
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell. Infect. Microbiol. , 29 July 2022
Sec. Microbiome in Health and Disease
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.964710
This article is part of the Research Topic Mechanism of Colonization and Persistence of Gut Commensal Microbiota and Pathogens View all 5 articles
The mammalian gut microbiota comprises a variety of commensals including potential probiotics and pathobionts, influencing the host itself. Members of the microbiota can intervene with host physiology by several mechanisms, including the secretion of a relatively well-reported set of metabolic products. Another microbiota influence mechanism is the use of secreted proteins (i.e., the secretome), impacting both the host and other community members. While widely reported and studied in pathogens, this mechanism remains understood to a lesser extent in commensals, and this knowledge is increasing in recent years. In the following minireview, we assess the current literature covering different studies, concerning the functions of secretable proteins from members of the gut microbiota (including commensals, pathobionts, and probiotics). Their effect on host physiology and health, and how these effects can be harnessed by postbiotic products, are also discussed.
Human microbiota research has become a relevant subject in microbiology in the last few decades. Moreover, the relationships between microbiota, health, and disease have gained increasing attention, given their potential implications in human physiology and applications in human medicine (Cullen et al., 2020). The human microbiota contains an enormous genetic content; it includes an estimated set of 22 million protein-coding genes for the intestinal microbiota (Tierney et al., 2019), an overwhelming amount compared with the approximately 21,000 protein-coding genes found in the human genome (Pertea et al., 2018). This gene repertoire is involved in microbial adaptation to the human body and how those microbes interact with their niches across the human body. This enormous gene set from the human microbiota, known as the human microbiome, can induce changes in the host by the action of their gene products.
One of the most relevant mechanisms of this influence effect is releasing a variety of molecules into their niches (Shine and Crawford, 2021). From fermentation products to antimicrobial peptides, these molecules are involved in various processes influencing the host and other community members. For example, short-chain fatty acids (SCFAs) potentially affect the host metabolism and signaling, with implications for health and disease (Ratajczak et al., 2019; Dalile et al., 2019; Sanna et al., 2019). The study of the role of the SCFA on the relation between human microbiota and human cells is a relatively well-covered topic covered in other reviews (Tan et al., 2014; Zheng et al., 2022). Moreover, other reviews cover the effect and role of the pleiad of secreted metabolites on the interaction between human microbiota and the host and the interaction between microbial communities themselves (e.g., (Shine and Crawford, 2021)). Among the repertoire of secreted compounds produced by the microbiota, an increasingly relevant group of study targets corresponds to the poorly studied secretable proteins known as the secretome (Tjalsma et al., 2000).
Several findings concerning the structure and function of secretion systems and secreted proteins have been made formerly from studies using pathogenic microbes (Green and Mecsas, 2016; Nicod et al., 2017). In several cases, pathways connecting the secretion of different proteins with changes in the host cellular program are also proposed (Hueck, 1998; Bumann et al., 2002; van Ulsen and Tommassen, 2006), suggesting that secreted protein effectors result from a very specialized relationship between specific pathogens and their host. Likewise, there is a possibility that several commensal-associated proteins are involved in a particular relationship with the host. Therefore, the potential role of secreted proteins is gaining importance in understanding the human microbiome-host relationship dynamics. However, there is little evidence about the repertoire of secreted proteins produced by gut commensals. In the following minireview, we assess the current literature covering the main studies of secreted or exposed proteins as effectors on the host physiology in the gut microbiome. We also cover the known implications on host health and diseases and discuss future research projections in this field.
In order to produce any effect on the host, secreted proteins require certain systems for their release. Some mechanisms involved in protein secretion among gut commensals and probiotic microbes are currently reported. For example, several gut commensals contain genes for different bacterial secretion systems, being protein complexes involved in the transport of proteins from the cytoplasm to the extracellular side of the cell (Green and Mecsas, 2016). Genes encoding components of the Type VI secretion system (T6SS) are widely distributed among Proteobacteria and Bacteroidetes (Silverman et al., 2012), and they were recovered from some members of the gut microbiota, such as Bacteroides (Russell et al., 2014), with even the presence of some genomic islands associated with those systems in strains of the common commensal Bacteroides fragilis (Coyne et al., 2016). The main roles of the effect proteins using these secretion systems are related to ecological competition between related microorganisms (Chen et al., 2019; Wood et al., 2020). T6SS may also have a modulatory role: a study on Helicobacter hepaticus, a pathobiont from the gut microbiota, showed that T6SS limited the ability of this microbe to colonize and induce inflammation, promoting a balanced relationship with the host (Chow and Mazmanian, 2010); the breakdown of this balancing role may have a role in certain diseases such as inflammatory bowel disease and colon cancer.
Another system for protein secretion in gut commensals, and some probiotic organisms, consists of extracellular vesicles (EVs) or outer membrane vesicles (OMVs). EVs are lipid-closed bodies produced by cells and released into the extracellular spaces (Doyle and Wang, 2019). Previous studies have shown that some gut commensals can produce EVs (Cortés et al., 2021; Díaz-Garrido et al., 2021; Ñahui Palomino et al., 2021). For example, a study analyzing EVs obtained in fecal samples from patients in different colorectal cancer (CRC) stages and healthy conditions (Park et al., 2021) could suggest the taxonomic origin for those EVs; vesicles from members of the Rikenellaceae and Acidaminococcaceae families were enriched in late CRC subjects, whereas Lactobacillaceae-derived EVs were relatively enriched in early CRC subjects. Another recent study (De Saedeleer et al., 2021), utilizing a methodological framework for the extraction of microbiome-derived, extracellular components, identified several extracellular proteins, mostly associated with Bacteroidetes and Proteobacteria, previously identified in OMVs; some of those proteins include ribosomal proteins, chaperones, outer membrane proteins, and some enzymes (glutamine synthetase, recombinase A, and formate acetyltransferase), as well as a variety of proteins involved in metabolite transport. Another study could isolate EVs from an enriched culture of porcine intestinal microbiota incubated with beta-mannan (Lagos et al., 2020), showing that several proteins enriched in those EVs were associated with translation, energy metabolism, and amino acid/carbohydrate transport. These studies confirm the occurrence and importance of EVs/OMVs in the intestinal niche from human and non-human hosts (Cecil et al., 2019) and the relevance of those vesicles as a powerful new approach for therapeutic approaches (Morishita et al., 2021).
There is an increasing number of reports about the presence and function of different microbial secreted proteins and enzymes in the mammalian gut, with implications for health and disease (Figure 1). The first functions of secreted proteins observed in gut commensals were enzymatic. Gut commensals are constantly secreting different proteins to their niche to break different compounds. Among them, secreted carbohydrate-active enzymes (CAEs) are important proteins utilized by gut microbiota to degrade carbohydrates that the host cannot consume (El Kaoutari et al., 2013). Gut microbiota can also produce secreted protease activities influencing the activity of the host, as seen in early studies with a set of Bacteroides strains, which could reduce maltase and sucrase activities from human brush-border preparations by degradation (Riepe et al., 1980). This secreted bacterial protease activity could be involved in the brush border damage observed during bacterial overgrowth syndromes. More recently, meta-omic studies showed a functional connection between this secretable protease activity and the severity of ulcerative colitis (UC) (Mills et al., 2022).
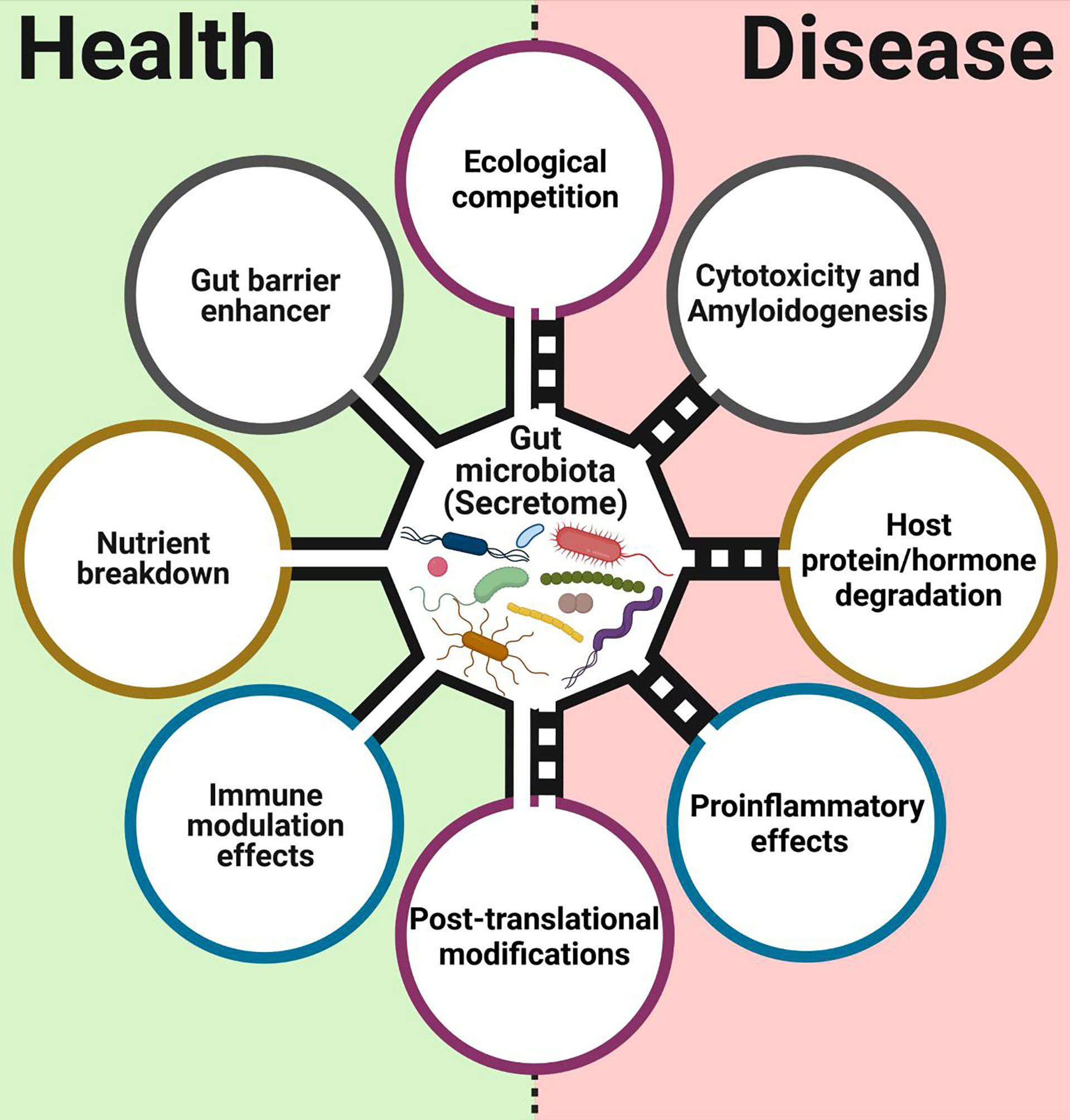
Figure 1 Main functions associated with proteins, enzymes, and surface layer proteins from commensals and probiotics bacterial intestinal. This figure was created with BioRender.com.
Despite being more researched in pathogens, some commensals were also analyzed in their capability to create adhesion structures from their secreted proteins. Veilonella parvula, a potential roleplayer in developing infant gut immunity, can secrete an autotransporter to auto-aggregate during biofilm formation to compete with other members of the microbial community (Béchon et al., 2020).
Some secreted proteins can produce posttranslational modifications to host proteins. O-linked N-acetylglucosaminylation (O-GlcNAcylation) by either the O-GlcNAcase (OGA) or the O-GlcNAc transferase (OGT) enzymes is predicted as a widely distributed feature in among members of Bacteroidetes and Firmicutes phyla, and depleted in the metagenome of subjects with UC (He et al., 2021). Moreover, microbial OGAs can influence the NF-κB signaling pathway, as observed in in-vitro studies, suggesting that those microbial enzymes can also influence the host (He et al., 2021). Some proteins from the microbiome can degrade peptide hormones: another in-vitro study showed that a protein called GelE, isolated from Enterococcus faecalis, is able to degrade the glucagon-like peptide-1 (GLP-1), a gastrointestinal hormone responsible for the regulation of appetite and glucose homeostasis (LeValley et al., 2020). This evidence points to a higher perspective about how microorganisms can interact directly with host proteins.
Another role observed from secreted proteins is their function as regulators of microbial communities. It has been observed that some organisms can induce inter- or intra-species antagonisms via the effect of some secreted proteins, like in the well-known case of the bacteriocins, peptides isolated from several gut commensals, a matter that has been addressed in other reviews (Dicks et al., 2018; Garcia-Gutierrez et al., 2019); several probiotics also contain bacteriocins, which some authors consider this as a probiotic trait (Deliorman Orhan, 2021). Some authors have even hypothesized that bacteriocins may pass the gut–blood barrier (Dicks et al., 2018), a feature with important implications for human infectious diseases. In the case of the commensals, strains from different gut organisms such as Enterococcus faecium, E. faecalis, Escherichia coli, Lactobacillus salivarius, L. acidophilus, L. gasseri, L. johnsonii, L. reuteri, and L. rhamnosus were found to produce different bacteriocins, with verified antimicrobial activity over members of the same or different species (Garcia-Gutierrez et al., 2019). In members of Bacteroidales, the role of secreted antimicrobial proteins (called BSAPs) has been observed even from genomic, experimental, and metagenomic data, suggesting that those proteins are toxic to members of the same species, mediating strain-level competition (Roelofs et al., 2016). Additionally, in B. fragilis strain 638R, the secretion of a highly expressed eukaryotic-like ubiquitin protein (named BfUbb), could inhibit growth from other related strains, suggesting that this protein is also a competition factor with an influence on the community composition (Chatzidaki-Livanis et al., 2017).
Secreted proteins are also associated with pathobionts. For example, in the case of the potential link between bacterial toxins and neurodegenerative diseases (Zhao et al., 2017; Zhao et al., 2021), some strains of B. fragilis secrete some proteins such as proteolipids, endo- and exotoxins, and fragilysin, a zinc-metalloproteinase with high neurotoxic activity. This latter toxin is also connected with potential effects in cancer (Haghi et al., 2019) and even with epigenetic changes (Allen et al., 2019). Other secreted proteins may contain amyloid-like properties, with implications for cancer and neurodegenerative disorders (Zhao et al., 2017), among other diseases. For example, FadA, secreted by Fusobacterium nucleatum, was associated with microbe reprogramming from commensal to pathogen phenotype during colorectal cancer development (Huang et al., 2022). Moreover, other amyloid-like proteins produced by some gut microorganisms may promote the formation of α-synuclein amyloids, a key etiologic factor for Parkinson’s disease (Wittung-Stafshede, 2022). One reported case of these amyloidogenic proteins is the curli protein CsgA from E. coli strain MC4100 (Sampson et al., 2020). A protein of Bacillus subtilis, called subtilisin, can translocate the gut epithelial barrier and cleave the human transthyretin (hTTR), generating a fragmented version with amyloidogenic properties with a potential role in senile systemic amyloidosis (SSA), a known factor involved in cardiovascular diseases (Peterle et al., 2020). Moreover, the protein fraction of the conditioned medium (i.e., the cell-free medium containing biologically active components derived from previous cell growth) from the potential probiotic Akkermansia muciniphila, cultivated in mucin-depleted conditions, could induce a set of phenotypic changes that could lead to alpha-synuclein aggregation, a pivotal feature associated Parkinson’s Disease development (Amorim Neto et al., 2022). This result raises the question of how potentially probiotic members of the gut microbiota may generate negative outcomes due to the effect of their secretable proteins.
Finally, as previously mentioned in the case of bacteriocins, there are discovered several secreted proteins found in probiotic microorganisms. Probiotics are defined as “live microorganisms which, when administered in adequate amounts, confer a health benefit on the host” (Hill et al., 2014; Reid, 2016). Probiotics have an extended repertoire of functions supporting their role in host health, and the secretion of bioactive proteins is one of those probiotic properties. A repertoire of proteins and enzymes secreted from some strains of commensals and probiotics has been discovered and studied (Table 1), and the compilation of their functions in the human gut has been previously reviewed (Sánchez et al., 2010). The main functions associated with the secreted proteins are the protection of the gut barrier (with concomitant reduction of barrier permeability), nutrient degradation (via catabolism of dietary polysaccharides), immunomodulation (especially in an anti-inflammatory manner), cell growth arrest, stimulation of hormones involved in metabolism regulation and competition against pathogens, among other roles (Table 1). It is worth to notice that several species contain characterized proteins with the reported effect produced by their probiotic sources (Table 1). A remarkable example is Akkermansia muciniphila, with the proteins Amuc_1100 and P9, with immunoregulatory and metabolic-regulating properties, aspects already reported for the role of this organism in murine models and human subjects (Everard et al., 2013; Depommier et al., 2019). It is also worth to indicate that microbial protein secretion also can be induced by host cells: a previous in-vitro study showed that the presence of colonic epithelial cell-derived vesicles could induce Lactobacillus rhamnosus GG, a widely studied probiotic strain, to secrete a protein named P40, a protecting agent of intestinal epithelial cells (Yang et al., 2019). This observation suggests a strong connection between some probiotic microbes (formerly isolated from human samples), and human cellular products.
Furthermore, some experiments additionally suggest that there are actually several unidentified secretable proteins involved in the probiotic effect of some commonly used agents (Sánchez et al., 2010). Those unidentified mixtures showed improving effects on gut barrier integrity (Ewaschuk et al., 2008), or amelioration of enteropathogenic infection (Broekaert et al., 2007), among other effects.
The use of the term “postbiotic” has increased in recent years (Wegh et al., 2019). A postbiotic agent can be defined as a “preparation of inanimate microorganisms and, or, their components conferring a health benefit on the host” (Salminen et al., 2021). Secreted proteins enter the scope of this concept. Some studies have already made connections between the role of some beneficial secreted proteins and their potential use as postbiotics, such as the case of the protein HM0539 from Lactobacillus rhamnosus GG (Gao et al., 2019). Postbiotics can be discovered by evaluating the effect of dead probiotic cells on the host. For example, the role of the Amuc_1100 protein (Table 1) was investigated after it was observed that pasteurized extracts from A. muciniphila could produce even a higher effect than the live organism (Plovier et al., 2017). The use of specific secretable proteins as postbiotics may exhibit some advantages compared with the use of probiotics (Nataraj et al., 2020). For example, probiotics involve the use of living and independent organisms that must be controlled and directed to specific objectives, often in a strain-dependent manner. Due to safety reasons (Sanders et al., 2010), the use of genetically-modified probiotics in human subjects is subject to controversies (Plavec and Berlec, 2020). Postbiotic agents can overcome this limitation: as well as several secretable proteins can be purified, and several of them exhibit a similar effect directly; in comparison with the live microbial source, the use of the purified secreted protein can offer new standards for safety, with the same beneficial effects for human health. Moreover, the production of isolated proteins as postbiotics also involves easier storage and generation strategies than the storage and production of living organisms, with concomitant effects on the scalability of mass production at an industrial level.
The evidence presented in this minireview shows that secreted proteins produced by the gut microbiota may influence the host in positive and negative manners. In the first case, the discovery of new secreted proteins utilized by pathobionts can help to find new therapeutic targets to attack during certain diseases. On the other hand, the discovery of beneficial proteins secreted by commensals or probiotics may lead to the development of novel postbiotic products.
Today, there are several methods available to discover new secreted proteins. In addition to the classic “conditioned medium” experiments, utilized in the past to discover single proteins or undefined mixtures (Sánchez et al., 2010), the arrival of comparative genomics, transcriptomics, and metagenomics have become powerful allies in the discovery and characterization of more secreted proteins. For example, in-silico reconstruction of the secretome has been made from the pangenome of Lactobacillus rhamnosus (Kant et al., 2014), or for the Bifidobacterium genus (Lugli et al., 2018), relevant groups of the human gut microbiota organisms including probiotic agents, formulating the concept of the pansecretome. This type of reconstruction can also be made from metagenomes or even metatranscriptomes. For example, a study using a set of obese and normal-weight Mexican children metatranscriptomes detected the set of potentially secreted proteins using an in-silico approach from the content of those RNA sequenced samples (Gallardo-Becerra et al., 2020). This latter study formulated the concept of secrebiome.
By the use of transcriptome analyses, the expression pattern of some gut microbiota members can be also directed to the search for valuable secretable protein production patterns. A transcriptomic study comparing Akkermansia muciniphila expression in the presence or absence of mucin (Shin et al., 2019) showed the upregulation of 79 genes encoding secreted protein candidates during mucin depletion (including the postbiotic agent Amuc_1100). Those transcriptome changes were consistent with the previous observation that A. muciniphila grown under mucin-depleted conditions is capable of reducing obesity and improving intestinal barrier integrity in high-fat diet-induced diabetic mice.
Furthermore, metagenomics is also useful for discovering previously unknown genes with the potential to be secreted proteins. A massive (1,773 samples) metagenomic screening searching for novel small genes (Sberro et al., 2019) showed that near 30% of a set of ~4,000 novel gene families were predicted to have a signal peptide or have transmembrane regions, suggesting that a considerable proportion of novel gene families in the human microbiome could be targeted to be secreted.
Finally, by either using classical biochemical/molecular techniques or by combining those strategies with “-omic” disciplines, the ability to discover new secreted proteins opens the possibility of more functional secreted proteins that may influence the host.
JC conceived the study; BV-V, DG, and JC gathered the data. BV-V made the table and the figure. JC wrote the paper. All the authors read the manuscript and approved the content.
JC research is supported by Fondecyt ANID project #11200209. BV-V is supported by ANID Grant For Doctorate Studies in Chile (ANID Doctorado Nacional 2021-21211564).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Allen, J., Hao, S., Sears, C. L., Timp, W. (2019). Epigenetic changes induced by Bacteroides fragilis toxin. Infect. Immun. 87, e00447–18. doi: 10.1128/IAI.00447-18
Amorim Neto, D. P., Bosque, B. P., Pereira de Godoy, J. V., Rodrigues, P. V., Meneses, D. D., Tostes, K., et al (2022). Akkermansia muciniphila induces mitochondrial calcium overload and α -synuclein aggregation in an enteroendocrine cell line. iScience 25, 103908. doi: 10.1016/j.isci.2022.103908
An, B. C., Hong, S., Park, H. J., Kim, B.-K., Ahn, J. Y., Ryu, Y., et al (2019). Anti-colorectal cancer effects of probiotic-derived p8 protein. Genes (Basel) 10, 624. doi: 10.3390/genes10080624
Ankaiah, D., Esakkiraj, P., Perumal, V., Ayyanna, R., Venkatesan, A. (2017). Probiotic characterization of Enterococcus faecium por1: Cloning, over expression of enterocin-a and evaluation of antibacterial, anti-cancer properties. J. Funct. Foods 38, 280–292. doi: 10.1016/j.jff.2017.09.034
Ankaiah, D., Palanichamy, E., Antonyraj, C. B., Ayyanna, R., Perumal, V., Ahamed, S. I. B., et al (2018). Cloning, overexpression, purification of bacteriocin enterocin-b and structural analysis, interaction determination of enterocin-A, B against pathogenic bacteria and human cancer cells. Int. J. Biol. Macromol. 116, 502–512. doi: 10.1016/j.ijbiomac.2018.05.002
Béchon, N., Jiménez-Fernández, A., Witwinowski, J., Bierque, E., Taib, N., Cokelaer, T., et al (2020). Autotransporters drive biofilm formation and autoaggregation in the diderm firmicute Veillonella parvula. J. Bacteriol. 202, e00461–20. doi: 10.1128/JB.00461-20
Bernardo, D., Sánchez, B., Al-Hassi, H. O., Mann, E. R., Urdaci, M. C., Knight, S. C., et al (2012). Microbiota/host crosstalk biomarkers: regulatory response of human intestinal dendritic cells exposed to Lactobacillus extracellular encrypted peptide. PLoS One 7, e36262. doi: 10.1371/journal.pone.0036262
Broekaert, I. J., Nanthakumar, N. N., Walker, W. A. (2007). Secreted probiotic factors ameliorate enteropathogenic infection in zinc-deficient human Caco-2 and T84 cell lines. Pediatr. Res. 62, 139–144. doi: 10.1203/PDR.0b013e31809fd85e
Bumann, D., Aksu, S., Wendland, M., Janek, K., Zimny-Arndt, U., Sabarth, N., et al (2002). Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect. Immun. 70, 3396–3403. doi: 10.1128/IAI.70.7.3396-3403.2002
Cecil, J. D., Sirisaengtaksin, N., O’Brien-Simpson, N. M., Krachler, A. M. (2019). Outer membrane vesicle-host cell interactions. Microbiol. Spectr. 7, 7–1. doi: 10.1128/microbiolspec.PSIB-0001-2018
Chatzidaki-Livanis, M., Coyne, M. J., Roelofs, K. G., Gentyala, R. R., Caldwell, J. M., Comstock, L. E. (2017). Gut symbiont Bacteroides fragilis secretes a eukaryotic-like ubiquitin protein that mediates intraspecies antagonism. MBio 8, e01902–17. doi: 10.1128/mBio.01902-17
Chen, C., Yang, X., Shen, X. (2019). Confirmed and potential roles of bacterial T6SSs in the intestinal ecosystem. Front. Microbiol. 10, 1484. doi: 10.3389/fmicb.2019.01484
Chow, J., Mazmanian, S. K. (2010). A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7, 265–276. doi: 10.1016/j.chom.2010.03.004
Cortés, A., Sotillo, J., Rinaldi, G., Cantacessi, C. (2021). Gut-microbiota-derived extracellular vesicles: Overlooked mediators in host-helminth interactions? Trends Parasitol. 37, 690–693. doi: 10.1016/j.pt.2021.05.009
Coyne, M. J., Roelofs, K. G., Comstock, L. E. (2016). Type VI secretion systems of human gut bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17, 58. doi: 10.1186/s12864-016-2377-z
Cullen, C. M., Aneja, K. K., Beyhan, S., Cho, C. E., Woloszynek, S., Convertino, M., et al (2020). Emerging priorities for microbiome research. Front. Microbiol. 11, 136. doi: 10.3389/fmicb.2020.00136
Dalile, B., Van Oudenhove, L., Vervliet, B., Verbeke, K. (2019). The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 16, 461–478. doi: 10.1038/s41575-019-0157-3
Deliorman Orhan, D. (2021). “Bacteriocins produced by probiotic microorganisms,” in Advances in probiotics Dhanasekaran, D., Sankaranarayanan, A. eds. (Amsterdam, NL: Elsevier), 277–291. doi: 10.1016/B978-0-12-822909-5.00018-6
Depommier, C., Everard, A., Druart, C., Plovier, H., Van Hul, M., Vieira-Silva, S., et al (2019). Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 25, 1096–1103. doi: 10.1038/s41591-019-0495-2
De Saedeleer, B., Malabirade, A., Ramiro-Garcia, J., Habier, J., Trezzi, J.-P., Peters, S. L., et al (2021). Systematic characterization of human gut microbiome-secreted molecules by integrated multi-omics. ISME Commun. 1, 82. doi: 10.1038/s43705-021-00078-0
Díaz-Garrido, N., Badia, J., Baldomà, L. (2021). Microbiota-derived extracellular vesicles in interkingdom communication in the gut. J. Extracell. Vesicles 10, e12161. doi: 10.1002/jev2.12161
Dicks, L. M. T., Dreyer, L., Smith, C., van Staden, A. D. (2018). A review: The fate of bacteriocins in the human gastro-intestinal tract: Do they cross the gut-blood barrier? Front. Microbiol. 9, 2297. doi: 10.3389/fmicb.2018.02297
Doyle, L. M., Wang, M. Z. (2019). Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 8, 727. doi: 10.3390/cells8070727
Du, M., Xie, X., Yang, S., Li, Y., Jiang, T., Yang, J., et al (2021). Lysozyme-like protein produced by Bifidobacterium longum regulates human gut microbiota using In vitro models. Molecules 26, 6480. doi: 10.3390/molecules26216480
El Kaoutari, A., Armougom, F., Gordon, J. I., Raoult, D., Henrissat, B. (2013). The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11, 497–504. doi: 10.1038/nrmicro3050
Everard, A., Belzer, C., Geurts, L., Ouwerkerk, J. P., Druart, C., Bindels, L. B., et al (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110, 9066–9071. doi: 10.1073/pnas.1219451110
Ewaschuk, J. B., Diaz, H., Meddings, L., Diederichs, B., Dmytrash, A., Backer, J., et al (2008). Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 295, G1025–G1034. doi: 10.1152/ajpgi.90227.2008
Fang, K., Jin, X., Hong, S. H. (2018). Probiotic escherichia coli inhibits biofilm formation of pathogenic e. coli via extracellular activity of DegP. Sci. Rep. 8, 4939. doi: 10.1038/s41598-018-23180-1
Fernández-Tomé, S., Marin, A. C., Ortega Moreno, L., Baldan-Martin, M., Mora-Gutiérrez, I., Lanas-Gimeno, A., et al (2019). Immunomodulatory effect of gut microbiota-derived bioactive peptides on human immune system from healthy controls and patients with inflammatory bowel disease. Nutrients 11, 2605. doi: 10.3390/nu11112605
Gallardo-Becerra, L., Cornejo-Granados, F., García-López, R., Valdez-Lara, A., Bikel, S., Canizales-Quinteros, S., et al (2020). Metatranscriptomic analysis to define the secrebiome, and 16S rRNA profiling of the gut microbiome in obesity and metabolic syndrome of Mexican children. Microb. Cell Fact. 19, 61. doi: 10.1186/s12934-020-01319-y
Gao, J., Li, Y., Wan, Y., Hu, T., Liu, L., Yang, S., et al (2019). A novel postbiotic from Lactobacillus rhamnosus GG with a beneficial effect on intestinal barrier function. Front. Microbiol. 10, 477. doi: 10.3389/fmicb.2019.00477
Garcia-Gutierrez, E., Mayer, M. J., Cotter, P. D., Narbad, A. (2019). Gut microbiota as a source of novel antimicrobials. Gut Microbes 10, 1–21. doi: 10.1080/19490976.2018.1455790
Green, E. R., Mecsas, J. (2016). Bacterial secretion systems: an overview. Microbiol. Spectr. 4, 4–1. doi: 10.1128/microbiolspec.VMBF-0012-2015
Haghi, F., Goli, E., Mirzaei, B., Zeighami, H. (2019). The association between fecal enterotoxigenic B. fragilis with colorectal cancer. BMC Cancer 19, 879. doi: 10.1186/s12885-019-6115-1
He, X., Gao, J., Peng, L., Hu, T., Wan, Y., Zhou, M., et al (2021). Bacterial O-GlcNAcase genes abundance decreases in ulcerative colitis patients and its administration ameliorates colitis in mice. Gut 70, 1872–1883. doi: 10.1136/gutjnl-2020-322468
Hering, N. A., Richter, J. F., Fromm, A., Wieser, A., Hartmann, S., Günzel, D., et al (2014). TcpC protein from E. coli nissle improves epithelial barrier function involving PKCζ and ERK1/2 signaling in HT-29/B6 cells. Mucosal Immunol. 7, 369–378. doi: 10.1038/mi.2013.55
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al (2014). Expert consensus document. the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Huang, A., Torres, A., Patel, R., Saxena, A., Patel, M. (2022). Fusobacterium nucleatum as a marker for epithelial to mesenchymal transition in colorectal cancer. FASEB J. 36 Suppl 1. doi: 10.1096/fasebj.2022.36.S1.L7993
Hueck, C. J. (1998). Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62, 379–433. doi: 10.1128/MMBR.62.2.379-433.1998
Kant, R., Rintahaka, J., Yu, X., Sigvart-Mattila, P., Paulin, L., Mecklin, J.-P., et al (2014). A comparative pan-genome perspective of niche-adaptable cell-surface protein phenotypes in Lactobacillus rhamnosus. PLoS One 9, e102762. doi: 10.1371/journal.pone.0102762
Kim, B., Wang, Y.-C., Hespen, C. W., Espinosa, J., Salje, J., Rangan, K. J., et al (2019). Enterococcus faecium secreted antigen a generates muropeptides to enhance host immunity and limit bacterial pathogenesis. eLife 8, e45343. doi: 10.7554/eLife.45343
Lagos, L., Leanti La Rosa, S., Ø Arntzen, M., Ånestad, R., Terrapon, N., Gaby, J. C., et al (2020). Isolation and characterization of extracellular vesicles secreted in vitro by porcine microbiota. Microorganisms 8, 983. doi: 10.3390/microorganisms8070983
LeValley, S. L., Tomaro-Duchesneau, C., Britton, R. A. (2020). Degradation of the incretin hormone glucagon-like peptide-1 (GLP-1) by Enterococcus faecalis metalloprotease GelE. mSphere 5, e00585–19. doi: 10.1128/mSphere.00585-19
Li, Q., Hu, W., Liu, W.-X., Zhao, L.-Y., Huang, D., Liu, X.-D., et al (2021). Streptococcus thermophilus inhibits colorectal tumorigenesis through secreting β-galactosidase. Gastroenterology 160, 1179–1193.e14. doi: 10.1053/j.gastro.2020.09.003
Lugli, G. A., Mancino, W., Milani, C., Duranti, S., Turroni, F., van Sinderen, D., et al (2018). Reconstruction of the bifidobacterial pan-secretome reveals the network of extracellular interactions between bifidobacteria and the infant gut. Appl. Environ. Microbiol. 84, e00796–18. doi: 10.1128/AEM.00796-18
Mills, R. H., Dulai, P. S., Vázquez-Baeza, Y., Sauceda, C., Daniel, N., Gerner, R. R., et al (2022). Multi-omics analyses of the ulcerative colitis gut microbiome link bacteroides vulgatus proteases with disease severity. Nat. Microbiol. 7, 262–276. doi: 10.1038/s41564-021-01050-3
Morishita, M., Horita, M., Higuchi, A., Marui, M., Katsumi, H., Yamamoto, A. (2021). Characterizing different probiotic-derived extracellular vesicles as a novel adjuvant for immunotherapy. Mol. Pharm. 18, 1080–1092. doi: 10.1021/acs.molpharmaceut.0c01011
Ñahui Palomino, R. A., Vanpouille, C., Costantini, P. E., Margolis, L. (2021). Microbiota-host communications: Bacterial extracellular vesicles as a common language. PLoS Pathog. 17, e1009508. doi: 10.1371/journal.ppat.1009508
Nataraj, B. H., Ali, S. A., Behare, P. V., Yadav, H. (2020). Postbiotics-parabiotics: the new horizons in microbial biotherapy and functional foods. Microb. Cell Fact. 19, 168. doi: 10.1186/s12934-020-01426-w
Nicod, C., Banaei-Esfahani, A., Collins, B. C. (2017). Elucidation of host-pathogen protein-protein interactions to uncover mechanisms of host cell rewiring. Curr. Opin. Microbiol. 39, 7–15. doi: 10.1016/j.mib.2017.07.005
Park, J., Kim, N.-E., Yoon, H., Shin, C. M., Kim, N., Lee, D. H., et al (2021). Fecal microbiota and gut microbe-derived extracellular vesicles in colorectal cancer. Front. Oncol. 11. doi: 10.3389/fonc.2021.650026
Pertea, M., Shumate, A., Pertea, G., Varabyou, A., Breitwieser, F. P., Chang, Y.-C., et al (2018). CHESS: a new human gene catalog curated from thousands of large-scale RNA sequencing experiments reveals extensive transcriptional noise. Genome Biol. 19, 208. doi: 10.1186/s13059-018-1590-2
Peterle, D., Pontarollo, G., Spada, S., Brun, P., Palazzi, L., Sokolov, A. V., et al (2020). A serine protease secreted from Bacillus subtilis cleaves human plasma transthyretin to generate an amyloidogenic fragment. Commun. Biol. 3, 764. doi: 10.1038/s42003-020-01493-0
Petrova, M. I., Imholz, N. C. E., Verhoeven, T. L. A., Balzarini, J., Van Damme, E. J. M., Schols, D., et al (2016). Lectin-like molecules of Lactobacillus rhamnosus GG inhibit pathogenic Escherichia coli and Salmonella biofilm formation. PLoS One 11, e0161337. doi: 10.1371/journal.pone.0161337
Plavec, T. V., Berlec, A. (2020). Safety aspects of genetically modified lactic acid bacteria. Microorganisms 8, e00796–18. doi: 10.3390/microorganisms8020297
Plovier, H., Everard, A., Druart, C., Depommier, C., Van Hul, M., Geurts, L., et al (2017). A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113. doi: 10.1038/nm.4236
Quévrain, E., Maubert, M. A., Michon, C., Chain, F., Marquant, R., Tailhades, J., et al (2016). Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in crohn’s disease. Gut 65, 415–425. doi: 10.1136/gutjnl-2014-307649
Rangan, K. J., Pedicord, V. A., Wang, Y.-C., Kim, B., Lu, Y., Shaham, S., et al (2016). A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science 353, 1434–1437. doi: 10.1126/science.aaf3552
Ratajczak, W., Rył, A., Mizerski, A., Walczakiewicz, K., Sipak, O., Laszczyńska, M. (2019). Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim. Pol. 66, 1–12. doi: 10.18388/abp.2018_2648
Reid, G. (2016). Probiotics: definition, scope and mechanisms of action. Best Pract. Res. Clin. Gastroenterol. 30, 17–25. doi: 10.1016/j.bpg.2015.12.001
Riepe, S. P., Goldstein, J., Alpers, D. H. (1980). Effect of secreted Bacteroides proteases on human intestinal brush border hydrolases. J. Clin. Invest. 66, 314–322. doi: 10.1172/JCI109859
Roelofs, K. G., Coyne, M. J., Gentyala, R. R., Chatzidaki-Livanis, M., Comstock, L. E. (2016). Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. MBio 7, e01055–16. doi: 10.1128/mBio.01055-16
Russell, A. B., Wexler, A. G., Harding, B. N., Whitney, J. C., Bohn, A. J., Goo, Y. A., et al (2014). A type VI secretion-related pathway in bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–236. doi: 10.1016/j.chom.2014.07.007
Salminen, S., Collado, M. C., Endo, A., Hill, C., Lebeer, S., Quigley, E. M. M., et al (2021). The international scientific association of probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 18, 649–667. doi: 10.1038/s41575-021-00440-6
Sampson, T. R., Challis, C., Jain, N., Moiseyenko, A., Ladinsky, M. S., Shastri, G. G., et al (2020). A gut bacterial amyloid promotes α-synuclein aggregation and motor impairment in mice. eLife 9, e53111. doi: 10.7554/eLife.53111
Sánchez, B., Urdaci, M. C., Margolles, A. (2010). Extracellular proteins secreted by probiotic bacteria as mediators of effects that promote mucosa-bacteria interactions. Microbiology (Reading Engl) 156, 3232–3242. doi: 10.1099/mic.0.044057-0
Sanders, M. E., Akkermans, L. M. A., Haller, D., Hammerman, C., Heimbach, J., Hörmannsperger, G., et al (2010). Safety assessment of probiotics for human use. Gut Microbes 1, 164–185. doi: 10.4161/gmic.1.3.12127
Sanna, S., van Zuydam, N. R., Mahajan, A., Kurilshikov, A., Vich Vila, A., Võsa, U., et al (2019). Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 51, 600–605. doi: 10.1038/s41588-019-0350-x
Sberro, H., Fremin, B. J., Zlitni, S., Edfors, F., Greenfield, N., Snyder, M. P., et al (2019). Large-Scale analyses of human microbiomes reveal thousands of small, novel genes. Cell 178, 1245–1259.e14. doi: 10.1016/j.cell.2019.07.016
Shine, E. E., Crawford, J. M. (2021). Molecules from the microbiome. Annu. Rev. Biochem. 90, 789–815. doi: 10.1146/annurev-biochem-080320-115307
Shin, J., Noh, J.-R., Chang, D.-H., Kim, Y.-H., Kim, M. H., Lee, E. S., et al (2019). Elucidation of Akkermansia muciniphila probiotic traits driven by mucin depletion. Front. Microbiol. 10, 1137. doi: 10.3389/fmicb.2019.01137
Silverman, J. M., Brunet, Y. R., Cascales, E., Mougous, J. D. (2012). Structure and regulation of the type VI secretion system. Annu. Rev. Microbiol. 66, 453–472. doi: 10.1146/annurev-micro-121809-151619
Tan, J., McKenzie, C., Potamitis, M., Thorburn, A. N., Mackay, C. R., Macia, L. (2014). The role of short-chain fatty acids in health and disease. Adv. Immunol. 121, 91–119. doi: 10.1016/B978-0-12-800100-4.00003-9
Tierney, B. T., Yang, Z., Luber, J. M., Beaudin, M., Wibowo, M. C., Baek, C., et al (2019). The landscape of genetic content in the gut and oral human microbiome. Cell Host Microbe 26, 283–295.e8. doi: 10.1016/j.chom.2019.07.008
Tjalsma, H., Bolhuis, A., Jongbloed, J. D., Bron, S., van Dijl, J. M. (2000). Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64, 515–547. doi: 10.1128/MMBR.64.3.515-547.2000
Tomaro-Duchesneau, C., LeValley, S. L., Roeth, D., Sun, L., Horrigan, F. T., Kalkum, M., et al (2020). Discovery of a bacterial peptide as a modulator of GLP-1 and metabolic disease. Sci. Rep. 10, 4922. doi: 10.1038/s41598-020-61112-0
van Ulsen, P., Tommassen, J. (2006). Protein secretion and secreted proteins in pathogenic neisseriaceae. FEMS Microbiol. Rev. 30, 292–319. doi: 10.1111/j.1574-6976.2006.00013.x
Wegh, C. A. M., Geerlings, S. Y., Knol, J., Roeselers, G., Belzer, C. (2019). Postbiotics and their potential applications in early life nutrition and beyond. Int. J. Mol. Sci. 20, 4673. doi: 10.3390/ijms20194673
Wittung-Stafshede, P. (2022). Gut power: Modulation of human amyloid formation by amyloidogenic proteins in the gastrointestinal tract. Curr. Opin. Struct. Biol. 72, 33–38. doi: 10.1016/j.sbi.2021.07.009
Wood, T. E., Aksoy, E., Hachani, A. (2020). From welfare to warfare: The arbitration of host-microbiota interplay by the type VI secretion system. Front. Cell. Infect. Microbiol. 10, 587948. doi: 10.3389/fcimb.2020.587948
Yan, F., Cao, H., Cover, T. L., Whitehead, R., Washington, M. K., Polk, D. B. (2007). Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 132, 562–575. doi: 10.1053/j.gastro.2006.11.022
Yang, L., Higginbotham, J. N., Liu, L., Zhao, G., Acra, S. A., Peek, R. M., et al (2019). Production of a functional factor, p40, by lactobacillus rhamnosus GG is promoted by intestinal epithelial cell-secreted extracellular vesicles. Infect. Immun. 87, e00113-19. doi: 10.1128/IAI.00113-19
Yin, M., Yan, X., Weng, W., Yang, Y., Gao, R., Liu, M., et al (2018). Micro integral membrane protein (MIMP), a newly discovered anti-inflammatory protein of Lactobacillus plantarum, enhances the gut barrier and modulates microbiota and inflammatory cytokines. Cell. Physiol. Biochem. 45, 474–490. doi: 10.1159/000487027
Yoon, H. S., Cho, C. H., Yun, M. S., Jang, S. J., You, H. J., Kim, J.-H., et al (2021). Akkermansia muciniphila secretes a glucagon-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice. Nat. Microbiol. 6, 563–573. doi: 10.1038/s41564-021-00880-5
Zhao, Y., Jaber, V., Lukiw, W. J. (2017). Secretory products of the human GI tract microbiome and their potential impact on Alzheimer’s disease (AD): detection of lipopolysaccharide (LPS) in AD hippocampus. Front. Cell. Infect. Microbiol. 7, 318. doi: 10.3389/fcimb.2017.00318
Zhao, Y., Jaber, V., Lukiw, W. J. (2021). Gastrointestinal tract microbiome-derived pro-inflammatory neurotoxins in alzheimer’s disease. J. Aging Sci. 9, Suppl 5.
Keywords: secretome, gut microbiome, secretion systems, extracellular vesicles, probiotics, postbiotics
Citation: Vidal-Veuthey B, González D and Cárdenas JP (2022) Role of microbial secreted proteins in gut microbiota-host interactions. Front. Cell. Infect. Microbiol. 12:964710. doi: 10.3389/fcimb.2022.964710
Received: 08 June 2022; Accepted: 06 July 2022;
Published: 29 July 2022.
Edited by:
Marjorie Pizarro-Guajardo, Texas A&M University, United StatesReviewed by:
Almagul Kushugulova, Nazarbayev University, KazakhstanCopyright © 2022 Vidal-Veuthey, González and Cárdenas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Juan P. Cárdenas, anVhbi5jYXJkZW5hc0B1bWF5b3IuY2w=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.