- Laboratório de Bacteriologia, Instituto Butantan, São Paulo, Brazil
Enteroaggregative Escherichia coli (EAEC) is an important cause of diarrhea in children and adults worldwide. This pathotype is phenotypically characterized by the aggregative-adherence (AA) pattern in HEp-2 cells and genetically associated to the presence of the aatA gene. EAEC pathogenesis relies in different virulence factors. At least, three types of adhesins have been specifically associated with EAEC strains: the five variants of the aggregative adherence fimbriae (AAF), the aggregative forming pilus (AFP) and more recently, a fibrilar adhesin named CS22. Our study aimed to evaluate the presence of AAF, AFP and CS22-related genes among 110 EAEC strains collected from feces of children with diarrhea. The presence of aggR (EAEC virulence regulator) and genes related to AAFs (aggA, aafA, agg3A, agg4A, agg5A and agg3/4C), AFP (afpA1 and afpR) and CS22 (cseA) was detected by PCR, and the adherence patterns were evaluated on HeLa cells. aggR-positive strains comprised 83.6% of the collection; among them, 80.4% carried at least one AAF-related gene and presented the AA pattern. aggA was the most frequent AAF-related gene (28.4% of aggR+ strains). cseA was detected among aggR+ (16.3%) and aggR- strains (22.2%); non-adherent strains or strains presenting AA pattern were observed in both groups. afpR and afpA1 were exclusively detected among aggR- strains (77.8%), most of which (71.4%) also presented AA pattern. Our results indicate that AAF- and AFP-related genes may contribute to identify EAEC strains, while the presence of cseA and its importance as an EAEC virulence factor and genotypic marker needs to be further evaluated.
Introduction
Enteroaggregative Escherichia coli (EAEC) is an important worldwide spread intestinal pathogen that has been related to acute and persistent diarrhea in children and adults from both developed and developing countries, travelers’ diarrhea and outbreaks of diarrhea associated with ingestion of contaminated food and water (Estrada-Garcia and Navarro-Garcia, 2012; Hebbelstrup-Jensen et al., 2014; Gomes et al., 2016). This pathotype was first described by Nataro et al. (1987) after observing that E. coli strains isolated from Chilean children with diarrhea presented a stacked-brick-like pattern when adhered on the surface of HEp-2 cells and between them on the coverslip (Nataro et al., 1987) in adherence assays (Cravioto et al., 1979). Since then, this so-called aggregative adherence (AA) pattern has been used as a gold standard to phenotypically define the EAEC category.
A number of EAEC virulence factors, either chromosome- or plasmid-borne, are intimately related to the presence of aggR, a transcriptional regulator belonging to the Ara-C family present in the EAEC virulence plasmid called pAA (Morin et al., 2013). Among other important EAEC virulence factors, pAA harbors aap and aatA genes, which respectively encodes the protein dispersin and its translocator (Sheikh et al., 2002; Nishi et al., 2003). The latest, formerly known as the pCVD432 probe (Baudry et al., 1990), has been widely used to genetically characterize E. coli strains belonging to this category (Cerna et al., 2003; Robins-Browne et al., 2004; Sarantuya et al., 2004; Jenkins et al., 2006; Panchalingam et al., 2012; Houpt et al., 2014; Patzi-Vargas et al., 2015; Gomes et al., 2016; Hebbelstrup-Jensen et al., 2017).
Among EAEC virulence factors related to adherence and regulated by AggR, the aggregative adherence fimbriae (AAF), which belongs to the usher-chaperone fimbriae family, have been reported as responsible for the AA pattern. So far, five variants of AAF have been described (AAF/I to V) (Savarino et al., 1994; Czeczulin et al., 1997; Bernier et al., 2002; Boisen et al., 2008; Jønsson et al., 2015). Although there are notable differences between the major pillin-encoding genes of each AAF type, the other genes composing the operon (usher, chaperone and minor pilin) are highly conserved (Savarino et al., 1994; Boisen et al., 2008; Jønsson et al., 2017a). Whilst AAF types seem to occur individually among EAEC strains, the occurrence of strains simultaneously carrying genes for both AAF/III and AAF/V has been also reported (Jønsson et al., 2017b; Dias et al., 2020; Petro et al., 2020).
Recently, Petro et al. (2020) and Boisen et al. (2020) observed the presence of the CS22 related gene cseA associated with the presence of aggR in EAEC strains. This gene encodes the structural subunit of the fibrilar adhesin CS22, which has been firstly described as a colonization factor in enterotoxigenic E. coli (ETEC) (Pichel et al., 2000). Although none of the strains presenting cseA (cseA+) analyzed by Petro et al. (2020) and Boisen et al. (2020) presented the AA pattern in adherence assays using HEp-2 cells, Boisen et al. (2020) showed that the cseA+ strain C671-15 adhered to colonic organoid similarly to the EAEC prototype strain 042.
While analyzing a heteropathogenic enteroaggregative/enterohemorrhagic E. coli (EAEC/EHEC) strain showing AA pattern, Lang et al. (2018) identified a new plasmid lacking aggR which harbored the aatA and aap genes. This plasmid (pAFP) also carried a new adhesin named aggregative-forming pilus (AFP) encoded by the afp operon, which among its components carries an AraC-like regulator very similar to aggR, named afpR. AFP was shown to be involved in bacterial piliation, autoaggregation, adhesion and cytotoxicity (Lang et al., 2018) and was recently observed mediating AA in a hybrid enteroaggregative/uropathogenic E. coli (EAEC/UPEC) strain (Schüroff et al., 2021; Schüroff et al., 2022). Moreover, phylogenetic analyses have shown that AFP-positive strains carrying other virulence genes related to EAEC pathogenesis (aat operon, type 6 secretion system genes and aap), are clustered with other fecal EAEC strains (Schüroff et al., 2021).
The present study aims to evaluate the presence of genes related to AAF, CS22 and AFP in a collection of EAEC strains isolated from diarrheic feces and previously classified by the presence of aatA gene, in order to identify genetic marker(s) that will allow us to specifically define the EAEC category.
Materials and methods
Bacterial strains
A total of 110 E. coli strains, characterized as EAEC by the presence of aatA, were considered for the present study. These strains were previously obtained from fecal samples collected from children (from 1 to 10 years) with acute diarrhea, in an epidemiological study carried out in the city of Salvador, Brazil (Bueris et al., 2007). Strains were kept at -80˚C in Luria-Bertani (LB) broth containing 15% glycerol and were routinely cultivated on MacConkey or LB agar.
Adherence assays
Qualitative adherence-assays were performed according to the method described by Cravioto et al. (1979) with modifications. HeLa cells (ATCC CCL-2) were cultured in Dulbecco modified Eagle medium (DMEM) (Cultilab, Brazil) containing 10% fetal bovine serum (FBS), using 24-well plates (Corning, USA) containing a glass coverslip in each well, until reaching 70% confluency (~48 h). Bacterial strains statically grown for 18 h at 37˚C in LB broth were diluted (1:50) in DMEM containing 2% FBS and 1% D-mannose, and inoculated on HeLa cells. After two periods of 3 h of incubation at 37˚C in 5% CO2, including a medium change between these periods, each well was washed with phosphate-buffered saline (PBS), fixed with methanol, stained with May-Grunwald and Giemsa (Merck Millipore, USA), and examined by light microscopy.
DNA isolation
One colony isolated from LB agar culture grown for 18 h at 37°C was selected and mixed with 200 µL of ultra-pure water and incubated at 100˚C for 10 min. After boiling, the lysates were incubated in an ice-bath for 5 min and centrifuged at 1,300 x g for 5 min. The supernatants were collected and stored at -20˚C.
Gene detection
The detection of the targeted virulence genes was performed by polymerase chain reactions (PCR). Simplex reactions were used to detect the presence of aggR, encoding the master EAEC virulence regulator (Morin et al., 2013); agg5A, encoding the AAF/V major pilin subunit (Jønsson et al., 2015); cseA, encoding the ETEC colonization factor CS22 (Pichel et al., 2000); and afpA1 and afpR, which are part of the AFP-encoding operon (Lang et al., 2018). Multiplex reaction was used to detect the genes encoding the major pilin subunits of AAF/I to IV (aggA, aafA, agg3A and agg4A, respectively), and agg3/4C, corresponding to the usher of AAF/III, IV and V, (Savarino et al., 1994; Czeczulin et al., 1997; Bernier et al., 2002; Boisen et al., 2008; Jønsson et al., 2015). Table 1 shows primer sequences and concentrations, amplicon sizes, annealing temperatures and positive controls used in each reaction.
PCR was performed in a final volume of 25 µL containing the 20-35 pmol of each primer; dATP, dTTP, dCTP and dGTP (0.1 mM each); 1.5 U Taq DNA polymerase (Invitrogen, USA); 5.0 µL 10X PCR buffer (Invitrogen), MgCl2 (1.5-2.0 mM) and 1.0 µL of DNA template. Cycling was conducted as follows: 1 x 94˚C/5 min; 30 x (94˚C/1 min, primer specific annealing temperature presented in Table 1/1 min, and 72˚C/1 min), followed by a final cycle of 72˚C/5 min. Amplicons were analyzed by 0.7% (simplex PCR) or 2% (multiplex PCR) agarose gel electrophoresis in Tris-borate-EDTA buffer using the 1 kb ladder (Invitrogen) as marker. The gels were stained with UniSafe Dye (Uniscience) and the amplicons were visualized in a UV transiluminator (Alliance HD 6, Uvitec, UK). E. coli HB101 or DH5α were used as negative controls. Positive controls are listed in Table 1.
Results
The AA-pattern is presented by most of the aatA-positive strains
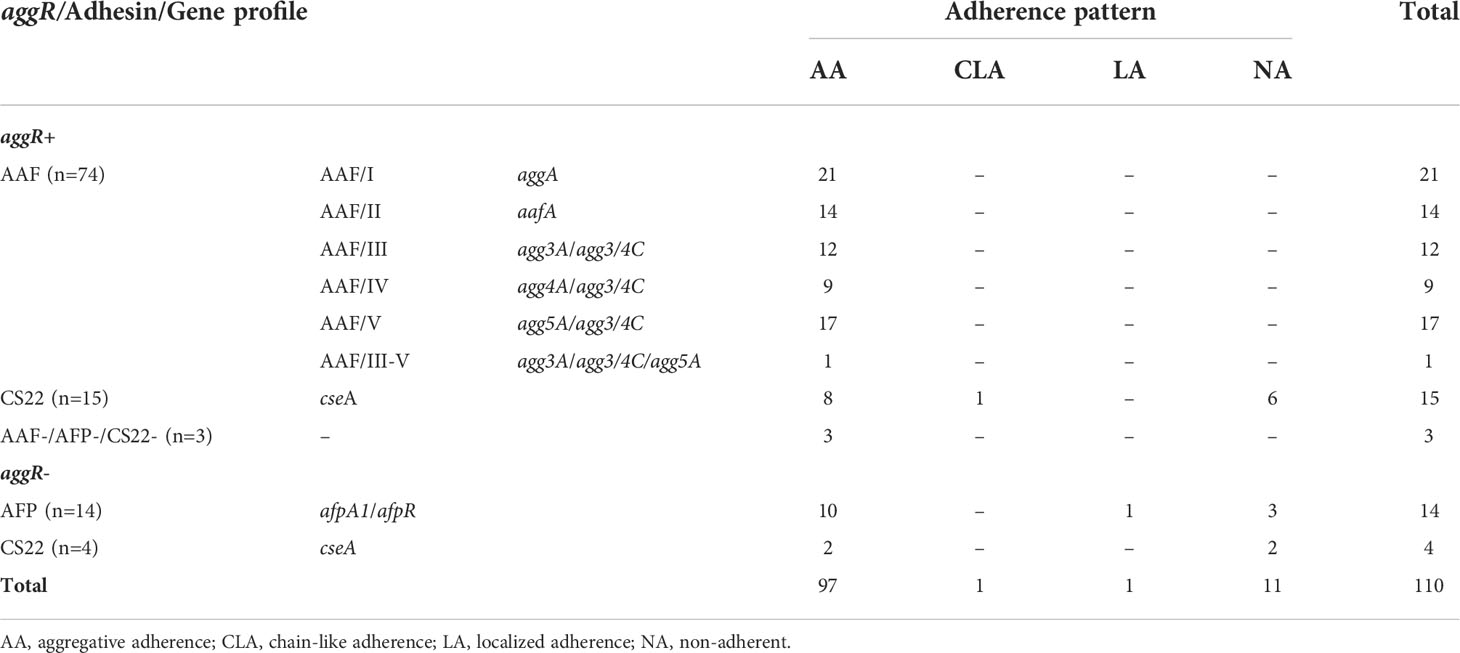
Among the 110 aatA+ EAEC strains submitted to adherence assays with HeLa cells, 97 (88.2%) presented the AA pattern. One strain presenting chain-like adherence (CLA) and one presenting localized adherence (AL) patterns were also observed. Eleven strains (10%) were non-adherent (NA) (Table 2).
Genetic characterization shows an important diversity of EAEC related genes
aggR was detected in 92 out of 110 (83.6%) aatA+ EAEC strains analyzed. Among the aggR+ strains, 74 (80.4%) presented one of the genes encoding the AAF major pilin subunits (Table 2). aggA was the most frequent among them, detected in 21 of these strains (28.4%), followed by agg5A (17 strains/22.9%), aafA (14 strains/18.9%), agg3A (12 strains/16.2%) and agg4A (9 strains/12.2%). Except for one strain (1.4%), harboring agg3A and agg5A, strains did not harbor more than one gene encoding the major pilin subunit. Finally, agg3/4C was detected in all strains harboring agg3A, agg4A and agg5A (39 strains/52.7%). cseA was detected in 15 of the 92 aggR+ strains (16.3%). The remaining three aggR+ strains (3.3%) did not carry either cseA or AAF-encoding genes. AFP-encoding genes were not detected among aggR+ strains.
Among the 18 aggR- strains, 14 (77.8%) carried both afpR and afpA1 genes, while four (22.2%) were cseA+. AAF-encoding genes were not detected among aggR- strains.
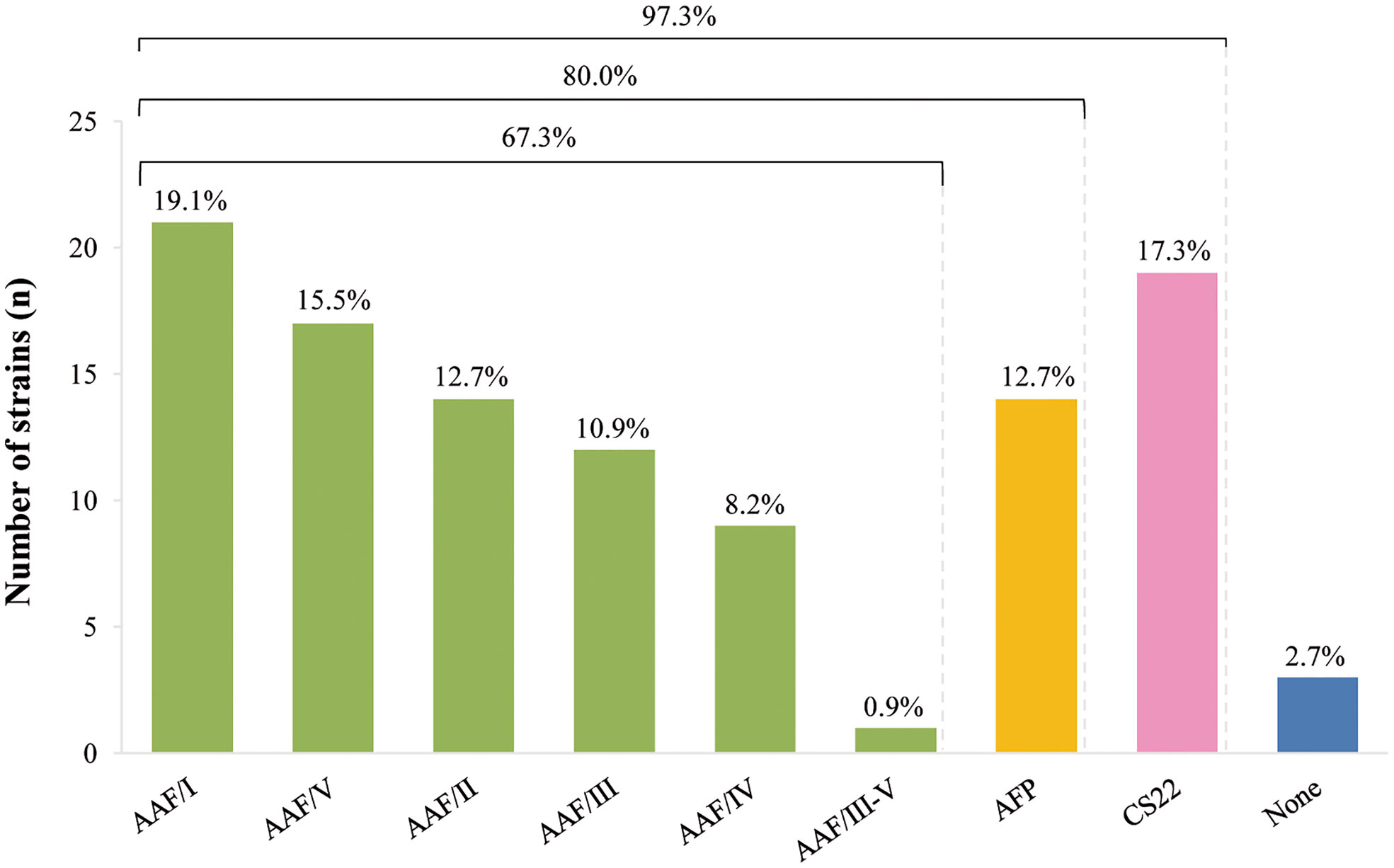
Finally, we emphasize that AAF, AFP and CS22-encoding genes were not simultaneously detected in any of the studied strains. Figure 1 shows the frequency of strains carrying genes related to AAF (67.3%), AFP (12.7%) and CS22 (17.3%) among the 110 aatA+ EAEC strains, independently of the presence of aggR. Only 2.7% were devoid of any of these genes.
Discussion
The 110 E. coli strains selected for this study were previously classified as EAEC by the presence of the aatA gene, using a multiplex PCR to directly detect strains belonging to four main diarrheagenic E. coli categories (enteropathogenic E. coli, EPEC; Shiga-toxin producing/enterohaemorrhagic E. coli, STEC/EHEC; ETEC and EAEC), during an epidemiological case-control study of the etiology of acute diarrhea (Bueris et al., 2007).
Adherence assays, which correspond to the phenotypic gold standard method used for EAEC characterization, were performed using HeLa cells in order to access the adherence pattern of aatA+ EAEC strains, and correlate it with the presence of different adhesins.
Among the 110 strains, 97 (88.2%) strains presented the AA pattern on HeLa cells showing that, in our collection, strains bearing the EAEC characteristic genetic marker aatA do not completely correspond to strains presenting AA pattern.
It is well known that aggR is present in the majority of EAEC strains (Hebbelstrup-Jensen et al., 2014; Rogawski et al., 2017; Lima et al., 2018; Boisen et al., 2020; Levine et al., 2020; Meza-Segura et al., 2020), and regulates different virulence factors in these strains. Although aggR has an important role in EAEC pathogenesis, it is also known that not all E. coli strains presenting the AA pattern harbor aggR. In fact, the classification of EAEC category in two different groups considering the presence or absence of aggR have been proposed: typical EAEC (tEAEC; AA pattern and aggR+) and atypical EAEC (aEAEC; AA pattern and aggR-) (Kaper et al., 2004; Harrington et al., 2006). According to this definition, our results showed that among 97 strains presenting AA pattern, 85 (87.6%) were tEAEC (aggR+), and, 12 (12.4%) were aEAEC (aggR-). The presence of aggR was also detected in one strain presenting CLA pattern (0.9%), and in other six (5.5%) NA aatA+ EAEC strains.
It has been suggested that the presence of aggR in tEAEC confers more virulence to these strains (Kaper et al., 2004; Estrada-Garcia and Navarro-Garcia, 2012; Hebbelstrup-Jensen et al., 2014). However, it is important to note that aEAEC strains have also been responsible for important diarrhea outbreaks (Čobeljić et al., 1996; Itoh et al., 1997). Guerrieri et al. (2019) also showed that in a Galleria mellonella infection model, aEAEC presented virulence levels comparable to the ones observed to tEAEC. Therefore, AA/aggR- (aEAEC) strains should also be considered as an important diarrheagenic agent despite the absence of the aggR regulon.
Among EAEC virulence factors regulated by aggR, the aggregative adherence fimbriae (AAF) have been associated with the AA pattern in different EAEC prototype strains (Savarino et al., 1994; Czeczulin et al., 1997; Bernier et al., 2002; Boisen et al., 2008; Jønsson et al., 2015). In the present work, genes related to at least one of five AAF types (AAF/I to V) described to date were detected in 74 out of 92 (80.4%) aggR+ strains. Among them, the AAF/I major pilin subunit aggA (28.4%) was the most frequent pilin-encoding gene found, followed by agg5A (22.9%), aafA (18.9%), agg3A (16.2%) and agg4A (12.2%). Although a heterogeneous frequency profile of AAF pilin-encoding genes have been reported among different EAEC strains collections, aggA also featured as the most frequent AAF pilin related gene in some studies, ranging from 17.8 - 26.4% (Jønsson et al., 2015; Jønsson et al., 2017b; Boisen et al., 2020; Petro et al., 2020). Other studies respectively pointed to aafA (21.8%) (Chattaway et al., 2017), agg4A (14.5%) (Hebbelstrup-Jensen et al., 2017), and agg5A (20%) (Dias et al., 2020) as the most frequent AAF pilin related genes. It is of notice that all AAF+ strains herein detected also presented the AA pattern on HeLa cells.
One strain harboring genes of the operons simultaneously encoding AAF/III and AAF/V (agg3A and agg5A, respectively) was also detected in our collection. Similar strains encoding AAF/III- and AAF/V-related genes have been reported in other studies (Jønsson et al., 2017b; Dias et al., 2020; Petro et al., 2020) and although transcription of both agg3A and agg5A has been shown in some of these strains (Jønsson et al., 2017b), further details on the fimbria structure and its regulation mechanisms remain unknown.
Boisen et al. (2020) recently described the presence of a CS22-like gene cluster among EAEC strains harboring aggR. This cluster is an operon highly homologous to the one encoding an ETEC colonization factor named CS22 (Pichel et al., 2000). A frequency of 3.1% among 97 EAEC (aatA+ and/or aaiC+) strains carrying a complete CS22-like gene cluster was reported by Boisen et al. (2020), while Petro et al. (2020) reported the occurrence of 16.1% among 56 EAEC (aatA+ and/or aaiC+) strains harboring cseA. These strains, however, were respectively minimal or not adherent to HEp-2 cells in adherence assays.
In the present work, 19 (17.3%) aatA+ EAEC strains harboring the cseA marker were detected. However, in contrast to what have been previously reported by Boisen et al. (2020) and Petro et al. (2020), where all the CS22+ strains were also aggR+, our study showed that only 15 out of 19 (78.9%) cseA+/aatA+ EAEC strains were aggR+. We also found in HeLa adherence assays that eight (53.3%) of these strains presented the AA pattern and one (6.7%) the CLA pattern; while the remaining six strains (40.0%) were NA. These results show that despite no correlation between presence of cseA and adherence pattern was found, some strains presented the AA pattern on HeLa cells. Likewise, we also found two cseA+/AA strains among four aggR- strains (two presenting the AA pattern and two NA). Although Boisen et al. (2020) reported that three cseA+ EAEC were non-adherent on HEp-2 cells, they also observed that one of these strains was able to adhere to human intestinal epithelium in a colonoid model. In this sense, the presence of CS22-encoding genes and their role in EAEC adherence, as well as their regulation by AggR are still unclear. In fact, CS22 is an important virulence factor involved in ETEC colonization (Pichel et al., 2000) and there are no reports concerning the presence of aggR (which belongs to the AraC regulator family) in ETEC, suggesting that CS22 related genes are not under exclusive aggR regulation.
The AFP biogenesis-related genes share high similarity with the genes composing the operon encoding the bundle-forming pili (BFP), a type IV pilus associated with localized adherence pattern of EPEC strains (Lang et al., 2018). Furthermore, Schüroff et al. (2021) showed that AFP are expressed as thin rigid structures, instead of the characteristic BFP bundles and mediates the AA pattern of a hybrid EAEC/uropathogenic E. coli (UPEC) strain isolated from a urinary infection case. In another study, Dias et al. (2020) also reported 15 aatA+/aggR- strains (6.8%) out of 220 aatA+ EAEC strains harboring AFP-related genes; among which, 12 also presented the AA pattern in HeLa cells adherence assays.
Among the 110 aatA+ EAEC strains studied, the present work detected 14 strains harboring AFP-encoding genes (afpA1 and afpR); all of them lacking aggR. Among these 14 aggR-/AFP+/aatA+ EAEC strains, 10 (71.4%) presented the AA pattern on HeLa cells, two were NA and one presented LA. As the LA pattern is a phenotypic characteristic of EPEC strains related to the production of BFP (Gomes et al., 2016), the occurrence of this adherence pattern in one aEAEC strain needs to be further studied.
Results obtained in this study draw our attention to the fact that among the 110 aatA+ EAEC strains studied, AAF, CS22 or AFP were exclusively present in 107 strains; respectively, 74 (67.3%), 19 (17.3%), and (14) 12.7%. Also important to note that 100% of these 74 AAF+ strains were aggR+ and presented AA pattern in HeLa cells adherence assays, while 100% of AFP+ strains were aggR-. While the role of AAF in pathogenesis and as a specific EAEC marker is well stablished, recent data showed that AFP has emerged as an important virulence trait in a subset of EAEC strains carrying aat, aai and aap (Lang et al., 2018; Dias et al., 2020; Schüroff et al., 2021; Schüroff et al., 2022). Considering our results, and the EAEC classification proposed by Kaper et al. (2004), we suggest that the search for AAF- and AFP-encoding genes may contribute to genotypically identify and discriminate EAEC strains. Regarding CS22, our results shows that it is not exclusively related to the presence of aggR, nor to the AA pattern. As mentioned earlier, CS22 has been described as a virulence factor involved in ETEC colonization (Pichel et al., 2000), but no other significant data on its role in EAEC pethogenesis has been reported. Unlike AAF and AFP, the role and importance of CS22 as a virulence factor in EAEC strains remains virtually unexplored and needs to be validated as an EAEC marker in other EAEC collections.
The present study also showed that the majority of our EAEC strains (80%) harbored either one AAF variant or AFP, highlighting their importance as target antigens to be used in EAEC diagnosis and/or prevention strategies. Further studies including aatA- EAEC strains are necessary to evaluate the specificity of theses gene markers.
Data availability statement
The raw data used as reference for our analyses are available in the Butantan Institute Repository (https://repositorio.butantan.gov.br/handle/butantan/4425).
Author contributions
WE and CA conceived and supervised the research. CF, BR, and CA conducted the experiments. CF, WE, and CA conducted the analyses, wrote and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grant 2018/04144-0 from São Paulo Research Foundation (FAPESP) to WE.
Acknowledgments
We would like to thank Dr Paulo Schüroff for designing the primers for afpA1 genes.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andrade, F. B., Gomes, T. A. T., Elias, W. P. (2014). A sensitive and specific molecular tool for detection of both typical and atypical enteroaggregative Escherichia coli. J. Microbiol. Methods 106, 16–18. doi: 10.1016/j.mimet.2014.07.030
Baudry, B., Savarino, S. J., Vial, P., Kaper, J. B., Levine, M. M. (1990). A sensitive and specific dna probe to identify enteroaggregative Escherichia coli, a recently discovered diarrheal pathogen. J. Infect. Dis. 161, 1249–1251. doi: 10.1093/infdis/161.6.1249
Bernier, C., Gounon, P., Le Bouguénec, C. (2002). Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect. Immun. 70, 4302–4311. doi: 10.1128/IAI.70.8.4302-4311.2002
Boisen, N., Østerlund, M. T., Joensen, K. G., Santiago, A. E., Mandomando, I., Cravioto, A., et al. (2020). Redefining enteroaggregative Escherichia coli (EAEC): genomic characterization of epidemiological EAEC strains. PLoS Negl. Trop. Dis. 14, 1–19. doi: 10.1371/journal.pntd.0008613
Boisen, N., Struve, C., Scheutz, F., Krogfelt, K. A., Nataro, J. P. (2008). New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect. Immun. 76, 3281–3292. doi: 10.1128/IAI.01646-07
Bueris, V., Sircilli, M. P., Taddei, C. R., Santos, M. F., Franzolin, M. R., Martinez, M. B., et al. (2007). Detection of diarrheagenic Escherichia coli from children with and without diarrhea in Salvador, Bahia, Brazil. Mem. Inst. Oswaldo Cruz. 102, 839–844. doi: 10.1590/S0074-02762007005000116
Cerna, J. F., Nataro, J. P., Estrada-Garcia, T. (2003). Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia coli strains. J. Clin. Microbiol. 41, 2138–2140. doi: 10.1128/JCM.41.5.2138-2140.2003
Chattaway, M. A., Day, M., Mtwale, J., White, E., Rogers, J., Day, M., et al. (2017). Clonality, virulence and antimicrobial resistance of enteroaggregative Escherichia coli from Mirzapur, Bangladesh. J. Med. Microbiol. 66, 1429–1435. doi: 10.1099/jmm.0.000594
Čobeljić, M., Miljkovic-Selimovic, B., Paunovic-Todosijevic, D., Velickovic, Z., Lepsanovic, Z., Zec, N., et al. (1996). Enteroaggregative Escherichia coli associated with an outbreak of diarrhoea in a neonatal nursery ward. Epidemiol. Infect. 117, 11–16. doi: 10.1017/S0950268800001072
Cravioto, A., Gross, R. J., Scotland, S. M., Rowe, B. (1979). An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Cur. Microbiol. 3, 95–99. doi: 10.1007/BF02602439
Czeczulin, J. R., Balepur, S., Hicks, S., Phillips, A., Hall, R., Kothary, M. H., et al. (1997). Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect. Immun. 65, 4135–4145. doi: 10.1128/iai.65.10.4135-4145.1997
Dias, R. C. B., Tanabe, R. H. S., Vieira, M. A., Cergole-Novella, M. C., Santos, L. F., Gomes, T. A. T., et al. (2020). Analysis of the virulence profile and phenotypic features of typical and atypical enteroaggregative Escherichia coli (EAEC) isolated from diarrheal patients in Brazil. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.00144
Elias, W. P., Suzart, S., Trabulsi, L. R., Nataro, J. P., Gomes, T. A. T. (1999). Distribution of aggA and aafA gene sequences among Escherichia coli isolates with genotypic or phenotypic characteristics, or both, of enteroaggregative e. coli. J. Med. Microbiol. 48, 597–599. doi: 10.1099/00222615-48-6-597
Estrada-Garcia, T., Navarro-Garcia, F. (2012). Enteroaggregative Escherichia coli pathotype: a genetically heterogeneous emerging foodborne enteropathogen. FEMS Immunol. Med. Microbiol. 66, 281–298. doi: 10.1111/j.1574-695X.2012.01008.x
Gomes, T. A. T., Elias, W. P., Scaletsky, I. C. A., Guth, B. E. C., Rodrgues, J. F., Piazza, R. M. F., et al. (2016). Diarrheagenic Escherichia coli. Braz. J. Microbiol. 47, 3–30. doi: 10.1016/j.bjm.2016.10.015
Guerrieri, C. G., Pereira, M. F., Galdino, A. C. M., Santos, A. L. S., Elias, W. P., Schuenck, R. P., et al. (2019). Typical and atypical enteroaggregative Escherichia coli are both virulent in the Galleria mellonella model. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01791
Harrington, S. M., Dudley, E. G., Nataro, J. P. (2006). Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol. Lett. 254, 12–18. doi: 10.1111/j.1574-6968.2005.00005.x
Hebbelstrup-Jensen, B., Olsen, K. E. P., Struve, C., Krogfelt, K. A., Petersen, A. M. (2014). Epidemiology and clinical manifestations of enteroaggregative Escherichia coli. Clin. Microbiol. Rev. 27, 614–630. doi: 10.1128/CMR.00112-13
Hebbelstrup-Jensen, B., Poulsen, A., Rasmussen, S. H. R., Struve, C., Engberg, J. H., Friis-Møller, A., et al. (2017). Genetic virulence profile of enteroaggregative Escherichia coli strains isolated from danish children with either acute or persistent diarrhea. Front. Cell. Infect. Microbiol. 7. doi: 10.3389/fcimb.2017.00230
Houpt, E., Gratz, J., Kosek, M., Zaidi, A. K. M., Qureshi, S., Kang, G., et al. (2014). Microbiologic methods utilized in the MAL-ED cohort study. Clin. Infect. Dis. 59, S225–S232. doi: 10.1093/cid/ciu413
Itoh, Y., Nagano, I., Kunishima, M., Ezaki, T. (1997). Laboratory investigation of enteroaggregative Escherichia coli O untypeable:H10 associated with a massive outbreak of gastrointestinal illness. J. Clin. Microbiol. 35, 2546–2550. doi: 10.1128/jcm.35.10.2546-2550.1997
Jønsson, R., Liu, B., Struve, C., Yang, Y., Jørgensen, R., Xu, Y., et al. (2017a). Structural and functional studies of Escherichia coli aggregative adherence fimbriae (AAF/V) reveal a deficiency in extracellular matrix binding. Biochim. Biophys. Acta Proteins Proteom. 1865, 304–311. doi: 10.1016/j.bbapap.2016.11.017
Jønsson, R., Struve, C., Boisen, N., Mateiu, R. V., Santiago, A. E., Jenssen, H., et al. (2015). Novel aggregative adherence fimbria variant of enteroaggregative Escherichia coli. Infect. Immun. 83, 1396–1405. doi: 10.1128/IAI.02820-14
Jønsson, R., Struve, C., Boll, E. J., Boisen, N., Joensen, K. G., Sørensen, C. A., et al. (2017b). A novel pAA virulence plasmid encoding toxins and two distinct variants of the fimbriae of enteroaggregative Escherichia coli. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.00263
Jenkins, C., Chart, H., Willshaw, G. A., Cheasty, T., Smith, H. R. (2006). Genotyping of enteroaggregative Escherichia coli and identification of target genes for the detection of both typical and atypical strains. Diagn. Microbiol. Infect. Dis. 55, 13–19. doi: 10.1016/j.diagmicrobio.2005.10.019
Kaper, J. B., Nataro, J. P., Mobley, H. L. T. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Lang, C., Fruth, A., Holland, G., Laue, M., Mühlen, S., Dersch, P., et al. (2018). Novel type of pilus associated with a shiga-toxigenic E. coli hybrid pathovar conveys aggregative adherence and bacterial virulence. Emerg. Microbes Infect. 7, 2–16. doi: 10.1038/s41426-018-0209-8
Levine, M. M., Nasrin, D., Acácio, S., Bassat, Q., Powell, H., Tennant, S. M., et al. (2020). Diarrhoeal disease and subsequent risk of death in infants and children residing in low-income and middle-income countries: analysis of the GEMS case-control study and 12-month GEMS-1A follow-on study. Lancet Glob. Health 8, 204–214. doi: 10.1016/S2214-109X(19)30541-8
Lima, A. A. M., Medeiros, P. H. Q. S., Havt, A. (2018). Enteroaggregative Escherichia coli subclinical and clinical infections. Curr. Opin. Infect. Dis. 31, 433–439. doi: 10.1097/QCO.0000000000000477
Meza-Segura, M., Zaidi, M. B., León, A. V., Moran-Garcia, N., Martinez-Romero, E., Nataro, J. P., et al. (2020). New insights into DAEC and EAECpathogenesis and phylogeny. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.572951
Morin, N., Santiago, A. E., Ernst, R. K., Guillot, S. J., Nataro, J. P. (2013). Characterization of the AggR regulon in enteroaggregative Escherichia coli. Infect. Immun. 81, 122–132. doi: 10.1128/IAI.00676-12
Nataro, J. P., Deng, Y., Maneval, D. R., German, A. L., Martin, W. C., Levine, M. M. (1992). Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect. Immun. 60, 2297–2304. doi: 10.1128/iai.60.6.2297-2304.1992
Nataro, J. P., Kaper, J. B., Robins-Browne, R., Prado, V., Vial, P., Levine, M. M. (1987). Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr. Infect. Dis. J. 6, 829–831. doi: 10.1097/00006454-198709000-00008
Nishi, J., Sheik, J., Mizuguchi, K., Luisi, B., Burland, V., Boutin, A., et al. (2003). The export of coat protein from enteroaggregative Escherichia coli by a specific ATP-binding cassette transporter system. J. Biol. Chem. 278, 45680–45689. doi: 10.1074/jbc.M306413200
Panchalingam, S., Antonio, M., Hossain, A., Mandomando, I., Ochieng, B., Oundo, J., et al. (2012). Diagnostic microbiologic methods in the GEMS-1 case/control study. Clin. Infect. Dis. 55, 294–302. doi: 10.1093/cid/cis754
Patzi-Vargas, S., Zaldi, M. B., Perez-Martinez, I., León-Cen, M., Michel-Ayala, A., Chaussabel, D., et al. (2015). Diarrheagenic Escherichia coli carrying supplementary virulence genes are an important cause of moderate to severe diarrhoeal disease in Mexico. PLoS Negl. Trop. Dis. 9, 1–18. doi: 10.1371/journal.pntd.0003510
Petro, C. D., Duncan, J. K., Seldina, Y. I., Allué-Guardia, A., Eppinger, M., Riddle, M. S., et al. (2020). Genetic and virulence profiles of enteroaggregative Escherichia coli (EAEC) isolated from deployed military personnel (DMP) with travelers’ diarrhea. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.00200
Pichel, M., Binsztein, N., Viboud, G. (2000). CS22, a novel human enterotoxigenic Escherichia coli adhesin, is related to CS15. Infect. Immun. 68, 3280–3285. doi: 10.1128/IAI.68.6.3280-3285.2000
Robins-Browne, R. M., Bordun, A. M., Tauschek, M., Bennet-Wood, V. R., Russell, J., Oppedisano, F., et al. (2004). Escherichia coli and community-acquired gastroenteritis, Melbourne, Australia. Emerg. Infect. Dis. 10, 1797–1805. doi: 10.3201/eid1010.031086
Rogawski, E. T., Guerrant, R. L., Havt, A., Lima, I. F. N., Medeiros, P. H. Q. S., Seidman, J. C., et al. (2017). Epidemiology of enteroaggregative Escherichia coli infections and associated outcomes in the MAL-ED birth cohort. PLoS Negl. Trop. Dis. 11, 1–17. doi: 10.1371/journal.pntd.0005798
Sarantuya, J., Nishi, J., Wakimoto, N., Erdene, S., Nataro, J. P., Sheikh, J., et al. (2004). Typical enteroaggregative Escherichia coli is the most prevalent pathotype among e. coli strains causing diarrhea in mongolian children. J. Clin. Microbiol. 42, 133–139. doi: 10.1128/JCM.42.1.133-139.2004
Savarino, S. J., Fox, P., Yikang, D., Nataro, J. P. (1994). Identification and characterization of a gene cluster mediating enteroaggregative Escherichia coli aggregative adherence fimbria I biogenesis. J. Bacteriol. 176, 4949–4957. doi: 10.1128/jb.176.16.4949-4957.1994
Schüroff, P. A., Abe, C. M., Silva, J. W., Coelho, C. P., Andrade, F. B., Hernandes, R. T., et al. (2022). Role of aggregate-forming pilus (AFP) in adherence and colonization of both intestinal and urinary tracts. Virulence. 13, 1423–1433. doi: 10.1080/21505594.2022.2112818
Schüroff, P. A., Salvador, F. A., Abe, C. M., Wami, H. T., Carvalho, E., Hernandes, R. T., et al. (2021). The aggregate-forming pili (AFP) mediates the aggregative adherence of a hybrid-pathogenic Escherichia coli (UPEC/EAEC) isolated from a urinary tract infection. Virulence. 12, 3073–3093. doi: 10.1080/21505594.2021.2007645
Sheikh, J., Czeczulin, J. R., Harrington, S., Hicks, S., Henderson, I. R., Le Bouguénec, C., et al. (2002). A novel dispersin protein in enteroaggregative Escherichia coli. J. Clin. Invest. 110, 1329–1337. doi: 10.1172/JCI16172
Keywords: enteroaggregative Escherichia coli, aggregative-adherence fimbriae, aggregative-forming pilus, AAF, AFP, CS22, adhesins
Citation: Freire CA, Rodrigues BO, Elias WP and Abe CM (2022) Adhesin related genes as potential markers for the enteroaggregative Escherichia coli category. Front. Cell. Infect. Microbiol. 12:997208. doi: 10.3389/fcimb.2022.997208
Received: 18 July 2022; Accepted: 19 October 2022;
Published: 08 November 2022.
Edited by:
Teresa Estrada-Garcia, Instituto Politécnico Nacional de México (CINVESTAV), MexicoReviewed by:
David Durand, Universidad Peruana Cayetano Heredia, PeruIruka N. Okeke, University of Ibadan, Nigeria
Copyright © 2022 Freire, Rodrigues, Elias and Abe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cecilia M. Abe, Y2VjaWxpYS5hYmVAYnV0YW50YW4uZ292LmJy
 Claudia A. Freire
Claudia A. Freire Beatriz O. Rodrigues
Beatriz O. Rodrigues Waldir P. Elias
Waldir P. Elias Cecilia M. Abe
Cecilia M. Abe