- 1Host Microbe Interaction Research Laboratory, Faculty of Bioresources and Environmental Sciences, Ishikawa Prefectural University, Nonoichi, Japan
- 2Laboratory of Food Immunology, Department of Science and Technology on Food Safety, Faculty of Biology-Oriented Science and Technology, Kindai University, Kinokawa, Japan
In recent years, with the development of non-cultivation approaches, it has become evident that intestinal bacteria have a significant impact on human health. However, because one-third of the genes cannot be annotated, it is difficult to elucidate the function of all intestinal bacteria by in silico analysis, and it is necessary to study the intestinal bacteria by culturing them. In addition, various media recommended for each individual bacterium have been used for culturing intestinal bacteria; however, the preparation of each medium is complex. To simultaneously culture many bacteria and compare bacterial phenotypes under the same conditions, a medium capable of culturing a wide range of bacteria is needed. In this study, we developed GAM + blood medium (GB medium), which consists of Gifu anaerobic medium containing 5% (v/v) horse blood; it is easy to prepare and it allowed the successful cultivation of 85% of the available predominant species in the human intestinal microbiota.
1 Introduction
Animals maintain a complex microbiota in their intestinal lumen, and it is becoming increasingly clear that the intestinal microbiota and health are closely related (Hsiao Elaine et al., 2013; Rosshart et al., 2017; Sharon et al., 2019; Buffington et al., 2021). Therefore, recent research in the field of gut microbes has focused on the function of intestinal microbiota as a community. Next-generation sequencing analysis of DNA and RNA in human feces has been performed since the early 21st century (Shendure and Ji, 2008). These non-cultivation methods have revealed the gene expression profile of the human intestinal microbiota, the catalogue of human intestinal microbial genes, and the predominant species of the intestinal microbiota (Qin et al., 2010; Nishijima et al., 2016). However, there are several undeveloped aspects of the information that can be obtained from next-generation sequencing analysis. Metagenomic gut microbiota analysis uses DNA extracted from feces for next-generation sequencing. However, it has been reported that the analysis of sequencing results vary greatly depending on that the used DNA extraction protocols (Costea et al., 2017). In addition, bias has been reported in amplicon-based library preparation due to sequencing primers (Gohl et al., 2016). Furthermore, even when amino acid sequences are revealed via next-generation sequencing, one-third of the genes cannot be annotated (Chang et al., 2015), and it is difficult to elucidate and regulate the function of the human intestinal microbiota based on their DNA sequences and microbiota composition.
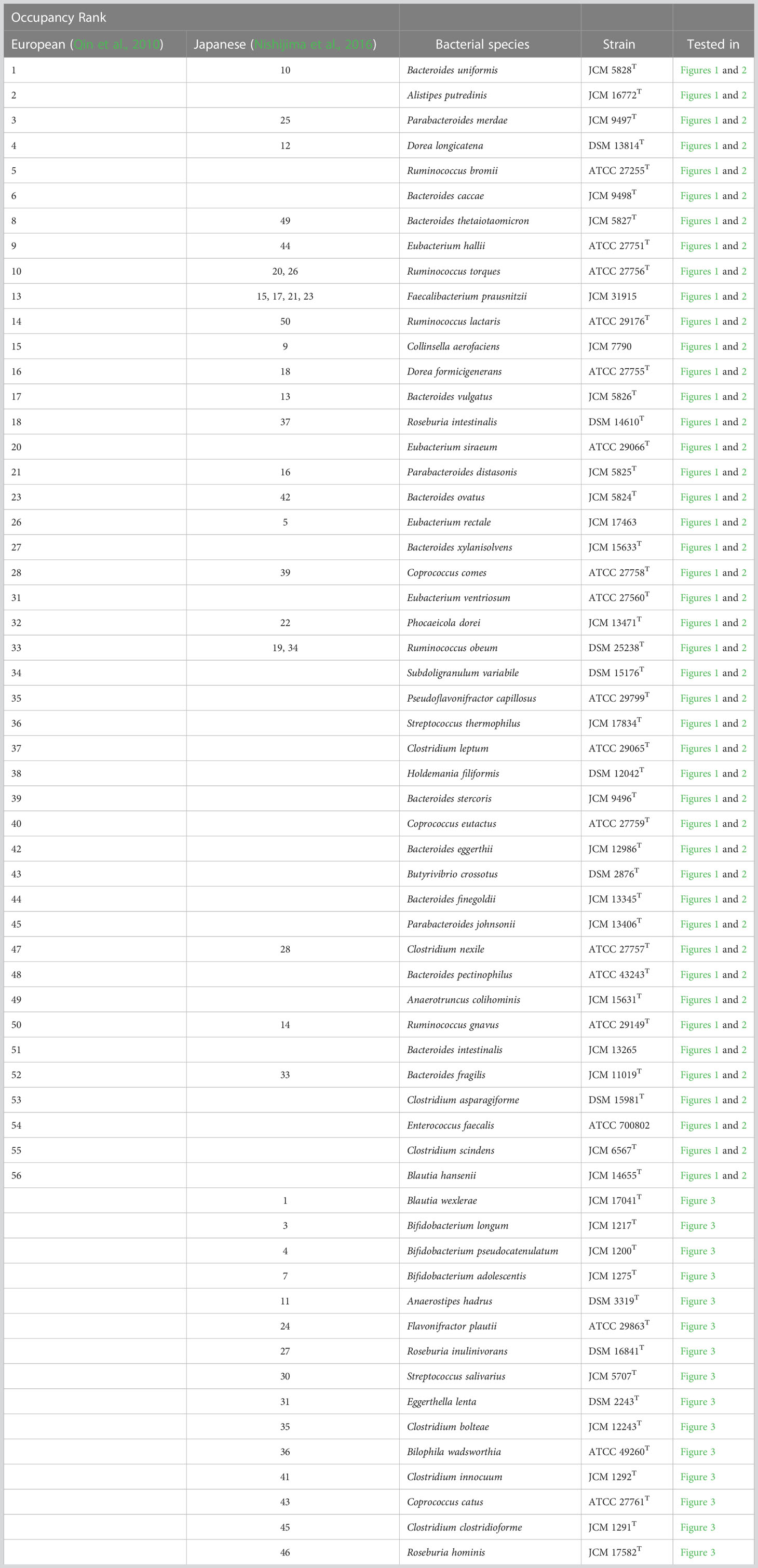
In 2010, the results of genome analysis using next-generation sequencing without cultivation reported 56 genomes that were predominant in the intestines of Europeans (Qin et al., 2010) (Table 1). Of the 56 genomes, 45 could be successfully assigned to cultured strains (white column in Table 1). For these 45 species, representative strains are available from the culture collections [such as the American Type Culture Collection (ATCC), the German Collection of Microorganisms and Cell Cultures GmbH (DSMZ), and the Japan Collection of Microorganisms (JCM)] (Table 1). However, the 11 genomes that could not be assigned to cultured bacteria cannot be investigated using live bacteria (gray rows in Table 1). Similarly, the 50 predominant strains in the gut of Japanese individuals have been reported (Nishijima et al., 2016), of which 41 are available (Table 2). To stably culture these species and strains, it is recommended to use the medium (Tables 1, 2) designated for each species by the respective distributing institution. However, when several intestinal bacterial taxa are cultured simultaneously, for example, in a 96-well plate, it is necessary to provide different media for each well. This results in greatly increased time, effort, and cost. In addition, different media hamper physiological comparisons among bacteria owing to differences in composition and accurate quantification of bacterial growth because of the presence or absence of precipitation in the media.
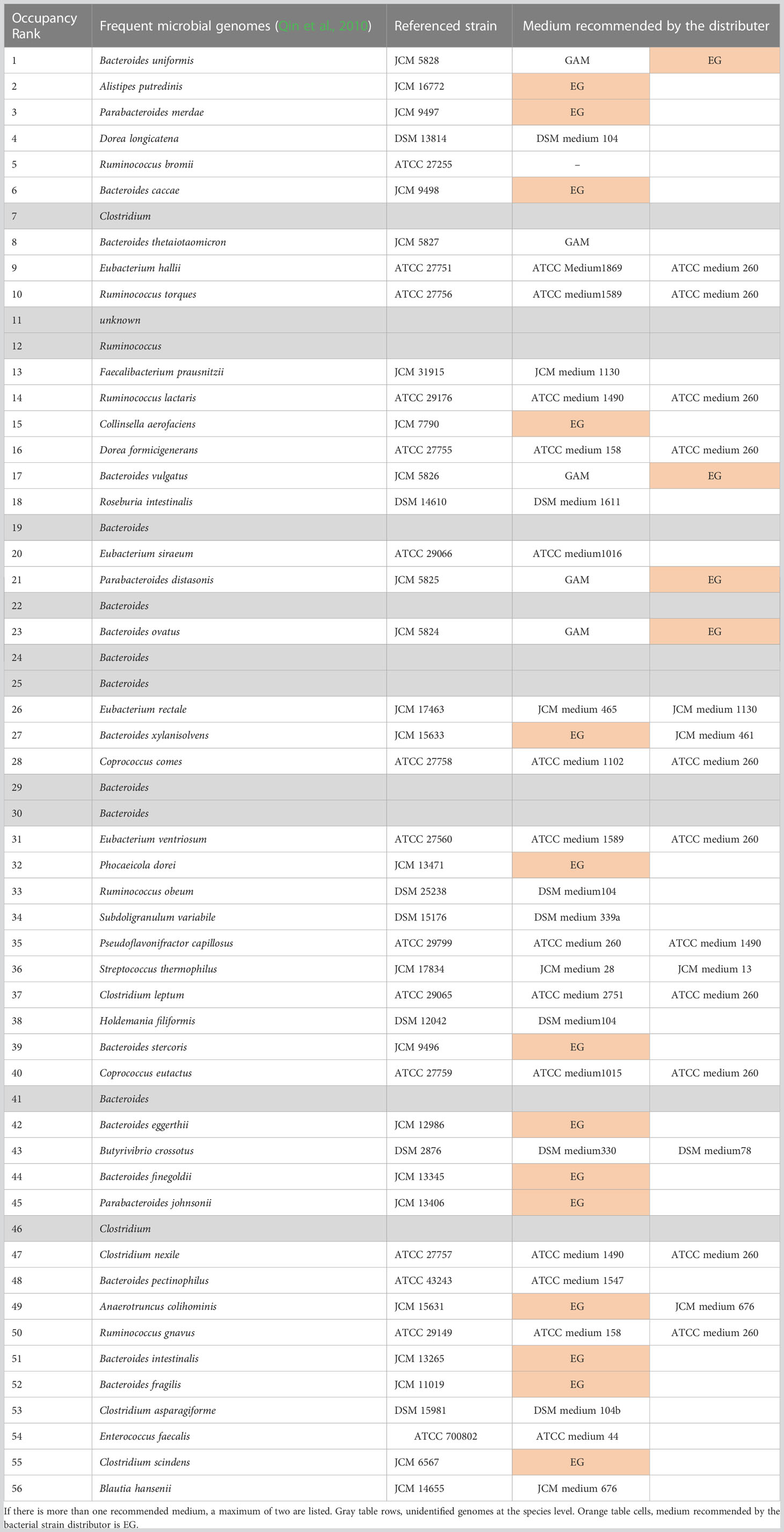
Table 1 Fifty-six most dominant species in the gut of Europeans (Qin et al., 2010) and their recommended medium.
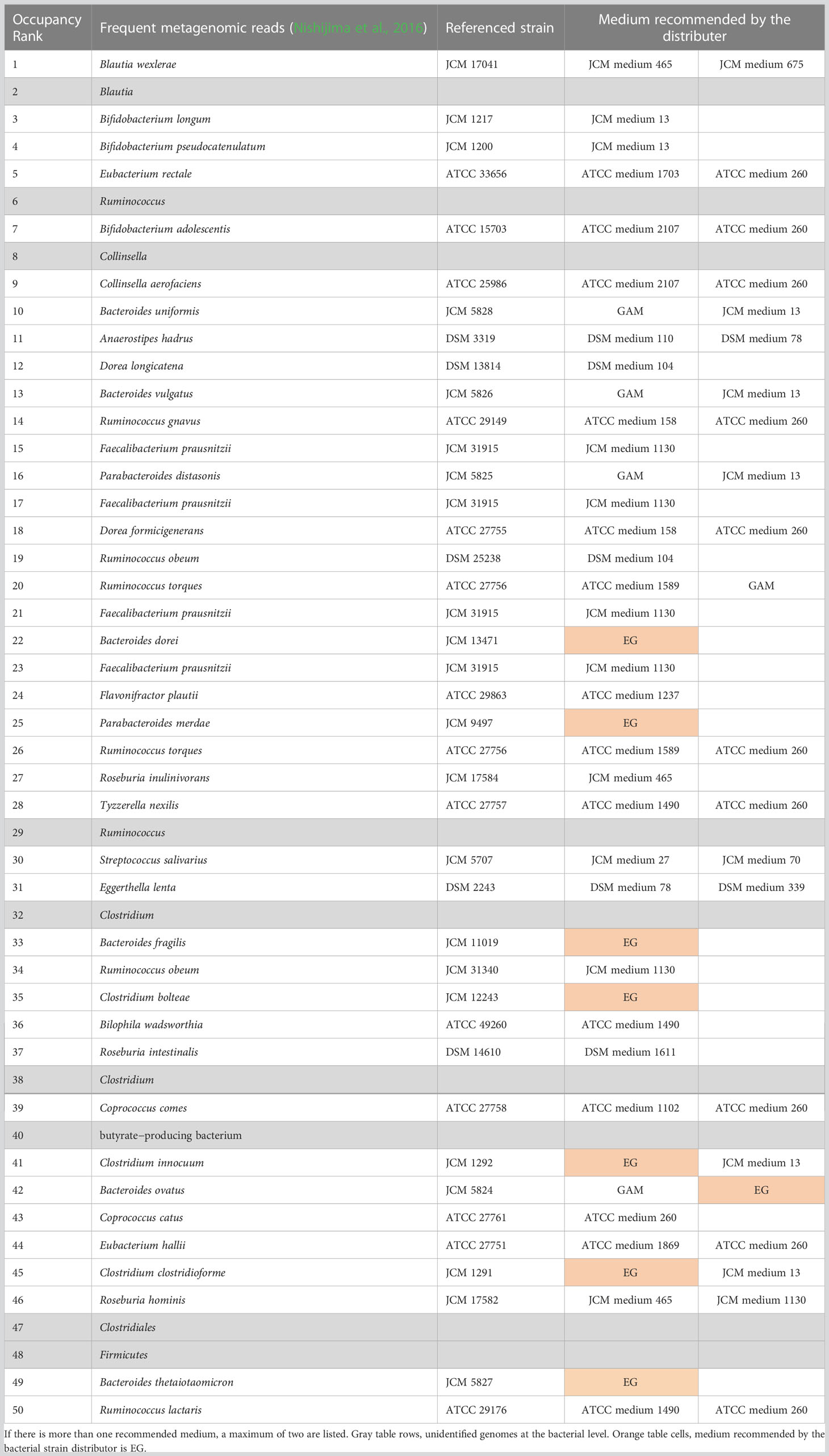
Table 2 Fifty most dominant species in the gut of Japanese (Nishijima et al., 2016) and their recommended medium.
Therefore, we set out to develop a method capable of culturing a large number of species of intestinal bacteria without producing precipitates, and found a method for utilizing Gifu anaerobic medium (GAM) for both pre-culture and main culture (Gotoh et al., 2017). Using this culturing method, 32 of the 44 predominant species of European gut microbiota available at the time were successfully cultured (Gotoh et al., 2017). Using this system, we previously reported five findings. First, we quantified polyamines in the predominant species of the human gut microbiota and reported the existence of many previously unknown metabolic and transport systems for polyamines (Sugiyama et al., 2017). Second, we used our system to screen for the oligosaccharide Gal-β1,4-Rha, which is not utilized by the predominant species of the human gut microbiota and is specifically utilized by bifidobacteria (Hirano et al., 2021). Third, we have also reported a comprehensive analysis of the growth inhibitory activity of medium-chain fatty acids on the predominant species in the gut of Europeans (Matsue et al., 2019). Fourth, we analyzed the effects of micronized “okara” on the growth and metabolic production of the predominant species (Nagano et al., 2020). Fifth, we analyzed phenylethylamine production by the predominant species in the gut of Europeans and found that phenethylamine from gut bacteria stimulated the production of colonic serotonin (Sugiyama et al., 2022). Thus, a system that can grow a wide range of gut microbiota under the same conditions facilitates cross-species comparisons and provides a variety of insights. However, because 32 species represent only 73% of the 44 species, the development of a culture method capable of culturing a wider variety of intestinal bacteria is desired.
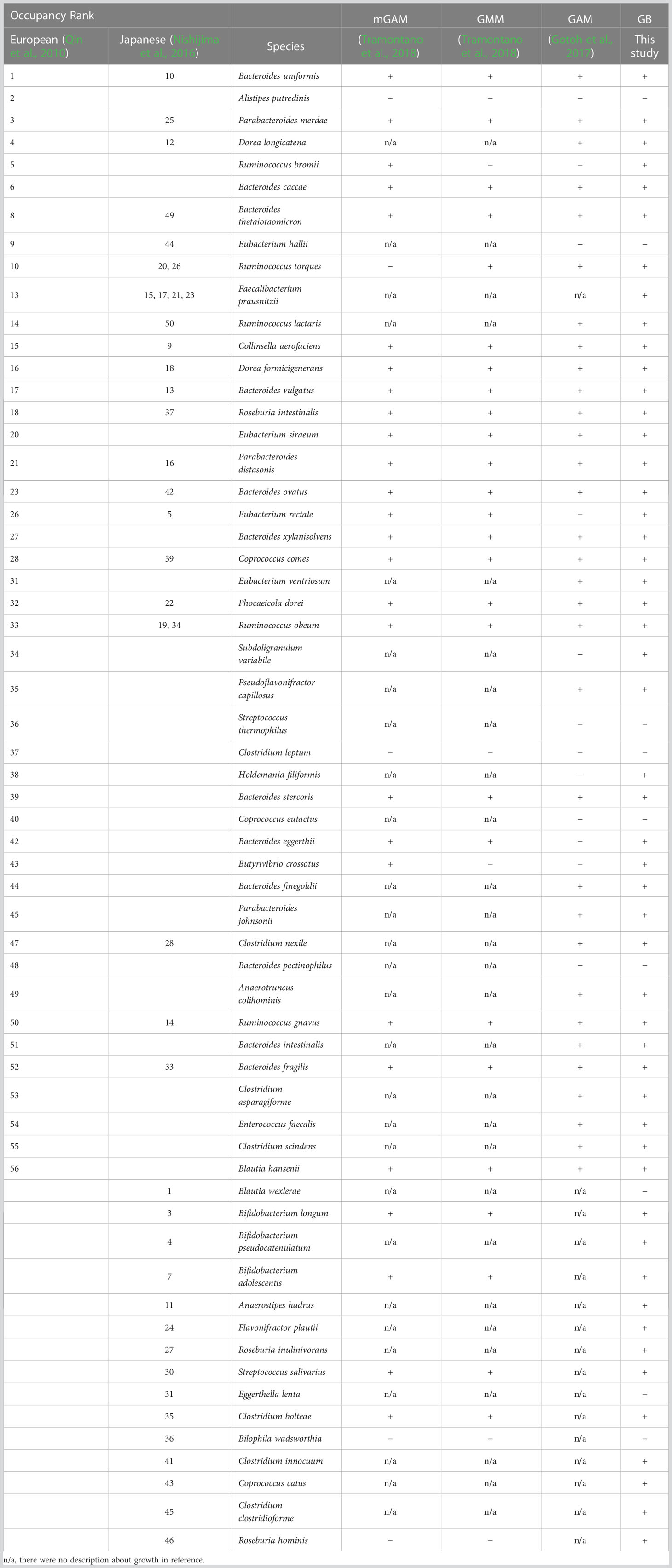
Some studies have reported culturing a wide variety of intestinal bacteria using different media. A modified Gifu anaerobic medium (mGAM) is lighter in color and more transparent and is useful in the isolation and cultivation of anaerobic bacteria and in drug susceptibility testing. A total of 45 species commonly occurring within the human population were inoculated into mGAM, and 34 species (76%) were able to grow (Tramontano et al., 2018). However, when bacteria were isolated from human feces using mGAM, 174 genera were detected by 16S rRNA gene analysis and, 48 genera were isolated, suggesting that many genera cannot be cultured using mGAM (Biclot et al., 2022). Although the gut microbiota medium (GMM) (Goodman et al., 2011) is a chemically defined medium, the number of isolated and cultured bacteria is 70% of the genera (Goodman et al., 2011) and 71% of the families (Rettedal et al., 2014) detected in fecal samples, and 33 of the 45 species commonly occurring within the human population (73%) are culturable (Tramontano et al., 2018). Thus, although attempts have been made to culture a wide range of bacteria, it is difficult to completely represent the gut microbiota.
In this study, we developed a culture medium and method that allows the cultivation of more intestinal bacteria, enables comprehensive and simple cultivation of the predominant species of human gut microbiota, and simplifies the subsequent analysis.
2 Materials and methods
2.1 Microbe strains
Bacteria were obtained from the American Type Culture Collection (ATCC), the German Collection of Microorganisms and Cell Cultures GmbH (DSMZ), and the Japan Collection of Microorganisms (JCM) (Table 3). Bacteria were cultured at 37°C in an anaerobic chamber (10% CO2, 10% H2, and 80% N2; InvivO2 400; Ruskinn Technology, Bridgend, UK).
2.2 Preparation of GAM + Eggerth–Gagnon medium (GE)
GAM (Nissui Pharmaceutical, Tokyo, Japan) was autoclaved (115°C for 15 min), immediately placed in a closed container with Aneropack Kenki (Mitsubishi Gas Chemical Company, Tokyo, Japan), and allowed to stand overnight to remove oxygen. Eggerth–Gagnon (EG) medium (composition: proteose peptone No. 3, yeast extract, Na2HPO4, glucose, soluble starch, l-cystine, l-cysteine ·HCl·H2O, and horse blood) was prepared according to the JCM’s instructions1. Materials other than blood were autoclaved, placed in a closed container together with Aneropack Kenki, and allowed to stand overnight to remove dissolved oxygen. Horse blood (horse whole blood defibrinated sterile; Nippon Bio-Supp. Center, Tokyo, Japan) stored anaerobically with Aneropack Kenki was added to the GAM at 5% (v/v) in an anaerobic chamber. GAM and EG medium were mixed in a 1:1 (v/v) ratio.
2.3 Preparation of GAM supplemented with Blood medium (GB)
GAM was autoclaved (115°C, 15 min), immediately placed in a closed container together with Aneropack Kenki, and allowed to stand overnight to remove dissolved oxygen. Horse blood that was stored anaerobically with Aneropack Kenki was then added to GAM at 5% (v/v) in an anaerobic chamber. To prepare GBsheep, sheep blood (Japan Bio Serum, Tokyo, Japan) was added instead of horse blood using the same procedure, and for GBhuman, human blood (Tennessee Blood Services, Tennessee, US) was added instead of horse blood, using the same procedure.
2.4 Culturing system
The experimental procedure is shown in Figure 1. Bacteria were cultured in an anaerobic chamber. First, bacterial strains were inoculated from frozen glycerol stock in 500 μL or 3 mL of media in 96-deep well plates or vials, respectively, and incubated at 37°C for 24-96 hours. GAM (Figure 1B), GE (Figure 1C) or GB (Figures 1D, 2, 3, Supplementary Figures S1, S2) were used as the medium for pre-culture. For pre-culturing in vials, 500 µL of the pre-culture solution was transferred to a 96-well plate before using a copy stand. Approximately 2 µL of the respective culture collection was inoculated in 500 μL of GAM in 96-deep well plates using a copy plate stand (Tokken, Chiba, Japan). After 48 hours (Figure 3; Supplementary Figures S1, S2) or 96 hours (Figure 1) of anaerobic incubation, growth was measured as the optical density at 600 nm (OD600) using Thermo Scientific™ Multiskan™ GO (Thermo Fisher Scientific, Waltham, MA). For Figure 2, measurements were taken over time up to 96 hours. The possibility of culture contamination was eliminated by 16S rDNA sequencing using previously described procedures (Gotoh et al., 2017) (Supplementary Tables S1, S2).
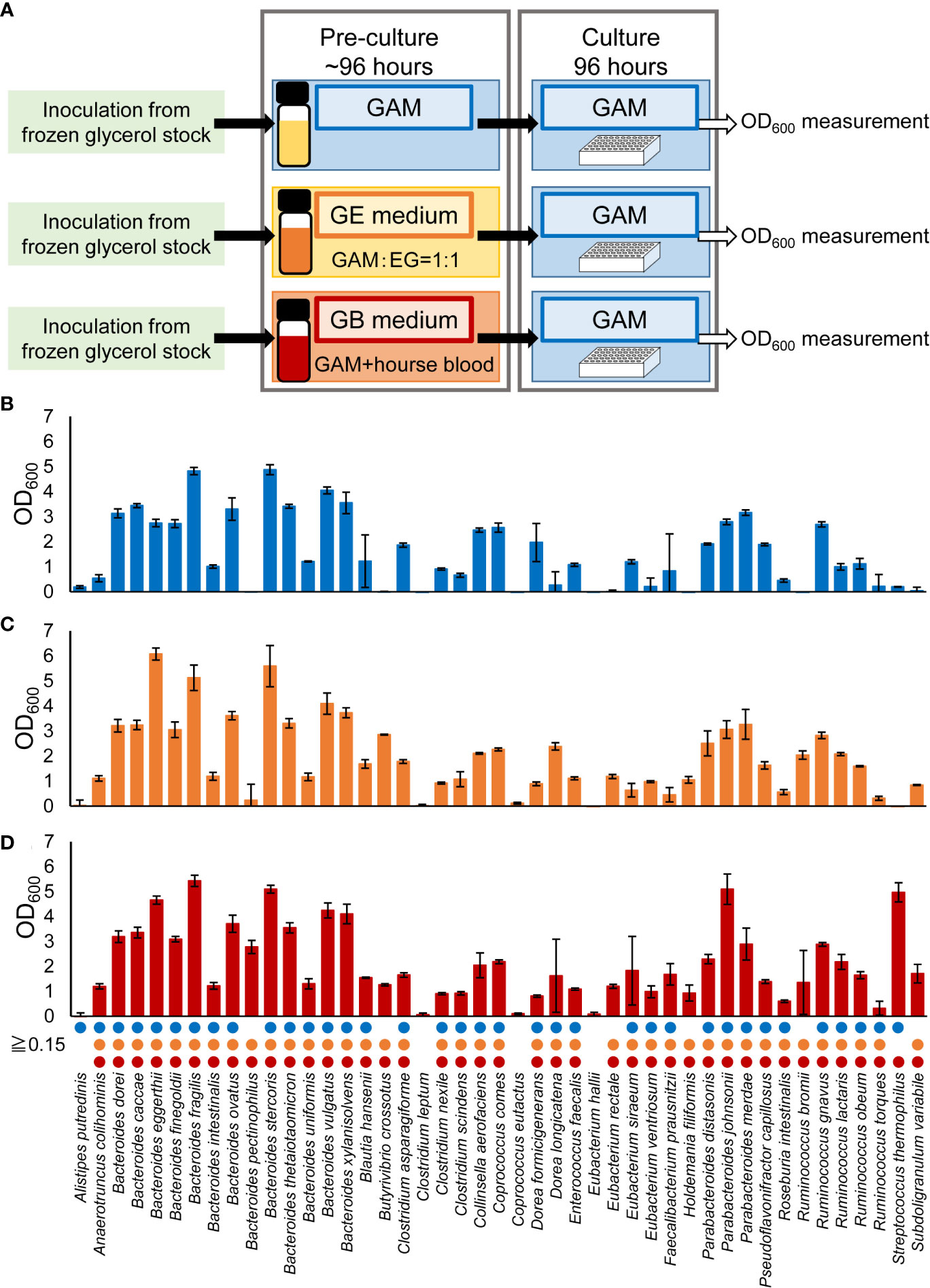
Figure 1 Comparison of pre-culture medium conditions required for culturing a wide range of predominant species of intestinal bacteria. (A) Experimental overview. (B–D) After pre-culturing in GAM (blue bars in B), GE medium (orange bars in C), or GB medium (red bars in D), the bacterial species were cultured in GAM for 96 hours. Bacterial growth was measured by determining the OD600. Circles indicate bacteria with an OD600 ≥ 0.15. Data are presented as the mean ± standard deviation (n = 3). (B) The growth was confirmed using 16S rDNA sequencing in cases where bacteria that did not grow in previous reports (Gotoh et al., 2017) using the GAM in pre-culture were grown in this study (Supplementary Table S1).
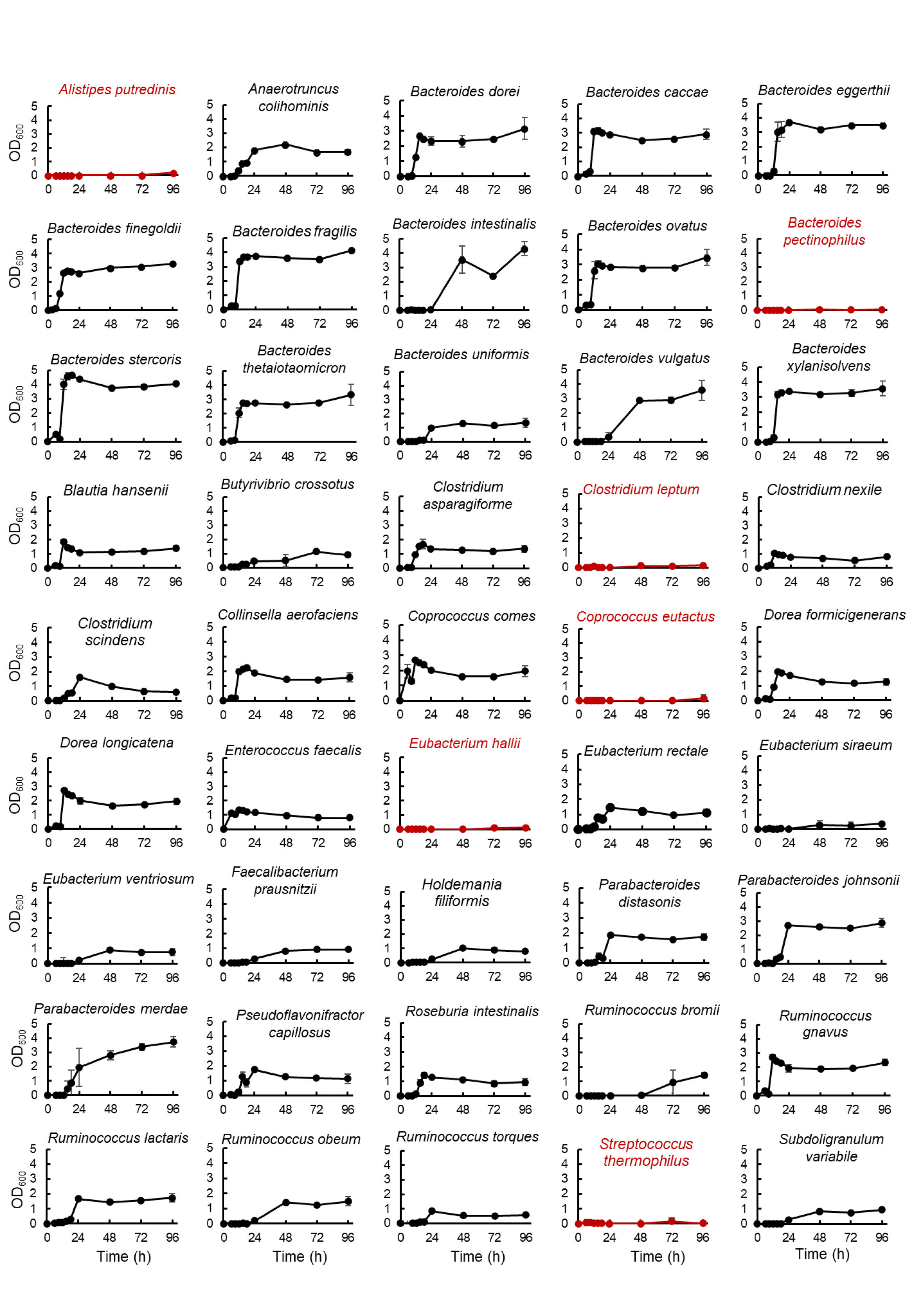
Figure 2 Growth curve of predominant species of intestinal bacteria grown using the developed culture system. After pre-culturing in GB medium, bacterial species were cultured in GAM for 96 hours. Growth was tracked by measuring OD600 over time. Bacteria with two or more points with an OD600 greater than 0.15 are shown on the graph in black. The growth of these bacteria was confirmed using 16S rDNA sequencing (Supplementary Table S2). Bacteria with one or zero points with an OD greater than 0.15 are indicated by red graphs. Data are presented as the mean ± standard deviation (n = 3).
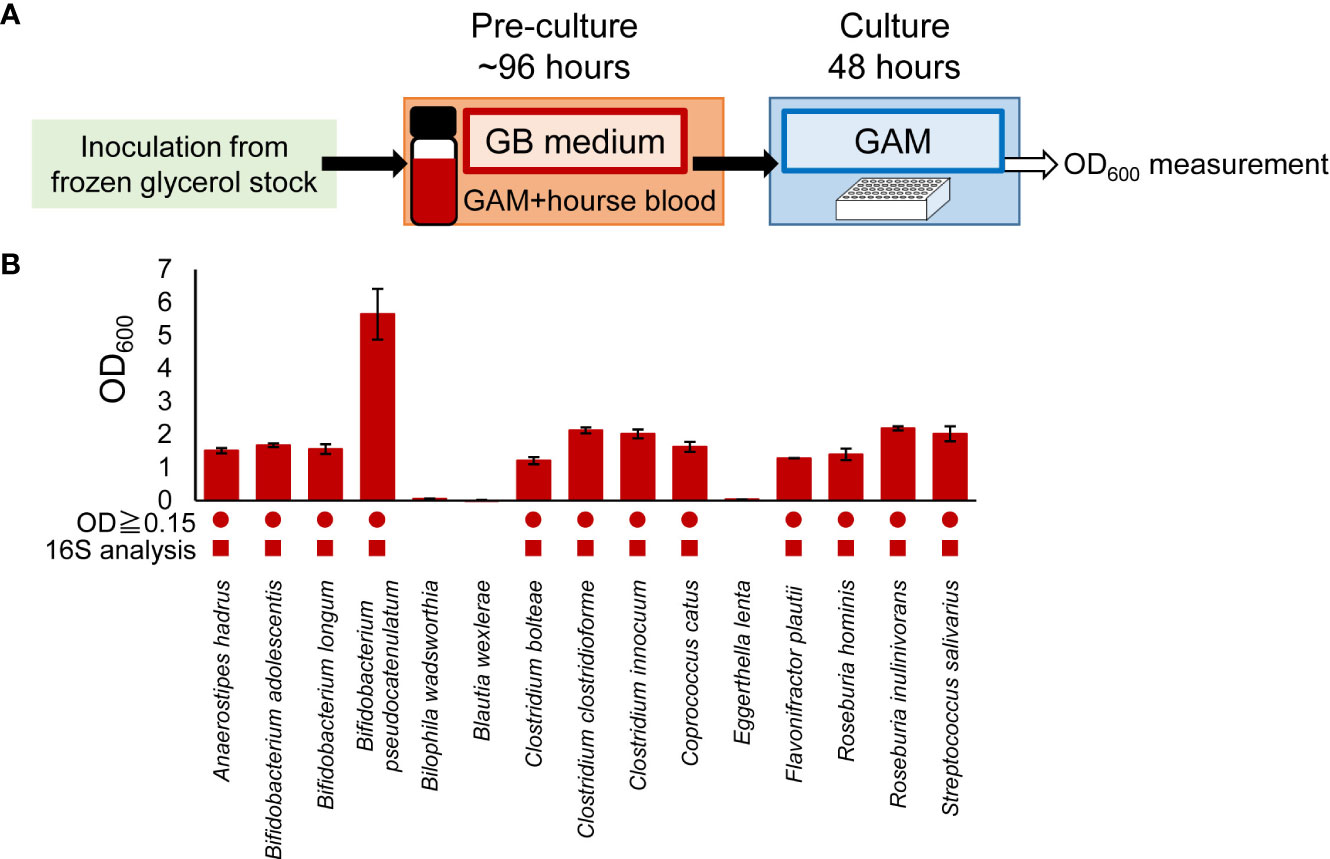
Figure 3 Adaptability of GB medium for predominant species in other gut microbiome projects. (A) Experimental overview. (B) After pre-culturing in GB medium, bacterial species were cultured in GAM for 48 hours. Bacterial growth was measured by determining the OD600. Circles indicate bacteria OD600 ≥ 0.15. Square symbols indicate growth confirmed by 16S rDNA sequencing analysis (Supplementary Table S2). Data represent means ± standard deviation (n = 3).
3 Results
3.1 Development of culture media capable of culturing a wide range of bacteria
We previously reported that 32 of the 56 predominant species in the human gut microbiota can be cultured in GAM (Gotoh et al., 2017). As with GAM, EG medium is recommended for numerous gut microbes (Table 1). Therefore, GE medium, a 1:1 (v/v) mixture of GAM and EG, was prepared. GAM + blood medium (GB medium) was also prepared by adding 5% (v/v) of horse blood to GAM, with reference to the fact that horse blood was supplemented to the EG medium at a final concentration of 5% (v/v). Of the 56 predominant species of European intestinal commensal microbiota identified using non-culture methods, 45 species available from the culture collection were pre-cultured in GAM, GE, and GB (Figure 1A). It was difficult to measure OD600 in GE and GB because of the turbidity derived from the added horse blood; therefore, the pre-culture was inoculated into GAM and cultivated anaerobically for 96 h at 37°C to test the growth of bacterial species by measuring the OD600 value (Figure 1A). The presence or absence of growth was determined using a threshold of OD600 = 0.15, as previously described (Tramontano et al., 2018). The number of bacterial species whose growth in GAM exceeded 0.15 was 36 (80% of the tested strains) when the pre-culture was performed on GAM (Figure 1B), 40 (89% of the tested strains) when the pre-culture was performed on GE medium (Figure 1C), and 41 (91% of the tested strains) when the pre-culture was performed on GB medium (Figure 1D). Compared to pre-culture using the conventional method of GAM (Gotoh et al., 2017), the number of species that could be grown was increased using our newly prepared GE or GB media for pre-culture. GB was chosen for subsequent experiments because it was able to culture the greatest number of species.
3.2 Stable growth of a wide range of bacteria using GB medium
To verify the stability of the culture of 45 bacterial species, which were confirmed to be growing when pre-cultured in GB and primarily cultured in GAM (Figure 1D), the same culturing method was used to culture these 45 bacterial species and measure their growth over time for 96 hours (Figure 2). A total of 39 species showed continuous growth (Figure 2), and 16S rDNA analysis of the bacterial cultures confirmed the species of the growing bacteria (Supplementary Table S2). Four species that did not grow in the conditions described in Figure 1D (Alistipes putredinis, Eubacterium hallii, Clostridium leptum, and Coprococcus eutactus) also did not grow in the conditions described in Figure 2. Streptococcus thermophilus and Bacteroides pectinophilus grew in the conditions described in Figure 1D; however, continuous growth was unstable (Figure 2). Thus, 39 of the 45 (87%) available species of the predominant species of European intestinal microbiota can be stably cultured using GB for pre-culture and GAM for the main culture.
3.3 Application of culture methods using GB medium to gut microbiota most dominant species derived from different human populations
It is becoming clear that the predominant species of bacteria vary in different human populations. Therefore, to investigate whether the culturing methods developed in this study could be applied to the predominant species in other gut microbiome projects, we cultured the predominant species of intestinal microbiota in the Japanese population (Nishijima et al., 2016) using GB for pre-culture and GAM for main culture (Figure 3). The predominant species of intestinal microbiota in the Japanese population were as diverse as the predominant species in Europeans (Table 2). As with Europeans, there was also a wide variety of recommended media (Table 2). Of the 50 predominant species of intestinal microbiota in Japanese individuals, 41 species publicly available from distributors such as the JCM, the ATCC, and the DSMZ were selected for examination (Table 3). Twenty-six strains were excluded from the study because they were identical to the predominant species in Europeans (Table 3). Consequently, 15 bacterial species (Table 3) were newly cultured in GB for pre-culture and GAM for the main culture (Figure 3). Because most predominant bacterial species of the European gut microbiota reached the stationary phase at 48 h of culture (Figure 2), the culture was not cultivated further (Figure 3A). Of the 15 strains, 12 grew sufficiently with an OD600 greater than 0.15, and contamination of the culture was excluded by 16S rDNA sequencing (Figure 3B and Supplementary Table S2). Together with the results of Figure 2, 32 of the 36 species (89%) intestinal microbiota in Japanese were cultured. These results indicate that GB is a potential medium for growing a wide range of bacterial species, the existence of which has been suggested in numerous human gut microbiome projects without culturing.
3.4 Effect of replacement of horse blood with other mammal’s blood on bacterial growth
Next, growth was tested when the horse blood added to the GB medium was replaced by blood from other mammals. Fifty-one species grown in GB medium containing horse blood were cultured in GBsheep or GBhuman medium prepared using sheep or human blood, respectively, instead of horse blood. The results show that 48 (94% of the tested 51 strains successfully cultured in GB) and 45 (88% of the tested 51 strains successfully cultured in GB) strains grew in GBsheep (Supplementary Figure S1) and GBhuman (Supplementary Figure S2), respectively.
4 Discussion
In this study, we succeeded in developing a new method for culturing a wide range of intestinal bacteria under the same conditions using an easily prepared GB medium, which can be prepared from only two materials thereby reducing the time and effort required for culturing. Using GB medium, 51 of 60 strains (85%) of European- and Japanese-predominant species were successfully cultured. Some of the predominant species, such as Subdoligranulum variabile and Roseburia hominis, which were previously unculturable in GMM, mGAM, or GAM, were cultured in GB (Table 4).
We have cultured Flavonifractor plautii many times using this system, but the cultivation is not always successful. In this report, we have provided data from a successful culture. There is a need for further improved culturing methods for better reproducibility.
In this study, we selected and cultured representative strains of each species. Bacterial strains, even those of the same species, vary in their characteristics, and these differences may affect human health (Yan et al., 2020). Since it is unclear whether other strains of the same species can be cultured using the method described in this study, it needs to be attempted in the future.
In addition, we have yet to attempt to isolate bacteria from feces using the GB medium. Additional experiments are needed to use the methods described in this study for the isolation of unknown bacteria from human feces. In the future, we plan to determine how many of the fecal bacteria (as detected from fecal DNA information by non-cultivation approaches) can be isolated using GB media.
A liquid growth medium was used in this study to simultaneously culture many bacterial species at the same time. It is difficult to simultaneously culture dozens of different bacteria on solid media because of the large space required for culturing. However, culturing on solid media is necessary to isolate bacteria. Moreover, cell growth can be directly confirmed by colony formation when cultured on solid media.
In a previous report, in which GAM was used in the pre-culture and GAM in the main culture, 32 species were grown (Gotoh et al., 2017). In this study, 36 species were successfully grown (Figure 1B). This may be attributed to the extended incubation time of the main culture up to 96 h (this study) compared to the previous 48 h (previous report).
Remarkably, the number of culturable species increased when GB was used in the pre-culture (Figure 1D) compared to when GAM was used (Figure 1B), even though the main culture had the same GAM. In this culture system, approximately 2 µL was brought into the main culture from the pre-culture medium, which was only 0.4% of the volume. In bacterial culture, it is suggested that if the pre-culture is carefully devised, the subsequent successional culture can grow well, even if the medium and bacteria are somewhat nutritionally incompatible. In the food industry, starter culture, which is equivalent to pre-culture, is used in the production of fermented foods. Starter culture may be defined as “a preparation or material containing large numbers of variable microorganisms, which may be added to accelerate a fermentation process” (Holzapfel, 2002). Starter cultures are used to manufacture foods such as cheese (Somerville et al., 2022), yogurt (Chen et al., 2017), sake (Yamashita, 2021), and wine (Capozzi et al., 2015), for example. Although initiation of spontaneous fermentation requires a relatively long time, using a starter culture can shorten this time (Holzapfel, 2002). The pre-culture may be used to improve subsequent growth. Indeed, it has been reported that two-stage cultures, including a pre-culture to promote growth, were used to isolate bacteria from feces and helped successfully culture a multitude of new species (Lagier et al., 2016). Thus, pre-culturing is an important step for bacterial analysis via culturing.
Notably, in this study, we successfully cultured Faecalibacterium prausnitzii JCM 31915. F. prausnitzii is reduced in the gut microbiota of donors with type 2 diabetes (Qin et al., 2012), Crohn’s disease (Fujimoto et al., 2013) and cirrhosis (Qin et al., 2014) compared to healthy donors. F. prausnitzii is very difficult to culture, and the preparation of the recommended medium, JCM 1130 medium (YCFA medium), requires a mixture of more than 20 ingredients. It has also been reported that F. prausnitzii can be cultured in mGAM-CRI medium, which is prepared by supplementing mGAM with bovine rumen, cellobiose, and inulin (Bellais et al., 2022). In this study, we found that F. prausnitzii JCM 31915 could be cultured on GB medium, which is more easily prepared and has fewer ingredients than other media. In addition, several phylogenetic groups exist in F. prausnitzii and have recently been reclassified into the following four species (Sakamoto et al., 2022): F. prausnitzii (type strain ATCC 27768T), Faecalibacterium duncaniae (type strain JCM 31915T tested in this study), Faecalibacterium hattorii (type strain JCM 39210T), and Faecalibacterium gallinarum (type strain JCM 17207T). Using our method, it may be possible to culture three other species (F. prausnitzii, F. hattorii, and F. gallinarum).
It has been estimated that there are more than 1,000 uncultured bacterial species in the human gut based on metagenomic analysis (Almeida et al., 2019). Our culturing method using GB medium, which is easy to prepare, may be applicable to the culture of bacteria whose functions and ecology are unknown and should be tested in the future. Culturomics, a culturing approach using bacterial culture, MALDI-TOF mass spectrometry, and 16S rRNA sequencing, have been developed for the cultivation and identification of unknown bacteria (Lagier et al., 2018). In culturomics, there are reports of successful analysis of new bacterial species by improving the culture medium (Lagier et al., 2012; Lagier et al., 2016). Furthermore, the use of GB media, combined with techniques such as culturomics, would also help in the analysis of unknown bacteria.
Administration of antibiotics and prebiotics can significantly modify the gut microbiota; however, they may also affect non-targeted bacteria (Hirano et al., 2021; Maier et al., 2021). Therefore, it is necessary to analyze the effects of certain molecules on individual bacteria. As our method makes it possible to culture a wide range of commensal intestinal bacteria under the same conditions, it may be useful for future research on agents that improve the intestinal microbiota.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
Conceptualization, RH and SK; Data curation, RH; Methodology, RH, IN, RS, and SK; Investigation, RH, IN, RN, AB, and RS; Validation, AB; Resources, SK; Writing – Original Draft, RH and SK; Writing – Review and Editing, RH, and SK; Visualization, RH; Funding Acquisition, SK; Project Administration, SK; Supervision, SK. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a Grant-in-Aid for Scientific Research (B) 20H02908, Grant-in-Aid from the Mitani Foundation for Research, and Grant-in-Aid for Challenging Research (Exploratory) 26660071.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2023.1056866/full#supplementary-material
Footnotes
References
Almeida, A., Mitchell, A. L., Boland, M., Forster, S. C., Gloor, G. B., Tarkowska, A., et al. (2019). A new genomic blueprint of the human gut microbiota. Nature 568, 499–504. doi: 10.1038/s41586-019-0965-1
Bellais, S., Nehlich, M., Ania, M., Duquenoy, A., Mazier, W., van den Engh, G., et al. (2022). Species-targeted sorting and cultivation of commensal bacteria from the gut microbiome using flow cytometry under anaerobic conditions. Microbiome 10, 24. doi: 10.1186/s40168-021-01206-7
Biclot, A., Huys, G. R. B., Bacigalupe, R., D’hoe, K., Vandeputte, D., Falony, G., et al. (2022). Effect of cryopreservation medium conditions on growth and isolation of gut anaerobes from human faecal samples. Microbiome 10, 80. doi: 10.1186/s40168-022-01267-2
Buffington, S. A., Dooling, S. W., Sgritta, M., Noecker, C., Murillo, O. D., Felice, D. F., et al. (2021). Dissecting the contribution of host genetics and the microbiome in complex behaviors. Cell 184, 1740–56.e16. doi: 10.1016/j.cell.2021.02.009
Capozzi, V., Garofalo, C., Chiriatti, M. A., Grieco, F., Spano, G. (2015). Microbial terroir and food innovation: the case of yeast biodiversity in wine. Microbiological Res. 181, 75–83. doi: 10.1016/j.micres.2015.10.005
Chang, Y.-C., Hu, Z., Rachlin, J., Anton, B. P., Kasif, S., Roberts, R. J., et al. (2015). COMBREX-DB: an experiment centered database of protein function: knowledge, predictions and knowledge gaps. Nucleic Acids Res. 44, D330–D3D5. doi: 10.1093/nar/gkv1324
Chen, C., Zhao, S., Hao, G., Yu, H., Tian, H., Zhao, G. (2017). Role of lactic acid bacteria on the yogurt flavour: a review. Int. J. Food Properties 20, S316–SS30. doi: 10.1080/10942912.2017.1295988
Costea, P. I., Zeller, G., Sunagawa, S., Pelletier, E., Alberti, A., Levenez, F., et al. (2017). Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 35, 1069–1076. doi: 10.1038/nbt.3960
Fujimoto, T., Imaeda, H., Takahashi, K., Kasumi, E., Bamba, S., Fujiyama, Y., et al. (2013). Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn's disease. J. Gastroenterol. Hepatol. 28, 613–619. doi: 10.1111/jgh.12073
Gohl, D. M., Vangay, P., Garbe, J., MacLean, A., Hauge, A., Becker, A., et al. (2016). Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 34, 942–949. doi: 10.1038/nbt.3601
Goodman, A. L., Kallstrom, G., Faith, J. J., Reyes, A., Moore, A., Dantas, G., et al. (2011). Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. 108, 6252–6257. doi: 10.1073/pnas.1102938108
Gotoh, A., Nara, M., Sugiyama, Y., Sakanaka, M., Yachi, H., Kitakata, A., et al. (2017). Use of gifu anaerobic medium for culturing 32 dominant species of human gut microbes and its evaluation based on short-chain fatty acids fermentation profiles. Bioscience Biotechnology Biochem. 81, 2009–2017. doi: 10.1080/09168451.2017.1359486
Hirano, R., Sakanaka, M., Yoshimi, K., Sugimoto, N., Eguchi, S., Yamauchi, Y., et al. (2021). Next-generation prebiotic promotes selective growth of bifidobacteria, suppressing Clostridioides difficile. Gut Microbes 13, 1973835. doi: 10.1080/19490976.2021.1973835
Holzapfel, W. H. (2002). Appropriate starter culture technologies for small-scale fermentation in developing countries. Int. J. Food Microbiol. 75, 197–212. doi: 10.1016/S0168-1605(01)00707-3
Hsiao Elaine, Y., McBride Sara, W., Hsien, S., Sharon, G., Hyde Embriette, R., McCue, T., et al. (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463. doi: 10.1016/j.cell.2013.11.024
Lagier, J. C., Armougom, F., Million, M., Hugon, P., Pagnier, I., Robert, C., et al. (2012). Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infection 18, 1185–1193. doi: 10.1111/1469-0691.12023
Lagier, J.-C., Dubourg, G., Million, M., Cadoret, F., Bilen, M., Fenollar, F., et al. (2018). Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 16, 540–550. doi: 10.1038/s41579-018-0041-0
Lagier, J.-C., Khelaifia, S., Alou, M. T., Ndongo, S., Dione, N., Hugon, P., et al. (2016). Culture of previously uncultured members of the human gut microbiota by culturomics. Nat. Microbiol. 1, 16203. doi: 10.1038/nmicrobiol.2016.203
Maier, L., Goemans, C. V., Wirbel, J., Kuhn, M., Eberl, C., Pruteanu, M., et al. (2021). Unravelling the collateral damage of antibiotics on gut bacteria. Nature 599, 120–124. doi: 10.1038/s41586-021-03986-2
Matsue, M., Mori, Y., Nagase, S., Sugiyama, Y., Hirano, R., Ogai, K., et al. (2019). Measuring the antimicrobial activity of lauric acid against various bacteria in human gut microbiota using a new method. Cell Transplant. 28, 1528–1541. doi: 10.1177/0963689719881366
Nagano, T., Hirano, R., Kurihara, S., Nishinari, K. (2020). Improved effects of okara atomized by a water jet system on α-amylase inhibition and butyrate production by Roseburia intestinalis Bioscience Biotechnology Biochem. 84, 1467–1474. doi: 10.1080/09168451.2020.1741337
Nishijima, S., Suda, W., Oshima, K., Kim, S.-W., Hirose, Y., Morita, H., et al. (2016). The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 23, 125–133. doi: 10.1093/dnares/dsw002
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. doi: 10.1038/nature11450
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Qin, N., Yang, F., Li, A., Prifti, E., Chen, Y., Shao, L., et al. (2014). Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64. doi: 10.1038/nature13568
Rettedal, E. A., Gumpert, H., Sommer, M. O. A. (2014). Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat. Commun. 5, 4714. doi: 10.1038/ncomms5714
Rosshart, S. P., Vassallo, B. G., Angeletti, D., Hutchinson, D. S., Morgan, A. P., Takeda, K., et al. (2017). Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171, 1015–28.e13. doi: 10.1016/j.cell.2017.09.016
Sakamoto, M., Sakurai, N., Tanno, H., Iino, T., Ohkuma, M., Endo, A. (2022). Genome-based, phenotypic and chemotaxonomic classification of Faecalibacterium strains: proposal of three novel species Faecalibacterium duncaniae sp. nov., Faecalibacterium hattorii sp. nov. and Faecalibacterium gallinarum sp. nov. Int. J. Systematic Evolutionary Microbiol. 72, 005379. doi: 10.1099/ijsem.0.005379
Sharon, G., Cruz, N. J., Kang, D.-W., Gandal, M. J., Wang, B., Kim, Y.-M., et al. (2019). Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell 177, 1600–18.e17. doi: 10.1016/j.cell.2019.05.004
Shendure, J., Ji, H. (2008). Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145. doi: 10.1038/nbt1486
Somerville, V., Berthoud, H., Schmidt, R. S., Bachmann, H.-P., Meng, Y. H., Fuchsmann, P., et al. (2022). Functional strain redundancy and persistent phage infection in Swiss hard cheese starter cultures. ISME J. 16, 388–399. doi: 10.1038/s41396-021-01071-0
Sugiyama, Y., Mori, Y., Nara, M., Kotani, Y., Nagai, E., Kawada, H., et al. (2022). Gut bacterial aromatic amine production: aromatic amino acid decarboxylase and its effects on peripheral serotonin production. Gut Microbes 14, 2128605. doi: 10.1080/19490976.2022.2128605
Sugiyama, Y., Nara, M., Sakanaka, M., Gotoh, A., Kitakata, A., Okuda, S., et al. (2017). Comprehensive analysis of polyamine transport and biosynthesis in the dominant human gut bacteria: potential presence of novel polyamine metabolism and transport genes. Int. J. Biochem. Cell Biol. 93, 52–61. doi: 10.1016/j.biocel.2017.10.015
Tramontano, M., Andrejev, S., Pruteanu, M., Klünemann, M., Kuhn, M., Galardini, M., et al. (2018). Nutritional preferences of human gut bacteria reveal their metabolic idiosyncrasies. Nat. Microbiol. 3, 514–522. doi: 10.1038/s41564-018-0123-9
Yamashita, H. (2021). Koji starter and koji world in Japan. J. Fungi 7, 569. doi: 10.3390/jof7070569
Keywords: standard medium, predominant intestinal bacteria, culture, GB medium, intestinal bacteria
Citation: Hirano R, Nishita I, Nakai R, Bito A, Sasabe R and Kurihara S (2023) Development of culture methods capable of culturing a wide range of predominant species of intestinal bacteria. Front. Cell. Infect. Microbiol. 13:1056866. doi: 10.3389/fcimb.2023.1056866
Received: 29 September 2022; Accepted: 16 June 2023;
Published: 13 July 2023.
Edited by:
Parisa Shabani, Northeast Ohio Medical University, United StatesReviewed by:
Sabrina Naud, Karolinska University Hospital, SwedenMaryam Tidjani Alou, Aix Marseille Université, France
Copyright © 2023 Hirano, Nishita, Nakai, Bito, Sasabe and Kurihara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shin Kurihara, c2t1cmloYXJhQHdha2Eua2luZGFpLmFjLmpw
 Rika Hirano
Rika Hirano Izumi Nishita1
Izumi Nishita1 Ayaka Bito
Ayaka Bito Shin Kurihara
Shin Kurihara