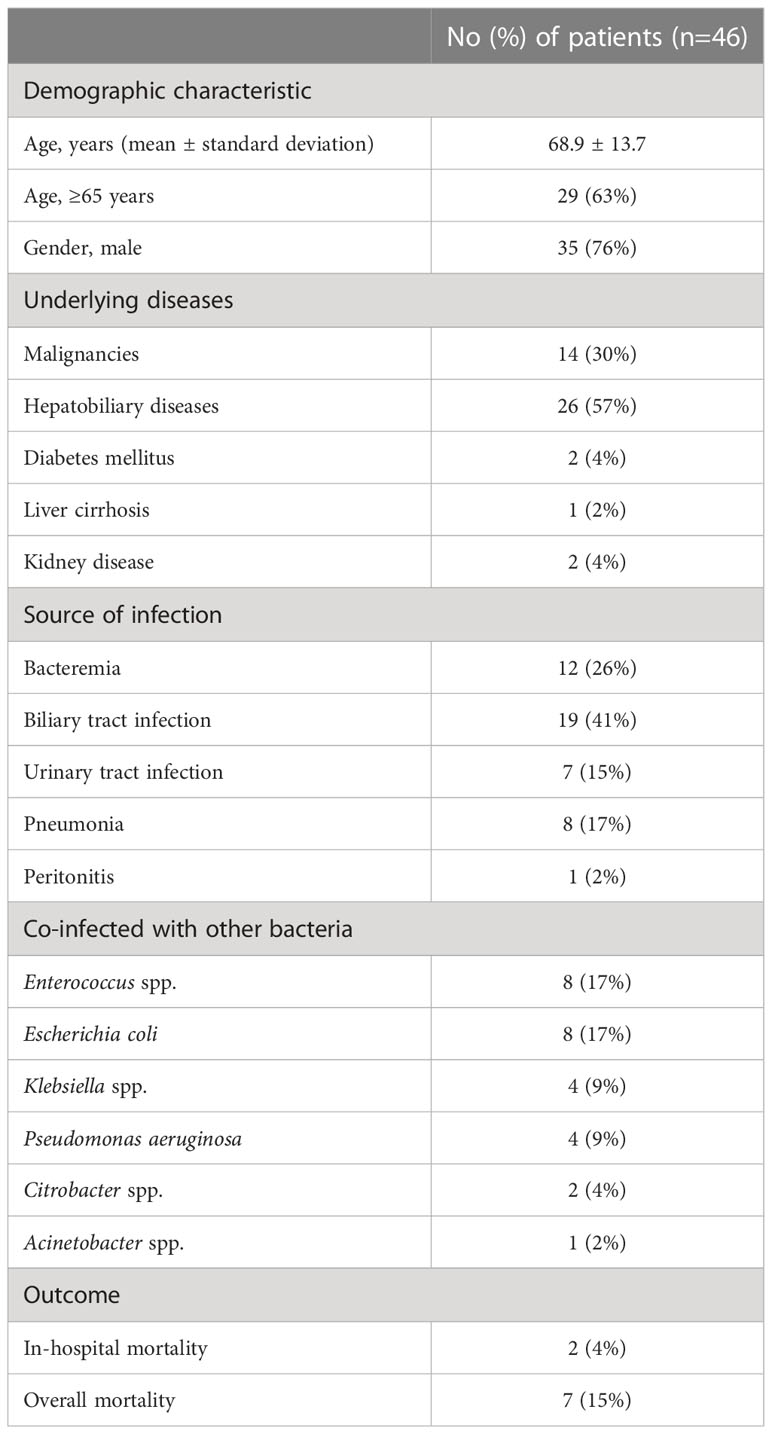
Table 1 Epidemiological data of 46 patients with extra-intestinal A. caviae infection from 2017 to 2020.
Antimicrobial susceptibility profiles
Antimicrobial susceptibility testing revealed the resistance rates to antibiotics were 38% (18/47) for cefoxitin, 34% (16/47) for cefuroxime, 32% (15/47) for cefotaxime, 30% (14/47) for tetracycline, 26% (12/47) for ceftazidime, 23% (11/47) for chloramphenicol, 21% (10/47) for cefepime, and 21% (10/47) for trimethoprim-sulfamethoxazole Most strains were susceptible to aminoglycosides and carbapenems (Table 2). 36% (17/47) of the strains showed susceptibility to all antimicrobial drugs tested. 40% (19/47) of the isolates were identified as MDR strains, of which 11 were from bile, 5 were from urine, 2 were from sputum, and 1 was from bloodstream. Moreover, we identified two strains resistant to carbapenems (Table S2).
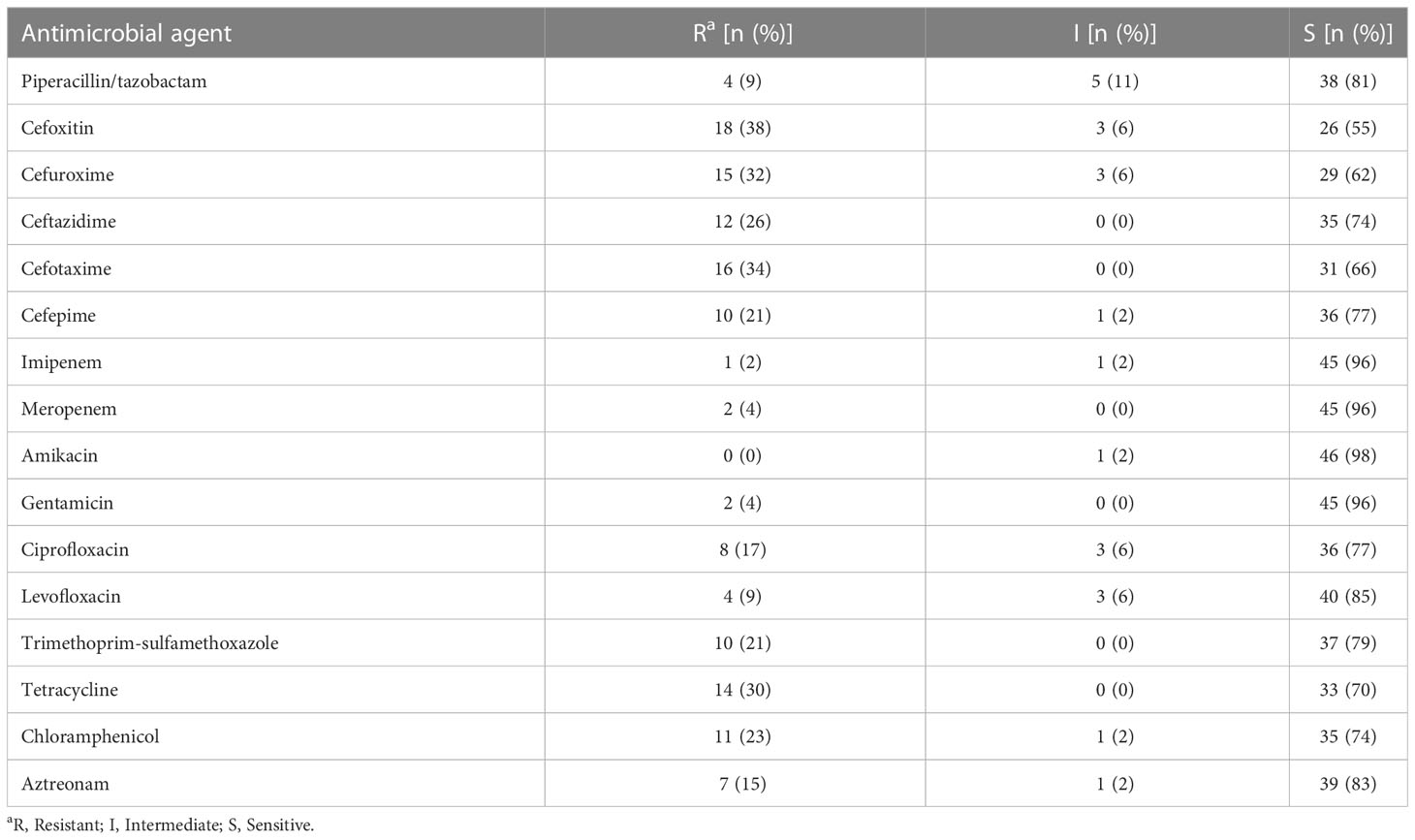
Table 2 Antimicrobial susceptibility patterns of 47 A. caviae strains iaolated from extra-intestinal infections.
Antibiotic resistance genotypic and phenotypic characteristic
The antimicrobial resistance genes of A. caviae were shown in Figure 1 and Table S2. 24 out of 47 isolates were found to be non-susceptible to cephalosporins and cephamycins, and all of which carried β-lactamase gene, including blaMOX, blaPER-3, blaOXA, blaNDM, and blaCphA. Most of the isolates (94%, 44/47) carried blaMOX, and 7 isolates carried blaPER-3. However, the AmpC β-lactamase gene, blaAQU-2, was present only in S169. The metallo-β-lactamase (MBL) gene, blaCphA and blaBRP, was detected in S175 and S189, respectively. Another MBL gene, blaNDM, was present in S169 and S189, which exhibited carbapenem resistance. However, blaCphA gene present in S175 was not associated with carbapenem resistance. Class A extended-spectrum β-lactamases (ESBLs) genes, blaCTX-M-3 and blaTEM-1, were simultaneous present in S225. Oxacillin-hydrolyzing (OXA)-type Class D β-lactamase encoding genes, blaOXA-278, blaOXA-724, blaOXA-1, blaOXA-10, which confer resistance to cephalosporins, were detected in S162, S175, S189, S225, respectively. The plasmid-mediated quinoloneresistance genes qnrS2, qnrVC, aac(6’)-Ib-cr were detected in 7 isolates, and the presence of both genes aac(6’)-Ib-cr and qnrS2 was observed in S189. Most of the isolates (86%, 6/7) carrying quinolone resistance genes manifested corresponding resistance phenotypes. Tetracycline resistance genes tet(A), tet(E), tet(31) were detected in 14 isolates, all of which manifested tetracycline resistance phenotypes. All 7 strains harbored sulfonamide resistance genes dfrA1, dfrA12, dfrA14, dfrA15b manifested resistance to trimethoprim-sulfamethoxazole. 15 strains harbored the chloramphenicol resistance genes floR, catB3, catII, and catI, 80% (12/15) of which were non-susceptible to chloramphenicol.

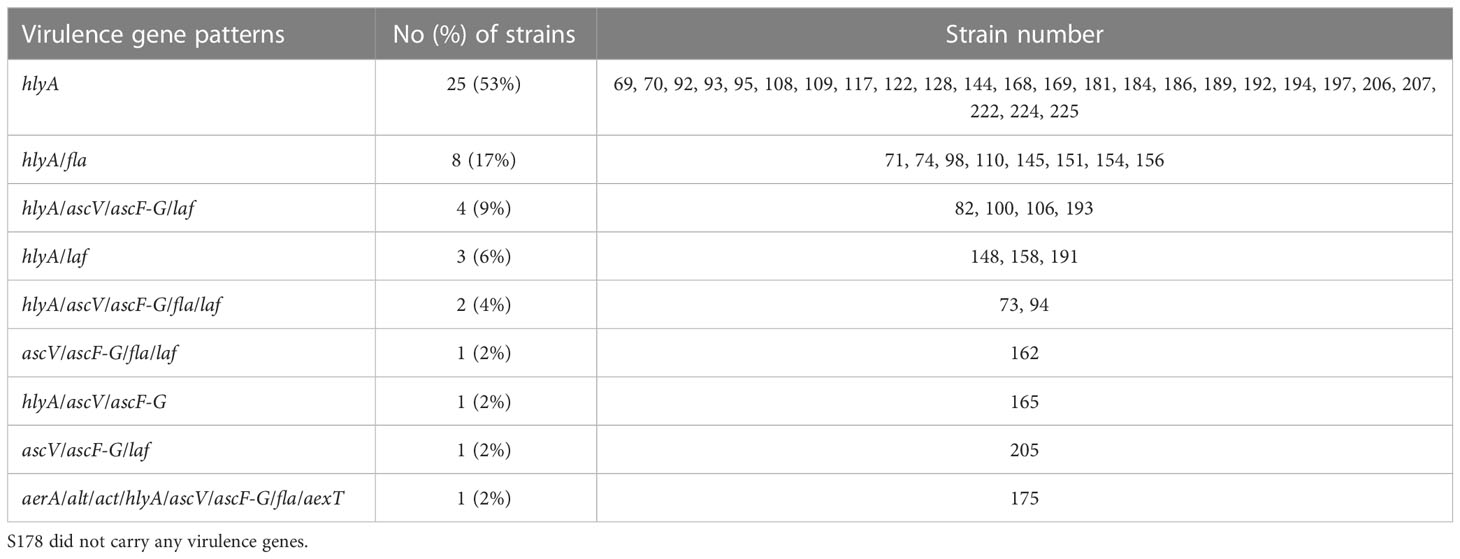
Figure 1 Construction of phylogenetic tree of 47 A. caviae strains. Antimicrobial resistance (AMR) genes were detected with more than 70% coverage and 80% identity in each genome using BLAST. Virulence genes were identified by blasting the VFDB database. Red and blue represented the antimicrobial resistance genes and virulence genes, respectively.
Distribution of virulence genes in A.caviae strains
We investigated representative 13 virulence genes in these clinical strains, including aerolysin (aerA), heat-stable cytotonic enterotoxin (ast), heat-labile cytotonic enterotoxin (alt), cytotoxic enterotoxin (act), hemolysin (hlyA), phospholipase (lip), type III secretion system components (ascV and ascF-G), flagellin (fla), elastase (ela), lateral flagella (laf), ADP-ribosyltransferase toxin (aexT), collagenase (col). The distribution of virulence genes in A. caviae was shown in Figure 1 and Table S3. Most of the strains (98%, 46/47) carried at least one of the virulence genes. Of these strains, 94% (44/47) carried hlyA, followed by fla (26%, 12/47), laf (23%, 11/47), ascV (21%, 10/47), ascF-G (21%, 10/47), and three or more virulence genes were detected in 21% (10/47) of clinical isolates. The genes encoding ast, lip, ela, and col were not detected in the studied strains. Analysis of the gene profiles based on the distribution of the 13 virulence genes revealed 9 virulence patterns summarized in Table 3. The most predominant patterns observed were hlyA (53%, 25/47) and hlyA/fla (17%, 8/47). Only S175 carried several virulence genes, including aerA, alt, act, hlyA, ascV, ascF-G, fla, and aexT.
Phylogenetic analysis
Phylogenetic analysis was performed on a total of 568 strains, including 521 strains reported between 2014 and 2021 from NCBI database (Table S4) and 47 clinical strains in our study, and a phylogenetic tree with 243,480 SNP loci was generated (Figure 2). All 568 strains were divided into four major clusters, of which L1 cluster had a low genetic relationship, L2 cluster contained less drug resistance genes, L3 and L4 cluster contained more drug resistance genes. Phylogenetic analysis showed that 47 clinical strains were distributed among the four clusters (Figure 2 and Table S5). The ANI between S108 and S109, which were isolated with a separation interval of 14 days from different departments on the same floor, was 100%.
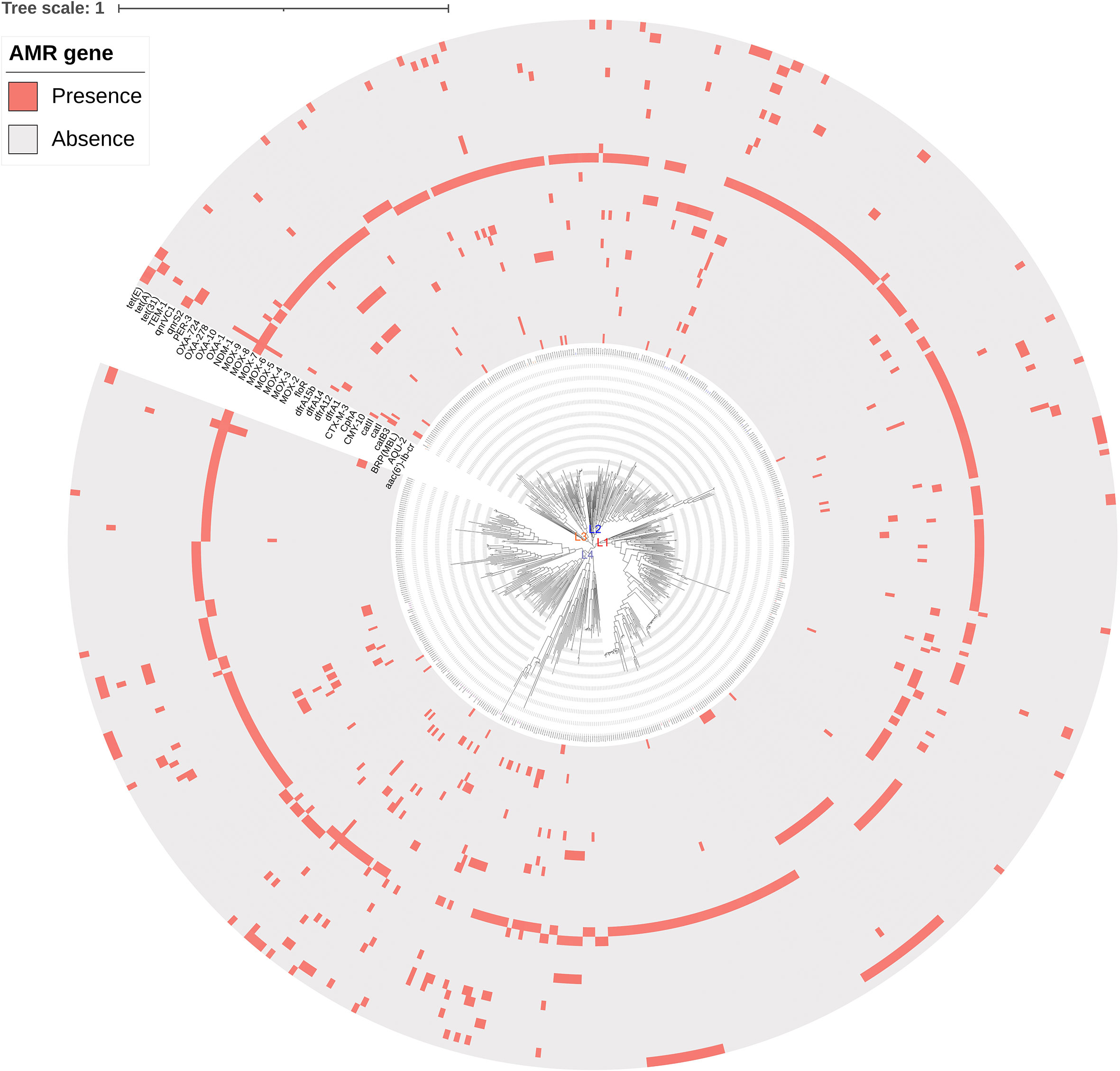
Figure 2 Phylogenetic tree of 568 A. caviae strains. Maximum likelihood phylogenetic tree was constructed using RAxML (v8.2.4). The 47 isolates in L1, L2, L3, and L4 clusters were marked in red, blue, orange, and purple color, respectively. AMR gene: antimicrobial resistance gene.
Discussion
In this study, it was found that more than 60% of extra-intestinal infections caused by A. caviae developed in elderly patients aged ≥ 65 years, most having immunocompromised conditions or underlying diseases, including hepatobiliary diseases, malignancies, diabetes mellitus, and renal disease. Furthermore, over 50% of patients with A. caviae infection had coexisting hepatobiliary diseases, including biliary stones, obstructive biliary disease, gallbladder or biliary tract tumor, and liver cancer, of which 8 patients suffered from bloodstream infections, 16 patients had biliary tract infections, and 1 patient had biliary tract infection followed by bloodstream infection. The high occurrence of infection with A. caviae may be attributable to biliary tract obstruction or stasis with increased intraductal pressure during hepatobiliary diseases. Moreover, translocation of A. caviae present in the gastrointestinal tract into intrahepatic and extrahepatic bile ducts, as well as hepatic veins and lymphatics, remain a possible cause of biliary tract infection and bacteremia (An et al., 2021). Polymicrobial infections occurred in 14 of 19 patients with A. caviae biliary tract infection, demonstrating that patients with biliary tract infection have a higher risk of developing polymicrobial infections. Malignant tumor patients who have received chemotherapy are more vulnerable to foodborne infections due to the disruption of their intestinal mucosal barrier, and the ensuing neutropenia makes the host susceptible to opportunistic infections (Baden et al., 2016). Consistent with previous studies, malignancy was found to be a common underlying disease in our study (Wu et al., 2015; Chen et al., 2021). Furthermore, liver cirrhosis has been recognized as a predisposing condition associated with Aeromonas bacteremia in regions with a high prevalence of chronic hepatitis (Tang et al., 2014). However, none of the 12 patients with A. caviae bacteremia had liver cirrhosis, and only one patient with liver cirrhosis developed urinary tract infection. Although A. caviae has been considered the primary culprit of bacteremia and biliary tract infections, A. caviae has also been reported in a few cases of urinary tract infections (Chao et al., 2012). In our study, more than 15% (7/46) of patients suffered from urinary tract infection, 5 of which are immunocompromised or undergoing invasive therapeutical procedures. Therefore, A. caviae should be considered a possible pathogen in immunocompromised patients, individuals suffering from underlying diseases, elderly patients, and patients undergoing invasive medical procedures.
CLSI has recommended third and fourth generation cephalosporins, fluroquinolones, and trimethoprim-sulfamethoxazole for the treatment of infections with Aeromonas spp. To date, there are only a few studies on the antimicrobial resistance of extra-intestinal A. caviae isolates, with one study reporting 15%-30% resistance rates for ceftazidime, cefepime, and ceftriaxone from 2012 to 2017 (Yang et al., 2019). Results of our study demonstrated that the resistance rate for cefotaxime was over 30%, and the resistance rates for ceftazidime, cefepime and trimethoprim-sulfamethoxazole were over 20%. Although the incidence of MDR reached to 40%, resistance rates to several antibiotics remained below 10%, including piperacillin-tazobactam, levofloxacin, gentamicin, amikacin, imipenem, and meropenem. These findings provide guidance for selecting appropriate empirical treatment before obtaining AST results.
In the present study, blaMOX was detected in more than 90% of isolates. This result concurs with previous findings that AmpC β-lactamases genes are species-specific to Aeromonas spp. and that all A. caviae isolates carry blaMOX (Fosse et al., 2003; Wu et al., 2015). However, only 50% (22/44) of the strains carrying blaMOX exhibited the cephalosporins or cephamycins non-susceptibility. Thus, the presence of the blaMOX gene was not associated with corresponding antibiotics non-susceptibility as reported previously (Walsh et al., 1997).
The class A extended-spectrum β-lactamases (ESBLs) gene blaPER-3 shared 99% identity with blaPER-1 and was initially identified within an isolate of Aeromonas punctata in France adjacent to a copy of ISCR1 (insertion sequence common regions, ISCRs), and then detected in two A.caviae isolates from a medical center in Taiwan and A.veronii isolated from chicken cloaca (Toleman et al., 2006; Wu et al., 2011; Wang et al., 2020). We found 7 blaPER-3-producing A. caviae isolates, all of which were MDR strains. Previous research revealed that the horizontal transfer of genetic elements, such as plasmids and integrons, could lead to an increased incidence of MDR among environmental Aeromonas isolates (Jacobs & Chenia, 2007). The spread of Enterobacteriaceae carrying the blaPER-1 gene as a chromosomal insert has been reported in Europe (Perilli et al., 2007). Therefore, caution should be exercised to prevent transmission of drug resistance genes between Aeromonas spp. by genetic elements.
Carbapenem resistance has been detected in A.hydrophila and A.veronii isolates, but is rarely found in A. caviae strains (Chen et al., 2012). However, recent studies have found that plasmid-encoded or non-plasmid encoded blaNDM-1, blaOXA-181, blaVIM, and blaKPC-2 genes contribute to carbapenem resistance in clinical A. caviae isolates, indicating the possibility of evolution and transmission of resistance genes in clinical strains (Adler et al., 2014; Anandan et al., 2017; Tang et al., 2020; Luo et al., 2022; Xu et al., 2022). In this study, two strains isolated from urine harbored blaNDM-1 and exhibited carbapenem resistance, one of which co-harbored of blaCTX-M-3 and class D β-lactamases gene blaOXA-1, which conferred resistance to third or fourth generation cephalosporins. The emergence of MDR strains poses a threat that challenges the diagnosis, clinical treatment, and control of infectious diseases. Studies on the genetic characteristics of these two strains are being conducted to elucidate the role of mobile genetic elements, such as plasmids and integrons, in the transmission of resistance.
According to one study, the AmpC β-lactamase gene blaAQU-2 was found only in A. hydrophila and A. jandaei strains isolated from chicken rinse (Wang et al., 2021). The MBL gene blaCphA, which is involved in the intrinsic resistance of Aeromonas to carbapenems, was found in the majority of A.hydrophila isolates but rarely detected in A. caviae strains (Wu et al., 2015; Wu et al., 2019). In our study, one blaCphA-carrying A. caviae, which co-harbored both blaAQU-2 and the class D β-lactamases gene blaOXA-724, conferred resistance to third generation cephalosporins, but did not exhibit carbapenem resistance in vitro, possibly attributed to the difficulty in detecting carbapenemase activity of carbapenemase hydrolyzing Aeromonas (CphA) through conventional AST, or genetic modifications that alter the expression of CphA (Wu et al., 2012).
To date, the virulence and pathogenic mechanism of Aeromonas remain obscure. Many virulence factors, including cytotoxins, enterotoxins, hemolysins, cell surface structures, lipases, proteases, aerolysins, and secretory systems, contributed to survival, environmental adaptation, and disease pathogenesis (Li et al., 2015; Wu et al., 2019; Zhou et al., 2019; Chen et al., 2021; Sun et al., 2021). Previous studies observed lower virulence and lower fatality rates in A. caviae compared to other Aeromonas spp. (Wu et al., 2015; Wu et al., 2019). However, findings from several studies on virulence genes and virulence phenotype of A. caviae remain controversial. In the present study, we revealed 9 virulence patterns composed of one to eight genes. Additionally, we found that the majority of the A. caviae strains carried at least one of the virulence genes, and three or more virulence genes were detected in more than 20% of these isolates. Pablos et al. reported that A. caviae infrequently carried aerA, hlyA, and ast genes (Pablos et al., 2010). Wu et al. found that the major genotype in clinical A. caviae isolates was lip, col, and ela (Wu et al., 2019). However, our finding contradicts the results of previous studies. More than 90% of isolates carried hlyA, and none carried lip, col, or ela. In accordance with previous research, our study showed that ast was absent in clinical A. caviae isolates (Aguilera-Arreola et al., 2007). Research has demonstrated that flagella glycosylation in A.hydrophila plays a vital role in Hep-2 cell adhesion and biofilm formation (Merino et al., 2014). Previous study showed that fla and laf were highly prevalent in A. caviae isolated from human faeces (Santos et al., 2011). In a recent study, 92.3% of A. caviae isolated from clinical specimens harbored fla (Miyagi et al., 2021). This study deteced fla and laf in only 26% and 23% of extra-intestinal A. caviae isolates, respectively. The type III secretion system (TTSS) of Aeromonas has been proved to play an essential role in pathogenicity (Yu et al., 2004). Studies found ascV and ascF-G in only a few extra-intestinal A. caviae isolates (14.3%, 2/14) (Chacon et al., 2004). Chen et al. demonstrated that infection with ascF-G Aeromonas was associated with mortality (Chen et al., 2021). We found that more than 20% of extra-intestinal A. caviae isolates (21%, 10/47) possesed these TTSS encoding genes, but only one patient, who was infected with A. caviae harboring ascV and ascF-G, died in the hospital.
One patient (case 46) infected with A. caviae carrying 8 virulence genes was cured after 14 days of hospitalization. However, another patient (case 18) infected with A. caviae possessing only hemolysin encoding gene hlyA died 45 days after admission. The significant difference between the two patients was that the latter had liver cancer. Our work showed that the overall mortality of extra-intestinal A. caviae infection was 15% (7/46). Additionally, we found that most of the strains (86%, 6/7) isolated from the 7 patients who died harbored only one virulence coding gene hlyA, and 5 of the 7 patients suffered from cancer. It is possible that host factors, such as underlying diseases, prolonged hospitalization, and not the virulence of A. caviae itself, are the main causes of death. We did not find an association between virulence genes and the pathogenicity of extra-intestinal A. caviae stains, suggesting the need for further research to identify specific virulence gene and the mechanism of pathogen-host interaction in extra-intestinal A. caviae infection.
In this retrospective study, we analyzed the genomic evolutionary characteristics of A. caviae based on 47 clinical isolates and 521 public strains available in the NCBI database. The genetic relationships of the 47 strains were identified, and cluster analysis indicated differences in SNPs among the 47 strains. These isolates had low genetic relationships, which might be due to the long separation interval of these strains. Genomic evolutionary analysis showed that stains in L3 and L4 contained more drug resistance genes than those in L2, presumably because changes in some key SNPs determined the subsequent evolution, and some drug resistance genes were inserted during the evolution. A high degree of homology between S108 and S109, but low homology between other stains indicated that there existed a risk of nosocomial transmission, but no clustering of hospital-onset infections was noted.
Conclusion
The epidemiological characteristics, antimicrobial resistance profiles, virulence genes, and phylogenetic traits described in this study strengthen our understanding of A.caviae strains that cause extra-intestinal infections. It may contribute to the management of extra-intestinal infections as well as the prevention and control of drug resistance.
Data availability statement
The data presented in the study are deposited in the National Library of Medicine repository (https://www.ncbi.nlm.nih.gov/sra/PRJNA902936), accession number PRJNA902936.
Ethics statement
This study involving human participants was reviewed and approved by the Ethics Committee of First Medical Center of Chinese PLA General Hospital.
Author contributions
YS, L-FW, D-XS, and YC designed the study. KZ, SL, LG, L-YY, and JG performed the experiments and interpreted the data. YS, YC, and L-FW wrote the first draft of the paper. YS, YC, L-FW and D-XS reviewed and approved the final report. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (No. 81472012 and No.31200142).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1084352/full#supplementary-material
References
Adler, A., Assous, M. V., Paikin, S., Shulman, A., Miller-Roll, T., Hillel, S., et al. (2014). Emergence of VIM-producing aeromonas caviae in Israeli hospitals. J. Antimicrob. Chemother. 69, 1211–1214. doi: 10.1093/jac/dkt505
Aguilera-Arreola, M. G., Hernandez-Rodriguez, C., Zuniga, G., Figueras, M. J., Garduno, R. A., Castro-Escarpulli, G. (2007). Virulence potential and genetic diversity of aeromonas caviae, aeromonas veronii, and aeromonas hydrophila clinical isolates from Mexico and Spain: A comparative study. Can. J. Microbiol. 53, 877–887. doi: 10.1139/W07-051
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
An, Z., Braseth, A. L., Sahar, N. (2021). Acute cholangitis: Causes, diagnosis, and management. Gastroenterol. Clin. North Am. 50, 403–414. doi: 10.1016/j.gtc.2021.02.005
Anandan, S., Gopi, R., Devanga Ragupathi, N. K., Muthuirulandi Sethuvel, D. P., Gunasekaran, P., Walia, K., et al. (2017). First report of blaOXA-181-mediated carbapenem resistance in aeromonas caviae in association with pKP3-a: Threat for rapid dissemination. J. Glob Antimicrob. Resist. 10, 310–314. doi: 10.1016/j.jgar.2017.07.006
Baden, L. R., Swaminathan, S., Angarone, M., Blouin, G., Camins, C. B., Casper, C., et al. (2016). Prevention and treatment of cancer-related infections, version 2.2016, NCCN clinical practice guidelines in oncology. J. Natl. Compr. Canc Netw. 14, 882–913. doi: 10.6004/jnccn.2016.0093
Chacon, M. R., Soler, L., Groisman, E. A., Guarro, J., Figueras, M. J. (2004). Type III secretion system genes in clinical aeromonas isolates. J. Clin. Microbiol. 42, 1285–1287. doi: 10.1128/JCM.42.3.1285-1287.2004
Chao, C. M., Gau, S. J., Lai, C. C. (2012). Aeromonas genitourinary tract infection. J. Infect. 65, 573–575. doi: 10.1016/j.jinf.2012.06.012
Chao, C. M., Lai, C. C., Tang, H. J., Ko, W. C., Hsueh, P. R. (2013). Biliary tract infections caused by aeromonas species. Eur. J. Clin. Microbiol. Infect. Dis. 32, 245–251. doi: 10.1007/s10096-012-1736-1
Chen, J. S., Hsu, G. J., Hsu, B. M., Yang, P. Y., Kuo, Y. J., Wang, J. L., et al. (2021). Prevalence, virulence-gene profiles, antimicrobial resistance, and genetic diversity of human pathogenic aeromonas spp. from shellfish and aquatic environments. Environ. pollut. 287, 117361. doi: 10.1016/j.envpol.2021.117361.
Chen, P. L., Ko, W. C., Wu, C. J. (2012). Complexity of beta-lactamases among clinical aeromonas isolates and its clinical implications. J. Microbiol. Immunol. Infect. 45, 398–403. doi: 10.1016/j.jmii.2012.08.008
Chen, Y. W., Su, S. L., Li, C. W., Tsai, C. S., Lo, C. L., Syue, L. S., et al. (2021). Pancreaticobiliary cancers and aeromonas isolates carrying type secretion system genes ascF-ascG are associated with increased mortality: An analysis of 164 aeromonas infection episodes in southern Taiwan. Front. Cell Infect. Microbiol. 11, 749269. doi: 10.3389/fcimb.2021.749269
Chuang, H. C., Ho, Y. H., Lay, C. J., Wang, L. S., Tsai, Y. S., Tsai, C. C. (2011). Different clinical characteristics among aeromonas hydrophila, aeromonas veronii biovar sobria and aeromonas caviae monomicrobial bacteremia. J. Korean Med. Sci. 26, 1415–1420. doi: 10.3346/jkms.2011.26.11.1415
Dwivedi, M., Mishra, A., Prasad, A., Azim, A., Singh, R. K., Baronia, A. K., et al. (2008). Aeromonas caviae septicemia in immunocompetent gastrointestinal carriers. Braz. J. Infect. Dis. 12, 547–548. doi: 10.1590/S1413-86702008000600023
Fernandez-Bravo, A., Figueras, M. J. (2020). An update on the genus aeromonas: Taxonomy, epidemiology, and pathogenicity. Microorganisms 8 (1), 1-39. doi: 10.3390/microorganisms8010129
Fosse, T., Giraud-Morin, C., Madinier, I. (2003). [Phenotypes of beta-lactam resistance in the genus aeromonas]. Pathol. Biol. (Paris) 51, 290–296. doi: 10.1016/S0369-8114(03)00027-0
Hoel, S., Vadstein, O., Jakobsen, A. N. (2019). The significance of mesophilic aeromonas spp. in minimally processed ready-to-Eat seafood. Microorganisms 7 (3), 91, 1-25. doi: 10.3390/microorganisms7030091
Institute CaLS (2015). Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated of fastidious bacteria. CLSI document M45-A3 3rd ed, 16–18.
Jacobs, L., Chenia, H. Y. (2007). Characterization of integrons and tetracycline resistance determinants in aeromonas spp. isolated from south African aquaculture systems. Int. J. Food Microbiol. 114, 295–306. doi: 10.1016/j.ijfoodmicro.2006.09.030
Jamal, W., Albert, M. J., Rotimi, V. O. (2014). Real-time comparative evaluation of bioMerieux VITEK MS versus bruker microflex MS, two matrix-assisted laser desorption-ionization time-of-flight mass spectrometry systems, for identification of clinically significant bacteria. BMC Microbiol. 14, 289. doi: 10.1186/s12866-014-0289-0
Janda, J. M., Abbott, S. L. (2010). The genus aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09
Kimura, M., Araoka, H., Yoneyama, A. (2013). Aeromonas caviae is the most frequent pathogen amongst cases of aeromonas bacteremia in Japan. Scand. J. Infect. Dis. 45, 304–309. doi: 10.3109/00365548.2012.737474
Lamy, B., Kodjo, A., Laurent, F. (2009). Prospective nationwide study of aeromonas infections in France. J. Clin. Microbiol. 47, 1234–1237. doi: 10.1128/JCM.00155-09
Li, F., Wang, W., Zhu, Z., Chen, A., Du, P., Wang, R., et al. (2015). Distribution, virulence-associated genes and antimicrobial resistance of aeromonas isolates from diarrheal patients and water, China. J. Infect. 70, 600–608. doi: 10.1016/j.jinf.2014.11.004
Li, R., Zhu, H., Ruan, J., Qian, W., Fang, X., Shi, Z., et al. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272. doi: 10.1101/gr.097261.109
Luo, X., Mu, K., Zhao, Y., Zhang, J., Qu, Y., Hu, D., et al. (2022). Emergence of bla NDM- 1-carrying aeromonas caviae K433 isolated from patient with community-acquired pneumonia. Front. Microbiol. 13, 825389. doi: 10.3389/fmicb.2022.825389
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Merino, S., Wilhelms, M., Tomas, J. M. (2014). Role of aeromonas hydrophila flagella glycosylation in adhesion to hep-2 cells, biofilm formation and immune stimulation. Int. J. Mol. Sci. 15, 21935–21946. doi: 10.3390/ijms151221935
Miyagi, K., Shimoji, N., Shimoji, S., Tahara, R., Uechi, A., Tamaki, I., et al. (2021). Comparison of species, virulence genes and clones of aeromonas isolates from clinical specimens and well water in Okinawa prefecture, Japan. J. Appl. Microbiol. 131, 1515–1530. doi: 10.1111/jam.15038
Pablos, M., Remacha, M. A., Rodriguez-Calleja, J. M., Santos, J. A., Otero, A., Garcia-Lopez, M. L. (2010). Identity, virulence genes, and clonal relatedness of aeromonas isolates from patients with diarrhea and drinking water. Eur. J. Clin. Microbiol. Infect. Dis. 29, 1163–1172. doi: 10.1007/s10096-010-0982-3
Perilli, M., De Santis, F., Mugnaioli, C., Rossolini, G. M., Luzzaro, F., Stefani, S., et al. (2007). Spread of enterobacteriaceae carrying the PER-1 extended-spectrum beta-lactamase gene as a chromosomal insert: a report from Italy. J. Antimicrob. Chemother. 59, 323–324. doi: 10.1093/jac/dkl487
Santos, P. G., Santos, P. A., Bello, A. R., Freitas-Almeida, A. C. (2011). Association of aeromonas caviae polar and lateral flagella with biofilm formation. Lett. Appl. Microbiol. 52, 49–55. doi: 10.1111/j.1472-765X.2010.02965.x
Sun, Y., Zhao, Y., Xu, W., Fang, R., Wu, Q., He, H., et al. (2021). Taxonomy, virulence determinants and antimicrobial susceptibility of aeromonas spp. isolated from bacteremia in southeastern China. Antimicrob. Resist. Infect. Control 10, 43. doi: 10.1186/s13756-021-00911-0
Tang, L., Huang, J., She, J., Zhao, K., Zhou, Y. (2020). Co-Occurrence of the bla KPC-2 and mcr-3.3 gene in aeromonas caviae SCAc2001 isolated from patients with diarrheal disease. Infect. Drug Resist. 13, 1527–1536. doi: 10.2147/IDR.S245553
Tang, H. J., Lai, C. C., Lin, H. L., Chao, C. M. (2014). Clinical manifestations of bacteremia caused by aeromonas species in southern Taiwan. PloS One 9, e91642. doi: 10.1371/journal.pone.0091642
Toleman, M. A., Bennett, P. M., Walsh, T. R. (2006). ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70, 296–316. doi: 10.1128/MMBR.00048-05
Walsh, T. R., Stunt, R. A., Nabi, J. A., MacGowan, A. P., Bennett, P. M. (1997). Distribution and expression of beta-lactamase genes among aeromonas spp. J. Antimicrob. Chemother. 40, 171–178. doi: 10.1093/jac/40.2.171
Wang, Y., Hou, N., Rasooly, R., Gu, Y., He, X. (2021). Prevalence and genetic analysis of chromosomal mcr-3/7 in aeromonas from U.S. animal-derived samples. Front. Microbiol. 12, 667406. doi: 10.3389/fmicb.2021.667406
Wang, X., Zhai, W., Wang, S., Shen, Z., Wang, Y., Zhang, Q. (2020). A novel transposon, Tn6518, mediated transfer of mcr-3 variant in ESBL-producing aeromonas veronii. Infect. Drug Resist. 13, 893–899. doi: 10.2147/IDR.S239865
Wu, C. J., Chen, P. L., Hsueh, P. R., Chang, M. C., Tsai, P. J., Shih, H. I., et al. (2015). Clinical implications of species identification in monomicrobial aeromonas bacteremia. PloS One 10, e0117821. doi: 10.1371/journal.pone.0117821
Wu, C. J., Chen, P. L., Wu, J. J., Yan, J. J., Lee, C. C., Lee, H. C., et al. (2012). Distribution and phenotypic and genotypic detection of a metallo-beta-lactamase, CphA, among bacteraemic aeromonas isolates. J. Med. Microbiol. 61, 712–719. doi: 10.1099/jmm.0.038323-0
Wu, C. J., Chuang, Y. C., Lee, M. F., Lee, C. C., Lee, H. C., Lee, N. Y., et al. (2011). Bacteremia due to extended-spectrum-beta-lactamase-producing aeromonas spp. at a medical center in southern Taiwan. Antimicrob. Agents Chemother. 55, 5813–5818. doi: 10.1128/AAC.00634-11
Wu, C. J., Ko, W. C., Lee, N. Y., Su, S. L., Li, C. W., Li, M. C., et al. (2019). Aeromonas isolates from fish and patients in tainan city, Taiwan: Genotypic and phenotypic characteristics. Appl. Environ. Microbiol. 85 (21), e01360-19. doi: 10.1128/AEM.01360-19
Xu, S., Tu, J., Zhang, L., Chen, Y., Dong, X., Chi, X., et al. (2022). Detection of NDM-1-Positive aeromonas caviae from bacteremia by using whole-genome sequencing. Infect. Drug Resist. 15, 2835–2841. doi: 10.2147/IDR.S360353
Yang, S., He, T., Sun, J., Sun, S. (2019). Distinct antimicrobial resistance profiling of clinically important aeromonas spp. in southwest China: A seven-year surveillance study. Infect. Drug Resist. 12, 2971–2978. doi: 10.2147/IDR.S216926
Yu, H. B., Rao, P. S., Lee, H. C., Vilches, S., Merino, S., Tomas, J. M., et al. (2004). A type III secretion system is required for aeromonas hydrophila AH-1 pathogenesis. Infect. Immun. 72, 1248–1256. doi: 10.1128/IAI.72.3.1248-1256.2004
Keywords: Aeromonas caviae, phylogenetic analysis, antimicrobial resistance, extra-intestinal infections, virulence
Citation: Song Y, Wang L-f, Zhou K, Liu S, Guo L, Ye L-y, Gu J, Cheng Y and Shen D-x (2023) Epidemiological characteristics, virulence potential, antimicrobial resistance profiles, and phylogenetic analysis of Aeromonas caviae isolated from extra-intestinal infections. Front. Cell. Infect. Microbiol. 13:1084352. doi: 10.3389/fcimb.2023.1084352
Received: 30 October 2022; Accepted: 13 February 2023;
Published: 24 February 2023.
Edited by:
Yolanda López-Vidal, Faculty of Medicine, National Autonomous University of Mexico, MexicoReviewed by:
Etinosa Igbinosa, University of Benin, NigeriaAndrea Micke Moreno, Faculty of Veterinary Medicine and Animal Science, University of São Paulo, Brazil
Copyright © 2023 Song, Wang, Zhou, Liu, Guo, Ye, Gu, Cheng and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ding-xia Shen, NDg3NTY3NjlAcXEuY29t; Yan Cheng, Y2hlbmd5YW4wOTIwMDZmcmVlQGFsaXl1bi5jb20=
†These authors have contributed equally to this work
 Yang Song1†
Yang Song1† Ling Guo
Ling Guo Jiang Gu
Jiang Gu Yan Cheng
Yan Cheng