- 1Department of Microbiology, ESIC Medical College & Hospital, Faridabad, HR, India
- 2Department of Biochemistry, Bose Institute, Kolkata, WB, India
Background: With the rapid increase in COVID-19 cases and the discovery of new viral variants within India over multiple waves, the expensive reagents and time-consuming sample pretreatment required for qPCR analysis have led to slower detection of the disease. The vast Indian population demands an inexpensive and competent sample preparation strategy for rapid detection of the disease facilitating early and efficient containment of the disease.
Methods: In this study, we have surveyed the spread of COVID-19 infection over Faridabad, Haryana, India for 6 months. We also devised a simple single-step method for total RNA extraction using a single tube and compared its efficacy with the commercially available RNA isolation kits.
Findings: Our findings suggest that determining Ct values for samples subjected to the One Pot RNA extraction method was as efficient as the commercially available kits but delivers a subtle advantage in a way, by minimizing the cost, labor and sample preparation time.
Conclusion: This novel crude RNA extraction method is stable and capable of operating in developing countries like India for low resource settings, without the use of expensive reagents and instruments. Additionally, this method can be further adapted to pooling samples strategies owing to its high sensitivity.
Introduction
Coronaviruses (CoVs) existed throughout human history, and are known to cause diseases ranging from mild respiratory illnesses to severe diseases like SARS (Severe Acute Respiratory Syndrome) and MERS (Middle East Respiratory Syndrome) in humans (Ye et al., 2020). The co-evolution with the host immune system bestows the CoVs capability of evading the protective responses resulting in transmission from one host to the other (Schultze and Aschenbrenner, 2021). In December 2019, a new member of the CoV family emerged in China; later termed as Severe Acute Respiratory Syndrome (SARS) associated with CoV-2 (Halaji et al., 2020). SARS-CoV-2 causes a severe form of respiratory illness called COVID-19 (World Health Organization, 1988). In contrast to SARS or Middle East Respiratory Syndrome (MERS), the SARS-CoV-2 has shown unpredicted spread over the world where 518 million individuals (approximately) contracted the infection with about 6 million fatalities (WHO Situation Report, 18th May 2022) (World Health Organization, 2022). The current COVID -19 pandemic due to widespread human-to-human transmission is now rising at an alarming rate (World Health Organization, 1988). The variable immune response in the host has displayed a spectrum of clinical patterns ranging from mild to severe cases (García, 2020). Besides symptomatic cases, the increase in the number of cases may be due to contact with the asymptomatic individual leading to rapid spread in the community (Oran and Topol, 2020; Antonucci et al., 2022). Therefore, to control the spreading of infections from one community to another and to provide accurate treatment to patients, correct diagnosis of the pathogen is the first line of requirement. Although many different diagnostic approaches have been introduced, there remains scope for rapid and cost-effective approaches (Kellner et al., 2019; Paz et al., 2020; Azhar et al., 2021). Its effective implementation would offer immense possibilities for rapid and widespread testing that has so far proven to be necessary for measuring the progression of the disease in multiple countries. The quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) has been widely used by several laboratories as a gold standard test for routine diagnosis of COVID-19 infection, developed by the USA-CDC (Carter et al., 2020). However, inadequate access to reagents and equipment due to high cost and the time-consuming sample pre-treatment has slowed the disease detection and impeded efforts to prevent viral spread (Vandenberg et al., 2021). With clinical samples, fast, inexpensive and sensitive RNA isolation methods may help in point-of-care (POC) pathogen detection. The objective of this study was to develop a simple, rapid and inexpensive one-tube testing method to diagnose SARS-CoV-2 which can be performed in low-resource settings.
Materials and methods
Study population
The study was carried out in ESIC Medical College and Hospital, Faridabad located in the Haryana district of India. The geographical map of the exact location is provided in (Figure 1). In this study, we surveyed the COVID-19 infections spread over Haryana for a period of 6 months (May 2020 to October 2020). This study was approved by the Indian government and the institutional review boards at participating institutions. Patients with severe acute respiratory illness (SARI) or influenza-like illness (ILI) i.e., clinically symptomatic population are recruited as per the guidelines of the Institutional Ethics Committee. The demographic characteristics like age, gender, travel history like migrant laborers, etc., place of residence, source of income, and co-morbidities if any was determined by taking a detailed history of the patient. The study excluded those individuals who refused to give written consent. Also, hospitalized patient or children with symptoms not related to COVID-19 infection was excluded from this study.
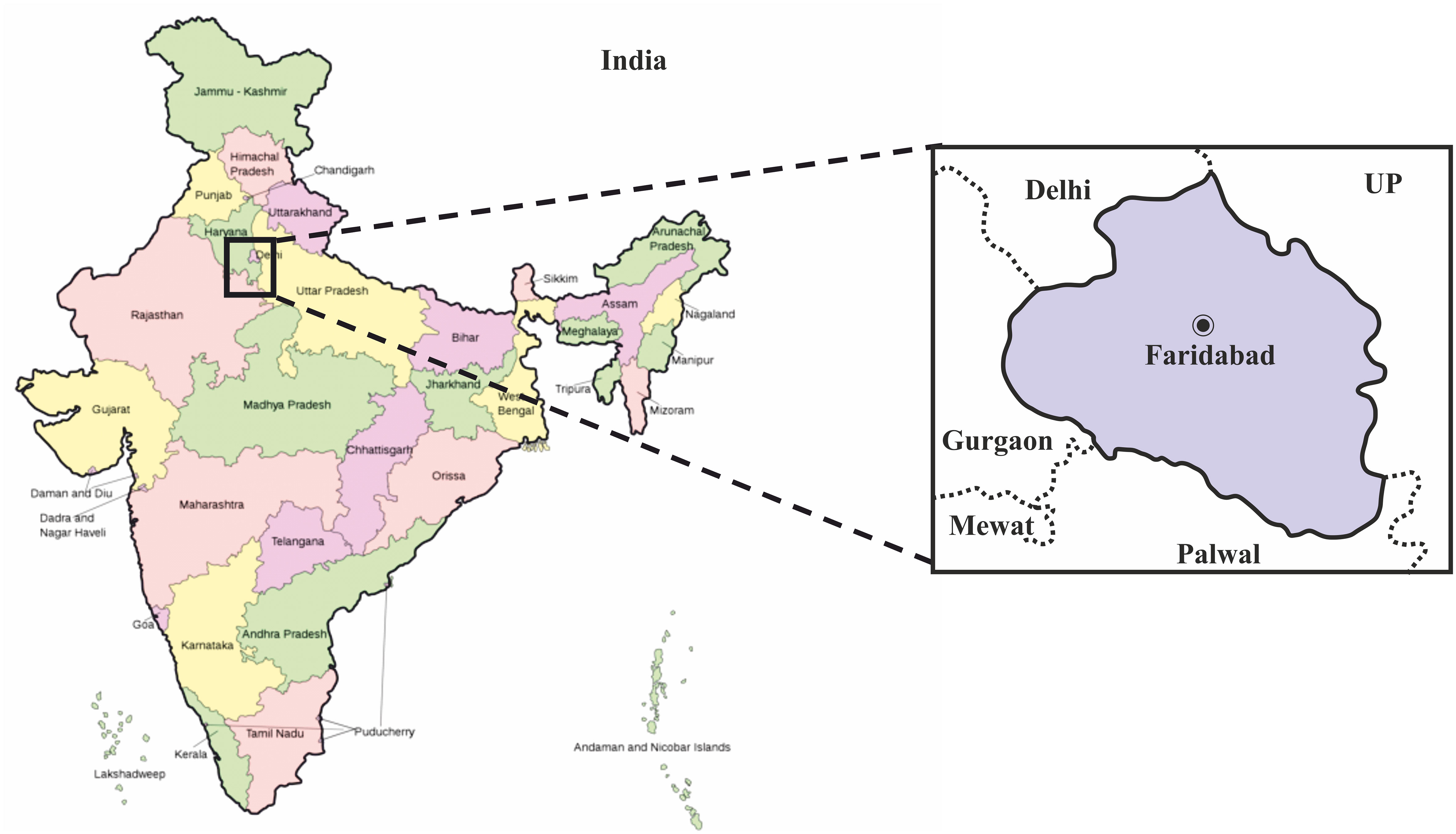
Figure 1 Geographical map of India showing the exact location of Faridabad district in the state of Haryana, where this study was carried out. The map of India was obtained from the online wikimedia commons website (URL -https://commons.wikimedia.org/wiki/File:India-map-en.svg).
Sample collection and transportation:
Before taking samples, informed consent was obtained from the patients or parents/guardians of the children. The samples were collected according to the Indian Council of Medicinal Research (ICMR) 2020 COVID-19 test protocol. Nasopharyngeal (NP) and oropharyngeal (OP) swab samples were collected from the consecutive population using sterile nylon flocked swabs (Himedia) and were immediately placed in HiViralTM Transport Medium (Himedia). The transport medium contains Hanks balanced salt solution, a protective protein, antibiotics to inhibit microbial growth and buffers to control the pH. The swab samples were further labeled and transported on the ice at the earliest to the laboratory for detailed investigations.
Study design:
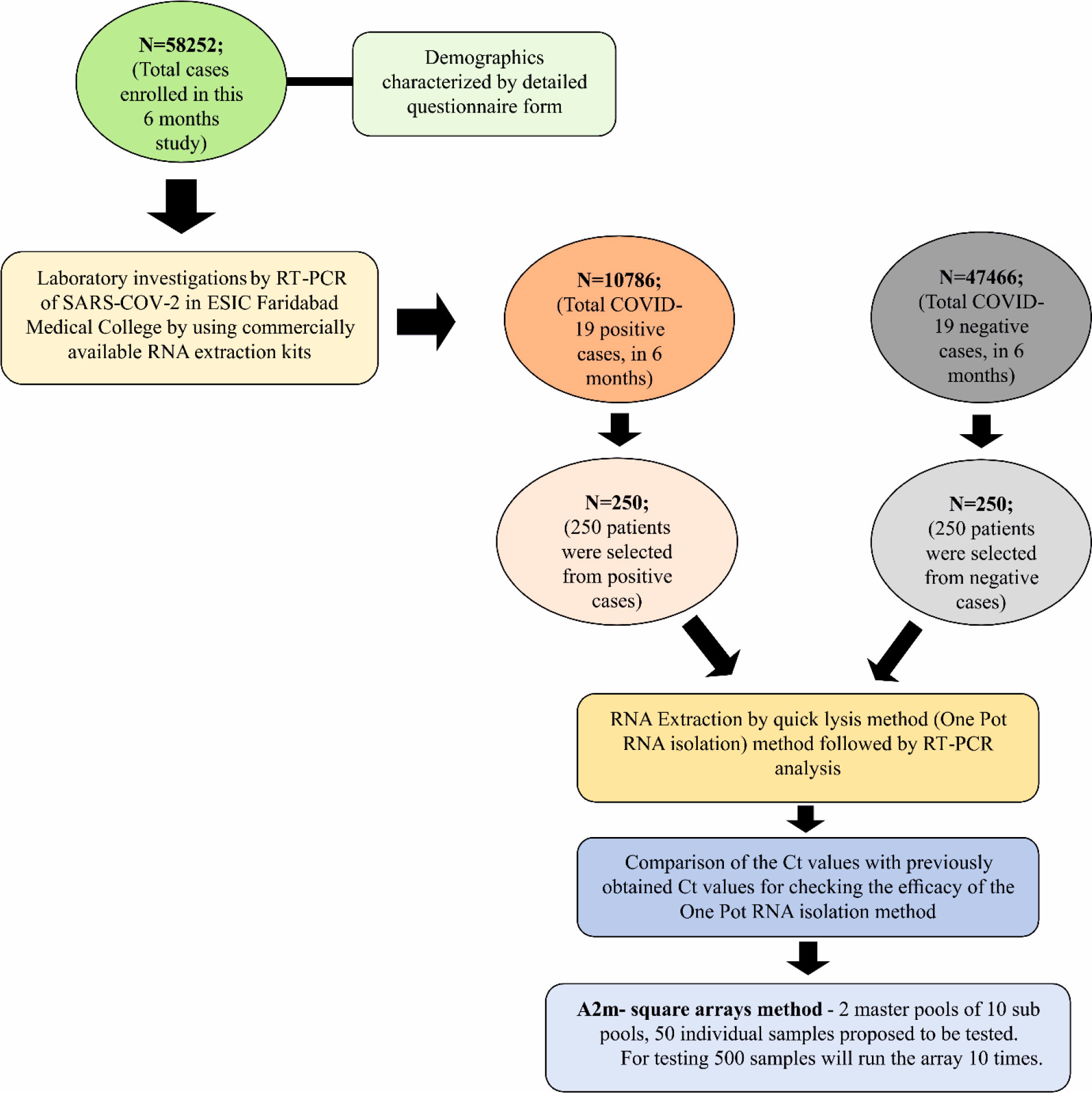
It is a retrospective study where out of a total of 58252 cases, only 500 cases were selected; 250 NP/OP aliquots from a previously tested COVID-19 positive group of individuals and 250 NP/OP aliquots from previously tested COVID-19 negative group of individuals were taken (Figure 2). The viral RNA was extracted from all these NP/OP swabs by employing our newly devised One-Pot RNA isolation method. The positive and negative results for SARS-COV-2 virus genes were determined by the Ct value obtained from qRT-PCR tests, using kits validated by ICMR.
RT-PCR for different genes of SARS-Cov-2
For the qRT-PCR analysis, the kits approved by ICMR (Indian Council of Medical Research) were employed for preparing samples as per the manufacturer’s instructions. The amplification of SARS-COV-2 genes was carried out by targeting the, (N) Nucleocapsid gene, (E) Envelope gene, (RdRp) RNA-dependent RNA polymerase gene (Cho et al., 2020; Udugama et al., 2020). The samples with a Cycle threshold (Ct) value ≤ 40 were interpreted as positive, while Ct value >40 or N.A. were interpreted as negative. The samples which were positive for N gene, E gene and RdRp gene after qRT-PCR analysis were considered as COVID-19 positive. Each time the qRT-PCR results were validated by positive amplification of the Internal Control (IC) gene provided in the kit. The PCR amplification was carried out following the kit’s instruction using the ABI7500 Real-time PCR system (Applied Biosystems USA)
RNA extraction by quick lysis method
The previous Acid-guanidinium-phenol-chloroform (AGPC) method of RNA extraction by Chomczynki and Sacchi was further modified for extracting viral RNA from the patient’s nasopharyngeal swab samples (Chomzynski and Sacchi, 1987). We devised a single tube quick-lysis method for a fast and efficient yield of crude viral RNA. This One-Pot RNA extraction method includes the use of the following materials and reagents: a) Quick Lysis solution (4M guanidinium thiocyanate, 0.5% (wt/vol) N-lauryl sarcosine, b) Chloroform, c) Glycogen solution (3.5mg/ml stock) d) 100% cold ethanol and e) 2 ml microcentifuge tubes.
Several microcentrifuge tubes (2ml) containing 500 µl Quick-Lysis solution and 300 µl chloroform were kept as ready-to-use stocks. While testing, 200 µl of NP/OP swab from VTM (Viral transport media) was directly added to the 800 µl of ready-to-use stock in the microcentrifuge tube followed by the addition of 5 µl of glycogen solution. The mixture was then vortexed and incubated at 65°C for 15 mins. Further, it was centrifuged at 24104g for 5 mins allowing phase separation (Figure 3). The upper yellow layer was discarded and 1 ml of ice-cold ethanol solution was added to the bottom layer followed by again centrifugation at 24104g for 5 mins. Finally, the supernatant was discarded followed by air drying of the tubes for 10 mins (to remove residual ethanol). The RNA pellet was then further resuspended in 20 µl of DEPC-treated water, quantified and subsequently used as a template in qRT-PCR analysis for detecting viral load in NP/OP swabs of affected patients. The Cycle threshold (Ct) values obtained after extracting RNA using this method were compared to the Ct values attained using a commercial RNA isolation kit (HiPurA™ Viral RNA Purification Kit from Himedia) simultaneously at the same time since RNA extraction can be affected by the storage time of the samples. We performed non-parametric Mann -Whitney tests to determine whether differences between the Ct values of the sample prepared using a commercial RNA extraction kit and the new One-pot RNA isolation method were significant for the three genes tested. Significance level was based on 2-tailed α (significance level) = 0.05. We also performed Two one sided t-test (TOST) for assessing the true equivalence between the Ct values obtained.
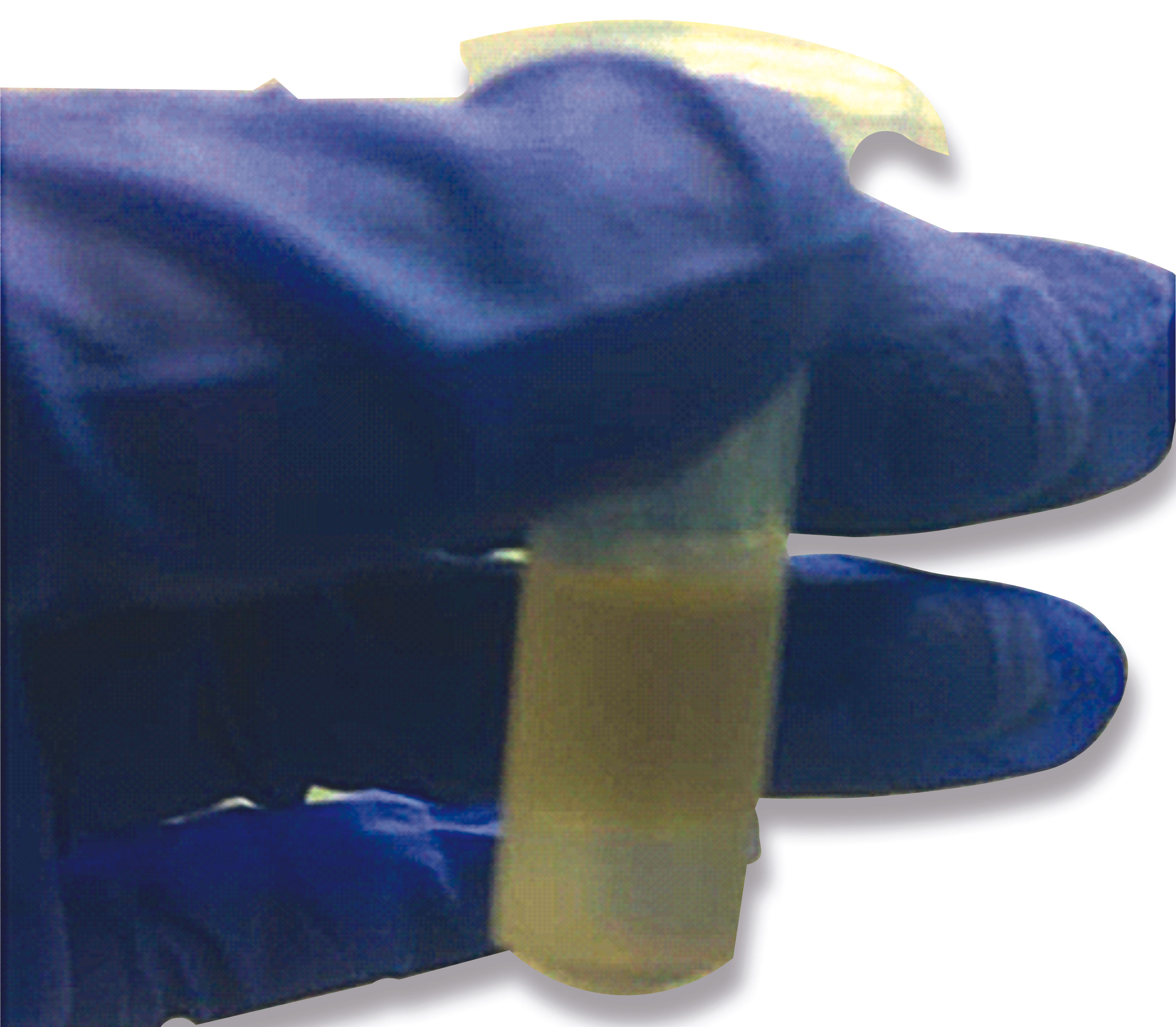
Figure 3 A microfuge tube showing two-phase separation after addition of Quick lysis solution, chloroform, and glycogen to VTM sample containing the NP/OP swab followed by brief heating at 65 °C and subsequent centrifugation at high speed. The lower phase contains the viral RNA precipitated by the glycogen that is being used in the Quick Lysis solution.
Pooling testing strategies:
The Ministry of Health and Family Welfare, Govt. of India has made provision for a one-time pool sample of 25 to be tested for Covid-19 district-level surveillance. Therefore, the modified A2m- square arrays method with two different master pools was applied in this study which generally provides better efficiency and positive predictive value (PPV) (Taylor et al., 2010). This pooling strategy is a two-dimensional array (“A2m”) with master pool testing. The first test was carried out for the master pool; if the master pool tests positive, then pools consisting of all samples in each row and column of the array are tested at the same time. Retesting is done for all the samples at points of intersection between positive rows and positive columns. In the situation of a positive row but no positive columns, all samples in the positive row are tested. As testing row and column pools occur in a single step of the testing process, thus A2m is a three-stage pooling algorithm. Only A2m algorithms with the similar number of rows and columns will be taken into account. In testing this algorithm, the number of samples included in each pool depends on the expected prevalence rate of the disease under study and the characteristics of the diagnostic test. Further, it also depends on dilution effects in the master pool, the length of the sensitivity depends on both the sensitivity/lower limit of detection of the assay and viral load in acute infection (Westreich et al., 2008). The optimal pool size required for testing was calculated with an available online calculator (http://www.bios.unc.edu/_mhudgens/optimal.pooling.b.htm). Therefore, according to the pooling size strategy mentioned above, the master pool size was 25. Two master pools of 10 sub-pools and 50 individual samples were tested in this algorithm. So, for testing 500 samples we have to run the array 10 times. For quality control, 5% of negative samples were randomly tested by standard qRT-PCR to check the false-negative rate.
Results
COVID-19 prevalence over Faridabad, Haryana, India
For the period of 6 months, a total of n=58252 samples were collected from different individuals who came to visit ESIC Medical college and hospital, Faridabad, Haryana for routine clinical checkups and testing. The RT-PCR testing of samples collected from their NP/OP swabs indicated 10786 individuals were positive for COVID-19 while the rest of 47466 were tested negative for COVID-19 indicating a prevalence rate of 18.52%. Out of the total n=58252 samples: 35184 samples were from males; 22805 samples were collected from females while 262 samples were from neonates. A total of 7086 males were found to be positive for COVID-19 while 3700 females were positive for COVID-19 (Table 1). However, all the neonates tested negative for COVID-19. The percentage of occurrence of COVID-19 was calculated to be 20.1% and 16.2% in the case of males and females respectively for the period of 6 months of study. The infected percentage obtained (Figure 4A) for the different age groups are as follows: 0-17 years – 9.7%; 18-35 years- 16.2%; 36-53 years- 23.8%; 54-71 years- 25.4% and 72-89 years- 28.8% (Table 1). Thus, our data indicate that SARS-CoV-2 is highly affecting individuals belonging to higher age groups. Among those who were positive for SARS-COV-2, the most predominant comorbid conditions were diabetes (37.2%), hypertension (30%), Respiratory disorder (24%), renal disorder (14.7%) and obesity (11.2%) (Table 1). Amongst the COVID-19 affected patients, most of the patients had Ct values within 25.01 to 35.00 for the N gene (Figure 4B) while, Ct values within 20.01 to 35.00 were observed for both the RdRp and E gene. Thus, from our data, we can speculate that the relative abundance of the viral RdRp and E gene is much higher compared to the N gene in the affected patient samples.

Table 1 Table showing the percentage of COVID-19 infected patients and the total number of COVID-19 infected patients (n) in each of the different groups.
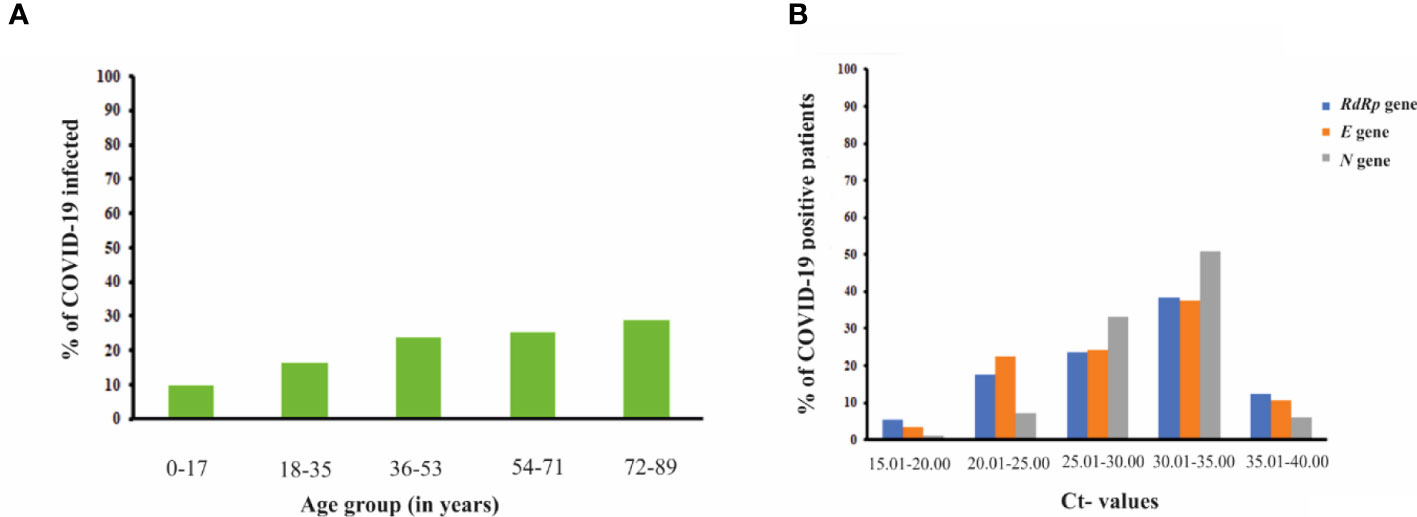
Figure 4 (A) Bar graph representation showing the percentage of COVID-19 infected patients from different age groups. The percentage of COVID-19 infected patients raised with the increase in the age group of the individuals (B) Graph showing correlation of Ct-values of different viral genes (RdRp gene, E gene and N gene) with the percentage of COVID-19 positive patients.
One-pot RNA isolation method was effective and sensitive in extracting viral RNA from NP/OP swabs
This One-pot RNA isolation method employed simple denaturing and a precipitation solution, with a centrifugation method to extract the crude RNA which can be directly used for RT-PCR testing. The Quick lysis solution was able to lyse the SARS-COV-2 Virus coating within a very short time. Also, heating at a higher temperature reduces the risk of handling positive samples without compromising the integrity of the viral RNA. Similar to other commercially available RNA isolation kits, this new RNA isolation method shows the Ct-value and Internal Control (IC) perfectly. The Ct values obtained after RT-PCR analysis using this new method of RNA isolation were almost similar to those obtained after RT-PCR analysis utilizing the commercial method of RNA isolation (Figure 5A). Previously tested samples negative for COVID-19 were found to be negative (data not shown) and COVID-19 positive samples were positive with this new RNA isolation method (Figure 5A). Statistical analysis TOST proved that there were no significant differences between the Ct values obtained after PCR amplification employing both method of RNA extraction (Supplementary Information: Figures S1-S3). The sensitivity and specificity of this One Pot method were calculated using a previously described method (Baratloo et al., 2015).

Figure 5 (A) Box-plot showing no significant change in the Ct values of 250 COVID-19 infected patients (for RdRp gene, E gene and N gene) obtained from using the One-pot (OP) RNA isolation method when compared to Ct values acquired using commercially available RNA-isolation kits. (n.s.: not significant; Mann-Whitney test and TOST). (B) Graph showing similar sensitivity in viral gene detection (N gene, Rdrp gene and E gene) when RNA extraction by One-Pot (OP) method was compared to commercial RNA kit method (Statistical analysis: Mann-Whitney test).
The One Pot method exhibited 100% sensitivity (true positives divided by the true positive plus false negatives) and 98.8% specificity (true negatives divided by the true negative plus false positive). We have also performed the pooling strategy where samples were pooled and then tested. The positive result obtained from the highly diluted pooled samples marked the uncompromised sensitivity of this newly devised RNA-isolation method. Viral RNA was extracted from a positive patient using both the commercial kit and One Pot method and was further diluted in the following proportion gradient: 1:1, 1:5, 1:10, 1:15, 1:20, and 1:25. The viral load was detected for each of the dilutions thrice. At higher dilution, the Ct values for N gene, Rdrp gene, and E gene were found to be almost similar in the case of both RNA-extraction methods (Figure 5B). Mann-Whitney t-test confirmed that there exists no significant difference between the Ct values obtained using both the RNA extraction methods. Viral RNA can still be detected from 25 times diluted RNA samples suggesting the at par sensitivity of the RNA extraction by One Pot method to that of the commercial kit method.
Discussion
Globally, as on, 11 January 2022, there have been 308,458,509 confirmed cases of COVID-19, counting 5,492,595 deaths, reported to WHO (World Health Organization) (World Health Organization, 2022). However, in India, from 3rd January 2020 to 11 January 2022, there have been 35,875,790 confirmed cases of COVID-19 with 484,213 deaths, according to WHO (World Health Organization, 2022). From these 6 months studies, the prevalence rate of Covid-19 infection was found to be 18.52% in 2020 in Faridabad, Haryana India. Like previous data showing men are more susceptible to infection than women, our data also revealed that males are more predisposed to COVID-19 infection than females (Bwire, 2020). However, it was also reported by Jian-Min Jin et al. that men and women have the same Covid-19 prevalence rate but in the case of men, the risk of complications and death is much more, independent of age (Jin et al., 2020). Although previous studies pointed to the fact that COVID-19 prevalence increases in adolescents and youth compared to older adults, our data revealed that the rate of infection increased with the increasing age group of the individual suggesting that older people are more prone to COVID-19 infection than the younger ones (Rumain et al., 2021). The reason for this observation can be attributed to age-related gradual weakening of immune system function (immunosenescence) or maybe due to hyperactive yet ineffective immune system (inflammaging) in older people leading to systemic inflammation (Franceschi et al., 2006; Aw et al., 2007). This study used data from relatively early in the pandemic (pre-vaccines), and it’s notable that children were much less likely to be infected pre-Delta strain.
To address various issues, several POCs have been established to enable the COVID-19 diagnosis outside the centralized testing facilities to hasten the clinical decision-making within a short window of time. Like, CRISPR-Cas9 enzymology-based diagnostic platform-specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) provides ultra-high specific amplification of nucleic acid sequences from clinical samples with the use of an isothermal cycler (Kellner et al., 2019). Also, another CRISPR-based, FnCas9, Editor Linked Uniform Detection Assay (FELUDA) has been designed in India for rapid and accurate detection of SARS-CoV-2 nucleic acids at a very low cost (Azhar et al., 2021). On the other hand, the use of a Rapid Antigen Test (RAT) kit provides rapid detection of the infection without the need for going to the lab for testing (Khandker et al., 2021). Nevertheless, the test possesses a lower accuracy for patients having no symptoms and can also give false negative results (Khandker et al., 2021). In addition, a sensitive RNA-isolation protocol exploiting Trizol to purify viral RNA from various types of clinical specimens with nominal BSL-1 precautions has been formulated recently (Paz et al., 2020). However, this RNA-isolation method included many transfer steps and was time-consuming.
Although the recent advent of the cartridge-based Xpert Xpress SARS-CoV-2 test (Cepheid) provides a runtime of only 45 min, this may not be a perfect choice, when testing thousands of patient samples per day (Goldenberger et al., 2020). Also, the costs per sample of a single-use cartridge which can be only used in the GeneXpert machine (Cepheid) are significantly much higher compared to our One Pot method. The GeneXpert machine (Cepheid) is very sophisticated and expensive and is not possible to use in small setup diagnostics in India. On the other hand, our One Pot method is user-friendly and can be used in every PCR machine in any setup with minimal cost.
To facilitate rapid, inexpensive and efficient testing of COVID-19 infected samples, we have tried to develop one tube RNA-isolation method which is capable of detecting SARS-CoV-2 RNA in the NP/OP swabs. The efficacy of this new method was tested by comparing the Ct values of the clinical samples with the Ct values obtained by employing commercially available RNA-isolation kits. The use of this new method is highly recommended when dealing with a huge number of clinical samples in low-resource settings like small mohalla clinics, primary health care centers, etc. This One-pot RNA-isolation method is highly cost-effective with a cost of approximately Rs. 35/sample (in Indian currency) when compared to commercially available RNA isolation kits with a cost of approximately Rs 200/sample. For isolating RNA, the time consumed by other commercial kits with the column method is more than 1 hour, but with this new method, the time required is approximately 30 mins. Thus, the use of this One-pot RNA isolation kit would highly benefit mankind by saving cost, time and labor thereby facilitating rapid and early detection of the disease. This study also implies the application of a resource-conserving testing algorithm employing sample pooling for clinical health diagnosis.
Conclusion
This study from India characterizes the disease pattern and the possible outcomes associated with it. In this study, we have tried to investigate the spread of COVID-19 infection over the region of Faridabad, Haryana (northern India) over 6 months (May 2020-October 2020) implementing our newly devised crude RNA extraction method. The rate of infection was found to be higher in the case of individuals belonging to higher age groups. Also, we have successfully developed an RNA isolation method using a single tube which can be employed for testing a large number of samples in a low resource setting, without compromising the qRT-PCR results. This quick, sensitive and inexpensive RNA-isolation method can be a better-suited alternative to most of the expensive, time-consuming commercially available RNA isolation kits.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving human participants were reviewed and approved by Ethical committee of ESIC medical college and hospital, Faridabad. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
Conceptualization, SJ. Formal analysis, SJ and AB. Funding acquisition, AP. Investigation, SJ. Methodology, SJ, AB and AP. Project administration, SJ and AP. Supervision, SJ and AP. Writing – original draft, SJ and AB. Writing – review & editing, SJ, AB and AP. All authors contributed to the article and approved the submitted version.
Funding
This work is funded by the intramural grant of ESIC Medical College & Hospital, Faridabad, Haryana, India.
Acknowledgments
The authors would like to acknowledge Dr. (Prof.) Asim Kumar Das, Dean of ESIC Hospital and Medical college, Faridabad, Haryana, India for his immense support and providing the intramural funds and laboratory space to conduct this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.962057/full#supplementary-material
Supplemental Information | The statistical analysis of the Ct-value comparison of N gene, E gene and Rdrp gene between the One-Pot RNA extraction and commercial kit method is provided as charts and equivalent test plot.
Abbreviations
SARS-CoV-2, Severe Acute Respiratory Syndrome- Coronavirus-2; CoVs, Coronaviruses; COVID-19, Corona Virus Disease-19; MERS, Middle East Respiratory Syndrome; VTM, Viral Transport Medium; POC, Point of Care; DEPC, Diethyl Pyrocarbonate; NP/OP, Nasopharyngeal/Oropharyngeal; WHO, World Health Organization; CRISPR, Clustered Regularly Interspaced Short Palindromic Repeats; ICMR, Indian Council of Medical Research; SARI, Severe Acute Respiratory Illness; ILI, Influenza Like Illness; Ct-value, Cycle threshold value; BSL-1, Biosafety Level-1; USA-CDC, United States of America- Centers for Disease Control and Prevention; qRT-PCR, quantitative Reverse Transcription-Polymerase Chain Reaction; N gene, Nucleocapsid gene; RdRp gene, RNA dependent RNA polymerase gene; E gene, viral Envelope gene; TOST, Two one-sided t-test.
References
Antonucci, F., Fiore, J. R., de Feo, L., Granato, T., di Stefano, M., Faleo, G., et al. (2022). Increased SARS-CoV-2 seroprevalence in healthy blood donors after the second pandemic wave in south-Eastern Italy: evidence for asymptomatic young donors as potential virus spreaders. Infect. Dis. 54, 241–246. doi: 10.1080/23744235.2021.2003856
Aw, D., Silva, A. B., Palmer, D. B. (2007). Immunosenescence: emerging challenges for an ageing population. Immunology 120, 435–446. doi: 10.1111/j.1365-2567.2007.02555.x
Azhar, M., Phutela, R., Kumar, M., AH, A., Rauthan, R., Gulati, S., et al. (2021). Rapid and accurate nucleobase detection using FnCas9 and its application in COVID-19 diagnosis. Biosens Bioelectron 183, 113207. doi: 10.1016/j.bios.2021.113207
Baratloo, A., Hosseini, M., Negida, A. (2015). El ashal g. part 1: Simple definition and calculation of accuracy, sensitivity and specificity. Emerg. (Tehran) 3, 48–49.
Bwire, G. M. (2020). Coronavirus: Why men are more vulnerable to covid-19 than women? SN Compr. Clin. Med. 2, 874–876. doi: 10.1007/s42399-020-00341-w
Carter, L. J., Garner, L. V., Smoot, J. W., Li, Y., Zhou, Q., Saveson, C. J., et al. (2020). Assay techniques and test development for COVID-19 diagnosis. ACS Cent Sci. 6, 591–605. doi: 10.1021/acscentsci.0c00501
Cho, H., Jung, Y. H., Cho, H. B., Kim, H., Kim, K. (2020). Positive control synthesis method for COVID-19 diagnosis by one-step real-time RT-PCR. Clinica Chimica Acta 511, 149–153. doi: 10.1016/j.cca.2020.10.001
Chomzynski, P., Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate–Phenol–Chloroform extraction. Anal. Biochem. 162, 156–159. doi: 10.1006/abio.1987.9999
Franceschi, C., Bonafè, M., Valensin, S., Olivieri, F., De Luca, M., Ottaviani, E., et al. (2006). Inflamm-aging: An evolutionary perspective on immunosenescence. Ann. N Y Acad. Sci. 908, 244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x
García, L. F. (2020). Immune response, inflammation, and the clinical spectrum of COVID-19. Front. Immunol. 11. doi: 10.3389/fimmu.2020.01441
Goldenberger, D., Leuzinger, K., Sogaard, K. K., Gosert, R., Roloff, T., Naegele, K., et al. (2020). Brief validation of the novel GeneXpert xpress SARS-CoV-2 PCR assay. J. Virol. Methods 284, 113925. doi: 10.1016/j.jviromet.2020.113925
Halaji, M., Farahani, A., Ranjbar, R., Heiat, M., Dehkordi, F. S. (2020). Emerging coronaviruses: first SARS, second MERS and third SARS-CoV-2: epidemiological updates of COVID-19. Infez Med. 28, 6–17.
Jin, J.-M., Bai, P., He, W., Wu, F., Liu, X.-F., Han, D.-M., et al. (2020). Gender differences in patients with COVID-19: Focus on severity and mortality. Front. Public Health 8. doi: 10.3389/fpubh.2020.00152
Kellner, M. J., Koob, J. G., Gootenberg, J. S., Abudayyeh, O. O., Zhang, F. (2019). SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 14, 2986–3012. doi: 10.1038/s41596-019-0210-2
Khandker, S. S., Nik Hashim, N. H. H., Deris, Z. Z., Shueb, R. H., Islam, M. A. (2021). Diagnostic accuracy of rapid antigen test kits for detecting SARS-CoV-2: A systematic review and meta-analysis of 17,171 suspected COVID-19 patients. J. Clin. Med. 10, 3493. doi: 10.3390/jcm10163493
Oran, D. P., Topol, E. J. (2020). Prevalence of asymptomatic SARS-CoV-2 infection. Ann. Intern. Med. 173, 362–367. doi: 10.7326/M20-3012
Paz, S., Mauer, C., Ritchie, A., Robishaw, J. D., Caputi, M. (2020). A simplified SARS-CoV-2 detection protocol for research laboratories. PloS One 15, e0244271. doi: 10.1371/journal.pone.0244271
Rumain, B., Schneiderman, M., Geliebter, A. (2021). Prevalence of COVID-19 in adolescents and youth compared with older adults in states experiencing surges. PloS One 16, e0242587. doi: 10.1371/journal.pone.0242587
Schultze, J. L., Aschenbrenner, A. C. (2021). COVID-19 and the human innate immune system. Cell 184, 1671–1692. doi: 10.1016/j.cell.2021.02.029
Taylor, S. M., Juliano, J. J., Trottman, P. A., Griffin, J. B., Landis, S. H., Kitsa, P., et al. (2010). High-throughput pooling and real-time PCR-based strategy for malaria detection. J. Clin. Microbiol. 48, 512–519. doi: 10.1128/JCM.01800-09
Udugama, B., Kadhiresan, P., Kozlowski, H. N., Malekjahani, A., Osborne, M., Li, V. Y. C., et al. (2020). Diagnosing COVID-19: The disease and tools for detection. ACS Nano 14, 3822–3835. doi: 10.1021/acsnano.0c02624
Vandenberg, O., Martiny, D., Rochas, O., van Belkum, A., Kozlakidis, Z. (2021). Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 19, 171–183. doi: 10.1038/s41579-020-00461-z
Westreich, D. J., Hudgens, M. G., Fiscus, S. A., Pilcher, C. D. (2008). Optimizing screening for acute human immunodeficiency virus infection with pooled nucleic acid amplification tests. J. Clin. Microbiol. 46, 1785–1792. doi: 10.1128/JCM.00787-07
World Health Organization (1988). Office of library and health literature services (Geneva Switzerland: World Health Organization).
Keywords: SARS-CoV-2, COVID-19, one pot method, RNA isolation, ct value
Citation: Jain S, Bhowmick A and Pandey AK (2022) A rapid One-Pot RNA isolation method for simplified clinical detection of SARS-COV-2 infection in India. Front. Cell. Infect. Microbiol. 12:962057. doi: 10.3389/fcimb.2022.962057
Received: 19 July 2022; Accepted: 16 September 2022;
Published: 04 October 2022.
Edited by:
Léanie Kleynhans, Stellenbosch University, South AfricaReviewed by:
Mariantonietta Di Stefano, University of Foggia, ItalyJeffrey A. Lewis, University of Arkansas, United States
Copyright © 2022 Jain, Bhowmick and Pandey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anil Kumar Pandey, ZHJwYW5kZXlha0B5YWhvby5jby5pbg==; Sonia Jain, c3d0c29uaWFqYWluQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Sonia Jain
Sonia Jain Arghya Bhowmick
Arghya Bhowmick Anil Kumar Pandey1*
Anil Kumar Pandey1*