
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 02 March 2023
Sec. Clinical Microbiology
Volume 13 - 2023 | https://doi.org/10.3389/fcimb.2023.1082347
This article is part of the Research Topic Molecular diagnostic methods for bacteria and fungi detection View all 10 articles
Background: In this study, we used real-time quantitative PCR (RQ-PCR) to rapidly detect Mucorales and Aspergillus in formalin-fixed, paraffin-embedded (FFPE) samples, targeting 18SrRNA gene and 28SrRNA gene. Identification of Mucorales and Aspergillus was analysed by combining Mucorales RQ-PCR (Mucorales18SrRNA and Mucorales28SrRNA) with Aspergillus RQ-PCR (Aspergillus18SrRNA and Aspergillus28SrRNA).
Objectives: The aims of this study were to compare the diagnostic performances of four RQ-PCR assays as single and combined diagnostic and identification tools.
Methods: We collected 12 control group samples and 81 experimental group samples diagnosed by histopathology, including mucormycosis (19 patients, 21 FFPE samples), aspergillosis (54 patients, 57 FFPE samples) and mucormycosis with aspergillosis (3 patients, 3 FFPE samples). All samples were detected by four RQ-PCR tests to compare and analyze diagnostic performance.
Results: The sensitivities of Mucorales18SrRNA and Mucorales28SrRNA were both 75%, with the tests having specificities of 97.10% and 94.20%. The sensitivities of Aspergillus18SrRNA and Aspergillus28SrRNA were 73.33% and 65%, with the tests having specificities of 87.88% and 81.82%. The values of the evaluation indexes of the combined detection of Mucorales28SrRNA and Aspergillus18SrRNA (M28A18) were the highest with a kappa coefficient value of 0.353, followed by M18A18. M28A18 had a sensitivity of 67.90% and a specificity of 100%.
Conclusions: We recommend using the combination of Mucorales RQ-PCR and Aspergillus RQ-PCR as a screening tool to detect samples suspected of mucormycosis and/or aspergillosis.
Over the past few decades, the incidence of invasive mold disease (IMD) has increased significantly and the fungal spectrum of IMD has broadened. According to literature, over 100,000 IMD cases occur each year, and these are associated with high morbidity and mortality in immunocompromised patients who have hematological malignancy and transplantation (Ruiz-Camps and Jarque, 2014; Ruhnke et al., 2015; Pegorie et al., 2017; Springer et al., 2018).
The members of the order Mucorales and genus Aspergillus are the most common opportunistic pathogens of IMD (Oren and Paul, 2014). Because of the significantly different antifungal susceptibilities and the complexity of the population of patients at risk, management of patients with IMD which lacks typical clinical manifestations has become increasingly complex. Therefore, early and reliable diagnostic methods are necessary for effective treatment.
Currently, the gold standard for the diagnosis of invasive fungal infections depends on histopathology and culture. However, culture is time-consuming and may fail if the potential microbial causes are not considered during sample collection, so formalin-fixed, paraffin-embedded (FFPE) bioptic material is collected for subsequent histological diagnosis. FFPE tissues obtained from patients with proven IMDs are frequently used to detect the etiology of invasive mycoses (Tarrand et al., 2003; Munoz-Cadavid et al., 2010; Babouee Flury et al., 2014). While histopathology can prove invasive fungal infections, the analytical correctness of histological findings is no more than 79% (Sangoi et al., 2009). Therefore, preliminary histological results should be interpreted cautiously (Guarner and Brandt, 2011) and supported by the culture whenever possible. In addition, histopathological observations of fungal shape and arrangement may not be sufficient for accurate identification of the Mucorales and Aspergillus if only a limited quantity of fungal hyphae is present.
In recent years, considerable efforts have been made to develop more sensitive and specific tools and protocols for IMD diagnosis. It is reported that PCR-based techniques, including conventional, semi-nested and real-time PCR, can be used to identify fungal agents in FFPE tissue (Bialek et al., 2005; Rickerts et al., 2006; Walsh et al., 2011; Springer et al., 2016a). RQ-PCR is very suitable for detecting the DNA of FFPE samples which are easily degraded. There are reports using the 18SrRNA gene and the 28SrRNA gene regions to detect and distinguish mucormycosis and invasive aspergillosis (Bialek et al., 2005; Walsh et al., 2011; Springer et al., 2016a; Gade et al., 2017).
The objective of this study was to evaluate RQ-PCR protocols by the use of TaqMan technology for detection and identification of Mucorales and Aspergillus in FFPE samples, targeting the 18SrRNA gene and the 28SrRNA gene. Identification of Mucorales and Aspergillus was analyzed by the combination of Mucorales RQ-PCR (Mucorales18SrRNA and Mucorales28SrRNA) and Aspergillus RQ-PCR (Aspergillus18SrRNA and Aspergillus28SrRNA).
Ethical approval was obtained from the West China Hospital Ethics Committee of Sichuan University. According to local ethics, we have applied for exemption from written informed consent. We collected 81 experimental group samples (from 76 patients) in the Department of Pathology of West China Hospital from January 2015 to January 2018 with positive histopathology results, including mucormycosis (19 patients, 21 FFPE samples), aspergillosis (54 patients, 57 FFPE samples) and mucormycosis with aspergillosis (3 patients, 3 FFPE samples).
In addition, 12 FFPE tissue specimens from patients were used as controls including 6 without IMDs and 6 with other fungal infections. All of the slides including hematoxylin-eosin (H&E), periodic acid-Schiff (PAS) and/or Gomori-methenamine-silver (GMS) from each patient were reviewed and confirmed according to European Organization for Research on Treatment of Cancer and the Mycoses Study Group (EORTC/MSG) (De Pauw et al., 2008) by two professional and experienced pathologists with consistent diagnosis independently and in duplicate.
Isolates of laboratory strains included Aspergillus flavus, Aspergillus fumigatus, Rhizomucor miehei, Candida albicans, Cryptococcus neoformans, and Fusarium oxysporum. All isolated strains were provided by the Clinical Microbiology Laboratory, West China Hospital of Sichuan University.
Four FFPE tissue sections with 4-to-5-um from each specimen were used for DNA extraction. Each section was cut at a different position of the disposable knife of the microtome to prevent DNA cross-contamination due to attached cells at that position of the knife from one section to the next. If the sample surface was exposed to air, discard the first 2–3 sections. For deparaffinization, the sections were put into 1.5 mL tubes. 1 ml of xylene was added, centrifuged at full speed for 2 minutes at room temperature and discarded the supernatant by pipetting. Then, 1 ml ethanol was added, centrifuged and discarded like xylene. After incubation of the tissue at 37°C to evaporate the remains of the ethanol, DNA extraction was performed according to the manufacturer’s instructions for the QIAamp DNA FFPE tissue kit (Qiagen, Germany) with the following modifications. All FFPE tissue samples were incubated over night in proteinase K and ATL buffer at 56°C. Fungal cell walls were lysed by Arthrobacter luteus lyticase L2524 (Sigma, USA) for 45minutes at 37°C. The DNA was eluted with 100 µl Buffer ATE and stored at -20°C.
DNA extraction of laboratory strains was performed according to the manufacturer’s instructions for the QIAamp DNA Mini kit (Qiagen, Germany) with the following modifications. Fungal cell walls were lysed by Arthrobacter luteus lyticase L2524 (Sigma, USA) for 30 minutes at 30°C, incubated 1 to 3 hours in proteinase K and ATL buffer at 56°C. The DNA was stored at -20°C.
Mucorales RQ-PCR primers and probes targeting the 18SrRNA gene and the 28SrRNA gene were described by Springer et al. (Springer et al. 2016a). Aspergillus RQ-PCR primers and probes targeting the 18SrRNA gene were described by Walsh et al. (Walsh et al., 2011). The protocols of RQ-PCR amplifications were performed as described previously (Walsh et al., 2011; Springer et al. 2016a) with the exception of the design of a new primer pair. For optimal design of new primers and probes, multiple sequence alignments of Aspergillus 28SrRNA gene (National Center for Biotechnology Information [NCBI] public genetic database [GenBank]) were created using Geneious software (Biomatters, Auckland, New Zealand). Primer Express 3.0 (Applied Biosystems, Foster City, CA) was used to help select primers and probes of optimal melting temperatures (Table 1). The primers and probes of Aspergillus 28SrRNA were verified for its specificity by six laboratory isolated strains and normal human DNA, which thermocycling protocol are the same as Aspergillus 18SrRNA. The locations of real-time PCR assay targets are shown in Figure 1.
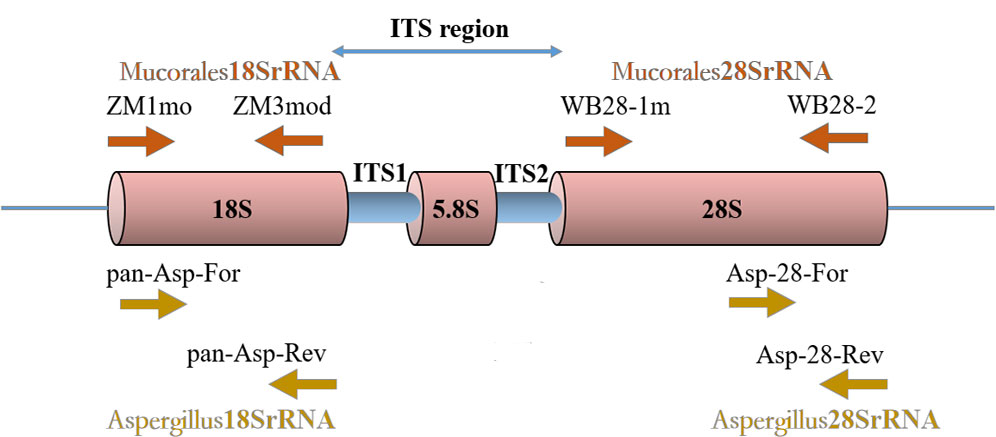
Figure 1 Locations of four real-time PCR assays target. An rDNA single repeat is shown. The primers ZM1mo and ZM3mod of Mucorales18SrRNA assay target 18SrRNA gene region. The primers WB28-1m and WB28-2 of Mucorales28SrRNA assay target 28SrRNA gene region. The primers pan-Asp-For and pan-Asp-Rev of Aspergillus18SrRNA assay target 18SrRNA gene region. The primers Asp-28-For and Asp-28-Rev of Aspergillus28SrRNA assay target 28SrRNA gene region.
We test all samples in triplicate. Amplification had to be reproducible, occurring in all 3 replicate wells, for a sample to be considered RQ-PCR positive. The positivity cutoff of Mucorales18SrRNA and Mucorales28SrRNA was defined as both wells having Cq values of <36. The positivity cutoff of Aspergillus18SrRNA and Aspergillus28SrRNA was defined as both wells having Cq values of <35 and <33, respectively. To validate the presence of amplifiable DNA and absence of inhibitory substances, a PCR in FFPE samples was performed using the primer set G1 and G2 targeting the human β globin gene (Bialek et al., 2005). When the result was negative, DNA extraction was repeated if enough material was available. All primers and probes were synthesized in Sangon Biotech (Sangon, Shanghai, China).
For all assays, RQ-PCR amplifications were performed in a 25 ul mixture using a StepOnePlus thermocycler (Applied Biosystems). Each reaction mixture contained 12.5 ul TaqMan Universal PCR Master Mix (Applied Biosystems), 900 nM forward and reverse primer, 200 nM probe and 5 ul extracted DNA. The DNA extracted from Rhizomucor miehei and Aspergillus flavus were serially diluted and tested for each RQ-PCR assay to determine the limit of detection (LoD). In each run, negative (FFPE tissue specimen without IMD) and positive (isolated strains of Rhizomucor miehei and Aspergillus flavus) controls were included.
Identification of Mucorales and Aspergillus was analyzed by Mucorales RQ-PCR in combination with Aspergillus RQ-PCR, including Mucorales18SrRNA and Aspergillus18SrRNA (M18A18), Mucorales18SrRNA and Aspergillus28SrRNA (M18A28), Mucorales28SrRNA and Aspergillus18SrRNA (M28A18), and Mucorales28SrRNA and Aspergillus28SrRNA (M28A28). True positives of Mucorales RQ-PCR in combination with Aspergillus RQ-PCR were defined as cases proven according to the following criteria: for mucormycosis samples, Mucorales RQ-PCR positivity and Aspergillus RQ-PCR negativity; for mucormycosis with aspergillosis samples, both positivity; for aspergillosis samples, Aspergillus RQ-PCR positivity and Mucorales RQ-PCR negativity. Other results were regarded as negatives.
All samples, including 81 experimental group samples and 12 control group samples, were detected by four RQ-PCR tests to compare and analyze the diagnostic performance, including negative predictive value (NPV), positive predictive value (PPV), sensitivity and specificity with likelihood ratios (LRs), and 95% confidence intervals (CIs) were calculated for NPV, PPV, sensitivity, and specificity. Cohen’s kappa coefficient was calculated to measure the agreement between any two assays. Statistical analysis was performed using SPSS, version 20 (SPSS, Chicago, IL, USA).
The study involved 76 patients (F/M, 42/34; age, 50.95 ± 17.31 years) with following comorbidities: 23 (30.26%) had diabetes, 16 (21.05%) had hypertension, 10 (13.16%) had bronchiectasis, 8 (10.53%) had solid tumor, 7 (9.21%) had Chronic obstructive pulmonary disease (COPD), 5 (6.58%) had hematological malignancy, 5 (6.58%) had tuberculosis, and 16 (21.05%) had others. Slides stained with PAS or GMS were considered, respectively, positive if magenta or brown-black fungal hyphae with morphological features were observed (Figure 2). Positive special staining with GMS and/or PAS were 24/29 (82.76%). Positive culture cases were 12/52 (23.08%). Positive 1, 3-beta-D-glucan assay cases were 9/40 (22.50%). Positive Galactomannan assay cases were 12/42 (28.57%). The principal site of infections was in lungs (61 cases), next were in other sites (including one ileum, three nasal cavity, four maxillary sinus, two trachea, one external auditory canal, one toe and three main bronchus). CT of the chest was obtained in 61 patients with pulmonary infection. There was a wide spectrum of radiological findings, with the most common being 26 nodules, followed by 20 mass, 16 cavitation, 7 consolidation, 5 pleural effusion, and 3 air crescent sign. Result of bronchofibroscopy were obtained in 56/61 patients with pulmonary infection: 18 patients were normal, and other patients were mainly necrosis, luminal stenosis, and purulent secretion.
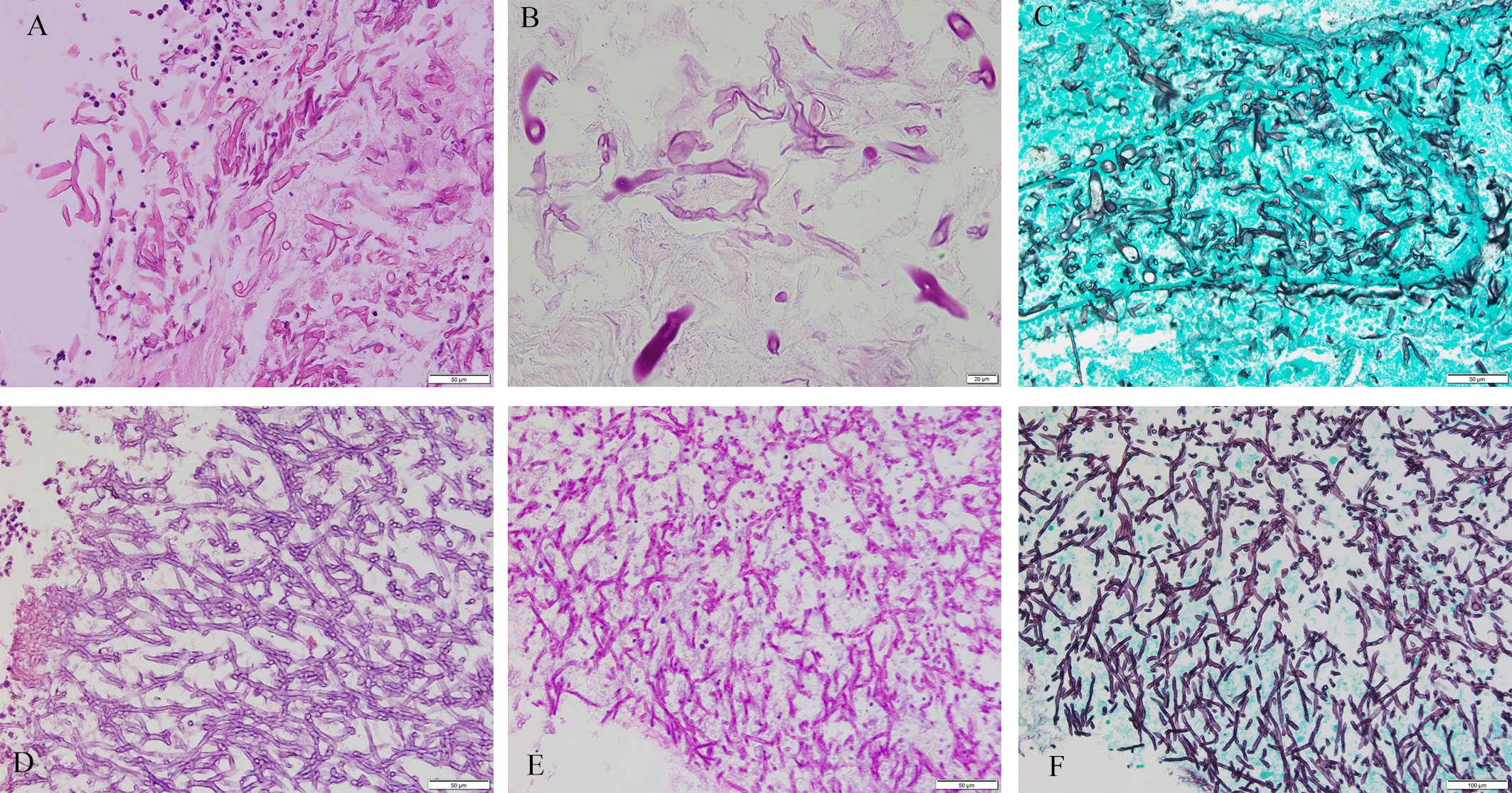
Figure 2 Mucorales and Aspergillus. Cytomorphology of Mucorales and Aspergillus in FFPE samples processed using the HE staining, PAS staining and GMS staining. The mycelium appeared magenta after PAS staining and brown-black after GMS staining. Magnification, 400×. (A) Mucorales with HE; (B) Mucorales with PAS; (C) Mucorales by GMS; (D) Aspergillus with HE; (E) Aspergillus with PAS; (F) Aspergillus with GMS. Lack of images of tissues with both Aspergillus and Mucorales infection.
The LOD of Mucorales18SrRNA and Mucorales28SrRNA was 10-1copies/ul in Rhizomucor miehei DNA. In AspergillusRQ-PCR assays, the LOD between Aspergillus 18SrRNA and Aspergillus28SrRNA was different (100copies/ul vs. 101copies/ul) for Aspergillus flavus DNA. The analytical specificity of Aspergillus28SrRNA assays was tested by adding genomic DNA from the six isolated strains. No cross-reactivity with non-Mucorales species or human DNA was observed. The specificity of Mucorales18SrRNA, Mucorales28SrRNA and Aspergillus18SrRNA have been tested in the previous articles (Walsh et al., 2011; Springer et al., 2016a).
Ninety-three different samples from 88 patients were analysed by the four different real-time PCR assays (Figure 3; Table 2). All control group samples were negative using the four different RQ-PCR assays. Thirteen experimental group samples were negative in all RQ-PCR assays.
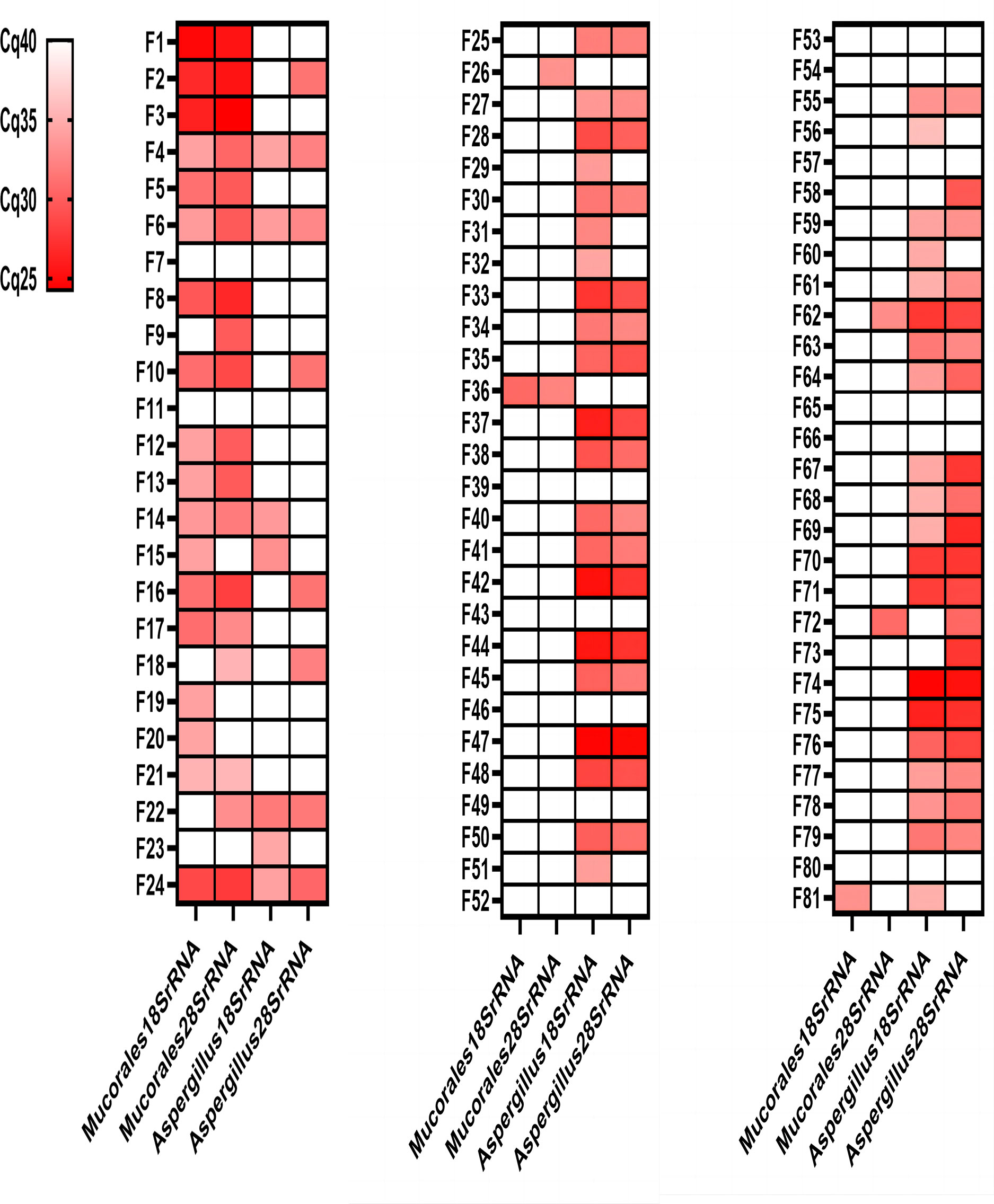
Figure 3 Genetic test results of the four RQ-PCR assays for 81 experimental group samples. Mucormycosis samples (F1-F21); Mucormycosis with Aspergillosis samples (F22-F24); Aspergillosis samples (F25-F81). Red: positive; White: negative.
The Mucorales18SrRNA assay was positive in 17 of 21 mucormycosis samples (80.95%), 1 of 3 mucormycosis with aspergillosis samples (33.33%), and 2 of 57 aspergillosis samples (3.51%). The Mucorales28SrRNA assay behaved similarly, detecting 16 out of 21 mucormycosis samples (76.19%), 2 out of 3 mucormycosis with aspergillosis samples (66.67%), and 4 out of 57 aspergillosis samples (7.02%). Only two samples were negative by two different Mucorales RQ-PCR despite positive histology showed fungal hyphae (F7 and F11). The Aspergillus18SrRNA assay was positive in 41 of 57 aspergillosis samples (71.93%), 3 of 3 mucormycosis with aspergillosis samples (100%), and 4 of 21 mucormycosis samples (19.05%). The Aspergillus28SrRNA assay behaved similarly, detecting 37 out of 57 aspergillosis samples (64.91%), 2 out of 3 mucormycosis with aspergillosis samples (66.67%), and 6 out of 21 mucormycosis samples (28.57%). 12 control samples were negative in each assay. A concomitant infection was diagnosed in three samples by histopathology but verified by Mucorales RQ-PCR and Aspergillus RQ-PCR in two (F22 and F24). There were some “abnormal results” in our study, e.g., 2 of aspergillosis samples were positive by Mucorales18SrRNA (F36 and F81); 4 of the aspergillosis samples were positive by Mucorales28SrRNA (F26, F36, F62 and F72); 4 of mucormycosis samples were positive by Aspergillus18SrRNA (F4, F6, F14 and F15); 6 of mucormycosis samples were positive by Aspergillus28SrRNA (F2, F4, F6, F10, F16 and F18).
The sensitivity, specificity, PPV, NPV, positive and negative LRs of all four RQ-PCR assays are shown in Table 3. The sensitivities of Mucorales18SrRNA and Mucorales 28SrRNA were both 75%, with specificity, PPV, NPV, positive LR, and negative LR of Mucorales18SrRNA being 97.10%, 90.00%, 91.78%, 25.86, and 0.26, respectively. Mucorales 28SrRNA assay showed specificity of 94.20%, PPV of 81.82%, NPV of 91.55%, positive LR of 12.93, and negative LR of 0.27. The sensitivity, specificity, PPV and NPV of Aspergillus28SrRNA assays were lower than Aspergillus18SrRNA assays, for Aspergillus18SrRNA assays: 73.33%, 87.88%, 91.67%, and 64.54%, respectively; for Aspergillus28SrRNA assays: 65%, 81.82%, 86.67%, and 56.25%, respectively. The positive LR and negative LR of Aspergillus18SrRNA assays and Aspergillus28SrRNA assays were 6.05 and 0.30 vs. 3.58 and 0.43.
The true positive results of M18A18, M18A28, M28A18, and M28A28 were as follows: for mucormycosis samples, 13, 12, 13, and 10, respectively; for mucormycosis with aspergillosis samples, 1, 1, 2, and 2, respectively; for aspergillosis samples, 40, 37, 40, and 35, respectively. All combined tests were negative in control group samples (Table 4).
A pairwise comparison of the tests showed that the highest level of agreement was M28A18, with a kappa coefficient value of 0.353. All other pairs of biomarkers showed less agreement (Table 5). Any combinations of Mucorales RQ-PCR assays and Aspergillus RQ-PCR assays had the same specificity (100%), PPV (100%), and positive LR (Infinity). The sensitivity, NPV, and negative LR were as follows: for M28A18, 67.90%, 31.58%, and 0.32 respectively; for M18A18, 66.67%, 30.77%, and 0.33, respectively; for M18A28, 61.73%, 27.91%, and 0.38, respectively; for M28A28, 58.02%, 26.09%, and 0.42, respectively. In all the samples, the values of the evaluation indexes of the combined detection of M28A18 were the highest, followed by M18A18.
The detection and identification of Mucorales and Aspergillus from FFPE samples played an important role in the diagnosis and management of aspergillosis and mucormycosis, whereas microscopy, serology, and culture were restricted by several disadvantages (Jensen et al., 1997; Frater et al., 2001; Rickerts et al., 2007; Hofman et al., 2010; Hamilos et al., 2011). Numerous RQ-PCR assays have been described for detection of Mucorales (Bernal-Martinez et al., 2013; Millon et al., 2013; Lengerova et al., 2014) and Aspergillus (Lass-Florl et al., 2011; Fricke et al., 2012; Paholcsek et al., 2014; Pini et al., 2015). In this study, we evaluated two Mucorales RQ-PCR assays, two Aspergillus RQ-PCR assays, and four combined tests to rapidly detect and identify Mucorales and Aspergillus.
More recently, the RQ-PCR of Mucorales from 268 serum samples and 12 FFPE samples was a promising test method with sensitivity of 91% targeting 18SrRNA gene (Springer et al., 2016b) and 86% targeting 28SrRNA gene (Springer et al., 2016a). In this study, two Mucorales RQ-PCR only had a sensitivity of 75%. The lower sensitivity may be relevant to small sample size and different sample types. The RQ-PCR detecting Mucorales also had a specificity of 100% in FFPE samples but a lower sensitivity of 56% by Hata et al. (Hata et al., 2008). The specificity of our two Mucorales RQ-PCR assays was 97.1% and 94.20%, respectively. As reported in the literature, the RQ-PCR specificity was 87.5% (Springer et al., 2016a). In Mucorales RQ-PCR assays, the diagnostic parameters and LoD values for the different assays indicated that Mucorales18SrRNA would provide the best diagnostic accuracy. These results support the findings of Springer et al. (Springer et al., 2016a; Springer et al., 2016b).
In our study, the specificity and sensitivity of Aspergillus18S rRNA were 87.88% and 73.33%, respectively, which is consistent with the results of previously published studies (Hadrich et al., 2011; Luong et al., 2011). The MycAssay™ Aspergillus real-time PCR kit was tested on tissues by the manufacturer with 15 different Aspergillus spp., including multiple strains of Aspergillus fumigatus, Aspergillus niger, Aspergillus terreus, and Aspergillus nidulans, having a sensitivity of 82% and a specificity of 79% (Lass-Florl et al., 2011). In our study, the specificity of two Aspergillus RQ-PCR assays was elevated, whereas sensitivity was reduced.
However, some limitations and several considerations indicate that it has some drawbacks in the Mucorales RQ-PCR or Aspergillus RQ-PCR alone. Our studies have evaluated the utility of detection of Mucorales RQ-PCR and Aspergillus RQ-PCR. Our finding, combining the results of these two tests gave optimal specificity and PPV, could be used to detect and identify Mucorales and Aspergillus and may provide a solution when gold standard tests were conflicting. Given the ubiquity of Aspergillus and Mucorales in the environment, combining Mucorales RQ-PCR with Aspergillus RQ-PCR would give clinicians greater confidence in detecting and identifying them at the same time and reduce the false positive and/or negative rate. The best combination was the M28A18, with a sensitivity of 67.90%, a specificity of 100%, which had the highest diagnostic potential with FFPE samples. This is inconsistent with the conclusion that Mucorales18SrRNA is better than Mucorales28SrRNA in diagnostic significance, which may be related to the small sample size. In all samples, the number of true positives of M18A18 was very similar to M28A18 (54 VS. 55). Their diagnostic significance needs to be further evaluated in future studies with larger sample sizes.
This study has several limitations, which may be the cause of some “abnormal results”. First, the sample size is too small, especially that the mucormycosis sample is only 21 cases from 19 patients. Due to the limited number of FFPE samples and strains, the very low number of Aspergillus and Mucorales species was tested, the number of Aspergillus and Mucorales species that can be detected by these primers needs to be further evaluated. Second, RQ-PCR assays require strictly positive and negative controls. The limitations of false positive and false negative errors due to amplification and contamination for assessing the value of a molecular diagnostic test have been eloquently highlighted (Mies, 1994; Cataloluk et al., 2003). In some samples, fluorescence signals higher than the positive Cq cutoff was detected, which may be caused by false negatives due to the low number of fungal hyphae. Third, the use of mold-active drugs may affect detection result. There are conflicting reports about the effect of antifungal therapy on the performance of tests. Antifungal therapy has been reported to both decrease (Reinwald et al., 2012) and increase (Musher et al., 2004) the diagnostic performance of RQ-PCR. Furthermore, “abnormal results” may be caused by a condition other than Mucormycosis and Aspergillosis or by drug treatment. Fourthly, Formalin fixed and paraffin wax embedded tissues can cause DNA degradation, only short sequences can be amplified from this type of tissue (Bonin et al., 2003). Although the amplified sequences of the four RQ-PCRs in this study were less than 200bp, which weakened the influence of DNA fragmentation, it may still reduce the sensitivity. Fifth, due to funding reasons, we did not use commercialized kits for the detection of Mucorales and Aspergillus DNA in clinical samples. Finally, Misclassification using these inconsistent criteria of RQ-PCR can occur for many reasons, e.g., detection of pathogenic fungi may be missed due to the diversity of fungal species and test samples, and there are multiple laboratorial protocols. Each suffers from different disadvantages such as vulnerability to contamination or limited detection of selected species or genera.
In conclusion, this preliminary study showed that the two Aspergillus RQ-PCR assays and two Mucorales RQ-PCR assays had high potential for the diagnosis of Mucorales and Aspergillus in FFPE samples. We envisage Aspergillus RQ-PCR and Mucorales RQ-PCR combination approach as a nearpatient test, allowing an immediate detection and identification of Mucorales and Aspergillus, with RQ-PCR results being available within a short time for samples of mucormycosis with aspergillosis. This combination approach can provide useful information when a small number of fungi are present, or the histological diagnosis is difficult in mucormycosis with aspergillosis samples. In the future, these assays may be used as a screening tool to detect other types of samples suspected of having mucormycosis and/or aspergillosis, such as serum, bronchoalveolar lavage fluid (BALF), and cytological samples. The results of our study should be validated in multicenter studies to develop tests for this clinical application.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by West China Hospital Ethics Committee of Sichuan University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
YJ and FY and conceived and designed the experiments. XJ analyzed the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work is supported by Chengdu Science and Technology Program (2019-YF05-00324-SN), West China Hospital of Sichuan University Scientific and Technological Achievements Transformation Fund (CGZH21011) and 1·3·5 project for disciplines of excellence–Clinical Research Incubation Project, West China Hospital, Sichuan University (No. 2020HXFH024).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Babouee Flury, B., Weisser, M., Prince, S. S., Bubendorf, L., Battegay, M., Frei, R., et al. (2014). Performances of two different panfungal PCRs to detect mould DNA in formalin-fixed paraffin-embedded tissue: What are the limiting factors? BMC Infect. Dis. 14, 692. doi: 10.1186/s12879-014-0692-z
Bernal-Martinez, L., Buitrago, M. J., Castelli, M. V., Rodriguez-Tudela, J. L., Cuenca-Estrella, M. (2013). Development of a single tube multiplex real-time PCR to detect the most clinically relevant mucormycetes species. Clin. Microbiol. Infect. 19 (1), E1–E7. doi: 10.1111/j.1469-0691.2012.03976.x
Bialek, R., Konrad, F., Kern, J., Aepinus, C., Cecenas, L., Gonzalez, G. M., et al. (2005). PCR based identification and discrimination of agents of mucormycosis and aspergillosis in paraffin wax embedded tissue. J. Clin. Pathol. 58 (11), 1180–1184. doi: 10.1136/jcp.2004.024703
Bonin, S., Petrera, F., Niccolini, B., Stanta, G. (2003). PCR analysis in archival postmortem tissues. Mol. Pathol. 56 (3), 184–186. doi: 10.1136/mp.56.3.184
Cataloluk, O., Cakmak, E. A., Buyukberber, N., Barlas, O. (2003). Formalin fixing and paraffin embedding may lead to extra band development in PCR. New microbiologica 26 (2), 193–198.
De Pauw, B., Walsh, T. J., Donnelly, J. P., Stevens, D. A., Edwards, J. E., Calandra, T., et al. (2008). Revised definitions of invasive fungal disease from the European organization for research and treatment of Cancer/Invasive fungal infections cooperative group and the national institute of allergy and infectious diseases mycoses study group (EORTC/MSG) consensus group. Clin. Infect. Dis. 46 (12), 1813–1821. doi: 10.1086/588660
Frater, J. L., Hall, G. S., Procop, G. W. (2001). Histologic features of zygomycosis: emphasis on perineural invasion and fungal morphology. Arch. Pathol. Lab. Med. 125 (3), 375–378. doi: 10.1043/0003-9985(2001)125<0375:HFOZ>2.0.CO;2
Fricke, S., Fricke, C., Oelkrug, C., Blatz, R., Schonfelder, U., Niederwieser, D., et al. (2012). A real-time PCR for the detection and characterisation of aspergillus species. Mycoses 55 (5), 416–425. doi: 10.1111/j.1439-0507.2011.02161.x
Gade, L., Hurst, S., Balajee, S. A., Lockhart, S. R., Litvintseva, A. P. (2017). Detection of mucormycetes and other pathogenic fungi in formalin fixed paraffin embedded and fresh tissues using the extended region of 28S rDNA. Med. Mycol 55 (4), 385–395. doi: 10.1093/mmy/myw083
Guarner, J., Brandt, M. E. (2011). Histopathologic diagnosis of fungal infections in the 21st century. Clin. Microbiol. Rev. 24 (2), 247–280. doi: 10.1128/CMR.00053-10
Hadrich, I., Mary, C., Makni, F., Elloumi, M., Dumon, H., Ayadi, A., et al. (2011). Comparison of PCR-ELISA and real-time PCR for invasive aspergillosis diagnosis in patients with hematological malignancies. Med. Mycol 49 (5), 489–494. doi: 10.3109/13693786.2010.540724
Hamilos, G., Samonis, G., Kontoyiannis, D. P. (2011). Pulmonary mucormycosis. Semin. Respir. Crit. Care Med. 32 (6), 693–702. doi: 10.1055/s-0031-1295717
Hata, D. J., Buckwalter, S. P., Pritt, B. S., Roberts, G. D., Wengenack, N. L. (2008). Real-time PCR method for detection of zygomycetes. J. Clin. Microbiol. 46 (7), 2353–2358. doi: 10.1128/JCM.02331-07
Hofman, V., Dhouibi, A., Butori, C., Padovani, B., Gari-Toussaint, M., Garcia-Hermoso, D., et al. (2010). Usefulness of molecular biology performed with formaldehyde-fixed paraffin embedded tissue for the diagnosis of combined pulmonary invasive mucormycosis and aspergillosis in an immunocompromised patient. Diagn. Pathol. 5, 1. doi: 10.1186/1746-1596-5-1
Jensen, H. E., Salonen, J., Ekfors, T. O. (1997). The use of immunohistochemistry to improve sensitivity and specificity in the diagnosis of systemic mycoses in patients with haematological malignancies. J. Pathol. 181 (1), 100–105. doi: 10.1002/(SICI)1096-9896(199701)181:1<100::AID-PATH100>3.0.CO;2-O
Lass-Florl, C., Follett, S. A., Moody, A., Denning, D. W. (2011). Detection of aspergillus in lung and other tissue samples using the MycAssay aspergillus real-time PCR kit. Can. J. Microbiol. 57 (9), 765–768. doi: 10.1139/w11-064
Lengerova, M., Racil, Z., Hrncirova, K., Kocmanova, I., Volfova, P., Ricna, D., et al. (2014). Rapid detection and identification of mucormycetes in bronchoalveolar lavage samples from immunocompromised patients with pulmonary infiltrates by use of high-resolution melt analysis. J. Clin. Microbiol. 52 (8), 2824–2828. doi: 10.1128/JCM.00637-14
Luong, M. L., Clancy, C. J., Vadnerkar, A., Kwak, E. J., Silveira, F. P., Wissel, M. C., et al. (2011). Comparison of an aspergillus real-time polymerase chain reaction assay with galactomannan testing of bronchoalvelolar lavage fluid for the diagnosis of invasive pulmonary aspergillosis in lung transplant recipients. Clin. Infect. Dis. 52 (10), 1218–1226. doi: 10.1093/cid/cir185
Mies, C. (1994). Molecular biological analysis of paraffin-embedded tissues. Hum. Pathol. 25 (6), 555–560. doi: 10.1016/0046-8177(94)90218-6
Millon, L., Larosa, F., Lepiller, Q., Legrand, F., Rocchi, S., Daguindau, E., et al. (2013). Quantitative polymerase chain reaction detection of circulating DNA in serum for early diagnosis of mucormycosis in immunocompromised patients. Clin. Infect. Dis. 56 (10), e95–101. doi: 10.1093/cid/cit094
Munoz-Cadavid, C., Rudd, S., Zaki, S. R., Patel, M., Moser, S. A., Brandt, M. E., et al. (2010). Improving molecular detection of fungal DNA in formalin-fixed paraffin-embedded tissues: comparison of five tissue DNA extraction methods using panfungal PCR. J. Clin. Microbiol. 48 (6), 2147–2153. doi: 10.1128/JCM.00459-10
Musher, B., Fredricks, D., Leisenring, W., Balajee, S. A., Smith, C., Marr, K. A. (2004). Aspergillus galactomannan enzyme immunoassay and quantitative PCR for diagnosis of invasive aspergillosis with bronchoalveolar lavage fluid. J. Clin. Microbiol. 42 (12), 5517–5522. doi: 10.1128/JCM.42.12.5517-5522.2004
Oren, I., Paul, M. (2014). Up to date epidemiology, diagnosis and management of invasive fungal infections. Clin. Microbiol. Infect. 20 Suppl 6, 1–4. doi: 10.1111/1469-0691.12642
Paholcsek, M., Leiter, E., Markovics, A., Biro, S. (2014). Novel and sensitive qPCR assays for the detection and identification of aspergillosis causing species. Acta Microbiol. Immunol. Hung 61 (3), 273–284. doi: 10.1556/AMicr.61.2014.3.3
Pegorie, M., Denning, D. W., Welfare, W. (2017). Estimating the burden of invasive and serious fungal disease in the united kingdom. J. Infect. 74 (1), 60–71. doi: 10.1016/j.jinf.2016.10.005
Pini, P., Bettua, C., Orsi, C. F., Venturelli, C., Faglioni, L., Forghieri, F., et al. (2015). Clinical performance of a commercial real-time PCR assay for aspergillus DNA detection in serum samples from high-risk patients: comparison with a galactomannan enzyme immunoassay. Eur. J. Clin. Microbiol. Infect. Dis. 34 (1), 131–136. doi: 10.1007/s10096-014-2211-y
Reinwald, M., Hummel, M., Kovalevskaya, E., Spiess, B., Heinz, W. J., Vehreschild, J. J., et al. (2012). Therapy with antifungals decreases the diagnostic performance of PCR for diagnosing invasive aspergillosis in bronchoalveolar lavage samples of patients with haematological malignancies. J. Antimicrob. Chemother. 67 (9), 2260–2267. doi: 10.1093/jac/dks208
Rickerts, V., Just-Nubling, G., Konrad, F., Kern, J., Lambrecht, E., Bohme, A., et al. (2006). Diagnosis of invasive aspergillosis and mucormycosis in immunocompromised patients by seminested PCR assay of tissue samples. Eur. J. Clin. Microbiol. Infect. Dis. 25 (1), 8–13. doi: 10.1007/s10096-005-0078-7
Rickerts, V., Mousset, S., Lambrecht, E., Tintelnot, K., Schwerdtfeger, R., Presterl, E., et al. (2007). Comparison of histopathological analysis, culture, and polymerase chain reaction assays to detect invasive mold infections from biopsy specimens. Clin. Infect. Dis. 44 (8), 1078–1083. doi: 10.1086/512812
Ruhnke, M., Groll, A. H., Mayser, P., Ullmann, A. J., Mendling, W., Hof, H., et al. (2015). Estimated burden of fungal infections in Germany. Mycoses 58 Suppl 5, 22–28. doi: 10.1111/myc.12392
Ruiz-Camps, I., Jarque, I. (2014). [Invasive mould disease in haematological patients]. Rev. Iberoam Micol 31 (4), 249–254. doi: 10.1016/j.riam.2014.06.002
Sangoi, A. R., Rogers, W. M., Longacre, T. A., Montoya, J. G., Baron, E. J., Banaei, N. (2009). Challenges and pitfalls of morphologic identification of fungal infections in histologic and cytologic specimens: a ten-year retrospective review at a single institution. Am. J. Clin. Pathol. 131 (3), 364–375. doi: 10.1309/AJCP99OOOZSNISCZ
Springer, J., Goldenberger, D., Schmidt, F., Weisser, M., Wehrle-Wieland, E., Einsele, H., et al. (2016a). Development and application of two independent real-time PCR assays to detect clinically relevant mucorales species. J. Med. Microbiol. 65 (3), 227–234. doi: 10.1099/jmm.0.000218
Springer, J., Lackner, M., Ensinger, C., Risslegger, B., Morton, C. O., Nachbaur, D., et al. (2016b). Clinical evaluation of a mucorales-specific real-time PCR assay in tissue and serum samples. J. Med. Microbiol. 65 (12), 1414–1421. doi: 10.1099/jmm.0.000375
Springer, J., White, P. L., Kessel, J., Wieters, I., Teschner, D., Korczynski, D., et al. (2018). A comparison of aspergillus and mucorales PCR testing of different bronchoalveolar lavage fluid fractions from patients with suspected invasive pulmonary fungal disease. J. Clin. Microbiol. 56 (2), e01655–17. doi: 10.1128/JCM.01655-17
Tarrand, J. J., Lichterfeld, M., Warraich, I., Luna, M., Han, X. Y., May, G. S., et al. (2003). Diagnosis of invasive septate mold infections. a correlation of microbiological culture and histologic or cytologic examination. Am. J. Clin. Pathol. 119 (6), 854–858. doi: 10.1309/EXBV-YAUP-ENBM-285Y
Walsh, T. J., Wissel, M. C., Grantham, K. J., Petraitiene, R., Petraitis, V., Kasai, M., et al. (2011). Molecular detection and species-specific identification of medically important aspergillus species by real-time PCR in experimental invasive pulmonary aspergillosis. J. Clin. Microbiol. 49 (12), 4150–4157. doi: 10.1128/JCM.00570-11
Keywords: mucorales, aspergillus, FFPE, RQ-PCR, 18SrRNA gene, 28SrRNA gene
Citation: Jiang X, Jiang Y and Ye F (2023) Detection and identification of Mucorales and Aspergillus in paraffin-embedded samples by real-time quantitative PCR. Front. Cell. Infect. Microbiol. 13:1082347. doi: 10.3389/fcimb.2023.1082347
Received: 28 October 2022; Accepted: 17 February 2023;
Published: 02 March 2023.
Edited by:
Christoph Gabler, Freie Universität Berlin, GermanyReviewed by:
Eman A. Gouda M. Youssef, Lundquist Institute for Biomedical Innovation, United StatesCopyright © 2023 Jiang, Jiang and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong Jiang, aW1wbGFzbWFjZWxsQHNjdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.