
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 12 October 2022
Sec. Clinical Microbiology
Volume 12 - 2022 | https://doi.org/10.3389/fcimb.2022.953443
This article is part of the Research Topic Pathogenesis, Diagnostics, Treatments of Mycobacterium tuberculosis and Its co-Infection with HIV or SARS-CoV-2 View all 10 articles
 Jitendra Singh1,2
Jitendra Singh1,2 Niti Singh3
Niti Singh3 Gayatri Suresh3
Gayatri Suresh3 Rahul Srivastava3
Rahul Srivastava3 Upasna Aggarwal3
Upasna Aggarwal3 Digamber Behera3,4
Digamber Behera3,4 Murali Munisamy2
Murali Munisamy2 Anvita Gupta Malhotra5
Anvita Gupta Malhotra5 Sarman Singh1,5,6*
Sarman Singh1,5,6*Setting: Tuberculosis Research Laboratory, Division of Clinical Microbiology and Molecular Medicine, Department of Laboratory Medicine, All India Institute of Medical Sciences, and the National Institute of Tuberculosis and Respiratory Diseases (NITRD), both situated in New Delhi.
Objectives: We aimed to identify the distribution of various genotypes of M. tuberculosis among HIV-positive and HIV-negative patients suspected of having Tuberculosis, seen at the National Institute of Tuberculosis and Respiratory Diseases, New Delhi, which is a tertiary care dedicated TB hospital.
Patients and methods: Genotyping by Spoligotyping and 24 loci MIRU-VNTR was performed and analyzed using SITVITWEB and MIRU-VNTRplus. Drug susceptibility patterns were also analyzed.
Results: A total of 503 subjects who were PTB/EPTB suspected were recruited and 287 were culture positive. Among them, 276 had growth of Mycobacterium tuberculosis (MTB) and in 11 patients non-tuberculous mycobacteria (NTM) were grown. The isolation rate of NTM was predominantly from HIV positive [10 of 130 (7.6%)] patients. Of the total isolates of MTB, 156 (56.5%) were from HIV negative patients and 120 (43.5%) were from HIV positive patients. All 276 M. tuberculosis isolates were genotyped and tested for drug susceptibility patterns. The CAS genotype was most predominant [153 (55.4%)], followed by Beijing lineage [44 (15.9%)], East African India [25 (9.1%)] and others [54 (19.6%)]. Beijing genotype was significantly more common in HIV positive patients (22.5%) than in HIV negative patients (10.9%). In MIRU-VNTR analysis, clustering was found to be more frequent in CAS strains irrespective of HIV status. In the HIV positive group, spoligotyping could differentiate various genotypes in 90% of isolates and MIRU-VNTR analysis in 84.2% of isolates. The clustering of various MTB strains was more associated with drug resistance.
Conclusion: The Beijing lineage was predominant in HIV-TB coinfected cases, even though the Central Asian Strain (CAS) was overall more predominant in the region.
Tuberculosis (TB) and Human immunodeficiency virus (HIV) infections are lethal chronic infections and are among the leading causes of mortality globally. In 2019, an estimated 1.2 million TB deaths occurred among HIV-negative people and 208,000 deaths amongst HIV-positive people (Global tuberculosis report, 2020). As per a WHO 2020 report, 69% of notified TB patients had documented HIV test results, as compared to 64% in 2018. The burden of HIV-associated TB is the highest in the WHO African region, where 86% of TB patients had a documented HIV test result. Globally, 88% of the TB-HIV co-infected patients were on antiretroviral therapy (ART), while this proportion was 95% in India (WHO, 2016).
Although both TB and HIV are deadly by themselves, they have an additive pathogenic influence when they come together, thereby making the HIV-TB co-infection a “double trouble” for society (Munawwar and Singh, 2012). Coordinated pathogenesis of HIV and Mycobacterium tuberculosis poses a major medical challenge as HIV provides an enormous opportunity for M. tuberculosis to multiply rapidly within the intracellular setting, and M. tuberculosis provides a favorable environment for HIV to replicate unhindered via TB-associated protein malnutrition and increased depletion or non-activation of T lymphocytes (Friedland et al., 2007; Munawwar and Singh, 2012).
Molecular genotyping reveals the different levels of genetic polymorphisms of M. tuberculosis and is applicable in monitoring disease transmission, detecting outbreaks, and confirming laboratory cross-contaminations and clonal spread of dominant clones. Under certain circumstances, it is used to distinguish between members of the M. tuberculosis complex that have critical clinical importance (Allix-Béguec et al., 2010). Due to its clonal structure (Sreevatsan et al., 1997), the comparative genotype analysis of MTCs from different human populations can give unique insights into the dissemination dynamics and evolutionary genetics of this pathogen; therefore, spoligotyping has been used to understand the emerging problem of multidrug-resistant (MDR) TB and the virulence of certain epidemic strains of M. tuberculosis (for example; the Beijing strain), as well as to better understand the epidemiology of TB and TB-HIV co-infection (Cadmus et al., 2011).
Spoligotyping is based on polymorphism in the direct repeat (DR) locus and is used as a primary genotyping method in combination with molecular typing methods based on variable number tandem repeats (VNTRs) of the 24 loci DNA elements known as mycobacterial interspersed repetitive units (MIRU) (Supply et al., 2006). MIRU-VNTR in combination with spoligotyping has become a striking alternative to the traditional IS6110-RFLP fingerprinting method, with maximum discriminatory power (Pitondo-Silva et al., 2013).
Information on the genotypes of M. tuberculosis isolated from India, in comparison with those obtained globally, are important for understanding the global spread and phylogeographical specificity of the predominant circulating clones of tubercle bacilli. In addition, M. tuberculosis molecular genotyping data obtained from India so far have not addressed the issue of HIV-TB co-infected patients, a subpopulation involved in the spread of the disease in Asia in general and India in particular (Gutierrez et al., 2006; Sharma et al., 2008; Thomas et al., 2011; Vadwai et al., 2012; Singh et al., 2015). We have previously analyzed the molecular genotypes of M. tuberculosis among extrapulmonary and pulmonary tuberculosis patients irrespective of their HIV status (Sankar et al., 2013; Singh et al., 2015). Here, we aimed to investigate the spoligotyping and 24 loci MIRU-VNTR based population structure of M. tuberculosis clinical isolates from HIV negative and MTB positive patients and HIV-TB co-infected patients, and to compare the patterns obtained with those available in the SIT-VIT web database of the Pasteur Institute of Guadeloupe (Demay et al., 2012).
This study was performed at the Tuberculosis Research Laboratory, Division of Clinical Microbiology and Molecular Medicines, All India Institute of Medical Sciences (AIIMS), New Delhi. The total duration of the study was three years from April 2012 to March 2015. All the patients were recruited at the National Institute of Tuberculosis and Respiratory Diseases (NITRD), New Delhi. As a standard protocol in India, all TB suspected cases are tested for HIV as per the standard guidelines issued by the National AIDS Control Organization and WHO. The inclusion criteria were all HIV-positive patients suspected of concomitant tuberculosis infection and clinically suspected patients with pulmonary tuberculosis (PTB) and/or extra-pulmonary tuberculosis (EPTB). Informed consent was obtained from all the recruited cases. However, we excluded patients taking anti-tuberculous treatment (ATT) for more than two weeks at the time of recruitment, patients under immune-suppressive treatment, and pregnant female patients. This study was approved by the Institutional Ethics Committees of the All India Institute of Medical Sciences, and the National Institute of Tuberculosis and Respiratory Diseases, New Delhi.
The clinical samples were transported on a daily basis to the Clinical Microbiology Laboratory at the All India Institute of Medical Sciences, New Delhi, where these were inoculated into BACTEC MGIT™ 960 as described earlier (Sankar et al., 2013). Flashed positive cultures were confirmed as M. tuberculosis by species specific multiplex PCR (m-PCR), which amplifies hsp65, esat6, and its regions of the mycobacterial genome (Sankar et al., 2013).
DNA from the cultures was isolated as previously described (Sankar et al., 2013). After checking the quality of the DNA, spoligotyping was performed using a commercially available kit (Mapmygenome India Limited) as per the manufacturer’s instructions (Thomas et al., 2011; Vadwai et al., 2012; Singh et al., 2015). Twenty-four loci MIRU-VNTR was performed by PCR amplification of individual loci using specific primers, as described previously (Supply et al., 2006).
The susceptibility pattern to Isoniazid (INH), Rifampicin (RIF), Streptomycin (SM), and Ethambutol (EMB) was determined using the BACTEC MGIT™ 960 (Becton- Dickinson, Sparks, USA) following the manufacturer’s instructions, as described earlier (Singh et al., 2015).
Genetic data analysis was performed using standard methods as described earlier (Hanekom et al., 2008; Narayanan et al., 2008), wherein spoligotypes were identified and analyzed as character types. The obtained spoligotyping patterns were compared with those available in the SITVIT_WEB database (http://www.pasteur-guadeloupe.fr:8081/SITVIT2). The Hunter-Gaston discriminatory index (HGDI) was used as a numerical index for MIRU-VNTR discriminatory power (Hunter and Gaston, 1988). The allelic diversity of the loci was identified as highly discriminant if the HGI was >0.6, moderately discriminant if the HGI was between 0.3 and 0.6, and poorly discriminant if the HGI was less than 0.3.
Differences among the lineages of the isolates and drug susceptibility patterns were analyzed by the chi-squared test and Fisher’s exact test as appropriate using STATA 9.0. The adjusted odds ratio (OR) and 95 percent confidence interval (CI) were calculated. Statistical significance was defined as a p value of 0.05 or less.
A total of 503 subjects who were suspected to have PTB and/or EPTB, were recruited from the Antiretroviral Therapy (ART) center and OPDs and wards of NITRD, New Delhi. Most of the patients were residents of Delhi (n=215, 74.9%), and the rest were from neighboring states in India, including Haryana (29, 10.1%), Uttar Pradesh (27, 9.4%), and Bihar (16, 5.6%), as these patients had visited the NITRD, New Delhi for their diagnosis and treatment (Table 1).
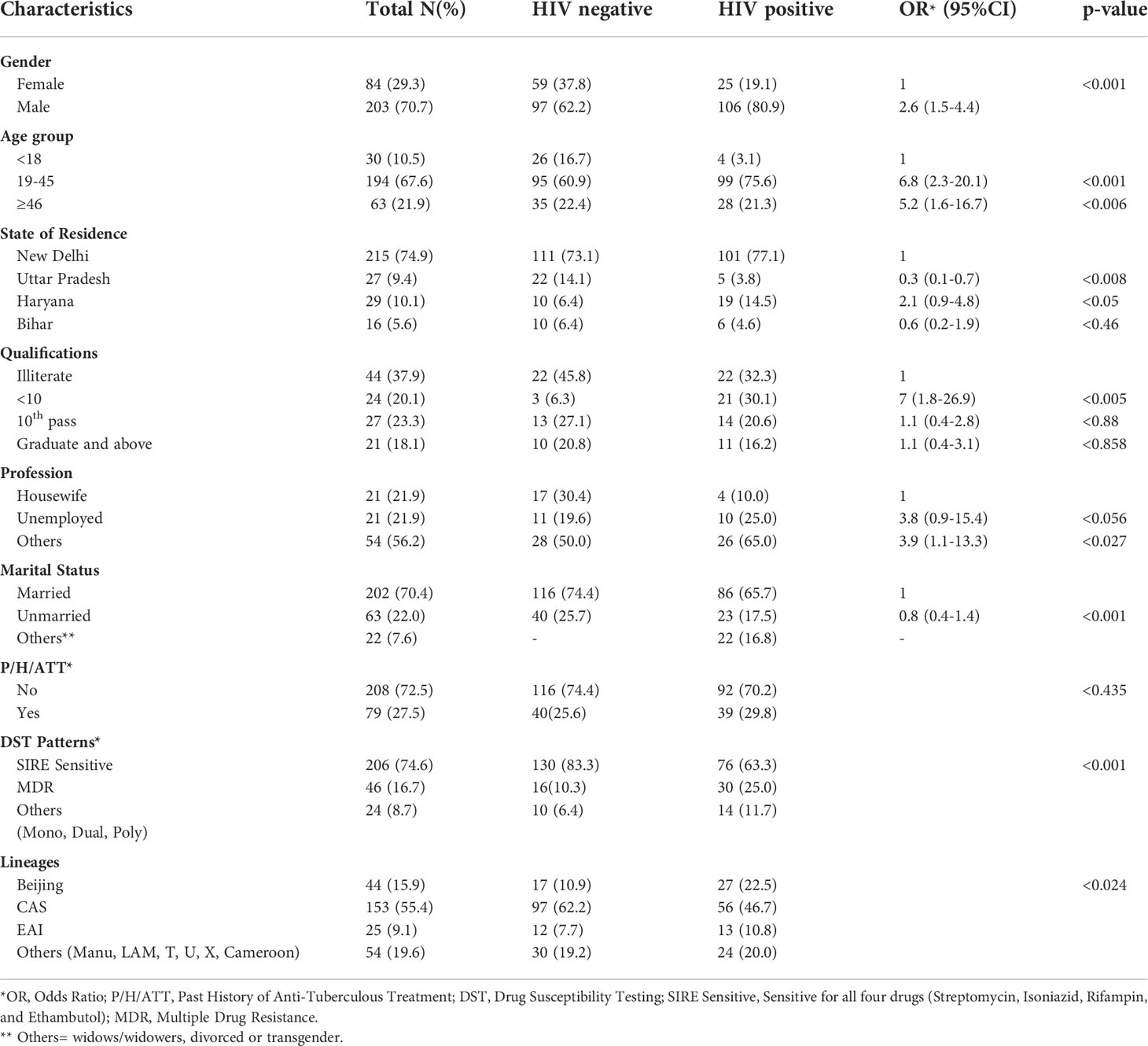
Table 1 Demographic details of the patients, drug resistance patterns, and phylogenetic lineages of the MTB isolates.
These 503 patients were divided into two groups based on their HIV status. In the HIV/AIDS positive group, there were 249 of 503 (49.5%) patients who had HIV-TB coinfection, either PTB and/or EPTB. Their mean age was 35.6 ± 11.8 years. Of these, 130 (52.2%) subjects were mycobacterial culture positive and their mean age was 36.6 ± 12.5 years. In the HIV-negative group, we had 254 of 503 (50.5%) patients who had PTB and or EPTB. Their mean age was 32.8 ± 13.8 years. Of these patients, 156 (61.4%) were found to be mycobacterial culture positive with a mean age of 33.0 ± 14.0 years. This difference was statistically insignificant. Most patients were in the age group of 19-45 years with a p-value <0.001.
Of the 249 HIV seropositive patients, 220 (88.4%) were suspected of PTB and 29 (11.6%) were EPTB cases, while in 254 were HIV seronegative patients, 244 (96.1%) were PTB suspected, and the remaining 10 (3.9%) were EPTB suspected cases. Thus the prevalence of EPTB in HIV positive and HIV-negative was statistically highly significant (Table 1).
Overall, 287 patients were culture positive patients, of them 203 (70.7%) were male and 84 (29.3%) female. We found that male preponderance was common in both HIV-negative (62.2%) as well as in HIV-positive (80.9%) patients. However, this male preponderance was significantly (p <0.001) more in HIV-positive cases. All mycobacterial cultures were subjected to m-PCR to differentiate MTB and non-tuberculous mycobacteria (NTM). Samples from 11 (3.8%) patients were found to be NTM (all HIV positive) and the remaining 276 were MTB and subjected to genotyping studies (Table 2). Only one HIV-negative patient had NTM while in 10 HIV-positive patients (7.6%) NTM were isolated.
The socio-economic and educational data revealed that in the HIV positive group, most of the patients were literate but not matriculate. Patients in the HIV-TB group were likely to be unemployed or have their own small businesses. Of the patients, 202 (70.4%) were married while 63 (22.0%) were unmarried. The remaining 22 (7.65) were widows/widowers, divorcees, or transgender.
A history (6 months or more) of taking anti-tuberculous treatment (ATT) showed that 208 out of 287 (72.5%) of all the mycobacterial culture positive patients had a previous history of ATT. The probability of having a past history of tuberculosis was similar irrespective of HIV status. (Table 1).
Before drug susceptibility testing and genotyping, all the culture isolates were checked by in-house multiplex PCR for species identification. Out of 287 isolates, 276 (96.2%) were identified as MTB, and the remaining 11 (3.8%) were identified as NTM. Ten (90.9%) NTM were isolated from HIV-positive patients and one (9.1%) from a HIV-negative patient.
Among 276 MTB isolates, 206 (74.6%) were sensitive to first line drugs and 46 (16.7%) were MDR. The remaining 24 (8.7%) were mono and dual resistant. Of the HIV-negative patients, 130 (83.3%) were SIRE sensitive while in HIV-positive cases, 76 (63.3%) were sensitive to first line drugs. Of the HIV-negative patients, 16 (10.3%) MDR cases were observed, while 30 (25.0%) MDR cases were observed in HIV-positive patients. The p-value was <0.001 (Table 1).
Of the 276 culture positive MTB isolates, 120 were HIV-positive and 156 were HIV-negative. The lineage distribution of these populations in pulmonary and extrapulmonary tuberculosis infection is illustrated in Table 2.
The molecular characterization of the 276 MTB isolates revealed that the Central Asian Strain (CAS) genotype was found to be most predominant [153 (55.4%)], followed by Beijing lineage [44 (15.9%)], East African Indian [25 (9.1%)], and other lineages [54 (19.6%)]. In HIV-negative patients, the CAS genotype was predominant (62.2%) followed by Beijing (10.9%), EAI (7.7%), and others (19.25%). The distribution of various genotypes in HIV-positive patients was not significantly different and CAS genotypes remained predominant (Tables 1, 2).
Based on the spoligotyping pattern and 24 loci MIRU-VNTR analysis, an Un-weighted pair group method with arithmetic mean (UPGMA) phylogenetic tree was generated (Figures 1A, B) using MIRU-VNTR Plus online tool. Accordingly, in HIV-negative patients, a total of 19 STs comprising 140 (89.7%) isolates were identified while the remaining 16 (10.3%) isolates had unique/unidentified ST patterns. The lineages and corresponding spoligotype patterns of 156 isolates are shown in Supplementary Table 1. Of the 19 different STs observed, ST26 of CAS1_DEL had a maximum of 51.3% strains, followed by ST1 of Beijing with 10.9%, ST25 of CAS1_DEL with 3.2%, ST22 with 0.6%, ST53 of T1 with 7.7%, EAI3_IND with 4.5% and the remaining belonged to other STs. While in the 120 HIV-positive patients, the CAS family consisted of six STs (Supplementary Table 2). Among the STs of the CAS family, 37.5% of strains belonged to ST26, 3.3% each belonged to ST289 and ST1343, while 0.8% belonged to ST25 and ST2419 each.
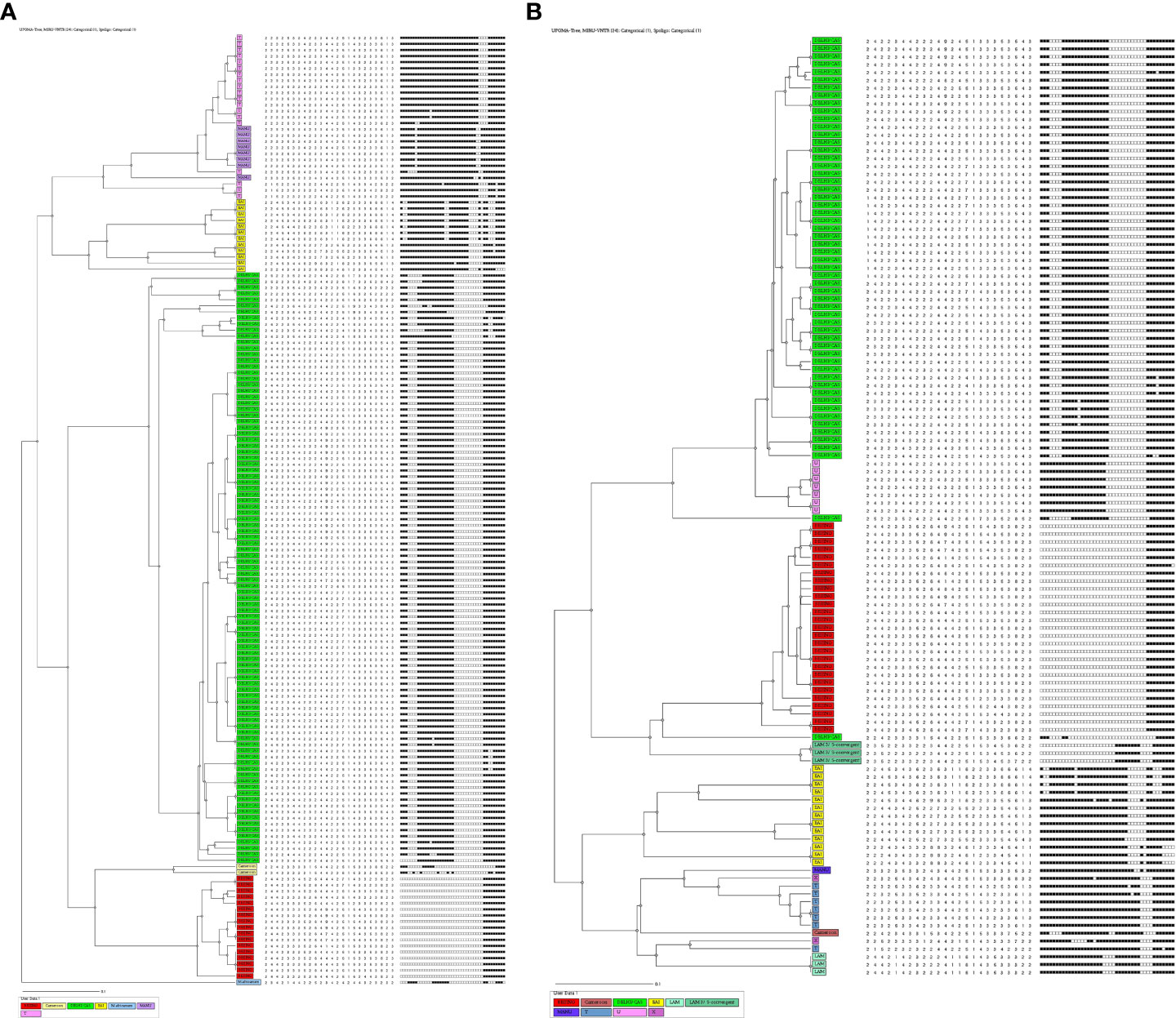
Figure 1 (A) UPGMA-dendrogram representing MIRU-VNTR and spoligotype patterns of 156 isolates from HIV-negative cases. (B) UPGMA-dendrogram representing MIRU-VNTR and spoligotype patterns of 120 isolates from HIV Positive cases.
In the EAI lineage, a total of 3 different STs were identified comprising 8 isolates of EAI, in which ST458 was found in 4 (3.3%) isolates followed by ST138 in 3 (2.5%) and ST1970 in 1 (0.8%) isolate. The single most dominant ST of the Beijing family was ST1 in 27 (22.5%) strains. Whereas other strains belonged to less predominant STs (Supplementary Table 2).
The distribution of the lineages in HIV positive and HIV negative patients were analyzed using the chi-square test which showed a significant (p=0.00816) difference in lineage distribution that was observed in both the study population (Table 3).
The combination of different epidemiological and phylogeographical techniques is important for better interpretation of molecular results. When the clustering rate by spoligotyping was determined, we found that Beijing was the most (100%) clustered lineage followed by CAS with 96.7% (89/92) in HIV-negative and 94.6% (53/56) in HIV-positive patients. The EAI lineage showed 87.5% clustering in HIV-negative while in HIV-positive patients it showed a 75% clustering rate. In HIV-negative cases, Manu was predominant with an 87.5% (7/8) clustering rate, followed by T lineage with 88.9% (16/18).
MIRU-VNTR revealed that 90.7% (88/97) and 96.4% (54/56) isolates of the largest cluster belonged to the CAS family in HIV-negative and HIV-positive patients respectively. In Beijing lineage, the clustering rate was 70.6% in HIV-negative and 63% in HIV-positive cases which was much lower than spoligotyping. Using MIRU-VNTR we identified 131 different genotypes in 34 clusters and 25 orphan isolates with a clustering rate of 84% in HIV negative cases, whereas in HIV positive cases 101 various genotypes were recognized in 28 clusters with an 84.2% clustering rate (Table 4).
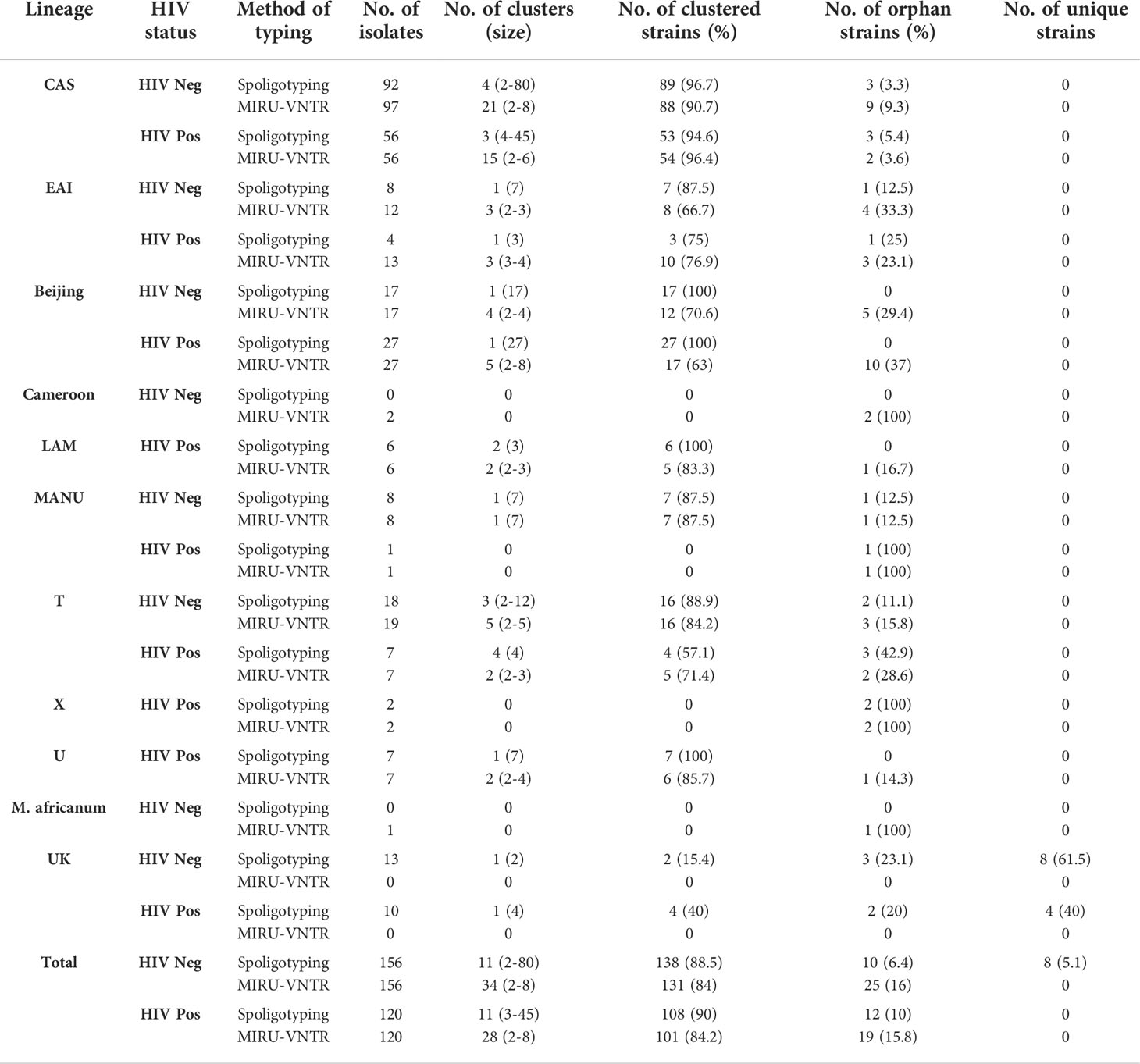
Table 4 Distribution and clustering patterns of various lineages of M. tuberculosis in HIV seronegative and HIV seropositive patients.
In HIV-negative cases, the largest and smallest cluster comprised 80 and 2 isolates respectively by spoligotyping. However, in HIV-positive cases, the largest cluster consisted of 45 isolates, while the smallest cluster had 3 isolates. When orphan strains were checked by both methods in all lineages, we found 6.4% (10/156) of orphan strains by spoligotyping in HIV-negative cases, while using MIRU-VNTR, the rate was 16% (25/156). In HIV-positive cases, spoligotyping showed 10% (12/120) isolates as an orphan while MIRU-VNTR revealed 15.8% (19/120) (Table 5). We did not find any significant difference with regard to the demographic background of the patients as most of them were from North India only (Table 6). The results of 24 -loci MIRU-VNTRs analysis showed that loci QUB-26, Mtub21, ETR-c, and MIRU-26 were highly discriminant in MTB isolates from HIV-negative as well as HIV-positive cases (Figures 2A, B).
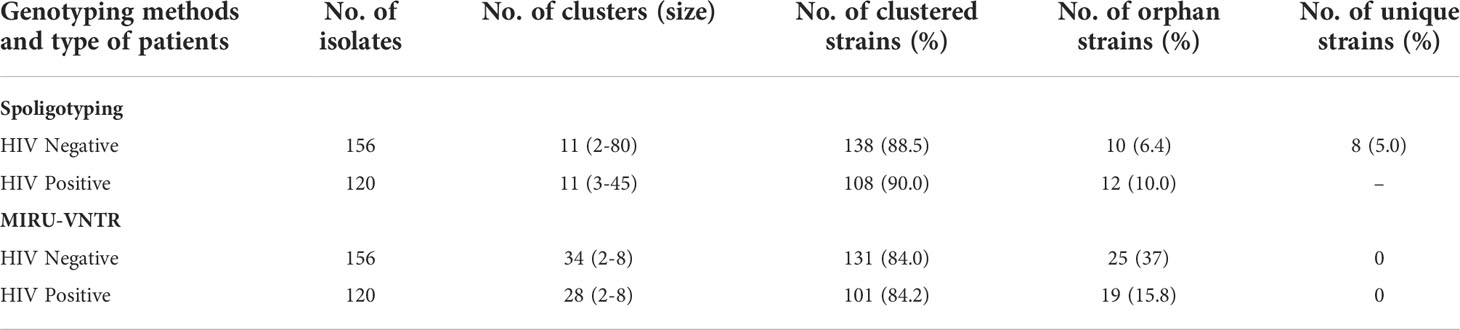
Table 5 Overall clustering patterns of MTB isolates by both the molecular techniques in HIV-negative and HIV-positive patients.
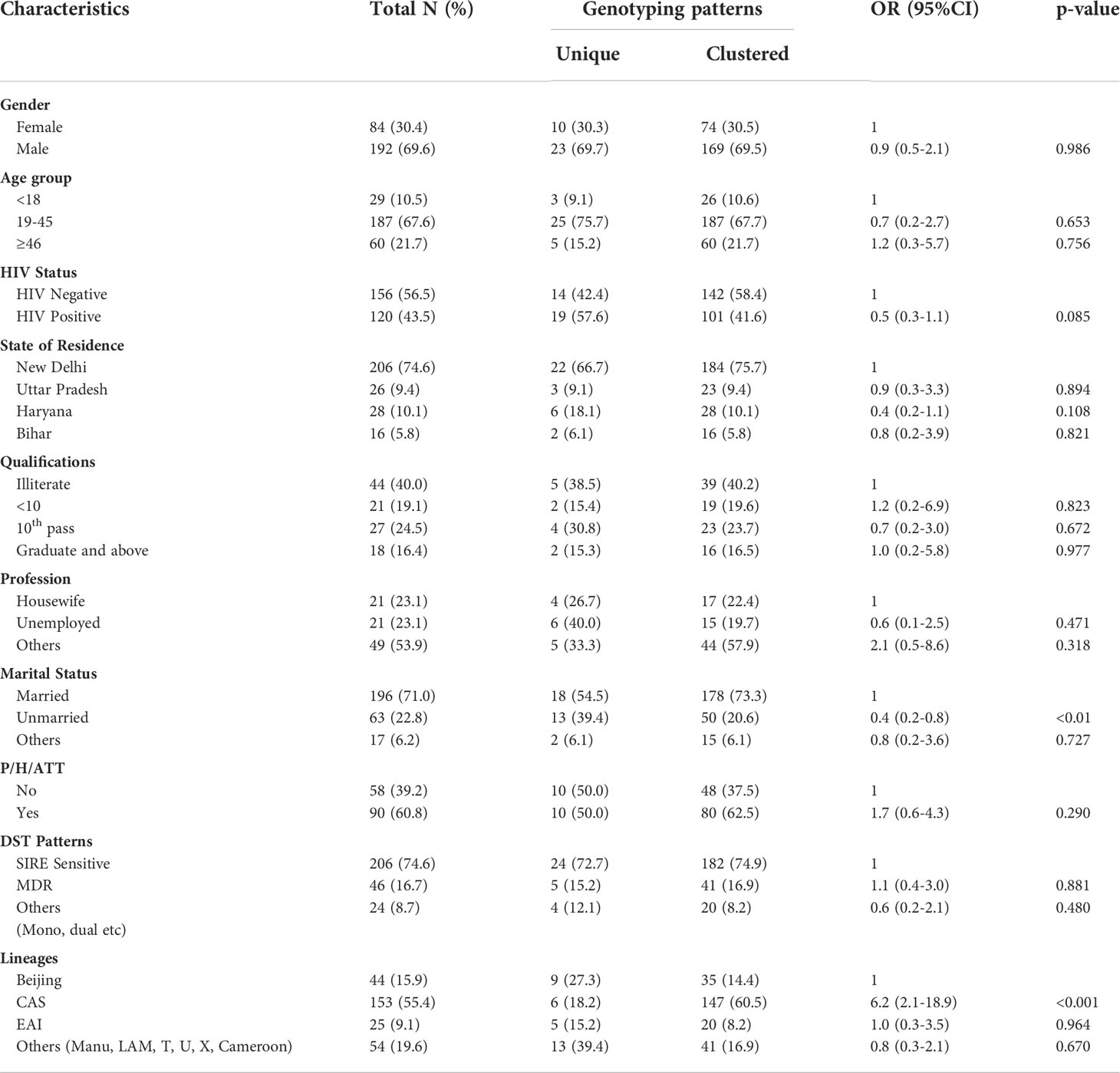
Table 6 Co-relation of various demographical and other factors between Clustered and Unique MTB strains.
The minimum Spanning Tree (MST) analysis was done by using MIRU-VNTRplus. The various SITs amongst MTB isolates from HIV-negative patients (Figure 2A) and HIV-positive patients (Figure 2B) showed predominant SITs and the evolutionary relationship between the lineages and their SITs.
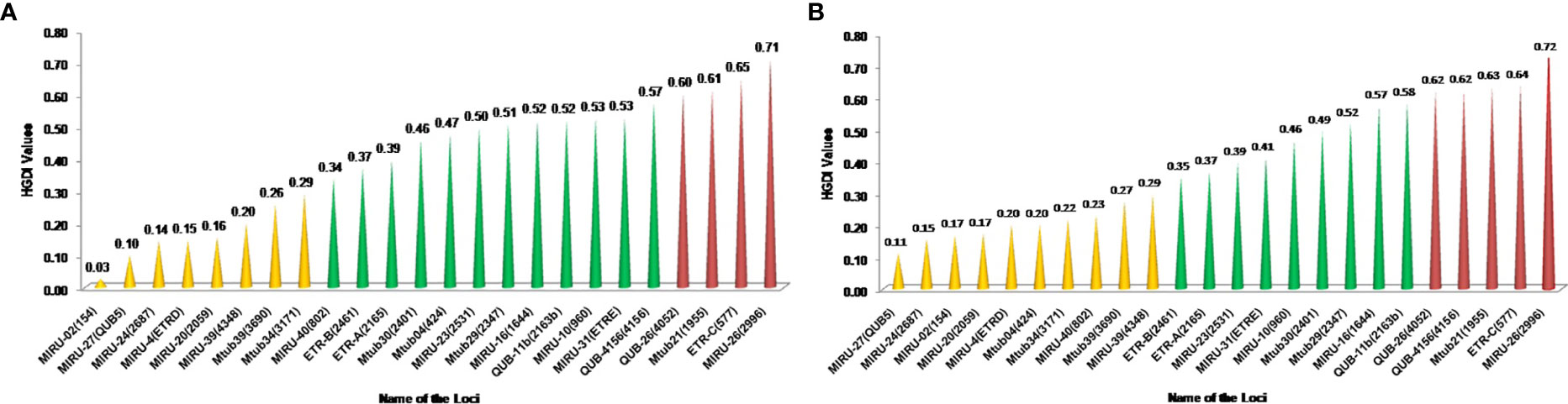
Figure 2 (A) Minimum spanning trees of MTC isolates from HIV-negative patients typed with both spoligotyping and MIRU-VNTR. Each circle represents a genotype. The distance between circles represents how closely related the different genotypes are to each other. MST connects each genotype based on the degree of changes required to go from one allele to another. The configuration of the tree is represented by branches i.e. continuous versus dotted lines and circles representing each pattern. The length of the branches denotes the distance between patterns whereas the intricacy of the lines indicates the number of spacers altered between two patterns. The thicker lines represent one change while thinner ones indicate 2 or 3. The size of the circle is comparable to the total number of MTB isolates in this study. The color of the circles represents the phylogenetic lineage to which the specific pattern belongs. Beijing patterns are circled in red. Patterns colored in green indicate Delhi/CAS strains. EAI strains are depicted in yellow. (B) Minimum spanning trees of MTC isolates from HIV-positive patients typed with both spoligotyping and MIRU-VNTR. Each circle represents a genotype. The distance between circles represents how closely related the different genotypes are to each other (detailed description as mentioned in Figure 3A).
This study describes the genetic diversity in M. tuberculosis isolated from HIV-TB co-infected and HIV uninfected TB cases. Most of our patient population was from the North Indian states of Delhi’s national capital region, Haryana and Uttar Pradesh. Therefore, the incidence and prevalence rates of HIV and TB were in line with other studies. However, in our study, we compared the drug resistance pattern and genotypes side-by-side isolated from these two patient groups under a controlled single laboratory study.
As reported in various other studies, we found that CAS was the predominant genotype followed by Beijing and EAI (Devi et al., 2015; Singh et al., 2015; Singh et al., 2021). Interestingly, the same pattern was seen in our patient populations irrespective of their HIV status. This could be explained based on the circulation of these genotypes in the environment and the ethnic make-up of North Indian patients (Sankar et al., 2013). In our earlier studies, we also found similar patterns in various genotypes (Sankar et al., 2013; Singh et al., 2015). Similar results were also found in the southern part of India (Narayanan et al., 2008). However, in this present study, the Beijing genotype was more prevalent in HIV seropositive patients rather than HIV-negative patients (Table 1). Similar results were also reported in a study conducted on HIV-positive patients with TB meningitis (Caws et al., 2006). Narayanan et al. (Narayanan et al., 2008) and Gupta et al. (2014) also reported Beijing genotype only in HIV-infected patients and not in HIV-uninfected patients. et alVeigas et al. (Viegas et al., 2013) suggested that the association of Beijing strain with HIV seropositive status could be due to the combination of the increased virulence of the strain, and higher susceptibility of HIV positive patients. However, a study from Western Maharashtra, India found no association between HIV status and spoligotypes (Chatterjee et al., 2010).
Generally, the prevalence of different genotypes of MTB is dependent on the geographical location and the ethnicities of the population, as reported in our previous studies (Sankar et al., 2013; Singh et al., 2015). In the present study, Beijing genotypes were also significantly more common in HIV-positive patients and CAS lineage was more common in HIV-negative cases, which could be explained by the fact that CAS lineage is the most common lineage circulating in northern parts of India (Sankar et al., 2013), and the same pattern was seen in this study. A population-based investigation for 20 years in rural Malawi reported ST129 (EAI5) to be the most common lineage associated with HIV status (Glynn et al., 2010). A study from Ethiopia found that in HIV-positive subjects, the T family was the most predominant (38.5%) (Mihret et al., 2012). This also suggests that the frequency of various lineages depends more on geographical locations rather than HIV status.
Anti-TB drug resistance can develop after acquiring the infection (secondary), but it can also be a baseline (primary) when the person is infected with drug-resistant strains of M. tuberculosis. In the present study, we observed an overall drug resistance (resistance to any drug) in 25.4% and MDR in 16.7% isolates (Table 1). A higher rate of MDR was found in isolates from HIV-positive patients as compared to isolates from HIV-negative patients. This may be attributed to the previous history of TB in a higher number of patients from the HIV-positive group, as well as the lower socio-economic status of these patients. Saldanha et al. (2019) also reported a high prevalence of MDR-TB in HIV-positive patients and associated this with the treatment of previous TB infection with sub-optimal anti-TB regimens. Similarly, Sethi et al. (2013) reported that TB was more prevalent in patients from lower socioeconomic backgrounds and that there was a higher number of MDR-MTB (27.3%) in HIV seropositive subjects compared to HIV seronegative subjects (15.4%). A prior study carried out by Isaakidis et al. (2014) also confirmed the very high burden of DR-TB in HIV-positive patients (38%) at an ART center in Mumbai. In the current study, most of our patients were from Delhi and its adjoining states. Our findings may not necessarily represent the entire country, as the socio-economic conditions of patients, HIV prevalence, and medical practices differ significantly from region to region. Clustering is often used to group strains with similar genotypic traits. On the analysis of clustering amongst the various phylogenetic lineages, we found clustering in 88.8% and 86.6% HIV-uninfected and HIV-infected patient groups. This reflects the high transmission rate within these population groups (Odhiambo et al., 1999). In addition to sub-lineage analysis, we also observed that CAS1_DEL (ST26) and Beijing (ST1) clades were predominant in both the study populations with no significant difference. These findings are in line with previous publications (Singh et al., 2004; Arora et al., 2009; Sankar et al., 2013; Singh et al., 2015).
Nutrition and timely medical care are important determinants of disease outcome. In India, even though TB is more prevalent in male patients and a poor disease outcome is more likely in females due to various socio-economic factors (Singh et al., 2012) and the finding of our study were on similar lines.
There was no significant difference in the prevalence of clustered or unique isolates among different age groups. The percentage of clustered isolates based on 24 loci MIRU-VNTR was, however, higher among married patients. This could be due to better opportunities for the circulation of these strains in the families, though it may an overestimation (García de Viedma et al., 2011). We found a higher percentage of clustered isolates of CAS lineage which showed a significantly higher clustering rate than other lineages. This may be because CAS is more prevalent in North India and most of the isolates from both groups are from north India (Arora et al., 2009; Singh et al., 2015). We also observed unique isolates with lineages like Manu, LAM, T, U X, and Cameroon, demonstrating an assortment of strains and newly emerging strains in these patients, and that this trend should be considered seriously by the program managers.
As per our findings in the HIV-negative group as well as in those in the HIV-positive group, MIRU26, ETRC, Mtub21, QUB4156, and QUB26 were highly discriminating, but MIRU2, MIRU24, MIRU20, and ETRD were feebly discriminating (Figures 3A, B). Earlier we also found that QUB4156, QUB26, and MIRU26 were highly discriminating and MIRU2, MIRU4 or ETRD and MIRU24 were poorly discriminating (Sankar et al., 2013).
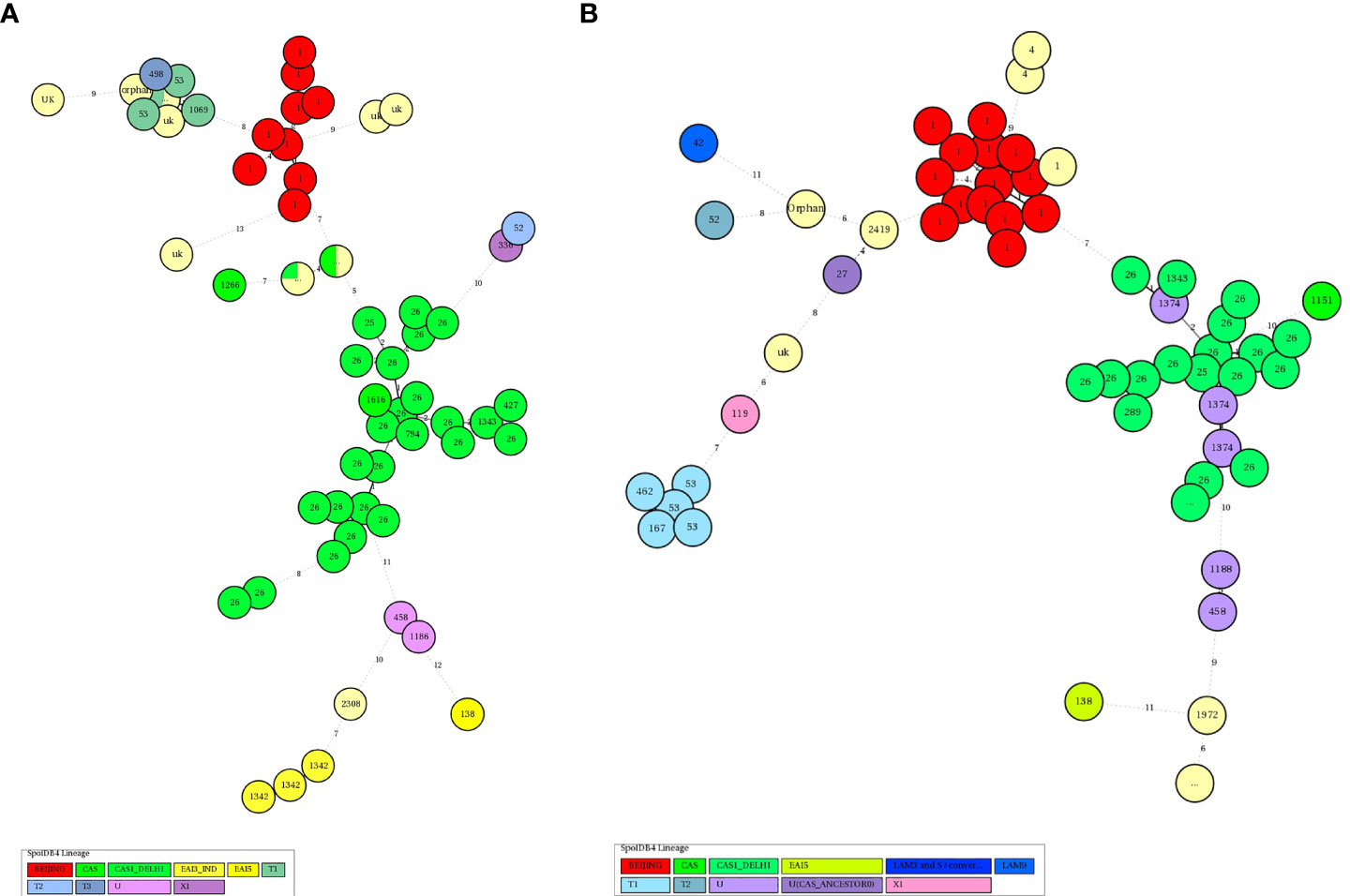
Figure 3 (A) The Hunter-Gaston discriminatory index of MIRU-VNTR alleles of TB isolates from HIV-Negative patients MIRU: Mycobacterial Interspersed Repetitive Units, VNTR, Variable Number Tandem Repeats; HGDI, Hunter-Gaston Diversity index. The allelic diversity of the loci was classified as highly discriminant [Hunter-Gaston Index (HGI) >0.6], moderately discriminant (HGI ≥0.3 to ≤0.6), and poorly discriminant (HGI < 0.3). (B) The Hunter-Gaston discriminatory index of MIRU-VNTR alleles of TB isolates from HIV- TB co-infected patients.
It might be expected that some lineages and clones of MTB are more prevalent or have severe disease outcomes in HIV-positive patients. However, in the present study, we found that though all clones could be isolated from HIV-positive patients, the incidence of the Beijing genotype was more frequent in this group of patients. It is important to note that in North India, even though the CAS lineage is more prevalent, in HIV-infected patients CAS lineage had a low incidence compared to Beijing lineage. We are not able to explain fully why the EPTB in the HIV-infected patients in our study was not as common as that reported by others (Yang et al., 1995; Giri et al., 2013), but it was significantly more common (11.6%) than in HIV-negative patients (3.9%). This could be because our patient recruiting center is a dedicated TB hospital and EPTB cases are more commonly referred to other general care hospitals for want of wider scope for surgical and medical management.
The present study provides important molecular genotypic data of M. tuberculosis isolates from HIV-negative and HIV-positive patients of Northern India. Central Asian lineage despite being the most predominant isolate circulating in the study area and both HIV-positive and HIV-negative TB cases, the Beijing genotype showed a statistically significant high incidence rate in HIV-TB cases. However, the clustering rate of predominant genotypes was similar in both HIV-positive and HIV-negative TB cases, which shows the high transmissibility of these genotypes in the community. Probability of having extrapulmonary tuberculosis and isolation of non-tuberculous mycobacterial were also significantly more in HIV-infected patients.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The studies involving human participants were reviewed and approved by Institutional Ethical Committee, All India Institute of Medical Sciences, New Delhi - 110029. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
SS, NS, DB, and UA designed the study. JS, NS, RS, and GS collected clinical samples and patient clinical information. JS performed genotyping analysis. JS, RS, and GS performed the experiments. SS and JS analysed data. JS, NS, and SS wrote the manuscript. Final modifications in manuscript done by MM and AM. SS arranged for financial support. All authors contributed to the article and approved the submitted version.
The study received financial support from Indian Council of Medical Research (ICMR), New Delhi (ICMR) Sanction Order Number: 5/8/5/15/10-ECD-I) and publication support from Indo-U.S. Science and Technology Forum (IUSSTF) (Award Letter No. IUSSTF/USISTEF/8th Call/HI-051/2017/2018-19).
We are thankful to all the patients who cooperated in this study. We are also grateful to Indian Council of Medical Research (ICMR), New Delhi (ICMR Sanction Order Number: 5/8/5/15/10-ECD-I) and Indo-U.S. Science and Technology Forum (IUSSTF) (Award Letter No. IUSSTF/USISTEF/8th Call/HI-051/2017/2018-19) for financial support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.953443/full#supplementarymaterial
TB, Tuberculosis; PTB, Pulmonary Tuberculosis; EPTB, Extrapulmonary Tuberculosis; HIV, Human Immunodeficiency Virus; MIRU-VNTR, Mycobacterial Interspersed Repetitive Unite - Variable Number Tandem Repeat; DST-SIRE, Drug Susceptibility Testing for Streptomycin (STR), Isoniazid (INH), Rifampin (RIF), and Ethambutol (EMB); CAS, Central Asian; EAI, East-African-Indian.
Allix-Béguec, C., Fauville-Dufaux, M., Stoffels, K., Ommeslag, D., Walravens, K., Saegerman, C., et al. (2010). Importance of identifying mycobacterium bovis as a causative agent of human tuberculosis. Eur. Respir. J. 35 (3), 692–694. doi: 10.1183/09031936.00137309
Arora, J., Singh, U. B., Suresh, N., Rana, T., Porwal, C., Kaushik, A., et al. (2009). Characterization of predominant mycobacterium tuberculosis strains from different subpopulations of India. Infect. Genet. Evol. 9 (5), 832–839. doi: 10.1016/j.meegid.2009.05.008
Global tuberculosis report. (2020) Geneva: World Health Organization 2020 Licence: CC BY-NC-SA 3.0 IGO. Available at: https://www.who.int/publications-detail-redirect/9789240013131" https://www.who.int/https://www.who.int/publications-detail-redirect/9789240013131"publications-detail-redirect/9789240013131.
Cadmus, S., Hill, V., van, S. D., Rastogi, N. (2011). Spoligotype profile of mycobacterium tuberculosis complex strains from HIV-positive and -negative patients in Nigeria: a comparative analysis. J. Clin. Microbiol. 49 (1), 220–226. doi: 10.1128/JCM.01241-10
Caws, M., Thwaites, G., Stepniewska, K., Lan, N. T. N., Duyen, N. T. H., Phuong, N. T., et al. (2006). Beijing Genotype of mycobacterium tuberculosis is significantly associated with human immunodeficiency virus infection and multidrug resistance in cases of tuberculous meningitis. J. Clin. Microbiol. 44 (11), 3934–3939. doi: 10.1128/JCM.01181-06
Chatterjee, A., D’Souza, D., Vira, T., Bamne, A., Ambe, G. T., Nicol, M. P., et al. (2010). Strains of mycobacterium tuberculosis from western maharashtra, India, exhibit a high degree of diversity and strain-specific associations with drug resistance, cavitary disease, and treatment failure. J. Clin. Microbiol. 48 (10), 3593–3599. doi: 10.1128/JCM.00430-10
Demay, C., Liens, B., Burguière, T., Hill, V., Couvin, D., Millet, J., et al. (2012). SITVITWEB–a publicly available international multimarker database for studying mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect. Genet. Evol. 12 (4), 755–766. doi: 10.1016/j.meegid.2012.02.004
Devi, K. R., Bhutia, R., Bhowmick, S., Mukherjee, K., Mahanta, J., Narain, K. (2015). Genetic diversity of mycobacterium tuberculosis isolates from Assam, India: Dominance of Beijing family and discovery of two new clades related to CAS1_Delhi and EAI family based on spoligotyping and MIRU-VNTR typing. Mokrousov I editor. PloS One 10 (12), e0145860. doi: 10.1371/journal.pone.0145860
Friedland, G., Churchyard, G. J., Nardell, E. (2007). Tuberculosis and HIV coinfection: current state of knowledge and research priorities. J. Infect. Dis. 196 Suppl 1, S1–S3. doi: 10.1086/518667
García de Viedma, D., Mokrousov, I., Rastogi, N. (2011). Innovations in the molecular epidemiology of tuberculosis. Enferm Infecc Microbiol. Clin. 29 Suppl 1, 8–13. doi: 10.1016/s0213-005x(11)70012-x
Giri, P. A., Deshpande, J. D., Phalke, D. B. (2013). Prevalence of pulmonary tuberculosis among HIV positive patients attending antiretroviral therapy clinic. N Am. J. Med. Sci. 5 (6), 367–370. doi: 10.4103/1947-2714.114169
Glynn, J. R., Alghamdi, S., Mallard, K., McNerney, R., Ndlovu, R., Munthali, L., et al. (2010). Changes in mycobacterium tuberculosis genotype families over 20 years in a population-based study in northern Malawi. PloS One 175 (8), e12259. doi: 10.1371/journal.pone.0012259
Gupta, A., Kulkarni, S., Rastogi, N., Anupurba, S. (2014). A study of mycobacterium tuberculosis genotypic diversity & drug resistance mutations in varanasi, north India. Indian J. Med. Res. 139 (6), 892–902.
Gutierrez, M. C., Ahmed, N., Willery, E., Narayanan, S., Hasnain, S. E., Chauhan, D. S., et al. (2006). Predominance of ancestral lineages of mycobacterium tuberculosis in India. Emerging Infect. Dis. 12 (9), 1367–1374. doi: 10.3201/eid1209.050017
Hanekom, M., van der Spuy, G. D., Gey van Pittius, N. C., McEvoy, C. R. E., Hoek, K. G. P., Ndabambi, S. L., et al. (2008). Discordance between mycobacterial interspersed repetitive-unit-variable-number tandem-repeat typing and IS6110 restriction fragment length polymorphism genotyping for analysis of mycobacterium tuberculosis Beijing strains in a setting of high incidence of tuberculosis. J. Clin. Microbiol. 46 (10), 3338–3345. doi: 10.1128/JCM.00770-08
Hunter, P. R., Gaston, M. A. (1988). Numerical index of the discriminatory ability of typing systems: an application of simpson’s index of diversity. J. Clin. Microbiol. 26 (11), 2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988
Isaakidis, P., Das, M., Kumar, A. M. V., Peskett, C., Khetarpal, M., Bamne, A., et al. (2014). Alarming levels of drug-resistant tuberculosis in HIV-infected patients in metropolitan Mumbai, India. PloS One 9 (10), e110461. doi: 10.1371/journal.pone.0110461
Mihret, A., Bekele, Y., Loxton, A. G., Jordan, A. M., Yamuah, L., Aseffa, A., et al. (2012). Diversity of mycobacterium tuberculosis isolates from new pulmonary tuberculosis cases in Addis Ababa, Ethiopia. Tuberc Res. Treat. 2012, 892079. doi: 10.1155/2012/892079
Munawwar, A., Singh, S. (2012). AIDS associated tuberculosis: a catastrophic collision to evade the host immune system. Tuberculosis (Edinb). 92 (5), 384–387. doi: 10.1016/j.tube.2012.05.006
Narayanan, S., Gagneux, S., Hari, L., Tsolaki, A. G., Rajasekhar, S., Narayanan, P. R., et al. (2008). Genomic interrogation of ancestral mycobacterium tuberculosis from south India. Infect. Genet. Evol. 8 (4), 474–483. doi: 10.1016/j.meegid.2007.09.007
Odhiambo, J. A., Borgdorff, M. W., Kiambih, F. M., Kibuga, D. K., Kwamanga, D. O., Ng’ang’a, L., et al. (1999). Tuberculosis and the HIV epidemic: increasing annual risk of tuberculous infection in Kenya, 1986-1996. Am. J. Public Health 89 (7), 1078–1082. doi: 10.2105/AJPH.89.7.1078
Pitondo-Silva, A., Santos, A. C. B., Jolley, K. A., Leite, C. Q. F., Darini AL da, C. (2013). Comparison of three molecular typing methods to assess genetic diversity for mycobacterium tuberculosis. J. Microbiol. Methods 93 (1), 42–48. doi: 10.1016/j.mimet.2013.01.020
Saldanha, N., Runwal, K., Ghanekar, C., Gaikwad, S., Sane, S., Pujari, S. (2019). High prevalence of multi drug resistant tuberculosis in people living with HIV in Western India. BMC Infect. Dis. 819, 391. doi: 10.1186/s12879-019-4042-z
Sankar, M. M., Singh, J., Angelin Diana, S. C., Singh, S. (2013). Molecular characterization of mycobacterium tuberculosis isolates from north Indian patients with extrapulmonary. Tuberculosis. 93 (1), 75–83. doi: 10.1016/j.tube.2012.10.005
Sethi, S., Mewara, A., Dhatwalia, S. K., Singh, H., Yadav, R., Singh, K., et al. (2013). Prevalence of multidrug resistance in mycobacterium tuberculosis isolates from HIV seropositive and seronegative patients with pulmonary tuberculosis in north India. BMC Infect. Dis. 13 (1), 137. doi: 10.1186/1471-2334-13-137
Sharma, P., Chauhan, D. S., Upadhyay, P., Faujdar, J., Lavania, M., Sachan, S., et al. (2008). Molecular typing of mycobacterium tuberculosis isolates from a rural area of kanpur by spoligotyping and mycobacterial interspersed repetitive units (MIRUs) typing. Infect. Genet. Evol. 8 (5), 621–626. doi: 10.1016/j.meegid.2008.05.002
Singh, J., Sankar, M. M., Kumar, P., Couvin, D., Rastogi, N., Singh, S., et al. (2015). Genetic diversity and drug susceptibility profile of mycobacterium tuberculosis isolated from different regions of India. J. Infect. 71 (2), 207–219. doi: 10.1016/j.jinf.2015.04.028
Singh, S., Singh, J., Kumar, S., Gopinath, K., Balooni, V., Singh, N., et al. (2012). Poor performance of serological tests in the diagnosis of pulmonary tuberculosis: Evidence from a contact tracing field study. Pai M editor. PloS One 7 (7), e40213. doi: 10.1371/journal.pone.0040213
Singh, A. V., Singh, S., Yadav, A., Kushwah, S., Yadav, R., Sai, D. K., et al. (2021). Genetic variability in multidrug-resistant mycobacterium tuberculosis isolates from patients with pulmonary tuberculosis in north India. BMC Microbiol. 21 (1), 123. doi: 10.1186/s12866-021-02174-6
Singh, U. B., Suresh, N., Bhanu, N. V., Arora, J., Pant, H., Sinha, S., et al. (2004). Predominant tuberculosis spoligotypes, Delhi, India. Emerging Infect. Dis. 10 (6), 1138–1142. doi: 10.3201/eid1006.030575
Sreevatsan, S., Pan, X., Stockbauer, K. E., Connell, N. D., Kreiswirth, B. N., Whittam, T. S., et al. (1997). Restricted structural gene polymorphism in the mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. U.S.A. 94 (18), 9869–9874. doi: 10.1073/pnas.94.18.9869
Supply, P., Allix, C., Lesjean, S., Cardoso-Oelemann, M., Rüsch-Gerdes, S., Willery, E., et al. (2006). Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of mycobacterium tuberculosis. J. Clin. Microbiol. 44 (12), 4498–4510. doi: 10.1128/JCM.01392-06
Thomas, S. K., Iravatham, C. C., Moni, B. H., Kumar, A., Archana, B. V., Majid, M., et al. (2011). Modern and ancestral genotypes of mycobacterium tuberculosis from andhra pradesh, India. PloS One 6 (11), e27584. doi: 10.1371/journal.pone.0027584
Vadwai, V., Shetty, A., Supply, P., Rodrigues, C. (2012). Evaluation of 24-locus MIRU-VNTR in extrapulmonary specimens: study from a tertiary centre in Mumbai. Tuberculosis (Edinb). 92 (3), 264–272. doi: 10.1016/j.tube.2012.01.002
Viegas, S. O., Machado, A., Groenheit, R., Ghebremichael, S., Pennhag, A., Gudo, P. S., et al. (2013). Mycobacterium tuberculosis Beijing genotype is associated with HIV infection in Mozambique. PloS One 8 (8), e71999. doi: 10.1371/journal.pone.0071999
WHO (2016) Global tuberculosis report. Available at: http://apps.who.int/medicinedocs/en/d/Js23098en/.
Yang, Z. H., Mtoni, I., Chonde, M., Mwasekaga, M., Fuursted, K., Askgård, D. S., et al. (1995). DNA Fingerprinting and phenotyping of mycobacterium tuberculosis isolates from human immunodeficiency virus (HIV)-seropositive and HIV-seronegative patients in Tanzania. J. Clin. Microbiol. 33 (5), 1064–1069. doi: 10.1128/jcm.33.5.1064-1069.1995
Keywords: spoligotyping, genotypes, HIV, Mycobacerium tuberculosis, molecular epidemiology
Citation: Singh J, Singh N, Suresh G, Srivastava R, Aggarwal U, Behera D, Munisamy M, Malhotra AG and Singh S (2022) A comparative analysis of molecular genotypes of Mycobacterium tuberculosis isolates from HIV-positive and HIV-negative patients. Front. Cell. Infect. Microbiol. 12:953443. doi: 10.3389/fcimb.2022.953443
Received: 26 May 2022; Accepted: 30 August 2022;
Published: 12 October 2022.
Edited by:
Divakar Sharma, University of Delhi, IndiaReviewed by:
Mariza Gonçalves Morgado, Oswaldo Cruz Foundation (Fiocruz), BrazilCopyright © 2022 Singh, Singh, Suresh, Srivastava, Aggarwal, Behera, Munisamy, Malhotra and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarman Singh, c2FybWFuX3NpbmdoQHlhaG9vLmNvbQ==; c2FybWFuLnNpbmdoQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.