- 1Department of Stomatology, Beijing Friendship Hospital, Capital Medical University, Beijing, China
- 2Immunology Research Center for Oral and Systemic Health, Beijing Friendship Hospital, Capital Medical University, Beijing, China
Objectives: To investigate the pathogenic effect of Porphyromonas gingivalis cultured media on the esophagus and the mechanism underlying the effect.
Background: Periodontitis is strongly associated with esophageal squamous cell carcinoma (ESCC). The cultured media of P. gingivalis may act on healthy esophagus to trigger a malignant transformation; however, this has not been confirmed.
Methods: Cell migration assays and cell cycle measurements were performed on normal human esophageal epithelial cells in the presence or absence of P. gingivalis cultured media. The esophagi of healthy adult C57BL/6J mice were isolated and cultured in-vitro. Hematoxylin-eosin and immunohistochemical staining using antibodies against proliferating cell nuclear antigen (PCNA), Claudin 1 and Claudin 4 were performed to detect dysplasia in specific tissues. Total mRNA was extracted to determine transcriptional dysregulation. A specific inhibitor of Sonic hedgehog signaling, cyclopamine, was used to confirm the underlying molecular mechanism.
Results: In the presence of P. gingivalis cultured media, proliferation and migration of normal human esophageal epithelial cells were up-regulated, and aneuploid cells appeared. Compared with control cells, the arrangement of mouse esophageal epithelial cells became disordered, the percentage of PCNA-positive cells increased, and the positive staining of Claudin 1 and Claudin 4 became weak. In addition, the expression of cancer-related pathway genes was up-regulated but tight junction-related gene expression was down-regulated. The Sonic hedgehog pathway was abnormally activated, and its inhibition reduced the pathogenic effect of P. gingivalis cultured media.
Conclusions: We revealed that the cultured media of the key periodontal pathogen, P. gingivalis, can induce the malignant transformation of normal esophageal epithelium through the Sonic hedgehog pathway.
1 Introduction
Periodontitis is a common oral inflammatory disease with a high prevalence of 45% to 50% and is the sixth most common human disease. The average worldwide prevalence of severe periodontitis has been estimated to be 11% (Kassebaum et al., 2014; Slots, 2017; Sanz et al., 2020) and recent studies support association between periodontitis and several systemic diseases, including diabetes, cardiovascular diseases, chronic obstructive pulmonary disease, and digestive cancers, such as esophageal cancer (Chapple et al., 2013; Linden et al., 2013; Zhou and Lou, 2019; Sanz et al., 2020).
Esophageal cancer ranks as the eighth most common cancer worldwide and the 5-year overall survival rate is only 10% because of its aggressive nature and the difficulty of early diagnosis (Pennathur et al., 2013; Domper Arnal et al., 2015; Gupta and Kumar, 2017; Zhou and Lou, 2019). Esophageal cancer consists of two main histological subtypes, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). In developing countries, ESCC is the predominant subtype. Epidemiological studies show that patients with periodontitis may have a significantly higher risk of ESCC, even after controlling for confounding variables, such as smoking (Abnet et al., 2001; Hiraki et al., 2008; Chen et al., 2015; Chen et al., 2016; Nwizu et al., 2017). These findings indicate that oral bacterial infections may be significantly related to ESCC susceptibility.
Porphyromonas gingivalis, one of the “red complex” bacterial species in the oral cavity, is a representative pathogen of periodontitis. Accumulating evidence indicates that P. gingivalis is strongly associated with ESCC. The abundance of P. gingivalis in saliva and dental plaque is associated with an increased occurrence and poor prognosis of ESCC, and high levels of P. gingivalis-specific antibodies in serum is an independent risk factor for poor ESCC prognosis (Ahn et al., 2012; Peters et al., 2017; Gao et al., 2018; Chen et al., 2019; Kageyama et al., 2019; Meng et al., 2019; Chen et al., 2021; Kawasaki et al., 2021). Additionally, the detection rates of P. gingivalis antigen, the secreted gingipain protease, and specific 16S rDNA are significantly higher in ESCC tissue compared with rates in adjacent tissue or in healthy controls (Gao et al., 2016; Yuan et al., 2017). Few studies have investigated the mechanism for the association between P. gingivalis and ESCC. Based on studies of ESCC patients, animal cancer models, and ESCC cell lines, P. gingivalis promotes the progression of the malignant phenotype and reduces therapeutic efficacy. The chronic inflammatory response, immune invasion, and epithelial-mesenchymal transition represent key links between the presence of P. gingivalis and ESCC development (Yuan et al., 2019; Qi et al., 2020; Chen et al., 2021).
However, the studies above focused on the effect of P. gingivalis on damaged esophagus in which cancer was already established. It is not known whether the cultured media of P. gingivalis can affect healthy esophageal epithelium and trigger dysplasia. Therefore, the aim of this study was to investigate the effect of P. gingivalis cultured media on healthy esophagus and to explore the underlying mechanism of any effect.
2 Materials and methods
2.1 Preparation of P. gingivalis strain cultured media
P. gingivalis liquid medium consisted of 30g/L tryptone soybean broth, 1 mg/mL yeast extract, 1 mg/L vitamin K1 and 5 mg/L hemin. For culture plates, 0.2 g/L agar and 5% sterile defibrinated sheep blood were added.
A standard strain of P. gingivalis, ATCC 33277 (kindly provided by Prof. Huanxin Meng, Peking University School and Hospital of Stomatology, China), was inoculated on plates and cultured at 37°C under anaerobic conditions (AnaeroPouch, MGC, Japan). A monoclonal colony was picked and amplified in the liquid medium. When the bacteria grew to the exponential phase, a density of 0.6 (OD600), the bacterial suspension was centrifuged at a speed of 10,000 × g for 10 minutes at 4°C to collect the cultured media. The cultured media was filtered through a 0.22-μm filter (Millipore, Ireland) before adding to cell or organ culture media at the required concentration.
2.2 Cell culture
Normal human esophageal epithelial Het-1A cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher, USA) at 37°C in 5% CO2.
2.3 Cell migration assay
Het-1A cells were assigned to three groups: cultured in medium supplemented with 5% P. gingivalis blank medium (0% CM), cultured in medium supplemented with 0.5% P. gingivalis cultured media (0.5% CM), and cultured in medium supplemented with 5% P. gingivalis cultured media (5% CM). After culture for 24 h, Het-1A cells from each group were trypsinized and resuspended in 500 μL serum free medium, and seeded at a density of 2 × 105 cells/mL in the upper wells of transwell inserts with a polyethylene terephthalate membrane pore size of 8.0 µm (MCEP24H48, Millipore, USA) in a 24-well plate. The lower chamber contained 750 μL culture medium containing 10% FBS. Cells were cultured in the transwell at 37°C with 5% CO2 for 24 hours. Then, culture medium was replaced by PBS in both chambers. After that, cells attached on the upper surface of the membrane were wiped off with a cotton swab, and cells that had migrated across the transwell membrane were fixed with methyl alcohol for 5 minutes, stained with DAPI, and counted under a fluorescence microscope.
2.4 Cell cycle measurement
The cell cycle of esophageal epithelial cells stained by propidium iodide (PI) was analyzed by flow cytometry. Het-1A cells (1×106 cells) were digested with 0.25% trypsin to make a single cell suspension, washed twice with PBS, and then resuspended in 250 μL PBS. The cells were then fixed overnight at 4°C by the addition of 750 μL anhydrous ethanol. The next day, the cells were centrifuged and washed with PBS. One milliliter of DNA staining solution (containing 0.2 mg/ml RNase, Cell Cycle Staining Kit, MultiSciences, China) was added to 106 cells, vortex-mixed for 5 to 10 seconds and incubated in the dark at room temperature for 30 minutes. The lowest sample loading speed was selected for the flow cytometer (Attune NxT, Thermo Fisher Scientific, USA). Flow cytometry was used to examine the cell cycle in accordance with standard procedures. Cell cycle distribution was analyzed by ModFit, and the proportions of cells in G0/G1, S and G2/M were calculated.
2.5 Organ culture in vitro
Fifteen healthy adult C57BL/6J mice were kept in the Institute of Materia Medica, Chinese Academy of Medical Sciences & Peking Union Medical College under standard conditions, and randomly assigned to the following groups: “0% CM” (treated with 5% P. gingivalis blank medium as control, n = 3), “0.5% CM” (treated with 0.5% P. gingivalis cultured media, n = 3), “5% CM” (treated with 5% P. gingivalis cultured media, n = 3), “0.5% CM + cyclopamine (CPM)” (treated with 0.5% P. gingivalis cultured media and 5 μM cyclopamine to inhibit SHH signaling (CPM, MCE, USA), n = 3) and “5% CM + CPM” (treated with 5% P. gingivalis cultured media and 5 μM CPM, n = 3). At a minimum, biological triplicates were analyzed. The organ culture system was described previously (Jia et al., 2018).
2.6 Immunohistochemical and immunofluorescence staining
Sections were prepared as previously reported (Jia et al., 2018). Antibodies against the following were used for immunohistochemical analysis: proliferating cell nuclear antigen (PCNA, mouse or rabbit monoclonal; 1:50, Santa Cruz Biotechnology), Claudin 1 (CLDN1, Proteintech), Claudin 4 (CLDN4, Proteintech), SHH (1:50, Abcam), GLI1 (1:50, Abcam). Corresponding secondary antibodies were used (1:100, ZSGB-BIO). For immunohistochemical staining, reactions were developed with diaminobenzidine (DAB, ZSGB-BIO). The percentage of PCNA positive cells per 400 times microscopic field was counted in a minimum of 3 sections. The Image J software calculates the staining area and the percentage of positive area of CLDN1, CLDN4 and SHH staining sections. For immunofluorescence staining, the sections were incubated with fluorescent secondary antibodies [Fluorescein-Conjugated AffiniPure Goat Anti-Mouse IgG (H+L), ZF-0312, ZSGB-BIO, China] at 37°C for 1 hour in the dark, washed with PBS, mounted with fluorescent mounting medium containing DAPI (ZLI-9557, ZSGB-BIO, China) and observed by fluorescence microscopy (FV1000, Olympus, Japan).
2.7 RNA isolation and sequencing
Total RNA from Het-1A cells of the 0% CM and 5% CM groups was extracted and purified in accordance with the manufacturer’s protocol. Sequencing libraries were generated using the VAHTS mRNA-seq v2 Library Prep Kit for Illumina in accordance with the manufacturer’s recommendations, and index codes were added to attribute sequences to each sample. The libraries were sequenced on an Illumina NovaSeq platform to generate 150-bp paired-end reads following the manufacturer’s instructions. Raw data (raw reads) were first processed through primary quality control to obtain clean data (clean reads), which were used for downstream analyses. Paired-end clean reads were aligned to the reference genome using TopHat. HTSeq was used to count the reads mapped to each gene. Differential expression analysis between two conditions was performed using the DEGSeq R package (1.20.0). Differentially expressed genes (DEGs) were defined as those for which the adjusted P-value was below 0.05 and the log2(fold change) was more than 1. Gene Ontology (GO) and KEGG enrichment analyses of DEG sets were performed using GOseq R and KOBAS 3.0, respectively. GO terms with adjusted P-value below 0.05 were considered significantly enriched by DEGs.
2.8 Real-time quantitative polymerase chain reaction
Total RNA was isolated from Het-1A cells of 0% CM, 0.5% CM, and 5% CM groups using TRNzol (DP424, TIANGEN BIOTECH, China) in accordance with the manufacturer’s instructions. First-strand cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (K1622, Thermo, USA) in accordance with the manufacturer’s protocol. SYBR® Premix Ex Taq™ II (Tli RNase H Plus, RR82LR, TaKaRa) was used to perform RT-qPCR. The qPCR thermocycling conditions were: 95°C for 5 min; 40 cycles of 95°C for 10 seconds, and 60°C for 35 seconds. After the amplification reaction, 70 cycles (60°C to 95°C for 10 sec) were performed to establish the melting curve of the PCR product. The 2−ΔΔCt method was applied to calculate relative mRNA levels. The primer sequences used in the study were: 1) GAPDH forward, 5′-AAATCAAGTGGGGCGATGCT-3′ and reverse, 5′-CAAATGAGCCCCAGCCTTCT-3′; 2) HHIP forward, 5′-AATGCAGAGCCACGGTACAA-3′ and reverse, 5′-CAGAGGCACTTGTTCGGTCT-3′.
2.9 Statistical analysis
Statistical analyses were performed using SPSS software version 25. Data are expressed as means ± SD of at least three independent experiments. The conduct of the experiment and the data assessment were performed by different researchers. Statistical differences of PCNA, SHH, CLDN1, CLDN4 positive rates and migration cell numbers among groups were analyzed by one-way ANOVA. The CLDN1/CLDN4 positive area percentages were compared between the samples with CPM addition and the samples without blocking the Shh pathway by paired t-test. P values below 0.05 were considered statistically significant.
3 Results
3.1 P. gingivalis cultured media leads to malignant transformation of esophageal epithelial cells
3.1.1 Cell culture results
The mean numbers of cells that migrated across the transwell membrane were significantly different among groups (61 cells in the 0% CM group, 145 cells in the 0.5% CM group, and 199 cells in the 5% CM group, P < 0.0001). Representative pictures and the corresponding statistics of transwell assays are presented in Figures 1A, B. The number of migration cells increased significantly in 0.5% CM group than in the 0% CM group (P = 0.0098). Similarly, the number of migration cells in 5% CM group was also significantly higher than that in 0% CM group (P = 0.0054). The effect of P. gingivalis cultured media on cell cycle progression was also determined. The addition of P. gingivalis cultured media for 24 h did not change the proportion of cells in S phase (still 50% approximately); however, a large number of aneuploid cells appeared as shown in Figure 1C. The aneuploid cells accounted for 23.66% of cells in the 0.5% CM group and for 16.51% in the 5% CM group (Figure 1C). The cell cycle distribution did not show statistically significant differences among groups (P > 0.05).
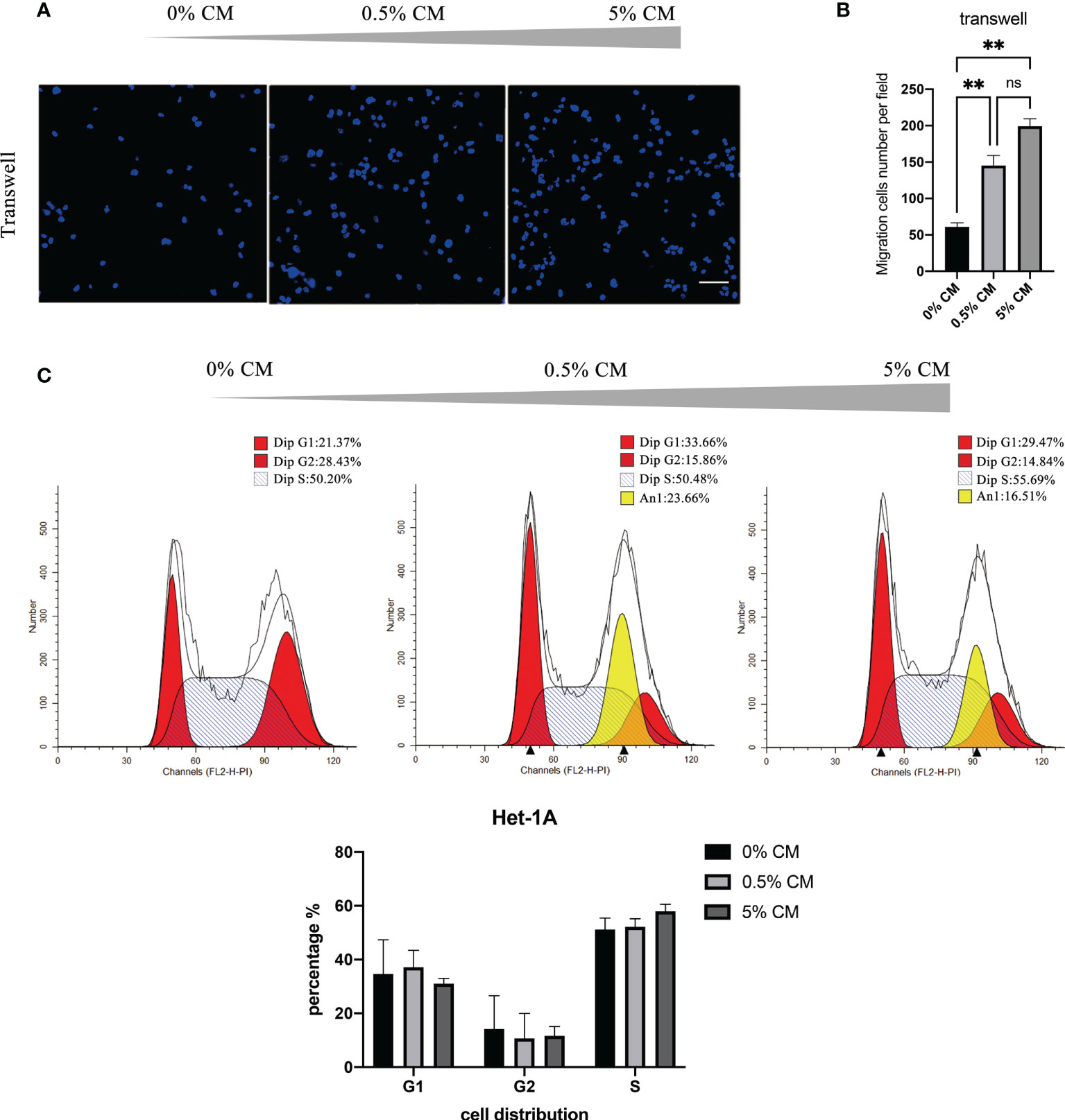
Figure 1 P. gingivalis cultured media promotes migration and proliferation of Het-1A cells. (A) Pictures of typical transwell assays. (B) Statistical analysis of transwell assay results. (C) Cell cycle analysis with propidium iodide staining. Scale bar: 100 μm. **P < 0.01, ns, no statistical significance.
3.1.2 Organ culture results
Treatment of normal mouse esophagus with P. gingivalis cultured media for 7 days significantly increased the number of stratified epithelial cell layers and the cell arrangement became disordered as shown in Figure 2A (white arrows). A statistically significant increase in the percentage of PCNA-positive cells was detected, from 51.8% to 65.9% in the 0.5% CM group (P = 0.010). In the 5% CM group, this percentage increased even further to 73.4% (P = 0.005, Figure 2B). PCNA results indicated that cellular proliferative capacity was increased by P. gingivalis cultured media.
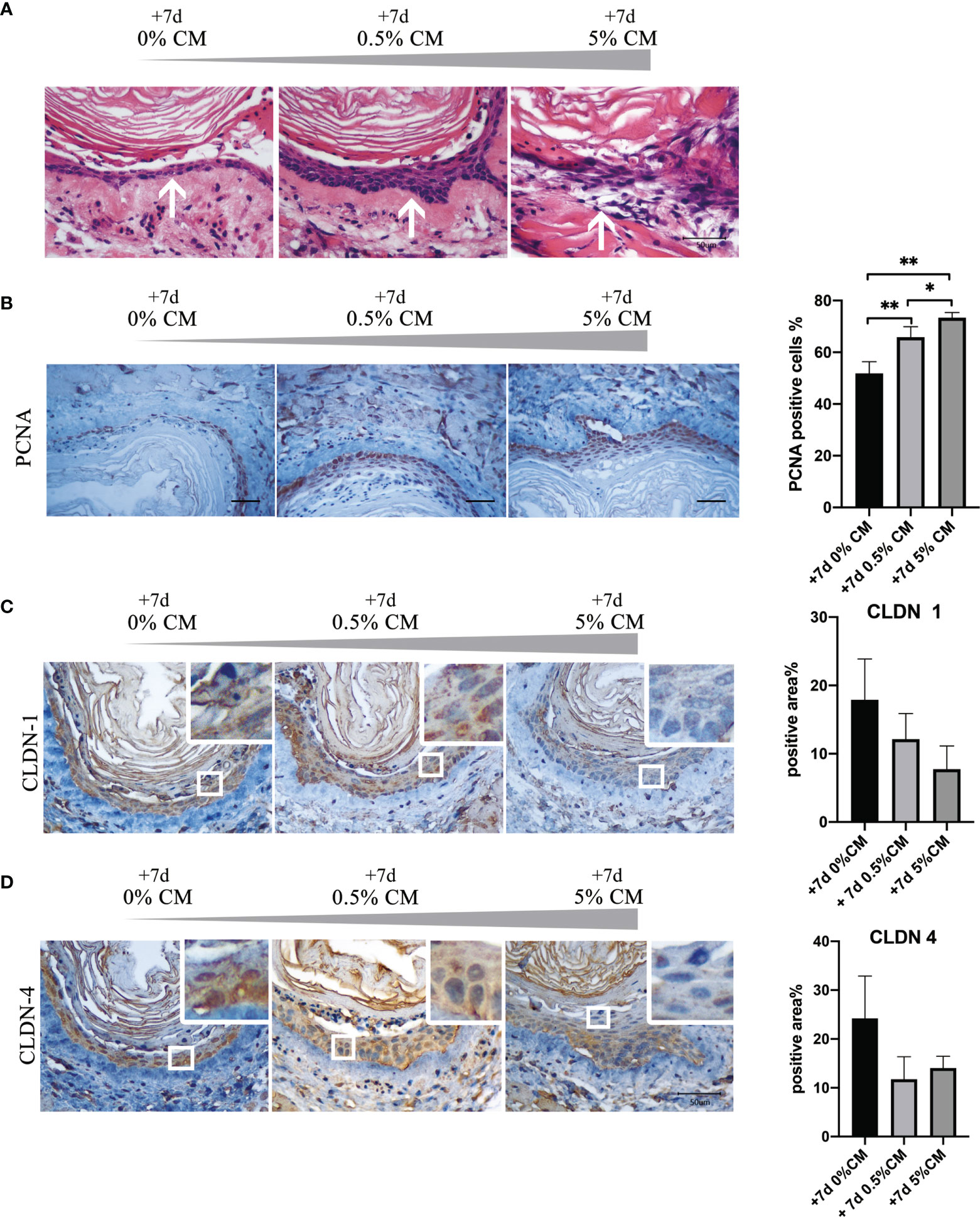
Figure 2 Addition of P. gingivalis cultured media to normal mouse esophagus in vitro for 7 days leads to malignant transformation. (A) The number of esophageal epithelial cell layers increased after treatment with 0.5% and 5% P. gingivalis cultured media and the cell arrangement was disordered in the 5% CM group (white arrows). (B) The rate of PCNA-positive cells was significantly in 0.5% CM and 5% CM groups compared with the control group. (C, D) The expression of CLDN1 and CLDN4 was down-regulated gradually as the concentration of P. gingivalis cultured media increased compared with the control esophageal epithelium. Scale bar: 50 μm. *P < 0.05; **P < 0.01.
However, the expression of the cell tight junction markers, CLDN1 and CLDN4, decreased with the increase in P. gingivalis cultured media concentration as shown in Figure 2C, indicating esophageal epithelial barrier was impaired. The percentage of positive area in CLDN1 immunohistochemical staining section was much lower in 5% CM group, although the statistically significant difference did not exist (P = 0.066). With respect to CLDN4, the percentage of positive area was also much lower with the addition of P. gingivalis cultured media (0% CM group vs. 0.5% CM group, P = 0.093; 0% CM group vs. 5% CM group, P = 0.197).
3.2 RNA-seq data indicated up-regulation of genes related to cancer and down-regulation of genes related to tight junctions
To profile global gene expression related to the malignant transformation of esophageal cells after treatment with P. gingivalis cultured media, total mRNA of Het-1A cells from the 0% CM and 5% CM groups was extracted and sequenced. A total of 2832 DEGs were detected between 0% CM and 5% CM groups. Addition of 5% P. gingivalis cultured media for 24 hours promoted overexpression of 1688 genes and down-regulated expression of 1144 genes (Figure 3A).
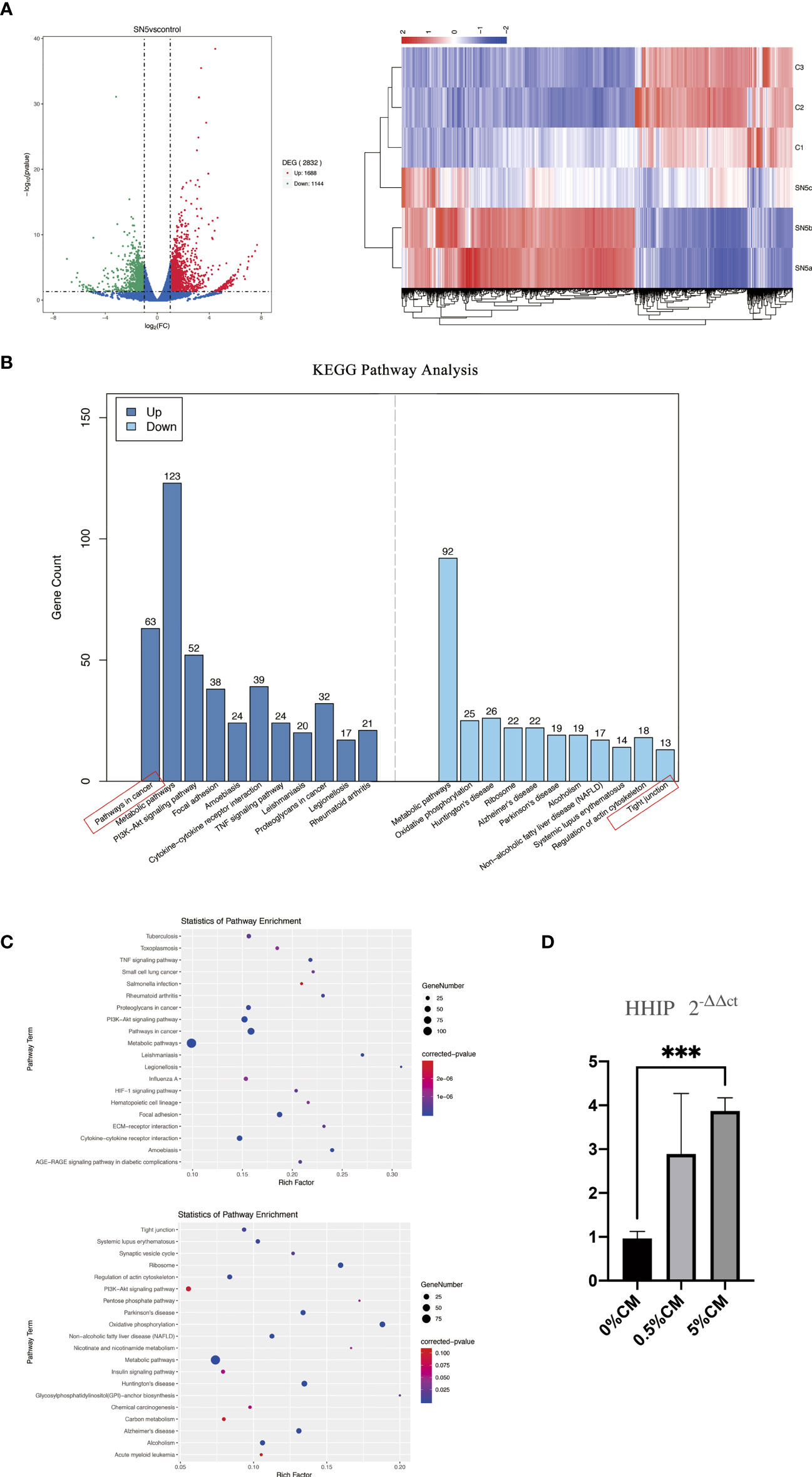
Figure 3 Comparison of the transcriptome profiles of cultured esophagi from 0% CM and 5% CM groups. (A) Volcano plot presenting the number, fold change, and statistical significance of DEGs and the cluster analysis of DEGs. Rows represent samples, and columns represent genes. (B) Up- and down-regulated pathways according to KEGG pathway analysis. (C) Numbers of DEGs and corresponding P values in terms related to up- and down-regulated pathways. (D) Differences in HHIP expression among 0% CM, 0.5% CM and 5% CM groups were determined by RT-qPCR. ***P < 0.001.
KEGG pathway analysis showed the primary up-regulated pathway to be in cancer, followed by metabolic pathways, PI3K-Akt signaling, focal adhesion, amoebiasis, cytokine-cytokine receptor interaction, and TNF signaling. The pathway associated with tight junctions was down-regulated (Figure 3B). The expression levels of genes associated with the cancer pathway, such as WNT3A, WNT2B, PTK2, PTGS2, CSF1R, ITGA2, MMP9, LPAR3, LPAR1, MMP2 and Hedgehog-interacting protein (HHIP) were up-regulated. Among these up-regulated genes, HHIP attracted our attention because it is a target gene of the SHH pathway, which is silent in normal mature esophageal cells, but is closely linked to cell proliferation and mobility. Up-regulation of HHIP was confirmed by RT-qPCR, P < 0.0001.
We then examined the expression of SHH of in vitro-cultured mouse esophagus. Immunohistochemistry indicated that, in normal mouse esophageal epithelial cells, SHH was nearly absent. However, with P. gingivalis cultured media treatment, strong positive expression of SHH was detected (Figure 4, red arrows). Compared with expression at the time of sacrifice and 7 days after culture, the amount of SHH positive staining increased markedly in 0.5% CM group (+7d 0% CM group vs. +7d 0.5% CM group, P = 0.0929) and in 5% CM group (+7d 0% CM group vs. +7d 5% CM group, P = 0.1421), although the statistically significant difference did not exist. Consistent with SHH expression pattern, the expression levels of GLI1, a marker for Hh-responding cells, were barely detectable in in vitro-cultured mouse esophagus. With P. gingivalis cultured media treatment for 7 days, GLI1 expression became positive and was transported from the cell cytoplasm into the nucleus, which is critical to indicate activation of the SHH pathway.
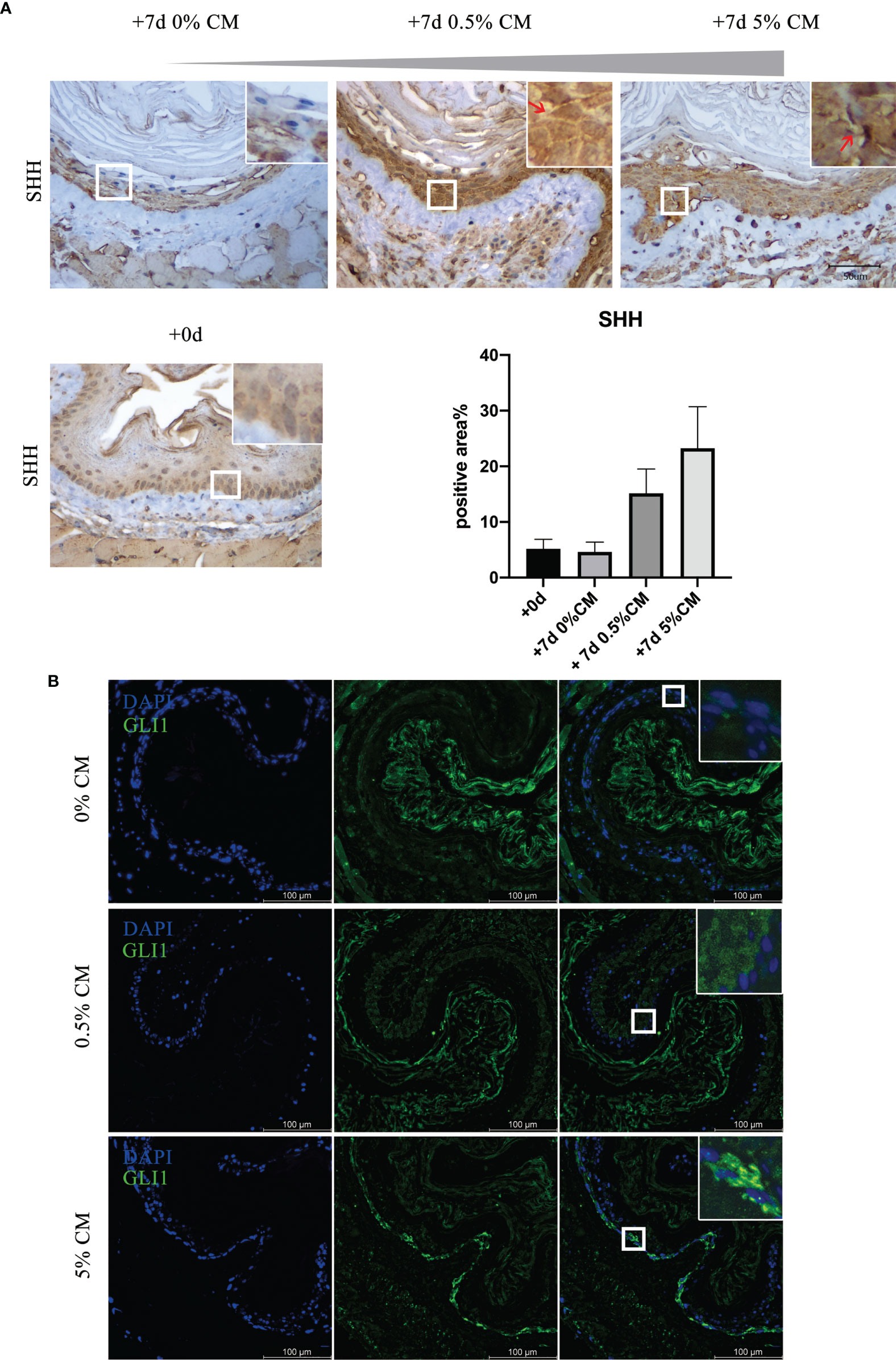
Figure 4 The SHH pathway was activated in P. gingivalis cultured media-treated mouse esophagi. (A) Overexpression of SHH protein in epithelial cells in 0.5% CM and 5% CM groups as indicated by red arrows. (B) GLI1 expression increased with 0.5% or 5% P. gingivalis cultured media treatment for 7 days, and the GLI protein was translocated from the cytoplasm into the nucleus of epithelial cells, especially in the 5% CM group. Scale bars: 50 μm (A); 100 μm (B).
3.3 Inhibition of the SHH pathway reduces the pathogenic effect of P. gingivalis cultured media
To investigate the role of SHH during the malignant transformation of normal esophageal epithelium by treatment with P. gingivalis cultured media, 5 μM of a specific SHH signaling inhibitor, CPM, was added to the culture medium for 7 days. GLI1 protein was predominately localized in the cytoplasm in samples treated with P. gingivalis cultured media plus CPM (Figure 5A), indicating disturbed Shh signaling. In the 0.5% P. gingivalis cultured media-treated samples, the percentage of PCNA-positive cells decreased significantly with the addition of 5 μM CPM (P<0.001), and this difference also existed between 5% CM and 5% CM + CPM groups (P<0.0001, Figure 5B).
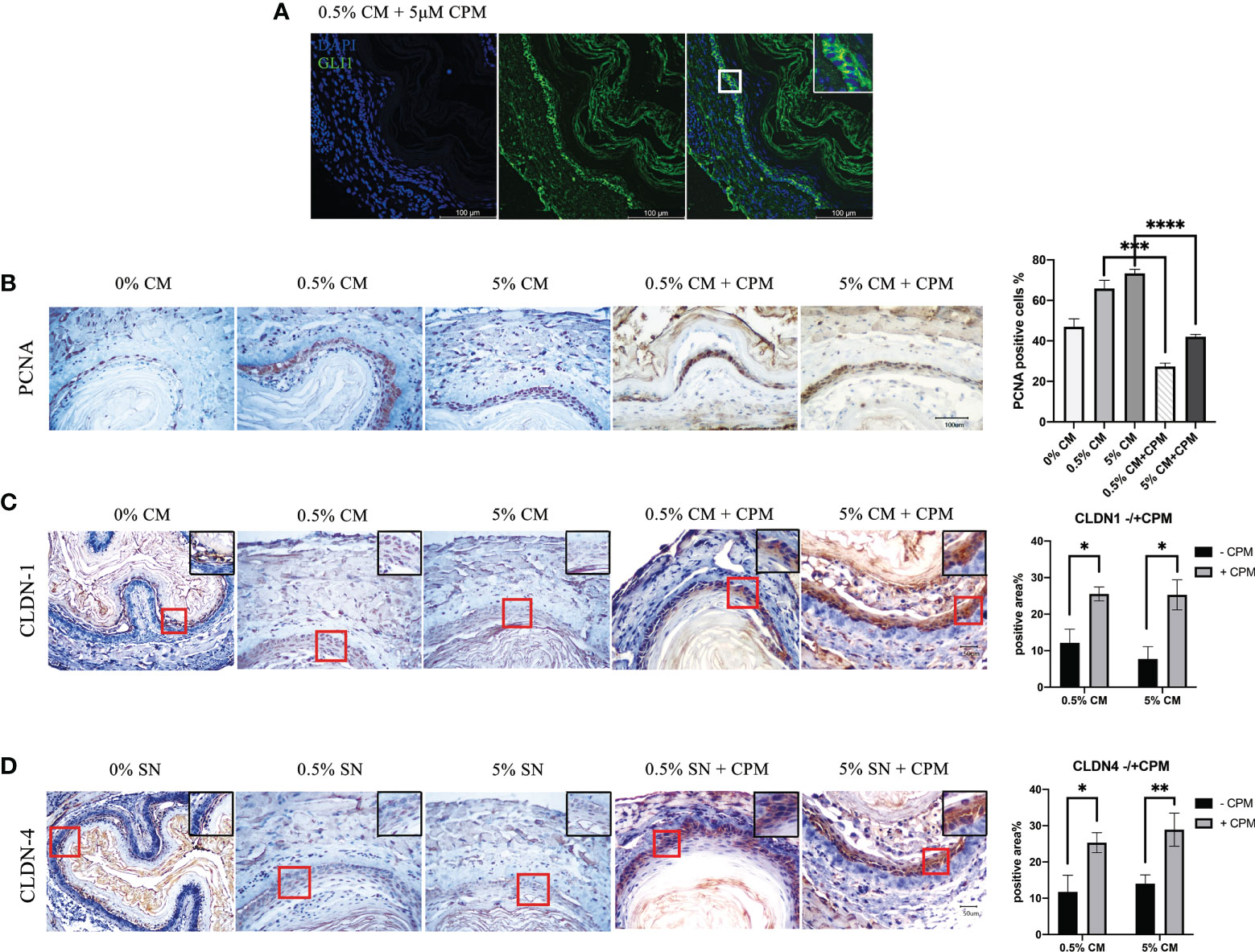
Figure 5 SHH inactivation reduced the percentage of PCNA-positive cells, which was increased in esophagi by P. gingivalis cultured media treatment, and recovered the expression of tight junction markers. (A) Predominately cytoplasmic localization of GLI1 in mouse esophageal epithelial cells cultured in CPM-supplemented medium. (B) Percentages of PCNA-positive cells in 0.5% CM and 5% CM groups were significantly decreased when CPM was added. (C) CLDN1 expression was decreased in 0.5% CM and 5% CM groups and was increased back to normal in the esophageal epithelium with the addition of CPM. (D) Accumulation of CLDN4 in esophageal epithelial cells in the presence of CPM, compared with culture without CPM. Scale bars: 100 μm (A); 50 μm (B–D). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
In addition, CLDN1 expression in the esophageal epithelial cells became markedly positive in the presence of CPM (0.5% CM group vs. 0.5% CM + CPM group, P = 0.027; 5% CM group vs. 5% CM + CPM group, P = 0.027; Figure 5C). The percentage of positive area in CLDN4 immunohistochemical staining section was also significantly higher in the presence of CPM (0.5% CM group vs. 0.5% CM + CPM group, P = 0.026; 5% CM group vs. 5% CM + CPM group, P = 0.009; Figure 5D). The expression patterns of CLDN1 and CLDN4 indicated recovery of tight junctions and intactness of the epithelial barrier after blocking the SHH pathway in esophageal epithelial cells.
4 Discussion
Transition of the esophagus from a normal to a cancerous state has been intensively investigated. ESCC arises in a sequential manner, beginning with inflammation, flowed by hyperplasia, dysplasia, carcinoma in situ, and invasive carcinoma (Chen et al., 2017). Accumulating evidence supports a close correlation between periodontal pathogens and esophageal carcinoma stage. In a prospective study, bacteria in oral wash samples were assessed. After adjusting for body mass index, smoking and alcohol habits, the abundance of P. gingivalis marginally trended with a higher risk of ESCC (OR = 1.30; P = 0.09) (Peters et al., 2017). Analysis of 190 ESCC patients for a median follow-up of 5-years showed that P. gingivalis in the cytoplasm of ESCC cells and stroma was positively correlated with invasion depth, lymphatic metastasis, tumor-node metastasis stage, and shorter overall survival (Qi et al., 2020).
The molecular mechanism underlying the intracellular invasion of P. gingivalis to promote the aggressive progression of ESCC is poorly understood. Qi et al. revealed that activation of TGFβ-dependent Smad/YAP/TAZ signaling may be involved (Qi et al., 2020). Meng et al. found that enhanced proliferation and motility of ESCC cells induced by P. gingivalis infection in vitro were accompanied by activation of NF-κB signaling (Meng et al., 2019). Moreover, Chen et al. found that P. gingivalis could up-regulate the glucose uptake rate of ESCC cells and increase the incidence of invasive carcinoma in a mouse esophageal tumor model induced by 4-nitroquinoline 1-oxide. In addition, P. gingivalis can also promote epithelial-mesenchymal transition and the recruitment of myeloid suppressor cells by up-regulating interleukin 6 (IL6) (Chen et al., 2021). Yuan et al. identified epigenetic alteration in ESCC patients with intracellular P. gingivalis infection, and blocking B7-H4 LD5B could improve the protective immunity against P. gingivalis (Yuan et al., 2019).
However, almost all of the above studies focused on the effect of P. gingivalis in the carcinoma phase, not on earlier stages of the oncogenesis process, which has important clinical significance. In this study, we paid close attention to the causal relationship between the cultured media of P. gingivalis and the squamous dysplasia stage, which is a critical pathological stage during the oncogenesis process of ESCC.
Dysplasia is defined by the histopathological appearance of nuclear atypia, loss of normal cell polarity, and abnormal tissue maturation without invasion through the basement membrane (Dawsey et al., 1994; Dawsey et al., 1994). In our study, P. gingivalis cultured media promoted the proliferation and migration of normal esophageal epithelial Het-1A cells. More importantly, aneuploid cells appeared after treatment for 24 h (Figure 1). Additionally, after treatment of normal mouse esophagus with P. gingivalis cultured media in vitro, the number of epithelial cell layers increased, the cell arrangement became disordered, the rate of cells positive for the proliferation marker, PCNA, was significantly increased, and the positive staining of the tight junction markers, CLDN1 and CLDN4, became weak (Figure 2). Consistent with this phenotype, KEGG pathway analysis of human esophageal epithelial cell transcriptome sequencing showed that after treatment with P. gingivalis cultured media, cancer pathway-related genes were up-regulated, and tight junction pathway-related genes were down-regulated (Figure 3).
These results indicated that P. gingivalis cultured media can directly induce dysplasia of esophageal epithelial cells. This important finding may explain why a high abundance of P. gingivalis in saliva has an increased risk of ESCC; that is, before epithelial barrier destruction, the harmful cultured media has already been acting on the healthy esophagus (Peters et al., 2017). In 2020, Qi et al. suggested that P. gingivalis alone might not be tumorigenic during the onset of ESCC because NE6 cells, an immortal cell line, failed to form colonies in soft agar colony formation assays after P. gingivalis infection and because subcutaneous xenografts from P. gingivalis-infected NE6 cells grew slowly and gradually disappeared (Qi et al., 2020). Our findings do not exactly correspond to those of Qi et al.; however, this difference is reasonable because of the complicated influence of many factors and the long process from the dysplasia stage to the carcinoma stage.
In this study, esophageal epithelial dysplasia was accompanied by activation of the SHH pathway, as shown in Figures 3, 4. SHH is a significant embryonic development-related gene, which is exclusively expressed in epithelial cells. The encoded SHH protein acts on mesenchymal cells through a paracrine pathway. A large number of studies showed the SHH pathway to be silent in normal mature esophageal epithelial cells, but in dysplasia and cancer, the SHH pathway is activated (Zhu et al., 2011; Okumura et al., 2014; Wadhwa et al., 2017). The SHH pathway inhibitor, CPM, has been used in the clinical treatment of cancer (Berman et al., 2003; Yu et al., 2019). We previously confirmed that SHH can regulate the proliferation and differentiation capacity of mouse esophageal epithelial cells. In K14-Cre;Shhfl/fl mice, the number of esophageal epithelial cells decreased and proliferation was disrupted compared with wild-type littermates. There were also significant differences in the expression of genes related to the cell cycle and cell motility between mutant and control samples, indicating a relationship of the SHH pathway with malignant transformation (Jia et al., 2018).
To further investigate the role of SHH activation in the P. gingivalis cultured media-induced phenotype, we added 5 μM CPM to the culture medium to block SHH signaling (Figure 5A). In the CPM-treated samples, P. gingivalis cultured media failed to increase PCNA-positive rates or to break down tight junctions (Figure 5), indicating that the SHH pathway may be the molecular mechanism underlying P. gingivalis cultured media-induced esophageal epithelial dysplasia. This finding may be of significance in the search for new targets to prevent esophageal epithelial dysplasia. Further studies are required to confirm the pathogenic effect in vivo and to identify the key pathogenic components in P. gingivalis cultured media.
5 Conclusions
We show that the cultured media of the key periodontal pathogen, P. gingivalis, can initiate the malignant transformation of normal esophageal epithelium through the SHH pathway. Our study provides direct causal evidence for the carcinogenic effect of a periodontal pathogen on normal esophagus. These findings will help identify new therapeutic targets for ESCC and inform the development of measures for the early prevention of ESCC.
Data availability statement
The data presented in the study are deposited in the Gene Expression Omnibus repository, accession number GSE210408.
Ethics statement
The animal study was reviewed and approved by the Biomedical Ethics Committee, Beijing Friendship Hospital, Capital Medical University.
Author contributions
XJ and XH conceived the study and designed the research; XJ, JL, and YH performed the experiments; JL and YH contributed to data analysis and interpretation; XJ and XH wrote the manuscript; XH contributed to manuscript review. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (to XJ, No. 82101005), the National Natural Science Foundation of China (to XH, No. 82071141), and the Natural Science Foundation of Beijing Municipality (to XH, No.7202036).
Acknowledgments
We thank Prof. Huanxin Meng (Peking University School and Hospital of Stomatology, China) for the generous gift of the standard strains of P. gingivalis, ATCC 33277. We acknowledge Xiaoqian Xu (Beijing Friendship Hospital, Capital Medical University, China) for assistance on the statistical analysis. We thank Jeremy Allen, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing a draft of this manuscript. In addition, the authors would like to appreciate Qiping Luo for his kindly support on figure preparation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abnet, C. C., Qiao, Y. L., Mark, S. D., Dong, Z. W., Taylor, P. R., Dawsey, S. M. (2001). Prospective study of tooth loss and incident esophageal and gastric cancers in China. Cancer Causes Control 12, 847–854. doi: 10.1023/a:1012290009545
Ahn, J., Segers, S., Hayes, R. B. (2012). Periodontal disease, Porphyromonas gingivalis serum antibody levels and orodigestive cancer mortality. Carcinogenesis 33, 1055–1058. doi: 10.1093/carcin/bgs112
Berman, D. M., Karhadkar, S. S., Maitra, A., De. Oca, R., Gerstenblith, M. R., Briggs, K., et al. (2003). Widespread requirement for hedgehog ligand stimulation in growth of digestive tract tumours. Nature 425, 846–851. doi: 10.1038/nature01972
Chapple, I. L., Genco, R., Working group 2 of joint EFP/AAP workshop (2013). Diabetes and periodontal diseases: Consensus report of the joint EFP/AAP workshop on periodontitis and systemic diseases. J. Periodontol. 40 (Suppl 14), S106–S112. doi: 10.1902/jop.2013.1340011
Chen, Y., Chen, X., Yu, H., Zhou, H., Xu, S. (2019). Oral microbiota as promising diagnostic biomarkers for gastrointestinal cancer: A systematic review. Onco. Targets Ther. 12, 11131–11144. doi: 10.2147/OTT.S230262
Chen, M. F., Lu, M. S., Hsieh, C. C., Chen, W. C. (2021). Porphyromonas gingivalis promotes tumor progression in esophageal squamous cell carcinoma. Cell. Oncol. (Dordr). 44, 373–384. doi: 10.1007/s13402-020-00573-x
Chen, H., Nie, S., Zhu, Y., Lu, M. (2015). Teeth loss, teeth brushing and esophageal carcinoma: a systematic review and meta-analysis. Sci. Rep. 5, 15203. doi: 10.1038/srep15203
Chen, Q. L., Zeng, X. T., Luo, Z. X., Duan, X. L., Qin, J., Leng, W. D. (2016). Tooth loss is associated with increased risk of esophageal cancer: Evidence from a meta-analysis with dose-response analysis. Sci. Rep. 6, 18900. doi: 10.1038/srep18900
Chen, X. X., Zhong, Q., Liu, Y., Yan, S. M., Chen, Z. H., Jin, S. Z., et al. (2017). Genomic comparison of esophageal squamous cell carcinoma and its precursor lesions by multi-region whole-exome sequencing. Nat. Commun. 8, 524. doi: 10.1038/s41467-017-00650-0
Dawsey, S. M., Lewin, K. J., Liu, F. S., Wang, G. Q., Shen, Q. (1994). Esophageal morphology from linxian, china. Squamous histologic findings in 754 patients. Cancer 732027–2037. doi: 10.1002/1097-0142(19940415)73:8<2027::aid-cncr2820730803>3.0.co;2-3
Dawsey, S. M., Lewin, K. J., Wang, G. Q., Liu, F. S., Nieberg, R. K., Yu, Y., et al. (1994). Squamous esophageal histology and subsequent risk of squamous cell carcinoma of the esophagus. A prospective follow-up study from linxian, China. Cancer 741686–1692. doi: 10.1002/1097-0142(19940915)74:6<1686::aid-cncr2820740608>3.0.co;2-v
Domper Arnal, M. J., Ferrández Arenas, Á., Lanas Arbeloa, Á. (2015). Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World. J. Gastroenterol. 21, 7933–7943. doi: 10.3748/wjg.v21.i26.7933
Gao, S., Li, S., Ma, Z., Liang, S., Shan, T., Zhang, M., et al. (2016). Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect. Agent. Cancer 11, 3. doi: 10.1186/s13027-016-0049-x
Gao, S. G., Yang, J. Q., Ma, Z. K., Yuan, X., Zhao, C., Wang, G. C., et al. (2018). Preoperative serum immunoglobulin G and a antibodies to Porphyromonas gingivalis are potential serum biomarkers for the diagnosis and prognosis of esophageal squamous cell carcinoma. BMC. Cancer 18, 17. doi: 10.1186/s12885-017-3905-1
Gupta, B., Kumar, N. (2017). Worldwide incidence, mortality and time trends for cancer of the oesophagus. Eur. J. Cancer Prev. 26, 107–118. doi: 10.1097/CEJ.0000000000000249
Hiraki, A., Matsuo, K., Suzuki, T., Kawase, T., Tajima, K. (2008). Teeth loss and risk of cancer at 14 common sites in Japanese. Cancer Epidemiol. Biomarkers. Prev. 17, 1222–1227. doi: 10.1158/1055-9965.EPI-07-2761
Jia, X., Min, L., Zhu, S., Zhang, S., Huang, X. (2018). Loss of sonic hedgehog gene leads to muscle development disorder and megaesophagus in mice. FASEB. J. 32, 5703–5715. doi: 10.1096/fj.201701581R
Kageyama, S., Takeshita, T., Takeuchi, K., Asakawa, M., Matsumi, R., Furuta, M., et al. (2019). Characteristics of the salivary microbiota in patients with various digestive tract cancers. Front. Microbiol. 101780. doi: 10.3389/fmicb.2019.01780
Kassebaum, N. J., Bernabé, E., Dahiya, M., Bhandari, B., Murray, C. J., Marcenes, W. (2014). Global burden of severe periodontitis in 1990-2010: A systematic review and meta-regression. J. Dent. Res. 93, 1045–1053. doi: 10.1177/0022034514552491
Kawasaki, M., Ikeda, Y., Ikeda, E., Takahashi, M., Tanaka, D., Nakajima, Y., et al. (2021). Oral infectious bacteria in dental plaque and saliva as risk factors in patients with esophageal cancer. Cancer 127 (4), 512–519. doi: 10.1002/cncr.33316
Linden, G. J., Lyons, A., Scannapieco, F. A. (2013). Periodontal systemic associations: Review of the evidence. J. Clin. Periodontol. 40 (Suppl 14), S8–S19. doi: 10.1111/jcpe.12064
Meng, F., Li, R., Ma, L., Liu, L., Lai, X., Yang, D., et al. (2019). Porphyromonas gingivalis promotes the motility of esophageal squamous cell carcinoma by activating NF-κB signaling pathway. Microbes Infect. 21, 296–304. doi: 10.1016/j.micinf.2019.01.005
Nwizu, N. N., Marshall, J. R., Moysich, K., Genco, R. J., Hovey, K. M., Mai, X., et al. (2017). Periodontal disease and incident cancer risk among postmenopausal women: results from the women's health initiative observational cohort. Cancer Epidemiol. Biomarkers. Prev. 26, 1255–1265. doi: 10.1158/1055-9965.EPI-17-0212
Okumura, H., Uchikado, Y., Setoyama, T., Matsumoto, M., Owaki, T., Ishigami, S., et al. (2014). Biomarkers for predicting the response of esophageal squamous cell carcinoma to neoadjuvant chemoradiation therapy. Surg. Today 44, 421–428. doi: 10.1007/s00595-013-0580-y
Pennathur, A., Gibson, M. K., Jobe, B. A., Luketich, J. D. (2013). Oesophageal carcinoma. Lancet 381, 400–412. doi: 10.1016/S0140-6736(12)60643-6
Peters, B. A., Wu, J., Pei, Z., Yang, L., Purdue, M. P., Freedman, N. D., et al. (2017). Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 77, 6777–6787. doi: 10.1158/0008-5472.CAN-17-1296
Qi, Y. J., Jiao, Y. L., Chen, P., Kong, J. Y., Gu, B. L., Liu, K., et al. (2020). Porphyromonas gingivalis promotes progression of esophageal squamous cell cancer via TGFβ-dependent Smad/YAP/TAZ signaling. PLoS. Biol. 18, e3000825. doi: 10.1371/journal.pbio.3000825
Sanz, M., Marco Del Castillo, A., Jepsen, S., Gonzalez-Juanatey, J. R., D'Aiuto, F., Bouchard, P., et al. (2020). Periodontitis and cardiovascular diseases: Consensus report. J. Clin. Periodontol. 47, 268–288. doi: 10.1111/jcpe.13189
Slots, J. (2017). Periodontitis: Facts, fallacies and the future. Periodontol. 2000. 75, 7–23. doi: 10.1111/prd.12221
Wadhwa, R., Wang, X., Baladandayuthapani, V., Liu, B., Shiozaki, H., Shimodaira, Y., et al. (2017). Nuclear expression of gli-1 is predictive of pathologic complete response to chemoradiation in trimodality treated oesophageal cancer patients. Br. J. Cancer 117, 648–655. doi: 10.1038/bjc.2017.225
Yuan, X., Liu, Y., Kong, J., Gu, B., Qi, Y., Wang, X., et al. (2017). Different frequencies of Porphyromonas gingivalis infection in cancers of the upper digestive tract. Cancer Lett. 404, 1–7. doi: 10.1016/j.canlet.2017.07.003
Yuan, X., Liu, Y., Li, G., Lan, Z., Ma, M., Li, H., et al. (2019). Blockade of immune-checkpoint B7-H4 and lysine demethylase 5B in esophageal squamous cell carcinoma confers protective immunity against P. gingivalis infection. Cancer Immunol. Res. 7, 1440–1456. doi: 10.1158/2326-6066.CIR-18-0709
Yu, J., Wu, R., Wang, Z., Chen, S., Chen, S., Guo, G., et al. (2019). Cyclopamine suppresses human esophageal carcinoma cell growth by inhibiting glioma-associated oncogene protein-1, a marker of human esophageal carcinoma progression. Med. Sci. Monit. 25, 1518–1525. doi: 10.12659/MSM.912858
Zhou, Y., Luo, G. H. (2019). Porphyromonas gingivalis and digestive system cancers. World. J. Clin. Cases 7, 819–829. doi: 10.12998/wjcc.v7.i7.819
Keywords: periodontal-systemic disease interactions, microbiome, porphyromonas gingivalis, esophagus, dysplasia, sonic hedgehog
Citation: Jia X, Liu J, He Y and Huang X (2022) Porphyromonas gingivalis secretion leads to dysplasia of normal esophageal epithelial cells via the Sonic hedgehog pathway. Front. Cell. Infect. Microbiol. 12:982636. doi: 10.3389/fcimb.2022.982636
Received: 30 June 2022; Accepted: 16 September 2022;
Published: 03 October 2022.
Edited by:
Aditi Chopra, Manipal College of Dental Sciences, IndiaReviewed by:
Tianfan Cheng, The University of Hong Kong, Hong Kong SAR, ChinaAkihiro Yoshida, Matsumoto Dental University, Japan
Copyright © 2022 Jia, Liu, He and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofeng Huang, aHVhbmd4ZjE5OThAMTYzLmNvbQ==
 Xueting Jia
Xueting Jia Jinan Liu1,2
Jinan Liu1,2